Abstract
The CRISPR/Cas system is widely used for genome editing. However, robust and targeted insertion of a DNA segment remains a challenge. Here, we present a fusion nuclease (Cas9-N57) to enhance site-specific DNA integration via a fused DNA binding domain of Sleeping Beauty transposase to tether the DNA segment to the Cas9/sgRNA complex. The insertion was unidirectional and specific, and DNA fragments up to 12 kb in length were successfully integrated. As a test of the system, Cas9-N57 mediated the insertion of a CD19-specific chimeric antigen receptor (CD19-CAR) cassette into the AAVS1 locus in human T cells, and induced intrahepatic cholangiocarcinoma in mice by simultaneously mediating the insertion of oncogenic KrasG12D into the Rosa26 locus and disrupting Trp53 and Pten. Moreover, the nuclease-N57 fusion proteins based on AsCpf1 (AsCas12a) and CjCas9 exhibited similar activity. These findings demonstrate that CRISPR-associated nuclease-N57 protein fusion is a powerful tool for targeted DNA insertion and holds great potential for gene therapy applications.
INTRODUCTION
The CRISPR/Cas system confers acquired immunity against foreign genetic elements via guide RNA-dependent DNA or RNA nuclease activity in bacteria and archaea (1). CRISPR effectors such as Cas9 and Cas12a/Cpf1 have become widely used tools for genome engineering, including DNA sequence change, gene expression regulation and genomic loci labeling (2–4). Recently, prime-editing, a ‘search-and-replace’ genome editing technology, has been developed to mediate targeted short deletions and insertions, all 12 possible base-to-base conversions, and combinations thereof, which enables any specific change around a target site (5). However, efficient targeted insertion of a large exogenous DNA fragment remains a challenge.
CRISPR-mediated targeted transgene integration is generally based on homology-directed repair (HDR) (6,7), non-homologous end-joining (NHEJ) (8,9), microhomology-mediated end-joining (MMEJ) (10–12), or homology-mediated end-joining (HMEJ) (13–16). HDR-based integration is precise without any insertions and deletions (indels) at the junction sites, but restrained in dividing cells with low efficiency (8,17–19). NHEJ-, MMEJ- and HMEJ-based approaches promote targeted insertion by in vivo cleaving transgene donors without homology arms (HAs) or with short (5–20 bp) or long (∼800 bp) HAs. Tild-CRISPR, an HMEJ-based method, achieves robust DNA knock-in by in vitro cleaving donors with long HAs (∼800 bp) in mouse and human embryos (14). SATI, another HMEJ-based method, allows for efficient targeted integration by in vivo cleaving DNA donors with a single HA (20). Easi-CRISPR, a method based on single-strand annealing (SSA), boosts DNA knock-in using a single-stranded DNA donor with medium-sized HAs (55–105 bp) in mouse embryos (21). Among these strategies, CRISPR/Cas9-mediated homology-independent targeted integration (HITI), an NHEJ-based approach, achieves robust targeted knock-in by in vivo cleaving DNA donor without HAs in both dividing and non-dividing cells (9). Once the donor DNA is inserted in the desired direction, the Cas9 target site is disrupted and further Cas9 cutting is prevented. When the donor DNA is inserted in the reverse direction, the Cas9 target site will remain intact and the inserted transgene will be cut out by a second round of Cas9 cutting (9). Thus, the direction of HITI-mediated transgene insertion is predetermined.
Recruiting the DNA donor to the vicinity of double-strand DNA break sites promotes HR-mediated transgene insertion. Cas9 fused with avidin/streptavidin or SNAP can tether biotin-labeled or O6-benzylguanine (BG)-labeled ssODN/dsDNA donors to editing sites and achieve robust knock-in in mammalian cells and mouse embryos (22–25). HUH endonucleases form covalent bonds with specific sequences of ssODN and bring ssODN donor to target sites when fused with Cas9 (26). Donor DNA can be linked covalently to sgRNA to form a RNA-DNA hybrid and mediate efficient targeted insertion (27). Additionally, multivalent display of ssODN on Cas9 increases the efficiency of knock-in and has been used successfully to engineer pancreatic β cells to secrete interleukin-10 (28). In yeast, a LexA-Fkh1p fusion protein facilitates Cas9-induced knock-in as LexA binds to specific DNA sequences and Fkh1p binds to phosphothreonines on multiple proteins involved in HR-mediated DNA repair (29).
In addition to the CRISPR system, transposons can be used to insert DNA segments into genome without introducing DNA double-strand breaks. Natural CRISPR-associated transposases and engineered dCas9-Homo sapiens mariner 1 transposase fusion can mediate targeted integration both in vitro and in Escherichia coli cells (30–33). Recently, the proof-of-concept that guide RNA-directed dCas9-transposase fusions mediate targeted insertion has been reported (34). A piggyBac transposase mutant with reduced DNA-binding activity is fused to a catalytically dead Cas9 mutant with high fidelity (dCas9-HF1), and the fusion protein can introduce targeted insertion of donor DNA at the CCR5 safe harbor site in human cells although with low efficiency (34).
To develop a robust and site-specific DNA integration strategy, we fused Sleeping Beauty transposase (SB) to dCas9, but the fusion protein failed to mediate targeted insertion. Then, we combined the HITI approach with the idea of tethering the DNA donor to the editing site with the Cas9/sgRNA complex. By fusing the DNA-binding domain of SB to Cas9, the DNA donor containing the SB corresponding binding sequence can be recruited to the target site and increase transgene integration in both cell cultures and mice. This approach exhibits high genome-wide specificity and can be used with a variety of natural and engineered Cas nucleases and thus is a promising tool for future genome engineering applications.
MATERIALS AND METHODS
Plasmid construction
The human codon-optimized SpCas9 (Addgene # 52962), eCas9 (Addgene # 104172), AsCpf1 (Addgene #69982) and CjCas9 (Addgene # 68338) plasmids were obtained from Addgene. sgRNA sequences were designed through https://www.benchling.com/ and are listed in Supplementary Table S1. A detailed description of the construction of Cas9 fusion proteins, nuclease-SB, nuclease-N57 and corresponding donor and sgRNA plasmids is provided in the Supplementary Methods. Cas9-donor-M2rtTA, pBlue-AAVS1-AAVS1 and pBlue-AAVS1-Puro-Cas9:p300-M2rtTA-AAVS1 plasmids were described previously (35).
Cell culture and transfection
All cell lines were obtained from ATCC. MCF7 and Jurkat E6–1 cell lines were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. NIH3T3 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% FBS and 1% penicillin/streptomycin. All cell lines were cultured at 37°C in a humidified atmosphere with 5% CO2.
MCF7 and NIH3T3 cells were transfected with polyethylenimine (PEI, Sigma). A total of 500 ng plasmid was used in each reaction. Approximately 1 × 105 cells were plated in 24-well plates 1 day before transfection. Jurkat E6–1 cells were transfected with the Cell Line Nucleofector® Kit V, program X-001 (for Nucleofector® II, Lonza). A total of 2 μg plasmid was used in each reaction. After 5 days, the original media were replaced with media containing 0.75, 2 and 4 μg/ml puromycin for Jurkat E6–1, MCF7 and NIH3T3 cells, respectively, for a period of 3 days. Sixty cells were seeded into a whole 96-well plate (less than one cell per well), and after 2 weeks the clones were harvested and identified by polymerase chain reaction (PCR) and sequencing.
