Abstract
目的
探讨表没食子儿茶素没食子酸酯(EGCG)诱导CHD5基因去甲基化促进KG-1、THP-1细胞凋亡的相关机制。
方法
设实验组及正常对照组,实验组:不同浓度(25、50、75、100、150 μg/mL)EGCG处理作用于KG-1、THP-1细胞,正常对照组:不加EGCG处理。MSP法检测KG-1、THP-1细胞CHD5基因甲基化;MTT法检测作用48 h后细胞增殖;流式细胞术检测作用48 h后细胞周期和细胞凋亡状况;RT-qPCR、Western blot检测DNMT1、CHD5、p19Arf、p53、p21Cip1基因和蛋白表达。
结果
EGCG呈现剂量依赖性逆转了KG-1、THP-1细胞CHD5基因高甲基化;EGCG以剂量依赖性的方式抑制KG-1、THP-1细胞增殖(P < 0.05);EGCG诱导细胞周期停滞于G1期,促进细胞凋亡;EGCG以剂量依赖性的方式下调DNMT1的mRNA和蛋白表达,上调CHD5、p19Arf、p53、p21Cip1的mRNA和蛋白表达(P < 0.05)。
结论
EGCG可通过下调DNMT1降低KG-1、THP-1细胞CHD5基因高甲基化,恢复CHD5基因表达,从而上调p19Arf、p53、p21Cip1表达诱发细胞凋亡。
Keywords: 表没食子儿茶素没食子酸, CHD5基因, 甲基化, p19Arf-p53-p21Cip1信号通路, 急性髓系白血病
Abstract
Objective
To investigate the mechanism by which epigallocatechin gallate (EGCG) induces CHD5 gene demethylation and promotes the apoptosis of acute myeloid leukemia KG-1 and THP-1 cell lines.
Methods
KG-1 and THP-1 cells treated with 25, 50, 75, 100 or 150 μg/mL EGCG for 48 h were examined for CHD5 gene methylation using MSP and for cell proliferation using MTT assay. The changes in cell cycle and apoptosis of the two cell lines after treatment with EGCG for 48 h were detected using flow cytometry. The mRNA and protein expressions of DNMT1, CHD5, p19Arf, p53 and p21Cip1 in the cells were detected using RT-quantitative PCR and Western blot.
Results
EGCG dose-dependently reversed hypermethylation of CHD5 gene and reduced the cell viability in both KG-1 and THP-1 cells (P < 0.05). EGCG treatment caused obvious cell cycle arrest in G1 phase, significantly increased cell apoptosis, downregulated the expression of DNMT1 and upregulated the expressions of CHD5, p19Arf, p53 and p21Cip1 in KG-1 and THP-1 cells (P < 0.05).
Conclusion
EGCG reduces hypermethylation of CHD5 gene in KG-1 and THP-1 cells by downregulating DNMT1 to restore its expression, which results in upregulated expressions of p19Arf, p53 and p21Cip1 and induces cell apoptosis.
Keywords: epigallocatechin gallate, CHD5 gene, methylation, p19Arf-p53-p21Cip1 signaling pathway, acute myleoid leukemia
急性髓系白血病(AML)是一种异质性克隆血液恶性肿瘤,以髓样祖细胞不受控制的增殖为特征,以及分化成熟血细胞的能力受到抑制导致造血功能不全[1-2]。AML发生机制与表观遗传学改变有关,研究表明抑癌基因高甲基化与白血病的发生密切相关[3-4]。因此,降低抑癌基因高甲基化状态是白血病的一种新的治疗策略。
DNA甲基转移酶(DNMT)在DNA甲基化中起重要作用,DNMT抑制剂降低DNA链的高甲基化,恢复沉默的抑癌基因表达[5]。目前常用的抑制剂如5-aza-2'-脱氧胞苷等缺乏靶点特异性,机体稳定性差且对健康细胞具有高毒性[6]。与这些合成抑制剂相比,小分子天然产物具有毒性低,副作用小且经济等特性成为新兴的靶向表观遗传调节剂。
绿茶是一种风靡全球的天然饮品,研究显示绿茶主要的化学物质成分:表没食子儿茶素-3-没食子酸酯(EGCG),可通过诱导肿瘤细胞凋亡或阻断肿瘤细胞周期,抑制肿瘤细胞增殖[7-8]。EGCG与DNMT的催化部位中Glu1265、Arg1310、Arg1311和LYS1482四个氨基酸残基形成氢键,直接抑制DNMT功能,降低抑癌基因的甲基化状态,从而抑制肿瘤细胞的增殖[9-10]。Sheng等[11]发现EGCG通过降低DNMT逆转SCUBE2的甲基化,恢复SCUBE2的表达,抑制乳腺癌细胞增殖。因此,EGCG可能对抑癌基因甲基化起靶向调控作用。然而目前EGCG调控AML抑癌基因甲基化恢复其表达研究较少[12],抑癌基因甲基化影响细胞增殖的具体分子机制尚不清楚,EGCG是否可通过调节抑癌基因甲基化影响AML细胞增殖和凋亡也值得进一步探讨。
