Abstract
MicroRNAs are known to be dysregulated in prostate cancer. These small noncoding RNAs can function as biomarkers and are involved in the biology of prostate cancer. The canonical mechanism for microRNAs is post-transcription regulation of gene expression via binding to the 3′ untranslated region of mRNAs, resulting in RNA degradation and/or translational repression. Thus, oncogenic microRNAs, also known as oncomiRs, often have high expression in prostate cancer and target the mRNAs of tumor suppressors. Conversely, tumor-suppressive microRNAs have reduced expression in cancer and typically target oncogenes. Some microRNAs function outside the classical mechanism and serve to stabilize their mRNA targets. Herein, we review contemporary studies that demonstrate oncogenic and tumor-suppressive activity of microRNAs in prostate cancer.
Keywords: MicroRNAs, Prostate cancer, OncomiRs
Introduction
Prostate cancer (PCa) is the second leading cause of cancer-related deaths and the most common cancer diagnosis among men in the United States. PCa is a significant medical burden, as 1 in 9 men are diagnosed with PCa in their lifetime (www.pcf.org). PCa begins as a localized disease that is typically treated with active surveillance, radical prostatectomy, or radiation therapy. Metastatic PCa spreads to the lymph nodes and bone, often causing osteolytic lesions. Metastatic PCa is initially androgen-responsive, termed castrate-sensitive PCa (mCSPC), and reduced with androgen deprivation therapies. PCa that adapts to androgen deprivation therapies becomes castrate-resistant PCa (mCRPC) [1]. MicroRNAs (miRs) have been implicated in all stages of PCa from localized disease to mCRPC.
Over the past two years, miR research in the PCa field remains focused on the characterization of differentially expressed miRs. MiRs are small noncoding RNAs, 18–22 base pairs in length, which regulate the proteome of a cell by causing post-transcriptional degradation or translational inhibition of target mRNAs by binding to the 3′ untranslated region (UTR). This canonical activity of miRs is to reduce protein levels of their targets by either blocking translation or inducing degradation of the target mRNA, via imperfect or perfect complementary binding to the 3′ UTR. MiRs are defined as oncogenic or tumor suppressive based on their targets and differential expression in PCa. Hypothesis generation for such studies often use publicly available databases (e.g. The Cancer Genome Atlas), which can be interrogated to identify dysregulated miRs. However, miRs can also be identified through RNA sequencing, real-time quantitative PCR, or array expression profiling of in-house cohorts. These miRs are then studied for their biological functions, which will be the focus of this review, and utility as PCa biomarkers, which has been reviewed elsewhere [2]. The gene targets of miRs can be confirmed by different assays with varying degrees of evidence. Assays that provide strong evidence include reporter assays (luciferase, etc.), Western blot, and quantitative PCR; assays that provide weaker evidence include microarrays, next-generation sequencing, and pulsed stable isotope labeling by amino acids in cell culture (pSILAC) [3]. This review was limited to 55 references, with emphasis on publications in high-impact journals, inclusion of patient samples, inclusion of genetic mouse models (GEMs), and noncanonical miR actions (Figure 1).
Figure 1.
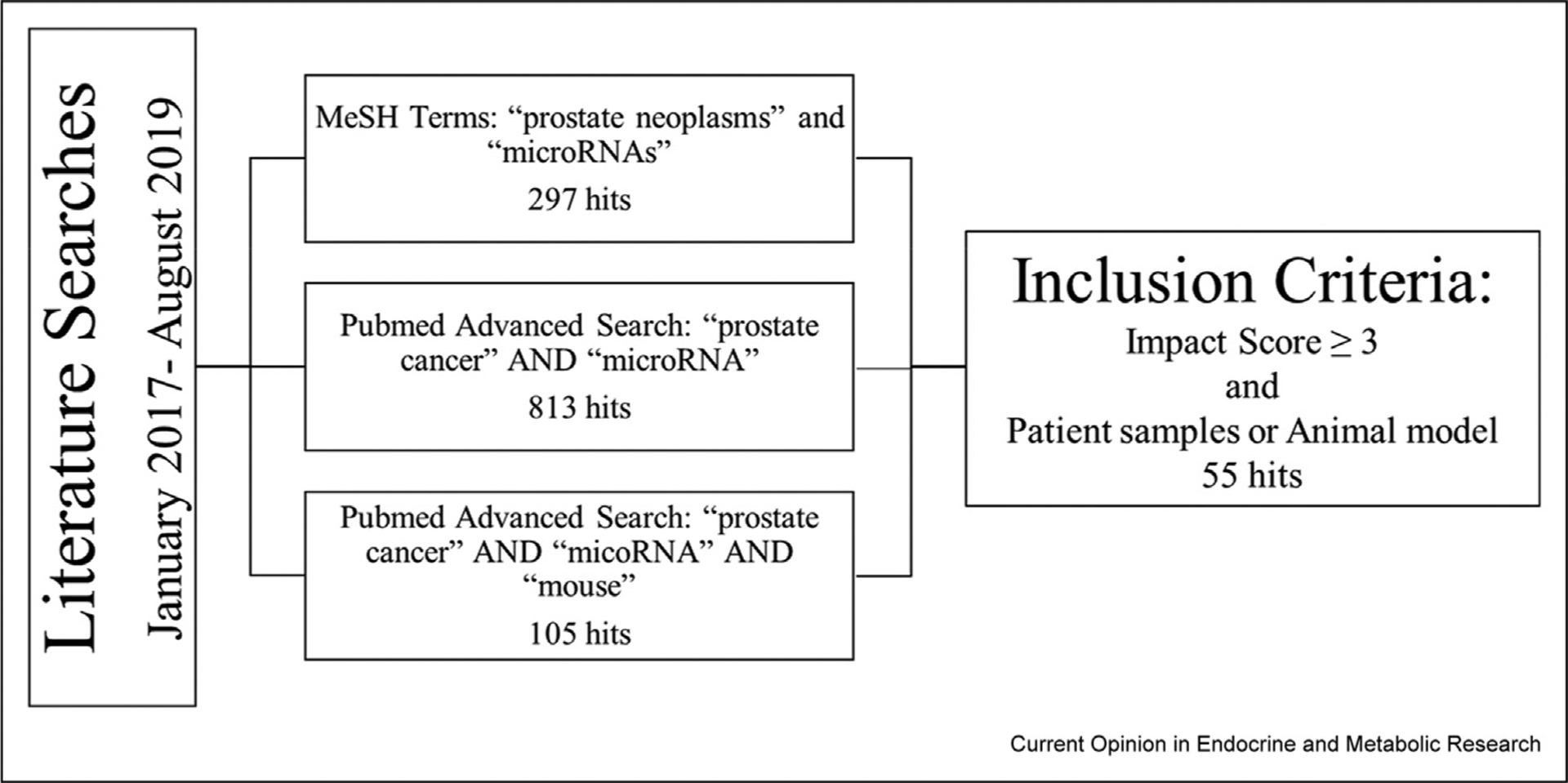
Literature Search and Inclusion Criteria for Review: The Pubmed database and MeSH terms were queried for specific key words relating to this review. Publications discussing the biological function of specific microRNAs in prostate cancer were compiled, and those that utilized patient samples or animal models were selected for potential inclusion. Author discretion was used for final literature selection.
