Abstract
ABC ATPases form one of the largest clades of P-loop NTPase fold enzymes that catalyze ATP-hydrolysis and utilize its free energy for a staggering range of functions from transport to nucleoprotein dynamics. Using sensitive sequence and structure analysis with comparative genomics, for the first time we provide a comprehensive classification of the ABC ATPase superfamily. ABC ATPases developed structural hallmarks that unambiguously distinguish them from other P-loop NTPases such as an alternative to arginine-finger-based catalysis. At least five and up to eight distinct clades of ABC ATPases are reconstructed as being present in the last universal common ancestor. They underwent distinct phases of structural innovation with the emergence of inserts constituting conserved binding interfaces for proteins or nucleic acids and the adoption of a unique dimeric toroidal configuration for DNA-threading. Specifically, several clades have also extensively radiated in counter-invader conflict systems where they serve as nodal nucleotide-dependent sensory and energetic components regulating a diversity of effectors (including some previously unrecognized) acting independently or together with restriction-modification systems. We present a unified mechanism for ABC ATPase function across disparate systems like RNA editing, translation, metabolism, DNA repair, and biological conflicts, and some unexpected recruitments, such as MutS ATPases in secondary metabolism.
INTRODUCTION
The nucleotide triphosphates (NTPs), predominantly ATP and GTP, are used both as substrates for biosynthetic reactions and as the ‘energy currency’ to perform mechanical work in cells. Out of the large number of NTP-binding protein folds at least 10 distinct folds have evolved to use the free energy of NTP binding/hydrolysis to drive biologically useful work. Of these, the P-loop NTPase fold (named after their eponymous NTP-binding sequence motif) is by far the most widespread and diverse, being present in 10–18% of the proteins with globular domains encoded by the genomes of cellular life-forms (1–5). P-loop NTPases catalyze the hydrolysis of the β-γ phosphate bond of a bound NTP. The majority of members of this vast assemblage of proteins use the free energy of this reaction to drive conformational changes in themselves and other interacting molecules across all cellular processes (2–5). Depending on the context of the conformational changes and the rate of NTP hydrolysis, the P-loop NTPases act either as switches that ensure quality control of macromolecular assembly pathways or as molecular motors/engines (2–7).
In the past two decades, comparative genomic studies have shown that the major monophyletic lineages of the P-loop NTPases can be traced to the last universal common ancestor (LUCA) of all extant cellular life forms. This indicates that founding members of the higher-order groups of the P-loop NTPases had already emerged prior to the LUCA (2–5). Comparative sequence and structure analyses have shown that the deepest evolutionary split in the P-loop NTPases divides them into two major divisions: the kinase-GTPase (KG) division and the ASCE (The additional strand, catalytic E) division (3,5). The ASCE division includes several major clades: namely, the ABC, AAA+, PilT, HerA-FtsK, helicase (superfamily 1/2; SF1/2), and RecA-ATP-synthase, along with some smaller lineages (e.g. KAP) (2–5,8,9).
The ABC (ATP-binding cassette) ATPases, which form one of the largest of these clades of the ASCE division are major drivers of energetic processes in cellular systems across all superkingdoms of life (10). These include: (i) transport of molecules across membranes (ABC-type transporters) (11,12); (ii) the assembly of iron-sulfur clusters (the SufC ATPase) (13,14); (iii) mismatch repair (e.g. MutS-MutL complex) (15,16); (iv) nucleotide-excision repair (e.g. UVRA) (17); (v) DNA-looping and other DNA-strand manipulations (18,19); (vi) regulation of chromosome architecture (SMC ATPases) (20); (vii) control of different steps of translation and ribosome assembly (21–23); (viii) assembly of complexes related to mRNA editing (MRB1590/RNA-ABC; hereinafter RNA-ABC) (24).
Since the discovery of the transporter ABC ATPases in the early 1970s (11,25), a wealth of structural and biochemical studies has greatly enhanced our knowledge of these enzymes. More recently, several crystal structures of transporter and non-transporter ABC ATPases with NTP and nucleic acid substrates elucidate their unified mode of operation (17,21–24,26–30), by illustrating: (i) their open (ATP-free) and closed conformations (ATP-bound); (ii) how ATP-binding induces conformational changes aiding their dimerization; (iii) the role of inserts like the zinc (Zn)-hook and the SMC hinge domain in coupling the parts of the P-loop ATPase domain separated by long coiled-coil segments (31). Additionally, several recent studies implementing single-molecule imaging techniques have helped visualize the action of non-transporter representatives such as the Rad50–Mre11 complex or the eukaryotic cohesin and condensin complexes featuring SMC ABC ATPases (32,33).
Over the past two decades, we have systematically surveyed all the major branches of the P-loop NTPases, barring the ABC ATPases, using comparative genomics, sequence and structure analysis (2–5,34). Thus, we have been able to provide a foundation for further biochemical studies on these enzymes and also reconstruct some of the earliest events in the evolution of life, given their extensive diversification before the LUCA. Until relatively recently much of the focus on ABC ATPases has been on the transporters (10,11,35); however, the recent flood of data on other members of the superfamily, especially those involved in nucleic acid dynamics, prompted us to address outstanding questions pertaining to the evolution, biochemistry and the underappreciated diversity of the biology of ABC ATPases. Accordingly, we performed comprehensive sequence, structure, phylogenetic and comparative genomic analyses of the less-understood ABC ATPases and the molecular systems centered on them. Consequently, we provide here: (i) a unified definition of the ABC clade and clarify their higher-order relationships; (ii) identification of previously-undetected lineages; (iii) prediction of previously unreported roles for ABC ATPases; (iv) the trends in the colonization of various functional niches including a major expansion in the context of diverse biological conflict systems.
ABC ATPases ARE MEMBERS OF THE ASCE-DIVISION OF THE P-loop NTPase FOLD
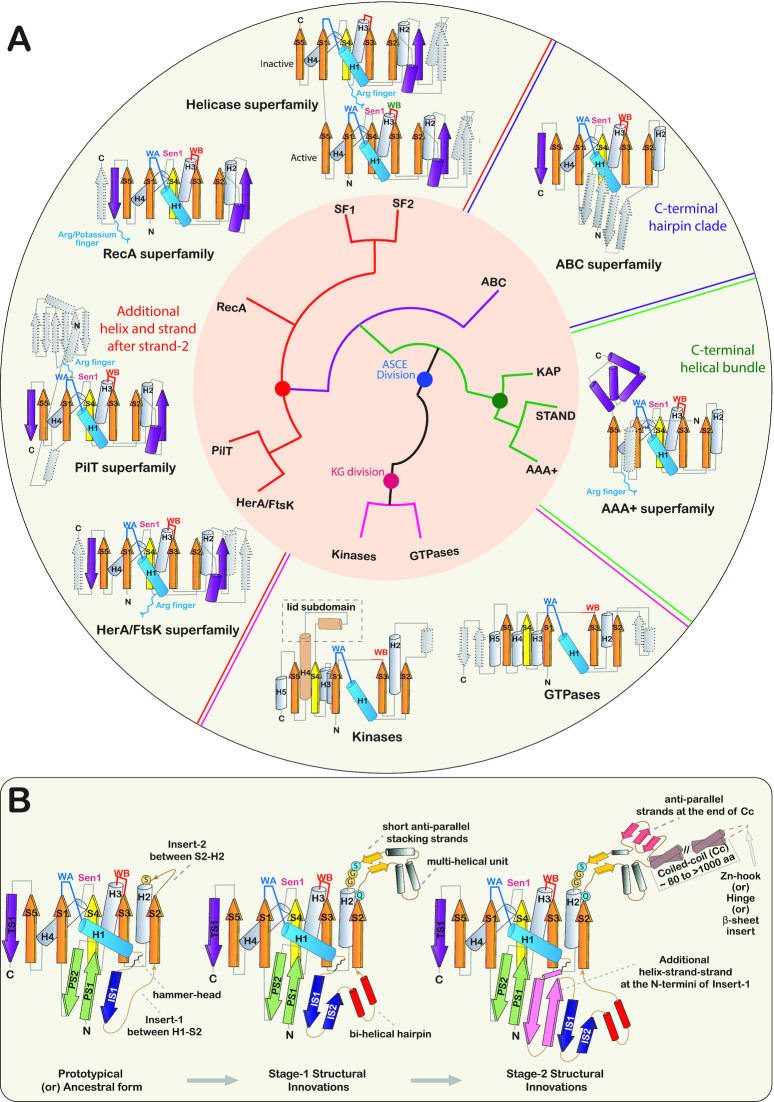
The P-loop NTPase fold is a three-layered α/β sandwich containing regularly recurring α-β units with the β-strands forming a central, mostly parallel sheet, which is sandwiched between α-helices on both sides (2–5) (Figure 1A). The active site is located at the C-terminal end of the core β-strands of the central sheet. The catalytically active P-loop NTPases are characterized by two strongly conserved sequence motifs, the P-loop (phosphate-binding loop; also known as Walker A) and Walker B, which respectively bind the β and γ phosphate moieties of the NTP, and a catalytic Mg2+ cation (6,36). The P-loop, which lies between the first strand and helix of the fold, typically displays the signature GxxxxGK [ST] and wraps around the polyphosphate moiety of the bound nucleotide. The Walker B motif is composed of a conserved aspartate (less often glutamate) at the C-terminus of a hydrophobic strand and provides a bond for the coordination of an Mg2+ cation. The octahedral coordination sphere of the Mg2+ cation is completed by ligands in the form of the β and γ-phosphate moieties of the bound NTP, and the conserved C-terminal serine or threonine residue of the P-loop motif (2–6,36).
Figure 1.
(A) Topology diagrams depicting the major divisions of the P-loop NTPases and accompanying cladogram depicting higher-order relationships. The cladogram depicts the two major divisions: the kinase-GTPase (KG) division and the ASCE (The additional strand, catalytic E division) division. Strands and helices forming the core of ASCE P-Loop NTPase domain are numbered and colored. Core strands and helices are in orange and grey-white, respectively, with central strand S4 in yellow. Synapomorphies shared across different lineages are colored violet, elements not conserved across lineages are colored gray and outlined in dotted lines. Abbreviations: WA, Walker A; WB, Walker B and Sen1, sensor-1. (B) Topology diagrams depicting the structural developments in the evolution of ABC ATPases. The elaborations of the insert regions are depicted.
Based on structural landmarks (Figure 1A), the KG division, which includes the kinases and GTPases, can be distinguished from the rest of the P-loop NTPases by several similarities, most prominently the adjacent placement of the strand preceding Walker A and the Walker B strand (3–5). In contrast, the ASCE division to which the ABC ATPases belong is characterized by (i) an additional strand in the core sheet between the P-loop strand and the Walker B strand; (ii) a conserved proton-abstracting acidic residue (usually glutamate) which primes a water molecule for the nucleophilic attack on the γ-phosphate group of ATP (3,5). In most ASCE clades, including the ABC ATPases, this residue occurs immediately downstream of the Walker B aspartate; (iii) a typical preference for ATP over other NTPs as a substrate.
To objectively define the distinctive features of the ABC ATPases we collated structures of all known representatives of the ABC clade and used them as queries to run DALI searches against the Protein Data Bank (PDB) database. Thus, we retrieved divergent and unannotated representatives including inactive members. We then built a structure-informed multiple sequence alignment of all these representatives of the ABC superfamily to map major sequence landmarks on to the basic template of the ASCE division (Figure 1A). In this mapping, we refer to the core strands of the ASCE group of the P-loop fold as S1–S5 and the helices as H1-H4 (Figure 1A-B). We denote the strands and helices occurring in the insert regions within the P-loop domain unique to the ABC clade as insert strands/helices (IS or IH), those that lie N-terminal to the core domain as ‘preceding strand/helix’ (PS/PH) and those to the C-terminus of the core as ‘terminal strand/helix’ (TS). These are numbered separately from N- to the C-terminus (Figure 1, Supplementary Table S1).
In the ABC ATPase superfamily, the Walker A motif typically takes the form Gx[N/S/T]GxGK[ST][S/T/N] (Supplementary Table S2), with an additional polar residue beyond the core C-terminal alcoholic residue and another conserved polar residue before the second glycine. The Walker B motif retains the ancestral condition inferred for the ASCE clade (12,35,37). However, in a few instances the glutamate C-terminal to the Walker B aspartate is replaced by an aspartate, glutamine, or serine/threonine (see Supplementary Table S2). Of the ASCE NTPases, the core sheet of the ABC ATPases is closest to the AAA+ superfamily in lacking additional strands to the ‘‘right’’ of S2 (Figure 1A) and in this respect differs from the other superfamilies of the ASCE division such as RecA-F1, helicases and PilT ATPases (2,4). It was earlier proposed that the adenoviral packaging ATPase IVA2 might share certain features with the ABC ATPases (9). However, the availability of new sequence data indicates that they are members of the viral and transposon encoded family of the FtsK-HerA clade of the ASCE division rather than being related to the ABC ATPases. ABC ATPases also share with the rest of the ASCE group the Sensor-1 motif defined by a polar residue at the end of S4 (2–4,38), which recognizes the γ-phosphate of the bound ATP and may help hold it in place as a ‘linchpin residue’ (2–4,39). Instead of the S/T/N/D typical of other superfamilies of the ASCE group, ABC ATPases contain a histidine at this position (termed the Sensor-H; Figure 1A, 1B and Supplementary Table S2); hence, the loop downstream of S4 has often been termed the ‘H-loop’ or the switch region (12,35,37). In ABC ATPases, this residue also acts as a ‘gatekeeper’ regulating the access of water to the active site (39).
THE DISTINGUISHING STRUCTURAL AND CATALYTIC FEATURES OF ABC ATPases
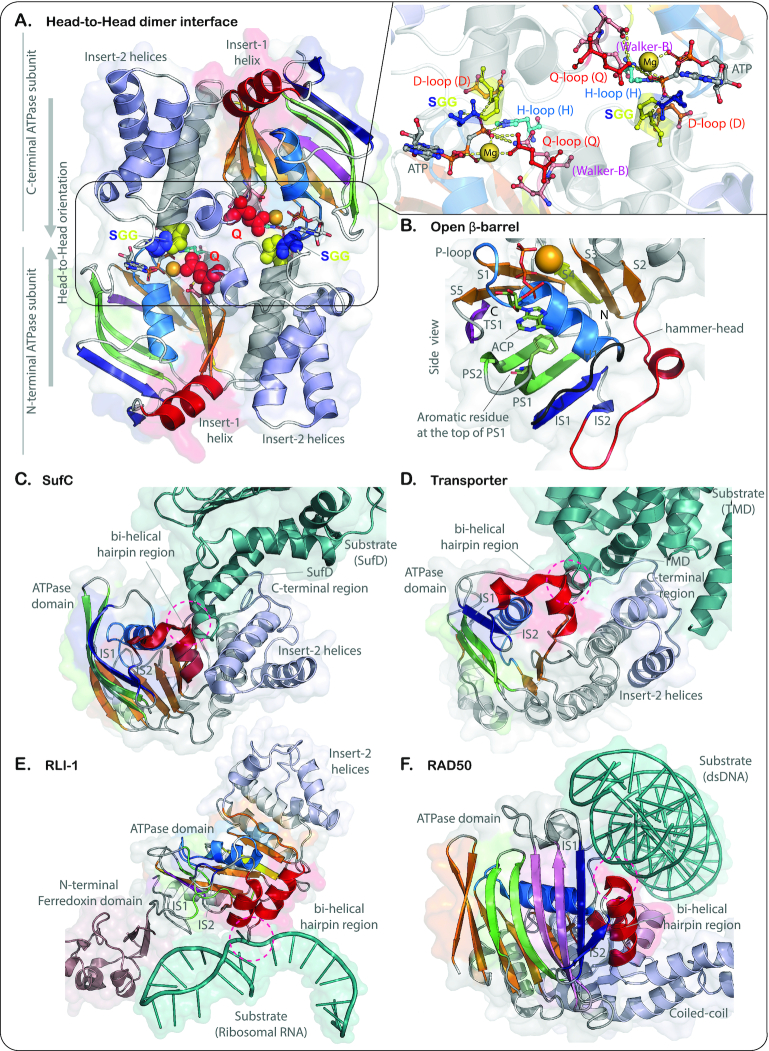
ABC ATPases have several shared derived characters (synapomorphies) setting them apart from the rest of the ASCE division: (i) a conserved glutamine in the loop C-terminal to S2 which is referred to as the Q-loop signature; (ii) a conserved serine that marks the beginning of H2 (or the end of the Q-loop) (12,37) and forms a hydrogen bond with the γ-phosphate of the ATP. In many of the ABC families, this residue is part of a characteristic SGG motif (Figure 1, Supplementary Tables S1 and S2). (iii) the D-loop signature associated with the loop downstream of the Walker-B strand. This loop displays a conserved aspartate (DExxxxxD) that is frequently but not always conserved in ABC ATPases (Supplementary Table S2). (iv) ABC ATPases possess two strands N-terminal to the core domain (PS1 and PS2) that form a β-hairpin (Figure 1). In the loop of this hairpin, a majority of the ABCs possess a conserved aromatic residue (Supplementary Tables S1 and S2), which usually participates in π–π stacking interactions with the nucleobase of the bound ATP and positions it in the active site. (iii) Within the core P-loop domain, all ABCs have an insert between H1 and S2, which we term ‘insert-1’; it varies considerably in length and structural elaboration (Figure 1B). (iv) Similarly, the core ABC P-loop fold shows a second variable insert between S2 and H2, which we term ‘insert-2’. Insert-2 varies in length from less than ten residues to more than a thousand residues and assumes different structural forms (see below, Figure 1B). (v) They also possess a C-terminal strand (TS1), that forms a hairpin with S5 from the core (Figure 1B). Strikingly, the C-terminal S5-TS1 hairpin and the N-terminal PS1–PS2 hairpin stack together with the extended elements from insert-1 to convert the core P-loop sheet into an open β-barrel-like structure through which the H1 is threaded (Figure 2A and B). Such a barrel configuration is exclusive to the ABC ATPases and is not observed in any other superfamily of P-loop NTPases.
Figure 2.
Structural depictions of ABC-ATPase showing: (A) head-to-head dimeric interface (PDB ID: 5X40). (B) open barrel-like structure through which the H1 is threaded (PDB ID: 3FVQ). (C–F) helical element of insert-1 forming a major binding interface for either partner proteins (e.g. SufC (PDB ID: 2ZU0) and transporters (PDB ID: 1L7V)) or nucleic acids (RLI-1 (PDB ID: 5LL6) and Rad50 (PDB ID: 5DNY)). The coloring scheme is as follows: Core strands (S1-S5) (orange); additional strand (S4) within the core-sheet of the ASCE clade (yellow); preceding strands (PS1-PS2) forming the barrel (pale-green); core helices (H1–H5) (gray); P-loop signature (marine); insert-1 strands IS1 and IS2 (dark blue); insert-1 helix or the bi-helical hairpin (red); hammer-head loop (black); Insert-2 helices (blueish white); the partner proteins or nucleic acids (green).
While the three-step reaction cycle of the ABC ATPases broadly resembles other members of the ASCE clade it also shows some unique features. In the first step, the polarized lytic water adopts a bridging position between the general base (glutamate in Walker B) and the γ-phosphate (39). This is followed by hydrolysis resulting in the formation of ADP, HPO42– and a protonated glutamate. Structural and mechanistic studies suggest that the direct transfer of the proton to the catalytic glutamate in ABC ATPases differs from several other KG and ASCE class P-loop NTPases, which might use a second water or the substrates in proton-relay chains (39). In the second step, the proton transfer from the catalytic glutamate to the freed γ-phosphate yields H2PO4– and resets the general base. The third step appears to involve a conformational change in the ABC-specific sensor-H resulting in it moving away to allow increased hydration of the active site, transient change in coordination of the active site Mg2+ and release of the end-products.
Our systematic analysis also affirmed that almost all ABC ATPases, including the newly identified clades, are distinguished from most other members of the ASCE clade, such as AAA+, HerA/FtsK, RecA-ATP synthase, the helicases and PilT, in lacking a key active site feature, the arginine finger (2–4,40). The arginine finger increases the efficiency of ASCE NTP hydrolysis through the stabilization of a negative charge of the γ-phosphate in the pentavalent transition intermediate (transition state of the reaction) (41,42) in both the KG and ASCE divisions of P-loop NTPases. Hence, its absence is puzzling, given that the ABC ATPases efficiently hydrolyze ATP. The answer to this conundrum comes from extensive structural evidence which shows these enzymes to function as obligate ‘head-to-head’ dimers (Figure 2A). This obligate head-to-head dimer allows the conserved serine (from the SGG-like signature) of one subunit to align with the ATP-binding site of the opposite subunit and vice versa. Thus, the sidechain hydroxyl group of the serine and backbone amide groups of the two downstream residues from one subunit are positioned to mold the conformation of the phosphates of the bound ATP of the opposite subunit in the dimer. Earlier studies have hypothesized that this conserved serine might deliver the positive charge of the H2 helix-dipole (43,44). Thus, in the head-head dimeric configuration, this positive charge from the helix-dipole might take the place of the arginine finger to stabilize the reaction intermediate.
