Abstract
目的
探讨LINC01106在结直肠癌中的表达及其对结直肠癌细胞增殖及凋亡能力的影响。
方法
分析公共数据库结直肠癌患者的肿瘤组织及正常组织LINC01106表达水平及其对患者预后的影响;构建LINC01106敲减及过表达结直肠癌细胞株;采用CCK-8实验检测LINC01106敲减及过表达对结直肠癌细胞增殖能力的影响;流式细胞术检测LINC01106敲减对结直肠癌细胞凋亡水平的影响;免疫印迹检测LINC01106敲减对结直肠癌细胞p-STAT3、STAT3、Bcl-2蛋白表达的影响;建立裸鼠移植瘤模型观察LINC01106表达水平对结直肠肿瘤体内生长的影响,随机分为2组,9只/组。对照组:接种SW480细胞,敲减组:接种LINC01106敲减的SW480细胞。
结果
TCGA数据库分析结果显示结直肠癌肿瘤组织LINC01106的表达水平明显高于正常组织,且LINC01106的表达水平与结直肠癌患者预后相关,差异具有统计学意义(P < 0.05);敲减LINC01106可以明显抑制SW480结直肠癌细胞增殖,促进凋亡(P < 0.05),过表达LINC01106可以明显促进SW480结直肠癌细胞增殖;敲减LINC01106可以抑制p-STAT3和Bcl-2蛋白表达水平;下调LINC01106可以抑制裸鼠移植瘤体内生长。
结论
LINC01106在结直肠癌患者肿瘤组织中表达显著上调,且与患者预后相关。LINC01106可以通过STAT3/Bcl-2信号调控SW480结直肠癌细胞增殖及凋亡;LINC01106有望成为结直肠癌诊断及预后评估的潜在标志物。
Keywords: 结直肠癌, LINC01106, 信号转导与转录激活因子3, 增殖, 凋亡
Abstract
Objective
To investigate the expression of LINC01106 in colorectal cancer and its role in regulating the proliferation and apoptosis of colorectal cancer cells.
Methods
We analyzed the data of LINC01106 expression levels in tumor tissues and normal tissues of patients with colorectal cancer in TCGA database and explored the association of LINC01106 expression level with the prognosis of the patients. Colorectal cancer SW480 cell lines with LINC01106 knockdown or overexpression were established, and their proliferation and apoptosis relative to the parental cells were evaluated using CCK-8 assay and flow cytometry, respectively. The expressions of p-STAT3, STAT3, and Bcl-2 in the cells were detected by immunoblotting. Nude mouse models bearing xenografts of SW480 cells with LINC01106 knockdown or naïve SW480 cells were established to observe the effect of LINC01106 knockdown on the growth of SW480 cells in vivo.
Results
Analysis of the data from TCGA database showed that the expression level of LINC01106 was significantly higher in colorectal cancer tissues than in normal tissues, and LINC01106 expression level was significantly related to the prognosis of the patients (P < 0.05). Knockdown of LINC01106 significantly inhibited the proliferation and promoted apoptosis of SW480 cells (P < 0.05), while LINC01106 overexpression significantly promoted proliferation of the cells. LINC01106 knockdown in SW-480 cells obviously lowered the expressions of p- STAT3 and Bcl-2 and suppressed the growth of the xenograft in nude mice.
Conclusion
LINC01106 is significantly up-regulated in colorectal cancer tissue and is related to the prognosis of the patients. LINC01106 can regulate the proliferation and apoptosis of SW480 cells through STAT3/Bcl-2 signaling and may serve as a potential marker for the diagnosis and prognostic evaluation of colorectal cancer.
