Abstract
COVID-19 morbidity and mortality are increased via unknown mechanisms in patients with diabetes and kidney disease. SARS-CoV-2 uses angiotensin-converting enzyme 2 (ACE2) for entry into host cells. Because ACE2 is a susceptibility factor for infection, we investigated how diabetic kidney disease and medications alter ACE2 receptor expression in kidneys. Single cell RNA profiling of kidney biopsies from healthy living donors and patients with diabetic kidney disease revealed ACE2 expression primarily in proximal tubular epithelial cells. This cell-specific localization was confirmed by in situ hybridization. ACE2 expression levels were unaltered by exposures to renin-angiotensin-aldosterone system inhibitors in diabetic kidney disease. Bayesian integrative analysis of a large compendium of public -omics datasets identified molecular network modules induced in ACE2-expressing proximal tubular epithelial cells in diabetic kidney disease (searchable at hb.flatironinstitute.org/covid-kidney) that were linked to viral entry, immune activation, endomembrane reorganization, and RNA processing. The diabetic kidney disease ACE2-positive proximal tubular epithelial cell module overlapped with expression patterns seen in SARS-CoV-2–infected cells. Similar cellular programs were seen in ACE2-positive proximal tubular epithelial cells obtained from urine samples of 13 hospitalized patients with COVID-19, suggesting a consistent ACE2-coregulated proximal tubular epithelial cell expression program that may interact with the SARS-CoV-2 infection processes. Thus SARS-CoV-2 receptor networks can seed further research into risk stratification and therapeutic strategies for COVID-19–related kidney damage.
Keywords: ACE inhibitors, acute kidney injury, COVID-19, diabetic nephropathy, molecular networks, proximal tubules, SARS-CoV-2, scRNAseq
Graphical abstract
Editor’s Note.
This is one of several articles we think you will find of interest that are part of our special issue of Kidney International addressing the challenges of dialysis and transplantation during the COVID-19 pandemic. Please also find additional material in our commentaries and letters to the editor sections. We hope these insights will help you in the daily care of your own patients.
Translational Statement.
To understand the overwhelming burden of kidney disease in coronavirus disease 2019 (COVID-19), we mapped the expression of the severe acute respiratory syndrome coronavirus 2 receptor, ACE2, in healthy kidney, early diabetic kidney diseases (DKDs), and COVID-19–associated kidney diseases. Single-cell RNA sequencing of 111,035 cells identified ACE2 predominantly in proximal tubular epithelial cells. ACE2 upregulation was observed in DKD but was not associated with renin-angiotensin-aldosterone system (RAAS) inhibition, arguing against an increased risk of COVID-19 among patients taking RAAS inhibitors. Molecular network analysis linked ACE2 expression to innate immune response and viral entry machinery, thereby revealing potential therapeutic strategies against COVID-19.
Coronavirus disease 2019 (COVID-19) disproportionally affects people with diabetes, hypertension, and kidney disease.1, 2, 3, 4, 5 The underlying molecular and physiologic causes of this association could be as varied as drugs used to treat these conditions, disease biology,6, 7, 8 direct infection of relevant organs by the virus9, 10, 11 and consequent tissue destruction, and the cytokine storm that occurs secondary to infection.3 Upper- and lower-airway tissues are likely the primary sites of infection, but recent data indicate that the viral tropism includes kidney tissue.10 , 12 Furthermore, patients with kidney disease have high mortality rates from COVID-19.10 , 13, 14, 15 Understanding the disease-specific molecular processes in COVID-19 relative to kidney disease and diabetes can have a significant impact on public health.
COVID-19 develops from infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a betacoronavirus with a single-stranded RNA genome. It gains entry into specific cell types through interaction of the viral surface spike protein with a cell surface receptor.16 As with the coronavirus (SARS-CoV)17 , 18 that caused severe acute respiratory syndrome in the early 2000s, angiotensin-converting enzyme 2 (ACE2) is the primary cell-entry receptor for SARS-CoV-2.16 , 19 , 20 ACE2 expression levels correlate with higher risk of SARS-CoV-2 infection.9 , 21 , 22
ACE2, a membrane-bound metallopeptidase, is a master regulator of the renin-angiotensin-aldosterone system (RAAS).23 SARS-CoV and SARS-CoV-2 bind ACE2 on the cell surface.24 , 25 Proteolytic cleavage of the coronavirus spike protein enables fusion with host membranes.26 TMPRSS2 appears to be the primary protease responsible for this cleavage in lung epithelial cells.27 However, SARS-CoV-2 can also be internalized and the S protein can be cleaved in the endosomal compartment by acid-activated proteases such as cathepsin-L and dipeptidyl peptidase 4, both of which are involved in glucose metabolism and immune systems.8 , 20 , 28, 29, 30
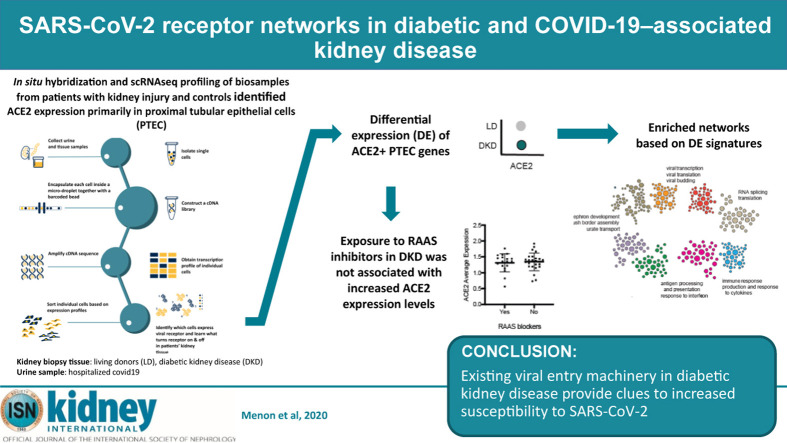
Single-cell RNA sequencing (scRNAseq) of SARS-CoV-2 target tissues provides a way to identify specific cell types with enhanced ACE2 expression and determine whether these cells possess molecular machinery that facilitate viral entry and subsequent virus-induced cytotoxicity. Characterization of these molecular processes could greatly accelerate the identification and development of novel therapeutic options for SARS-CoV-2 infection and noninvasive means for stratifying individuals at risk for COVID-19. Therefore, in the present study, we explored the expression and associated biological processes of ACE2 and other SARS-CoV-2 entry factors in kidney cells from healthy living donors (LDs), patients with diabetic kidney disease (DKD), and COVID-19 patients that required hospitalization (COV). Using in situ hybridization and scRNAseq techniques, we (i) localized the cellular expression of ACE2 in kidneys, (ii) characterized the cellular programs associated with ACE2 expression in DKD and COV, (iii) mapped the ACE2-associated changes to emerging data on SARS-CoV-2–induced cellular responses, and (iv) tested associations with DKD phenotypic characteristics (Figure 1 ). This study included a DKD cohort because COVID-19 disproportionally affects individuals with diabetes and kidney disease.3 , 5 , 8 , 14 , 31 , 32 Side-by-side analysis of the DKD and COV cohorts were aimed at identifying ACE2-associated mechanisms in DKD shared with COVID-19 associated kidney disease.
Figure 1.
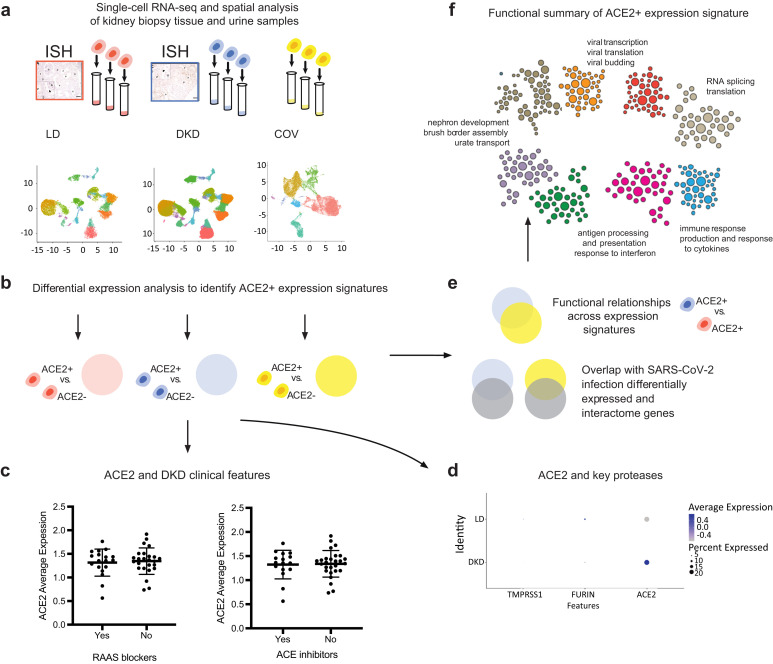
Study overview. To understand how the kidney may be affected by coronavirus disease 2019 (COVID-19) we performed spatial, systems, and clinical association analyses of angiotensin-converting enzyme 2 (ACE2) and other severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) host factors in kidney biopsies from living donors (LD) and patients with diabetic kidney disease (DKD) and kidney cells isolated from the urine of hospitalized COVID-19 patients (COV). (a) Biopsy samples from DKD and LD were processed for in situ hybridization (ISH) and single-cell RNA sequencing (scRNAseq) profiling. scRNAseq of LD, DKD, and urine cell pellets from COV samples were analyzed to determine cell type expression specificity of ACE2 in healthy and disease states. (b) For each scRNAseq dataset, ACE2+ differential expression signatures were identified. (c) Association of ACE2 expression levels in DKD with clinical characteristics were evaluated, including exposure to renin-angiotensin-aldosterone system (RAAS) blockers and ACE inhibitors. (d) Expression of ACE2 and key proteases between LD and DKD proximal tubule epithelial cells (PTECs) were compared. (e) ACE2 expression signatures across datasets identified aspects induced in PTECs expressing DKD samples compared to LD. These gene sets significantly overlapped those reported to be affected by direct SARS-CoV-2 infection. (f) The biological processes in ACE2+ expression signatures were characterized by projecting these signature genes onto PTEC-specific functional networks at HumanBase (https://hb.flatironinstitute.org/covid-kidney). These networks represent genes and their interactions in biological processes and pathways active in PTECs.