Flow cytometry
Five days after transfection, cells were treated with puromycin for 3 days and expanded in media without puromycin for 2–3 weeks. Then cells were isolated and resuspended in 500 μl FACS buffer and loaded onto a flow cytometer (BD Fortessa, CA, USA) for the detection of EGFP+ and mNeonGreen+ cells.
Western blotting
Western blotting was performed according to previously described standard protocols with minor alteration (36). Briefly, cells of iKA-CRISPR clone #17 and MCF cells transfected with Cas9, Cas9-SB, Cas9-N123 or Cas9-N57 were lysed, boiled for 10 min at 95°C, separated on a 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis gel, transferred to a nitrocellulose membrane and detected with sequential blotting with primary antibodies against GAPDH (6004–1-Ig, Proteintech), Cas9 (ab191468, Abcam), Flag tag (66008–3, Proteintech) and anti-mouse IgG HRP secondary antibody (7076S, Cell Signaling Technology). Protein expression was detected by chemiluminescence according to the manufacturer's instructions (WBKLS0500, Millipore, MA, USA).
T7 endonuclease I (T7E1) assay
T7E1 assay was performed as described previously (35). Briefly, targeted regions were amplified by PCR and purified with a PCR Purification Kit (Cat. No. 28106, Qiagen). The purified DNA (400 ng) was denatured and cooled for heteroduplex formation in NEB buffer 2 with a thermocycler (Thermo Fisher Scientific, Singapore). The heteroduplex PCR products were incubated with T7 endonuclease I (M0302L, NEB) for 2 h at 37°C and then subjected to electrophoresis on 2% agarose gels. The primers for the T7E1 assay to assess the editing efficiency at the HBG1 site were described previously (35).
RT-qPCR
RT-qPCR was performed as described previously (35). Briefly, total RNA was isolated from cells and reverse-transcribed into cDNA with a Prime Script RT Reagent Kit (Cat. No. RR047A, TAKARA). qPCR was performed on a LightCycler 96 System (Roche, Switzerland) with SYBR Premix Ex Taq II (Cat. No. RR820, TAKARA) as follows: 95°C for 30 s, followed by 45 cycles of 95°C for 5 s and 60°C 34 s. The primers for IL-1RN and HBG1 were described previously (35). All other primers used are listed in Supplementary Table S1.
Tracking of indels by decomposition (TIDE) assay
The tracking of indels by decomposition (TIDE) assay is a bioinformatics tool to estimate the frequency of insertions and deletions (indels) in a pool of cells (37). We used two PCR amplicons from control and test samples with the same set of primers; all parameters were set to the online default (http://tide.nki.nl). All primers used are listed in Supplementary Table S1.
NovaSeq and analysis
NovaSeq (30 times genome coverage) was performed by Novogene (Beijing, China). Illumina paired-end reads were trimmed with Trimmomatic v.0.36 (38), and read quality was assessed with FastQC v.0.11.8. The plasmid sequence was then appended to the human genome sequence downloaded from GENCODE (GRCh38/hg38) (39), and reads were aligned with the modified human genome sequence with BWA 0.7.17 (40). Quality control on the raw alignment data was performed by filtering unqualified reads, including unmapped reads and multi-mapped reads, with SAMtools v.1.7.2 (41). PCR-duplicated reads were identified and removed with Picard v.2.16.0 (42). Paired-end reads with one read located on the plasmid and the mate read located on human chromosomes were retrieved from the alignment results, and Integrative Genomics Viewer (IGV) v.2.3.34 (43) was used to evaluate the mapping quality of these soft-clipped reads. To determine the direction of insertion, forward and reverse insertion reference sequences were constructed in chr7:5527490–5527491 for the alignment of paired-end reads.
Hematoxylin and eosin (H&E) staining, immunofluorescence staining and immunohistochemistry
C57BL/6 mice were euthanized through CO2 asphyxiation. Livers were fixed overnight in 4% paraformaldehyde at 4°C, embedded in paraffin, sliced into 5 μm sections and subjected to H&E staining. Immunofluorescence staining was performed as described previously (35). The GFP antibody was purchased from Abcam (Cat. No. ab6556). Nuclei were counterstained with DAPI (H3570, Invitrogen). Images were captured on a Zeiss LSM 880 confocal microscope (Germany).
Liver sections were deparaffinized with xylene and rehydrated via serial ethanol dilutions. For antigen retrieval, the sections were boiled for 6 min in a pressure cooker. Endogenous peroxidases were inactivated for 10 min and then blocked with 5% bovine serum albumin for 20 min. The sections were incubated first overnight with a primary antibody against Ck19 (1:100, ab133496, Abcam) at 4°C, then with a secondary HRP-conjugated anti-rabbit IgG antibody (7074S, Cell Signal Technology) for 1 h at room temperature, and subsequently with the chromogenic substrate for 20 min. Then the sections were counterstained with hematoxylin, dehydrated sequentially via serial dilutions with ethanol and xylene, and sealed with neutral resins for long-term storage. Images were obtained with a Nikon 11R laser scanning microscope with a × 20 objective (Japan).
Hydrodynamic tail vein injection
A total of 30 μg Cas9/Cas9-SB/Cas9-N57 plasmids, 15 μg DNA donor and 6 μg sgRNA per target site were mixed with 1.5–1.8 ml 0.9% sterile saline (10% of body weight) and then delivered to each mouse by hydrodynamic tail vein injection lasting 7 s. After 4–5 weeks, the mice were euthanized for liver assessment.
Statistics
Fisher's exact test was used to assess targeted insertion rate based on PCR experiments of single cell clones, and one-way ANOVA test for other experiments.
RESULTS
Cas9-N57 catalyzes robust targeted DNA integration
To insert an exogenous DNA fragment into the genome without DNA double-strand breaks, we developed a hybrid Cas9 and transposase system, aiming to capitalize on the site-specific targeting ability of Cas9 and the DNA insertion capability of transposase. SB100X (henceforth ‘SB’) is a hyperactive transposase (44), and dCas9 is a nuclease-dead Cas9 mutant. In principle, the dCas9-SB fusion protein can be directed to the target site by a guide RNA, and SB can excise the gene of interest (GOI) from a donor plasmid and insert it into the target site (Supplementary Figure S1A and B). However, the system failed when tested at the AAVS1 locus in MCF7 cells (Supplementary Figure S1C and D).