我们前期研究发现AML患者CHD5基因甲基化率高于正常对照组,且存在CHD5基因甲基化的病例CHD5 mRNA表达水平相应降低[13]。CHD5位于染色体1p36位点,被认为是一种潜在的肿瘤抑制基因,可调节p19Arf-p53-p21Cip1等抑制肿瘤增殖相关信号通路,诱导细胞凋亡[14]。研究表明在多种人类癌症中存在CHD5启动子高甲基化,导致表观遗传沉默[15-16]。本课题组前期证实CHD5基因甲基化的AML患者外周血p19Arf,p53,p21Cip1 mRNA和蛋白均呈现低表达。在本研究的初步实验中,发现KG-1、THP-1细胞的CHD5基因存在高甲基化,但EGCG是否可调节CHD5基因甲基化恢复CHD5表达未见报道,p19Arf-p53-p21Cip1信号通路介导CHD5基因甲基化影响AML细胞增殖凋亡的作用尚不明确。因此我们拟在前期研究基础上,以KG-1、THP-1细胞为研究对象,明确EGCG是否诱导CHD5基因启动子去甲基化进而影响CHD5表达,通过调控p19Arf-p53-p21Cip1信号通路从而促进细胞凋亡。本研究旨在分析EGCG是否可通过调节CHD5甲基化影响细胞增殖和凋亡,探讨EGCG在AML中的相关表观遗传机制,为EGCG应用于AML治疗提供进一步的科学依据。
1. 材料和方法
1.1. 细胞培养
KG-1、THP-1细胞购自中科院典型培养物保藏委员会上海细胞库,培养于含10%胎牛血清的RPMI 1640完全培养基中,培养温度为37 ℃,CO2浓度为5%,传代间隔时间为36~48 h,细胞中常规培养,取对数生长期细胞进行实验。
1.2. MSP法检测CHD5基因甲基化
1×105KG-1、THP-1细胞接种到12孔板,用25、50、75、100、150 μg/mL EGCG处理48 h,设空白对照组。细胞基因组DNA由TIANamp Genomic DNA kit(TIANGEN)提取,使用NanoDrop微量分光光度计(Thermo)检测基因组DNA纯度。亚硫酸盐修饰按照EZ DNA Methylation-Gold Kit(ZYMO)操作说明进行,修饰并纯化后的基因组DNA为模板上PCR仪扩增。CHD5甲基化上游引物为5'-CGGGGAGTAGGTTAAGGC-3',下游引物为5'-TACCAACCTTAACCCGTACG-3';CHD5未甲基化上游引物为5'-TGTGGGGAGTAGG TTAAGGT-3',下游引物为5'-ATACCAACCTTAACC CATACAC-3'。PCR体系设置为50 μL;反应条件为:95 ℃预变性3 min,94 ℃ 30 s,54 ℃ 30 s,72 ℃ 1 min,40个循环,然后72 ℃ 7 min。配制1.5%琼脂糖凝胶,上样后放置电泳槽检测。
1.3. MTT法检测细胞增殖
制备1×105/mL的KG-1、THP-1细胞悬液,取100 μL/孔接种到96孔板,分别加入25、50、75、100、150 μg/mL EGCG(TCI),并设空白对照组及调零组,每组设5个复孔。于37 ℃培养箱孵育细胞48 h后,取5 mg/mL的MTT溶液20 μL加入每孔,继续37 ℃培养4 h。1000 r/min离心10 min,吸弃上清,PBS清洗1次,加入150 μL DMSO,放置摇床上温和震荡10 min,待甲瓒结晶溶解后,上酶标仪(Thermo)于570 nm波长处测定光密度值(A570 nm),根据下列公式计算细胞增殖率:细胞增殖率=(实验组A570 nm-调零组A570 nm)/(空白对照组A570 nm-调零组A570 nm)×100%。实验重复2次。
1.4. 流式细胞术检测EGCG处理后KG-1、THP-1细胞周期和细胞凋亡
1×105 KG-1、THP-1细胞接种到6孔板,用25、50、75、100、150 μg/mL EGCG处理48 h,1200 r/min离心5 min收集细胞,冰冷PBS洗涤并重悬2次,加入70%乙醇在4 ℃下冷却过夜,1500 r/min离心5 min,小心吸弃乙醇,预冷PBS清洗2次。细胞重悬在PI染色液(KeyGEN)离心5 min(20 mg/mL PI,0.2 mg/mL RNase),37 ℃孵育30 min,过筛网后采用流式细胞仪分析细胞周期分布(BD)。按异硫氰酸荧光素(FITC)细胞凋亡检测试剂盒(KeyGEN)操作说明检测各孔凋亡情况,FITC标记的Annexin V和PI室温下固定细胞15 min,上机检测细胞早期和晚期凋亡率。
1.5. RT-qPCR检测DNMT1、CHD5、p19Arf、p53、p21Cip1 mRNA表达水平
1×105 KG-1、THP-1细胞接种到12孔板,用25、50、75、100、150 μg/mL EGCG处理48 h,设空白对照组。1200 r/min离心5 min收集细胞,冰冷PBS洗涤,应用TRIzol试剂(Takara)提取总RNA,离心样品加入氯仿,转移上清并加入预冷异丙醇。16 000 r/min离心10 min,弃上清加入冰冷乙醇清洗2次,离心后小心吸弃上清,产物以焦碳酸二乙酯水溶解。RNA定量采用NanoDrop微量分光光度计。取1 μg RNA用于合成cDNA,按PrimeScript RT Reagent Kit with gDNA Eraser(Takara)操作说明进行。qPCR按照SYBR Green qPCR Master Mix(Thermo)操作说明进行,所用引物见表 1。扩增循环数设为40个,变性温度设为95 ℃,时间30 s,各基因退火温度见表 2,时间为30 s,延伸温度和时间分别为72 ℃,30 s。相对mRNA表达水平采用2-ΔΔCt计算,β-actin作为参照基因。所有实验均重复至少3次。
1.
