Abstract
YoeB–YefM, the widespread type II toxin–antitoxin (TA) module, binds to its own promoter to autoregulate its transcription: repress or induce transcription under normal or stress conditions, respectively. It remains unclear how YoeB–YefM regulates its transcription depending on the YoeB to YefM TA ratio. We find that YoeB–YefM complex from S.aureus exists as two distinct oligomeric assemblies: heterotetramer (YoeB–YefM2–YoeB) and heterohexamer (YoeB–YefM2–YefM2–YoeB) with low and high DNA-binding affinities, respectively. Structures of the heterotetramer alone and heterohexamer bound to promoter DNA reveals that YefM C-terminal domain undergoes disorder to order transition upon YoeB binding, which allosterically affects the conformation of N-terminal DNA-binding domain. At TA ratio of 1:2, unsaturated binding of YoeB to the C-terminal regions of YefM dimer forms an optimal heterohexamer for DNA binding, and two YefM dimers with N-terminal domains dock into the adjacent major grooves of DNA to specifically recognize the 5′-TTGTACAN6AGTACAA-3′ palindromic sequence, resulting in transcriptional repression. In contrast, at TA ratio of 1:1, binding of two additional YoeB molecules onto the heterohexamer induces the completely ordered conformation of YefM and disassembles the heterohexamer into two heterotetramers, which are unable to bind the promoter DNA optimally due to steric clashes, hence derepresses TA operon transcription.
INTRODUCTION
Toxin–antitoxin (TA) systems, the widespread genetic modules in bacterial genomes and plasmids, are emerging as key players in stress responses (1–4). TA gene loci were originally found on plasmids that provided growth advantage for bacteria with TA-containing plasmids and killed plasmid-free bacteria via a mechanism called post-segregation killing (PSK) (5,6). Since then, TA operons were been found not only in plasmids, but also on the chromosomes of most free-living bacteria to facilitate bacteria cell survival under various stress conditions (7). TA system consists of a gene pair encoding a stable toxin that impedes the cell growth by interfering with the vital cellular processes such as transcription, translation, DNA replication and membrane homeostasis, and an unstable antitoxin that counteracts the toxin activity (8,9). Currently, six classes of TA system have been reported on the basis of mechanism for neutralizing the biological effect of toxins (10,11).
Type II TA system is the most widely studied module found in most of the free-living bacteria (12–15). In the type II TA systems, toxins are made of proteins which inhibit replication by disrupting DNA gyrase or translation by cleaving mRNA, tRNA and rRNA, inactivating ribosome elongation factors and glutamyl-tRNA synthetase (GltX) (10,16). Most of the type II toxins are endoribonucleases that adopt microbial RNase fold (17). Antitoxins are regulatory proteins that are typically comprised of two domains, i.e. N-terminal DNA binding domain and C-terminal toxin neutralizing domain that is intrinsically disordered (10,18). The DNA binding domain binds their own operator to regulate transcription of the TA operon. Some of type II TA modules are subject to transcriptional autoregulation to ensure that toxins are repressed transcriptionally under normal growth condition, and are induced only when cells are under stress (16,19,20). The most prevalent mode of transcriptional autoregulation, termed ‘conditional cooperativity’, has been found in a number of type II systems including phd/doc, ccdAB and relBE. The core concept of conditional cooperativity is that the level of TA operon transcription is controlled by the ratio of toxin to antitoxin, and toxin acts as a co-repressor or de-repressor for the antitoxin at lower or higher molar ratio to either suppress or derepress transcription (10,21–23). Although some of type II TA modules have conditional cooperativity as a general mechanism to regulate transcription, the underlying molecular/structural mechanism differs significantly among individual type II TA modules (4,10).
YoeB–YefM TA complex is the widespread type II TA system among archaea and bacteria including major pathogens such as Streptococcus pneumoniae, Mycobacterium tuberculosis and Staphylococcus aureus (24–26). Previous research demonstrated that the YoeB–YefM complex plays a key role in processes such as biofilm formation and response to oxidative stress (27). YoeB–YefM from Escherichia coli was first identified as a homolog of axe-txe from Enterococcus faecium (28). The toxin YoeB, an atypical microbial ribonuclease (RNase), belongs to the superfamily ParE/RelE that could bind the ribosome at A-site, followed by cleaving the mRNA and inhibiting the translation (24,29). The antitoxin YefM is a structural homolog of Phd antitoxin and neutralizes the YoeB toxicity (30). Kamada and Hanaoka determined the structure of YoeB–YefM complex from E. coli as the heterohexamer in the asymmetric unit, i.e. asymmetric disordered-ordered C-terminal region of YefM dimer bound YoeB monomer forms heterotrimer (YefM2–YoeB1), followed by interaction of two heterotrimers to establish heterohexamer (31). In addition, the structure of YoeB in complex with 30S and 70S ribosomes shed light to its mechanism of mRNA cleaving recently (24,29). Like other type II systems, the YoeB–YefM complex binds to its own promoter to autoregulate transcription (32). However, the underlying structural mechanism remains elusive: i.e. it is unclear how the YoeB–YefM complex with different toxin to antitoxin ratio affects its oligomeric state, promoter DNA binding, and ultimately TA gene transcription.
In S. aureus, a major human pathogen that causes a wide range of clinical infections, two chromosomally encoded yefM-yoeB paralogs (YefMSa1–YoeBSa1 and YefMSa2–YoeBSa2) have been identified. The first paralogue YefMSa1 and YoeBSa1 share high sequence identity with YefM (42%) and YoeB (51%) from E. coli, therefore hereafter termed as YoeB–YefM for simplicity. In the present work, we find S. aureus YoeB–YefM complex exists in two different oligomeric states: heterotetramer and heterohexamer in solution. Biochemical analysis reflected heterohexamer as the higher affinity DNA-binding state (∼200-fold) than heterotetramer. We further determined the crystal structures of heterotetramer (YoeB–YefM2–YoeB) alone and heterohexamer (YoeB–YefM2–YefM2–YoeB) bound to its promoter DNA at 2.40 and 2.35 Å resolutions to understand the structural mechanism of transcriptional autoregulation. Interestingly, the structures clearly show YoeB toxin binding induced the intrinsic disordered to ordered transition of YefM C-terminal domain. Moreover, structural analysis explained why the heterohexamer state can optimally bind to the promoter DNA whereas the heterotetramer cannot. Our work provides detail molecular insights into understanding YoeB–YefM complex mediated transcriptional autoregulation, which will help target this type II TA system to overcome antibiotics resistance in S. aureus and other pathogenic bacteria.
MATERIALS AND METHODS
Plasmid constructions
The genes encoding antitoxin yefM (SAOUHSC_02692) and toxin yoeB (SAOUHSC_02691) were amplified from the S. aureus (NCTC8325 strain) genomic DNA, and cloned into pET28a and pET22b vectors (Novagen) to create pET28a-yefM, pET28a-yoeB, pET22b-yefM and pET22b-yoeB expression constructs. Mutant plasmids were derived from the corresponding plasmids via site-directed mutagenesis. All plasmids used in this study are listed in the Supplementary Table S3.
Protein expression and purification
The plasmid containing the full-length yefM was transformed into E. coli BL21 (DE3) for expression of the recombinant YefM protein. Different combination of plasmids were co-transformed into E. coli BL21 (DE3) to express the YoeB–YefM complex as list in Supplementary Table S3. The heterohexamer complex was obtained by co-expression of pET22b-yefM (no tag) and pET28a-yoeB (N-terminal 6xHis-tag); while the heterotetrameric protein complex was achieved by co-expression of pET28a-yefM (N-terminal 6xHis tag) and pET22b-yoeB (no tag). The same strategy was also applied for the expression of site-mutation heterohexameric complex. Co-expression of pET28a-yefM (D23A and D44A) and pET22b-yoeB (K29A and R63A) plasmids gained the heterotrimer. The transformants were grown in LB medium at 37°C until the OD600 reached approximately 0.8, followed by induction with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 16°C for 18 h. Cell was harvested by centrifugation at 8000 rpm during 6 min and frozen at −80°C. The cell pellets were resuspended in buffer A (50 mM Tris–HCl pH 8.0, 500 mM NaCl), followed by lysis on ice via the ultrasonicator (Qsonica, USA). After centrifugation (12 000 rpm for 30 min at 4°C), supernatant was poured onto the Ni2+ affinity chromatographic column (GE Healthcare, USA), followed by further purification with size-exclusion chromatography (Superdex 16/200; GE Healthcare, USA).