Oncogenic and tumor suppressor microRNAs identified in localized prostate cancer
Putative oncomiRs and tumor suppressor miRs are identified by differential expression in benign tissue versus PCa in radical prostatectomy samples from localized disease. In these localized tumors, the pathways and processes regulated by miRs include epithelial–mesenchymal transition (EMT), microenvironment, metabolism, and proliferation (Table 1). Gene targets of miRs are predicted by sequence-based prediction algorithms and often through effects of miR overexpression on the 3’ UTR.
Table 1.
MiR dysregulated in localized PCa.
| Local PCa OncomiR | PCa phenotype and target assays | Pathway/mRNA target | Patient samples | References |
|---|---|---|---|---|
| miR-30d | SQX, L | Angiogenesis/MYPT1 | N = 12, matched frozen RP; N = 225 PCa and N = 25 benign FFPE RP; MSKCC and TCGA | [19] |
| miR-182 | SQX | Wnt pathway | N = 25, paired fresh RP | [17] |
| miR-183 cluster | SQX | Metabolism/ZIP1 | N = 2 PrE cells | [24] |
| miR-193a-5p | SQX, L | EMT/PCHDA | N = 62, FFPE RP; N = 35, benign, FFPE TURP | [8] |
| miR-486-5p | SQX, L | TGFβ/SMAD2 | N = 23, paired FFPE RP; GSE36802, GSE8126, GSE23022, MSKCC, GSE54010, GSE45604 | [12] |
| miR-543 | OX, L | EMT/RKIP | N = 42, fresh and FFPE RP | [5] |
| miR-590-3p | SQX, L | INPP4B | N = 28, paired biopsy | [27] |
| Local PCa TS miR | PCa phenotype and target assays | Pathway/mRNA target | Patient samples | References |
| miR-124-3p, miR-129-5p, miR-378 | L | Metabolism/CPT1A, CACT, CRAT | N = 30, FFPE RP LCM | [25] |
| miR-33a | L | Metabolism/PIM1 | N = 18, paired frozen RP; MSKCC | [26] |
| miR-129 | SQX, L | EMT/Growth/EST1 | N = 30, paired frozen RP | [7] |
| miR-143 | SQX with nanoparticle miR delivery, L | UPAR | N = 26, frozen RP | [33] |
| miR-150 | SQX, L | EMT/WNT/TRPM4 | N = 87, paired frozen RP; GSE30994, GSE38241, GSE46602, GSE55945, GSE103512, TCGA | [18] |
| miR-200c-3p | SQX, L | EMT/ZEB2 | N = 22, paired RP; TCGA | [6] |
| miR-218 | SQX, rabbit corneal assay, L | Angiogenesis/RICTOR | GSE21036 | [20] |
| miR-199a-5p | L | Angiogenesis/HIF-1A | N = 134, PCa; N = 69 benign FFPE needle biopsy and transurethral resection; N = 6 frozen RP | [21] |
| miR-338-3p | SQX, L | Growth/RAB23 | N = 24, paired frozen RP; GSE21036 | [35] |
| miR-384 | SQX, L | HOXB7 | [34] | |
| miR-487a-3p | SQX, L | Growth/CCND1 | GSE8126, GSE6956; PCa tissue chip | [29] |
| miR-212 | SQX, L | Proliferation/EN2 | N = 58, paired frozen RP | [31] |
| miR-455-3p | SQX, L | Proliferation/eIF4E | N = 47, PCa; N = 18 benign; N = 10 paired frozen RP | [30] |
| miR-491-5p | SQX, L | PDGFRA | N = 18, paired frozen RP | [28] |
| miR-10a | SQX, L | YAP pathway/KDM4A | N = 34, paired RP | [32] |
| miR-539 | SQX, L | EMT/TGFβ/DLX1 | N = 134, RP; GSE55945, GSE45016, GSE38241, TCGA | [11] |
| miR-198 | SQX | MIB1 | CPC-GENE | [36] |
PDX = patient-derived xenograft; OX = orthotopic xenograft; SQX = subcutaneous xenograft; L = luciferase; ICI = intracardiac injection; CAM = chick chorioallantoic membrane assay; TVI = tail vein injection; ZEM = zebrafish extravasation assay; RP = radical prostatectomy; BPH = benign prostatic hyperplasia; PCa = prostate cancer; PrE = primary epithelial cell; TURP = transurethral resection of the prostate; TCGA = The Cancer Genome Atlas; CPC-GENE = The Canadian Prostate Cancer Genome Network; MSKCC = Memorial Sloan Kettering Cancer Center data set (GSE21032); GSE# = NCBI GEO accession ID.
MiRs have been shown to regulate EMT by phenotypic assays and via direct targets known to be involved in the EMT pathway. In PCa, EMT is stimulated by multiple pathways (transforming growth factor β [TGFβ], inflammation, cytokines, etc.) with a wide array of downstream effectors which all converge on characteristic EMT changes: decreased E-cadherin and increased Vimentin, N-cadherin, and Snail1 protein levels [4]. The oncomiR-543 induced a switch from high E-cadherin to high Vimentin, a hallmark of EMT [5]. On the other hand, tumor-suppressive miR-200c-3p and miR-129 increased E-cadherin by targeting ZEB2 [6] and ETS1 [7], respectively. The oncomiR-193a-5p stands out among the EMT regulatory miRs because it was identified by an indirect approach through a circular RNA (circRNA) [8*]. circAMOTL1L expression is low in high-grade PCa and functions as a miR-193a-5p ‘sponge’ with 6 binding sites [8]. Expression of circAMOTL1L and a miR-193a-5p inhibitor resulted in smaller PC3 xenografts than either alone.
TGFβ and Wnt pathways are key regulators of the tumor microenvironment and EMT [9]. TGFβ stimulation results in the activation of SMAD proteins which causes specific transcriptional changes as well as increased processing of pri-miRs to mature miRs [10]. OncomiR-486-5p and tumor suppressor miR-539 have opposing effects on the TGFβ pathway and downstream-activated SMAD transcription factors. miR-539 suppresses the pathway by inhibiting DLX1, an activator of SMAD4 [11]. Somewhat paradoxical, SMAD2 is the direct target of an oncomiR miR-486-5p [12]. However, tumor-suppressive functions of SMAD2 have been reported by others [13,14], and TGFβ is known to have tumor-suppressing activity in early PCa, the phenomenon known as the ‘TGFβ paradox’ [15]. Similarly, oncomiR-182 and tumor suppressor miR-150 have opposing effects on the Wnt pathway. Wnt pathway activation results in the translocation of β-catenin to the nucleus to induce transcriptional changes and EMT [16]. Several Wnt pathway inhibitors are direct targets of miR-182, resulting in larger PCa xenografts with miR overexpression [17], while miR-150 targeted TRPM4, an activator of Wnt signaling [18].