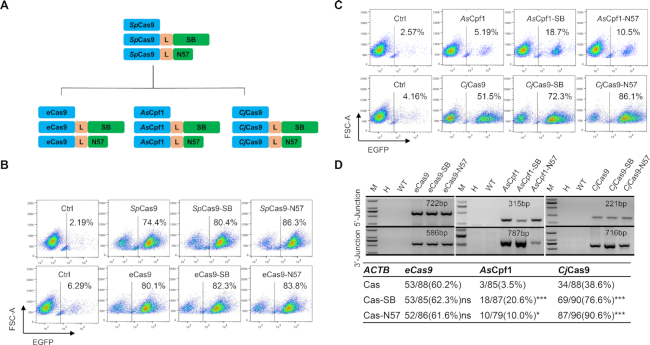
We noticed that SB contains two DNA-binding domains, PAI and RED, as well as a DDE domain that is involved in cleavage and strand transfer (45) (Supplementary Figure S2A). Therefore, we hypothesized that the DNA-binding ability of SB could be used to recruit the exogenous DNA segment close to the cutting site of Cas9 and facilitate insertion. Given that HITI is a currently used strategy for efficient targeted DNA insertion (9), we tested whether the DNA-binding capability of SB could further improve the efficiency of HITI. To this end, we constructed three plasmids by fusing Cas9 with full-length SB (Cas9-SB), the PAI and RED domains of SB (Cas9-N123) and the PAI domain of SB (Cas9-N57; Figure 1A and Supplementary Figure S2A and B).
Figure 1.
Cas9-N57 catalyzes robust-targeted DNA integration. (A) Schematic of Cas9-SB, Cas9-N123 and Cas9-N57 for site-specific DNA integration. (B) Schematic of the IRES-Puro-pEF1α-EGFP DNA donor and targeting strategy at the ACTB locus. Filled and open boxes indicate the coding and 3′-untranslated regions of the human ACTB gene, respectively. Numbers inside the boxes indicate the serial numbers of the exons. Black and gray arrows indicate the outer and inner direct repeats in the inverted repeats (IRs), respectively. g indicates the sgACTB target site in the donor. IRES, internal ribosome entry site; puro, puromycin resistant gene; PA, polyA; EGFP, enhanced GFP; EF1α, EF1-alpha promoter. Primer positions for genomic PCR are shown. (C) Results of FACS analysis of the frequencies of EGFP+ cell populations in MCF7 cells transfected with the indicated plasmids. Ctrl, cells transfected with gRNA and DNA donor. (D) Genomic PCR and statistics of targeted single clones generated with Cas9, Cas9-SB, Cas9-N123 and Cas9-N57. M, Marker; H, H2O; WT, wild-type. Primers for the 5′-junction, 3′-junction and WT were ACTB-5-F/5-R, 3-F/ACTB-3-R and ACTB-5-F/ACTB-3-R, respectively. Correct knock-in clones/total checked clones (percentage). ns, not significant; **P < 0.01, ***P < 0.001, Fisher's exact test. (E) Sequence analysis of 5′- and 3′-junctions in representative targeted clones. The Ref seq is the predicted junction sequence. Genomic and insertion regions are indicated with different-colored bars. Black arrowheads indicate junction sites. (F) Efficiency of knock-in, indicated by the percentage of targeted single clones. Numbers above each bar denote total clones checked. *P < 0.05, **P < 0.01, ***P < 0.001, Fisher's exact test.
To test the system, a DNA cassette containing IRES-Puro-pEF1α-EGFP and SB-binding DNA sequence (IR) was designed to be inserted downstream of the stop codon of the actin beta (ACTB) gene (Figure 1B). MCF7 breast cancer cells were transfected with the fusion proteins and donor plasmids at a transfection frequency of about 15–18%. The cells were selected with puromycin for 3 days and then cultured for two more weeks. FACS analysis showed that the frequency of EGFP+ cells was about 1.5-fold in cells transfected with fusion proteins than in cells transfected with Cas9 (Figure 1C). Furthermore, genomic PCR demonstrated that the donor fragment was inserted into the ACTB site in the transfected cell populations (data not shown). To more precisely assess the efficiency of insertion, genomic PCR was performed in single clones. Ratios of clones with correct insertion to total grown-up clones were 54/96 (56.3%) for Cas9, 49/96 (51.0%) for Cas9-SB, 75/96 (78.1%) for Cas9-N123 and 80/96 (83.3%) for Cas9-N57 (Figure 1D). Sanger sequencing of the genomic PCR amplicons showed that the junction sites might contain small indels (Figure 1E). Besides the ACTB site, similar phenomena were observed at the GAPDH, PGK1 and AAVS1 sites (Figure 1F and Supplementary Figure S3). Statistical analyses indicated that Cas9-N57 significantly increased the efficiency of targeted integration across all the tested sites. Therefore, we focused on Cas9-N57 in the following experiments. These results demonstrated that the targeted integration mediated by Cas9-N57 was more efficient than that mediated by Cas9.
Cas9-N57 exhibits high genome-wide specificity
To examine the genome-wide specificity of Cas9-N57, unbiased whole-genome sequencing (WGS) was performed with MCF7 cells transfected with Cas9, Cas9-SB and Cas9-N57 that targeted the ACTB locus. As shown in Figure 2A, targeted insertion was detected in all three samples. Ratios of targeted reads to total reads at the ACTB site were 18/26 (69.2%) for Cas9, 23/30 (76.7%) for Cas9-SB and 22/24 (91.7%) for Cas9-N57, consistent with the results of FACS analysis and single-clone identification (Figures 1C–F and 2A). Indels were also detected at the junction sites, 3/18 (16.7%) for Cas9, 6/23 (26.1%) for Cas9-SB and 2/22 (9.1%) for Cas9-N57, with the most abundant indel being a 1-bp insertion (Figure 2B). Whole-genome analysis revealed that the insertion in each sample mapped predominantly to the target site, and ratios of targeted insertion reads to total insertion reads were 18/19 (94.7%) for Cas9, 23/25 (92.0%) for Cas9-SB and 22/24 (91.7%) for Cas9-N57 (Figure 2C). Additionally, no repeated tandem insertion events were observed in the WGS data. Similar WGS results were observed at the GAPDH locus (Figure 2A–C), and one off-target site in a GAPDH pseudogene in chromosome X was confirmed by junction PCR and amplicon sequencing (data not shown). Together, these results implied that Cas9-N57 exhibits high genome-wide specificity.
Figure 2.
Cas9-N57 exhibits high genome-wide specificity. (A) IGV analysis showing junction sites at the ACTB and GAPDH loci. Arrows indicate junction sites. Genomic and insertion regions are indicated with different-colored bars. (B) Comparison of the position and length of insertions/deletions (indels) around junction sites among the Cas9, Cas9-SB and Cas9-N57 groups. The indel ratio (indel reads/total reads) is shown in the inset. (C) Targeted integration and potential off-target integration. The x-axis and y-axis denote chromosomal position and insertion percentage, respectively.
