Abstract
Circular RNAs (circRNAs) encompass a widespread and conserved class of RNAs, which are generated by back-splicing of downstream 5′ to upstream 3′ splice sites. CircRNAs are tissue-specific and have been implicated in diseases including cancer. They can function as sponges for microRNAs (miRNAs) or RNA binding proteins (RBPs), for example. Moreover, some contain open reading frames (ORFs) and might be translated. The functional relevance of such peptides, however, remains largely elusive. Here, we report that the ORF of circZNF609 is efficiently translated when expressed from a circZNF609 overexpression construct. However, endogenous proteins could not be detected. Moreover, initiation of circZNF609 translation is independent of m6A-generating enzyme METTL3 or RNA sequence elements such as internal ribosome entry sites (IRESs). Surprisingly, a comprehensive mutational analysis revealed that deletion constructs, which are deficient in producing circZNF609, still generate the observed protein products. This suggests that the apparent circZNF609 translation originates from trans-splicing by-products of the overexpression plasmids and underline that circRNA overexpression constructs need to be evaluated carefully, particularly when functional studies are performed.
INTRODUCTION
Large parts of the human genome produce non-coding RNAs such as microRNAs (miRNAs), long non-coding RNAs (lncRNAs) and various other classes (1). One of these classes are circRNAs, which have been discovered decades ago but attracted attention only recently when it was found that these RNAs are abundantly and widely expressed, particularly in neuronal tissues (2–4). CircRNAs are generated by splicing of a downstream splice donor to an upstream splice acceptor resulting in a circularized RNA lacking a 5′ and a 3′ end (5,6). This process is commonly referred to as back-splicing (7,8). Depending on the splice sites that are used, circRNAs can contain one or several exons and also intronic sequences. Although circRNA biogenesis is not yet fully understood, some aspects of this process have been unraveled. Specific hallmarks of loci that produce circRNAs are ‘back-folding’ of intronic sequences that position upstream and downstream splice sites close to each other allowing for back-splicing. Such structures can be achieved by complementary intronic sequences that hybridize to each other. In agreement with this model, inverted repeats allowing for direct base pairing are often found next to the splice sites that are engaged in back-splicing (9–13). Alternatively, RNA binding proteins (RBPs) can contribute to back-splicing by simultaneous binding to both sites flanking the circularized exons. RBPs might dimerize and thus loop out the pre-mRNA sequences that will be circularized. The tissue-specific RBPs Muscleblind (MBL) in Drosophila and Quaking (QKI) in human have been shown to function as trans-acting factors promoting back-splicing and circular RNA production (14,15).
The molecular functions of circRNAs appear to be rather diverse and are in most cases only poorly understood. Initial studies on the circRNA ciRS-7 revealed that it contains >70 conserved binding sites for the microRNA miR-7 and it has been demonstrated that ciRS-7 functions as sponge that negatively regulates miR-7-guided gene regulation in the brain (2,3). Interestingly, ciRS-7 also contains a binding site for miR-671, which is sufficiently complementary to allow for Ago2-mediated cleavage of ciRS-7. Thus, expression of miR-671 liberates large quantities of miR-7 leading to a robust regulatory output. ciRS-7 has recently been genetically inactivated in mice and the regulatory network containing miR-7, miR-671 and ciRS-7 was largely confirmed. Mice lacking ciRS-7 show alterations in sensorimotor gating and synaptic transmission suggesting a role in neuronal functions and a potential link to associated diseases (16). Further analysis showed that ciRS-7 forms a highly sophisticated RNA-centered regulatory network involving miR-7, miR-671, ciRS-7 and the long non-coding RNA Cyrano, highlighting the importance of balancing gene expression programs in brain function (17). In addition to miRNA sponges, circRNAs have evolved different functions, which are not well understood, yet. For example, circRNAs may serve as sponges for RBPs and thus negatively regulate their functions. In addition, circRNAs may also engage in RNA-RNA interactions although direct evidence for such a scenario remains rather scarce (18).
Several circRNAs contain start AUGs followed by open reading frames (ORFs) that could serve as templates for translation. Indeed, it has been reported that circRNAs can be translated and give rise to protein products (19,20). Since circRNAs do not contain 5′ cap structures, cap-independent initiation is required. Several studies have characterized internal ribosomal entry sites (IRES), which help recruiting ribosomes to circRNAs. Other studies found highly specialized translation initiation factors such as eIF4G2 that recruits YTHDF3, an m6A reader protein that directly interacts with the modified circRNA sequences leading to translation initiation (21–23). Although a number of publications have reported on circRNA translation, endogenous protein products clearly originating from a circRNA are difficult to detect (24–26,27). Furthermore, a comprehensive computational and experimental study concluded that AUG-containing circRNAs are not generally templates for translation (28). In contrast, a characterization of the translatome of the human heart identified a large number of uncharacterized micropeptides and several of them originate from circRNAs (29). These findings suggest that circRNA translation might be more prevalent in primary tissues.
To better understand the molecular mechanism of circRNA translation, we used circZNF609 translation as a test case. This circRNA appears physiologically important since it is upregulated during primary neuron differentiation (4) and aging (30) of the mouse brain. Furthermore, human circZNF609 (also called myocardial infarction-associated circular RNA or MICRA in this study) is lower in blood samples from patients with myocardial infarction compared to healthy control samples, which could even be used for prediction of left ventricular dysfunction (31). In human and mice, circZNF609 is downregulated during muscle differentiation and may encode for a protein (19). It was proposed that circZNF609 and its putative protein products might function in regulating myogenesis. In addition, it was recently reported that it regulates the transition of G1 to S phase in rhabdomyosarcoma (32).
Here, we have investigated circZNF609 translation in detail. Our work demonstrates that our circZNF609 constructs are indeed translated and encode for a larger panel of proteins. This process appears to be independent of the m6A-generating METTL3/METTL14 complex. In addition, comprehensive mutagenesis studies reveal that circZNF609 does not contain obvious IRES sequences. Surprisingly, we finally find that these proteins are efficiently produced even in the absence of circRNA production. Since we did not find evidence for endogenous proteins originating from circZNF609, we conclude that the observed translation results from linear splicing by-products and circZNF609 is most likely not translated from such constructs.
MATERIALS AND METHODS
Plasmids
pcDNA3.1(+) ZKSCAN1 MCS-WT Split GFP (Addgene plasmid #69908) was a gift from Jeremy Wilusz (University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA). pcDNA3.1(+) ZKSCAN1 MCS-WT Split GFP was used as backbone to clone pcDNA3.1(+) ZKSCAN1-circZNF609 and its derivation. pcDNA3.1(+)-circZNF609i and pcDNA3.1(+)-circZNF609i-3xFlag were a gift from Irene Bozzoni (Sapienza University of Rome, Rome, Italy). Briefly, pcDNA3.1(+)-circZNF609i were generated by cloning the sequence of murine ZNF609 circRNA exon and its upstream 860 nts intron into pcDNA3.1(+) vector. 200 nts intron downstream of ZNF609 circRNA exon was added to the plasmids. Finally, the inverted sequence of the upstream 860 nts intron was cloned downstream of this 200 nts. pcDNA-Cas9-GFP and psg-RFP were generated as described (33). Two gRNAs targeting METTL3 were cloned into the psg-RFP resulting psg-RFP-g1 and psg-RFP-g2. Flag/HA tagged METTL3 (FH-METTL3) and Flag/HA-tagged (FH-METTL14) were cloned and described previously (34). pGEM-3E5-T7t was a gift from So Umekage (Toyohashi University of Technology, Toyohashi, Aichi, Japan) as described previously (35). pGEM-3E5-T7t was used as backbone to clone pGEM-3E5-T7t-circZNF609 and its derivation for production of circZNF609 RNA in vitro. All the sequence of the oligos can be found at Supplementary Table S1.
Antibodies
Following antibodies were used for western blot: mouse-anti-HA (monoclonal, 1:1000, Covance, clone HA.11), rabbit-anti-GAPDH (polyclonal, clone FL-335, 1:500, Santa Cruz Biotechnology), rabbit-anti-GAPDH (monoclonal, clone 14C10, Cell Signaling mAb #2118, 1:1000), mouse-anti-beta-Actin (monoclonal, clone AC15, 1:10 000, Abcam), Rat-anti-m6A (monoclonal, clone 9B7 (36)), mouse-anti-Flag (monoclonal, clone M2, 1:1000, Sigma-Aldrich), mouse-anti-beta-Tubulin (polyclonal, 1:1000, Abcam), rabbit-anti-METTL3 (polyclonal, 15073-1-AP, 1:1000, proteintech). Goat-anti-rabbit/mouse/rat IRDye 680RD or goat-anti-rabbit/mouse/rat/guinea pig IRDye 800CW antibodies (Li-Cor Biosciences) were used as secondary antibodies.
Cell lines, cell culture and generation of stable cell lines
HEK293T (ATCC), N2a (ATCC), C643 (CLS, RRID:CVCL_5969) cells were cultivated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and Penicillin/Streptomycin (P/S) antibiotics mix (all from Sigma).