Results
Cohort studied for SARS-CoV-2 receptor expression and regulation
Kidney cell expression profiles were obtained from early DKD (n = 44) and LD (n = 18) kidney biopsies and from COV urine samples (n = 13). Clinical characteristics at the time of sample collection are provided for the DKD and COV (Table 1 ) cohorts. Additional details are provided in the Methods and the Supplementary Methods).
Table 1.
Clinical characteristics of DKD and LD cohorts at time of biopsy and COV cohort at time of urine sample collection
| Characteristic | DKD at biopsy (n = 44) | COV at sample collection (n = 13) | LD at biospy (n = 18) |
|---|---|---|---|
| Age, yr | 41 ± 11 | 50 ± 17 | 45 ± 10 |
| Male, % | 32 | 54 | 49 |
| Race, % | |||
| White | 0 | 38 | 88 |
| Black | 0 | 54 | 6 |
| American Indian | 100 | 0 | 0 |
| Other | 0 | 8 | 6 |
| Diabetes duration, yr | 12.2 ± 7.5 | – | – |
| Body mass index, kg/m2 | 36.9 (7.3) | 34.3 | – |
| HbA1c, % | 9.2 ± 2.4 | 7.9 (N = 4) | – |
| Diastolic blood pressure, mm Hg | 72 ± 10 | – | – |
| ACR, mg/g, median [IQR] | 18 [9–53] | – | – |
| iGFR, ml/min | 159 ± 58 | – | 101 ± 17 |
| Use of antihypertensives, % | 43 | 54 | – |
| Use of RASS blockers, % | 43 | 8 | – |
| Use of angiotensin receptor blockers, % | 7 | 8 | – |
| Use of ACE inhibitors, % | 36 | 15 | – |
| Baseline CKD, % | – | 46 | – |
| History of kidney transplant, % | – | 23 | – |
| History of DM, % | 100 | 54 | – |
| On immunosuppressant, % | – | 23 | – |
| Time since first positive COVID-19 test, d [IQR] | – | 11 [5–29] | – |
| AKI by KDIGO, % | – | 62 | – |
| KDIGO stage 1 | – | 25 | – |
| KDIGO stage 2 | – | 0 | – |
| KDIGO stage 3 | – | 75 | – |
| Renal recovery, % | – | 38 | – |
| COVID-19 treatment features, % | – | – | |
| Need for RRT | – | 23 | – |
| Indwelling urinary catheter | – | 69 | – |
| ICU admission | – | 85 | – |
| Intubation | – | 85 | – |
| ECMO | – | 23 | – |
ACE, angiotensin converting enzyme; ACR, albumin/creatinine ratio; AKI, acute kidney injury; CKD, chronic kidney disease; COV, coronavirus disease 2019 cohort; COVID-19, coronavirus disease 2019; DKD, diabetic kidney disease; DM, diabetes mellitus; ECMO, extracorporeal membrane oxygenation; ICU, intensive care unit; iGFR, iothalamate-measured glomerular filtration rate; KDIGO, Kidney Disease: Improving Global Outcomes; LD, living donor; RAAS, renin-angiotensin-aldosterone system; RRT, renal replacement therapy.
Continuous variables are presented as mean (SD).
scRNAseq-based definition of SARS-CoV-2 receptor expression in kidney tissue
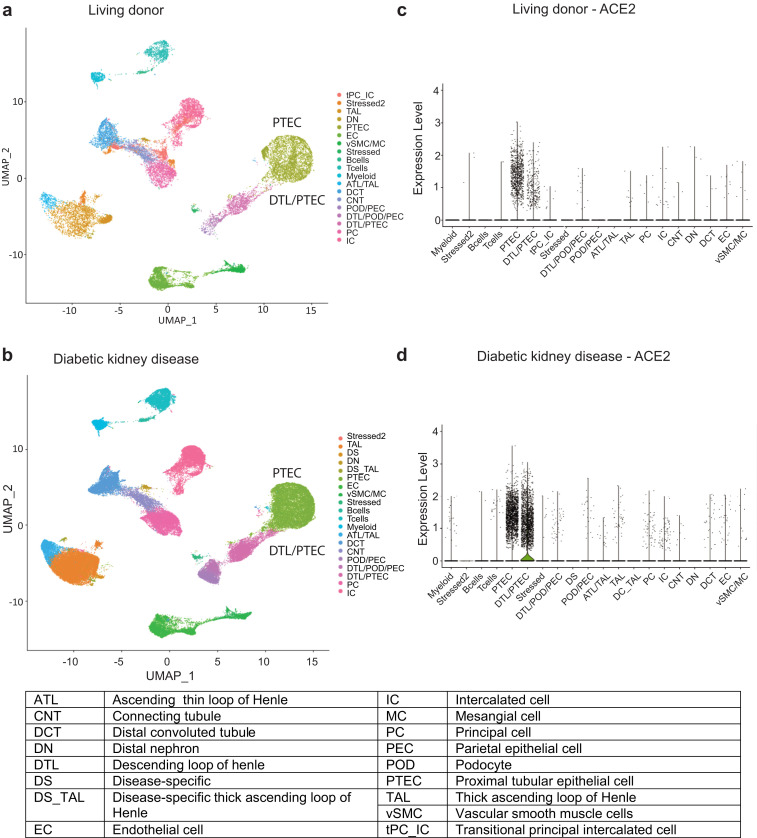
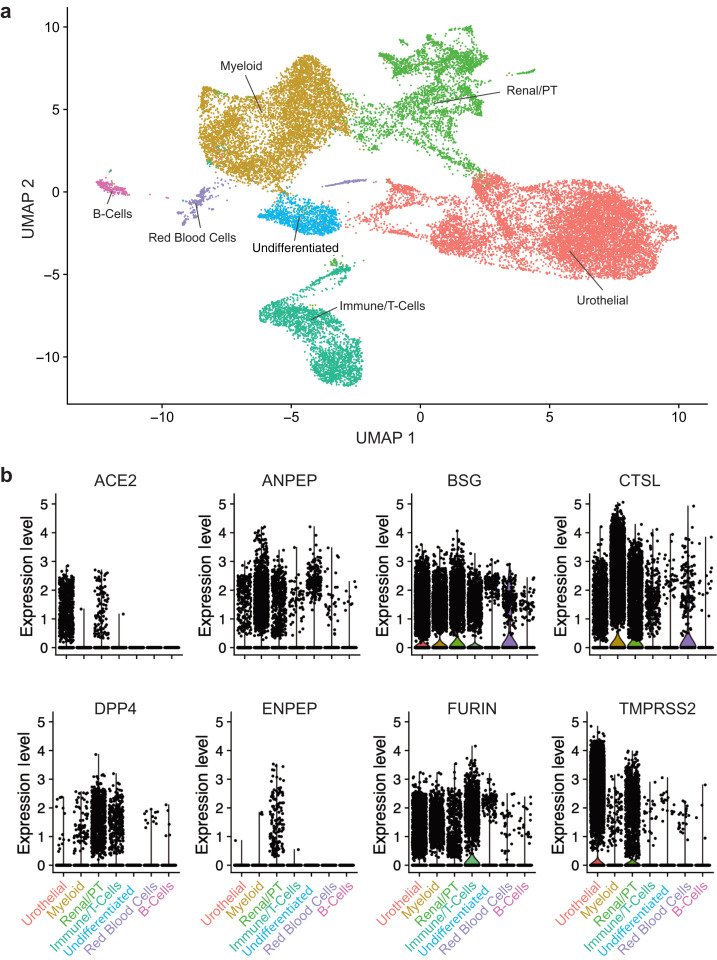
To define the expression pattern of ACE2, cell populations obtained by scRNAseq analysis of kidney biopsies (44 DKD and 18 LD) were clustered based on the transcription profiles of individual cells. A combined analysis of 111,035 cells from the 2 data sets with a resolution (clustering granularity) of 0.6 defined 21 cell clusters (Figure 2a and b, UMAP plots). Cells from both sample sources populated 18 of these, indicating the presence of cells with similar identity in both sample sources (Supplementary Figure S1). The 18 cell clusters covered the entire spectrum of kidney cell types found along the nephron- and tissue-resident immune cells. Furthermore, we identified 2 DKD-specific clusters (disease-specific and disease-specific thick ascending loop of Henle) and 1 LD-specific cluster (transitional principal cell-intercalated cluster). The violin plots in Figure 2c and d show a restricted cell-type-specific mRNA expression of ACE2 in proximal tubular epithelial cells (PTECs), identified using standard transcript markers including cubilin.33 ACE2, in both LD and DKD, was predominantly expressed in cells expressing cubilin, found in 2 clusters: PTEC and a hybrid cluster containing both distal limb of loop of Henle and PTEC. For further analysis, we focused on the cubilin-positive PTEC in these 2 clusters.
Figure 2.
Unsupervised clustering of cells from living donors (LD) and patients with diabetic kidney disease (DKD). UMAP plots showing the distributions in unsupervised clustering of (a) 25,163 LD kidney cells into 19 clusters and (b) 85,872 cells from DKD into 21 clusters; 22% of the total cells in both DKD and LD were cubilin-positive proximal tubular epithelial cells (PTECs). The violin plots for (c) LD and (d) DKD show cell type specificity of ACE2 expression. DTL, distal limb of loop of Henle.
