Abstract
Prokaryotic CRISPR–Cas immune systems are classified into six types based on their effector complexes which cleave dsDNA specifically (types I, II and V), ssRNA exclusively (type VI) or both ssRNA via a ruler mechanism and ssDNA unspecifically (type III). To date, no specific cleavage of ssDNA target has been reported for CRISPR–Cas. Here, we demonstrate dual dsDNA and ssDNA cleavage activities of a subtype I-D system which carries a type III Cas10-like large subunit, Cas10d. In addition to a specific dsDNA cleavage activity dependent on the HD domain of Cas10d, the helicase Cas3′ and a compatible protospacer adjacent motif (PAM), the subtype I-D effector complex can cleave ssDNA that is complementary in sequence to the crRNA. Significantly, the ssDNA cleavage sites occur at 6-nt intervals and the cleavage is catalysed by the backbone subunit Csc2 (Cas7), similar to the periodic cleavage of ssRNA by the backbone subunit of type III effectors. The typical type I cleavage of dsDNA combined with the exceptional 6-nt spaced cleavage of ssDNA and the presence of a type III like large subunit provide strong evidence for the subtype I-D system being an evolutionary intermediate between type I and type III CRISPR–Cas systems.
INTRODUCTION
Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR associated (Cas) genes, CRISPR–Cas, constitutes an important anti-viral system present in approximately 40% of bacteria and 85% of archaea (1). Three main phases are involved in CRISPR–Cas immunity: adaptation, expression and interference. Adaptation involved the insertion of a short sequence of the invader genome (protospacer) into the CRISPR DNA array where it becomes a spacer. The expression stage includes transcription and maturation of CRISPR RNA (crRNA) which then assembles with Cas proteins to form the effector complex. Interference is guided by sequence complementarity between crRNA and the invading nucleic acid, leading to target recongnition and then cleavage by the effector complex. Adaptation and interference generally require the presence of a protospacer adjacent motif (PAM) (2).
To date, CRISPR–Cas systems have been classified into classes 1 and 2, each of which is divided into three major types. Class 1 (types I, III and IV) effector complexes carry multiple protein subunits whereas those of class 2 (types II, V and VI) carry a single multidomain protein, in addition to crRNA (3). Type I, II and V systems primarily target double-stranded (ds) DNA whereas type VI systems degrade single-stranded (ss) RNA and type III systems exhibit dual RNA and ssDNA cleavage activities (4). The substrate(s) of type IV interference remain to be identified. The type III effectors can cleave target transcripts at 6 nucleotides (nt) intervals via the Csm3/Cmr4 backbone subunit (5,6). Moreover, effector binding to the target RNA activates two activities of the large subunit Cas10: unspecific ssDNA cleavage by the histidine aspatate (HD) nuclease domain and synthesis of secondary messenger cyclic oligo adenylates by the Palm polymerase domain (cyclase) (7–9).
The signature protein of type I systems, Cas3, carries a helicase domain (Cas3′) for unwinding the target dsDNA and an endonuclease HD domain (Cas3″) for nicking the non-target strand (NTS) and degrading the DNA (10–12). Exceptionally, the type I-D system encodes both a type I signature helicase protein, Cas3′, and a type III Cas10-like large subunit, Cas10d, in addition to Csc1 (Cas5) and the backbone protein Csc2 (Cas7) (13) (Figure 1A). Moreover, a Cas11-like small subunit protein (SS) was predicted to be encoded at the 3′ end of cas10d initiating from an internal AUG codon (14). Type I-D CRISPR–Cas is abundant in cyanobacteria and many archaeal species, but very little is known about its interference mechanism(s). Here, we demonstrate that type I-D effector complex of Sulfolobus islandicus LAL14/1 cleaves dsDNA substrates in vitro in a manner similar to that of other type I subtype systems, and exceptionally, it also cleaves ssDNA specifically by a ruler–like mechanism that is similar to the protospacer transcript cleavage of type III systems.
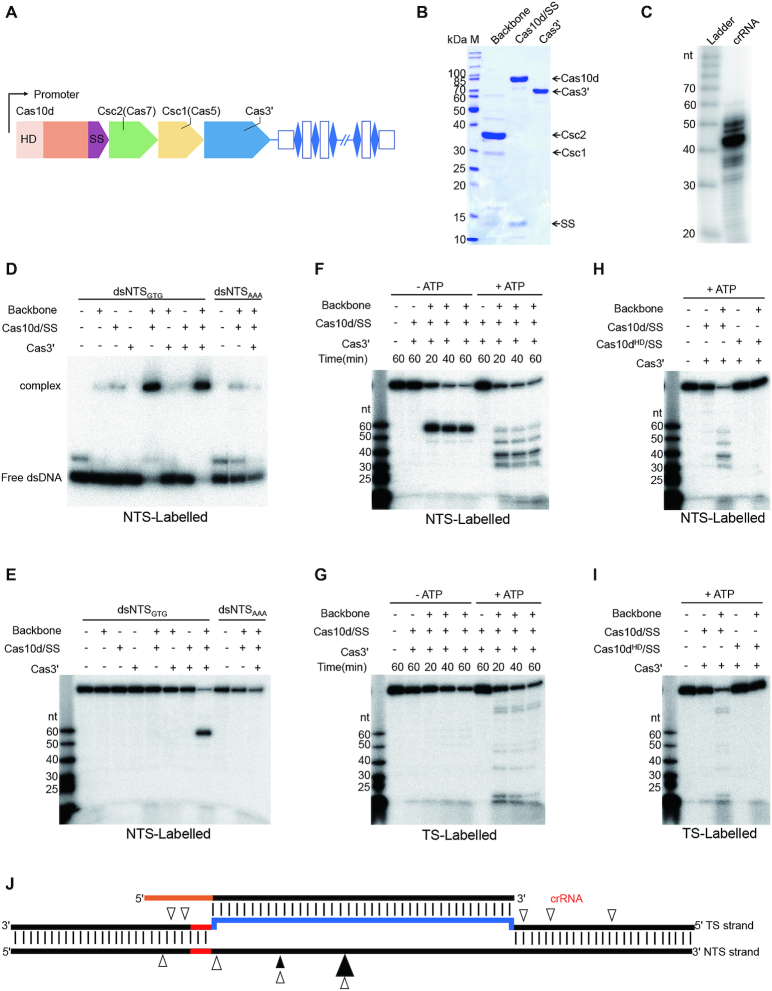
Figure 1.
