Abstract
The chloroplasts are a crucial part of photosynthesizing plant cells and are extensively utilized in phylogenetic studies mainly due to their maternal inheritance. Characterization and analysis of complete plastome sequences is necessary to understand their diversity and evolutionary relationships. Here, a panel of thirteen plastomes from various potato taxa are presented. Though they are highly similar with respect to gene order and content, there is also a great extent of SNPs and InDels between them, with one of the Solanum bukasovii plastomes (BUK2) having the highest number of SNPs and InDels. Five different potato plastome types (C, S, A, W, W2) are present in the panel. Interestingly, the S. tuberosum subsp. tuberosum (TBR) accession has a W-type plastome, which is not commonly found in this species. The S-type plastome has a conserved 48 bp deletion not found in other types, which is responsible for the divergence of the S-type from the C-type plastome. Finally, a phylogenetic analysis shows that these plastomes cluster according to their types. Congruence between the nuclear genome and the plastome phylogeny of these accessions was seen, however with considerable differences, supporting the hypothesis of introgression and hybridization between potato species.
Introduction
The chloroplasts are an essential part of all photosynthesizing plant cells. Chloroplast DNA, the plastome, is maternally inherited and has a higher degree of conservation than nuclear or mitochondrial DNA, in terms of gene order and content [1, 2]. The plastome is composed of a single circular DNA molecule, typically ranging between 115 and 165 kb in land plants [3]. Potato (Solanum sp.) plastomes range from 154 to 156 kb, with a typical quadripartite structure including one large single copy (LSC) region, one small single copy (SSC) region, and two copies of inverted repeat regions (IRa and IRb). The inverted repeats are ~25 kb in length and separated by the LSC (~86 kb) and SSC (~18 kb) regions [3, 4]. Despite the degree of plastome conservation in most land plants, many structural and single nucleotide variations have been observed. These variations include interspecies deletions [3], insertions and deletions (InDels), single nucleotide polymorphisms (SNPs), genome expansions, and polymorphic single sequence repeats (SSRs) [4, 5].
The taxonomy of wild and cultivated Solanum species is very complex and is continuously debated. The Solanum genus includes ornamentals and nightshades as well as food crops such as tomato, potato, and eggplant. A recent taxonomy study on potato—members of the tuber-forming clade of Solanum—reported that there are 107 wild and 4 cultivated potato species [6], instead of the 232 species previously proposed [7]. The plastome was used extensively in previous studies as phylogenetic markers [8], however, the plastome phylogeny does not necessarily follow the nuclear genome phylogeny [9]. Nonetheless, generating both phylogenies is useful in terms of studying cross-hybridization and introgression in potato [8].
Five basic plastome types (A, S, C, T, and W) were previously described in potato based on a restriction fragment length polymorphism (RFLP) analysis [10]. The W-type is considered the most primitive [10], since its restriction fragment pattern is closest to those of Mexican diploids as well as non-tuber-bearing Solanum species. The majority of the plastome types are derived from the W-type through various point mutations with the exception of the T-type, which is derived by a single deletion of 241 bp from the W-type [11]. The additional W1, W2, and W3 types were derived from the W-type by a single nucleotide change shown in the PvuII restriction fragment pattern, whereas the C-type was derived by a single change that led to a difference in the BamHI restriction fragment pattern [10, 11]. The A and S-type originated from the C-type through single nucleotide changes, evidenced through changes in the BamHI and HindIII restriction fragment patterns, respectively [10]. A diverse set of plastomes has been found among potato species as a result of successive domestication, polyploidization and selection [11, 12].
Just like mitochondrial DNA, plastid DNA can be used to study maternal genetic lineage. However, the use of partial plastome sequences was shown to be insufficient to classify closely related species [13], and it is therefore necessary to characterize complete plastomes to deduce phylogenetic relationships and evolution of species.
In this study, the plastomes from a panel of 13 potato accessions were characterized. The nuclear genomes of 12 of these accessions were previously studied for structural variation [14, 15], and in the present study we present their complete plastome sequences and from an additional individual wild potato accession. Furthermore, SNPs, InDels, repeat sequences present as well as the type of each plastome are identified based on polymorphisms. The topology obtained from a phylogenetic analysis of the complete plastomes of 16 accessions/species (13 from the panel and three reference sequences) is compared with the previously published topology obtained using the nuclear genomes.
Materials and methods
Plant materials, DNA library preparation, and sequencing
DNA was extracted from plants of twelve Peruvian potato accessions: Solanum stenotomum subsp. goniocalyx (GON1—CIP 702472), S. stenotomum subsp. goniocalyx (GON2—CIP 704393), S. phureja (PHU—CIP 703654), S. xajanhuiri (AJH—CIP 703810), S. stenotomum subsp. stenotomum (STN—CIP 705834), S. bukasovii (BUK1—CIP 761748), S. bukasovii (BUK2 –CIP 761748), S. tuberosum subsp. andigena (ADG1—CIP 700921), S. tuberosum subsp. andigena (ADG2—CIP 702853), S. curtilobum (CUR—CIP 702937), S. juzepczukii (JUZ—CIP 706050), S. chaucha (CHA- CIP 707129) and one Chilean accession: S. tuberosum subsp. tuberosum (TBR—CIP 705053) and sequenced using Illumina PE Technology as described in [14]. The genome of a second individual from the S. bukasovii (BUK2 –CIP 761748) accession, BUK2, was sequenced with 10X Genomics’ GemCode technology (https://www.10xgenomics.com/).
de novo assembly and annotation
Raw reads obtained from Illumina sequencing were initially processed using Trimmomatic v0.36 to remove adapter sequences and low-quality bases [16]. TruSeq3 paired end Illumina adapters, low-quality bases, and reads less than 60 bp in length were removed. The parameters used for the Trimmomatic were ILLUMINACLIP: TruSeq3-PE. fa: 2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW: 4:15 MINLEN:60. The reads of BUK2 obtained from the 10X Genomics’ GemCode technology were run through LongRanger Basic to perform read trimming and barcode error correction (https://support.10xgenomics.com/genome-exome/software/pipelines/latest/installation). The filtered reads of each landrace were assembled into a complete plastome using NOVOPlasty: de novo organellar genome assembler [17], which uses a seed-and-extend algorithm to de novo construct a complete plastome from whole genome sequencing (WGS) data. A seed sequence of 1000 bp was randomly selected from the doubled monoploid S. tuberosum Group Phureja clone DM1-3 plastome [18]. The expected genome range was set to 145 kb-165 kb, and other parameters set to default. Inverted repeat regions were detected in assembled sequences using REPuter software [19]. The assembled sequences were annotated using GeSeq [20] with S. commersonii (GenBank accession NC_028069.2) [4] and S. tuberosum (GenBank accession DQ231562.1) [3] species as references. The annotations were manually curated to adjust start and stop codons using Blastn searches [21]. The circular maps of all the plastomes were drawn using OGDRAW v1.3.1 [22]. The inverted repeats and gene structure in junction sites of all the plastomes were visualized using IRscope [23].
Variant detection
SNPs and InDels were determined from the multiple sequence alignment of the thirteen plastomes. Multiple sequence alignments (MSA) were performed using MAFFT v7 with 1PAM/k = 2 scoring matrix [24]. SNPs were detected using SNP-sites [25]. InDels were detected using a custom python script and Geneious Prime 2019 (Geneious Prime 2019.2 https://www.geneious.com). From the MSA, a consensus sequence was obtained and annotated to determine the genes harboring the variant sites. Furthermore, at each variant site any base with a frequency of more than 50% was considered a reference allele. Gapped annotations were obtained from Geneious Prime 2019, and the variants were annotated using gapped annotations with BEDTools v2.28 [26].
Repeat identification
Simple sequence repeats (SSRs) were identified using PHOBOS v3.3.12 [27] with a minimum number of repeat units as follows: 10 for mononucleotide repeats, 5 for dinucleotide repeats, 4 for trinucleotide repeats, and 3 for tetranucleotide repeats, pentanucleotide repeats, and hexanucleotide repeats. Tandem repeats were detected using Tandem Repeat Finder v4.09 [28]. Forward, palindromic, reverse, and complemented repeats present in all the plastomes were identified using REPuter software with minimum repeat size of 30 bp and a hamming distance of 3 [19].