Localization of ACE2 in kidneys
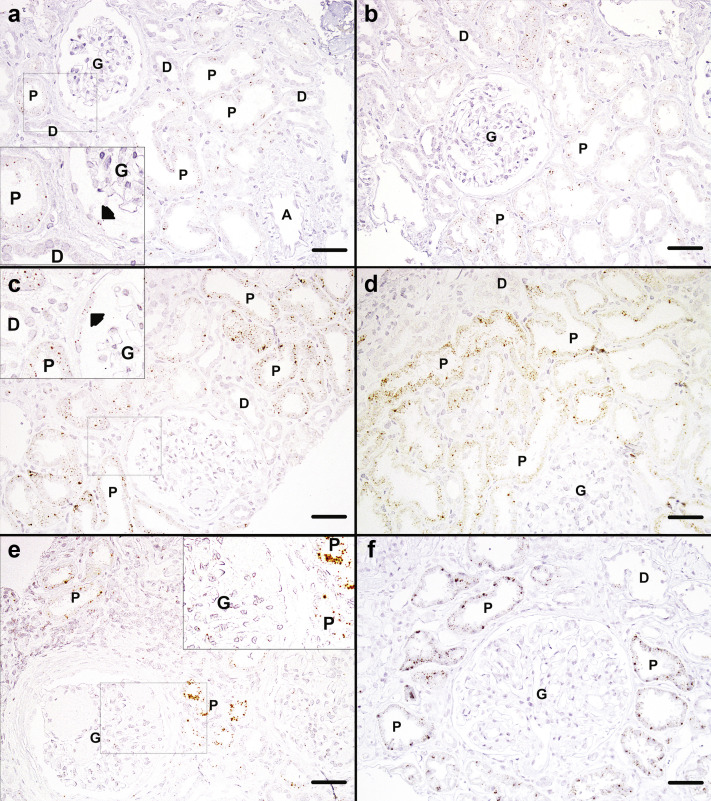
Cellular localization of ACE2 transcripts via in situ hybridization in the kidney is shown in Figure 3 and is consistent with ACE2 expression only in proximal tubules and focally in parietal cells. Representative images from 2 control kidney biopsies (taken from 2 LD kidneys at time of transplantation) after in situ hybridization with the use of ACE2 specific probes revealed punctate signal of ACE2 expression in PTECs, but not in distal tubular cells, glomeruli, or arterioles (Figure 3a and b). A few parietal cells also showed positive signal (arrowhead in inset of Figure 3a). A clear increase in ACE2 signal in PTEC was evident in representative images from 2 kidney biopsies with mild features of DKD (Figure 3b and c) and 2 with advanced DKD (Figure 3d and e). Focally parietal cells also showed increased signal (Figure 3c inset), while other cell types remained negative. Loss of signal was seen in areas of interstitial fibrosis and tubular atrophy (Figure 3e), concomitant with loss of proximal cells in advanced DKD.
Figure 3.
In situ detection of angiotensin-converting enzyme 2 (ACE2) in deidentified living donor (LD) and diabetic kidney disease tissue.In situ hybridization for ACE2 mRNA in (a,b) 2 LD reference control biopsies, (c,d) 2 renal biopsies from patients with mild diabetic nephropathy, and (e,f) 2 cases of advanced diabetic nephropathy. Small brown dots representing ACE2 mRNA transcripts are seen scattered at low density in proximal tubules (PTs) of control biopsies (a,b) and at higher density in mild (c,d) and advanced (e,f) diabetic nephropathy. Small brown dots can also be seen in parietal cells in control biopsies (a) and mild diabetic nephropathy (c). No ACE2 transcript signal is seen in distal tubules (D), glomeruli (G), or arterioles (A). Original magnification ×200. Bars = 50 μm. Arrowheads indicate parietal cells. To optimize viewing of this image, please see the online version of this article at www.kidney-international.org/.
Defining proximal-tubule ACE2 coregulated gene programs
To define the functional context of ACE2 expression in PTECs, we first identified genes specifically expressed in PTECs that express ACE2 (ACE2+). To this end, we first compared the expression profiles of ACE2+ with PTECs with no detectable ACE2 (ACE2−) in LD and DKD. The resulting gene expression signatures (ACE2+ signatures) consisted of genes with increased expression in ACE2+ compared with ACE2− in LD (LD ACE2+ signature; Supplementary Table S1) and DKD (DKD ACE2+ signature; Supplementary Table S2).
To define the DKD-specific component of the ACE2+ signature, we next identified disease-induced genes, i.e., those upregulated in DKD ACE2+ compared with LD ACE2+ (DKD-induced ACE2+ signature; Supplementary Table S3). Table 2 provides a summary of the key ACE2+ gene signatures defined in this study.
Table 2.
ACE2+ signatures defined from single-cell sequencing data
| Signature | Definition | Figure | Table |
|---|---|---|---|
| LD ACE2+ signature | Genes with increased expression in ACE2+ vs. ACE2− PTECs in LD | Not shown | Supplementary Table S1 |
| DKD ACE2+ signature | Genes with increased expression in ACE2+ vs. ACE2− PTECs in DKD | Figure 4a | Supplementary Table S2 |
| DKD induced ACE2+ signature | Genes with increased expression in DKD ACE2+ PTECs compared with LD ACE2+ PTECs | Supplementary Figure S2 | Supplementary Table S3 |
| COV ACE2+ signature | Genes with increased expression in ACE2+ vs. ACE2− PTECs in COV | Figure 7a | Supplementary Table S10 |
ACE2+, angiotensin-converting enzyme 2–positive; COV, coronavirus disease 2019–associated disease that required hospitalization; DKD, diabetic kidney disease; LD, living donor; PTEC, proximal tubular epithelial cell.
Functional characterization of the ACE2-coregulated gene programs in proximal tubular epithelial cells
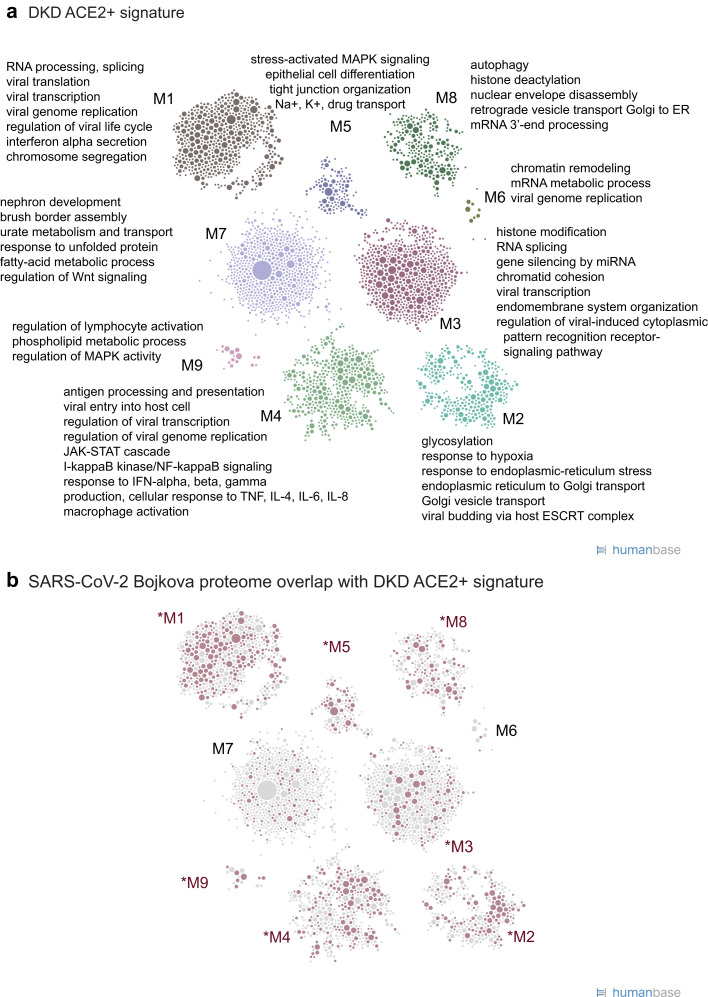
To functionally characterize the molecular machinery expressed in ACE2 + cells in DKD, we projected the DKD ACE2+ signature genes onto the HumanBase functional network that represents biological processes and pathways active in PTECs. Intuitively, this functional network is constructed by probabilistically integrating a large compendium of thousands of public omics datasets to predict the likelihood of 2 genes acting together in processes in PTECs.34 , 35 We clustered the DKD ACE2+ signature genes within this PTEC functional network (Figure 4a36 ; Supplementary Table S2), and the resulting modules (Figure 4a; Supplementary Table S4) contained key processes of tubular function and failure (M7) as well as disease signals (M3–M5, M7). Importantly, we also identified key processes important to both host response and viral replication, including viral entry and genome replication (M1, M4), viral gene transcription (M1, M3), and endomembrane organization and transport (M2–M4, M7). Immune processes were significantly enriched across multiple modules and encompassed innate immune responses (M1, M4) and both production and response to tumor necrosis factor, interferons, and cytokine and macrophage activation (M4). Thus, this signature contains both key elements of documented DKD pathophysiology and processes generally associated with viral infections.37 , 38
Figure 4.
Functional summary of diabetic kidney disease (DKD) angiotensin-converting enzyme 2–positive (ACE2+) expression signature. (a) ACE2+ coexpression signatures were used for community clustering in a positive proximal tubular epithelial cell–specific functional network to identify enriched processes and pathways in DKD biopsy samples. (b) Highlight of ACE2+ signature genes (red circles) shared with the set of proteins that increase expression in response to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection (Bojkova proteome36; Supplementary Table S6). Red text indicates modules with the strongest overlap (P < 0.05), highlighting modules and processes in the ACE2+ signature relevant to SARS-CoV-2 infection.
The signature above describes processes in ACE2+ compared with ACE2− PTECs in DKD. To discover which aspects of DKD could be linked to a more severe COVID-19 disease progression, we identified genes upregulated in ACE2+ DKD compared with LD (Supplementary Table S3). Analysis of these genes demonstrated prominent signals reflecting induction of processes such as inflammation and immune signaling, regulation of kidney size, and morphogenesis of kidney epithelium, which are consistent with reported impacts of diabetic pathology on proximal tubules and PTECs (Supplementary Table S5; Supplementary Figure S2A). Notably, many genes and processes connected to RNA splicing and several aspects related to viral biology were also induced in DKD.
We compared the DKD ACE2+ signature genes with published SARS-CoV-2–relevant gene sets20 , 36 , 39 , 40 (see Methods). In each comparison, a significant fraction of each of the SARS-CoV-2 gene sets was shared with the DKD ACE2+ signature (Table 3 ), suggesting a set of common responses to SARS-CoV-2 infection shared with the DKD ACE2+ signature. Specifically, a significant fraction of the proteins that change expression in response to SARS-CoV-2 infection36 are also DKD ACE2+ signature genes (40%; P < 2.2 × 10−16), showing consistency at both levels of regulation (Figure 4b; Table 3; Supplementary Table S6). Processes in nearly all of the modules of the DKD ACE2+ signature contained genes that were perturbed in SARS-CoV-2–infected cells (Supplementary Table S7). However, the module most relevant to kidney biology (M7) was not enriched in SARS-CoV-2 processes (1.17-fold enrichment; P = 0.048), reflecting the kidney specificity of the DKD ACE2+ signature. The transcripts in the DKD-induced ACE2+ signature (Supplementary Figure S2A; Supplementary Table S3) also significantly overlapped with specific SARS-CoV-2 infection gene sets (Table 3). The shared signals are focused on translation, antiviral responses, and antigen presentation and related signaling (as evidenced by significant enrichment of the SARS-CoV-2 infection gene set from Bojkova et al. 36 in individual modules M1, M2, and M5 in Supplementary Figure S2B [red nodes] and Supplementary Table S8). Taken together, these functional analyses suggest that expression programs active in ACE2-expressing PTECs in DKD could interact with viral infection and modulate host response.
