Abstract
The recently characterized mammalian writer (methyltransferase) and eraser (demethylase) of the DNA N6-methyladenine (N6mA) methyl mark act on single-stranded (ss) and transiently-unpaired DNA. As YTH domain-containing proteins bind N6mA-containing RNA in mammalian cells, we investigated whether mammalian YTH domains are also methyl mark readers of N6mA DNA. Here, we show that the YTH domain of YTHDC1 (known to localize in the nucleus) binds ssDNA containing N6mA, with a 10 nM dissociation constant. This binding is stronger by a factor of 5 than in an RNA context, tested under the same conditions. However, the YTH domains of YTHDF2 and YTHDF1 (predominantly cytoplasmic) exhibited the opposite effect with ∼1.5–2× stronger binding to ssRNA containing N6mA than to the corresponding DNA. We determined two structures of the YTH domain of YTHDC1 in complex with N6mA-containing ssDNA, which illustrated that YTHDC1 binds the methylated adenine in a single-stranded region flanked by duplexed DNA. We discuss the hypothesis that the writer-reader-eraser of N6mA-containining ssDNA is associated with maintaining genome stability. Structural comparison of YTH and SRA domains (the latter a DNA 5-methylcytosine reader) revealed them to be diverse members of a larger family of DNA/RNA modification readers, apparently having originated from bacterial modification-dependent restriction enzymes.
INTRODUCTION
DNA methylation in bacteria and archaea is common, occurring at ring carbon C5 of the cytosine (yielding 5-methylcytosine, 5mC), or the exocyclic amino groups of either cytosine at N4 (yielding N4-methylcytosine, N4mC) or adenine at N6 (yielding N6-methyladenine, N6mA) (1,2). The great majority (if not all) of bacterial and archaeal DNA methyltransferases (MTases) are associated with a set of conserved sequence motifs important for three essential functions: binding methyl donor S-adenosyl-l-methionine (SAM), binding the DNA substrate, and catalyzing the methyl transfer between the donor and substrate (3–5). It was these predictive motifs that aided in the discovery of mammalian DNA cytosine-C5-specific Dnmt1 (6) and Dnmt3 (7).
The existence of 5mC was first reported in DNA from ox spleen ∼70 years ago (8), whereas detection of N6mA in mammalian DNA was reported only recently, in parallel with development and application of ultrasensitive mass spectrometry. There are three debatable issues regarding N6mA in mammalian DNA: (i) whether low-level N6mA can be accurately detected, (ii) how N6mA is generated and (iii) what the potential functions of N6mA might be.
First, low levels of N6mA have been reported in DNA from mouse (9,10), human (11,12), and human malignant brain tumor glioblastoma (13). Other studies have failed to detect N6mA in several mouse tissues and mitochondrial DNA from human placenta (14), DNA isolated from mouse embryonic stem cells or brain and liver tissue (15) or cultured human cells (16). Nevertheless, more recent reports present evidence for low levels of N6mA in DNA from cultured human and mouse cells; however, that modified adenosine is incorporated (at least in part) by DNA polymerase using deoxy-N6mA converted from RNA-derived ribo-N6mA, via the nucleotide salvage pathway (17,18).
Second, there is also uncertainty regarding the mammalian enzyme(s) (suggested to include HemK2/N6AMA1, MettL3-MettL14 complex and MettL4) responsible for generating N6mA in mammalian DNA (10,13,19–21). Human HemK2, which was thought to be a DNA N6mA MTase (11), is actually a protein MTase active on glutamine and lysine (19,20,22,23). Guided by the sequential order of conserved sequence motifs unique to Class β amino (N6mA and N4mC) MTases (4,24,25), we identified human MettL3–MettL14 as a DNA adenine MTase active on single-strand and unpaired DNA in vitro (26). Previous to that, MettL3–MettL14 had been characterized extensively as generating N6mA in RNA ((27,28) and references therein). Murine MettL4 is responsible for N6mA deposition in genic elements associated with transcriptional silencing (10). Intriguingly, recombinant human MettL4 expressed in human embryonic kidney (HEK293T) cells has in vitro enzymatic activity on mitochondrial DNA (21), whereas recombinant human MettL4 purified from Escherichia coli has RNA MTase activity (29).
Third, having N6mA in DNA might be a double-edged sword. One potential advantage of having N6mA in DNA might help to reduce the mutagenic potential of cellular 8-oxo-2′-deoxyguanosine (8-oxo-dGTP) (30). Opposite to the template adenosine, 8-oxo-dGTP can be misincorporated into DNA using Hoogsteen pairing that involves the exocyclic amino group of adenosine. Methylation of that adenosine amino group makes N6mA:8-oxo-dG significantly less stable than the A:8-oxo-dG Hoogsteen pair in the DNA duplex region, resulting in less efficient misincorporation of 8-oxo-dGTP opposite N6mA by human DNA polymerase β (30). This observation might be related to the association between MettL3–MetL14 recruitment to DNA damaged sites, and its methyltransfer activity being required for the repair (31). On the other hand, one potential disadvantage of using N6mA (or N4mC) in DNA, compared to 5mC, is that the methylated amino group of N6mA (or N4mC) is involved in normal hydrogen-bonding for Watson-Crick base pairing, while the cytosine C5 position is not. Indeed, incorporation of N6mA into a DNA template causes RNA polymerase Pol II pausing, associated with the lower stability and slower kinetics of base pairing, and the stability of N6mA:U base pair is weakened (32). Similar site-specific pausing at N6mA, by a modified phage DNA polymerase inserting T across from a template N6mA, underlies the ability of PacBio SMRT sequencers to detect methylated adenines (33), and there is additional evidence for weaker N6mA:T than A:T base pairs (34–36).
Human and mouse homologs (Alkbh1 and Alkbh4) of the E. coli repair enzyme AlkB can function as sequence-independent DNA N6mA demethylases (9,10,13), and at least the mouse Alkbh1 prefers locally-unpaired DNA substrates (37). It is intriguing that both the writer (MTase) and eraser (demethylase) of the N6mA methyl mark act on single-stranded and transiently unpaired DNA. We wondered whether there are N6mA DNA-binding domains in mammals. For this reason, we investigated the YTH domain of YTHDC1, as YTH domain-containing proteins bind N6mA-containing RNA in mammalian cells (reviewed in (38–40) and references therein). There are five YTH domain-containing proteins in humans (DC1, DC2, DF1, DF2 and DF3), and among them YTHDC1 is known to localize in the nucleus (41), where it plays important roles in gene regulation via exon selection during pre-mRNA splicing (42). In contrast, YTHDF1 is predominantly cytoplasmic and YTHDF2 migrates to mitotic chromatin in human induced pluripotent stem cells (43). Aside from their conserved YTH domains, DC1 and DC2 are unrelated to each other and unrelated to DF1–3, based on amino acid sequence, size, and overall domain organization. The three paralogs DF1–3, on the other hand, share high amino acid identity over their entire length (38). Notably, in addition to the conserved YTH domain located in the C-terminal region of DF1–3 and DC2 and an internal region of DC1, DF1–3 and DC1 contain low-complexity sequences (P/Q/N-rich) throughout and lack recognizable modular protein domains.
Here we show that the YTH domain of YTHDC1 binds to single-stranded (ss) DNA containing N6mA, with a 10 nM dissociation constant. Under the same conditions, this YTH binding affinity for N6mA in a DNA context is stronger by a factor of 5 than such binding in an RNA context. However, the YTH domains of YTHDF1 and YTHDF2 exhibited the opposite effect with ∼1.5–2× stronger binding to N6mA in ssRNA than in the corresponding DNA. We determined two structures of the YTH domain of YTHDC1 binding the methylated adenosine in ssDNA, and compared these structures to that of a previously characterized RNA-bound YTH domain of YTHDC1. Finally, we discuss the relationship between mammalian DNA base modification reader domains and bacterial modification-dependent restriction enzymes.
MATERIALS AND METHODS
Protein expression and purification
The protein fragments used are YTHDC1 residues 345–509 of UniProtKB/Swiss-Prot Q96MU7, YTHDF1 residues 361–559 of Q9BYJ9 and for YTHDF2 residues 391–579 of Q9Y5A9. YTHDC1 (pXC2129) and YTHDF1 (pXC2130) constructs were purchased from Addgene (plasmid #64652 and #64653) containing N-terminal 6xHis-tag. The YTH domain of human YTHDF2 as a GST-tagged fusion in a pGEX-4T1 vector was synthesized by Biomatik. Each plasmid was transformed into BL21-CodonPlus cells for expression.
Each protein was purified from 6 L of LB inoculated with overnight starter culture and grown to A600 of ∼1 at 37°C; after which the samples were cooled to 16°C and induced by 1 mM isopropyl thiogalactopyranoside for 12 h. Cells were harvested at 4°C and then suspended into buffer A (20 mM Tris pH 7.5, 400 mM NaCl, 5% glycerol) to a final volume of 30 ml before undergoing sonication at 75% amplitude (3 s on and 9 s off for 2 min). The sonicated samples were centrifuged at 25 000 rpm at 4°C, for 2 h.