Identification of chloroplast DNA types
Two methods were used to identify plastome types. Method 1: Seven microsatellite markers and their PCR product sizes were used to determine the types [12, 29]. Marker analysis was conducted using primer sequences of these seven markers with an in silico PCR “reaction” [30]. Method 2: In silico restriction endonuclease analysis: the digestion sites for five restriction enzymes BamHI, HindIII, KpnI, PvuII, and XhoI were used for an in silico restriction digest reaction. The digestion was performed in silico using RestrictionMapper (http://www.restrictionmapper.org/). Custom scripts were used to parse the output and compare the results amongst 13 plastomes to determine the length polymorphisms. These restriction fragment patterns were compared with the previous studies to determine the types [1, 10].
Phylogenetic analyses
To understand the phylogenetic position of these plastomes, a maximum parsimony phylogeny was constructed using PAUP* version 4.0a167 with 1000 bootstrap replicates [31]. Published plastomes of S. tuberosum group Phureja DM1-3 516 R44 (DM) [18], S. commersonii (GenBank accession NC_028069.2) [4], and S. tuberosum (GenBank accession DQ231562.1) [3] were also included in the phylogeny. One inverted repeat region from each plastome was removed before constructing the phylogeny to avoid data duplication.
Results and discussion
Structure and organization of potato plastomes
Whole genomic DNA was previously sequenced from a panel of 12 potato accessions (Table 1) [14]. In this study, another S. bukasovii genome (BUK2) from the same accession as BUK1 was added to the panel, and their combined 13 plastomes were assembled and investigated to assess the diversity among them. For each potato accession, the entire plastome was assembled into a single circular sequence using trimmed WGS reads, with a typical quadripartite structure of a plastid DNA molecule: one large single copy (LSC), one small single copy (SSC), and two inverted repeat regions (IRa and IRb) (Fig 1). The sequencing coverage of each plastome ranges from 9,348 to 25,856 X. The size of the plastomes ranges between 155,486 bp (AJH) and 155,584 bp (BUK2) (Table 2), and they are all highly identical in structure and genome organization. Inverted repeat (IR) regions of 25,593 bp are found in 12 of the plastomes, while the plastome of BUK2 differs with 25,596 bp of IR regions (3 bp more per IR). The size of the LSC and SSC regions ranges from 85,929 bp to 86,005 bp and 18,363 bp to 18,394 bp respectively. The GON1, GON2, PHU, STN, and CUR plastomes have the same size and boundaries. Similarly, the CHA and ADG2 plastomes have the same size (Table 2). The GC content in the AJH and ADG1 plastomes is 37.8%, while it is 37.9% in the rest of them.
Table 1. A panel of 13 potato plastomes.
| Potato Accession | Label | Nuclear ploidy level | CIP Accession identifier | Sequence coverage (X) on plastome | GenBank accession # |
|---|---|---|---|---|---|
| Solanum stenotomum subsp. goniocalyx | GON1 | Diploid | 702472 | 14,576 | MT120855 |
| Solanum stenotomum subsp. goniocalyx | GON2 | Diploid | 704393 | 25,856 | MT120856 |
| Solanum xajanhuiri | AJH | Diploid | 703810 | 15,388 | MT120857 |
| Solanum phureja | PHU | Diploid | 703654 | 15,995 | MT120858 |
| Solanum stenotomum subsp. stenotomum | STN | Diploid | 705834 | 12,028 | MT120859 |
| Solanum bukasovii | BUK1 | Diploid | 761748 | 25,459 | MT120860 |
| Solanum bukasovii | BUK2 | Diploid | 761748 | 9,348 | MT120867 |
| Solanum juzepczukii | JUZ | Triploid | 706050 | 14,319 | MT120863 |
| Solanum chaucha | CHA | Triploid | 707129 | 11,628 | MT120864 |
| Solanum tuberosum subsp. andigena | ADG1 | Tetraploid | 700921 | 10,177 | MT120861 |
| Solanum tuberosum subsp. andigena | ADG2 | Tetraploid | 702853 | 12,789 | MT120862 |
| Solanum tuberosum subsp. tuberosum | TBR | Tetraploid | 705053 | 9,873 | MT120865 |
| Solanum curtilobum | CUR | Pentaploid | 702937 | 9,815 | MT120866 |
The plastomes from a panel of 13 potato accessions with various nuclear ploidy levels including seven diploid, two triploid, three tetraploid, and one pentaploid were assembled. Two individuals, BUK1 and BUK2, from the same accession of the wild S. bukasovii were sequenced.
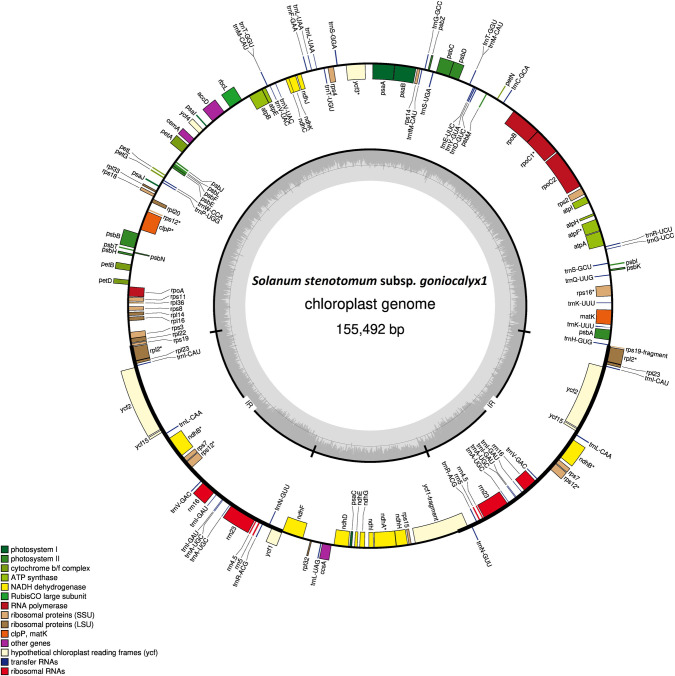
Fig 1. Gene structure of the Solanum stenotomum subsp. goniocalyx 1 plastome.
Structure and gene content of the GON1 plastome. A panel of 13 potato plastomes all have similar gene content and structure. Genes that lie outside the circle are transcribed clockwise, genes inside the circle are transcribed counter-clockwise. The inner circle represents GC content.
Table 2. Plastome size and range of individual segments.
| Potato accession | Size (bp) | LSC (size bp) | IRb (size bp) | SSC (size bp) | IRa (size bp) |
|---|---|---|---|---|---|
| GON1 | 155,492 | 1–85,930 (85,930) | 85,931–111,523 (25,593) | 111,524–129,899 (18,376) | 129,900–155,492 (25,593) |
| GON2 | 155,492 | 1–85,930 (85,930) | 85,931–111,523 (25,593) | 111,524–129,899 (18,376) | 129,900–155,492 (25,593) |
| AJH | 155,486 | 1–85,937 (85,937) | 85,938–111,530 (25,593) | 111,531–129,893 (18,363) | 129,894–155,486 (25,593) |
| PHU | 155,492 | 1–85,930 (85,930) | 85,931–111,523 (25,593) | 111,524–129,899 (18,376) | 129,900–155,492 (25,593) |
| STN | 155,492 | 1–85,930 (85,930) | 85,931–111,523 (25,593) | 111,524–129,899 (18,376) | 129,900–155,492 (25,593) |
| BUK1 | 155,491 | 1–85,929 (85,929) | 85,930–111,522 (25,593) | 111,523–129,898 (18,376) | 129,899–155,491 (25,593) |
| BUK2 | 155,584 | 1–85,998 (85,998) | 85,999–111,594 (25,596) | 111,595–129,988 (18,394) | 129,989–155,584 (25,596) |
| JUZ | 155,532 | 1–85,981 (85,981) | 85,982–111,574 (25,593) | 111,575–129,939 (18,365) | 129,940–155,532 (25,593) |
| CHA | 155,518 | 1–85,968 (85,968) | 85,969–111,561 (25,593) | 111,562–129,925 (18,364) | 129,926–155,518 (25,593) |
| ADG1 | 155,530 | 1–85,971 (85,971) | 85,972–111,564 (25,593) | 111,565–129,937 (18,373) | 129,938–155,530 (25,593) |
| ADG2 | 155,518 | 1–85,968 (85,968) | 85,969–111,561 (25,593) | 111,562–129,925 (18,364) | 129,926–155,518 (25,593) |
| TBR | 155,564 | 1–86,005 (86,005) | 86,006–111,598 (25,593) | 111,599–129,971 (18,373) | 129,972–155,564 (25,593) |
| CUR | 155,492 | 1–85,930 (85,930) | 85,931–111,523 (25,593) | 111,524–129,899 (18,376) | 129,900–155,492 (25,593) |
The plastomes were assembled into a complete circular molecule with four segments included, i.e. one large single copy region (LSC), one small single copy region (SSC), and two inverted repeats (IRa and IRb). The overall size ranges from 155,486 to 155,584 bp. GON1, GON2, PHU, STN, and CUR have the same size and boundaries, and ADG2 and CHA have the same. The other six plastomes vary in size and boundaries.