In addition to the SGG-like motif, the cooperativity in ATP binding and hydrolysis between the two subunits is also enhanced by the aspartate downstream of Walker-B (‘D-loop’ D), which when present, acts in trans by coordinating a water molecule in the active site of the opposite subunit of the dimer (45,46). Further, studies on multiple ABC ATPases suggest that this cooperativity is key for the performance of mechanical work across diverse systems (7). Thus, the work cycle of the ABC ATPases is envisaged as being initiated by the partner molecules which induce the initial dimerization and cooperative ATP-binding by the ABC dimer. This ATP binding rather than ATP hydrolysis drives the power-stroke of the ABC ATPases (7,39) by inducing conformational changes that may then be relayed as translational motion in the partner molecules. This is followed by ATP-hydrolysis, whose primary role is in reinitiating the work cycle upon release of the reaction products (7,39). ATP hydrolysis by the two subunits appears to occur in an alternating fashion enhancing the kinetics of the work cycle (7).
STRUCTURAL DEVELOPMENTS IN THE EVOLUTION OF ABC ATPases
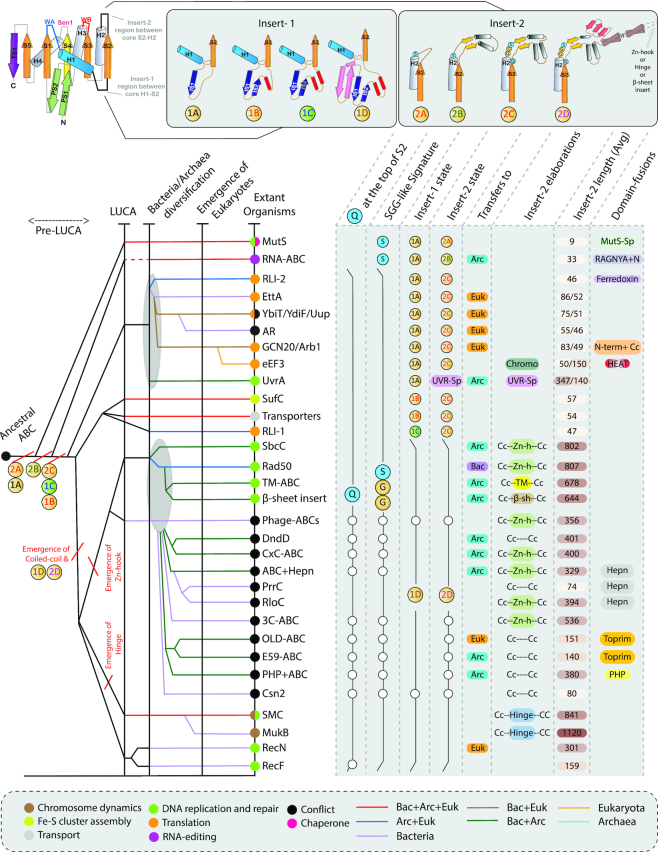
The information gleaned from structure and sequence analysis allowed us to reconstruct the ancestral form of the ABC ATPases (Figure 3). This prototypical form can be visualized as containing the N-terminal PS1-PS2 hairpin, followed by the core domain with the β-sheet (in S5–S1–S4–S3–S2 order) and sandwiching interleaved helices (H1–H4) and the C-terminal TS1 (Figure 1B). The insert-1 and insert-2 are predicted as being in a largely rudimentary form, each likely of less than 10 residues (Figure 1B). In the ancestral state, insert-1 was characterized by a ‘hammer-head’ like loop (47) downstream of H1 followed by the strand IS1 before connecting to the core S2 (Figure 2A). Insert-2 featured two short extended regions flanking a central loop. Further, we posit that the basic dimerization mode, which obviated the need for an arginine finger, was already present in the ancestral ABC ATPase with the conserved serine at the end of H2 playing a key role in the coupling of the two subunits (Figures 1B, 2A). Among the extant members of the ABC superfamily, a form closest to this inferred ancestral state is retained in the MutS family of ABC ATPases (Figure 3). The version of the domain in RNA-ABC and one of the two ATPase domains of the EttA-like families, UVRA, EF3 and RLI-1 (21,22,27,48) are also relatively close to the inferred primitive state (Figure 3).
Figure 3.
Inferred evolutionary history of ABC ATPases. The figure shows several relative temporal epochs associated with certain major transitions marked by vertical black lines. Individual lineages, labeled to the right, are traced to their maximal inferred evolutionary depth by horizontal lines, which are colored by extant phyletic distributions. Broken horizontal lines indicate a lineage cannot be traced beyond that point. Panels on the right show the sequence and structural synapomorphies. Fast-evolving clades with minor deviations in the conserved features are highlighted using white circles and black outlines. Panels on the top show the states of the insert region elaborations. Slanting red lines mark key structural and/or functional transitions.
Our analysis demonstrates that major developments in course of the evolution of the ABC superfamily likely occurred in two distinct stages. The first stage was marked by innovations in insert-1 where an additional short strand (IS2) emerged forming a β-hairpin with IS1. Further, a helical element with a single α-helix (transporter-like ATPases) or two helices forming a bi-helical hairpin (one of the subunits of RLI, RecN/RecF, the Rad50/SbcC -like assemblage and SMCs) emerged downstream of the above elements in insert-1 (Figures 1B and 3). This helical element forms a major binding interface for either partner proteins (e.g. transporters and SufC) or nucleic acids (e.g. one of the domains of RLI, Rad50/SbcC, SMCs) (Figure 2C–F). Likewise, in the first stage, a more elaborate form of insert-2 emerged, characterized by a central unit with 2–4 helices formed by a progression of α-helical hairpins (Figures 1B and 3). The elaboration of insert-2 also provided an interface for the interaction with target proteins or nucleic acids in conjunction with the helical region in insert-1. Notably, these structural developments were also coupled with the incorporation of the conserved serine at C-terminus of H2 into an SGG-like motif and the emergence of a conserved glutamine just downstream of core strand S2 (the Q-loop; Figures 1B and 3, Supplementary Tables S1 and S2) (12,35,37). Given that this glutamine interacts with the SGG motif of the second subunit in the dimer and participates in the coordination of Mg2+ it marked the origin of an enhanced interface for cooperative ATP binding and hydrolysis by the two subunits (35) (Figure 2A). Hence, this set of innovations appears to have aided the efficient performance of mechanical work on proteins or nucleic acids.
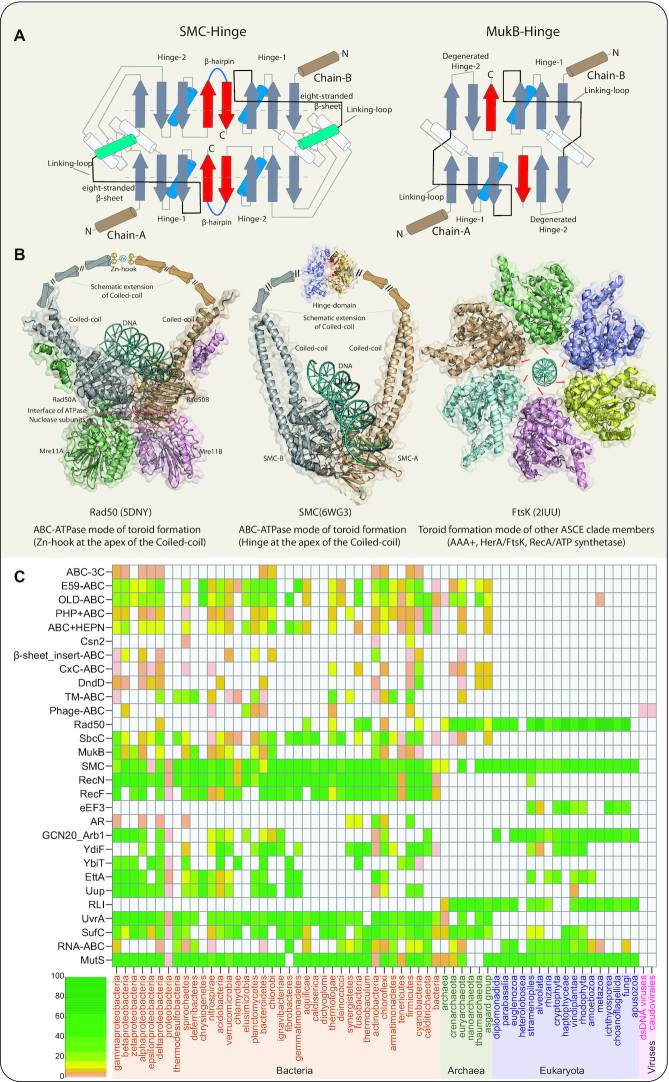
The second major stage of structural innovation was marked by even more dramatic elaboration of the two inserts specifically in the RecN/RecF, Rad50/SbcC and SMC clades (Figures 1B and 3. See below for definitions). The insert-1 region developed an N-terminal ‘helix-strand-strand’ element. The β-hairpin from this element was incorporated into the barrelized sheet of the ABC P-loop domain (Figure 1B). Insert-2 developed a giant insert in the form of a coiled-coil arm comprised of two anti-parallel helices, followed by 3 terminal strands (Figures 1B and 3). Additional lineage-specific elaborations are seen in the apex of this coiled-coil insert (Figure 1B). A Zn-hook module (a split Zn-ribbon domain, ZnR) is inserted into it in the Rad50/SbcC-like clade. In the active dimeric state, each Zn-hook motif of the two monomers contributes two cysteines to reconstitute the tetrahedral coordination of a Zn2+ ion typical of Type-1 ZnRs (49). In the SMC-like clade, the ‘hinge region’ is inserted in this region, and similarly contributes to their dimer interface via hinge-hinge interactions (25,44). In the classic SMC ATPases, the hinge region has two duplicated domains, hinge-I and II, each with a three-stranded β-sheet and two α-helices (31,50). C-terminal to the last strand of the β-sheet, there is a further β-hairpin that is structurally similar to those found in Zn-hooks (Figure 4A). This suggests that the hinge might have developed via insertion of the two hinge domains into a precursor with a Zn-hook, followed by incorporation of that ancestral Zn-hook hairpin to form the five-stranded β-sheet of hinge-II. In the dimer, the interaction of hinge-I and -II from opposite monomers allows it to assume a doughnut-shaped structure with the hinges forming two eight-stranded β-sheets (Figure 4A, left). Based on structural analysis and phyletic patterns determined in this study (see below), we could confidently infer that the bacteria-specific MukB family (51) of the SMC-like clade has undergone a degeneration of the hinge region from the ancestral state (Figure 3) with the degradation of the hinge domain II (Figure 4A). Beyond the Zn-hook and the hinge domains, we also identified other inserts in the same region in some of the novel clades identified in this study; these will be discussed further in the context of functional predictions for the newly identified clades.
Figure 4.
(A) Topological diagrams showing the differences between the SMC hinge domains and the MukB. The insert region between the core strand 1 and 2 is shown in gray. Loops connecting domains 1 and 2 are shown in black. Domain-2 of MukB hinge displays a degenerate version. (B) Structural and(or) schematic representations showing the different modes of toroid formation in the ABC ATPases (first two panels) and the rest of the ASCE clade (Right panel). The arginine fingers situated on the side opposite to the active site are shown as red lines with the nucleic acid being threaded through the central pore of the torus (right panel). (C) Phyletic patterns of the families within the ABC ATPase superfamily color-coded according to the percentage of representation within the specified taxa. Individual boxes correspond to individual taxa grouped by superkingdom, e.g. firmicutes and actinobacteria in the bacterial superkingdom. The taxa names belonging to the 3 superkingdoms of life and the viruses are provided on the X-axis, whereas the Y-axis shows ABC ATPase families/clades.
These inserts in the coiled-coil region are found in the ABC clades specializing in DNA manipulations and represent a convergent evolutionary solution to the problem of forming a hoop-like structure around double-helical DNA (Figure 4B). In the case of RecA, AAA+ and FtsK/HerA superfamilies, a topologically equivalent solution has been attained via the formation of multimeric rings with the help of arginine fingers situated on the opposite side of the active site with the nucleic acid being threaded through the central pore of the ring (2–4) (Figure 4B). Given that the ABC ATPases have a very different head-to-head dimerization mode, they have instead achieved the DNA-encircling hoop-like state via a dimer interface in the coiled-coil insert while retaining the ancestral dimerization mode and coupled ATP-binding of the core P-loop domain (Figure 4B).
IDENTIFICATION OF NOVEL MEMBERS OF THE ABC SUPERFAMILY USING SEQUENCE SEARCHES
ABC ATPases represent one of the most populous clades of ASCE group P-loop NTPases. Typically, free-living organisms have a minimum of 15–20 transporter ABC ATPases. Moreover, nearly all organisms have at least one each of a SufC-like, a translation-related ABC-ATPase and a Rad50/SbcC-like representative. Further, nearly all eukaryotes have at least six SMC proteins per organism. Not surprisingly, much attention has been devoted both in terms of biochemical and structural studies on these universal and abundant representatives. However, previous studies hinted a vast radiation of other ABC ATPases with more restricted phyletic patterns, performing roles other than in transport. For instance, the RNA-ABC clade with a mRNA editing role was only recently characterized in kinetoplastids (24). Hence, we carried out a systematic search for the ABC ATPases with a focus on identifying novel clades with more restricted or sporadic phyletic patterns using iterative sequence profile searches with the PSI-BLAST and JACKHMMER programs and profile-profile comparisons using HHpred. Representatives with structures were then used as seeds for initial iterative searches against the NCBI non-redundant (nr) database; the searches were then transitively expanded using newly identified members. All the candidates recovered in these sequence searches were compared to the above structure-informed multiple alignment to detect the features that distinguished them from other ASCE NTPases and confirm their membership in the ABC clade.
For example, a PSI-BLAST search initiated with an archaeal Rad50 (ARM75002.1: Acidianus manzaensis, crenarchaeota) against the NCBI nr database recovered several bacterial Rad50-like proteins at significant e-values (OGM94115.1, e-value: 4e–19; KPJ83571.1, e-value: 7e–17). These were confirmed with sequence signatures, profile-profile searches and gene-neighborhood analysis as unambiguous members of the Rad50 clade in bacteria as opposed to its bacterial ortholog SbcC. Conversely, transitive searches (a search protocol where a chain of new searches is reinitiated using the most divergent true positives recovered in the first search) initiated with SbcC from bacteria (WP_081412283.1: Tuberibacillus calidus, firmicutes) recovered novel versions of SbcC (e.g. WP_083758457.1, PKL53170.1, PSG97727.1, e-value: 5e–47) from euryarchaeota, crenarcheota and asgardarchaea that were distinct from its archaeal ortholog Rad50. Thus, we were able to identify examples of likely lateral transfers of Rad50 from archaea to bacteria and SbcC in the reverse direction. Further searches using SbcC (WP_030014645.1) and Rad50 (BAL52404.1) as seeds recovered a broad collection of ABC-ATPases with novel features distributed across a wide range of bacteria, containing a conserved cysteine at the end of S4 (see below). As another example, separate searches using a representative of the DndD family of ABC ATPases (WP_056497450.1: Sphingomonas sp,) as query recovered several previously-unidentified versions (ABE52444.1, 3e–68; ABQ26658.1, 3e–09) that, in contrast to the DndD, possess a zinc-hook, typified by an atypical ‘CxC’ signature (Figure 3, Supplementary Table S2).
We ran such searches exhaustively till no new ABC ATPase domains were recovered. All retrieved proteins were then clustered using the BLASTCLUST program and clusters belonging to previously known ABC ATPase groups were separated from the clusters that were not unified to any of the known groups. Multiple sequence alignments were constructed for each of these clusters and their sequence signatures were recorded (Supplementary Table S2, Data). Subsequent structure predictions were performed, and the structural features were utilized to further unify the clusters with known clades or define new ones. Thus, we identified at least 11 previously unidentified clades and also several distinct divergent subclades of previously known clades (Figure 3, Supplementary Table S2).
HIGHER-ORDER RELATIONSHIPS AND EVOLUTIONARY CLASSIFICATION OF THE ABC ATPase SUPERFAMILY
We used the information obtained from the newly identified clades and the above-described structural inferences to establish the higher-order relationships between ABC ATPases based on synapomorphies defining specific groupings. These higher-order groups were also completely reproduced in a dendrogram constructed by average linkage clustering from a distance matrix based on pairwise DALI Z-scores for structure similarity (Supplementary Material). We also combined this information with the phyletic patterns of the clades (Figure 4C) to infer the possible temporal sequence of their emergence (Figure 3).
The basal branches: MutS and RNA-ABC
The version of the ABC ATPase domain found in the MutS clade (15,48) is closest to the ancestral version of the ABC ATPases. It is widely distributed in all three superkingdoms of life suggesting that it was already present in the LUCA. It lacks elaborations in the insert regions and also lacks the glutamine downstream of S2 and has only the conserved serine of the SGG-like motif (Figure 3, Supplementary Tables S1 and S2). These together suggest that it was the first clade to split off from the ancestral ABC ATPase. The next split marks the separation of the RNA-ABC ATPase, which like MutS, lacks any major developments in insert-1 and the glutamine downstream of S2 (Figure 3, Supplementary Tables S1 and S2, Data). However, the insert-2 region of the RNA-ABC ATPase is characterized by two short-antiparallel strands interleaved by two helices which it shares with the remaining ABC ATPases (Figure 3). Interestingly, it exhibits a sporadic distribution across bacteria, archaea and eukaryotes indicative of high evolutionary mobility (Figure 4C). This sporadic pattern of retention might relate to a distinctive self-contained RNA-related role (24) outside of the core RNA processing systems of both eukaryotes and prokaryotes.
The translation factors and UvrA
The next major branch to split off radiated into a clade of translation factor ABC ATPases, which includes the second ATPase domain of RLI (RLI-2), both the domains of Etta-like families, GCN20/ARB1 and eEF3. While none of these have phyletic patterns spanning all three superkingdoms their complementary patterns indicate that they descend from a single ancestral member that was likely present in the LUCA (Figures 3 and 4C). Another member of this assemblage is the bacteria-specific DNA-repair protein UvrA (17,52). These ABC ATPases are unified by the development in the insert-2 region that contains three or more helices forming a multi-helical unit. This assemblage of ABC ATPases also shows a canonical SGG motif in H2, a Q downstream of S2 and a conserved D immediately after the sensor-H (Figure 3, Supplementary Table S2). This suggests that all these well-known features first emerged in the common ancestor they shared with the ‘classical ABC ATPases’.
The ‘classical ABC ATPases’
The remaining members of the superfamily form the ‘classical ABC ATPase’ assemblage which encompasses several major clades namely SufC, the ABC transporters, RLI-1 (the first ATPase domain of RLI), RecF, RecN, SMC-like families and the Rad50/SbcC-like clades. These are unified by a synapomorphy in insert-1, typified by a short strand IS2 after IS1, forming an β-hairpin with it, and a further downstream helix or a bi-helical hairpin (Figure 3). Within this vast assemblage, RLI-1, SufC and the transporters likely separated first from the rest of the assemblage as the helical element in their insert-1 is not strongly elaborated. SufC and likely a single ancestral ABC transporter can be inferred as being present in the LUCA (Figure 3). The rest, which includes RecF, RecN, SMC-like families and the Rad50/SbcC-like families are unified as the coiled-coil clade by the characteristic coiled-coil segment followed by three strands within insert-2. Of these, the RecN and RecF families are the most primitive as they have relatively short coiled-coil segments and lack any elaboration in the apex of the coiled-coil. The remaining families of the coiled-coil clade fall into two major clades: the Zn-hook clade, which includes SbcC, Rad50 and several other related families (some of which secondarily lost the Zn-hook), and the Hinge clade containing the SMC and MukB families. The predominantly bacterial phyletic pattern of SbcC together with the predominantly archaeo-eukaryotic phyletic pattern of its ortholog Rad50 points to a single common ancestor of these being present in the LUCA. The hinge-clade prototyped by the SMC family has representatives in all three superkingdoms of life again indicating a presence in the LUCA (Figure 3).
RECRUITMENT OF ABC ATPases ACROSS DIVERSE FUNCTIONAL THEMES
We combined the above natural classification with previously known functional information, where available, and new functional predictions based on a systematic analysis of conserved gene-neighborhoods and domain architectures of the ABC ATPases. In the following sections, we present this functional mapping along with the logic used for the new functional inferences. The functions of ABC ATPases can be broadly categorized as related to (i) reorganization of protein complexes; (ii) transport; (iii) dynamics of nucleic acids and nucleoprotein complexes. While across these roles the ABC ATPases utilize a strongly conserved work cycle, they have been recruited to different biological functions through combination with other domains in the same polypeptide or independent functional partners.
Thus, certain ABC ATPases have been recruited independently on several occasions to similar functional themes. In other cases, they have captured different functional niches within the same general functional theme such as translation termination. However, some roles, like transmembrane transport, have remained more or less fixed since the earliest period of their appearance. First, we only briefly survey the role of ABC ATPases in the structural reorganization of protein complexes and transport as these functions have been previously considered in multiple studies (7,10,11,37). We elaborate on the nucleic acid-related functions of ABC ATPases as this functional category encompasses several new findings. We focus primarily on the novel inferences that emerged from the systematic analysis in this study and discuss them grouped by function. However, if a closely-related, newly-identified subclade has acquired an entirely new function, we still describe it alongside its better-studied paralogs.