Due to the YoeB cytotoxicity for E. coli, recombinant YoeB protein was purified by refolding method (33). Briefly, the purified protein of heterohexamer was used to denature in buffer B (20 mM Tris–HCl pH 8.0, 7 M Guanidine-HCl), and the His-YoeB protein was purified with Ni2+ affinity chromatography, followed by refolding with buffer C (20 mM Tris–HCl pH 8.0, 500 mM NaCl, 5% glycerol) at 16°C utilizing gradient dialysis method. The refolded YoeB protein was further purified using size-exclusion chromatography.
Crystallization and structure determination
Crystallization was performed using sitting-drop vapor diffusion method at 16°C by mixing equal volume (1 μl) of protein (∼12 mg/ml) and reservoir solution. The crystals of heterotetramer were grown in the condition containing 16% (w/v) polyethylene glycol 3350, 0.07 M citric acid and 0.03 M Bis-Tris propane, pH 3.4. To obtain the crystals of YoeB–YefM complex with promoter DNA, the heterohexameric complex was incubated with 26-bp duplex promoter DNA (5′-TTATTGTACAGATATTTGTACAATTG-3′) at 1:1.2 molar ratio for 2 h at 4°C, followed by mixing equal volume of complex and reservoir solution. Optimized crystals were obtained in 10% (w/v) polyethylene glycol 8000, 8% (v/v) ethylene glycol and 0.1 M HEPES pH 7.5. The crystal of YoeB-dimer was obtained in the condition of 25% (w/v) polyethylene glycol 3350, 0.1 M citric acid pH 3.5, with 1/5 volume additive of 20% 1.0 M ammonium sulfate.
The crystals were cryoprotected in 25% glycerol and flash-cooled in liquid nitrogen. All diffraction data were collected at the Shanghai Synchrotron Radiation Facility (SSRF) at the beamlines BL17U1 and BL18U1 and processed with the XDS package (34). Molecular replacement method was used to determine the initial phase with Phenix.phaser (35). The structure of heterotetramer (YoeB2–YefM2) was solved with the dimer (YoeB1–YefM1) from E. coli (PDB: 2A6Q) (31) as a search model. The segment (YoeB and YefM (residues of 40–83)) of heterotetramer was used as search model to solve the structure of heterohexamer-DNA, and the model was further completed after several iterations of automatic and manual building with Phenix.autobuild and Coot (36), respectively. The YoeB monomer from heterotetramer was used to solve the structure of YoeB-dimer. Refinement was performed by combining Phenix.refine and manual building with coot. The data collection and structure refinement statistics are summarized in Supplementary Table S1.
Small-angle X-ray scattering (SAXS) measurement
Solution SAXS data were collected at the Shanghai Synchrotron Radiation Facility (SSRF) at the beamline BL19U2. The samples were prepared as three gradient concentrations (1, 3 and 5 mg/ml) in buffer C (20 mM Tris–HCl pH 8.0, 500 mM NaCl, 5% glycerol). For each measurement, 20 consecutive frames of 1 s exposure time were recorded. Similarly, the background data were recorded in the same buffer and were subtracted from the protein patterns. Data were processed using ATSAS software suites (37), the primusqt program was used for data average, subtraction and comparison with the theoretical scattering curves from the structural models using the program crysol.
Electrophoretic mobility shift assay
Interaction of protein with DNA was validated with the Chemiluminescence EMSA kit (Beyotime Biotechnology, China), as per manufacturers instruction. Briefly, the DNA fragments containing the promoter region were generated by annealing two complementary oligonucleotides labeled with biotin at the 5′ end of forward strand (5′-TTATTGTACAGATATTTGTACAATTG-3′). The constant (2 nM) biotin-labeled DNA fragment was incubated with the increasing amount (2 nM - 400 nM) of purified protein at 25°C for 20 min. Samples were then loaded onto 6.5% native polyacrylamide gel in 0.5× TBE buffer at 80 V for 80 min. The DNA was transferred to a nylon membrane and subsequently UV-cross-linked at 302 nm for 15 min. Chemiluminescence was detected by conjugation of Streptavidin-HRP according to the manufacturer's instructions. The images were obtained using ImageQuant LAS 4000 mini (GE, Piscataway, NJ, USA).
Size exclusion chromatography/multi-angle light scattering
The molecular weight in solution of samples was measured using size exclusion chromatography coupled with multi-angle light scattering (SEC-MALS). Protein samples (2 mg/ml, 100 μl) in buffer A (50 mM Tris–HCl pH 8.0, 500 mM NaCl) were injected into the analytical Superdex 200 Increase 10/300 GL column separating and detecting by the AKTA Purifier System (GE Healthcare, USA) coupled with MALS instrument (DAWN HELEOS 8; Wyatt Technologies, USA) with 0.5 ml/min flow ratio. ASTRA 7 software suite was used to calculate the average weight molecular mass from the intercept of the Debye plot using Zimm's model as implemented in Wyatt's ASTRA software.
Isothermal titration calorimetry (ITC) assays
ITC experiments were performed at 10°C using Microcal PEAQ-ITC equipment (MicroCal Inc. USA). Duplex DNA (600–1200 μM) was dissolved in the reaction buffer A (50 mM Tris–HCl pH 8.0, 500 mM NaCl) and titrated against protein sample (50–100 μM) in the same buffer. The first injection (0.5 μl) was followed by 19 injections of 2 μl with the stirring rate of 750 r.p.m. Data were fitted to one-site binding model via the MicroCal PEAQ-ITC analysis software (MicroCal Inc. USA).
RESULTS
YoeB–YefM complex possess two oligomeric states in solution
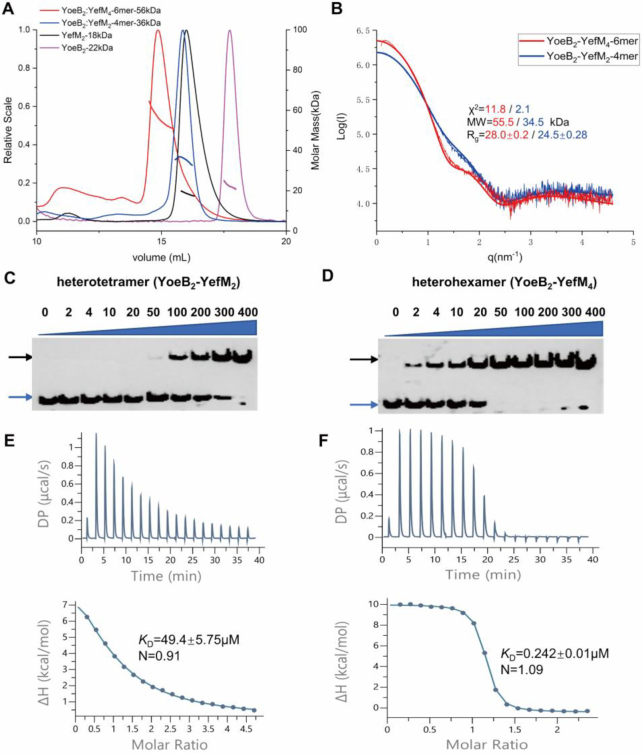
The YoeB–YefM complex co-expressed in E. coli BL21 (DE3) was eluted at two different elution volumes via size-exclusion chromatography. It suggested that the samples may present in different oligomeric states in solution. To evaluate the molecular weight and stoichiometry of the corresponding samples, size-exclusion chromatography coupled with multi-angle light scattering (SEC-MALS) experiments were performed. The apparent molecular weight of YefM, YoeB and two different YoeB–YefM complexes were approximately 18, 22, 36 and 56 kDa, respectively (Figure 1A). Given the theoretical molecular weight of YoeB toxin (10.4 kDa) and YefM antitoxin (9.4 kDa), we conclude that YefM and YoeB exist as a dimer in solution. In addition, the 36 kDa complex peak could be heterotetramers (YoeB2–YefM2, 39.6 kDa) consisting of two YoeB and two YefM molecules while the 56 kDa sample peak could be heterohexamers (YoeB2–YefM4, 60.4 kDa) comprised of two YoeB and four YefM molecules. Small-angle X-ray scattering (SAXS) experiment also calculated molecular weights of 56 and 35 kDa for the two YoeB–YefM protein complexes, which is consistent with the SEC/MALS results (Figure 1B). Taken together, these results supported that YoeB–YefM complex exists as two different oligomeric states in solution: i.e. heterotetramer (YoeB2–YefM2) and heterohexamer (YoeB2–YefM4).
Figure 1.