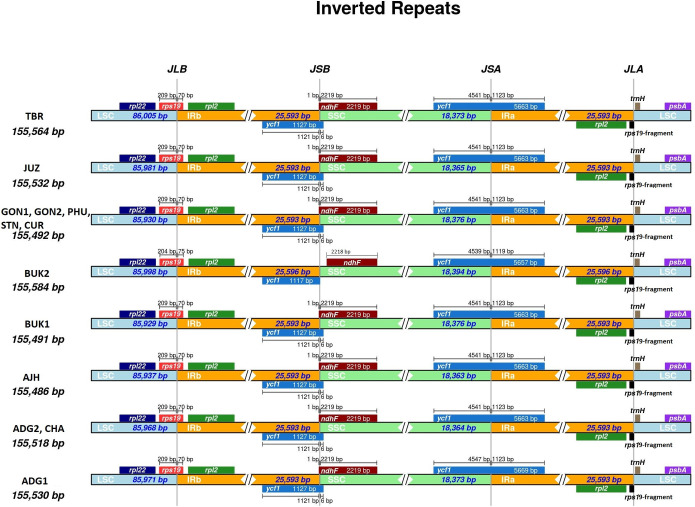
Each plastome has 143 genes in total, of which 20 are present in both of the IR regions and five are duplicated in the LSC region (Table 3). Overall, 115 unique (single copy) genes are present in all the plastomes, of which 81 are protein coding genes, 30 are tRNAs, and four are the rRNAs. A total of 14 intronic regions are present in nine genes, of which five are found in the LSC region, three in IR regions, and one in the SSC region. The circular map of the GON1 plastome with the gene structure is shown in Fig 1. The gene structure at the inverted repeat junction sites is similar in all plastomes assembled here, with the exception of BUK2. The JLA (LSC-IRa junction site) is located upstream of the rps19-fragment and downstream of the trnH-GUG gene. The JLB (junction between LSC-IRb) is situated within the rps19 gene. The junction between SSC and IRb is located within the ycf1 and ndhF genes in all of them except in BUK2, where it is located in the intergenic region between these two genes. Furthermore, the JSA (SSC-IRa junction) is located within the ycf1 gene (Fig 2). These 13 plastomes share similar gene structure, organization and gene boundaries with the previously sequenced S. commersonii, and S. tuberosum plastomes [3, 4]. However, the 13 plastomes each have more genes than those of S. commersoni and S. tuberosum, likely due to the usage of different annotation tools and curation methods. Nonetheless, the core set of genes and the unique protein coding genes, tRNAs, and rRNAs are the same in all of them.
Table 3. Distribution of genes in the plastomes.
| Genome | Total Genes | Genes in LSC | IRb | SSC | IRa | Genes in junction sites | tRNAs | rRNAs | Introns |
|---|---|---|---|---|---|---|---|---|---|
| GON1 | 143 | 87 | 20 | 11 | 21 | 4 | 30 | 4 | 14 |
| GON2 | 143 | 87 | 20 | 11 | 21 | 4 | 30 | 4 | 14 |
| AJH | 143 | 87 | 20 | 11 | 21 | 4 | 30 | 4 | 14 |
| PHU | 143 | 87 | 20 | 11 | 21 | 4 | 30 | 4 | 14 |
| STN | 143 | 87 | 20 | 11 | 21 | 4 | 30 | 4 | 14 |
| BUK1 | 143 | 87 | 20 | 11 | 21 | 4 | 30 | 4 | 14 |
| BUK2 | 143 | 87 | 21 | 12 | 21 | 2 | 30 | 4 | 14 |
| JUZ | 143 | 87 | 20 | 11 | 21 | 4 | 30 | 4 | 14 |
| CHA | 143 | 87 | 20 | 11 | 21 | 4 | 30 | 4 | 14 |
| ADG1 | 143 | 87 | 20 | 11 | 21 | 4 | 30 | 4 | 14 |
| ADG2 | 143 | 87 | 20 | 11 | 21 | 4 | 30 | 4 | 14 |
| TBR | 143 | 87 | 20 | 11 | 21 | 4 | 30 | 4 | 14 |
| CUR | 143 | 87 | 20 | 11 | 21 | 4 | 30 | 4 | 14 |
All the plastomes have 143 genes in total, with 87 genes in the LSC region, 20 in the IRb, 11 in the SSC, and 21 in the IRa, and four in junction sites, except BUK2. The BUK2 plastome has 21 genes in the IRb, 12 in the SSC region and only two genes in the junction sites. All the plastomes have 30 unique tRNA and four unique rRNA, and 14 intron regions.
Fig 2. Representation of junction-sites in 13 potato plastomes.
Four junction sites are present in all the plastomes, JLB (LSC-IRb junction), JSB (IRb-SSC junction), JSA (SSC-IRa junction), JLA (IRa-LSC junction). The BUK2 plastome differs from the other by the locations of the junctions.
Presence of variants
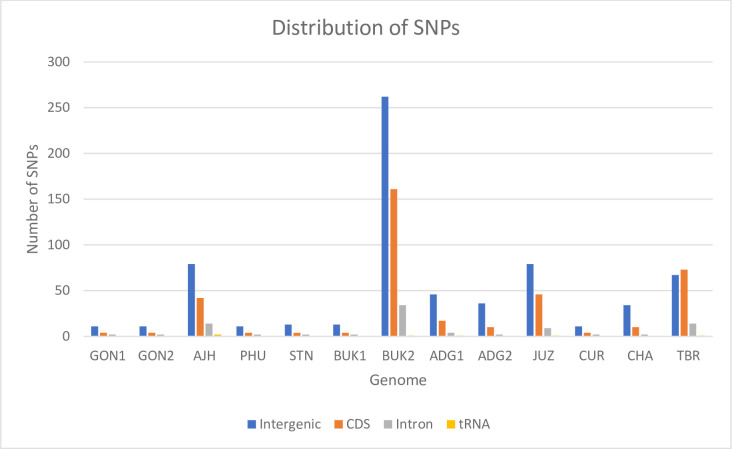
Despite the highly conserved nature of the plastomes, noticeable variations are present in closely related species. The multiple sequence alignment of the 13 complete chloroplast sequences reveals a great number of single nucleotide polymorphisms (SNPs) and insertions/deletions (InDels), even though they are structurally highly similar. A total of 746 SNP sites are detected and out of these, 563 are singleton variable sites (two variants), 178 are parsimony-informative sites (two variants), two are singleton variable sites (three variants), and three are parsimony-informative sites (three variants). Among the 13 plastomes, BUK2 has the highest number of SNPs compared to a majority call reference with 458 SNP sites, followed by the TBR, AJH, and JUZ, whose plastomes have 155, 137, and 135 SNPs, respectively. More than 50% of the SNPs are present in intergenic and intron regions (Fig 3). The genes harboring SNPs are listed in Table 4. Out of 746 overall SNP sites, only 57 SNPs are located in IR regions indicating that IR regions are more conserved than the LSC region with 519 SNPs and the SSC region with 170 SNPs. These results are in agreement with other studies [5, 8]. Overall, 57 genes in total have SNPs, and a few genes have more SNPs than others. The ycf1 gene has the highest number of SNPs (55 SNPs), followed by the ndhA gene with 30 SNPs, clpP and ndhF genes with 18 and 16 SNPs respectively (S1 Fig).