dsDNA binding and cleavage by the type I-D effector. (A) The S. islandicus LAL14/1 CRISPR–Cas type I-D locus. (B) SDS-PAGE of type I-D backbone, Cas10d/SS and Cas3′ subunits, protein bands corresponding to the five subunits are indicated on the right. SS: small subunit, M: protein mass marker. (C) crRNA co-purified with type I-D backbone complex. RNA extracted from the backbone complex was 5′ end radiolabelled and subjected to electrophoresis in a 12% denaturing PAGE gel. (D) Electrophoretic mobility shift assay showing complex formation between radiolabelled dsDNA with the type I-D effector; dsNTSGTG: dsDNA carrying GTG PAM with 5′-end of NTS radiolabelled; dsNTSAAA: same as dsNTSGTG but with GTG replaced by AAA. Samples were run in a 8% native PAGE. (E) dsDNA cleavage by type I-D effector requiring all the components (backbone, Cas10d/SS, Cas3′) and a right PAM (GTN). dsDNA as used in (D) was incubated with a single, two or all the three components at 65 °C for 1 h before electrophoresis in a 12% denaturing PAGE. (F, G) Cleavage of dsDNA with GTG PAM by type I-D effector in the absence (+) or presence (+) of ATP, backbone, Cas10d/SS and Cas3′. The radiolabelled strand, NTS or TS, is indicated at the bottom of each panel. Time duration (minutes) is shown. (H, I) dsDNA cleavage by type I-D effector requiring the HD domain of Cas10d. Cas10dHD: HD mutant of Cas10d. The reactions were performed in the presence of ATP. The absence (-) or presence (+) of backbone, Cas10d/SS, Cas10dHD and Cas3′ is indicated. The same radiolabled oligo size ladder (nt) is shown on the left in (E to I). (J) Schematic presentation of the cleavage sites, in the presence (empty triangles) or absence (filled triangles) of ATP, on the dsDNA target. Larger triangles indicate the sites where the most prominent cleavage was observed in (F).
MATERIALS AND METHODS
Strains, media and growth conditions
Sulfolobus islandicus LAL14/1 and derivatives were grown at 78°C with 150 rpm/min shaking in basic salt medium (15) supplemented with 0.2% sucrose, 0.2% casamino acids, 0.005% yeast extract and a vitamin mixture (SCVY). Escherichia coli strains DH5α and Rosetta were propagated in Lysogeny broth (LB) medium at 37°C with 200 rpm/min shaking. If applicable, antibiotics were added to E. coli cultures as the following: ampicillin (100 μg/ml, Sigma-Aldrich), kanamycin (25 μg/ml, Sigma-Aldrich) and chloramphenicol (10 μg/ml, Sigma-Aldrich). The S. islandicus LAL14/1 derivative devoid of the type I-A and type III-B systems but retaining the I-D effector module (I-D strain), was constructed from the CRISPR array-deficient strain arrays as described previously (16).
Plasmid construction
PCR primers for the construction of the plasmids were listed in Supplementary Table S1. To construct the plasmid for the purification of I-D backbone complex from Sulfolobus, the csc1 gene encoding a C-terminal His-tagged Csc1 (csc1-his) was amplified from S. islandicus LAL14/1 genome and cloned into pEXA2 (17) via NdeI and NheI, giving rise to pEXA2-Csc1. Artificial CRISPR arrays with 11 (spacer 5–6) or 9 (spacer 4–17) repeat spacer units were generated by PCR as described (18) and fused to an arabinose promoter by overlap PCR and cloned into the plasmid pEXA2-Csc1 via NheI and NotI, yieldding pEXA2-Csc1-S5–6 and pEXA2-Csc1-S4–17, respectively. The gene encoding Cas3′ with a C-terminal His-tag was amplified from LAL14/1 genome by PCR and inserted into the E. coli expression plasmid pMAL-TEV (derived from pMAL-c5X, NEB) by overlap PCR, generating pMAL-TEV-Cas3′. To co-purify Csc1 and Csc2 (WT and mutants) from E. coli, csc1-his and csc2 were amplified from LAL14/1 genome and inserted into plasmid pETDuet-1 (Sigma-Aldrich) by overlap PCR successively, generating pETDuet-Csc1-Csc2 (Supplementary Table S1). Constructs encoding the Cas10d HD mutants and Csc2 mutants were obtained by the one-step site-directed mutagenesis protocol (19). All plasmid constructs were verified by DNA sequencing (GATC Biotech).
I-D CRISPR–Cas effector purification
The I-D strain containing either pEXA2-Csc1-S5–6 or pEXA2-Csc1-S4–17 (Supplementary Table S1) was used to purify two different backbone complexes (Csc1, Csc2 and crRNA) carrying spacers 5–6 and 4–17-derived crRNA, respectively. Cells were cultivated in 12 litres SCVY medium to OD600 of 0.7–0.9, collected by centrifugation at 8000 g and the cell pellet was resuspended in lysis/binding buffer (20 mM HEPES, pH 7.5, 300 mM NaCl, 20 mM imidazole). Following lysis by sonication and French press, cell extract was separated from cell debris by centrifugation at 13000 g for 20 min at 4°C. The extract was loaded on a Histrap column (GE Healthcare) for purification and eluted with the elution buffer (20 mM HEPES, pH 7.5, 300 mM NaCl, 200 mM imidazole). The fractions were pooled and concentrated to 1 ml, followed by size exclusion chromatography (SEC) with Superdex 200 column using the chromatography buffer (20 mM HEPES, pH 7.5, 300 mM NaCl).