Different oligomeric states and DNA-binding affinity of YoeB–YefM complex. (A) Size-exclusion chromatography coupled with multiangle light scattering (SEC-MALS) analysis of heterohexamer (red), heterotetramer (blue), YefM (black) and YoeB (magenta). Elution profiles and the calculated molar masses are represented by the corresponding lines. (B) Comparison of the experimental small-angle X-ray scattering (SAXS) curves with the theoretical scattering curves calculated from the structures of heterohexamer (red line) and heterotetramer (blue line). (C) and (D) Electrophoretic mobility shift assay (EMSA) of heterotetramer and heterohexamer with the promoter DNA. The increasing amount (2–400 nM) of proteins was incubated with the constant biotin labeled 26-bp DNA duplex (2 nM). Blue and black arrows denote positions of unbound oligonucleotide and protein-DNA complex. (E and F) Measurement of binding affinities of complexes against the promoter DNA utilizing ITC. (E) Promoter DNA (1.2 mM) titrated against heterotetramer (50 μM) and; (F) DNA (0.6 mM) against heterohexamer (50 μM).
The heterotetramer and heterohexamer complexes have different promoter DNA-binding affinities
In Staphylococcus aureus, the promoter of yefM-yoeB constitutes 8-bp palindromic sequence with 6-bp spacer distance (5′-TTATTGTACAGATATTTGTACAATTG-3′) located at –35 and –10 regions. To examine the specific binding of DNA fragment with YefM and the two oligomeric states of YoeB–YefM, we performed electrophoretic mobility shift assays (EMSA) using 26-bp duplex DNA labeled with 5′ biotin label. The results showed that both heterohexamer and heterotetramer are able to bind the promoter DNA (Figure 1C and D). However, heterohexamer has much higher promoter binding affinity than heterotetramer. In contrast, YefM dimer failed to bind the promoter DNA sequence in this setting (Supplementary Figures S1A and B). In order to obtain the quantitative values for DNA binding affinity, isothermal titration calorimetry (ITC) was employed. The interaction of protein complexes with the DNA is an endothermic reaction, resulting in an entropy-driven binding interaction. The equilibrium dissociation constant (KD) value for heterohexamer (0.242 ± 0.01 μM) was ∼200-fold higher than heterotetramer (49.4 ± 5.75 μM) (Figure 1E and F, Supplementary Table S2).
The distinct DNA-binding affinity of different oligomeric states clearly indicated that the heterohexamer with lower ratio of YoeB to YefM (1:2) has much stronger binding affinity for promoter DNA whereas the heterotetramer with equal amount of YoeB and YefM has neglectable DNA binding affinity. These biochemical results are compatible with the notion that YoeB–YefM complex with different TA ratio autoregulate transcription through differential DNA binding activities.
Structure of the heterotetramer (YoeB–YefM2–YoeB) complex
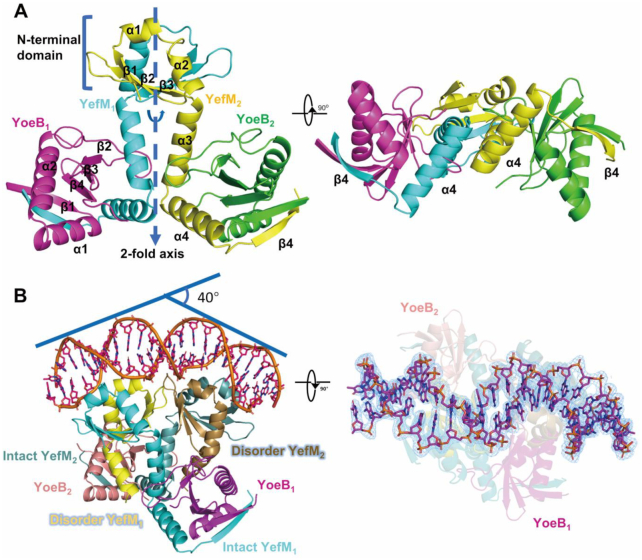
To obtain the structural information for different oligomeric states of YoeB–YefM, both protein complexes were subjected to crystallization experiments but only crystals for heterotetramer were obtained. The structure was determined at the resolution of 2.40 Å and refined to Rwork/Rfree of 18.97%/22.34% (Supplementary Table S1). The crystal belongs to space group P61 and contains two copies of heterotetramer per asymmetric unit. Each heterotetramer is composed of two YoeB and two YefM molecules, which is consistent with the heterotetramer state in solution. The final model covers all residues of YoeB (residues 2–88 of 88) and YefM (residues 2–83 of 83) molecules except the first Met and His-tag are disordered.
The overall structure of heterotetramer assembles into inverted T-shaped architecture and has internal non-crystallographic 2-fold symmetry along the YefM dimeric interface (Figure 2A). The two N-terminal domains (residues 1–40) of YefM form a compact homodimer comprising of central six-stranded β sheets (β1↑, β2↑, β3↓, β3′↑, β2′↓, β1′↓) and four α helices (α1, α2, α1′, α2′). The dimerization is mediated by hydrogen bond between β3 and β3′ as well as hydrophobic packing interactions through Leu13, Leu16, Val20, Val27 and Val29 from each monomer. Additionally, the two α3 helices (residues 41–57) from each monomer also contribute to the dimerization via hydrophobic contacts. The interface of the homodimer buried ∼1288 Å2 accessible surface area as determined by PDB PISA server (38). Both C-terminal domains (residues 41–81) of YefM are well-folded with two α-helices (α3–α4) and one β-strand (β4), which is significantly different with the structure from E.coli where two C-terminal domain is intrinsically disordered (31). Moreover, the C-terminal domain of YefM forms U-shaped structure tightly wrapped around the YoeB monomer burying the accessible surface area of 1650 Å2. Each YoeB monomer adopts a microbial RNase fold (31). The catalytic core is located at the surface of β-sheet consisted of β1, β4, β3 and β2 strands, and is completely blocked by the C-terminal domain of YefM.
Figure 2.
Structures of heterotetramer and heterohexamer-DNA complex. (A) Overall structure (left) and orientations rotated by 90° (right) of the heterotetramer (YoeB–YefM2-YoeB) complex are illustrated in cartoon, YefM are shown in cyan and yellow while YoeB in magenta and green. (B) Overall structure of the heterohexamer–DNA (YoeB–YefM2–YefM2–YoeB–DNA) complex lateral (left) and top view (right). Two intact YefM molecules are shown in cyan/teal and while other two disordered YefM in yellow/brown. Two YoeB molecules are illustrated by magenta/salmon. DNA is shown in orange. The 2Fo – Fc electron density map is displayed at the level of 1.0σ around DNA molecule.
Structure of the heterohexamer (YoeB–YefM2–YefM2–YoeB) in complex with its promoter DNA
Since EMSA and ITC experiments showed higher affinity of heterohexamer for promoter DNA binding, we tried to co-crystallize the heterohexamer complex with the 26-bp duplex DNA to stabilize the heterohexamer complex and promote crystal formation. We succeeded in obtaining the heterohexamer–DNA complex crystals. The crystal belongs to space group P43212 and contains two heterohexamers bound promoter DNA complexes (heterohexamer-DNA) in the asymmetric unit. Each heterohexamer comprises of four YefM and two YoeB molecules, which is in agreement with its heterohexameric state in solution. The structure was refined to Rwork/Rfree of 19.71%/22.25% at 2.35 Å resolution (Supplementary Table S1). In the final model, two YoeB (residues 2–88 of 88) and two YefM (residues 1–83) molecules were completely modeled. However, the electron density maps for the C-terminal domain (residues 58–83) of other two YefM molecules are disordered. The DNA molecules were modelled with core 22–23 bp fragments.
The structure of heterohexamer-DNA complex has an internal pseudo-dyad axis which intersects the DNA at center and is perpendicular to the DNA double helix. The heterohexamer forms a unique YoeB–YefM2–YefM2–YoeB arrangement (Figure 2B). Four YefM molecules form two homodimers in which the N-terminal domains (residues 1–40) constitute two positively charged surface docked into the major grooves of the DNA with the total buried surface area of ∼1955 Å2. Two YoeB molecules individually interact with the two intact YefM C-terminal domains (residues 40–83), while the other two YefM C-terminal domains (residues 55–83) are structurally disordered in the absence of YoeB binding.
The heterohexamer structure in the complex can be divided into two heterotrimeric structures (YoeB–YefM2), the architecture of which is similar to the previously determined DNA-free YoeB–YefM complex structure from E.coli (PDB: 2A6Q) (RMSD of 1.26 Å for 217 Cα) (31). However, the relative positions of two N-terminal DNA-binding domains (DBDs) of YefM are significantly different between two structures with 5.9° deviation that is likely induced by DNA binding (Supplementary Figures S2A and B). It suggested that the relative positions of two DBDs are further adjusted upon DNA binding. In addition, the theoretical scattering curve calculated from the heterohexamer (it was isolated from the heterohexamer-DNA) fits the SAXS experimental data poorly, i.e. χ2 value of 11.8 (Figure 1B). The reasons behind this might be different conformations of apo and DNA-bound states, and the model lacked of the two intrinsically disordered C-terminal regions of YefM in the heterohexamer structure.