METTL3-knockout cells were generated using the CRISPR/Cas9 genome editing as previously described (33). Briefly, C643 cells were transfected with pcDNA-Cas9-GFP and two psg-RFP plasmids expressing guide RNAs by using Lipofectamine 2000, according to the manufacturer's instructions. Control clones were generated by transfecting the Cas9 nuclease only. Forty-eight hours post-transfection GFP- (control) and GFP/RFP-positive single cells were sorted in 96 well-plates with a FACSAria II cell sorter (BD Biosciences) and cultured. Knockout clones were tested by western blotting. Positive clones were confirmed by sequencing of the genomic region targeted by the gRNAs. For generation of stable C643 WT OE (C643 WT cells expressing FH-METTL3 and FH-METTL14) and C643 M3-KO Rescue (C643-M3 KO cells expressing FH-METTL3 and FH-METTL14) cell line expressing both FH-METTL3 and FH-METTL14 constructs (34), cells were transfected with plasmids in 6-well format. Cells were split one day post-transfection into one 10 cm plate and selection was started 2 days post-transfection with 300 μg/ml Hygromycin B. After verification of the positive clones, the stable clones were maintained in the same medium as used for selection.
For generation of stable inducible Flp-In T-REx 293 cell lines, the cells were co-transfected with pOG44 and pcDNA5-FRT/TO in 6-well format. One day post-transfection, the cells were split into one 10 cm plate. Selection was started 2 days post-transfection using 15 μg/ml Blasticidin and 200 μg/ml Hygromycin B (Life Technologies). Clones were picked two weeks later and tested for expression levels of circZNF609 RNA by northern blot. Expression was induced for 48 hours with final concentration of 1 μg/ml Doxycycline (Sigma–Aldrich). Stable clones were maintained in the same medium as used for selection.
SDS-PAGE and western blotting
For performing a western blot, samples were shortly heated at 95°C for 5 min and then loaded onto a 10–15% SDS-PAGE gel. After separation, the proteins were blotted onto an Amersham Protran Premium 0.45 µm membrane (GE Healthcare) using 1x Towbin buffer for 1 min/kDa constant at 2 mA/cm2. The membrane was blocked in TBS containing 0.1% Tween-20 and 5% milk for 1h and subsequently incubated with the primary antibody overnight at 4°C. After three washing steps with TBS containing 0.1% Tween-20, the secondary antibody was added for 1 h at RT. After three washing steps with TBS containing 0.1% Tween-20, the membrane was scanned on a LI-COR reader.
Preparation of RNA
RNA extraction from mammalian cells has been done with TRIzol reagent (Life Technologies) or NucleoSpin RNA Kit (Macherey-Nagel) following the manufacturer's protocol.
mRNA (poly(A)+ RNA) was isolated using Oligo d(T)25 Magnetic beads (NEB) according to manufacturer's protocol. The flow-through was collected and served as poly(A)- RNA. After the first isolation, the poly(A)+ RNA was used for the second round of isolation with new beads to deplete most ribosomal RNA. The RNA after isolation was collected and cleaned up from the magnetic beads using NucleoSpin RNA Kit (Macherey-Nagel). The quality of poly(A)+ RNA was confirmed using TapeStation (Agilent) before subsequent HPLC analysis of nucleosides.
RNase R treatment was performed using RNase R (Biozym) according to manufacturer's protocol. The reaction was cleaned up using NucleoSpin RNA Kit (Macherey-Nagel).
RNA in vitro transcription (IVT) using T7 polymerase
The reaction requires a DNA template containing the T7 promoter upstream of the sequence as indicated in Figure 5A. In order to perform the in vitro transcription, the DNA templates were first amplified using PCR. 2 μg of the PCR product was then used in a 500 μl transcription reaction containing final concentration of 30 mM Tris, pH 8.0, 10 mM DTT, 0.01% Triton X-100, 25 mM MgCl2, 2 mM spermidine, 5 mM ATP, 5 mM CTP, 5 mM UTP, 5 mM GTP (all from Thermo Scientific), 0.4 U/ml thermostable inorganic pyrophosphatase (NEB) and 0.1 mg/ml T7 polymerase (produced from our lab) at 37°C for at least 3 h. For increasing self-splicing reaction of circRNA, the reaction was shifted to 55°C for 30 min. Afterwards, the reaction was digested with 1 μl of 1 U/μl DNase I (Thermo Scientific) for 15 min at 37°C. The RNA extraction from in vitro transcription was collected in 750 μl TRIzol-LS reagent (Life Technologies, Carlsbad, USA) following TRIzol-LS manual. After isolation, the RNA was loaded on 4–6% UREA gel or 1% MOPS Agarose gel for seperation. Of note, we have never obtained good separation of IVT RNA (for big RNA as circZNF609) using UREA gel. Only in 1% MOPS Agarose gel, we obtained good separation of the IVT RNA. Desired RNA was isolated from MOPS Agarose gel using Zymoclean Gel RNA Recovery Kit (Zymo Research) following the manufacturer's protocol.
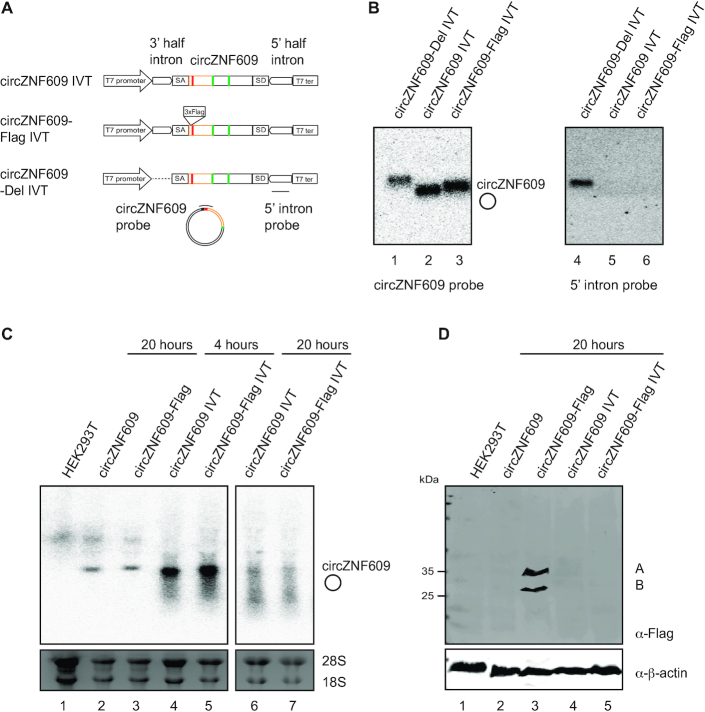
Figure 5.
No evidence for exogenous in vitro transcribed (IVT) circZNF609 translation. (A) Schematic overview of constructs for production of circZNF609 in vitro using a self-splicing permutated intron–exon (PIE) system. The circZNF609 exon is cloned in between the 3′ half intron and the 5′ half intron of the self-splicing group I intron of the T4 phage Td gene. All constructs include splice acceptors (SA, CUACC) and splice donors (SD, GGGU). These sequences (9 nts) are necessary for the self-splicing reaction and will be included into the sequence of circZNF609 after ligation. The construct circZNF609-Del lacks the 3′ half-intron and serve as a negative control for the self-splicing reaction. (B) Northern blot for confirmation of IVT circZNF609. 10 ng of isolated IVT circZNF609 was loaded on 1% MOPS Agarose gel. Northern blot was performed with a probe spanning the circZNF609 splice junction (lanes 1–3) or a probe targeting the 5′ intron (lanes 4–6). (C) Northern blot of HEK293T transfected with plasmids for overexpression of circZNF609 (lanes 2, 3) and IVT circZNF609 (lanes 4–7). For examination of transfected IVT circZNF609, RNA was harvested 4 h (lanes 4, 5) and 20 hours post transfection (lanes 6, 7). (D) Western blot of HEK293T cells transfected with plasmids for overexpression of circZNF609 (lane 2, 3) and IVT circZNF609 (lane 4, 5) 20 h post transfection.
Quantitative real-time PCR (qPCR)
qPCR was done with Sso Fast Eva Green Mix (Bio-Rad, Hercules, USA) using 0,5 μM forward and 0,5 μM reverse primer and cDNA from 10 ng RNA as template. qPCR was run on a CFX96 cycler (Bio-Rad, Hercules, USA) using the standard program as given in the SsoFast EvaGreen SuperMix manual. Data were evaluated using the ΔΔCt method and normalized to control sample.
Digestion of RNA and subsequent HPLC analysis of nucleosides
After Poly(A)+ purification of total RNA using oligo d(T)25 Magnetic beads (NEB), the RNA (total RNA, poly(A)+ RNA and poly(A)- RNA) was digested into single nucleosides by incubating with an enzyme mix of Benzonase (Sigma No. E8263), Phosphodiesterase I (Worthington Cat. No. LS003926) and FastAP (Thermo Scientific) for 3 h at 37°C (37).
Subsequently, the digested RNA-sample was loaded onto a Superdex Peptide column (GE Healthcare, 0.4 M Ammoniumacetate, 1 ml/min) to remove the enzymes and the pyrimidines. The adenosine (+m6A) containing fraction, which eluted between 36.7 min and 40.5 min, was collected and evaporated to dryness in a speedvac. The remaining pellet was then dissolved in H2O and directly applied to a HPLC system (WellChrom from Knauer) equipped with a Hypercarb-column (5 μm, 100 × 2.1; Thermo Scientific), Pump K-1001, Diode Array Detector K-2800, column oven and a Vacuum Degasser from Techlab GmbH (Germany).
For the separation of the nucleosides a gradient of the buffers A (50 mM NH4Ac, pH 5.0) and B (20% 50 mM NH4Ac, pH 5.0/80% acetonitrile) was applied at a flow rate of 0.25 ml/min and a column temperature of 55°C.
The detection of the nucleosides was performed simultaneously at two different wavelengths 260 nm and 280 nm and the resulting chromatograms were analysed with the software ChromGate Client/Server Version 3.1.7. After normalizing the m6A peak-area to the corresponding adenosine peak-area, the relative m6A-amount between the different samples was used.