ABC ATPases IN THE ASSEMBLY OF PROTEINS WITH FE-S CLUSTERS AND TRANSPORT
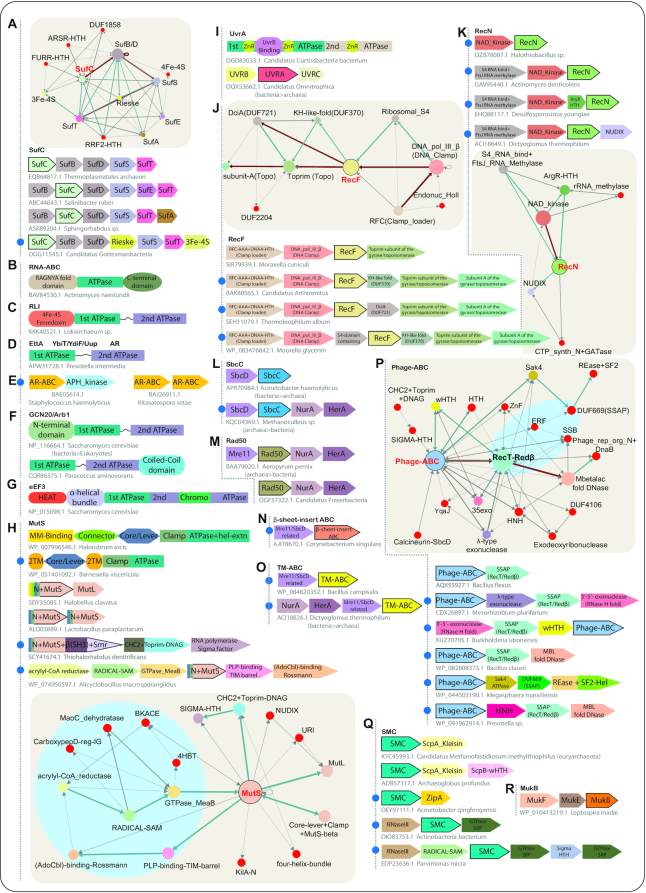
Iron-sulfur (Fe–S) clusters are ancient and versatile inorganic cofactors used across the three superkingdoms of life. Different protein machineries, namely NIF/ISC and SUF, have evolved to mediate the assembly of Fe–S clusters into target proteins (13,53). The components of the SUF system include a two-subunit cysteine desulfurase complex of SufS (a pyridoxal 5′-phosphate-dependent desulfurase), the sulfur shuttle protein SufE (54,55), the homologous parallel-β helix domain subunits, SufB and SufD (14) and the ABC-ATPase SufC (56). These together form a SufB–(SufC)2–SufD complex onto which the Fe–S clusters are transiently assembled (13,14) before transfer to the substrate. The SufB–(SufC)2–SufD complex then transfers the Fe–S cluster to Fe–S carrier proteins such as SufA or SufT that then deliver it to the target (57,58).
Previous studies had shown that the SufC ABC ATPase along with genomically-associated SufB and SufD are widely conserved across bacteria and archaea (13) (Figure 5A). We could also recover the SufC-SufB association in some photosynthetic eukaryotes such as stramenopiles, Rhodophyta and cryptophyta. The phyletic profile of SufC is consistent with its presence in the LUCA, which, along with SufC and the paralogous SufB and SufD, constituted one of the ancestral Fe-S cluster-loading systems. While SufC’s gene-neighborhood association with SufB and SufD is the most widely observed linkage, we also observed linkages to other components of the system such as SufS and SufT in various prokaryotic lineages and sporadic linkages to SufE and SufA (Figure 5A). Further, our analysis recovered conserved gene-neighborhood associations with a gene encoding a Rieske-type ferredoxin domain that coordinates a 2Fe–2S cluster (59) and occasionally another gene coding for a 3Fe–4S type ferredoxin (Figure 5A, Supplementary Material). It is conceivable that these function as alternative Fe–S carrier proteins of the SUF system.
Figure 5.
(A–R) Representative depictions of domain architectures, gene neighborhoods and the corresponding contextual network of ABC-ATPases across diverse functional themes of ‘house-keeping’ and repair. Domain architectures only depict globular domains and the proteins are not drawn to scale. Gene neighborhoods are depicted as box arrows with the multiple domains in each product individually colored. All contexts are labeled with the organism name and NCBI accession. Blue dots denote newly identified systems in this study or systems containing a previously unrecognized component. All domain and organism expansions are provided in Supplementary Material.
In evolutionary terms, the SufC ATPase is specifically related to the ABC transporters, unified by the unambiguous structural synapomorphies in insert-1 (IS1–IS2–H) and insert-2 (multi-helical unit containing three helices) and other shared sequence features (Figure 3, Supplementary Tables S1 and S2). Further, the SufC ATPase interacts with its protein substrates in a manner similar to the interaction of the ABC transporters with their transmembrane subunits (or domains) (Figure 2C and D). This suggests that the basic mechanism of the ATP-driven conformational change in a protein partner was already in place in the common ancestor of the transporters and the SufC family. The exact temporal phase of the origin of the transporter ABCs is difficult to determine due to extensive dissemination of their genes across the genomes of the extant organisms by lateral transfers and rapid lineage-specific evolutionary adaptions for the transport of specific substrates such as xenobiotics and protein toxins. However, as ABC transporters are used by organisms across the three superkingdoms of life (35), it is conceivable that they were already present in the LUCA. Several Fe–S cluster proteins are part of the electron transport chain and operate close to membranes. Hence, it is possible that both SufC and ABC transporters emerged from an ancestral ATPase that catalyzed conformational changes in proteins close to the membrane.
ABC ATPases IN RIBONUCLEOPROTEIN COMPLEXES
The ABC ATPases were recruited independently on at least two distinct occasions to roles in ribonucleoprotein complexes. The first of these is one of the basal branches of the ABC superfamily, the RNA-ABC. The second was the common ancestor of the RLI-EttA, GCN20/Arb1/eEF3 clade. Of these, the translation termination factor RLI family (also called ABCE1 or HP68) is conserved throughout the archaeo-eukaryotic branch of life and the translation elongation regulator EttA and its related families evince a pan-bacterial distribution. Hence, it is conceivable that the LUCA already possessed an ABC ATPase of the RLI-EttA, GCN20/Arb1/eEF3 clade, which had a ribosome-associated function. GCN20 and Arb1 have more complex phyletic patterns indicative of a major role for lateral transfer events in their spread (see below), whereas eEF3 shows a restricted phyletic pattern suggestive of later derivation from one of the more ancient lineages of this clade (Figure 3). While having a role related to the ribosome, the specific functions of RLI, EttA, GCN20, Arb1 and eEF3 are distinct suggesting that they diversified in the different superkingdoms of life to occupy multiple translation-related niches. We briefly discuss below the functional aspects of the multiple recruitments of ABC ATPases to ribonucleoprotein complexes and the notable evolutionary aspects of their radiation in this context.
The RNA-ABC clade
The sporadic phyletic pattern of RNA-ABC across the three superkingdoms of life is indicative of both extensive lateral transfer and gene-loss making its ultimate provenance unclear. However, as noted above, in structural terms it is close to the inferred prototypical ABC protein pointing to an early origin for this clade. Its presence in the basal eukaryotes such as the parabasalid and kinetoplastids suggests that it was probably present in early eukaryotes followed by losses in multiple lineages (Supplementary Data). It shows a unique, strongly conserved domain architecture with fusions to N- and C-terminal domains (24), which have been respectively demonstrated to participate in RNA-binding and dimerization. Through an examination of the structure of the N-terminal domain, we show that it represents an unreported version of the RAGNYA fold (Figures 3 and 5B, Supplementary Data), a domain found in several other nucleic acid-binding contexts (60). Given that the RNA-ABC protein shows no other domain fusions or operonic linkages it is predicted to function as a standalone engine for the ATP-dependent reorganization of ribonucleoprotein complexes. Further, its wide distribution in diverse organisms suggests that it is likely to bring this role to bear in biological functional contexts beyond the specific reported role in kinetoplastid mRNA editing (24,61).
The RLI (ABCE) family
This family shows two ABC ATPase domains connected by a distinctive flexible linker region (Figure 5C). Further, it is distinguished by the fusion to an N-terminal iron-sulfur (4Fe–4S)-cluster-binding ferredoxin domain (Figures 3, 5C) that has been recruited for an unusual role. RLI splits the 80S ribosomes into 60S and 40S subunits (62,63), either after canonical termination by release factors (eRF1/aRF1) or upon recognition of stalled ribosomes by quality-control systems that use the catalytically inactive Dom34/Pelota as a release factor (see reviews: (62,64). Besides this, RLI also engages the 30S post-splitting complex participating in the subsequent steps of ribosome recycling (65). Key to those above functions are the 4Fe–4S ferredoxin domain and the inter-ABC linker: the former undergoes a large structural movement to interact with the 60S protein uL14 and interferes with its role as an inter-subunit bridge (23,62,66). The latter might couple the ferredoxin domain with rRNA interaction (23,26) (Figure 2E). Interestingly, RLI mirrors other release factors in showing a distinction between the archaeo-eukaryotic and bacterial lineages (58,60): bacteria lack it and instead use the evolutionarily unrelated ribosome recycling factor (RRF), which contains a conserved domain shared with several amino-acyl tRNA synthetases (58,60). Indeed, the proliferation of ferredoxin domains in archaea (67) might have allowed the acquisition of this domain by RLI and its specific role in translation termination in the archaeo-eukaryotic lineage.
EttA and related families
Bacterial EttA (also called ABCF) is a regulator of the 70S ribosome in the translation elongation cycle (22), which is characterized by an inter-ABC linker even more developed than in RLI (Figure 5D). Notably, EttA has been lost in cyanobacteria, firmicutes and chlorobi. This is particularly striking because we identified EttA in several eukaryotes, primarily those with photosynthetic capacity, such as chlorophyta, stramenopiles and rhodophyta. Given that EttA is predicted to function in the plastids of these organisms and its absence in cyanobacteria (from which the plastids were derived in early eukaryotes), it appears likely that it was acquired from a distinct bacterial endosymbiont. This provides further evidence for the hypothesis that a second endosymbiont facilitated the establishment of the cyanobacterial endosymbiont as the plastid in the early evolution of photosynthetic eukaryotes (e.g. a chlamydial endosymbiont) (68,69). EttA senses the ATP/ADP ratio in the cells and allows the formation of the peptide bond only when the ATP concentrations are high enough that it can occupy the active site of EttA; if EttA is instead predominantly bound to ADP, it stalls the peptide bond formation and causes termination (22,70). Thus, while EttA does not function as the house-keeping termination factor, it does facilitate the termination of protein synthesis based on ATP availability. This reinforces the proposal that RLI and EttA acquired their specific roles from an ancestral ABC ATPase that had a more generic role in regulating the termination of translation.
Three further, poorly characterized families specifically related to EttA to the exclusion of others were recently described, the Uup, YbiT and YdiF families (71), with Uup and EttA on one side and YbiT and YdiF on the other forming higher-order groupings. Like EttA, YdiF is observed in plastid-containing eukaryotic lineages, representing a second transfer from this group of families to the eukaryotes (71). Unlike EttA, this transfer likely proceeded through an early endosymbiont of the cyanobacteria lineage, given its strong representation in cyanobacteria. In terms of domain architectures, YdiF and Uup to the exclusion of others contain a C-terminal helical extension after the second ATPase domain. The overlapping yet not-mutually-exclusive phyletic distributions of these families with EttA have two key implications: (i) while no single family (with the possible exception of Uup) can be confidently traced to the last bacterial common ancestor (LBCA), taken as a whole at least one representative was present perhaps in the LBCA, which underwent subsequent duplications early in the bacteria. (ii) The uncharacterized families are likely involved in comparable and even potentially overlapping functional roles in translation termination.
There are several other smaller EttA-like families (71,72), with predominant phyletic concentration either in the actinobacteria or firmicutes. Notable among these are the set of ribosomal antibiotic protection or antibiotic resistance families, which have been documented in ATP-dependent ejection of antibiotics that inhibit peptide-bond formation at the ribosome peptidyl-transferase center (PTC) (73–76). Their mode of action resembles EttA in that they bind to the E-site and sense the occupancy of the PTC, thus either directly interacting with and dislodging antibiotics or inducing a change in ribosome conformation resulting in their expulsion (77). Our analysis proposes the monophyly for these families, wherein an initial version was recruited to such a role from the YdiF family (Figure 3). We observed a gene-neighborhood association with an APH-like kinase (Figure 5E), which has been documented to inactivate antibiotics via phosphorylation (78,79). We also observe instances of clustering of multiple copies of these antibiotic resistance families on the genome (Figure 5E). These observations indicate that these families and the associated genes might be part of a multipronged antibiotic resistance system in the bacteria that possess them. Further, the proliferation of numerous paralogous EttA-like families in bacteria might have occurred, as termination of translation was probably a beneficial response in various contexts including metabolite limitation, environmental stresses and biological conflicts (including antibiotics and other ribosome-targeting effectors) (64).
GCN20 (ABCF3), Arb1 (ABCF2) and eEF3
The three translation factors, Arb1, GCN20 and eEF3, form a distinct family within the translation clade with ribosome-associated functions (Figure 3). Of these GCN20 and Arb1 are two close paralogous eukaryote-specific lineages. Their bacterial orthologs are the YheS-like ABCs (71), which are distinguished from their eukaryotic counterparts by the fusion to a C-terminal coiled-coil domain (Figures 3 and 5F). The eukaryotic Arb1 and GCN20 instead show a highly variable N-terminal domain, which in the case of GCN20 interacts with the ribosome-associated protein kinase GCN2 that phosphorylates eIF2 (80,81) (Figure 5F). Almost all bacterial species with GCN20/Arb1 cognates show at least two close paralogous copies. The bacterial GCN20/Arb1-like proteins were likely derived from the pool of EttA-like families in bacteria, which subsequently underwent a late duplication. This was followed by the eukaryotes acquiring two copies of this family, potentially via the alphaproteobacterial mitochondrial progenitor, which then persisted as GCN20 and Arb1. In eukaryotes, GCN20 functions in conjunction with the kinase GCN2 to regulate translation in response to amino acid starvation (82,83). In contrast, Arb1 acts as one of the Ribosome-associated quality control (RQC) factors at the ribosomal E-site to stimulate RQC-specific release factor Vms1 for the release of stalled ribosomes (84,85). However, we observed that the bacterial versions show no specific correlation with either Vms1 homologs or serine/threonine kinases. Hence, it is likely they function via distinct interactions mediated by the C-terminal coiled-coil region.
eEF3 and the related New1 (86) are restricted to eukaryotes and show a sporadic distribution with a presence in chromalveolates, chlorophyta, rhodophyta and opisthokonts. eEF3 is distinguished from other members of this clade by an N-terminal fusion to HEAT repeats and an α-helical bundle and the insertion of a chromo domain within the insert-2 region of the second ATPase domain (21) (Figures 3 and 5G). EF3 acts at the E-site to expel the tRNA after peptide-bond formation during translation elongation and, like RLI, can also separate the ribosome subunits. Further, similar to the ferredoxin domain in RLI (Figure 2E), the N-terminal domains of eEF3 undergo a major structural reorientation and make contacts with the protein SX2 in the head of the 40S ribosomal subunit and the helix 39 of the rRNA in the 60S subunit (21,66), whereas the chromo domain interacts with 60S ribosomal protein rpL5/11 and 5S ribosomal RNA (21,66,70). This functional overlap with RLI has probably led to eEF3’s secondary loss in various eukaryotic lineages such as metazoans. However, in terms of sequence and structure features and a role at the E-site, eEF3 is closer to Arb1 and GCN20; hence, it is likely it was derived as a eukaryote-specific divergent branch from one of those two lineages rather than from RLI.
ABC ATPases IN DNA REPAIR, RECOMBINATION AND DYNAMICS
ABC ATPases have been recruited for roles in core DNA repair and recombination systems on at least three independent occasions. The first of these is represented by the basal-most clade of the ABC ATPases, MutS, which recognizes base mismatches as part of a post-DNA-replication error correction process. The second recruitment, UvrA, occurred in the context of nucleotide-excision-repair. The third recruitment occurred at the base of the coiled-coil clade likely in the pre-LUCA era. Within the latter clade, SbcC/Rad50 and some newly identified clades show a strong association with double-strand break and recombinational repair and are always coupled with a DNase such as SbcD/Mre11. The Hinge clade, with representatives such as SMC, acquired a major role in other aspects of DNA dynamics such as chromosomal organization. We briefly describe below the salient features and genomic associations that went with each of these recruitment events to DNA-related functions.
The MutS clade in DNA repair
The MutS clade is widely distributed across the three superkingdoms of life and certain nucleocytoplasmic large DNA viruses infecting unicellular eukaryotes. The phylogenetic tree topologies suggest the presence of a distinct archaeo-eukaryotic branch and a bacterial branch; however, there have also been several lateral transfers between the superkingdoms (Supplementary Material). A subset of MutS proteins has the GHKL superfamily ATPase MutL as a functional partner (Figure 5H). Our analysis indicates that the MutS–MutL pair is primarily prevalent in the archaeo-eukaryotic lineage and is restricted to the firmicutes and few other lineages among the bacteria. Therefore, the MutS–MutL combination likely originated in the archaeo-eukaryotic lineage and was transferred to bacteria where it spread further through lateral transfers. Such an inference is congruent with the overall phylogenetic tree topology of the MutS clade (Supplementary Material). Notably, the MutS versions that co-occur with MutL in the genome show a distinctive domain fusion to the so-called MM (mismatch-recognition) domain, and the predominantly α-helical connector, core/lever and clamp regions N-terminal to the ABC domain (Figure 5H). In these proteins, the dimeric MutS ABC domain catalyzes the formation of a clamp around DNA for the recognition of the mismatch (16,87). This clamp then recruits MutL to form the MutS-MutL complex that mediates strand discrimination followed by an endonucleolytic cut either via a standalone endonuclease (e.g. MutH) or the unique MutL-associated endonuclease domain (15,48,88).
Beyond the above subclade, there are multiple distinct subclades with other conserved gene-neighborhood associations. One of these combines two genes with MutS ABC domains and is widespread across various prokaryotic lineages. Both the MutS ABC domains encoded by this gene-dyad show several unique sequence features distinguishing them from the MutL-associated versions: while they lack the N-terminal MM-binding and connector domains, they have the so-called core-lever/clamp suggesting that they still form a clamp around dsDNA. This dyad likely forms a distinct heterodimeric complex similar to those seen in the eukaryotes such as MutSα (a heterodimer of MSH2 and MSH6) (89,90) and MutSβ (heterodimer of MSH2 and MSH3). Like the latter complex, they could recognize other forms of DNA-damage such as longer insertions and deletions (91).
A further subclade of the MutS clade (sometimes termed MutS2) is also characterized by the lack of the N-terminal MM-binding and the connector domains but in this case, the ABC domain is fused to a C-terminal β-barrel SH3 fold domain followed by a Smr (small MutS-related) domain. It is conceivable that their SH3-fold and Smr domains might play a lesion-recognition role analogous to the MM-binding domain. The Smr domain has been claimed to possess endoDNase activity (92) while a recent study has shown that this remains uncertain as several versions lack the conserved metal-binding residues that are necessary for such an activity. Nonetheless, several Smr domains have been shown to possess metal-independent RNase activity (93), raising the possibility that this domain might remove RNA primers or mis-incorporated ribonucleotides. Consistent with this proposal, a subset of these occur in a widely distributed conserved gene-neighborhood across bacteria which links MutS to genes encoding a DnaG-like Toprim domain primase and an RNA polymerase Sigma factor (94) (Figure 5H). Further, the DnaG primase might initiate re-synthesis of the DNA during lesion repair. The Sigma factor from this operon might either initiate transcription in response to specific conditions or might facilitate the coupling of this repair activity with transcription by RNA polymerase. Certain MutS2 representatives have also been shown to promote homologous recombination and DNA repair in bacteria (95). Thus, the activity of the MutS ABC ATPase appears to have been recruited for at least 4 distinct DNA repair processes.
A novel membrane-associated MutS subclade
One of the previously unrecognized MutS subclades that we uncovered in this study is widely but sporadically distributed across bacterial lineages such as bacteroidetes, chloroflexi, deferribacteres, fibrobacteres and firmicutes. It is typified by the lack of the MM-binding and connector domains of the classical MutS proteins. Notably, these versions start with a two-transmembrane (2TM) domain just before the core-lever domain and contain a second 2TM domain just N-terminal to the clamp domain (Figure 5H). Based on the location of the pair of 2TM domains we predict that the core MutS ATPase and accessory domains are likely to form an intracellular clamp such that its aperture opens on one side towards the plane of the membrane. We propose that this version represents a novel adaptation of MutS perhaps as a DNA pump that could function in a specialized DNA transfer process or membrane-proximal DNA repair.