The recombinant Cas10d with a C-terminal His-tag was purified from E. coli as described (20) and was copurified with small subunit (SS). Csc1 and Csc2 (WT and mutants) complex was purified from E. coli Rosetta carrying the plasmid pETDuet-Csc1-Csc2 or a derivative. Cell cultures were induced with 0.3 mM IPTG overnight, centrifuged at 8000 g for 10 min at 4°C and the supernatant was heat-treated at 70°C for 15 minutes followed by centrifugation at 13 000 g for 15 min. Proteins in the supernatant were purified by HisTrap and SEC as described above. crRNA carrying the spacer 5–6 was purchased (IDT Integrated DNA Technologies) and added to Csc1 and Csc2 to form the backbone complex.
The recombinant Cas3′ with a C-terminal His-tag fused at the N-terminus of maltose binding protein (MBP) linked with the TEV protease recognition site was overexpressed from pMAL-TEV-Cas3′ in E. coli Rosetta with 0.3 mM IPTG overnight. Then, the fusion protein was purified by HisTrap column and SEC as described above. The SEC fractions were pooled, and TEV protease was added and kept in 4°C overnight. The solution was again purified by HisTrap column, and the elution was pooled to 1 ml and further purified by SEC using Superdex 200 column with the chromatography buffer (20 mM HEPES, pH 7.5, 300 mM NaCl).
RNA extraction and sequencing
The native type I-D backbone complex was purified as described above from S. islandicus LAL14/1 carrying plasmid pEXA2-Csc1. Following purification, RNA was extracted with TRIzol (Thermo Fisher Scientific) and about 1 μg was subjected to Illumina sequencing which generated 1 Gb clean data (BGI, China). The sequence reads were analysed using the public Galaxy server (21). FastaQ files were mapped to the LAL14/1 genome with Bowtie2 (22) version 2.3.4.1. Mapped reads were counted using the feature Counts (23) version 1.6.0.3 tool with a reference file defining positions of all CRISPR spacers from all LAL14/1 arrays. The individual crRNA content of the complex was determined as percent of total crRNA reads.
Labelling of crRNA and DNA Substrates
DNA oligonucleotides were purchased from IDT and purified by recovering the corresponding bands from 12% denaturing polyacrylamide gel (PAGE). crRNA and ssDNA was 5′ labeled with [γ-32P]-ATP (PerkinElmer) and T4 polynucleotide kinase (New England Biolabs). dsDNA targets were generated by overlap PCR using one 5′-labelled primer and the other unlabelled primers (Supplementary Table S1), and purified with 12% native PAGE.
DNA cleavage assays
dsDNA cleavage assays were conducted in 10 μl of reaction containing 0.02 mg/ml backbone complex purified from Sulfolobus or 0.05 mg/ml Csc1 and Csc2 complex purified from E. coli plus 200 nM crRNA ordered from IDT, 0.03 mg/ml Cas10d, 0.01 mg/ml Cas3′ and 20 nM dsDNA substrate in the cleavage buffer (20 mM MES pH 6, 10 mM MgCl2, 1 mM DTT) at 65°C for 1 h, or for a different time span as indicated. Reactions were stopped by adding 1 μl 6 M guanidinium thiocyanate and 12 μl 2x RNA loading dye (New England Biolabs). For electrophoresis, samples were heated for 3 min at 95°C and analysed on a 12% denaturing PAGE. For ssDNA cleavage, the reaction containing the same amount of backbone complex as aforementioned, 0.03 mg/ml Cas10d and 20 nM ssDNA substrate in the cleavage buffer (20 mM MES pH 6, 1 mM DTT) was incubated at 65°C for 1 h. Samples were analysed on a 18% denaturing PAGE. Results were recorded by phosphor imaging with a Typhoon FLA-7000. Relative ssDNA cleavage was estimated by image quantification of bands on denaturing PAGE using the accessory analysis tool equipped with Typhoon FLA 7000.
Electrophoretic mobility shift assays (EMSA)
Each reaction contains the same amount of backbone complex as aforementioned, 0.03 mg/ml Cas10d and 20 nM dsDNA or ssDNA substrates in the binding buffer (20 mM MES pH 6, 10 mM MgCl2, 1 mM DTT for dsDNA and 20 mM MES pH 6, 1 mM DTT for ssDNA) at 65°C for 30 min. 0.01 mg/ml Cas3′ was added when applicable. Then, the same volume of 2 × native loading buffer (0.02% bromophenol blue, 40% glycerol, v/v) was added and samples were loaded to an 8% native PAGE. Electrophoresis was carried out at 200 V using 40 mM Tris, 20 mM acetic acid (pH 8.4 at 25°C) as running buffer. Results were recorded by phosphor imaging with Typhoon FLA-7000.
Structure modelling
Structural model for Csc2 was generated by automatic model-building with I-Tasser (24). All structure figures were exhibited using PyMOL (http://www.pymol.org/).
RESULTS
crRNA content of the endogenous backbone complex
The S. islandicus LAL14/1 genome encodes five CRISPR arrays; arrays 1 and 2, adjacent to type I-A cas gene module, carry identical repeat sequences whereas arrays 3 to 5, located close to the type I-D cas gene module, share repeat sequences that are different from those of arrays 1 and 2 (25) (Supplementary Figure S1A). To analyse the crRNA content of the I-D effector complex, a plasmid-borne csc1 (cas5) carrying a His-tag coding sequence was introduced into LAL14/1 and the C-terminally tagged Csc1 protein was purified. Following size exclusion chromatography (SEC) the type I-D subcomplex containing the His-tagged Csc1 (Cas5), Csc2 (Cas7) and crRNA (backbone complex) was identified (Supplementary Figure S1B–D). The absence of Cas10d and Cas3′ in the complex suggest a weak or no association of the two with the backbone. The crRNAs extracted from the SEC fractions (Supplementary Figure S1D) were then pooled, sequenced, and the resulting sequence reads were analysed. The predominant crRNA sizes were 48, 49 and 50 nt (Supplementary Figure S1E). Mapping the reads revealed that 4.90, 36.59 and 58.31% of the crRNA contents derived from arrays 3, 4 and 5, respectively. This correlated with their genomic association to the I-D system (Supplementary Figure S1A and S1F). Importantly, the reads from spacer 6 of array 5 (spacer 5–6) constituted 31% of the entire library (Supplementary Figure S2). Furthermore, spacer 5–6 matches gp18 of the virus SIRV2 (16) and was shown to be effective in in vivo CRISPR-targeting (20) and, therefore, it was employed in the in vitro cleavage assays (see below). The processed crRNAs from arrays 3, 4 and 5 have a 9 nucleotide 5′-repeat handle and a trimmed 3′-end with no or a very short repeat sequence.
