Key Points
Conditional deletion of Tip60 from the murine hematopoietic system leads to HSC loss, in both the fetal and adult stages.
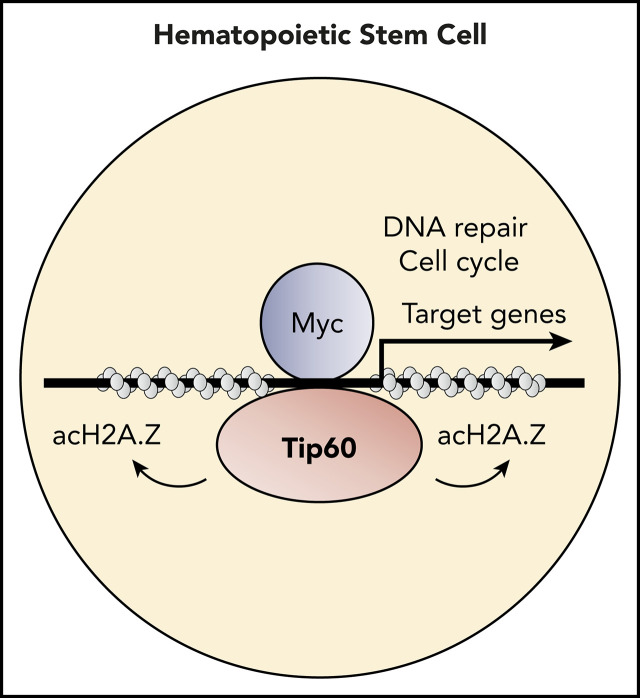
Tip60 regulates genes that are involved in critical biological processes for HSC maintenance through acetylation of H2A.Z.
Abstract
Hematopoietic stem cells (HSCs) have the potential to replenish the blood system for the lifetime of the organism. Their 2 defining properties, self-renewal and differentiation, are tightly regulated by the epigenetic machineries. Using conditional gene-knockout models, we demonstrated a critical requirement of lysine acetyltransferase 5 (Kat5, also known as Tip60) for murine HSC maintenance in both the embryonic and adult stages, which depends on its acetyltransferase activity. Genome-wide chromatin and transcriptome profiling in murine hematopoietic stem and progenitor cells revealed that Tip60 colocalizes with c-Myc and that Tip60 deletion suppress the expression of Myc target genes, which are associated with critical biological processes for HSC maintenance, cell cycling, and DNA repair. Notably, acetylated H2A.Z (acH2A.Z) was enriched at the Tip60-bound active chromatin, and Tip60 deletion induced a robust reduction in the acH2A.Z/H2A.Z ratio. These results uncover a critical epigenetic regulatory layer for HSC maintenance, at least in part through Tip60-dependent H2A.Z acetylation to activate Myc target genes.
Visual Abstract
Introduction
Hematopoietic stem cells (HSCs) possess 2 defining properties: self-renewal and multilineage differentiation, under various tightly regulated epigenetic mechanisms.1 Chromatin-modifying enzymes play crucial roles in regulating gene expression for HSC maintenance.2-5 Histone acetylation, a reversible covalent posttranslational modification (PTM), is one of the most studied chromatin modifications, catalyzed by lysine acetyltransferases (KATs) and mediates unique and specific effects on gene transcription by altering the degree of chromatin condensation.6,7 According to structural homology and acetylation mechanisms, KATs are classified into 5 representative families: GNAT, MYST, p300/CBP, SRC, and TAF1.8 The MYST family is defined by the proteins containing a C2HC-type zinc finger and an acetyl-CoA binding domain and consists of 5 members: Tip60/Kat5, Moz/Kat6a, Morf/Kat6b, Hbo1/Kat7, and Mof/Kat8. Previous studies of various KAT deletions in mice detailed their importance in the maintenance or differentiation of HSCs. Homozygous deletion and catalytic mutant Moz/Kat6a mice revealed its essential role in both fetal and adult HSC maintenance as a repressor of p16 expression to prevent HSC senescence.5,9 Mof/Kat8 is critical for adult, not fetal, HSC maintenance, and the catalytically inactive mutant neither restores H4K16ac levels nor rescues colony-forming ability in adult hematopoiesis.10 The Hbo1-Brd1 complex is necessary for transcription of genes that regulate erythroid development.11 Each lysine acetyltransferase has a specific regulatory role in hematopoiesis, despite some redundancy in their substrate specificities.12
Lysine acetyltransferase 5, (Kat5, also known as tat-interactive protein-60 [Tip60]) plays a key role in DNA damage response and repair, as well as in gene-specific transcriptional regulation.13 It is part of the evolutionarily conserved nucleosomal acetyltransferase of the H4 (NuA4) protein complex, which acetylates the nucleosomal histone H3, H4, H2A, and H2A variants.14 Homozygous global Tip60 deletion in mice leads to preimplantation lethality at embryonic day (E)3.5,15 indicating its requirement for embryonic development. In the hematopoietic system, a conditional deletion of Tip60 in regulatory T (Treg) cells impairs their function in peripheral immune organs by suppressing the transcriptional activity of FOXP3.16 However, the role of Tip60 in HSCs is largely unknown. In this study, using murine conditional Tip60 deletion models, we demonstrated a critical role for Tip60 in murine HSC maintenance. Genome-wide transcriptome and chromatin profiling revealed that Tip60 and c-Myc colocalized at active chromatin loci to activate transcription of their target genes. Notably, Tip60 deletion reduced the acetylation level of H2A.Z at the target gene promoters. We thus propose a new epigenetic mechanism in HSC maintenance: Tip60-mediated H2A.Z acetylation for the activation of Myc target genes.
Materials and methods
Mice
Tip60flox mice were generated by inserting the LoxP sites flanking introns 2 and 11 of the mouse Tip60 gene. Cre-recombinase–mediated excision was designed to remove exons 3 to 11, which includes the chromo-finger, zinc finger, and acetyl Co-A–binding domains. Embryonic stem cell clones with correct homologous recombination were injected into C57/Bl6 blastocysts, which transmitted the targeted allele via germline after implantation (supplemental Figure 1A-B, available on the Blood Web site). Additional details are described in the supplemental Data. All mice were housed in a sterile barrier facility within the Comparative Medicine Facility at the National University of Singapore. All experiments performed in mice were approved by the Institutional Animal Care and Use Committee.
Inducible Tip60 deletion
For in vivo Tip60 deletion, Mx1-cre; Tip60f/f mice were injected with 300 μg polyinosinic:polycytidylic acid (pIpC; GE Healthcare) pIpC per body for 3 consecutive days. For in vitro Tip60 deletion, LSK or c-Kit+ cells were sorted from the fetal liver of Rosa26 Cre-ERT2; Tip60f/f embryos (E13.5-15.5) by FACS Aria (BD Biosciences, San Jose, CA); cultured in Stemline II (MilliporeSigma, Burlington, MA) supplemented with 5% fetal bovine serum, murine recombinant SCF 100 ng/μL, IL-3 6 ng/μL, IL-6 10 ng/μL, and IL-11, 20 ng/μL (Peprotech, Cranbury, NJ) and 4-hydroxytamoxifen (4-OHT; MilliporeSigma) 400 nM for 72 hours; and collected for analysis.
Flow cytometry
Single-cell suspensions were analyzed by flow cytometry. Cells stained with antibodies were analyzed or sorted using the LSRII Flow Cytometer or FACS Aria (BD Biosciences). Flow Jo 7.5 (Tree Star, Ashland, OR) was used for data analysis. The antibodies are described in the supplemental Data.
Retroviral transduction and Tip60 KAT rescue
FLAG-tagged human TIP60 wild-type and catalytically inactive mutant constructs17 were cloned into the MSCV vector (TIP60wt and TIP60mut, respectively). TIP60wt and TIP60mut retroviruses were produced in BOSC23 cells. FACS-sorted LSK cells obtained from Tip60f/f; Vav-iCre embryos (CD45.2+) at E13.5 were seeded in RetroNectin-coated plates (Takara, Mountain View, CA) containing retroviral supernatants for 24 hours. Transduced cells (2 × 103) were injected into lethally (900 rads) irradiated congenic mice (CD45.1+), along with whole bone marrow (WBM) cells from congenic mice (CD45.1+ CD45.2+). Transduction of plasmid DNAs were verified by Sanger sequencing of the PCR-amplified genomic products amplified by the following primers: Tip60KAT-forward, 5′-GTGTTTCCTTGACCATAAGACACTGTACTA T-3′, and Tip60KAT-reverse, 5′-GGTCTGGGACCAGTAGCTTCGATA G-3′.
Bone marrow transplantation assay
For the competitive transplantation assay, unfractionated fetal liver cells (1 × 106 cells) from Tip60f/+, Tip60+/Δ, and Tip60Δ/Δ E14.5 embryos were injected into lethally irradiated (900 rads) congenic mice, along with competitor WBM cells (1 × 105 cells). WBM cells (1 × 106 cells) from Tip60f/f and Tip60f/f; Mx1-Cre mice, which had been injected with 800 μg pIpC per body every other day for 7 days, were collected 10 days after the last injection and injected into lethally irradiated congenic mice, along with competitor WBM cells (2 × 105 cells). For reciprocal transplantation assays, unfractionated WBM cells from Mx1-Cre; Tip60f/f and Tip60f/f mice were injected into lethally irradiated (900 rads) congenic mice along with competitor WBM cells (1 × 106 and 2 × 105; 5 × 105 and 5 × 105 cells, respectively), and the recipients were injected with 800 μg pIpC per animal every other day for 4 days after engraftment. To exclude the cell-extrinsic effect caused by Tip60 deletion, unfractionated CD45.1+ WBM cells (1 × 106 cells) were injected into lethally irradiated Tip60f/f and Tip60f/f; Mx1-Cre mice. After transplantation, chimerism analysis was performed as described in the supplemental Data.
