Abstract
Prime editors (PEs) enable targeted precise editing, including the generation of substitutions, insertions and deletions, in eukaryotic genomes. However, their genome-wide specificity has not been explored. Here, we developed Nickase-based Digenome-seq (nDigenome-seq), an in vitro assay that uses whole-genome sequencing to identify single-strand breaks induced by CRISPR (clustered regularly interspaced short palindromic repeats)-Cas9 (CRISPR-associated protein 9) nickase. We used nDigenome-seq to screen for potential genome-wide off-target sites of Cas9 H840A nickase, a PE component, targeted to nine human genomic sites. Then, using targeted amplicon sequencing of off-target candidates identified by nDigenome-seq, we showed that only five off-target sites showed detectable PE-induced modifications in cells, at frequencies ranging from 0.1 to 1.9%, suggesting that PEs provide a highly specific method of precise genome editing. We also found that PE specificity in human cells could be further improved by incorporating mutations from engineered Cas9 variants, particularly eSpCas9 and Sniper Cas9, into PE.
INTRODUCTION
Genome engineering has been achieved by either non-homologous end joining (NHEJ) or homology-directed repair (HDR) following the generation of DNA double-stranded breaks (DSBs) by programmable nucleases in eukaryotic cells (1–4). NHEJ-mediated knock-out of a wide range of target genes has been induced by CRISPR technology with high efficiency (5). Precise genome editing is achievable by HDR in the presence of designed donor DNAs (6–10), but HDR efficiency still remains unsatisfactory (11,12). Moreover, HDR leaves uncorrected indel sequences at the DSB sites and potentially leads to the integration of donor DNAs at untargeted sites (11,13,14). Significant improvements in DNA modification efficiencies have been achieved by base editing systems, which use a fusion between deaminase and Cas9 nickase or catalytically inactive dead Cas9 protein to deaminate cytosine (15–18) and adenine bases (19,20). However, there were several limitations in the full-fledged use of base editors for gene corrections: for example, base editors had relatively broad editing windows, limiting their utility for creating precise DNA substitutions. In addition, only ‘A-T to G-C’ and ‘C-G to T-A’ conversions were achievable, and other types of conversions, such as ‘A-T to T-A’ or ‘G-C to T-A’ conversions, remained hard to achieve. Furthermore, precise deletions, insertions, or transversion mutations, which are necessary for the treatment of 54% of human genetic disorders, were beyond the scope of base editing systems (21,22). Recently, prime editing technology was developed as a powerful tool to overcome the low efficiency, low specificity, or limited spectrum of possible gene corrections (23).
Prime editors (PEs) consist of Cas9 H840A nickase fused to an engineered M-MLV reverse transcriptase (RT) and a prime editing guide RNA (pegRNA) that carries the primer binding sequence (PBS) and an RT template sequence with the desired edit (23). PEs can potentially lead to all types of genetic modifications including transition and transversion mutations, insertions, and deletions, as well as combinations of these mutations, in eukaryotic cells without inducing DSBs. Prime editing is achieved by five distinct steps: i) the generation of a single-stranded break (SSB) in the non-target strand via Cas9 H840A nickase, ii) DNA/RNA hybridization between the PBS in the pegRNA and the 3′-region of the nicked strand, iii) RT-mediated reverse transcription of the nicked strand according to the RT template sequence, which generates a 3'-flap containing the edit, iv) incorporation of the 3'-flap sequence in the DNA following ligation and v) final incorporation of the edited sequence in both strands through DNA repair.
The initial PE study, in which mutation frequencies were measured at several known Cas9 off-target sites, suggested that PEs have higher specificity compared with the canonical CRISPR-Cas9 system (23). However, genome-wide PE specificities have yet to be determined; this information is essential for clinical and biotechnological applications. In this study, we developed nickase-based Digenome-seq (nDigenome-seq), which effectively profiles DNA SSBs induced by Cas9 H840A nickase on a genome-wide scale. The levels of off-target effects (expected and unexpected edits) at each potential site were quantified by targeted amplicon sequencing analysis during the validation step. This method tactically utilizes the fact that a Cas9 H840A-mediated nick is essential for achieving PE-mediated genetic modifications. Herein, we report that PE-mediated genome editing is quite precise, with high genome-wide specificities.
MATERIALS AND METHODS
Plasmid construction
The pCMV-PE2 (Addgene plasmid #132775) and pU6-pegRNA-GG-acceptor (Addgene plasmid #132777) plasmids were a gift from David Liu. pegRNA-encoding plasmids were generated by Golden Gate assembly using a custom acceptor plasmid according to methods described in a previous paper. (23) pCMV-PE2 was modified to generate a set of plasmids encoding the PE2 variants, which included pCMV-PE2-HF (N497A, R661A, Q695A and Q926A), pCMV-ePE2 (K848A, K1003A and R1060A), pCMV-Evo-PE2 (M495V, Y515N, K526E and R661Q), pCMV-Hypa-PE2 (N692A, M694A, Q695A and H698A) and pCMV-Sniper-PE2 (F539S, M763I and K890N), by incorporating mutations via NEBuilder® HiFi DNA Assembly Master Mix (New England Biolabs) with gblocks containing mutations (IDT) and site-directed mutagenesis (Q5 Site-Directed Mutagenesis Kit, New England Biolabs).
Human cell culture and transfection
HEK293T cells (ATCC CRL-11268) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin and verified using an STR profile. HEK293T cells were seeded onto a 24-well plate and transfected with appropriate plasmids [for PE2 experiments, the PE2- or PE2 variant-encoding plasmid (1500 ng) and the pegRNA-encoding plasmid (500 ng); for PE3 experiments, the PE2-encoding plasmid (1500 ng), the pegRNA-encoding plasmid (500 ng) and sgRNA-encoding plasmid (167 ng); for Cas9 experiments, the Cas9-encoding plasmid (1500 ng) and the sgRNA-encoding plasmid (500 ng)] using 3 μl of Lipofectamine 2000 (Life Technologies). After 96 h, gDNA was isolated with a DNeasy tissue kit (Qiagen) according to the manufacturer's instructions.
Expression and purification of Cas9 H840A nickase
The plasmid encoding the His6-Cas9 H840A nickase was generated by site-directed mutagenesis of a pET28 plasmid encoding His6-Cas9 nuclease. The recombinant Cas9 H840A protein containing a nuclear localization signal, an HA epitope and an His-tag at the N terminus was expressed and purified according to methods described previously (ABE paper). Briefly, expression of the recombinant Cas9 H840A protein was induced in BL21 Star cells using 0.1 mM IPTG, after which the cells were lysed by sonication. The recombinant protein in the soluble lysate obtained after centrifugation was purified using Ni-NTA agarose beads (Qiagen) and heparin agarose beads (Heparin Sepharose 6 Fast Flow; GE Healthcare). The resulting protein fractions were concentrated using an Ultracel 100K cellulose column (Millipore), and the purity and concentration of the Cas9 protein were analyzed by sodium dodecyl sulphate-polyacrylamide gelelectrophoresis.
Cas9 nickase-mediated in vitro digestion of gDNA
gDNA was isolated from HEK293T cells using a DNeasy tissue kit (Qiagen) according to the manufacturer's instructions. Recombinant Cas9 H840A nickase (100 nM) and sgRNA (300 nM) were pre-incubated at RT for 10 min and mixed with 20 μg of gDNA in a reaction buffer (100 mM NaCl, 50 mM Tris–HCl, 10 mM MgCl2, 100 μg/ml bovine serum albumin, at pH 7.9) for 8 h at 37°C. Digested gDNA was treated with RNase A (50 μg/ml) and protease K to degrade the sgRNA and recombinant Cas9 H840A nickase and then purified again with a DNeasy tissue kit (Qiagen).