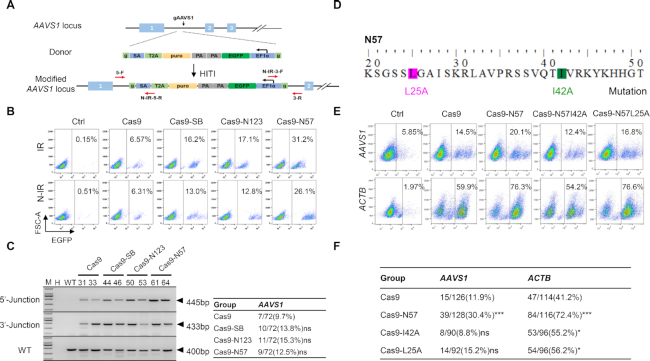
Cas9-N57 mediates integration of large DNA fragments
Efficient targeted insertion of long DNA segments is challenging. To evaluate whether Cas9-N57 could catalyze the integration of a large DNA fragment, a 12-kb DNA donor was designed to be inserted into the AAVS1 site in MCF7 cells (Figure 3A). The donor contained the EGFP coding sequence and an iKA-CRISPR system, consisting of a doxycycline-inducible Cas9-p300 expression cassette that can be used to activate gene transcription and cleave genomic DNA (35). FACS analysis of transfected cells showed that the frequency of EGFP+ cells was 11.4% with Cas9, 14.7% with Cas9-SB and 23.6% with Cas9-N57 (Figure 3B). Targeted insertion was verified in single clones with sequencing of genomic PCR amplicons (Figure 3C and D). Next, a single clone was selected to test the function of the iKA-CRISPR system (Figure 3E). Western blotting results confirmed that expression of Cas9-p300 was dependent on the doxycycline treatment (Figure 3F). We found that 14-bp-long sgRNAs targeting the promoter regions of IL1RN and HBG1 activated the transcription of these genes, whereas 20 bp-long sgRNAs targeting HBG1 cleaved genomic DNA (Figure 3G and H). Collectively, these data indicated that Cas9-N57 could successfully mediate targeted integration of a large DNA fragment.
Figure 3.
Cas9-N57 mediates integration of large DNA fragments. (A) Schematic of the large DNA donor and the strategy for targeted integration at the AAVS1 locus. SA, splice acceptor; T2A, 2A self-cleaving peptides from Thosea asigna virus 2A; TRE, tetracycline-responsive element-regulated promoter; Cas9/p300, Cas9/p300-core-domain fusion protein; M2rtTA, Tet-On advanced transactivator. Primer positions for genomic PCR are shown. (B) Results of FACS analysis of the frequencies of EGFP+ MCF7 cell populations transfected with the indicated plasmids. (C) Genomic PCR and statistics of targeted single clones generated with Cas9, Cas9-SB, Cas9-N123 and Cas9-N57. Primers for the 5′-junction and 3′-junction were 5-F/5-R and 3-F/3-R, respectively. **P < 0.01, ***P < 0.001, Fisher's exact test. (D) Sequence analysis of 5′- and 3′-junctions in representative targeted clones. (E) Results of FACS analysis of the iKA clone 17#. (F) Western blotting analysis showing the time-course expression of doxycycline-inducible Cas9-p300. (G) Inducible activation of gene transcription in iKA clone 17#. Relative mRNA expression of IL1RN and HBG1 was detected by RT-qPCR in MCF7 cells transfected with 14-bp gRNAs targeting the promoter regions of IL1RN and HBG1. Mean values are presented with SD, n = 3. ***P < 0.001, one-way ANOVA test. (H) T7EI analysis showing inducible DNA cleavage in iKA clone 17#. A single 20-bp gRNA was used for the HBG1 locus.
Targeted insertion of the chimeric antigen receptor in T cells with Cas9-N57
Chimeric antigen receptor (CAR)-T cells are a powerful immunotherapeutic tool (46). Genetic cargo delivery systems, including CRISPR/Cas, transposons and viral vectors, have been employed to induce stable DNA integration in T cells (47–49). However, the efficiency of site-specific integration with these systems is relatively low. To address this limitation, CD19-specific CAR was inserted into the AAVS1 ‘safe harbor’ locus in T cells using Cas9-N57 (Figure 4A). The CAR cassette, comprising T2A-Puro-pEF1α-CAR(CD19)-T2A-mNeonGreen, was electroporated into Jurkat T cells and the cells were analyzed by FACS after 30 days. The frequency of mNeonGreen+ cells was 1.70% with the control, 9.74% with Cas9, 16.4% with Cas9-SB and 21.6% with Cas9-N57 (Figure 4B). Consistent with these observations, RT-qPCR revealed that expression of CD19-CAR was higher in Cas9-N57-electroporated cells than in cells electroporated with Cas9 (Figure 4C). As shown in Figure 4D, genomic PCR of single cell clones revealed that Cas9-SB and Cas9-N57 increased the insertion efficiency from 24.1 to 36.3% and to 33.8%, respectively, although only marginal significance was observed (P-values were 0.0495 and 0.1151 for Cas9-SB and Cas9-N57, respectively. Fisher's exact test). Integration at the AAVS1 site was also verified in single clones by sequencing of genomic PCR-generated amplicons (Figure 4E). Together, these results suggested that Cas9-N57 can moderately mediate efficient site-specific insertion of CAR in human T cells.
Figure 4.
Targeted insertion of the chimeric antigen receptor in T cells with Cas9-N57. (A) Schematic of the T2A-puro-pEF1α-CAR(CD19)-T2A-mNeonGreen donor and targeting strategy at the AAVS1 locus. CAR-CD19, anti-CD19 chimeric antigen receptor. Primer positions for genomic PCR are shown. (B) Results of FACS analysis of the frequencies of mNeonGreen+ cell populations in human Jurkat T cells 30 days after electroporation. (C) RT-qPCR of CAR-CD19 expression. Mean values are presented with SD, n = 3. *P < 0.05, ***P < 0.001, one-way ANOVA test. (D) Genomic PCR and statistics of single clones generated with Cas9-N57 electroporation. Primers for the 5′-junction, 3′-junction and WT were 5-F/5-R, 3-F/3-R and 5-F/3-R, respectively. ns: not significant; *P < 0.05, Fisher's exact test. (E) Sequence analysis of 5′- and 3′-junctions in representative clones.
Cancer modeling in mice with Cas9-N57
The above results demonstrated that Cas9-N57 could work in cell culture. Next, the functionality of Cas9-N57 was tested in vivo. We first tested whether Cas9-N57 could work in mouse cells. The DNA cassette T2A-Puro-pEF1α-EGFP was designed to be inserted into the Gt(ROSA)26Sor (Rosa26) locus (Supplementary Figure S4A), a well-documented safe site for DNA insertion (50). FACS analysis revealed that mouse NIH3T3 cells transfected with Cas9-N57 had more EGFP+ cells than those transfected with Cas9 (Supplementary Figure S4B). Next, we used hydrodynamic tail vein injection, a simple, convenient and efficient method of transgene expression (51,52), to deliver Cas9-N57, EGFP reporter donor and sgRosa26 plasmids into the mouse liver (Supplementary Figure S4C). Five weeks after injection, immunofluorescence staining revealed that the injection of Cas9-N57 resulted in more EGFP+ cells than Cas9 (Supplementary Figure S4D and E). These data demonstrated that Cas9-N57 could function in mice.