M6A meRIP (Methylated RNA Immunoprecipitation)
Total RNA was isolated from C643 WT and C643 M3-KO cells by TRIzol. 20 μg of anti-m6A antibody (Clone 9B7 (36)) was pre-bound to protein G magnetic beads (Pierce-Thermo Scientific) in 500 μl IP buffer (25 mM Tris pH 7.5, 150 mM KCl, 0.5% NP-40, 2 mM EDTA) for 1 h at room temperature or at least 2 h at 4°C. Afterward, the beads were washed two time with IP buffer before adding 10 μg of total RNA in a final volume of 500 μl of IP buffer. The mixture of RNA and Protein G beads was incubated for 2 hours at 4°C. Samples were washed twice with IP buffer. After the first wash, the beads were transferred to a new eppendorf tube. Then the beads were washed four times with wash buffer (20 mM Tris pH 7.5, 300 mM KCl, 0.5% NP-40, 1mM MgCl2). After the final wash, the beads were transferred to a new eppendorf tube and RNA was isolated from the beads using TRIzol. After purification, RNA was reverse-transcribed using First strand cDNA synthesis kit (Thermo Scientific) with random hexamers, and enrichment of m6A-containing transcripts was determined by quantitative PCR.
Digestion of plasmids with restriction enzymes
5 μg of plasmids was digested in a 50 μl reaction containing FastDigest Green Buffer (Thermo Scientific), 3 μl FastDigest restriction enzyme (Thermo Scientific) and 1 μl FastAP Thermosensitive Alkaline Phosphatase (Thermo Scientific) (1 U/μl) for 30 minutes at 37°C. After separation on 1% agarose gel, the desired fragments were isolated using NucleoSpin Gel and PCR Clean-up (Macherey-Nagel).
Transfection of RNA to HEK293T cells
2 ml of 400 000 HEK293T cells/ml were seeded into six-wells plates. 500 ng of in vitro transcribed RNA was transfected into six-well plates using Lipofectamine 2000. Four hours post-transfection, the medium was removed and replaced with 2 ml of fresh complete medium. The cells were incubated for 16 h. The cells were harvested 4 and 20 h post-transfection for northern blot and western blot analysis.
Northern blotting
Northern blot for circRNAs was prepared with 1% MOPS agarose gel. 2× RNA loading dye (45% Formamide, 1x MOPS, 2% Formaldehyde, 5% Glycerol, 0.01% Bromophenol Blue) was added to 20 μg of RNA samples. After incubating at 65°C for 10 min, samples were chilled on ice. 2.3 μl of EtBr (400 μg/μl) were added directly into the samples before loading into the gel. Once the gel was checked under UV light, it was soaked and shaked into Milli-Q water for 5 minutes followed by 50 mM NaOH for 30 min, 50 mM Tris pH 7.5 for 30 min and 20× SSC for 30 min at room temperature. Afterwards, the transfer was set up by disposing a glass plate on top two square dishes filled with 20× SSC. Two strips of Whatman paper were soaked into 2× SSC and laid on a plate in order to hang both ends into the 20× SSC of the dishes. After removing bubbles, the gel was placed facedown. The area around the gel was sealed with parafilm to prevent 20× SSC from ‘short-circuiting’. The nitrocellulose membrane (Amersham Hybond-N) of the size of the gel was soaked into Milli-Q water and 2x SSC and it was then placed on the gel. After removing all bubbles, three 2× SSC-soaked pieces of Whatman paper were placed on the membrane. A stack of towel papers was put on top with a second glass plate and an additional weight of 500 g. The transfer was set at room temperature overnight, preferably for 18 h, after which the success of transfer was verified by visualizing the membrane under UV light. The RNA was crosslinked to the membrane using the UV Stratalinker at 254 nm. Afterward, 20–30 ml hybridization solution (5x SSC, 20 mM NaPi pH 7.2, 7% SDS, 0.02% Albumin fraction V, 0.02% Ficoll 400, 0.02% Polyvinylpyrrolidone K30) was put into a cylindrical glass bottle, in which the membrane was already put into with the RNA facing inwards, and then placed in a hybridization oven at 50–60°C for 60 min.
The oligo sequence for northern blot was listed in Supplementary Table S1. Probe labeling was done using 20 μM DNA oligonucleotide with 20 μCi of γ 32P-ATP (Hartmann Analytics) using T4 PNK enzyme (Life Technologies) according to the manufacturer's protocol. The reaction was incubated for 60 min at 37°C. The reaction was mixed with 30 μl 30 mM EDTA to stop PNK reaction.
The membrane was washed twice with wash solution 1 (5× SSC, 1% SDS) and once with wash solution 2 (1× SSC, 1% SDS), incubating each wash step for 10 min on a turning wheel at the same temperature as hybridization. After the last step, the liquid was discarded and the membrane was wrapped in saran for exposure to a phosphor screen and then signals were scanned by Personal Molecular Imager System (Bio-Rad).
Statistical analyses
Statistical analyses have been performed for the data presented in Figure 2C using Graphpad Prism 5. Two-sample assuming equal variances t-test was used for testing the significant of the graphs. P values less than 0.05 were considered to be significant. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ns: P > 0.05. Graphs and error bars reflect means ± s.d. (standard deviation).
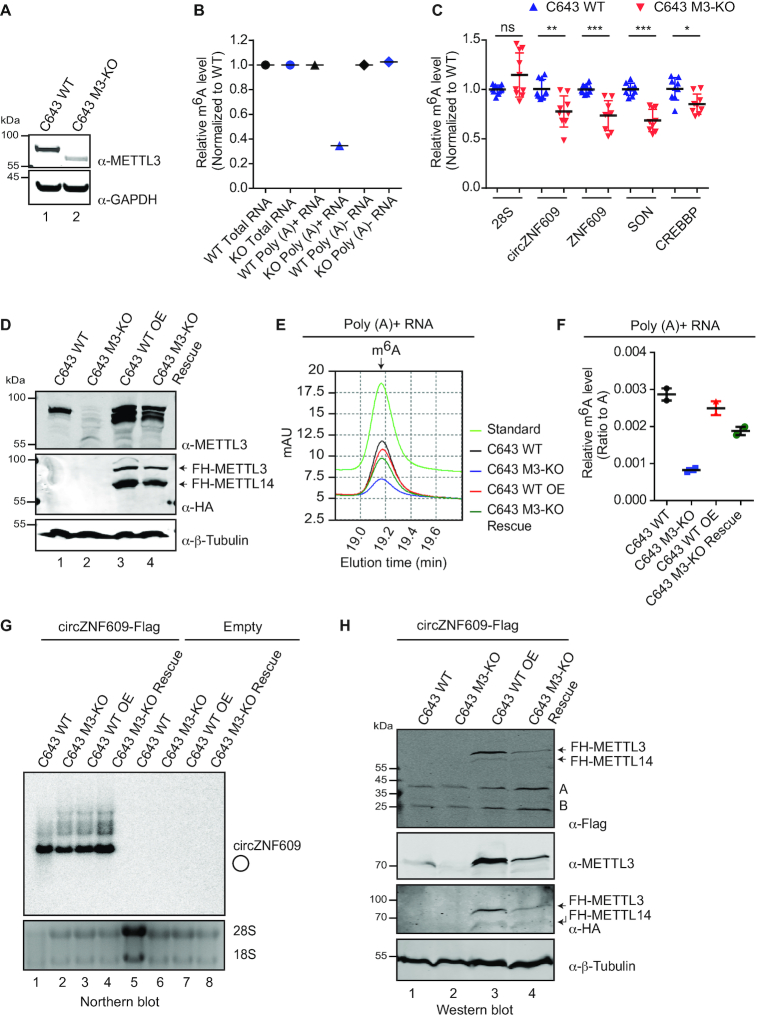
Figure 2.
circZNF609 is translated independently of METTL3 activity. (A) Western blot of the METTL3 knockout C643 cell line (C643 M3-KO) using an anti-METTL3 antibody. (B) HPLC analysis of the relative expression of m6A expression of total RNA, poly(A)+ and poly(A)-RNA from the C643 M3-KO cell line compared to C643 WT cells. (C) m6A meRIP of total RNA from C643 WT and C643 M3-KO cells using anti-m6A antibody. Relative enrichment of different transcripts after m6A meRIP was first calculated to input and then normalized to WT. 28S served as negative control while SON and CREBBP served as positive controls. The experiment was performed with three biological replicates. Graphs and error bars reflect means ± s.d. (standard deviation). ns: P > 0.05, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. (D) Western blot of C643 wildtype (C643 WT), METTL3 knock out cells (C643 M3-KO), C643 WT cells overexpressing FH-METTL3/14 (C643 WT OE) and C643 METTL3 knock out cells overexpressing FH-METTL3/14 (C643 M3-KO Rescue). (E) HPLC analysis of m6A level from poly(A)+ RNA of C643 WT (black), C643 M3-KO (blue), C643 WT OE (red) and C643 M3-KO Rescue (dark green). m6A standard nucleoside is indicated (green). The m6A peak is detected after about 19.1 minutes of the HPLC run. (F) Relative m6A level of poly(A)+ RNA of C643 WT (black), C643 M3-KO (blue), C643 WT OE (red) and C643 M3-KO Rescue (dark green). The ratio of m6A to A was used for the graph. The experiment was performed with two biological replicates. (G) Northern blot of the C643 cell lines (C643 WT, C643 M3-KO, C643 WT OE, C643 M3-KO Rescue) transfected with plasmids for overexpression of circZNF609-Flag. The signal was detected with a probe spanning the splice junction of circZNF609. (H) Western blot experiments of C643 cell lines (C643 WT, C643 M3-KO, C643 WT OE, C643 M3-KO Rescue) transfected with circZNF609-Flag constructs for analysis of circZNF609 translation.
