Abstract
Ikaros family zinc finger 1(IKZF1) encodes a lymphoid-restricted zinc finger transcription factor named Ikaros that regulates lymphocyte differentiation and proliferation as well as self-tolerance. Increasing evidence indicates that IKZF1 could contribute to the pathogenesis of autoimmune diseases. Recent research has provided evidence that IKZF1 might correlate with Systemic lupus erythematosus (SLE), but no clear definition has been made yet. In this study, we focus on the relationship between IKZF1 polymorphisms and SLE susceptibility, cytokine levels, and clinical characteristics in the Chinese Han population.
One thousand seventy-six subjects, including 400 SLE patients and 676 healthy controls, were included in this study. Three single nucleotide polymorphisms within IKZF1 containing rs4917014, rs11980379, and rs4132601 were genotyped in all subjects by an improved multiplex ligation detection reaction technique. 143 subjects from SLE patients were randomly selected for testing the levels of serum cytokines. The clinical characteristics of SLE patients were gathered and collated from medical records. The data were analyzed mainly using SPSS20.0 (SPSS lnc., Chicago, IL).
Significant relationships were observed between rs4132601 and SLE susceptibility, CD40 ligand, and malar rash (P < .001, P = .04, and P = .01, respectively). In addition, significant relationships were observed between rs4917014 and susceptibility, granzyme B level, and hematological disorder in SLE (P = .005, P = .03 and P = .005, respectively).
The results further support that IKZF1 may have an important role in the development and pathogenesis of SLE. Allele G of rs4132601 and rs4917014 is related to a decreased risk of SLE occurrence and associated with clinical features in SLE patients, including CD40 ligand level, granzyme B level, malar rash, and hematological disorder, which play important roles in disease progression.
Keywords: CD40 ligand, granzyme B, hematological disorder, ikaros family zinc finger 1, malar rash, single nucleotide polymorphisms, systemic lupus erythematosus
1. Introduction
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease characterized by a large number of autoantibodies generation, complement activation, and immune complex deposition, which results in multisystem microvascular inflammation, tissue damage, and potentially life-threatening multi-organ failure.[1] Comparing with the general population, SLE patients have nearly a 3-fold increased risk of death.[2] Although SLE poses a severe threat to human health, the exact etiopathology of SLE is not clear.
Genetic factors play essential roles in the etiology and development of SLE.[3] Numerous genes associated with SLE have been identified, mainly influencing lymphoid cell function, dendritic-cell function and Interferon (IFN) signaling.[4] Ikaros, encoded by ikaros family zinc finger 1(IKZF1), is one of the most substantial transcription factors demonstrated in the control of lymphocyte specification and differentiation. It widely expresses in hematopoietic precursors and regulates lymphocyte differentiation, proliferation, and self-tolerance through regulation of B-cell, T-cell, and dendritic cell signaling, which are important to maintain the immune system stability and avoid the occurrence of autoimmune diseases.[5–8] With the development of genetic studies, IKZF1 has been identified as risk loci for hematological malignancies, solid tumors, and autoimmune diseases.[9,10]
Meanwhile, several single nucleotide polymorphisms (SNPs) such as rs11980379 and rs4132601 have also been identified as the susceptible variants for some immune diseases such as inflammatory bowel disease, Crohn disease and primary Sjoègren syndrome.[11–13] Previous data implicated that the expression levels of IKZF1 mRNA in SLE patients decreased significantly, making IKZF1 a considerable factor in the characteristic cascade of SLE.[14] Moreover, some studies suggested that IKZF1 might be a candidate gene for SLE.[15,16] Meanwhile, IKZF1 rs4917014 may affect the genetic predisposition to SLE.[15,17] In short, a reliable basis of data from different research is accumulating, which provides support for the potential role of IKZF1 underlying SLE. Nevertheless, the exact role of IKZF1 in SLE is remained to be further explored.
Therefore, IKZF1 is a promising topic of researches to understand immune deficiency and autoimmunity diseases. Besides, the hematological disorder is the most common clinical symptom of SLE in China. Considering the vital role of IKZF1 in the hematological system regulation, IKZF1 may contribute to the etiology and clinical manifestation of SLE. In the present study, we aim to explore the association between the IKZF1 polymorphisms rs4917014, rs11980379 and rs4132601, and SLE in the Chinese Han population.
2. Methods
2.1. Participants
We recruited 400 SLE patients and 676 gender- and age-matched health controls from 2014 to 2018 in West China Hospital. All the 1076 subjects are Chinese and signed informed consents. The included patients were first diagnosed in our hospital with fulfilling SLE criteria 1997 released by the American College of Rheumatology classification.[18] Drug-induced SLE patients and patients who had received regular treatment with lupus-related drugs were excluded. All included controls lived in the same area with the patients and randomly invited to participate in the study. Participants were considered healthy and included in the control group when they did not disclose any autoimmune disorders, chronic diseases, and endemic infectious diseases and had normal blood test results. The blood tests included routine blood tests, blood biochemical indexes tests, tumor marker tests, and immune indexes tests. All the individual participants were uniquely numbered to ensure complete information and personal privacy. This study was approved by the Ethics Committee of West China Hospital (No. 81601830) and conducted in the light of the 1975 Declaration of Helsinki. Informed consent was obtained from every participant.
2.2. Patient information
We gathered and collated the patients’ information from medical records including general information (age, gender) and clinical characteristics (including disease duration, disease activity (determined by the SLE disease activity index 2000 (SLEDAI-2k))[19] and main symptoms such as malar rash, hematological disorder (excluding drug factors, platelet (PLT) < 100∗10^9/L or white blood cell (WBC) < 3∗10^9/L) and proteinuria). All the patients were in the active stage with SLEDAI > 4.