RT-qPCR使用的引物
Primers used for gene expression analysis (RT-qPCR)
| Primer | Sequence | Annealing temp (℃) |
| DNMT1-F | 5'-AAGAATGGTGTTGTCTACCGAC-3' | 55 |
| DNMT1-R | 5'-CATCCAGGTTGCTCCCCTTGGATGG-3' | 55 |
| CHD5-F | 5'-TCCGCAAGCAGGTCAACTACA-3' | 56 |
| CHD5-R | 5'-CTCATCCTCAGAGCCAATGGAA-3' | 56 |
| p19Arf-F | 5'-CCATGTGGACCTGTCACTGT-3' | 60 |
| p19Arf-R | 5'-AAGATGTAGAGCGGGCCTTT-3' | 60 |
| p53-F | 5'-CTGGGGAGTCTTGAGGGACC-3' | 59 |
| p53-R | 5'-CAGGTTGTCTAAATTCCTAG-3' | 59 |
| p21Cip1-F | 5'-CTTGTGGAGCCGGAGCT-3' | 58 |
| p21Cip1-R | 5'-TGGTGTCTCGGTGACAAAGT-3' | 58 |
| β-actin-F | 5'-TGGCACCCAGCACAATGAA-3' | 60 |
| β-actin-R | 5'-CTAAGTCATAGTCCGCCTAGAAGCA-3' | 60 |
1.6. Western blot检测DNMT1、CHD5、p19Arf、p53、p21Cip1蛋白表达
2×105 KG-1、THP-1细胞接种到6孔板,用25、50、75、100、150 μg/mL EGCG处理24 h,并设空白对照组。收集细胞PBS清洗2次后采用蛋白提取试剂盒(Sangon)提取蛋白,蛋白浓度采用BCA定量试剂盒测定(Sangon)。取50 μg蛋白质经SDS-PAGE凝胶电泳,随后转移至PVDF膜(Santa Cruz),5%脱脂奶粉封闭2 h,加入DNMT1、CHD5、p19Arf、p53、p21Cip1一抗(Santa Cruz)于4 ℃温度条件下振荡孵育过夜,膜经清洗3次后加入二抗(Santa Cruz,)室温孵育2 h,洗膜3次采用ECL发光试剂盒显色(Sangon),曝光X胶片,显影并定影,扫描胶片,光灰度值采用Image J软件分析。
1.7. 统计分析
数值资料以均数±标准差表示,采用SPSS软件进行单因素方差分析,P < 0.05表示差异有统计学意义。
2. 结果
2.1. KG-1、THP-1细胞CHD5启动子甲基化状态
MSP检测结果发现在KG-1、THP-1细胞中,CHD5基因启动子表现出明显的甲基化(图 1A),以正常人血液DNA作为非甲基化阳性对照,通用甲基化DNA作为甲基化阳性对照,以H2O为阴性对照。
1.
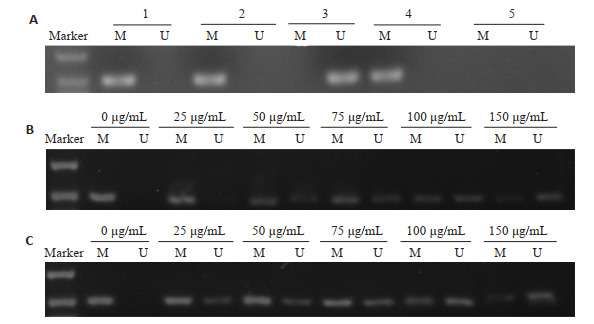
KG-1、THP-1细胞CHD5基因的甲基化状态及EGCG的去甲基化作用
Methylation status of CHD5 gene in KG-1 and THP-1 cells and CHD5 gene demethylation induced by EGCG. M: Methylation-specific PCR. U: Unmethylation-specific PCR. A: Methylation status of CHD5 gene in KG-1 and THP-1 cells. Line 1: Methylation status of CHD5 gene in KG-1 cells. Line 2: Methylation status of CHD5 gene in THP-1 cells. Line 3: Unmethylation positive control. Line 4: Methylation positive control. Line 5: Negative control; B: Demethylation in KG-1 cells; C: Demethylation in THP-1 cells.
2.2. EGCG逆转了KG-1、THP-1细胞CHD5基因的高甲基化状态
为明确EGCG是否对KG-1、THP-1细胞的CHD5基因产生去甲基化作用,我们使用25、50、75、100、150 μg/mL EGCG处理细胞。结果发现(图 1B、C),与未处理EGCG对照组比较,CHD5基因的甲基化特异性条带逐渐减弱,呈现剂量依赖性;而非甲基化特异性条带则呈现剂量性增强趋势。
2.3. EGCG对KG-1、THP-1细胞的增殖抑制作用
本实验使用25、50、75、100、150 μg/mL EGCG处理KG-1、THP-1细胞,培育48 h后,酶标仪检测EGCG对KG-1、THP-1细胞是否有增殖抑制作用。结果表明(图 2),25、50、75、100、150 μg/mL EGCG浓度组与未处理的对照组比较,KG-1、THP-1细胞增殖率均呈现剂量依赖性显著降低(P < 0.05)。
2.
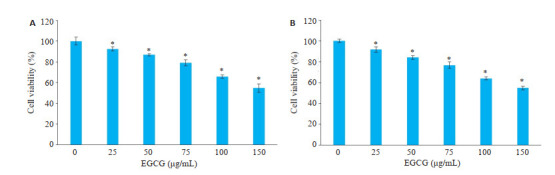
EGCG对KG-1、THP-1细胞的增殖抑制作用
Inhibition of EGCG on proliferation of KG-1 (A) and THP-1 (B) cells (n=5). *P < 0.05 vs control group.
2.4. EGCG对KG-1、THP-1细胞周期和凋亡的影响
不同浓度EGCG处理KG-1、THP-1细胞48 h后,加入PI单染或Annexin V和PI双染检测细胞周期分布的变化及对细胞凋亡率的改变。如图 3所示,EGCG处理导致细胞阻滞在G1期,S期或G2期细胞比率则呈现下降趋势。Annexin V和PI双染结果显示(图 4):与对照组比较,EGCG诱导KG-1、THP-1细胞凋亡增加(P < 0.05),且表现出剂量依赖性。
3.