Interactions between YoeB and YefM
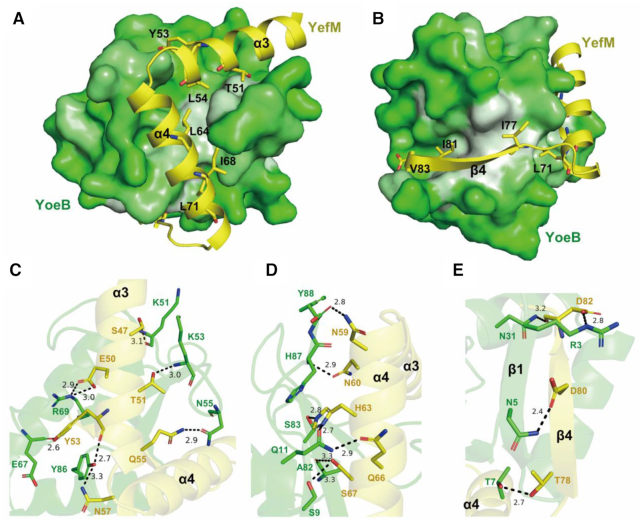
YoeB uses a large and highly complementary interface to recognize the YefM disordered region. The β sheet consisted of β1, β4, β3 and β2 of YoeB creates a concave surface, in which exposed hydrophobic residues (Leu52, Leu56, Tyr59, Val71, Ala82 and Tyr86) make extensive hydrophobic interactions with the hydrophobic residues (Thr51, Tyr53, Leu54, Leu64, Ile68 and Leu71) at α3, α4 helices in YefM (Figure 3A). Moreover, the hydrogen bond network facilitates the specific interactions of YefM and YoeB. The highly conserved residues Glu50 of YefM and Arg69 of YoeB form a salt bridge interaction. Polar residues (Ser47, Glu50, Thr51, Tyr53, Gln55, Asn57, Asn59, Asn60, His63, Gln66 and Ser67) of YefM form hydrogen bonds with the YoeB adjacent residues at either backbone or sidechain (Figure 3C and D).
Figure 3.
Interactions between YoeB and YefM. (A and B) The hydrophobic interaction between YoeB and YefM. The hydrophobic surface of YoeB are presented in white, while the hydrophobic residues of YefM are shown as sticks. (C–E) The detailed hydrogen-bonds interaction of YoeB (green) and YefM (yellow) (α3 (C), α4 (D) and β4 (E)) are presented as blank dashed lines.
The functional oligomer state of YoeB is homodimer that bind at A-site of ribosome to cleave RNA (39,40), while it exists as monomer in the YefM-YoeB complex. Compared with YoeB dimer structure, the last three residues and first β1 strand of YoeB exhibit significant conformational rearrangement (Supplementary Figure S3). The last three residues (Tyr86, His87 and Tyr88) of YoeB adopt a flexible conformation in the apo structure, but are stabilized by the residues located on helices α3 and α4 of YefM via hydrogen bonds in the complex structure. The tyrosyl group of Tyr86 of YoeB forms hydrogen bond with the backbone oxygen of Tyr53 from YefM (Figure 3C). The residues His87 of YoeB stacks pairs with the imidazole ring of the His63 of the YefM (Figure 3D). The hydroxyl group in tyrosyl of Tyr88 forms hydrogen bond with the Tyr53 of YefM and the carboxyl terminal is stabilized by the side chain of Asn57 and Asn59 of YefM via two pair of hydrogen bonds (Figure 3D). The first β1-strand (residues 2–8) of YoeB and the β4-strand (residues 77–83) of YefM fold into an antiparallel β-sheet (Figure 3E). The hydrophobic residues (Ile77, Ile81 and Val83) at β4 of YefM are oriented towards the hydrophobic cavity and interact hydrophobically with Leu4, Ile6, Phe13, Leu34, Ile79 and Ile81 of YoeB (Figure 3B). Compared with the dimeric structure of YoeB, the interactions between YefM and YoeB likely impede YoeB homodimerization: i.e. the interactions between β4 of YefM and β1 of YoeB extended and twisted towards the dimeric interface of YoeB (Figure 3D and Supplementary Figures S3A and S3B). Therefore, YefM suppress the toxicity of YoeB via preventing its dimerization and blocking the active site.
Structural basis of promoter DNA binding and recognition by heterohexamer
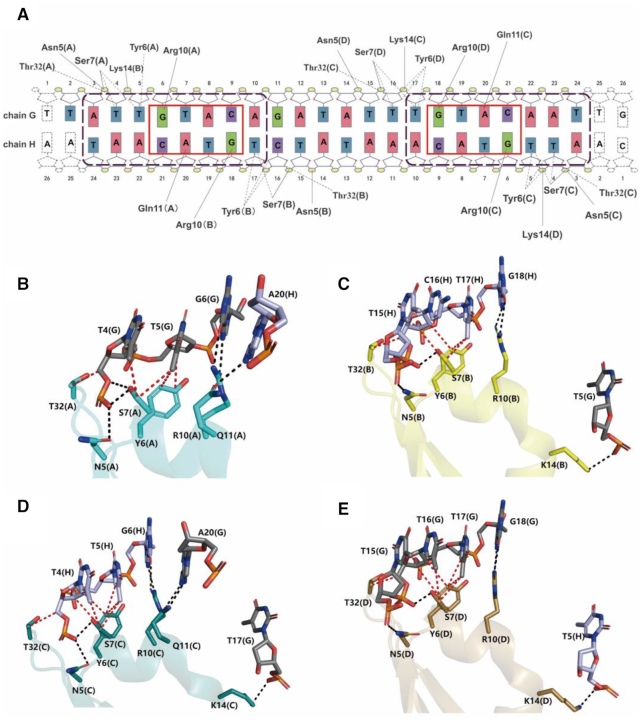
The duplex promoter DNA in heterohexamer-DNA complex adopts an overall B-DNA like conformation. However, the DNA molecule has undergone noticeable bending of 40° toward the heterohexamer protein (Figure 2B). Analysis of the DNA conformation with w3DNA server (41) revealed the angles of propeller twist and roll exhibit much greater than B-form DNA at the major groove of sequence of 5′-GTAC-3′ region while reduce at the AT-rich minor groove, the helical twist angle (∼30°) at the major groove deviates from the value of ideal B-form DNA (36.0° ± 1.01) (Supplementary Figures S4B-S4D). These local deviations result major and minor groove widths are significantly distorted: AT-rich minor groove is compressed to 8.7 Å (compared to 12.5 Å for B-DNA) while the major groove, is expanded to 22.2 Å compared to 16.6 Å for B-DNA (Supplementary Figure S4A).
The heterohexamer recognizes palindromic sequence of the promoter DNA at adjacent major grooves using two symmetrical DNA-binding domains, each comprised of residues coming from a dimer of YefM. The main interactions with major groove of DNA are mediated by the antiparallel HTH (α1-turn-α2) motifs inserted into the major groove of DNA to recognize 5′-TTGTACA-3′ sequence, and the winged loops between β2 and β3 provide additional minor groove contacts. Sequence-specific base interactions are mediated by highly conserved residue Arg10 at the antiparallel HTH motif with the guanine bases of 5′-TTGTACA-3′ palindromic sequence. The residue Gln11 forms hydrogen bond interaction with the adenine of the 3′- TAACATGT -5′ sequence (Figure 4A-E). Thus, the Arg10-Gln11 pair specifically recognizes the core GTAC box (5′-ATTGTACA-3′) of palindromic sequence. Indeed, mutation of the core sequence 5′-ATTGTACA-3′ to 5′-ATTTTAAA-3′ completely abolished the protein-DNA interaction (Supplementary Figure S1C). In addition, the thymine bases flanked in the core GTAC box are also important for protein-DNA interaction, that contact the residues of Tyr6 and Ser7 as Van der Waals interactions. Substitution of 5′-ATTGTACA-3′ to 5′-AGCGTACG-3′ resulted in the loss of interaction (Supplementary Figure S1D). In addition to the base-specific interactions, the residues (Asn5, Ser7, Lys14 and Thr32) participate in hydrogen bond interactions with the phosphate backbone of DNA (Figure 4A-E). Furthermore, the mutational studies were performed to confirm the interaction, the results showed that various mutants of residues in YefM (R10A, Q11A, R10A/Q11A, Y6A, S7A, Y6A/S7A and N5A/K14A/T32A) decreased and abolished the binding affinity to the promoter DNA. (Supplementary Figure S8).
Figure 4.
DNA recognition and binding. (A) Schematic overview of the interactions between heterohexamer and DNA. Solid and dashed lines depict the interactions of hydrogen bond and Van der Waals interaction interactions, respectively. Red and purple boxes presented the core GTAC box and palindromic sequence, respectively. (B–E) Interactions of YefM with promoter DNA. The hydrogen bonds and Van der Waals interactions are presented as blank and red dashed lines, respectively. The chain number of YefM and DNA are presented in parentheses.