dsDNA cleavage characteristic of type I CRISPR systems by the I-D effector complex
To determine dsDNA binding and cleavage by the type I-D effector complex in vitro, we purified recombinant Cas10d and Cas3′ from Escherichia coli and a backbone complex from a LAL14/1 subclone carrying a single species of crRNA, derived from spacer 5–6 (Figure 1B and C). Consistent with the prediction (14) and the structural similarity between the C-terminal region of Cas10d (residues 720–847) and Cas11-like small subunit proteins (Manav MC, Van LB, JL, AF, XP and Brodersen DE, in preparation), a small protein of 14 kDa was co-purified with the His-tagged Cas10d (Figure 1B) and was confirmed to be the C-terminal part of Cas10d (Supplementary Figure S3). The Cas10d preparation was then termed Cas10d/SS. In contrast, no SS was detected in the backbone complex (Figure 1B). The combination of Cas10d/SS and the backbone complex was necessary, and sufficient, for binding to the dsDNA target (115 bp), while neither alone was able to bind to the DNA (Figure 1D). Replacing the PAM GTN with AAA completely abolished binding (Figure 1D) which suggested that Cas10d or the backbone recognises the PAM but only establishes a stable interaction with the target when both are present to form an R-loop.
Next, we incubated the backbone complex, Cas10d/SS and Cas3′ individually and in different combinations with the dsDNA target and separated the samples on a denaturing PAGE to assay for cleavage of the 5′-end radiolabeled non-target strand (NTS) (Figure 1E). In the absence of ATP, the effector complex produced mainly a single cut on the NTS DNA, and the cleavage appeared specific and required the entire effector complex including Cas3′. The specificity was further confirmed by changing the PAM GTN to AAA which abolished the single cut (Figure 1E). In the presence of ATP which provides energy for the helicase activity, three additional cleavage products from NTS were observed indicating that cleavage was extended beyond the protospacer (Figure 1F). To examine possible cleavage of the target strand (TS), the dsDNA substrate was radiolabelled at the TS 5′-end and subjected to the same cleavage assays as performed for the NTS. In the absence of ATP, the target strand remained almost uncleaved except for very weak cleavages in the protospacer region (Figure 1G left panel), but the presence of ATP promoted TS cleavage beyond the protospacer in both directions (Figure 1G, right panel). The HD nuclease domain is part of the large subunit Cas10d in contrast to other type I systems where it is usually found in the Cas3 or Cas3″ subunit (3). To determine its function, the HD residues in the domain were substituted with alanines and the mutant Cas10d protein was subjected to cleavage assays as above. Replacing the wildtype Cas10d with the mutant clearly abolished dsDNA cleavage (Figure 1H and I), establishing that the Cas10d HD nuclease domain is responsible for dsDNA cleavage in the type I-D CRISPR system.
Overall, dsDNA cleavage by the I-D effector complex requires a PAM, a helicase Cas3′ and an HD nuclease (Cas3″) embedded in Cas10d (Figure 1E – I). The cleavage patterns for both NTS and TS (Figure 1J) resemble those observed for other type I systems (11,26). We conclude that the type I-D system exhibits dsDNA cleavage activity characteristic of other type I CRISPR–Cas systems.
Periodic ssDNA cleavage by the I-D effector is comparable with target RNA cleavage by type III systems
The rudivirus SIRV2 infects LAL14/1 and produces a high level of ssDNA replicative intermediates (27). Therefore we investigated whether the type I-D effector complex can cleave ssDNA specifically. First we examined binding to the target strand ssDNA by the backbone complex carrying a crRNA derived from spacer 5–6. Whereas Cas10d/SS alone did not bind to ssDNA, the backbone complex formed a stable complex with the ssDNA substrate (Figure 2A). Adding Cas10d/SS to the reaction produced a further shift, suggesting that the backbone complex bound to the ssDNA and that Cas10d/SS was then recruited by the backbone complex. In contrast to dsDNA binding which initiates at the PAM site (28,29) (Figure 1D), binding to ssDNA here appeared to be facilitated by base-pairing between crRNA and the complementary TS ssDNA and therefore did not require a PAM (Figure 2A). Next, we examined ssDNA cleavage by the effector complex. After 1 hr incubation of the 5′-labeled 60 nt TS ssDNA substrate with one, two or three of the components (backbone complex, Cas10d/SS and Cas3′) at 65°C, samples were separated by denaturing PAGE (Figure 2B). The combination of backbone complex and Cas10d/SS cleaved the TS ssDNA at four sites producing 4 major products (Figure 2B). A mixture of Cas3′ and Cas10d/SS triggered some unspecific ssDNA cleavage (Figure 2B). Consistent with the result from the binding assay (Figure 2A), ssDNA cleavage by the backbone and Cas10d/SS required neither a PAM sequence nor Cas3′ (Figure 2B). An estimation of the sizes of the ssDNA cleavage products suggests a ruler mechanism (Figure 2B). To identify the precise cleavage sites on the TS, we ran the cleavage products in a denaturing gel along with 5′-labelled oligo markers of different lengths. This demonstrated that cleavage of the TS indeed occurred at 6 nt intervals producing products of 38, 32, 26 and 20 nt (Supplementary Figure S4).