Angiogenesis is regulated by oncomiR-30d and tumor suppressor miR-218 via opposing effects on the vascular endothelial growth factor A (VEGFA) pathway. miR-30d–overexpressing LNCaP and DU145 xenografts had significantly higher microvessel density than miR-30de knockdown cells [19]. Conversely, CWR22Rv1 xenografts overexpressing miR-218 formed smaller tumors with less CD31+ tumor cells, and in a rabbit cornea model, these cells did not produce a neovascular response [20]. Vascular endothelial growth factor (VEGF) expression was reduced by miR-199a-5p, a miR lower in PCa biopsies than in benign prostatic hyper-plasia and benign prostate tissue, via targeting of hypoxia-inducible factor 1-alpha (HIF-1α) [21].
Metabolism in the prostate is distinct from that in other organs because of a block in the tricarboxylic acid cycle (TCA) cycle that provides high citrate for the seminal fluid [22]. PCa is metabolically unique compared with other cancers because the normal prostate favors glycolysis for energy production but switches to the TCA and oxidative phosphorylation during localized disease [23]. Stable expression of the miR-183 family cluster (miR-183, miR-96, and miR-182) in RWPE1 benign prostate cells altered the metabolism by inhibition of ZIP1, a zinc transporter responsible for high levels of zinc in benign prostate that suppresses the TCA cycle [24]. miR-183 cluster–overexpressing PC3 xenografts were larger than controls. The carnitine cycle and fatty acid metabolism are regulated by the tumor-suppressive miR-124-3p, miR-129-5p, and miR-378 [25*], whose reduced levels in PCa leads to increased expression of their target mRNAs, CPT1A, CACT, and CrAT, allowing PCa cells to favor fatty acid oxidation for energy production. Along these same lines, reduced expression of miR-33a promoted beta-oxidation of fatty acids through the upregulation of PIM1 and other targets [26].
Proliferation is a common endpoint in phenotypic assays that is often regulated by miRs. miR-590-3p was found to increase proliferation by targeting INPP4B [27], while the tumor suppressor miR-491-5p, miR-487a-3p, miR-455-3p, miR-212, and miR-10a reduced proliferation by targeting PDGFRA [28], CCND1 [29], eIF4E [30], EN-2 [31], and KDM4A [32], respectively, in PCa cell lines and mouse xenografts. MiR mimics were used to overexpress these miRs in mouse xenografts, and miR-590-3p increased tumor size, while the tumor suppressor miR-overexpressing cells resulted in smaller tumors. miR-143, miR-384, miR-338-3p, and miR-198 all have a tumor suppressor phenotype mediated through a confirmed target (UPAR [33], HOXB7 [34], RAB23 [35], and MIB1 [36], respectively), but the exact pathway was not determined. In further validation of the tumor-suppressive role of miR-143, Szczyrba et al. [37] found this miR to be the most downregulated (~fourfold) in PCa compared with noncancerous prostate tissue by deep sequencing, and Wach et al. [38] had similar findings in a microarray data set.
Oncogenic and tumor suppressor microRNAs in advanced prostate cancer
PCa metastases require distinct transcriptional activity from localized PCa to support metastasis and seeding of extraprostatic tissues such as bone and lung [39]. To identify miRs with a role in advanced PCa, many studies compared miR expression in mCSPC and mCRPC with that in localized PCa. The miRs in metastases effect similar pathways to those of localized disease, and studies use specialized metastasis models, such as chick chorioallantoic membrane (CAM) assays, zebrafish xenografts, mouse intracardiac injections, tail vein injections, and subcutaneous and orthotopic xenografts (Table 2).
Table 2.
MiRs in metastatic and advanced PCa.
| Metastasis OncomiR | PCa phenotype and target assays | Pathway/mRNA target | Patient samples | References |
|---|---|---|---|---|
| miR-96 | ICI, L | EGF/ETV6 | N = 14, PCa; N = 55, PCa TMA; MSKCC | [61] |
| miR-149-3p | ICI, zebrafish xenografts, L | Angiogenesis/NF-kβ/DAB2IP | TCGA, GSE21032 | [54] |
| miR-194 | CAM, TVI, OX, L | EMT/SOCS2 | N = 44, RP; TCGA, MSKCC, GSE35988, GSE46691, GSE62116, GSE62667, GSE72291, GSE79956, GSE79957 | [40] |
| miR-22 | MTA1 transgenic mouse, L | EMT/CDH1 | N = 10, fresh RP; TCGA | [46] |
| miR-210-3p | ICI, L | EMT/NF-kβ/TNIP1, SOCS1 | N = 140, PCa; N = 9, metastatic tissue; TCGA | [55] |
| miR-652 | SQX, L | EMT/NED/PPP2R3A | N = 585, RP | [47] |
| miR-210-3p | ICI (PICK1 OE cells), L | TGFβ signaling/PICK1 | N = 198 PCa, N = 10 bone met FFPE, N = 51 frozen RP or biopsy, TCGA, GSE46602 | [50] |
| Metastasis TS miR | PCa phenotype and target assays | Pathway/mRNA target | Patient samples | References |
| miR-133a-3p | ICI, TVI, L | Growth/EGFR, FGFR1, IGF1R, MET | N = 20 paired; N = 225 PCa, N = 48 benign, frozen needle biopsy or surgery; TCGA | [60] |
| miR-141 | SQX, OX, L | Stemness/CD44, EZH2, RAC1, CDC42, ARPC5, CDC42EP3 | N = 21 RP, N = 26 primary PCa cell lines | [58] |
| miR-141-3p | ICI, L | NF-kβ/TRAF5/6 | N = 141 surgery or needle biopsy, TCGA | [56] |
| miR-203 | SQX, TVI | Angiogenesis/SNAI2 | TCGA | [53] |
| miR-221-5p | ZEM, OX | EMT | GSE21036 | [42] |
| miR-338-5p, miR-421 | CAM, SQX, L | SPINK1 | N = 20, frozen PCa; N = 238, FFPE TMA; TCGA, GSE45604, MSKCC | [45] |
| miR-19a-3p | Tibial injection | TGFβ/SMAD2/4 | N = 121 frozen surgery or needle biopsy; TCGA | [49] |
| miR-383 | SQX, ICI, L | Stemness/CD44 | N = 122, FFPE PCa; TCGA | [59] |
| miR-449a | SQX, L | Angiogenesis/PrLZ | N = 38, frozen RP | [44] |
| miR-466 | OX, ICI, L | Bone metastasis/RUNX2 | N = 48, matched LCM RP; TCGA | [65] |
| miR-1247 | TVI | EMT/NRP1 | GDS2547 | [41] |
| miR-3622a | OX, ICI, L | EMT/ZEB1, SNAI2, CTNNB1 | N = 138, paired microdissected FFPE RP; TCGA | [43] |
| CRPC miR | PCa phenotype and target assays | Pathway/mRNA target | Patient samples | References |
| miR-100-5p | PDX with androgen ablation | Survival/MTOR | N = 12, PDX | [66] |