Non-DNA repair roles for members of the MutS clade
Strikingly, the second novel subclade we uncovered in this survey points, for the first time, to a non-DNA-related role for a member of the MutS clade. Unlike the above versions, these occur in a conserved six-gene locus found in certain bacteroidetes, elusimicrobia, tenericutes, thermotogae, proteobacteria and firmicutes. In order, this operon codes for: (i) an acrylyl-CoA reductase that is otherwise found as one of the domains of the multidomain propionyl-CoA synthase (PCS) complex that has been recently shown to sequester a toxic intermediate during CO2 fixation (96); (ii) a pyridoxal-5′-phosphate (PLP), S-adenosyl-l-methionine (SAM), and [4Fe–4S]-dependent radical-SAM enzyme predicted to function as a lysine-2,3-aminomutase; (iii) a metallochaperone MeaB GTPase, which is involved in the assembly of the vitamin B12-dependent enzymes (97,98); (iv) MutS; (v) a PLP-binding TIM barrel enzymatic domain; (vi) An adenosylcobalamin (AdoCbl)-binding Rossmann fold domain protein (Figure 5H). The latter two proteins are known to constitute a hetero-tetrameric lysine 5,6-aminomutase complex (99,100). These proteins are likely involved in the SAM, PLP and AdoCbl-dependent synthesis of an amino acid (likely lysine)-derived metabolite where the GTPase acts as a metallochaperone for loading the B12 cofactor on to the subunit of the lysine 5,6-aminomutase. By analogy to its role in DNA-repair, we propose that the MutS protein in this system might play the role of an assembly factor of the whole multi-protein enzymatic complex in an ATP-dependent manner.
UvrA
While nucleotide-excision-repair (NER) is seen in all three superkingdoms of life, their core components have evolved independently in each of the superkingdoms (101,102). The bacterial NER system is comprised of the UvrABC complex containing the UvrA ABC ATPase, the UvrB SF-2 helicase and UvrC with two distinct endonuclease domains. An operon combining the genes for these three proteins is conserved throughout most bacterial lineages and has also been transferred sporadically to few archaea (Figure 5I, Supplementary Material). UvrA has two tandem ABC ATPase domains and has acquired inserted domains in the multihelical element of the insert-2 region (17,52,103) (Figures 3, 5I). In the first of the two ABC-ATPase domains, there are two sets of inserted domains in this region, i.e., the α/β fold UvrB-binding domain, which is itself flanked by two type-I Zn-ribbon domains. The insert-2 region of the second ABC ATPase lacks the UvrB-binding domain but has one Zn-ribbon. In addition, our sequence analysis reveals that the inserted domains of the first ABC unit were lost multiple times suggesting that the UvrB-binding domain might be dispensable for the core function in NER. Our searches also uncovered hitherto unreported versions of UvrA, with four ABC ATPase domains within the same polypeptide in several prokaryotic lineages. These lack operonic linkages with UvrB and UvrC raising the possibility that they function independently of the UvrABC complex.
RecF-mediated DNA repair systems
RecF is one of the most primitive of lineages within the coiled-coil insert-containing clade. Its pan-bacterial distribution without conserved representation in archaea or eukaryotes raises the possibility that it emerged along with RecN (see below) from a basal pre-LUCA member of the coiled-coil clade (Figure 3). These appear to have evolved as part of a radiation of early systems for the recombinational repair of DNA breaks and gaps that probably became important with growing genome size (104–106). RecF is part of a repair system along with RecO and RecR (an inactive Toprim domain protein). This system repairs ssDNA gaps that might arise from incomplete DNA replication on faulty templates (reviewed in (107,108)) by facilitating recombination (109). RecF has also been demonstrated to operate in the repair of DSBs in bacteria that lack RecBCD homologs (107). RecF-mediated homologous recombination and subsequent NER component-based repair have also been implicated in relieving stalled replication forks caused by DNA ADP-Ribosylating toxins (110). RecF is part of a conserved gene-neighborhood widespread across various bacterial lineages, which additionally codes for the clamp loader AAA+ ATPase, responsible for loading the DNA sliding clamp (DNA polymerase III β subunit) (2), and a Toprim subunit of the gyrase/topoisomerase family (Figure 5J). Further, in these neighborhoods, RecF might be linked to three distinct genes coding for proteins with DUF370, DUF721 domains and a single-strand nucleic acid binding S4 domain protein (Figure 5H). Our analysis showed that the DUF370 has a KH-like fold implicated in single-stranded nucleic acid binding while the DUF721 is the recently characterized DciA (dna[CI] antecedent) found to be a widespread bacterial protein required for loading the replicative helicase onto DNA (111). Of these, DciA has the most widespread association with RecF found across various bacterial lineages, while S4 and DUF370 were more restricted (Supplementary Material). It is conceivable that the DUF370 and S4 domains might bind single-stranded DNA or an RNA primer after DciA facilitates assembly of the functionally linked replicative helicase and it unwinds the DNA.
RecN-mediated repair systems
RecN shows a pan-bacterial distribution and our survey also uncovered transfers to eukaryotes such as stramenopiles and the viridiplantae. Its phyletic distribution is comparable to RecF – indeed, both RecF and RecN might have emerged via a duplication event from a common precursor in the ancestral bacterium (Figure 3). Consistent with this, RecN interacts with both the RecFOR- and RecBCD-dependent bacterial recombinational repair pathways and is among the first responders to DSBs (19). RecN also physically interacts with RecA and stimulates RecA’s DNA strand invasion via its ATPase activity (106). We identified an almost absolutely conserved genomic association of RecN with the NAD+ kinase (NADK) (Figure 5K) across numerous bacterial lineages suggesting that it is most likely an ancient association going back to the ancestral bacterium. In addition to the core RecN-NADK association, in several bacterial lineages, it may also show a further operonic association with the FtsJ RNA methylase with an S4 RNA-binding domain. Other less conserved associations might extend this operonic linkage to include the ArgR-family of HTH domains and a Nudix phosphoesterase also implicated in NAD+ processing (Figure 5K). These associations are enigmatic because, unlike RecN, none of these proteins have a direct role in DNA repair. NADK phosphorylates NAD+ to synthesize the metabolite NADP+ whereas FtsJ catalyzes the 2′-O-methylation of the sugar of the universally conserved U2552 in 23S rRNA (112,113). It is conceivable that the linkage of these housekeeping functions to a DNA repair protein RecN plays a role in sensing stress conditions that might pose challenges to genomic integrity.
SbcC/Rad50-dependent double-strand break repair systems
In extant organisms, double-strand DNA breaks are repaired by either homologous recombination (114), or one of several non-homologous (microhomology dependent) end-joining pathways (115). The mainstay of the recombinational repair system in the LUCA can be confidently reconstructed as having a RecA-like recombinase, i.e., the precursor of RecA, RadA, Rad51 and DMC1, which was involved in homologous recombination, and the hetero-tetrameric DNA-end resection complex comprised of an SbcC/Rad50-like ABC ATPase and SbcD/Mre11 nuclease of the calcineurin-like phosphoesterase superfamily (116) (Figures 3, 5L-M). The descendants of this ancestral ABC ATPase with a Zn-hook in the apex of the coiled-coil region are nearly universally conserved across the three superkingdoms of life (Figure 3, Supplementary Material). The Rad50–Mre11 complex acts as a double-strand break sensor in archaea and eukaryotes (18,117). A characteristic feature of this complex is its structural reorganization in conjunction with end-recognition which results in the Mre11 dimer being positioned to process the DNA end (118). Likewise, the bacterial SbcC-SbcD forms a complex that performs comparable roles in DNA-double strand break repair, especially when the bacterial RecBCD system is not functional (119,120). In archaea, the Rad50–Mre11 system shows a genomic linkage to the genes coding for the ATPase HerA and the nuclease NurA that generate 3′ overhangs in DNA required for the subsequent repair steps (121) (Figure 5M).
As noted above, we found examples of transfers of the Rad50–Mre11 system to bacteria and the SbcC-SbcD system to various archaea. The affinities of these transferred versions were confirmed with a phylogenetic tree that includes comprehensive representation of Rad50 and SbcC (Supplementary Material: Tree file). In the former case (e.g. in cyanobacteria, proteobacteria, spirochaetes, thermobaculum, and chloroflexi) the transferred Rad50–Mre11 element additionally includes genes for the HerA–NurA dyad suggesting that a complex similar to that in archaea is also formed in the bacteria with these genes. Interestingly, the SbcC–SbcD system transferred to the archaea acquired associations with the HerA–NurA dyad in archaea and these together were transferred again back and disseminated across several lineages of bacteria (Figure 4C). Taken together, these findings suggest back and forth transfers between bacteria and archaea in the SbcC–Rad50 clade after their initial separation and that both subclades were able to constitute complexes with HerA–NurA in both the archaeal and bacterial superkingdoms (Figure 5L and M, Supplementary Material).
The β-sheet insert and TM clades: novel DSB repair systems
We identified two related, novel clades that share specific sequence and structure features with the SbcC/Rad50 clade (Supplementary Table S2) including a similarly sized coiled-coil region of around 650 amino acids residues (Figure 3). The two clades are united by a modified sensor motif (H-loop) usually lacking a histidine and typically of the form [T/S]C, which also distinguishes them from the canonical SbcC/Rad50 proteins. Moreover, they also show displacement of the Zn-hook at the apex of the coiled-coil by either of two distinct domains respectively in the two clades. The first clade displays a β-strand rich domain (the β-sheet insert clade) (Figure 5N) and the second a 2TM-domain (TM-clade) (Figure 5O). They are both sporadically but widely distributed across diverse bacteria and are also in a few archaeal lineages – a pattern suggesting extensive lateral mobility. Like SbcC/Rad50, both of these clades retain a strong operonic linkage to a SbcD/Mre11-related nuclease. Beyond the two-gene core of these operons, a few representatives of the TM clade from archaea (crenarchaeota and thaumarchaeota) and bacteria (dictyoglomi) are encoded in more extended operons with further associations with genes for the NurA nuclease and the HerA ATPase (Figure 5O). These associations suggest that these clades probably evolved via rapid divergence from the more widespread Rad50 of archaeo-eukaryotic provenance (Figure 3). Based on their contextual associations and relationship to the SbcC/Rad50 clade, we predict that these clades might also function in DNA double-strand break repair. The insertion of the 2TM suggests that the TM clade might specifically mediate repair in a membrane-anchored fashion, perhaps during chromosome segregation or acquisition of DNA via transformation or conjugation.
ABC ATPases mediating homologous DNA-recombination in bacteriophages
As members of the higher-order Zn-hook clade, most of them possess a Zn-hook (Figure 3); however, this domain has degenerated to a non-Zn-binding state in some representatives. They are found in large DNA bacteriophages of the caudovirales assemblage or their prophage derivatives in certain bacterial genomes (122). They are typically encoded close to the viral replication origins and are characterized by operonic associations with single-strand DNA annealing proteins (SSAPs) that play a key role in homologous DNA recombination (122,123) (Figure 5P). The most common of these gene-neighborhoods includes a two-gene core coding for an ABC ATPase and an SSAP of the RecT/Redβ family. A set of operons lack the RecT/Redβ association; here the ABC gene is instead linked to genes encoding DUF669 and the Sak4 ATPase of the RecA family (124). A closer examination suggests that the DUF669 is a SSAP with a permutated version of the RNase H fold comparable to the RecT/Redβ domains (Supplementary Material). This ABC ATPase–SSAP dyad may be combined with several alternative genes in different viruses coding for distinct nucleases such as (Figure 5P): (i) a λ-type exonuclease with the REase fold (122,125). (ii) A 3′-5′-exonuclease typified by the Escherichia coli ExoI, a member of the RNase H fold (126). These gene-neighborhoods additionally encode a winged HTH (wHTH) protein that is elsewhere fused to the FtsK–HerA superfamily ATPases (127). (iii) A metallobetalactamase (MBL) fold DNase (128). A further association with a potential DNase of the HNH fold is sometimes observed across these nuclease types (Figure 5P). Despite their diversity, these systems are predicted to be functionally similar as they are specifically found in phages with linear chromosomes that face the DNA-end problem following replication due to removal of the RNA primer. The ABC ATPase with the associated nuclease and SSAPs help solve this problem via recombination, with the SSAPs promoting single-strand annealing often in conjunction with the associated RecA-like Sak4 ATPase by binding to the 3′' DNA overhangs generated by the associated nucleases (129). These nucleases likely form a complex with the cognate ABC ATPase, as observed in the case of Rad50–Mre11, and with the latter bridging the homologous chromosomes with its coiled-coil region (Figure 4B).
The Hinge clade: ABC ATPases in chromosome organization
With the growth in genome size, one of the major problems faced by cells was the packaging of the genomic DNA into a submicron space. ABC ATPases of the Hinge clade, which appear to have emerged by the LUCA (Figure 3), were key players in the solution to this problem (130–132). Prokaryotes code for a protein that is likely close to the ancestral version of this clade in the form of the single SMC protein found across archaea and several bacterial lineages. This single SMC ABC ATPase underwent proliferation to give rise to six paralogs in the ancestral eukaryote. In prokaryotes, a common genomic association links the SMC gene with that coding for its partner protein ScpA, the archetypal member of the kleisin superfamily, which contains a helical N-terminal domain linked to a C-terminal wHTH domain (133,134) (Figure 5Q). In some cases, there is a further association with a gene coding for a second partner protein ScpB (135,136), which contains a tandem duplication of a wHTH domain (137–139) (Figure 5Q). This suggests that even in the LUCA the SMC ABC ATPase likely formed a complex with a kleisin, which is still retained in the form of the single SMC complex of the prokaryotes. The eukaryotic paralogs form heterodimeric cores of complexes with different roles such as cohesin (Smc1–Smc3) involved in chromosomal cohesion, condensin (Smc2–Smc4) involved in chromosome compaction and the Smc5-Smc6 complex maintaining genome stability through replication and segregation (20,130,134). These combine with similarly diversified members of the kleisin superfamily: Scc1 with cohesin, Brn1 with condensin and Nse4 for Smc5-Smc6 complexes (20,140).
Apart from ScpA and ScpB, the SMC ABC-ATPases are often genomically linked in several bacterial lineages with one or both GTPase subunits of the signal recognition particle (SRP) ribonucleoprotein (141) and RNaseIII (Figure 5Q). The significance of this linkage is not immediately apparent due to the functional disparity of the SRP and RNaseIII with the SMCs. However, it is conceivable that association with the SRP may play a role in anchoring the SMC complex to the membrane during chromosome segregation. The associated RNaseIII might have a role in the processing of the SRP RNA. This association with cell-division and segregation is also reinforced by the proteobacteria-specific genomic association with ZipA (Figure 5Q), which is a membrane protein with a large cytosolic C-terminal region (142) involved in the formation of a proto-ring which marks the septum site during cell-division (142).
Several gammaproteobacteria have a second SMC paralog, MukB (Figure 3), and we found an extensive but sporadic presence of MukB homologs in several other bacterial lineages (Figure 4C, Supplementary Material). MukB shows an operonic association with two other genes MukE and MukF which code for wHTH domain proteins and function as partners of MukB (143,144) analogous to the kleisins and ScpB (145,146) (Figure 5R). Notably, MukB and its partners are faster evolving relative to the SMCs and show degeneration of one of two copies of the Hinge domain in their hinge regions (Figure 4A). Moreover, they show extensive dissemination via lateral transfer. These features suggest that MukB branched off from the more conserved and widespread SMCs with which they sometimes co-occur in conserved gene-neighborhoods. Accordingly, we propose that the MukBEF system might be a mobile ‘selfish’ version of the SMC system that might have caused an ‘addiction’ for their maintenance in certain bacteria by taking over a role in chromosome segregation.
ABC ATPases IN BIOLOGICAL CONFLICT SYSTEMS
Comparative genomics and follow-up biochemical and biological studies in the past two decades have revealed a remarkable diversity of biological conflict systems. These may be broadly defined as systems that are deployed in conflicts that occur at all levels of biological organization including: (i) within genomes between integrated selfish elements and the genomes they reside in; (ii) between different competing genomes in the same cell; (iii) between different organisms of the same species; (iv) between organisms of different species. The last category spans the entire range of interactions from host–parasite to predator-prey interactions. Common to all these levels of biological conflicts is the use of protein armaments typically termed effectors, that target other biomolecules in conflict situations as part of regulated larger biomolecular assemblies (147). These are usually enzymatic (catalytic effectors (148,149) or sometimes non-catalytic (membrane targeting effectors which form pores (150,151). Examples of such include the toxin-antitoxin (TA) involved intra-genomic conflicts, restriction-modification (RM), CRISPR/Cas systems and diverse nucleotide-activated effector systems deployed against invasive nucleic acids, polymorphic- and related toxin systems used in inter-organismal conflicts. A key feature of these systems is an extraordinary array of regulatory components that either prevent the premature deployment of the effectors or their deployment against self-targets or their recycling after deployment.
It has become increasingly clear that the P-loop NTPases of practically every higher-order assemblage have been deployed either as effectors or as regulatory components of biological conflict systems (9,64,122,148,149,152–156). Some of these roles include (i) toxic effectors (e.g. P-loop kinases); (ii) recycling chaperones, e.g. the classical AAA+ ATPases in the Type-6 and PVC secretion systems; (iii) ATPase components involved in DNA-protein dynamics in RM and CRISPR/Cas systems, e.g. SF1 and SF2 helicases, AAA+ and FtsK/HerA ATPases; (iv) regulators of effectors, e.g. AAA+ of the STAND subclade and certain GTPases; (v) transporters, e.g. ABC transporters and FtsK/HerA ATPases which are the ATP-dependent engines of effector transport respectively in the Type-1 and Type-7 secretory systems.
Notable among these conflict systems are the RM systems, which are typified by a modification component that modifies the self-DNA (usually via methylation of adenine or cytosine) and a restriction component, usually an endonuclease, that cleaves unmodified non-self or foreign parasitic DNA (e.g. viruses or plasmids) to protect the host cell against their deleterious consequences (157,158). It has become clear in the past decade that ABC ATPases play a more direct role in RM and standalone restriction-only or modification-only systems as remodelers of DNA-protein complexes and as sensory switches that regulate RM-associated RNA-targeting effectors, e.g. PrrC and RloC (154,155,159–161). Our systematic analysis revealed that these systems are just the tip of the iceberg of an extensive radiation of conflict-related ABC ATPases participating in various aspects of the defense against invasive nucleic acids and also self-nucleic-acid-targeting responses that serve as a backup for the failure of mainline defensive mechanisms like the RM or CRISPR/Cas systems. Notably, all of the ABC ATPases in conflict systems have been recruited from the coiled-coil clade and within it, most representatives can be clearly shown to belong to the Zn-hook clade. This raises the possibility that the vast radiation ABC ATPases in conflict systems probably derives from a single or few ancestral versions. We describe below the numerous recruitments of ABC ATPases to conflict systems.
Inactive ABC ATPases facilitate the acquisition of spacer DNA in type-II CRISPR/Cas systems
The Csn2 protein is one of several accessory proteins that are found as part of certain CRISPR/Cas systems, which are prokaryotic adaptive restriction systems that target invasive nucleic acids using complementary guide RNAs (crRNAs/tracrRNA) synthesized from DNA segments acquired from genomes of invaders (spacers) (162,163). Csn2 is specifically encoded by type-II CRISPR/Cas systems, where it occurs along with the genes for the characteristic Cas1 and Cas2 proteins of CRISPR/Cas systems and the diagnostic Cas9 restriction component of type-II CRISPR/Cas systems with RNase H- and HNH- superfamily nuclease domains. Within these gene-neighborhoods, Csn2 is specifically linked to Cas1–Cas2 with a strong preserved ‘Cas1-Cas2-Csn2’ gene order (Figure 6A). Indeed, a complex formed by Cas1–Cas2–Csn2 (the spacer-acquisition complex) interacts with the restriction component Cas9 via Csn2 to form a single multimeric complex (164,165). However, Csn2 has been shown to be primarily involved in the spacer-acquisition (immunization) stage along with the core components Cas1 and Cas2 and is dispensable for the maturation of the crRNAs and the cleavage of the target DNA (166,167). In this context, Csn2 forms a homotetrameric toroid around DNA, facilitating the integration of spacers by end-joining upon cleavage of the integration site by Cas1 integrase (168).
Figure 6.
(A–C) Representative depictions of gene neighborhoods of diverse biological conflict systems centered on Csn2, DndD/CxC and 3C-ABC-ATPases. The corresponding contextual networks based on the gene-neighborhoods are shown. The system of labeling is as in Figure 5.
Csn2 is confidently predicted to be an inactive ABC ATPase because of the complete erosion of the sequence features such as the P-loop, Walker-B and sensor-H motifs that are characteristic of the ABC superfamily (169,170). However, it retains the diagnostic structural features of the ABC superfamily such as the characteristic N-terminal preceding strands and insert-1 region (containing IS1–IS2 and a helix; Figure 3, Supplementary Tables S1 and S2). The recently solved crystal structure of Csn2 confirmed our proposal that Csn2 is an ABC ATPase, which specifically belongs to the coiled-coil clade (Figure 3) (165,169). These observations suggest that like the cellular coiled-coil ABC ATPases (SbcC/Rad50) or their phage counterparts (see above), the Csn2 marks and possibly bridges double-strand breaks for repair. However, being inactive, Csn2 is merely a passive DNA-end-binding component like the Ku or the SRAP domains (171) and does not perform any ATP-dependent manipulation of the DNA. Notably, we identified examples of active coiled-coil clade ABC ATPase systems embedding within CRISPR/Cas systems (Figure 7C, see below). These systems may act as backups in the event of failure of the main effector system. This indicates that Csn2 was probably recruited to type-II CRISPR/Cas systems from an active DNA end-binding ABC ATPase precursor. Further, related ATPases (e.g. PrrC and RloC) are also seen in biological conflict systems like RM systems as sensors of nucleotide triphosphates (see below) associated with viral replication (154,155,159–161). Hence, it cannot be ruled out that Csn2 additionally plays such a sensory role in the type-II CRISPR/Cas systems.
