Abstract
The N4-methylation of cytidine (m4C and m42C) in RNA plays important roles in both bacterial and eukaryotic cells. In this work, we synthesized a series of m4C and m42C modified RNA oligonucleotides, conducted their base pairing and bioactivity studies, and solved three new crystal structures of the RNA duplexes containing these two modifications. Our thermostability and X-ray crystallography studies, together with the molecular dynamic simulation studies, demonstrated that m4C retains a regular C:G base pairing pattern in RNA duplex and has a relatively small effect on its base pairing stability and specificity. By contrast, the m42C modification disrupts the C:G pair and significantly decreases the duplex stability through a conformational shift of native Watson-Crick pair to a wobble-like pattern with the formation of two hydrogen bonds. This double-methylated m42C also results in the loss of base pairing discrimination between C:G and other mismatched pairs like C:A, C:T and C:C. The biochemical investigation of these two modified residues in the reverse transcription model shows that both mono- or di-methylated cytosine bases could specify the C:T pair and induce the G to T mutation using HIV-1 RT. In the presence of other reverse transcriptases with higher fidelity like AMV-RT, the methylation could either retain the normal nucleotide incorporation or completely inhibit the DNA synthesis. These results indicate the methylation at N4-position of cytidine is a molecular mechanism to fine tune base pairing specificity and affect the coding efficiency and fidelity during gene replication.
INTRODUCTION
RNA chemical modifications have been increasingly recognized as one of nature's general strategies to define, diversify, and regulate RNA structures and functions in numerous biological processes. To date, over 160 post-transcriptional modifications have been identified in all types of RNAs in the three domains of life (1). Many of these modifications have been demonstrated to play critical roles in both normal and diseased cellular functions and processes such as development, circadian rhythms, embryonic stem cell differentiation, meiotic progression, temperature adaptation, stress response, and tumorigenesis, etc (2). Similar to DNA and protein epigenetic markers, these RNA modifications, also termed as ‘epitranscriptome’, can be dynamically and reversibly regulated by specific reader, writer, and eraser enzymes, representing a new layer of gene regulation (3). Accordingly, these modification-associated enzymes, as an important research frontier toward RNA-based drug discovery, have become useful molecular tools and drug targets (4).
Methylation has been known as the most abundant RNA chemical modification since the first methylated nucleobase was discovered over 70 years ago (5). These methylated nucleotides in different types of RNAs play diverse and key roles in cells, ranging from the stabilization of tRNA structure, reinforcement of the codon-anticodon interaction, regulation of wobble base pairing, and prevention of frameshift errors, to the RNA quality control and localization (6,7). For example, two forms of dimethylated adenosines, N6-dimethyladenosine (m62A) and 2,8-dimethyladenosine (m2,8A), in ribosomal RNA (rRNA) can result in the multi-drug resistance in many bacteria (8,9). Many methylated nucleobases like 5-methylcytidine (m5C), N1-methylguanidine (m1G), N1-methyladenosine (m1A), N3-methylcytidine (m3C), N7-methylguanidine (m7G), and 2′-O-methylated sugar (2′-Nm) in the anticodon stem loops of transfer RNA (tRNA) are directly involved in the codon recognition and can induce or inhibit the frameshifting mutations during translation (10,11). N6-methyladenosine (m6A), the most commonly found internal modification in eukaryotic mRNAs and some long noncoding RNAs, is actively working in mRNA stability, structure switches, miRNA processing, protein synthesis, and epigenetic inheritance (12,13). The oxidative demethylation of this mRNA m6A is catalyzed by several dioxygenases such as FTO, AlkBH1, AlkBH3, and AlkBH5 (6,7,14–16), further bridging this methylation with a wider range of cellular functions and disease states.
Compared to other methylated nucleosides, the N4-methylcytidine (m4C) has been much less investigated. It is known that m4C is common in prokaryotic DNAs and plays significant roles in bacterial evolution and epigenetic gene regulation. The methylation leads to the structural disruption of DNA major groove as well as the protein recognition and binding. Recently, m4C in Helicobacter pylori, which is a Gram-negative, spiral-shaped microaerophilic bacterium causing various diseases including gastric cancer (17), was found to act as a global epigenetic regulator and affect the transcription, ribosome assembly and overall pathogenesis of this bacterium (18). In RNA, the m4C has been confirmed as a major methylated base in both cytoplasmic and mitochondrial rRNAs of bacterial and eukaryotic cells (19–21). The working enzyme responsible for m4C in Escherichia coli rRNA is RsmH (also known as mraW), an S-adenosyl methionine (SAM)-dependent methyltransferase (22,23), which can further methylate m4C to N4,N4-dimethylcytidine (m42C). The presence of m4C in particular was speculated to stabilize the rRNA folding in mitochondrial small ribosomal subunits. Very recently, METTL15, a member of the mammalian methyltransferase-like (METTL) enzyme family and a sequence orthologue of the E. coli RsmH protein, has been identified to introduce m4C into human mitochondrial 12S rRNA and is required for efficient mitochondrial protein synthesis and mitoribosome biogenesis, providing a potential new drug target for the treatment of mitochondrial disorders (24). More interestingly, m42C has been uniquely detected in the viral RNAs from ZIKV and HCV virions and the cells infected by these virus (25).
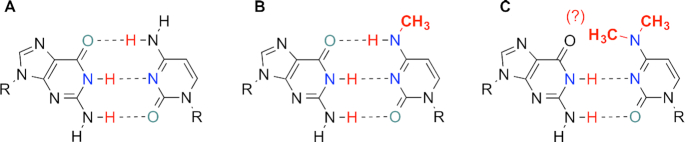
One of the direct molecular consequences of these methylated nucleobases is the effect on base pairing stability and specificity. Since the N4-position directly participates the Watson-Crick pairing, as shown in Figure 1, the single methylation of m4C might be able to either retain or disrupt the hydrogen bonding between C and G, depending on the conformation of the methyl group, while the dimethylated m42C, which is generated from m4C by RsmH, seems to disrupt the C:G pair with a potential wobble-like or other pairing patterns and thus reduce the base pairing fidelity of cytosine. In addition, the methyl groups might also affect the enzyme recognition modes. Therefore, we hypothesize that the methylation at N4-position of cytidine is a potential molecular mechanism not only to modify RNA structures, but also to fine tune the base pairing specificity and affect the efficiency and fidelity of gene replication during transcription and reverse-transcription, which could result in the increased viral gene mutation rates. Toward this goal, here we report the chemical synthesis of m4C and m42C phosphoramidite building blocks and their incorporation into RNA oligonucleotides. The RNAs containing either m4C or m42C residues were used in base pairing stability and specificity studies, crystal structure and molecular dynamic simulation studies, as well as their biological function studies in reverse transcription with different enzymes.
Figure 1.
Watson-Crick pairing patterns of RNAs containing guanine with native and methylated cytidines. (A) Canonical G:C base pair. (B) G:m4C pair. (C) G:m42C pair with unknown methyl conformations.
MATERIALS AND METHODS
Materials and general procedures of synthesis
Anhydrous solvents were used and redistilled using standard procedures. All solid reagents were dried under a high vacuum line prior to use. Air sensitive reactions were carried out under argon. RNase-free water, tips and tubes were used for RNA purification, crystallization and thermodynamic studies. Analytical TLC plates pre-coated with silica gel F254 (Dynamic Adsorbents) were used for monitoring reactions and visualized by UV light. Flash column chromatography was performed using silica gel (32–63 μm). All 1H, 13C and 31P NMR spectra were recorded on a Bruker 400 spectrometer. Chemical shift values are in ppm. 13C NMR signals were determined by using APT technique. High-resolution MS were achieved by ESI at University at Albany, SUNY. The NMR and MS spectra of the modified nucleosides are shown in Supplementary Figure S2–S19.
Synthesis of m42C phosphoramidite
1-(2′-O-tert-Butyldimethylsilyl-3′,5′-O-di-tert-butylsilylene-beta-d-ribofuranosyl)-N4,N4-dimethylcytidine 2. To a solution of compound 1 (1.5 g, 3.0 mmol) in THF (30 mL) was added NaH (0.6 g, 15 mmol, 60% dispersion in mineral oil) in portions at 0°C under Ar. After 15 min, MeI (0.75 ml, 12 mmol) was added. The reaction mixture was warmed to room temperature and stirred for 24 h. The mixture was quenched with water (50 ml) and extracted with Ethyl Acetate (3 × 50 ml). The organic layer was dried by Na2SO4, filtered and evaporated under reduced pressure. The residue was purified by silica gel chromatography to give compound 2 (1.3 g, 2.5 mmol, 82% yield) as a white solid. TLC Rf = 0.3 (DCM:MeOH = 20:1). 1H NMR (500 MHz, CDCl3) δ 7.33 (d, J = 7.5 Hz, 1H), 5.77 (d, J = 7.5 Hz, 1H), 5.62 (s, 1H), 4.47 (dd, J = 5.5, 9.5 Hz, 1H), 4.33 (d, J = 9.5 Hz, 1H), 4.21–4.15 (m, 1H), 3.94 (dd, J = 9.0, 10.5 Hz, 1H), 3.83 (dd, J = 4.5, 9.5 Hz, 1H), 3.16 (s, 3H), 3.01 (s, 3H), 0.99 (s, 9H), 0.98 (s, 9H), 0.90 (s, 9H), 0.21 (s, 3H), 0.12 (s, 3H). 13C NMR (125 MHz, CDCl3) δ 163.5, 155.0, 139.7, 94.5, 91.2, 75.8, 75.2, 74.4, 67.9, 27.5, 27.0, 26.0, 22.7, 20.3, 18.2, –4.4, –4.8. HRMS (ESI-TOF) [M+H]+ = 526.3130 (calc. 526.3132). Chemical formula: C25H47N3O5Si2.
