Abstract
Background
Real-time reverse transcription-PCR (rRT-PCR) has been the most effective and widely implemented diagnostic technology since the beginning of the COVID-19 pandemic. However, fuzzy rRT-PCR readouts with high Ct values are frequently encountered, resulting in uncertainty in diagnosis.
Methods
A Specific Enhancer for PCR-amplified Nucleic Acid (SENA) was developed based on the Cas12a trans-cleavage activity, which is specifically triggered by the rRT-PCR amplicons of the SARS-CoV-2 Orf1ab (O) and N fragments. SENA was first characterized to determine its sensitivity and specificity, using a systematic titration experiment with pure SARS-CoV-2 RNA standards, and was then verified in several hospitals, employing a couple of commercial rRT-PCR kits and testing various clinical specimens under different scenarios.
Findings
The ratio (10 min/5 min) of fluorescence change (FC) with mixed SENA reaction (mix-FCratio) was defined for quantitative analysis of target O and N genes, and the Limit of Detection (LoD) of mix-FCratio with 95% confidence interval was 1.2≤1.6≤2.1. Totally, 295 clinical specimens were analyzed, among which 21 uncertain rRT-PCR cases as well as 4 false negative and 2 false positive samples were characterized by SENA and further verified by next-generation sequencing (NGS). The cut-off values for mix-FCratio were determined as 1.145 for positive and 1.068 for negative.
Interpretation
SENA increases both the sensitivity and the specificity of rRT-PCR, solving the uncertainty problem in COVID-19 diagnosis and thus providing a simple and low-cost companion diagnosis for combating the pandemic.
Funding
Detailed funding information is available at the end of the manuscript.
Keywords: COVID-19, SARS-CoV-2, rRT-PCR, CRISPR diagnosis, Cas12a, SENA
1. Introduction
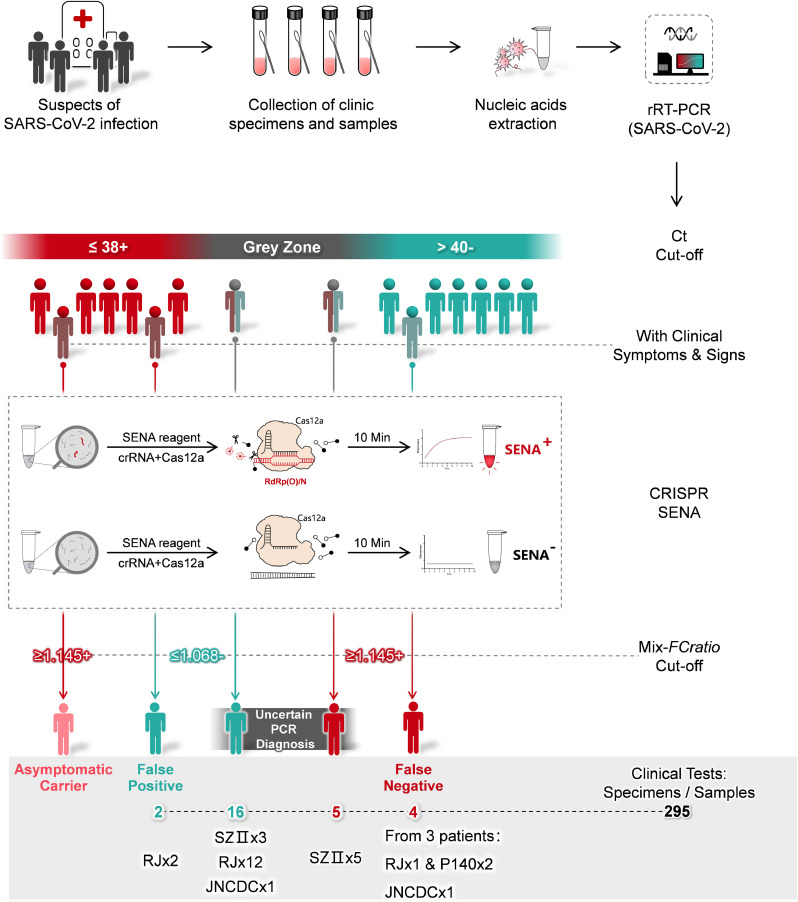
Since December 2019, theoutbreak of COVID-19, caused by the infection of the SARS-CoV-2, has rapidly spread throughout the world, and is now a global pandemic [1]. Till June 1, 2020, the outbreak has affected 216 countries, areas and territories, infected 6 million people, and caused more than 370 thousand of death [2]. One of the greatest public health concerns in combating the pandemic is a prompt response to the urgent demand for rapid and accurate diagnosis of the virus. Currently, nucleic acid amplification-based molecular diagnostics (MDx) is the most accurate, fast and affordable and thus the preferred method for diagnosis of SARS-CoV-2 infection, and the real-time reverse transcription PCR (rRT-PCR) kits have been successfully developed by quite a few laboratories and commercial companies [3]. However, since its clinical application at least six months ago in China, the diagnostic performance of rRT-PCR for SARS-CoV-2 has brought some urgent challenges, particularly the uncertain negative or positive readouts associated with the frequently encountered high Ct-value “grey zones” [[4], [5], [6], [7], [8]]. Besides of “human error” factors such as misconducted sampling, unqualified reagents and uncalibrated diagnostic equipment, inefficient RT reaction and PCR amplification of clinical samples with very low viral loads are likely the major intrinsic causative factors for the fuzzy rRT-PCR readouts and uncertain diagnosis. Although repetitive sampling and assays are implemented for final confirmation of the diagnosis, these “trouble shooting” efforts are time-consuming and may still fail to detect the low viral load samples from some mild or asymptomatic patients, or from the recovering patients, resulting in false-negative diagnosis that may cause serious public concerns in battling against the pandemic (Fig. 1).
Research in context.