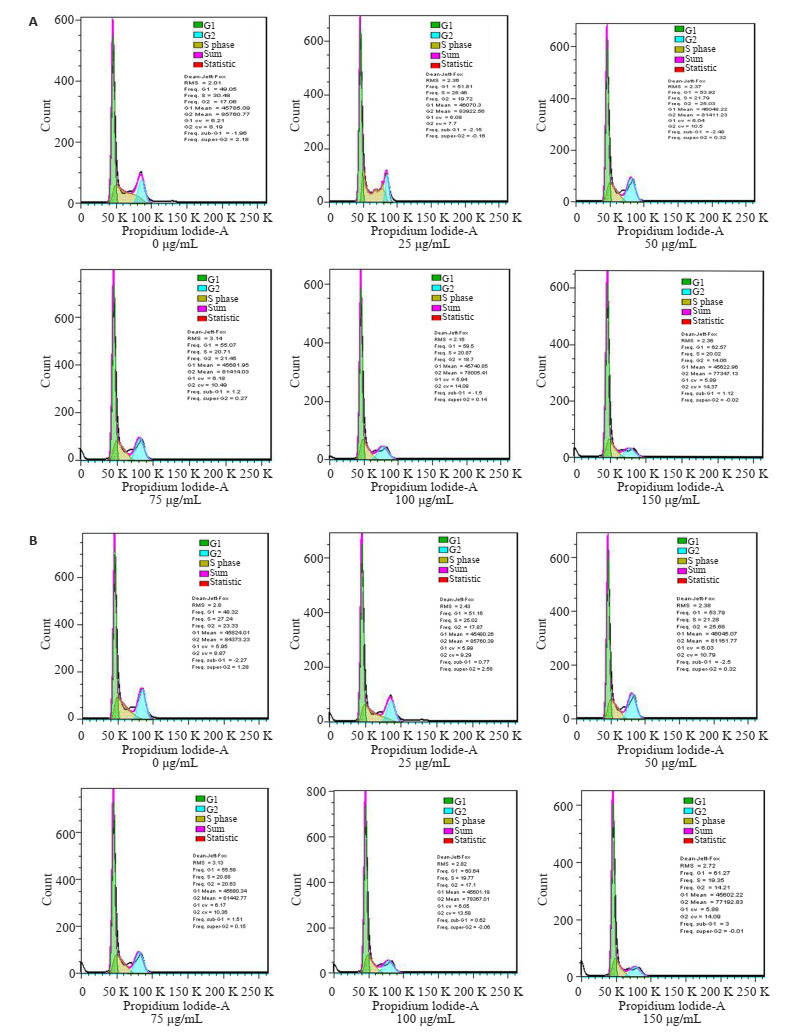
EGCG对KG-1、THP-1细胞周期的作用
Effect of EGCG on cell cycle of KG-1 (A) and THP-1 (B) cells.
4.
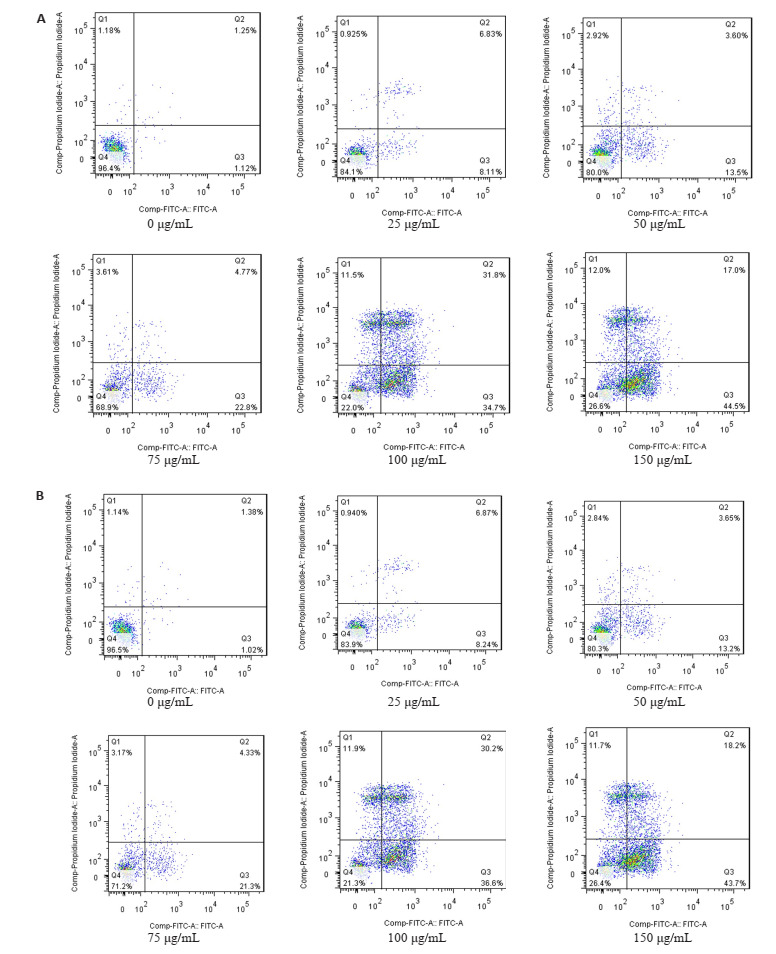
EGCG对KG-1、THP-1细胞凋亡的作用
Effect of EGCG on apoptosis of KG-1 (A) and THP-1 (B) cells.
2.5. EGCG对KG-1、THP-1细胞CHD5、DNMT、p19Arf、p53、p21Cip1 mRNA的表达影响
如图 5所示,EGCG处理KG-1、THP-1细胞后,DNMT1 mRNA表达程度呈现剂量依赖性下降趋势,25、50、75、100、150 μg/mL EGCG浓度组DNMT1 mRNA表达量与EGCG未处理的细胞相比有显著差异(P < 0.05);CHD5、p19Arf、p53、p21Cip1的mRNA水平的表达水平增加,具有剂量依赖型,与EGCG未处理的细胞相比,CHD5、p19Arf、p53、p21Cip1的mRNA表达量呈现出统计学差异(P < 0.05)。
5.
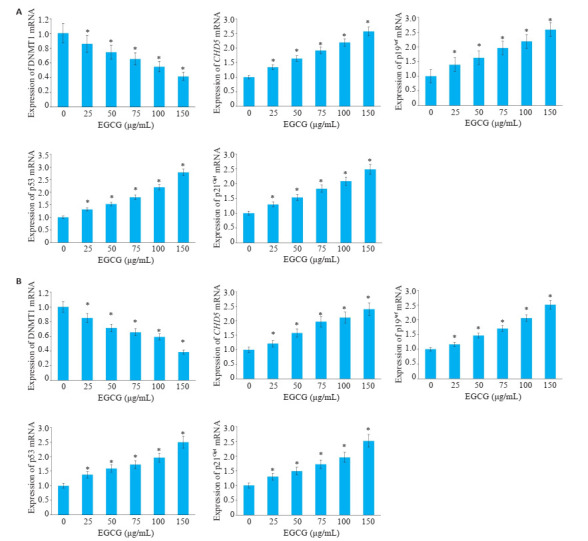
EGCG对KG-1、THP-1细胞DNMT1,CHD5,p19Arf,p53,p21Cip1 mRNA表达的作用
mRNA expressions of DNMT1, CHD5, p19Arf, p53 and p21Cip1 in KG-1 (A) and THP-1 (B) cells treated with EGCG. *P < 0.05 vs control group (n=3).