Fig 3. Distribution of SNPs in a panel of 13 potato plastomes.
The number of SNPs and their distribution across different regions show that the majority of the SNPs are present in intergenic regions. However, the BUK2, AJH, JUZ, and TBR plastomes also have many SNPs in the CDS regions.
Table 4. Genes harboring SNPs and InDels.
| Genes harboring SNPs, and InDels | |||
|---|---|---|---|
| SNPs | Insertions | Deletions | |
| GON1 | atpB, atpF, ndhA, rps3, rps4, ycf1 | - | - |
| GON2 | atpB, atpF, ndhA, rps3, rps4, ycf1 | - | - |
| AJH | atpA, atpB, atpE, atpF, ccsA, cemA, clpP, matK, ndhA, ndhC, ndhD, ndhF, ndhG, psaA, psbB, psbC, rbcL, rpl14, rpl22, rpoB, rpoC1, rps11, rps16, rps3, trnQ-UUG, trnR-UCU, ycf1, ycf3 | - | clpP |
| PHU | atpB, atpF, ndhA, rps3, rps4, ycf1 | - | - |
| STN | atpB, atpF, ndhA, rps3, rps4, ycf1 | - | - |
| BUK1 | atpB, atpF, ndhA, rps3, rps4, ycf1 | - | - |
| BUK2 | accD, atpA, atpB, atpF, atpH, atpI, ccsA, clpP, matK, ndhA, ndhC, ndhD, ndhF, ndhG, ndhH, ndhJ, petA, petB, petD, psaA, psaB, psaC, psbA, psbC, psbH, psbI, psbT, psbZ, rbcL, rpl14, rpl16, rpl20, rpl22, rpoA, rpoB, rpoC1, rpoC2, rps11, rps15, rps16, rps19, rps2, rps3, rps4, rps8, trnR-UCU, ycf1, ycf2, ycf3 | atpF, clpP, ndhF, rps19-fragment | matK, ycf3, ycf1 |
| ADG1 | atpH, ccsA, clpP, ndhA, ndhC, rpoB, rpoC1, rpoC2, rps4, trnL-UAG, ycf1 | ndhA, ycf1 | ClpP |
| ADG2 | ccsA, cemA, ndhA, ndhF, rpoB, rps15, ycf1, ycf2 | - | clpP |
| JUZ | atpB, atpF, ccsA, cemA, clpP, ndhA, ndhC, ndhD, ndhF, ndhG, ndhH, petA, psaA, psbB, psbC, rbcL, rpl14, rpl22, rpoB, rpoC1, rps11, rps16, rps3, trnR-UCU, ycf1, ycf3 | - | clpP |
| CUR | atpB, atpF, ndhA, rps3, rps4, ycf1 | - | - |
| CHA | ccsA, cemA, ndhA, ndhF, rpoB, rps15, ycf1, ycf2 | - | clpP |
| TBR | atpA, atpB, ccsA, clpP, matK, ndhA, ndhC, ndhD, ndhF, ndhG, ndhH, psaA, psbA, psbC, psbI, rbcL, rpl14, rpl20, rpl22, rpoB, rpoC1, rpoC2, rps11, rps15, rps3, rps8, rrn23, trnR-UCU, ycf1, ycf1-fragment, ycf2, ycf3 | clpP | clpP |
The SNPs and InDels were annotated. The BUK2, TBR, AJH, and JUZ plastomes have higher number of genes with SNPs compared with the other plastomes. The GON1, GON2, PHU, ST, BUK, and CUR plastomes lack InDels. Comparatively, the SNPs are more common than the InDels in all plastomes.
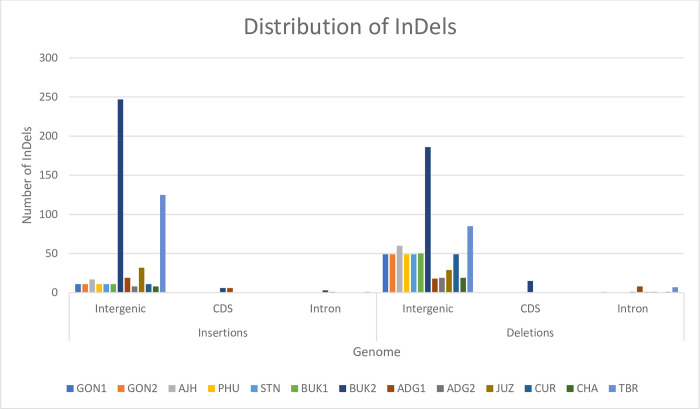
Similarly, InDels were detected from the multiple sequence alignment of the plastomes. Overall, 790 InDels are present of which 413 are insertions and 377 are deletions. There are more deletions than insertions in the plastomes of GON1, GON2, AJH, PHU, STN, BUK1, ADG2, CHA, AND CUR, while those of TBR, JUZ, and BUK2 contain more insertions than deletions. Overall, BUK2 and TBR have more InDels than the others with 458 and 218 InDels, respectively. Annotation shows that more than 90% of these InDels are present in intergenic and intron regions, which may have significant impact on gene expression regulation. Only a small number of InDels are present in genes (Fig 4 and Table 4). From a total of 790 InDels, only four InDels are present in IR regions, 714 in the LSC region, and 72 InDels in the SSC region. This further confirms the highly conserved nature of IR regions. In total, only four genes have deletions and six genes have insertions. The clpP gene has the highest number of InDels with 17 InDels in its intron region. Also, the ycf1 and the matK genes have 12 and nine InDels, respectively (S2 Fig). High divergence of the ycf1 gene has been observed in the plastomes of many plant species and this has led to it being used as a DNA barcode in land plants [5, 32].
Fig 4. Distribution of InDels in a panel of 13 potato plastomes.
Comparatively, BUK2 and TBR have the greatest number of InDels of all the plastomes. More than 90% of the InDels are present in intergenic regions.
A few length mutations are also present, some of them conserved in multiple plastomes. A 48 bp deletion is present in the GON1, GON2, PHU, STN, BUK1, and CUR plastomes and this is the largest deletion found in the panel. It is present in the intergenic region between rps16 and trnQ-UUG genes in the LSC region. Similarly, a 10 bp deletion is present in AJH, ADG1, ADG2, JUZ, CHA, and TBR in the intergenic region between ndhG and ndhI genes in the SSC region. Likewise, the second largest deletion of 30 bp is present in the AJH plastome in the intergenic region between the trnS-GCU and trnG-UCC genes in the LSC region. BUK2 also has 30 and 18 bp deletions in the intergenic region of LSC, and a 9 bp and 6 bp deletion in the matK and ycf1 genes, respectively.
Moreover, a 7 bp insertion is located in the intergenic region between rbcL and accD genes in GON1, GON2, PHU, STN, BUK1, and CUR. Similarly, AJH, JUZ, TBR, and BUK2 have a 7 bp insertion in the intergenic region between psaA and ycf3 genes. The largest insertion of 55 bp is in the BUK2 plastome in the intergenic region between the rpl20 and rps12 genes. The TBR plastome has 30 bp and 27 bp inserted in the intergenic region between trnG-UCC, trnR-UCU and rps16, trnQ-UUG genes, respectively. Many of these length mutations are present in the intergenic regions, hence they have not altered the gene content nor the overall structure of the 13 plastomes.