Whole-genome and nDigenome sequencing
gDNA (1 μg), which had been previously treated with Cas9 H840A nickase, was sheared to obtain fragments of about 500 bp in size using a Covaris system (Life Technologies) and blunt-ended using End Repair Mix (illumina). The fragmented DNA was adenylated to prevent self-ligation and ligated with adapters to produce libraries via TruSeq DNA Library Prep Kits (illumina), which were then subjected to whole-genome sequencing (WGS) using a HiSeq X Ten Sequencer (Illumina) at Macrogen. WGS was performed at a sequencing depth of 30–40×. Sequencing reads were aligned to a reference genome sequence (hg19 from UCSC) using Isaac aligner with the following parameters: base quality cutoff, 15; keep duplicate reads, yes; variable read length support, yes; realign gaps, no; and adaptor clipping, yes (adaptor: AGATCGGAAGAGC*,*GCTCTTCCGATCT). After separating the aligned sequences into the forward and reverse strands using SAMtools, DNA SSB sites were identified using the original Digenome programs, which are used to calculate the count of the sequence reads with their 5′ ends starting at the same nucleotide position and the sequencing depth at each nucleotide position. The source code of the original version of Digenome used in this manuscript is available at https://github.com/snugel/digenome-toolkit. gDNA that included the on-target and potential off-target sites was amplified using KAPA HiFi HotStart DNA polymerase (Roche) according to the manufacturer's instructions. The resulting polymerase chain reaction amplicons containing Illumina TruSeq HT dual index adapter sequences were subjected to 150-bp paired-end sequencing using Illumina iSeq 100. Substitutions and indel frequencies were calculated by MAUND, which is available at https://github.com/ibs-cge/maund.
Statistical analyses
All results from experiments with three independent replicates were expressed as mean ± s.e.m. P-values were calculated by the two-tailed Student's t-test.
RESULTS
Editing efficiency of PE2 in human cells
First, we compared the editing efficiencies of PE2, designed to induce six different types of edits (1-nt insertions or deletions, 3-nt insertions or deletions, or two different kinds of 1-nt substitutions), with the indel frequencies generated by Cas9 nuclease at seven endogenous target sites (Supplementary Figure S1a–d). We transfected plasmids encoding PE2 or Cas9 nuclease into HEK293T cells together with those encoding a corresponding pegRNA or sgRNA, respectively. Although the PE2 efficiency is lower than that of Cas9 nuclease, we found that PE2s induce the expected edit with a broad range of efficiencies (0.4–48.6%; average, 11.7 ± 1.7%) depending on the target site and the type of desired mutation (targeted insertion, deletion and substitution efficiencies averaging 9.1 ± 2.1%, 10.5 ±3.4 % and 15.6 ± 3.3%, respectively) with very low frequencies of undesired indels (without expected edit; 0.16 ± 0.05%) (Supplementary Figure S1a–c), suggesting that PE2-mediated editing results in a higher purity of the desired product compared with Cas9 nuclease-mediated HDR, which induces higher frequencies of unwanted byproducts such as indels (15). We also found that the editing efficiencies of PE2 were lower than those of Cas9 nuclease and were independent of the Cas9 nuclease-mediated indel frequencies at a given site (Supplementary Figure S1c).
Tolerance of PE2 and Cas9 for mismatched pegRNAs and sgRNAs
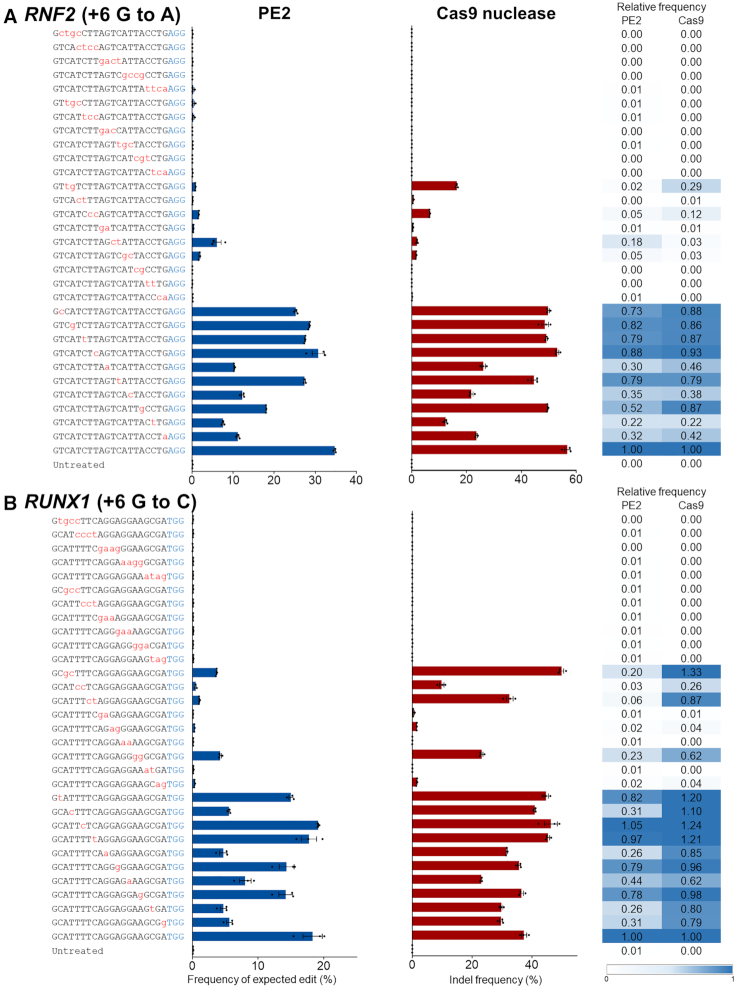
To appraise the specificity of PE2, we examined its tolerance for mismatches in the pegRNA spacer sequence and compared it to that of Cas9 nuclease guided by mismatched sgRNAs. To this end, we transfected HEK293T cells with plasmids encoding PE2 or Cas9 along with plasmids encoding the accompanying pegRNA or sgRNA, respectively, each of which carried zero to four mismatches in the spacer sequences. Then, we determined the frequencies of expected edit and indel frequencies at three endogenous target sites (Figure 1 and Supplementary Figure S2). Overall, PE2 and Cas9 nuclease tolerated most of the single nucleotide mismatches but showed almost complete loss of activities when transfected with a guide RNA with a three or four nucleotide mismatch. Although PE2 has a slightly higher level of tolerance for certain mismatches compared to Cas9 nuclease, for instance, pegRNA containing two mismatches (at positions 11–12, numbered 1–20 in the 5′ to 3′ direction) in the RNF2 site (the relative frequency of edits and indels induced by PE2 and Cas9 at the mismatched versus matched sites was 0.18 and 0.03, respectively), PE2 generally showed a lower tolerance for mismatched targets compared to the Cas9 nuclease (Figure 1 and Supplementary Figure S2). Collectively, these results indicate that PE2 enables precise genome editing with less tolerance for mismatches between the target and pegRNA.
Figure 1.
Tolerance of PE2 and Cas9 nuclease for mismatched pegRNAs or sgRNAs, respectively. (A and B) Together with plasmids encoding PE2 or Cas9 nuclease, plasmids encoding pegRNA or sgRNA that differed from the RNF2 (A) and RUNX1 (B) target sequences by 1–4 nt were respectively transfected into HEK293T cells. Editing frequencies and indel frequencies were measured by targeted deep sequencing. The PAM is shown in blue and the mismatched nucleotides are shown in red lowercase letters. Error bars indicate the s.e.m. (three biologically independent samples). Relative frequencies were calculated by dividing the frequency of edits or indels obtained using mismatched pegRNAs or sgRNAs by the frequency of the on-target edits.