1-(2′-O-tert-Butyldimethylsilyl-beta-d-ribofuranosyl)-N4,N4-dimethylcytidine 3. To a solution of compound 2 (1.3 g, 2.5 mmol) in THF (20 ml) at 0°C was added a solution of hydrogen fluoride-pyridine complex (hydrogen fluoride ∼70%, pyridine ∼30%; 0.5 mL) in pyridine (3 ml). After 1 h at 0°C the reaction was complete and pyridine (7.5 ml) was added. The mixture was diluted with DCM (200 ml) and washed with sat. NaHCO3 and brine. The organic layer was dried over Na2SO4 and evaporated. The residue was purified by silica gel chromatography to give compound 3 (700 mg, 1.8 mmol, 73% yield) as a white solid. TLC Rf = 0.5 (DCM:MeOH = 10:1). 1H NMR (500 MHz, CDCl3) δ 7.54 (d, J = 7.5 Hz, 1H), 5.81 (d, J = 8.0 Hz, 1H), 5.29 (d, J = 4.5 Hz, 1H), 4.88–4.85 (m, 1H), 4.84–4.80 (m, 1H), 4.10–4.08 (m, 1H), 3.89–3.86 (m, 1H), 3.73–3.68 (m, 1H), 3.14 (s, 3H), 3.02 (s, 3H), 0.83 (s, 9H), 0.03 (s, 3H), 0.01 (s, 3H). 13C NMR (125 MHz, CDCl3) δ 163.5, 155.5, 144.2, 96.1, 91.8, 85.9, 73.3, 70.9, 62.2, 25.7, 17.9, –4.8, –5.2. HRMS (ESI-TOF) [M+H]+ = 386.2111 (calc. 386.2111). Chemical formula: C17H31N3O5Si.
1-(2′-O-tert-Butyldimethylsilyl-5′-O-4,4′-dimethoxytrityl-5′-beta-d-ribofuranosyl)- N4,N4-dimethylcytidine 4. To a solution of compound 3 (700 mg, 1.8 mmol) in dry pyridine (10 mL) was added 4,4′-Dimethoxytrityl chloride (1.25 g, 3.6 mmol) under Ar. The resulting solution was stirred at room temperate overnight. The reaction was quenched with methanol (1 ml) and stirred for another 5 min. The reaction mixture was then concentrated to dryness under vacuum. The residue was purified by silica gel chromatography to give compound 4 (1.1 g, 1.6 mmol, 89% yield) as a white solid. TLC Rf = 0.6 (ethyl acetate). 1H NMR (500 MHz, CDCl3) δ 8.13 (d, J = 7.5 Hz, 1H), 7.46–7.43 (m, 2H), 7.36–7.23 (m, 8H), 6.87–6.84 (m, 4H), 5.88 (d, J = 1.0 Hz, 1H), 5.34 (d, J = 7.5 Hz, 1H), 4.39–4.33 (m, 1H), 4.32–4.30 (m, 1H), 4.07–4.04 (m, 1H), 3.80 (s, 6H), 3.60 (dd, J = 2.0, 11.0 Hz, 1H), 3.51 (dd, J = 3.0, 11.5 Hz, 1H), 3.20 (s, 3H), 2.96 (s, 3H), 0.94 (s, 9H), 0.36 (s, 3H), 0.22 (s, 3H). 13C NMR (125 MHz, CDCl3) δ 163.6, 158.63, 158.62, 155.4, 144.6, 140.8, 135.6, 135.4, 130.3, 130.2, 128.3, 128.0, 127.0, 113.24, 113.23, 90.9, 90.4, 86.9, 82.8, 76.6, 69.1, 61.5, 55.2, 25.9, 18.1, –4.3, –5.5. HRMS (ESI-TOF) [M+H]+ = 688.3415 (calc. 688.3418). Chemical formula: C38H49N3O7Si.
1-(2′-O-tert-Butyldimethylsilyl-3′-O-(2-cyanoethyl-N,N-diisopropylamino)phosphoramidite-5′-O-4,4′-dimethoxytrityl-5′-beta-d-ribofuranosyl)-N4,N4-dimethylcytidine 5. To a solution of compound 4 (225 mg, 0.33 mmol) in DCM (5 ml) was added N,N-di-iso-propylethylamine (0.24 ml, 1.32 mmol), 1-methyl-1H-imidazole (27 μl, 0.33 mmol) and 2-cyanoethyl N,N-diisopropylchlorophosphoramidite (0.17 ml, 0.66 mmol). The resulting solution was stirred at room temperature overnight under Ar. The reaction was quenched with water and extracted with ethyl acetate. After drying the organic layer over Na2SO4 and evaporation, the residue was purified by silica gel chromatography to give compound 5 (200 mg, 0.23 mmol, 68% yield) as a white solid. TLC Rf = 0.6 (ethyl acetate). 1H NMR (500 MHz, CDCl3) δ 8.24–8.22 (m, 1H), 7.48–7.45 (m, 2H), 7.37–7.22 (m, 9H), 6.86–6.83 (m, 4H), 5.77 (d, J = 0.5 Hz, 1H), 5.28 (d, J = 8.0 Hz, 1H), 4.33–4.23 (m, 3H), 3.79 (s, 6H), 3.74–3.73 (m, 1H), 3.65–3.42 (m, 5H), 3.18 (s, 3H), 2.93 (s, 3H), 2.38 (t, J = 6.5 Hz, 2H), 1.15 (s, 3H), 1.13 (s, 3H), 1.11 (s, 3H), 1.09 (s, 3H), 0.90 (s, 9H), 0.28 (s, 3H), 0.14 (s, 3H).31P NMR (202 MHz, CDCl3) δ 150.06, 148.89. HRMS (ESI-TOF) [M+H]+ = 888.4490 (calc. 888.4497). Chemical formula: C47H66N5O8PSi.
Synthesis and purification of m4C and m42C containing RNA oligonucleotides
All oligonucleotides were chemically synthesized at 1.0 μmol scales by solid phase synthesis using the Oligo-800 synthesizer. The m4C and m42C-phosphoramidite were dissolved in acetonitrile to a concentration of 0.1 M. I2 (0.02 M) in THF/Py/H2O solution was used as an oxidizing reagent. Coupling was carried out using 5-ethylthio-1H-tetrazole solution (0.25 M) in acetonitrile for 12 min, for both native and modified phosphoramidites. 3% trichloroacetic acid in methylene chloride was used for the 5′-detritylation. Synthesis was performed on control-pore glass (CPG-500) immobilized with the appropriate nucleoside through a succinate linker. All the reagents used are standard solutions obtained from ChemGenes Corporation. The oligonucleotide was prepared in DMTr off form. After synthesis, the oligos were cleaved from the solid support and fully deprotected with 1:1 v/v ammonium hydroxide solution (28% NH3 in H2O) and methylamine (40% w/w aqueous solution) at 65°C for 45 min. The solution was evaporated to dryness by Speed-Vac concentrator. The solid was dissolved in 100 μl DMSO and was desilylated using a triethylamine trihydrogen fluoride (Et3N•3HF) solution at 65°C for 2.5 h. Cooled down to room temperature the RNA was precipitated by adding 0.025 ml of 3 M sodium acetate and 1 ml of ethanol. The solution was cooled to –80°C for 1 h before the RNA was recovered by centrifugation and finally dried under vacuum.
The oligonucleotides were purified by IE-HPLC at a flow rate of 1 ml/min. Buffer A was 20 mM Tris–HCl, pH 8.0; buffer B 1.25 M NaCl in 20 mM Tris–HCl, pH 8.0. A linear gradient from 100% buffer A to 70% buffer B in 20 min was used to elute the oligos. The analysis was carried out by using the same type of analytical column with the same eluent gradient. All the modified-oligos were checked by MALDI-TOF MS. The 31-mer RNA template oligonucleotides were purified on a preparative 20% denaturing polyacrylamide gel (PAGE). The MS-spectra, HPLC purification profiles and the gel image are shown in Supplementary Figure S20–S34.