For His-tagged YTH domains of YTHDC1 and YTHDF1, the supernatant was passed through a 3.4 μm filter and diluted to a volume of 100 ml to a final concentration of 15 mM imidazole using buffer B (300 mM imidazole, 20 mM Tris pH 7.5, 400 mM NaCl, 5% glycerol). The filtered supernatant was loaded onto a 5 ml GE HisTrap column by a Bio-Bad NGC™ chromatography system using a flowrate of 2 ml/min and then washed for 15× column volumes with 15 mM imidazole. Each protein eluted over a broad peak starting at 60 mM imidazole. The estimated protein yields were ∼65 mg per 6 L culture. The pooled fractions (∼28.5 ml at 2.3 mg/ml) to which 2 mg of TEV protease was added to cleave 6xHis tag for 15 h at 4°C while undergoing two rounds of dialysis in 3.5 kDa Snakeskin dialysis tubing (Thermo Scientific). The cleaved products (with an additional N-terminal glycine) were passed through a subtractive HisTrap column. YTHDC1 interacted with the column and was eluted with a low concentration of imidazole, whereas YTHDF1 was collected in the flow through. The proteins were concentrated to 2 ml and loaded onto a Superdex S75 10/300 GL equilibrated with buffer C (20 mM Tris pH 7.5, 150 mM NaCl) with 1 mM tris(2-carboxyethyl)phosphine (TCEP). The samples eluted as a symmetrical peak at an elution volume consistent with a monomer.
For GST-tagged YTH domain of YTHDF2, the clarified supernatant after cell lysate was passed through a gravity fed GST column with a volume of 10 ml at room temperature, washed with 170 ml of buffer A and the GST tag was cleaved on column with thrombin protease at room temperature for 2 h before elution. The cleaved product has no additional residue at the N-terminus but have an additional five residues at the C-terminus (RPHRD). The samples were concentrated and loaded onto a S75 size exclusion column as described above. Supplementary Figure S1D shows examples of purified YTH domains used in the study.
Binding assays of protein-nucleic acids interaction
DNA and RNA oligonucleotides containing N6mA or FAM tagged oligos were synthesized by B. Baker of New England Biolabs, Inc., and unmodified oligos were purchased from Integrated DNA Technologies. Nucleic acid samples were suspended in buffer C (without TCEP) to final stock concentrations of 4 mM (ss oligo) and then some annealed using a thermocycler to 2 mM (ds oligo). We used three binding assays to characterize the binding affinity of YTHDC1 to methylated oligodeoxynucleotides: (i) isothermal titration calorimetry (ITC), (ii) MicroScale Thermophoresis (MST) and (iii) electrophoretic mobility shift.
For ITC, protein samples (100 μM in buffer C) were injected from the syringe into sample cells containing 10 μM of the 11-mer oligonucleotides in the same buffer (DNA: 5′-CGCGGACTCTG-3′ or RNA: 5′-CGCGGACUCUG-3′ where A = N6mA), using the automated PEAQ-ITC (Malvern Instrument Ltd). The experiments were conducted at 25°C with 19 injections at 2 μl each, or 13 injections (2.9 μl) if more heat was required, with a reference setting of 10 μcal/s. Binding constants were calculated by fitting the data using the ITC data analysis module ‘one set of sites’ supplied by the manufacturer.
MST experiments were conducted using a Monolith NT.115 microscale thermophoresis instrument from NanoTemper Technologies. A 2-fold serial dilution of protein was produced starting at 20 μM in buffer C with 0.05% (v/v) added Tween 20 (Sigma-Aldrich). Varied protein concentrations (total of 16 samples) in 0.2 ml PCR tubes were mixed with an equal volume of the 6-carboxy-fluorescein (FAM)-labeled 11-mer oligonucleotide at 20 nM in buffer C and matching 0.05% Tween 20. Approximately 10 μl of the mixed samples, at half their respective starting concentrations, were loaded into Monolith specific capillary tubes. The signals had below 5% error among samples with regards to the fluorescence and a fluorescence intensity of 500 counts using 40% power on a medium MST setting. The MST data was fitted using Mo.Affinity Analysis software (NanoTemper) included with the instrument.
Electrophoretic mobility shift assays (EMSA) were performed with the same set of samples used in the MST assays. An aliquot of 5 μl of complex was loaded into 6% native PAGE gels, and ran at 100 V for 30 min at 4°C in 0.5× TBE buffer. The images were collected using a BioRad ChemiDoc imager in fluorescence mode.
Crystallography
The Art Robbins Gryphon Crystallization Robot was used to set up 0.4-μl sitting drops (0.2 μl of complex plus 0.2 μl of well solution) at ∼19°C of the protein–DNA complexes. A crystallization hit was observed using the 11 nt oligo 5′-CGCGGACTCTG-3′ (A = N6mA) in a complex with YTH domain of YTHDC1 (10 mg/ml in 1:2 molar ratio of protein to DNA) in 0.1 M Bis–Tris pH 6.0, 0.2 M ammonium sulfate and 29% (w/v) polyethylene glycol (PEG) 3350. The crystals appeared after ∼15 days incubation at 19°C. The crystal shape was a consistent flat parallelogram, and suffered from minor crystal layering (Supplementary Figure S1A).
At molar ratios of 2:1 YTH to DNA, protein only crystals appeared within 24 h with 0.1 M ammonium sulfate, 0.1–0.15 M Bis–Tris pH 5.5 and 23–29% PEG 3350. The protein only crystals were very birefringent (Supplementary Figure S1B). Because the YTH domain contains a hydrophobic cage consisting of two tryptophan residues interacting with methylated adenine (see Results), the crystals of the protein-DNA complex had no UV fluorescence from tryptophan (perhaps due to quenching) and little to no birefringence, while the protein only crystals had strong Trp UV fluorescence.
The second crystallization attempt used a set of three oligos from 9–10 nt with varied 3′ sequence. A 10 nt 5′-CGCGGACTTC-3′ (A = N6mA) incubated with YTH crystallized within 12 h at 19°C as heavily layered stacks with time became circular discs. This complex (in 1:2 ratio of protein to DNA) was readily crystallizable under hundreds of conditions. The problem of heterogeneous nucleation was resolved by generating a screen using the optimal crystallization condition (0.1 M Bis–Tris pH 7.0, 0.24 M ammonium sulfate, 23% PEG 3350) with increasing concentration of glycerol as an additive. Individual crystals emerged starting at ∼10% glycerol and with an eventual single crystal per crystallization drop at 20% (Supplementary Figure S1C).
Single crystals were flash frozen in liquid nitrogen by equilibrating in a cryoprotectant buffer containing the crystallization solution and 25% (v/v) ethylene glycol. X-ray diffraction data were collected at the SER-CAT beamline 22ID of the Advanced Photon Source at Argonne National Laboratory. Crystallographic datasets were processed with HKL2000 (44). Molecular replacement was performed with the PHENIX PHASER module (45) by using the known structure of the human YTHDC1 YTH domain (PDB ID 4R3H) as a search model. Structure refinement was performed with PHENIX Refine (46) with 5% randomly chosen reflections for the validation by the Rfree value. COOT (47) was used for the manual building of the structure model and corrections between refinement rounds. DNA models were built into difference density during the first several rounds of refinement for the two complex structures. Structure quality was analyzed during PHENIX refinements and finally validated by the PDB validation server. Molecular graphics were generated by using PyMol (Schrödinger, LLC).
RESULTS
YTH binding to single-strand DNA containing N6mA
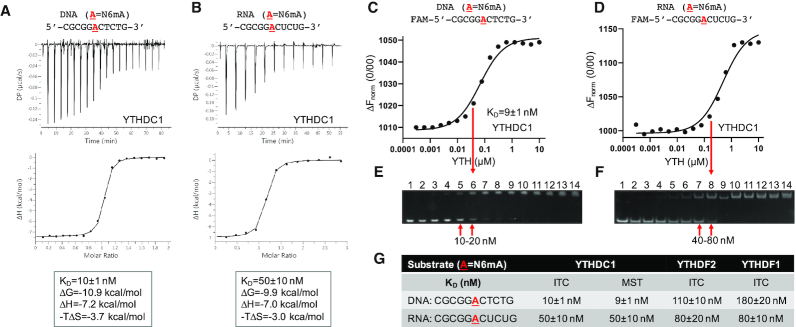
We used three independent methods, all of which revealed that the binding of human YTHDC1′s YTH domain, to a 11-nt single-strand (ss) oligodeoxynucleotide (DNA oligo) containing a single N6mA in the context of GGACT, had higher affinity than its binding to the corresponding ssRNA of the same length and sequence (containing GGACU). The DNA GGACT and RNA GGACU were chosen as they are the equivalent DNA or RNA recognition sequence of MettL3-MettL14 (26,48). We first used ITC to quantitatively measure the dissociation constants (KD). The YTH domain bound the methylated DNA oligo with a KD of 10 nM (Figure 1A). Under the same conditions, the modified RNA oligo exhibited 5-fold reduced binding affinity (Figure 1B). Because this observation was somewhat unexpected, we repeated the binding experiments with another biophysical technique, MST, to measure the strength of interaction between the YTH and oligos. Very similar to the ITC measurements, we observed a 9 nM binding affinity for the modified DNA oligo, with 5.5× reduced affinity for the corresponding RNA oligo (Figure 1C, D). The same samples used for the MST assays were then used for an electrophoretic mobility shift assay (EMSA), and again confirmed the enhanced affinity for the methylated DNA oligo relative to the RNA control (Figure 1E, F). Under the same conditions, we observed no measurable binding to unmodified ssDNA or ssRNA oligos, or to double-stranded (ds)DNA or RNA/DNA hybrid oligos containing N6mA on one strand (Supplementary Figure S2A).