Keywords: colorectal cancer, LINC01106, STAT3, proliferation, apoptosis
结直肠癌是常见的消化道恶性肿瘤,据全球数据统计,结直肠癌发病率及死亡率均位居肿瘤疾病的第2位[1]。在中国,结直肠癌的发病率和死亡率均位居肿瘤疾病的第5位[2]。随着人们生活方式和饮食结构的改变,结直肠癌的发病呈上升趋势。由于早期筛查的缺乏,大多数结直肠癌患者在确诊时就已经处于中晚期并伴随转移。然而,转移和复发正是结直癌治疗失败的主要原因[3-4]。结直肠癌的治疗目前仍然是以手术切除为主,放化疗为辅[5-6]。因此,展开肿瘤诊断及预后标志物的研究,探索结直肠癌发病的分子机制是当前的热点。
长链非编码RNA(LncRNA)是调节型非编码RNA的成员之一,LncRNA一度被认为是与基因表达无关的噪音,近年来研究表明,LncRNA在基因表达过程中发挥复杂精确的调控功能,其异常表达能影响癌细胞的增殖、凋亡、迁移、侵袭、转移,从而促进癌症的发生发展[7-8]。研究表明,长链非编码RNA LINC01106介导多种肿瘤的生长和转移[9],但LINC01106在结直肠癌中的表达及作用尚未见报道。本研究通过分析公共数据库中LINC01106表达水平与结直肠癌预后的关系,同时在通过调控LINC01106的表达观察其对结直肠癌细胞生长及转移的影响。
1. 材料和方法
1.1. 材料
1.1.1. 细胞株
结直肠癌SW480细胞,由蚌埠医学院药学院生化药理实验室冻存。
1.1.2. 试剂
DMEM培养基、胰蛋白酶购自美国Gibco公司;胎牛血清(浙江四季青);STAT3、p-STAT3、Bcl-2抗体(Abcam);双染试剂盒(凯基);裸鼠(维通利华)。
1.2. 实验方法
1.2.1. TCGA数据库分析LINC01106在结直肠癌及正常组织中的表达水平
通过TCGA数据库分析275例结直肠癌组织样本和349例正常组织样本中LINC01106表达水平,并且对270例样本(分别为135例LINC01106高表达组和135例LINC01106低表达组)进行生存期分析。
1.2.2. 过表达及敲减LINC01106结直肠癌细胞株构建
将SW480结直肠癌细胞接种于6孔细胞培养板中,置37 ℃培养箱中培养24 h,当细胞密度达70%~80%时进行转染,使用Lipo2000转染试剂(Invitrogen)将LINC01106 siRNA、过表达质粒及各自相对应的阴性对照(上海吉玛制药技术有限公司)转染SW480结直肠癌细胞株,置于37 ℃培养箱中继续培养48 h,分别将肿瘤细胞分为过表达组、过表达对照组、敲减组和敲减对照组。
1.2.3. RT- qPCR检测RNA表达水平
使用TRAzol(Invitrogen)提取结直肠癌细胞RNA,紫外分光光度计测其浓度,按逆转录试剂盒说明书(Mir-XTM miRNA First-Strand Synthesis Kit, TAKARA)操作进行反转录将RNA反转录为cDNA,RT-qPCR试剂盒(SYBR Premix Ex TaqTM Ⅱ,TAKARA)进行定量PCR扩增,重复40个循环。相对表达量用2-ΔΔCT法计算,每个样本按照相同条件检测3次。
1.2.4. CCK-8检测过表达及敲减LINC01106对结直肠癌细胞增殖的影响
取SW480细胞转染操作24 h,以1×104/孔接种于96孔板中,分别设置过表达组、过表达对照组、敲减组和敲减对照组,每组设置5个复孔,置于37 ℃培养箱中培养48 h后,向每孔加入10 μL CCK-8溶液(Biosharp),注意避免产生气泡,置于37 ℃培养箱培养2 h后,酶标仪测定吸光度A450 nm,实验重复3次。综合实验结果分析细胞的活力。
1.2.5. Transwell检测过表达及敲减LINC01106对结直肠癌侵袭转移的影响
取结直肠癌细胞株SW480进行细胞侵袭试验。1:8稀释50 mg/L的Matrigel后,加50 μL稀释后的Matrigel包被Transwell小室底部膜,待其风干。胰酶消化细胞、离心后去上清,用无血清DMEM培养液重悬细胞至单细胞悬液,设置敲减组和对照组,以1×105/100 μL的细胞悬液接种到上室,下室加500 μL含10%FBS的DMEM培养液,48 h后取出小室,去培养液,用棉签擦除上室细胞,4%多聚甲醛固定细胞15 min,结晶紫染色10 min,PBS漂洗3次,镜下观察穿过小室的细胞数并拍照。
1.2.6. 流式细胞仪检测过表达及敲减对LINC01106细胞凋亡的影响