2.2.1. IKZF1 polymorphisms genotyping
1 to 2 mL EDTA anti-coagulant blood was obtained from subjects, of which 200uL was used for Genomic DNA extraction by QuickGene DNA whole blood kit S (QuickGene, FujiFilm, Japan). An improved multiplex ligation detection reaction (iMLDR) technique (Genesky Biotechnologies Inc., Shanghai, China) was used for all SNPs (rs4917014, rs11980379, rs4132601) genotyping. The PCR program was set as follows: 95 °C 2 minute, 11 cycles × (94 °C 20 second, 65 °C to 0.5 °C/cycle 40 second, 72 °C 1 minute), 24 cycles × (94 °C 20 second, 59 °C 30 seconds, 72 °C 1.5 minute), 72 °C 2 minute, 4 °C after that. The ligation cycling program was set as below: 95 °C 2 minute; 38 cycles × (94 °C 1 minute, 56 °C 4 minute), 4 °C hold. ABI 3730XL was conducted for genotyping by loading a 0.5uL ligation product. Then GeneMapper 4.1 (AppliedBiosystems, Foster City, CA) was applied to analyze the initial data. Meanwhile, 10% of total DNA samples were randomly selected to sequence for confirming the iMLDR results.
2.3. Antibodies and cytokines testing
One hundred forty-three subjects from SLE patients were randomly selected for testing the levels of serum cytokines including interleukin-6, interleukin-17A, IFN-γ, tumor necrosis factor-α, CD40 ligand (CD40L), and granzyme B. All the 400 SLE patients were tested for other markers, including anti-nuclear antibody (ANA), anti-double-stranded DNA antibodies, anti-Smith antibodies, anti-ribonuclear protein antibody, anti-SSA/ anti-SSB antibody, anti-ribosomal protein antibody, and routine blood test (including Red blood cell (RBC), hemoglobin (HGB), PLT, and WBC).
2.3.1. Laboratory assays
R&D Human Inflammation Assays were used for cytokines detection. Autoantibodies of all the SLE patients were tested by indirect immunofluorescence reagent kits (Euroimmun, Germany). Routine blood test (including RBC, HGB, PLT, and WBC) was analyzed by full-automatic blood analyzer (Sysmex, Japan). All the tests were conducted based on manufacturers’ instruction.
2.4. Statistical analysis
The software “PS: Power and Sample Size Calculation” was applied for Hardy–Weinberg equilibrium appraising for each polymorphism. Haploview software package (version 4.2, Mark Daly's lab, Boston) was used for polymorphisms linkage disequilibrium exploring.[20] Statistical Package for the Social Sciences version 20.0 (SPSS, SPSS Inc., Chicago, IL) was applied for all statistical analyses. The student t test or Mann–Whitney U test was appropriately used to compare demographic and clinical data between groups. For the allele case-control comparisons, Pearson Chi-squared test or Fisher exact test was used. The odds ratio (OR) and 95% Confidence Interval (CI) were figured out to estimate the association of SNPs with susceptibility and development. When the 2-sided P-value was less than .05, the results were considered to be statistically significant.
For the case and control comparison, we used some analytic methods: dominant model (TG/TC+GG/CC vs TT), recessive model (GG/CC vs TT+TG/TC), allele frequency distribution (allele T vs allele G/C).
3. Results
3.1. Characteristics of the subjects
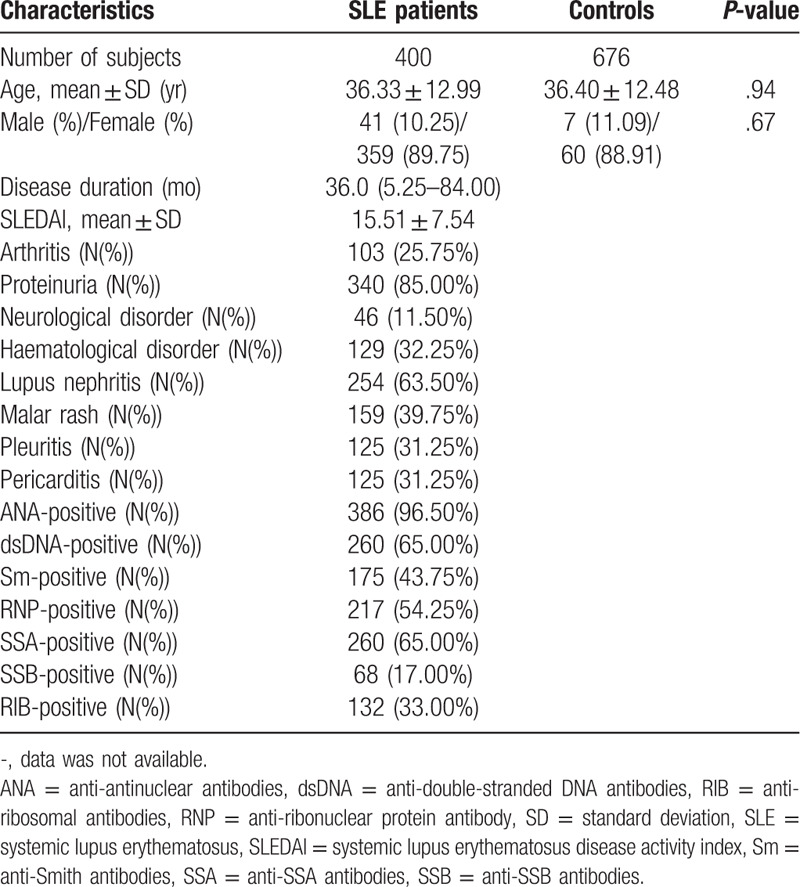
Table 1 presents the primary demographic and clinical characteristics of all participants. Two groups of subjects, namely SLE patients, and controls with 400 and 676 subjects, were interviewed. The mean ages were 36.33 and 36.40 years, respectively, in SLE patients and controls. The female percentages were 89.75 in SLE patients and 88.91 in controls. The 2 groups matched adequately in age and gender (P = .935 and P = .666, respectively). The disease-related information was shown for the patients. The disease duration ranged from 5.25 to 84 months, and the mean of SLEDAI score that responses disease activity was 15.51. The positive percentages of various marker antibodies were: anti-nuclear antibody (96.50%), anti-double-stranded DNA antibodies (65.00%), anti-Smith antibodies (43.75%), ribonuclear protein antibody (54.25%), anti-SSA antibodies (65.00%), anti-SSB antibodies (17.00%), anti-ribosomal antibodies (33.00%). In addition, the patients had multiple clinical symptoms, the proportion of which appeared as: arthritis (25.75%), proteinuria (85.00%), neurological disorder (11.50%), haematological disorder (35.25%), lupus nephritis (63.50%), Malar rash (39.75%), pleuritic (31.25%), pericarditis (31.25%).
