Abstract.
Molecular data are required to improve our understanding of the epidemiology of leptospirosis in Africa and to identify sources of human infection. We applied molecular methods to identify the infecting Leptospira species and genotypes among patients hospitalized with fever in Tanzania and compared these with Leptospira genotypes detected among animals in Tanzania to infer potential sources of human infection. We performed lipL32 real-time PCR to detect the presence of pathogenic Leptospira in acute-phase plasma, serum, and urine samples obtained from study participants with serologically confirmed leptospirosis and participants who had died with febrile illness. Leptospira blood culture was also performed. In positive specimens, we performed species-specific PCR and compared participant Leptospira secY sequences with Leptospira reference sequences and sequences previously obtained from animals in Tanzania. We detected Leptospira DNA in four (3.6%) of 111 participant blood samples. We detected Leptospira borgpetersenii (one participant, 25.0%), Leptospira interrogans (one participant, 25.0%), and Leptospira kirschneri (one participant, 25.0%) (one [25%] undetermined). Phylogenetic comparison of secY sequence from the L. borgpetersenii and L. kirschneri genotypes detected from participants was closely related to but distinct from genotypes detected among local livestock species. Our results indicate that a diverse range of Leptospira species is causing human infection. Although our analysis suggests a close relationship between Leptospira genotypes found in people and livestock, continued efforts are needed to obtain more Leptospira genetic material from human leptospirosis cases to help prioritize Leptospira species and genotypes for control.
INTRODUCTION
Leptospirosis is a zoonotic disease, with an estimated annual incidence of up to approximately 100 cases per 100,000 people in Tanzania.1 Previous studies in Tanzania have identified contact with cattle and working in rice fields as risk factors for acute human leptospirosis,2,3 but further data are needed to understand transmission pathways and to confirm sources of Leptospira infection for people. Pathogenic Leptospira have been detected by culture or nucleic acid amplification methods in a number of animal hosts in Tanzania including cattle, goats, sheep, and rodents.4,5 Sequence-based species identification of Tanzanian animal isolates and DNA indicates that Leptospira borgpetersenii, Leptospira interrogans, and Leptospira kirschneri are circulating in animal populations in Tanzania. However, the contributions, and relative importance, of these Leptospira species to human disease in Tanzania are unclear.
Direct detection of Leptospira by culture can be challenging in humans as Leptospira are fastidious organisms that may take weeks to grow in culture, limiting the utility of this approach to provide a timely diagnosis for an acutely unwell patient.6 Because of the challenges of Leptospira culture in vitro, molecular diagnostic assays based on nucleic acid amplification tests (NAAT) including real-time PCR of the lipL32 gene have been developed to aid diagnosis. PCR techniques have also been used to directly determine the species of infecting Leptospira for some of the more common pathogenic Leptospira species. This has been achieved using PCR-based amplification of secY and ompL1 genes using species-specific primers and probes to show variation between Leptospira species.7 The sensitivity of these assays has not been fully determined but when used in a two-step algorithm with a lipL32 assay was able to identify four common pathogenic Leptospira species (L. borgpetersenii, L. interrogans, L. kirschneri, and Leptospira noguchii).7 Sequence-based typing schemes of selected gene targets (e.g., 16S rRNA rrs, secY, and lfb1)8–10 have also been developed for Leptospira. Amplification of a ∼435-bp fragment of the secY gene has been shown to have good phylogenetic discrimination between pathogenic Leptospira species and has been widely used in the East African region.4,8,11 Sequence-based approaches have not only been applied to Leptospira isolates8 but can also be applied directly to clinical samples to determine the infecting species and genotype, and to investigate links between human and animal Leptospira infection.12
In this study, we aimed to characterize the genetic diversity of human Leptospira infection in northern Tanzania and to infer possible sources of human infection. We applied direct pathogen detection methods including culture and NAAT to detect Leptospira infection and determine the infecting Leptospira species among febrile patients in northern Tanzania. We then used sequence-based analysis of the secY gene to compare pathogenic Leptospira genotypes detected in patients with those previously reported in animals in Tanzania to advance our understanding of the relationship between human and animal Leptospira infection.
METHODS
Study setting.
Our study was performed among patients presenting to the Kilimanjaro Christian Medical Centre (KCMC), a 450-bed zonal referral hospital, and to Mawenzi Regional Referral Hospital (MRRH), a 300-bed regional referral hospital. Both hospitals are located in the Moshi municipal district in northern Tanzania. Moshi (population ∼180,000) is the administrative capital of the Kilimanjaro Region (population ∼1.6 million), which is located in the north of Tanzania.
Study procedures and participants.
To detect the presence of Leptospira among febrile patients, we used plasma, serum, and urine collected from patients enrolled in two fever etiology studies that have been previously described.2,13 Briefly, we enrolled adult and pediatric inpatients admitted with a fever at the KCMC and MRRH from September 2007 through August 2008 (study 1) and from February 2012 through May 2014 (study 2). A questionnaire was administered to each participant that included questions on the duration of illness and any antibacterial treatment received before presentation at the hospital. We used participant questionnaire responses to estimate the duration of their fever (median and range) before sample collection.