取对数期生长的SW480细胞,胰酶消化后,重悬细胞至单细胞悬液,调整细胞终浓度为1× 105 cells/mL,设置敲减组和对照组,接种于12孔板,每孔接种细胞1 mL。按照实验设计方案处理后,用预冷的PBS(0.01 mol/L,pH7.2)漂洗1次,胰酶消化,离心去上清后,PBS漂洗2次。收集细胞约1~5×104个。重悬细胞使其悬浮于300 μL Binding Buffer中,加入3 μL AnnexinV-FITC混匀后加入3 μL PI混匀,室温避光反应10 min。流式细胞仪检测各组细胞凋亡情况。
1.2.7. 免疫印迹检测蛋白表达水平
收集稳定表达细胞,使用细胞裂解液冰上裂解30 min,提取细胞总蛋白,BCA蛋白定量法检测细胞总蛋白浓度,用细胞裂解液稀释各组蛋白至等浓度。100 ℃煮沸5 min变性。等量上样于10% SDS-PAGE凝胶电泳后转膜,使用5%脱脂牛奶室温封闭2 h;PBS洗净,一抗室温孵育2 h后洗净,二抗室温孵育2 h,TBST洗膜,曝光,用Bio Imaging system(Gene Genius)对图像进行灰度扫描分析。
1.2.8. LINC01106敲减对结直肠癌细胞移植瘤体内生长的影响
从维通利华公司购买18只6周雌性裸鼠,在动物中心提前饲养1周左右,以使其适应新的生长环境并随机分为对照组及LINC01106敲减组。将处于生长对数期的SW480细胞及LINC01106敲减SW480细胞分别接种于裸鼠皮下,每只裸鼠皮下接种0.2 mL细胞悬液,对裸鼠进行标记以及初次称重。待皮下肿瘤长至100 mm3开始记录瘤体积。每天观察皮下移植瘤生长情况,每3 d检测裸鼠体质量及瘤体积,并做好记录。待裸鼠皮下移植瘤生长到一定体积(约1000 mm3),采用颈椎脱臼法处死裸鼠,解剖分离瘤体以及各脏器。部分瘤体及脏器用4%多聚甲醛固定备用,剩余瘤体及脏器冻存于液氮中。用医疗废物专用袋将裸鼠尸体装好,送至动物中心处理。
1.2.9. 免疫组化
取小鼠移植瘤组织,制作石蜡切片。切片常规脱蜡至水,Hydrogen Peroxide Block中孵育10 min,滴加Ultra V Block室温孵育5 min封闭非特异性背景染色。缓冲液清洗后,一抗孵育2 h,二抗孵育30 min。显色剂显色7 min,自来水充分冲洗,复染,脱水,透明,封片。
1.3. 统计学方法
数据分析采用SPSS 24.0统计软件,计量资料以均数±标准差表示,两组间均数的比较用独立样本t检验,P < 0.05为差异有统计学意义。
2. 结果
2.1. LINC01106在SW480结直肠癌组织中高表达并且与患者不良预后相关
通过分析TCGA数据库中临床样本数据,我们发现LINC01106在结直肠癌组织中的表达水平明显高于正常组织,组间差异具有统计学意义(P < 0.05,图 1)。进一步分析数据发现,LINC01106高表达结直肠癌患者预后较LINC01106低表达患者差。
1.
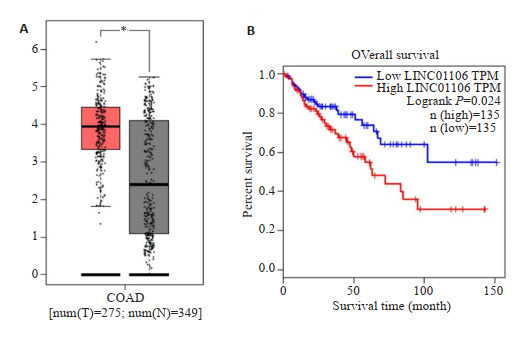
TCGA数据库分析LINC01106表达与预后的关系
Relationship between expression of LINC01106 and prognosis of patients with colorectal cancer. A: Expressions of LINC01106 in colorectal cancer tissue and normal tissue; B: Kaplan-Meier survival curves of patients with high and low expressions of LINC01106. *P < 0.05 for comparison between tumor group and normal group.
2.2. SW480结直肠癌细胞LINC01106过表达和敲减验证
RT- qPCR实验结果显示,LINC01106过表达组SW480结直肠细胞的LINC01106表达水平明显高于阴性对照组;LINC01106敲减组SW480结直肠癌细胞的LINC01106表达水平明显低于阴性对照组(图 2,P < 0.05)。
2.
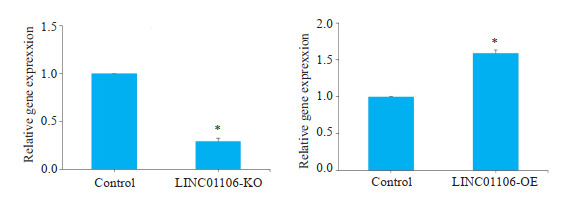
RT-qPCR验证LINC01106过表达及敲减
Verification of LINC01106 overexpression and knockdown in SW-480 cells by RT-qPCR. *P < 0.05 vs control.