RNA/ChIP-sequencing
RNA-sequencing (RNA-seq) libraries were prepared using Illumina Tru-Seq Stranded Total RNA with Ribo-Zero Gold Kit protocol, according to the manufacturer’s instructions (Illumina, San Diego, CA). Libraries were validated with a Bioanalyzer (Agilent Technologies, Palo Alto, CA), diluted, and applied to an Illumina flow cell by using the Illumina Cluster Station. Chromatin immunoprecipitation sequencing (ChIP-seq) libraries were prepared with the NEBNext ChIP-Seq Library Prep Reagent Set and multiplexed (New England Biolabs, Ipswich, MA). Each library was sequenced on an Illumina Hiseq2000 sequencer. The details of chromatin immunoprecipitation and sequence analysis are described in the supplemental Data.
ChIP-seq similarity analysis
A similarity analysis was performed on normalized pointwise mutual information (NPMI).18 After normalization, NPMI ranged from 1 for complete co-occurrence (correlation limit), to 0 for independent peak profiles, to −1 when peaks did not occur together (anticorrelation limit). NPMI values were clustered using Euclidean distance and Ward linkage in R (R Project for Statistical Computing; https://www.r-project.org/). All data sets except Tip60 profiling are accessible through an intuitive Web browser interface (CODEX; http://codex.stemcells.cam.ac.uk/).
Statistical analysis
The statistical significances were assessed by Student t test using GraphPad Prism software (La Jolla, CA), unless otherwise specified.
Results
Tip60 deletion leads to rapid HSC loss in both fetal and adult stages
To study the role of Tip60 in HSCs, we generated mice in which a loxP-flanked Tip60 allele (exons 3-11, which includes the chromo, zinc finger, and acetyl Co-A binding domains; supplemental Figure 1A-B)16 was conditionally deleted by improved Cre (iCre) recombinase, the expression of which was under the control of the Vav1 promoter (Tip60f/f; Vav-iCre) (supplemental Figure 2A-C).19 Tip60f/+; Vav-iCre and Tip60 f/f mice were crossed, and among the 76 viable progenies obtained, none of the 19 (25%) expected Tip60f/f; Vav-iCre (Tip60Δ/Δ) mice was born (Figure 1A). In a retrospective analysis, Tip60Δ/Δ embryos were viable at E14.5; however, they became anemic at E17.5 (Figure 1B), suggesting that the cause of the lethality of the Tip60Δ/Δ embryos was hematopoietic failure, mainly anemia (supplemental Figure 2D). Tip60 deletion led to a decrease in the size of the fetal liver, accompanied by six- to ninefold reductions in the number of fetal liver cells at E14.5 and E17.5 (Figure 1C; supplemental Figure 2E). The subpopulations of hematopoietic stem and progenitor cells (HSPCs) were dramatically reduced (Figure 1D-E). To assess the repopulating ability, we performed competitive transplantation, using whole fetal liver cells of E14.5 embryos, which demonstrated complete loss of hematopoietic reconstitution by Tip60Δ/Δ cells (Figure 1F; supplemental Figure 2F-G). There were few Tip60Δ/Δ HSCs present in the bone marrow (BM) of the recipients at 16 weeks after transplantation, suggesting impairment of the long-term repopulating capability of fetal Tip60Δ/Δ HSCs (Figure 1G). Notably, heterozygous deletion of Tip60 (Tip60+/Δ, Tip60f/+; Vav-iCre) did not affect hematopoietic reconstitution, which indicated that there was no gene dosage effect of Tip60 in HSC function.
Figure 1.
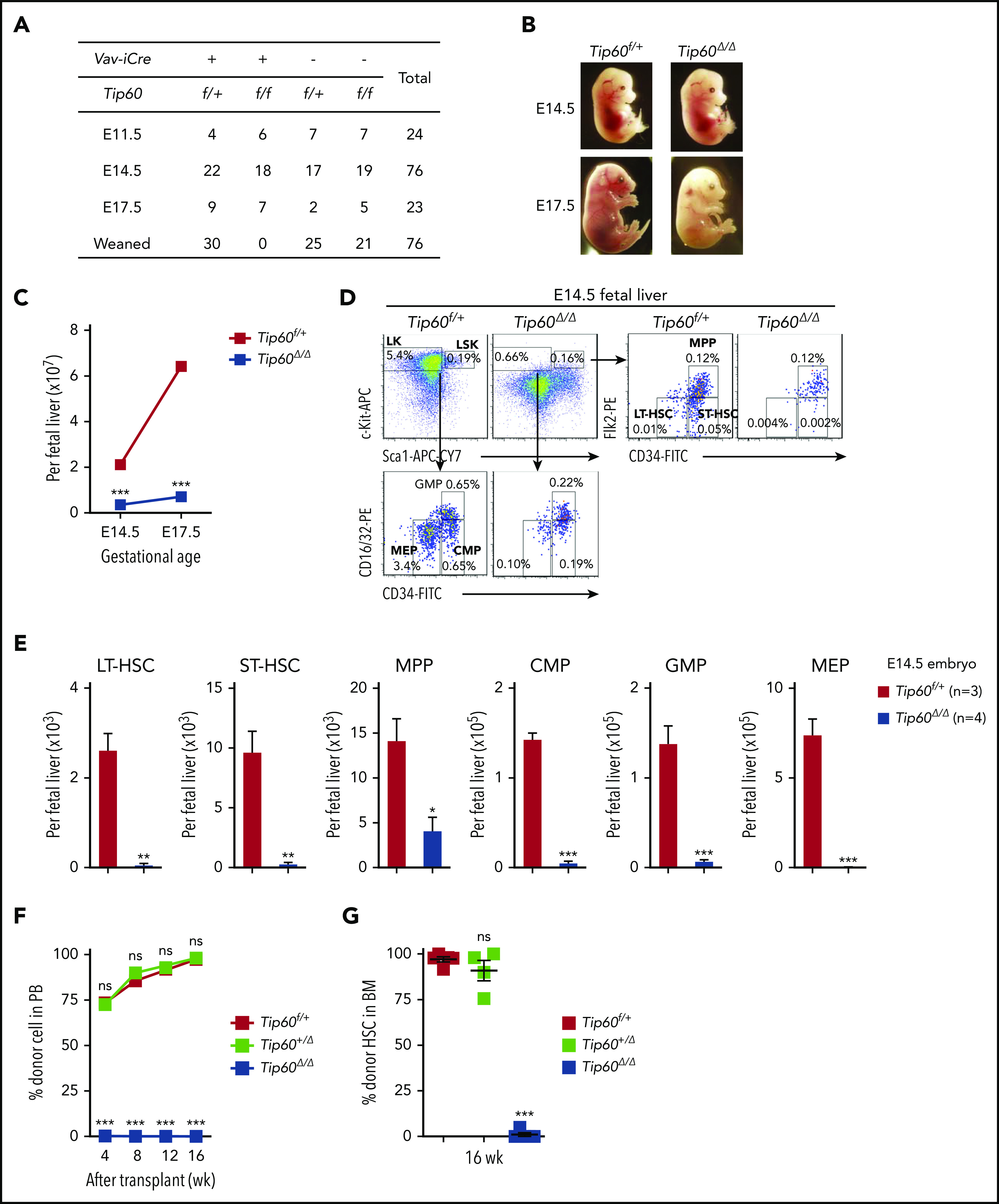
Genetic deletion of Tip60 leads to fetal hematopoietic failure. (A) The genotype distribution of offspring and embryos derived from extensive Tip60f/f × Tip60f/+; Vav-iCre breeding. (B) Representative images of Tip60f/+ and Tip60Δ/Δ (Tip60f/f; Vav-iCre) embryos at E14.5 and E17.5. (C) Average absolute number of nucleated cells from control and Tip60Δ/Δ fetal liver at different gestational ages (n = 3-5 embryos for each genotype for each gestational age). (D) Representative dot plots of flow cytometry analysis of control and Tip60Δ/Δ E14.5 fetal liver cells and frequencies of the gated populations. (E) Absolute number of HSPC subpopulations from control and Tip60Δ/Δ E14.5 embryos from the same littermates: LSK (Lin−c-Kit+Sca-1+), LK (Lin−c-Kit+Sca-1−), LT-HSC (CD34−Flk2−LSK), ST-HSC (CD34+Flk2−LSK), MPP (CD34+Flk2+LSK), GMP (CD34+CD16/32+LK), CMP (CD34+CD16/32−LK), and MEP (CD34−CD16/32−LK). (F) Whole fetal liver cells (1 × 106) from Tip60f/+, Tip60+/Δ, and Tip60Δ/Δ E14.5 embryos were transplanted into lethally irradiated congenic mice (CD45.1+), along with 1 × 105 congenic WBM (CD45.1+CD45.2+) cells. Donor chimerism of recipient PB is shown. (G) Donor chimerism of recipient BM HSCs (CD34+ Flk2−LSK) at 16 weeks after the transplantation. The values are presented as means ± standard error of the mean. Statistical analyses were performed vs Tip60f/+. *P < .05; **P < .01; ***P < .001, ns; not significant.
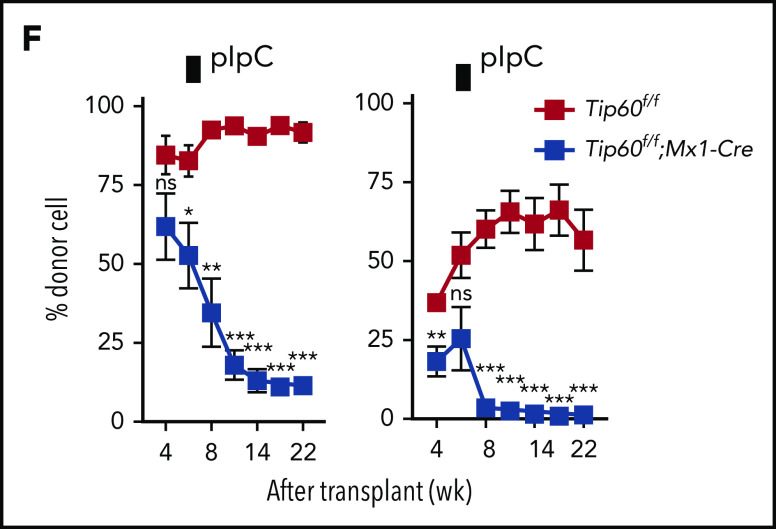
To determine the role of Tip60 in adult HSCs, we generated Tip60f/f and Tip60f/+ mice wherein expression of Cre is driven by the interferon-inducible promoter of the gene encoding Mx1. In this model, the Tip60 gene was excised by injecting pIpC, leading to a reduction of Tip60 protein in HSPCs (supplemental Figure 3A-C).20 Tip60 deletion induced a dramatic decrease in cellularity and number of HSPCs in the BM (Figure 2A-B), whereas heterozygous Tip60 deletion did not, again demonstrating the absence of a gene dosage effect of Tip60 in adult hematopoiesis as well. In competitive transplantations, Tip60Δ/Δ WBM cells did not reconstitute hematopoiesis (Figure 2C; supplemental Figure 3D), with few or no Tip60Δ/Δ HSCs detected in the BM 16 weeks after the transplantation (Figure 2D), indicating an impaired long-term repopulating capability in the adult stage, as well. Short-term in vivo homing assays demonstrated no significant difference in engraftment efficiency (Figure 2E).