Table 1.
Demographic and clinical characteristics of subjects.
3.2. Genotyping and linkage disequilibrium evaluation
IMLDR method was used for the 3 SNPs (rs11980379, rs4132601, rs4917014) genotyping of all subjects in this study. All the detections obtained effective results, which were in complete accordance with the results from the randomly selected samples’ direct sequencing. No significant deviation was found in the analysis of Hardy–Weinberg equilibrium (HWE).
Haploview was applied for Linkage disequilibrium evaluation, which shows that rs11980379 and rs4132601 were in complete linkage disequilibrium (Fig. 1). Accordingly, to avoid repetitive results, the analyses except susceptibility analysis were not carried out on the rs11980379.
Figure 1.
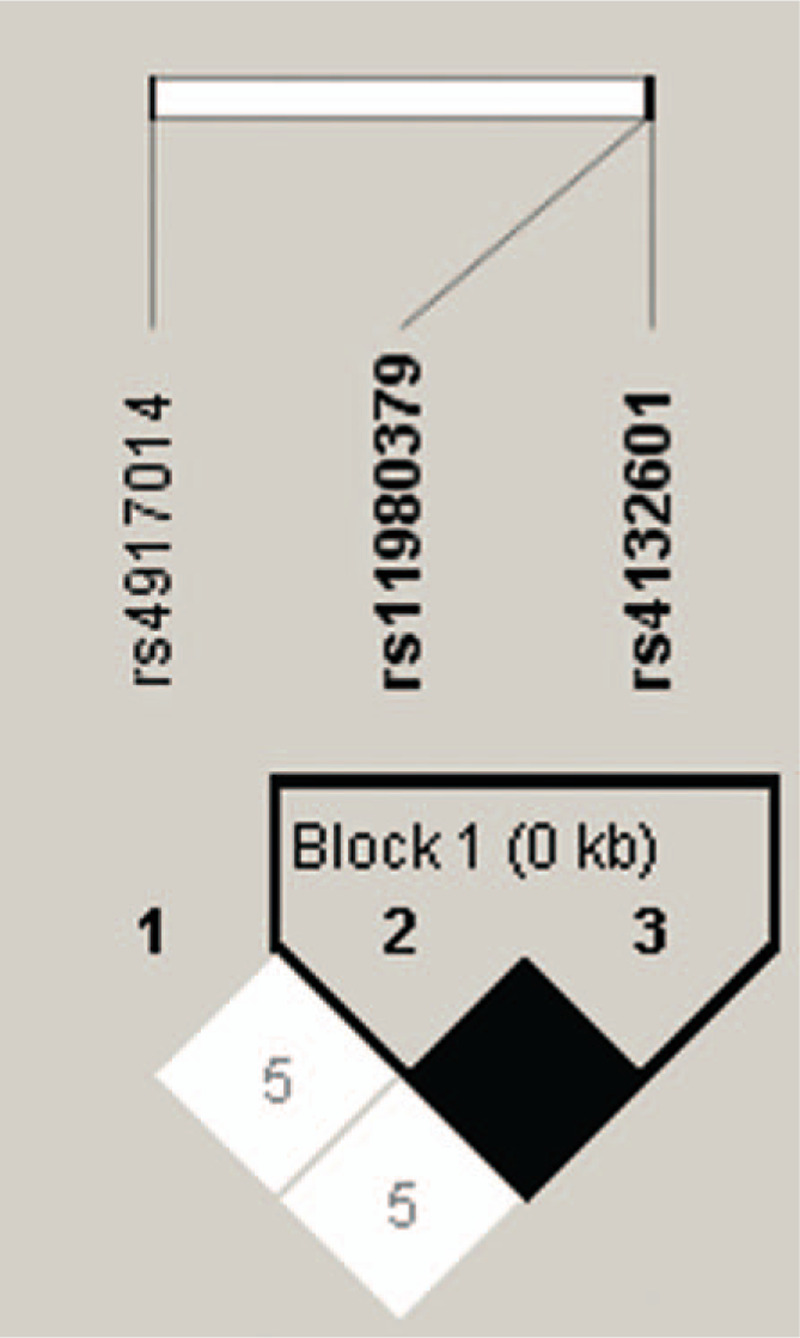
Linkage disequilibrium evaluation for ikaros family zinc finger 1 single nucleotide polymorphisms in 1076 individuals. The linkage disequilibrium plot shows r2 values between each pair of single nucleotide polymorphisms. Rs11980379 and rs4132601 were in complete linkage disequilibrium (black squares, r2 = 1.00).