Evidence before this study
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has exploded all over the world and is already a global pandemic. rRT-PCR technique is the current gold standard for molecular diagnosis of COVID-19 and is recommended by both the Centers for Disease Control and Prevention (CDC) and World Health Organization (WHO). However, the diagnostic performance of rRT-PCR for SARS-CoV-2 has been challenged by the frequently encountered “grey zone” problem that is caused by high Ct values. Usually, sample re-collection and retesting are required, which is time-consuming and labor-intensive, and a simple and low-cost technology for solving this “grey zone” problem and correcting false positive or false negative diagnosis is thus urgently needed.
Added value of this study
We here created a CRISPR-based diagnostic technology, namely the Specific Enhancer for PCR-amplified Nucleic Acid (SENA), and proved that the LoD per reaction of SENA is at least two copies less than that of rRT-PCR. Besides, the mix-FCratio was defined for quantitative analysis of SENA readouts and the cut-off values for mix-FCratio were also determined. Then, SENA was verified with 295 clinical specimens with 4 false negative and 2 false positive rRT-PCR diagnosis identified. Moreover, SENA also resolved the uncertainty problems of 21 specimens that fell in the rRT-PCR “grey zone”.
Implications of all the available evidence
SENA is a safe, simple, stable, quick and low-cost diagnostic tool with no need of extra instruments. More importantly, it may efficiently eliminate the uncertainty problems of PCR-based diagnosis and thus, may have great potential in applications such as COVID-19 diagnosis and many other clinical scenarios.
Alt-text: Unlabelled box
Fig. 1.
Schematic description of SENA and its application as a confirmation diagnosis for rRT-PCR diagnosis of COVID-19. Generally, nucleic acids are extracted from the clinical specimens such as pharyngeal swabs of the suspects of SARS-CoV-2 infection and then subject to rRT-PCR analysis. The diagnostic reports are based on the Ct cut-off values guided by the supplier of rRT-PCR kits. However, high Ct-value designated “grey zone” associated uncertain fuzzy readouts are often encountered. Besides, some probably false-positive or false-negative cases may be indicated by their atypical clinical symptoms or signs. For all these cases, the corresponding rRT-PCR products can be sent to another physically isolated room for SENA analysis and the ambiguity may be clarified by SENA with its positive and negative cut-off mix-FCratio. The real-life data related to these scenarios revealed in this study are shown in the figure and details are illustrated in the text. RJ, JNCDC and SZII are the names of the hospitals and the number indicates the overall number of patients identified. While P140 was a patient in DF hospital, and two distinct samples from P140 were identified to be false-negative. For details, please ref to Supplementary Table 3.
With the characterization of non-specific trans-cleavage activities against single-stranded nucleic acids in several CRISPR-associated (Cas) proteins, e.g., Cas12, Cas13 and Cas14 [[9], [10], [11], [12], [13], [14], [15]], Clustered Regularly Interspaced Short Palindromic Repeats Diagnostics (CRISPR-Dx) technology [16, 17] was established and has been developing rapidly. The underline mechanism for CRISPR-Dx, as illustrated by the Cas12a-based HOLMES system for example [18], is based on the efficient trans-cleavage activity against a fluorophore quencher (FQ)-labeled single-stranded DNA reporter by Cas12a triggered upon target DNA recognition, which is guided by a specific CRISPR RNA (crRNA), generating exponentially increasing fluorescence signal within several minutes. With this mechanism, here, we design a Specific Enhancer for detection of PCR-amplified Nucleic Acids (SENA) to improve both the detection sensitivity and specificity against the pre-amplified targeted SARS-CoV-2 genomic fragments (Fig. 1). Briefly, the COVID-19 clinical samples are firstly analyzed by the well-established rRT-PCR assays and amplicons with uncertain readouts are then verified by SENA in a physically isolated space, avoiding contamination of the PCR laboratory during pipetting.
2. Methods
2.1. rRT-PCR reactions
Reactions were conducted in a 25 μl reaction mixture following the instructions offered by the commercial suppliers of the reaction kits (ref to Supplementary Table 1a). Usually, the reaction cycle parameters were set as reverse transcription at 50°C for 10 min, denaturation at 95°C for 5 min, then followed by 45 cycles of amplification, i.e., 95°C for 10 s and 55°C for 40 s.
2.2. SENA reagents and detection
2.2.1. Preparation of SENA reaction reagents
Candidate crRNA guide sequences with the “TTN” PAM sequences were designed as shown in Supplementary Table 1b, and the crRNAs were prepared following the procedures previously described [11]. Except the target DNA, the 2 × SENA reagent comprises of 2 × NEB buffer 3.1, 500 nM LbCas12a (Tolo Biotech.), 1 μM synthesized crRNAs (for each specified target and thus varies according to the rRT-PCR kit supplier, Supplementary Table 1b), 1 μM FQ-reporter, and 1 U/μl RNase inhibitor (TaKaRa).
2.2.2. SENA detection
To avoid the aerosol contamination of the MDx laboratory, after the rRT-PCR reaction, their products must be transferred to a physically isolated room to perform SENA detection. It is also important to choose proper SENA detection reagents corresponding to the rRT-PCR kits. To prepare a 20 μl SENA reaction system, with corresponding positive and negative controls, 2 μl PCR products and 8 μl RNase-free H2O were mixed with 10 μl 2 × SENA reagent, and the mixture was then measured on an appropriate fluorescence reader with FAM fluorescence collected following the programs: 48 °C 30 s per cycle, 20 cycles. Both the slope and the Fluorescence Change (FC) can be calculated at any time points as desired.
2.3. Next generation sequencing (NGS)
The rRT-PCR products were purified by AMPure XP beads (Beckman Coulter Life Sciences, US), and libraries were then constructed following the procedures of end repair, dA-tailing and adaptor ligation, with the StepWise DNA Lib Prep Kit for Illumina (ABclonal, China). After PCR amplification, samples were sequenced on Illumina Miniseq to produce 2 × 150 bp paired-end reads. After adaptor trimming and quality trimming, the clean reads were mapped to the reference genome of SARS-CoV-2 (MN908947.3) using Bowtie2 [19].