Figure 2.
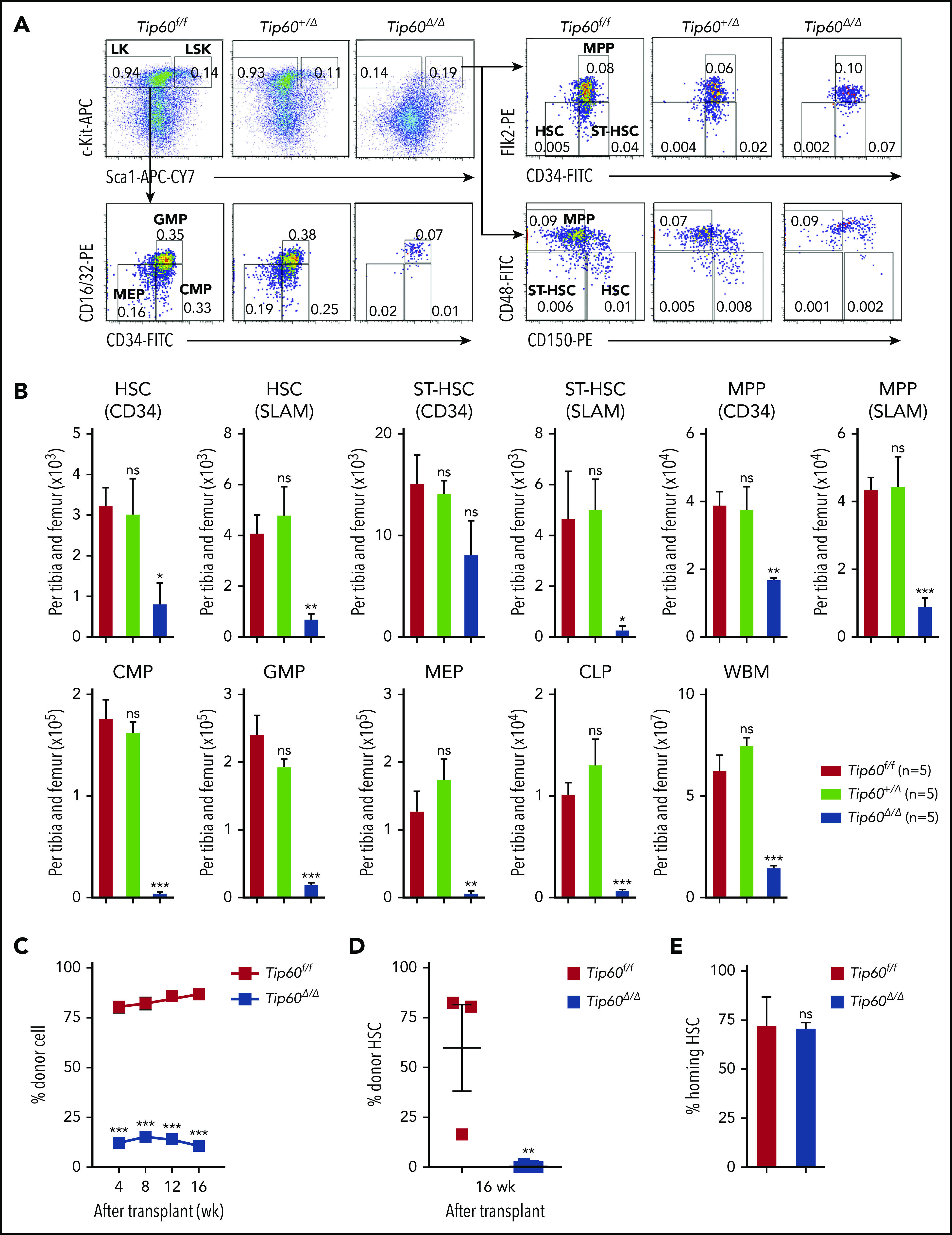
Tip60 is essential for adult HSC maintenance in a cell-intrinsic manner. (A) Male 6- to 10-week-old Tip60f/f, Tip60f/+; Mx1-Cre, and Tip60f/f; Mx1-Cre mice were injected with pIpC for 3 consecutive days. BM cells were analyzed for the frequency of HSPCs by flow cytometry 5 days after the last injection: LSK (Lin−c-Kit+Sca-1+), LK (Lin−c-Kit+Sca-1−), HSC (CD34, CD34−Flk2−; SLAM, CD150+CD48−LSK), ST-HSC (CD34, CD34+Flk2−; SLAM, CD150−CD48−LSK), MPP (CD34, CD34+Flk2+; SLAM, CD150−CD48+LSK), GMP (CD34+CD16/32+LK), CMP (CD34+CD16/32−LK), MEP (CD34−CD16/32−LK), and CLP (Lin−IL7R+c-Kit+Sca1+Flk2+). Representative dot plots and frequencies of the HSPC subpopulation from Tip60f/f, Tip60+/Δ, and Tip60Δ/Δ mice are shown. (B) The absolute number of HSPCs and WBM cells from Tip60f/f, Tip60+/Δ, and Tip60Δ/Δ mice is shown. (C) WBM cells (1 × 106) from control Tip60f/f and Tip60Δ/Δ mice (CD45.2+) were transplanted into lethally irradiated recipient mice (CD45.1+), along with 2 × 105 congenic WBM cells (CD45.1+CD45.2+). Donor chimerism of recipient PB is shown. (D) Donor chimerism of recipient BM HSCs (CD150+CD48−LSK) at 16 weeks after transplantation. (E) CFSE-labeled HSCs were injected into lethally irradiated congenic mice, and homing efficiency was analyzed 16 hours after the injection (n = 3). (F) WBM cells (1 × 106) from Tip60f/f or Tip60f/f; Mx1-Cre mice (CD45.2+) (left) were transplanted into lethally irradiated recipient mice (CD45.1+) along with 2 × 105 congenic WBM cells (CD45.1+CD45.2+). WBM cells (5 × 105) from Tip60f/f or Tip60f/f;Mx1-Cre mice (CD45.2+) (right) were transplanted into lethally irradiated recipient mice (CD45.1+), along with 5 × 105 congenic WBM cells (CD45.1+CD45.2+). pIpC was injected into the recipients 6 weeks after transplantation. Donor chimerism in all nucleated cells of recipient PB are shown. Values are presented as means ± standard error of the mean. Statistical analyses were performed vs Tip60f/f. *P < .05; **P < .01; ***P < .001; ns; not significant
Tip60 is a critical cell-intrinsic regulator of HSCs
Because Cre under the Mx1 promoter is activated by pIpC in various tissues,20 reciprocal competitive transplantations were conducted to exclude a cell-extrinsic effect of Tip60 deletion. First, WBM cells from Tip60f/f; Mx1-Cre or Tip60f/f mice were transplanted into lethally irradiated congenic mice, and pIpC was injected into the recipients 6 weeks after transplantation. Peripheral blood (PB) chimerism analysis revealed a continuous decrease in Tip60f/f; Mx1-Cre cells in all lineages over time, but not in control (Tip60f/f) cells (Figure 2F; supplemental Figure 3E). Second, WBM cells from congenic mice were transplanted into lethally irradiated recipient Tip60f/f; Mx1-Cre and Tip60f/f mice, and the mice were subsequently injected with pIpC. The chimerism of donor-derived cells was sustained in both groups for 8 weeks. However, all the Tip60f/f; Mx1-Cre recipients died within 3 months (supplemental Figure 3F), with clinical presentations of severe dermatitis, hair loss, and deteriorating general condition. PB analysis demonstrated stable blood counts, which were derived from donor mice (data not shown), suggesting that the hematopoietic defect was not the cause of death. Given that Tip60 plays a critical role in response to DNA double-strand breaks (DSBs),17,21 Tip60 deletion may have exacerbated the genotoxicity of irradiation, leading to fatal failure of nonhematopoietic organs wherein Tip60 is deleted. In summary, we conclude that Tip60 is a critical cell-intrinsic regulator of HSCs.
Tip60 acetyltransferase activity is essential for HSC function
Because our conditional deletion model ablated all the functional domains of Tip60, we investigated whether its acetyltransferase activity is essential for HSC function. We overexpressed both wild-type TIP60 and acetyltransferase-defective TIP60 (Q377E/G380E)17 in HEK293T cells, transducing FLAG-TIP60 wild-type (TIP60wt) and FLAG-TIP60 mutant (TIP60mut) retrovirally, and confirmed that both proteins were expressed at similar levels (supplemental Figure 4A). Tip60Δ/Δ LSK cells purified from E13.5 fetal livers were transduced with TIP60wt, TIP60mut, or empty vector (EV) and subsequently transplanted into lethally irradiated congenic mice, along with congenic WBM cells (Figure 3A; supplemental Figure 4B). The Tip60Δ/Δ LSK cells transduced with EV did not recover long-term hematopoiesis, whereas the transduction of TIP60wt rescued the defect successfully (Figure 3B). Notably, the transduction of TIP60mut did not rescue the defect to any detectable level. Furthermore, we confirmed that both TIP60wt and TIP60mut interacted with key components of the NuA4 complex, including p400, RUVBL1, RUVBL2, and TRRAP (supplemental Figure 4C-E), suggesting that the catalytic mutations in TIP60 did not affect its binding to the complex. However, we cannot fully exclude the possibility that the TIP60 mutant could have some loss or gain of additional protein interactions that may account for its inability to rescue HSC regenerative capacity. Collectively, these results demonstrate a specific requirement of Tip60 acetyltransferase activity for the regeneration of HSCs.