3.3. Association between IKZF1 SNPs and SLE susceptibility
Table 2 provides the allele and genotype frequencies of IKZF1 SNPs (rs4132601, rs11980379, and rs4917014) in the SLE patients and controls.
Table 2.
Genotype distributions of ikaros family zinc finger 1 in systemic lupus erythematosus patients and controls.
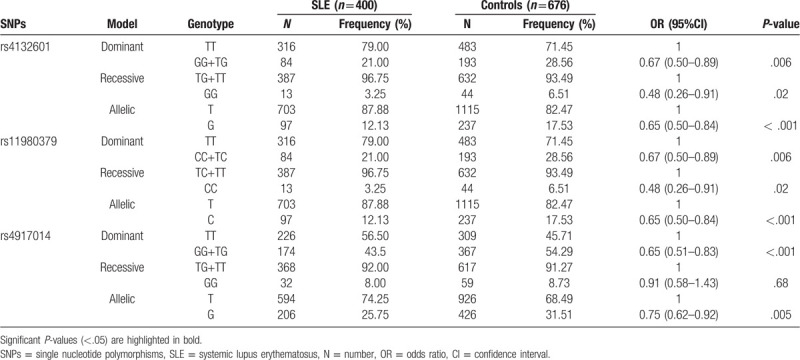
There were statistical differences between SLE patients and healthy subjects in genotype distributions of rs4132601 and rs11980379 (dominant model, OR = 0.67, 95%CI = 0.50–0.89, P = .006; recessive model, OR = 0.48, 95%CI = 0.26–0.91, P = .02; allelic model, OR = 0.65, 95%CI = 0.50–0.84, P < .001). Significant difference was also found in the genotype distribution of SNP rs4917014 between the 2 groups (dominant model, OR = 0.65, 95%CI = 0.51–0.83, P < .001; allelic model, OR = 0.75, 95%CI = 0.62–0.92, P = .005).
3.4. Association between IKZF1 SNPs and cytokines in SLE patients
Figures 2 and 3 illustrate the correlation analyses between IKZF1 polymorphisms and cytokine levels in 143 SLE patients. For rs4132601, there were 98 TT genotype carriers, 32 GT genotype carriers, and 13 GG genotype carriers. A significant difference was found in the serous level of CD40L of different genotype carriers, and GG genotype carriers had the highest level (P = .04). The analytical results also showed that the IKZF1 rs4917014 TT genotype correlated with a reduced level of granzyme B in SLE patients (P = .03). No significant difference was found between IKZF1 SNPs (rs4132601 and rs4917014) and the other cytokine levels in SLE patients (P > .05).
Figure 2.
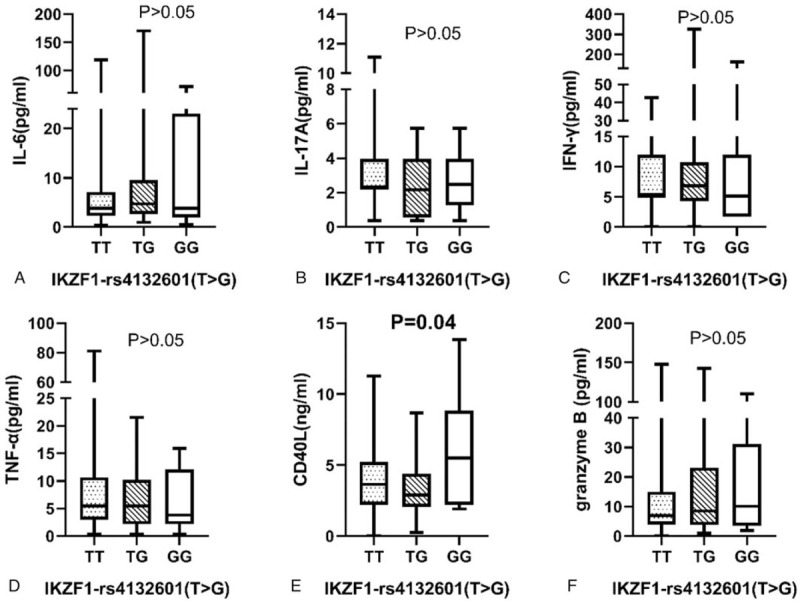
Association between ikaros family zinc finger 1-rs4132601 genotype distribution and cytokines in SLE patients. Cytokines in 143 systemic lupus erythematosus patients with different genotype of ikaros family zinc finger 1-rs4132601 (98 TT, 32 GT and 13 GG). (A) IL-6 (pg/mL): TT genotype 3.81 (2.3, 7.14), TG genotype 4.72(2.60,9.54) and GG genotype 3.21(1.55,13.98), P = .48. (B) IL-17A (pg/mL): TT genotype 2.33 (2.18, 3.96), TG genotype 2.18 (0.55, 3.96) and GG genotype 2.48 (2.18, 3.96), P = .69. (C) IFN-γ (pg/mL): TT genotype 5.16 (5.16, 11.99), TG genotype 6.87 (1.75, 10.28) and GG genotype 5.16 (1.75, 11.14), P = .48. (D) TNF-α (pg/mL): TT genotype 5.44 (2.97, 10.57), TG genotype 5.44 (2.13, 10.18) and GG genotype 4.22 (2.55, 9.18), P = .71. (E) CD40L(ng/mL): TT genotype 3.64 (2.20, 5.24), TG genotype 2.89 (2.06, 4.34) and GG genotype 5.46 (3.17, 7.61), P = .04. (F) Granzyme B (pg/mL): TT genotype 6.85 (3.95, 15.02), TG genotype 8.49 (3.74, 23.13) and GG genotype 6.85 (4.36, 23.11), P = .69. The results represent the median and interquartile of cytokines levels.