Cas9-N57 was then employed to model the development of cancer in mice. Because mutations in KRAS (16–18% of cases), TP53 (26–44%) and PTEN (∼3.7%) are the main drivers of intrahepatic cholangiocarcinoma (ICC) (53), we used Cas9-N57 to simultaneously mediate targeted insertion of KrasG12D (a hotspot diver mutant (53)) into the Rosa26 site and disrupt Trp53 and Pten in the mouse liver. To this end, a DNA donor construct that contained a KrasG12D expression cassette, as well as eight sgRNAs targeting the coding sequence of Trp53 and Pten (four for each), were generated. The formation of tumors in the liver was examined 4 weeks after the hydrodynamic tail vein injection of these constructs and Cas9-N57 plasmids (Figure 5A and B). Liver weights were 0.94 ± 0.14 g with the control injection (with all plasmids excluding the nucleases Cas9, Cas9-SB and Cas9-N57), 1.44 ± 0.46 g with Cas9, 2.20 ± 0.86 g with Cas9-SB and 2.60 ± 1.22 g with Cas9-N57 (Figure 5C). The number of surface liver tumors was 3.7 ± 3.0 with Cas9, 8.2 ± 5.5 with Cas9-SB and 12.4 ± 9.5 with Cas9-N57 (Figure 5C). To estimate the targeted insertion rate, quantitative genomic PCR assays were performed but failed. The reason was possibly that the in vivo delivery rate was extremely low, only <5% liver cells were EGFP positive (Supplementary Figure S4D and E) and much less cells were inserted with delivered DNA. Since a tumor could raise from a single mutated cell, tumor growth could reflect the targeted insertion rate of KrasG12D. The grownup tumors exhibited pathological features of bile duct differentiation and were positive for cytokeratin 19 (Ck19), a marker of ICC (Figure 5D). Sequencing of genomic PCR amplicons from tumor nodules demonstrated targeted integration of KrasG12D into the Rosa26 locus and disruption of Trp53 and Pten (Figure 5E and F; Supplementary Figure S5). Collectively, these data demonstrated that Cas9-N57 performed better than Cas9 at simultaneously mediating site-specific KrasG12D insertion and inducing mutations in Trp53 and Pten in the mouse liver, which supported the use of Cas9-N57 for modeling cancer in vivo.
Figure 5.
Cancer modeling in mice with Cas9-N57. (A) Experimental design for liver cancer modeling in vivo. (B) Macroscopic analysis of mouse liver tumors harvested 4 weeks after injection. (C) Quantification of liver weight and surface liver tumor number per mouse. Error bars represent SD. n = 11–13 mice. **P < 0.01, one-way ANOVA test. (D) Representative images of H&E and IHC staining of Ck19 in mouse liver tumors. Scale bar, 100 μm. n = 11–13 mice. (E) Genomic PCR of targeted integration in tumors generated through Cas9, Cas9-SB and Cas9-N57 injection. Primers for the 5′-junction and 3′-junction were R26–5-F/R26–5-R and R26–3-F/R26–3-R, respectively. (F) Sequence analysis of 5′- and 3′-junctions in tumors.
Multiple nuclease-N57 versions based on other CRISPR systems
The Cas9-N57 described in the previous sections was based on SpCas9. To expand the potential applications of this new tool, we generated three new fusion proteins based on eCas9, AsCpf1 (Acidaminococcus sp. BV3L6) and CjCas9 (Campylobacter jejuni) and tested their activity at the ACTB and AAVS1 loci in MCF7 cells (Figure 6A). eCas9 is an enhanced specificity SpCas9 mutant that reduces off-target effects and maintains robust on-target cleavage (54). CRISPR/Cpf1, a class 2 type V CRISPR system, recognizes a T-rich protospacer-adjacent motif (PAM) via a self-processed single crRNA and cleaves the DNA, leaving sticky ends (3). CjCas9 is considerably smaller than SpCas9, which facilitates therapeutic delivery into a wide range of cell types via lentiviral or adeno-associated viral (AAV) vectors (55). For the ACTB site, results of FACS analysis and PCR of single cell clones revealed that eCas9 exhibited no significant effect (Figure 6B and D), while the activity of both AsCpf1-N57 and CjCas9-N57 was clearly increased compared to that of AsCpf1 and CjCas9 alone, respectively (Figure 6C and D). For the AAVS1 site, eCas9-N57 exhibited an increase trend although without statistical significance (Fisher's exact test), and the insertion rates were too low for AsCpf1- and CjCas9-based fusions to be power enough to prove statistical significance although an increase trend was also observed (Supplementary Figure S6). Together, these observations indicated that AsCpf1-N57 and CjCas9-N57 could mediate robust targeted insertion of exogenous DNA, suggesting that this tool has broad potential applications.
Figure 6.
AsCpf1-N57 and CjCas9-N57 enhance targeted integration at the ACTB locus. (A) The schematic structures of Cas-N57, including SpCas9-N57, eCas9-N57, AsCpf1-N57 and CjCas9-N57. (B andC) Results of FACS analysis of the frequencies of EGFP+ cell populations in MCF7 cells 21 days after transfection with the indicated plasmids. (D) Genomic PCR and statistics of targeted single clones generated with the indicated nuclease-N57 plasmids. Primers for 5′-junction and 3′-junction were AS-CJ-ACTB-SB-F2/5-R and AS-CJ-ACTB-SB-3-F6/AS-CJ-ACTB-SB-3-R6 for AsCpf1-N57, AS-CJ-ACTB-SB-F2/5-R and AS-CJ-ACTB-SB-3-F5/ AS-CJ-ACTB-SB-3-R5 for CjCas9-N57. ns: not significant; *P < 0.05, ***P < 0.001, Fisher's exact test.
Mechanism of Cas9-N57
Finally, we explored how Cas9-N57 induced highly efficient targeted integration. Western blotting assay showed that expression of Cas9-N57 was comparable to that of Cas9 (Supplementary Figure S7A). The TIDE assay is a simple and accurate assay to precisely determine the frequency of targeted mutations induced by the CRISPR/Cas9 system (37), and it showed that Cas9-N57 and Cas9 exhibited similar DNA cleavage activity (Supplementary Figure S7B). Thus, it was of minimal possibility that Ca9-N57 achieved robust insertion via increasing protein expression or nuclease activity. N57, through its interaction with the IR of donor DNA, might recruit the transgene close to the target locus to increase knock-in. To test this, donor DNA cassettes containing the T2A-Puro-pEF1α-EGFP fragment with or without IR were designed to be inserted into the AAVS1 locus (Figure 7A). The number of EGFP+ cells decreased when the donor without IR was used: frequencies were 6.57% for Cas9, 16.2% for Cas9-SB, 17.1% for Cas9-N123 and 31.2% for Cas9-N57 in the group with IR and 6.31% for Cas9, 13.0% for Cas9-SB, 12.8% for Cas9-N123 and 26.1% for Cas9-N57 in the group without IR (Figure 7B). Targeted insertion of the donors without IR was further verified in single clones by junction PCR and DNA sequencing of the amplicons, and statistical analyses showed no significant difference between each Cas9 fusion variant and Cas9 alone, although the fusion variants retained relatively high efficiency (Figure 7C). Furthermore, to test the donor recruitment model, we mutated two amino acid residues (I42A and L25A) in the N57 domain that are essential for DNA binding (56,57) (Figure 7D). Cas9 fusion proteins with the two mutated N57 domains had similar protein expression and similar DNA cleavage activity to the WT fusion protein (Supplementary Figure S7C and D). For integration at the AAVS1 and ACTB sites, FACS and PCR of single cell clones analyses revealed that the two mutants obviously and significantly decreased the enhancement effect although residual effect remained at the ACTB site (Figure 7E and F). Collectively, all the above data demonstrated that N57 tethering the donor to the editing site played a major role and other currently unidentified mechanisms might also play a minor role.