RESULTS
Apparent translation of overexpressed circZNF609
CircZNF609 shares the start codon with its cognate mRNA ZNF609 and contains a stop codon generating an ORF upon circularization that includes one additional amino acid which is not present in the full-length protein translated from the ZNF609 mRNA (Figure 1A, Supplementary Figure S1A). During the course of this study, it was reported that circZNF609 associates with ribosomes and can be translated (19,22). We therefore used circZNF609 as a test case to study the underlying mechanisms.
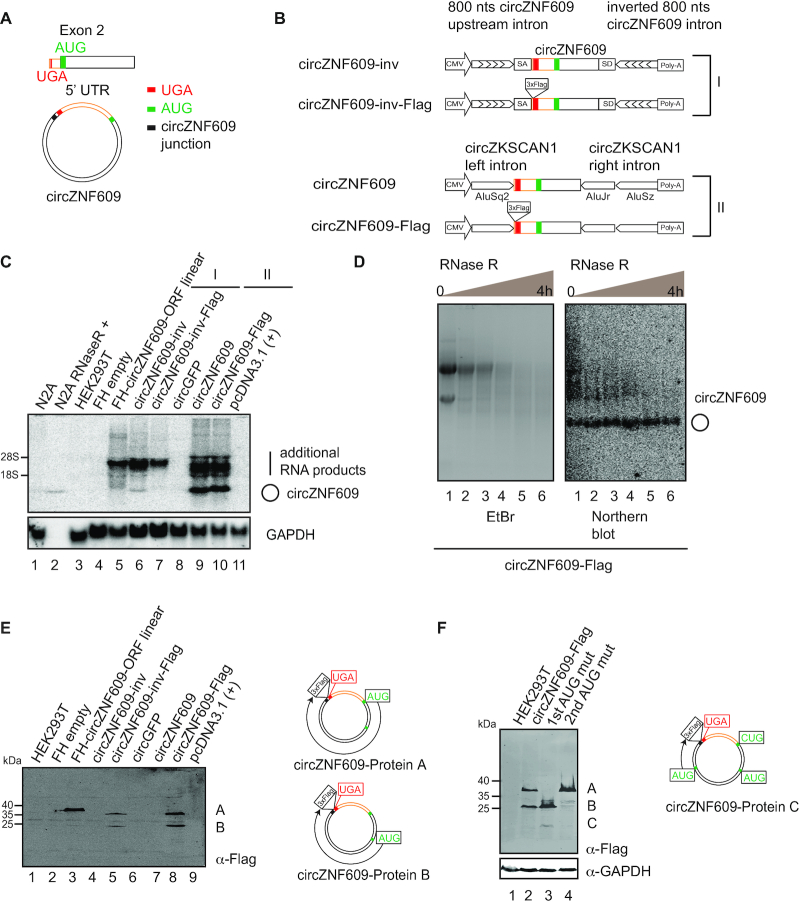
Figure 1.
Overexpressed circZNF609 is translated. (A) circZNF609 contains an ORF upon circularization. The stop codon (UGA, red) is located in front of the start codon (AUG, green) in the linear transcript. Upon circularization, circZNF609 forms a complete ORF and a 5′ UTR (yellow) is located between the stop and start codon. (B) Schematic representation of the constructs generated for overexpression of circZNF609 (circZNF609-inv (I) and circZNF609 (II)). 3xFlag was inserted in front of the stop codon of circZNF609 to generate circZNF609-inv-Flag (I) and circZNF609-Flag (II) plasmids. (C) HEK293T cells transfected with different overexpression constructs were analyzed by northern blotting using a probe spanning the splice junction of circZNF609. (D) RNA isolated from HEK293T cells transfected with circZNF609-Flag was isolated for RNase R digestion at different time points. Left panel: RNA after digestion with RNase R was loaded on 1% MOPS Agarose gel. Right panel: Northern blot of the corresponding membrane transferred from MOPS Agarose gel of RNase R-digested RNA was detected with a probe spanning circZNF609 junction. (E) Western blot of HEK293T cells transfected with different constructs from (B, C) detected by anti-Flag antibody. Analysis of circZNF609 sequence predicts two AUGs in-frame with 3xFlag and thus could be detected by anti-Flag antibodies (circZNF609-protein A and B). (F) Western blot of HEK293T cells transfected with different overexpressed circZNF609 AUG mutants. The third protein products appeared when the first AUG is mutated to CUG (lane 3). Scheme of the third AUG which can be used to generate the circZNF609-protein C.
We used two different circRNA overexpression systems allowing for the in frame fusion to a C-terminal 3× FLAG tag that can be detected by western blotting (Figure 1B, I, (2); II, (38)). Northern blotting confirmed the presence of endogenous circZNF609 in N2a cells treated with RNase R (Figure 1C, lane 2), which serves as size standard for our overexpression experiments. Plasmids from both expression systems (I, II) were transfected into HEK293T cells and the generated products were analyzed by northern blotting. In both cases, a number of potentially linear transcripts but also the circular RNAs were detected (Figure 1C, Supplementary Figure S1B). To further confirm the circular nature of circZNF609, we digested the overexpressed RNAs with RNase R at different time points and observed that circZNF609 remained stable while other RNA products were digested (Figure 1D, Supplementary Figure S1C).
To investigate translation from our constructs, we transfected plasmids encoding linear (N-terminal Flag/HA (FH) tagged) and circular forms of the circZNF609 ORF into HEK293T cells and performed anti-FLAG western blotting (Figure 1E). While the ORF in its linear form produced a protein band migrating at the expected size, all circular variants generated two protein bands. The expression system II appeared to be more efficient and was thus used for all following studies. Interestingly, a closer examination of the ORF revealed that the two bands (A and B) could be readily explained by the alternative use of two start codons present on the circular RNA (Figure 1E). To validate this hypothesis, we mutated the first AUG as well as the second AUG independently of each other (Figure 1F). Indeed, when the first start codon was mutated, only the second band appeared and vice versa the larger protein was made only when the second AUG was mutated. Northern blotting indicated that all constructs produced equal amounts of circular RNA (Supplementary Figure S1D). Close examination of the western blots identified a third albeit less strong band migrating much faster than the other two protein products (band C). Strikingly, a third AUG can be found within the ORF and mutating this AUG abolishes production of this very short peptide (data not shown). However, protein C appears to be stronger when the first AUG is mutated suggesting potential competition between the start codons. Of note, the protein bands were also detectable when the linear forms of the circZNF609 ORF are transfected as C-terminal Flag/HA tagged versions that use the circZNF609 AUGs for translation initiation instead of the AUGs from the expression vector (Supplementary Figure S1E-F). Taken together, circZNF609 can be translated and produces at least three different proteins.
CircZNF609 is translated in a METTL3 knockout cell line
How translation is initiated on circRNAs remains largely elusive. For example, it has been suggested that m6A methylation of circRNAs helps to recruit the translation machinery (21–23). To address this possibility in more detail, we deleted METTL3, the catalytic enzyme generating internal m6A modifications on mRNAs, from the thyroid carcinoma cell line C643 (Figure 2A, Supplementary Figure S2A). The knockout was confirmed by western blotting using anti-METTL3 antibodies and by HPLC analysis of total, poly(A)+ and poly(A)- RNA (Figure 2A, B). Of note, the m6A peak is only reduced in the poly(A)+ fraction because our RNA preparation still contained non-coding RNAs that are m6A modified by other enzymes. In addition, a shorter protein band is recognized in the knockout cells, which might represent a truncated version of METTL3 but in case this is true, is most likely non-functional because m6A levels on poly(A)+ RNA are clearly reduced (Figure 2B). Next, we confirmed m6A modification of endogenous circZNF609 in C643 WT and METTL3 KO cells (C643 M3-KO) (Figure 2C). m6A-modified RNA was immunoprecipitated from total RNA using anti-m6A antibodies and circZNF609 relative enrichment was analyzed by qPCR (Figure 2C). CircZNF609 enrichment in anti-m6A immunoprecipitation was moderately decreased in C643 M3-KO cells compared to C643 WT cells. Interestingly, the full length ZNF609 mRNA was also decreased in METTL3 KO cells suggesting that it was m6A modified as well. Similarly, the positive controls SON and CREBBP (39) were also reduced. 28S rRNA served as negative control and was not reduced since m6A modification on rRNA is not catalyzed by METTL3. Our data is therefore in agreement with reports showing that circZNF609 is efficiently m6A methylated (19,22).
To further corroborate these observations, we performed rescue experiments (Figure 2D–H). We generated stable cell lines overexpressing FH-METTL3 and its essential co-factor FH-METTL14 (Figure 2D). Simultaneous expression of FH-METTL3 and FH-METTL14 in C643 WT (C643 WT OE) and C643 M3-KO (C643 M3-KO Rescue) cells resulted in increased m6A levels indicating successful functional rescue (Figure 2D–F).
To assess the requirement of METTL3 activity for circZNF609 translation, the circZNF609 construct was transfected into C643 WT, C643 M3-KO, C643 WT OE and C643 M3-KO Rescue cells (Figure 2G, H). Northern blotting revealed similar expression of circZNF609 in all cell lines (Figure 2G). Western blotting using antibodies against the FLAG-tag detected similar expression levels in all cell lines tested (Figure 2H). Because we did not observe a reduced expression of the two circZNF609-generated proteins A and B in METTL3-lacking cells, we conclude that at least METTL3-generated m6A modification might be rather dispensable for circZNF609 translation in the used cell line. Nevertheless, it is still possible that other m6A-modification pathways could be involved.