Importance of nicking in PE2 activity
To test the possibility that unexpected genome editing occurs through Cas9 nickase-independent incorporation of a 3′ extension of the pegRNA (PBS), we performed an insilico search for endogenous off-target sites that contain an identical PBS (HEK3, RNF2 and RUNX1) but lack a Cas9 protospacer adjacent motif (PAM) sequence. Targeted amplicon sequencing analysis revealed, however, that none of these sites displayed a Cas9 nickase-independent mutation (Supplementary Table S1). In addition, to confirm the requirement for nicking in PE2 activity, we generated a catalytically inactive dead PE2 (dPE2) that consists of a dead Cas9 (incorporation of D10A and H840A mutations) and an engineered M-MLV RT. Transfection of the dPE2 module with pegRNA into HEK293T cells did not induce any mutations at any of the seven endogenous target sites (Supplementary Figure S1d). These results establish that Cas9 H840A nickase-mediated nicking is essential for PE2-mediated genome editing.
Nickase-based Digenome-seq (nDigenome-seq)
To evaluate the genome-wide specificity of PE2, which can be measured as the genome-wide pattern of nicks formed by Cas9 H840A nickase, which is essential to PE2-mediated genome editing, genomic DNA (gDNA) isolated from HEK293T cells was incubated with ribonucleoproteins (RNPs) consisting of purified Cas9 H840A nickase protein (100 nM) and in vitro transcribed sgRNA (300 nM). The treated gDNA was subjected to whole genome sequencing following DNA fragmentation, end repair and adaptor ligation (Figure 2A). The genome-wide specificity was investigated by measuring the frequencies of off-target edits obtained with pegRNAs targeted to nine endogenous sites.
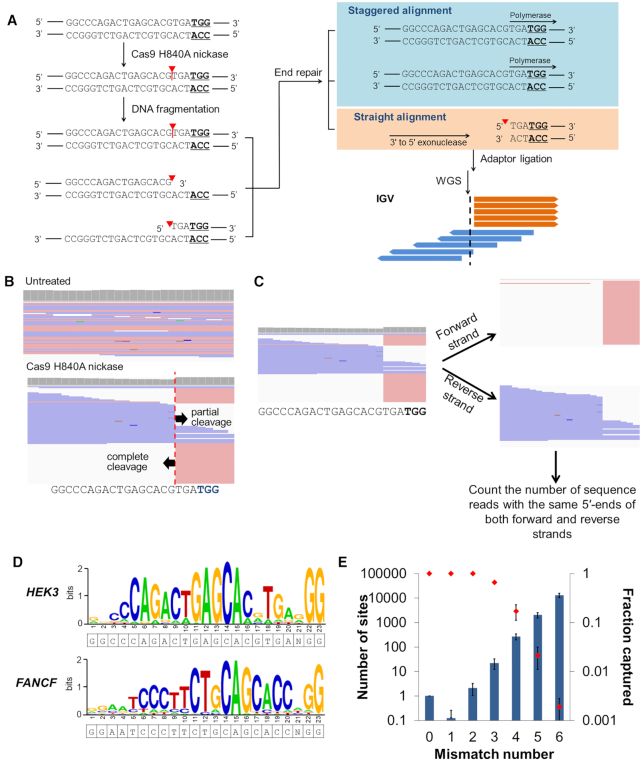
Figure 2.
nDigenome-seq to identify SSB sites generated in vitro in the human genome by Cas9 H840A. (A) Overview of nDigenome-seq. gDNA is prepared from cells and treated with Cas9 H840A nickase and sgRNA, resulting in a nick in the protospacer of on-target and off-target sites. The fragmentation step, done by sonication, renders a library of ∼500 bp fragments with random ends. Treatment with a polymerase with 3′ to 5′ exonuclease activity leads to end repair, during which the 3′-end resulting from the nick (that is, fragments with a 5′-overhang) is filled. As a result, the filled strand shows a random end sequence due to the random fragmentation, thereby exhibiting staggered alignment in an IGV file. However, the fragments with a 3′-overhang are trimmed by 3′ to 5′ exonuclease activity, resulting in fragments with blunt ends. Thus, the trimmed fragments show a straight alignment pattern in an IGV file. In summary, a straight alignment pattern along with a staggered alignment pattern indicates the occurrence of a nick induced by Cas9 H840A nickase at both on-target and off-target sites. We used a TruSeq DNA Library Prep Kit (illumina) for adaptor ligation and to generate a sequencing library. Forward and reverse sequence reads are shown in pink and blue, respectively. Red triangles and vertical lines represent cleavage positions. (B) An IGV image of sequence reads that reveals complete and partial cleavage of the non-target and target strands, respectively, at the HEK3 on-target site. (C) Aligned sequence data were separated into forward and reverse strands, and the number of sequence reads with the same 5′ end at each position were counted to identify in vitro SSB sites. (D) Representative sequence logos for the HEK3 and FANCF sites were obtained from WebLogo using DNA sequences at the sites captured by nDigenome (sequence read counts starting at this position ≥10 and count/depth ≥20%). (E) The number of potential off-target sites with homologous sequences (left y-axis; bars) increased exponentially as the number of mismatches with the nine Cas9 H840A on-target sites tested increased. In contrast, the fractions of captured sites (right y-axis, red dots) decreased sharply. Error bars indicate the s.e.m (n = 9).
First, we investigated the cleavage pattern formed by Cas9 H840A nickase RNPs at the on-target sites. Integrative Genomics Viewer (IGV) images revealed that Cas9 H840A nickase induced complete cleavage of the non-target strand (the strand containing the PAM). However, the target strand was also, although only partially, cleaved by the Cas9 nickase (Figure 2B). Based on this result, we considered profiling in vitro SSB sites that were generated in the non-target strand of Cas9 target sites without consideration of the SSB in the target strand. Herein, we propose using nDigenome-seq for the determination of genome-wide specificity, which uses the following procedure to probe genome-wide SSB sites generated by Cas9 H840A nickase. First, we separated the sequencing reads into forward and reverse strands using SAMtools (Figure 2C). Second, the number of sequence reads with the same 5′ end and sequence depth at each nucleotide position were determined (Figure 2C and Supplementary Figure S3a). Third, to identify in vitro SSB sites, we computationally screened for sites where the count of the sequence reads with 5′ ends starting at the same nucleotide position was 10 or more and straight alignments account for at least 20% of the sequence reads (Figure 2C and Supplementary Figure S3a). Finally, we screened for off-target sites that accompany canonical PAM (5′-NGG-3′)-containing sites with six or fewer mismatches or non-canonical PAM (5′-NGH-3′ or 5′-NHG-3′; H is A, C, or T)-containing sites with five or fewer mismatches compared to the on-target sequence.
Based on this nDigenome-seq method, we identified 39–536 in vitro SSB sites (average number, 210 ± 71) associated with the nine on-target sites (Figure 2D; Supplementary Figures S3b-S4 and Table S2). In contrast, we could not detect any in vitro SSB sites in the untreated gDNA, confirming the validity of the nDigenome-seq method. As expected, all 9 of the on-target and all 19 of the off-target sites with one or two mismatches containing an NGG PAM sequence were captured by nDigenome-seq (Figure 2E). However, as the number of mismatches increased from three to six, the fraction of in vitro SSB sites decreased exponentially from 0.66 to 0.0019, which was similar to the pattern obtained with the Cas9-based Digenome-seq method (Figure 2E) (24,25).
Comparison of in vitro cleavage sites of Cas9 H840A nickase and Cas9 nuclease targeted to the same sites
Next, although Cas9 nuclease-mediated Digenome-seq and Cas9 H840A nickase-mediated nDigenome-seq use different parameters to define off-target loci, we compared the off-target in vitro cleavage sites identified by Cas9 nuclease (24) and Cas9 H840A nickase targeted to five genomic sites (HEK3, FANCF, RNF2, EMX1 and HEK4 sites). When targeted to the FANCF, RNF2 and HEK4 sites, Cas9 and Cas9 H840A nickase exhibited similar off-target profiles. In contrast, when targeted to the HEK3 and EMX1 sites, a significant portion of the off-target sites recognized by Cas9 and Cas9 H840 were distinct (Supplementary Figure S5). This result requires an in-depth investigation of Cas9 H840A nickase-induced off-target effects that are independent of Cas9. Nonetheless, it is interesting to note that the great majority of off-target sites detected by Digenome-seq in vitro and validated in vivo for Cas9 (49/52 = 94%) (24) were also identified by Cas9 H840A nickase.