2.6. EGCG对KG-1、THP-1细胞CHD5、DNMT、p19Arf、p53、p21Cip1蛋白表达的影响
不同浓度EGCG(25、50、75、100、150 μg/mL)处理KG-1、THP-1细胞24 h后,检测蛋白表达水平,结果显示(图 6)随着EGCG浓度升高,DNMT1蛋白表达水平呈现降低趋势;CHD5、p19Arf、p53、p21Cip1蛋白表达水平则呈现剂量依赖性增加,与未处理对照组比较,EGCG浓度组出现显著差异(P < 0.05)。
6.
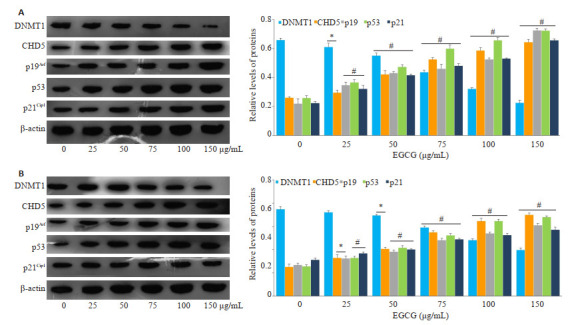
EGCG对KG-1、THP-1细胞DNMT1,CHD5,p19Arf,p53,p21Cip1蛋白表达的作用
Protein expressions of DNMT1, CHD5, p19Arf, p53 and p21Cip1 were detected in KG-1 (A) and THP-1 (B) cells treated with EGCG. *P < 0.05 vs control group (n=3), #P < 0.01 vs control group (n=3).
3. 讨论
近年来,大量研究表明表观遗传修饰在癌症生理学和病因学中起着至关重要的作用[17-18]。表观遗传修饰指独立于基因序列改变调节功能基因的表达,包括DNA甲基化、RNA和组蛋白的修饰。其中,DNA甲基化最为常见,现已发现多种抑癌基因的异常DNA甲基导致基因沉默是癌症的形成和进展的重要因素之一。如何等[19]在肝癌中发现SOCS3启动子区异常甲基化修饰导致SOCS3转录抑制而造成表达水平下降,进一步影响细胞凋亡。DNA甲基化改变是可逆的,可通过去甲基化恢复沉默的抑癌基因表达,一些合成表观遗传修饰剂已经通过癌症治疗的临床试验。如DNMT抑制剂氮杂替丁和癸二胺(胞苷类似物)已被美国食品药品监督管理局批准用于慢性粒细胞白血病和骨髓增生异常综合征治疗[20]。然而,合成DNMT抑制剂缺乏靶点特异性,机体稳定性差以及对健康细胞具有较高的毒性。因此,具有抗肿瘤作用的天然小分子产物引起广泛的研究兴趣。
目前已有多种天然小分子产物被证实具有表观遗传调节因子作用,可用于癌症的预防和治疗,如芹菜素、叶酸、染料木素、番茄红素、杨梅素、柚皮素、韧皮素、原儿茶酸、槲皮素、迷迭香酸、芥子酸和萝卜硫素等,具备口服给药,副作用少,且药效维持时间长等特点[21]。其中绿茶的主要成分EGCG被证实通过抑制DNMT功能,减少多种抑癌基因甲基化。如Morris等[22]用EGCG处理结肠癌细胞后降低DNMT表达活性,降低了RXRα启动子甲基化并恢复RXRα基因的表达;Sheng等[11]将EGCG加入MCF-7和MDA-MB-231细胞孵育后检测DNMT活性,与EGCG未处理组活性比较显著降低;Assumpção等[23]发现EGCG与DNMT1的MTAse结构域相互作用,使BT-549细胞中RASSF1A的启动子区脱甲基化。前期研究结果表明AML患者外周血CHD5甲基化率较高,CHD5 mRNA表达水平下调[12],因此推测CHD5甲基化可能导致了CHD5表达的沉默。本研究采用EGCG处理人急性髓系白血病细胞系,探讨EGCG调节CHD5基因甲基化的表观遗传作用。MSP实验结果表明EGCG可降低KG-1、THP-1细胞CHD5基因启动子甲基化,并呈现出剂量依赖性,实时定量RT-PCR结果也显示KG-1、THP-1细胞CHD5 mRNA呈现低表达,EGCG处理后可恢复CHD5 mRNA表达,降低DNMT1 mRNA的表达,提示EGCG可降低DNMT逆转CHD5基因高甲基化,恢复沉默CHD5基因的表达。
除了抑制DNMT活性外,EGCG还被证实具有诱导凋亡、抑制表皮生长因子受体介导的信号通路、破坏血管生成、抑制端粒酶和DNMT以及减少氧化应激水平等作用[24-26]。本研究采用MTT法证实EGCG对KG-1、THP-1细胞有明显的增殖抑制作用,尤其在75、100、150 μg/mL浓度时,细胞活力受到明显抑制。Zhang等[27]发现EGCG可诱导肺癌A549细胞周期停滞于G1期,G2和S期细胞比率明显低于对照组;Zan等[28]也证实EGCG对乳腺癌MCF-7细胞的增殖活力抑制作用有明显的时间依赖性和剂量依赖性,流式细胞术分析表明EGCG在G2/M期诱导细胞凋亡,破坏细胞周期进程。本研究也发现,EGCG可阻滞KG-1、THP-1细胞生长周期于G1期,剂量依赖性地促进细胞早期凋亡。CHD5,编码一个染色质重塑构蛋白,已被报道为多种人类癌症中一个新的肿瘤抑制基因,如肾细胞癌、胃癌和乳腺癌等[29-32]。研究表明,CHD5促进细胞凋亡可通过调控p19Arf-p53-p21Cip1信号通路对细胞增殖起抑制作用。前期研究结果也表明AML患者体内p19Arf,p53,p21Cip1 mRNA和蛋白水平较对照组降低,进一步检测发现,CHD5基因启动子在AML患者与未甲基化AML患者和对照组比较甲基化率较高,存在CHD5基因甲基化的AML患者p19Arf,p53,p21Cip1 mRNA和蛋白均低表达。本研究在前期基础上探寻EGCG调控CHD5甲基化及促进细胞凋亡的分子机制,将不同浓度EGCG作用于KG-1、THP-1细胞,发现EGCG可对CHD5基因产生去甲基化作用,恢复CHD5 mRNA和蛋白表达,同时也增加p19Arf,p53,p21Cip1 mRNA和蛋白表达,促进细胞凋亡,进一步证实EGCG可作为治疗AML的靶向表观遗传药物。
本研究首次报道EGCG可以通过降低DNMT的表达逆转KG-1、THP-1细胞DNA甲基化,重新恢复CHD5基因的表达,进而激活p19Arf-p53-p21Cip1信号通路,促进白血病细胞凋亡。我们的研究表明对EGCG可作为靶向功能基因的表观遗传修饰治疗药物,为EGCG治疗急性髓系白血病的临床应用提供参考。然而,EGCG作为表观遗传修饰药物的具体调节分子机制以及相应的体外和临床试验需进一步探讨。综上所述,本研究揭示了EGCG对CHD5基因产生去甲基化作用,通过调控p19Arf-p53-p21Cip1信号通路促进细胞凋亡,为EGCG作为新型表观遗传药物提供科学依据。
Biographies
吴明彩,博士,副教授,E-mail: williyia@wnmc.edu.cn
蒋明,主治医师,E-mail: 954967767@qq.com
Funding Statement
国家自然科学基金(31971199);安徽高校自然科学研究项目重点项目(KJ2018A0262);安徽省高等学校省级质量工程教学研究重点项目(2019jyxm025);皖南医学院博士科研启动基金项目(WYRCQD2018001);皖南医学院青年骨干人才项目(wyqnyx201906);安徽省大学生创新训练项目(S201910368088,201810368063,201810368064)
Supported by National Natural Science Foundation of China (31971199)
Contributor Information
吴 明彩 (Mingcai WU), Email: williyia@wnmc.edu.cn.