Sequence alignment of YefM depicted that the DNA interaction residues (Tyr6Ser7, Arg10Gln11) are relatively conserved in diverse range of bacteria. In addition, their promoter region contains the GATC core box with 8 bp spacer while different flanking region sequences (most harbor with T/A bases) (Supplementary Figure S5). Hence, this interaction fashion between YefM and DNA might be conserved among diverse bacteria.
Structural basis of differential DNA binding affinities between heterotetramer and heterohexamer complexes
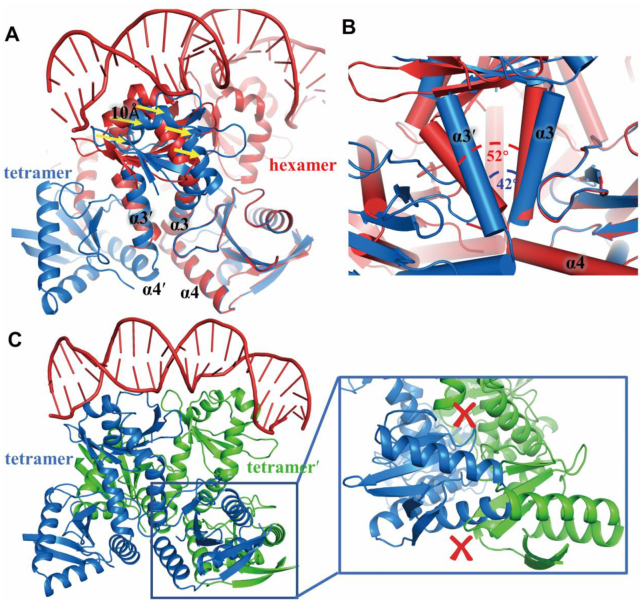
To understand why the heterohexamer possess higher DNA-binding affinity than heterotetramer and how these two oligomeric states differ, structural comparison was performed. When the heterotetrametric structure is superposed onto the structure of heterohexamer-DNA using the common YoeB molecules, the C-terminal region (residues 57–83, α4β4) of YefM completely overlap whereas the N-terminal regions (residues 1–56, β1α1α2β2β3α3) of heterotetramer swing away from the major groove of DNA about 10 Å measured by the displacement of Cα of Arg10 (Figure 5A and Supplementary Figure S6A and S6C). The helix α3 of YefM act as a hinge between the N- and C-terminal domain, and is crucial for this conformational change. The two helices α3 from each monomer of YefM dimer cross at different angles of 42° and 52° in heterotetramer and heterohexamer, respectively (Figure 5B). In heterotetramer, saturated binding of YefM molecules with YoeB enables C-terminal domain to form ordered conformation (α3α4β4). To accommodate two ordered α4 helices, the two α4 arrange in a parallel manner to avoid collision (Supplementary Figure S6D). Consequently, the helix α3 of YefM in heterotetramer rotates about 10° compared with heterohexamer, resulting in the rotation of whole N-terminal domain at ∼10 Å away from the DNA major groove (Supplementary Figure S6B). Both ordered conformation of C-terminal domains of YefM dimer upon binding of YoeB constrains the movement of N-terminal DNA-binding domain, which is less optimal for DNA binding. In contrast, the asymmetric disorder-order conformation of YefM dimer in the heterohexamer results in more flexible N-terminal DNA-binding domains to accommodate optimal DNA binding.
Figure 5.
Structural comparison of heterotetramer and heterohexamer-DNA complex. (A) Superposed the structures of heterotetramer (blue) and heterohexamer-DNA (red) based on the mutual YoeB molecule. The C-terminal regions (residues 57–83, α4β4) of YefM completely overlap, while the N-terminal domain of YefM has a shift (10 Å) presented by the yellow arrows. (B) Two helices (α3-α3′) from each monomer of YefM dimer cross at different angles of 42° and 52° in heterotetramer and heterohexamer, respectively. (C) Structural model for binding of two heterotetramers to DNA simultaneously. The crosses in red presented steric clashes in the C-terminal region of YefM and YoeB molecules.
In the structure of heterohexamer-DNA complex, the N-terminal DNA-binding domains of YefM dimer insert into adjacent major grooves of promoter DNA. If the two heterotetramer structures are superimposed with the heterohexamer structure using the N-terminal DNA-binding domains, severe steric clashes would occur at the C-terminal regions of YefM and YoeB molecule as illustrated in Figure 5C. This analysis is consistent with the ITC data that the heterotetramer bind DNA at a stoichiometry of 1:1 (N = 0.909). Therefore, two heterotetramers are unable to bind the promoter DNA simultaneously. Taken together, these results supported that heterohexamer is the optimal state to bind the promoter DNA, whereas heterotetramer binds promoter DNA with much weaker affinity due to the steric clashes, that prevent two heterotetramers to form an octamer complex. Therefore, the comparative structural analysis explained why the heterotetramer has a low affinity for promoter DNA-binding than heterohexamer.
Although, upon binding the additional YoeB makes the conformational change in N- and C-terminal domain of YefM, most residues in YefM keep the same orientation in the heterohexamer and heterotetramer. However, only the Glu50 and Tyr53 residues undergo slight rotation (Supplementary Figure S6E). Interestingly, the YefM disordered conformation begins at Tyr53 residue, suggesting Tyr53 of YefM might be important for initial recognition by the additional YoeB molecule.
The heterohexamer is the optimal oligomeric state for promoter DNA-binding
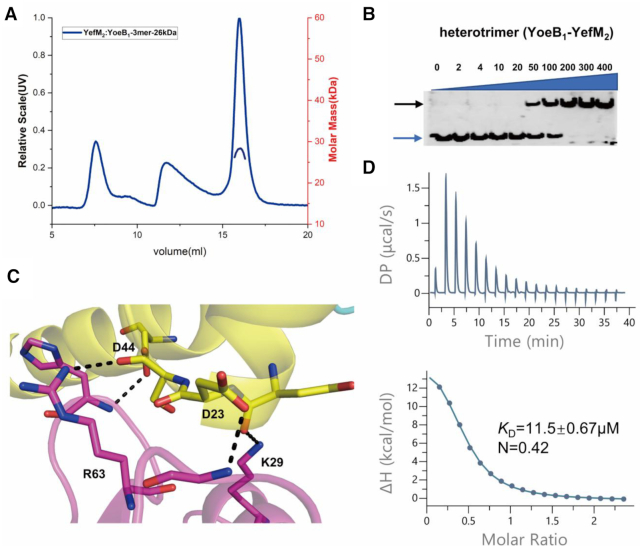
YoeB–YefM complex from Staphylococcus aureus could adopt heterohexameric state both in solution and crystal structure. Analysis of the heterohexameric structure (YoeB–YefM2–YefM2–YoeB) revealed that two heterotrimers (YoeB–YefM2/YoeB′–YefM′2) manly interact with each other through hydrogen bonds. The side chain of Asp23 and Asp44 residues of YefM forms hydrogen bonds with the backbone amide atoms of Gly47 and His66 from YoeB′. The side chain of Lys29 and Arg63 of YoeB′ participates in the hydrogen bond interactions with the backbone carbonyl group of Asn21 and Ser24 of YefM, respectively (Figures 6C). The similar interactions are also found in the YoeB–YefM structure (PDB code: 2A6Q) from E. coli (Supplementary Figures S7). These key residues might mediate the formation of heterohexamer assembly. These four residues (Asp23 and Asp44 of YefM, Lys29 and Arg63 of YoeB) corresponding to the binding site for the heterohexamer formation were abbreviated as D23D44K29R63 binding site for simplicity. Consistent with the structure, mutation of these residues to alanine essentially abolished the formation of heterohexamer as demonstrated by SEC-MALS assays: the average molecular mass of the mutant is about 26 kDa, which represents the heterotrimeric state (Figure 6A). Additionally, results from EMSA and ITC assays showed significant reduction (KD = 11.5±0.67 μM, ∼47-fold decrease) in the DNA binding affinity of D23D44K29R63 mutant as compared with the wild type heterohexameric complex (Figure 6B and D). Therefore, these results demonstrated that the D23D44K29R63 site is critical for the heterohexameric state formation and optimal DNA-binding affinity.
Figure 6.