Validation of in vitro cleavage sites
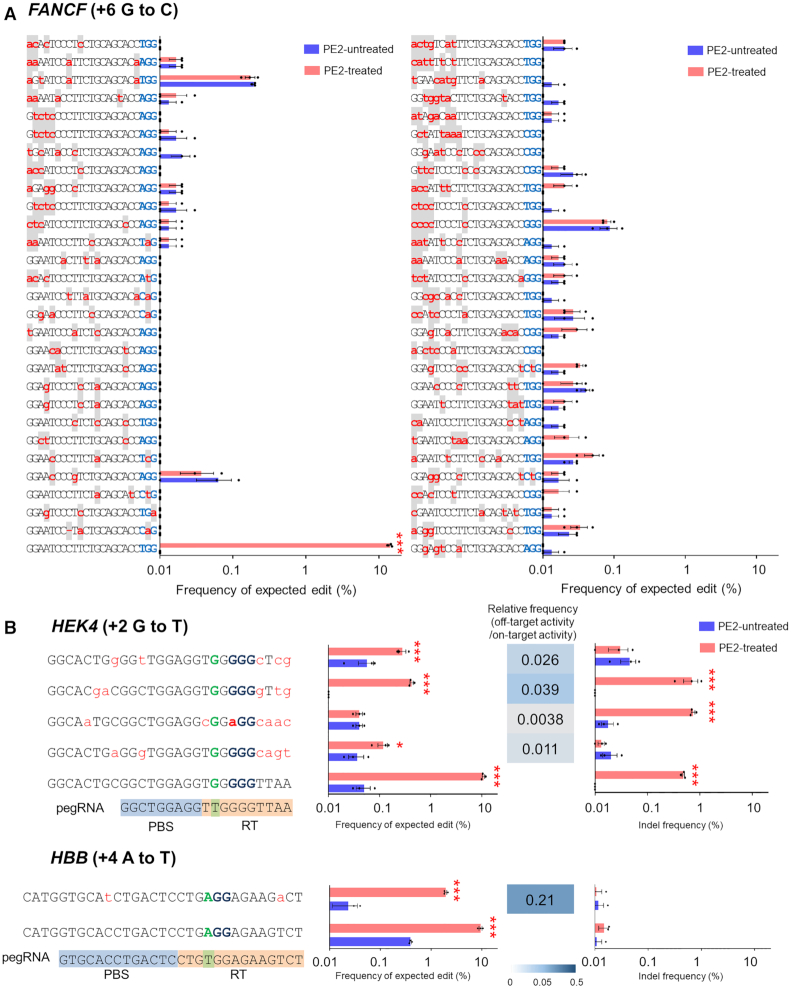
To check if the off-target sites identified by nDiginome-seq show unintended, PE2-induced mutations in vivo, we transfected plasmid DNA encoding PE2 and pegRNAs respectively targeting nine human genomic sites (installing at position +1 a CTT insertion (ins) (HEK3), +6 G to C (FANCF), +6 G to A (RNF2), +5 G to T (EMX1), +6 G to C (DNMT1), +6 G to C (RUNX1), +5 G to T (VEGFA), +2 G to T (HEK4) and +4 A to T (HBB) mutations), and the editing frequencies were measured at the 283 in vitro cleavage sites identified by Cas9 H840A nickase-mediated nDigenome-seq. Surprisingly, no detectable mutations at the in vitro cleavage sites predicted by nDigenome-seq were identified by the targeted deep sequencing for seven of the nine targets (Figure 3A and Supplementary Table S3). PE2 coupled with pegRNA targeting HEK4 or HBB caused mutations at five off-target sites, with frequencies ranging from 0.1 to 1.9% (Figure 3B). Considering that 3–20 validated off-targets (10 ± 3) were identified by Cas9 nuclease-mediated Digenome-seq for the EMX1, FANCF, RNF2, HEK3 and HEK4 sites (24). These result suggests that PE2 provides a highly specific programmable editing system. In particular, PE2 did not induce mutations at the DNMT1-06 site, which contains two nucleotide mismatches in the spacer sequence containing an NGG PAM sequence, 1 nt mismatch in the PBS, and three nucleotide mismatches in the RT sequence, or the HEK4-017 site, which contains three nucleotide mismatches in the PAM distal region of the spacer sequence containing an NGG PAM sequence, no mismatches in the PBS and 3 nt mismatches in the RT sequence (Supplementary Figure S6). In contrast, Cas9 nuclease induced off-target mutations at both the DNMT1-06 and HEK4-017 sites. The reason for the high specificity of PE2 may be explained by the speculation that although SSBs are generated at the off-targets by Cas9 H840A nickase, these sites do not necessarily share an identical sequence with the on-target sequence in the RT template region of the pegRNA. It is likely that reverse-transcribed edits are not applied to the gDNA because of poor hybridization of the 3′-flap with the opposite strand.
Figure 3.
Validation of PE2 off-target sites identified by Cas9 H840A nickase-mediated nDigenome-seq. (A) In vitro SSB sites captured by nDigenome-seq after treatment with Cas9 H840A nickase targeting FANCF (installing +6 G to C mutation) were subjected to validation by targeted deep sequencing using gDNA extracted from HEK293T cells transfected with plasmids encoding PE2 and pegRNA. The mismatched nucleotides are shown in red lowercase letters on a gray background, and PAM sequences are shown in blue. Error bars indicate the s.e.m. (three biologically independent samples). (B) On-target and validated off-target sites identified in human cells using targeted deep sequencing. The mismatched nucleotides are shown in red lowercase letters and PAM sequences are shown in blue. Precise edit and indel frequencies at the validated off-target sites for Cas9 H840A nickase targeted to HEK4 and HBB. PBS and RT template sequences are highlighted with blue and orange backgrounds, respectively. Error bars indicate the s.e.m. (three biologically independent samples). P-values were calculated by Student's t-test between PE2-untreated and PE2-treated. P -value, *; <0.05, **; <0.01, ***; <0.001.
Tolerance of PE2 to mismatches in the RT and PBS regions
To take a closer look at how the RT template and PBS in the pegRNA impose specificity on PE2, we constructed various pegRNA-encoding templates that carry up to four mismatched nucleotides in either the PBS or RT region. Using those mismatched pegRNAs, we investigated the frequencies of expected edits at RUNX1 (installing +6 G to C mutation), HEK3 (installing +1 CTT insertion) and RNF2 (installing +6 G to A mutation) in HEK293T cells (Figure 4 and Supplementary Figure S7). One mismatch in either the PBS or RT region did not affect the precise editing efficiency significantly. However, two or more mismatches sometimes resulted in a significant decrease in editing efficiency, depending on the mismatch site (Figure 4 and Supplementary Figure S7). In particular, mismatches in nick-adjacent PBS and RT template regions critically affected the editing efficiency. We next found a representative target with an associated off-target site that had an identical protospacer sequence and a one base mismatch in the RT sequence. A measurement of prime editing efficiency revealed that the expected edit frequency in the off-target site decreased up to 65% (6.5–65.1%; average, 40.5 ± 8.8%) compared to the on-target activity (Supplementary Figure S8). These results suggest that a bilateral checkpoint in the RT template and PBS confers specificity to the PE system. In another respect, they imply that the sequence similarity in the RT template and PBS regions (that is, a target sequence) should be considered in the identification of off-target sites for the PE system.
Figure 4.