Repeat analyses
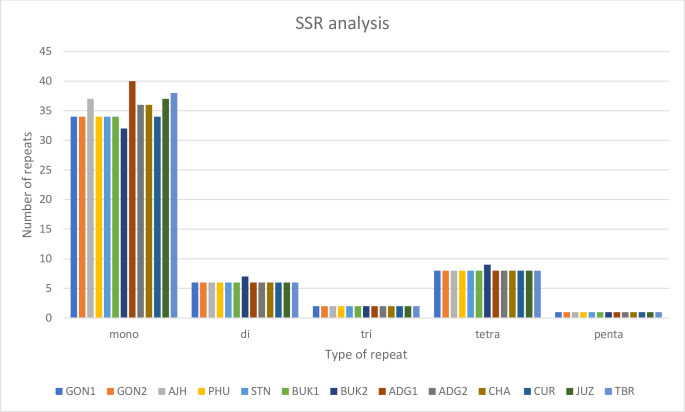
Simple sequence repeats (SSRs) are important molecular genetic markers that are extensively used in populations genetics, evolutionary studies, diversity assessments, and ecological studies [13]. Between 51 to 57 SSRs are present in each of the 13 plastomes, ranging from mononucleotide repeats to pentanucleotide repeats. The mononucleotide repeat abundance is the highest amongst the SSRs for all the plastomes (Fig 5). The GON1, GON2, PHU, STN, BUK1, BUK2, and CUR plastomes each have 51 SSRs in total, of which 34 (67%) are mono-repeats. While all of them have the same number of di-, tri-, tetra-, and pentanucleotide repeats (except for BUK2, which has one extra di- and tetra- repeat), they differ in mono-repeats. BUK2 and ADG1 have the lowest and highest number of mono-repeats containing 32 and 40 repeats, respectively. Similarly, all of the dinucleotide repeats are composed of AT/TA repeats, and 50% of trinucleotide repeats are TTA, indicating an abundance of AT bases in chloroplast SSR sequences.
Fig 5. Analysis of SSRs in a panel of 13 potato plastomes.
Five SSRs were identified in all the plastomes, namely mononucleotide, dinucleotide, trinucleotide, tetranucleotide, and pentanucleotide repeats. Mononucleotide repeats are the most abundant.
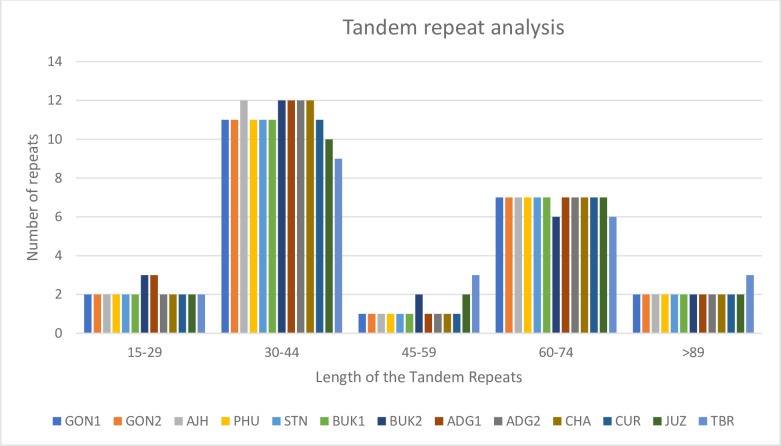
Also, 23–25 tandem repeats are present in all of them and the lengths of tandem repeats range from 26 bp to 91 bp. Overall, GON1, GON2, PHU, STN, BUK1, JUZ, TBR, and CUR have 23 tandem repeats, AJH, CHA, and ADG2 have 24 tandem repeats and ADG1, BUK2 have 25 repeats. The length of these repeat sequences is mostly in the range of 30–44 bp. Three tandem repeat sequences in TBR are 90 bp large and the other 12 plastomes have two 90 bp tandem repeat sequences (Fig 6). Similarly, 31–35 repeat sequences are present in all of them, including forward, palindromic, reverse, and complementary repeat sequences. Overall, 31 direct and palindromic repeat sequences are present in GON1, GON2, AJH, PHU, STN, BUK1, ADG1, ADG2, CHA, and CUR. Similarly, 32, 33, and 35 direct and palindromic repeat sequences are present in JUZ, TBR and BUK2 respectively. However, BUK2 is the only one that has two reverse and one complementary repeat sequences. All plastomes have 16 palindromic repeats, and they range from 56 bp to 30 bp, while the forward repeats range from 61 bp to 30 bp.
Fig 6. Analysis of tandem repeats in a panel of 13 potato plastomes.
Overall, 23 to 25 tandem repeats are present in all the plastomes and their sizes range from 26 bp to 91 bp. The size of the repeats is divided into five different ranges and number of repeats present in that particular range are presented here. Most of the repeats have a size in the range of 30–44 bp.
Identification of chloroplast DNA types
Potato has a diverse cytoplasm with different plastome types, which have been previously demonstrated to affect agronomic traits [33]. Some of the cytoplasm types in potato have been shown to induce higher percentages of tuberization, higher tuber yields, and earlier vine maturity [33]. In order to study the plastome diversity among the different accessions in the genome panel, their types were identified. Plastome specific markers and an in silico restriction enzyme analysis were used to identify plastome types for potato and the results were compared with previous studies [12, 29]. The plastome types of the GON1, GON2, PHU, STN, BUK1, CUR, ADG1, AJH, and JUZ accessions were positively identified, whereas the rest of the plastomes could not be clearly identified with the marker analysis due to the lack of supporting data from previous studies. The GON1, GON2, PHU, STN, BUK1, and CUR plastomes are of S-type, which is the most commonly found in Solanum stenotomum species, whereas ADG1, AJH, and JUZ are found to be of the C-type (Table 5). Both S and C-types are rarely found in modern potato cultivars, and instead are predominantly found in landraces or wild species [33]. Our results confirm these earlier findings.
Table 5. Identifying plastome types from marker analysis.
| Genome | NTCP6 | NTCP7 | NTCP8 | NTCP9 | NTCP12 | NTCP14 | NTCP18 | cp_type |
|---|---|---|---|---|---|---|---|---|
| GON1 | 127 | 173 | 251 | 289 | 129 | 150 | 186 | S |
| GON2 | 127 | 173 | 251 | 289 | 129 | 150 | 186 | S |
| PHU | 127 | 173 | 251 | 289 | 129 | 150 | 186 | S |
| STN | 127 | 173 | 251 | 289 | 129 | 150 | 186 | S |
| BUK1 | 127 | 173 | 251 | 289 | 129 | 150 | 186 | S |
| CUR | 127 | 173 | 251 | 289 | 129 | 150 | 186 | S |
| ADG1 | 173 | 174 | 251 | 289 | 126 | 150 | 187 | C |
| ADG2 | 174 | 174 | 250 | 289 | 127 | 151 | 186 | C/A |
| CHA | 174 | 174 | 250 | 289 | 127 | 151 | 186 | C/A |
| AJH | 174 | 174 | 249 | 289 | 127 | 151 | 186 | C |
| JUZ | 174 | 174 | 250 | 289 | 126 | 150 | 186 | C |
| TBR | 174 | 174 | 255 | 310 | 125 | 150 | 188 | inconclusive |
| BUK2 | 174 | 174 | 253 | 249 | 128 | 154 | 186 | inconclusive |
The in silico PCR product size for each marker used are presented here. The respective plastome type was determined after comparison with previous studies. The ADG2 and CHA plastomes are identified as ambiguous between C/A type cpDNAs due to lack of supporting data from the previous marker analysis studies, while TBR and BUK2 are inconclusive.
The chloroplast DNA types of ADG2, CHA, TBR, and BUK2 were instead identified by in silico restriction endonuclease analysis. The restriction fragment patterns found were compared with the previous studies to identify plastome types [10]. GON1, GON2, PHU, STN, BUK1, CUR, ADG1, AJH, JUZ, CHA, and ADG2 plastomes gained a 3769 bp fragment when digested with BamHI, which indicates that these plastomes are either S-type, C-type, or the A-type. Loss of a 48 bp fragment, when digested with HindIII, confirms GON1, GON2, PHU, STN, BUK1, and CUR as the S-type. Similarly, loss of a ~300 bp fragment when digested with BamHI confirms ADG2 and CHA as A-type. No loss of any fragment when digested with BamHI and HindIII confirms ADG1, AJH, and JUZ as C-type plastome. Since none of these changes were observed in TBR, it is a W-type. Similarly, since BUK2 also does not have any of these changes except for a gain of a 20.9 kb fragment when digested with PvuII, suggests BUK2 has a W2-type chloroplast DNA (Table 6).