2.4. Systematic titration and regression analyses
2.4.1. Systematic titration experimentation
2.4.1.1. The standard RNA templated
The SARS-CoV-2 RNA standards were purchased from Genewell (Shenzhen, China). According to the supplier's information, three plasmids containing the whole sequences of N and E genes, and partial sequence of the Orf1ab, i.e., from 13,237 to 13,737 of the SARS-CoV-2 complete genome (MN908947.3), were transcribed in vitro individually. The RNA products were mixed with equal molar, aliquoted with addition of 1 μg of human RNA per tube, and subject to lyophilization and subsequent quantification with digital PCR. This SARS-CoV-2 RNA standard dry powder containing 1808 copies of O gene, 1795 copies of N gene and 1160 copies of E gene was dissolved with 10 μl RNase-free water to obtain the original stock solution (estimated 180.8 copies/μl of O, 179.5 copies/μl of N and 116 copies/μl of E).
2.4.1.2. The preparation of the serially diluted RNA templates
Pharyngeal swab samples were collected from 40 adult patients in Shenzhen Second Peoples’ Hospital by the Clinical Diagnosis Laboratory and the nucleic acids of each sample were extracted with the pre-packaged nucleic acid extraction kit (Da'An Gene., Ltd., Guangzhou, China), according to the manufacturer's instructions, ended up with 55 μl extracts per sample. After rRT-PCR assays employing 5 μl of the extracts from each sample, all of the samples were shown to be SARS-CoV-2 negative. The remaining 50 μl extracts of each sample were mixed together and 5 μl of the mixture was once again analyzed by rRT-PCR and confirmed to be SARS-CoV-2 negative. Then, the mixed nucleic acid extract was used as the dilution buffer (totally about 2 ml) for serial dilution of the SARS-CoV-2 RNA standard stocks, generating desired concentrations (i.e., 5, 2, 1, 0.8, 0.6, 0.4, 0.2, 0.1, 0.05, 0.025, 0.01, 0.005 copies/μl), and 5 μl of each of the diluted solutions were used as templates for rRT-PCR analysis, forming gradient template concentrations (i.e., 25, 10, 5, 4, 3, 2, 1, 0.5, 0.25, 0.125, 0.05, 0.025 copies/Rx).
Replica setting: We analyzed 9 replicas for each of the concentrations of 1 and 0.5 RNA template copies/Rx while 6 replicas for each of the rest concentrations (Supplementary Fig. 1). rRT-PCR reactions: rRT-PCR reaction kit was supplied by BG (Supplementary Table 1a), who follows the primer sets recommended by Chinese CDC. Instead of the recommended 40 cycles of PCR amplification, we set 45 cycles as routine aiming at recording the maximum exact Ct values if possible.
SENA detection: Every amplicon of the rRT-PCR reactions was subjected to SENA detection. Specially, for this experimentation, three sets of SENA reagents were individually used, O-SENA contains the crRNA targeting the O sequence, N-SENA contains the N-targeting crRNA, and the mix-SENA contains crRNAs for both sequences.
2.4.2. Choice of FCratio as the standard readout for SENA detection and mix-SENA as the standard reagent for clinical application
Three readout parameters were compared as shown in Supplementary Fig. 1 (original data in Supplementary Table 2). The slope (increase of fluorescence/min) represents the reaction rate of Cas12a trans-cleavage activity, but in this experiment, it represents neither the initial rate of the enzyme under limited substrate condition nor the pure first-order reaction rate varies according to the substrate concentration, particularly, demonstrated in cases of high template concentration, in which, the slope goes down along with the increase of the template concentration. In addition, when the substrate concentration is low, the slope of SENA is hard to be distinguished from that of the negative control, ending with ambiguous cut-offs. The FC (the fold of change of fluorescence between that of the sample over that of the negative control at certain time point, usually 5–30 min) does show clear differences between the positive amplicons from that of negative control, and it also shows certain quantitation character particularly at the low concentration templates cases. Because of these properties, FC has been a parameter used by a few users of CRISPR-Dx [20]. However, it seems that the absolute value of the FC usually varies along the reaction time and sometimes it is influenced by the change of the fluorescence signal of the negative control. Besides, it is difficult to determine the “best choice” of the FC recorded at certain time points, which may cause confusing in clinical applications. It is clear, we need a stable readout which reflects the dynamic process and the quantitative correlation of SENA reaction with the low concentration of the templates on one hand and should be robust and accurate for clinical diagnosis on the other hand. We defined FCratio, which is the ratio between FC's of SENA detection at 10 min versus 5 min after the beginning of the fluorescence reading. It not only measures the fluorescence change of SENA against the negative control background so that the quantity of the amplicons, particularly at the low concentration range may be represented, but also normalizes the slope of the fluorescence change of SENA to eliminate the complex background differences. As shown in Supplementary Fig. 2, FCratio significantly amplified the positive signals and represents the quantitation of the amplicons at low template concentrations to certain extent.
As shown in Supplementary Fig. 2, the capacity of the three SENA reactions are compared. It is obvious that N-SENA is the least sensitive one, while although O-SENA seems much more sensitive than that of N-SENA and largely comparable to that of mix-SENA, its signal at the very low template concentration range seems uncertain in some cases. Therefore, for clinical application, mix-SENA is the best choice.