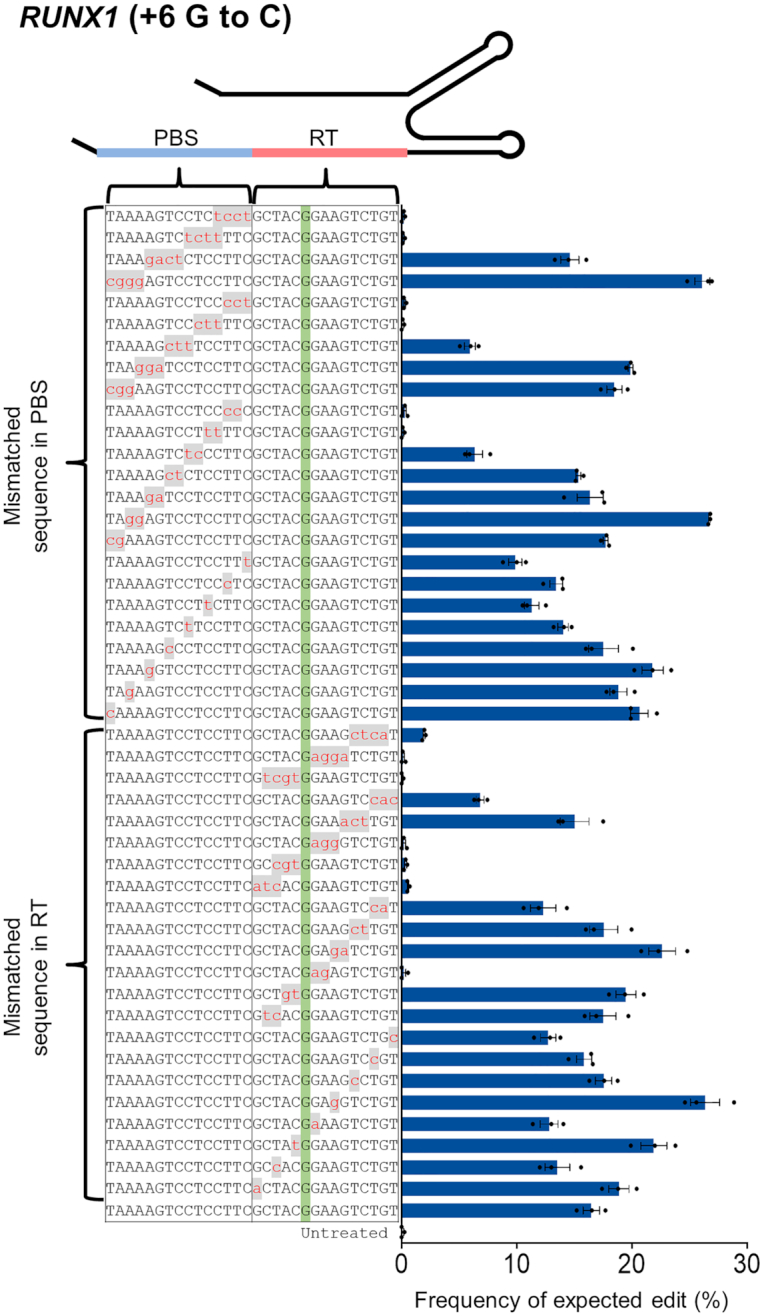
Tolerance of PE2 for mismatches in the PBS or RT template region of the pegRNA and enhanced specificity with engineered PE2. Frequency of targeted edits induced by PE2 using pegRNAs containing one to four mismatched nucleotides in the PBS or RT template. The PAM is shown in blue, and the mismatched nucleotides are shown in red lowercase letters on a gray background. Error bars indicate the s.e.m. (three biologically independent samples).
Effect of the PBS and RT template lengths on PE2 specificity
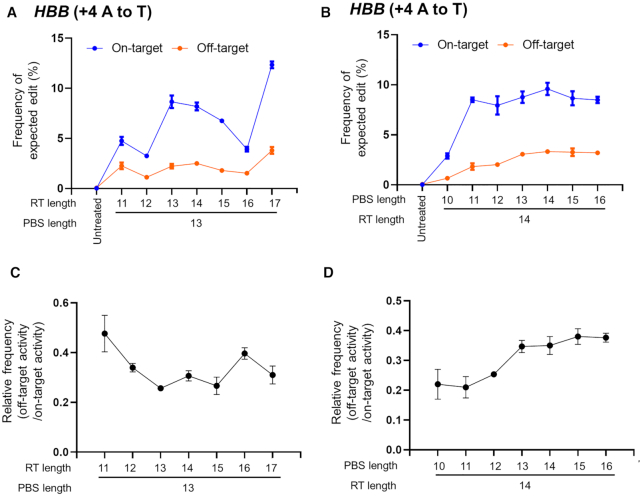
To investigate whether the PBS and RT template lengths affect PE2 specificity, we transfected HEK293T cells with plasmids encoding PE2 and pegRNAs (installing +4 A to T (HBB) mutation) carrying different PBS or RT-template lengths and measured the editing frequency at on-target and off-target sites (Figure 5A–D). Overall, changing the RT template length did not affect PE2 specificity (Figure 5C); a relatively short PBS template (11–13 nt) was associated with higher specificity than a long RT template (14–17 nt) (Figure 5D).
Figure 5.
Effect of the lengths of the RT template and PBS in the pegRNA on the efficiency of on-target and off-target editing in HEK293T cells. (A and B) The dependence of the PE2 specificity on the lengths of the PBS or RT template was investigated using HBB-targeted pegRNAs with variable PBS lengths (10–16 bp) or RT lengths (11–17 bp), respectively (A and B). (C andD) Representative graphs showing relative frequencies (off-target activity/on-target activity) depending on the PBS length (C) or the RT template sequence length (D). Error bars indicate the s.e.m. (three biologically independent samples).
Specificity enhancement of PE2 via engineering of Cas9 H840A
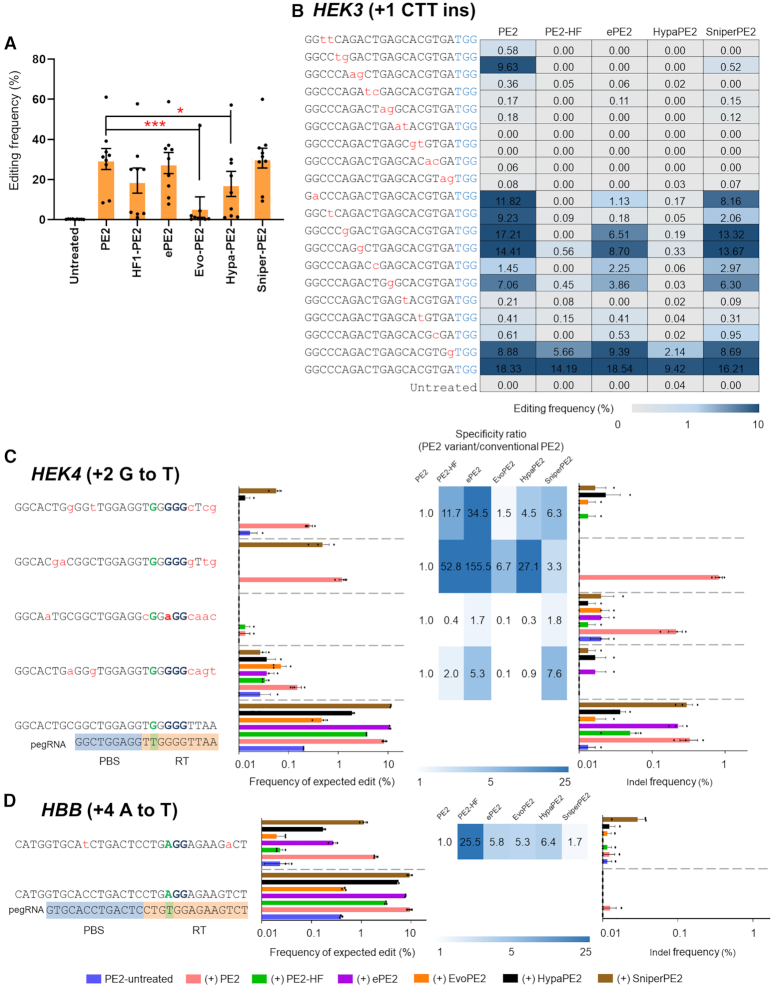
Next, we hypothesized that engineering the Cas9 H840A portion of PE2 might improve PE specificity. Cas9 variants, including eCas9 (26), Cas9-HF (27), HypaCas9 (28), EvoCas9 (29) and SniperCas9 (30), are known to have enhanced specificity compared to Cas9. Thus, we generated PE2 variants that included the Cas9 variant mutations in the Cas9 H840A domain. Then, we transfected HEK293T cells with the plasmids encoding the PE2 variants (PE2, ePE2, PE2-HF, HypaPE2, EvoPE2 and SniperPE2) with respective pegRNAs (installing +1 CTT ins (HEK3), +6 G to C (FANCF), +6 G to A (RNF2), +5 G to T (EMX1), +6 G to C (DNMT1), +6 G to C (RUNX1), +5 G to T (VEGFA), +2 G to T (HEK4) and +4 A to T (HBB) mutations) and measured the precise editing frequency at the nine endogenous target sites (Figure 6A).