Table 6. Restriction endonuclease analysis and polymorphisms in fragment length.
| Genome | BamHI | HindIII | KpnI | PvuII | XhoI | cp type |
|---|---|---|---|---|---|---|
| GON1, GON2, PHU, STN, BUK, CUR | 15.6 kb + 3.76 kb → 19.4 kb | 2.58 kb—48 bp → 2.54 kb | - | - | - | S |
| ADG2, CHA | 15.6 kb + 3.76 kb → 19.4 kb & 3.88 kb—297 bp → 3.59 kb | - | - | - | - | A |
| ADG1, AJH, JUZ | 15.6 kb + 3.76 kb → 19.4 kb | - | - | - | - | C |
| TBR | - | - | - | - | - | W |
| BUK2 | - | - | - | 2 X 20.9 kb → 41.8 kb | - | W2 |
Different polymorphisms were observed in fragment lengths when digested with five restriction enzymes. These length polymorphisms were compared with previous studies to identify plastome types [1, 10]. The RFLP analysis confirms the findings from the marker analysis and also identify the types of ADG2, CHA, BUK2, and TBR.
The cultivated diploid species GON1, GON2, PHU, and STN all have the S-type plastome. The same classification was observed for a majority of these accessions in prior studies [1, 10, 12]. However, we show that BUK1 and BUK2, the two plastomes from the wild diploid species (same accession), have S-type and W2-type cpDNAs, respectively. The majority of Solanum bukasovii accessions studied previously were shown to have the C-type, though four accessions of S. bukasovii were previously demonstrated to have the S-type [11, 34]. A W- type was shown in two accessions of S. bukasovii [11, 34], but a W2-type of S. bukasovii cpDNA has not been mentioned in previous studies. Also, the W2-type has been seen in other wild accessions from potato species [1]. This diversity of plastome types in Solanum bukasovii is likely due to the successful domestication and parallel differentiation from time to time and place to place from ancestral species [11].
S. curtilobum, CUR, which is a cultivated pentaploid, has an S-type plastome like the cultivated diploids. This is in agreement with previous studies [10, 12]. CHA and ADG2 have an A-type plastome, while ADG1 has the C-type. The differences in types of the two S. tuberosum subsp. andigena accessions (ADG1 and ADG2) were reported, having either A, C, or an S-type [1]. Similarly, the A-type was found previously in a few accessions of Solanum chaucha (CHA) [12]. The cultivated diploid AJH and the triploid JUZ also have the C-type plastome. These results are in agreement with the previous studies where S. xajanhuiri (AJH) and S. juzepczukii (JUZ) species were shown to have the C-type [12, 34]. Finally, the tetraploid TBR has a W-type cpDNA, which was previously only observed in one variety of S. tuberosum subsp. tuberosum [10]. The T-type plastome is more commonly found in the cultivated Chilean potato S. tuberosum subsp. tuberosum. It is interesting that the accession of S. tuberosum subsp. tuberosum in our panel has a W-type plastome, and it may partially explain its close relatedness with wild potato species [14].
Polymorphisms responsible for the divergence of chloroplast DNA types
According to the initial study that presented the method of classifying plastome types by restriction endonuclease analysis [10], the W-type is the most primitive type and other plastome types are derived from this. It was shown that other plastome types were derived by a single change in the restriction fragment pattern, except for the T-type. Here, we discuss in detail the point mutations that are responsible for the type divergence. It was shown in vitro that the W2-type (sometimes shown as W”) was derived from the W-type with a single change in the PvuII restriction fragment pattern that led to a 21.2 kb fragment [1, 10]. Here, we show that it is the single point mutation in the PvuII restriction site present in the rps11 gene in the LSC region that is responsible for the origin of the W2-type. The BUK2 plastome has a change of A to G in the PvuII restriction site, which made BUK2 gain a fragment of 20.9 kb (S3 and S4 Figs). Likewise, gain of a 3.66 kb fragment was observed in A, S, and C-type plastomes when in vitro digested with BamHI [10]. Here, we show that a single polymorphism in the BamHI restriction site at the intergenic region between cemA and petA genes in the LSC region is responsible for this gain (S3 Fig). A gain of 3.76 kb fragment was observed in GON1, GON2, AJH, PHU, STN, BUK, CUR, ADG1, ADG2, CHA, and JUZ plastomes with a change of nucleotide from G to A, indicating either A, S, or C-type (S4 Fig).
Furthermore, a single site change (G to C) in the coding region of the ccsA gene in the SSC region forms a second BamHI restriction pattern (ggatcc), as observed in ADG2 and CHA (S3 Fig). This led to these plastomes losing a ~300 bp fragment (S4 Fig). Similarly, a loss of ~350bp fragment was used to distinguish type-A from type-C when digested with BamHI [10]. Finally, a 48 bp deletion mentioned above in GON1, GON2, PHU, STN, BUK1, and CUR in the LSC region makes the HindIII digested fragment 48 bp shorter (S3 and S4 Figs). This loss of 48 bp distinguishes the S-type from the C-type, as mentioned in the previous studies [10]. Also, the PCR product size of the NTCP6 marker can distinguish the S-type from C-type—a product of 127 bp was observed for the S-type plastomes, corresponding to products of 173 and 174 bp in other types. Hence, it clearly shows that a deletion of ~48 bp in the above-mentioned region confirms a plastome to be an S-type. No T-type plastome was found in these 13 potato accessions, but a 241 bp deletion is commonly used to identify this type [11]. These particular polymorphic regions will greatly aid in plastome type identification (Fig 7).
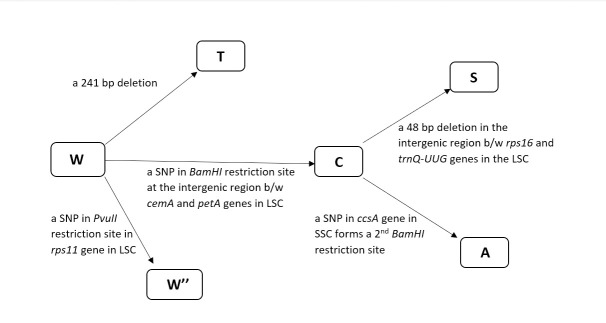
Fig 7. Schematic representation of evolution of cpDNA types in potato.
The W-type is the primitive type of potato plastome from which other types were derived. A 241 bp deletion in the W-type gave rise to the T-type cpDNA [11]. The W2 (W”) is derived from W due to a single nucleotide change. Similarly, the C-type is derived from the W-type by a single nucleotide change which further formed the S- and A-type plastomes. The A-type derived from the C-type due to the formation of a second restriction site by an SNP, while a 48 bp deletion in the C-type led to the S-type.