蒋 明 (Ming JIANG), Email: 954967767@qq.com.
章 尧 (Yao ZHANG), Email: zhangyao@ahedu.gov.
References
- 1.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374(23):2209–21. doi: 10.1056/NEJMoa1516192. [Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic classification and prognosis in acute myeloid leukemia[J]. N Engl J Med, 2016, 374(23): 2209-21.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen HL, Liu T, Liu J, et al. Circ-ANAPC7 is upregulated in acute myeloid leukemia and appears to target the MiR-181 family. Cell Physiol Biochem. 2018;47(5):1998–2007. doi: 10.1159/000491468. [Chen HL, Liu T, Liu J, et al. Circ-ANAPC7 is upregulated in acute myeloid leukemia and appears to target the MiR-181 family[J]. Cell Physiol Biochem, 2018, 47(5): 1998-2007.] [DOI] [PubMed] [Google Scholar]
- 3.Younesian S, Shahkarami S, Ghaffari P, et al. DNA hypermethylation of tumor suppressor genes RASSF6 and RASSF10 as independent prognostic factors in adult acute lymphoblastic leukemia. Leuk Res. 2017;61:33–8. doi: 10.1016/j.leukres.2017.08.016. [Younesian S, Shahkarami S, Ghaffari P, et al. DNA hypermethylation of tumor suppressor genes RASSF6 and RASSF10 as independent prognostic factors in adult acute lymphoblastic leukemia[J]. Leuk Res, 2017, 61: 33-8.] [DOI] [PubMed] [Google Scholar]
- 4.Zhou JD, Zhang TJ, Li XX, et al. Methylation-independent CHFR expression is a potential biomarker affecting prognosis in acute myeloid leukemia. J Cell Physiol. 2018;233(6):4707–14. doi: 10.1002/jcp.26253. [Zhou JD, Zhang TJ, Li XX, et al. Methylation-independent CHFR expression is a potential biomarker affecting prognosis in acute myeloid leukemia[J]. J Cell Physiol, 2018, 233(6): 4707-14.] [DOI] [PubMed] [Google Scholar]
- 5.Pathania R, Ramachandran S, Mariappan G, et al. Combined inhibition of DNMT and HDAC blocks the tumorigenicity of cancer stem-like cells and attenuates mammary tumor growth. Cancer Res. 2016;76(11):3224–35. doi: 10.1158/0008-5472.CAN-15-2249. [Pathania R, Ramachandran S, Mariappan G, et al. Combined inhibition of DNMT and HDAC blocks the tumorigenicity of cancer stem-like cells and attenuates mammary tumor growth[J]. Cancer Res, 2016, 76(11): 3224-35.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tyagi T, Treas JN, Mahalingaiah PK, et al. Potentiation of growth inhibition and epigenetic modulation by combination of green tea polyphenol and 5-aza-2'deoxycytidine in human breast cancer cells. Breast Cancer Res Treat. 2015;149(3):655–68. doi: 10.1007/s10549-015-3295-5. [Tyagi T, Treas JN, Mahalingaiah PK, et al. Potentiation of growth inhibition and epigenetic modulation by combination of green tea polyphenol and 5-aza-2'deoxycytidine in human breast cancer cells [J]. Breast Cancer Res Treat, 2015, 149(3): 655-68.] [DOI] [PubMed] [Google Scholar]
- 7.Braicu C, Gherman CD, Irimie A, et al. Epigallocatechin-3-gallate (EGCG) inhibits cell proliferation and migratory behaviour of triple negative breast cancer cells. J Nanosci Nanotechnol. 2013;13(1):632–7. doi: 10.1166/jnn.2013.6882. [Braicu C, Gherman CD, Irimie A, et al. Epigallocatechin-3-gallate (EGCG) inhibits cell proliferation and migratory behaviour of triple negative breast cancer cells[J]. J Nanosci Nanotechnol, 2013, 13(1): 632-7.] [DOI] [PubMed] [Google Scholar]
- 8.李 彬彬, 万 郑, 孔 霞, et al. 表没食子儿茶素没食子酸酯通过调控P53/ miR-34a抑制鼻咽癌细胞的增殖. 中国病理生理杂志. 2015;31(9):1557–62. doi: 10.3969/j.issn.1000-4718.2015.09.004. [李彬彬, 万郑, 孔霞, 等.表没食子儿茶素没食子酸酯通过调控P53/ miR-34a抑制鼻咽癌细胞的增殖[J].中国病理生理杂志, 2015, 31 (9): 1557-62.] [DOI] [Google Scholar]
- 9.Gan RY, Li HB, Sui ZQ, et al. Absorption, metabolism, anti-cancer effect and molecular targets of epigallocatechin gallate (EGCG): An updated review. Crit Rev Food Sci Nutr. 2018;58(6):924–41. doi: 10.1080/10408398.2016.1231168. [Gan RY, Li HB, Sui ZQ, et al. Absorption, metabolism, anti-cancer effect and molecular targets of epigallocatechin gallate (EGCG): An updated review[J]. Crit Rev Food Sci Nutr, 2018, 58(6): 924-41.] [DOI] [PubMed] [Google Scholar]