Study personnel drew blood from each participant for serology, and participants were asked to return 4–6 weeks after enrollment for collection of a convalescent serum sample (both studies). We also collected acute urine samples from participants in study 1 and acute blood samples for Leptospira culture from participants enrolled in study 2.
Leptospira culture.
For Leptospira culture, three drops of whole blood were inoculated immediately into 6-mL Ellinghausen–McCullough–Johnson–Harris culture media supplemented with 5-fluorouracil. Inoculated culture media were batch-shipped to the U.S. CDC where they were kept at 30°C for up to 6 months and monitored weekly for Leptospira growth by darkfield microscopic examination.
Selection of participants for real-time PCR.
We selected participants for real-time PCR testing based on predetermined serological case definitions. In both studies, microscopic agglutination testing (MAT) was performed on acute and convalescent serum samples using a panel of 20 serovars belonging to 17 serogroups following standard laboratory procedures as previously described (Supplemental Table).13–15 Participants who had either a ≥ 4-fold rise in MAT titers between acute and convalescent serum samples or a single reciprocal MAT titer ≥ 800 to any serovar on acute or convalescent Leptospira were considered to be positive for acute leptospirosis and were selected for real-time PCR.16 The predominant reactive serogroup for each was defined as the serogroup containing the serovar with the highest agglutinating titer. In addition, we also selected study participants who had died before study follow-up for inclusion in our study (subsequently referred to as decedent participants). Such participants had provided only an acute-phase serologic sample, and as MAT is considered insensitive during the acute phase of illness, we included these samples to allow for missed cases within the decedent participant group.17
Molecular detection of Leptospira DNA.
We performed real-time PCR testing to detect pathogenic Leptospira DNA in archived acute plasma and urine samples for participants recruited from study 1, and in acute serum samples for participants recruited from study 2. First, we extracted DNA from plasma and serum using the QIAamp® DNA Blood Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions with a final elution volume of 100 μL to increase DNA concentration. DNA was extracted from urine using QIAamp DNA Blood Mini Kit with a pretreatment step designed to maximize the yield of bacterial DNA from biological samples.18–20 Briefly, up to 1,000 μL of urine was centrifuged at 10,000 × g for 10 minutes. The resulting pellet was washed in 200 μL Tris-EDTA (TE) buffer (Biotechnology grade, pH 8.0; VWR International Ltd., Lutterworth, United Kingdom) and then resuspended in an enzyme mix containing 50 μL lysozyme (10 mg/mL), 50 μL mutanolysin (1 mg/mL), and 4 μL lysostaphin (1 mg/mL) made up to a final volume of 200 μL with TE buffer, and incubated at 37°C for 1 hour. Subsequently, 180 μL lysis buffer (Buffer AL, Qiagen) and 20 μL proteinase K were added to each sample, and extraction was completed following the standard spin column extraction protocol.
To detect pathogenic Leptospira DNA in participant samples, we used a real-time PCR assay targeting the lipL32 gene following previously published protocols,21,22 using the ABI 7500 real-time PCR system (Thermo Fisher Scientific, Waltham, MA). Assays were performed at National Collaborating Centre for Reference and Research on Leptospirosis, Dutch Royal Tropical Institute (now Amsterdam Medical Centre), Amsterdam, the Netherlands (Study 1), and at the Bacterial Special Pathogens Laboratory, U.S. CDC, Atlanta, GA (study 2). Primers and probes are shown in Table 1. Inhibition was evaluated using an endogenous internal positive control (human rnaseP) (study 1) or an exogenous positive control (Applied Biosystems). Each real-time reaction run included DNA extracted from a pure culture of L. interrogans as a positive control (study 1: L. interrogans serovar Copenhagenii strain Wijnberg supplied by the National Leptospirosis Reference Laboratory, Amsterdam, the Netherlands; study 2: L. interrogans serovar Icterohaemorrhagiae strain RGA from the American Type Culture Collection [ATCC] number 43642).24 In study 1, the master mix contained (per reaction) 1.25 μL of each primer, used at a concentration of 10 μM; 0.25 μL probe (10 μM concentration); 0.5 μL of ROX (1:10 dilution); 4.25 μL nuclease-free water; 12.5 μL Platinum quantitative PCR (qPCR) Supermix-UDG (Thermo Fisher Scientific, Waltham, MA); and 5 μL of the sample. Inhibition was evaluated in a separate qPCR reaction run on the same plate. The master mix for this reaction contained (per reaction) 1 μL of each primer (10 μM concentration), 0.3 μL probe (10 μM concentration), 0.5 μL ROX (1:10 dilution), 4.7 μL nuclease-free water, 12.5 μL Platinum qPCR Supermix-UDG (Thermo Fisher Scientific), and 5 μL of the sample. For study 2, the master mix contained (per reaction) 0.9 μL of each primer (20 μM concentration), 0.5 μL probe (5 μM concentration), 0.3 μL nuclease-free water, 10 μL PerfeCTa® qPCR ToughMix®, Low ROX™ (Quanta Biosciences, Gaithersburg, MD), 2 μL 10X Exo IPC Mix, 0.4 μL 50X Exo IPC DNA, and 5 μL of the sample. For both studies, the amplification protocol consisted of 2 minutes at 50°C and 10 minutes at 95°C, followed by 45 cycles of amplification (95°C for 15 seconds and 60°C for 60 seconds [study 1] or 58°C for 60 seconds [study 2]). PCR-grade water was used as a non-template, negative control, and included in duplicate in each PCR run. Samples were tested in duplicate. Reaction runs were considered valid when both replicates of the L. interrogans control amplified with cycle threshold (Ct) values < 40 and when all replicates of the negative controls showed no evidence of amplification. Samples with a Ct value < 40 were considered positive for pathogenic Leptospira infection.