Figure 6.
Enhancing PE2 specificities with PE2 variants. (A) Precise edit frequencies achieved by PE2, PE2-HF, ePE2, Evo-PE2, Hypa-PE2 and Sniper-PE2 at nine selected endogenous target sites in HEK293T cells. Error bars indicate the s.e.m. (n = 9). P -values were calculated by Student's t-test. P-value, *; <0.05, **; <0.01, ***; <0.001. (B) Plasmids encoding mismatched pegRNAs that differed from the HEK3 site (installing +1 CTT ins) by one to 2 nt were transfected into HEK293T cells. Precise editing frequencies were measured by targeted deep sequencing. (C andD) Improved specificity of PE2 variants as validated at off-target sites with a high sequence similarity with the HEK4 (C) and HBB (D) on-target sites. Specificity ratios were calculated by dividing the specificity of conventional PE2 (on-target frequency/off-target frequency) by the specificity of the PE2 variant (on-target frequency/off-target frequency). The heatmap represents the relative specificity of the PE2 variants compared to that of the canonical PE2. The mismatched nucleotides are shown in red lowercase letters, and the PAM sequences are shown in blue. PBS and RT template sequences are highlighted with blue and orange backgrounds, respectively. Error bars indicate the s.e.m. (three biologically independent samples).
Prime editing efficiency is governed by gene-targeting activity in the first stage and by the mutation efficiency, involving RT, in the second stage. That is, decreased efficiency in either of the two stages would result in reduced prime editing activity. A previous study revealed that evoCas9, HypaCas9 and Cas9-HF show lower editing activity compared to eCas9 and Sniper-Cas9 (31,32). Here we present experimental data that show that ePE2 and Sniper-PE2 exhibited an on-target editing efficiency comparable to that of the canonical PE2 (Figure 6A). In contrast, PE2-HF, HypaPE2 and EvoPE2 showed a markedly reduced prime editing efficiency (Figure 6A). To investigate the specificities of the PE2 variants, we transfected HEK293T cells with plasmids encoding each PE2 variant (except for EvoPE2, which exhibits an on-target efficiency that is very low), together with a plasmid encoding a pegRNA carrying up to two-bp mismatches in the spacer sequence and measured the editing frequency at the target site (Figure 6B). In general, all of the PE2 variants showed increased specificities compared to the canonical PE2. For instance, the canonical PE2 system was highly tolerant of pegRNAs containing a 2-bp mismatch at position 5–6, editing the target site with an efficiency of 9.6%. However, ePE2, PE2-HF, HypaPE2 and SniperPE2 showed efficiencies of <0.01, <0.01, <0.01 and 0.52, respectively (Figure 6B). These results show that the PE2 variants have a higher specificity compared to the canonical PE2.
Next, we transfected plasmids encoding each PE2 variant and a pegRNA targeting the HEK4 site (installing +2 G to T mutation) or HBB (installing +4 A to T mutation) and measured the expected edit levels and indel frequencies at the on-target and validated off-target sites. ePE2 and SniperPE2 showed reduced expected edit and indel frequencies at the off-target sites, including four validated off-target sites for HEK4 and one off-target site for HBB, while retaining on-target prime-editing efficiency (Figure 6C and D). In contrast, the PE2-HF and HypaPE2 systems showed a concomitant loss in their on-target activity, which may compromise the utility of these systems for prime editing (Figure 6C and D). PE2 variants, particularly ePE2 and SniperPE2, await further investigations of their genome-wide specificity before their use in prime editing.
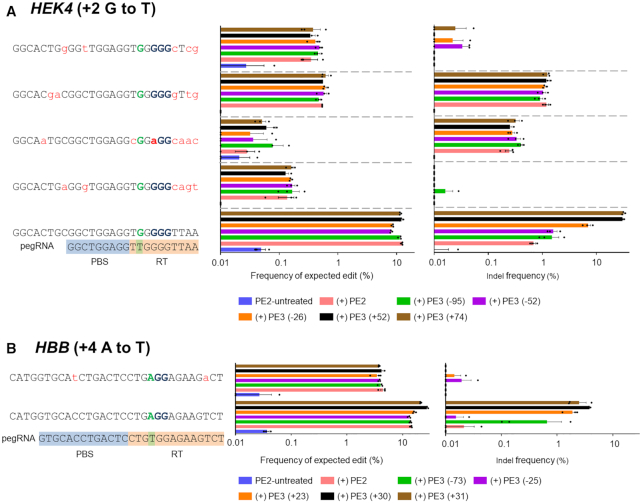
Off-target effects of PE3
PE3 consists of PE2, pegRNA and an sgRNA that is complementary to the PBS (or RT) strand. The Cas9 H840A nickase of PE2 and the sgRNA induce a nick in the non-edited strand, which is intended to enhance the prime editing activity. To investigate the off-target effects of PE3, we transfected plasmids encoding PE2, pegRNA targeting the HEK4 site (installing +2 G to T mutation) or HBB (installing +4 A to T mutation) and five different sgRNAs into HEK293T cells and measured the frequency of expected edits and indels at the on-target and validated off-target sites. First, we measured the PE3-induced frequency of DNA modifications at on- and off-target HEK4 sites. In line with results from a previous report (23), PE3 modules targeting the HEK4 site did not increase the editing frequency at either the on-target or off-target sites relative to PE2 (Figure 7A). On the contrary, the editing efficiency at the HBB on-target site was increased by up to 1.9-fold by the addition of sgRNA (+30; the number refers to the number of nucleotides from the PE2 nick site, +; 3′ downstream, −; 5′ upstream) compared to that of PE2 (Figure 7B). Next, the off-target effects of PE2 and PE3 were compared at the validated off-target sites. Despite the increased on-target editing efficiency of PE3, the editing efficiencies of PE2 and PE3 at the off-target sites were the same (Figure 7A and B). It is noteworthy that, for PE3, the frequencies of indels increased even at the on-target sites, which is also in line with the previous report (23). Thus, the possible occurrence of unwanted byproduct indels must be considered when the PE3 system is used. Taken together, these results suggest that the risk of off-target effects are not increased for PE3 compared to PE2. One possible explanation would be that, except when paralogous genes are present, it is highly likely that the sgRNA used as part of PE3 would have no recognizable protospacer sequence near the off-target sites.
Figure 7.
Specificity of PE3. (A and B) Plasmids encoding PE2, pegRNA and sgRNA were transfected into HEK293T cells and the frequencies of precise edits and unwanted indels were measured using targeted deep sequencing at the HEK4 (A) and HBB (B) on-target and validated off-target sites. The mismatched nucleotides are shown in red lowercase letters and the PAM sequences are shown in bold font. PBS and RT template sequences are highlighted with blue and orange backgrounds, respectively. Error bars indicate the s.e.m. (three biologically independent samples).