Table 3.
Overlap between ACE2 expression signature lists and previously reported SARS-CoV-2–associated gene sets
| No. of genes in overlap | DKD ACE2+ signature (3734) | DKD-induced ACE2+ (839) | COV ACE2+ signature (2895) |
|---|---|---|---|
| Bojkova_proteome (2666) | 1073 (40%) | 185 (7%) | 809 (30%) |
| Gordon_interactome (332) | 123 (37%) | 4 (1%) | 84 (25%) |
| Zhou_interactome (119) | 53 (44.5%) | 3 (3%) | 33 (28%) |
| Blanco-Melo_NHBE (553) | 126 (23%) | 37 (7%) | 76 (14%) |
| Fold enrichment | |||
| Bojkova_proteome (2666) | 2.16 | 1.65 | 2.10 |
| Gordon_interactome (332) | 1.98 | 0.29 | 1.75 |
| Zhou_interactome (119) | 2.39 | 0.60 | 1.92 |
| Blanco-Melo_NHBE (553) | 1.22 | 1.59 | 0.95 |
| P value | |||
| Bojkova_proteome (2666) | <2.2 × 10−16 | 1.01 × 10−12 | <2.2 × 10−16 |
| Gordon_interactome (332) | 1.78 × 10−15 | 0.9996 | 1.21 × 10−7 |
| Zhou_interactome (119) | 7.95 × 10−11 | 0.88 | 0.00012 |
| Blanco-Melo_NHBE (553) | 0.0079 | 0.0036 | 0.71 |
ACE2, angiotensin-converting enzyme 2–positive; COV, coronavirus disease 2019; DKD, diabetic kidney disease; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
The size of each gene set is indicated in parentheses with its name.
Association of ACE2 mRNA expression in proximal tubular epithelial cells with clinical features
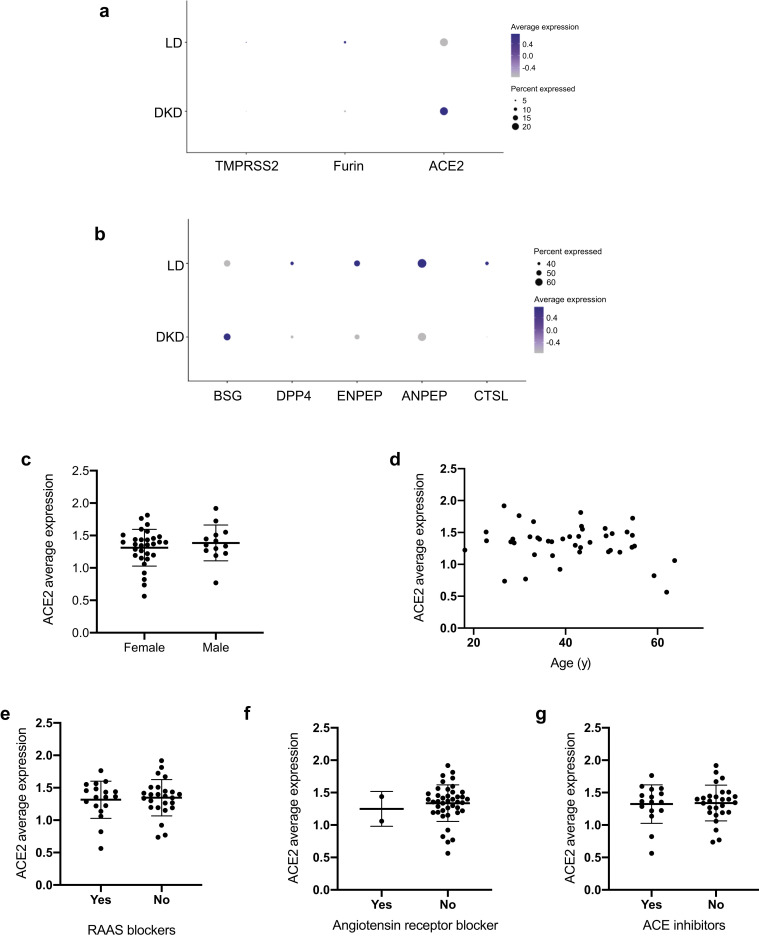
We analyzed the association between ACE2 gene expression levels and clinical measures of DKD to identify potential contributors to elevated ACE2 expression. Higher expression of ACE2 was observed in cubilin-positive PTECs in DKD compared with LD (Wilcoxon rank sum test, P < 0.009; log fold change = 0.05; Figure 5 a). Meanwhile, proteases implicated in coronavirus infection,39 , 41 including ANPEP, BSG, cathepsin-L, dipeptidyl peptidase 4, ENPEP, FURIN, and TMPRSS2, also were detected at varying levels in PTECs in LD and DKD (Figure 5b). In testing for individual-level associations with clinical characteristics, ACE2 expression levels in PTECs per participant was associated with neither baseline characteristics in the DKD cohort (e.g. age and sex; Figure 5c and d) nor treatment exposures to RAAS inhibitors (Figure 5e–g). No consistent relationships were found between ACE2 expression in PTECs and parameters of structural injury across the spectrum of DKD available to us in this cohort.
Figure 5.
Severe acute respiratory syndrome coronavirus 2–related host gene expression in diabetic kidney disease (DKD) biopsy samples and relationships to clinical phenotypes. (a,b) Expression of angiotensin-converting enzyme 2 (ACE2) and related proteases in proximal tubular epithelial cells (PTECs). Dot plots showing the expression of ACE2 and proteases in PTECs from DKD and LD tissue according to single-cell analysis. Color indicates expression level, and the size of the dot indicates the percentage of cells expressing the gene. Proteases were grouped into 2 sets based on the overall expression pattern in proximal cells (a, expression in <30% of the cells; b, expression in ≥30% of the cells). (c–g) Clinical associations of ACE2 expression in DKD. Relationships between ACE2 expression in PTECs and (c) biological sex, (d) age, (e) exposure to any renin-angiotensin-aldosterone-system (RAAS) blockers, (f) exposure to angiotensin receptor blockers, and (g) exposure to ACE inhibitors were examined. No significant correlations between clinical factors and ACE2 expression were found in the DKD cohort.
Transcriptional profile of urine-derived PTECs from COVID-19 patients
Finally, to determine the COVID-19–induced cellular responses in kidneys, we used scRNAseq to analyze urine-derived PTECs from patients hospitalized for COVID-19. Urine from 13 COV patients yielded 25,791 cells that passed our quality control threshold. Unsupervised clustering produced 7 clusters: a PTEC/kidney cell, 3 immune cell types, a red blood cell, a urothelial cell, and an undifferentiated cell cluster (Figure 6 a; Supplementary Figure S3; Supplementary Table S9 for cluster markers). Of the cells in the PTEC cluster, 13% expressed ACE2, and proteases including ANPEP, BSG, cathepsin-L, dipeptidyl peptidase 4, ENPEP, FURIN, and TMPRSS2 also were expressed in this cluster (Figure 6b).
Figure 6.
Single-cell analysis of cells isolated from the urine of patients hospitalized with coronavirus disease 2019. (a) UMAP plot showing the 7 clusters identified from 25,791 cells. (b) Violin plots of severe acute respiratory syndrome coronavirus 2–associated proteases ACE2, ANPEP, BSG, CTSL, DPP4, ENPEP, FURIN, and TMPRSS2 in the 7 identified cell types.
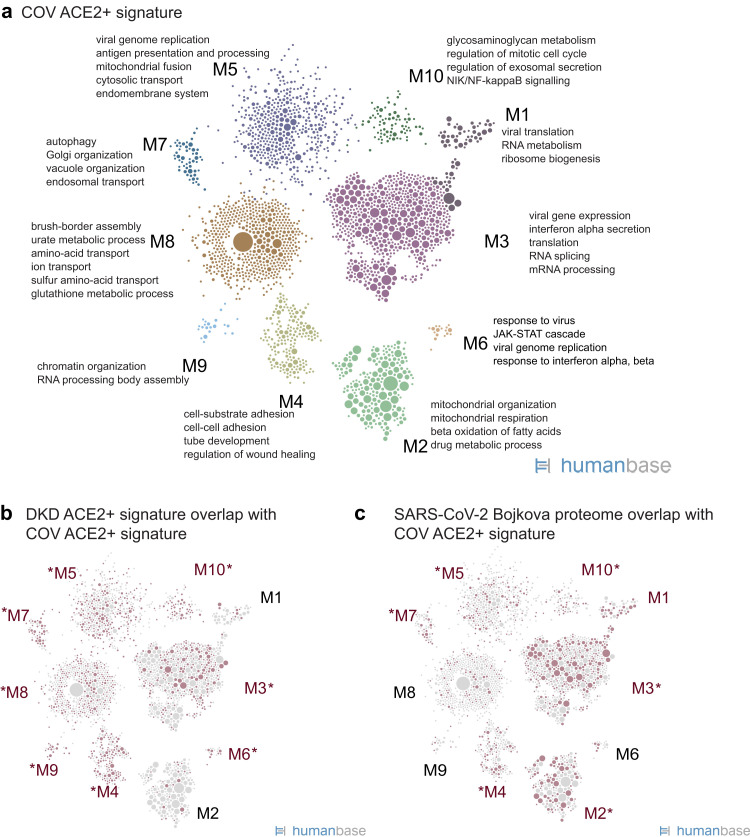
Functional network analysis of the COV ACE2+ signature identified 10 modules, with M3, M5, and M6 enriched for viral processes, including viral genome replication and viral gene expression (Figure 7 a36; Supplementary Tables S10 and S11). Cellular functions in the modules also reported to be critical for coronavirus infections were identified, such as translation, endomembrane system remodeling, and RNA metabolism pathways (M1, M3, M5, M7). Modules enriched in mitochondrial pathways indicative of cell stress were also present in the signature (M2, M7), and PTEC-specific processes (M8).