Table 1.
Real-time PCR methods used to target genes of pathogenic Leptospira spp. among patients hospitalized with fever, Tanzania, 2007–2008 and 2012–2014
| Genus/species | Target gene | Primer/probe name | Sequence (5′-3′) | Annealing temp (°C) | Cycles |
|---|---|---|---|---|---|
| Pathogenic Leptospira spp. | lipL3222 | LipL32-45F | AAG CAT TAC CGC TTG TGG TG | 60 | 40 |
| LipL32-286R | GAA CTC CCA TTT CAG CGA TT | ||||
| LipL32-189P | FAM-AA AGC CAG GAC AAG CGC CG-BHQ1 | ||||
| Leptospira borgpetersenii | ompL17 | F_bpn | GAT TCG GGT TAC AAT TAG ACC | 65 | 45 |
| R_bpn1 | TTG ATC TAA CCG GAC CAT AGT | ||||
| Leptospira interrogans | secY7 | PFLint2 | CTT GAG-CCT GCG CGT TAY C | 63 | 45 |
| PRLint2 | CCG ATA ATT CCA GCG AAG ATC | ||||
| TaqLint2 | TET-CTC ATT TGG TTA GGA GAA CAG ATC A-BHQ1 | ||||
| Leptospira kirschneri | secY7 | F_nery | CTG GCT TAA TCA ATG CTT CTG | 60 | 45 |
| R_nery | CTC TTT CGG TGA TCT GTT CC | ||||
| TqM_nery | Texas Red-CAG TTC CAG TTG TAA TAG ATA AGA TTC-BHQ2 | ||||
| Leptospira noguchii | secY7 | FLnog2 | TCA GGG TGT AAG AAA GGT TC | 63 | 45 |
| RLnog2 | CAA AAT TAA AGA AGA AGC AAA GAT | ||||
| TaqLnog | FAM-CGA TTG GCT TTT TGC TTG AAC CAT C-BHQ1 | ||||
| TqM_bpn | Cy5.5 (Quasar 705)-TAC TAA GGA TGG TTT GGA CGC TGC-BHQ2 | ||||
| Pan Leptospira spp. | SecY23 | SecYFd | 5′-ATG CCG ATC ATY TTY GCT TC-3′ | 52°C | 45 |
| secYR3 | 5′-TTC ATG AAG CCT TCA TAA TTT CTC A-3′ |
Identification of infecting Leptospira species and genotype.
For samples that were positive for pathogenic Leptospira DNA by lipL32 real-time PCR assay, we performed real-time PCR using sets of Leptospira species–specific probes and flanking primers designed for the direct detection of common Leptospira species in clinical samples as shown in Table 1. Primers and probes targeting the secY gene were used for the detection of L. interrogans, L. kirschneri, and Leptospira noguchi. Primers and probes targeting the ompL1 gene were used for the detection of L. borgpetersenii.7 In each run, we included negative controls and species-specific positive controls (L. borgpetersenii strain Ballum Mus 127 DMSO 4/21/15; L. interrogans strain Icterohaemorrhagiae RGA, ATCC 43462; L. kirschneri serovar Cynopteri strain 3522C, ATCC 49945; and L. noguchi strain 2001034031).
For samples that were positive by lipL32 real-time PCR, we also amplified and sequenced a ∼435-base pair (bp) fragment of the secY gene to investigate the infecting genotype within each Leptospira species. PCR amplification was performed using previously published protocols. PCR amplicons were purified using the QIAquick PCR Purification Kit (Qiagen), and Sanger sequencing was performed by Source Bioscience Ltd. (Nottingham, United Kingdom).
Phylogenetic analysis of the secY gene.
We performed phylogenetic analysis of the secY gene using MEGA7.0 software.25 Final sequences were aligned and compared with sequences from reference serovars accessed through GenBank8,26 and with sequences from other studies in Tanzania4,27 using the ClustalW algorithm. Notably, included in the comparison were sequences of Leptospira detected in Tanzanian livestock in the Moshi municipal district during the same time period as study 2 (2013–2014). This included 17 Leptospira sequences derived from cattle samples, one sequence derived from a goat sample, and one derived from a sheep sample.4 We also included published sequences from Leptospira reference serovars,8 and from serovars previously isolated in Tanzania in our final alignment.27 The model test function in MEGA7.0 was used to select the most appropriate nucleotide substitution model for the aligned sequences. The final phylogenetic tree was generated using a maximum likelihood method with 500 bootstraps repeats as a test of relationships between the aligned sequences.
Research ethics.