DISCUSSION
Programmable nickases such as Cas9 nickases and zinc finger nickases are widely used, not only to improve specificity through paired nicking (33–35), but also as fusions with functional domains including cytosine deaminases (15,16), adenosine deaminases (19), Rad51 (36) and engineered M-MLV RT (23). Although several methods, such as IDLV (Integrase deficient lentiviral vectorcapture) capture (37), GUIDE-seq (genome-wide, unbiased identification of DSBs evaluated by sequencing) (38), HTGTS (high-throughput, genome-wide translocation sequencing) (39), BLISS (breaks labeling in situ and sequencing) (40), CIRCLE-seq (circularization for invitro reporting of cleavage effects by sequencing) (41), SITE-seq (selective enrichment and identification of tagged genomicDNA ends by sequencing) (42) and DISCOVER-Seq (discovery of insitu Cas off-targets and verification by sequencing) (43) were developed to define genome-wide off-target effects of Cas9 nucleases, these methods cannot identify nickase-mediated genome-wide off-target effects. In a previous study, we successfully identified genome-wide off-target effects of cytosine base editors (CBEs) (44) and adenine base editors (ABEs) (45), which are fusions between Cas9 D10A nickase and a deaminase. However, in that study, we identified genome-wide off-target effects by detecting DSBs that were induced via CBEs and USER (uracil-specific excision reagent) or ABEs and Endonuclease V.
In this study, we developed nDigenome-seq to assess genome-wide SSBs generated in vitro. Using this method, we identified genome-wide SSB sites induced by Cas9 H840A nickase, a PE component, in vitro. We then tested whether these sites could be validated as off-target PE sites by targeted amplicon sequencing of gDNA extracted from HEK293T cells transfected with plasmids encoding PE2 and pegRNA. These experiments indicated that PE is highly specific, consistent with results of a previous study published by David Liu's group (23). In addition, the tolerance of PE2 to mismatches in various regions of the pegRNA, including the spacer sequence, RT template and PBS, show the importance of RT template and PBS for PE specificity. Unlike Cas9 nuclease specificity, which is affected by one checkpoint, the spacer sequence, PE specificity is additionally affected by the RT template and PBS, which serve as bilateral checkpoints. Accordingly, PE2 specificity is higher than that of Cas9 nuclease. For instance, Cas9 nuclease induces high frequencies of indels (12.1 and 38.3%, respectively) at the FANCF-5 and EMX1-3 off-target sites, which each possess 2 nt mismatches in the spacer sequence relative to the on-target site (The indel frequency at the FANCF and EMX1 on-target sites is 44.5 and 61.6%, respectively) (44). However, the frequency of PE2-induced mutations was below noise levels (<0.05%) at the FANCF-5 and EMX1-3 off-target sites (Supplementary Table S3). Therefore, to effectively design specific pegRNAs, we should monitor not only the spacer sequence but also the RT template and PBS at potential off-target sites.
An additional increase in PE specificity was achieved by the use of Cas9 variants. Sniper-Cas9 and eCas9 enabled prime editing with reduced frequencies of off-target effects (Figure 6). Further incremental increases could be obtained by the use of sophisticated designs for the pegRNAs. For instance, a pegRNA with an elongated PBS resulted in increased prime editing at HBB off-target sites (Figure 5C). An alteration in the RT template length may also affect specificity (Figure 5D). However, these results are not directly applicable to all target genes and should be determined empirically.
nDigenome-seq is also expected to be useful for profiling the genome-wide specificity of CBE, ABE and hRad51-Cas9, as well as that of PE. However, although nicking induced by Cas9 H840A nickase is essential in PE2-assisted genome engineering, Cas9 H840A nickase-mediated nDigenome-seq is an indirect method for profiling the genome-wide specificity of PE. A more direct method, such as genome-wide off-target analysis by two-cell embryo injection (GOTI) as reported previously (46) or transcriptome sequencing (47), would complement the nDigenome-seq-based description of the PE specificity landscape.
CONCLUSION
Prime editing was developed to overcome the low efficiency of HDR-mediated precise gene correction and to expand CRISPR technology into treatments for a wider range of genetic disorders. Given the high efficiency of PE-mediated gene correction, it is quite demanding to investigate possible off-target sites of a prime editing system. In summary, our data, obtained using the nDigenome-seq method, revealed the high specificity with which precise, PE-mediated genome editing can be achieved. The data suggest that the RT template and PBS sequences serve as a dual checkpoint for precise editing and that dissimilarity in either of these two sequences prevents unintended off-target activity. We also found that PE2 specificity was further improved by the incorporation of mutations found in Cas9 variants, especially evident with ePE2 and SniperPE2, which were much more sensitive than PE2 to even one-base mismatches with the on-target sequence. Although further studies, including the investigation of pegRNA-independent off-target effects, will be required to understand PE specificities fully, our results based on the nDigenome-seq method should promote the broader use of PEs in clinical and biotechnological applications.
DATA AVAILABILITY
Sequencing data was deposited in the NCBI Sequence Read Archive (SRA) database with BioProject accession code PRJNA659391.
CODE AVAILABILITY
The source code of the version of Digenome used in this manuscript is available at https://github.com/snugel/digenome-toolkit.
Supplementary Material
ACKNOWLEDGEMENTS
Author contributions: D.K., Y.-S.K. and J.-H.K. supervised the research. D.K., D.Y.K. and S.B.M. performed the experiments and bioinformatics analysis. D.K. and Y.-S.K. wrote the manuscript. All authors approved the manuscript.
Contributor Information
Do Yon Kim, Genome Editing Research Center, Korea Research Institute of Bioscience and Biotechnology (KRIBB), Daejeon 34141, Republic of Korea; KRIBB School of Bioscience, Korea University of Science and Technology (UST) , Daejeon 34141, Republic of Korea.
Su Bin Moon, Genome Editing Research Center, Korea Research Institute of Bioscience and Biotechnology (KRIBB), Daejeon 34141, Republic of Korea; KRIBB School of Bioscience, Korea University of Science and Technology (UST) , Daejeon 34141, Republic of Korea.
Jeong-Heon Ko, Genome Editing Research Center, Korea Research Institute of Bioscience and Biotechnology (KRIBB), Daejeon 34141, Republic of Korea; KRIBB School of Bioscience, Korea University of Science and Technology (UST) , Daejeon 34141, Republic of Korea.
Yong-Sam Kim, Genome Editing Research Center, Korea Research Institute of Bioscience and Biotechnology (KRIBB), Daejeon 34141, Republic of Korea; KRIBB School of Bioscience, Korea University of Science and Technology (UST) , Daejeon 34141, Republic of Korea; GenKOre, Daejeon 34141, Republic of Korea.
Daesik Kim, Genome Editing Research Center, Korea Research Institute of Bioscience and Biotechnology (KRIBB), Daejeon 34141, Republic of Korea.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
KRIBB Research Initiative Program [KG-20200023 to Y.-S.K.,and D.K.]; The Ministry of Science and ICT [NRF-2016M3A9B6903343 to Y.-S.K.]; National Research Council of Science & Technology [CAP-15-03-KRIBB to Y.-S.K., D.K.]; Institute for Basic Science [IBS-R021-D1 to D.K.]. Funding for open access charge: National Research Council of Science & Technology [CAP-15-03-KRIBB].
Conflict of interest statement. D.Y.K., S.B.M., J.-H.K., Y.-S.K. and D.K. have filed a patent application based on this work. Y.-S.K. is a founder of, and holds stock in, GenKOre, Inc.