Figure 3.
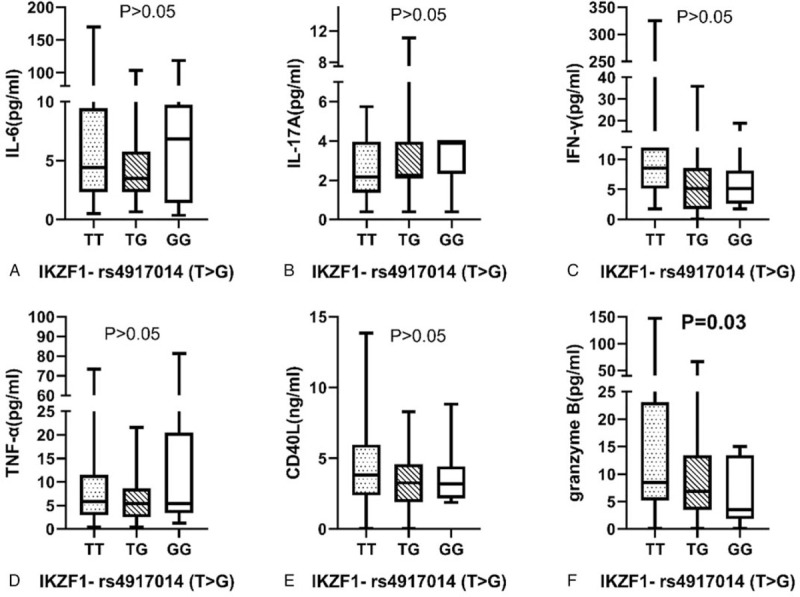
Association between ikaros family zinc finger 1-rs4917014 genotype distribution and cytokines in systemic lupus erythematosus patients. Cytokines in 143 SLE patients with different genotype of ikaros family zinc finger 1-rs4917014 (86 TT, 46 GT and 11 GG). (A) IL-6 (pg/mL): TT genotype 4.41 (2.30, 9.47), TG genotype 3.51 (2.30, 5.78) and GG genotype 6.84 (1.40, 9.74), P = .36. (B) IL-17A (pg/mL): TT genotype 2.18 (1.37, 3.96), TG genotype 2.18 (2.18, 3.96) and GG genotype 3.96 (2.33, 3.96), P = .62. (C) IFN-γ (pg/mL): TT genotype 5.16 (5.16, 11.99), TG genotype 5.16 (2.61, 8.58) and GG genotype 8.58 (3.46, 13.70), P = .40. (D) TNF-α (pg/mL): TT genotype 5.03 (2.13, 12.21), TG genotype 5.84 (3.71, 7.93) and GG genotype 4.63 (2.13, 5.44), P = .82. (E) CD40L (ng/mL): TT genotype 3.56 (2.06, 4.70), TG genotype 4.13 (2.62, 6.14) and GG genotype 3.04 (2.09, 4.96), P = .40. (F) Granzyme B (pg/mL): TT genotype 8.49 (5.19, 23.13), TG genotype 6.85 (3.53, 13.4) and GG genotype 3.53 (1.85, 13.4), P = .03. The results represent the median and interquartile of cytokines levels.
3.5. Association between IKZF1 SNPs and clinical characteristics in SLE patients
Table 3 presents the analysis results about the influence of IKZF1 SNPs on clinical symptoms in SLE patients. Significant associations were found both in rs4132601 and malar rash (P = .01) and rs4917014 and hematological disorder (P = .005). The patients with the GG genotype of rs4917014 had a lower frequency of hematological disorder than the patients with TG and TT genotype. No Statistical difference was found between IKZF1 SNPs and other clinical characteristics, including lupus nephritis, neurological disorder, arthritis, proteinuria, pleuritis, pericarditis in SLE patients (all P > .05).
Table 3.
Association between ikaros family zinc finger 1 polymorphisms and clinical characteristics in systemic lupus erythematosus patients.

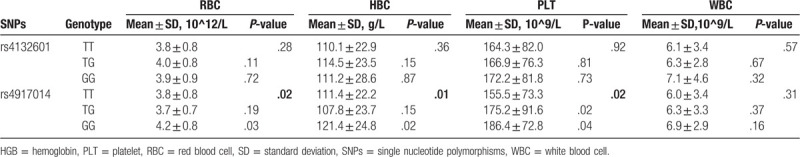
Moreover, the analysis of the association of the SNPs with specific hematological parameters is depicted in Table 4. IKZF1 rs4917014 was significantly associated with the level of RBC (P = .02), HGB (P = .01), and PLT (P = .02), but not related to the level of WBC (P > .05). No significant difference was observed between rs4132601 and all hematological index (P > .05).
Table 4.
Association between ikaros family zinc finger 1 polymorphisms and hematological index in systemic lupus erythematosus patients.