UV-melting temperature (Tm) study
Solutions of the duplex RNAs (1.5 μM) were prepared by dissolving the purified RNAs in sodium phosphate (10 mM, pH 7.0) buffer containing 100 mM NaCl. The solutions were heated to 95°C for 5 min, then cooled down slowly to room temperature, and stored at 4°C for 2 h before Tm measurement. Thermal denaturation was performed in a Cary 300 UV–Visible Spectrophotometer with a temperature controller. The temperature reported is the block temperature. Each denaturizing curve was acquired at 260 nm by heating and cooling from 5 to 80°C for four times in a rate of 0.5°C/min. All the melting curves were repeated for at least four times. The thermodynamic parameters of each strand were obtained by fitting the melting curves in the Meltwin software.
Crystallization
Crystallization was carried out by vapor diffusion hanging drop method. The crystallization conditions of CCGG(m4C)GCCGG (300 μM) were: 10% v/v (+/–)-2-methyl-2,4-pentanediol (MPD), 0.040 M sodium cacodylate trihydrate pH 7.0, 0.012 M spermine tetrahydrochloride, 0.08 M potassium chloride, 0.02 M magnesium chloride hexahydrate. The CCGG(m42C)GCCGG (300 μM) was crystallized in two conditions: (i) 10% v/v (+/–)-2-methyl-2,4-pentanediol (MPD), 0.040 M sodium cacodylate trihydrate pH 7.0, 0.012 M spermine tetrahydrochloride, 0.08 M sodium chloride, and (2) 10% v/v (+/–)-2-methyl-2,4-pentanediol, 0.040 M sodium cacodylate trihydrate pH 6.0, 0.012 M spermine tetrahydrochloride, 0.012 M sodium chloride and 0.080 M potassium chloride. Crystals were cryoprotected by 35% of MPD prior to freezing in liquid nitrogen.
Diffraction data collection
The diffraction data for each determined structure were collected from a single crystal at the SER-CAT 22-ID beamline at the Advanced Photon Source (APS), Argonne National Laboratory, USA. The diffraction data were processed with XDS (26) or HKL3000 (27) and truncated with STARANISO (http://staraniso.globalphasing.org/cgi-bin/staraniso.cgi) using anisotropic diffraction limits. The anisotropic cut-off surface for the data of CCGG(m4C)GCCGG has been determined from 1.93 Å (best diffraction limit) to 2.29 Å (lowest cut-off diffraction limit). In the case of CCGG(m42C)GCCGG, the anisotropic diffraction limits for the data collected from P212121 crystal were between 1.65 Å and 1.91 Å. Diffraction limits for the data from R32 crystal were between 1.81 Å and 2.75 Å. Supplementary Table S3 lists detailed statistics of the data processing. Coordinates and structure factors were deposited in the PDB under the accession numbers 6WY2 [CCGG(m4C)GCCGG], 6WY3 [CCGG(m42C)GCCGG-P212121], and 6Z18 [CCGG(m42C)GCCGG-R32].
Structure determination and refinement
Our previously deposited structure of the native CCGGCGCCGG RNA duplex (PDB ID: 4MS9) (28) was used as an initial model for the phase determination of the structure of CCGG(m4C)GCCGG RNA in Phaser (29). The model was then taken for the subsequent steps of manual and automatic refinement with Coot (30) and Phenix (31). TLS parameters (32) were applied at the later stages of the structure refinement. In the case of CCGG(m42C)GCCGG-P212121 structure the initial search model contained a part of the 4MS9 structure. The initial search in Phaser included 4 copies of the CCGG duplex from 4MS9. Then, the missing part of the structure was manually built in Coot. The starting model for the CCGG(m42C)GCCGG-R32 structure was an ideal CCGGCGCCGG duplex generated in Coot. The refinement of both structures was analogous to the CCGG(m4C)GCCGG RNA structure. Rwork, Rfree factors (33) and geometric parameters were controlled during refinement which was carried out until the difference electron density maps, geometry, and refinement statistics were satisfactory. The quality of refined structures was tested using MolProbity (34). The final refinement statistics are given in Supplementary Table S3. The geometrical restraints for m4C and m42C were generated in Sketcher from the CCP4 package (35).
Molecular simulation
To study the m4C and m42C nucleotides in the context of the RNA duplex in MD simulations, we developed AMBER (36) type force-field parameters for the atoms of the modified nucleoside. We used the AM1-BCC (37) charge model to calculate the atomic charges, which is developed as a fast yet accurate alternate for ESP-fit using Hartree-Fock theory and 6-31G* basis-sets (38). AMBER99 force-field parameters were used for bonded interactions, and AMBER99 parameters with Chen-Garcia corrections (39) for the bases and Bergonzo-Cheatham corrections (40) for the backbone were used for LJ interactions. The unmodified RNA duplex was constructed in A-form using make-na server that automates the Nucleic Acid Builder (NAB) suite of AMBER, and mutated to create the modifications.
Molecular dynamics simulations were performed using Gromacs-2018 package (41). The simulation system included the RNA duplex in a solution of 0.1 M NaCl solution in a 3D periodic box. The box size was 4.5 × 4.3 × 5.5 nm containing 24 Na+ ions, 6 Cl− ions and 3130 water molecules. The system was subjected to energy minimization to prevent any overlap of atoms, followed by a 1 ns equilibration run. The equilibrated system was then subjected to a 500 ns production run. The MD simulations incorporated leap-frog algorithm with a 2 fs timestep to integrate the equations of motion. The system was maintained at 300 K and 1 bar, using the velocity rescaling thermostat (42) and Parrinello-Rahman barostat (43), respectively. The long-ranged electrostatic interactions were calculated using particle mesh Ewald (PME) (44) algorithm with a real space cut-off of 1.2 nm. LJ interactions were also truncated at 1.2 nm. TIP4PEw model (45) was used represent the water molecules, and LINCS (46) algorithm was used to constrain the motion of hydrogen atoms bonded to heavy atoms. Co-ordinates of the RNA molecule were stored every 20 ps for further analysis.
Reverse transcription (RT) assays
RT assays were performed with AMV RT (ThermoFisher) and HIV-1 RT (AS ONE Corp.) in 20 μl total solution containing 10X reverse transcription buffer: 50 mM Tris (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM DTT. Final reaction mixtures contained RNA template (5 μM), DNA FAM-primer (2.5 μM) and dNTP (1 mM). After addition of Rnase inhibitor (20 U) and each RTs: AMV RT (10 U), HIV-1 RT (4 U), the mixtures were incubated at 37°C for 1 h. The reactions were quenched with stop solution [98% formamide, 0.05% xylene cyanol (FF), and 0.05% bromophenol blue], heated to 90°C for 5 min and then cooled to 0°C in ice-bath, and analysed by 15% PAGE with 8 M urea at 250 V for 1–1.5 h. The fluorescent and UV gel imaging were taken on a Bio-Rad Gel XR+ imager.
RESULTS AND DISCUSSION
Chemical synthesis of m4C and m42C-phosphoramidite building blocks
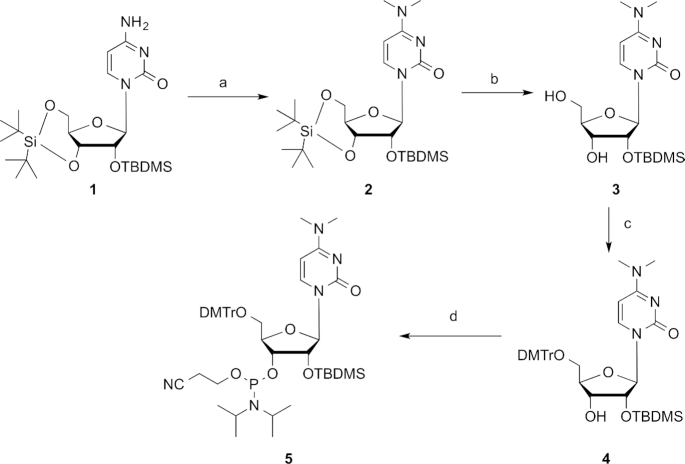
The N4-methylcytidine (m4C) phosphoramidite was synthesized according to the literature procedure starting from the silylated uridine (Supplementary Figure S1) (47). The activation of C-4 position with 2,4,6-triisopropylbenzene sulfonyl chloride (TPSCl), followed by the treatment with aqueous methylamine solution and the acetylation using acetic anhydride provided compound S3, which was selectively desilylated by hydrogen fluoride in pyridine (HF·Py), tritylated with trityl chloride at the 5′-position and finally converted to the m4C phosphoramidite building block for the subsequent oligonucleotide solid-phase synthesis. Similarly, we started the synthesis of m42C phosphoramidite from the silylated cytidine 1 (Figure 2). The dimethylation of 1 using methyl iodide in the presence of sodium hydride gave compound 2 in a high yield, which was selectively desilylated by hydrogen fluoride, 5′-tritylated with trityl chloride and converted to the final product 5 through regular 3′-phophitylation reaction. Although the m42C modified RNA strands could also be achieved through post-oligo conversion strategy (48), our phosphoramidite building block provides a direct, more efficient and high-quality method to make these modified RNAs.