2.3. 过表达和敲减LINC01106对结直肠癌细胞SW480增殖及凋亡的影响
CCK-8实验结果显示,在结直肠癌细胞SW480中敲减LINC01106后,SW480细胞增殖能力明显受到抑制;相反,LINC01106过表达组细胞增殖能力明显增强(图 3,P < 0.05)。PI和Annexin V双染实验结果显示,LINC01106敲减组SW480细胞凋亡增多(图 4,P < 0.05)。
3.
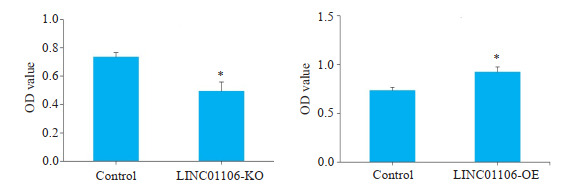
CCK-8检测LINC01106对SW480细胞增殖的影响
Effects of LINC01106 overexpression or knockdown on the proliferation of SW480 cells (assessed by CCK-8 assay at 48 h). *P < 0.05 vs control.
4.
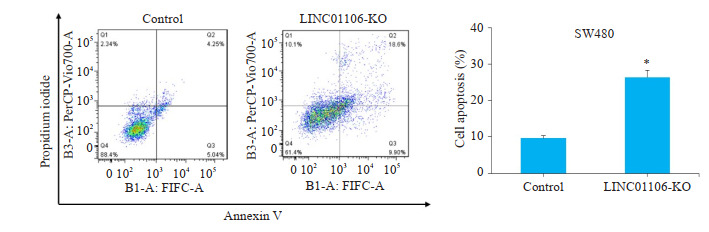
PI & Annexin V检测LINC01106对SW480细胞凋亡的影响
Effect of LINC01106 knockdown on proliferation of SW480 cells analyzed by flow cytometry. *P < 0.05 vs control.
2.4. LINC01106通过调控STAT3信号通路抑制SW480结直肠癌细胞凋亡
为了进一步探究LINC01106影响肿瘤细胞凋亡的分子机制,我们检测了STAT3及其下游分子Bcl-2蛋白表达水平。Western blot实验结果显示,在SW480细胞敲减LINC01106后,p-STAT3和Bcl-2蛋白表达水平明显下降(图 5,P < 0.05)。
5.
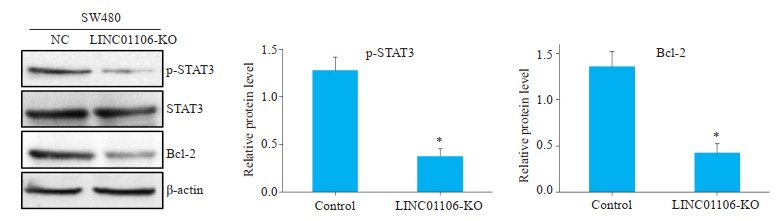
免疫印迹检测LINC01106对蛋白表达的影响
Effects of LINC01106 knockdown on expressions of STAT3, p-STAT3 and Bcl-2 in SW480 cells detected by Western blotting. *P < 0.05 vs control.
2.5. LINC01106敲减抑制SW480细胞体内生长
SW480细胞移植瘤实验结果显示,LINC01106敲减组小鼠移植瘤生长速度明显比阴性对照组慢,移植瘤体积增加速度明显减弱(图 6)。我们进一步对移植瘤免疫组化,结果表明LINC01106敲减后移植瘤Cleaved Caspase-3和Cleaved Caspase-9表达水平上升(图 7)。
6.
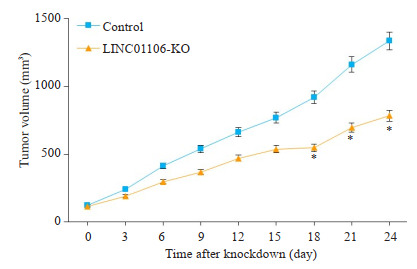
LINC01106敲减对移植瘤体内生长的影响
Effect of LINC01106 knockdown on growth of SW-480 cell xenografts in nude mice. *P < 0.05 vs control.
7.