Figure 7.
Functional summary of angiotensin-converting enzyme 2–positive (ACE2+) expression signature in kidney cells isolated from the urine of hospitalized corona virus disease 2019 (COVID-19) patients (COV). (a) Community clustering and enriched processes of the ACE2+ expression signature in proximal tubule epithelial cells (PTECs) isolated from COV. (b) Highlight of genes (red circles) from the DKD ACE2+ signature (Figure 4a; Supplementary Table S2) that overlap the ACE2+ signature in PTEC from COVID-19 patients. Red text indicates modules with the strongest gene overlap (P < 0.0105), thus indicating ACE2+-associated processes shared between PTECs in diabetic kidneys and PTECs in the context of COVID-19. (c) Highlight of ACE2+ signature genes (red circles) shared with the set of proteins that increase expression in response to SARS-CoV-2 infection (Bojkova proteome36; Supplementary Table S6).
Overall, the COV ACE2+ signature significantly overlapped the DKD ACE2+ signature (P < 2.22 × 10−16; 30% of DKD ACE2+ signature genes are also in COV ACE2+ signature) and was functionally consonant with processes critical for viral infection and immune response (processes represented in starred modules showed significant enrichments in Figure 7b and Supplementary Tables S12 and S13). A module specific to the COV ACE2+ signature was enriched in mitochondrial pathways (M2), which may reflect a stress signal in kidney cells shed in the urine (Supplementary Table S14). A more extensive set of interferon receptors, interferon-stimulated genes, and cytokines were differentially expressed in the COV ACE2+ signature (Supplementary Table S10) compared with the DKD ACE2+ signature. In addition, the COV ACE2+ signature also had significant concordance with other reported SARS-CoV-2 datasets20 , 36 , 39 , 40 (Table 3), sharing key viral processes such as viral regulation, interferon response, stress signaling, and endomembrane organization and transport (as evidenced by significant enrichments of the SARS-CoV-2 infection gene list36 in COV ACE2+ signature modules M3, M5, M6, and others, as depicted in Figure 7c and listed in Supplementary Table S14). This is further confirmed by examination of the processes that are enriched when the PTEC network is clustered with only the shared genes (intersection of COV ACE2+ signature and the SARS-CoV-2 infection set from Bojkova et al. 36 (Supplementary Table S15).
Discussion
With the pandemic spread of SARS-CoV-2 and the increased morbidity and mortality of COVID-19–associated disease in patients with diabetes and kidney disease, it is imperative to define the underlying mechanisms that would promote rapid development of risk reduction strategies. Autopsy studies of patients with COVID-19 as well as susceptibility and infection of ex vivo kidney cultures are consistent with a direct SARS-CoV-2 infection of the kidney tissue with viral tropism being limited to specific renal cell types.10 , 15 , 41 ACE2, the primary SARS-CoV-2 receptor, governs RAAS and associated pathways critical to kidney function. Therefore, we studied the cell types expressing ACE2 in kidney and the corresponding activated functional programs with the goal of uncovering mechanisms that underlie viral susceptibility.
ACE2 expression was localized, in kidneys, predominately to PTECs in LD and DKD. The only other nephron segment with low ACE2 expression were parietal glomerular epithelial cells, which is consistent with their shared developmental origin with PTEC. In contrast to published murine data,42, 43, 44, 45, 46, 47 no significant expression was seen in glomerular podocytes or endothelial cell compartments. Consistent with our findings, PTEC appear to be the main cell type for SARS-CoV-2 infection in the kidney.10 , 12 , 15 Unlike in other organ systems, little coexpression of ACE2 and TMPRSS2 was observed, but robust coexpression of other cell surface proteases, including cathepsin-L, which is associated with viral infection and uptake, were detected.15 , 41 The viral mRNA signal in the glomerular compartment detected by Puelles et al. may represent intracellular viral particles gaining access to glomerular cells via an ACE2-independent mechanism.12
Even though none of the DKD cohort were infected with SARS-CoV-2 at the time of biopsy, our pathway analysis showed that processes including viral infection, protein processing, and antigen presentation were enriched in ACE2-expressing PTECs in DKD. ACE2 is coexpressed in PTECs with a set of genes that also function in establishing viral replication, host responses, and innate immunity. These results and the consistent functional themes observed between DKD and COV ACE2+ signatures suggest that the PTEC gene sets coexpressed with ACE2 in DKD may establish a cellular program that interacts with processes induced by viral infection and host immune response. The ACE2-associated pathways could interact in 2 ways with SARS-CoV-2 infections: (i) The upregulation of viral infection pathways in DKD could explain the higher susceptibility of this patient population; and (ii) if viral infection of ACE2+ PTECs further activates pathways already increased in diabetes, this cumulative and exacerbating activation might lead to kidney damage.
The ACE2 program presents several testable hypotheses for studying the cytopathology of infection and the influence of commonly used diabetes and hypertension drugs. For example, ACE2+ signature genes that are also reported to change expression in response to SARS-CoV-2 infection (highlighted in Figure 4c and listed in Supplementary Table S6 [“Bojkova proteome list”]) may directly interface with SARS-CoV-2 infection in kidney PTECs. We clustered the PTEC functional network with this overlap gene list and identified enriched pathways (Supplementary Table S16). Genes involved in endomembrane transport and viral gene expression could facilitate viral entry, replication, and exit48 (see the Supplementary Discussion for more details). Inflammation and interferon response genes in the list include IFNGR1, ILF3, TNFAIP2, and TNFAIP8, and successful host defense against SARS-CoV-2 may hinge on their activity. It is intriguing that a number of the ACE2+ DKD signature genes encode for proteins that may interact with SARS-CoV-2 proteins,39 including the membrane “M” protein (which suppresses type 1 interferon and cytokine responses) and others (Supplementary Discussion). If these physical interactions occur during infection and diminish host defense, ACE2+ PTECs could be impaired in their ability to mount an appropriate host response, providing one explanation for increased susceptibility. A number of commonly used drugs may influence the ACE2+ coexpression signature. Therapeutic targets in the DKD ACE2+ signature (Figure 4a) include insulin receptor (M4), insulin-like growth factor 1 receptor (M4), and dipeptidyl peptidase 4 (M7). Dipeptidyl peptidase 4 is a protease associated with the entry of other coronaviruses and a key factor in glucose metabolism.8 , 30 Studies in the kidney organoid model confirm expression of ACE2 in the correct functional context of tubular cell clusters, and support ACE2-mediated SARS-CoV-2 infection and subsequent reduced cytotoxicity on addition of soluble ACE2.49 Combining this organoid system with the human interaction maps generated in this study can form an experimental framework for mechanistic studies on the cytopathology of infection and influence of commonly used drugs.
The inflammatory signals observed in the DKD PTECs, expected based on prior studies in this and other cohorts,50 , 51 could be maladaptive for viral infection. Recent results from COVID-19 pulmonary samples described impaired immune responses,52 , 53 and work on SARS-CoV-2–infected cultured lung epithelial cells specifically showed a blunted interferon response despite robust cytokine production.40 Thus, there may be immune hallmarks associated with SARS-CoV-2 pathology. If a similarly asymmetric inflammatory response is already present in DKD kidneys at time of infection, viral infection could amplify the stress responses, increasing cytopathology and enabling further viral propagation. PTECs shed in urine from COV, presented here, showed an interferon activation and response signature, indicating that interferon response to SARS-CoV-2 infection can be robust in the kidney.
As data emerged on the role of ACE2 as a receptor for SARS-CoV-2, significant concerns were raised about risks of RAAS inhibitors, frequently prescribed in patients with diabetes and chronic kidney diseases. Based on nonhuman animal models, RAAS inhibitors were predicted to increase ACE2 levels thereby elevating COVID-19 morbidity and mortality.31 , 54 , 55 However, our findings in this DKD cohort counter such predictions by providing evidence for an absence of RAAS inhibition–associated ACE2 expression induction in PTECs. Meanwhile, a series of case-control, database, and electronic health record studies also did not find any association of RAAS inhibitors with poor outcomes in 3 independent cohorts of patients with COVID-19 and RAAS exposures.56, 57, 58
Studies on regulation of ACE2 gene expression in kidney disease, to date, have been inconclusive.44 , 59, 60, 61 A strength of the present study is the localization of the ACE2 expression to the cellular context of PTECs in kidney disease and COVID-19, consistent with published work from reference kidney tissue12 , 62 , 63 and the Human Protein Atlas64 (https://www.proteinatlas.org; Supplementary Figure S4). The resolution from single-cell studies is critical because disease-associated loss of PTECs can confound bulk tissue analysis.
This study was limited to assessing mRNA levels, which capture only one of several levels of regulation of ACE2 function. Our analyses, therefore, focused on the changes in transcriptional programs as a functional readout and not protein activity. However, our mRNA findings for ACE2 expression are consistent with protein expression specificity from the Human Protein Atlas and the ACE2 signatures with reported SARS-CoV-2 infection gene sets at both mRNA and protein levels. We restricted our analysis to ACE2, the best characterized SARS-CoV-2 receptor. As additional receptors or mechanisms of SARS-CoV-2 cellular entry are discovered, our data sets can be mined for regulation and coexpression in a disease-specific context.
In summary, the present work identifies the regulation and associated cellular machinery of ACE2 and associated SARS-CoV-2 coreceptors in PTECs in kidney health and metabolic and viral disease.
The SARS-CoV-2 receptor associated networks are now available (https://hb.flatironinstitute.org/covid-kidneyhb.flatironinstitute.org/covid-kidney) to seed further research into urgently needed therapeutic strategies for COVID-19.
Methods
Study population
Diabetic kidney disease biopsy cohort
An early DKD cohort was selected as the discovery cohort, given the impact of COVID-19 on individuals with diabetes and kidney disease.3 , 5 , 8 , 14 , 31 , 32 Patient characteristics are provided in Table 1 and the Supplementary Methods.