This study was conducted in accordance with the World Medical Association Declaration of Helsinki. It was approved by the KCMC Research Ethics Committee (#295), the Tanzania National Institutes for Medical Research National Ethics Coordinating Committee (NIMR/HQ/R.8cNo1. 11/283 and NIMR/HQ/R.8a/Vol.IX/1499), Duke Health Institutional Review Board (IRB#Pro00016134), the University of Otago Human Ethics Committee (Health) (H15/055), and the University of Glasgow College of Medical, Veterinary, and Life Sciences Ethics Committee (Project No. 200120020). Written informed consent was obtained from all participants. Study data are available at http://dx.doi.org/10.5525/gla.researchdata.881.
RESULTS
In total, 1,849 participants who were recruited into the two fever etiology studies had blood drawn for leptospirosis serology, and 1,294 (70.0%) had Leptospira blood culture. MAT serological testing for leptospirosis was performed on paired acute and convalescent samples for 1,225 (66.3%) participants, and there were 108 decedents. A total of 109 (5.9%) participants met our selection criteria for real-time PCR testing for Leptospira and had samples available for testing. These included samples from 81 participants who met our serologic case definition of leptospirosis and 28 decedent participants. Of 109 participants, 106 (98.1%) participants had blood derivatives, including plasma in 58 (53.2%) and serum in 48 (44.0%) available for testing, and 30 (27.5%) had urine available for testing.
Data on fever duration and self-reported antibacterial use were available from 108 (99.1%) participants undergoing PCR testing. The median (range) reported duration of the fever before sample collection was 7 (1–366) days. Of participants, 64 (58.7%) reported antibacterial use before enrollment.
Pathogenic Leptospira testing and speciation.
Leptospira culture was negative on all 1,294 participants. By real-time PCR, we detected pathogenic Leptospira DNA in three (3.7%) of 81 participants with serologically confirmed acute leptospirosis and one (3.6%) of 28 decedent participants (Table 2). The decedent participant did not meet our serologic case definition of leptospirosis (low titer in the acute sample and no convalescent sample available). All positive PCR reactions were from serum samples. Of four real-time PCR-positive samples, species-specific real-time PCR identified one (25%) as positive for L. borgpetersenii, one (25%) L. interrogans, and one (25%) L. kirschneri, each in a single study participant (Table 2). In the single decedent sample (25%) that was lipL32 real-time PCR positive, we were unable to amplify DNA using the species-specific real-time PCR reactions.
Table 2.
LipL32 and species-specific RT-PCR CT values, and MAT predominant serogroup in participants with Leptospira DNA detected, Tanzania, 2012–2014
| Participant number | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| PCR | ||||
| LipL32 CT value | 38.7 | 36.3 | 32.3 | 38.4 |
| L. borgpetersenii CT value | ND | 37.8 | ND | ND |
| Leptospira interrogans CT value | 37.4 | ND | ND | ND |
| L. kirschneri CT value | ND | ND | 37.8 | ND |
| Leptospira noguchii CT value | ND | ND | ND | ND |
| Leptospira species (secY sequencing) | NA | L. borgpetersenii | L. kirschneri | NA |
| MAT | ||||
| Predominant MAT serogroup | Sejroe | Pyrogenes | Sejroe | Pyrogenes |
| Acute reciprocal titer | 0 | 0 | 0 | 400 |
| Convalescent reciprocal titer | 200 | 800 | 12,800 | NA |
CT = cycle threshold; L. borgpetersenii = Leptospira borgpetersenii; L. kirschneri = Leptospira kirschneri; MAT = microscopic agglutination testing; NA = not available; ND = not detected.
Comparison between genetic and serological results.
When compared with the MAT serological results, all three participants with real-time PCR-positive samples met the case definition for acute leptospirosis by virtue of a 4-fold rise in MAT titer between the acute and convalescent samples. The predominant reactive serogroup was Sejroe for the participants with L. interrogans and L. kirschneri infection, and was Pyrogenes for the participant with L. borgpetersenii infection (Table 2). For the real-time PCR-positive decedent, a single titer of 1:400 against serogroup Pyrogenes was obtained by MAT.
Leptospira phylogenetic analysis.
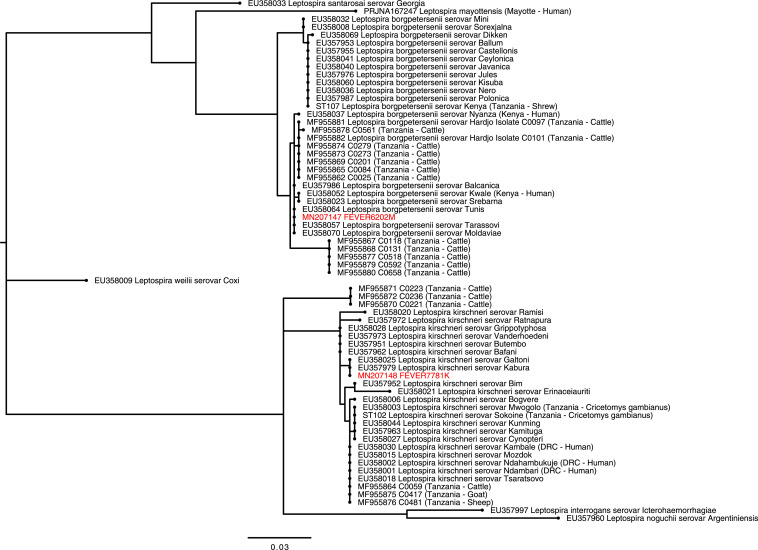
SecY sequences were obtained from two (50.0%) of four participants whose samples were positive by lipL32 real-time PCR. Comparison with published secY reference sequences8 confirmed the infecting Leptospira species as L. borgpetersenii and L. kirschneri (Figure 1, Table 2). When compared with reference serovars, L. borgpetersenii secY sequence from a participant in our study was 100% identical to reference sequences from L. borgpetersenii serovar Balcanica (EU357986), Moldaviae (EU358070), Tarassovi (EU358057), and Tunis (EU358064). L. kirschneri secY sequence generated in our study was 100% identical to sequences obtained from reference serovars including L. kirschneri serovar Galtoni (EU358025), and serovar Kabura (EU 357979).