CircZNF609 peptides are not produced from cis-spliced by-products
Translation of circRNAs could also be caused by linear splicing by-products, which is often not sufficiently controlled for. Therefore, we designed a series of experiments to assess whether overexpressed circZNF609 is indeed translated from the circular RNA. We first addressed potential cis-splicing events (Figure 3A). When the transfected plasmid containing the circZNF609 overexpression cassette is transcribed, it is conceivable that the polymerase may occasionally read-through termination signals and produces a rolling circle RNA. Such an RNA would be capped and contain multiple copies of the circZNF609 overexpression cassette (concatemers). Since 5′ and 3′ splice sites are present, such RNAs could undergo splicing and generate a transcript that could be translated in a cap-dependent manner (Figure 3A). To rule out such a scenario, we cut the plasmid in two halves using different restriction enzymes and transfected the linear DNA, which cannot result in rolling circle transcription (Figure 3B and C). Moreover, DNA fragments were de-phosphorylated to prevent potential re-ligation of the fragments occurred. As expected, circZNF609 was still generated since circRNA splicing of the circRNA expression cassette is still possible from the linearized templates (Figure 3D, Supplementary Figure S3A). Consistently, western blotting against the FLAG-tag clearly shows that the two protein bands are produced also from the plasmid fragments containing the circRNA expression cassette, ruling out rolling circle transcription and potential cis-splicing events (Figure 3E). When a digestion strategy that removes the promoter of the circRNA cassette (K-X) was chosen as negative control, neither a circRNA nor the corresponding proteins were detected (Figure 3B-E, Supplementary Figure S3A).
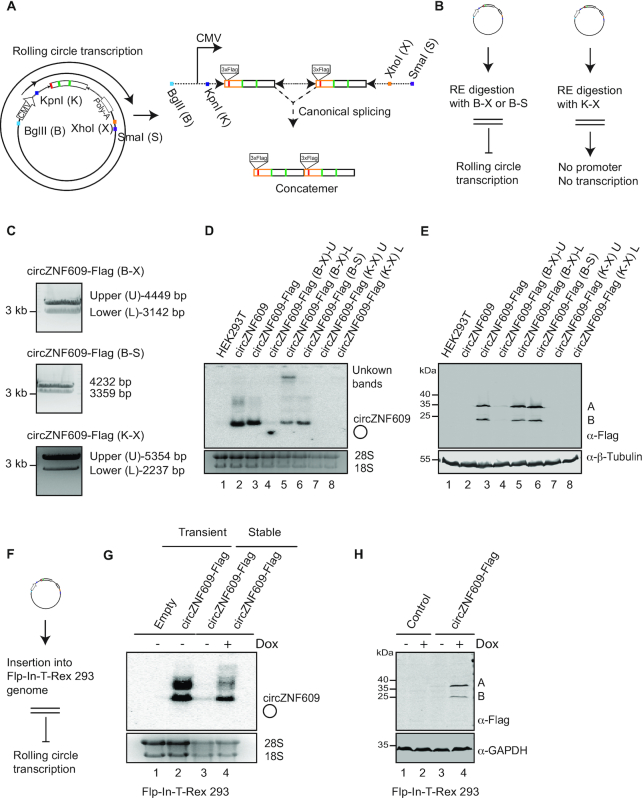
Figure 3.
circZNF609 is not translated from linear cis-splicing RNA products. (A) Schematic overview of the proposed rolling circle transcription and generation of a linear concatemers by canonical splicing. Four restriction enzymes (RE) were used for digestion. BglII (B) is located upstream while KpnI (K) downstream of the CMV promoter. Both XhoI (X) and SmaI (S) are located downstream of the poly(A) signal. (B) Digestion of the plasmids with both BglII (B) and XhoI (X) or BglII (B) and SmaI (S) prevents rolling circle transcription. RE digestion of the plasmids with both KpnI (K) and XhoI (X) generates circZNF609 fragments without a promoter and therefore transcription from both fragments is abolished. (C) Agarose gel of the circZNF609-Flag plasmids digested with either Bglll and XhoI (B-X), Bglll and SmaI (B-S) or KpnI and XhoI (K-X) restriction enzymes. B-X digestion results in two fragments: the upper (U) fragment does not contain circZNF609-Flag while the lower (L) fragment contains circZNF609-Flag with its promoter and poly(A) signal. B-S digestion results in two fragments similar to B-X digestion. However, the two fragments were not separated well so both fragments were combined in one reaction. K-X digestion gave two fragments: the upper (U) fragment contains CMV promoter without circZNF609-Flag while the lower (L) fragment contains circZNF609-Flag without its CMV promoter. (D) Northern blot of the HEK293T cell lines transfected with plasmids for overexpression of circZNF609-Flag and the fragments isolated from C). The signal was detected with a probe spanning junction of circZNF609. (E) Western blot of the HEK293T cells transfected with different digested circZNF609-Flag plasmid fragments using anti-Flag antibodies. (F) Schematic overview of the strategy for insertion a single copy of circZNF609-Flag construct into Flp-In-T-rex 293 genome. This will not allow for rolling circle transcription and therefore prevents potential unwanted cis-splicing products. (G) Northern blot of the Flp-In-T-rex 293 cell lines transiently transfected with plasmids for overexpression of circZNF609-Flag and of Flp-In-T-rex 293 stable cell lines expressing circZNF609-Flag in an inducible manner by Doxycycline (Dox). A probe targeting the Flag sequence was used for circZNF609 detection. (H) Western blot of the corresponding samples from G) was performed using anti-Flag antibodies. GAPDH served as loading control.
Genomic integration of the circRNA cassette into the genome would also rule out cis-splicing from rolling circle transcripts. Therefore, we next generated an inducible Flp-In-T-rex 293 cell line that contains this construct stably integrated into the genome (Figure 3F). Comparing transient expression from overexpressed plasmids with stably integrated constructs showed that both systems produce circZNF609 equally well. Of note, stable integration seems to produce less additional RNA bands (Figure 3G, Supplementary Figure S3B). When analyzing the produced protein bands by western botting against the FLAG-tag, we observed translation and production of the two circZNF609 peptides A and B (Figure 3H), demonstrating that rolling circle transcription, cis-splicing and translation from a linear transcript is not the basis for the observed circZNF609 translation.
circZNF609 lacks a classical IRES structure and various AUGs are pervasively used
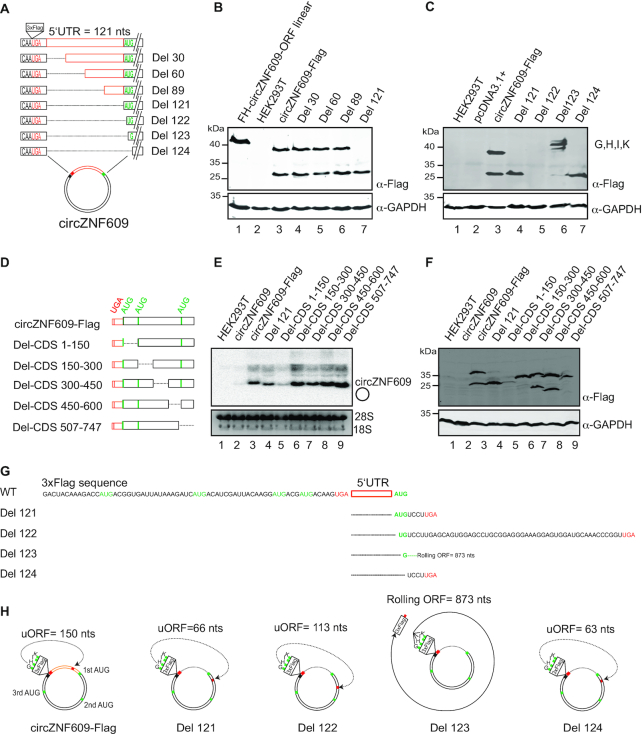
During cap-independent translation initiation, specific IRES elements help to position the ribosomal subunits to the correct start codon (40). To search for potential IRESs, we investigated sequences upstream of the first AUG (referred to as 5′ UTR) of circZNF609 since this is the region where IRES elements are often located (Figure 4A). The sequence was systematically shortened and the plasmids were transfected into HEK293T cells (Figure 4A–C). The transfected constructs were equally expressed (Supplementary Figure S4A-B) and deletion of 89 nts of the 121 nts 5′ UTR did not result in any reduction of the two protein bands (lanes 3–6). Complete deletion of the 5′ UTR (Del 121) led to the loss of the higher molecular weight protein (protein A), which is initiated from the first AUG. The second, downstream AUG, however, is still efficiently used (lane 7) suggesting that fusing the stop codon directly to the first AUG inhibits initiation but the 5′ UTR does not contain a specific IRES element. We next reasoned that an IRES might form within the CDS and therefore systematically deleted the CDS (Figure 4D–F). All deletion constructs (Figure 4D) were transfected into HEK293T cells and RNA and the translated proteins were analyzed by northern blotting (Figure 4E) and western blotting (Figure 4F). All truncated constructs resulted in proteins of the expected size initiated from the first and the second AUG and therefore, an IRES element within the CDS is also unlikely. Of note, deletion of the 3′ terminal fragment (Del-CDS 507–747) resulted in only a single peptide starting from the first AUG. The protein produced from the second AUG gave a very faint band. Whether this sequence indeed affects initiation from the second AUG, however, needs to be further investigated.