Figure 2.
Synthesis of m42C phosphoramidite 5. Conditions: (a) MeI, NaH, THF; (b) HF.Py, THF; (c) DMTrCl, Py; (d) (i-Pr2N)2P(Cl)OCH2CH2CN, (i-Pr)2NEt, 1-methylimidazole, DCM.
Both of the phosphoramidite building blocks were well compatible with the regular solid-phase RNA synthesis conditions, including trichloroacetic acid (TCA) and oxidative iodine treatments, and thus, the coupling yields were very similar to those of the commercially available native counterparts. They were also found to be stable under basic cleavage from the solid-phase beads and Et3N·3HF treatment to remove the TBDMS groups during deprotection and HPLC purification of the RNA oligonucleotides. As a demonstration, different RNA sequences containing these two modifications were synthesized and their molecular mass have been confirmed by ESI or MALDI-TOF MS, as shown in Supplementary Table S1.
Thermal denaturation and base pairing studies of m4C and m42C RNA duplexes
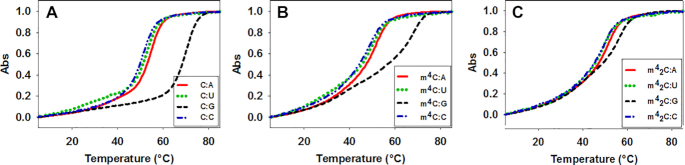
We synthesized two sets of RNA oligonucleotides to investigate the thermodynamic properties and base pairing specificity of m4C and m42C in RNA duplexes. The normalized Tm curves of the native and modified RNA duplexes, [5′-GGACUXCUGCAG-3′ & 3′-CCUGAYGACGUC-5′] with Watson-Crick and other non-canonical base pairs (X pairs with Y), are shown in Figure 3. The detailed melting temperature data are summarized in Table 1. Compared to the native counterparts, both m4C and m42C-modified RNA duplexes showed decreased thermal stability. In the native C:G paired 12mer duplexes (compare entry 1, 5 and 9 in Table 1), the m4C decreases the Tm by 2.0°C, while the m42C dramatically decreases the Tm by 15.5°C, corresponding to a △G° reduction of 6.4 and 9.5 kcal/mol respectively. Similarly, the non-canonical base paired (ex. C:A, C:U and C:C) duplexes containing these two modifications also showed significantly lower melting temperatures. In the case of m4C, the Tm drops by 4.1°C in the C:A mismatched duplex (entry 2 versus 6), 3.5°C in the C:U mismatched one (entry 3 versus 7) and 3.6°C for the C:C mismatched one (entry 4 versus 8), corresponding to the △G° reduction of 2.8, 2.8 and 2.0 kcal/mol respectively. While with the m42C residue, the Tm drops by 4.2°C in the C:A mismatched duplex (entry 2 versus 10), 5.3°C in the C:U mismatched one (entry 3 versus 11) and 2.9°C for the C:C mismatched one (entry 4 versus 12), corresponding to the △G° reduction of 2.6, 2.7 and 1.8 kcal/mol respectively.
Figure 3.
Normalized UV-melting curves of RNA duplexes. (A) Native sequence 5′-GGACUCCUGCAG-3′) pairs with matched and mismatched strands. (B) m4C modification sequence (5′-GGACUm4CCUGCAG-3′) pairs with matched and mismatched sequences. (C) m42C modification sequence (5′-GGACUm42CCUGCAG-3′) pairs with matched and mismatched sequences.
Table 1.
Duplex stability and base pairing specificity of m4C and m42C in a 12-mer RNA duplex [5′-GGACUXCUGCAG-3′ & 3′-CCUGAYGACGUC-5′] (X pairs with Y)
| Base Pairs | T m | △Tm | –△G° | ||
|---|---|---|---|---|---|
| Entry | X | Y | (°C)a | (°C)b | (kcal/mol)c |
| 1 | C | G | 69.6 | 20.6 | |
| 2 | C | A | 54.2 | –15.4 | 14.0 |
| 3 | C | U | 52.9 | 16.7 | 14.3 |
| 4 | C | C | 50.7 | 18.9 | 12.4 |
| 5 | m4C | G | 67.6 | 14.2 | |
| 6 | m4C | A | 50.1 | 17.5 | 11.2 |
| 7 | m4C | U | 49.4 | 18.2 | 11.5 |
| 8 | m4C | C | 47.1 | 20.5 | 10.4 |
| 9 | m42C | G | 54.1 | 11.1 | |
| 10 | m42C | A | 50.0 | 4.1 | 11.4 |
| 11 | m42C | U | 47.6 | 6.5 | 10.6 |
| 12 | m42C | C | 47.8 | 6.3 | 10.6 |
aThe Tms were measured in sodium phosphate (10 mM, pH 7.0) buffer containing 100 mM NaCl, Tm values reported are the averages of four measurements.
b△Tm values are relative to the duplexes with only Watson–Crick pairs.
cObtained by non-linear curve fitting using Meltwin 3.5 (49).
These results indicate that although the m4C has a relatively small effect on its base pairing stability, the regular C:G base pairing in the context of RNA duplex was still perturbed by the methylation to certain extent. The additional methyl group in m42C significantly disrupts the C:G pair and the overall duplex stability, which is consistent with the pairing pattern proposed in Figure 1. Indeed, when we compared these two modifications with the native C in a self-complementary 10-mer duplex context (CCGGC*GCCGG)2, where two consecutive m4C:G and m42C:G pairs are introduced in the middle of the duplex, the Tm drops by 7.7 and 32.2°C respectively, as shown in Supplementary Figure S35 and Table S2. On the other hand, the comparison of the base pairing specificity in each duplex system indicated different effects of these two modifications. When directly comparing the Tms of each normal Watson-Crick base paired duplex with its own mismatched ones, as shown in the △Tm column (Table 1), the m4C retains similar pairing specificity as C, while the m42C significantly decreases the discrimination between C:G pair and other mismatched C:A, C:U and C:C pairs.
Crystal structure studies of RNA duplexes containing m4C and m42C
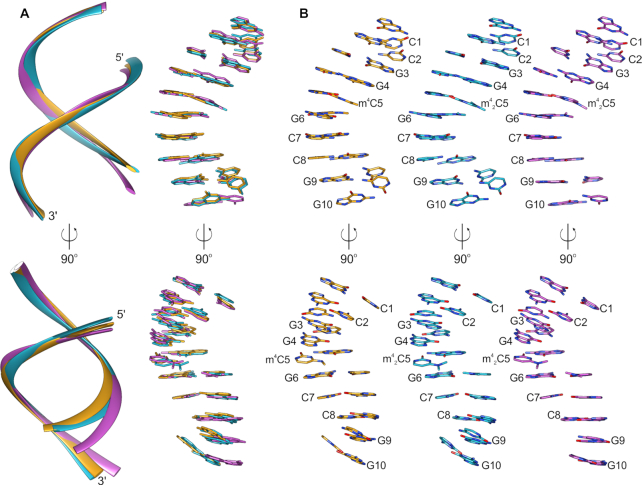
To gain further structural insights into these two methylated cytidines, we obtained three crystal structures using the self-complementary 10mer duplex (CCGGC*GCCGG)2 as the model system with two consecutive m4C:G or m42C:G pairs in the middle. The study included one structure of m4C-10mer and two structures of m42C-10mer in two different crystal forms. The diffraction data collection and final structure refinement statistics are summarized in Supplementary Table S3. Overall, all the three structures show A-type RNA duplexes with regular 3′-endo sugar pucker conformation, as shown in Figure 4.
Figure 4.
Crystal structures of RNA 10-mers carrying m4C and m42C modifications. (A) Superposition of the three determined structures presented only as backbone (cartoon, left) and nucleobases (sticks, right); (B) individual structures of RNA 10-mers CCGG(m4C)GCCGG (orange), CCGG(m42C)GCCGG crystallized in the P212121 space group (cyan) and CCGG(m42C)GCCGG crystallized the R32 space group (violet) shown in the same orientation as in panel A.
Overall duplex comparison
The structure with m4C5 modification presents the closest structural analogy to the structure of native 10-mer duplex that we solved previously (PDB ID: 4MS9) (28). All the five strands in the asymmetric unit of this structure are highly similar to each other (rmsd of the superposed backbone atoms of the single strands is no greater than 0.68 Å) and they also show high similarity to the native duplex (highest rmsd between single strands superposition is 0.77 Å). In addition, the backbone distance between P1 and P9 in the m4C structure (the shortest 25.1 Å in chain A and the longest 26.5 Å in chain E) is very close to the native one (25.4 Å). In the m4C structure, there are two Mg2+ ions bound in the major groove of the duplex, each coordinated by six water molecules (Supplementary Figure S36A). Three of these water molecules create hydrogen bonds with G3 and G4. In addition, a potassium ion is also observed in the structure, which contributes to the inter-duplex stabilization. It is worth noting that, especially for this structure, the anisotropic truncation of the data with STARANISO made a huge improvement of the electron density maps (Supplementary Figure S36A) in comparison to the maps obtained with spherical truncation of the data (Supplementary Figure S36B). This step was crucial for the interpretation of the structure and identification of not only bound ions but also for the analysis of the methyl modification of m4C5.