- 10.Mereles D, Hunstein W. Epigallocatechin-3-gallate (EGCG) for Clinical Trials: More pitfalls than promises? Int J Mol Sci. 2011;12(9):5592–603. doi: 10.3390/ijms12095592. [Mereles D, Hunstein W. Epigallocatechin-3-gallate (EGCG) for Clinical Trials: More pitfalls than promises[J]? Int J Mol Sci, 2011, 12(9): 5592-603.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheng J, Shi WL, Guo H, et al. The inhibitory effect of (-)- Epigallocatechin-3-gallate on breast cancer progression via reducing SCUBE2 methylation and DNMT activity. Molecules. 2019;24(16):E2899. doi: 10.3390/molecules24162899. [Sheng J, Shi WL, Guo H, et al. The inhibitory effect of (-)- Epigallocatechin-3-gallate on breast cancer progression via reducing SCUBE2 methylation and DNMT activity[J]. Molecules, 2019, 24(16): E2899.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu DS, Shen JZ, Yu AF, et al. Epigallocatechin-3-gallate and trichostatin a synergistically inhibit human lymphoma cell proliferation through epigenetic modification of p16INK4a. Oncol Rep. 2013;30(6):2969–75. doi: 10.3892/or.2013.2734. [Wu DS, Shen JZ, Yu AF, et al. Epigallocatechin-3-gallate and trichostatin a synergistically inhibit human lymphoma cell proliferation through epigenetic modification of p16INK4a[J]. Oncol Rep, 2013, 30(6): 2969-75.] [DOI] [PubMed] [Google Scholar]
- 13.吴 明彩, 蒋 明, 董 婷, et al. CHD5基因启动子甲基化调控p19Arf/p53/ p21Cip1信号通路促进急性髓系白血病发病的研究. 中国实验血液学杂志. 2019;27(4):1001–7. doi: 10.19746/j.cnki.issn.1009-2137.2019.04.002. [吴明彩, 蒋明, 董婷, 等. CHD5基因启动子甲基化调控p19Arf/p53/ p21Cip1信号通路促进急性髓系白血病发病的研究[J].中国实验血液学杂志, 2019, 27(4): 1001-7.] [DOI] [PubMed] [Google Scholar]
- 14.Bagchi A, Papazoglu C, Wu Y, et al. CHD5 is a tumor suppressor at human 1p36. Cell. 2007;128(3):459–75. doi: 10.1016/j.cell.2006.11.052. [Bagchi A, Papazoglu C, Wu Y, et al. CHD5 is a tumor suppressor at human 1p36[J]. Cell, 2007, 128(3): 459-75.] [DOI] [PubMed] [Google Scholar]
- 15.Mulero-Navarro S, Esteller M. Chromatin remodeling factor CHD5 is silenced by promoter CpG island hypermethylation in human cancer. Epigenetics. 2008;3(4):210–5. doi: 10.4161/epi.3.4.6610. [Mulero-Navarro S, Esteller M. Chromatin remodeling factor CHD5 is silenced by promoter CpG island hypermethylation in human cancer[J]. Epigenetics, 2008, 3(4): 210-5.] [DOI] [PubMed] [Google Scholar]
- 16.Ma ZL, Song JL, Liu SM, et al. Decreased expression of the CHD5 gene and its clinicopathological significance in breast cancer: Correlation with aberrant DNA methylation. Oncol Lett. 2016;12(5):4021–6. doi: 10.3892/ol.2016.5147. [Ma ZL, Song JL, Liu SM, et al. Decreased expression of the CHD5 gene and its clinicopathological significance in breast cancer: Correlation with aberrant DNA methylation[J]. Oncol Lett, 2016, 12 (5): 4021-6.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nebbioso A, Tambaro FP, Dell'Aversana C, et al. Cancer epigenetics: Moving forward. PLoS Genet. 2018;14(6):e1007362. doi: 10.1371/journal.pgen.1007362. [Nebbioso A, Tambaro FP, Dell'Aversana C, et al. Cancer epigenetics: Moving forward[J]. PLoS Genet, 2018, 14(6): e1007362.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukuoka K, Mamatjan Y, Tatevossian R, et al. Clinical impact of combined epigenetic and molecular analysis of pediatric low grade gliomas. Neuro Oncol. 2020:noaa077. doi: 10.1093/neuonc/noaa077. [Fukuoka K, Mamatjan Y, Tatevossian R, et al. Clinical impact of combined epigenetic and molecular analysis of pediatric low grade gliomas[J]. Neuro Oncol, 2020: noaa077.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.何 靖炀, 刘 秋英, 魏 玲, et al. BORIS通过表观修饰对人肝癌细胞SOCS3表达的调控. 四川大学学报:医学版. 2018;49(1):1–7. [何靖炀, 刘秋英, 魏玲, 等. BORIS通过表观修饰对人肝癌细胞SOCS3表达的调控[J].四川大学学报:医学版, 2018, 49(1): 1-7.] [Google Scholar]
- 20.Navada SC, Steinmann J, Lubbert M, et al. Clinical development of demethylating agents in hematology. J Clin Invest. 2014;124(1):40–6. doi: 10.1172/JCI69739. [Navada SC, Steinmann J, Lubbert M, et al. Clinical development of demethylating agents in hematology[J]. J Clin Invest, 2014, 124(1): 40-6.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schnekenburger M, Diederich M. Epigenetics offer new horizons for colorectal cancer prevention. Curr Colorectal Cancer Rep. 2012;8(1):66–81. doi: 10.1007/s11888-011-0116-z. [Schnekenburger M, Diederich M. Epigenetics offer new horizons for colorectal cancer prevention[J]. Curr Colorectal Cancer Rep, 2012, 8 (1): 66-81.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris J, Moseley VR, Cabang AB, et al. Reduction in promotor methylation utilizing EGCG (epigallocatechin-3-gallate) restores RXRα expression in human colon cancer cells. Oncotarget. 2016;7(23):35313–26. doi: 10.18632/oncotarget.9204. [Morris J, Moseley VR, Cabang AB, et al. Reduction in promotor methylation utilizing EGCG (epigallocatechin-3-gallate) restores RXRα expression in human colon cancer cells[J]. Oncotarget, 2016, 7(23): 35313-26.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Assumpção JHM, Takeda AAS, Sforcin JM, et al. Effects of propolis and phenolic acids on triple-negative breast cancer cell lines: Potential involvement of epigenetic mechanisms. Molecules. 