| miR-196b-3p | Allograft SQX with castration | NFkB/MEIS2 | N = 80, PCa tissues | [57] |
| miR-302/367 cluster (302a, 302b, 302d, 367) | OX and SQX with castration, L | Hippo-YAP pathway/LATS2 | N = 29, PCa; N = 22, benign fresh RP | [63] |
| miR-744 | SQX, L | Wnt/NKD1 | N = 10, ADPC; N = 10, CRPC, frozen RP and TURP; MSKCC | [51] |
| miR-1205 | SQX, L | EGLN3 | N = 12, paired CRPC, RP LCM FFPE; TCGA | [62] |
PDX = patient-derived xenograft; OX = orthotopic xenograft; SQX = subcutaneous xenograft; L = luciferase; ICI = intracardiac injection; CAM = chick chorioallantoic membrane assay; TVI = tail vein injection; ZEM = zebrafish extravasation assay; RP = radical prostatectomy; BPH = benign prostatic hyperplasia; PCa = prostate cancer; PrE = primary epithelial cell; TURP = transurethral resection of the prostate; TCGA = The Cancer Genome Atlas; CPC-GENE = The Canadian Prostate Cancer Genome Network; MSKCC = Memorial Sloan Kettering Cancer Center data set (GSE21032); GSE# = NCBI GEO accession ID.
For EMT, miR-194 [40] increased mesenchymal protein markers in PC3 and 22Rv1 cells, while the tumor-suppressive miR-1247 [41], miR-221-5p [42], miR-3622a [43], miR-449a [44], and miR-338-5p/421 [45] increased epithelial markers. miR-22 directly targets and reduces E-cadherin to promote EMT [46]. Das et al. [40*] observed that serum levels of miR-194 were also significantly higher in patients with metastatic castrate-resistant prostate cancer (CRPC) than in those with localized PCa. When miR-194–overexpressing PC-3 cells were injected orthotopic into nude mice, 6 of 9 miR-overexpressing xenografts developed metastases, whereas all control tumors remained localized to the prostate [40*], supporting potent oncogenic activity of this miR.
The repression of tumor-suppressive miRs in metastatic disease occurs by both genomic loss and promoter hypermethylation. Bucay et al. [43] observed that miR-3622a had low expression in 70% of CRPC because of both homozygous/heterozygous deletion at the locus (8p21) (50% of patients) and epigenetic silencing with hypermethylation (55% of PCa tumors). Two SPINK1-targeting tumor suppressor miRs, miR-338-5p and miR-421, are also silenced in PCa by promoter hypermethylation [45]. Restoration of miR-338-5p and miR-421 suppressed metastatic bone, liver, and lung lesions of 22Rv1 subcutaneous xenografts [45]. Nam et al. [47] showed that miR-652 targets PPP2R3A, leading to oncogenic activity at several stages of PCa progression via induction of EMT (PC3 cells) and neuroendocrine-like differentiation in LNCaP cells. These phenotypes were mediated by different downstream pathways in each cell type, activation of the PI3K-AKT and ERK-1/2 pathways in PC3 cells and activation of the Wnt pathway in LNCaP cells [47], demonstrating that the transcriptional background of cells is important for the phenotype.
The tumor microenvironment of advanced PCa is also influenced by miRs [48]. miR-19a-3p was downregulated in metastatic compared with nonmetastatic PCa tissue and inhibited bone metastasis after intratibial injections of SCID mice by targeting SMAD2 and SMAD4, which are downstream mediators of TGFβ signaling [49]. miR-210-3p augmented TGFβ signaling by targeting PICK1, a tumor suppressor of bone metastasis [50]. Oncogenic miR-744 activated Wnt signaling in androgen-independent PCa cells. In patient tissues, miR-744 was highly expressed in mCRPC compared with mCSPC and its expression associates with recurrence-free survival [51]. Conversely, tumor-suppressive miR-1 [52] suppresses Wnt signaling through androgen receptor (AR) regulation.
Angiogenesis is a critical process during PCa metastasis. Tumor-suppressive miR-203 and miR-149-3p inhibit angiogenesis by a complementary mechanism. miR-203–overexpressing DU145 subcutaneous xenografts had smaller tumors with less blood microvessels than controls [53] and conditioned media from miR-203–overexpressing cells caused a decrease in human umbilical vein endothelial cells (HUVEC) tube formation [53]. Overexpression of miR-149-3p in PC3 also inhibited angiogenesis, but by inhibiting HUVEC proliferation and migration [54].
Several miRs target inflammatory pathways in mCSPC and mCRPC. miR-210-3p [55] and miR-141-3p [56] both promote bone metastases by increasing NF-kβ signaling. miR-210-3p is highly expressed in bone lesions compared with localized PCa and behaves similar to an oncogene by targeting TRAF5/6, which is part of the TNFα pathway leading to NF-kβ activation [55]. AntagomiR-210-3p was able to reduce bone metastasis and prolong survival of nude mice that received intracardiac PC3 cells. The miR-210-3p locus was amplified in 19 of 68 bone metastases from patients compared with only 1 of 81 nonbone metastases, suggesting the requirement of this miR for only a subset of metastases. On the other hand, miRs can be regulated by the constitutive NF-kβ signaling that often occurs in mCRPC. miR-196b-3p is one such miR that was shown to function as an oncogene using the Myc-CaP mouse. miR-196b3p–overexpressing Myc-CaP tumors were orders of magnitude larger than controls in immune-competent castrated mice [57].
miR-141 and miR-383 are involved in the putative PCa stem cell population. miR-141 is low in the CD44+ population of patient PCa tumors, and inducible miR-141 inhibited lung metastasis of PC3, DU145, and LAPC9 orthotopic xenografts, compared with controls [58**]. Huang et al. [56] independently showed that miR-141-3p suppressed metastasis in PCa, supporting the results of Liu et al. [58**]. miR-383 targets CD44 directly, and miR-383–overexpressing PC3 injected into nude mice had decreased CD44 protein levels and reduced metastases compared with controls. The miR-383 locus (8p22) is deleted in ~48% of PCa cases in The Cancer Genome Atlas data set, confirmed by their own patient cohort, similar to this group’s miR-3622a finding discussed earlier [59].