Figure 1.
YTH binding methylated adenine in DNA. (A, B) ITC measurements of YTHDC1 binding methylated ssDNA (panel A) or methylated ssRNA (panel B). (C, D) MST measurements and (E, F) electrophoretic mobility shift assays of YTHDC1 binding to oligos containing a single N6mA in DNA or RNA. (G) Summary of KD values of three YTH domains (see Supplementary Figure S2). Data represent the mean ± SD of N independent determinations (N = 2 for ITC and N = 3 for MST).
For comparison, we also purified the corresponding YTH domains from YTHDF1 and YTHDF2 and measured their binding affinities to the same sets of DNA or RNA oligos by ITC (Figure 1G and Supplementary Figure S2B-D). These three proteins have distinct but overlapping binding specificities on RNA (49). The binding affinities for the methylated ssRNA by the three YTH domains were similar, in the range of 50–80 nM (Figure 1G). However, the three YTH domains bound to the DNA molecule very differently. While YTHDC1 bound DNA 5× more strongly than to RNA, YTHDF1 and YTHDF2 exhibited the opposite effect by binding DNA 1.4–2.2× weaker than to RNA. Significantly, YTH domain of YTHDC1 bound N6mA-containing DNA more strongly by a factor of 11 or 18, respectively, than did the corresponding YTH domains of YTHDF1 and YTHDF2.
Overall structure of YTHDC1 YTH domain bound to methylated DNA
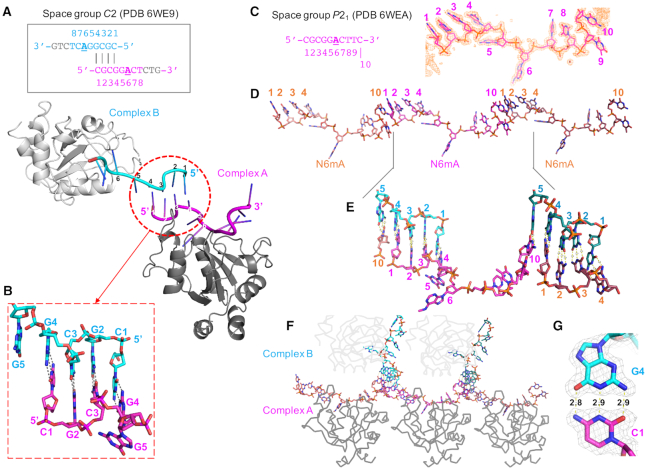
We next sought to understand how the YTH domain binds methylated DNA, and why it recognizes N6mA preferentially in the context of DNA over RNA. Accordingly, we co-crystallized the YTH domain of YTHDC1, with the same 11-nt ssDNA oligo (5′-CGCGGACTCTG-3′) containing a centrally located N6mA that we had used for the binding studies. The complex crystallized in space group C2, resulting in a structure determined to resolution of 1.59 Å (Supplementary Table S1). In the crystallographic asymmetric unit, there are two complexes (A and B) with each YTH domain binding a piece of ssDNA (Figure 2A). Interestingly, with the 11-nt utilized for crystallization, the 5′ end CGCG sequence base pairs with the corresponding neighboring DNA molecule, forming a 4-base pair duplex (Figure 2B). However, three nucleotides at the 3′ end have poor electron density for the bases in complex A or are completely disordered (no visible density) in complex B due to the lack of direct crystallographic packing interactions.
Figure 2.
Overall structures of YTH-DNA complexes in two space groups. (A) Two complexes of YTH-11mer DNA in space group C2 (PDB 6WE9). (B) The 5′ CGCG of two neighboring DNA molecules form 4-bp duplex. (C) Ten nucleotides of DNA used in space group P21 (PDB 6WEA), superimposed with an omit Fo-Fc electron density map (orange) contoured at 4σ above the mean by omitting the entire 10-nt DNA. (D) A long pseudo ssDNA formed by stacking the 3′ cytosine C10 to the 5′ end of neighboring molecule. (E) The DNA (cyan) bound with molecule B provides the five pairing nucleotides for the joint sequence of DNA (magenta) bound with molecule A. (F) Crystal lattice in space group P21 showing YTH (in gray) binds the methylated adenine in the single-strand region flanked with 5-base pair duplexes. (G) Example of G4:C1 base pair superimposed with an omit Fo – Fc electron density map (light gray) contoured at 4σ above the mean. The numerical numbers are interatomic distances in Å.
To improve the quality of electron density for the 3′ end, we designed a series of ssDNA oligos varying in length and/or sequence, and screened crystallizations for different space groups with different morphology (Supplementary Figure S1C). A complex formed with a 10-nt ssDNA oligo (5′-CGCGGACTTC-3′) crystallized in space group P21, where we observed ordered electron density for all 10 nucleotides (Figure 2C). There are two copies of each complex (A and B), resulting in a total of four complexes within the P21 asymmetric unit. The ssDNA bound by molecule A stacks head-to-tail with neighboring molecules on both ends, forming a pseudo-continuous ssDNA molecule with a joint sequence of (5′-C10-C1G2C3G4-3′) (Figure 2D). The DNA bound by molecule B provides the five pairing nucleotides (5′-CGCGG-3′) for the joint sequence (Figure 2E), thus generating a continuous lattice in the crystal with a 5-base pair duplex flanking the single-strand region where YTH binds the methylated adenine (Figure 2F). Whereas the phosphate group at the joint of the two DNA molecules, connecting C10 and C1, is of course missing, the five G:C base pairs form three classic Watson–Crick hydrogen bonds, respectively, that are 2.8–2.9 Å apart (Figure 2G).
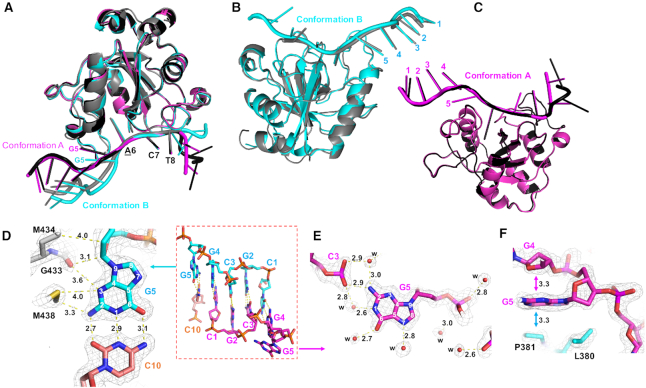
Two structural conformations of YTH-bound DNA
The protein components of the four complexes (A and B in both space groups) are highly similar, with root-mean-square deviations of 0.1–0.5 Å across 135 pairs of Cα atoms (Figure 3A). The largest deviation involves complex B in space group C2, where the 3′ nucleotides are disordered and not modeled due to lack of electron density (colored grey in Figure 3B). Among the four bound DNA molecules, only three nucleotides adopt the same conformations: N6mA at position 6, and the two 3′ nucleotides at positions 7 and 8 (C7 and T8) (Figure 3A). The remaining nucleotides exhibit varied conformations, depending on whether they are involved in base pairing. The two complexes can be distinguished by the guanosine at position 5 (G5) and whether it is involved in base pairing (conformation B) or not (conformation A) (Figure 3B, C).
Figure 3.
Two YTH-bound DNA conformations. (A) Superimposition of four complexes, A and B (black and dark gray) in space group C2 (PDB: 6WE9) and A and B (magenta and cyan) in space group P21 (PDB: 6WEA). (B) Superimposition of two B complexes in space groups C2 (gray) and P21 (cyan). (C) Superimposition of two A complexes in space groups C2 (black) and P21 (magenta). (D) Guanine G5 of complex B involves in base pairing. (E, F) Guanine G5 of complex A involves in intra-molecular interaction and protein–DNA interaction. The omit Fo – Fc electron density map (light grey) contoured at 4σ above the mean. Panels D–F are depicted from the structure (PDB: 6WEA).
In conformation B, guanosine G5 pairs with cytidine C10 of the neighboring DNA molecule (Figure 3D). Three protein residues (Met438, Gly433 and Met434) make van der Waals contacts with N2 and N3 atoms of the guanosine G5 base and its ribose ring (Figure 3D). In conformation A, the guanosine G5 base makes intra-molecular interactions with the phosphate group bridging between nucleotides G2 and C3, the N2 atom of the guanosine G5 makes a direct hydrogen bond with one of the phosphate oxygen atoms, and there are two water-mediated interactions with the same phosphate group (Figure 3E). In addition, the guanosine ring of G5 stacks with two 5-membered rings–the ribose of the preceding guanosine G4 and the proline ring of Pro381 (Figure 3F). While the interactions with the guanosine G5 seen in the two conformations do not provide base specific recognition, whether or not G5 is stacked with the 5′ bases, these interactions with YTH residues presumably enhance the binding affinity.