Figure 4.
Analysis of circZNF609 UTR and ORF sequences. (A) Schematic overview of the 121 nts of the circZNF609 5′ UTR in between the stop codon UGA and the first start codon AUG. Truncated mutants of circZNF609 5′ UTR are indicated. (B and C) Western blot of HEK293T cells transfected with all 5′ UTR deletion mutants. Anti-Flag antibody was used to detect the proteins while GAPDH served as loading control. (D) Schematic overview of different coding sequence (CDS) mutants of the circZNF609-Flag construct. (E) Northern blot of HEK293T cells transfected with the different CDS mutants. The signal was detected with a probe targeting Flag and therefore circZNF609 without Flag was not detected (lane 2). (F) Western blot of HEK293T cells transfected with the different CDS mutants. (G) The 3xFlag sequence contains 4 AUGs, which can be presumably used for translation of an upstream open reading frame (uORFs). (H) circZNF609-Flag WT contains an uORF from AUGs of 3xFlag sequence. Deletion of circZNF609 UTR could lead to disruption of these uORF and eventually results in a panel of different products. Scheme of circZNF609 5′ UTR mutagenesis of each one nucleotide from Del 121 to Del 124 for understanding the usage of uORFs.
The puzzling observation that the complete deletion of the 5′ UTR (Del 121, Figure 4B) inhibits initiation from the first AUG prompted us to further investigate the sequence environment upstream of the first start codon. We generated several mutant constructs (Figure 4A) and northern blotting was performed to verify the equal expression of all mutant circZNF609 (Supplementary Figures S4A and B). We started with construct Del 121 and shortened it by one nt (Del 122, Figure 4A, C). Unexpectedly, this construct did not efficiently translate protein A or B (Figure 4B and C). Further truncations led to even more surprising results. Deleting one nucleotide more resulted in the generation of four peptides with higher molecular weights than peptide A (Del 123, Figure 4C, peptides G, H, I, K). A closer examination of this construct revealed that the triple FLAG-tag that we inserted contains four additional AUGs and the 5′ UTR of circZNF609 contains a stop codon in frame with the FLAG-AUGs (Figure 4G, H). This upstream ORF (uORF) is most likely translated in all constructs but due to its short product escaped detection in western blots (Figure 4H). In construct Del 121, a stop codon in the ORF is used and products from the FLAG-AUGs are not visible (Figure 4G, H). When we now change the frame by deleting single nt, this stop codon becomes out of frame and FLAG-AUG-driven initiation is now terminated at a stop codon further downstream (Figure 4G, Del 122). Deleting one more nt, this stop codon becomes out of frame and the entire circle is translated until it reaches a stop right after the FLAG tag (Figure 4G and H). Further shortening by one nt changed the frame again but now a stop codon immediately downstream of the first AUG is used and again the very short and invisible product is generated. Thus, due to this uORF only the second AUG is used and peptide B is made. This was observed for all mutants in this frame (Del 121 and Del 124). Of note, these two start codons are independent of the frames of the uORF and peptides A and B are therefore always present. However, they are less prominent when the longer peptides G–K are synthesized, probably due to ribosome occupancy that might interfere with initiation (Del 123). This might explain why neither A nor B was made anymore for Del 122 (Figure 4C, G and H).
Taken together, our systematic investigations demonstrate that translation from exogenous circZNF609 is a multifaceted process and many different AUGs are probably even randomly utilized. Thus, we conclude that our constructs may attract ribosomes rather pervasively.
In vitro transcribed circZNF609 is not translated in cells
To further corroborate our finding that circZNF609 can be translated from several AUGs and to control that the observed translation is caused by the overexpression construct, we transcribed circZNF609 in vitro using a self-splicing expression system (35) (Figure 5A, Supplementary Figure S5A). We cloned untagged and FLAG-tagged circZNF609 into the vector containing all necessary elements of a group I intron for self-splicing. As control, we deleted the 3′ part of the flanking intron, which should abrogate expression of the circRNA (Figure 5A). RNA from in vitro transcription reaction was loaded on an agarose gel (Supplementary Figure S5B, lane 1–3). This revealed a number of different bands corresponding to the predicted products from self-splicing reactions (Supplementary Figure S5A). Northern blotting using probes against the 5′ intron and circZNF609 (Supplementary Figure S5B) indicated that circZNF609 migrates at the size of product 3 (Supplementary Figure S5A-B). This band was cut out and purified. Northern blot using a probe spanning circZNF609 junction (Figure 5B, lanes 1–3) and a probe targeting the 5′ intron (Figure 5B, lanes 4–6) confirmed that we obtained pure circZNF609 from the in vitro transcription reaction. Furthermore, RNase R digestion confirmed that the purified bands were circular (Supplementary Figure S5C). The purified circZNF609 was transfected into HEK293T cells (Figure 5C, D). However, protein bands A and B were only detectable when the overexpression construct but not the RNA was transfected (Figure 5D). To test whether circZNF609 is indeed transfected, we extracted RNA 4 h and 20 h after transfection and performed northern blotting. Indeed, circZNF609 can be readily detected after 4 h and is efficiently degraded after 20 h suggesting that circZNF609 is not trapped in unsolved or aggregated lipid structures caused by transfection that are inaccessible for cellular proteins and enzymes (Figure 5C). These findings are consistent with previous observations (19) and we suggest that in vitro transcribed circZNF609 is not translated. In agreement, we also did not observe translation in a reticulocyte-based in vitro translation system (data not shown).
circZNF609 protein products are generated from splicing by-products
Since we did not find evidence for IRES sequences and translation initiation appears rather random and in vitro transcribed circZNF609 is not translated, we further examined protein and RNA products generated from our overexpression constructs.
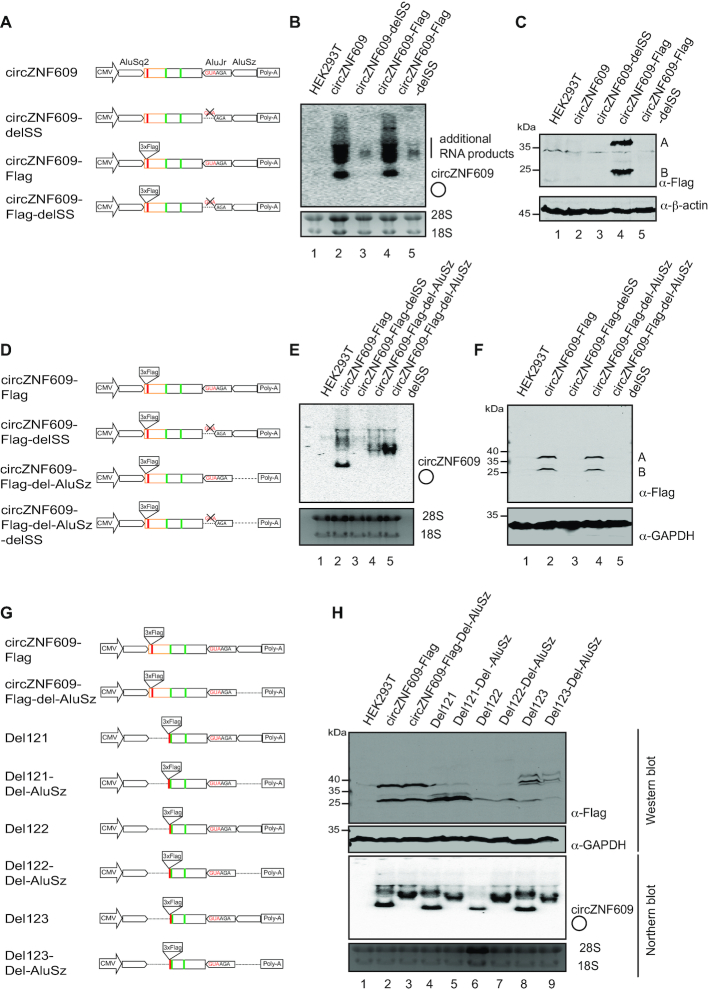
Splicing and thus the presence of 5′ and 3′ splice sites flanking circZNF609 are essential for circRNA biogenesis. To test whether splicing is also required for the translated proteins, we deleted the 3′ splice site from our FLAG-tagged and untagged constructs and expressed them in HEK293T cells (Figure 6A). As expected, when the splice site is deleted, circZNF609 is not generated (Figure 6B). Of note, also other RNA bands and by-products are strongly reduced suggesting that splicing of these constructs generates several unwanted RNAs in addition to the circRNA. When analyzing translation by western botting against the FLAG-tag, we did not observe FLAG-tagged protein products when the splice site is deleted (Figure 6C).
Figure 6.
circZNF609 protein products are most likely generated from splicing by-products. (A) The schematic overview represents deletion of the splice sites (SS) for disruption of circZNF609 production. Specifically, 3 nts (SS, GUA) in the ZKSCAN1 intron downstream of circZNF609 were deleted. (B) Northern blot of HEK293T cells transfected with the delSS mutant. A probe spanning the circZNF609 splice junction was used to detect the signal. Of note, deletion of splice site abrogates circZNF609 production as well as the by-products. (C) Western blot of HEK293T transfected with delSS mutants detected by anti-Flag antibodies. Beta-actin served as loading control. (D) Schematic overview of the circZNF609-Flag constructs deleted of the splice site (SS) and the AluSz located in the ZKSCAN1 intron downstream of circZNF609. (E) Northern blot of HEK293T cells transfected with different deletion mutants. The signal was detected with a probe spanning the circZNF609 splice junction. (F) Western blot of HEK293T cells transfected with different deletion mutants detected with the anti-Flag antibody. GAPDH served as loading control. (G) Schematic overview of circZNF609-Flag constructs and the 5′ UTR deletion constructs (Del 121, Del 122, Del 123) with deletion of AluSz located in the ZKSCAN1 intron downstream of circZNF609. (H) Lower panel: Northern blot of HEK293T cells transfected with different deletion mutants. The signal was detected with a probe spanning the circZNF609 splice junction. Upper panel: Western blot of HEK293T cells transfected with different deletion mutants detected with the anti-Flag antibody. GAPDH served as loading control.