Figure 7.
The mechanisms of Cas9-N57-catalyzed targeted DNA insertion. (A) Schematic of the non-inverted repeat (N-IR) donor and targeting strategy at the AAVS1 locus. (B) Results of FACS analysis of the frequencies of EGFP-positive cell populations in MCF7 cells. IR/N-IR, donors with/without IR. (C) Genomic PCR analysis and statistics of single clones generated with Cas9, Cas9-SB, Cas9-N123 and Cas9-N57 with N-IR donor. The primers for 5′-junction, 3′-junction and WT were 5-F/N-IR-5-R, N-IR-3-F/3-R and 5-F/3-R, respectively. ns: not significant, Fisher's exact test. (D) Schematic of the mutations that disrupt the DNA binding activity of N57. (E) Results of FACS analysis of the frequencies of EGFP-positive cell populations in MCF7 cells transfected with the indicated plasmids at the AAVS1 and ACTB loci. (F) Statistics of single clones generated with Cas9, Cas9-N57, Cas9-N57I42A and Cas9-N57L25A. ns: not significant; *P < 0.05, ***P < 0.001, Fisher's exact test.
DISCUSSION
In the study, we present a strategy to tether the DNA donor template to Cas9, which could enhance the efficiency of HITI-mediated transgene insertion. Our results indicate that Cas9-N57 can catalyze efficient targeted DNA insertion in a unidirectional, site-specific and homology-independent manner in both cultured cells and mice (Supplementary Figure S8).
A variety of targeted integration methods mediated by the CRISPR system have been reported (Supplementary Table S2). Most methods are based on HDR and thus achieve precise gene editing without any indels at the junction sites. However, two major disadvantages limit the application of HDR-based methods. One is their well-known low efficiency. The other is that these methods are not accessible to non-dividing cells because HDR is used primarily in the S/G2 cell-cycle phases (58). Donor manipulation is widely used to increase the efficiency of HDR-based methods, such as using ssDNA instead of dsDNA, chemically modifying donor DNA, linearizing the donor in vivo and recruiting the donor to the insertion site (21–22,25,59–60). Efficiency is indeed increased by donor manipulation, but it is still lower than that of NHEJ-based methods. In the current work, we combined the NHEJ-based method HITI with donor linearization in vivo and recruitment of the donor to the insertion site. We demonstrated that the combination of these three strategies achieved highly targeted integration in diverse cell types.
Because HITI can achieve robust targeted DNA integration in a wide variety of adult cell types (including dividing and non-dividing cells) (9), Cas9-N57 might further increase editing efficiency in HITI-based applications. For example, in this study, we used Cas9-N57 to establish reporter cell lines by inserting an IRES-reporter-polyA DNA fragment downstream of the stop codon of endogenous genes or by inserting a reporter gene with an upstream splicing accept site in an intron. Cas9-N57 might also be used to improve the efficacy of gene therapy for retinitis pigmentosa in rat models by inserting a functional copy of exon 2 to restore the function of the Mertk gene, which has potential clinical applications (9,61). Additionally, Cas9-N57 has DNA-cleaving capability and thus can execute knock-in and knockout simultaneously, thereby allowing for more complex applications, like modeling of cancer development in vivo, as proved in this study. Moreover, Cpf1and small Cas9 have been used successfully to construct variants of nuclease-N57, thereby endowing nuclease-N57 with distinctive characteristics. Future optimization of nuclease-N57 using the engineering approaches reported for CRISPR/Cas systems, such as the use of split Cas9, a photochemical inducible system and PAM-changed nucleases, would further expand its potential application (62–64).
For most experiments, Cas-N57 and Cas-SB were used side by side (Figure 1D and F, Supplementary Figure S3C, Figures 3C, 4D and 6D). We noted that Cas-N57 outperformed Cas-SB at most tested sites in different cell lines. In addition, N57 is smaller than SB and thus it is easier for delivery, particularly for lentivirus and adeno-associated virus package. Compared to SB, N57 lacks transposase activity and thus has no chance to introduce potential transposon-mediated off-target insertion. Therefore, Cas-N57 would be a better choice than Cas-SB when targeted insertion is needed.
Cas9-N57 contains the PAI DNA-binding domain of SB, which binds to the IR sequence in donor DNA. Our data in Figure 7 and Supplementary Figure S7 demonstrate that the high efficiency mainly results from the physical drawing of the donor DNA close to the target site. We noted that, although it was not statistically significant, the insertion efficiency of donor DNA without IR mediated by Cas9-N57 was still marginally higher than that mediated by Cas9 alone (Figure 7C). Additionally, we found that N57 mutants without DNA-binding activity almost completely lost the enhancement effect at the AAVS1 site but still retained marginal effect at the ACTB sites when assessed by single clone PCR assay (Figure 7F). These observations demonstrate that recruiting the donor to the target site is the major mechanism and other currently unidentified mechanisms might also be involved to a less extent.
In summary, the fusion protein Cas9-N57 elicits highly efficient targeted DNA integration in both cultured cells and mice and can be used for genome engineering in vertebrate species for basic research purposes and potential clinical gene therapy.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Penghui Zhou from Sun Yat-sen University for providing the CD19-CAR plasmid. We also thank every member of Dr Ying Lin and Dr Zhili Rong's lab for their helpful discussion and suggestions.
Contributor Information
Shufeng Ma, Department of Nephrology, Shenzhen Hospital, Southern Medical University, Shenzhen 518110, China; Cancer Research Institute, School of Basic Medical Sciences, Southern Medical University, Guangzhou 510515, China.
Xinlong Wang, Cancer Research Institute, School of Basic Medical Sciences, Southern Medical University, Guangzhou 510515, China.
Yongfei Hu, Dermatology Hospital, Southern Medical University, Guangzhou 510091, China.
Jie Lv, Cancer Research Institute, School of Basic Medical Sciences, Southern Medical University, Guangzhou 510515, China.
Chengfang Liu, Cancer Research Institute, School of Basic Medical Sciences, Southern Medical University, Guangzhou 510515, China.
Kaitong Liao, Cancer Research Institute, School of Basic Medical Sciences, Southern Medical University, Guangzhou 510515, China.
Xiaohua Guo, Department of Nephrology, Shenzhen Hospital, Southern Medical University, Shenzhen 518110, China.
Dong Wang, Dermatology Hospital, Southern Medical University, Guangzhou 510091, China; Department of Bioinformatics, School of Basic Medical Sciences, Southern Medical University, Guangzhou 510515, China.
Ying Lin, Cancer Research Institute, School of Basic Medical Sciences, Southern Medical University, Guangzhou 510515, China.