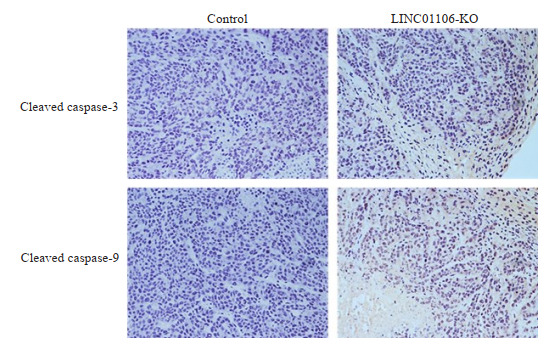
免疫组化检测移植瘤cleaved caspase-3和cleaved caspase-9表达情况
Immunohistochemistry for detecting the expression of cleaved caspase-3 and cleaved caspase-9 in xenografts of SW480 cells with LINC01106 knockdown (Original magnification: ×200).
3. 讨论
结直肠癌是临床常见的消化道恶性肿瘤之一,其发病机制跟生活、饮食以及遗传密切相关。越来越多的证据表明结直肠癌的发生发展涉及表观遗传学,而LncRNA在表观遗传中的作用使得其与结直肠癌进展有着千丝万缕的关联[7-8, 10]。LncRNA通过在转录前水平、转录水平以及转录后水平三个层次调控基因的表达,并且LncRNA的异常表达可以介导促癌基因的激活,从而促进肿瘤的生长[11]。随着分子生物学技术的进步,LncRNA功能的解密进展迅速。研究表明,LncRNA参与众多细胞生物学过程,包括转录调控、细胞周期调控、表观遗传调控、基因组印迹等[12]。LncRNA可以与DNA、RNA以及蛋白质发生相互作用,形成复合体,调控基因的表达过程[13-14];也可以通过与转录因子、染色质修饰因子、核糖蛋白结合影响生物学作用[15];甚至可以影响通过影响DNA甲基化在转录后水平调节mRNA的加工、剪接、翻译[16-17]。此外,LncRNA可作为促癌或抑癌基因在多种肿瘤的发生发展中发挥重要作用并与癌症患者的预后具有相关性,其中包括乳腺癌、结直肠癌、肺癌、卵巢癌等[18]。因此,LncRNA有望成为新的肿瘤诊断及预后标志物。
LncRNA不仅参与结直肠癌细胞的生长和转移,还与结直肠癌患者预后密切相关[19]。随着基因测序的发展,越来越多的LncRNA被证明参与结直肠癌的发生发展过程。研究表明,LINC00152在结直肠癌中呈高表达状态并且能明显促进结直肠癌细胞的增殖、侵袭及转移能力[20-21]。Wnt/β-catenin信号通路是结直肠癌经典的信号传导通路,该信号的激活也是促进结直肠癌进展的原因之一[22]。研究表明,LncRNA CCAL可以通过调控Wnt/β-catenin通路促进结直肠癌细胞生长[23]。除此之外LncRNA可以通过调控转录因子发挥促结直肠癌的作用,如LncRNA DUXAP10可以通过调控JAK-STAT的激活刺激结直肠癌的生长[24]。
本研究中,我们分析了LINC01106在结直肠癌和正常组织中的相对表达水平,发现LINC01106在结直肠癌组织中高表达,并且LINC01106的高表达与结直肠癌患者不良预后相关。结果提示我们,LINC01106在结直肠癌的进展中扮演重要角色。为了进一步探讨LINC01106与结直肠癌的关系,我们分别构建LINC01106敲减的结直肠癌细胞株和过表达的结直肠癌细胞株,发现敲减LINC01106可显著抑制SW480结直肠癌细胞的增殖,而过表达LINC01106则可促进细胞增殖。为了探讨敲减LINC01106抑制结直肠癌细胞增殖的可能原因,我们通过PI和Annexin V双染实验观察敲减LINC01106对SW480结直肠癌细胞凋亡的影响,发现敲减LINC01106可以促进细胞凋亡。随后我们探讨了LINC01106促癌的可能机制,Western blot实验结果表明,敲减LINC01106可以抑制p-STAT3和Bcl-2蛋白表达。STAT3是信号传导与转录激活因子,负责调控多种细胞生物学过程,如增殖、凋亡、侵袭转移等。正常组织中,STAT3的激活是十分短暂的,只能持续几分钟到几小时。然而,在癌组织中STAT3往往呈现持续激活状态。目前,STAT3已经是公认的癌基因,通过调控STAT3的表达和激活可以促进肿瘤的发生发展[25-27]。Bcl-2是抗凋亡家族的成员之一,也是经典的STAT3下游信号[28]。通过上述结果我们发现,LINC01106可以影响STAT3蛋白激活以及Bcl-2蛋白表达,LINC01106可能通过影响STAT3信号通路的传导发挥促癌作用。此外,我们将LINC01106敲减的SW480细胞接种于裸鼠皮下,与阴性对照组相比敲减组瘤体积明显较小并且免疫组化结果表明Cleaved caspase-3和Cleaved caspase-9表达水平上升,提示凋亡信号被激活。
综上所述,LINC01106在结直肠癌中表达上调与其恶性进展相关,LINC01106可以作为一个促癌基因调控结直肠癌细胞增殖和凋亡,其可能通过影响STAT3信号传导发挥促癌作用,但LINC01106影响STAT3激活的分子机制仍需深入研究。探究LINC01106在结直肠癌中的作用及分子机制可为结直肠癌的治疗提供新的思路,同时LINC01106也可能成为结直肠癌诊断及预后标志物。
Biography
谷昱琛,硕士,药师,E-mail: 469399704@qq.com
Funding Statement
国家自然科学基金(81903684);蚌埠医学院第一附属医院优秀青年科学基金(2019byyfyyq06)
Supported by National Natural Science Foundation of China (81903684)
Contributor Information
谷 昱琛 (Yuchen GU), Email: 469399704@qq.com.