Living donor biopsy cohort
Reference healthy tissue, 18 LD biopsies, were obtained before perfusion and before placement in the recipient. Participant mean age was 45.1 ± 10.2 years (range 30–66), 11 of the 18 donors were female (61%), mean spot urine protein-to-creatinine ratio was 0.08 ± 0.04 g/g with range (0.03–0.20), and mean iothalamate glomerular filtration rate (GFR) was 100.6 ± 16.9 ml/min per 1.73 m2 (range 81–144). Fifteen of 17 donors were white, 1 of Hispanic descent, and 1 of African ancestry.
COVID-19 cohort
The procurement protocol of urinary cells for single-cell analysis is described in the Supplementary Methods, and baseline characteristics of the study participants are summarized in Table 1.
Kidney biopsy sample processing for single cell
Single-cell transcriptomes were generated with 2–3 mg of the biopsy core samples from 62 CryoStor (Stemcell Technologies) preserved DKD and LD biopsies. Tissue processing and single-cell isolation were performed according to our published protocol65 and as detailed in the Supplementary Methods and at https://www.kpmp.org/for-researchers#protocols.
Urine cell preparation for scRNAseq in patients with COVID-19
Urine single-cell preparation followed the protocol published by Arazi et al. 66 with various modifications as detailed in the Supplementary Methods.
scRNAseq data generation and analysis
scRNAseq data generation followed methods developed for the Kidney Precision Medicine Project (KPMP) and are described in detail in the Supplementary Methods and at https://www.kpmp.org/for-researchers#protocols.65 , 67
In situ hybridization of ACE2 in control and diabetic kidney biopsies
In situ detection of ACE2 mRNA transcripts was performed with the use of RNAscope Probe Hs-ACE2 (Advanced Cell Diagnostics, catalog no. 848151) according to the manufacturer’s protocol by kidney pathologists able to identify cell types in kidney tissue. Housekeeping gene ubiquitin C (RNAscope Positive Control Probe Hs-UBC; Advanced Cell Diagnostics, catalog no. 310041) was used as an internal mRNA control and the bacteria DapB (RNAscope Negative Control Probe DapB, Advanced Cell Diagnostics, catalog no. 310043) as a negative control gene.
Identification of differentially expressed ACE2+ coregulated gene signatures
Differentially expressed genes were identified between ACE2+ and ACE2− PTECs in DKD and COV samples with the use of the FindAllMarkers Seurat function. For LD, DKD, and DKD-induced signatures, all genes with Bonferroni-adjusted P < 0.05, positive log fold change, and found in ≥10% of DKD ACE2+ PTEC cells were selected. For the COV ACE2+ signature, all genes with nominal unadjusted P < 0.05, positive log fold change, and found in ≥10% of ACE2+ PTEC cells were selected. (Supplementary Tables S1–S3 and S10).
Overlap of differentially expressed ACE2+ coregulated gene signatures with SARS-CoV-2 relevant gene sets
SARS-CoV-2–relevant gene sets were compiled from multiple published sources: (i) Bojkova_proteome is a list of differentially expressed proteins in the Caco-2 cell line after SARS-Cov-2 infection (P < 0.05 at any time point)36; (ii) Gordon_interactome is a set of host proteins identified as physically interacting with SARS-CoV-2 viral proteins in HEK-293T cells39; (iii) Zhou_interactome is a literature-curated list of genes related to diverse coronaviruses20; and (iv) Blanco-Melo_NHBE is a list of differentially regulated genes in response to SARS-CoV-2 infection in normal human bronchial epithelial cells.40 ACE2 was removed from ACE2+ coexpressed gene sets before computing overlaps. P values were computed with the use of the hypergeometric test with a count of 20,000 genes used as background. Specifically, P values were computed with the R function 1 − phyper(s − 1, g1, N − g1, g2), where s is the number of genes shared between the 2 gene sets, N is the number of background genes, g1 is the number of genes in gene set 1, and g2 is the number of genes in the second gene set. Fold enrichment was computed as (s/g1)/(g2/N).
Functional network analysis
To determine the biological processes and pathways in the ACE2+ differentially expressed gene sets, we performed functional network clustering in the PTEC gene functional network derived from GIANT 2.0.34 , 35 Community clustering in the network was performed to identify tightly connected sets of genes with the use of HumanBase.io module detection function68 (https://hb.flatironinstitute.org/covid-kidney). The network was clustered with each set of differentially expressed genes constituting each ACE2+ signature: LD, DKD, DKD-induced, and COV (Supplementary Tables S1–S3 and S10). See the Supplementary Methods for details. The Gene Ontology enrichment outputs are provided in Supplementary Tables S4, S5, S11, S13, S15, and S16 and can be interactively explored at https://hb.flatironinstitute.org/covid-kidney.
Statistical analysis of clinical associations
Spearman correlation was used to evaluate the association of the steady-state average PTEC-specific gene expression levels of ACE2 and age at time of biopsy for the 44 DKD samples. The Mann-Whitney nonparametric test was applied to evaluate sex and treatment difference of ACE2 expression in the DKD cohort.
Data access
LD single-cell data sets are searchable at http://nephrocell.miktmc.org. A dynamic user-friendly interface at HumanBase (https://hb.flatironinstitute.org/covid-kidney) is available for researchers to explore the functional networks of gene expression signatures.
Disclosure
MK reports grants from the National Institutes of Health (NIH) and nonfinancial support from the University of Michigan during the conduct of the study as well as grants from JDRF, Astra-Zeneca, NovoNordisk, Eli Lilly, Gilead, Goldfinch Bio, Merck, Chan Zuckerberg Initiative, Janssen, Boehringer-Ingelheim, Moderna, Chinook, amfAR, Angion, RenalytixAI, Retrophin, the European Union Innovative Medicine Initiative, and Certa outside of the submitted work, and has licensed patent PCT/EP2014/073413 “Biomarkers and methods for progression prediction for chronic kidney disease.” OT reports serving on the scientific advisory boards of Caris Life Sciences and GoldFinch Bio, both outside of the submitted work. LHM reports grants from NIH–National Institute of Diabetes and Digestive and Kidney Diseases and the National Center for Advancing Translational Sciences during the conduct of the study and other research funding from Boehringer Ingelheim, serves on the Reata Pharmaceutical CKD Advisory Board, and has received honoraria for American Society of Nephrology Board Review. SE has served as coinvestigator on grants funded by Gilead Sciences, NovoNordisk, Astra-Zeneca, Jannsen, and Eli Lilly. ASN reports funding from the Michigan Institute of Clinical Health Research, Novartis, and the National Institute of Allergy and Infectious Diseases.
Author Contributions
MK, LM, ASN, RGN, RM, and OGT designed the study; SLD, CCL, ASN, LHM, and RSo enrolled participants and collected data; RGN, HCL, and ASN provided kidney biopsy tissue samples from respective studies; CCB, EAO, BG, and SLD developed protocol and processed samples for single cell analysis; the in situ hybridization was accomplished by JBH, JL, YY; RM, VN, SE, DF, JH, EAO, RSe, AKW, CLT, XC, YW, and AB were involved in data analysis; MK, RGN, ASN, JAS, LHM, and HCL provided clinical input; PMK, JZS, and CEW provided scientific input for interpretation of results; RM, LS, VN, CLT, AKW, RSe, RGN, OGT, and MK wrote and compiled the manuscript.
Study Approval
All studies of the DKD population were approved by the Institutional Review Board of the National Institute of Diabetes and Digestive and Kidney Diseases under protocol 13-DK-N151. Transplant biopsies were obtained after review and approval of the Transplant Transcriptomic Atlas study under HUM00150968; DKD kidney biopsy gene expression analysis was performed under HUM00002468; COVID-19 urine samples were obtained after review and approval under HUM00004729, all reviewed by the University of Michigan Medical School Institutional Review Boards.
Acknowledgements
The authors thank Lois Jones, RN, Enrique Diaz, RN, Bernadine Waseta, and Camille Waseta for performing the studies in the diabetes cohort and the University of Michigan Advanced Genomics Core for providing expert technical assistance with single-cell processing and sequencing. They also thank Dr. Carmen Mirabelli for reviewing this manuscript and providing valuable feedback.