Figure 7.
(A–C) Representative depictions of gene neighborhoods of diverse biological conflict systems centered on HEPN, PHP and Toprim containing ABC-ATPases. The corresponding contextual networks based on the gene-neighborhoods are shown. The system of labeling is as in Figure 5.
Decoding the functions of the DndD and CxC clades
The ABC ATPase DndD is a component of the Dnd DNA-modification system. As noted above, we found a further clade related to DndD, the CxC clade, which we named after the unusual signature of its Zn-hook motif (Figure 3, Supplementary Material). The Dnd system catalyzes the modification of self-DNA by replacing the non-bridging oxygen in the phosphate backbone of the DNA by sulfur (forming a phosphorothioate) (172,173). The well-characterized core of the Dnd system is comprised of the enzymes DndA, a PLP-dependent cysteine desulphurase (169), which abstracts a sulfur from cysteine and the PP-loop ATP pyrophosphatase DndC, which incorporates the sulfur into the DNA backbone via an adenylated intermediate (174,175). While it is not usually associated with a restriction component, the system encoded by the DndFGHI operon has been identified as a restriction component for unmodified DNA in certain Dnd systems (176,177).
Aside from these components, Dnd systems contain three conserved but poorly understood components DndB, DndD and DndE (Figure 6B). Our earlier analysis had shown DndB to be a member of the ParB superfamily of enzymes that possess both nuclease and NTPase activity (178). While the ABC ATPase DndD has been shown to possess ATPase activity (179), its role in the Dnd system has remained enigmatic. For instance, DndD has also been erroneously described as an AAA+ ATPase and was not recognized as an ABC ATPase (180,181). We identified two tandem copies of the DNA-binding ribbon‐helix‐helix (RHH) domain (139) in DndE, which is consistent with it preferentially binding nicked double-stranded DNA substrates in a sequence-independent manner; however, its role in these systems has also been enigmatic (182,183). Our analysis recovered numerous divergent DndE homologs and revealed that DndD shows a nearly universal linkage to DndE but is not always linked to the other genes of the Dnd system. Further, the related CxC clade of ABC ATPases also shows a comparable linkage to a gene coding for a DndE homolog. Given that DndE is a non-specific DNA-binding protein, based on the precedence of other members of the coiled-coil clade such as the SMCs, which operate along with non-specific DNA-binding partners such as the kleisins and ScpB (Figure 5Q), we propose that ABC-ATPase-DndE complex forms a similar DNA-manipulating complex.
This complex might be utilized both in the DNA-modifying Dnd system as well as the below systems which we uncovered for the first time in this study. In the case of the Dnd system, we propose that the backbone modification requires the manipulation of the strands by the DndB–DndE complex for the catalytic DndA–DndC complex to access the modification sites. Indeed, the ParB superfamily DndB protein could play a further role in this process by acting as a nuclease that allows easier access for the backbone-modifying machinery, or as an ATPase that regulates the action of DndC. Consistent with the nuclease proposal, we found that there are multiple Dnd-like systems wherein, instead of DndB, there are genes coding for an endoDNases of the HKD restriction enzyme superfamily. We also found an unexpected twist to the Dnd system in the form of a conserved gene-neighborhood-association linking DndC and DndD to a DndE homolog that is further fused to an FtsK-like ATPase (Figure 6B). HerA/FtsK superfamily ATPases are involved in DNA translocation (127); hence, it is conceivable that the action of these systems involves a DNA-pumping activity akin to the functional coupling of the homologous ATPase HerA to several ABC ATPases of the Rad50–SbcC-like clades.
Outside of the Dnd systems, we found strong parallel operonic associations for the DndD and CxC clades (along with their usual partner DndE), conserved across various bacterial and archaeal lineages, with a predicted endoDNase of the HKD superfamily typically fused to an SF-2 helicase module that is related to restriction enzymes from previously characterized Type I/III-like ATP-dependent RM systems (184) (Figure 6B). Additionally, these operons may be coupled to one or both modification methylases respectively of the DNA cytosine methylase and the circularly permuted cytosine-N4-methylase families and a previously uncharacterized globular domain found in several RM systems (MTaX1: methylase associated X1; Figure 6B). Distinct from classical RM systems, these systems are also associated with a gene coding for a small, rapidly evolving protein of around 60–70 amino acids that is predicted to have a mostly disordered structure. Based on the precedence of TA systems, we predict that this small protein (analogous to an antitoxin) potentially acts as a negative regulatory component that might sense the presence of invaders by acting as a binding factor or substrate for invader-encoded enzymes like viral peptidases.
The similar recruitment of the DndD and CxC clade ABC ATPases both in Dnd and RM-like operons, and their coupling to DndE, strongly indicates that the two related ATPases likely perform equivalent roles. In the recently described Dnd-like ‘sspABCD-sspE’ systems, sspA, sspD and sspE are equivalents of DndA, DndC and DndB of the Dnd system (180). Interestingly, these systems lack a direct homolog of the DndD or CxC clade ABC ATPases; instead, we find that the ABC ATPase has been displaced by an ATPase with a distinct C-terminal domain, sspC, belonging to the STAND-Orc-CDC6 clade of AAA+ NTPases. Likewise, instead of DndE these systems contain sspB which contains a triple wHTH domain related to the DNA-binding domain of the restriction endonuclease FokI (185). This suggests that the core modification components are mixed and matched with different ATPase and DNA-binding components which might perform comparable roles in these DNA-modification-based conflict systems.
A novel Zn-hook-containing ABC clade associated with diverse effectors
Our study uncovered an ABC clade widely distributed across all major bacterial lineages and on rare occasions in archaea, which typically occurs as part of a mobile three-gene operon. Accordingly, we named this clade the ABC-3C (for ABC-three component) clade (Figure 3). The ABC gene is always the third or the 3′-most gene in these operons. Of the two other genes in the operon, the 5′-most gene codes a protein marked by a variable multidomain architecture, whereas the middle gene codes for a distinctive single domain protein which we term MC (for middle component) (Figure 6C). The diversity in the first multidomain component is primarily in the variable N-terminal domain, which we predict to be the effector module of the system. These include (i) multiple related subfamilies of REase fold domains; (ii) a trypsin superfamily peptidase; (iii) an enzyme of C-N hydrolase superfamily. In some cases, an inactive version of this is combined with a further N-terminal active REase fold component; (iv) A HNH superfamily domain; (v) An α/β-hydrolase; (vi) A TIR domain; (vii) a double-Rossmann domain dehydrogenase related to the NAD+-dependent dihydropyrimidine dehydrogenases. Additionally, when the trypsin domain is present there may be further effector components encoded by another 5′ standalone gene in the form of a metallobetalactamase or calcineurin-like superfamily nuclease or an HKD superfamily endoDNase with a C-terminal SAD(SRA) domain (Figure 6C). The effector component additionally contains one of at least 12 distinct conserved regions C-terminal to the above catalytic effector domains. These C-terminal regions contain one or more conserved domains, which are either α+β or all α-helical and with distinct patterns of charged and polar residues. We did not detect a relationship of these to any other previously characterized domains. Corresponding to these types of C-terminal regions in the effector component, there are also at least eight types of MCs (Figure 6C), each defined by a distinctive pattern of conserved charged residues (Supplementary Material). At least three of these MCs can be confidently unified with the HTH fold, although others appear unrelated to any other known domain (Figure 6C, Supplementary Material).
There are multiple striking aspects of these systems: (i) the effectors encompass enzymes that are predicted to target a range of cellular components such as phosphodiester bonds in nucleic acids (REase, Metallobetalactamase, calcineurin), potentially N-glycosidic linkages in nucleic acids or nucleotides (CN-hydrolase, TIR), possibly nucleobases (the dihydropyrimidine dehydrogenases-like Rossmann domains), membranes (α/β hydrolase), proteins (trypsin-like peptidase) and NAD+ (TIR). This diversity is not typical of RM systems where the effectors primarily target nucleic acids. (ii) Unlike RM systems, even when the effector domains are predicted to target DNA, there are no corresponding modification enzymes or modified DNA ‘reader’ domains in these systems that might help in self-non-self discrimination. Rather, they resemble the recently described nucleotide-regulated effector systems where diverse effectors are coupled to the sensing of nucleotide signals (152). (iii) In addition to their tight linkage, the three components also co-evolve, i.e. related types of effector component C-terminal regions come with corresponding versions of MCs. This is also reflected in the subtypes of 3C clade ABC ATPases associating with each variety of MC and C-terminal regions of the effector component (Figure 6C).
Due to these features, we propose that these define novel biological conflict systems that function as a multimeric complex comprised of the effector component, MC and the 3C-ABC ATPase. This complex is presumed to be in an inactive state by default. Based on the precedence of the PrrC and RloC systems (159,161,186–188), we suggest that the ABC ATPases respond to the presence of invasive entities, such as DNA viruses. This sensing event is predicted to reconfigure the multimeric complex in an ATP-dependent manner to unleash the effector. As at least some versions are HTH domains, MCs could generally function akin to kleisins as DNA-binding partners for the ABC ATPases. Due to the typically large size of the effector component C-terminal regions relative to the actual effector and MC domains we propose that it serves as the platform on which the remaining two components assemble. In this proposal, the conformational change transmitted by the ABC ATPase would result in an unfurling and activation of the effector. Finally, given that the effector domains include those that might target membranes or NAD+, we propose that some of them could act on cellular targets in a ‘suicidal’ mode to limit viral spread to kin.
ABC ATPases coupled to a diverse array of nuclease and other effector domains
Our study uncovered a vast radiation of such ABC ATPases that are closely coupled, either in the same polypeptide or in tightly linked gene-neighborhoods with a diverse array of effector domains (Figures 3 and 7A–C). We describe the entire gamut of these systems below followed by a unified account of their possible mode of action.
Systems with ABC-ATPases linked to diverse RNase domains of the HEPN superfamily
Several distinct clades of the HEPN superfamily of RNase domains are found fused to ABC ATPases. These occur across prokaryotes as mobile standalone genes or embedded in RM systems (e.g. Escherichia PrrC) or as part of gene-neighborhoods of increasing complexity (Figure 7A). Beyond the standalone versions, the simplest of the gene-neighborhoods feature two genes respectively coding for an ABC ATPase with a C-terminal HEPN domain and a standalone HEPN domain protein. A further conserved two-gene neighborhood combines a gene for an ABC ATPase+HEPN protein with a gene coding for a three-domain protein with a ParB nuclease/NTPase, an HNH endoDNase and a novel C-terminal domain which might recognize modified DNA. More elaborate neighborhoods feature three genes respectively coding for an ABC ATPase with a C-terminal HEPN and Zn-ribbon domain, a protein with a standalone URI domain nuclease (189), and a small protein with an RHH DNA-binding domain (139,190). Notably, all these neighborhoods combine the ABC ATPase+HEPN with either a second predicted RNase (HEPN) or one of multiple DNases that consistently lack an associated modification component (Figure 7A).
Systems with ABC ATPases associated with PHP and histidine kinase domains
The core of these mobile systems found across prokaryotes is a protein combining the ABC ATPase domain with a PHP (Polymerase and histidinol phosphatase) domain (116) (Figures 3, 7B). The PHP domains are TIM-barrel fold phosphoesterases, versions of which also cleave nucleic acids (191,192). The ABC ATPase+PHP genes might show associations with genes coding for a REase fold restriction enzyme fused to an SF2 helicase and an m6A methylase with TRD domains (Figure 7B). These are likely counterparts of the ABC ATPase+HEPN systems, where the ABC ATPase+PHP gene is embedded in bona fide Type I-like R-M systems (Figure 7B). Closely related ABC ATPases also occur in certain conserved gene-neighborhoods independently of the PHP domain. These versions are typically linked to a gene coding for a minimal histidine kinase domain (193) fused to a Schlafen-family RNase domain (194). The latter element with the minimal histidine kinase domain also occurs as an independent element of diverse conflicts system in various effectors(195). In a variant of the above theme, the ABC ATPase gene is combined with two genes respectively coding for a histidine kinase and a protein with a receiver domain fused to a C-terminal S/T/Y/small-molecule kinase domain (Figure 7B).
Systems with ABC ATPases fused to Toprim domains
Two sister families of ABC ATPases, OLD (196–200), and E59 (201,202), which display a coiled-coil insert but lack a Zn-hook, are typically associated with a Toprim domain (Figure 3). Recent crystal structures of the OLD proteins emphatically support their membership in the coiled-coil clade (199,200). The OLD (Overcoming Lysogenization Defect) ABC-ATPase was originally identified in the lysogenic bacteriophage P2 as a gene coding for a product with potential toxicity to lysogenization defective (lyd) Escherichia coli mutants and a role in defense against other lysogenic phages such as λ that compete for the same host species (197,198). Likewise, the E59 (Early59, abbreviated as Ea59 or E59), originally identified in phage λ, is implicated in inter-phage conflict and bacterial counter-phage defense mechanisms (201–203). These sister families share the same structural inserts and similar coiled-coil segments (∼150 residues) but are distinguished by several sequence features (Supplementary Table S2 and alignments provided in the supplementary material). These include frequent deviations from the ABC signatures in the OLD family such as a commonly observed histidine in place of the aspartate in the D-loop and a basic residue in place of the glutamine in the Q-loop. Notably, these unusual features are required for OLD ATPase activity and its toxic effect on RecBC− hosts (199).
The Toprim domains of the archetypal OLD protein and of others such as RNase M5 have been shown to have both RNase and DNase activity (196,204,205) (Figure 7C). Both families are part of highly mobile gene neighborhoods that are widespread in the two prokaryotic superkingdoms. The OLD family has been transferred independently to a unicellular holozoan (the filasterian Capsaspora owczarzaki) and the bivalve mollusc Crassostrea where it has undergone lineage-specific expansions. The simplest of these neighborhoods couple genes for either the OLD or the E59 family and a standalone Toprim gene, suggesting that these likely retain the ancestral condition of these families (Figure 7C). In other neighborhoods, the Toprim domain is fused to the C-terminus of either of these ABC ATPases and they are found as part of conventional Type I RM systems with a REase fold restriction enzyme fused to an SF2 helicase and an m6A DNA methylase with TRD domains. In some of these neighborhoods of the OLD family, a HEPN domain is fused to the REase fold endoDNase domains and in these instances, the Toprim associated with the ABC-ATPase is inactive (Figure 7C). This supports potential functional equivalence between HEPN and at least a subset of Toprim domains. In the E59 family, there may also be associations with RM systems coding for a cytosine C5 methylase instead of an m6A DNA methylase (Figure 7C). Either independently of conventional Type I RM systems or sometimes co-occurring with them, is a notable gene neighborhood, which combines the ABC ATPase+Toprim of the OLD family with a gene coding for a UvrD-like helicase, which is further fused to a 3′-5′ exonuclease domain of the RNase H fold (Figure 7C). These have recently been described as ‘class-2’ OLD systems (199).
Systems related to the above where the Toprim component is displaced
Both the OLD and E59 ABC ATPase components occur as part of gene-neighborhoods where the Toprim module has been displaced by one of several other modules. Both the OLD and E59 families show parallel combinations with an NTPase/nuclease of the ParB superfamily (178) (Figure 7C). Further, the OLD ABC ATPase alone is combined with a gene for a previously unreported version of the REase fold, whereas the E59 family is combined with genes encoding either of two endonucleases respectively displaying the HNH fold (widely distributed) or the REase fold (less common). As with the Toprim-containing versions, this core neighborhood might be combined with conventional type-I-like RM systems with REase+SF2 helicase domains (occasionally with a further fusion to a HEPN domain) and m6A methylases with TRDs (Figure 7C). An unusual set of such HNH-associated systems display N-terminal fusions of the E59 ABC ATPase to (i) a further HNH domain and another helical domain; (ii) an m6A DNA methylase. Interestingly, both these fused domains are predicted to be catalytically inactive due to the loss of their active site residues (Figure 7C). Strikingly, a subset of gene-neighborhoods featuring the second fusion additionally codes for a patatin-like phospholipase A related to the bacterial toxin ExoU (206), with N-terminal transmembrane helices. Likewise, another subset of these is linked to an α/β hydrolase gene coding for a TLE1 toxin-like lipase (207).
Notably, both the OLD and E59 families often show conserved C-terminal α-helical extensions (199,200) fused to either the ABC ATPase or the nuclease component. Crystal structures have revealed a relationship to the bacterial controller (C) proteins from RM systems (200). In the case of the OLD family, the versions without the helical extension were recently classified as class-1 and those with the helical extension as class-2 (199,200). In the class-2 homologs, this C-terminal α-helical extension contributes conserved residues to the Toprim active site and is required for DNA nicking and cleavage activity (200). Structural modeling also points to a possible role for it in binding and orienting the substrate DNA (200). This latter function may more generally hold for the comparable helical extensions of the E59 family, which are more fast-evolving, and lack the conserved residues contributed to the nuclease active site by the OLD class-2 helical inserts. Some members of the E59 clade lacking the Toprim domain have a conserved N-terminal domain predicted to possess an α+β fold. While there are no indications that this domain is a catalytic domain, it might play a role in interactions with the components of partner systems like RM.
ABC ATPases and linked effectors as back-ups for mainline anti-invader systems
Studies on the classic PrrC and RloC systems have revealed a remarkable interplay between restriction of invasive DNA and attacks on self-RNA in the context of the defense against coliphages like T4 (154,155,159–161,208). In these systems, the ABC ATPase is fused to a C-terminal RNase domain of the HEPN superfamily (209–213) (Figures 3, 7A) which targets the anticodon loops of tRNAs. PrrC is genomically linked to the Type Ic R-M system PrrI, whose three core RM components (hsdMSR) negatively regulate the PrrC by keeping it catalytically inactive (154,160,214). However, when phages like T4 deploy antirestriction peptides, such as Stp, which inhibit the restriction endoDNase of the PrrI RM system, it activates the associated PrrC evidently by interacting with the PrrC-restriction endoDNase interface (208). This unleashes the C-terminal RNase domain, which is further augmented by GTP hydrolysis and binding of dTTP (159), to act as the second line of defense against the virus by cleaving the anticodon loop of self-tRNAs (154,160,214). Unlike PrrC, the RloC is not linked to the RM systems and its HEPN RNase domain is kept inactive by its N-terminal ABC-ATPase domain under normal conditions. RloC responds to DNA damage, which could be induced by viral infections, and dTTP which could act as a signal related to viral replication, to unleash its HEPN RNase through conformational changes in the ATPase domain. These self-tRNA targeting responses inhibit translation and can have deleterious consequences on the replicating virus (215).
Thus, the ABC ATPases are combined with a multiplicity of C-terminal nuclease domains, such as HEPN, PHP, PIN and Toprim. These domains are known to have either RNase or DNase activity or both. Further, we find multiple independent associations of these systems with bona fide RM systems. Taken together, our observations point to a general functional principle for the combination of ABC ATPases with nuclease domains. This suggests that upon sensing of signals indicating the failure of front-line defense systems, the ABC ATPase unmasks associated nuclease domains to either attack self-RNAs (likely those associated with translation, e.g. tRNAs) or DNA in an indiscriminate fashion as suggested by the role of the Toprim DNase activity in the killing of recBC-Escherichia coli (199). Other examples of the above systems also show characteristic features of DNA-targeting enzymes such as the RHH DNA-binding domain found in systems with the URI endonuclease, or the HNH+ParB combination related to type IV RM systems, versions of which have been shown to cleave DNA (216–218).
The RloC-like systems (described above) have been shown to not just cleave the anticodon loop but also excise the wobble base from the cleaved loop (186,219). Consistent with this, we find multiple instances of a second RNase component such as a second HEPN or Schlafen domain in these systems. Accordingly, we posit that these second RNases might likewise catalyze an additional cleavage. Further, the systems which contain a UvrD-like helicase with a 3′-5′ exonuclease domain might also catalyze exonucleolytic degradation of the cleaved nucleic acids. In either case, such double-nuclease action can considerably limit repair, especially of the targeted RNAs by ligation (155). Additionally, there are coupled effectors that might not target nucleic acids, such as the two distinct lipases, which likely target the membrane, and the histidine kinase-receiver domain two-component systems, which may activate a kinase effector (Figure 7C). Absence of linked DNA-modification enzymes or other inhibitory components (like immunity proteins) in these cases suggests that these enzymes, just as the associated nucleases, are likely kept inactive in the default state in a complex with the ABC ATPase component. Together, these examples indicate that in addition to or in place of the attack on nucleic acids, the line of defense mediated by the ABC ATPases might also trigger responses via linked effectors, which might include a suicidal attack on the cell-membrane or phosphorylation of particular substrates.