刘 哲 (Zhe LIU), Email: liuzhecpu@outlook.com.
王 路路 (Lulu WANG), Email: wanglulu@njglyy.com.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2018, 68(6): 394-424.] [DOI] [PubMed] [Google Scholar]
- 2.Chen WQ, Zheng RS, Baade PD, et al. Cancer statistics in China, 2015. CACancer J Clin. 2016;66(2):115–32. doi: 10.3322/caac.21338. [Chen WQ, Zheng RS, Baade PD, et al. Cancer statistics in China, 2015[J]. CACancer J Clin, 2016, 66(2): 115-32.] [DOI] [PubMed] [Google Scholar]
- 3.Shen XN, Hu X, Mao JY, et al. The long noncoding RNA TUG1 is required for TGF-β/TWIST1/EMT-mediated metastasis in colorectal cancer cells. Cell Death Dis. 2020;11(1):65. doi: 10.1038/s41419-020-2254-1. [Shen XN, Hu X, Mao JY, et al. The long noncoding RNA TUG1 is required for TGF-β/TWIST1/EMT-mediated metastasis in colorectal cancer cells[J]. Cell Death Dis, 2020, 11(1): 65.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D'Andrea V, Panarese A, Tonda M, et al. Cancer stem cells as functional biomarkers. Cancer Biomark. 2017;20(3):231–4. doi: 10.3233/CBM-151176. [D'Andrea V, Panarese A, Tonda M, et al. Cancer stem cells as functional biomarkers[J]. Cancer Biomark, 2017, 20(3): 231-4.] [DOI] [PubMed] [Google Scholar]
- 5.Thanikachalam K, Khan G. Colorectal cancer and nutrition. Nutrients. 2019;11(1):164. doi: 10.3390/nu11010164. [Thanikachalam K, Khan G. Colorectal cancer and nutrition[J]. Nutrients, 2019, 11(1): 164.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geng F, Wang Z, Yin H, et al. Molecular targeted drugs and treatment of colorectal cancer: recent progress and future perspectives. Cancer Biother Radiopharm. 2017;32(5):149–60. doi: 10.1089/cbr.2017.2210. [Geng F, Wang Z, Yin H, et al. Molecular targeted drugs and treatment of colorectal cancer: recent progress and future perspectives[J]. Cancer Biother Radiopharm, 2017, 32(5): 149-60.] [DOI] [PubMed] [Google Scholar]
- 7.Bommer GT, Gerin I, Feng Y, et al. P53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17(15):1298–307. doi: 10.1016/j.cub.2007.06.068. [Bommer GT, Gerin I, Feng Y, et al. P53-mediated activation of miRNA34 candidate tumor-suppressor genes[J]. Curr Biol, 2007, 17 (15): 1298-307.] [DOI] [PubMed] [Google Scholar]
- 8.Isin M, Dalay N. LncRNAs and neoplasia. Clin Chim Acta. 2015;444:280–8. doi: 10.1016/j.cca.2015.02.046. [Isin M, Dalay N. LncRNAs and neoplasia[J]. Clin Chim Acta, 2015, 444: 280-8.] [DOI] [PubMed] [Google Scholar]
- 9.Sun FF, Liang WW, Tang KJ, et al. Profiling the lncRNA-miRNAmRNA CeRNA network to reveal potential crosstalk between inflammatory bowel disease and colorectal cancer. PeerJ. 2019;7:e7451. doi: 10.7717/peerj.7451. [Sun FF, Liang WW, Tang KJ, et al. Profiling the lncRNA-miRNAmRNA CeRNA network to reveal potential crosstalk between inflammatory bowel disease and colorectal cancer[J]. PeerJ, 2019, 7: e7451. DOI:10.7717/peerj.7451.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, Ma SQ, Huang J, et al. Roles of long noncoding RNAs in colorectal cancer metastasis. Oncotarget. 2017;8(24):39859–76. doi: 10.18632/oncotarget.16339. [Li H, Ma SQ, Huang J, et al. Roles of long noncoding RNAs in colorectal cancer metastasis[J]. Oncotarget, 2017, 8(24): 39859-76.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krijger I, Mekenkamp LJ, Punt CJ, et al. MicroRNAs in colorectal cancer metastasis. J Pathol. 2011;224(4):438–47. doi: 10.1002/path.2922. [Krijger I, Mekenkamp LJ, Punt CJ, et al. MicroRNAs in colorectal cancer metastasis[J]. J Pathol, 2011, 224(4): 438-47.] [DOI] [PubMed] [Google Scholar]