The heterohexamer is the optimum state for promoter DNA-binding. (A) SEC-MALS characterization of the heterohexamer after mutation. Data analysis indicated the molecular mass of complex (26 kDa) consistent with the mass of heterotrimeric state. (B) EMSA analysis of the promoter DNA with the heterotrimer (after mutation of heterohexamer). The increasing concentration (2 nM - 400 nM) of protein incubated with 2 nM DNA. Blue and black arrows denote positions of unbound oligonucleotide and protein–DNA complex. (C) Detail interactions at the D23D44K29R63 site. Dash lines indicated the hydrogen bond between YefM and YoeB. (D) Measurement of binding affinity for promoter DNA (1.2 mM) and heterotrimer (100 μM) utilizing ITC. The dissociation constant (KD) and the binding stoichiometry (N) are shown.
DISCUSSION
Bacterial TA systems play a key role in adaption to stress conditions. A hallmark of TA operons is autoregulation of its transcription through a mechanism known as conditional cooperativity. In the present study, we used YoeB–YefM TA complexes from S. aureus as a model system to study the underlying molecular mechanism. The YefM dimer on its own is unable to bind the DNA, whereas the YoeB–YefM complexes have differential DNA-binding affinities depending on the TA molecular ratio. Biochemical data supported that the ratios of YoeB and YefM are determinant in fine-tune its own transcription. We found that YoeB–YefM can form two different oligomeric states including heterotetramer (YoeB2–YefM2) and heterohexamer (YoeB2–YefM4) in solution and exhibit distinct promoter DNA binding affinities. In the meantime, the same tetrameric structure (PDB:7BWF) from Staphylococcus aureus was reported (42), which is agree with our observations in this study. This suggests that YoeB–YefM forms distinct oligomeric complexes at different TA molecular ratio to regulate TA transcription. At TA ratio of 1:2, heterohexamer complex binds to promoter DNA with high affinity to repress TA transcription under normal growth conditions. Under stress conditions, the SOS response induces Lon protease, which preferentially cleaves unstable antitoxin YefM (43). This will cause an increase in the TA molar ratio. Indeed at the TA ratio of 1:1, heterotetramer complex is formed, which binds to promoter DNA with neglectable affinity and causes derepression of TA operon transcription. Hence, the TA ratio alteration is linked to the changes in the TA oligomeric state to regulate gene transcription. Different oligomeric states exist in many TA systems covered various toxins with diverse activities and folds, and antitoxins belong to different classes, such as Phd/Doc (21), relBE (23), AtaR-AtaT (44,45), CcdAB (22,46) and VapBC (47,48). In contrast, some TA systems formed only single oligomeric state, such as DinJ/YafQ (49,50) and MqsRA (51), which might achieve the transcription autoregulation through different mechanisms.
What is the underlying structural mechanism for different oligomeric states to regulate TA gene transcription? So far, no crystal structures of repressed states bound operator DNA and derepressed state alone complexes were determined, simultaneously. Through detailed structural analysis, we now understand that the YoeB–YefM operon is regulated by YoeB–YefM complexes at two different levels. The first level is regulated by conformational change and allostery through the molecular switch YefM protein. The unstable antitoxin YefM uses its unstructured C-terminal domain as a sensor to recognize the toxin YoeB and allosterically regulate the conformation of N-terminal DNA-binding domain. YefM shows significant structural differences in the absence and presence of toxin YoeB. Binding of YoeB makes the YefM C-terminal intrinsic disorder region form structurally ordered conformation and this allosteric effect extends to the N-terminal domain to affect the DNA binding affinity. This allosteric coupling might be due to the small hydrophobic core shielded by the aromatic side chains of the helix α3 of YefM, which was also described in the other Phd/doc TA system (21). Therefore, the C-terminal intrinsic disorder region of YefM is a fundamental structural element of antitoxin coupling toxin binding to DNA recognition through helix α3. The second level is through forming different oligomeric states to further regulate DNA binding: i.e. each heterotrimer (YoeB–YefM2) is a DNA recognition unit recognizing promoter DNA with an affinity of ∼11.5 μM. Heterohexamer formation brings two such units together with optimal spacing to bind simultaneously to the promoter DNA, which increases DNA binding affinity to 0.242 μM through avidity effect. Mutation of D23D44K29R63 residues at the heterohexamer interface reduced the hexamer into two trimers. Consistent with the avidity effect, this results in ∼47-fold decrease in the DNA-binding affinity as compared to heterohexamer (11.5 μM versus 0.242 μM). More or less the same binding site (Low affinity, L site) was also found in CcdAB (46) and Phd/Doc (21) systems. Binding of two additional molecules of YoeB to the heterohexamer disrupts DNA binding also at two levels: triggering conformational changes within the YefM dimer to affect N-terminal DNA binding domain allosterically; dissembling the hexamer into two tetramers with much lower DNA binding affinity (∼49.4 μM) due to the steric clashes.
The crystal structure of heterohexamer-DNA complex revealed that four N-terminal domains of YefM form two DNA-binding units that insert into two adjacent major grooves of promoter DNA. The promoter of yefM-YoeB constitutes 8-bp palindromic sequence (5′-TTATTGTACAGATATTTGTACAATTG-3′). The core palindromic sequence (5′-GTAC(N)8GTAC-3′) was also found for the yefM-yoeB promoter region among the diverse microbial species such as Escherichia coli K12, Shigella boydii and Streptococcus agalactiae (32) (Supplementary Figure S5B). Based on the DNA structural analysis and biochemical assay, the conserved residues (Arg10–Glu11) on the antiparallel HTH motif of YefM specifically recognize the guanine and adenine bases of the core palindromic sequence (5′-GTAC(N)8GTAC-3′). In addition, the flanking sequence are crucial for specific recognition via the hydrophobic interaction. Two YefM–YoeB paralogs from S. aureus (YefMSa1–YoeBSa1 and YefMSa2–YoeBSa2) do not cross-talk with each other and are transcriptionally autoregulated by their own cognate antitoxin (52). The YefMSa1 and YefMSa2 paralogs share conserved residue (Arg10) and sequence identity of 26.4%. The promoter for both paralogs harbors the same 5′-GTAC(N)8GTAC-3′ DNA core sequence but different flanking sequences (Supplementary Figure S5B). Further comparative structural work is necessary to fully understand their DNA binding specificity, which is likely controlled by specific interaction with the flanking DNA sequence. Moreover, the space between the core sequence may be correlated with the DNA bending angle: i.e. more bending angle with the increase in DNA spacing. Comparative analysis of YefM-YoeB and Phd/Doc systems, which have distance of 8- and 9-bp between core GTAC box, showed that binding of the YefM-YoeB or Phd/Doc complexes to the duplex DNA caused DNA to bend about 40° and 60°, respectively (53). These results indicated that the DNA bending angle depends on the distance between the core DNA sequence.
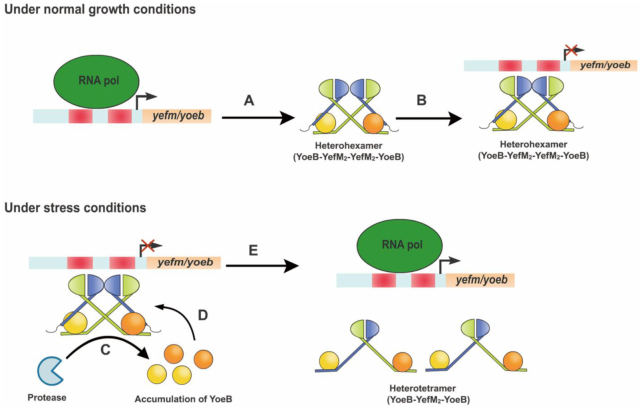
The main objective of this study was to explore the molecular mechanism of conditional cooperativity of YefM-YoeB system. Combined with our structural and corresponding biochemical data in this study and others, we proposed a model for YoeB–YefM transcriptional autoregulation (Figure 7). Under normal growth condition, the transcription of the TA operon is strictly repressed. This repression is achieved through a complex of YoeB and YefM where YoeB toxin acts as a corepressor with a TA ratio of 1:2. Each YoeB molecule interacts with one of asymmetric YefM dimer through the C-terminal intrinsic disordered region to form trimeric YoeB–YefM2 complex. Two trimeric complexes are closely associated with each other via D23D44K29R63 to form heterohexamer with greater DNA binding affinity through avidity effect and strongly bind promoter DNA to repress TA operon. During the stress conditions, preferential proteolytic degradation of YefM in DNA-loaded heterohexamer by Lon protease releases more YoeB toxins (43,54), which leads to partial activation of transcription. YefM is rapidly degraded by the protease, resulting the accumulation of YoeB, which further bind to the two unoccupied YefM molecules in the heterohexamer complex. Binding of two extra YoeB molecules changes the conformation of YefM dimer in the heterohexamer, which dissembles heterohexamer into two heterotetramers with much lower DNA binding affinity (∼49 μM). This results in the transcriptional derepression of TA operon. Consequently, the un-neutralized YoeB toxin is able to inhibit the translational initiation and cellular growth by its own ribosome-dependent RNase activity. In conclusion, variation in the ratio of YoeB to YefM is manifested as different oligomeric states of YoeB–YefM complex which dynamically regulates its own transcription to adjust the environmental cues.