Figure 3.
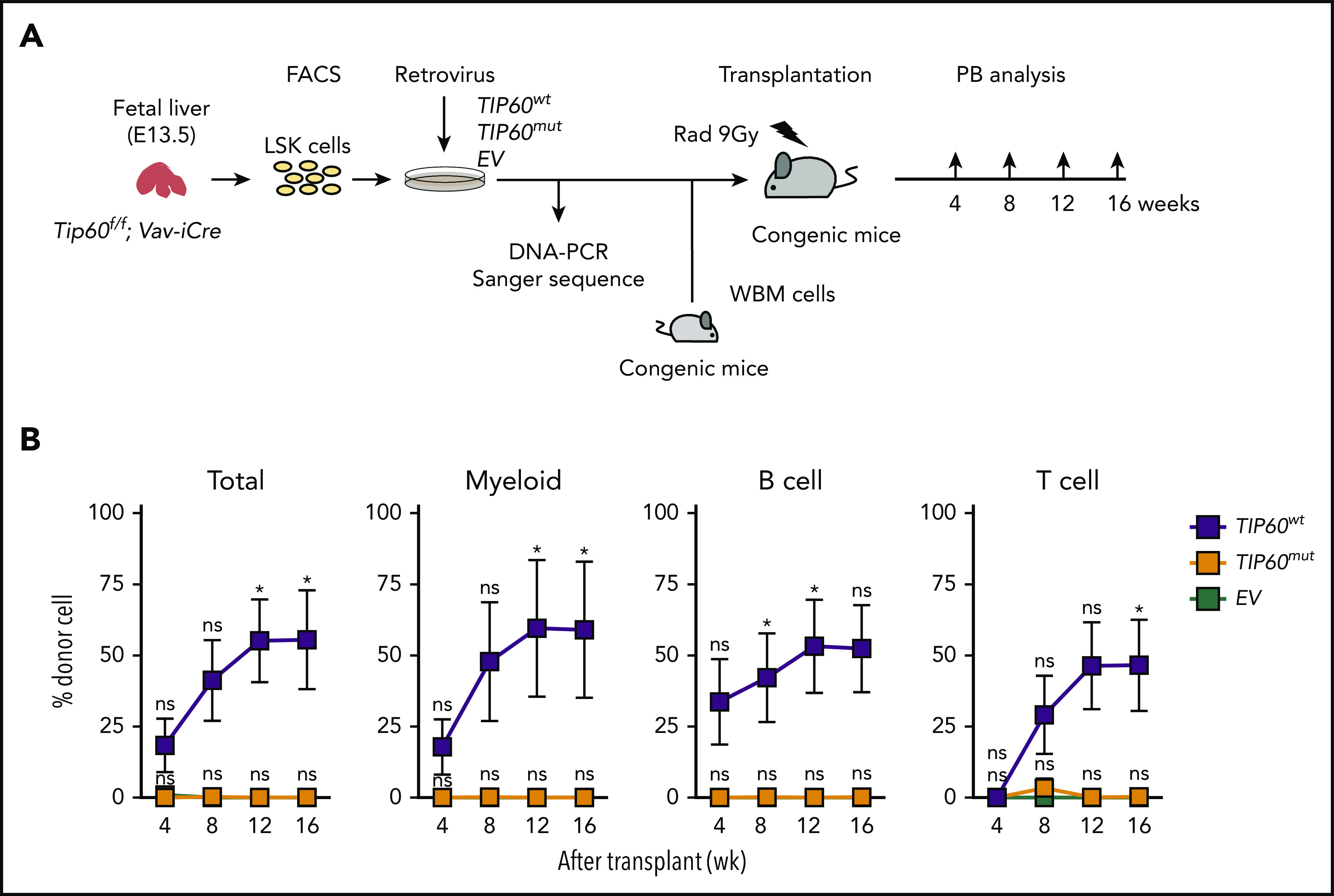
Tip60 acetyltransferase activity is critical for HSC function. (A) The experimental scheme for in vivo repopulating assays: Tip60Δ/Δ (Tip60f/f; Vav-iCre) LSK cells at E13.5 (CD45.2+) were transduced with TIP60wt, TIP60mut (supplemental Figure 4B), and EV plasmid retrovirally, and transplanted into lethally irradiated recipient mice (CD45.1+), along with congenic WBM cells (CD45.1+CD45.2+). Transduction of plasmid DNA was verified by Sanger sequencing of the PCR-amplified genomic products. (B) Chimerism of recipient PB was monitored at different times for 16 weeks after the transplantation. Percentages of donor cells in total (all nucleated cell), myeloid (Gr1+Mac1+), B cells (CD19+B220+), and T cells (CD3+CD4+) of recipient PB are shown. Values are presented as means ± standard error of the mean. Statistical analyses were performed vs EV. *P < .05, ns; not significant.
Tip60 maintains HSC genome integrity
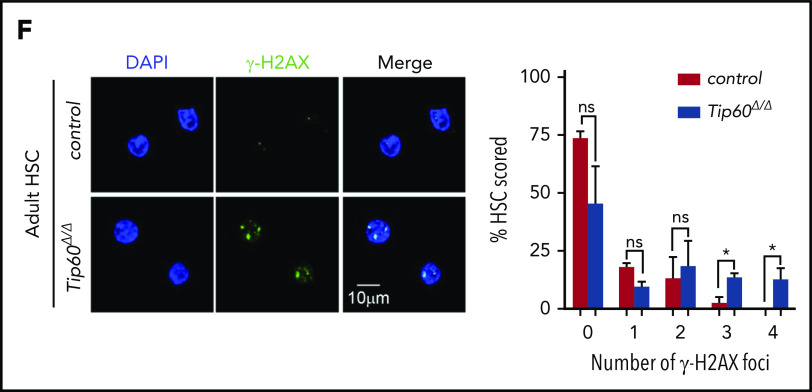
To elucidate the cellular mechanisms underlying the loss of Tip60Δ/Δ HSCs, we assessed apoptosis and cell-cycle status. Tip60 deletion induced apoptosis in fetal CD45+ cells and adult HSCs, exhibiting increased annexin V+DAPI− (4′,6-diamidino-2-phenylindole) cells and cleaved caspase-3 levels (Figure 4A-B). Cell-cycle analysis revealed fewer quiescent Tip60Δ/Δ HSCs (G0: Ki67−Hoeschst−) with a concordant increased percentage of cells in the cycling phase (S-G2/M: Ki67+Hoeschst+) compared with the control (Figure 4C). Moreover, DNA content analysis, using fetal LSK cells of Tip60f/f; Rosa26-CreERT2 and Tip60f/f embryos at E14.5, subsequently treated with 4-OHT in culture for 5 days, revealed an increase in cells with ≥4N DNA content (Figure 4D). Such an increase was not seen in LSK cells from Rosa26-CreERT2 embryos, suggesting that this effect is specific to the deletion of Tip60 (supplemental Figure 4F). Microscopic examination revealed that Tip60Δ/Δ cells grew in size and became multinucleated, exhibiting abnormal nuclear morphology, including multilobulated nuclei and micronuclei (supplemental Figure 4G). Given that the Tip60 complex has been shown to be important for the DNA DSB repair process,17,21 we investigated DNA damage in Tip60Δ/Δ HSCs. Remarkably, an alkaline comet assay demonstrated increased tail moment in Tip60Δ/Δ HSPCs (Figure 4E), suggesting that Tip60 deletion impaired DNA repair, which was further verified by the accumulation of γH2AX signals, a marker for unrepaired DSB loci (Figure 4F). Collectively, these results indicate that Tip60 deletion induces aberrant cell-cycle progression and apoptosis that is possibly caused by DNA breaks in HSCs.
Figure 4.