2.4.3. Quality analyses of the rRT-PCR titration data
The quality of the Ct values vs the concentration of the standard templates for rRT-PCR of the systematic titration experiment were analyzed both empirically and statistically. Firstly, as shown in Supplementary Table 2 and Supplementary Fig. 1, the amplification efficiencies of the two genes represented by the valuable Ct readouts were different. The apparently lower sensitivity of the N gene amplification is in contrast with the clinical experiences and might due to the difference in the property of the templates used in different experiments (laboratory standard RNA template vs clinical real viral template). Secondly, although linear regression can be readily made between the Ct values and the log2 (conc) (Supplementary Fig. 3a) as that of the previously published tests, the quality of the regression as judged by the R2s (Supplementary Fig. 3a) and the residues (Supplementary Fig. 3b) are clearly suboptimal likely due to the limited number of replicas in the experimentation. On the other hand, this titration was designed with taking at least the two most fundamental limitation factors about the sensitivity of COVID-19 rRT-PCR diagnosis into consideration, i.e., the sampling ambiguity and the influence of the biological/chemical contaminants from the clinical samples. Therefore, the data will be used for determination of the LoD for rRT-PCR and SENA (Methods 4.4 and Supplementary Fig. 2), and the regression function will be used to estimate the “apparent Ct” values of the samples with Ct values greater than 45 (no amplification signal) but their SENA detection is positive (Methods 4.5 and Supplementary Fig. 4).
2.4.4. Determination of limit of detection (LoD) for rRT-PCR and SENA
The LoD values for rRT-PCR (N-Ct and O-Ct) and SENA (N-FCratio, O-FCratio and mix-FCratio) were estimated based on the systematic titration employing standard RNA templates (Supplementary Fig. 1 and Supplementary Table 2). The fractions of positive replicates versus the number of target molecules (copies) per reaction for N and O gene of COVID-19 were plotted and used the sigmoidal functions (1) to fit the data via R (software version 3.5.0). The 95% confidence intervals were derived by bootstrapping the model residues and were visualized by R (software version 3.5.0) with built-in ggplot2 library [21].
2.4.5. Regression of rRT-PCR Ct values and the SENA FCratio versus the concentration of the templates employing the data from the systematic titration
2.4.5.1. Regression of rRT-PCR Ct values with the concentration of the templates (copies/Rx)
Since the PCR product increased exponentially with the initial concentration of the sample (x), and the Ct value of rRT-PCR parameter (y) was inversely correlated with the initial concentration, especially in the range of low copy number (low template concentration) samples, the power function equation (a<1) should be suitable for the data fitting. However, some of the experimental groups included very low initial sample concentrations (<1 copy/Rx), those amplification efficiencies should be different (particularly affected by sampling ambiguity) from that of the groups with high initial template concentration. Therefore, the power function formula with four parameters (Y=aXn+bXm) was used to match all the experimental group data to obtain a more accurate data model (Functions 1, Supplementary Fig. 4).
2.4.5.2. Regression of SENA FCratio with the concentration of the templates (copies/Rx)
The exponential function (first order association kinetics of the interaction between a substrate and an enzyme, Y=a+b(1-ecX)) is used to fit the data of FCratio against the concentration of the templates. At low concentration (especially when the concentration is less than 2 copies/Rx), the FCratio is positively correlated with the template concentrations. However, when the template concentration reaches to 2 copies/Rx and more, the FCratio does not increase accordingly and the curve tend to be flatted out. In addition, as FCratio has already been normalized by the fluorescence signal of the negative background, it is stable and, in this case, we give the parameter Y0 being set as a constant value between 0.9 to 1 (Functions 2 in Supplementary Fig. 5).
2.4.5.3. Regression of rRT-PCR Ct values with the SENA mix-FCratio values
In practice, quite significant portions of the clinical positive samples detected by mix-SENA with their FCratio readings higher than the positive cut-off, but with a negative PCR Ct value (40 - 45, depending on the scenario). Under certain circumstances, people may be interested to learn the copy number of the templates for the corresponding rRT-PCR assays or even the “probable” Ct values of these assays. With the aid of the above-mentioned two regression functions (1 in Supplementary Fig. 4 and 2 in Supplementary Fig. 5), these data could be estimated. One may firstly substitute the Y in Functions 2 by the measured FCratio value and the corresponding X can be calculated representing the “estimated concentration of the template”. Then this X value can be used to estimate the corresponding “estimated Ct-value” as the Y of Functions 1. We estimated all the ambiguous Ct values of the positive amplicons and plotted them against their corresponding mix-FCratio (Fig. 4). All the real and estimated Ct values for both N and O genes are plotted against the corresponding FCratio values of mix-SENA as X axis. An exponential decay function (with X0 = 1; When X≤X0, Ct=∞; otherwise, one phase decay) fits well to all the data (R2=0.9238) and is used for analyzing the clinical data and adjust the cut-off values accordingly (Fig. 4).
Fig. 4.
Apparent correlation plot of the rRT-PCR Ct values against the SENA mix-FCratios in SARS-CoV-2 detection. All the data of the systematic titration experiment with low concentrations of standard RNA templates (Supplementary Table 2a) and the data of clinical tests employing samples with ambiguous rRT-PCR readouts are used in this plot. In case the Ct values are too high to be detected by the rRT-PCR assay, i.e., Ct>40~45, depending on the scenarios, the mix-FCratio-correlated “apparent Ct values” may be estimated via the template concentration-related regression functions (Methods 4.5); however, majority of the extremely high “apparent Ct values” in real negative samples are arbitrary and are adopted merely to simplify the presentation. The positive cut-off of the mix-SENA detection (mix-FCratio=1.145) is defined by the C3 and C6 samples of the systematic titration experiment (Fig. 2a), while the negative cut-off of the mix-SENA detection (mix-FCratio=1.068) is defined by the P278–1 sample of the clinical tests (Supplementary Table 3). This plot confirmed the cut-off Ct values of rRT-PCR test provided by the kit supplier (BG, ref to all detectable Ct values shown as solid dots). Meanwhile, with the aid of mix-SENA, the sensitivity of rRT-PCR was increased up to the detected level of O-Ct=43.3 in samples of D2 and D4 and estimated level of O-Ct=49.5 in samples of C6 and P280. In addition, false positives were detected with O-Ct values as low as 39 (P278–1), which was also the key test to define the negative cut-off of mix-SENA.