Figure 7.
The model for transcriptional autoregulation of YoeB–YefM TA system. Under normal growth conditions, the transcription of the operon is strictly repressed. (A) The YefM antitoxin and YoeB toxin are transcribed as bicistronic operons by an autoregulated promoter. (B) Since YefM antitoxin appear to be more efficiently translated than YoeB toxin, they tend to form heterohexamer (YoeB–YefM2–YefM2–YoeB) for binding of the promoter at low ratio of YoeB:YefM, resulting in repression of the transcription. During stress condition, the operon is activated by the protease and excess YoeB. (C) The proteolytic degradation of YefM in DNA-loaded heterohexamer induces partial activation of transcription and release YoeB molecules. (D) The excess YoeB molecules can act as a derepressor to bind the remaining two free C termini of YefM in the heterohexamer complex. (E) Binding of two extra YoeB molecules changes the conformation of YefM dimer in the heterohexamer, which dissembles heterohexamer into two heterotetramers (YoeB–YefM2–YoeB) with much lower DNA binding affinity, resulting in the transcriptional derepression of the operon.
The knowledge generated from the current study provides clue for strategies of disrupting conditional cooperativity in YoeB–YefM type II TA systems to overcome antibiotic resistance. TA system is used as a weapon to promote bacterial survival under stress conditions: i.e. subpopulation of persister bacteria can survive the stress condition and regrow the whole population when stress is relieved. It is conceivable that selective disruption of YefM antitoxin translation by CRISPR gene editing may cause deregulation of YoeB–YefM TA system. In this case both YoeB and YefM can be transcribed into mRNA but YefM cannot be translated into a functional protein so that YoeB will be constitutively expressed to suppress bacterial growth as a suicide mechanism. Such CRISPR enzyme can be delivered into antibiotic resistant bacteria through bacteriophage vehicles. Alternatively, small molecule modulators of the YoeB toxin, which selectively disrupt its interaction with YefM (cause derepression of YoeB–YefM TA system) but not affect its ribosome-dependent RNase activity can also be used as drugs to target YoeB–YefM TA system to overcome antibiotic resistance.
DATA AVAILABILITY
The atomic coordinates have been deposited in the Protein Data Bank (PDB) (codes: 6L8E for heterohexamer-DNA (YoeB2-YefM4-DNA); 6L8F for heterotetramer (YoeB2–YefM2); 7CUA for YoeB dimer).
Supplementary Material
ACKNOWLEDGEMENTS
We would appreciate Prof. Tengchuang Jin for assistance with critical comments and manuscript revision. We thank the staff at beamline BL17U1, BL18U1 and BL19U1 of the Shanghai Synchrotron Radiation Facility for help with data collection.
Author contributions: Lu Xue and Jian Yue performed the experiments, analyzed data and wrote paper; Jiyuan Ke modified the manuscript, Muhammad Hidayatullah Khan provided advice for the experimental process and modified the article; Wen Wen and Baolin Sun provided some interpretation during the experiments; Zhongliang Zhu and Liwen Niu supervised the project.
Contributor Information
Lu Xue, Hefei National Laboratory for Physical Sciences at the Microscale, Division of Molecular and Cellular Biophysics, University of Science and Technology of China, Hefei, Anhui 230026, China; School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230026, China.
Jian Yue, Hefei National Laboratory for Physical Sciences at the Microscale, Division of Molecular and Cellular Biophysics, University of Science and Technology of China, Hefei, Anhui 230026, China; School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230026, China.
Jiyuan Ke, Lead Discovery Department, H3 Biomedicine Inc, 300 Technology Square FL 5, Cambridge, MA 02139, USA.
Muhammad Hidayatullah Khan, Hefei National Laboratory for Physical Sciences at the Microscale, Division of Molecular and Cellular Biophysics, University of Science and Technology of China, Hefei, Anhui 230026, China; School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230026, China.
Wen Wen, School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230026, China.
Baolin Sun, School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230026, China.
Zhongliang Zhu, Hefei National Laboratory for Physical Sciences at the Microscale, Division of Molecular and Cellular Biophysics, University of Science and Technology of China, Hefei, Anhui 230026, China; School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230026, China.
Liwen Niu, Hefei National Laboratory for Physical Sciences at the Microscale, Division of Molecular and Cellular Biophysics, University of Science and Technology of China, Hefei, Anhui 230026, China; School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230026, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31621002 to L.N., U1632124 to L.N. and 31270770 to Z.Z.]; Ministry of Science and Technology of China [2017YFA0504903 to L.N.]; Hefei National Science Center Pilot Project Funds (in part). Funding for open access charge: National Natural Science Foundation of China [31621002 to L.N.]; Ministry of Science and Technology of China [2017YFA0504903 to L.N.]; Hefei National Science Center Pilot Project Funds (in part).
Conflict of interest statement. None declared.
REFERENCES
- 1. Gerdes K., Christensen S.K., Lobner-Olesen A.. Prokaryotic toxin–antitoxin stress response loci. Nat. Rev. Microbiol. 2005; 3:371–382. [DOI] [PubMed] [Google Scholar]
- 2. Yamaguchi Y., Inouye M.. Regulation of growth and death in Escherichia coli by toxin–antitoxin systems. Nat. Rev. Microbiol. 2011; 9:779–790. [DOI] [PubMed] [Google Scholar]
- 3. Van Melderen L. Toxin-antitoxin systems: why so many, what for. Curr. Opin. Microbiol. 2010; 13:781–785. [DOI] [PubMed] [Google Scholar]
- 4. Harms A., Brodersen D.E., Mitarai N., Gerdes K.. Toxins, targets, and Triggers: An overview of Toxin-Antitoxin biology. Mol. Cell. 2018; 70:768–784. [DOI] [PubMed] [Google Scholar]
- 5. Gerdes K., Rasmussen P.B., Molin S.. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. U.S.A. 1986; 83:3116–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ogura T., Hiraga S.. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc. Natl. Acad. Sci. U.S.A. 1983; 80:4784–4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Leplae R., Geeraerts D., Hallez R., Guglielmini J., Dreze P., Van Melderen L.. Diversity of bacterial type II toxin–antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Res. 2011; 39:5513–5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hayes F., Van Melderen L.. Toxins-antitoxins: diversity, evolution and function. Crit. Rev. Biochem. Mol. Biol. 2011; 46:386–408. [DOI] [PubMed] [Google Scholar]
- 9. Deter H.S., Jensen R.V., Mather W.H., Butzin N.C.. Mechanisms for differential protein production in Toxin-Antitoxin systems. Toxins (Basel). 2017; 9:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Page R., Peti W.. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 2016; 12:208–214. [DOI] [PubMed] [Google Scholar]
- 11. Unterholzner S.J., Poppenberger B., Rozhon W.. Toxin-antitoxin systems: biology, identification, and application. Mob Genet. Elem. 2013; 3:e26219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang Q.E., Walsh T.R.. Toxin-antitoxin systems and their role in disseminating and maintaining antimicrobial resistance. FEMS Microbiol. Rev. 2017; 41:343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Makarova K.S., Wolf Y.I., Koonin E.V.. Comprehensive comparative-genomic analysis of type 2 toxin–antitoxin systems and related mobile stress response systems in prokaryotes. Biol. Direct. 2009; 4:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xie Y., Wei Y., Shen Y., Li X., Zhou H., Tai C., Deng Z., Ou H.Y.. TADB 2.0: an updated database of bacterial type II toxin–antitoxin loci. Nucleic Acids Res. 2018; 46:D749–D753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rocker A., Meinhart A.. Type II toxin: antitoxin systems. More than small selfish entities. Curr. Genet. 2016; 62:287–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fraikin N., Goormaghtigh F., Van Melderen L.. Type II toxin-antitoxin systems: evolution and revolutions. J. Bacteriol. 2020; 202:e00763-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ramisetty B.C.M., Santhosh R.S.. Endoribonuclease type II toxin–antitoxin systems: functional or selfish. Microbiology. 2017; 163:931–939. [DOI] [PubMed] [Google Scholar]