miR-133a-3p and miR-96 were found to have opposing effects on proliferation and metastatic potential; miR-133a-3p targeted multiple receptors, including epidermal growth factor receptor (EGFR), fibroblast growth factor receptor 1 (FGFR1), and hepatocyte growth factor receptor (MET), which would normally inhibit phosphatidylinositol 3-kinase/protein kinase B (PI3K/AKT) signaling [60], and miR-96 was found to be upregulated by EGFR signaling [61]. Both the downregulation of miR-133a-3p and the upregulation of miR-96 promoted PCa proliferation and metastasis, as expected. miR-1205 was also found to enhance PCa proliferation. The copy number of the locus containing miR-1205 was increased in CRPC tumors and cell lines compared with androgen-dependent cancers [62*]. CRISPR/Cas9 was used to knock out miR-1205 in androgen-independent PC3 cells, leading to decreased proliferation and significantly smaller tumors with less Ki67 expression than in controls when subcutaneously injected into nude mice.
The miR-302/367 cluster is oncogenic and expressed at high levels in localized PCa patient tissues as well as the PTENflox/flox GEM model [63*]. Stable overexpression of this miR cluster in LNCaP androgen-dependent PCa cells increased proliferation and decreased apoptosis under androgen-deprived culture conditions. miR-302/367–overexpressing orthotopic LNCaP and PC3 xenografts had increased emergence of CRPC compared with controls after castration. Conversely, knockdown miR-302/367 PC3 subcutaneous grafts had reduced formation incidence, growth rate, and tumor size compared with controls. Hippo pathway kinase LATS2 was ultimately identified as a target gene that could reverse miR-302/367 effects on tumor growth.
There are several unique pathways in metastatic PCa that are partially regulated by miRs [64]. Colden et al. found that miR-466 targets the osteogenic transcription factor RUNX2, resulting in decreased spontaneous bone metastases after intracardiac injections of miR-466–overexpressing PC3 cells [65]. miR-100-5p was upregulated in patient-derived xenografts after surgical castration and CRPC relapse [66*]. Likewise, in vitro, anti-miR-100-5p treatment of androgen-deprived LNCaP cells, or androgen-independent DU145 cells, increased apoptosis, but the targets of miR-100-5p were not explored.
MiRs that directly regulate androgen signaling
Androgens regulate normal prostate growth and that of PCa. Multiple miRs have been shown to regulate androgen activity and contribute to PCa survival and growth [67,68]. These miRs contribute to constitutive ligand-independent activation of ARs or through other mechanisms [69] (Table 3).
Table 3.
MiRs involved in AR signaling.
| AR miR | PCa phenotype and target assays | Pathway/mRNA target | Patient samples | References |
|---|---|---|---|---|
| miR-1 | SQX, L | AR signaling/TCF7 | N = 40 biopsy; MSKCC | [52] |
| miR-32 | miR-32 transgenic mice alone and with Pten+/− | Growth/RAC2 | [75] | |
| miR-130b | ICI, OX, L | AR | N = 19, PCa; N = 16 healthy donor serum, N = 12 PCa serum; N = 4 PrE, N = 14 PCa PrE; N = 4 bone metastases tissue; MSKCC | [73] |
| miR-181c-5p | OX with enzalutamide treatment, L | enzalutamide resistance/ARv7 | N = 13, BPH; N = 26, PCa | [70] |
| miR-346, miR-361-3p, miR-197-3p | CRPC PDX, L | AR | N = 1, PDX; TCGA, GSE34932 | [72] |
PDX = patient-derived xenograft; OX = orthotopic xenograft; SQX = subcutaneous xenograft; L = luciferase; ICI = intracardiac injection; CAM = chick chorioallantoic membrane assay; TVI = tail vein injection; ZEM = zebrafish extravasation assay; RP = radical prostatectomy; BPH = benign prostatic hyperplasia; PCa = prostate cancer; PrE = primary epithelial cell; TURP = transurethral resection of the prostate; TCGA = The Cancer Genome Atlas; CPC-GENE = The Canadian Prostate Cancer Genome Network; MSKCC = Memorial Sloan Kettering Cancer Center data set (GSE21032); GSE# = NCBI GEO Accession ID.
miR-1 and miR-181c-5p are both tumor suppressor miRs that act at different points in the androgen axis. miR-1 is induced by the AR [52], and miR-181c-5p represses ARV7, a splice variant of AR that supports mCRPC development. In a regulatory mechanism similar to miR-193a-5p described earlier, miR-181c-5p binds a circRNA, circRNA17 [70*]. However, circRNA17 stabilizes and supports the activity of miR-181c-5p, rather than serving as an inhibitory sponge. This was discovered with PCa orthotopic xenografts that only responded to combined expression of miR-181c-5p with circRNA17, but had no effect with miR-181c-5p alone.
MiRs have also been implicated in diverse mechanisms of drug resistance in the treatment of PCa [71]. Fletcher et al. found that miR-346, miR-361-3p, and miR-197-3p act noncanonically by binding and stabilizing the 3′ UTR of ARs, resulting in increased AR protein [72]. Xenografts derived from patients with CRPC displayed increased expression of miR-197 and miR-361-3p after enzalutamide treatment. Cannistraci et al. [73] found that miR-130b was an oncomiR that targets ARs, leading to its degradation. They determined that the c-Met oncogene leads to the overexpression of miR-130b as a mechanism of hormone therapy resistance and that patients with recurrent PCa or CRPC expressed higher levels of miR-130b in their primary tumors than in nonrecurrent patient tumors.
miR-32 is an androgen-regulated miR that is highly expressed in CRPC and increases PCa cell growth in vitro [74]. Latonen et al. [75**] developed a probasin-driven transgenic miR-32 mouse model to assess the effects of miR-32 in prostate epithelium. The miR-32+/+ mice had an increased proliferative index by Ki67 and PCNA in all prostate lobes and increased goblet cell metaplasia and PIN compared with wild-type mice, indicating weak oncogenic activity. In addition, no further oncogenic activity was observed when the mice were crossed with heterozygous Pten mice (Pten+/−) to create mir-32+/+ x Pten ± mice, suggesting that this miR is not sufficient to induce cancer but does induce precancerous lesions.