4. Discussion
As one of the most prototypic and complex autoimmune diseases, SLE is characterized by various autoantibody production, complement activation, immune complex deposition, and tissue damage. It is a pathological condition that can lead to incapacitation and decrease life expectancy. Comparing with the general population, people suffering from SLE have nearly a 3-fold increased risk of death.[1,2] Although SLE's pathogenesis is unclear, the view that environmental, and genetic factors influence the disease has been widely accepted.[21] In the past few decades, a variety of genetic factors associated with SLE have been identified. Notably, more than 20 robust susceptibility loci and genes of SLE have been identified by numerous genome-wide association studies in European populations.[22–25] Meanwhile, the incidence and prevalence of SLE vary markedly among different ethnic or geographical people, which suggests the genetic heterogeneity is vital for SLE risk.[26,27] Therefore, the study about SLE susceptibility genes and loci in non-European populations is necessary. In this study, we identified 2 SNPs, rs4132601, and rs4917014 of IKZF1, associating significantly with SLE susceptibility in the southwest China Han population. According to the results, the allele G of both 2 SNPs correlated with reduced susceptibility to SLE. As far as we know, no report demonstrating the association between these IKZF1 SNPs and SLE in the Chinese Han population has been made. Many previous studies have reported that some IKZF1 SNPs were associated mainly with Leukemia[28,29] and some autoimmune diseases such as inflammatory bowel disease, Crohn disease and primary Sjoègren syndrome.[11–13] In this study, we observed that IKZF1 is one of SLE susceptibility genes in the southwest China Han population, providing the support that IKZF1 may correlate with many autoimmune diseases susceptibility.
Meanwhile, significant associations were observed between rs4132601 and the serum level of CD40L and malar rash in SLE patients. Comparing to TT and TG genotype carriers, the patients with GG genotype had higher CD40L concentration. As a member of the tumor necrosis factor superfamily, CD40L participates in B cell differentiation, isotype switching and germinal center formation by engaging with its receptor CD40 on B cells, which plays a central role in the induction of immune responses.[30] CD40 and CD40L interactions are thus considered to be one of the central mechanisms in autoimmune development. In SLE, B cells and monocytes express CD40L aberrantly. During the active stage of SLE, CD40L is abnormally overexpressed by CD4+T and CD8+T cells.[31] Preclinical studies showed that transgenic mice ectopically expressing CD40L on B cells developed lupus-like disease. Meanwhile, lupus-prone NZB/W mice delayed the onset of symptoms. They improved the survival by treating with an anti-CD40L monoclonal antibody, suggesting the suppression of CD40 and CD40L interactions probably contribute to the amelioration of lupus-like disease.[32,33] Considering that the patients with rs4132601 GG genotype had a significantly higher CD40L concentration, IKZF1 might affect SLE's development by maintaining high levels of CD40L. It probably provides new targets and ideas for the treatment. It is somewhat interesting that rs4132601G seems associated with an increased risk of development but a decreased risk of SLE occurrence. Some other researches showed similar phenomena. For instance, the female is more susceptible to SLE, while female patients of SLE commonly suffered less severe disease state than male patients of SLE. Some studies deduced that higher accumulations of genetic load are required for males than females in disease occurrence.[34,35] The different faces of IKZF1-rs4132601 in SLE do not have a clear definition. Whether the phenomenon is associated with complex immune microenvironment and gene-gene interactions is worth studying further.
What's more, a statistical correlation was found between rs4917014 and the serum level of granzyme B and hematological disorder in SLE. The patients carrying GG genotype had a significantly lower granzyme B level than patients with TT and TG genotypes. Extracellular granzyme B is thought to participate in SLE's pathogenesis by cleaving autoantigens resulting in the formation of immunogenic neoepitopes and leading to the formation of pathogenic autoantibodies.[36,37] Since rs4917014 allele G was associated with a lower granzyme B level and a decreased risk of SLE susceptibility, we presume that IKZF1 rs4917014 may influence disease occurrence by preventing the overexpression of granzyme B.
The result that the rs4917014 minor allele G was associated with a decreased risk of hematological disorder for SLE patients was notable. IKZF1 was known for encoding ikaros which regulate immune cell development and homeostasis.[8] In adults, ikaros Expression is restricted mostly to lymphopoietic tissues such as spleen, thymus, and peripheral blood leukocytes.[38,39] Further analysis in this study showed that the genotype of rs4917014 was significantly associated with the level of RBC, HGB, and PLT, providing support for that IKZF1 rs4917014 might participate in the development of SLE by influencing the hematological indicator levels. Other evidence suggested that ikaros is essential in the regulation of critical pathways. For example, it regulates numerous signal activators and transducers, including IFN-α producers, playing important roles in immune regulation.[16] At the same time, some studies have identified the rs4917014 is one of the expression quantitative trait loci of IKZF1. It contributes to the down-regulation of complement gene and up-regulation of the type 1 interferon gene, both of which are marks in SLE pathophysiology.[40] Therefore, IKZF1 rs4917014 polymorphisms may influence the occurrence and development of SLE by regulation of type 1 IFN and complement. It is almost certain that a compelling genetic association runs between these transcription factors and the immune dysfunction associated with SLE.[41–45]
However, there are several potential limitations to this study. Firstly, this is a single-center study with a limited number of cases and population. Secondly, although only the patients diagnosed for the first time were included, the influence of previously administered medications on clinical manifestations could not be completely excluded. Lastly, this study focused on the relationships between IKZF1 polymorphisms and susceptibility as well as clinical symptoms of SLE, but no more in-depth study on the mechanism or pathway. Given these limitations, what we have found in this study should be interpreted prudently. Furthering studies in larger sample sizes and other ethnic populations are required to confirm the observations in this study. The study concentrated on the mechanism of IKZF1 genetic variants in SLE'S pathogenesis should also be conducted in vitro and in vivo.