The two structures carrying dimethylated m42C5 were solved in two different crystal forms. Interestingly, these two orthogonal and rhombohedral crystals of m42C-10mer grew in nearly identical crystallization conditions; they sometimes even appeared together in the same crystallization drops. This may suggest that the double methylation introduces more structural perturbations in the duplex structure and the modified RNA adopts more than one conformation to compensate the m42C5 modification. This is even more visible when the structures are superposed onto each other (Figure 4A). There are two duplexes (A–B and C–D) in the CCGG(m42C)GCCGG-P212121 structure; rmsds of their superposed single strands vary from 0.96 Å (chains A and B) up to 1.87 Å (chains B and C). In the CCGG(m42C)GCCGG-R32 structure (one duplex in the asymmetric unit), superposed chains present rmsd of 1.44 Å. The backbone distances between P1 and P9 of the strands in CCGG(m42C)GCCGG-P212121 structure vary between 25.7 Å (chain B) and 30.6 Å (chain C), while for the strands of the CCGG(m42C)GCCGG-R32 structure, these distances are 25.8 Å (chain A) and 28.9 Å (chain B). In the duplex-to-duplex comparison, the duplexes A–B and C–D of the CCGG(m42C)GCCGG-P212121 show rmsd of 1.45 Å when they are superposed onto each other. The rmsd is 1.68 and 1.85 Å for the superposition of duplexes A-B and C-D of CCGG(m42C)GCCGG-P212121 with the duplex from the CCGG(m42C)GCCGG-R32 structure. Overall, the introduction of m42C5 modification into the RNA 10-mer causes much more significant structural perturbations to the RNA helix than the m4C5 modification.
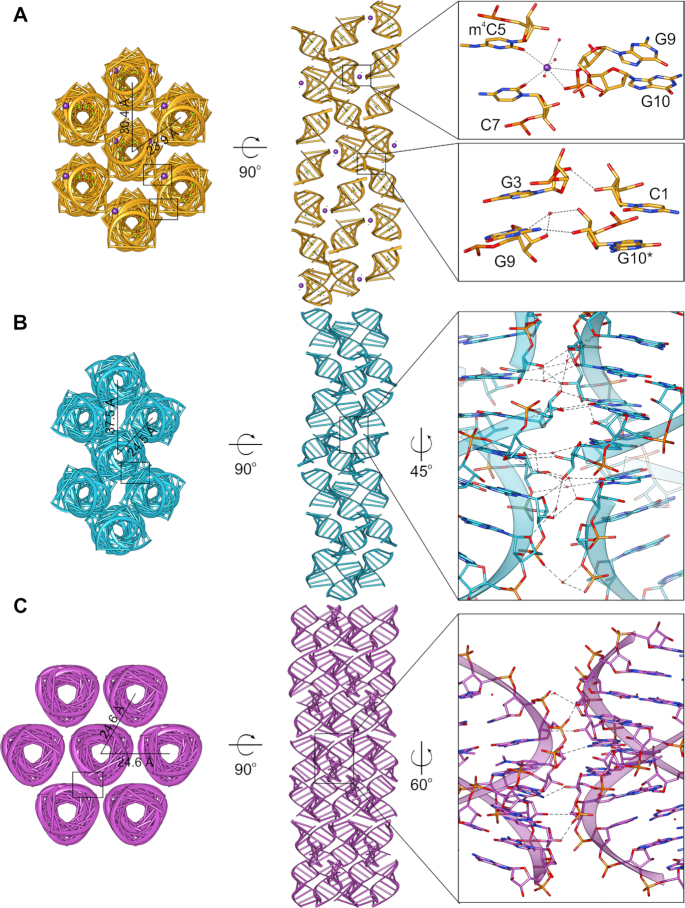
Crystal packing and helix–helix interactions analysis
In the crystal lattice, all duplexes create infinite helices by stacking of the terminal bases. Helix axis of the duplex with m4C5 modification runs along the longest face diagonal of the unit cell (Supplementary Figure S37A) while in both crystal forms of the m42C5-duplexes, infinite helix direction is parallel to the longest unit cell axes (Supplementary Figure S37B and C). The helix axis of the m42C5 structure crystallized in the space group R32 is very close to the straight line, while helices in the other two structures are locally bent and resemble an S-shape. These features are further determined by inter-helix packing in the crystal lattice (Figure 5). Overall, the tightest helices packing is in the m42C5 rhombohedral crystals, where axis-to-axis distance of the neighbouring helices is 24.6 Å (Figure 5C). In this crystal form, each helix makes a direct contact with six other helices, where the duplexes are arranged on the same level in the crystal lattice. This packing is very similar to the packing of the native duplex, except for the neighbouring duplexes in the m42C5 rhombohedral crystals that are rotated by 60° to one another and they present a few inter-helix contacts along the minor groove. In the other two structures (m4C5 and m42C5 in P212121 space group), helices interact with only four neighouring helices (Figure 5A, B) and the axis-to-axis distances of the interacting helices are 23.9 and 24.5 Å, respectively. The distances of the axes of the distant helices are 30.4 Å and 37.5 Å in m4C5 and m42C5 in P212121 space group, respectively. Therefore, they are not in the proximity to create direct inter-helix contacts, and the closest distance is no nearer than ∼7 Å (m4C5) and ∼11 Å (m42C5 in P212121 space group). In these two crystal forms, due to the wavy shape of the helices, the inter-helix contacts are significantly different. In the m4C5 structure (Figure 5A), potassium ions participate in the inter-helix stabilization by coordinating with the backone oxygen atoms of G9 and G10 from one helix, O2 atoms of m4C5 and C7, and three water molecules. Other stabillizing interactions involve C1 and G10 from the consecutive duplexes within one helix, which are H-bonded with G3 and G9 from the neighbouring helix. The same interaction pattern between two helices is repeated every two and a half duplex (every asymmetric unit). On the other hand, the m42C5 in P212121 space group seems to present the most developed hydrogen-bonding network from all three determined structures. Such stronger crystal contacts are consistent with the better diffraction properties of the m42C5 orthorombic crystals.
Figure 5.
Crystal packing of the three solved 10-mer structures. (A) CCGG(m4C)GCCGG, (B) CCGG(m42C)GCCGG in the P212121 space group and (C) CCGG(m42C)GCCGG in the R32 space group. Left and center panels show the views from the top and along the axis of the seven neighboring RNA helices in the crystal lattice; distances between the helices axes are provided. Right panels are close-up views of the crystal contacts between duplexes; black rectangles indicate locations of the zoomed regions.
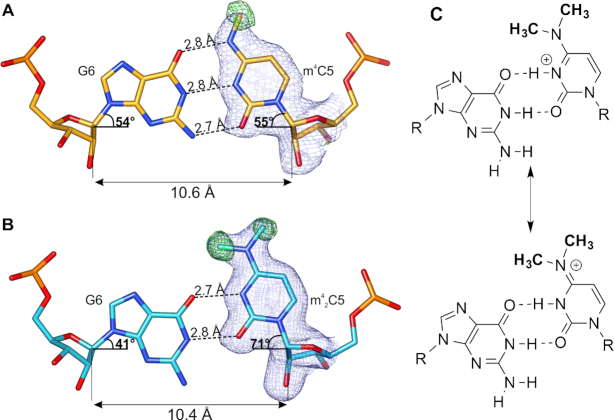
Influence of the m4C and m42C on base pairing
The electron density maps confirmed the m4C5 and m42C5 methylations and clearly showed the positions of methyl groups in the structures (Figure 6). In both modified bases, methyl groups are placed almost ideally in-plane with the C5 base plane. Single methylation of cytosine has a minor effect on the geometry of the m4C5:G6 pairing and does not disturb the Watson-Crick pairing (Figure 6A). N4 is still able to form the hydrogen bond with O6 of G6. On average, the C1′ atoms of m4C5:G6 are placed 10.6 Å away and the λ angles of m4C5 and G6 are 54° and 55°, similar to the geometry of the unmodified C:G pair. Nonetheless, the presence of methyl group in m4C5 disables the N4 from being a partner in another hydrogen bond from the side of the major groove, which could be vital for RNA-protein recognition.
Figure 6.