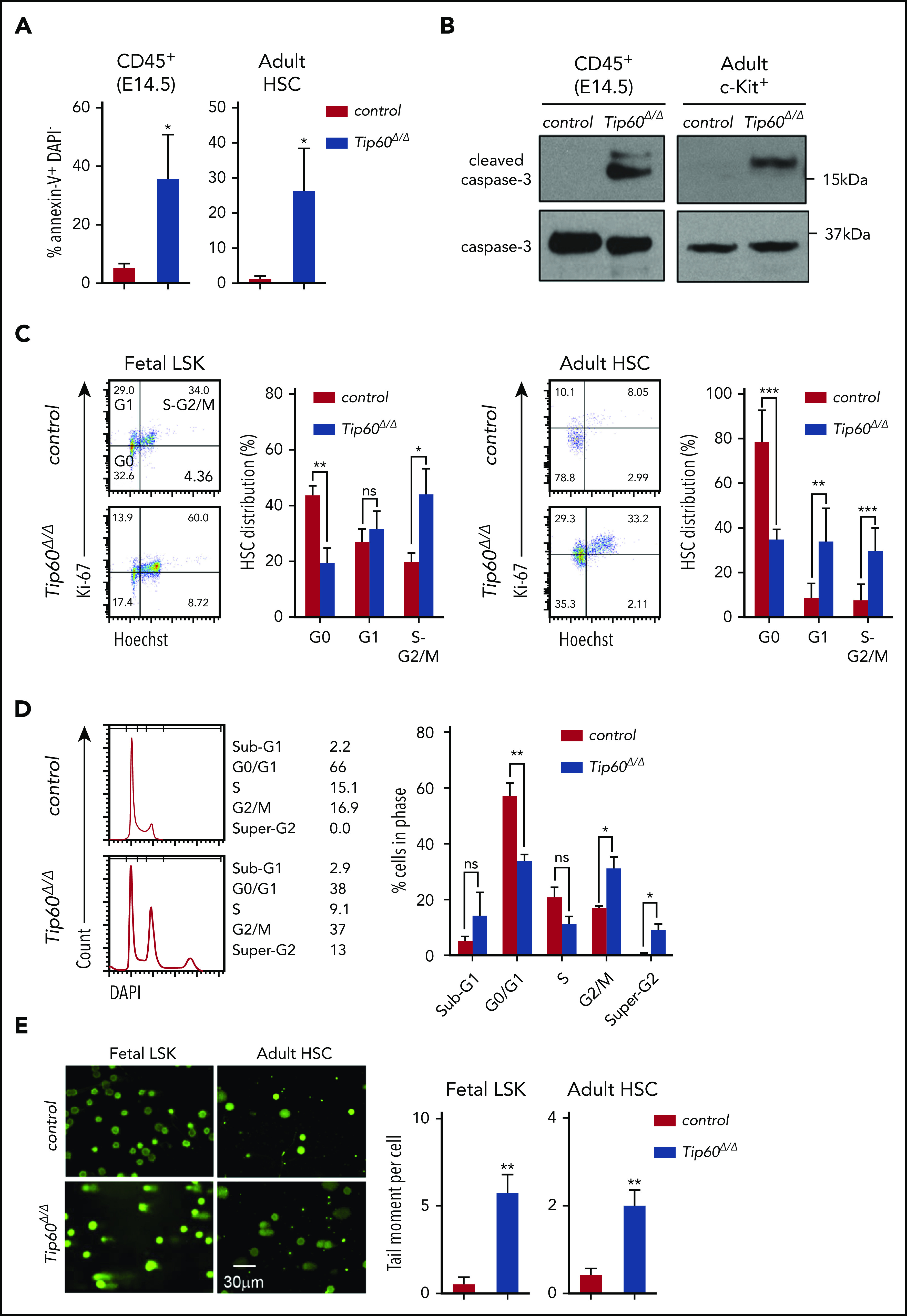
Tip60 loss leads to apoptosis and impairs cell cycle progression in HSCs. (A) Annexin V and DAPI staining of CD45+ cells from E14.5 embryos of control (Tip60f/+; n = 3), Tip60Δ/Δ (Tip60f/f; Vav-iCre) (n = 4), and BM CD150+CD48−LSK cells from control (Tip60f/f; n = 3) and Tip60Δ/Δ (Tip60f/f;Mx1-Cre) mice (n = 3) (pIpC injection on days 1-3, and analysis on day 5). Percentages of annexin V+DAPI− cells are shown. (B) Immunoblot analysis of caspase-3 and cleaved caspase-3 using CD45+ cells from E14.5 embryos of control and Tip60Δ/Δ and BM c-Kit+ cells from control (Tip60f/+) and Tip60Δ/Δ mice. (C) Cell-cycle status of fetal LSK (E14.5) (left) and adult CD150+CD48− LSK cells (right) was analyzed by flow cytometry. Representative dot plots (left), with average percentages of cells in each phase (right). (D) DNA content analysis by DAPI staining of LSK cells purified from Tip60f/f and Tip60f/f; Rosa26-CreERT2 embryos at E14.5, treated with 4-OHT in culture for 5 days (left). Percentages of cells in each phase (right). (E) Assessment of DNA breaks by alkaline comet assay. Images of control (Tip60f/+) and Tip60Δ/Δ fetal LSK cells and control (Tip60f/f) and Tip60Δ/Δ adult CD150+CD48−LSK cells (left). Quantitative values of tail DNA moment (right). (F) Images of immunostaining of γ-H2AX in control (Tip60f/f) and Tip60Δ/Δ CD150+CD48−LSK cells (left). Percentages of cells (y-axis) that have a corresponding number of γ-H2AX foci (x-axis) are graphed (right). The values are presented as means ± standard error of the mean. *P < .05; **P < .01; ***P < .001; ns, not significant
Tip60 is essential for expression of Myc target genes
To identify the genes regulated by Tip60 in a cell-intrinsic manner, we performed RNA-seq using LSK cells purified from Tip60f/f; Rosa26-CreERT2 and Tip60f/f embryos that were treated with 4-OHT for 72 hours (supplemental Figure 5A-B). A total of 1278 genes exhibited a significant change in expression levels, with 847 genes upregulated and 431 genes downregulated (log2-fold change >0.5; false discovery rate [FDR] <0.05) in Tip60Δ/Δ cells compared with the control (Tip60f/f; supplemental Figure 5C). Gene set enrichment analysis (GSEA), using the Molecular Signature Database MSigDB, revealed that E2F and Myc target genes were downregulated in Tip60Δ/Δ cells (supplemental Figure 5D). Given that Tip60 functions mainly as a transcriptional coactivator for several sequence-specific transcription factors,22-24 it is plausible that a variety of transcription factors are involved in the underlying mechanisms for the rapid loss of Tip60Δ/Δ HSCs. Hence, an open-ended GSEA was conducted on all available gene sets of transcription factor targets. Interestingly, multiple Myc-associated gene sets were specifically enriched in the control (Figure 5A-B). Although c-Myc at RNA and protein levels were not suppressed (supplemental Figure 5E-F), most Myc target genes were transcriptionally inactivated upon Tip60 deletion. Among the 124 differentially expressed Myc target genes (supplemental Figure 5G), we observed a significant overrepresentation of Tip60 targets in the downregulated group compared with the upregulated group (P < 10−5; Fisher’s exact test; supplemental Figure 5H). Our results demonstrate that Tip60 acts as a coactivator of Myc target genes.
Figure 5.
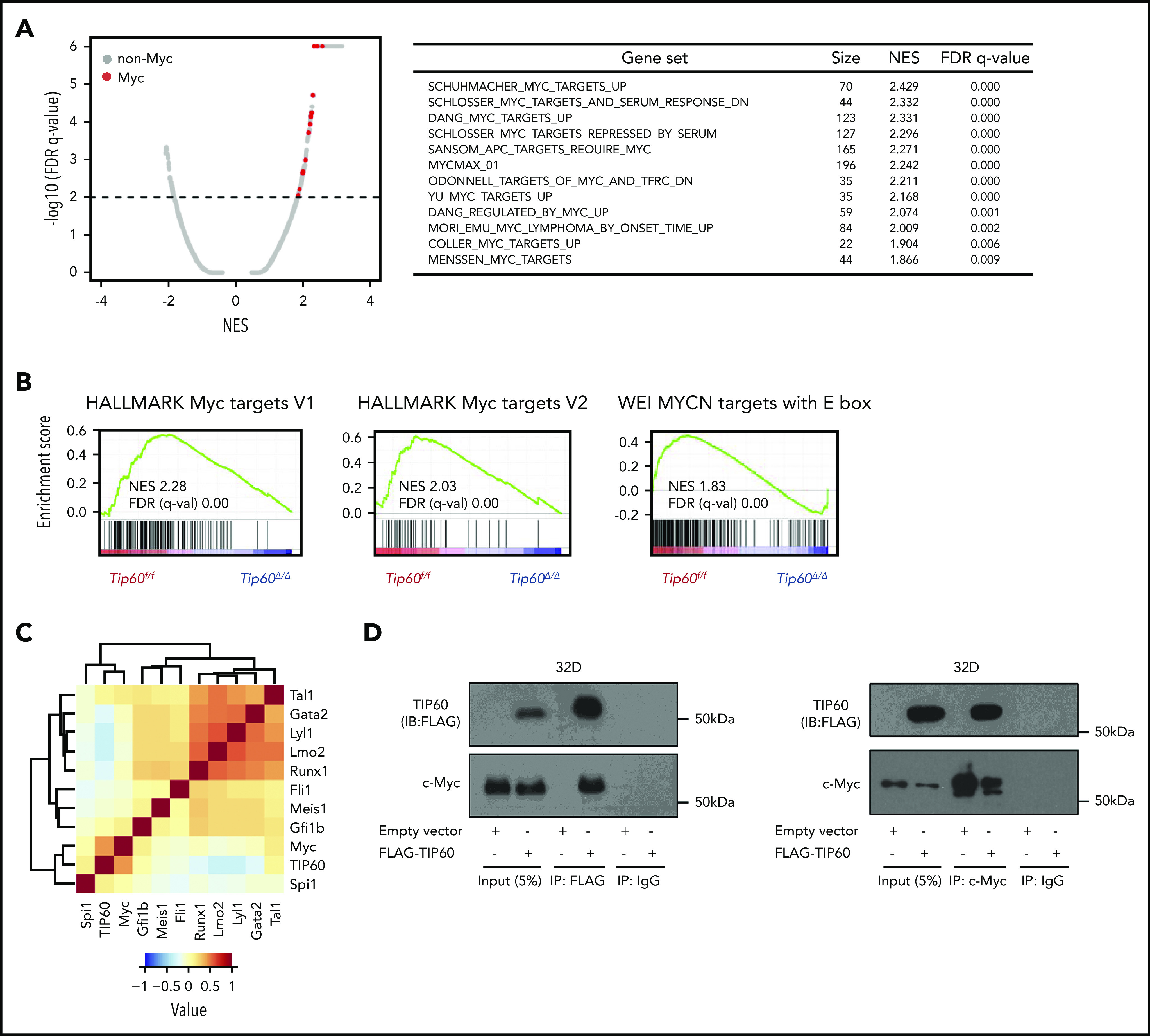
Tip60 regulates Myc target genes. (A) A plot (left) of FDR vs normalized enrichment score (NES) based on GSEA from RNA-seq data of control (Tip60f/f) vs Tip60Δ/Δ LSK cells. Distribution of all transcription factor target gene sets obtained from the MSigDB (c2 and c3) is shown. The red dots indicate the gene sets for Myc and the gray dots, the non-Myc transcription factors. The dashed line represents the FDR cutoff. List of enriched Myc-associated gene sets (right). (B) Representative GSEA plots for Myc target gene sets. (C) Correlation analysis of genome-wide occupancy for Tip60 and 10 transcriptional factors (c-Myc, Tal1, Gata2, Lyl1, Lmo2, Runx1, Fli1, Meis1, Gfi1b, and Spi1) in a murine hematopoietic progenitor cell line, HPC-7, using NPMI. The color intensity represents the correlation efficiency of each 2 transcriptional factors. A total of 3651 c-Myc binding peaks, as well as 4 347 Tip60, 7 596 Tal1, 2 796 Gata2, 3 432 Lyl1, 5 202 Lmo2, 5 992 Runx1, 18 796 Fli1, 7 690 Meis1, 3 279 Gfi1b, and 18 249 Spi1 (PU.1) peaks were identified. (D) Cells of the hematopoietic progenitor cell line 32D were transduced with FLAG-TIP60 plasmid (left). Cell lysates were subjected to immunoprecipitation with FLAG-M2 beads. Proteins present in immunoprecipitates or cell lysates (input) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotted with antibodies for FLAG and c-Myc. Interaction was confirmed by reciprocal coimmunoprecipitation (right).