Despite these limitations, our study identified that 2 common variants (rs4132601 and rs4917014) of IKZF1 are associated with the susceptibility to SLE in the Chinese Han population. Furthermore, these 2 SNPs correlated with the inflammatory cytokines and clinical manifestations in SLE patients, which probably contribute to the development of the disease. According to the results, we illustrated that allele G of IKZF1 rs4132601 was associated with decreased SLE's susceptibility and increased risk of SLE's development. The allele G of IKZF1 rs4917014 correlated with decreased SLE's susceptibility and reduced risk of hematological disorder in SLE patients. This study may provide evidence for the amelioration of our understanding of the exact role of IKZF1 in the pathogenesis of autoimmune diseases.
Acknowledgments
Thanks to Dr H. Tejeda Mora from Erasmus MC Rotterdam in the Netherlands for the language modification that bettered our manuscript.
Author Contributions
L.C., Q.N., and J.L.Z. designed the study; Z.C.H., B.Y., and Y.K.W. performed experiments; L.C., Q.N., Z.C.H., and J.L.Z. statistical analyses and interpreted results; L.C., Q.N., and J.L.Z. wrote the manuscript. All authors reviewed the manuscript.
Conceptualization: Lin Chen.
Data curation: Zhuochun Huang, Bin Yang, Yongkang Wu.
Formal analysis: Lin Chen, Qian Niu, Zhuochun Huang.
Funding acquisition: Bin Yang.
Investigation: Qian Niu, Zhuochun Huang, Bin Yang, Yongkang Wu.
Methodology: Qian Niu, Zhuochun Huang.
Project administration: Lin Chen.
Writing – original draft: Lin Chen.
Writing – review & editing: Qian Niu.
Glossary
Abbreviations: CD40L = CD40 ligand, HGB = hemoglobin, IFN = Interferon, IKZF1 = ikaros family zinc finger 1, iMLDR = improved multiplex ligation detection reaction, PLT = platelet, RBC = Red blood cell, SLE = systemic lupus erythematosus, SLEDAI = systemic lupus erythematosus disease activity index, SNPs = single nucleotide polymorphisms, WBC = white blood cell.
References
- [1].Rahman A, Isenberg DA. Systemic lupus erythematosus. N Engl J Med 2008;358:929–39. [DOI] [PubMed] [Google Scholar]
- [2].Yurkovich M, Vostretsova K, Chen W, et al. Overall and cause-specific mortality in patients with systemic lupus erythematosus: a meta-analysis of observational studies. Arthritis Care Res 2014;66:608–16. [DOI] [PubMed] [Google Scholar]
- [3].Rullo OJ, Tsao BP. Recent insights into the genetic basis of systemic lupus erythematosus. Ann Rheum Dis 2013;72: (Suppl 2): ii56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Tsokos GC. Systemic lupus erythematosus. N Engl J Med 2011;365:2110–21. [DOI] [PubMed] [Google Scholar]
- [5].Agnihotri P, Robertson NM, Umetsu SE, et al. Lack of Ikaros cripples expression of Foxo1 and its targets in naive T cells. Immunology 2017;152:494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Schwickert TA, Tagoh H, Gultekin S, et al. Stage-specific control of early B cell development by the transcription factor Ikaros. Nat Immunol 2014;15:283–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cytlak U, Resteu A, Bogaert D, et al. Ikaros family zinc finger 1 regulates dendritic cell development and function in humans. Nat Commun 2018;9: 1239–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fan Y, Lu D. The ikaros family of zinc-finger proteins. Acta Pharm Sin B 2016;6:513–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chen Q, Shi Y, Chen Y, et al. Multiple functions of ikaros in hematological malignancies, solid tumor and autoimmune diseases. Gene 2019;684:47–52. [DOI] [PubMed] [Google Scholar]
- [10].Hoshino A, Okada S, Yoshida K, et al. Abnormal hematopoiesis and autoimmunity in human subjects with germline IKZF1 mutations. J Allergy Clin Immunol 2017;140:223–31. [DOI] [PubMed] [Google Scholar]
- [11].Franke A, McGovern DP, Barrett JC, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet 2010;42:1118–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012;491:119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Qu S, Du Y, Chang S, et al. Common variants near IKZF1 are associated with primary Sjogren's syndrome in Han Chinese. PLoS One 2017;12: e0177320–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hu W, Sun L, Gao J, et al. Down-regulated expression of IKZF1 mRNA in peripheral blood mononuclear cells from patients with systemic lupus erythematosus. Rheumatol Int 2011;31:819–22. [DOI] [PubMed] [Google Scholar]
- [15].Dang J, Shan S, Li J, et al. Gene-gene interactions of IRF5, STAT4, IKZF1 and ETS1 in systemic lupus erythematosus. Tissue Antigens 2014;83:401–8. [DOI] [PubMed] [Google Scholar]
- [16].Hu SJ, Wen LL, Hu X, et al. IKZF1: a critical role in the pathogenesis of systemic lupus erythematosus? Mod Rheumatol 2013;23:205–9. [DOI] [PubMed] [Google Scholar]
- [17].Bentham J, Morris DL, Graham DSC, et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat Genet 2015;47:1457–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 1997;40: 1725–5. [DOI] [PubMed] [Google Scholar]
- [19].Gladman DD, Ibanez D, Urowitz MB. Systemic lupus erythematosus disease activity index 2000. J Rheumatol 2002;29:288–91. [PubMed] [Google Scholar]
- [20].Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263–5. [DOI] [PubMed] [Google Scholar]