The m4C and m42C pairing with G. (A) The m4C5:G6 pair (from chain C and B, respectively) of the CCGG(m4C)GCCGG duplex and (B) m42C5:G6 pair (from chain B and A, respectively) of the CCGG(m42C)GCCGG structure in P212121 space group; dashed lines indicate hydrogen bonds; blue mesh represents 2Fo – Fc electron density map (contoured at 1σ) for m4C and m42C; green mesh is the omit Fo – Fc map (contoured at 3σ) calculated only for methyl groups of the modified nucleotides. (C) Two possible forms of the m42C5:G6 pair.
Introduction of a second methyl group on m42C5 causes much more severe perturbations in the duplex structure. Because N4 in m42C5 is not a hydrogen bond donor to O6 atom of G6, the m42C5:G6 pairing is very different from the canonical Waston-Crick pair. To accommodate the two methyl groups, the hydrogen bonds are shifted to a wobble-like pairing pattern (Figure 6B). As a result, only two H-bonds are formed in m42C5:G6 pair: (i) between O2 of m42C5 and N1 of G6, and (ii) between N3 of m42C5 and O6 of G6. This pattern indicates that the dimethylated m42C5 residue in the structure might exist as a protonated form, similar to the one observed in i-DNA base pairing (50). In the meantime, the two electron-donating methyl groups might be able to enhance the electron resonance within the N3-C4-N4 atoms and result in an equilibrium between the protonated N3-form and an ‘iminium’ form with cation on the N4 position (Figure 6C). With the current resolution limitation of the two structures, the unrestrained refinement is not effective enough to differentiate the two tautomers with more precise assignment of bond lengths in this aromatic system. Of course, it is also possible that the charged cation forms are accompanied by a neutral pairing form containing only one hydrogen bond with relatively lower occupancy in the crystal lattice; our MD simulation supports the existence of a form of m42C5:G6 pair with a single H-bond (see below).
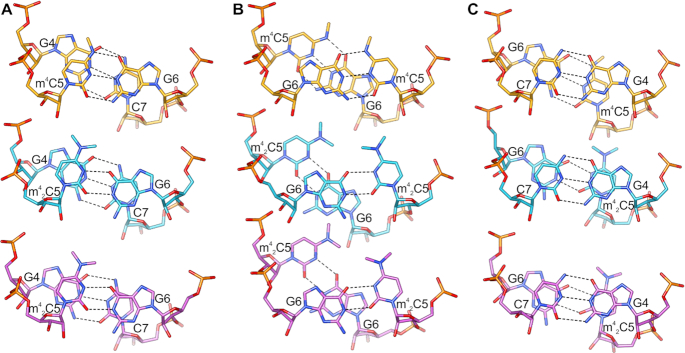
The shift from canonical H-bond pattern in m42C5:G6 also leads to the dramatic conformational change: the average λ angles of m42C5 and G6 are now 71° and 41°, respectively (Figure 6B). The distance between C1′ atoms slightly decreases to the average of 10.4 Å. Consequently, the stacking interactions of the base pair steps with m42C5 are highly perturbed in comparison to the native duplex and to the duplex carrying the single m4C5 methylation (Figure 7). These perturbations most likely introduce a higher tendency of the m42C5 duplex to adopt various conformations in order to avoid a steric clash between methyl group of m42C5 and G6. This intrinsic flexibility can also explain the differences between particular duplexes in CCGG(m42C)GCCGG-P212121 and CCGG(m42C)GCCGG-R32 structures.
Figure 7.
Base pair steps overlap. (A) G4-m4C/m42C5 step; (B) m4C/m42C5:G6 and (C) G6-C7 step in the 10-mer RNA duplexes CCGG(m4C)GCCGG (orange), CCGG(m42C)GCCGG in the P212121 space group (cyan), and CCGG(m42C)GCCGG in the R32 space group (violet).
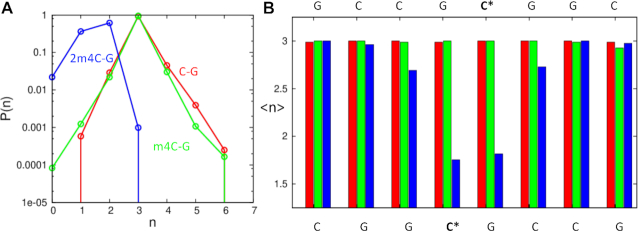
Molecular simulation studies
To investigate the dynamic property of the hydrogen bonding patterns in the structure, we conducted MD simulations studies. The ensemble of structures obtained from the simulations were used to calculate the difference in hydrogen bonding between the modified cytosine and the complementary guanidine nucleobases. Figure 8A shows the distribution of the number of H-bonds between the aforementioned bases. The unmodified base-pair shows the characteristic peak at n = 3, corresponding to the three hydrogen bonds observed in a canonical C:G base pair. As expected, the distribution remains unperturbed when this canonical pair is mutated to m4C:G, implying that the single methylation can be well accommodated in the base pairing pattern and has little impact on the pairing dynamics. However, for the double methylated m42C:G pair, the peak in the distribution moves significantly to the left, yielding an average of ∼1.5 H-bonds. This result indicates that the double methylation cannot be accommodated in the canonical base-pairing orientation due to the steric hindrances of two methyl groups of m42C with the O6 of the pairing G. Moreover, even the wobble-type pairing, where two hydrogen bonds seem to be formed, is not very stable probably due to the capability of the deprotonation of N3 position. Therefore, it is very likely that the m42C residue exists as a mixed form in the duplex context. On the other hand, the average number of hydrogen bonds obtained for all the base-pairs in the duplex is shown in Figure 8B, indicating the structural perturbation caused by the m42C is mainly local to the modified bases, except for one neighboring base-pair, which also shows an average decrease of one H-bond. This is also consistent with our structural studies.
Figure 8.
Molecular simulation studies of the 10mer-RNA duplexes containing C:G (red), m4C:G (green) and m42C:G (blue) pairs. (A) The distribution of H-bond numbers between the above-mentioned bases. (B) The average number of hydrogen bonds of all the base-pairs in the duplexes.
Reverse transcription studies of m4C and m42C in primer extension reactions
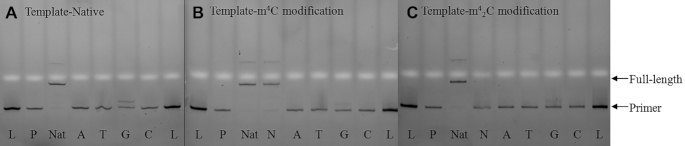
In order to further investigate the potential molecular consequences of the base pairing discrimination induced by the methylation of cytidine in RNA, we conducted the template directed primer extension reactions as the reverse transcription model. As shown in Figure 9, the 5′-end of DNA primer was labeled with fluorescent FAM group and the two 31nt-long modified RNAs were synthesized as the templates with either m4C or m42C on the starting site of the replication reaction, which represents a direct and effective way to explore the enzymatic compatibility and coding property of modified residues. The reverse transcription yields or fidelity with different base pairing substrates in the presence of two different reverse transcriptase, AMV-RT and HIV-1-RT, were quantitated by the fluorescence gel images with single-nucleotide resolution.
Figure 9.

Primer extension reaction as the reverse transcription model.
When the Avian Myeloblastosis Virus Reverse Transcriptase (AMV-RT), which is an RNA-directed DNA polymerase widely applied in RT-PCR and RNA sequencing (51), was used in the system, the reverse transcription reaction completes in the presence of all the natural dNTPs with native RNA template (Figure 10A, lane Nat). In the presence of different dNTP substrates, only dGTP but no other dNTPs can be incorporated against the starting C residue on the native template (lane A, T, G, C). With m4C modified RNA template (Figure 10B), although the dGTP can still be incorporated, the overall yield is dramatically reduced from the initial 48.4% (lane G in Figure 10A) to 18.2% (lane G in Figure 10B). On the other hand, in the presence of all natural dNTPs, the full-length product could still be obtained with comparably high yield to the native system (lane Nat vs N). Furthermore, with m42C modified RNA template (Figure 10C), the incorporation yield of dGTP is further decreased to less than 5% (lane G). However, with m42C residue, no full-length product could be observed in the presence of all natural dNTPs (lane N), indicating that the double methylation completely inhibits the AMV-RT activity in this reverse transcription process.
Figure 10.
Fluorescent gel images of standing-start primer extension reactions with AMV-RT using native (A), m4C-modified (B) and m42C-modified (C) RNA strands as templates. Lanes: L, reference DNA 20mer ladder; P, primer; Nat, natural template with all four dNTPs as positive controls in each gel; A, T, G, and C, reactions in the presence of the respective dNTP only; N, reactions in the presence of all four dNTPs.