2.5. SENA detection of clinical samples
Nucleic acids are extracted from the total 295 clinical specimens, such as the pharyngeal swabs and nasopharyngeal swabs of the suspects of SARS-CoV-2 infection, which collected in Ruijin Hospital, Shenzhen Second People's Hospital, and Jinan Center for Disease Control and Prevention, and then subject to rRT-PCR analysis. rRT-PCR reactions were conducted in a 25 μl reaction mixture following the instructions offered by the commercial suppliers of the reaction kits (refer to Supplementary Table 1a). Then, the corresponding rRT-PCR products were sent to another physically isolated room and 2 μl PCR products were carefully transferred to new tubes for SENA analysis (refer to Methods 2.2) and NGS (refer to Methods 2.3).
2.6. Detection of antibodies
The detection of anti-SARS-CoV2 antibodies was executed by the point-of-care microfluidic platform integrating a home-made fluorescence detection analyzer (Suxin, Shanghai, China). A total of 10 μl plasma was added into the loading chamber of microchip followed by the addition of 70 μl sample dilution buffer. After incubation for 15 min at room temperature, the microchips were loaded onto the fluorescence detection analyzer, and fluorescence signal was detected from the analyzer, following the manufacturer's instruction.
2.7. Ethics statement
All experiments using human material were performed in accordance with the instructional guidelines and agreement of the Ethical Committee of Ruijin Hospital (#2018–48), and Shenzhen Second People's Hospital (#202003009005). Written informed consent was waived given the context of emerging infectious diseases.
3. Results
3.1. Development and characterization of SENA
To prepare appropriate crRNAs for SENA detection, we firstly determined the amplicon sequences from several commercial rRT-PCR kits used in China and then designed specific crRNAs corresponding to each of the distinct amplicons (Supplementary Table 1a). Candidate crRNAs were prepared and analyzed individually in a SENA system, which comprised of, in addition to the crRNAs, Cas12a, FQ-reporter and the rRT-PCR products using templates of either the positive or negative controls. The apparently most appropriate crRNAs, i.e., the lowest fluorescence with the negative control and highest with the positive control, were chosen for the final formulation of the SENA assay reagents (data not shown). In general, four formulations of regents were prepared employing crRNAs against corresponding assay targets, i.e., Orf1ab (abbreviated as O), E or N genes, individually, and the mixture of O and N (abbreviated as mix).
The performance of SENA was quantitatively characterized via a systematic titration upon rRT-PCR amplicons employing pure SARS-CoV-2 RNA standards comprised of the O and N fragments, individually or mixed, as the templates. As it is aware that the viral nucleic acids extracted from patients’ samples such as nasopharyngeal swabs usually contain some biological and chemical contaminants that might inhibit the enzyme activities for reverse transcription and PCR reactions and is likely one of the causal effects attributed to the low efficiency of rRT-PCR in clinical analysis [22]. In order to mimic the clinical sampling for the titration experimentation, the RNA standards were serially diluted in buffer prepared by mixing the nucleic acid extracts from 40 COVID-19 negative people, generating RNA templates ranging from 0.025 to 25 copies per reaction (Rx).
Due to the Poisson distribution property of sampling, replica variations become extremely significant when the template copies in individual reaction are designed to be low, i.e., less than 3–4 copies/Rx, near the limit of detection (LoD) for rRT-PCR [21, 23], and extremely low, i.e., equal to and less than 1 copy/Rx. To overcome this sampling ambiguity problem, we performed 9 replicas for groups with 1 and 0.5 RNA template copies/Rx while 6 replicas for each of the rest concentrations. In addition, although the rRT-PCR assay supplier, BioGerm (BG, Shanghai, China), who follows the Chinese CDC recommended primer sets (Supplementary Table 1a), recommends 40 cycles of PCR amplification, we set 45 cycles as routine aiming at recording maximum exact Ct values if possible. After rRT-PCR reaction, all amplicons were subjected to 3 individual SENA reactions, i.e., N-SENA, O-SENA and mix-SENA with crRNAs targeting O gene, N gene and both, respectively.
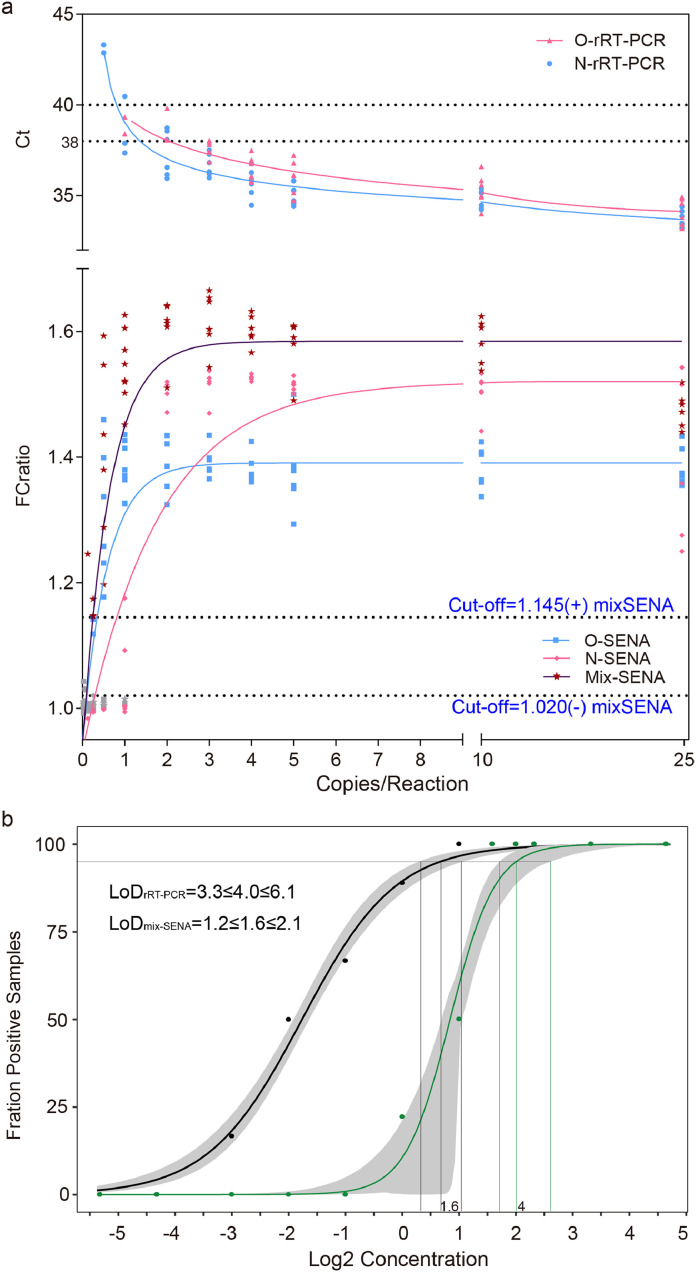