Next, we reasoned that complementary Alu sequences within the constructs are essential as well since they need to fold back on each other to allow for back-splicing. We deleted the Alu sequence downstream of circZNF609 and analyzed circRNA generation by northern blotting (Figure 6D and E). CircZNF609 is not expressed anymore, when the downstream Alu element is deleted (Figure 6E, lane 4). Surprisingly, when we analyzed protein synthesis from these constructs by western blotting, we can still readily detect protein bands A and B (Figure 6F, lane 4). This was fully unexpected since circZNF609 is not present as the northern blots clearly indicate. These findings suggest that the protein A and B might be expressed from a linear splicing concatemer. To test this, we simultaneously deleted the splice sites and the Alu element and found that protein A and B are not translated anymore (Figure 6F, lane 5). These results lead to the conclusion that circZNF609 proteins A and B are not produced from a circular RNA. Although unlikely, our results cannot exclude that peptides A and B are simultaneously produced from circZNF609 and linear splicing products. Thus, although circZNF609 expression is lost, proteins generated from the linear transcripts are still detected.
Since we had observed that circZNF609 construct can randomly initiate and pervasively translate (Figure 4C), we tested whether these effects are also present in the Alu-deleted constructs (Figure 6G). All Alu deletion mutants do not produce circRNAs but protein products are still readily detected (Figure 6H). Notably, all protein products are not detectable when we deleted further the splice site (Supplementary Figure S6A–C). Thus, we conclude that the proteins are not produced from the circular RNAs but from linear transcripts and circRNA overexpression systems need to be treated with caution when functional studies are performed and interpreted.
DISCUSSION
CircRNAs have been described decades ago (41,42) but a potential role as regulatory RNAs have only been recognized recently, when RNAseq was combined with bioinformatics strategies to identify reads from back-splicing of 5′ and 3′ ends of exons on a transcriptome-wide scale (3,9,43). Since then, circRNAs have been profiled in various tissues including several types of cancer (44,45). CircZNF609 has initially been found in human myoblasts where it controls cell proliferation (19). Interestingly, circZNF609 is upregulated in rhabdomyosarcoma, a skeletal muscle-derived pediatric cancer where it regulates cell cycle progression from G1 to S phase and thus is important for cancer cell proliferation (32). circZNF609 is highly abundant circRNA in brain and associated with neuronal differentiation and aging (4,30). Moreover, circZNF609 expression is correlated with several cancers and might play important roles in tumor development and metastasis (46). In agreement with these reported functions, we identify circZNF609 as a potential oncogene important for colorectal cancer progression (data not shown).
Although many circRNAs have been identified, their individual functions are often only poorly understood. Since a large number of circRNAs contain ORFs, it has been shown that circRNAs possess coding potential and can indeed be translated (19,20,29). Some of these peptides have even been implicated in diseases such as cancer (24,25,47,48). In contrast, a comprehensive study on AUG-containing circRNAs did not find evidence for circRNA translation (28). Since circRNAs do not contain 5′ m7G caps, potential translation initiation is cap-independent. In such cases, IRES elements can be utilized that recruit ribosomal subunits to the cognate AUG. Our truncation studies, however, revealed that all sequences can be deleted and translation is still detectable suggesting an IRES-independent alternative initiation mechanism. It has been shown that circRNAs are m6A-modified (49) and this modification is critical for translation initiation (21,22). The m6A reader protein YTHDF3 interacts with circular RNAs and recruits the initiation factor variant eIF4G2, which facilitates initiation (21,22). Although circZNF609 is m6A-modified in our assays, knock out of METTL3 did not lead to reduced translation suggesting an METTL3-independent initiation mechanism in our cellular system. Alternatively, other m6A-generating enzymes could be involved in circZNF609 methylation, which cannot be ruled out in our assays.
Since endogenous protein products are difficult to find, it is not clear if circZNF609 is mainly a non-coding or a functional protein coding RNA. To shed more light on the proposed mechanisms of translation of circZNF609, we extensively tested numerous constructs and conditions and concluded that translation of our overexpressed constructs can start at various AUGs. A major concern about circRNA overexpression systems is the generation of linear transcripts as well as cis and trans splicing events that are rarely controlled for in cell biological experiments. Thus, we attempted to carefully delineate circZNF609 translation from our overexpression constructs. Using restriction digestion and integration of the plasmid into the genome, we found that circZNF609 is apparently translated and thus not produced from concatemers generated by cis-splicing of rolling circle transcripts. Moreover, splicing plays an important role in producing circZNF609 proteins because splice site deletion results in a complete loss of the protein products. However, when one Alu element was deleted and circRNA splicing was completely abolished, the protein products were still readily detectable suggesting a trans splicing event leading to a linear transcript that is translated. The molecular basis for the observed trans splicing is unclear and needs to be investigated further. Translation from a linear by-product is also corroborated by the finding that in vitro transcribed and transfected circRNAs do not lead to detectable protein products (Figure 5) (19). On the other hand, it is also conceivable that circRNAs need to go through cellular biogenesis in order to be licensed for efficient translation as has been suggested (19). Taken together, it is tempting to speculate that circRNA overexpression constructs may undergo trans-splicing and thus such tools need to be evaluated carefully. Although not tested yet, it is likely that similar systems such as using other flanking introns might result in similar trans-splicing effects and should be controlled carefully as well.
What could be the reasons for the association of circZNF609 with ribosomes as has been observed previously (19) if it is not translated? Ribosome-associated lncRNAs are more sensitive to nonsense-mediated decay and are often less stable and degraded at ribosomes (50,51). It is conceivable that ribosome binding to circRNAs could also lead to enhanced turnover of circRNAs or to the initiation of nonsense-mediated decay, which has not been studied so far. On the other hand, circZNF609 was not detected on ribosomes in HEK293T cells (52). This suggests condition- or cell type-specific effects that need to be considered.
Supplementary Material
ACKNOWLEDGEMENTS
We thank S. Ammon, C. Friederich and Annamaria Dalfino Spinelli for technical support. We thank Jeremy Wilusz, Irene Bozzoni, Ivano Legnini and So Umekage for providing reagents.
Contributor Information
Hung Ho-Xuan, Regensburg Center for Biochemistry (RCB), Laboratory for RNA Biology, University of Regensburg, Regensburg, Germany.
Petar Glažar, Laboratory for Systems Biology of Gene Regulatory Elements, Berlin Institute for Medical Systems Biology, Max-Delbrück Center for Molecular Medicine, Berlin, Germany.
Claudia Latini, Regensburg Center for Biochemistry (RCB), Laboratory for RNA Biology, University of Regensburg, Regensburg, Germany.
Kevin Heizler, Regensburg Center for Biochemistry (RCB), Laboratory for RNA Biology, University of Regensburg, Regensburg, Germany.
Jacob Haase, Institute of Molecular Medicine, Section for Molecular Cell Biology, Martin-Luther-University Halle-Wittenberg, Charles Tanford Protein Center, 06120 Halle, Germany.
Robert Hett, Regensburg Center for Biochemistry (RCB), Laboratory for RNA Biology, University of Regensburg, Regensburg, Germany.
Marvin Anders, Department of Surgery, University Hospital Regensburg, Regensburg, Germany.
Franziska Weichmann, Regensburg Center for Biochemistry (RCB), Laboratory for RNA Biology, University of Regensburg, Regensburg, Germany.
Astrid Bruckmann, Regensburg Center for Biochemistry (RCB), Laboratory for RNA Biology, University of Regensburg, Regensburg, Germany.
Debbie Van den Berg, Department of Cell Biology, Erasmus MC, Wytemaweg 80, 3015 CN, Rotterdam, Netherlands.
Stefan Hüttelmaier, Institute of Molecular Medicine, Section for Molecular Cell Biology, Martin-Luther-University Halle-Wittenberg, Charles Tanford Protein Center, 06120 Halle, Germany.
Nikolaus Rajewsky, Laboratory for Systems Biology of Gene Regulatory Elements, Berlin Institute for Medical Systems Biology, Max-Delbrück Center for Molecular Medicine, Berlin, Germany.
Christina Hackl, Department of Surgery, University Hospital Regensburg, Regensburg, Germany.