Figure 1.
Phylogenetic tree showing the relationship between Leptospira secY gene (434-bp fragment) derived from human infections in Tanzania with sequence from reference serovars8 and previously published sequences from human and animal infection in Tanzania4,27 The phylogenetic tree was constructed using the maximum likelihood method using the Tamura 3-parameter (T92) nucleotide substitution model with a discrete gamma distribution.44 The tree with the highest log likelihood is shown. Sequences from our study are labeled with unique identifiers (FEVERXXXX) and GenBank accession numbers and highlighted in red. Sequences generated from reference Leptospira serovars and from other studies in Tanzania are shown and labeled with GenBank accession numbers and ST types, respectively.45 Expanded clades show reference serovars most closely related to the study genotypes. Clades of more distantly related species are collapsed and labeled with species names only. Country locations and host are shown in parenthesis for East African studies. DRC = Democratic Republic of Congo; sv = serovar; ST = sequence types. This figure appears in color at www.ajtmh.org.
Sequences obtained from participants in our study showed not only a high degree of similarity but also some minor distinctions when compared with Leptospira secY sequences obtained from animals in Tanzania (Figure 1).4,27 Leptospira borgpetersenii sequences were highly similar (99.8% of 434 bp) to sequences of L. borgpetersenii serovar Hardjo (Hardjo-bovis) isolated from cattle in the Moshi municipal district, but with a single nucleotide substitution in the human-derived sequence. L. kirschneri sequences were also highly similar (99.1% of 434 bp) but differed by four nucleotide substitutions from genotypes detected in cattle, sheep, and goats sampled in the Moshi municipal district.
DISCUSSION
We detected Leptospira DNA in four participants with acute febrile illness in northern Tanzania, including one patient who died with acute leptospirosis. Despite the small number of real-time PCR-positive samples, our results show a striking amount of diversity in infecting Leptospira species. Three Leptospira species—L. borgpetersenii, L. interrogans, and L. kirschneri—were detected in participants with acute febrile illness. Sequence-based analysis showed a high degree of similarity between secY sequence obtained from L. borgpetersenii and L. kirschneri genotypes infecting people and L. borgpetersenii and L. kirschneri genotypes detected in local cattle, goats, and sheep also sampled in the Moshi municipal district. The findings of our study corroborate previous serological findings that detected multiple Leptospira types are circulating in Tanzania and support the hypothesis that livestock are a potential source of human infection.2,4,13
To date, little information has been available regarding the predominant Leptospira species implicated in human disease in Tanzania.28 PerfeCTa qPCR ToughMix, Low ROX interrogans, and L. kirschneri have previously been implicated as a cause of human infection elsewhere in Tanzania although the methods used in the earlier study lacked sufficient resolution to discriminate to the species level.27 Our approach, which used a combination of species-specific PCR and sequence-based typing, has generated to our knowledge the first species-level identification of pathogenic Leptospira associated with human disease and provides robust evidence that multiple Leptospira types are contributing to human leptospirosis in Tanzania.
Phylogenetic analysis of a partial fragment of the secY gene enabled comparison with L. borgpetersenii and L. kirschneri genotypes identified in local livestock species. Sequences of the Leptospira secY gene obtained from our study participants showed a high degree of similarity to secY sequences obtained previously from livestock in Tanzania. In particular, L. borgpetersenii is commonly carried by cattle in northern Tanzania,4 and the genotype derived from a patient in our study was highly similar to that of L. borgpetersenii sequences obtained from cattle in Tanzania that were sampled during the same time period and same geographic region.4 However, a high degree of similarity was also seen in sequences from three Leptospira reference serovars including L. borgpetersenii serovar Balcanica, previously detected in cattle in Zimbabwe29; L. borgpetersenii serovar Tunis, previously detected from pigs in Tunisia30; and L. borgpetersenii serovar Tarassovi, which has been detected among a wide variety of wildlife and livestock from a broad geographical distribution.31–36 Likewise, the L. kirschneri genotype derived from a human participant in our study was similar to but distinct from sequences derived from livestock in Tanzania.4 This finding suggests a less recent common ancestor between the L. kirschneri genotypes detected in our study and those previously detected in livestock than for L. borgpetersenii genotypes.