- 12.Saha P, Verma S, Pathak RU, et al. Long noncoding RNAs in mammalian development and diseases. Adv Exp Med Biol. 2017;1008:155–98. doi: 10.1007/978-981-10-5203-3_6. [Saha P, Verma S, Pathak RU, et al. Long noncoding RNAs in mammalian development and diseases[J]. Adv Exp Med Biol, 2017, 1008: 155-98.] [DOI] [PubMed] [Google Scholar]
- 13.Szcześniak MW, Makałowska I. lncRNA-RNA Interactions across the Human Transcriptome. PLoS One. 2016;11(3):e0150353. doi: 10.1371/journal.pone.0150353. [Szcześniak MW, Makałowska I. lncRNA-RNA Interactions across the Human Transcriptome[J]. PLoS One, 2016, 11(3): e0150353.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi Y, Zhao Y, Huang Y, et al. A brief review of RNA-protein interaction database resources. Noncoding RNA. 2017;3(1):E6. doi: 10.3390/ncrna3010006. [Yi Y, Zhao Y, Huang Y, et al. A brief review of RNA-protein interaction database resources[J]. Noncoding RNA, 2017, 3(1): E6.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guttman M, Rinn JL. Modular regulatory principles of large noncoding RNAs. Nature. 2012;482(7385):339–46. doi: 10.1038/nature10887. [Guttman M, Rinn JL. Modular regulatory principles of large noncoding RNAs[J]. Nature, 2012, 482(7385): 339-46.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang H, Liu P, Zhang J, et al. Long noncoding RNA MIR31HG exhibits oncogenic property in pancreatic ductal adenocarcinoma and is negatively regulated by miR-193b. Oncogene. 2016;35(28):3647–57. doi: 10.1038/onc.2015.430. [Yang H, Liu P, Zhang J, et al. Long noncoding RNA MIR31HG exhibits oncogenic property in pancreatic ductal adenocarcinoma and is negatively regulated by miR-193b[J]. Oncogene, 2016, 35 (28): 3647-57.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi XF, Sun M, Wu Y, et al. Post-transcriptional regulation of long noncoding RNAs in cancer. Tumour Biol. 2015;36(2):503–13. doi: 10.1007/s13277-015-3106-y. [Shi XF, Sun M, Wu Y, et al. Post-transcriptional regulation of long noncoding RNAs in cancer[J]. Tumour Biol, 2015, 36(2): 503-13.] [DOI] [PubMed] [Google Scholar]
- 18.Zhang Q, Su M, Lu GM, et al. The complexity of bladder cancer: long noncoding RNAs are on the stage. Mol Cancer. 2013;12(1):101. doi: 10.1186/1476-4598-12-101. [Zhang Q, Su M, Lu GM, et al. The complexity of bladder cancer: long noncoding RNAs are on the stage[J]. Mol Cancer, 2013, 12(1): 101.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun JF, Ding CH, Yang Z, et al. The long non-coding RNA TUG1 indicates a poor prognosis for colorectal cancer and promotes metastasis by affecting epithelial-mesenchymal transition. J Transl Med. 2016;14:42. doi: 10.1186/s12967-016-0786-z. [Sun JF, Ding CH, Yang Z, et al. The long non-coding RNA TUG1 indicates a poor prognosis for colorectal cancer and promotes metastasis by affecting epithelial-mesenchymal transition[J]. J Transl Med, 2016, 14: 42.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bian ZH, Zhang JW, Li M, et al. Correction: Long non-coding RNA LINC00152 promotes cell proliferation, metastasis, and confers 5- FU resistance in colorectal cancer by inhibiting miR-139-5p. Oncogenesis. 