Phylogenetic analysis
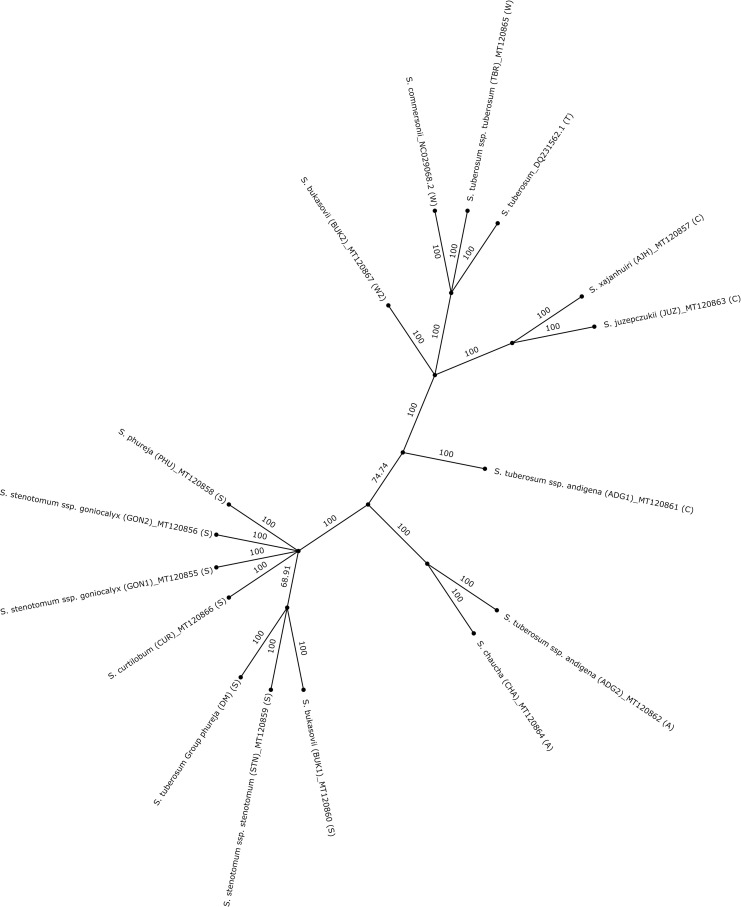
Twelve of the potato accessions in the panel (BUK2 is the exception), were previously classified by their nuclear genomes [14]. Here, the complete sequences of their plastomes (with the addition of BUK2) were analyzed to understand their phylogenetic relationships relative to the nuclear genome classification. The IRb region was omitted to reduce redundancy and bias. In addition, the plastomes of DM, S. commersonii, and S. tuberosum [3, 4, 18], all of which are publicly available, were also included in the phylogeny (Fig 8). All the plastomes group together according to their type. Most of the branches and nodes in the phylogeny are strongly supported by the bootstrap analysis, while the nodes at DM, BUK1, and STN, and the node separating S-type plastome with other types have moderate support. All the cultivated diploids with an S-type plastome are grouped into a single clade, where also BUK1 groups, despite being a wild species. It is interesting to see that the plastome of DM, a double monoploid derived from S. tuberosum group Phureja clone, is in the same clade as that of PHU [18] (Fig 8). The plastome of a cultivated pentaploid species, CUR, is grouped together with the diploid species. Similar results were observed previously but the reason for this is not known, however it can be presumed that S. curtilobum could be derived from a cross between S. juzepcuzkii as a male parent and a cultivated diploid species as a female parent [10]. The plastomes of ADG1 and ADG2 are present in different clades and this is clearly due to the difference in their type. However, the plastome of CHA, a triploid species, is closely related to that of ADG2. This is reasonable since Solanum chaucha (CHA) is believed to be derived from an andigena female and a stenotomum male [10]. The plastomes of another cultivated diploid, AJH and the triploid JUZ are placed close to each other. S. xajanhuiri (AJH) is a bitter potato derived from S. megistacrolobum, which also has a C-type plastome [12]. Finally, BUK2, TBR, S. tuberosum, and S. commersonii plastomes are clustered together in a single clade. This clade contains the W, W2, and T-types, and previous studies have shown that they (W, and T type) group in the same clade [12]. BUK2, a diploid wild species, is close to S. commersonii, which is also a wild diploid species. Not surprising was the fact that the plastome of S. commersonii was found to be phylogenetically distinct from the other cultivated potatoes as this was previously reported in a study of analyses of plastome restriction sites [35]. Consistent with these analyses, S. commersonii and S. tuberosum (which are clustered together) are sexually compatible [36].
Fig 8. Phylogenetic tree of 16 plastomes.
A phylogenetic tree was constructed using complete plastomes of sixteen potato accessions (without the IRb repeat). Plastome types of three other potato species were also determined (types are shown in brackets at the end). It is interesting to see that all the plastomes are grouped together according to their types.
Comparison between the plastome phylogeny and nuclear genome CNV-based classification
A copy number variation (CNV) based classification of the nuclear genomes from this panel of species (except BUK2) was previously presented [14]. That study shows that GON1, GON2, PHU, STN, and ADG1 cluster together with ADG2, with CHA being close to this cluster. A similar relationship is observed in our plastome phylogeny, where ADG2, and CHA cluster closer to the cultivated diploids, GON1, GON2, PHU, and STN (Fig 8). The bitter potatoes JUZ, CUR, and AJH clustered together in the CNV based classification. However, here only JUZ and AJH were in a single cluster, but CUR is closely related to the cultivated diploids. Moreover, TBR and M6 were clustered together and BUK1 remained an outlier as observed in the CNV classification. BUK1 here is grouped together with the other diploids, whereas TBR clustered with other wild species including BUK2. Despite that plastome classification does not necessarily follow species boundaries in potato (many potato species can readily cross) [9], a strong similarity was observed between the CNV based classification [14] and the plastome classification of the selected accessions in this study. Except for the CUR and BUK1, the rest of the phylogeny is in congruence with the CNV based classification of these accessions.
Conclusions
A panel of 13 potato plastomes were assembled and annotated. All of them are highly similar in gene content and genomic structures. This was also previously found for 202 plastomes of Solanum accessions in the section Petota [8], indicating a high degree of conservation. Nonetheless, variation is present in all the plastomes, with BUK2 having the highest number of SNPs and InDels among them. Overall, the ycf1 gene has a greater number of variations compared to the other genes. In addition, identified SSRs, along with SNPs and InDels, can be used as genetic markers in phylogenetic analyses. Most importantly, the plastome types of all accessions were identified. Though BUK1 and BUK2 are from the same accession of the wild species S. bukasovii, they have different plastome types, which is likely due to the accession being collected as a natural population. This also illustrates S. bukasovii being a natural outcrossing wild species, and the importance of plastome analyses for better understanding of potato genetic resources. Moreover, the phylogeny of 16 complete plastome sequences reveal clustering according to the specific types. Even though some congruency has been observed between plastome and nuclear genome clustering of the accessions in the panel, organelle phylogeny does not entirely correspond to the nuclear phylogeny, which is also known from literature. Finally, a detailed description of the polymorphisms responsible for the divergence of plastome types is beneficial for quickly identifying the potato plastome types. A 48 bp deletion was found in the GON1, GON2, PHU, STN, BUK1, and CUR plastomes that is associated only with the S-type plastome. The findings from this panel of potato accessions will be helpful in future research to effectively identify plastome types in potato species, as source for information in breeding programs, as well as to further the understanding of the evolutionary history of the potato.
Supporting information
(PDF)
(PDF)
(A) A point mutation (A to G) in the PvuII restriction site of the BUK2 plastome. (B) Mutation from G to A in the BamHI restriction site in the GON1, GON2, PHU, STN, BUK1, ADG1, ADG2, AJH, CUR, JUZ, and CHA plastomes. (C) A single nucleotide change from G to C in ADG2, and CHA genomes forms a BamHI restriction site. (D) A 48 bp deletion in the GON1, GON2, PHU, STN, CUR, and BUK1 plastomes.
(PDF)
The restriction fragment patterns of five different plastome types found in the 13 accessions are shown here, one accession per plastome type is represented here. (A) PvuII restriction fragment patterns, + indicates a gain of 20.9 kb in the BUK2 plastome. (B) BamHI restriction fragment patterns; × indicates a gain of 3.76 kb in all the plastomes except TBR, and BUK2. ≠ indicates a loss of ~300 bps from a 3.88 kb fragment. (C) HindIII restriction patterns;♦indicates a loss of 48 bps in the 2.58 kb fragment. The circles indicate loss of fragments in BUK2 plastome which were never reported.
(PDF)
Acknowledgments
The authors would like to express their appreciation to Rene Gomez for his support and expertise in selecting the type accessions of each potato taxon to sequence.
Data Availability
The plastome sequences are available from NCBI (accession numbers MT120855-MT120866).