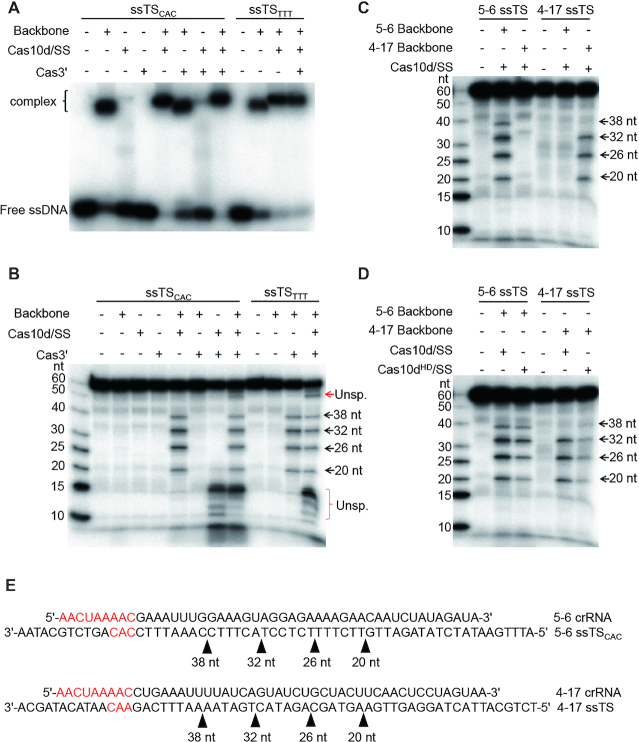
Figure 2.
Single-stranded target DNA binding and periodic cleavage by the I-D effector. (A) Electrophoretic mobility shift assay showing complex formation between radiolabelled ssDNA and the type I-D effector; ssTSCAC: target ssDNA with CAC at PAM position; ssTSTTT: target ssDNA with TTT at PAM position. (B–D) ssDNA cleavage by type I-D effector. The radiolabelled ssDNA targets are shown on top of each gel under which the presence (+) or absence (+) of the type I-D components is indicated. The sizes of cleavage products are depicted on the right side of each gel. Unsp.: Unspecific cleavage. Cas10dHD: HD mutant of Cas10d. (E) Schematic depiction of the cleavage sites on ssDNA targets.
Furthermore, to examine the specificity of ssDNA cleavage by the I-D system and to verify the ruler cleavage mechanism, we constructed a second backbone complex which carries a crRNA derived from spacer 4–17. As spacers 5–6 and 4–17 differ in length, the corresponding 4–17 TS ssDNA was then designed to have the same length from its 5′-end to the distal end of the protospacer region as for the 5–6 TS ssDNA. Remarkably, both ssDNAs were cut resulting in products of the same lengths, although the band corresponding to the largest product is weak for 5–6 TS, and barely visible for 4–17 (Figure 2C). The specificity of the ssDNA cleavage was demonstrated by incubating the effectors with a non-cognate ssDNA target which produced no cleavage (Figure 2C). Moreover, when assayed with Cas10d carrying the HD domain mutation, the cleavage pattern remained unaffected (Figure 2D). Nevertheless, Cas10d/SS must be present for the complex to effect cleavage (Figure 2B), possibly playing a structural role. No specific RNA cleavage activity was detected for the type I-D effector complex (Supplementary Figure S5), further supporting the specificity of ssDNA cleavage by the type I-D system.
Taken together, these data indicate that the type I-D effector complexes cleave ssDNA targets at 6 nt intervals within the protospacer region (Figure 2E), similarly to the mechanism by which protospacer transcripts are cleaved by the backbone subunit of type III effectors (5,6,30,31).
The backbone subunit Csc2 is the catalytic subunit for the periodic ssDNA cleavage
In type III systems, ss RNA rather than DNA is first recognized and bound by the effector and it has been well established that the bound RNA is cleaved periodically by multiple Csm3 (III-A) or Cmr4 (III-B) subunits in the backbone (5,6,30,31). Given the similar cleavage pattern, we reasoned that the ruler-based ssDNA cleavage observed here was catalysed by multiple Csc2 subunits. To examine this possibility, we mutated the conserved residues of Csc2 (Supplementary Figure S6A), which, through structure modelling (Supplementary Figure S7), align with the loop region carrying the catalytic residue of Csm3/Cmr4. Owing to the difficulties of mutagenesis in Sulfolobus, the Csc2 mutants were expressed and purified from E. coli (see Materials and Methods). Alanine substitution mutations pertaining to the residues E55, E58, D66 and E68 showed no changes in the ssDNA binding or cleavage compared to native Csc2 (Supplementary Figure S6C and D) but mutations involving the residues H54 and D59 affected both the binding and cleavage. Importantly, the decrease in cleavage appeared to be due to the weakened binding (Supplementary Figure S6C). Next we examined five conserved residues which, based on the structural model, align with the putative ‘thumb region’ adjacent to the loop. All the mutant proteins were expressed effectively in E. coli and were purified to similar yields (Figure 3A). Four of the five mutants (D181N, T184A, T186A and T187A) demonstrated a dramatic decrease in both ssDNA binding and cleavage. Therefore it remained inconclusive as to whether these residues are directly involved in ssDNA cleavage. In contrast, the E182Q mutation decreased the ssDNA cleavage to ∼25% relative to the WT setting but did not significantly affect ssDNA binding (Figure 3B and C). Importantly, none of the five mutations affected dsDNA cleavage (Figure 3D) which suggested that the mutations did not affect the structural or functional integrity required for dsDNA cleavage. Taken together, we conclude that the backbone subunit Csc2 is the catalytic subunit responsible for the periodic ssDNA cleavage.
Figure 3.