This work was supported in part by the Intramural Research Program at the National Institute of Diabetes and Digestive and Kidney Diseases (DK069062 to HCL and RGN; DK083912, DK082841, DK020572, and DK092926 to RGK), the extramural research program of the National Institute of Diabetes and Digestive and Kidney Diseases R24 DK082841 “Integrated Systems Biology Approach to Diabetic Microvascular Complications” and P30 DK081943 “University of Michigan O'Brien Kidney Translational Core Center” to MK, via the “Kidney Precision Medicine Project” (KPMP, funded by the following grants from the NIDDK: U2C DK114886, UH3DK114861, UH3DK114866, UH3DK114870, UH3DK114908, UH3DK114915, UH3DK114926, UH3DK114907, UH3DK114920, UH3DK114923, UH3DK114933, and UH3DK114937, with U2C DK114886 and UH3 DK114907) to MK and OGT, via the Chan Zuckerberg Initiative “Human Cell Atlas Kidney Seed Network” to MK and OGT, JDRF 5-COE-2019-861-S-B “JDRF and M-Diabetes Center of Excellence at the University of Michigan” to MK, and a COVID grant from amFAR, the Foundation for AIDS Research, to MK. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We acknowledge the Kidney Precision Medicine Project represented by the following members: American Association of Kidney Patients, Tampa, FL: Richard Knight; Beth Israel Deaconess, Boston, MA: Stewart Lecker, Isaac Stillman; Boston University, Boston, MA: Sushrut Waikar; Brigham and Women's Hospital, Boston, MA: Gearoid McMahon, Astrid Weins, Samuel Short; Broad Institute, Cambridge, MA: Nir Hacohen, Paul Hoover; Case Western Reserve, Cleveland, OH: Mark Aulisio; Cleveland Clinic, Cleveland, OH: Leslie Cooperman, Leal Herlitz, John O’Toole, Emilio Poggio, John Sedor, Stacey Jolly; Columbia University, New York, NY: Paul Appelbaum, Olivia Balderes, Jonathan Barasch, Andrew Bomback, Pietro A. Canetta, Vivette D. d’Agati, Krzysztof Kiryluk, Satoru Kudose, Karla Mehl, Jai Radhakrishnan, Chenhua Weng; Duke University, Durham, NC: Laura Barisoni; European Molecular Biology Laboratory, Heidelberg, Germany: Theodore Alexandrov; Indiana University, Indianapolis, IN: Tarek Ashkar, Daria Barwinska, Pierre Dagher, Kenneth Dunn, Michael Eadon, Michael Ferkowicz, Katherine Kelly, Timothy Sutton, Seth Winfree; John Hopkins University, Baltimore, MD: Steven Menez, Chirag Parikh, Avi Rosenberg, Pam Villalobos, Rubab Malik, Derek Fine, Mohammed Atta, Jose Manuel Monroy Trujillo; Joslin Diabetes Center, Boston, MA: Alison Slack, Sylvia Rosas, Mark Williams; Mount Sinai, New York, NY: Evren Azeloglu, Cijang (John) He, Ravi Iyengar, Jens Hansen; Ohio State University, Columbus, OH: Samir Parikh, Brad Rovin; Pacific Northwest National Laboratories, Richland, WA: Chris Anderton, Ljiljana Pasa-Tolic, Dusan Velickovic, Jessica Lukowski; Parkland Center for Clinical Innovation, Dallas, TX: George (Holt) Oliver; Patient Partners: Joseph Ardayfio, Jack Bebiak, Keith Brown, Taneisha Campbell, Catherine Campbell, Lynda Hayashi, Nichole Jefferson, Robert Koewler, Glenda Roberts, John Saul, Anna Shpigel, Edith Christine Stutzke, Lorenda Wright, Leslie Miegs, Roy Pinkeney; Princeton University, Princeton, NJ: Rachel Sealfon, Olga Troyanskaya; Providence Medical Research Center, Providence Health Care, Spokane, WA: Katherine Tuttle; Stanford University, Palo Alto, CA: Dejan Dobi, Yury Goltsev; University of California San Diego, La Jolla, CA: Blue Lake, Kun Zhang; University of California San Francisco, San Francisco, CA: Maria Joanes, Zoltan Laszik, Andrew Schroeder, Minnie Sarwal, Tara Sigdel; University of Michigan, Ann Arbor, MI: Ulysses Balis, Victoria Blanc, Oliver He, Jeffrey Hodgin, Matthias Kretzler, Laura Mariani, Rajasree Menon, Edgar Otto, Jennifer Schaub, Becky Steck, Chrysta Lienczewski, Sean Eddy; University of Pittsburgh, Pittsburgh, PA: Michele Elder, Daniel Hall, John Kellum, Mary Kruth, Raghav Murugan, Paul Palevsky, Parmjeet Randhawa, Matthew Rosengart, Sunny Sims-Lucas, Mary Stefanick, Stacy Stull, Mitchell Tublin; University of Washington, Seattle, WA: Charles Alpers, Ian de Boer, Ashveena Dighe, Jonathan Himmelfarb, Robyn Mcclelland, Sean Mooney, Stuart Shankland, Kayleen Williams, Kristina Blank, Jonas Carson, Frederick Dowd, Zach Drager, Christopher Park; UT Health San Antonio, Center for Renal Precision Medicine, San Antonio, TX: Kumar Sharma, Guanshi Zhang, Shweta Bansal, Manjeri Venkatachalam; UT Southwestern Medical Center, Dallas, TX: Asra Kermani, Simon Lee, Christopher Lu, Tyler Miller, Orson Moe, Harold Park, Kamalanathan Sambandam, Francisco Sanchez, Jose Torrealba, Toto Robert, Miguel Vazquez, Nancy Wang; Washington University, St. Louis, MO: Joe Gaut, Sanjay Jain, Anitha Vijayan; and Yale University, New Haven, CT: Randy Luciano, Dennis Moledina, Ugwuowo Ugochukwu, Francis Perry Wilson, Sandy Alfano.
Footnotes
see commentary on page 1404
Supplementary Discussion.
Supplementary Methods.
Supplementary References.
Figure S1. UMAP plot showing the distribution of LD and DKD cells.
Figure S2. Functional summary of DKD-induced ACE2+ signature.
Figure S3. Heatmap of cluster markers used to identify cell types in COV.
Figure S4. ACE2 immunostaining in human kidney tissue from the Human Cell Atlas.
Table S1. Differential expression gene list for the LD ACE2+ signature.
Table S2. Differential expression gene list for the DKD ACE2+ signature (Figure 4a).
Table S3. Differential expression gene list for the DKD-induced ACE2+ signature genes (Supplementary Figure S2A).
Table S4. The Gene Ontology enrichments for DKD ACE2+ signature network modules (Figure 4a).
Table S5. The Gene Ontology enrichments for the DKD-induced ACE2+ signature network modules (Supplementary Figure S2A).
Table S6. SARS-CoV-2 infection gene sets20 , 36 , 39 , 40 with overlaps indicated for both the DKD ACE2+ signature genes and the COV ACE2+ signature genes (Supplementary Figure S2B).
Table S7. A table of P values indicating the significance of the genes shared between the Bojkova proteome36 gene list and each module of the DKD ACE2+ signature (Figure 4b).
Table S8. A table of P values indicating the significance of the genes shared between the Bojkova proteome36 gene list and each module of the DKD-induced ACE2+ signature (Supplementary Figure S2).
Table S9. Cluster markers for single cell analysis of the COV samples (Figure 6).
Table S10. Differential expression gene list for the COV ACE2+ signature (Figure 7a).
Table S11. The Gene Ontology enrichments for network modules of COV ACE2+ signature (Figure 7a).
Table S12. COV and DKD ACE2+ signature overlap gene set (Figure 7b).
Table S13. The Gene Ontology enrichments for network modules of clustered genes shared by COV and DKD ACE2+ signatures (Figure 7b).
Table S14. A table of P values indicating the significance of the genes shared between the Bojkova proteome36 gene list and each module of the COV ACE2+ signature (Figure 7c).
Table S15. The Gene Ontology enrichments for network modules for the network clustering of genes shared between the Bojkova proteome36 gene list and the COV ACE2+ signature (Figure 7c).
Table S16. The Gene Ontology enrichments for network modules for the network clustering of genes shared between the Bojkova proteome36 gene list and the DKD ACE2+ signature (Figure 4b).
Supplementary Material
References
- 1.Argenziano M.G., Bruce S.L., Slater C.L., et al. Characterization and clinical course of 1000 patients with coronavirus disease 2019 in New York: retrospective case series. BMJ. 2020;369:m1996. doi: 10.1136/bmj.m1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apicella M., Campopiano M.C., Mantuano M., et al. COVID-19 in people with diabetes: understanding the reasons for worse outcomes. Lancet Diabetes Endocrinol. 2020;8:782–792. doi: 10.1016/S2213-8587(20)30238-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muniyappa R., Gubbi S. COVID-19 pandemic, corona viruses, and diabetes mellitus. Am J Physiol Endocrinol Metab. 2020;318:E736–E741. doi: 10.1152/ajpendo.00124.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shahid Z., Kalayanamitra R., McClafferty B., et al. COVID-19 and older adults: what we know. J Am Geriatr Soc. 2020;68:926–929. doi: 10.1111/jgs.16472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emami A., Javanmardi F., Pirbonyeh N., et al. Prevalence of underlying diseases in hospitalized patients with COVID-19: a systematic review and meta-analysis. Arch Acad Emerg Med. 2020;8 e35–e35. [PMC free article] [PubMed] [Google Scholar]
- 6.Pal R., Bhansali A. COVID-19, diabetes mellitus and ACE2: the conundrum. Diabetes Res Clin Pract. 2020;162:108132. doi: 10.1016/j.diabres.2020.108132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye M., Wysocki J., William J., et al. Glomerular localization and expression of angiotensin-converting enzyme 2 and angiotensin-converting enzyme: implications for albuminuria in diabetes. J Am Soc Nephrol. 2006;17:3067–3075. doi: 10.1681/ASN.2006050423. [DOI] [PubMed] [Google Scholar]
- 8.Drucker D.J. Coronavirus infections and type 2 diabetes—shared pathways with therapeutic implications. Endocr Rev. 2020;41:bnaa011. doi: 10.1210/endrev/bnaa011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muus C., Luecken, Eraslan G., et al. Integrated analyses of single-cell atlases reveal age, gender, and smoking status associations with cell type-specific expression of mediators of SARS-CoV-2 viral entry and highlights inflammatory programs in putative target cells. bioRxiv. 2020 doi: 10.1101/2020.04.19.049254. Accessed April 21, 2020. [DOI] [Google Scholar]
- 10.Braun F., Lütgehetmann M., Pfefferle S., et al. SARS-CoV-2 renal tropism associates with acute kidney injury. Lancet. 2020;396(10251):597–598. doi: 10.1016/S0140-6736(20)31759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding Y., He L., Zhang Q., et al. Organ distribution of severe acute respiratory syndrome (SARS) associated coronavirus (SARS-CoV) in SARS patients: implications for pathogenesis and virus transmission pathways. J Pathol. 2004;203:622–630. doi: 10.1002/path.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puelles V.G., Lütgehetmann M., Lindenmeyer M.T., et al. Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med. 2020 doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henry B.M., Lippi G. Chronic kidney disease is associated with severe coronavirus disease 2019 (COVID-19) infection. Int Urol Nephrol. 2020;52:1193–1194. doi: 10.1007/s11255-020-02451-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng Y., Luo R., Wang K., et al. Kidney impairment is associated with in-hospital death of COVID-19 patients. medRxiv. 2020;2002(2018):20023242. [Google Scholar]