Discussion: challenges and future directions in the PCa miR field
Many miRs have recently been implicated in all stages of PCa progression and may represent promising therapeutic targets or biomarkers to monitor disease. However, challenges exist in the miR field, regardless of the disease model. First, to validate the mRNA targets of miRs, a variety of methods are used with intrinsically different evidence strength levels. Targets can be validated by assays that provide strong evidence for regulation by a miR (reporter assay, Western blot, and qPCR) or less strong evidence (microarray, next-generation sequencing, pSILAC, etc.) [3]. Second, miRs are promiscuous regulators that are able to bind to and target multiple mRNAs, resulting in widespread effects on cell functions. Finally, complicating things further, different miRs can bind to and regulate the same mRNA and potentially have opposing effects on mRNA stability (as shown for miR-346, miR-361-3p, and miR-197-3p on ARs) [72]. These aspects of miR biology make studying them in isolation difficult and potentially biased. The typical gold standard to determine the function of genes is a transgenic knockout model; however, because miRs have overlapping target genes and pathways, they can compensate for one another. Thus, the functions of a miR determined in vitro may not be observed in a knockout model. This highlights the importance of the miR-32 study conducted by Latonen et al. [75], as they were able to successfully represent the miR function through the use of a transgenic miR mouse model. A particular challenge to studying miRs in PCa is the limited number of commonly used PCa cell lines, which were all derived from metastasis. MiRs found higher in localized disease are routinely assessed by overexpression in a PCa cell line derived from metastasis, which is not the appropriate transcriptional background.
MiRs are being studied for disease mechanisms and clinical utility at all stages of PCa. The challenge lies in determining how the collective miRNome functions in different diseased transcriptional backgrounds to promote PCa progression and the potential to manipulate miRs for therapeutic benefit.
Acknowledgements
The authors were supported by the grants Department of Defense Prostate Cancer Research Program Idea Development Award PC150495 (Nonn and Zenner) and the Urology Care Foundation Research Scholar Award Program from the North Central Section of the American Urological Association (Baumann).
Footnotes
Conflict of interest statement
Nothing declared.
References
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
* * of outstanding interest
- 1.Karantanos T, Corn PG, Thompson TC: Prostate cancer progression after androgen deprivation therapy: mechanisms of castrate resistance and novel therapeutic approaches. Oncogene 2013, 32:5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cozar J, et al. : The role of miRNAs as biomarkers in prostate cancer. Mutat Res Rev Mutat Res 2019, 781:165–174. [DOI] [PubMed] [Google Scholar]
- 3.Huang HY, , et al. miRTarBase: Updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res 2020, 48:D148–D154. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Culig Z: Epithelial mesenchymal transition and resistance in endocrine-related cancers. Biochim Biophys Acta Mol Cell Res 2019, 1866:1368–1375. [DOI] [PubMed] [Google Scholar]
- 5.Du Y, et al. : MiR-543 promotes proliferation and epithelial-mesenchymal transition in prostate cancer via targeting RKIP. Cell Physiol Biochem 2017, 41:1135–1146. [DOI] [PubMed] [Google Scholar]
- 6.Zhang J, et al. : MicroRNA-200c-3p/ZEB2 loop plays a crucial role in the tumor progression of prostate carcinoma. Ann Transl Med 2019, 7:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu S, et al. : MiR-129 inhibits cell proliferation and metastasis by targeting ETS1 via PI3K/AKT/mTOR pathway in prostate cancer. Biomed Pharmacother 2017, 96:634–641. [DOI] [PubMed] [Google Scholar]
- 8.*.Yang Z, et al. : Dysregulation of p53-RBM25-mediated circAMOTL1L biogenesis contributes to prostate cancer progression through the circAMOTL1L-miR-193a-5p-Pcdha pathway. Oncogene 2019, 38:2516–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]; MiR-193a-5p was found to be upregulated due to a downregulation of a circular RNA, CircAMOTL1L, which normally functions as a sponge for this miR. PC3 cells overexpressing circAMOTL1L and depleted of miR-193a-5p resulted in reduced xenograft tumor volumes.
- 9.Zhang J, Tian X-J, Xing J: Signal transduction pathways of EMT induced by TGF-β, SHH, and WNT and their crosstalks. J Clin Med 2016, 5:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butz H, et al. : Crosstalk between TGF-beta signaling and the microRNA machinery. Trends Pharmacol Sci 2012, 33: 382–393. [DOI] [PubMed] [Google Scholar]
- 11.Sun B, et al. : MicroRNA-539 functions as a tumour suppressor in prostate cancer via the TGF-beta/Smad4 signalling pathway by down-regulating DLX1. J Cell Mol Med 2019, 23: 5934–5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Y, et al. : The miR-486-5p plays a causative role in prostate cancer through negative regulation of multiple tumor suppressor pathways. Oncotarget 2017, 8: 72835–72846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoot KE, et al. : Keratinocyte-specific Smad2 ablation results in increased epithelial-mesenchymal transition during skin cancer formation and progression. J Clin Invest 2008, 118: 2722–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ju W, et al. : Deletion of Smad2 in mouse liver reveals novel functions in hepatocyte growth and differentiation. Mol Cell Biol 2006, 26:654–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boguslawska J, et al. : TGF-beta and microRNA interplay in genitourinary cancers. Cells 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kharaishvili G, et al. : Wnt signaling in prostate development and carcinogenesis. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 2011, 155:11–18. [DOI] [PubMed] [Google Scholar]
- 17.Wang D, et al. : MiR-182 promotes prostate cancer progression through activating Wnt/beta-catenin signal pathway. Biomed Pharmacother 2018, 99:334–339. [DOI] [PubMed] [Google Scholar]
- 18.Hong X, Yu JJ: MicroRNA-150 suppresses epithelial-mesenchymal transition, invasion, and metastasis in prostate cancer through the TRPM4-mediated beta-catenin signaling pathway. Am J Physiol Cell Physiol 2019, 316: C463–C480. [DOI] [PubMed] [Google Scholar]
- 19.Lin ZY, et al. : MicroRNA-30d promotes angiogenesis and tumor growth via MYPT1/c-JUN/VEGFA pathway and predicts aggressive outcome in prostate cancer. Mol Canc 2017, 16:48. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.*.Guan B, et al. : Tumor-suppressive microRNA-218 inhibits umor angiogenesis via targeting the mTOR component RICTOR in prostate cancer. Oncotarget 2017, 8:8162–8172. [DOI] [PMC free article] [PubMed] [Google Scholar]; MiRs-124-3p, 129-5p, and 378 were found to be downregulated in PCa, resulting in the increased expression of their targets: CPT1A, CACT, and CrAT. The increase in these targets led to more effective fatty acid metabolism in PCa cell lines, thereby participating in the dysregulation of metabolism in PCa.