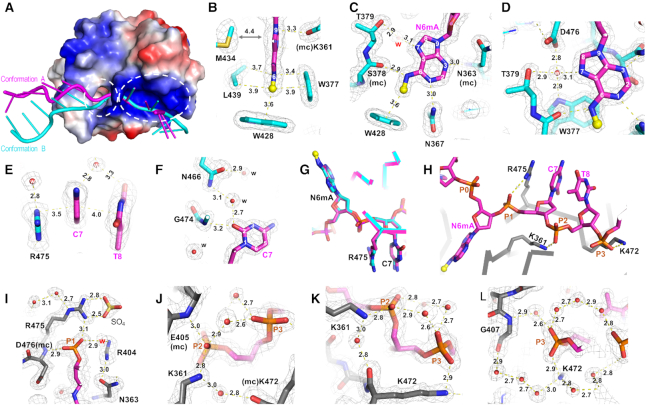
Interactions with N6mA-containing DNA
The methylated adenosine N6mA is inserted into a hydrophobic pocket (Figure 4A), where the base ring is stacked between Trp377 on one side and Leu439 and Met434 on the other side (Figure 4B). At the bottom of the pocket lies the indole ring of Trp428, juxtaposed to the methyl group of N6mA (Figure 4B). The adenosine ring is involved in four hydrogen bonds with, respectively, the side-chain of Asn367 (via Ade N1), the main-chain amide nitrogen atoms of Asn363 (via Ade N3), the main-chain carbonyl oxygen atom of Ser378 (via Ade N6 amino group), and a water molecule (via Ade N7) (Figure 4C). The water molecule is trapped at the protein-DNA interface, tetrahedrally coordinated with the Ade N7, the indole nitrogen of Trp377, the hydroxyl oxygen of Thr379, and one of the carboxylate oxygen atoms of Asp476 (Figure 4D). This pattern of interactions fully saturates the hydrogen-bonding capacity of polar atoms on the adenosine ring and, together with van der Waals contacts with the methyl group, defines the specificity for the methylated adenosine in the binding pocket.
Figure 4.
YTH interaction with N6mA (PDB 6WEA). (A) YTH domain contains a hydrophobic pocket (circled in dashed line) next to a basic surface. The surface charge at neutral pH is displayed as blue for positive, red for negative, and white for neutral. (B) An aromatic cage for binding N6mA. (C) Interactions with N6mA in the pocket. (D) A trapped water molecule has tetrahedral coordination. (E) Arg475 stacks with cytosine C7. (F) Interaction with cytosine C7. (G) Arg475 occupies the position where the N6mA would be normally located if it stacked with the neighboring nucleotide. (H) Three phosphate groups (P1, P2 and P3) 3′ to N6mA interaction with protein. (I) Arg475 interaction with the P1 phosphate group and a SO4 molecule used in crystallization. (J) Lys361 interaction with the P2 phosphate group. (K) Lys472 interaction with the P3 phosphate group. (L) An example of a water network surrounds phosphate groups. The 2Fo – Fc electron density map (light gray) is contoured at 2σ above the mean.
The next nucleotide, cytidine C7, is stacked in-between the side-chain of Arg475 and the following thymidine T8 of the same strand, both of which are solvent exposed (Figure 4E). In addition, cytidine C7 makes a direct but weak hydrogen bond (3.2 Å) with the main-chain amide nitrogen atom of Gly474, and a water-mediated interaction with the side chain of Asn466 (Figure 4F). Like guanosine G5, the interactions with cytidine C7 are not base-specific recognition. Interestingly, the guanidine group of Arg475 occupies the position where the N6mA would normally be located if it stacks with neighboring nucleotides (Figure 4G).
We note that numerous genome-wide studies did not produce an agreeable consensus DNA sequence containing N6mA from mammalian cells: sequences reported include TTTTTAGAAGC or TACA[A/G]GA in mouse ES cells (9,10), [G/C]AGG[C/T] in a human genome (especially in the mitochondria) (11), TGGATGGA in human glioblastoma (13), and CT[T/C/A]ATC in human mitochondria (21). From the face value of these studies, it seems that the sequences immediately before and after N6mA are both variable. In the current structures with bound ssDNA, the nucleotides (G5 and C7) before and after N6mA are not engaged in base specific interactions with YTH. We modeled the other three nucleotides (G/A/T) at position 7 and calculated binding interfaces between YTH and modeled nucleotides (Supplementary Figure S3). The differences in the binding interface are negligible.
YTH–DNA phosphate interactions
There are direct protein–DNA phosphate contacts concentrated on the three phosphates 3′ to the N6mA (labeled as P1, P2 and P3 in Figure 4H). Three positively-charged residues (Arg475, Lys361 and Lys472) lie on the highly-basic surface (Figure 4A), and interact directly with the three respective negatively-charged phosphate groups (Figure 4I-K). In addition, the main chain amide nitrogen atoms of Asp476 and Glu405 form hydrogen bonds with, respectively, one of the non-bridging oxygen atoms of the P1 and P2 phosphate groups (Figure 4I, J). In addition to these direct protein–DNA interactions, a complex network of ordered water molecules interconnects bases, phosphate groups, and amino acids. In the case of P3, protein atoms might function as one of the connecting groups in a water network surrounding the charged phosphate group (Figure 4L).
Evolutionarily-conserved interaction with N6mA
Interestingly, the residues involved in the contacts to N6mA are more highly conserved than the other DNA contacts (Supplementary Table S2). While there were no obvious orthologs for YTHDC1, YTHDF1 or YTHDF2 in Agnatha (jawless fish), the other six vertebrate classes had clear orthologs for all three proteins, with some positions conserved across both classes and YTH proteins. Of the DNA contacts in YTHDC1 YTH domain, 4/8 (50%) that contact N6mA involve positions highly conserved across the three proteins in six classes (Ser362, Asn363, Asn367, Trp377, Ser378, Thr379, Trp428 and Asp476; invariant residues in bold), while this is true for just 3/12 (25%) that make other DNA contacts (Lys361, Leu380, Pro381, Glu405, Gly433, Met434, Met438, Leu439, Asn466, Lys472, Gly474 and Arg475). The invariant residues concentrated in construction of the binding site provide direct binding to the methyl group of N6mA (Trp377 and Trp428; Figure 4B), the trapped water molecule (Asp476; Figure 4D) and the cytidine immediately 3′ to N6mA (Lys361, Asn466 and Arg475; Figure 4E–H). This suggests that recognition of the methylated base and binding of the 3′ cytosine are particularly well-conserved among YTH proteins, and are consistent with the extensive structural recognition and binding of the two nucleotides (N6mA6-C7) described earlier in this section.
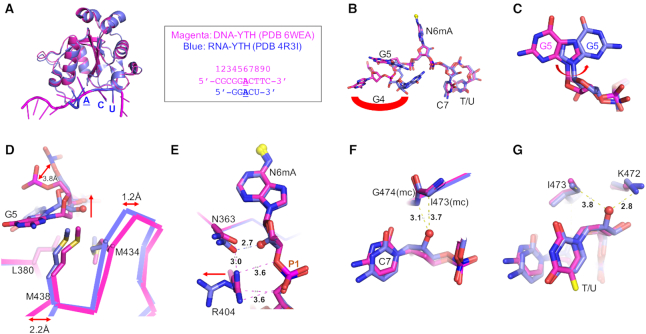
Comparison between DNA and RNA bound YTH domains
The YTH domain of YTHDC1 had been structurally characterized in complex with a 5-nt RNA (5′-GGACU-3′ where the underlined A is methylated at the N6 atom) (PDB 4R3I (50)). [YTHDC1 also binds RNA containing A methylated at the N1 atom (51) but, as this does not occur normally in DNA, we have not studied it here.] Superimposition of RNA- and DNA-bound YTH domains resulted in highly similar structures, having a RMSD of just 0.25 Å across 130 pairs of Cα toms (Figure 5A). For the RNA component, the 3′ ACU is superimposable with the corresponding DNA ACT, whereas the 5′ guanosine (G4) points in completely different directions in the two structures (Figure 5B). The internal (Ade-adjacent) guanosine, corresponding to G5 of the DNA used in our study, has a similar backbone conformation in both structures, but the guanosine ring is rotated ∼180°C along the glycosidic bond between the base and sugar group (Figure 5C). The conformational changes in RNA between these two 5′ guanosine nucleotides (G4 and G5) resulted in the loss of protein interactions described above for the DNA conformations (Figure 3), presumably partly responsible for reduced affinity of the RNA binding.
Figure 5.
Comparison between DNA- and RNA-bound YTH. (A) Superimposition of complex A in space group P21 and RNA-YTH complex (PDB: 4R3I). The DNA ACT (magenta) and RNA ACU (blue) are superimposable. (B) Difference in guanine G4 binding. (C) Difference in guanine G5 binding. (D) The effect of 2′-OH group of G5 pushes RNA molecule away. (E) The effect of 2-OH group of N6mA interrupts an Arg404-Asn363 interaction. (F, G) The 2-OH groups of cytosine C7 and uracil U8 have no visible impact on YTH binding.