Consistent with the theoretical analysis [23] and the rigorous experimentation [21], along with the decrease of the RNA templates to less than 3 copies/Rx, the rRT-PCR Ct values in some replicas, primarily that corresponding to the N gene, passed 38 (the cut-off for positive as recommended by the rRT-PCR kit suppliers) but were less than 40, which should be considered as entering the “grey zone”. The Ct values increased steadily when the concentration of the RNA templates further decreased, with more and more replicas showing one or both Ct values entering the “grey zone” and eventually all became “negative”, i.e., greater than 40 or even 45 (Fig. 2a, Supplementary Table 2 and Supplementary Fig. 1). Employing Ct=38 as the cut-off for “positive” detection, we estimated the LoD for O and N genes with 95% confidence interval (CI) of this set of rRT-PCR assay as 3.3≤4.0≤6.1 and 4.0≤4.1≤4.4, respectively (Supplementary Fig. 2). Most likely due to the influence of the complex combination of the targeted viral genomic fragments and the clinical sampling background, the LoD determined in this study was clearly higher than the published value of 2.0≤2.5≤3.7, which analyzed single target in a pure system [21].
Fig. 2.
Determination of the cut-off values for SENA detection (a) and the LoD values with 95% CI for both rRT-PCR (O-Ct, green dots) and SENA (mix-FCratio, black dots) (b) based on the systematic titration assays. All the experimental and analytical details are described in the text. Notice that the SENA negative cut-off was set as mix-FCratio=1.020 in this figure on the basis of the titration assay of the standard RNA templates but was adjusted to 1.068 along with the increase of the clinical applications (Figs. 1 and 4, Supplementary Table 3). LoD with 95% confidence interval was 1.2≤1.6≤2.1 with mix-SENA versus 3.3≤4.0≤6.1 with rRT-PCR.
The rRT-PCR amplicons were further analyzed by SENA detection with the measurement of the fluorescence signals for each corresponding replica. After comparison of the parameters of slope (increase of fluorescence/min) versus FC (the fold of change of fluorescence between that of the sample over that of the negative control at certain time point), we defined a parameter, FCratio, which is the ratio of the FC at 10 min to that at 5 min after the initiation of fluorescence reading (Supplementary Fig. 1 and Supplementary Table 2). We also found that in the cases with low concentrations of templates, the rRT-PCR efficiency of the two target genes (i.e., O and N) were different so as the SENA detection (Supplementary Figs. 1 and 2). In order to verify the existence of specific amplicons of SARS-CoV-2 nucleic acids in an individual rRT-PCR reaction, all of the amplicons of the replicas with RNA templates ranging from 0.125 to 2 copies/Rx were subjected to next generation sequencing (NGS) analysis. The results were found to be completely consistent with the perspective results of both O-SENA and mix-SENA. In addition, with mix-SENA, not only the signals are generally more significant than that of the O-SENA detection but also may resolve some of the ambiguity readouts found with N-SENA (Supplementary Fig. 1 and Supplementary Table 2). Based on these results, the mix-FCratio was demonstrated as the most sure-proof index for rRT-PCR confirmation, and we empirically estimated that mix-FCratio≥1.145 for positive cut-off, and mix-FCratio≤1.020 for negative cut-off (Fig. 2a). Of course, these two parameters are subject to further verification and adjustment along with the increase of tested samples. Because SENA is rRT-PCR based, the same methodology for determining the rRT-PCR LoD was used to estimate that of SENA by this set of data, corresponding to both individual O and N fragments (Supplementary Fig. 2) and in combination as indicated by the mix-SENA (Fig. 2b). As expected, the N-SENA LoD (3.7≤4.3≤4.8 with 95% CI) is very close to that of the N-Ct of rRT-PCR, while the LoD of O-SENA (1.1≤1.3≤1.7 with 95% CI) is significantly lower than that of O-Ct (Supplementary Fig. 2). Although the LoD of mix-SENA (1.2≤1.6≤2.1 with 95% CI) is slightly higher than that of O-SENA (Fig. 2b and Supplementary Fig. 2), it is apparently caused by its capable of confirming some of the ambiguous amplicons in the extremely low concentration cases (Supplementary Fig. 2) and thus, mix-FCratio is chosen for clinical applications.
3.2. Verification of SENA in different clinical scenarios
SENA was further verified in a few hospitals, testing various clinical specimens and samples under different scenarios (Fig. 1) and employing few more commercial rRT-PCR diagnosis kits in addition to BG which was used in the titration experiment (Supplementary Table 1a). Totally 295 clinical samples or specimens (mainly pharyngeal swabs) collected from 282 individuals were tested by rRT-PCR followed by SENA detection (Supplementary Table 3). Except for asymptomatic carriers, all the cases of uncertain analytic and false positive or negative readouts of rRT-PCR diagnosis were encountered and finally confirmed or corrected by SENA detection.