Thus, such an action by the ABC ATPases and the coupled effectors can provide a backup for the failure of the frontline defensive mechanisms. The host might either be able to tide over such inhibition as a period of dormancy (a known defense mechanism against phages (220), or if it undergoes suicide it can preempt viral spread to kin cells or other cells in a multicellular assemblage (154,155,159–161). As has been previously proposed, such ‘apoptotic’ responses are a widespread last line of defense against viruses across the superkingdoms of life (154,156,221). It is also notable that such systems have been acquired by viruses: e.g. OLD by the coliphage P2 and Burkholderia phage KS5 (200) and E59 by λ (201,202). These provide a counter against superinfection by other phages (197,198) or launch an attack on the host genome to preempt host counterattacks. Many of these occur in ‘mobile gene neighborhoods’; for instance, OLD family members frequently occur in neighborhoods enriched in transposons (200). This suggests that they have been repeatedly disseminated via lateral transfer and exchanged between bacterial and phage genomes. In addition to the above, multiple acquisitions and expansions of such ABC-Toprim enzymes in the ‘greater animal’ (holozoans) lineage suggests that such a mechanism might have also been adapted for defense against eukaryotic viruses.
GENERAL EVOLUTIONARY AND FUNCTIONAL CONSIDERATIONS
ABC ATPases emerged as part of the early radiation of the P-loop NTPases in association with nucleic acids
Based on their phyletic patterns we infer that at least 5 and up to 8 distinct versions of ABC ATPases spanning all the major higher assemblages of the superfamily and associated with different functions were already present in the LUCA (Figure 3). This suggests that the ABC ATPases had undergone a major radiation prior to the LUCA, as has been previously reported for another major P-loop NTPase superfamily from the ASCE-division, the AAA+ NTPases (Figure 1) (2). Smaller but comparable pre-LUCA radiations are also observed in the case of the helicase (SF-1 and SF-2) and RecA-ATP-synthase superfamilies of the ASCE-division of P-loop NTPases (222,223). Thus, not only were at least 5 distinct ASCE-division superfamilies (AAA+, ABC, RecA, Helicase and HerA/FtsK) present in the LUCA but they had radiated into at least one to six members each. Our study demonstrates that four of the five clades of ABC ATPases confidently inferred as being present in the LUCA (six of eight if we consider the larger, less-confidently inferred set in the LUCA) performed nucleic-acid-associated functions. In the case of the AAA+ ATPases three of six clades, in the case of the helicases 3 of 3 clades, in the case of HerA/FtsK ATPases one of one clade and in the case of the RecA-ATP synthase ATPases at least one of two clades traceable to the LUCA are inferred as having performed nucleic acid-related functions. Thus, much of the early radiation of the ASCE division, including the ABC ATPases, occurred in the context of the pre-LUCA emergence of systems associated with nucleic acids dynamics and biochemistry.
Further, among the ABC ATPases the ancestral MutS, Zn-hook, and SMC clades (total of 3) are inferred as performing DNA-related functions (Figure 3). A single ABC ATPase is confidently inferred as functioning in a translation-associated RNA-related context, while the second one, the RNA-ABC, might have performed an RNA-related role in the more inclusive reconstruction of the LUCA complement. In the case of the other ASCE-division superfamilies, at least 3 clades of AAA+ ATPases and at least 1 clade each of the helicases, HerA/FtsK and RecA-like superfamilies are inferred as having DNA-related roles in the LUCA (2,127,222,223). In contrast, in the KG-division, of the 13 clades of GTPases traceable to the LUCA, at least 8 perform RNA-related functions, mostly related to the translation apparatus, and only one might have doubtfully functioned in the context of DNA (3,5). This drastic difference in terms of the preferred nucleic acid-association between the ASCE and the KG divisions indicate that the former might have originally diversified with the fixation of DNA as the primary store of genetic information and the latter with the fixation of the core translation apparatus. Their radiation primarily in a DNA-related context suggests that this probably went together with the energy-intensive processes associated with DNA as the primary genetic material, such as unwinding of dsDNA, looping of DNA for compact packaging and bridging of DNA-ends during recombination.
The biochemical features of ABC ATPases are correlated with their unique functional niche
Our current study points to a basic mode of action that is common to all ABC ATPases. Consistent with this, we find that across diverse functional contexts there are only rare displacements of ABC ATPases by P-loop NTPases of any other superfamily. Structural and biochemical hallmarks of this uniqueness are the head-to-head dimerization mode (Figure 2A), barrelization of the core P-loop β-sheet (Figure 2B) and the absence of the arginine finger which has convergently evolved in the other KG and ASCE-division NTPases. The absence of the arginine finger is compensated by the distinctive, complementary Q-loop-SGG interaction between the two subunits (Figure 2A). This together with the mode of ATP-binding by the barrelized β-sheet facilitates an ATPase work-cycle that results in translational motion (Figure 2B). This contrasts with the rotatory motion typical of the other ASCE ATPases such as those of the AAA+, HerA/FtsK and RecA-like clades with corresponding multimeric toroidal assemblies or the asymmetric linear translocation of the helicases (224–226) (Figure 4B). The only non-P-loop ATPase domains that have displaced ABC ATPase domains in more than one functional context are the MORC-like clade of ATPases of the GHKL superfamily of ATPases. These include ATPases such as the MORCs, para-MORCs and MutL. MORCs occurs in counter-phage restriction systems in contexts equivalent to the ABC ATPases (193). Further, in eukaryotes certain MORCs have fused with the SMC hinge domains and a coiled-coil to generate a configuration equivalent to the SMCs (193). Hence, this clade of unrelated ATPases has likely convergently evolved the capacity to transduce mechanical work in a manner similar to the ABC ATPases.
Our natural classification of the ABC superfamily suggests that the dimer interface was augmented in multiple stages starting from a rudimentary form seen in the MutS family, through the initial developments in the RNA-ABC and translation factor-UvrA clade, to the more elaborate form of the classical ABC ATPases (Figures 1B and 3). It was this elaboration that allowed the emergence of a unique aspect of ABC function, i.e. active transport across the lipid bilayer, the key event which allowed cells to actively acquire nutrients, deliver toxin weaponry and extrude deleterious xenobiotics. Further, the developments in the dimer interface were accompanied by the development of inserts 1 and 2 that allowed more efficient coupling of the core ABC ATPase domain to substrates (Figure 2A–F). An important stage in this development was the emergence of progressively longer coiled-coil inserts (Figure 3). This resulted in a mechanism, largely unique among P-loop NTPases, where the Walker A and Walker B regions are conditionally decoupled by the coiled-coil (227,228). This allowed for the work cycle of the ATPase domain to be coupled with long-range bridging interactions that were dependent on the length of the coiled-coil (Figure 4B). This development appears to have gone along with the emergence of large DNA genomes because ABC ATPases of the coiled-coil clade performed two key roles, namely repair of dsDNA breaks and the packaging of chromosomes in a small space (229). Moreover, these recruitments were accompanied by the acquisition of special conserved partners, respectively the nucleases of the calcineurin-like phosphoesterase superfamily (Mre11/SbcD) and ScpB/Kleisins with wHTH domains (230,231).
ABC ATPases affirm deep evolutionary links between house-keeping aspects of nucleic acid biochemistry and conflict systems
A major aspect of this study is the identification of an extensive radiation of ABC ATPases in an array of diverse biological conflict systems. Notably, we show that all these ATPases have emerged from a single or few ancestral members of the coiled-coil clade. Just as certain house-keeping members of the coiled-coil clade function with a nuclease partner, these too function with one of several nuclease partners. Further, our identification of a pervasive link between these ATPases and RHH domains, such as DndE, suggests that, like the ScpB/kleisins of the SMC clade, several of these too might associate with comparable DNA-binding partners to constitute comparable DNA-encircling structures. However, in contrast to their relatives such as SbcC/Rad50 and SMCs, which are well-conserved ‘house-keeping proteins’, the versions from conflict systems are rapidly evolving and widely disseminated via lateral transfer. This prevents us from tying their origins to a precise temporal window; however, given the ubiquity of biological conflicts we suspect that they are likely to have emerged early (Figure 3).
Parallels to this situation are found in diverse nucleotide-activated conflict systems, which couple nucleotidyltransferases with a panoply of effector domains that likely target both self and non-self molecules (152). We had shown that the activating nucleotide-generating enzymes of these systems are ultimately related to highly conserved nucleotidyltransferases and polymerases involved in housekeeping roles such as tRNA maturation and replication. We had postulated that the sensing of damaged RNAs and genomic DNA to activate repair processes by nucleotidyltransferases and nucleic acid polymerases occurred as a response to the viral targeting of host-nucleic acids. This ancient, conserved repair process probably gave rise to the predecessors of the signaling nucleotides that activated anti-viral conflict systems as a byproduct. This eventually led to the fixation of dedicated nucleotidyltransferase paralogs of the housekeeping versions that activate anti-viral effectors based on nucleotide signals (152). In parallel, we postulate that the ABC ATPases which recognized dsDNA breaks, precursors and intermediates of viral replication were probably drafted as sensors for potential viral attacks to activate associated effectors. Indeed, given that several viruses themselves code for ABC ATPases that are part of the end-recombination apparatus, it is possible that the ABC ATPases in cellular conflict systems were recruited from such selfish replicons as they were likely pre-adapted to sense viral replication/recombination intermediates. Thus, these observations suggest the deriving of conflict-related sensors and switches from related enzymes that mediate and safeguard the flow of information in the ‘central-dogma’ (as pertaining to both cellular and selfish replicons) is likely to be a general phenomenon.
CONCLUSIONS
Other than the transporter ATPase clade, the ABC ATPases remained the only major superfamily of P-loop NTPases that were not previously comprehensively surveyed. Here we redress the situation with an in-depth analysis of the rest of this superfamily. Consequently, we show that, whereas the ABC ATPases initially radiated along with the ASCE-division NTPases primarily in DNA-related contexts, they have a mode of action that is distinct from all other P-loop NTPases. We show that this has been maintained through the evolution of this superfamily and augmented in multiple distinct stages to perform diverse translational motions in distinct cellular contexts. Notably, this allows us to understand ABC ATPase function under a common overarching structural framework.
Further, this work identifies several novel clades and clarifies the contextual linkages and relationships of several previously known but poorly understood clades. As a result, we present several testable hypotheses that would allow for the discovery of novel biochemistry. Examples of these include the role of MutS ATPases in biosynthetic complex assembly related to secondary metabolism and the possible mechanism of the DndD and related clades. Importantly, we identify several novel biological conflict systems wherein the ABC ATPases could play pivotal roles in the sensing of invasive entities and triggering a range of effector responses. Given the rising value of enzymes from biological conflict systems as potential biotechnological reagents, we believe that the systems reported here are likely to provide further opportunities in this regard.
Supplementary Material
ACKNOWLEDGEMENTS
This research was supported by the Intramural Research Program of the NIH, National Library of Medicine. Due to space limitations, we were regrettably unable to cite several research papers on ABC ATPases in the article; however, a more complete supplementary bibliography is provided in the supplementary material.
Contributor Information
Arunkumar Krishnan, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20894, USA.
A Maxwell Burroughs, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20894, USA.
Lakshminarayan M Iyer, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20894, USA.
L Aravind, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20894, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH postdoctoral visiting fellowship (to A.K.); intramural funds (to L.M.I., A.M.B., L.A.) of the National Library of Medicine at the National Institutes of Health, USA. Funding for open access charge: Intramural Funds of the National Library of Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1. Saraste M., Sibbald P.R., Wittinghofer A.. The P-loop–a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 1990; 15:430–434. [DOI] [PubMed] [Google Scholar]
- 2. Iyer L.M., Leipe D.D., Koonin E.V., Aravind L.. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 2004; 146:11–31. [DOI] [PubMed] [Google Scholar]
- 3. Leipe D.D., Wolf Y.I., Koonin E.V., Aravind L.. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 2002; 317:41–72. [DOI] [PubMed] [Google Scholar]
- 4. Leipe D.D., Koonin E.V., Aravind L.. STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J. Mol. Biol. 2004; 343:1–28. [DOI] [PubMed] [Google Scholar]
- 5. Leipe D.D., Koonin E.V., Aravind L.. Evolution and classification of P-loop kinases and related proteins. J. Mol. Biol. 2003; 333:781–815. [DOI] [PubMed] [Google Scholar]
- 6. Vetter I.R., Wittinghofer A.. Nucleoside triphosphate-binding proteins: different scaffolds to achieve phosphoryl transfer. Q. Rev. Biophys. 1999; 32:1–56. [DOI] [PubMed] [Google Scholar]
- 7. Higgins C.F., Linton K.J.. The ATP switch model for ABC transporters. Nat. Struct. Mol. Biol. 2004; 11:918–926. [DOI] [PubMed] [Google Scholar]
- 8. Aukema K.G., Kron E.M., Herdendorf T.J., Forest K.T.. Functional dissection of a conserved motif within the pilus retraction protein PilT. J. Bacteriol. 2005; 187:611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burroughs A.M., Iyer L.M., Aravind L.. Comparative genomics and evolutionary trajectories of viral ATP dependent DNA-packaging systems. Genome Dyn. 2007; 3:48–65. [DOI] [PubMed] [Google Scholar]
- 10. Dassa E. Natural history of ABC systems: not only transporters. Essays Biochem. 2011; 50:19–42. [DOI] [PubMed] [Google Scholar]
- 11. Theodoulou F.L., Kerr I.D.. ABC transporter research: going strong 40 years on. Biochem. Soc. Trans. 2015; 43:1033–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wilkens S. Structure and mechanism of ABC transporters. F1000Prime Rep. 2015; 7:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Garcia P.S., Gribaldo S., Py B., Barras F.. The SUF system: an ABC ATPase-dependent protein complex with a role in Fe-S cluster biogenesis. Res. Microbiol. 2019; 170:426–434. [DOI] [PubMed] [Google Scholar]
- 14. Hirabayashi K., Yuda E., Tanaka N., Katayama S., Iwasaki K., Matsumoto T., Kurisu G., Outten F.W., Fukuyama K., Takahashi Y. et al.. Functional dynamics revealed by the structure of the SufBCD complex, a novel ATP-binding Cassette (ABC) protein that serves as a scaffold for iron-sulfur cluster biogenesis. J. Biol. Chem. 2015; 290:29717–29731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang W. Structure and function of mismatch repair proteins. Mutat. Res. 2000; 460:245–256. [DOI] [PubMed] [Google Scholar]
- 16. Groothuizen F.S., Winkler I., Cristovao M., Fish A., Winterwerp H.H., Reumer A., Marx A.D., Hermans N., Nicholls R.A., Murshudov G.N. et al.. MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA. Elife. 2015; 4:e06744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jaciuk M., Nowak E., Skowronek K., Tanska A., Nowotny M.. Structure of UvrA nucleotide excision repair protein in complex with modified DNA. Nat. Struct. Mol. Biol. 2011; 18:191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Syed A., Tainer J.A.. The MRE11-RAD50-NBS1 complex conducts the orchestration of damage signaling and outcomes to stress in DNA replication and repair. Annu. Rev. Biochem. 2018; 87:263–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alonso J.C., Cardenas P.P., Sanchez H., Hejna J., Suzuki Y., Takeyasu K.. Early steps of double-strand break repair in Bacillus subtilis. DNA Repair (Amst.). 2013; 12:162–176. [DOI] [PubMed] [Google Scholar]
- 20. Uhlmann F. SMC complexes: from DNA to chromosomes. Nat. Rev. Mol. Cell Biol. 2016; 17:399–412. [DOI] [PubMed] [Google Scholar]
- 21. Andersen C.B., Becker T., Blau M., Anand M., Halic M., Balar B., Mielke T., Boesen T., Pedersen J.S., Spahn C.M. et al.. Structure of eEF3 and the mechanism of transfer RNA release from the E-site. Nature. 2006; 443:663–668. [DOI] [PubMed] [Google Scholar]
- 22. Boel G., Smith P.C., Ning W., Englander M.T., Chen B., Hashem Y., Testa A.J., Fischer J.J., Wieden H.J., Frank J. et al.. The ABC-F protein EttA gates ribosome entry into the translation elongation cycle. Nat. Struct. Mol. Biol. 2014; 21:143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Heuer A., Gerovac M., Schmidt C., Trowitzsch S., Preis A., Kotter P., Berninghausen O., Becker T., Beckmann R., Tampe R.. Structure of the 40S-ABCE1 post-splitting complex in ribosome recycling and translation initiation. Nat. Struct. Mol. Biol. 2017; 24:453–460. [DOI] [PubMed] [Google Scholar]
- 24. Shaw P.L., McAdams N.M., Hast M.A., Ammerman M.L., Read L.K., Schumacher M.A.. Structures of the T. brucei kRNA editing factor MRB1590 reveal unique RNA-binding pore motif contained within an ABC-ATPase fold. Nucleic Acids Res. 2015; 43:7096–7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Berger E.A., Heppel L.A.. Different mechanisms of energy coupling for the shock-sensitive and shock-resistant amino acid permeases of Escherichia coli. J. Biol. Chem. 1974; 249:7747–7755. [PubMed] [Google Scholar]
- 26. Karcher A., Buttner K., Martens B., Jansen R.P., Hopfner K.P.. X-ray structure of RLI, an essential twin cassette ABC ATPase involved in ribosome biogenesis and HIV capsid assembly. Structure. 2005; 13:649–659. [DOI] [PubMed] [Google Scholar]
- 27. Gupta S., Gellert M., Yang W.. Mechanism of mismatch recognition revealed by human MutSbeta bound to unpaired DNA loops. Nat. Struct. Mol. Biol. 2011; 19:72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pellegrino S., Radzimanowski J., de Sanctis D., Boeri Erba E., McSweeney S., Timmins J.. Structural and functional characterization of an SMC-like protein RecN: new insights into double-strand break repair. Structure. 2012; 20:2076–2089. [DOI] [PubMed] [Google Scholar]
- 29. Liu Y., Sung S., Kim Y., Li F., Gwon G., Jo A., Kim A.K., Kim T., Song O.K., Lee S.E. et al.. ATP-dependent DNA binding, unwinding, and resection by the Mre11/Rad50 complex. EMBO J. 2016; 35:743–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Diebold-Durand M.L., Lee H., Ruiz Avila L.B., Noh H., Shin H.C., Im H., Bock F.P., Burmann F., Durand A., Basfeld A. et al.. Structure of full-length SMC and rearrangements required for chromosome organization. Mol. Cell. 2017; 67:334–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kurze A., Michie K.A., Dixon S.E., Mishra A., Itoh T., Khalid S., Strmecki L., Shirahige K., Haering C.H., Lowe J. et al.. A positively charged channel within the Smc1/Smc3 hinge required for sister chromatid cohesion. EMBO J. 2011; 30:364–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Myler L.R., Gallardo I.F., Soniat M.M., Deshpande R.A., Gonzalez X.B., Kim Y., Paull T.T., Finkelstein I.J.. Single-molecule imaging reveals how Mre11-Rad50-Nbs1 initiates DNA break repair. Mol. Cell. 2017; 67:891–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Abramo K., Valton A.L., Venev S.V., Ozadam H., Fox A.N., Dekker J.. A chromosome folding intermediate at the condensin-to-cohesin transition during telophase. Nat. Cell Biol. 2019; 21:1393–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Aravind L., Iyer L.M., Leipe D.D., Koonin E.V.. A novel family of P-loop NTPases with an unusual phyletic distribution and transmembrane segments inserted within the NTPase domain. Genome Biol. 2004; 5:R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Locher K.P. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 2016; 23:487–493. [DOI] [PubMed] [Google Scholar]
- 36. Walker J.E., Saraste M., Runswick M.J., Gay N.J.. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982; 1:945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hopfner K.P. Invited review: Architectures and mechanisms of ATP binding cassette proteins. Biopolymers. 2016; 105:492–504. [DOI] [PubMed] [Google Scholar]
- 38. Neuwald A.F., Aravind L., Spouge J.L., Koonin E.V.. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999; 9:27–43. [PubMed] [Google Scholar]