We next examined effects of the presence (ribose) or absence (deoxyribose) of the 2′-hydroxyl group (OH) in the backbone sugar of the two equivalent sequences. The 2′-OH of guanosine G4 in RNA points toward the solvent and does not engage in any interactions. The addition of 2′-OH to G5 in RNA moves the ribose ring more than 1 Å away from the protein relative to its position in DNA (Figure 5D). In DNA, three hydrophobic residues (Leu380, Met434 and Met438) encompass the deoxyribose C2′ atom. A loop in the RNA complex, that contains Met434 and Met438, undergoes movement of between 1.2 Å (residue prior to Met434) and 2.2 Å (residue after Met438) relative to that of the DNA-bound conformation (Figure 5D). This loop movement is significant considering the overall RMSD of 0.25 Å between the two YDH complex structures. Furthermore, in the 1.18 Å-resolution structure of the apo-YTH domain (Supplementary Table S1), the corresponding loop in the absence of DNA is less ordered and contains discontinuous electron density in the main-chain between Gly433 and Lys437, as well as in the side chains of Met434 and Met438, suggesting that the loop is stabilized by the bound DNA substrate (Supplementary Figure S4A).
The inclusion of a 2′-OH on the methylated deoxy-adenosine (N6mA) disrupts the Arg404–Asn363 interaction, which runs in parallel and makes van der Waals contacts with the sugar-phosphate backbone in the DNA-bound conformation (Figure 5E). In the absence of DNA, the Arg404 interacts with a negatively-charged sulfate group (used for crystallization), which occupies an equivalent position of the P1 phosphate group (Supplementary Figure S4A). The comparison among the three structures—YTH alone, YTH-DNA and YTH-RNA—suggests that the Arg404–Asn363 bridge is unique to the deoxyribose of DNA-bound form. In contrast, the conserved aromatic cage for binding the methylated adenine base of DNA or RNA is rigid, and adopts the same conformation with and without bound substrate (Supplementary Figure S4B).
Finally, 2′-OH groups on cytidine C7 and uridine at position 8 in RNA ligands are accommodated without the need for protein conformational adjustment (Figure 5F and G). Taken together, the addition of 2′-OH groups in RNA resulted in three circumstances: (i) accommodation without changes in the protein conformation (at C7 and U8), (ii) disruption, unique to DNA, of intra-molecular interactions between two protein residues (at N6mA) and (iii) repulsion between protein and RNA (at G5). The differences, in the conformations of nucleotides G4, G5 (Figure 3D and F), and N6mA at position 6 (Figure 5E), and their varied interactions with the YTH domain of YTHDC1, might explain the limited but significant increase in binding affinity for DNA over RNA (Figure 1).
In addition, we compared YTHDC1-DNA interactions with that of YTHDF1-RNA (52). The similarity between the two sets of interactions lies in the binding of modified adenosine (N6mA) and its following nucleotide (Supplementary Figure S5). We note that the one of the major difficulties in the structural comparisons between DNA-bound and RNA-bound forms of YTH domains is due to the shorter nucleotides observed in the RNA-bound structures: by 5-nt in YTHDC1 and 4-nt in YTHDF1. Thus, the effect of longer strands on the geometry of nucleotides (i.e. secondary and tertiary RNA structure) surrounding N6mA cannot be addressed in the current comparison. We also note that our ITC data (Figure 1) do not support an enthalpy difference between the binding modes of DNA and RNA by the YTH domain of YTHDC1.
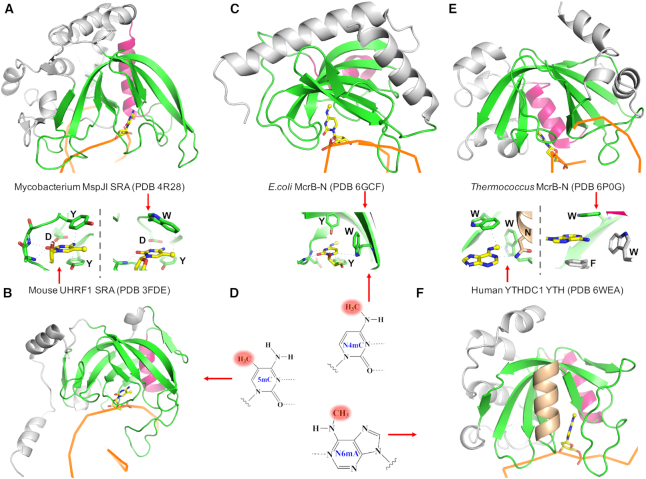
Connection of YTH to bacterial (modified cytosine restriction) McrBC
SET and RING finger-associated (SRA) domains (Figure 6A, B) were originally characterized in mammalian proteomes as readers of hemi-methylated CpG sequences – that is, sequences containing 5mC in only one strand, such as arise following DNA replication (53,54). SRA domains are also widespread in bacterial species, often associated with modification-dependent restriction endonucleases ((55,56) and references therein). We suggest that, like the SRA domain itself, the YTH domain may also have bacterial origins. McrBC (modified cytosine restriction) is an antiphage defense system of E. coli, which specifically cleaves invading DNA that is methylated in specific ways (57–59). The McrBC restriction system consists of two distinct subunits: McrB, comprising an N-terminal DNA binding domain (McrB-N) and a C-terminal AAA+ ATPase domain for GTP hydrolysis, and McrC, harboring an endonuclease domain (60). The McrB-N DNA binding domain recognizes DNA containing modified cytosine residues (61) and flips out of the duplex various modified cytosines including N4mC, 5mC and 5-hydroxymethylcytosine (5hmC) (58,59) (Figure 6C). This base flipping is independent of the sequence context of the target modified base. Interestingly McrB-N binds cytosine derivatives in the order of descending affinity of N4mC > 5mC > 5hmC (59). There is no mammalian protein domain currently known to bind N4mC; but MettL15 introduces N4mC in rRNA (62). However, the DUF55 domain of human thymocyte nuclear protein 1 (THYN1; also known as Thy28) shares a structural fold with the YTH domain (63,64), and has been suggested to bind DNA containing modified cytosine (65), while YTHDF2 binds 5mC in RNA (66). We note that the methyl groups of N4mC and N6mA share their location on the exocyclic amino groups of cytosine (at N4) and adenine (at N6) (Figure 6D), and the MTases responsible for these two types of amino (N4-cytosine and N6-adenine) methylation belong to the same MTase family group (4,25).
Figure 6.
The 5mC-binding SRA domain and N6mA-binding YTH domain are members of a larger family of modified nucleotide binding proteins associated with bacterial modification-dependent endonucleases. The conserved β-strands are in green and one conserved helix (colored in red) is behind the arch. The other α-helices are in gray. DNA strands are in ribbon with the flipped base in stick presentation. (A) MspJI SRA domain (with 5mC). (B) mouse UHRF1 SRA domain (with 5mC). (C) E. coli McrB-N DNA binding domain (with N4mC). (D) Three kinds of methylated DNA bases. (E) Thermococcus McrB-N in complex with a piece of dsDNA (with Ade). We note that a 19-bp duplex DNA was used for co-crystallization with Thermococcus McrB-N (67): a piece of 6-bp was observed that aligned tail-to-end in the crystal lattice and left no space for the missing two thirds of remaining oligo duplex. There are discrepancies of bases in the structure deposited in the PDB (6P0G) and that described in the publication (67): 5mC vs C on one strand and T versus C on the other strand. Nevertheless, the flipped base is an unmodified adenine. (F) Human YTHDC1 YTH domain (with N6mA) has a unique helix (colored in brown) in front of the arch. The modified bases are bound in a cage formed by 2–3 aromatic residues.
Recently, the structure of the McrB-N domain from the archaeon Thermococcus gammatolerans revealed a striking similarity to that of the eukaryotic YTH domain (67) (Figure 6E). This YTH-like DNA binding domain binds with about the same affinity to either double-stranded or single-stranded DNA containing N6mA, with an increased affinity relative to its binding to unmodified or 5mC-containing oligos. In contrast, it shows little affinity for RNA substrates, whether they are methylated or unmethylated. Looking at these various modification-dependent binding domains together, they share a common structural feature - a twisted anti-parallel β-sheet that forms an arch-like structure, with a variable number of helices packed against the outer surface (Figure 6). The inner surface of the arch, where DNA is bound, contains an aromatic pocket which defines the binding site of the modified nucleotide (Figure 6), which is flipped out from the DNA duplex if the substrates are double-strand DNA. A common helix lies in the back of the arch (colored red in Figure 6), perhaps functioning as a gatekeeper to control the access to the aromatic cage. The YTH domain of YTHDC1 contains an additional helix in front of the arch (colored brown in Figure 6F). The unique features for each domain lie in the number and length of their β-strands. Some strands are as long as 20-residues and curved, responsible for the arch-like appearance. A longer strand can be disrupted into two shorter strands linked by an inserted bulged segment. We consider the SRA and YTH domains as diverse members of a larger family of DNA/RNA modification readers.