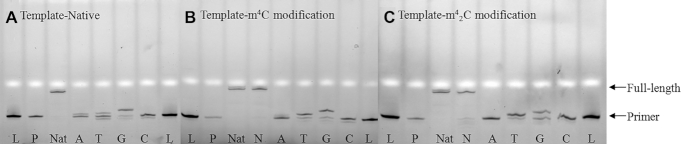
By contrast, when the HIV-1 reverse transcriptase, which has been known to have lower replication fidelity than AMV-RT, was applied in the system together with the native template, the incorporation yield of dGTP was largely increased (Figure 11A, lane G), and the mis-incorporations of dATP and dTTP could also be observed (lane A, T). In the presence of both m4C- and m42C-templates (Figure 11B, C), the full-length products were obtained with the presence of all natural dNTPs (lanes Nat and N), indicating the modifications do not inhibit the HIV-RT activity. Interestingly, the m4C modification significantly increases the dTTP incorporation efficiency from 23.2% in the native template to 72.9%, while retaining similar yield for the dGTP incorporation (lane T and G in Figure 11B). In the m42C template, the incorporation yield of dTTP is also increased to 83.6%, but the dGTP incorporation yield is decreased from the native 79.6% to 52% (lane T and G in Figure 11C). In addition, we further investigated the time course of this HIV-1 RT extended reaction with both m4C and m42C templates. Our gel image (Supplementary Figure S38) showed that the primer was completely consumed after 2 h with the m4C template and 1.5 h for the m42C one with quantitative yields of full-length products in the presence of all the natural dNTPs. In the case of m42C-containing RNA template, dTTP was the most efficiently incorporated nucleotide. After 0.5 h, 68.2% of dTTP incorporation was observed compared to the 25.7% of dGTP incorporation.
Figure 11.
Fluorescent gel images of standing-start primer extension reactions with HIV-1 RT using native (A), m4C-modified (B), and m42C-modified (C) RNA strands as templates. Lanes: L, reference DNA 20mer ladder; P, primer; Nat, natural template with all four dNTPs as positive controls in each gel; A, T, G and C, reactions in the presence of the respective dNTP only; N, reactions in the presence of all four dNTPs.
Base modifications have been known to have big impacts on the overall activity and fidelity of RNA polymerase and reverse transcriptase, and several widely studied modified bases such m6A, m5C, m5U, hm5U and pseudouridine have been evaluated in terms of the DNA or RNA synthesis error rates (52). It was reported that the HIV-1 RT as a low fidelity reverse transcriptase catalyzes nucleotide mismatch with an error frequency of 1/2000 to 1/4000 and a specificity of C:A pair over other mismatches, thus inducing a G to A mutation during HIV gene replication (53). Although the m4C was previously reported not to be G to A mutagenic (54), our results indicate that both mono- and di-methylated cytosine bases could instead specify the C:T pair and increase the G to T mutation during the reverse transcription of HIV-1 RT. Indeed, the plausible pairing patterns of the methylated C with other bases (Supplementary Figure S39) also show the m4C:T pair is the most stable one with two hydrogen bonds, and this pattern also exist in m42C:T pair with the protonated form. For other reverse transcriptase with higher fidelity like AMV-RT, the monomethylation m4C retains the normal nucleotide incorporation and the dimethylated m42C completely shuts down the DNA synthesis, which may provide an adaptive evolution mechanism for virus in responding to different selection stresses. In the meantime, the enzyme RsmH or its analogs that are responsible for the methylation processes in viral RNA genes might play important roles in virus mutation and the development of antiviral drug resistance, and be good potential molecular targets for new drug design and development.
CONCLUSIONS
In summary, we synthesized m4C and m42C phosphoramidites and a series of RNA oligonucleotides containing these two modifications. Our base-pairing and specificity studies showed that the m4C retains a regular C:G base pairing pattern in the context of RNA duplex and has a relatively small effect on its base pairing stability and specificity. The m42C modification disrupts the canonical C:G pairing geometry and significantly decreases the duplex stability, which also results in the loss of base pairing discrimination of C:G with C:A, C:T and C:C mismatched pairs. We also presented three crystal structures of RNA duplexes containing m4C and m42C residues, providing more detailed insights into the base pairing patterns and structural impacts of the methylated cytidines. The structures confirm that the mono-methylated C is well accommodated in paring to G with normal Watson-Crick pattern and does not affect the local and global structure conformations. On the other hand, the dimethylation induces a protonated cytidine in the structure and results in a significant conformational shift of C:G pair to a Wobble-like pairing pattern. Our molecular simulation studies on these two structures further indicates that the hydrogen bonds of m4C:G are quite stable while the ones in m42C:G pair are more dynamic and flexible. In addition, our investigation of the base methylation effects on the reverse transcription model showed that both mono- or di-methylated cytosine bases could specify the C:T pair and induce the G to T mutation during the reverse transcription by HIV-1 RT. For the reverse transcriptase with higher fidelity like AMV-RT, the methylation could either retain the normal nucleotide incorporation or completely shut down the DNA synthesis. This work provides detailed insights into the structure and importance of methylated cytidine modifications in RNA, and set up a knowledge foundation for further exploiting the biochemical and biomedical potentials of this methylation pathway towards the design and development of RNA based therapeutics.
DATA AVAILABILITY
Coordinates and structure factors were deposited in the PDB under the accession numbers 6WY2 [CCGG(m4C)GCCGG], 6WY3 [CCGG(m42C)GCCGG-P212121], and 6Z18 [CCGG(m42C)GCCGG-R32].
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Cen Chen and Prof. Zhen Huang for their help in MS-Spec experiments.
Diffraction data were collected at the Advanced Photon Source, Argonne National Laboratory, at the SER-CAT beamline 22-ID (supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under contract W-31-109-Eng-38). Structural work was supported by the Intramural Research Program of the National Cancer Institute, Center for Cancer Research.
Contributor Information
Song Mao, Department of Chemistry, University at Albany, State University of New York, 1400 Washington Ave. Albany, NY 12222, USA; The RNA Institute, University at Albany, State University of New York, 1400 Washington Ave. Albany, NY 12222, USA.
Bartosz Sekula, Synchrotron Radiation Research Section, Macromolecular Crystallography Laboratory, National Cancer Institute, Argonne, IL, USA.
Milosz Ruszkowski, Synchrotron Radiation Research Section, Macromolecular Crystallography Laboratory, National Cancer Institute, Argonne, IL, USA; Institute of Bioorganic Chemistry, Polish Academy of Sciences, Poznan, Poland.
Srivathsan V Ranganathan, The RNA Institute, University at Albany, State University of New York, 1400 Washington Ave. Albany, NY 12222, USA.
Phensinee Haruehanroengra, Department of Chemistry, University at Albany, State University of New York, 1400 Washington Ave. Albany, NY 12222, USA; The RNA Institute, University at Albany, State University of New York, 1400 Washington Ave. Albany, NY 12222, USA.
Ying Wu, Department of Chemistry, University at Albany, State University of New York, 1400 Washington Ave. Albany, NY 12222, USA; The RNA Institute, University at Albany, State University of New York, 1400 Washington Ave. Albany, NY 12222, USA.
Fusheng Shen, Department of Chemistry, University at Albany, State University of New York, 1400 Washington Ave. Albany, NY 12222, USA; The RNA Institute, University at Albany, State University of New York, 1400 Washington Ave. Albany, NY 12222, USA.
Jia Sheng, Department of Chemistry, University at Albany, State University of New York, 1400 Washington Ave. Albany, NY 12222, USA; The RNA Institute, University at Albany, State University of New York, 1400 Washington Ave. Albany, NY 12222, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NSF [CHE-1845486, MCB-1715234]; University at Albany, State University of New York. Funding for open access charge: [NSF-1715234].
Conflict of interest statement. None declared.