Source attribution for Leptospira infection requires a thorough characterization of local Leptospira serovars or sequence types as well as a good understanding of the animal host range and potential transmission pathways for each of these types.37,38 The relatively small amount of published Leptospira sequence data from Tanzania, combined with the limited amount of typing that can be performed for Leptospira in the absence of cultured isolates, means that we are unable to robustly infer the sources of infection for the febrile patients enrolled in our study. However, based on our current understanding of the epidemiology and biology of these Leptospira species, we think that it is plausible that the three different Leptospira spp. detected in patients in our study have distinct epidemiology and transmission pathways. First, the three different Leptospira spp. detected in our study have previously been identified in different animal hosts in Tanzania. To date, L. borgpetersenii has only been detected in cattle and rodents in Tanzania, whereas L. kirschneri appears to have a broader animal host range including cattle, sheep, goats, and rodents.4,27 In addition, other studies have demonstrated biological differences between Leptospira species. For example, L. borgpetersenii has reduced environmental survival compared with L. interrogans, and hence, relies more heavily on direct contact for host-to-host transmission.39 Therefore, we hypothesize that infection transmission may be occurring under different environmental conditions for each of the Leptospira species identified, with important implications for public health strategies to reduce the burden of infection.
Overall, our study was limited by the small number of PCR-positive samples from human participants available for analysis. Our PCR approach had relatively low sensitivity and detected fewer than 4% of acute leptospirosis cases, despite focusing our selection on acute blood and urine samples from participants that met international guidelines serological case definitions for acute leptospirosis.16 Testing the entire cohort with both MAT and PCR is likely to have detected additional cases as although MAT using paired serum is the reference standard for serological diagnosis for leptospirosis, it has imperfect sensitivity as some patients will not seroconvert.17,40–42 Resource limitations preclude testing all participants with both assays.
Many studies report that MAT testing of paired serum samples detected cases that were unable to be detected by PCR,17,40–42 but the proportion of serologically confirmed cases who tested positive by PCR was surprisingly low in our study. One possible explanation for the surprisingly low proportion positive by PCR was that at least half of participants in our studies reported a fever for greater than 7 days before sample collection. PCR has highest sensitivity during the bacteremic phase of leptospirosis during first 5–7 days of symptoms.43 The high reported prevalence of prior antimicrobial use may have further reduced the proportion positive by culture and PCR in our studies by sterilizing blood before enrollment preventing bacterial growth, or reducing leptospiral load below the lower limit of detection for the real-time PCR assay. In addition, we performed the PCR assays on serum and plasma, which may have lower sensitivity than whole blood.21 Despite this limitation, the small amount of molecular data that we obtained has helped to expand our understanding of the diversity of Leptospira present in northern Tanzania and also provided some intriguing insights into the relationship between infecting Leptospira species and serogroup in patients with acute febrile illness. For example, Sejroe was the predominant reactive serogroup in two patients who each were infected with different Leptospira species: L. borgpetersenii and L. interrogans. This suggests that molecular typing may reveal greater diversity than serology alone. Furthermore, the use of Leptospira real-time PCR allowed us to detect a fatal case of leptospirosis in our patient cohort that may otherwise have been undetected and demonstrates the potential value of PCR in understanding the prevalence of fatal leptospirosis.
Our study has provided some new insights into the complexity of Leptospira transmission pathways in Tanzania. Given the relatively small amount of data and the high incidence of human leptospirosis in Tanzania,1,15 we think that further work is needed to determine the diversity of Leptospira species and genotypes contributing to both human and animal infection, and to understand sources of infection and epidemiological transmission sources for human leptospirosis. This could in turn inform efforts to control and prevent human disease.
Based on our findings, we recommend the combined use of both serological and NAAT diagnostic approaches to diagnose and investigate the epidemiology of human leptospirosis. This is particularly important where delays in presentation to healthcare facilities are common, or when there is a high prevalence of prehospital antibacterial use. Additional efforts to obtain Leptospira genetic material from human leptospirosis cases are needed to help prioritize Leptospira species and genotypes for control and critical insights into the links between human and animal infection and help understand transmission of this complex but important pathogen.
Supplemental table
Acknowledgments:
We would like to thank those involved in recruitment, laboratory work, data management, and study administration, including Godfrey S. Mushi, Flora W. Mboya, Lilian E. Ngowi, Winfrida H. Shirima, Michael E. Butoyi, Anna H. Mwalla, Miriam L. Barabara, Ephrasia Mariki, Tumsifu G. Tarimo, Yusuf S. Msuya, Leila J. Sawe, Aaron E. Tesha, Luig J. Mbuya, Edward M. Singo, Isaac A. Afwamba, Thomas M. Walongo, Remigi P. Swai, Augustine M. Musyoka, Philoteus A. Sakasaka, O. Michael Omondi, Enoch J. Kessy, Alphonse S. Mushi, Robert S. Chuwa, Cynthia A. Asiyo, Frank M. Kimaro, and Francis P. Karia. In addition, we would like to thank the study participants as well as the clinical staff and administration at Kilimanjaro Christian Medical Centre and Mawenzi Regional Referral Hospital for their support during this study, and the climate scientists at the Tanzanian Production Company for provision of climate data.
Note: Supplemental table appears at www.ajtmh.org.