- 39. Priess M., Goddeke H., Groenhof G., Schafer L.V.. Molecular mechanism of ATP hydrolysis in an ABC transporter. ACS Cent Sci. 2018; 4:1334–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Donmez I., Patel S.S.. Mechanisms of a ring shaped helicase. Nucleic Acids Res. 2006; 34:4216–4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ogura T., Whiteheart S.W., Wilkinson A.J.. Conserved arginine residues implicated in ATP hydrolysis, nucleotide-sensing, and inter-subunit interactions in AAA and AAA+ ATPases. J. Struct. Biol. 2004; 146:106–112. [DOI] [PubMed] [Google Scholar]
- 42. Zhao Z., De-Donatis G.M., Schwartz C., Fang H., Li J., Guo P.. An arginine finger regulates the sequential action of asymmetrical hexameric ATPase in the double-stranded DNA translocation motor. Mol. Cell. Biol. 2016; 36:2514–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lebbink J.H., Fish A., Reumer A., Natrajan G., Winterwerp H.H., Sixma T.K.. Magnesium coordination controls the molecular switch function of DNA mismatch repair protein MutS. J. Biol. Chem. 2010; 285:13131–13141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lamers M.H., Georgijevic D., Lebbink J.H., Winterwerp H.H., Agianian B., de Wind N., Sixma T.K.. ATP increases the affinity between MutS ATPase domains. Implications for ATP hydrolysis and conformational changes. J. Biol. Chem. 2004; 279:43879–43885. [DOI] [PubMed] [Google Scholar]
- 45. Hopfner K.P., Karcher A., Shin D.S., Craig L., Arthur L.M., Carney J.P., Tainer J.A.. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000; 101:789–800. [DOI] [PubMed] [Google Scholar]
- 46. Hohl M., Hurlimann L.M., Bohm S., Schoppe J., Grutter M.G., Bordignon E., Seeger M.A.. Structural basis for allosteric cross-talk between the asymmetric nucleotide binding sites of a heterodimeric ABC exporter. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:11025–11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Alva V., Soding J., Lupas A.N.. A vocabulary of ancient peptides at the origin of folded proteins. Elife. 2015; 4:e09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lamers M.H., Perrakis A., Enzlin J.H., Winterwerp H.H., de Wind N., Sixma T.K.. The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature. 2000; 407:711–717. [DOI] [PubMed] [Google Scholar]
- 49. Kaur G., Iyer L.M., Subramanian S., Aravind L.. Evolutionary convergence and divergence in archaeal chromosomal proteins and Chromo-like domains from bacteria and eukaryotes. Sci. Rep. 2018; 8:6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Haering C.H., Lowe J., Hochwagen A., Nasmyth K.. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell. 2002; 9:773–788. [DOI] [PubMed] [Google Scholar]
- 51. Ku B., Lim J.H., Shin H.C., Shin S.Y., Oh B.H.. Crystal structure of the MukB hinge domain with coiled-coil stretches and its functional implications. Proteins. 2010; 78:1483–1490. [DOI] [PubMed] [Google Scholar]
- 52. Pakotiprapha D., Samuels M., Shen K., Hu J.H., Jeruzalmi D.. Structure and mechanism of the UvrA-UvrB DNA damage sensor. Nat. Struct. Mol. Biol. 2012; 19:291–298. [DOI] [PubMed] [Google Scholar]
- 53. Roland M., Przybyla-Toscano J., Vignols F., Berger N., Azam T., Christ L., Santoni V., Wu H.C., Dhalleine T., Johnson M.K. et al.. The plastidial Arabidopsis thaliana NFU1 protein binds and delivers [4Fe-4S] clusters to specific client proteins. J. Biol. Chem. 2020; 295:1727–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Loiseau L., Ollagnier-de-Choudens S., Nachin L., Fontecave M., Barras F.. Biogenesis of Fe-S cluster by the bacterial Suf system: SufS and SufE form a new type of cysteine desulfurase. J. Biol. Chem. 2003; 278:38352–38359. [DOI] [PubMed] [Google Scholar]
- 55. Ollagnier-de-Choudens S., Lascoux D., Loiseau L., Barras F., Forest E., Fontecave M.. Mechanistic studies of the SufS-SufE cysteine desulfurase: evidence for sulfur transfer from SufS to SufE. FEBS Lett. 2003; 555:263–267. [DOI] [PubMed] [Google Scholar]
- 56. Nachin L., Loiseau L., Expert D., Barras F.. SufC: an unorthodox cytoplasmic ABC/ATPase required for [Fe-S] biogenesis under oxidative stress. EMBO J. 2003; 22:427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gupta V., Sendra M., Naik S.G., Chahal H.K., Huynh B.H., Outten F.W., Fontecave M., Ollagnier de Choudens S.. Native Escherichia coli SufA, coexpressed with SufBCDSE, purifies as a [2Fe-2S] protein and acts as an Fe-S transporter to Fe-S target enzymes. J. Am. Chem. Soc. 2009; 131:6149–6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chahal H.K., Dai Y., Saini A., Ayala-Castro C., Outten F.W.. The SufBCD Fe-S scaffold complex interacts with SufA for Fe-S cluster transfer. Biochemistry. 2009; 48:10644–10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kolling D.J., Brunzelle J.S., Lhee S., Crofts A.R., Nair S.K.. Atomic resolution structures of rieske iron-sulfur protein: role of hydrogen bonds in tuning the redox potential of iron-sulfur clusters. Structure. 2007; 15:29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Balaji S., Aravind L.. The RAGNYA fold: a novel fold with multiple topological variants found in functionally diverse nucleic acid, nucleotide and peptide-binding proteins. Nucleic Acids Res. 2007; 35:5658–5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Travis B., Shaw P.L.R., Liu B., Ravindra K., Iliff H., Al-Hashimi H.M., Schumacher M.A.. The RRM of the kRNA-editing protein TbRGG2 uses multiple surfaces to bind and remodel RNA. Nucleic Acids Res. 2019; 47:2130–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nurenberg E., Tampe R.. Tying up loose ends: ribosome recycling in eukaryotes and archaea. Trends Biochem. Sci. 2013; 38:64–74. [DOI] [PubMed] [Google Scholar]
- 63. Simonetti A., Guca E., Bochler A., Kuhn L., Hashem Y.. Structural insights into the mammalian late-stage initiation complexes. Cell Rep. 2020; 31:107497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Burroughs A.M., Aravind L.. The origin and evolution of release factors: implications for translation termination, ribosome rescue, and quality control pathways. Int. J. Mol. Sci. 2019; 20:1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nurenberg-Goloub E., Kratzat H., Heinemann H., Heuer A., Kotter P., Berninghausen O., Becker T., Tampe R., Beckmann R.. Molecular analysis of the ribosome recycling factor ABCE1 bound to the 30S post-splitting complex. EMBO J. 2020; 39:e103788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gerovac M., Tampe R.. Control of mRNA translation by versatile ATP-Driven machines. Trends Biochem. Sci. 2019; 44:167–180. [DOI] [PubMed] [Google Scholar]
- 67. Anantharaman V., Koonin E.V., Aravind L.. Regulatory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains. J. Mol. Biol. 2001; 307:1271–1292. [DOI] [PubMed] [Google Scholar]
- 68. Huang J., Gogarten J.P.. Did an ancient chlamydial endosymbiosis facilitate the establishment of primary plastids. Genome Biol. 2007; 8:R99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Keeling P.J. The endosymbiotic origin, diversification and fate of plastids. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010; 365:729–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chen B., Boel G., Hashem Y., Ning W., Fei J., Wang C., Gonzalez R.L. Jr, Hunt J.F., Frank J.. EttA regulates translation by binding the ribosomal E site and restricting ribosome-tRNA dynamics. Nat. Struct. Mol. Biol. 2014; 21:152–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Murina V., Kasari M., Takada H., Hinnu M., Saha C.K., Grimshaw J.W., Seki T., Reith M., Putrins M., Tenson T. et al.. ABCF ATPases involved in protein synthesis, ribosome assembly and antibiotic resistance: structural and functional diversification across the tree of life. J. Mol. Biol. 2019; 431:3568–3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sharkey L.K., Edwards T.A., O’Neill A.J.. ABC-F proteins mediate antibiotic resistance through ribosomal protection. mBio. 2016; 7:e01975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Reynolds E.D., Cove J.H.. Resistance to telithromycin is conferred by msr(A), msrC and msr(D) in Staphylococcus aureus. J. Antimicrob. Chemother. 2005; 56:1179–1180. [DOI] [PubMed] [Google Scholar]
- 74. Singh K.V., Weinstock G.M., Murray B.E.. An Enterococcus faecalis ABC homologue (Lsa) is required for the resistance of this species to clindamycin and quinupristin-dalfopristin. Antimicrob. Agents Chemother. 2002; 46:1845–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Novotna G., Janata J.. A new evolutionary variant of the streptogramin A resistance protein, Vga(A)LC, from Staphylococcus haemolyticus with shifted substrate specificity towards lincosamides. Antimicrob. Agents Chemother. 2006; 50:4070–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hot C., Berthet N., Chesneau O.. Characterization of sal(A), a novel gene responsible for lincosamide and streptogramin A resistance in Staphylococcus sciuri. Antimicrob. Agents Chemother. 2014; 58:3335–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Crowe-McAuliffe C., Graf M., Huter P., Takada H., Abdelshahid M., Novacek J., Murina V., Atkinson G.C., Hauryliuk V., Wilson D.N.. Structural basis for antibiotic resistance mediated by the Bacillus subtilis ABCF ATPase VmlR. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:8978–8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sarwar M., Akhtar M.. Cloning of aminoglycoside phosphotransferase (APH) gene from antibiotic-producing strain of Bacillus circulans into a high-expression vector, pKK223-3. Purification, properties and location of the enzyme. Biochem. J. 1990; 268:671–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Trower M.K., Clark K.G.. PCR cloning of a streptomycin phosphotransferase (aphE) gene from Streptomyces griseus ATCC 12475. Nucleic Acids Res. 1990; 18:4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Garcia-Barrio M., Dong J., Ufano S., Hinnebusch A.G.. Association of GCN1-GCN20 regulatory complex with the N-terminus of eIF2alpha kinase GCN2 is required for GCN2 activation. EMBO J. 2000; 19:1887–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Masson G.R. Towards a model of GCN2 activation. Biochem. Soc. Trans. 2019; 47:1481–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Vazquez de Aldana C.R., Marton M.J., Hinnebusch A.G.. GCN20, a novel ATP binding cassette protein, and GCN1 reside in a complex that mediates activation of the eIF-2 alpha kinase GCN2 in amino acid-starved cells. EMBO J. 1995; 14:3184–3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sattlegger E., Hinnebusch A.G.. Polyribosome binding by GCN1 is required for full activation of eukaryotic translation initiation factor 2{alpha} kinase GCN2 during amino acid starvation. J. Biol. Chem. 2005; 280:16514–16521. [DOI] [PubMed] [Google Scholar]
- 84. Su T., Izawa T., Thoms M., Yamashita Y., Cheng J., Berninghausen O., Hartl F.U., Inada T., Neupert W., Beckmann R.. Structure and function of Vms1 and Arb1 in RQC and mitochondrial proteome homeostasis. Nature. 2019; 570:538–542. [DOI] [PubMed] [Google Scholar]
- 85. Verma R., Reichermeier K.M., Burroughs A.M., Oania R.S., Reitsma J.M., Aravind L., Deshaies R.J.. Vms1 and ANKZF1 peptidyl-tRNA hydrolases release nascent chains from stalled ribosomes. Nature. 2018; 557:446–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kasari V., Pochopien A.A., Margus T., Murina V., Turnbull K., Zhou Y., Nissan T., Graf M., Novacek J., Atkinson G.C. et al.. A role for the Saccharomyces cerevisiae ABCF protein New1 in translation termination/recycling. Nucleic Acids Res. 2019; 47:8807–8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Mardenborough Y.S.N., Nitsenko K., Laffeber C., Duboc C., Sahin E., Quessada-Vial A., Winterwerp H.H.K., Sixma T.K., Kanaar R., Friedhoff P. et al.. The unstructured linker arms of MutL enable GATC site incision beyond roadblocks during initiation of DNA mismatch repair. Nucleic Acids Res. 2019; 47:11667–11680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Toledo M., Sun X., Brieno-Enriquez M.A., Raghavan V., Gray S., Pea J., Milano C.R., Venkatesh A., Patel L., Borst P.L. et al.. A mutation in the endonuclease domain of mouse MLH3 reveals novel roles for MutLgamma during crossover formation in meiotic prophase I. PLos Genet. 2019; 15:e1008177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kolodner R.D. A personal historical view of DNA mismatch repair with an emphasis on eukaryotic DNA mismatch repair. DNA Repair (Amst.). 2016; 38:3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Graham W.J.t., Putnam C.D., Kolodner R.D.. The properties of Msh2-Msh6 ATP binding mutants suggest a signal amplification mechanism in DNA mismatch repair. J. Biol. Chem. 2018; 293:18055–18070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Iyer R.R., Pluciennik A., Napierala M., Wells R.D.. DNA triplet repeat expansion and mismatch repair. Annu. Rev. Biochem. 2015; 84:199–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Fukui K., Kosaka H., Kuramitsu S., Masui R.. Nuclease activity of the MutS homologue MutS2 from Thermus thermophilus is confined to the Smr domain. Nucleic Acids Res. 2007; 35:850–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Glover M.L., Burroughs A.M., Monem P.C., Egelhofer T.A., Pule M.N., Aravind L., Arribere J.A.. NONU-1 encodes a conserved endonuclease required for mRNA translation surveillance. Cell Rep. 2020; 30:4321–4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Helmann J.D. Where to begin? Sigma factors and the selectivity of transcription initiation in bacteria. Mol. Microbiol. 2019; 112:335–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Burby P.E., Simmons L.A.. MutS2 promotes homologous recombination in bacillus subtilis. J. Bacteriol. 2017; 199:e00682-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Bernhardsgrutter I., Vogeli B., Wagner T., Peter D.M., Cortina N.S., Kahnt J., Bange G., Engilberge S., Girard E., Riobe F. et al.. The multicatalytic compartment of propionyl-CoA synthase sequesters a toxic metabolite. Nat. Chem. Biol. 2018; 14:1127–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Lofgren M., Padovani D., Koutmos M., Banerjee R.. A switch III motif relays signaling between a B12 enzyme and its G-protein chaperone. Nat. Chem. Biol. 2013; 9:535–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Edwards T.E., Baugh L., Bullen J., Baydo R.O., Witte P., Thompkins K., Phan I.Q., Abendroth J., Clifton M.C., Sankaran B. et al.. Crystal structures of Mycobacterial MeaB and MMAA-like GTPases. J. Struct. Funct. Genomics. 2015; 16:91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Berkovitch F., Behshad E., Tang K.H., Enns E.A., Frey P.A., Drennan C.L.. A locking mechanism preventing radical damage in the absence of substrate, as revealed by the x-ray structure of lysine 5,6-aminomutase. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:15870–15875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Maity A.N., Chen Y.H., Ke S.C.. Large-scale domain motions and pyridoxal-5′-phosphate assisted radical catalysis in coenzyme B12-dependent aminomutases. Int. J. Mol. Sci. 2014; 15:3064–3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Rouillon C., White M.F.. The evolution and mechanisms of nucleotide excision repair proteins. Res. Microbiol. 2011; 162:19–26. [DOI] [PubMed] [Google Scholar]
- 102. Krishnan A., Burroughs A.M., Iyer L.M., Aravind L.. Unexpected evolution of lesion-recognition modules in eukaryotic NER and Kinetoplast DNA dynamics proteins from bacterial mobile elements. iScience. 2018; 9:192–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Rossi F., Khanduja J.S., Bortoluzzi A., Houghton J., Sander P., Guthlein C., Davis E.O., Springer B., Bottger E.C., Relini A. et al.. The biological and structural characterization of Mycobacterium tuberculosis UvrA provides novel insights into its mechanism of action. Nucleic Acids Res. 2011; 39:7316–7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Mehta A., Haber J.E.. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 2014; 6:a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Haber J.E. DNA Repair: The Search for Homology. Bioessays. 2018; 40:e1700229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Uranga L.A., Reyes E.D., Patidar P.L., Redman L.N., Lusetti S.L.. The cohesin-like RecN protein stimulates RecA-mediated recombinational repair of DNA double-strand breaks. Nat. Commun. 2017; 8:15282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Ayora S., Carrasco B., Cardenas P.P., Cesar C.E., Canas C., Yadav T., Marchisone C., Alonso J.C.. Double-strand break repair in bacteria: a view from Bacillus subtilis. FEMS Microbiol. Rev. 2011; 35:1055–1081. [DOI] [PubMed] [Google Scholar]
- 108. Rocha E.P., Cornet E., Michel B.. Comparative and evolutionary analysis of the bacterial homologous recombination systems. PLoS Genet. 2005; 1:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Pages V. Single-strand gap repair involves both RecF and RecBCD pathways. Curr. Genet. 2016; 62:519–521. [DOI] [PubMed] [Google Scholar]
- 110. Lawaree E., Jankevicius G., Cooper C., Ahel I., Uphoff S., Tang C.M.. DNA ADP-ribosylation stalls replication and is reversed by RecF-mediated homologous recombination and nucleotide excision repair. Cell Rep. 2020; 30:1373–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Brezellec P., Vallet-Gely I., Possoz C., Quevillon-Cheruel S., Ferat J.L.. DciA is an ancestral replicative helicase operator essential for bacterial replication initiation. Nat. Commun. 2016; 7:13271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Hager J., Staker B.L., Jakob U.. Substrate binding analysis of the 23S rRNA methyltransferase RrmJ. J. Bacteriol. 2004; 186:6634–6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Magni G., Orsomando G., Raffaelli N.. Structural and functional properties of NAD kinase, a key enzyme in NADP biosynthesis. Mini Rev. Med. Chem. 2006; 6:739–746. [DOI] [PubMed] [Google Scholar]
- 114. Piazza A., Heyer W.D.. Homologous recombination and the formation of complex genomic rearrangements. Trends Cell Biol. 2019; 29:135–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Chang H.H.Y., Pannunzio N.R., Adachi N., Lieber M.R.. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017; 18:495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Aravind L., Koonin E.V.. Phosphoesterase domains associated with DNA polymerases of diverse origins. Nucleic Acids Res. 1998; 26:3746–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Paull T.T. 20 Years of Mre11 biology: no end in sight. Mol. Cell. 2018; 71:419–427. [DOI] [PubMed] [Google Scholar]
- 118. Kashammer L., Saathoff J.H., Lammens K., Gut F., Bartho J., Alt A., Kessler B., Hopfner K.P.. Mechanism of DNA end sensing and processing by the Mre11-Rad50 complex. Mol. Cell. 2019; 76:382–394. [DOI] [PubMed] [Google Scholar]
- 119. Wendel B.M., Cole J.M., Courcelle C.T., Courcelle J.. SbcC-SbcD and ExoI process convergent forks to complete chromosome replication. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Dillingham M.S., Kowalczykowski S.C.. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 2008; 72:642–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Ahdash Z., Lau A.M., Byrne R.T., Lammens K., Stuetzer A., Urlaub H., Booth P.J., Reading E., Hopfner K.P., Politis A.. Mechanistic insight into the assembly of the HerA-NurA helicase-nuclease DNA end resection complex. Nucleic Acids Res. 2017; 45:12025–12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Iyer L.M., Koonin E.V., Aravind L.. Classification and evolutionary history of the single-strand annealing proteins, RecT, Redbeta, ERF and RAD52. BMC Genomics. 2002; 3:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Muyrers J.P., Zhang Y., Buchholz F., Stewart A.F.. RecE/RecT and Redalpha/Redbeta initiate double-stranded break repair by specifically interacting with their respective partners. Genes Dev. 2000; 14:1971–1982. [PMC free article] [PubMed] [Google Scholar]
- 124. Hutinet G., Besle A., Son O., McGovern S., Guerois R., Petit M.A., Ochsenbein F., Lecointe F.. Sak4 of phage HK620 is a RecA remote homolog with single-strand annealing activity stimulated by its cognate SSB protein. Front. Microbiol. 2018; 9:743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Aravind L., Makarova K.S., Koonin E.V.. SURVEY AND SUMMARY: holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 2000; 28:3417–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Lovett S.T. The DNA exonucleases of Escherichia coli. EcoSal Plus. 2011; 4:doi:10.1128/ecosalplus.4.4.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Iyer L.M., Makarova K.S., Koonin E.V., Aravind L.. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004; 32:5260–5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Dominski Z. Nucleases of the metallo-beta-lactamase family and their role in DNA and RNA metabolism. Crit. Rev. Biochem. Mol. Biol. 2007; 42:67–93. [DOI] [PubMed] [Google Scholar]
- 129. de Souza R.F., Iyer L.M., Aravind L.. Diversity and evolution of chromatin proteins encoded by DNA viruses. Biochim. Biophys. Acta. 2010; 1799:302–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Hassler M., Shaltiel I.A., Haering C.H.. Towards a unified model of SMC complex function. Curr. Biol. 2018; 28:R1266–R1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Hirano T. At the heart of the chromosome: SMC proteins in action. Nat. Rev. Mol. Cell Biol. 2006; 7:311–322. [DOI] [PubMed] [Google Scholar]
- 132. Makela J., Sherratt D.. SMC complexes organize the bacterial chromosome by lengthwise compaction. Curr. Genet. 2020; doi:10.1007/s00294-020-01076-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Schleiffer A., Kaitna S., Maurer-Stroh S., Glotzer M., Nasmyth K., Eisenhaber F.. Kleisins: a superfamily of bacterial and eukaryotic SMC protein partners. Mol. Cell. 2003; 11:571–575. [DOI] [PubMed] [Google Scholar]
- 134. Muir K.W., Li Y., Weis F., Panne D.. The structure of the cohesin ATPase elucidates the mechanism of SMC-kleisin ring opening. Nat. Struct. Mol. Biol. 2020; 27:233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Soppa J., Kobayashi K., Noirot-Gros M.F., Oesterhelt D., Ehrlich S.D., Dervyn E., Ogasawara N., Moriya S.. Discovery of two novel families of proteins that are proposed to interact with prokaryotic SMC proteins, and characterization of the Bacillus subtilis family members ScpA and ScpB. Mol. Microbiol. 2002; 45:59–71. [DOI] [PubMed] [Google Scholar]