2020;25(6):1289. doi: 10.3390/molecules25061289. [Assumpção JHM, Takeda AAS, Sforcin JM, et al. Effects of propolis and phenolic acids on triple-negative breast cancer cell lines: Potential involvement of epigenetic mechanisms[J]. Molecules, 2020, 25(6): 1289.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yokoyama M, Noguchi M, Nakao Y, et al. The tea polyphenol, (-)- epigallocatechin gallate effects on growth, apoptosis, and telomerase activity in cervical cell lines. Gynecol Oncol. 2004;92(1):197–204. doi: 10.1016/j.ygyno.2003.09.023. [Yokoyama M, Noguchi M, Nakao Y, et al. The tea polyphenol, (-)- epigallocatechin gallate effects on growth, apoptosis, and telomerase activity in cervical cell lines[J]. Gynecol Oncol, 2004, 92 (1): 197-204.] [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Man GCW, Chan TH, et al. A prodrug of green tea polyphenol (-)-epigallocatechin-3-gallate (Pro-EGCG) serves as a novel angiogenesis inhibitor in endometrial cancer. Cancer Lett. 2018;412:10–20. doi: 10.1016/j.canlet.2017.09.054. [Wang J, Man GCW, Chan TH, et al. A prodrug of green tea polyphenol (-)-epigallocatechin-3-gallate (Pro-EGCG) serves as a novel angiogenesis inhibitor in endometrial cancer[J]. Cancer Lett, 2018, 412: 10-20.] [DOI] [PubMed] [Google Scholar]
- 26.Crous-Masó J, Palomeras S, Relat J, et al. (-)-Epigallocatechin 3- gallate synthetic analogues inhibit fatty acid synthase and show anticancer activity in triple negative breast cancer. Molecules. 2018;23(5):1160. doi: 10.3390/molecules23051160. [Crous-Masó J, Palomeras S, Relat J, et al. (-)-Epigallocatechin 3- gallate synthetic analogues inhibit fatty acid synthase and show anticancer activity in triple negative breast cancer[J]. Molecules, 2018, 23(5): 1160.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang YW, Wang X, Han L, et al. Green tea polyphenol EGCG reverse cisplatin resistance of A549/DDP cell line through candidate genes demethylation. Biomed Pharmacother. 2015;69:285–90. doi: 10.1016/j.biopha.2014.12.016. [Zhang YW, Wang X, Han L, et al. Green tea polyphenol EGCG reverse cisplatin resistance of A549/DDP cell line through candidate genes demethylation[J]. Biomed Pharmacother, 2015, 69: 285-90.] [DOI] [PubMed] [Google Scholar]
- 28.Zan L, Chen Q, Zhang L, et al. Epigallocatechin gallate (EGCG) suppresses growth and tumorigenicity in breast cancer cells by downregulation of miR-25. Bioengineered. 2019;10(1):374–82. doi: 10.1080/21655979.2019.1657327. [Zan L, Chen Q, Zhang L, et al. Epigallocatechin gallate (EGCG) suppresses growth and tumorigenicity in breast cancer cells by downregulation of miR-25[J]. Bioengineered, 2019, 10(1): 374-82.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Du ZF, Li LL, Huang X, et al. The epigenetic modifier CHD5 functions as a novel tumor suppressor for renal cell carcinoma and is predominantly inactivated by promoter CpG methylation. Oncotarget. 2016;7(16):21618–30. doi: 10.18632/oncotarget.7822. [Du ZF, Li LL, Huang X, et al. The epigenetic modifier CHD5 functions as a novel tumor suppressor for renal cell carcinoma and is predominantly inactivated by promoter CpG methylation[J]. Oncotarget, 2016, 7(16): 21618-30.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu GR, Zhu HX, Zhang MH, et al. Histone deacetylase 3 is associated with gastric cancer cell growth via the miR-454-mediated targeting of CHD5. Int J Mol Med. 2018;41(1):155–63. doi: 10.3892/ijmm.2017.3225. [Xu GR, Zhu HX, Zhang MH, et al. Histone deacetylase 3 is associated with gastric cancer cell growth via the miR-454-mediated targeting of CHD5[J]. Int J Mol Med, 2018, 41(1): 155-63.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo XR, Xiong X, Shao Q, et al. The tumor suppressor interferon regulatory factor 8 inhibits β-catenin signaling in breast cancers, but is frequently silenced by promoter methylation. Oncotarget. 2017;8(30):48875–88. doi: 10.18632/oncotarget.16511. [Luo XR, Xiong X, Shao Q, et al. The tumor suppressor interferon regulatory factor 8 inhibits β-catenin signaling in breast cancers, but is frequently silenced by promoter methylation[J]. Oncotarget, 2017, 8(30): 48875-88.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hashimoto T, Kurokawa Y, Wada N, et al. Clinical significance of chromatin remodeling factor CHD5 expression in gastric cancer. Oncol Lett. 2020;19(1):1066–73. doi: 10.3892/ol.2019.11138. [Hashimoto T, Kurokawa Y, Wada N, et al. Clinical significance of chromatin remodeling factor CHD5 expression in gastric cancer[J]. Oncol Lett, 2020, 19(1): 1066-73.] [DOI] [PMC free article] [PubMed] [Google Scholar]