Gunter Meister, Regensburg Center for Biochemistry (RCB), Laboratory for RNA Biology, University of Regensburg, Regensburg, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [FOR2127]; European Union [ITN RNATRAIN, ERC grant 682291 ‘moreRNAs’]. The open access publication charge for this paper has been waived by Oxford University Press – NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1. Deveson I.W., Hardwick S.A., Mercer T.R., Mattick J.S.. The dimensions, dynamics, and relevance of the mammalian noncoding transcriptome. Trends Genet. 2017; 33:464–478. [DOI] [PubMed] [Google Scholar]
- 2. Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J.. Natural RNA circles function as efficient microRNA sponges. Nature. 2013; 495:384–388. [DOI] [PubMed] [Google Scholar]
- 3. Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. et al.. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013; 495:333–338. [DOI] [PubMed] [Google Scholar]
- 4. Rybak-Wolf A., Stottmeister C., Glazar P., Jens M., Pino N., Giusti S., Hanan M., Behm M., Bartok O., Ashwal-Fluss R. et al.. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell. 2015; 58:870–885. [DOI] [PubMed] [Google Scholar]
- 5. Li X., Yang L., Chen L.L.. The biogenesis, functions, and challenges of circular RNAs. Mol. Cell. 2018; 71:428–442. [DOI] [PubMed] [Google Scholar]
- 6. Ebbesen K.K., Hansen T.B., Kjems J.. Insights into circular RNA biology. RNA Biol. 2017; 14:1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016; 17:205–211. [DOI] [PubMed] [Google Scholar]
- 8. Ebbesen K.K., Kjems J., Hansen T.B.. Circular RNAs: identification, biogenesis and function. Biochim. Biophys. Acta. 2016; 1859:163–168. [DOI] [PubMed] [Google Scholar]
- 9. Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E.. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013; 19:141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang X.O., Wang H.B., Zhang Y., Lu X., Chen L.L., Yang L.. Complementary sequence-mediated exon circularization. Cell. 2014; 159:134–147. [DOI] [PubMed] [Google Scholar]
- 11. Ivanov A., Memczak S., Wyler E., Torti F., Porath H.T., Orejuela M.R., Piechotta M., Levanon E.Y., Landthaler M., Dieterich C. et al.. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015; 10:170–177. [DOI] [PubMed] [Google Scholar]
- 12. Dubin R.A., Kazmi M.A., Ostrer H.. Inverted repeats are necessary for circularization of the mouse testis Sry transcript. Gene. 1995; 167:245–248. [DOI] [PubMed] [Google Scholar]
- 13. Liang D., Wilusz J.E.. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014; 28:2233–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S.. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014; 56:55–66. [DOI] [PubMed] [Google Scholar]
- 15. Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A., Roslan S., Schreiber A.W., Gregory P.A., Goodall G.J.. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015; 160:1125–1134. [DOI] [PubMed] [Google Scholar]
- 16. Piwecka M., Glazar P., Hernandez-Miranda L.R., Memczak S., Wolf S.A., Rybak-Wolf A., Filipchyk A., Klironomos F., Cerda Jara C.A., Fenske P. et al.. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017; 357:eaam8526. [DOI] [PubMed] [Google Scholar]
- 17. Kleaveland B., Shi C.Y., Stefano J., Bartel D.P.. A network of noncoding regulatory RNAs acts in the mammalian brain. Cell. 2018; 174:350–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hentze M.W., Castello A., Schwarzl T., Preiss T.. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 2018; 19:327–341. [DOI] [PubMed] [Google Scholar]
- 19. Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M. et al.. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell. 2017; 66:22–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pamudurti N.R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L., Hanan M., Wyler E., Perez-Hernandez D., Ramberger E. et al.. Translation of CircRNAs. Mol. Cell. 2017; 66:9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., Jin Y., Yang Y., Chen L.L., Wang Y. et al.. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017; 27:626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Di Timoteo G., Dattilo D., Centron-Broco A., Colantoni A., Guarnacci M., Rossi F., Incarnato D., Oliviero S., Fatica A., Morlando M. et al.. Modulation of circRNA metabolism by m(6)A modification. Cell Rep. 2020; 31:107641. [DOI] [PubMed] [Google Scholar]
- 23. Zhao J., Lee E.E., Kim J., Yang R., Chamseddin B., Ni C., Gusho E., Xie Y., Chiang C.M., Buszczak M. et al.. Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat. Commun. 2019; 10:2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Begum S., Yiu A., Stebbing J., Castellano L.. Novel tumour suppressive protein encoded by circular RNA, circ-SHPRH, in glioblastomas. Oncogene. 2018; 37:4055–4057. [DOI] [PubMed] [Google Scholar]
- 25. Zhang M., Zhao K., Xu X., Yang Y., Yan S., Wei P., Liu H., Xu J., Xiao F., Zhou H. et al.. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun. 2018; 9:4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zheng X., Chen L., Zhou Y., Wang Q., Zheng Z., Xu B., Wu C., Zhou Q., Hu W., Wu C. et al.. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol. Cancer. 2019; 18:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang M., Huang N., Yang X., Luo J., Yan S., Xiao F., Chen W., Gao X., Zhao K., Zhou H. et al.. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018; 37:1805–1814. [DOI] [PubMed] [Google Scholar]
- 28. Stagsted L.V., Nielsen K.M., Daugaard I., Hansen T.B.. Noncoding AUG circRNAs constitute an abundant and conserved subclass of circles. Life Sci Alliance. 2019; 2:e201900398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van Heesch S., Witte F., Schneider-Lunitz V., Schulz J.F., Adami E., Faber A.B., Kirchner M., Maatz H., Blachut S., Sandmann C.L. et al.. The translational landscape of the human heart. Cell. 2019; 178:242–260. [DOI] [PubMed] [Google Scholar]
- 30. Gruner H., Cortes-Lopez M., Cooper D.A., Bauer M., Miura P.. CircRNA accumulation in the aging mouse brain. Sci. Rep. 2016; 6:38907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Salgado-Somoza A., Zhang L., Vausort M., Devaux Y.. The circular RNA MICRA for risk stratification after myocardial infarction. Int. J. Cardiol. Heart Vasc. 2017; 17:33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rossi F., Legnini I., Megiorni F., Colantoni A., Santini T., Morlando M., Di Timoteo G., Dattilo D., Dominici C., Bozzoni I.. Circ-ZNF609 regulates G1-S progression in rhabdomyosarcoma. Oncogene. 2019; 38:3843–3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Muller S., Glass M., Singh A.K., Haase J., Bley N., Fuchs T., Lederer M., Dahl A., Huang H., Chen J. et al.. IGF2BP1 promotes SRF-dependent transcription in cancer in a m6A- and miRNA-dependent manner. Nucleic Acids Res. 2019; 47:375–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Scholler E., Weichmann F., Treiber T., Ringle S., Treiber N., Flatley A., Feederle R., Bruckmann A., Meister G.. Interactions, localization, and phosphorylation of the m(6)A generating METTL3-METTL14-WTAP complex. RNA. 2018; 24:499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Umekage S., Kikuchi Y.. In vitro and in vivo production and purification of circular RNA aptamer. J. Biotechnol. 2009; 139:265–272. [DOI] [PubMed] [Google Scholar]
- 36. Weichmann F., Hett R., Schepers A., Ito-Kureha T., Flatley A., Slama K., Hastert F., Angstmann N., Cardoso C.M., Konig J.. Validation strategies for antibodies targeting modified ribonucleotides. RNA. 2020; doi:10.1261/rna.076026.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Su D., Chan C.T., Gu C., Lim K.S., Chionh Y.H., McBee M.E., Russell B.S., Babu I.R., Begley T.J., Dedon P.C.. Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. Nat. Protoc. 2014; 9:828–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kramer M.C., Liang D., Tatomer D.C., Gold B., March Z.M., Cherry S., Wilusz J.E.. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015; 29:2168–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G. et al.. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014; 505:117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Leppek K., Das R., Barna M.. Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol. 2018; 19:158–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Braun S., Domdey H., Wiebauer K.. Inverse splicing of a discontinuous pre-mRNA intron generates a circular exon in a HeLa cell nuclear extract. Nucleic Acids Res. 1996; 24:4152–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cocquerelle C., Mascrez B., Hetuin D., Bailleul B.. Mis-splicing yields circular RNA molecules. FASEB J. 1993; 7:155–160. [DOI] [PubMed] [Google Scholar]
- 43. Salzman J., Gawad C., Wang P.L., Lacayo N., Brown P.O.. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012; 7:e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kristensen L.S., Hansen T.B., Veno M.T., Kjems J.. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018; 37:555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Patop I.L., Kadener S.. circRNAs in Cancer. Curr. Opin. Genet. Dev. 2018; 48:121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. He Y., Huang H., Jin L., Zhang F., Zeng M., Wei L., Tang S., Chen D., Wang W.. CircZNF609 enhances hepatocellular carcinoma cell proliferation, metastasis, and stemness by activating the Hedgehog pathway through the regulation of miR-15a-5p/15b-5p and GLI2 expressions. Cell Death. Dis. 2020; 11:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liang W.C., Wong C.W., Liang P.P., Shi M., Cao Y., Rao S.T., Tsui S.K., Waye M.M., Zhang Q., Fu W.M. et al.. Translation of the circular RNA circbeta-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019; 20:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang Y., Gao X., Zhang M., Yan S., Sun C., Xiao F., Huang N., Yang X., Zhao K., Zhou H. et al.. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Inst. 2018; 110:304–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhou C., Molinie B., Daneshvar K., Pondick J.V., Wang J., Van Wittenberghe N., Xing Y., Giallourakis C.C., Mullen A.C.. Genome-Wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep. 2017; 20:2262–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zeng C., Fukunaga T., Hamada M.. Identification and analysis of ribosome-associated lncRNAs using ribosome profiling data. BMC Genomics. 2018; 19:414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Carlevaro-Fita J., Rahim A., Guigo R., Vardy L.A., Johnson R.. Cytoplasmic long noncoding RNAs are frequently bound to and degraded at ribosomes in human cells. RNA. 2016; 22:867–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ragan C., Goodall G.J., Shirokikh N.E., Preiss T.. Insights into the biogenesis and potential functions of exonic circular RNA. Sci. Rep. 2019; 9:2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.