Zhili Rong, Department of Nephrology, Shenzhen Hospital, Southern Medical University, Shenzhen 518110, China; Cancer Research Institute, School of Basic Medical Sciences, Southern Medical University, Guangzhou 510515, China; Dermatology Hospital, Southern Medical University, Guangzhou 510091, China; Bioland Laboratory (Guangzhou Regenerative Medicine and Health Guangdong Laboratory), Guangzhou 510005, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [NSFC 81872511 to Z.R., NSFC 81670093 to Z.R., in part to Y.L.]; National Science and Technology Major Project [2018ZX10301101 to Z.R., Y.L., in part]; Frontier Research Program Bioland Laboratory (Guangzhou Regenerative Medicine and Health Guangdong Laboratory) [2018GZR110105005 to Z.R., in part to Y.L.]; Program of Department of Science and Technology of Guangdong Province [2014B020212018 to Z.R.]; Natural Science Foundation of Guangdong Province [2017A030310331, 2018A030313455 to Y.L.]. Funding for open access charge: National Science and Technology Major Project of China [2018ZX10301101].
Conflict of interest statement. None declared.
REFERENCES
- 1. Makarova K.S., Wolf Y.I., Iranzo J., Shmakov S.A., Alkhnbashi O.S., Brouns S.J.J., Charpentier E., Cheng D., Haft D.H., Horvath P. et al.. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020; 18:67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hsu P.D., Lander E.S., Zhang F.. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014; 157:1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A. et al.. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015; 163:759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dominguez A.A., Lim W.A., Qi L.S.. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol. 2016; 17:5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anzalone A.V., Randolph P.B., Davis J.R., Sousa A.A., Koblan L.W., Levy J.M., Chen P.J., Wilson C., Newby G.A., Raguram A. et al.. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019; 576:149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang H., Wang H., Shivalila C.S., Cheng A.W., Shi L., Jaenisch R.. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013; 154:1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gonzalez F., Zhu Z., Shi Z.D., Lelli K., Verma N., Li Q.V., Huangfu D.. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell. 2014; 15:215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. He X., Tan C., Wang F., Wang Y., Zhou R., Cui D., You W., Zhao H., Ren J., Feng B.. Knock-in of large reporter genes in human cells via CRISPR/Cas9-induced homology-dependent and independent DNA repair. Nucleic Acids Res. 2016; 44:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Suzuki K., Tsunekawa Y., Hernandez-Benitez R., Wu J., Zhu J., Kim E.J., Hatanaka F., Yamamoto M., Araoka T., Li Z. et al.. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016; 540:144–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nakade S., Tsubota T., Sakane Y., Kume S., Sakamoto N., Obara M., Daimon T., Sezutsu H., Yamamoto T., Sakuma T. et al.. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat. Commun. 2014; 5:5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yao X., Wang X., Liu J., Hu X., Shi L., Shen X., Ying W., Sun X., Wang X., Huang P. et al.. CRISPR/Cas9-mediated precise targeted integration in vivo using a double cut donor with short homology arms. EBioMedicine. 2017; 20:19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aida T., Nakade S., Sakuma T., Izu Y., Oishi A., Mochida K., Ishikubo H., Usami T., Aizawa H., Yamamoto T. et al.. Gene cassette knock-in in mammalian cells and zygotes by enhanced MMEJ. BMC Genomics. 2016; 17:979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yao X., Wang X., Hu X., Liu Z., Liu J., Zhou H., Shen X., Wei Y., Huang Z., Ying W. et al.. Homology-mediated end joining-based targeted integration using CRISPR/Cas9. Cell Res. 2017; 27:801–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yao X., Zhang M., Wang X., Ying W., Hu X., Dai P., Meng F., Shi L., Sun Y., Yao N. et al.. Tild-CRISPR allows for efficient and precise gene knockin in mouse and human cells. Dev. Cell. 2018; 45:526–536. [DOI] [PubMed] [Google Scholar]
- 15. Xie L., Sun J., Mo L., Xu T., Shahzad Q., Chen D., Yang W., Liao Y., Lu Y.. HMEJ-mediated efficient site-specific gene integration in chicken cells. J. Biol. Eng. 2019; 13:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang J.P., Li X.L., Li G.H., Chen W., Arakaki C., Botimer G.D., Baylink D., Zhang L., Wen W., Fu Y.W. et al.. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol. 2017; 18:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chu V.T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K., Kuhn R.. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015; 33:543–548. [DOI] [PubMed] [Google Scholar]
- 18. Maruyama T., Dougan S.K., Truttmann M.C., Bilate A.M., Ingram J.R., Ploegh H.L.. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 2015; 33:538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Song J., Yang D., Xu J., Zhu T., Chen Y.E., Zhang J.. RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat. Commun. 2016; 7:10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Suzuki K., Yamamoto M., Hernandez-Benitez R., Li Z., Wei C., Soligalla R.D., Aizawa E., Hatanaka F., Kurita M., Reddy P. et al.. Precise in vivo genome editing via single homology arm donor mediated intron-targeting gene integration for genetic disease correction. Cell Res. 2019; 29:804–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Quadros R.M., Miura H., Harms D.W., Akatsuka H., Sato T., Aida T., Redder R., Richardson G.P., Inagaki Y., Sakai D. et al.. Easi-CRISPR: a robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Genome Biol. 2017; 18:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Savic N., Ringnalda F.C., Lindsay H., Berk C., Bargsten K., Li Y., Neri D., Robinson M.D., Ciaudo C., Hall J. et al.. Covalent linkage of the DNA repair template to the CRISPR-Cas9 nuclease enhances homology-directed repair. eLife. 2018; 7:e33761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ma M., Zhuang F., Hu X., Wang B., Wen X.Z., Ji J.F., Xi J.J.. Efficient generation of mice carrying homozygous double-floxp alleles using the Cas9-Avidin/Biotin-donor DNA system. Cell Res. 2017; 27:578–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gu B., Posfai E., Rossant J.. Efficient generation of targeted large insertions by microinjection into two-cell-stage mouse embryos. Nat. Biotechnol. 2018; 36:632–637. [DOI] [PubMed] [Google Scholar]
- 25. Carlson-Stevermer J., Abdeen A.A., Kohlenberg L., Goedland M., Molugu K., Lou M., Saha K.. Assembly of CRISPR ribonucleoproteins with biotinylated oligonucleotides via an RNA aptamer for precise gene editing. Nat. Commun. 2017; 8:1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Aird E.J., Lovendahl K.N., St Martin A., Harris R.S., Gordon W.R.. Increasing Cas9-mediated homology-directed repair efficiency through covalent tethering of DNA repair template. Commun. Biol. 2018; 1:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee K., Mackley V.A., Rao A., Chong A.T., Dewitt M.A., Corn J.E., Murthy N.. Synthetically modified guide RNA and donor DNA are a versatile platform for CRISPR-Cas9 engineering. eLife. 2017; 6:e25312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lim D., Sreekanth V., Cox K.J., Law B.K., Wagner B.K., Karp J.M., Choudhary A.. Engineering designer beta cells with a CRISPR-Cas9 conjugation platform. Nat. Commun. 2020; 11:4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Roy K.R., Smith J.D., Vonesch S.C., Lin G., Tu C.S., Lederer A.R., Chu A., Suresh S., Nguyen M., Horecka J. et al.. Multiplexed precision genome editing with trackable genomic barcodes in yeast. Nat. Biotechnol. 2018; 36:512–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bhatt S., Chalmers R.. Targeted DNA transposition in vitro using a dCas9-transposase fusion protein. Nucleic Acids Res. 2019; 47:8126–8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Klompe S.E., Vo P.L.H., Halpin-Healy T.S., Sternberg S.H.. Transposon-encoded CRISPR-Cas systems direct RNA-guided DNA integration. Nature. 2019; 571:219–225. [DOI] [PubMed] [Google Scholar]