Cell-cycle regulators play a pivotal role in HSC maintenance by tuning the balance between quiescence and self-renewal.25 The DNA repair process is critical as well, exemplified by recent studies that demonstrated that HSCs are susceptible to DNA damage caused by intrinsic and extrinsic stress factors, such as aging, replication, and genotoxic and oxidative stresses.26-28 Of note, the genes that were inactivated by Tip60 deletion are associated with cell-cycle and DNA repair. Therefore, we conclude that Tip60 maintains HSC functional integrity, mainly by cooperating with Myc family transcription factors to activate the multiple genes that are involved in essential cellular processes for HSC maintenance.
Tip60 and Myc coactivate target genes
To better understand the involvement of Tip60 in the active transcription of Myc target genes, we performed genome-wide ChIP-seq assays in the murine hematopoietic progenitor cell line HPC-7,29 using rabbit polyclonal antibodies that specifically recognize endogenous Tip60.23 Most of the 4347 high-confidence, Tip60-bound genomic loci identified (66.7%) were located at the proximal promoter regions (−1kb, +100 bp from the transcriptional start sites [TSSs]), including the promoters of previously reported Tip60 target genes (Ncl, Rps9, and Cdkn1b)23,30,31 (supplemental Figure 6A-B). To define the transcription factors that colocalize with Tip60, a correlation analysis of genome-wide Tip60 and 10 hematopoietic transcription factor occupancy (c-Myc, Tal1, Gata2, Lyl1, Lmo2, Runx1, Fli1, Meis1, Gfi1b, and Spi1) was performed, as previously described.18 Notably, c-Myc was the most enriched transcription factor in the Tip60-bound regions (Figure 5C), which corroborates the findings in our RNA-seq analysis. Given that Myc binding peaks are not more than those of other transcription factors, a specificity of Tip60 colocalization with c-Myc does not reflect an abundance of c-Myc binding peaks. Colocalization of c-Myc and Tip60 at the promoters of the target genes was validated by ChIP-qPCR (supplemental Figure 6C). Consistent with a previous study in nonhematopoietic cells,23 we demonstrated that TIP60 interacts with c-Myc by reciprocal coimmunoprecipitation experiments in 32D murine hematopoietic progenitor cells (Figure 5D).
To delineate the chromatin conformations, we generated heat maps of ChIP-seq peak signals between −5 and +5 kb from the TSS region for Tip60, active promoter histone mark H3K4me3, repressive histone mark H3K27me3, and DNase-1 hypersensitive sites (DHSs)18 and ranked the gene order based on expression levels (supplemental Figure 6D). Remarkably, the Tip60 binding intensity correlated with the RNA levels of the corresponding genes, and its occupancy overlapped with H3K4me3 enrichment and DHSs, but not with H3K27me3. Given that c-Myc and N-myc bind specifically to a common consensus sequence,32,33 we conclude that Tip60 colocalizes with Myc proteins at the transcriptional regulatory elements and active chromatin regions to activate genes in HSPCs.
Tip60 maintains acetylated H2A.Z at Myc target genes
Tip60 has been demonstrated to acetylate histone H3, H4, H2A, and H2A variants, as well as nonhistone proteins.13,17 H2A.Z is an evolutionarily conserved variant of canonical H2A. In Saccharomyces cerevisiae, ESA1, the yeast orthologue of Tip60, also mediates the acetylation of Htz1, the orthologue of H2A.Z.34,35 Drosophila Tip60 acetylates lysine 5 of H2Av, a functional homologue of the mammalian H2A.Z isoform.36 H2A.Z is enriched around TSS across the different species,37,38 whereas an acetylated form of H2A.Z (acH2A.Z) is enriched at the active promoter regions39 and confers nucleosome destabilization and open chromatin conformation.40 We evaluated global histone acetylation levels upon overexpression of TIP60 in the 32D hematopoietic progenitor cells and found increased H2A.Z acetylation levels, as well as H4K16, a known histone substrate of Tip60 (supplemental Figure 7A). Given that H2A.Z has been linked with actively transcribed regions, we further evaluated the changes in acH2A.Z along with an active enhancer and promoter mark (H3K27ac) and a repressive mark (H3K27me3), evoked by Tip60 deletion by performing ChIP-seq (supplemental Figure 7B). Consistent with the findings from a previous study,41 both acH2A.Z and H2A.Z were enriched at the active chromatin marked by H3K27ac (Figure 6A), and acH2A.Z exhibited a pronounced bimodal enrichment around the TSS at highly expressed genes, while exhibiting less enrichment at poorly expressed genes and no enrichment at nontranscribed genes (supplemental Figure 7C). Remarkably, we observed that the transcription levels of genes that were highly enriched in acH2A.Z and H2A.Z marks around the TSS are significantly higher than those of all transcribed genes (supplemental Figure 7D). We observed a similar trend for genes with H3K27ac enrichment, whereas those with H3K27me3 marks exhibited a significant decrease in their transcription levels. Moreover, the ratio of acH2A.Z/H2A.Z around the TSS correlated with RNA levels of the corresponding genes (Pearson’s correlation, R2 = 0.7), and most of the Tip60-bound genes (82.0%) were highly expressed and had a higher acH2A.Z/H2A.Z ratio (Figure 6B).
Figure 6.
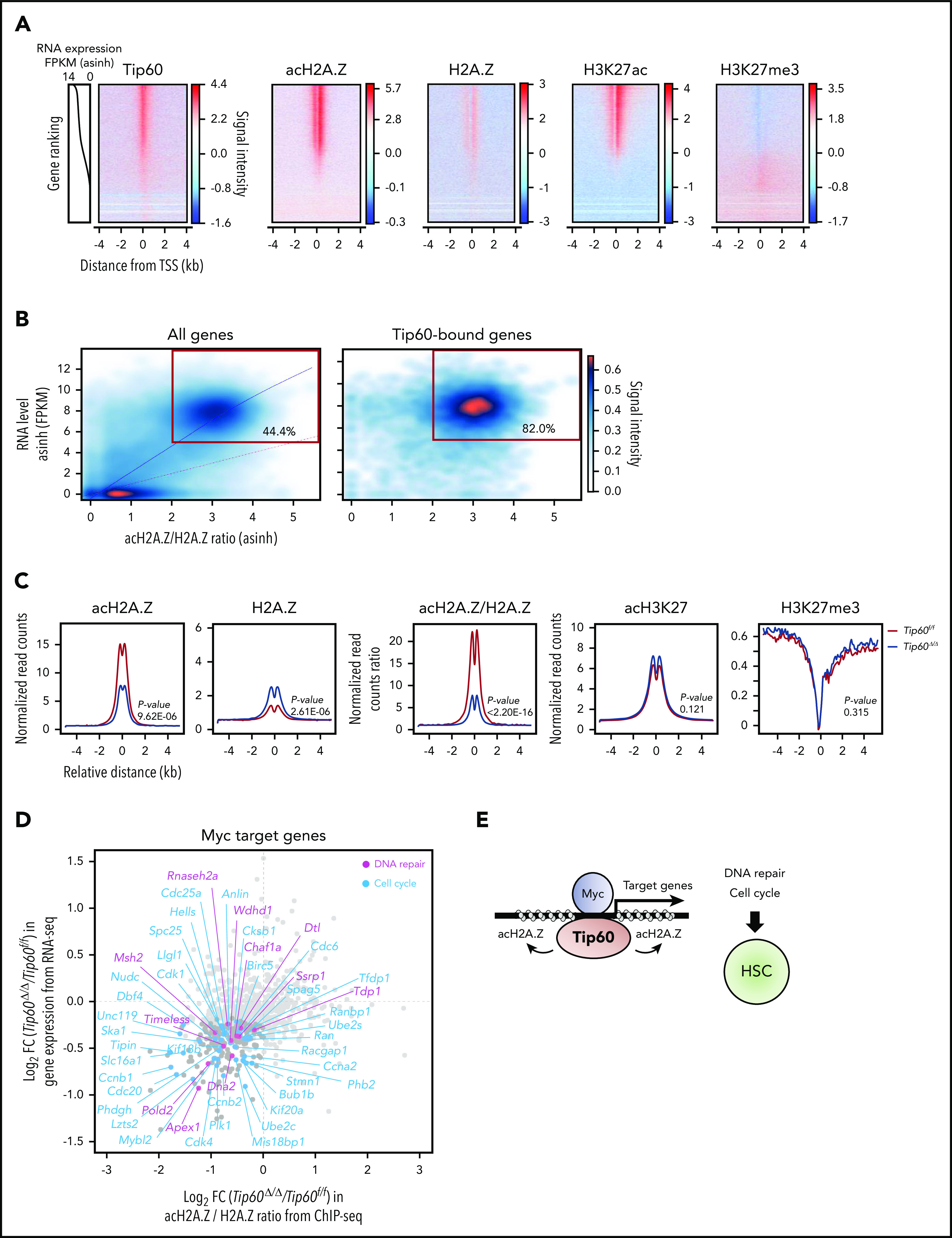
Tip60 maintains the acetylation of H2A.Z to activate Myc target genes. (A) An integrated view of Tip60 occupancy and acH2A.Z, H2A.Z, and H3K27me3 enrichment around TSS (TSS, ±4 kb) in ChIP-seq analysis using wild-type fetal c-Kit+ cells. Genes were ordered according to the expression levels (line plots) from high (top) to low (bottom). The color scale represents changes in signal intensity for each antibody. (B) Cloud plots represent all (left) and Tip60-bound (right) genes, according to the corresponding acH2A.Z/H2A.Z ratio from ChIP-seq and the gene expression level from RNA-seq. Correlation analysis between the acH2A.Z/H2A.Z ratio and the gene expression level is shown. The blue line represents the polynomial regression curve (R2 = 0.7). The red line represents a theoretical linear regression curve (y = x). Genes with a high acH2A.Z/H2A.Z ratio (>2) acH2A.Z and a high RNA level (log2 FPKM >5) are in red rectangles. Tip60-bound genes were identified based on Tip60 enrichment at the proximal promoter region (−1 kb, +100 bp from the TSS). (C) Mean plot of acH2A.Z, H2A.Z, acH2A.Z/H2A.Z ratio, acH3K27, and H3K27me3 enrichment based on relative distance from Tip60-bound proximal promoter regions in both control (purple) and Tip60Δ/Δ fetal c-Kit+ cells (red). Statistical analyses were performed using the Mann-Whitney Wilcoxon method. (D) Myc target genes (n = 816) were identified to be transcribed in both control and Tip60Δ/Δ fetal LSK cells in the RNA-seq analysis. The x-axis indicates log2-fold change in acH2A.Z/H2A.Z ratio at proximal promoter regions induced by Tip60 deletion and the y-axis indicates log2-fold change in gene expression level. DNA repair–related genes (blue) and cell-cycle–related genes (pink) with significant downregulation in their expression levels (P < .05) are labeled. Myc target genes were obtained from the gene sets described in Figure 5A. DNA repair– and cell-cycle–related genes were obtained from Gene Ontology categories GO:0006281 and GO:0007049, respectively. (E) Tip60 maintains HSCs through acetylation of H2A.Z, to activate Myc target genes that regulate cell-cycle and DNA repair processes. FPKM, fragments per kilobase per million.