REFERENCES
- 1. Boccaletto P., Machnicka M.A., Purta E., Piatkowski P., Baginski B., Wirecki T.K., de Crecy-Lagard V., Ross R., Limbach P.A., Kotter A. et al.. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018; 46:D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Machnicka M.A., Milanowska K., Osman Oglou O., Purta E., Kurkowska M., Olchowik A., Januszewski W., Kalinowski S., Dunin-Horkawicz S., Rother K.M. et al.. MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res. 2013; 41:D262–D267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Roundtree I.A., Evans M.E., Pan T., He C.. Dynamic RNA modifications in gene expression regulation. Cell. 2017; 169:1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jiang Q., Crews L.A., Holm F., Jamieson C.H.M.. RNA editing-dependent epitranscriptome diversity in cancer stem cells. Nat. Rev. Cancer. 2017; 17:381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Amos H., Korn M.. 5-Methyl cytosine in the RNA of Escherichia coli. Biochim. Biophys. Acta. 1958; 29:444–445. [DOI] [PubMed] [Google Scholar]
- 6. Yi C., Yang C.G., He C.. A non-heme iron-mediated chemical demethylation in DNA and RNA. Acc. Chem. Res. 2009; 42:519–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.M., Li C.J., Vagbo C.B., Shi Y., Wang W.L., Song S.H. et al.. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013; 49:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Giessing A.M., Jensen S.S., Rasmussen A., Hansen L.H., Gondela A., Long K., Vester B., Kirpekar F.. Identification of 8-methyladenosine as the modification catalyzed by the radical SAM methyltransferase Cfr that confers antibiotic resistance in bacteria. RNA. 2009; 15:327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lai C.J., Weisblum B.. Altered methylation of ribosomal RNA in an erythromycin-resistant strain of Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 1971; 68:856–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bjork G.R., Wikstrom P.M., Bystrom A.S.. Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science. 1989; 244:986–989. [DOI] [PubMed] [Google Scholar]
- 11. Ranasinghe R.T., Challand M.R., Ganzinger K.A., Lewis B.W., Softley C., Schmied W.H., Horrocks M.H., Shivji N., Chin J.W., Spencer J. et al.. Detecting RNA base methylations in single cells by in situ hybridization. Nat. Commun. 2018; 9:655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fu Y., Dominissini D., Rechavi G., He C.. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat. Rev. Genet. 2014; 15:293–306. [DOI] [PubMed] [Google Scholar]
- 13. Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G. et al.. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014; 505:117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Falnes P.O., Johansen R.F., Seeberg E.. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002; 419:178–182. [DOI] [PubMed] [Google Scholar]
- 15. Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y.G. et al.. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011; 7:885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Trewick S.C., Henshaw T.F., Hausinger R.P., Lindahl T., Sedgwick B.. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002; 419:174–178. [DOI] [PubMed] [Google Scholar]
- 17. Backert S., Neddermann M., Maubach G., Naumann M.. Pathogenesis of Helicobacter pylori infection. Helicobacter. 2016; 21(Suppl 1):19–25. [DOI] [PubMed] [Google Scholar]
- 18. Kumar S., Karmakar B.C., Nagarajan D., Mukhopadhyay A.K., Morgan R.D., Rao D.N.. N4-cytosine DNA methylation regulates transcription and pathogenesis in Helicobacter pylori. Nucleic Acids Res. 2018; 46:3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dubin D.T., Taylor R.H., Davenport L.W.. Methylation status of 13S ribosomal RNA from hamster mitochondria: the presence of a novel riboside, N4-methylcytidine. Nucleic Acids Res. 1978; 5:4385–4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Iwanami Y., Brown G.M.. Methylated bases of ribosomal ribonucleic acid from HeLa cells. Arch. Biochem. Biophys. 1968; 126:8–15. [DOI] [PubMed] [Google Scholar]
- 21. Bohnsack M.T., Sloan K.E.. The mitochondrial epitranscriptome: the roles of RNA modifications in mitochondrial translation and human disease. Cell. Mol. Life Sci. 2018; 75:241–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kimura S., Suzuki T.. Fine-tuning of the ribosomal decoding center by conserved methyl-modifications in the Escherichia coli 16S rRNA. Nucleic Acids Res. 2010; 38:1341–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wei Y., Zhang H., Gao Z.Q., Wang W.J., Shtykova E.V., Xu J.H., Liu Q.S., Dong Y.H.. Crystal and solution structures of methyltransferase RsmH provide basis for methylation of C1402 in 16S rRNA. J. Struct. Biol. 2012; 179:29–40. [DOI] [PubMed] [Google Scholar]
- 24. Van Haute L., Hendrick A.G., D'Souza A.R., Powell C.A., Rebelo-Guiomar P., Harbour M.E., Ding S., Fearnley I.M., Andrews B., Minczuk M.. METTL15 introduces N4-methylcytidine into human mitochondrial 12S rRNA and is required for mitoribosome biogenesis. Nucleic Acids Res. 2019; 47:10267–10281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McIntyre W., Netzband R., Bonenfant G., Biegel J.M., Miller C., Fuchs G., Henderson E., Arra M., Canki M., Fabris D. et al.. Positive-sense RNA viruses reveal the complexity and dynamics of the cellular and viral epitranscriptomes during infection. Nucleic Acids Res. 2018; 46:5776–5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kabsch W. Xds. Acta. Crystallogr. D. Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Minor W., Cymborowski M., Otwinowski Z., Chruszcz M.. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta. Crystallogr. D. Biol. Crystallogr. 2006; 62:859–866. [DOI] [PubMed] [Google Scholar]
- 28. Sheng J., Li L., Engelhart A.E., Gan J., Wang J., Szostak J.W.. Structural insights into the effects of 2′-5′ linkages on the RNA duplex. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:3050–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta. Crystallogr. D. Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta. Crystallogr. D. Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Winn M.D., Murshudov G.N., Papiz M.Z.. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003; 374:300–321. [DOI] [PubMed] [Google Scholar]
- 33. Brunger A.T. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992; 355:472–475. [DOI] [PubMed] [Google Scholar]
- 34. Chen V.B., Arendall W.B. 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C.. MolProbity: all-atom structure validation for macromolecular crystallography. Acta. Crystallogr. D. Biol. Crystallogr. 2010; 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A. et al.. Overview of the CCP4 suite and current developments. Acta. Crystallogr. D. Biol. Crystallogr. 2011; 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cornell W.D., Cieplak P., Bayly C.I., Gould I.R., Merz K.M., Ferguson D.M., Spellmeyer D.C., Fox T., Caldwell J.W., Kollman P.A.. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995; 117:5179–5197. [Google Scholar]
- 37. Jakalian A., Jack D.B., Bayly C.I.. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 2002; 23:1623–1641. [DOI] [PubMed] [Google Scholar]
- 38. Cornell W.D., Cieplak P., Bayly C.I., Kollman P.A.. Application of RESP charges to calculate conformational energies, hydrogen bond energies, and free energies of solvation. J. Am. Chem. Soc. 1993; 115:9620–9631. [Google Scholar]
- 39. Chen A.A., Garcia A.E.. High-resolution reversible folding of hyperstable RNA tetraloops using molecular dynamics simulations. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:16820–16825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bergonzo C., Cheatham T.E. 3rd. Improved force field parameters lead to a better description of RNA structure. J. Chem. Theory. Comput. 2015; 11:3969–3972. [DOI] [PubMed] [Google Scholar]
- 41. Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E.. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015; 1–2:19–25. [Google Scholar]
- 42. Bussi G., Donadio D., Parrinello M.. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007; 126:014101. [DOI] [PubMed] [Google Scholar]
- 43. Berendsen H.J.C., Postma J.P.M., van Gunsteren W.F., DiNola A., Haak J.R.. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984; 81:3684–3690. [Google Scholar]
- 44. Darden T.A., Pedersen L.G.. Molecular modeling: an experimental tool. Environ. Health Perspect. 1993; 101:410–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Horn H.W., Swope W.C., Pitera J.W., Madura J.D., Dick T.J., Hura G.L., Head-Gordon T.. Development of an improved four-site water model for biomolecular simulations: TIP4P-Ew. J. Chem. Phys. 2004; 120:9665–9678. [DOI] [PubMed] [Google Scholar]
- 46. Hess B., Bekker B., Berendsen H.J.C., Fraaije J.G.E.M.. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997; 18:1463–1472. [Google Scholar]
- 47. Lu J., Li N.S., Koo S.C., Piccirilli J.A.. Efficient synthesis of N4-methyl- and N4-hydroxycytidine phosphoramidites. Synthesis. 2010; 16:2708–2712. [Google Scholar]
- 48. Guennewig B., Stoltz M., Menzi M., Dogar A.M., Hall J.. Properties of N(4)-methylated cytidines in miRNA mimics. Nucleic Acid Ther. 2012; 22:109–116. [DOI] [PubMed] [Google Scholar]
- 49. McDowell J.A., Turner D.H.. Investigation of the structural basis for thermodynamic stabilities of tandem GU mismatches: solution structure of (rGAGGUCUC)2 by two-dimensional NMR and simulated annealing. Biochemistry. 1996; 35:14077–14089. [DOI] [PubMed] [Google Scholar]
- 50. Gehring K., Leroy J.L., Gueron M.. A tetrameric DNA structure with protonated cytosine.cytosine base pairs. Nature. 1993; 363:561–565. [DOI] [PubMed] [Google Scholar]
- 51. Myers J.C., Spiegelman S., Kacian D.L.. Synthesis of full-length DNA copies of avian myeloblastosis virus RNA in high yields. Proc. Natl. Acad. Sci. U.S.A. 1977; 74:2840–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Potapov V., Fu X., Dai N., Correa I.R. Jr., Tanner N.A., Ong J.L.. Base modifications affecting RNA polymerase and reverse transcriptase fidelity. Nucleic Acids Res. 2018; 46:5753–5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Preston B.D., Poiesz B.J., Loeb L.A.. Fidelity of HIV-1 reverse transcriptase. Science. 1988; 242:1168–1171. [DOI] [PubMed] [Google Scholar]
- 54. Suzuki T., Moriyama K., Otsuka C., Loakes D., Negishi K.. Template properties of mutagenic cytosine analogues in reverse transcription. Nucleic Acids Res. 2006; 34:6438–6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors were deposited in the PDB under the accession numbers 6WY2 [CCGG(m4C)GCCGG], 6WY3 [CCGG(m42C)GCCGG-P212121], and 6Z18 [CCGG(m42C)GCCGG-R32].