Specifically, samples from 139 patients of Ruijin Hospital (RJ, Shanghai, China) were assayed by rRT-PCR employing diagnostic kits of Liferiver (LR) and Beijing Genomics Institute (BGI), 137 of which had consistent readouts by all those of rRT-PCR kits, indicating two positive, 123 negative and 12 suspected that fell in the “grey zone” (Supplementary Table 3). SENA detection of these samples revealed not only the 12 suspected as negative but also identified one more positive among the original 123 negative individuals, clearly a case of false negative diagnosis (Supplementary Table 3). Besides, distinct rRT-PCR assay results, positive by BGI but negative by LR were shown for samples collected from 2 close contacts of COVID-19 patients and apparently asymptomatic (ref to Supplementary Table 3). However, the amplicons of both LR and BGI were shown as negative via SENA detection. All these ambiguous rRT-PCR amplicons (17 samples, ref to Supplementary Table 3) were finally analyzed by NGS, and the results were consistent with the SENA. Noticeably, the rRT-PCR false-negative COVID-19 patient was symptomatically mild at the point of admission with all the clinical laboratory tests negative but turned positive after 24 hours. On the other hand, although those 12 suspected patients had respiratory infection symptoms, they were finally excluded from COVID-19 according to the latest guideline for diagnosis and treatment from China National Health commission (the 6th edition). Similarly, in Shenzhen Second People's Hospital (SZII, Shenzhen, China), 5 uncertain rRT-PCR readouts for O gene were found among 139 individuals. Three of them had Ct value of 39.47, 39.7 and 40.56, respectively but the following SENA detection gave mix-FCratio values less than 1.0 for all of them, indicating all negative. The other two individuals had Ct values of 38.87 and 39.22, while their mix-FCratio values were 1.581 and 1609, respectively, indicating positive for both. In addition, there were another three individuals with Ct values larger than 40 for O gene and 36.09, 35.88 and 37.98 for N gene, respectively; however, the following SENA detection showed mix-FCratio values were 1.39, 1.55 and 1.21, respectively, indicating all positive. All these amplicons were further confirmed by NGS analysis (Supplementary Table 3), obtaining consistent results with those of SENA. Consistently, the three SENA-negative individuals were finally excluded from SARS-CoV-2 infection after being rechecked by rRT-PCR after 24 hours (Supplementary Table 3). Based on above data, it is clear, SARS-CoV-2 infection suspects with either rRT-PCR Ct values falling in the “grey zone” or with clear patient-contact epidemiological history but negative rRT-PCR tests, are strongly recommended to perform SENA detection to minimize the possibility of misdiagnosis. On the other hand, in case an rRT-PCR-positive suspect does not demonstrate any COVID-19 clinical symptoms and/or signs, SENA detection is also strongly recommended to eliminate either false-positive diagnosis or misdiagnosis of the so-called “asymptomatic carrier” or “asymptomatic patient”.
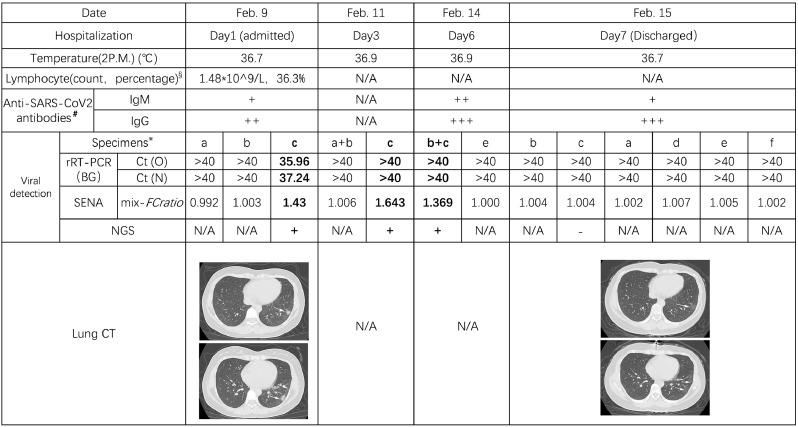
Besides of preventing false-negative or false-positive diagnosis, the highly sensitive property of SENA may also assist in providing evidence of viral clearance for COVID-19 recovering patients. A female patient in Dongfang Hospital (DF, Shanghai, China) was confirmed as COVID-19 positive by both rRT-PCR and CT scanning and showed ground-glass opacities mixed with consolidation along the subpleural area (Fig. 3). Accordingly, the SENA test was positive with the mix-FCratio of 1.43. After the hospitalization, the patient was further analyzed by rRT-PCR at two time points, obtaining all negative results with bilateralnasal and pharyngeal swab specimens. However, the mix-FCratios of SENA for some of her specimens were 1.64, 1.36 and 1.00, respectively, indicating that the virus was contained and yet to be cleared. On the seventh day, both rRT-PCR and the corresponding SENA detection for all of her specimens were negative and these results were confirmed by NGS and consistent with her normal CT scanning results (Fig. 3). Thus, she was discharged from the hospital and safely back to home. Similar cases were found in Jinan of Shandong Province, China, where the fecal samples from two recovering COVID-19 patients were tested negative by rRT-PCR but clearly positive by SENA (Supplementary Table 3). Considering a certain percentage of the recovered patients discharged from hospitals were reported to be re-detectable positive (RP) [24], the incomplete clearance of the SARS-CoV-2 virus ahead of discharge might be one of the possible causes. Therefore, it could be necessary to consider more sensitive detection approaches such as SENA as a potential index of viral clearance.
Fig. 3.
Schematic diagram of the hospitalization process of patient P140 (Shanghai DF Hospital). Detailed viral detection data are listed in Supplementary Table 3. Other clinical data indicated that P140 is a COVID-19 patient with mild clinical symptoms.
§Reference range for Lymphocyte count: 1.1–3.2 × 109/L; Reference range for the percentage of Lymphocyte: 20–50%.
༃Label of the antibodies: +, weak; ++, medium; +++, strong.
※Label of the specimens: a, pharyngeal swab; b, nasal (left) swab; c, nasal (right) swab; d, serum; e, plasma; f, fecal.