- 136. Mascarenhas J., Soppa J., Strunnikov A.V., Graumann P.L.. Cell cycle-dependent localization of two novel prokaryotic chromosome segregation and condensation proteins in Bacillus subtilis that interact with SMC protein. EMBO J. 2002; 21:3108–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Kim J.S., Shin D.H., Pufan R., Huang C., Yokota H., Kim R., Kim S.H.. Crystal structure of ScpB from Chlorobium tepidum, a protein involved in chromosome partitioning. Proteins. 2006; 62:322–328. [DOI] [PubMed] [Google Scholar]
- 138. Kwon S.Y., Kang B.S., Kim M.H., Kim K.J.. Cloning, expression, purification, crystallization and X-ray crystallographic analysis of ScpB (Rv1710) from Mycobacterium tuberculosis. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2007; 63:1058–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Aravind L., Anantharaman V., Balaji S., Babu M.M., Iyer L.M.. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 2005; 29:231–262. [DOI] [PubMed] [Google Scholar]
- 140. Wells J.N., Gligoris T.G., Nasmyth K.A., Marsh J.A.. Evolution of condensin and cohesin complexes driven by replacement of Kite by Hawk proteins. Curr. Biol. 2017; 27:R17–R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Wild K., Halic M., Sinning I., Beckmann R.. SRP meets the ribosome. Nat. Struct. Mol. Biol. 2004; 11:1049–1053. [DOI] [PubMed] [Google Scholar]
- 142. Pazos M., Natale P., Vicente M.. A specific role for the ZipA protein in cell division: stabilization of the FtsZ protein. J. Biol. Chem. 2013; 288:3219–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Rajasekar K.V., Baker R., Fisher G.L.M., Bolla J.R., Makela J., Tang M., Zawadzka K., Koczy O., Wagner F., Robinson C.V. et al.. Dynamic architecture of the Escherichia coli structural maintenance of chromosomes (SMC) complex, MukBEF. Nucleic Acids Res. 2019; 47:9696–9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Makela J., Sherratt D.J.. Organization of the Escherichia coli Chromosome by a MukBEF Axial Core. Mol. Cell. 2020; 78:250–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Woo J.S., Lim J.H., Shin H.C., Suh M.K., Ku B., Lee K.H., Joo K., Robinson H., Lee J., Park S.Y. et al.. Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions. Cell. 2009; 136:85–96. [DOI] [PubMed] [Google Scholar]
- 146. Zawadzka K., Zawadzki P., Baker R., Rajasekar K.V., Wagner F., Sherratt D.J., Arciszewska L.K.. MukB ATPases are regulated independently by the N- and C-terminal domains of MukF kleisin. Elife. 2018; 7:e31522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Galan J.E. Common themes in the design and function of bacterial effectors. Cell Host Microbe. 2009; 5:571–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Zhang D., de Souza R.F., Anantharaman V., Iyer L.M., Aravind L.. Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol. Direct. 2012; 7:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Zhang D., Burroughs A.M., Vidal N.D., Iyer L.M., Aravind L.. Transposons to toxins: the provenance, architecture and diversification of a widespread class of eukaryotic effectors. Nucleic Acids Res. 2016; 44:3513–3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Iacovache I., van der Goot F.G., Pernot L.. Pore formation: an ancient yet complex form of attack. Biochim. Biophys. Acta. 2008; 1778:1611–1623. [DOI] [PubMed] [Google Scholar]
- 151. Gilbert R.J. Pore-forming toxins. Cell. Mol. Life Sci. 2002; 59:832–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Burroughs A.M., Zhang D., Schaffer D.E., Iyer L.M., Aravind L.. Comparative genomic analyses reveal a vast, novel network of nucleotide-centric systems in biological conflicts, immunity and signaling. Nucleic Acids Res. 2015; 43:10633–10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Iyer L.M., Burroughs A.M., Anand S., de Souza R.F., Aravind L.. Polyvalent proteins, a pervasive theme in the intergenomic biological conflicts of bacteriophages and conjugative elements. J. Bacteriol. 2017; 199:e00245-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Anantharaman V., Makarova K.S., Burroughs A.M., Koonin E.V., Aravind L.. Comprehensive analysis of the HEPN superfamily: identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing. Biol. Direct. 2013; 8:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Burroughs A.M., Aravind L.. RNA damage in biological conflicts and the diversity of responding RNA repair systems. Nucleic Acids Res. 2016; 44:8525–8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Kaur G., Burroughs A.M., Iyer L.M., Aravind L.. Highly regulated, diversifying NTP-dependent biological conflict systems with implications for the emergence of multicellularity. Elife. 2020; 9:e52696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Vasu K., Nagaraja V.. Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol. Mol. Biol. Rev. 2013; 77:53–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Ershova A.S., Rusinov I.S., Spirin S.A., Karyagina A.S., Alexeevski A.V.. Role of restriction-modification systems in prokaryotic evolution and ecology. Biochemistry (Mosc.). 2015; 80:1373–1386. [DOI] [PubMed] [Google Scholar]
- 159. Klaiman D., Steinfels-Kohn E., Krutkina E., Davidov E., Kaufmann G.. The wobble nucleotide-excising anticodon nuclease RloC is governed by the zinc-hook and DNA-dependent ATPase of its Rad50-like region. Nucleic Acids Res. 2012; 40:8568–8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Meineke B., Shuman S.. Determinants of the cytotoxicity of PrrC anticodon nuclease and its amelioration by tRNA repair. RNA. 2012; 18:145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Klaiman D., Steinfels-Kohn E., Kaufmann G.. A DNA break inducer activates the anticodon nuclease RloC and the adaptive immunity in Acinetobacter baylyi ADP1. Nucleic Acids Res. 2014; 42:328–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Chylinski K., Makarova K.S., Charpentier E., Koonin E.V.. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res. 2014; 42:6091–6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Xiao Y., Ng S., Nam K.H., Ke A.. How type II CRISPR-Cas establish immunity through Cas1-Cas2-mediated spacer integration. Nature. 2017; 550:137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Ka D., Jang D.M., Han B.W., Bae E.. Molecular organization of the type II-A CRISPR adaptation module and its interaction with Cas9 via Csn2. Nucleic Acids Res. 2018; 46:9805–9815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Wilkinson M., Drabavicius G., Silanskas A., Gasiunas G., Siksnys V., Wigley D.B.. Structure of the DNA-bound spacer capture complex of a Type II CRISPR-Cas system. Mol. Cell. 2019; 75:90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Sapranauskas R., Gasiunas G., Fremaux C., Barrangou R., Horvath P., Siksnys V.. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011; 39:9275–9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Deltcheva E., Chylinski K., Sharma C.M., Gonzales K., Chao Y., Pirzada Z.A., Eckert M.R., Vogel J., Charpentier E.. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011; 471:602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Arslan Z., Wurm R., Brener O., Ellinger P., Nagel-Steger L., Oesterhelt F., Schmitt L., Willbold D., Wagner R., Gohlke H. et al.. Double-strand DNA end-binding and sliding of the toroidal CRISPR-associated protein Csn2. Nucleic Acids Res. 2013; 41:6347–6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Nam K.H., Kurinov I., Ke A.. Crystal structure of clustered regularly interspaced short palindromic repeats (CRISPR)-associated Csn2 protein revealed Ca2+-dependent double-stranded DNA binding activity. J. Biol. Chem. 2011; 286:30759–30768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Lee K.H., Lee S.G., Eun Lee K., Jeon H., Robinson H., Oh B.H.. Identification, structural, and biochemical characterization of a group of large Csn2 proteins involved in CRISPR-mediated bacterial immunity. Proteins. 2012; 80:2573–2582. [DOI] [PubMed] [Google Scholar]
- 171. Shukla V., Halabelian L., Balagere S., Samaniego-Castruita D., Feldman D.E., Arrowsmith C.H., Rao A., Aravind L.. HMCES functions in the alternative end-joining pathway of the DNA DSB repair during class switch recombination in B cells. Mol. Cell. 2020; 77:384–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Wang L., Jiang S., Deng Z., Dedon P.C., Chen S.. DNA phosphorothioate modification-a new multi-functional epigenetic system in bacteria. FEMS Microbiol. Rev. 2019; 43:109–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Chen S., Wang L., Deng Z.. Twenty years hunting for sulfur in DNA. Protein Cell. 2010; 1:14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. You D., Wang L., Yao F., Zhou X., Deng Z.. A novel DNA modification by sulfur: DndA is a NifS-like cysteine desulfurase capable of assembling DndC as an iron-sulfur cluster protein in Streptomyces lividans. Biochemistry. 2007; 46:6126–6133. [DOI] [PubMed] [Google Scholar]
- 175. Zhou X., He X., Liang J., Li A., Xu T., Kieser T., Helmann J.D., Deng Z.. A novel DNA modification by sulphur. Mol. Microbiol. 2005; 57:1428–1438. [DOI] [PubMed] [Google Scholar]
- 176. Xu T., Yao F., Zhou X., Deng Z., You D.. A novel host-specific restriction system associated with DNA backbone S-modification in Salmonella. Nucleic Acids Res. 2010; 38:7133–7141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. He W., Huang T., Tang Y., Liu Y., Wu X., Chen S., Chan W., Wang Y., Liu X., Chen S. et al.. Regulation of DNA phosphorothioate modification in Salmonella enterica by DndB. Sci. Rep. 2015; 5:12368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Maindola P., Raina R., Goyal P., Atmakuri K., Ojha A., Gupta S., Christie P.J., Iyer L.M., Aravind L., Arockiasamy A.. Multiple enzymatic activities of ParB/Srx superfamily mediate sexual conflict among conjugative plasmids. Nat. Commun. 2014; 5:5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Yao F., Xu T., Zhou X., Deng Z., You D.. Functional analysis of spfD gene involved in DNA phosphorothioation in Pseudomonas fluorescens Pf0-1. FEBS Lett. 2009; 583:729–733. [DOI] [PubMed] [Google Scholar]
- 180. Xiong X., Wu G., Wei Y., Liu L., Zhang Y., Su R., Jiang X., Li M., Gao H., Tian X. et al.. SspABCD-SspE is a phosphorothioation-sensing bacterial defence system with broad anti-phage activities. Nat. Microbiol. 2020; 5:917–928. [DOI] [PubMed] [Google Scholar]
- 181. Xiong L., Liu S., Chen S., Xiao Y., Zhu B., Gao Y., Zhang Y., Chen B., Luo J., Deng Z. et al.. A new type of DNA phosphorothioation-based antiviral system in archaea. Nat. Commun. 2019; 10:1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Yang Y., Xu G., Liang J., He Y., Xiong L., Li H., Bartlett D., Deng Z., Wang Z., Xiao X.. DNA backbone sulfur-modification expands microbial growth range under multiple stresses by its anti-oxidation function. Sci. Rep. 2017; 7:3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Yao P., Liu Y., Wang C., Lan W., Wang C., Cao C.. Investigating the interactions between DNA and DndE during DNA phosphorothioation. FEBS Lett. 2019; 593:2790–2799. [DOI] [PubMed] [Google Scholar]
- 184. Zaremba M., Toliusis P., Grigaitis R., Manakova E., Silanskas A., Tamulaitiene G., Szczelkun M.D., Siksnys V., Tamulaitiene G., Silanskas A. et al.. DNA cleavage by CgII and NgoAVII requires interaction between N- and R-proteins and extensive nucleotide hydrolysis. Nucleic Acids Res. 2015; 43:3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Wah D.A., Bitinaite J., Schildkraut I., Aggarwal A.K.. Structure of FokI has implications for DNA cleavage. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:10564–10569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Bitton L., Klaiman D., Kaufmann G.. Phage T4-induced DNA breaks activate a tRNA repair-defying anticodon nuclease. Mol. Microbiol. 2015; 97:898–910. [DOI] [PubMed] [Google Scholar]
- 187. Meineke B., Shuman S.. Structure-function relations in the NTPase domain of the antiviral tRNA ribotoxin Escherichia coli PrrC. Virology. 2012; 427:144–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Bacusmo J.M., Orsini S.S., Hu J., DeMott M., Thiaville P.C., Elfarash A., Paulines M.J., Rojas-Benitez D., Meineke B., Deutsch C. et al.. The t(6)A modification acts as a positive determinant for the anticodon nuclease PrrC, and is distinctively nonessential in Streptococcus mutans. RNA Biol. 2018; 15:508–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Aravind L., Koonin E.V.. Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res. 2001; 11:1365–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Schreiter E.R., Drennan C.L.. Ribbon-helix-helix transcription factors: variations on a theme. Nat. Rev. Microbiol. 2007; 5:710–720. [DOI] [PubMed] [Google Scholar]
- 191. Banos B., Lazaro J.M., Villar L., Salas M., de Vega M.. Editing of misaligned 3′-termini by an intrinsic 3′-5′ exonuclease activity residing in the PHP domain of a family X DNA polymerase. Nucleic Acids Res. 2008; 36:5736–5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Nakane S., Nakagawa N., Kuramitsu S., Masui R.. Characterization of DNA polymerase X from Thermus thermophilus HB8 reveals the POLXc and PHP domains are both required for 3′-5′ exonuclease activity. Nucleic Acids Res. 2009; 37:2037–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Iyer L.M., Abhiman S., Aravind L.. MutL homologs in restriction-modification systems and the origin of eukaryotic MORC ATPases. Biol. Direct. 2008; 3:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Yang J.Y., Deng X.Y., Li Y.S., Ma X.C., Feng J.X., Yu B., Chen Y., Luo Y.L., Wang X., Chen M.L. et al.. Structure of Schlafen13 reveals a new class of tRNA/rRNA- targeting RNase engaged in translational control. Nat. Commun. 2018; 9:1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Burroughs A.M., Ando Y., Aravind L.. New perspectives on the diversification of the RNA interference system: insights from comparative genomics and small RNA sequencing. Wiley Interdiscip. Rev. RNA. 2014; 5:141–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Aravind L., Leipe D.D., Koonin E.V.. Toprim–a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 1998; 26:4205–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Sironi G. Mutants of Escherichia coli unable to be lysogenized by the temperate bacteriophage P2. Virology. 1969; 37:163–176. [DOI] [PubMed] [Google Scholar]
- 198. Lindahl G., Sironi G., Bialy H., Calendar R.. Bacteriophage lambda; abortive infection of bacteria lysogenic for phage P2. Proc. Natl. Acad. Sci. U.S.A. 1970; 66:587–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Schiltz C.J., Adams M.C., Chappie J.S.. The full-length structure of Thermus scotoductus OLD defines the ATP hydrolysis properties and catalytic mechanism of Class 1 OLD family nucleases. Nucleic Acids Res. 2020; 48:2762–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Schiltz C.J., Lee A., Partlow E.A., Hosford C.J., Chappie J.S.. Structural characterization of Class 2 OLD family nucleases supports a two-metal catalysis mechanism for cleavage. Nucleic Acids Res. 2019; 47:9448–9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Gorbalenya A.E., Koonin E.V.. Superfamily of UvrA-related NTP-binding proteins. Implications for rational classification of recombination/repair systems. J. Mol. Biol. 1990; 213:583–591. [DOI] [PubMed] [Google Scholar]
- 202. Zhou X., Deng Z., Hopwood D.A., Kieser T.. Streptomyces lividans 66 contains a gene for phage resistance which is similar to the phage lambda ea59 endonuclease gene. Mol. Microbiol. 1994; 12:789–797. [DOI] [PubMed] [Google Scholar]
- 203. Benchimol S., Lucko H., Becker A.. A novel endonuclease specified by bacteriophage lambda. Purification and properties of the enzyme. J. Biol. Chem. 1982; 257:5211–5219. [PubMed] [Google Scholar]
- 204. Allemand F., Mathy N., Brechemier-Baey D., Condon C.. The 5S rRNA maturase, ribonuclease M5, is a Toprim domain family member. Nucleic Acids Res. 2005; 33:4368–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Hou L., Klug G., Evguenieva-Hackenberg E.. Archaeal DnaG contains a conserved N-terminal RNA-binding domain and enables tailing of rRNA by the exosome. Nucleic Acids Res. 2014; 42:12691–12706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Sato H., Frank D.W.. ExoU is a potent intracellular phospholipase. Mol. Microbiol. 2004; 53:1279–1290. [DOI] [PubMed] [Google Scholar]
- 207. Ma J., Pan Z., Huang J., Sun M., Lu C., Yao H.. The Hcp proteins fused with diverse extended-toxin domains represent a novel pattern of antibacterial effectors in type VI secretion systems. Virulence. 2017; 8:1189–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Penner M., Morad I., Snyder L., Kaufmann G.. Phage T4-coded Stp: double-edged effector of coupled DNA and tRNA-restriction systems. J. Mol. Biol. 1995; 249:857–868. [DOI] [PubMed] [Google Scholar]
- 209. O’Connell M.R. Molecular mechanisms of RNA targeting by Cas13-containing type VI CRISPR-Cas systems. J. Mol. Biol. 2019; 431:66–87. [DOI] [PubMed] [Google Scholar]
- 210. Peng S., Zhou K., Wang W., Gao Z., Dong Y., Liu Q.. High-resolution crystal structure reveals a HEPN domain at the C-terminal region of S. cerevisiae RNA endonuclease Swt1. Biochem. Biophys. Res. Commun. 2014; 453:826–832. [DOI] [PubMed] [Google Scholar]
- 211. Zhang B., Ye Y., Ye W., Perculija V., Jiang H., Chen Y., Li Y., Chen J., Lin J., Wang S. et al.. Two HEPN domains dictate CRISPR RNA maturation and target cleavage in Cas13d. Nat. Commun. 2019; 10:2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Pillon M.C., Goslen K.H., Gordon J., Wells M.L., Williams J.G., Stanley R.E.. It takes two (Las1 HEPN endoribonuclease domains) to cut RNA correctly. J. Biol. Chem. 2020; 295:5857–5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Garcia-Doval C., Schwede F., Berk C., Rostol J.T., Niewoehner O., Tejero O., Hall J., Marraffini L.A., Jinek M.. Activation and self-inactivation mechanisms of the cyclic oligoadenylate-dependent CRISPR ribonuclease Csm6. Nat. Commun. 2020; 11:1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Uzan M., Miller E.S.. Post-transcriptional control by bacteriophage T4: mRNA decay and inhibition of translation initiation. Virol J. 2010; 7:360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Snyder L. Phage-exclusion enzymes: a bonanza of biochemical and cell biology reagents. Mol. Microbiol. 1995; 15:415–420. [DOI] [PubMed] [Google Scholar]
- 216. Machnicka M.A., Kaminska K.H., Dunin-Horkawicz S., Bujnicki J.M.. Phylogenomics and sequence-structure-function relationships in the GmrSD family of Type IV restriction enzymes. BMC Bioinformatics. 2015; 16:336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Bair C.L., Black L.W.. A type IV modification dependent restriction nuclease that targets glucosylated hydroxymethyl cytosine modified DNAs. J. Mol. Biol. 2007; 366:768–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Iyer L.M., Zhang D., Aravind L.. Adenine methylation in eukaryotes: apprehending the complex evolutionary history and functional potential of an epigenetic modification. Bioessays. 2016; 38:27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Davidov E., Kaufmann G.. RloC: a wobble nucleotide-excising and zinc-responsive bacterial tRNase. Mol. Microbiol. 2008; 69:1560–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Jackson S.A., Fineran P.C.. Bacterial dormancy curbs phage epidemics. Nature. 2019; 570:173–174. [DOI] [PubMed] [Google Scholar]
- 221. Chopin M.C., Chopin A., Bidnenko E.. Phage abortive infection in lactococci: variations on a theme. Curr. Opin. Microbiol. 2005; 8:473–479. [DOI] [PubMed] [Google Scholar]
- 222. Gorbalenya A.E., Koonin E.V.. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. Curr. Opin. Struct. Biol. 1993; 3:419–429. [Google Scholar]
- 223. Aravind L., Walker D.R., Koonin E.V.. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999; 27:1223–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Yu X., West S.C., Egelman E.H.. Structure and subunit composition of the RuvAB-Holliday junction complex. J. Mol. Biol. 1997; 266:217–222. [DOI] [PubMed] [Google Scholar]
- 225. Gomis-Ruth F.X., Moncalian G., Perez-Luque R., Gonzalez A., Cabezon E., de la Cruz F., Coll M.. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature. 2001; 409:637–641. [DOI] [PubMed] [Google Scholar]
- 226. Yu X., Jezewska M.J., Bujalowski W., Egelman E.H.. The hexameric E. coli DnaB helicase can exist in different Quaternary states. J. Mol. Biol. 1996; 259:7–14. [DOI] [PubMed] [Google Scholar]
- 227. Soh Y.M., Burmann F., Shin H.C., Oda T., Jin K.S., Toseland C.P., Kim C., Lee H., Kim S.J., Kong M.S. et al.. Molecular basis for SMC rod formation and its dissolution upon DNA binding. Mol. Cell. 2015; 57:290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Minnen A., Burmann F., Wilhelm L., Anchimiuk A., Diebold-Durand M.L., Gruber S.. Control of Smc coiled coil architecture by the ATPase heads facilitates targeting to chromosomal ParB/parS and release onto flanking DNA. Cell Rep. 2016; 14:2003–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Soppa J. Prokaryotic structural maintenance of chromosomes (SMC) proteins: distribution, phylogeny, and comparison with MukBs and additional prokaryotic and eukaryotic coiled-coil proteins. Gene. 2001; 278:253–264. [DOI] [PubMed] [Google Scholar]
- 230. Cuylen S., Metz J., Haering C.H.. Condensin structures chromosomal DNA through topological links. Nat. Struct. Mol. Biol. 2011; 18:894–901. [DOI] [PubMed] [Google Scholar]
- 231. Wilhelm L., Burmann F., Minnen A., Shin H.C., Toseland C.P., Oh B.H., Gruber S.. SMC condensin entraps chromosomal DNA by an ATP hydrolysis dependent loading mechanism in Bacillus subtilis. Elife. 2015; 4:e06659. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.