DISCUSSION
Here we characterized, for the first time, the in vitro binding activity of the YTH domain of human YTHDC1 as preferentially targeting DNA, relative to RNA. YTH binds N6mA in the context of ssDNA, an activity comparable to that found in the human enzymatic activities of the MettL3–14 MTase complex and the demethylase Alkbh1, as they each act on damaged or unpaired DNA (26,37). Additional study will be required to address whether YTHDC1 is involved in the recognition of DNA adenine methylation in vivo and, if so, its impact on chromatin organization.
However, the following considerations suggest the hypothesis that genomic N6mA is associated with maintaining genome stability. First, upon ultraviolet irradiation, the human MTase complex MettL3-L14 is recruited within 2 min to the damaged sites, and MettL3 methylation activity is required for the DNA repair (31). Second, while DNA sequences are normally base paired in a canonical double helix, transient local unwinding of dsDNA occurs during the processes of transcription (forming a transcriptional bubble (68); Supplementary Figure S6A), replication (such as the single-strand regions between Okazaki fragments in the legging strand synthesis), recombination, and DNA repair. Furthermore, the existence of these transient non-B DNA structures is prolonged under physiological or biological stresses. For example, the accumulation of replication-associated ssDNA gaps has been observed in tumor cells (69–71). Accumulation of R-loops, a specific DNA–RNA hybrid with an unpaired ssDNA strand formed during transcription, is associated with disease in the context of cellular stress (72,73), and N6mA-associated R-loop accumulates during the cell cycle (43). Resolution of the four-way Holliday junction, the central intermediate of recombination, results in two dsDNA helices linked by a ssDNA region containing a single base gap (74) (Supplementary Figure S6B); and the gapped DNA is anisotropically bent (75). Perhaps cells respond to the inherent topological stress that arises from non-B DNA by coating ssDNA with protective protein complexes, that include the stress-induced writer-reader-eraser of N6mA-containing ssDNA. This would be consistent with the requirement of MettL3 methylation activity for UV-induced DNA repair (31). A single-nucleotide-resolution sequencing study revealed N6mA clusters associated with single-strand DNA binding protein on the human mitochondrial genome (12).
The appearance of ssDNA has been observed in E. coli as well as mammalian cells. For example, ssDNA can be induced by stress-induced DNA duplex destabilization (SIDD) in E. coli (76), and ssDNA is a common feature of the mammalian genome potentially involved in gene regulation (77). In E. coli, heterologous site-specific adenine methylation can induce the SOS DNA repair genes due to the action of the modification-specific endonuclease Mrr (modified DNA rejection and restriction) (78). In Caulobacter crescentus, the cell cycle-regulated DNA adenine MTase (CcrM) binds DNA by strand-separation of dsDNA and creates a bubble at its recognition site (79) (Supplementary Figure S6C). CcrM is active on both dsDNA and ssDNA as well as mismatches within or immediately outside of the recognition sequence (80,81).
Finally, we note that many nucleic acid-modifying enzymes are able to modify both DNA and RNA ((82) and references therein). This group of enzymes include members of the AlkB family, involved in the direct reversal of alkylation damage to both DNA and RNA (83), and members of the Apobec family of cytidine deaminases (84). Tet2, one of the ten-eleven translocation proteins initially discovered as DNA 5mC dioxygenases (85), mediates oxidation of 5mC in mRNA (86,87). Murine MettL4 was reported to be responsible for N6mA deposition in genic elements, corresponding with transcriptional silencing (10). Interestingly, recombinant human MettL4 expressed in HEK293T cells has in vitro enzymatic activity on mitochondrial DNA (21), whereas recombinant human MettL4 expressed in E. coli has RNA MTase activity (29). The former study showed that MettL4 localizes with mitochondria in all tested tissues (21) and the latter study found mainly nuclear localization of an exogenously introduced MettL4, and failed to identify appreciable levels of N6mA in mitochondrial DNA (29). The difference among these various studies illustrates the complex nature of DNA vs. RNA adenine methylation in mammalian genomes, and the accumulation of N6mA in DNA and/or RNA might reflect diverse cellular and mitochondrial stress responses under different laboratory conditions. The 5-fold difference in affinity, of the YTHDC1 YTH domain preference for DNA (Figure 1), therefore takes nothing away from the significance of YTH protein activities on RNA but suggests an additional layer of complexity involving mammalian DNA adenine methylation and its potential role in maintaining genome stability.
DATA AVAILABILITY
The X-ray structures (coordinates and structure factor files) of YTH domain with or without bound DNA have been submitted to PDB under accession number 6WEA (10-mer), 6WE9 (11-mer) and 6WE8 (no DNA).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Tao Wu of Baylor College of Medicine for discussion.
Authors contributions: C.B.W. performed protein purification, DNA binding assays and crystallization. J.R.H. performed X-ray data collection and structure determination. J.Z. helped with protein purification. M.T.B. made the initial suggestion for testing YTH binding methylated DNA and provided expression constructs. R.M.B. participated in discussion and performed analysis on sequence conservation and assisted in preparing the manuscript. X.Z. and X.C. organized and designed the scope of the study.
Contributor Information
Clayton B Woodcock, Department of Epigenetics and Molecular Carcinogenesis, University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
John R Horton, Department of Epigenetics and Molecular Carcinogenesis, University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Jujun Zhou, Department of Epigenetics and Molecular Carcinogenesis, University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Mark T Bedford, Department of Epigenetics and Molecular Carcinogenesis, University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Robert M Blumenthal, Department of Medical Microbiology and Immunology, and Program in Bioinformatics, The University of Toledo College of Medicine and Life Sciences, Toledo, OH 43614, USA.
Xing Zhang, Department of Epigenetics and Molecular Carcinogenesis, University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Xiaodong Cheng, Department of Epigenetics and Molecular Carcinogenesis, University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
U.S. National Institutes of Health (NIH) [R35GM134744 to X.C., R01GM126412 to M.T.B.]; Cancer Prevention and Research Institute of Texas (CPRIT) [RR160029 to X.C. who is a CPRIT Scholar in Cancer Research]. Funding for open access charge: MD Anderson Cancer Center.
Conflict of interest statement. M.T.B. is a cofounder of EpiCypher. Others declare no competing interests.
REFERENCES
- 1. Ehrlich M., Gama-Sosa M.A., Carreira L.H., Ljungdahl L.G., Kuo K.C., Gehrke C.W.. DNA methylation in thermophilic bacteria: N4-methylcytosine, 5-methylcytosine, and N6-methyladenine. Nucleic Acids Res. 1985; 13:1399–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sanchez-Romero M.A., Cota I., Casadesus J.. DNA methylation in bacteria: from the methyl group to the methylome. Curr. Opin. Microbiol. 2015; 25:9–16. [DOI] [PubMed] [Google Scholar]
- 3. Posfai J., Bhagwat A.S., Posfai G., Roberts R.J.. Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res. 1989; 17:2421–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Malone T., Blumenthal R.M., Cheng X.. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol. 1995; 253:618–632. [DOI] [PubMed] [Google Scholar]
- 5. Cheng X. Structure and function of DNA methyltransferases. Annu. Rev. Biophys. Biomol. Struct. 1995; 24:293–318. [DOI] [PubMed] [Google Scholar]
- 6. Bestor T., Laudano A., Mattaliano R., Ingram V.. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J. Mol. Biol. 1988; 203:971–983. [DOI] [PubMed] [Google Scholar]
- 7. Okano M., Xie S., Li E.. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 1998; 19:219–220. [DOI] [PubMed] [Google Scholar]
- 8. Wyatt G.R. Recognition and estimation of 5-methylcytosine in nucleic acids. Biochem. J. 1951; 48:581–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu T.P., Wang T., Seetin M.G., Lai Y., Zhu S., Lin K., Liu Y., Byrum S.D., Mackintosh S.G., Zhong M. et al.. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature. 2016; 532:329–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kweon S.M., Chen Y., Moon E., Kvederaviciute K., Klimasauskas S., Feldman D.E.. An adversarial DNA N(6)-methyladenine-sensor network preserves polycomb silencing. Mol. Cell. 2019; 74:1138–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xiao C.L., Zhu S., He M., Chen D., Zhang Q., Chen Y., Yu G., Liu J., Xie S.Q., Luo F. et al.. N(6)-Methyladenine DNA modification in the human genome. Mol. Cell. 2018; 71:306–318. [DOI] [PubMed] [Google Scholar]