REFERENCES
- 1.Biggs HM, Hertz JT, Munishi OM, Galloway RL, Marks F, Saganda W, Maro VP, Crump JA, 2013. Estimating leptospirosis incidence using hospital-based surveillance and a population-based health care utilization survey in Tanzania. PLoS Negl Trop Dis 7: e2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maze MJ, et al. 2018. Risk factors for acute human leptospirosis in northern Tanzania. PLoS Negl Trop Dis 12: e0006372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schoonman L, Swai ES, 2009. Risk factors associated with the seroprevalence of leptospirosis, amongst at-risk groups in and around Tanga city, Tanzania. Ann Trop Med Parasitol 103: 711–718. [DOI] [PubMed] [Google Scholar]
- 4.Allan KJ, et al. 2018. Assessment of animal hosts of pathogenic Leptospira in northern Tanzania. PLoS Negl Trop Dis 12: e0006444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mgode GF, Machang’u RS, Goris MG, Engelbert M, Sondij S, Hartskeerl RA, 2006. New Leptospira serovar sokoine of serogroup Icterohaemorrhagiae from cattle in Tanzania. Int J Syst Evol Microbiol 56: 593–597. [DOI] [PubMed] [Google Scholar]
- 6.Levett PN, 2001. Leptospirosis. Clin Microbiol Rev 14: 296–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferreira AS, Costa P, Rocha T, Amaro A, Vieira ML, Ahmed A, Thompson G, Hartskeerl RA, Inacio J, 2014. Direct detection and differentiation of pathogenic Leptospira species using a multi-gene targeted real time PCR approach. PLoS One 9: e112312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Victoria B, Ahmed A, Zuerner RL, Ahmed N, Bulach DM, Quinteiro J, Hartskeerl RA, 2008. Conservation of the S10-spc-alpha locus within otherwise highly plastic genomes provides phylogenetic insight into the genus Leptospira. PLoS One 3: e2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morey RE, Galloway RL, Bragg SL, Steigerwalt AG, Mayer LW, Levett PN, 2006. Species-specific identification of Leptospiraceae by 16S rRNA gene sequencing. J Clin Microbiol 44: 3510–3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merien F, Portnoi D, Bourhy P, Charavay F, Berlioz-Arthaud A, Baranton G, 2005. A rapid and quantitative method for the detection of Leptospira species in human leptospirosis. FEMS Microbiol Lett 249: 139–147. [DOI] [PubMed] [Google Scholar]
- 11.Dietrich M, Wilkinson DA, Soarimalala V, Goodman SM, Dellagi K, Tortosa P, 2014. Diversification of an emerging pathogen in a biodiversity hotspot: Leptospira in endemic small mammals of Madagascar. Mol Ecol 23: 2783–2796. [DOI] [PubMed] [Google Scholar]
- 12.Hamond C, Pestana CP, Medeiros MA, Lilenbaum W, 2016. Genotyping of Leptospira directly in urine samples of cattle demonstrates a diversity of species and strains in Brazil. Epidemiol Infect 144: 72–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biggs HM, et al. 2011. Leptospirosis among hospitalized febrile patients in northern Tanzania. Am J Trop Med Hyg 85: 275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goris MG, Hartskeerl RA, 2014. Leptospirosis serodiagnosis by the microscopic agglutination test. Curr Protoc Microbiol 32: Unit 12E 5. [DOI] [PubMed] [Google Scholar]
- 15.Maze MJ, et al. 2016. Comparison of the estimated incidence of acute leptospirosis in the Kilimanjaro Region of Tanzania between 2007–08 and 2012–14. PLoS Negl Trop Dis 10: e0005165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.CDC , 2013. Leptospirosis (Leptospira interrogans) 2013 Case Definition. Available at: https://wwwn.cdc.gov/nndss/conditions/leptospirosis/case-definition/2013/. Accessed October 21, 2015. [Google Scholar]
- 17.Limmathurotsakul D, Turner EL, Wuthiekanun V, Thaipadungpanit J, Suputtamongkol Y, Chierakul W, Smythe LD, Day NP, Cooper B, Peacock SJ, 2012. Fool’s gold: why imperfect reference tests are undermining the evaluation of novel diagnostics: a reevaluation of 5 diagnostic tests for leptospirosis. Clin Infect Dis 55: 322–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiagen , 2006. DNeasy® Blood & Tissue Handbook. Hilden, Germany: Qiagen. [Google Scholar]
- 19.Bag S, et al. 2016. An improved method for high quality metagenomics DNA extraction from human and environmental samples. Sci Rep 6: 26775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gill C, van de Wijgert JHHM, Blow F, Darby AC, 2016. Evaluation of lysis methods for the extraction of bacterial DNA for analysis of the vaginal microbiota. PLoS One 11: e0163148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galloway RL, Hoffmaster AR, 2015. Optimization of LipL32 PCR assay for increased sensitivity in diagnosing leptospirosis. Diagn Microbiol Infect Dis 82: 199–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stoddard RA, 2013. Detection of pathogenic Leptospira spp. through real-time PCR (qPCR) targeting the LipL32 gene. Methods Mol Biol 943: 257–266. [DOI] [PubMed] [Google Scholar]