REFERENCES
- 1. Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M.. RNA-guided human genome engineering via Cas9. Science. 2013; 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J.. RNA-programmed genome editing in human cells. eLife. 2013; 2:e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jiang W., Bikard D., Cox D., Zhang F., Marraffini L.A.. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013; 31:233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. et al.. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim H., Kim J.S.. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 2014; 15:321–334. [DOI] [PubMed] [Google Scholar]
- 6. Rouet P., Smih F., Jasin M.. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:6064–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chu V.T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K., Kuhn R.. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015; 33:543–548. [DOI] [PubMed] [Google Scholar]
- 8. Yao X., Wang X., Liu J., Hu X., Shi L., Shen X., Ying W., Sun X., Wang X., Huang P. et al.. CRISPR/Cas9—mediated precise targeted integration in vivo using a double cut donor with short homology arms. EBioMedicine. 2017; 20:19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yoshimi K., Kunihiro Y., Kaneko T., Nagahora H., Voigt B., Mashimo T.. ssODN-mediated knock-in with CRISPR-Cas for large genomic regions in zygotes. Nat. Commun. 2016; 7:10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang J.P., Li X.L., Li G.H., Chen W., Arakaki C., Botimer G.D., Baylink D., Zhang L., Wen W., Fu Y.W. et al.. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol. 2017; 18:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Paquet D., Kwart D., Chen A., Sproul A., Jacob S., Teo S., Olsen K.M., Gregg A., Noggle S., Tessier-Lavigne M.. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016; 533:125–129. [DOI] [PubMed] [Google Scholar]
- 12. Lin S., Staahl B.T., Alla R.K., Doudna J.A.. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife. 2014; 3:e04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zelensky A.N., Schimmel J., Kool H., Kanaar R., Tijsterman M.. Inactivation of Pol theta and C-NHEJ eliminates off-target integration of exogenous DNA. Nat. Commun. 2017; 8:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Merkle F.T., Neuhausser W.M., Santos D., Valen E., Gagnon J.A., Maas K., Sandoe J., Schier A.F., Eggan K.. Efficient CRISPR-Cas9-mediated generation of knockin human pluripotent stem cells lacking undesired mutations at the targeted locus. Cell Rep. 2015; 11:875–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R.. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016; 533:420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nishida K., Arazoe T., Yachie N., Banno S., Kakimoto M., Tabata M., Mochizuki M., Miyabe A., Araki M., Hara K.Y. et al.. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016; 353:aaf8729. [DOI] [PubMed] [Google Scholar]
- 17. Hess G.T., Fresard L., Han K., Lee C.H., Li A., Cimprich K.A., Montgomery S.B., Bassik M.C.. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat. Methods. 2016; 13:1036–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ma Y., Zhang J., Yin W., Zhang Z., Song Y., Chang X.. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat. Methods. 2016; 13:1029–1035. [DOI] [PubMed] [Google Scholar]
- 19. Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I., Liu D.R.. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 2017; 551:464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ryu S.-M., Koo T., Kim K., Lim K., Baek G., Kim S.-T., Kim H.S., Kim D.-e., Lee H., Chung E. et al.. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat. Biotechnol. 2018; 36:536–539. [DOI] [PubMed] [Google Scholar]
- 21. Rees H.A., Liu D.R.. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018; 19:770–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Koblan L.W., Doman J.L., Wilson C., Levy J.M., Tay T., Newby G.A., Maianti J.P., Raguram A., Liu D.R.. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 2018; 36:843–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Anzalone A.V., Randolph P.B., Davis J.R., Sousa A.A., Koblan L.W., Levy J.M., Chen P.J., Wilson C., Newby G.A., Raguram A. et al.. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019; 576:149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim D., Kim S., Kim S., Park J., Kim J.S.. Genome-wide target specificities of CRISPR-Cas9 nucleases revealed by multiplex Digenome-seq. Genome Res. 2016; 26:406–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim D., Bae S., Park J., Kim E., Kim S., Yu H.R., Hwang J., Kim J.I., Kim J.S.. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods. 2015; 12:237–243. [DOI] [PubMed] [Google Scholar]
- 26. Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F.. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016; 351:84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K.. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016; 529:490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen J.S., Dagdas Y.S., Kleinstiver B.P., Welch M.M., Sousa A.A., Harrington L.B., Sternberg S.H., Joung J.K., Yildiz A., Doudna J.A.. Enhanced proofreading governs CRISPR–Cas9 targeting accuracy. Nature. 2017; 550:407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Casini A., Olivieri M., Petris G., Montagna C., Reginato G., Maule G., Lorenzin F., Prandi D., Romanel A., Demichelis F. et al.. A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat. Biotechnol. 2018; 36:265–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee J.K., Jeong E., Lee J., Jung M., Shin E., Kim Y.-h., Lee K., Jung I., Kim D., Kim S. et al.. Directed evolution of CRISPR-Cas9 to increase its specificity. Nat. Commun. 2018; 9:3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim N., Kim H.K., Lee S., Seo J.H., Choi J.W., Park J., Min S., Yoon S., Cho S.R., Kim H.H.. Prediction of the sequence-specific cleavage activity of Cas9 variants. Nat. Biotechnol. 2020; doi:10.1038/s41587-020-0537-9. [DOI] [PubMed] [Google Scholar]
- 32. Schmid-Burgk J.L., Gao L., Li D., Gardner Z., Strecker J., Lash B., Zhang F.. Highly parallel profiling of Cas9 variant specificity. Mol. Cell. 2020; 78:794–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cho S.W., Kim S., Kim Y., Kweon J., Kim H.S., Bae S., Kim J.S.. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014; 24:132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mali P., Aach J., Stranges P.B., Esvelt K.M., Moosburner M., Kosuri S., Yang L., Church G.M.. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013; 31:833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ran F.A., Hsu P.D., Lin C.Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y. et al.. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013; 154:1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rees H.A., Yeh W.H., Liu D.R.. Development of hRad51-Cas9 nickase fusions that mediate HDR without double-stranded breaks. Nat. Commun. 2019; 10:2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gabriel R., Lombardo A., Arens A., Miller J.C., Genovese P., Kaeppel C., Nowrouzi A., Bartholomae C.C., Wang J., Friedman G. et al.. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat. Biotechnol. 2011; 29:816–823. [DOI] [PubMed] [Google Scholar]
- 38. Tsai S.Q., Zheng Z., Nguyen N.T., Liebers M., Topkar V.V., Thapar V., Wyvekens N., Khayter C., Iafrate A.J., Le L.P. et al.. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015; 33:187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Frock R.L., Hu J., Meyers R.M., Ho Y.J., Kii E., Alt F.W.. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol. 2015; 33:179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yan W.X., Mirzazadeh R., Garnerone S., Scott D., Schneider M.W., Kallas T., Custodio J., Wernersson E., Li Y., Gao L. et al.. BLISS is a versatile and quantitative method for genome-wide profiling of DNA double-strand breaks. Nat. Commun. 2017; 8:15058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tsai S.Q., Nguyen N.T., Malagon-Lopez J., Topkar V.V., Aryee M.J., Joung J.K.. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat. Methods. 2017; 14:607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cameron P., Fuller C.K., Donohoue P.D., Jones B.N., Thompson M.S., Carter M.M., Gradia S., Vidal B., Garner E., Slorach E.M. et al.. Mapping the genomic landscape of CRISPR-Cas9 cleavage. Nat. Methods. 2017; 14:600–606. [DOI] [PubMed] [Google Scholar]
- 43. Wienert B., Wyman S.K., Richardson C.D., Yeh C.D., Akcakaya P., Porritt M.J., Morlock M., Vu J.T., Kazane K.R., Watry H.L. et al.. Unbiased detection of CRISPR off-targets in vivo using DISCOVER-seq. Science. 2019; 364:286–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim D., Lim K., Kim S.T., Yoon S.H., Kim K., Ryu S.M., Kim J.S.. Genome-wide target specificities of CRISPR RNA-guided programmable deaminases. Nat. Biotechnol. 2017; 35:475–480. [DOI] [PubMed] [Google Scholar]
- 45. Kim D., Kim D.E., Lee G., Cho S.I., Kim J.S.. Genome-wide target specificity of CRISPR RNA-guided adenine base editors. Nat. Biotechnol. 2019; 37:430–435. [DOI] [PubMed] [Google Scholar]
- 46. Zuo E., Sun Y., Wei W., Yuan T., Ying W., Sun H., Yuan L., Steinmetz L.M., Li Y., Yang H.. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science. 2019; 364:289–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Grunewald J., Zhou R., Garcia S.P., Iyer S., Lareau C.A., Aryee M.J., Joung J.K.. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature. 2019; 569:433–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data was deposited in the NCBI Sequence Read Archive (SRA) database with BioProject accession code PRJNA659391.