Notably, Tip60 loss reduced acH2A.Z levels at the Tip60-bound promoters and distal enhancer regions, whereas neither H3K27ac nor H3K27me3 enrichment levels were altered (Figure 6C; supplemental Figure 7E). H2A.Z abundance within nucleosomes at the Tip60-bound promoters was not decreased; indicating that reduced acetylation of H2A.Z is not a direct consequence of H2A.Z eviction. Myc target genes (816 genes), which were obtained from Myc-associated gene sets (Figure 5A), demonstrated reduced acH2A.Z/H2A.Z ratios around TSS and significant downregulation of their RNA levels (217 genes), including various genes related to cell cycle and DNA repair (Figure 6D; supplemental Figure 7F). We conducted additional analysis for Tal1 and Spi1 (PU.1) target genes obtained from public database (supplemental Figure 7G). We chose Tal1 as a representative of the genes that showed minimum correlation with Tip60 in the genome-wide occupancy analysis and Spi1 (PU.1), as a representative of genes that showed high correlation (supplemental Figure 5C). A higher proportion of Myc target genes (69.8%) exhibited decreased acH2AZ/H2AZ ratio and gene expression compared with the Tal1 (33.02%) and Spi1 (PU.1) target genes (28.47%). We thus propose that the Tip60-acH2A.Z epigenetic axis contributes to HSC survival through activation of Myc target genes (Figure 6E).
Discussion
The lysine acetyltransferase Tip60 is the catalytic subunit of the multiprotein complex NuA4 and acts mainly in this large molecular complex. Previous studies revealed a critical requirement for 3 subunits of the complex in normal hematopoiesis; ablation of the gene encoding E1A-binding protein p400,42 transformation/transcription domain-associated protein (Trrap),43 and RuvB-like AAA ATPase 1 (Ruvbl1),44 resulted in rapid loss of adult HSCs from BM caused by apoptosis. These subunits are essential for the integrity, assembly, and activity of the complex,45-48 suggesting that the NuA4 complex is important for HSC maintenance. However, the role of Tip60, specifically the catalytic function, in normal hematopoiesis remains undefined. In the present study, we established Tip60 conditional knockout mice in 2 different Cre strains (Vav-iCre and Mx1-Cre) to study the role of Tip60 in both fetal and adult hematopoiesis. Importantly, we demonstrated that the acetyltransferase activity of Tip60 is critical for the long-term regenerating ability of murine HSCs.
The conditional deletion of Tip60 in murine hematopoietic cells evoked catastrophic DNA damage in HSCs in both the fetal and adult stages, thereby resulting in rapid apoptosis. It is noteworthy that conditional deletion of Tip60 in Treg cells increased their number in the thymus, whereas they were decreased in both the spleen and the lymph nodes.16 These results highlight that loss of Tip60 does not always result in cell lethality. Given that recent studies involving DNA repair–deficient mouse models indicate that HSCs are vulnerable to intrinsic and extrinsic DNA damage,26-28 Tip60 deletion provoked apoptosis in HSCs because of cellular susceptibility to DNA damage. Therefore, the essentiality of Tip60 for cell survival could be cell context dependent.
Tip60 deletion had a clear impact on functional HSCs loss. All the transplantation assays in this study were followed up for at least 16 weeks, thus excluding a contribution from progenitor cell–derived hematopoiesis. Indeed, bulk RNA-seq was performed in HSPCs; therefore, it is possible that gene expression changes in Myc and E2F targets are a consequence of a decrease in the number of proliferating cells. Nonetheless, the specific cobinding of Tip60 and Myc on the target gene promoters demonstrated by ChIP-seq analysis provides evidence of a direct role of Tip60 in the regulation of c-Myc target genes. Intriguingly, Tip60 deletion did not suppress c-Myc RNA and protein levels, indicating that downregulation of Myc target genes induced by Tip60 deletion is not a direct consequence of c-Myc levels. In line with a previous study,23 we also demonstrated interaction of Tip60 and c-Myc in hematopoietic progenitor cells. These results provide substantial evidence of a direct role of Tip60 in the regulation of Myc target genes. Noting that the HSC phenotype evoked by simultaneous c-myc and N-myc deletion in a previous study49 resembles those of the Tip60 conditional knockout mice, we conclude that Tip60 maintains HSC survival by interacting with c-Myc to coactivate target genes.
We demonstrated that the catalytically inactive TIP60 mutant did not rescue the impaired HSC function, although it maintained critical components of the NuA4 complex, indicating that Tip60 acetyltransferase activity was specifically required. H2A.Z has been validated to be a substrate of Tip6034-36 and has various functions in different species, including gene activation and repression, DNA repair, heterochromatin formation, and chromosome segregation.50-52 The diverse functions are influenced by PTMs, including acetylation, SUMOylation, ubiquitination, and methylation of lysines.53 Among them, acetylation is the most studied PTM as an active enhancer and promoter mark.39,40,54,55 We extended these findings to murine HSPCs by performing genome-wide ChIP-seq analysis to investigate how Tip60 impacts H2A.Z acetylation and chromatin structure. Notably, Tip60 deletion resulted in a global reduction in acH2A.Z/H2A.Z ratio around the TSS, as well as to some extent in distal enhancer regions bound by Tip60, and reduced expression of a portion of these corresponding genes, particularly Myc targets. These data indicate that Tip60-mediated H2A.Z acetylation may serve as a prerequisite for active gene transcription although how acetylated H2A.Z facilitates this machinery has remained largely enigmatic. Tip60 has also been described to acetylate H3, H4, and nonhistone proteins besides H2A.Z.13 Additional targets besides H2A.Z may be involved in the regulation of genes for HSC survival, and therefore further work is necessary to study the detailed mechanisms.
In addition, we cannot exclude the possibility that the Tip60-acH2A.Z epigenetic axis may involve other transcription factors besides c-Myc. Recent studies have suggested a putative association between H2A.Z and transcription factor E2F1 and coactivator Brd2 in active gene transcription.56,57 In future research, it will be of interest to identify and investigate how Tip60 cooperates with different transcription factors to facilitate gene transcription through acetylation of H2A.Z.
In summary, our study highlights the importance of Tip60 in maintaining proper cell-cycle progression and DNA repair in murine HSCs in both the fetal and adult stages, which is, at least in part, mediated by Tip60-dependent acetylation of H2A.Z, which activates Myc target genes.
Supplementary Material
The online version of this article contains a data supplement.
Acknowledgments
The authors thank Bruno Amati (Italian Institute of Technology, Milan, Italy) for providing rabbit polyclonal Tip60 antibodies; Issay Kitabayshi (National Cancer Research Center, Tokyo, Japan) and Chunaram Choudhary (The Novo Nordisk Foundation Center for Protein Research, Copenhagen, Denmark) for helpful discussions; Celestina Chin Ai Qi (CSI Singapore) for proofreading the manuscript; Cai Ping Koh (CSI Singapore) and members of the Tenen laboratory for technical support and helpful discussions; and the FACS facility and Quantitative Proteomics Core (CSI Singapore), Beijing Genomics Institute (Hong Kong, China), and Duke-NUS Genome Biology Facility (Singapore) for technical support.
This work was supported by the National Research Foundation, Singapore; the Singapore Ministry of Education; a Singapore Translational Research Award from the Singapore National Medical Research Council (NMRC/STaR/00018/2013) (D.G.T.); National Institutes of Health, National Cancer Institute grant R35CA197697 and National Heart, Lung and Blood Institute grant and P01HL131477 (D.G.T.); Cancer Research UK grant C1163/A21762 and Wellcome Trust grant 206328/Z/17/Z (B.G.). a research fellowship from the Uehara Memorial Foundation (A.N.), and funds from the Leukaemia and Blood Cancer New Zealand and the family of Marijana Kumerich (S.K.B.).
Footnotes
The RNA-Seq and ChIP-seq data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE120705).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.N., D.B., and D.G.T. undertook the concept and design of the study; D.B. and J. Li developed the murine models; A.N., H.S.K., and Q.-L.Z. performed the experiments, interpreted the data, and generated the figures; D.B., J.P., C.Q.W., V.K., and Y.Z. performed the experiments; Q.-L.Z., J. Lough, R.T.-M., V.E.A., R.H., and S.Z. performed bioinformatic analysis; H.Y. and T.B. supervised the bioinformatic analysis; D.R. performed the mass spectrometry experiments; A.N. and H.S.K. wrote the manuscript; R.S.W., M.O., S.J., B.G., and S.K.B. provided expert consultation; A.N., H.S.K., D.B., and D.G.T. reviewed the manuscript; and D.G.T. provided funding support.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Deepak Bararia, Miltenyi Biotec B.V. & Co. KG, Friedrich-Ebert-Str 68, 51429 Bergisch Gladbach, Germany; e-mail: deepakb@miltenyibiotec.de; and Daniel G. Tenen, Centre for Translational Medicine, National University of Singapore, 14 Medical Dr, #12-01 North Core, Singapore 117599, Singapore; e-mail: daniel.tenen@nus.edu.sg.