- 15.Farkash E.A., Wilson A.M., Jentzen J.M. Ultrastructural evidence for direct renal infection with SARS-CoV-2. J Am Soc Nephrol. 2020;31:1683–1687. doi: 10.1681/ASN.2020040432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gheblawi M., Wang K., Viveiros A., et al. Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system: celebrating the 20th anniversary of the discovery of ACE2. Circ Res. 2020;126:1456–1474. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia H.P., Look D.C., Shi L., et al. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J Virol. 2005;79:14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuba K., Imai Y., Ohto-Nakanishi T., et al. Trilogy of ACE2: a peptidase in the renin–angiotensin system, a SARS receptor, and a partner for amino acid transporters. Pharmacol Ther. 2010;128:119–128. doi: 10.1016/j.pharmthera.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo J., Huang Z., Lin L., et al. Coronavirus disease 2019 (COVID-19) and cardiovascular disease: a viewpoint on the potential influence of angiotensin-converting enzyme inhibitors/angiotensin receptor blockers on onset and severity of severe acute respiratory syndrome coronavirus 2 infection. J Am Heart Assoc. 2020;9 doi: 10.1161/JAHA.120.016219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou L., Niu Z., Jiang X., et al. Systemic analysis of tissue cells potentially vulnerable to SARS-CoV-2 infection by the protein-proofed single-cell RNA profiling of ACE2, TMPRSS2 and Furin proteases. bioRxiv. 2020 doi: 10.1101/2020.04.06.028522. Accessed April 16, 2020. [DOI] [Google Scholar]
- 21.Kumar A., Faiq M.A., Pareek V., et al. Relevance of enriched expression of SARS-CoV-2 binding receptor ACE2 in gastrointestinal tissue with pathogenesis of digestive symptoms, diabetes-associated mortality, and disease recurrence in COVID-19 patients. bioRxiv. 2020 doi: 10.1101/2020.04.14.040204. Accessed April 16, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Breidenbach J.D., Dube P., Ghosh S., et al. Impact of comorbidities on the expression of SARS-CoV-2 viral entry-related genes. J Pers Med. 2020;10:146. doi: 10.3390/jpm10040146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamming I., Cooper M.E., Haagmans B.L., et al. The emerging role of ACE2 in physiology and disease. J Pathol. 2007;212:1–11. doi: 10.1002/path.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diao B., Wang C., Wang R., et al. Human kidney is a target for novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. bioRxiv. 2020 doi: 10.1101/2020.03.04.20031120. Accessed April 22, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.South A.M., Diz D.I., Chappell M.C. COVID-19, ACE2, and the cardiovascular consequences. Am J Physiol Heart Circ Physiol. 2020;318:H1084–H1090. doi: 10.1152/ajpheart.00217.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shang J., Wan Y., Luo C., et al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ziegler C.G.K., Allon S.J., Nyquist S.K., et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoffmann M., Kleine-Weber H., Schroeder S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glowacka I., Bertram S., Müller M.A., et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qi F., Qian S., Zhang S., et al. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem Biophys Res Commun. 2020;526:135–140. doi: 10.1016/j.bbrc.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perico L., Benigni A., Remuzzi G. Should COVID-19 concern nephrologists? Why and to what extent? The emerging impasse of angiotensin blockade. Nephron. 2020;144:213–221. doi: 10.1159/000507305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Al Heialy S., Hachim M.Y., Senok A., et al. Regulation of angiotensin converting enzyme 2 (ACE2) in obesity: implications for COVID-19. bioRxiv. 2020 doi: 10.1101/2020.04.17.046938. Accessed April 21, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sahali D., Mulliez N., Chatelet F., et al. Characterization of a 280-kD protein restricted to the coated pits of the renal brush border and the epithelial cells of the yolk sac. Teratogenic effect of the specific monoclonal antibodies. J Exp Med. 1988;167:213–218. doi: 10.1084/jem.167.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greene C.S., Krishnan A., Wong A.K., et al. Understanding multicellular function and disease with human tissue-specific networks. Nat Genet. 2015;47:569–576. doi: 10.1038/ng.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong A.K., Krishnan A., Troyanskaya O.G. GIANT 2.0: genome-scale integrated analysis of gene networks in tissues. Nucleic Acids Res. 2018;46:W65–W70. doi: 10.1093/nar/gky408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bojkova D., Westhaus S., Costa R., et al. Sofosbuvir activates EGFR-dependent pathways in hepatoma cells with implications for liver-related pathological processes. Cells. 2020;9:1003. doi: 10.3390/cells9041003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Becker J.L., Miller F., Nuovo G.J., et al. Epstein-Barr virus infection of renal proximal tubule cells: possible role in chronic interstitial nephritis. J Clin Invest. 1999;104:1673–1681. doi: 10.1172/JCI7286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamanaka K., Oka K., Nakazawa S., et al. Immunohistochemical features of BK virus nephropathy in renal transplant recipients. Clin Transplant. 2012;26:20–24. doi: 10.1111/j.1399-0012.2012.01636.x. [DOI] [PubMed] [Google Scholar]
- 39.Gordon D.E., Jang G.M., Bouhaddou M., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e1039. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su H., Yang M., Wan C., et al. Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney Int. 2020;98:219–227. doi: 10.1016/j.kint.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tikellis C., Johnston Colin I., Forbes Josephine M., et al. Characterization of renal angiotensin-converting enzyme 2 in diabetic nephropathy. Hypertension. 2003;41:392–397. doi: 10.1161/01.HYP.0000060689.38912.CB. [DOI] [PubMed] [Google Scholar]
- 43.Nadarajah R., Milagres R., Dilauro M., et al. Podocyte-specific overexpression of human angiotensin-converting enzyme 2 attenuates diabetic nephropathy in mice. Kidney Int. 2012;82:292–303. doi: 10.1038/ki.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ye M., Wysocki J., Naaz P., et al. Increased ACE 2 and decreased ace protein in renal tubules from diabetic mice. Hypertension. 2004;43:1120–1125. doi: 10.1161/01.HYP.0000126192.27644.76. [DOI] [PubMed] [Google Scholar]
- 45.Riera M., Márquez E., Clotet S., et al. Effect of insulin on ACE2 activity and kidney function in the non-obese diabetic mouse. PLoS One. 2014;9 doi: 10.1371/journal.pone.0084683. e84683–e84683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bindom S.M., Lazartigues E. The sweeter side of ACE2: Physiological evidence for a role in diabetes. Mol Cell Endocrinol. 2009;302:193–202. doi: 10.1016/j.mce.2008.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Márquez E., Riera M., Pascual J., et al. Renin-angiotensin system within the diabetic podocyte. Am J Physiol Renal Physiol. 2014;308:F1–F10. doi: 10.1152/ajprenal.00531.2013. [DOI] [PubMed] [Google Scholar]
- 48.Klein S., Cortese M., Winter S.L., et al. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. bioRxiv. 2020 doi: 10.1101/2020.06.23.167064. Accessed August 18, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Monteil V., Kwon H., Prado P., et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913.e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nair V., Komorowsky C.V., Weil E.J., et al. A molecular morphometric approach to diabetic kidney disease can link structure to function and outcome. Kidney Int. 2018;93:439–449. doi: 10.1016/j.kint.2017.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmid H., Boucherot A., Yasuda Y., et al. Modular activation of nuclear factor-κB transcriptional programs in human diabetic nephropathy. Diabetes. 2006;55:2993–3003. doi: 10.2337/db06-0477. [DOI] [PubMed] [Google Scholar]
- 52.Chen H., Liu W., Liu D., et al. SARS-CoV-2 activates lung epithelia cell proinflammatory signaling and leads to immune dysregulation in COVID-19 patients by single-cell sequencing. medRxiv. 2020 doi: 10.1101/2020.05.08.20096024. Accessed May 20, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liao M., Liu Y., Yuan J., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 54.Ghosal S., Mukherjee J.J., Sinha B., et al. The effect of angiotensin converting enzyme inhibitors and angiotensin receptor blockers on death and severity of disease in patients with coronavirus disease 2019 (COVID-19): a meta-analysis. medRxiv. 2020 doi: 10.1101/2020.04.23.20076661. Accessed April 29, 2020. [DOI] [Google Scholar]
- 55.de Simone G., Mancusi C. Speculation is not evidence: antihypertensive therapy and COVID-19. Eur Heart J Cardiovasc Pharmacother. 2020;6:133–134. doi: 10.1093/ehjcvp/pvaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mehra M.R., Desai S.S., Kuy S., et al. Cardiovascular disease, drug therapy, and mortality in Covid-19. N Engl J Med. 2020;382(25) doi: 10.1056/NEJMoa2007621. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57.Mancia G., Rea F., Ludergnani M., et al. Renin-angiotensin-aldosterone system blockers and the risk of Covid-19. N Engl J Med. 2020;382:2431–2440. doi: 10.1056/NEJMoa2006923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reynolds H.R., Adhikari S., Pulgarin C., et al. Renin-angiotensin-aldosterone system inhibitors and risk of Covid-19. N Engl J Med. 2020;382:2441–2448. doi: 10.1056/NEJMoa2008975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mizuiri S., Ohashi Y. ACE and ACE2 in kidney disease. World J Nephrol. 2015;4:74–82. doi: 10.5527/wjn.v4.i1.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rao S., Lau A., So H.-C. Exploring diseases/traits and blood proteins causally related to expression of ACE2, the putative receptor of SARS-CoV-2: a Mendelian randomization analysis highlights tentative relevance of diabetes-related traits. Diabetes Care. 2020;43:1416–1426. doi: 10.2337/dc20-0643. [DOI] [PubMed] [Google Scholar]
- 61.Reich H.N., Oudit G.Y., Penninger J.M., et al. Decreased glomerular and tubular expression of ACE2 in patients with type 2 diabetes and kidney disease. Kidney Int. 2008;74:1610–1616. doi: 10.1038/ki.2008.497. [DOI] [PubMed] [Google Scholar]
- 62.Sungnak W., Huang N., Bécavin C., et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zou X., Chen K., Zou J., et al. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med. 2020;14:185–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Uhlén M., Fagerberg L., Hallström B.M., et al. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 65.Menon R., Otto E.A., Hoover P., et al. Single cell transcriptomics identifies focal segmental glomerulosclerosis remission endothelial biomarker. JCI Insight. 2020;5 doi: 10.1172/jci.insight.133267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arazi A., Rao D.A., Berthier C.C., et al. The immune cell landscape in kidneys of patients with lupus nephritis. Nat Immunol. 2019;20:902–914. doi: 10.1038/s41590-019-0398-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu H., Malone A.F., Donnelly E.L., et al. Single-cell transcriptomics of a human kidney allograft biopsy specimen defines a diverse inflammatory response. J Am Soc Nephrol. 2018;29:2069–2080. doi: 10.1681/ASN.2018020125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krishnan A., Zhang R., Yao V., et al. Genome-wide prediction and functional characterization of the genetic basis of autism spectrum disorder. Nat Neurosci. 2016;19:1454–1462. doi: 10.1038/nn.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.