Funding Statement
The authors acknowledge funding through a Nouvelles Initiatives (Project International) grant from the Centre SÈVE (Fonds de recherche du Québec - Nature et technologies (FRQ-NT) to M.V.S., N.A., D.E., and H.H.T.; Compute/Calcul Canada Resource Allocations for Research Portals and Platforms (The Potato Genome Diversity Portal), a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (NSERC) and a Génome Québec (GQ-AAC-2019-2) award to M.V.S.; A-base funding from Agriculture and Agri-Food Canada to H.H.T. from the Management Directed Genomics Call. The authors also gratefully acknowledge the support of the CGIAR Genebank Platform and appreciatively express thanks to the financial support for the sequencing by GIZ on behalf of the Federal Ministry of Economic Cooperation and Development, Germany. The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1.Hosaka K, Hanneman RE. Origin of chloroplast DNA diversity in the Andean potatoes. Theoretical and Applied Genetics. 1988;76(3):333–40. 10.1007/BF00265332 [DOI] [PubMed] [Google Scholar]
- 2.Thode VA, Lohmann LG. Comparative Chloroplast Genomics at Low Taxonomic Levels: A Case Study Using Amphilophium (Bignonieae, Bignoniaceae). Frontiers in Plant Science. 2019;10(796). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chung H-J, Jung JD, Park H-W, Kim J-H, Cha HW, Min SR, et al. The complete chloroplast genome sequences of Solanum tuberosum and comparative analysis with Solanaceae species identified the presence of a 241-bp deletion in cultivated potato chloroplast DNA sequence. Plant Cell Reports. 2006;25(12):1369–79. 10.1007/s00299-006-0196-4 [DOI] [PubMed] [Google Scholar]
- 4.Cho K-S, Cheon K-S, Hong S-Y, Cho J-H, Im J-S, Mekapogu M, et al. Complete chloroplast genome sequences of Solanum commersonii and its application to chloroplast genotype in somatic hybrids with Solanum tuberosum. Plant Cell Reports. 2016;35(10):2113–23. 10.1007/s00299-016-2022-y [DOI] [PubMed] [Google Scholar]
- 5.Liu E, Yang C, Liu J, Jin S, Harijati N, Hu Z, et al. Comparative analysis of complete chloroplast genome sequences of four major Amorphophallus species. Scientific Reports. 2019;9(1):809 10.1038/s41598-018-37456-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spooner DM, Ghislain M, Simon R, Jansky SH, Gavrilenko T. Systematics, Diversity, Genetics, and Evolution of Wild and Cultivated Potatoes. The Botanical Review. 2014;80(4):283–383. [Google Scholar]
- 7.Hawkes JG. The potato: evolution, biodiversity and genetic resources. London: Belhaven Press; 1990. viii + 259 pp. p. [Google Scholar]
- 8.Huang B, Ruess H, Liang Q, Colleoni C, Spooner DM. Analyses of 202 plastid genomes elucidate the phylogeny of Solanum section Petota. Scientific Reports. 2019;9(1):4454 10.1038/s41598-019-40790-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradshaw JE, Ramsay G. Chapter 1—Potato Origin and Production In: Singh J, Kaur L, editors. Advances in Potato Chemistry and Technology. San Diego: Academic Press; 2009. p. 1–26. [Google Scholar]
- 10.Hosaka K. Who is the mother of the potato?—restriction endonuclease analysis of chloroplast DNA of cultivated potatoes. Theoretical and Applied Genetics. 1986;72(5):606–18. 10.1007/BF00288998 [DOI] [PubMed] [Google Scholar]
- 11.Hosaka K. Successive domestication and evolution of the Andean potatoes as revealed by chloroplast DNA restriction endonuclease analysis. Theoretical and Applied Genetics. 1995;90(3):356–63. [DOI] [PubMed] [Google Scholar]
- 12.Sukhotu T, Kamijima O, Hosaka K. Nuclear and chloroplast DNA differentiation in Andean potatoes. Genome. 2004;47(1):46–56. 10.1139/g03-105 [DOI] [PubMed] [Google Scholar]
- 13.Bi Y, Zhang M-f, Xue J, Dong R, Du Y-p, Zhang X-h. Chloroplast genomic resources for phylogeny and DNA barcoding: a case study on Fritillaria. Scientific Reports. 2018;8(1):1184 10.1038/s41598-018-19591-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kyriakidou M, Achakkagari SR, Gálvez López JH, Zhu X, Tang CY, Tai HH, et al. Structural genome analysis in cultivated potato taxa. Theoretical and Applied Genetics. 2020;133(3):951–66. 10.1007/s00122-019-03519-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kyriakidou M, Anglin NL, Ellis D, Tai HH, Strömvik MV. Genome assembly of six polyploid potato genomes. Scientific Data. 2020;7(1):88 10.1038/s41597-020-0428-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dierckxsens N, Mardulyn P, Smits G. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Research. 2016;45(4):e18–e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.PGSC. Genome sequence and analysis of the tuber crop potato. Nature. 2011;475(7355):189–95. 10.1038/nature10158 [DOI] [PubMed] [Google Scholar]
- 19.Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Research. 2001;29(22):4633–42. 10.1093/nar/29.22.4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, et al. GeSeq –versatile and accurate annotation of organelle genomes. Nucleic Acids Research. 2017;45(W1):W6–W11. 10.1093/nar/gkx391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. NCBI BLAST: a better web interface. Nucleic Acids Research. 2008;36(suppl_2):W5–W9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greiner S, Lehwark P, Bock R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Research. 2019;47(W1):W59–W64. 10.1093/nar/gkz238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amiryousefi A, Hyvönen J, Poczai P. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 2018;34(17):3030–1. 10.1093/bioinformatics/bty220 [DOI] [PubMed] [Google Scholar]
- 24.Katoh K, Misawa K, Kuma Ki, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research. 2002;30(14):3059–66. 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Page AJ, Taylor B, Delaney AJ, Soares J, Seemann T, Keane JA, et al. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microbial Genomics. 2016;2(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–2. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraemer L, Beszteri B, Gäbler-Schwarz S, Held C, Leese F, Mayer C, et al. STAMP: Extensions to the STADEN sequence analysis package for high throughput interactive microsatellite marker design. BMC Bioinformatics. 2009;10(1):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Research. 1999;27(2):573–80. 10.1093/nar/27.2.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tiwari JK, Devi S, Chandel P, Ali N, Bhardwaj V, Singh BP. Organelle Genome Analysis in Somatic Hybrids Between Solanum tuberosum and S. pinnatisectum Revealed Diverse Cytoplasm Type in Potato. Agricultural Research. 2016;5(1):22–8. [Google Scholar]
- 30.San Millán RM, Martínez-Ballesteros I, Rementeria A, Garaizar J, Bikandi J. Online exercise for the design and simulation of PCR and PCR-RFLP experiments. BMC Research Notes. 2013;6(1):513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.PAUP* (Phylogenetic Analysis Using Parsimony (and Other Methods)). Dictionary of Bioinformatics and Computational Biology.
- 32.Dong W, Xu C, Li C, Sun J, Zuo Y, Shi S, et al. ycf1, the most promising plastid DNA barcode of land plants. Scientific Reports. 2015;5(1):8348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanetomo R, Gebhardt C. Cytoplasmic genome types of European potatoes and their effects on complex agronomic traits. BMC Plant Biology. 2015;15(1):162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hosaka K, Sanetomo R. Comparative differentiation in mitochondrial and chloroplast DNA among cultivated potatoes and closely related wild species. Genes & Genetic Systems. 2009;84(5):371–8. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez F, Wu F, Ané C, Tanksley S, Spooner DM. Do potatoes and tomatoes have a single evolutionary history, and what proportion of the genome supports this history? BMC Evolutionary Biology. 2009;9(1):191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnston SA, den Nijs TPM, Peloquin SJ, Hanneman RE. The significance of genic balance to endosperm development in interspecific crosses. Theoretical and Applied Genetics. 1980;57(1):5–9. 10.1007/BF00276002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(A) A point mutation (A to G) in the PvuII restriction site of the BUK2 plastome. (B) Mutation from G to A in the BamHI restriction site in the GON1, GON2, PHU, STN, BUK1, ADG1, ADG2, AJH, CUR, JUZ, and CHA plastomes. (C) A single nucleotide change from G to C in ADG2, and CHA genomes forms a BamHI restriction site. (D) A 48 bp deletion in the GON1, GON2, PHU, STN, CUR, and BUK1 plastomes.
(PDF)
The restriction fragment patterns of five different plastome types found in the 13 accessions are shown here, one accession per plastome type is represented here. (A) PvuII restriction fragment patterns, + indicates a gain of 20.9 kb in the BUK2 plastome. (B) BamHI restriction fragment patterns; × indicates a gain of 3.76 kb in all the plastomes except TBR, and BUK2. ≠ indicates a loss of ~300 bps from a 3.88 kb fragment. (C) HindIII restriction patterns;♦indicates a loss of 48 bps in the 2.58 kb fragment. The circles indicate loss of fragments in BUK2 plastome which were never reported.
(PDF)
Data Availability Statement
The plastome sequences are available from NCBI (accession numbers MT120855-MT120866).