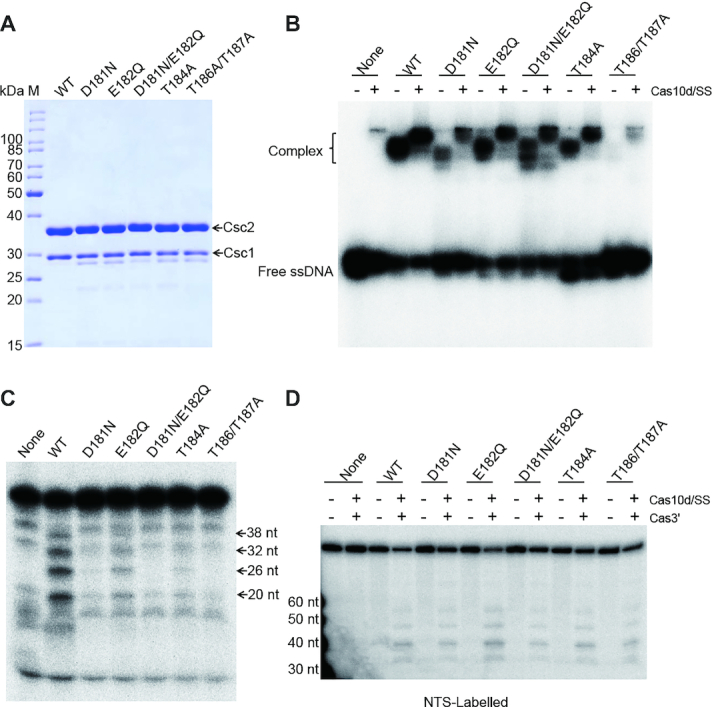
The backbone subunit Csc2 is the catalytic subunit for the periodic ssDNA cleavage. (A) SDS-PAGE of Csc1 and Csc2 (WT or mutated) co-purified from E. coli (see Methods), M: protein mass marker. ssDNA binding (B), ssDNA cleavage (C) and dsDNA cleavage (D) by type I-D effectors carrying the WT or one of the Csc2 mutants, Radiolabled ssTSCAC as in Figure 2A was used in (B) and (C); dsNTSGTG as in Figure 1 was used in (D). Reactions were performed using backbone complex containing the WT or mutant Csc2 indicated on top (B–D), in the presence (+) or absence (+) of Cas10d/SS (B, D) and Cas3′ (D). Cas10d/SS was included in all lanes in (C). None: No backbone complex added.
DISCUSSION
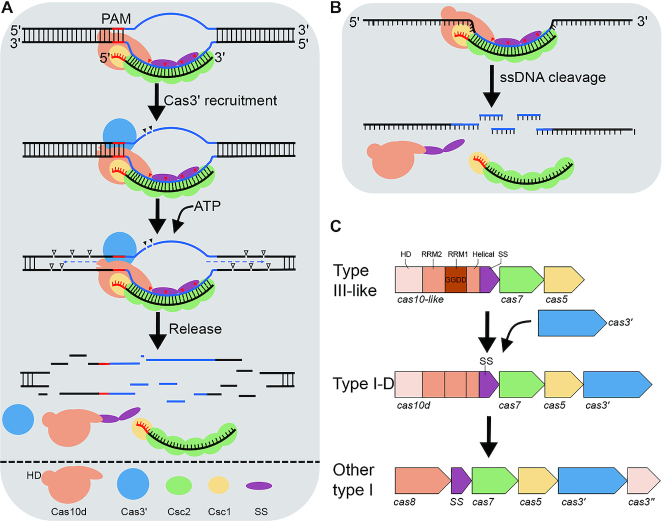
The type I-D effector module possesses mixed features of type I and type III modules. It carries the type I signature protein Cas3, split into the helicase Cas3′ and the nuclease Cas3″, with the latter merged at the N-terminus (residues 1–186) of the large subunit Cas10d. The major part of Cas10d (residues 187–719), on the other hand, structurally resembles more the large subunit Cas10 of type III systems (Manav MC, Van LB, JL, AF, XP and Brodersen DE, in preparation). On the basis of concatenated Cas protein sequences of all archaeal type I and type III effector modules, Vestergaard et al. infered that type I-D represented an intermediate between type I and type III effector modules (32). More recently, Makarova et al. proposed the effector module of type III systems to be the best candidate for the ancestral CRISPR–Cas state from which other types of CRISPR–Cas systems such as type IV, HRAMP and type I evolved (1). Here we provide the first biochemical data supporting type I-D system being an evolutionary intermediate between type I and type III systems.
The dsDNA cleavage by type I-D effector requires the helicase Cas3′, the HD nuclease domain at the N-terminus of Cas10d (Cas3″) and a functional PAM (Figure 4A), characteristic of type I dsDNA cleavage by other type I systems (11,26,33). DNA cleavage occurs also upstream and downstream of the protospacer suggesting a possible release of Cas10d/Cas3′ from the backbone after the cleavage of the protospacer region. In addition, the I-D effector exhibits specific ssDNA cleavage activity which is guided by crRNA (Figure 4B). The 6 nt intervalled cleavage catalysed by the backbone subunit Csc2 is reminiscent of target RNA cleavage by the type III effector which is also catalysed by the backbone subunit (5,6,30,31). In support of this, Csc2 has been shown to be evolutionarily grouped closer to Csm3/Cmr4 than to the canonical type I Cas7 proteins (34). Although crRNA base pairing with target DNA creates kinks every sixth nt in type I-E (35–37) and type I-F (38), as in type III systems, no periodic cleavage of a target has been reported for any of the two or other type I systems. Possibly the backbone subunit Cas7 has lost the catalytic acitivity during evolution, or the conformation of type I effectors is not favourable for the activity. Future work is needed to elucidate how the backbone in different effector complexes evolved to carry out periodic cleavage on ssRNA and ssDNA targets. Nevertheless, the high level of viral ssDNA replicative intermediates in SIRV2-infected LAL14/1 cells (27,39), and the wide presence of ssDNA viruses in both Bacteria and Archaea (40), strongly support the biological significance of the specific ssDNA targeting.
Figure 4.