- [21].Lewis MJ, Jawad AS. The effect of ethnicity and genetic ancestry on the epidemiology, clinical features and outcome of systemic lupus erythematosus. Rheumatology (Oxford) 2017;56: (Suppl 1): i67–77. [DOI] [PubMed] [Google Scholar]
- [22].Jiang SH, Athanasopoulos V, Ellyard JI, et al. Functional rare and low frequency variants in BLK and BANK1 contribute to human lupus. Nat Commun 2019;10: 2201–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Alarcon-Riquelme ME, Ziegler JT, Molineros J, et al. Genome-wide association study in an Amerindian ancestry population reveals novel systemic lupus erythematosus risk loci and the role of European admixture. Arthritis Rheumatol 2016;68:932–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Jarvinen TM, Hellquist A, Zucchelli M, et al. Replication of GWAS-identified systemic lupus erythematosus susceptibility genes affirms B-cell receptor pathway signalling and strengthens the role of IRF5 in disease susceptibility in a Northern European population. Rheumatology (Oxford) 2012;51:87–92. [DOI] [PubMed] [Google Scholar]
- [25].Graham RR, Cotsapas C, Davies L, et al. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat Genet 2008;40:1059–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ramos-Casals M, Brito-Zeron P, Kostov B, et al. Google-driven search for big data in autoimmune geoepidemiology: analysis of 394,827 patients with systemic autoimmune diseases. Autoimmun Rev 2015;14:670–9. [DOI] [PubMed] [Google Scholar]
- [27].Yang W, Ng P, Zhao M, et al. Population differences in SLE susceptibility genes: STAT4 and BLK, but not PXK, are associated with systemic lupus erythematosus in Hong Kong Chinese. Genes Immun 2009;10:219–26. [DOI] [PubMed] [Google Scholar]
- [28].Kuehn HS, Boisson B, Cunningham-Rundles C, et al. Loss of B Cells in patients with heterozygous mutations in IKAROS. N Engl J Med 2016;374:1032–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Churchman ML, Qian M, Te Kronnie G, et al. Germline genetic IKZF1 variation and predisposition to childhood acute lymphoblastic leukemia. Cancer Cell 2018;33:937e8–48e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Elgueta R, Benson MJ, de Vries VC, et al. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol Rev 2009;229:152–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Vukelic M, Li Y, Kyttaris VC. Novel treatments in lupus. Front Immunol 2018;9: 2658–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ma WT, Gao F, Gu K, et al. The role of monocytes and macrophages in autoimmune diseases: a comprehensive review. Front Immunol 2019;10: 1140–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Perper SJ, Westmoreland SV, Karman J, et al. Treatment with a CD40 antagonist antibody reverses severe proteinuria and loss of saliva production and restores glomerular morphology in murine systemic lupus erythematosus. J Immunol 2019;203:58–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gaudreau MC, Johnson BM, Gudi R, et al. Gender bias in lupus: does immune response initiated in the gut mucosa have a role? Clin Exp Immunol 2015;180:393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Alonso-Perez E, Suarez-Gestal M, Calaza M, et al. Lack of replication of higher genetic risk load in men than in women with systemic lupus erythematosus. Arthritis Res Ther 2014;16: R128–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Graham KL, Utz PJ. Sources of autoantigens in systemic lupus erythematosus. Curr Opin Rheumatol 2005;17:513–7. [DOI] [PubMed] [Google Scholar]
- [37].Kok HM, van den Hoogen LL, van Roon JAG, et al. Systemic and local granzyme B levels are associated with disease activity, kidney damage and interferon signature in systemic lupus erythematosus. Rheumatology (Oxford) 2017;56:2129–34. [DOI] [PubMed] [Google Scholar]
- [38].Mitchell JL, Seng A, Yankee TM. Expression and splicing of Ikaros family members in murine and human thymocytes. Mol Immunol 2017;87:1–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Frippiat JP, Crucian BE, de Quervain DJ, et al. Towards human exploration of space: The THESEUS review series on immunology research priorities. NPJ Microgravity 2016;2: 16040–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Westra HJ, Peters MJ, Esko T, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 2013;45:1238–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].He CF, Liu YS, Cheng YL, et al. TNIP1, SLC15A4, ETS1, RasGRP3 and IKZF1 are associated with clinical features of systemic lupus erythematosus in a Chinese Han population. Lupus 2010;19:1181–6. [DOI] [PubMed] [Google Scholar]
- [42].Cunninghame Graham DS, Morris DL, Bhangale TR, et al. Association of NCF2, IKZF1, IRF8, IFIH1, and TYK2 with systemic lupus erythematosus. PLoS Genet 2011;7: e1002341–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Leng RX, Wang W, Cen H, et al. Gene-gene and gene-sex epistatic interactions of MiR146a, IRF5, IKZF1, ETS1 and IL21 in systemic lupus erythematosus. PLoS One 2012;7: e51090–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lee HS, Bae SC. Recent advances in systemic lupus erythematosus genetics in an Asian population. Int J Rheum Dis 2015;18:192–9. [DOI] [PubMed] [Google Scholar]
- [45].Yin Q, Wu LC, Zheng L, et al. Comprehensive assessment of the association between genes on JAK-STAT pathway (IFIH1, TYK2, IL-10) and systemic lupus erythematosus: a meta-analysis. Arch Dermatol Res 2018;310:711–28. [DOI] [PubMed] [Google Scholar]