- 18. Park S.J., Son W.S., Lee B.J.. Structural overview of toxin–antitoxin systems in infectious bacteria: a target for developing antimicrobial agents. Biochim. Biophys. Acta. 2013; 1834:1155–1167. [DOI] [PubMed] [Google Scholar]
- 19. Mruk I., Kobayashi I.. To be or not to be: regulation of restriction-modification systems and other toxin–antitoxin systems. Nucleic Acids Res. 2014; 42:70–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cataudella I., Trusina A., Sneppen K., Gerdes K., Mitarai N.. Conditional cooperativity in toxin–antitoxin regulation prevents random toxin activation and promotes fast translational recovery. Nucleic Acids Res. 2012; 40:6424–6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Garcia-Pino A., Balasubramanian S., Wyns L., Gazit E., De Greve H., Magnuson R.D., Charlier D., van Nuland N.A., Loris R.. Allostery and intrinsic disorder mediate transcription regulation by conditional cooperativity. Cell. 2010; 142:101–111. [DOI] [PubMed] [Google Scholar]
- 22. Vandervelde A., Drobnak I., Hadzi S., Sterckx Y.G., Welte T., De Greve H., Charlier D., Efremov R., Loris R., Lah J.. Molecular mechanism governing ratio-dependent transcription regulation in the ccdAB operon. Nucleic Acids Res. 2017; 45:2937–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Boggild A., Sofos N., Andersen K.R., Feddersen A., Easter A.D., Passmore L.A., Brodersen D.E.. The crystal structure of the intact E. coli RelBE toxin–antitoxin complex provides the structural basis for conditional cooperativity. Structure. 2012; 20:1641–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Feng S., Chen Y., Kamada K., Wang H., Tang K., Wang M., Gao Y.G.. YoeB-ribosome structure: a canonical RNase that requires the ribosome for its specific activity. Nucleic Acids Res. 2013; 41:9549–9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cherny I., Gazit E.. The YefM antitoxin defines a family of natively unfolded proteins: implications as a novel antibacterial target. J. Biol. Chem. 2004; 279:8252–8261. [DOI] [PubMed] [Google Scholar]
- 26. Kang S.M., Kim D.H., Jin C., Lee B.J.. A systematic overview of type II and III Toxin-Antitoxin systems with a focus on druggability. Toxins (Basel). 2018; 10:515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chan W.T., Domenech M., Moreno-Cordoba I., Navarro-Martinez V., Nieto C., Moscoso M., Garcia E., Espinosa M.. The streptococcus pneumoniaeyefM-yoeB and relBE toxin-antitoxin operons participate in oxidative stress and biofilm formation. Toxins (Basel). 2018; 10:378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Grady R., Hayes F.. Axe-Txe, a broad-spectrum proteic toxin–antitoxin system specified by a multidrug-resistant, clinical isolate of Enterococcus faecium. Mol. Microbiol. 2003; 47:1419–1432. [DOI] [PubMed] [Google Scholar]
- 29. Pavelich I.J., Maehigashi T., Hoffer E.D., Ruangprasert A., Miles S.J., Dunham C.M.. Monomeric YoeB toxin retains RNase activity but adopts an obligate dimeric form for thermal stability. Nucleic Acids Res. 2019; 47:10400–10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Arbing M.A., Handelman S.K., Kuzin A.P., Verdon G., Wang C., Su M., Rothenbacher F.P., Abashidze M., Liu M., Hurley J.M. et al.. Crystal structures of Phd-Doc, HigA, and YeeU establish multiple evolutionary links between microbial growth-regulating toxin–antitoxin systems. Structure. 2010; 18:996–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kamada K., Hanaoka F.. Conformational change in the catalytic site of the ribonuclease YoeB toxin by YefM antitoxin. Mol. Cell. 2005; 19:497–509. [DOI] [PubMed] [Google Scholar]
- 32. Kedzierska B., Lian L.Y., Hayes F.. Toxin-antitoxin regulation: bimodal interaction of YefM-YoeB with paired DNA palindromes exerts transcriptional autorepression. Nucleic Acids Res. 2007; 35:325–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Singh S.M., Panda A.K.. Solubilization and refolding of bacterial inclusion body proteins. J. Biosci. Bioeng. 2005; 99:303–310. [DOI] [PubMed] [Google Scholar]
- 34. Kabsch W. Xds. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Franke D., Petoukhov M.V., Konarev P.V., Panjkovich A., Tuukkanen A., Mertens H.D.T., Kikhney A.G., Hajizadeh N.R., Franklin J.M., Jeffries C.M. et al.. ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Crystallogr. 2017; 50:1212–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Krissinel E., Henrick K.. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007; 372:774–797. [DOI] [PubMed] [Google Scholar]
- 39. Pavelich I.J., Maehigashi T., Hoffer E.D., Ruangprasert A., Miles S.J., Dunham C.M.. Monomeric YoeB toxin retains RNase activity but adopts an obligate dimeric form for thermal stability. Nucleic Acids Res. 2019; 47:10400–10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Maehigashi T., Ruangprasert A., Miles S.J., Dunham C.M.. Molecular basis of ribosome recognition and mRNA hydrolysis by the E. coli YafQ toxin. Nucleic Acids Res. 2015; 43:8002–8012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li S., Olson W.K., Lu X.J.. Web 3DNA 2.0 for the analysis, visualization, and modeling of 3D nucleic acid structures. Nucleic Acids Res. 2019; 47:W26–W34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Eun H.J., Lee K.Y., Kim D.G., Im D., Lee B.J.. Crystal structure of the YoeBSa1-YefMSa1 complex from Staphylococcus aureus. Biochem. Biophys. Res. Commun. 2020; 527:264–269. [DOI] [PubMed] [Google Scholar]
- 43. Christensen S.K., Maenhaut-Michel G., Mine N., Gottesman S., Gerdes K., Van Melderen L.. Overproduction of the Lon protease triggers inhibition of translation in Escherichia coli: involvement of the yefM-yoeB toxin–antitoxin system. Mol. Microbiol. 2004; 51:1705–1717. [DOI] [PubMed] [Google Scholar]
- 44. Jurenas D., Van Melderen L., Garcia-Pino A.. Mechanism of regulation and neutralization of the AtaR-AtaT toxin–antitoxin system. Nat. Chem. Biol. 2019; 15:285. [DOI] [PubMed] [Google Scholar]
- 45. Qian H., Yu H., Li P., Zhu E., Yao Q., Tai C., Deng Z., Gerdes K., He X., Gan J. et al.. Toxin-antitoxin operon kacAT of Klebsiella pneumoniae is regulated by conditional cooperativity via a W-shaped KacA-KacT complex. Nucleic Acids Res. 2019; 47:7690–7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. De Jonge N., Garcia-Pino A., Buts L., Haesaerts S., Charlier D., Zangger K., Wyns L., De Greve H., Loris R.. Rejuvenation of CcdB-poisoned gyrase by an intrinsically disordered protein domain. Mol. Cell. 2009; 35:154–163. [DOI] [PubMed] [Google Scholar]
- 47. Bendtsen K.L., Xu K., Luckmann M., Winther K.S., Shah S.A., Pedersen C.N.S., Brodersen D.E.. Toxin inhibition in C. crescentus VapBC1 is mediated by a flexible pseudo-palindromic protein motif and modulated by DNA binding. Nucleic Acids Res. 2017; 45:2875–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mate M.J., Vincentelli R., Foos N., Raoult D., Cambillau C., Ortiz-Lombardia M.. Crystal structure of the DNA-bound VapBC2 antitoxin/toxin pair from Rickettsia felis. Nucleic Acids Res. 2012; 40:3245–3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liang Y., Gao Z., Wang F., Zhang Y., Dong Y., Liu Q.. Structural and functional characterization of Escherichia coli toxin–antitoxin complex DinJ-YafQ. J. Biol. Chem. 2014; 289:21191–21202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ruangprasert A., Maehigashi T., Miles S.J., Giridharan N., Liu J.X., Dunham C.M.. Mechanisms of toxin inhibition and transcriptional repression by Escherichia coli DinJ-YafQ. J. Biol. Chem. 2014; 289:20559–20569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Brown B.L., Wood T.K., Peti W., Page R.. Structure of the Escherichia coli antitoxin MqsA (YgiT/b3021) bound to its gene promoter reveals extensive domain rearrangements and the specificity of transcriptional regulation. J. Biol. Chem. 2011; 286:2285–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schuster C.F., Bertram R.. Toxin-antitoxin systems of Staphylococcus aureus. Toxins (Basel). 2016; 8:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Garcia-Pino A., De Gieter S., Talavera A., De Greve H., Efremov R.G., Loris R.. An intrinsically disordered entropic switch determines allostery in Phd-Doc regulation. Nat. Chem. Biol. 2016; 12:490–496. [DOI] [PubMed] [Google Scholar]
- 54. Donegan N.P., Thompson E.T., Fu Z., Cheung A.L.. Proteolytic regulation of toxin–antitoxin systems by ClpPC in Staphylococcus aureus. J. Bacteriol. 2010; 192:1416–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates have been deposited in the Protein Data Bank (PDB) (codes: 6L8E for heterohexamer-DNA (YoeB2-YefM4-DNA); 6L8F for heterotetramer (YoeB2–YefM2); 7CUA for YoeB dimer).