- 32. Strecker J., Ladha A., Gardner Z., Schmid-Burgk J.L., Makarova K.S., Koonin E.V., Zhang F.. RNA-guided DNA insertion with CRISPR-associated transposases. Science. 2019; 365:48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Luo W., Galvan D.L., Woodard L.E., Dorset D., Levy S., Wilson M.H.. Comparative analysis of chimeric ZFP-, TALE- and Cas9-piggyBac transposases for integration into a single locus in human cells. Nucleic Acids Res. 2017; 45:8411–8422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hew B.E., Sato R., Mauro D., Stoytchev I., Owens J.B.. RNA-guided piggyBac transposition in human cells. Synth. Biol. 2019; 4:ysz018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ma S., Lv J., Sun J., Tang P., Li H., Zhou H., Zhang Z., Lin Y., Rong Z.. iKA-CRISPR hESCs for inducible and multiplex orthogonal gene knockout and activation. FEBS Lett. 2018; 592:2238–2247. [DOI] [PubMed] [Google Scholar]
- 36. Deng C.C., Zhu D.H., Chen Y.J., Huang T.Y., Peng Y., Liu S.Y., Lu P., Xue Y.H., Xu Y.P., Yang B. et al.. TRAF4 promotes fibroblast proliferation in keloids by destabilizing p53 via interacting with the deubiquitinase USP10. J. Invest. Dermatol. 2019; 139:1925–1935. [DOI] [PubMed] [Google Scholar]
- 37. Brinkman E.K., Chen T., Amendola M., van Steensel B.. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014; 42:e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Frankish A., Diekhans M., Ferreira A.M., Johnson R., Jungreis I., Loveland J., Mudge J.M., Sisu C., Wright J., Armstrong J. et al.. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019; 47:D766–D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. Genome Project Data Processing, S . The sequence alignment/map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Muramoto N., Oda A., Tanaka H., Nakamura T., Kugou K., Suda K., Kobayashi A., Yoneda S., Ikeuchi A., Sugimoto H. et al.. Phenotypic diversification by enhanced genome restructuring after induction of multiple DNA double-strand breaks. Nat. Commun. 2018; 9:1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Thorvaldsdottir H., Robinson J.T., Mesirov J.P.. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013; 14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mates L., Chuah M.K., Belay E., Jerchow B., Manoj N., Acosta-Sanchez A., Grzela D.P., Schmitt A., Becker K., Matrai J. et al.. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 2009; 41:753–761. [DOI] [PubMed] [Google Scholar]
- 45. Narayanavari S.A., Chilkunda S.S., Ivics Z., Izsvak Z.. Sleeping beauty transposition: from biology to applications. Crit. Rev. Biochem. Mol. Biol. 2017; 52:18–44. [DOI] [PubMed] [Google Scholar]
- 46. June C.H., O’Connor R.S., Kawalekar O.U., Ghassemi S., Milone M.C.. CAR T-cell immunotherapy for human cancer. Science. 2018; 359:1361–1365. [DOI] [PubMed] [Google Scholar]
- 47. Clauss J., Obenaus M., Miskey C., Ivics Z., Izsvak Z., Uckert W., Bunse M.. Efficient non-viral T-cell engineering by sleeping beauty minicircles diminishing DNA toxicity and miRNAs silencing the endogenous T-cell receptors. Hum. Gene Ther. 2018; 29:569–584. [DOI] [PubMed] [Google Scholar]
- 48. Eyquem J., Mansilla-Soto J., Giavridis T., van der Stegen S.J., Hamieh M., Cunanan K.M., Odak A., Gonen M., Sadelain M.. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017; 543:113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Marcucci K.T., Jadlowsky J.K., Hwang W.T., Suhoski-Davis M., Gonzalez V.E., Kulikovskaya I., Gupta M., Lacey S.F., Plesa G., Chew A. et al.. Retroviral and lentiviral safety analysis of gene-modified T cell products and infused HIV and oncology patients. Mol. Ther. 2018; 26:269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999; 21:70–71. [DOI] [PubMed] [Google Scholar]
- 51. Liu F., Song Y., Liu D.. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999; 6:1258–1266. [DOI] [PubMed] [Google Scholar]
- 52. Chen X., Calvisi D.F.. Hydrodynamic transfection for generation of novel mouse models for liver cancer research. Am. J. Pathol. 2014; 184:912–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ong C.K., Subimerb C., Pairojkul C., Wongkham S., Cutcutache I., Yu W., McPherson J.R., Allen G.E., Ng C.C., Wong B.H. et al.. Exome sequencing of liver fluke-associated cholangiocarcinoma. Nat. Genet. 2012; 44:690–693. [DOI] [PubMed] [Google Scholar]
- 54. Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F.. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016; 351:84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kim E., Koo T., Park S.W., Kim D., Kim K., Cho H.Y., Song D.W., Lee K.J., Jung M.H., Kim S. et al.. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017; 8:14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Izsvak Z., Khare D., Behlke J., Heinemann U., Plasterk R.H., Ivics Z.. Involvement of a bifunctional, paired-like DNA-binding domain and a transpositional enhancer in sleeping beauty transposition. J. Biol. Chem. 2002; 277:34581–34588. [DOI] [PubMed] [Google Scholar]
- 57. Yant S.R., Park J., Huang Y., Mikkelsen J.G., Kay M.A.. Mutational analysis of the N-terminal DNA-binding domain of sleeping beauty transposase: critical residues for DNA binding and hyperactivity in mammalian cells. Mol. Cell. Biol. 2004; 24:9239–9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Symington L.S., Gautier J.. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011; 45:247–271. [DOI] [PubMed] [Google Scholar]
- 59. Gutierrez-Triana J.A., Tavhelidse T., Thumberger T., Thomas I., Wittbrodt B., Kellner T., Anlas K., Tsingos E., Wittbrodt J.. Efficient single-copy HDR by 5′ modified long dsDNA donors. eLife. 2018; 7:e39468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yao X., Wang X., Liu J.L., Hu X.D., Shi L.Y., Shen X.W., Ying W.Q., Sun X.Y., Wang X., Huang P.Y. et al.. CRISPR/Cas9-mediated precise targeted integration in vivo using a double cut donor with short homology arms. EBioMedicine. 2017; 20:19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Suzuki K., Izpisua Belmonte J.C.. In vivo genome editing via the HITI method as a tool for gene therapy. J. Hum. Genet. 2018; 63:157–164. [DOI] [PubMed] [Google Scholar]
- 62. Zetsche B., Volz S.E., Zhang F.. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol. 2015; 33:139–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Adli M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018; 9:1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V.V., Nguyen N.T., Zheng Z., Gonzales A.P., Li Z., Peterson R.T., Yeh J.R. et al.. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015; 523:481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.