- 21.Zhong J, et al. : Downregulation of miR-199a-5p promotes prostate adeno-carcinoma progression through loss of its inhibition of HIF-1alpha. Oncotarget 2017, 8:83523–83538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costello LC, Franklin RB: The clinical relevance of the metabolism of prostate cancer; zinc and tumor suppression: connecting the dots. Mol Canc 2006, 5:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cutruzzolà F, et al. : Glucose metabolism in the progression of prostate cancer. Front Physiol 2017, 8:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dambal S, et al. : The miR-183 family cluster alters zinc homeostasis in benign prostate cells, organoids and prostate cancer xenografts. Sci Rep 2017, 7:7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valentino A, et al. : Deregulation of MicroRNAs mediated control of carnitine cycle in prostate cancer: molecular basis and pathophysiological consequences. Oncogene 2017, 36:6030. [DOI] [PubMed] [Google Scholar]
- 26.Karatas OF, et al. : miR-33a is a tumor suppressor microRNA that is decreased in prostate cancer. Oncotarget 2017, 8: 60243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen H, Luo Q, Li H: MicroRNA-590-3p promotes cell proliferation and invasion by targeting inositol polyphosphate 4-phosphatase type II in human prostate cancer cells. Tumour Biol 2017, 39 1010428317695941. [DOI] [PubMed] [Google Scholar]
- 28.Xu Y, et al. : MiR-491-5p negatively regulates cell proliferation and motility by targeting PDGFRA in prostate cancer. Am J Cancer Res 2017, 7:2545–2553. [PMC free article] [PubMed] [Google Scholar]
- 29.Wang M, et al. : MicroRNA-487a-3p functions as a new tumor suppressor in prostate cancer by targeting CCND1. J Cell Physiol 2019, 235:1588–1600. [DOI] [PubMed] [Google Scholar]
- 30.Zhao Y, et al. : MicroRNA-455-3p functions as a tumor suppressor by targeting eIF4E in prostate cancer. Oncol Rep 2017, 37:2449–2458. [DOI] [PubMed] [Google Scholar]
- 31.Zhou Y, et al. : The biological functions and mechanism of miR-212 in prostate cancer proliferation, migration and invasion via targeting Engrailed-2. Oncol Rep 2017, 38: 1411–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mu H, et al. : MiR-10a functions as a tumor suppressor in prostate cancer via targeting KDM4A. J Cell Biochem 2019, 120:4987–4997. [DOI] [PubMed] [Google Scholar]
- 33.Wach S, et al. : Exploring the MIR143-UPAR Axis for the inhibition of human prostate cancer cells in vitro and in vivo. Mol Ther Nucleic Acids 2019, 16:272–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong Z, et al. : MicroRNA-384 is lowly expressed in human prostate cancer cells and has anti-tumor functions by acting on HOXB7. Biomed Pharmacother 2019, 114:108822. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, Qin H: miR-338-3p targets RAB23 and suppresses tumorigenicity of prostate cancer cells. Am J Cancer Res 2018, 8:2564–2574. [PMC free article] [PubMed] [Google Scholar]
- 36.Ray J, et al. : MicroRNA198 suppresses prostate tumorigenesis by targeting MIB1. Oncol Rep 2019, 42:1047–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szczyrba J, et al. : The microRNA profile of prostate carcinoma obtained by deep sequencing. Mol Canc Res 2010, 8:529–538. [DOI] [PubMed] [Google Scholar]
- 38.Wach S, et al. : MicroRNA profiles of prostate carcinoma detected by multiplatform microRNA screening. Int J Canc 2012, 130:611–621. [DOI] [PubMed] [Google Scholar]
- 39.Rycaj K, Tang DG: Molecular determinants of prostate cancer metastasis. Oncotarget 2017, 8:88211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.*.Das R, et al. : MicroRNA-194 promotes prostate cancer metastasis by inhibiting SOCS2. Cancer Res 2017, 77: 1021–1034. [DOI] [PubMed] [Google Scholar]; In situ hybridization of localized PCa tissue showed that miR-194 expression associated with Gleason grade and that miR-194 was higher in the serum of men with mCRPC compared to those with localized disease. MiR-194-overexpressing PC3 cells used in tail vein injections and orthotopic xenografts resulted in increased metastases in both assays as well as decreased survival in the xenograft model.
- 41.Taddei ML, et al. : Stromal-induced downregulation of miR-1247 promotes prostate cancer malignancy. J Cell Physiol 2019, 234:8274–8285. [DOI] [PubMed] [Google Scholar]
- 42.Kiener M, et al. : miR-221-5p regulates proliferation and migration in human prostate cancer cells and reduces tumor growth in vivo. BMC Canc 2019, 19:627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bucay N, et al. : A novel microRNA regulator of prostate cancer epithelial-mesenchymal transition. Cell Death Differ 2017, 24:1263–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen W, et al. : Loss of miR-449a-caused PrLZ overexpression promotes prostate cancer metastasis. Int J Oncol 2017, 51: 435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhatia V, et al. : Epigenetic silencing of miRNA-338-5p and miRNA-421 drives SPINK1-positive prostate cancer. Clin Canc Res 2019, 25:2755–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dhar S, et al. : MTA 1-activated Epi-micro RNA-22 regulates E-cadherin and prostate cancer invasiveness. FEBS Lett 2017, 591:924–933. [DOI] [PubMed] [Google Scholar]
- 47.Nam RK, et al. : MicroRNA-652 induces NED in LNCaP and EMT in PC3 prostate cancer cells. Oncotarget 2018, 9: 19159–19176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rupaimoole R, et al. : miRNA deregulation in cancer cells and the tumor microenvironment. Canc Discov 2016, 6: 235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wa Q, et al. : Downregulation of miR19a3p promotes invasion, migration and bone metastasis via activating TGFbeta signaling in prostate cancer. Oncol Rep 2018, 39:81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dai Y, et al. : The TGF-beta signalling negative regulator PICK1 represses prostate cancer metastasis to bone. Br J Canc 2017, 117:685–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guan H, et al. : MicroRNA-744 promotes prostate cancer progression through aberrantly activating Wnt/beta-catenin signaling. Oncotarget 2017, 8:14693–14707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siu MK, et al. : TCF7 is suppressed by the androgen receptor via microRNA-1-mediated downregulation and is involved in the development of resistance to androgen deprivation in prostate cancer. Prostate Cancer Prostatic Dis 2017, 20: 172–178. [DOI] [PubMed] [Google Scholar]
- 53.Tian X, et al. : The miR-203/SNAI2 axis regulates prostate tumor growth, migration, angiogenesis and stemness potentially by modulating GSK-3beta/beta-CATENIN signal pathway. IUBMB Life 2018, 70:224–236. [DOI] [PubMed] [Google Scholar]
- 54.Bellazzo A, et al. : Cell-autonomous and cell non-autonomous downregulation of tumor suppressor DAB2IP by microRNA-149-3p promotes aggressiveness of cancer cells. Cell Death Differ 2018, 25:1224–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ren D, et al. : Oncogenic miR-210-3p promotes prostate cancer cell EMT and bone metastasis via NF-kappaB signaling pathway. Mol Canc 2017, 16:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang S, et al. : Downregulation of miR-141-3p promotes bone metastasis via activating NF-kappaB signaling in prostate cancer. J Exp Clin Canc Res 2017, 36:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jeong JH, et al. : A constitutive intrinsic inflammatory signaling circuit composed of miR-196b, Meis2, PPP3CC, and p65 drives prostate cancer castration resistance. Mol Cell 2017, 65:154–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.**.Liu C, et al. : MicroRNA-141 suppresses prostate cancer stem cells and metastasis by targeting a cohort of pro-metastasis genes. Nat Commun 2017, 8:14270. [DOI] [PMC free article] [PubMed] [Google Scholar]; MiR-141 was found to have lower expression in the CD44+ putative stem cell population in 21 primary human prostate tumors compared to the non-stem cell population. MiR-overexpressing LAPC9 CD44+ cells overexpressing miR-141 had significantly decreased subcutaneous xenograft growth compared to controls, and miR-141-overexpressing PC3 CD44+ cells had smaller orthotopic tumors and less lung metastases compared to controls. Furthermore, DOX-inducible miR-overexpressing LAPC9 cell orthotopic xenografts resulted in smaller primary tumors and less lung metastases. MiR-141 was found to directly target CD44 by the luciferase assay and further confirmed by other assays.