- 12. Koh C.W.Q., Goh Y.T., Toh J.D.W., Neo S.P., Ng S.B., Gunaratne J., Gao Y.G., Quake S.R., Burkholder W.F., Goh W.S.S.. Single-nucleotide-resolution sequencing of human N6-methyldeoxyadenosine reveals strand-asymmetric clusters associated with SSBP1 on the mitochondrial genome. Nucleic Acids Res. 2018; 46:11659–11670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xie Q., Wu T.P., Gimple R.C., Li Z., Prager B.C., Wu Q., Yu Y., Wang P., Wang Y., Gorkin D.U. et al.. N(6)-methyladenine DNA modification in glioblastoma. Cell. 2018; 175:1228–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ratel D., Ravanat J.L., Charles M.P., Platet N., Breuillaud L., Lunardi J., Berger F., Wion D.. Undetectable levels of N6-methyl adenine in mouse DNA: Cloning and analysis of PRED28, a gene coding for a putative mammalian DNA adenine methyltransferase. FEBS Lett. 2006; 580:3179–3184. [DOI] [PubMed] [Google Scholar]
- 15. Schiffers S., Ebert C., Rahimoff R., Kosmatchev O., Steinbacher J., Bohne A.V., Spada F., Michalakis S., Nickelsen J., Muller M. et al.. Quantitative LC-MS provides no evidence for m(6) dA or m(4) dC in the genome of mouse Embryonic stem cells and tissues. Angewandte Chemie. 2017; 56:11268–11271. [DOI] [PubMed] [Google Scholar]
- 16. Liu B., Liu X., Lai W., Wang H.. Metabolically generated stable isotope-labeled deoxynucleoside code for tracing DNA N(6)-methyladenine in human cells. Anal. Chem. 2017; 89:6202–6209. [DOI] [PubMed] [Google Scholar]
- 17. Musheev M.U., Baumgartner A., Krebs L., Niehrs C.. The origin of genomic N(6)-methyl-deoxyadenosine in mammalian cells. Nat. Chem. Biol. 2020; 16:630–634. [DOI] [PubMed] [Google Scholar]
- 18. Liu X., Lai W., Li Y., Chen S., Liu B., Zhang N., Mo J., Lyu C., Zheng J., Du Y.R. et al.. N(6)-methyladenine is incorporated into mammalian genome by DNA polymerase. Cell Res. 2020; doi:10.1038/s41422-020-0317-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Woodcock C.B., Yu D., Zhang X., Cheng X.. Human HemK2/KMT9/N6AMT1 is an active protein methyltransferase, but does not act on DNA in vitro, in the presence of Trm112. Cell Discov. 2019; 5:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li W., Shi Y., Zhang T., Ye J., Ding J.. Structural insight into human N6amt1-Trm112 complex functioning as a protein methyltransferase. Cell Discov. 2019; 5:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hao Z., Wu T., Cui X., Zhu P., Tan C., Dou X., Hsu K.W., Lin Y.T., Peng P.H., Zhang L.S. et al.. N(6)-deoxyadenosine methylation in mammalian mitochondrial DNA. Mol. Cell. 2020; 78:382–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Figaro S., Scrima N., Buckingham R.H., Heurgue-Hamard V.. HemK2 protein, encoded on human chromosome 21, methylates translation termination factor eRF1. FEBS Lett. 2008; 582:2352–2356. [DOI] [PubMed] [Google Scholar]
- 23. Metzger E., Wang S., Urban S., Willmann D., Schmidt A., Offermann A., Allen A., Sum M., Obier N., Cottard F. et al.. KMT9 monomethylates histone H4 lysine 12 and controls proliferation of prostate cancer cells. Nat. Struct. Mol. Biol. 2019; 26:361–371. [DOI] [PubMed] [Google Scholar]
- 24. Bujnicki J.M., Feder M., Radlinska M., Blumenthal R.M.. Structure prediction and phylogenetic analysis of a functionally diverse family of proteins homologous to the MT-A70 subunit of the human mRNA:m(6)A methyltransferase. J. Mol. Evol. 2002; 55:431–444. [DOI] [PubMed] [Google Scholar]
- 25. Woodcock C.B., Horton J.R., Zhang X., Blumenthal R.M., Cheng X.. Beta class amino methyltransferases from bacteria to humans: evolution and structural consequences. Nucleic Acids Res. 2020; doi:10.1093/nar/gkaa446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Woodcock C.B., Yu D., Hajian T., Li J., Huang Y., Dai N., Correa I.R. Jr, Wu T., Vedadi M., Zhang X. et al.. Human MettL3-MettL14 complex is a sequence-specific DNA adenine methyltransferase active on single-strand and unpaired DNA in vitro. Cell Discov. 2019; 5:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Frye M., Harada B.T., Behm M., He C.. RNA modifications modulate gene expression during development. Science. 2018; 361:1346–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu J., Dou X., Chen C., Chen C., Liu C., Xu M.M., Zhao S., Shen B., Gao Y., Han D. et al.. N (6)-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science. 2020; 367:580–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen H., Gu L., Orellana E.A., Wang Y., Guo J., Liu Q., Wang L., Shen Z., Wu H., Gregory R.I. et al.. METTL4 is an snRNA m(6)Am methyltransferase that regulates RNA splicing. Cell Res. 2020; 30:544–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang S., Song Y., Wang Y., Li X., Fu B., Liu Y., Wang J., Wei L., Tian T., Zhou X.. The m(6)A methylation perturbs the Hoogsteen pairing-guided incorporation of an oxidized nucleotide. Chem. Sci. 2017; 8:6380–6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xiang Y., Laurent B., Hsu C.H., Nachtergaele S., Lu Z., Sheng W., Xu C., Chen H., Ouyang J., Wang S. et al.. RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature. 2017; 543:573–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang W., Xu L., Hu L., Chong J., He C., Wang D.. Epigenetic DNA modification N(6)-methyladenine causes Site-Specific RNA polymerase II transcriptional pausing. J. Am. Chem. Soc. 2017; 139:14436–14442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Flusberg B.A., Webster D.R., Lee J.H., Travers K.J., Olivares E.C., Clark T.A., Korlach J., Turner S.W.. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods. 2010; 7:461–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Valinluck V., Liu P., Burdzy A., Ryu J., Sowers L.C.. Influence of local duplex stability and N6-methyladenine on uracil recognition by mismatch-specific uracil-DNA glycosylase (Mug). Chem. Res. Toxicol. 2002; 15:1595–1601. [DOI] [PubMed] [Google Scholar]
- 35. Lopez C.M., Lloyd A.J., Leonard K., Wilkinson M.J.. Differential effect of three base modifications on DNA thermostability revealed by high resolution melting. Anal. Chem. 2012; 84:7336–7342. [DOI] [PubMed] [Google Scholar]
- 36. Song Q.X., Ding Z.D., Liu J.H., Li Y., Wang H.J.. Theoretical study on the binding mechanism between N6-methyladenine and natural DNA bases. J. Mol. Model. 2013; 19:1089–1098. [DOI] [PubMed] [Google Scholar]
- 37. Zhang M., Yang S., Nelakanti R., Zhao W., Liu G., Li Z., Liu X., Wu T., Xiao A., Li H.. Mammalian ALKBH1 serves as an N(6)-mA demethylase of unpairing DNA. Cell Res. 2020; 30:197–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Patil D.P., Pickering B.F., Jaffrey S.R.. Reading m(6)A in the transcriptome: m(6)A-binding proteins. Trends Cell Biol. 2018; 28:113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liao S., Sun H., Xu C.. YTH domain: a family of N(6)-methyladenosine (m(6)A) readers. Genomics Proteomics Bioinformatics. 2018; 16:99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tong J., Flavell R.A., Li H.B.. RNA m(6)A modification and its function in diseases. Front Med. 2018; 12:481–489. [DOI] [PubMed] [Google Scholar]
- 41. Nayler O., Hartmann A.M., Stamm S.. The ER repeat protein YT521-B localizes to a novel subnuclear compartment. J. Cell Biol. 2000; 150:949–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xiao W., Adhikari S., Dahal U., Chen Y.S., Hao Y.J., Sun B.F., Sun H.Y., Li A., Ping X.L., Lai W.Y. et al.. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell. 2016; 61:507–519. [DOI] [PubMed] [Google Scholar]
- 43. Abakir A., Giles T.C., Cristini A., Foster J.M., Dai N., Starczak M., Rubio-Roldan A., Li M., Eleftheriou M., Crutchley J. et al.. N(6)-methyladenosine regulates the stability of RNA:DNA hybrids in human cells. Nat. Genet. 2020; 52:48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Otwinowski Z., Borek D., Majewski W., Minor W.. Multiparametric scaling of diffraction intensities. Acta Crystallogr. A. 2003; 59:228–234. [DOI] [PubMed] [Google Scholar]
- 45. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Headd J.J., Echols N., Afonine P.V., Grosse-Kunstleve R.W., Chen V.B., Moriarty N.W., Richardson D.C., Richardson J.S., Adams P.D.. Use of knowledge-based restraints in phenix.refine to improve macromolecular refinement at low resolution. Acta Crystallogr. D. Biol. Crystallogr. 2012; 68:381–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 48. Schibler U., Kelley D.E., Perry R.P.. Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. J. Mol. Biol. 1977; 115:695–714. [DOI] [PubMed] [Google Scholar]
- 49. Arguello A.E., Leach R.W., Kleiner R.E.. In vitro selection with a site-specifically modified RNA library reveals the binding preferences of N(6)-methyladenosine reader proteins. Biochemistry. 2019; 58:3386–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Xu C., Wang X., Liu K., Roundtree I.A., Tempel W., Li Y., Lu Z., He C., Min J.. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 2014; 10:927–929. [DOI] [PubMed] [Google Scholar]