Interference mechanisms and evolution of type I-D CRISPR–Cas system. (A) dsDNA interference by the type I-D effector. Cas10d recongnizes PAM, enabling base-pairing between guide sequence of crRNA and the target strand of protospacer, forming an R-loop. The Cas3′ helicase is recruited, Cas10d nicks the non-target strand of the protospacer. Next, Cas3′ unwinds the DNA at both directions of the protospcer, and Cas10d cuts ssDNA of both strands at multiple sites. Possibly, release of the cleavage products shuts off Cas10d activity, completing the DNA interference. Since neither Cas10d nor SS were detected in the backbone complex purified from Sulfolobus (Figure 1B), we infer that Cas10d/SS do not form a stable complex with the backbone in the absence of a target dsDNA. (B) Ruler-based ssDNA interference. The type I-D effector cleaves ssDNA targets at 6 nt intervals within the protospacer region, and no PAM is required. (C) Evolution model of type I CRISPR–Cas systems from a type III-like ancient system via a type I-D like intermediate. RRM: RNA recognition motif, SS: small subunit.
Based on both the earlier bioinformatic data (1,32) and the experimental evidence shown here, we propose a model on how type I effector modules evolved from a type III-like ancient CRISPR–Cas system via a type I-D like intermediate (Figure 4C). From the ancient system three events would be required to transform a type III-like effector module to a type I-D module: acquisition of a Cas3′ helicase; mutation of cas10 to acquire PAM recognition and binding activity; and inactivation of Cas10 GGDD Palm domain. Further evolution, involving three events, led to the formation of other, and more canonical, type I systems: separation of Cas3″ nuclease from the large subunit (and eventual merger with Cas3′), fission of the small subunit from the large subunit Cas10d, and loss of Cas7 enzymatic activity (Figure 4C). This proposed evolutionary pathway transformed a dormancy-based immune system (type III-like) into an immune system directly degrading invasive genomes (type I-like).
Supplementary Material
Contributor Information
Jinzhong Lin, Department of Biology, University of Copenhagen, Ole Maaløes Vej 5, DK-2200 Copenhagen N, Denmark.
Anders Fuglsang, Department of Biology, University of Copenhagen, Ole Maaløes Vej 5, DK-2200 Copenhagen N, Denmark.
Anders Lynge Kjeldsen, Department of Biology, University of Copenhagen, Ole Maaløes Vej 5, DK-2200 Copenhagen N, Denmark.
Kaiyan Sun, Department of Biology, University of Copenhagen, Ole Maaløes Vej 5, DK-2200 Copenhagen N, Denmark.
Yuvaraj Bhoobalan-Chitty, Department of Biology, University of Copenhagen, Ole Maaløes Vej 5, DK-2200 Copenhagen N, Denmark.
Xu Peng, Department of Biology, University of Copenhagen, Ole Maaløes Vej 5, DK-2200 Copenhagen N, Denmark.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Danish Council for Independent Research/Technology [DFF-7017-00060]; Novo Nordisk Foundation/Hallas MØller Ascending Investigator Grant [NNF17OC0031154 to X.P.]. Funding for open access charge: Basic funding from Copenhagen University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Makarova K.S., Wolf Y.I., Iranzo J., Shmakov S.A., Alkhnbashi O.S., Brouns S.J.J., Charpentier E., Cheng D., Haft D.H., Horvath P. et al.. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020; 18:67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mojica F.J.M., Diez-Villasenor C., Garcia-Martinez J., Almendros C.. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009; 155:733–740. [DOI] [PubMed] [Google Scholar]
- 3. Makarova K.S., Wolf Y.I., Alkhnbashi O.S., Costa F., Shah S.A., Saunders S.J., Barrangou R., Brouns S.J.J., Charpentier E., Haft D.H. et al.. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 2015; 13:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hille F., Richter H., Wong S.P., Bratovic M., Ressel S., Charpentier E.. The biology of CRISPR–Cas: backward and forward. Cell. 2018; 172:1239–1259. [DOI] [PubMed] [Google Scholar]
- 5. Ramia N.F., Spilman M., Tang L., Shao Y., Elmore J., Hale C., Cocozaki A., Bhattacharya N., Terns R.M., Terns M.P. et al.. Essential structural and functional roles of the Cmr4 subunit in RNA cleavage by the Cmr CRISPR–Cas complex. Cell Rep. 2014; 9:1610–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tamulaitis G., Kazlauskiene M., Manakova E., Venclovas C., Nwokeoji A.O., Dickman M.J., Horvath P., Siksnys V.. Programmable RNA shredding by the type III-A CRISPR–Cas system of Streptococcus thermophilus. Mol. Cell. 2014; 56:506–517. [DOI] [PubMed] [Google Scholar]
- 7. Tamulaitis G., Venclovas C., Siksnys V.. Type III CRISPR–Cas immunity: major differences brushed aside. Trends Microbiol. 2017; 25:49–61. [DOI] [PubMed] [Google Scholar]
- 8. Niewoehner O., Garcia-Doval C., Rostol J.T., Berk C., Schwede F., Bigler L., Hall J., Marraffini L.A., Jinek M.. Type III CRISPR–Cas systems produce cyclic oligoadenylate second messengers. Nature. 2017; 548:543–548. [DOI] [PubMed] [Google Scholar]
- 9. Kazlauskiene M., Kostiuk G., Venclovas C., Tamulaitis G., Siksnys V.. A cyclic oligonucleotide signaling pathway in type III CRISPR–Cas systems. Science. 2017; 357:605–609. [DOI] [PubMed] [Google Scholar]
- 10. Redding S., Sternberg S.H., Marshall M., Gibb B., Bhat P., Guegler C.K., Wiedenheft B., Doudna J.A., Greene E.C.. Surveillance and processing of foreign DNA by the Escherichia coli CRISPR–Cas system. Cell. 2015; 163:854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Westra E.R., van Erp P.B., Kunne T., Wong S.P., Staals R.H., Seegers C.L., Bollen S., Jore M.M., Semenova E., Severinov K. et al.. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol. Cell. 2012; 46:595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huo Y., Nam K.H., Ding F., Lee H., Wu L., Xiao Y., Farchione M.D. Jr, Zhou S., Rajashankar K., Kurinov I. et al.. Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation. Nat. Struct. Mol. Biol. 2014; 21:771–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Makarova K.S., Haft D.H., Barrangou R., Brouns S.J., Charpentier E., Horvath P., Moineau S., Mojica F.J., Wolf Y.I., Yakunin A.F. et al.. Evolution and classification of the CRISPR–Cas systems. Nat. Rev. Microbiol. 2011; 9:467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McBride T.M., Schwartz E.A., Kumar A., Taylor D.W., Fineran P.C., Fagerlund R.D.. Internal translation of large subunit transcripts drives small subunit synthesis in type I CRISPR–Cas interference complexes. 2020; bioRxiv doi:18 April 2020, preprint: not peer reviewed 10.1101/2020.04.18.045682. [DOI]