- 59.Bucay N, et al. : MicroRNA-383 located in frequently deleted chromosomal locus 8p22 regulates CD44 in prostate cancer. Oncogene 2017, 36:2667–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tang Y, et al. : Downregulation of miR-133a-3p promotes prostate cancer bone metastasis via activating PI3K/AKT signaling. J Exp Clin Canc Res 2018, 37:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsai YC, et al. : Epidermal growth factor receptor signaling promotes metastatic prostate cancer through microRNA-96-mediated downregulation of the tumor suppressor ETV6. Canc Lett 2017, 384:1–8. [DOI] [PubMed] [Google Scholar]
- 62.*.Wang Y, et al. : MicroRNA-1205, encoded on chromosome 8q24, targets EGLN3 to induce cell growth and contributes to risk of castration-resistant prostate cancer. Oncogene 2019, 38:4820–4834. [DOI] [PMC free article] [PubMed] [Google Scholar]; MiR-1205 was knocked out of PC3 cell using CRISPR/Cas9 technology, and these cells had reduced tumor growth and weight as well as decreased markers of proliferation compared to controls. These authors also performed miRNA-mRNA immunoprecipitation to confirm miR binding to its EGLN3 target.
- 63.*.Guo Y, et al. : miR-302/367/LATS2/YAP pathway is essential for prostate tumor-propagating cells and promotes the development of castration resistance. Oncogene 2017, 36: 6336–6347. [DOI] [PubMed] [Google Scholar]; * The miR-302/367 cluster was assessed for the ability to promote CRPC in subcutaneous and orthotopic xenograft models. This miR cluster was overexpressed in PCa cell xenografts and recipient mice were castrated two weeks post-xenograft. MiR cluster–overexpressing xenografts had increased and more rapid tumor growth compared to controls, and miR-knockdown xenografts had opposite results.
- 64.Weidle UH, et al. : The functional role of prostate cancer metastasis-related micro-RNAs. Canc Genom Proteomics 2019, 16:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Colden M, et al. : MicroRNA-466 inhibits tumor growth and bone metastasis in prostate cancer by direct regulation of osteogenic transcription factor RUNX2. Cell Death Dis 2017, 8, e2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.*.Nabavi N, et al. : miR-100-5p inhibition induces apoptosis in dormant prostate cancer cells and prevents the emergence of castration-resistant prostate cancer. Sci Rep 2017, 7:4079. [DOI] [PMC free article] [PubMed] [Google Scholar]; These authors treated PCa patient-derived xenografts with androgen deprivation therapy (ADT) to induce CRPC experimentally and found that miR-100-5p was upregulated post-ADT treatment.
- 67.Jackson BL, Grabowska A, Ratan HL: MicroRNA in prostate cancer: functional importance and potential as circulating biomarkers. BMC Canc 2014, 14:930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ostling P, et al. : Systematic analysis of microRNAs targeting the androgen receptor in prostate cancer cells. Cancer Res 2011, 71:1956–1967. [DOI] [PubMed] [Google Scholar]
- 69.Saraon P, et al. : Mechanisms of androgen-independent prostate cancer. Ejifcc 2014, 25:42. [PMC free article] [PubMed] [Google Scholar]
- 70.*.Wu G, et al. : Preclinical study using circular RNA 17 and micro RNA 181c-5p to suppress the enzalutamide-resistant prostate cancer progression. Cell Death Dis 2019, 10:37. [DOI] [PMC free article] [PubMed] [Google Scholar]; These authors found that AR binds to an androgen receptor enhancer site located upstream of PDLIM5, the host gene of circRNA17. CircRNA17 binds to and stabilizes miR-181c-5p, which targets ARV7, an important AR variant involved in CRPC. Mice treated with enzalutamide (anti-androgen therapy) post-orthotopic xenograft cell injection had the lowest tumor sizes and less metastatic foci with both circRNA17 and miR-181c-5p overexpression compared to either overexpressed alone. MiR-181c-5p overexpressed alone did not result in increased expression compared to the control vector, while miR-181c-5p and circRNA17 overexpression resulted in miR overexpression, indicating the stabilizing role of circRNA17.
- 71.Li F, Mahato RI: MicroRNAs and drug resistance in prostate cancers. Mol Pharm 2014, 11:2539–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fletcher CE, et al. : Androgen receptor-modulatory microRNAs provide insight into therapy resistance and therapeutic targets in advanced prostate cancer. Oncogene 2019, 38: 5700–5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cannistraci A, et al. : C-Met/miR-130b axis as novel mechanism and biomarker for castration resistance state acquisition. Oncogene 2017, 36:3718. [DOI] [PubMed] [Google Scholar]
- 74.Jalava SE, et al. : Androgen-regulated miR-32 targets BTG2 and is overexpressed in castration-resistant prostate cancer. Oncogene 2012, 31:4460–4471. [DOI] [PubMed] [Google Scholar]
- 75.**.Latonen L, et al. : In vivo expression of miR-32 induces proliferation in prostate epithelium. Am J Pathol 2017, 187: 2546–2557. [DOI] [PubMed] [Google Scholar]; This is the only contemporary paper that used a transgenic mouse model to assess the ability of a miR to drive prostate cancer. They found that miR-32 overexpression in transgenic mouse models caused increased proliferation in all prostate lobes as well as increased goblet cell metaplasia and increased prostatic intraepithelial neoplasia. They did not, however, observe any prostate cancer lesions.