- 51. Dai X., Wang T., Gonzalez G., Wang Y.. Identification of YTH domain-containing proteins as the readers for N1-methyladenosine in RNA. Anal. Chem. 2018; 90:6380–6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xu C., Liu K., Ahmed H., Loppnau P., Schapira M., Min J.. Structural basis for the discriminative recognition of N6-Methyladenosine RNA by the human YT521-B Homology domain family of proteins. J. Biol. Chem. 2015; 290:24902–24913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bostick M., Kim J.K., Esteve P.O., Clark A., Pradhan S., Jacobsen S.E.. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007; 317:1760–1764. [DOI] [PubMed] [Google Scholar]
- 54. Sharif J., Muto M., Takebayashi S., Suetake I., Iwamatsu A., Endo T.A., Shinga J., Mizutani-Koseki Y., Toyoda T., Okamura K. et al.. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007; 450:908–912. [DOI] [PubMed] [Google Scholar]
- 55. Zheng Y., Cohen-Karni D., Xu D., Chin H.G., Wilson G., Pradhan S., Roberts R.J.. A unique family of Mrr-like modification-dependent restriction endonucleases. Nucleic Acids Res. 2010; 38:5527–5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Loenen W.A., Raleigh E.A.. The other face of restriction: modification-dependent enzymes. Nucleic Acids Res. 2014; 42:56–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Stewart F.J., Panne D., Bickle T.A., Raleigh E.A.. Methyl-specific DNA binding by McrBC, a modification-dependent restriction enzyme. J. Mol. Biol. 2000; 298:611–622. [DOI] [PubMed] [Google Scholar]
- 58. Sukackaite R., Grazulis S., Tamulaitis G., Siksnys V.. The recognition domain of the methyl-specific endonuclease McrBC flips out 5-methylcytosine. Nucleic Acids Res. 2012; 40:7552–7562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zagorskaite E., Manakova E., Sasnauskas G.. Recognition of modified cytosine variants by the DNA-binding domain of methyl-directed endonuclease McrBC. FEBS Lett. 2018; 592:3335–3345. [DOI] [PubMed] [Google Scholar]
- 60. Nirwan N., Itoh Y., Singh P., Bandyopadhyay S., Vinothkumar K.R., Amunts A., Saikrishnan K.. Structure-based mechanism for activation of the AAA+ GTPase McrB by the endonuclease McrC. Nat. Commun. 2019; 10:3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kruger T., Wild C., Noyer-Weidner M.. McrB: a prokaryotic protein specifically recognizing DNA containing modified cytosine residues. EMBO J. 1995; 14:2661–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Van Haute L., Hendrick A.G., D'Souza A.R., Powell C.A., Rebelo-Guiomar P., Harbour M.E., Ding S., Fearnley I.M., Andrews B., Minczuk M.. METTL15 introduces N4-methylcytidine into human mitochondrial 12S rRNA and is required for mitoribosome biogenesis. Nucleic Acids Res. 2019; 47:10267–10281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yu F., Song A., Xu C., Sun L., Li J., Tang L., Yu M., Yeates T.O., Hu H., He J.. Determining the DUF55-domain structure of human thymocyte nuclear protein 1 from crystals partially twinned by tetartohedry. Acta Crystallogr. D. Biol. Crystallogr. 2009; 65:212–219. [DOI] [PubMed] [Google Scholar]
- 64. Luo S., Tong L.. Molecular basis for the recognition of methylated adenines in RNA by the eukaryotic YTH domain. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:13834–13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Spruijt C.G., Gnerlich F., Smits A.H., Pfaffeneder T., Jansen P.W., Bauer C., Munzel M., Wagner M., Muller M., Khan F. et al.. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013; 152:1146–1159. [DOI] [PubMed] [Google Scholar]
- 66. Dai X., Gonzalez G., Li L., Li J., You C., Miao W., Hu J., Fu L., Zhao Y., Li R. et al.. YTHDF2 Binds to 5-Methylcytosine in RNA and Modulates the Maturation of Ribosomal RNA. Anal. Chem. 2020; 92:1346–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hosford C.J., Bui A.Q., Chappie J.S.. The structure of the Thermococcus gammatolerans McrB N-terminal domain reveals a new mode of substrate recognition and specificity among McrB homologs. J. Biol. Chem. 2020; 295:743–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. He Y., Yan C., Fang J., Inouye C., Tjian R., Ivanov I., Nogales E.. Near-atomic resolution visualization of human transcription promoter opening. Nature. 2016; 533:359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Panzarino N.J., Krais J., Peng M., Mosqueda M., Nayak S., Bond S., Calvo J., Cong K., Doshi M., Bere M. et al.. Replication gaps underlie BRCA-deficiency and therapy response. 2019; bioRxiv doi:25 September 2019, preprint: not peer reviewed 10.1101/781955. [DOI] [PMC free article] [PubMed]
- 70. Nayak S., Calvo J.A., Cong K., Peng M., Berthiaume E., Jackson J., Zaino A.M., Vindigni A., Hadden K.M., Cantor S.B.. Inhibition of the translesion synthesis polymerase REV1 exploits replication gaps as a cancer vulnerability. Sci. Adv. 2020; 6:eaaz7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cong K., Kousholt A.N., Peng M., Panzarino N.J., Lee W.T.C., Nayak S., Krais J., Calvo J., Bere M., Rothenberg E. et al.. PARPi synthetic lethality derives from replication-associated single-stranded DNA gaps. 2019; bioRxiv doi:25 September 2019, preprint: not peer reviewed 10.1101/781989. [DOI]
- 72. Allison D.F., Wang G.G.. R-loops: formation, function, and relevance to cell stress. Cell Stress. 2019; 3:38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hegazy Y.A., Fernando C.M., Tran E.J.. The balancing act of R-loop biology: The good, the bad, and the ugly. J. Biol. Chem. 2020; 295:905–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Liu Y., Freeman A.D.J., Declais A.C., Wilson T.J., Gartner A., Lilley D.M.J.. Crystal structure of a Eukaryotic GEN1 resolving enzyme bound to DNA. Cell Reports. 2015; 13:2565–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Guo H., Tullius T.D.. Gapped DNA is anisotropically bent. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:3743–3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wang H., Noordewier M., Benham C.J.. Stress-induced DNA duplex destabilization (SIDD) in the E. coli genome: SIDD sites are closely associated with promoters. Genome Res. 2004; 14:1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kouzine F., Wojtowicz D., Baranello L., Yamane A., Nelson S., Resch W., Kieffer-Kwon K.R., Benham C.J., Casellas R., Przytycka T.M. et al.. Permanganate/S1 nuclease footprinting reveals Non-B DNA structures with regulatory potential across a Mammalian genome. Cell Syst. 2017; 4:344–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Heitman J., Model P.. Site-specific methylases induce the SOS DNA repair response in Escherichia coli. J. Bacteriol. 1987; 169:3243–3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Horton J.R., Woodcock C.B., Opot S.B., Reich N.O., Zhang X., Cheng X.. The cell cycle-regulated DNA adenine methyltransferase CcrM opens a bubble at its DNA recognition site. Nat. Commun. 2019; 10:4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Woodcock C.B., Yakubov A.B., Reich N.O.. Caulobacter crescentus cell cycle-regulated DNA methyltransferase uses a novel mechanism for substrate recognition. Biochemistry. 2017; 56:3913–3922. [DOI] [PubMed] [Google Scholar]
- 81. Reich N.O., Dang E., Kurnik M., Pathuri S., Woodcock C.B.. The highly specific, cell cycle-regulated methyltransferase from Caulobacter crescentus relies on a novel DNA recognition mechanism. J. Biol. Chem. 2018; 293:19038–19046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Forterre P., Grosjean H.. Grosjean H. The Interplay between RNA and DNA modifications. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. 2009; Austin: Landes Bioscience; 259–274. [Google Scholar]
- 83. Fedeles B.I., Singh V., Delaney J.C., Li D., Essigmann J.M.. The AlkB family of Fe(II)/alpha-ketoglutarate-dependent dioxygenases: repairing nucleic acid alkylation damage and beyond. J. Biol. Chem. 2015; 290:20734–20742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yang B., Li X., Lei L., Chen J.. APOBEC: From mutator to editor. J. Genet Genomics. 2017; 44:423–437. [DOI] [PubMed] [Google Scholar]
- 85. Ko M., An J., Bandukwala H.S., Chavez L., Aijo T., Pastor W.A., Segal M.F., Li H., Koh K.P., Lahdesmaki H. et al.. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature. 2013; 497:122–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Shen Q., Zhang Q., Shi Y., Shi Q., Jiang Y., Gu Y., Li Z., Li X., Zhao K., Wang C. et al.. Tet2 promotes pathogen infection-induced myelopoiesis through mRNA oxidation. Nature. 2018; 554:123–127. [DOI] [PubMed] [Google Scholar]
- 87. Guallar D., Bi X., Pardavila J.A., Huang X., Saenz C., Shi X., Zhou H., Faiola F., Ding J., Haruehanroengra P. et al.. RNA-dependent chromatin targeting of TET2 for endogenous retrovirus control in pluripotent stem cells. Nat. Genet. 2018; 50:443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The X-ray structures (coordinates and structure factor files) of YTH domain with or without bound DNA have been submitted to PDB under accession number 6WEA (10-mer), 6WE9 (11-mer) and 6WE8 (no DNA).