- 23.Ahmed A, Engelberts MF, Boer KR, Ahmed N, Hartskeerl RA, 2009. Development and validation of a real-time PCR for detection of pathogenic Leptospira species in clinical materials. PLoS One 4: e7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tindall BJ, 2014. ATCC 43642 replaces ATCC 23581 as the type strain of Leptospira interrogans (Stimson 1907) Wenyon 1926. Opinion 91. Judicial Commission of the International Committee on Systematics of prokaryotes. Int J Syst Evol Microbiol 64: 3584–3585. [DOI] [PubMed] [Google Scholar]
- 25.Kumar S, Stecher G, Tamura K, 2015. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benson DA, Karsch-Mizrachi I, Clark K, Lipman DJ, Ostell J, Sayers EW, 2011. GenBank. Nucleic Acids Res 40: D48–D53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mgode GF, Machang’u RS, Mhamphi GG, Katakweba A, Mulungu LS, Durnez L, Leirs H, Hartskeerl RA, Belmain SR, 2015. Leptospira serovars for diagnosis of leptospirosis in humans and animals in Africa: common Leptospira isolates and reservoir hosts. PLoS Negl Trop Dis 9: e0004251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muller SK, Assenga JA, Matemba LE, Misinzo G, Kazwala RR, 2016. Human leptospirosis in Tanzania: sequencing and phylogenetic analysis confirm that pathogenic Leptospira species circulate among agro-pastoralists living in Katavi-Rukwa ecosystem. BMC Infect Dis 16: 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feresu SB, Steigerwalt AG, Brenner DJ, 1999. DNA relatedness of Leptospira strains isolated from beef cattle in Zimbabwe. Int J Syst Bacteriol 49: 1111–1117. [DOI] [PubMed] [Google Scholar]
- 30.Bakoss P, Chadli A, 1965. The pig, reservoir of Leptospira mitis in Tunisia. Arch Inst Pasteur Tunis 42: 85–91. [Google Scholar]
- 31.Maze MJ, et al. 2017. The Global Distribution and Host Range of Leptospira Species and Serovars. 10th International Leptospirosis Society Meeting. Palmerston North, New Zealand. [Google Scholar]
- 32.Robinson AJ, Ramadass P, Lee A, Marshall RB, 1982. Differentiation of subtypes within Leptospira interrogans serovars Hardjo, Balcanica and Tarassovi, by bacterial restriction-endonuclease DNA analysis (BRENDA). J Med Microbiol 15: 331–338. [DOI] [PubMed] [Google Scholar]
- 33.Field PR, Santiago A, Chan SW, Patel DB, Dickeson D, Mitchell JL, Devine PL, Murphy AM, 2002. Evaluation of a novel commercial enzyme-linked immunosorbent assay detecting Coxiella burnetii-specific immunoglobulin G for Q fever prevaccination screening and diagnosis. J Clin Microbiol 40: 3526–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carmona-Gasca CA, Lara LL, Castillo-Sanchez LO, Ramirez-Ortega JM, Ko A, Palomera CL, de la Pena-Moctezuma A, 2011. Detection of Leptospira santarosai and L. kirschneri in cattle: new isolates with potential impact in bovine production and public health. Veterinaria Mexico 42: 277–288. [Google Scholar]
- 35.Aragon PR, Jacalne AV, Famatiga EG, 1965. Isolation of Leptospira from rats, dogs and pigs. Philippine J Sci 94: 45–54. [Google Scholar]
- 36.Chernukha YG, Isayeva RA, Mustafayeva NI, 1969. Antigenic properties of some strains of Leptospirae of the Tarassovi serological group. Systematic positiion of the strain Perepelicin and new serological type Vietnam. J Hyg Epidemiol Microbiol Immunol (Prague) 13: 118–125. [PubMed] [Google Scholar]
- 37.Guernier V, Allan KJ, Goarant C, 2017. Advances and challenges in barcoding pathogenic and environmental Leptospira. Parasitology 145: 595–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mather AE, Vaughan TG, French NP, 2015. Molecular approaches to understanding transmission and source attribution in nontyphoidal Salmonella and their application in Africa. Clin Infect Dis 61 (Suppl 4): S259–S265. [DOI] [PubMed] [Google Scholar]
- 39.Bulach DM, et al. 2006. Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. Proc Natl Acad Sci USA 103: 14560–14565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schreier S, Doungchawee G, Chadsuthi S, Triampo D, Triampo W, 2013. Leptospirosis: current situation and trends of specific laboratory tests. Expert Rev Clin Immunol 9: 263–280. [DOI] [PubMed] [Google Scholar]
- 41.Agampodi SB, Matthias MA, Moreno AC, Vinetz JM, 2012. Utility of quantitative polymerase chain reaction in leptospirosis diagnosis: association of level of leptospiremia and clinical manifestations in Sri Lanka. Clin Infect Dis 54: 1249–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thaipadungpanit J, et al. 2011. Diagnostic accuracy of real-time PCR assays targeting 16S rRNA and LipL32 genes for human leptospirosis in Thailand: a case-control study. PLoS One 6: e16236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haake DA, Levett PN, 2015. Leptospirosis in humans. Curr Top Microbiol Immunol 387: 65–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamura K, 1992. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol Biol Evol 9: 678–687. [DOI] [PubMed] [Google Scholar]
- 45.Jolley KA, Maiden MC, 2016. Leptospira spp. MLST Databases. Available at: http://pubmlst/org/leptospira. Accessed February 6, 2019. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.