- 15. Zillig W., Kletzin A., Schleper C., Holz I., Janekovic D., Hain J., Lanzendorfer M., Kristjansson J.K.. Screening for sulfolobales, their plasmids and their viruses in Icelandic solfataras. Syst. Appl. Microbiol. 1993; 16:609–628. [Google Scholar]
- 16. Bhoobalan-Chitty Y., Johansen T.B., Di Cianni N., Peng X.. Inhibition of type III CRISPR–Cas immunity by an archaeal Virus-Encoded Anti-CRISPR protein. Cell. 2019; 179:448–458. [DOI] [PubMed] [Google Scholar]
- 17. Gudbergsdottir S., Deng L., Chen Z.J., Jensen J.V.K., Jensen L.R., She Q.X., Garrett R.A.. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol. Microbiol. 2011; 79:35–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lin J., Feng M., Zhang H., She Q.. Characterization of a novel type III CRISPR–Cas effector provides new insights into the allosteric activation and suppression of the Cas10 DNase. Cell Discov. 2020; 6:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zheng L., Baumann U., Reymond J.L.. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004; 32:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. He F., Bhoobalan-Chitty Y., Van L.B., Kjeldsen A.L., Dedola M., Makarova K.S., Koonin E.V., Brodersen D.E., Peng X.. Anti-CRISPR proteins encoded by archaeal lytic viruses inhibit subtype I-D immunity. Nat. Microbiol. 2018; 3:461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Afgan E., Baker D., van den Beek M., Blankenberg D., Bouvier D., Cech M., Chilton J., Clements D., Coraor N., Eberhard C. et al.. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016; 44:W3–W10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 24. Yang J., Zhang Y.. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 2015; 43:W174–W181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jaubert C., Danioux C., Oberto J., Cortez D., Bize A., Krupovic M., She Q., Forterre P., Prangishvili D., Sezonov G.. Genomics and genetics of Sulfolobus islandicus LAL14/1, a model hyperthermophilic archaeon. Open Biol. 2013; 3:130010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Majumdar S., Terns M.P.. CRISPR RNA-guided DNA cleavage by reconstituted Type I-A immune effector complexes. Extremophiles. 2019; 23:19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Martinez-Alvarez L., Bell S.D., Peng X.. Multiple consecutive initiation of replication producing novel brush-like intermediates at the termini of linear viral dsDNA genomes with hairpin ends. Nucleic Acids Res. 2016; 44:8799–8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Blosser T.R., Loeff L., Westra E.R., Vlot M., Kunne T., Sobota M., Dekker C., Brouns S.J.J., Joo C.. Two distinct DNA binding modes guide dual roles of a CRISPR–Cas protein complex. Mol. Cell. 2015; 58:60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xiao Y.B., Luo M., Hayes R.P., Kim J., Ng S., Ding F., Liao M.F., Ke A.L.. Structure basis for directional R-loop formation and substrate handover mechanisms in type I CRISPR–Cas system. Cell. 2017; 170:48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jia N., Mo C.Y., Wang C., Eng E.T., Marraffini L.A., Patel D.J.. Type III-A CRISPR–Cas Csm complexes: assembly, periodic RNA cleavage, DNase activity regulation, and autoimmunity. Mol. Cell. 2019; 73:264–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. You L., Ma J., Wang J., Artamonova D., Wang M., Liu L., Xiang H., Severinov K., Zhang X., Wang Y.. Structure studies of the CRISPR-Csm complex reveal mechanism of Co-transcriptional interference. Cell. 2019; 176:239–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vestergaard G., Garrett R.A., Shah S.A.. CRISPR adaptive immune systems of Archaea. Rna Biol. 2014; 11:156–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Majumdar S., Ligon M., Skinner W.C., Terns R.M., Terns M.P.. Target DNA recognition and cleavage by a reconstituted Type I-G CRISPR–Cas immune effector complex. Extremophiles. 2017; 21:95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Makarova K.S., Aravind L., Wolf Y.I., Koonin E.V.. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR–Cas systems. Biol. Direct. 2011; 6:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jackson R.N., Golden S.M., van Erp P.B., Carter J., Westra E.R., Brouns S.J., van der Oost J., Terwilliger T.C., Read R.J., Wiedenheft B.. Structural biology. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science. 2014; 345:1473–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhao H., Sheng G., Wang J., Wang M., Bunkoczi G., Gong W., Wei Z., Wang Y.. Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli. Nature. 2014; 515:147–150. [DOI] [PubMed] [Google Scholar]
- 37. Mulepati S., Heroux A., Bailey S.. Structural biology. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science. 2014; 345:1479–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pausch P., Muller-Esparza H., Gleditzsch D., Altegoer F., Randau L., Bange G.. Structural variation of type I-F CRISPR RNA guided DNA surveillance. Mol. Cell. 2017; 67:622–632. [DOI] [PubMed] [Google Scholar]
- 39. Martinez-Alvarez L., Deng L., Peng X.. Formation of a viral replication focus in sulfolobus cells infected by the rudivirus sulfolobus islandicus Rod-Shaped virus 2. J. Virol. 2017; 91:e00486-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Koonin E.V., Dolja V.V., Krupovic M.. Origins and evolution of viruses of eukaryotes: The ultimate modularity. Virology. 2015; 479–480:2–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.