REFERENCES
- 1.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132(4):631-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma S, Gurudutta G. Epigenetic Regulation of Hematopoietic Stem Cells. Int J Stem Cells. 2016;9(1):36-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Konuma T, Oguro H, Iwama A. Role of the polycomb group proteins in hematopoietic stem cells. Dev Growth Differ. 2010;52(6):505-516. [DOI] [PubMed] [Google Scholar]
- 4.Cullen SM, Mayle A, Rossi L, Goodell MA. Hematopoietic stem cell development: an epigenetic journey. Curr Top Dev Biol. 2014;107:39-75. [DOI] [PubMed] [Google Scholar]
- 5.Katsumoto T, Aikawa Y, Iwama A, et al. MOZ is essential for maintenance of hematopoietic stem cells. Genes Dev. 2006;20(10):1321-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brownell JE, Zhou J, Ranalli T, et al. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84(6):843-851. [DOI] [PubMed] [Google Scholar]
- 7.Turner BM, Birley AJ, Lavender J. Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell. 1992;69(2):375-384. [DOI] [PubMed] [Google Scholar]
- 8.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012;11(5):384-400. [DOI] [PubMed] [Google Scholar]
- 9.Perez-Campo FM, Costa G, Lie-A-Ling M, Stifani S, Kouskoff V, Lacaud G. MOZ-mediated repression of p16(INK) (4) (a) is critical for the self-renewal of neural and hematopoietic stem cells. Stem Cells. 2014;32(6):1591-1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valerio DG, Xu H, Eisold ME, Woolthuis CM, Pandita TK, Armstrong SA. Histone acetyltransferase activity of MOF is required for adult but not early fetal hematopoiesis in mice. Blood. 2017;129(1):48-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mishima Y, Miyagi S, Saraya A, et al. The Hbo1-Brd1/Brpf2 complex is responsible for global acetylation of H3K14 and required for fetal liver erythropoiesis. Blood. 2011;118(9):2443-2453. [DOI] [PubMed] [Google Scholar]
- 12.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70(1):81-120. [DOI] [PubMed] [Google Scholar]
- 13.Squatrito M, Gorrini C, Amati B. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 2006;16(9):433-442. [DOI] [PubMed] [Google Scholar]
- 14.Doyon Y, Côté J. The highly conserved and multifunctional NuA4 HAT complex. Curr Opin Genet Dev. 2004;14(2):147-154. [DOI] [PubMed] [Google Scholar]
- 15.Hu Y, Fisher JB, Koprowski S, McAllister D, Kim MS, Lough J. Homozygous disruption of the Tip60 gene causes early embryonic lethality. Dev Dyn. 2009;238(11):2912-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao Y, Nagai Y, Deng G, et al. Dynamic interactions between TIP60 and p300 regulate FOXP3 function through a structural switch defined by a single lysine on TIP60. Cell Rep. 2014;7(5):1471-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ikura T, Ogryzko VV, Grigoriev M, et al. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102(4):463-473. [DOI] [PubMed] [Google Scholar]
- 18.Wilson NK, Schoenfelder S, Hannah R, et al. Integrated genome-scale analysis of the transcriptional regulatory landscape in a blood stem/progenitor cell model. Blood. 2016;127(13):e12-e23. [DOI] [PubMed] [Google Scholar]
- 19.de Boer J, Williams A, Skavdis G, et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur J Immunol. 2003;33(2):314-325. [DOI] [PubMed] [Google Scholar]
- 20.Kühn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269(5229):1427-1429. [DOI] [PubMed] [Google Scholar]
- 21.Murr R, Loizou JI, Yang YG, et al. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8(1):91-99. [DOI] [PubMed] [Google Scholar]
- 22.Bararia D, Trivedi AK, Zada AA, et al. Proteomic identification of the MYST domain histone acetyltransferase TIP60 (HTATIP) as a co-activator of the myeloid transcription factor C/EBPalpha. Leukemia. 2008;22(4):800-807. [DOI] [PubMed] [Google Scholar]
- 23.Frank SR, Parisi T, Taubert S, et al. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 2003;4(6):575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Legube G, Linares LK, Tyteca S, et al. Role of the histone acetyl transferase Tip60 in the p53 pathway. J Biol Chem. 2004;279(43):44825-44833. [DOI] [PubMed] [Google Scholar]
- 25.Rossi L, Lin KK, Boles NC, et al. Less is more: unveiling the functional core of hematopoietic stem cells through knockout mice. Cell Stem Cell. 2012;11(3):302-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beerman I, Seita J, Inlay MA, Weissman IL, Rossi DJ. Quiescent hematopoietic stem cells accumulate DNA damage during aging that is repaired upon entry into cell cycle. Cell Stem Cell. 2014;15(1):37-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rossi DJ, Bryder D, Seita J, Nussenzweig A, Hoeijmakers J, Weissman IL. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 2007;447(7145):725-729. [DOI] [PubMed] [Google Scholar]
- 28.Walter D, Lier A, Geiselhart A, et al. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature. 2015;520(7548):549-552. [DOI] [PubMed] [Google Scholar]
- 29.Pinto do O P, Kolterud A, Carlsson L. Expression of the LIM-homeobox gene LH2 generates immortalized steel factor-dependent multipotent hematopoietic precursors. EMBO J. 1998;17(19):5744-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134(1):162-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park JH, Sun XJ, Roeder RG. The SANT domain of p400 ATPase represses acetyltransferase activity and coactivator function of TIP60 in basal p21 gene expression. Mol Cell Biol. 2010;30(11):2750-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blackwell TK, Kretzner L, Blackwood EM, Eisenman RN, Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990;250(4984):1149-1151. [DOI] [PubMed] [Google Scholar]
- 33.Ma A, Moroy T, Collum R, Weintraub H, Alt FW, Blackwell TK. DNA binding by N- and L-Myc proteins. Oncogene. 1993;8(4):1093-1098. [PubMed] [Google Scholar]
- 34.Babiarz JE, Halley JE, Rine J. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev. 2006;20(6):700-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keogh MC, Mennella TA, Sawa C, et al. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 2006;20(6):660-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kusch T, Florens L, Macdonald WH, et al. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306(5704):2084-2087. [DOI] [PubMed] [Google Scholar]
- 37.Creyghton MP, Markoulaki S, Levine SS, et al. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell. 2008;135(4):649-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raisner RM, Hartley PD, Meneghini MD, et al. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123(2):233-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valdés-Mora F, Song JZ, Statham AL, et al. Acetylation of H2A.Z is a key epigenetic modification associated with gene deregulation and epigenetic remodeling in cancer. Genome Res. 2012;22(2):307-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishibashi T, Dryhurst D, Rose KL, Shabanowitz J, Hunt DF, Ausió J. Acetylation of vertebrate H2A.Z and its effect on the structure of the nucleosome. Biochemistry. 2009;48(22):5007-5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823-837. [DOI] [PubMed] [Google Scholar]
- 42.Fujii T, Ueda T, Nagata S, Fukunaga R. Essential role of p400/mDomino chromatin-remodeling ATPase in bone marrow hematopoiesis and cell-cycle progression. J Biol Chem. 2010;285(39):30214-30223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loizou JI, Oser G, Shukla V, et al. Histone acetyltransferase cofactor Trrap is essential for maintaining the hematopoietic stem/progenitor cell pool. J Immunol. 2009;183(10):6422-6431. [DOI] [PubMed] [Google Scholar]
- 44.Bereshchenko O, Mancini E, Luciani L, Gambardella A, Riccardi C, Nerlov C. Pontin is essential for murine hematopoietic stem cell survival. Haematologica. 2012;97(9):1291-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Auger A, Galarneau L, Altaf M, et al. Eaf1 is the platform for NuA4 molecular assembly that evolutionarily links chromatin acetylation to ATP-dependent exchange of histone H2A variants. Mol Cell Biol. 2008;28(7):2257-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elías-Villalobos A, Toullec D, Faux C, Séveno M, Helmlinger D. Chaperone-mediated ordered assembly of the SAGA and NuA4 transcription co-activator complexes in yeast. Nat Commun. 2019;10(1):5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitchell L, Lambert JP, Gerdes M, et al. Functional dissection of the NuA4 histone acetyltransferase reveals its role as a genetic hub and that Eaf1 is essential for complex integrity. Mol Cell Biol. 2008;28(7):2244-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jha S, Gupta A, Dar A, Dutta A. RVBs are required for assembling a functional TIP60 complex. Mol Cell Biol. 2013;33(6):1164-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laurenti E, Varnum-Finney B, Wilson A, et al. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell. 2008;3(6):611-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altaf M, Auger A, Covic M, Côté J. Connection between histone H2A variants and chromatin remodeling complexes. Biochem Cell Biol. 2009;87(1):35-50. [DOI] [PubMed] [Google Scholar]
- 51.Biterge B, Schneider R. Histone variants: key players of chromatin. Cell Tissue Res. 2014;356(3):457-466. [DOI] [PubMed] [Google Scholar]
- 52.Weber CM, Henikoff S. Histone variants: dynamic punctuation in transcription. Genes Dev. 2014;28(7):672-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sevilla A, Binda O. Post-translational modifications of the histone variant H2AZ. Stem Cell Res (Amst). 2014;12(1):289-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu G, Cui K, Northrup D, et al. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2013;12(2):180-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ku M, Jaffe JD, Koche RP, et al. H2A.Z landscapes and dual modifications in pluripotent and multipotent stem cells underlie complex genome regulatory functions. Genome Biol. 2012;13(10):R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Draker R, Ng MK, Sarcinella E, Ignatchenko V, Kislinger T, Cheung P. A combination of H2A.Z and H4 acetylation recruits Brd2 to chromatin during transcriptional activation. PLoS Genet. 2012;8(11):e1003047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Surface LE, Fields PA, Subramanian V, et al. H2A.Z.1 Monoubiquitylation Antagonizes BRD2 to Maintain Poised Chromatin in ESCs. Cell Rep. 2016;14(5):1142-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.