2018;7(8):63. doi: 10.1038/s41389-018-0067-1. [Bian ZH, Zhang JW, Li M, et al. Correction: Long non-coding RNA LINC00152 promotes cell proliferation, metastasis, and confers 5- FU resistance in colorectal cancer by inhibiting miR-139-5p[J]. Oncogenesis, 2018, 7(8): 63.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang YH, Fu J, Zhang ZJ, et al. LncRNA-LINC00152 downregulated by miR-376c-3p restricts viability and promotes apoptosis of colorectal cancer cells. Am J Transl Res. 2016;8(12):5286–97. [Zhang YH, Fu J, Zhang ZJ, et al. LncRNA-LINC00152 downregulated by miR-376c-3p restricts viability and promotes apoptosis of colorectal cancer cells[J].Am J Transl Res, 2016, 8(12): 5286-97.] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W, Zheng WH, Zhang L, et al. Long non-coding RNA ROR1- AS1 induces tumorigenesis of colorectal cancer by affecting Wnt/β- catenin signaling pathway. Biosci Rep. 2019;39(11):BSR20191453. doi: 10.1042/BSR20191453. [Wang W, Zheng WH, Zhang L, et al. Long non-coding RNA ROR1- AS1 induces tumorigenesis of colorectal cancer by affecting Wnt/β- catenin signaling pathway[J]. Biosci Rep, 2019, 39(11): BSR20191453.] [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Ma YL, Yang YZ, Wang F, et al. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/β-catenin signalling pathway via suppression of activator protein 2α. Gut. 2016;65(9):1494–504. doi: 10.1136/gutjnl-2014-308392. [Ma YL, Yang YZ, Wang F, et al. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/β-catenin signalling pathway via suppression of activator protein 2α[J]. Gut, 2016, 65(9): 1494-504.] [DOI] [PubMed] [Google Scholar]
- 24.Lian YF, Xu YT, Xiao CX, et al. The pseudogene derived from long non-coding RNA DUXAP10 promotes colorectal cancer cell growth through epigenetically silencing of p21 and PTEN. Sci Rep. 2017;7(1):7312. doi: 10.1038/s41598-017-07954-7. [Lian YF, Xu YT, Xiao CX, et al. The pseudogene derived from long non-coding RNA DUXAP10 promotes colorectal cancer cell growth through epigenetically silencing of p21 and PTEN[J]. Sci Rep, 2017, 7(1): 7312.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buettner R, Mora LB, Jove R. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin Cancer Res. 2002;8(4):945–54. doi: 10.1093/carcin/23.4.669. [Buettner R, Mora LB, Jove R. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention [J]. Clin Cancer Res, 2002, 8(4): 945-54.] [DOI] [PubMed] [Google Scholar]
- 26.Yu H, Jove R. The STATs of cancer: new molecular targets come of age. Nat Rev Cancer. 2004;4(2):97–105. doi: 10.1038/nrc1275. [Yu H, Jove R. The STATs of cancer: new molecular targets come of age[J]. Nat Rev Cancer, 2004, 4(2): 97-105.] [DOI] [PubMed] [Google Scholar]
- 27.Haura EB, Turkson J, Jove R. Mechanisms of disease: Insights into the emerging role of signal transducers and activators of transcription in cancer. Nat Clin Pract Oncol. 2005;2(6):315–24. doi: 10.1038/ncponc0195. [Haura EB, Turkson J, Jove R. Mechanisms of disease: Insights into the emerging role of signal transducers and activators of transcription in cancer[J]. Nat Clin Pract Oncol, 2005, 2(6): 315-24.] [DOI] [PubMed] [Google Scholar]
- 28.Reed JC. Mechanisms of apoptosis avoidance in cancer. Curr Opin Oncol. 1999;11(1):68–75. doi: 10.1097/00001622-199901000-00014. [Reed JC. Mechanisms of apoptosis avoidance in cancer[J]. Curr Opin Oncol, 1999, 11(1): 68-75.] [DOI] [PubMed] [Google Scholar]