To reconfirm and/or improve the cut-off values for SENA mix-FCratio, the ambiguous Ct values were re-estimated using the regression functions derived from the rRT-PCR assays with titrated standard RNA templates (Supplementary Figs. 4 and 5), and then the Ct values (both estimated and detected) were plotted against the corresponding mix-FCratios (Fig. 4). Combining the data from both RNA standards and clinical samples, it is clear that SENA detection is of both high sensitivity, identifying real positive samples with Ct values as high as more than 43 (approaching 50 as estimated), and high specificity, identifying real negative samples with Ct values as low as 39. Therefore, SENA can effectively eliminate uncertain diagnosis of rRT-PCR assays for SARS-CoV-2 infection. In addition, the cut-off value for SENA mix-FCratio remains unchanged as 1.145 for positive diagnosis while slightly increased to 1.068 for negative (Fig. 4), which is supposed to further increase along with the clinical applications.
4. Discussion
Instead of developing a closed CRISPR-Dx system, which ideally should be comprised of both target nucleic acids amplification and CRISPR-Cas-based trans-cleavage assays, SENA was created here to match the commercially available and widely applied rRT-PCR kits, and to solve the uncertainty challenge of the rRT-PCR “grey zone” in COVID-19 diagnosis. As expected, SENA specifically increases both the detection sensitivity (i.e., with the LoD of 1.6) and the specificity (i.e., with false positive and false negative samples detected) in COVID-19 diagnosis.
In a SENA detection system, the amplicons of rRT-PCR are taken as the target nucleic acids for the Cas12a-based trans-cleavage reaction, and the remaining single-stranded PCR probes that are fluorescently labelled may have influence on the SENA readouts. Firstly, the crRNAs were carefully designed to avoid targeting either the primers or the probes. Therefore, the remaining the probes will not trigger the trans-cleavage activities of Cas12a. Secondly, as only a small amount of the amplicons is added in the SENA reaction system, the remaining fluorophore will have no influence on the SENA results even if the rRT-PCR system uses the same fluorophore as that in SENA. On one hand, the addition of cleaved rRT-PCR probe increases only the background signal of SENA other than the FCratio. On the other hand, the un-cleaved rRT-PCR probe is also the target for Cas12a trans-cleavage, which is only 1/25 of the FQ-reporter in amount and will increase the saturation signal instead of the FCratio. Taken together, one may conclude that the remaining rRT-PCR probes have no influence on the SENA results. Considering the fact that detection of 5(6)-carboxyfluorescein (FAM) with its maximum wavelength of excitation and emission at 494 nm and 522 nm, respectively, is supported by most quantitative PCR machines, we simply chose FAM for labelling the SENA FQ-reporter. Of course, the SENA reporter can also be labelled with other fluorophores as previously reported [18].
Since the outbreak of COVID-19 pandemic, dozens of rRT-PCR diagnosis kits that use distinct PCR primer pairs and probes have been successfully developed and are commercially available. Here in this study, both LoD and cut-off values were determined for SENA with the employment of one of them, which is also commercially available in China. Although the LoD of SENA can be different for distinct rRT-PCR kits, the SENA cut-off values were shown to be consistent among the three tested rRT-PCR kits in this work (ref to Fig. 4 and Supplementary Table 3). For example, the cut-off values were empirically estimated as mix-FCratio≥1.145 for positive and mix-FCratio≤1.020 for negative with the employment of standard samples. Then, after the verification of SENA detection with 295 clinical specimens, using three different rRT-PCR kits (Supplementary Table 3), the cut-off value for positive remained unchanged and the cut-off value for negative slightly changed from 1.020 to 1.068. Therefore, when a mix-FCratio falls between 1.068 and 1.145, which is the “grey zone” of SENA detection at present, either NGS or clinical symptoms should be used to help diagnosis. Also, it is highly recommended that following researchers may precisely determine the LoD for a specific rRT-PCR kit as well as the cut-off values in future. With the increase of the samples tested, the SENA cut-off value for negative may become slightly larger, generating a smaller “grey zone” and consequently leading to less uncertainty in SENA diagnosis.
Besides, to precisely calculate the LoD of SENA, the quality of the rRT-PCR data in this study was also carefully evaluated before further SENA analysis. In general, most measuring techniques generate a signal response that is proportional to the amount of measurand present. For example, measured absorption is proportional to the concentration of the dissolved measurand as predicted by the Beer-Lambert law [25]. Regarding to rRT-PCR, the measured Ct values are logarithmic responses proportional to the log base 2 (log2) of the concentration of the measurand (the number of target molecules present). We thus plotted the Ct values against the log2 of their corresponding template concentrations, in which the Ct value decreased with the increase of the template concentration. Although the few most diluted samples were outside the linear range of rRT-PCR standard curve, which might be due to sampling background noise and limited number of replicates in the experiment (Supplementary Fig. 3), the data were on the whole fitted to a straight line. On the other hand, while linear data generated by linear measurements are normally distributed in a linear scale with the measurand concentration changing [26], quantitative real-time PCR data show normal distribution in a logarithmic scale [27]. To confirm it, we also showed the measured Ct values in residual plot relative to the fitted straight line (Supplementary Fig. 3b), and the plots showed how the spread of replicates increases with decreasing number of targets as that of the previously published tests [21]. Although other factors contribute to variation across replicates [28], sampling noise, which can be modeled by the Poisson distribution, is expected to dominate at very low copy numbers. In the future, it is recommended to increase the number of duplicates at very low copy numbers to further improve the data quality.
Taken together, considering the fact that rRT-PCR is the most popular MDx system and SENA is simple to operate, SENA has the potential to be widely used in various scenarios to solve the uncertainty problems of rRT-PCR as well as other nucleic acid amplification-based MDx. Of course, to minimize the possibility of aerosol contamination during opening of the rRT-PCR tubes and pipetting, physical separation of the SENA detection center from the clinical PCR laboratory is an absolute requisition. Abided by this rule, SENA has been demonstrated convenient and effective in several hospitals and centers for disease control and prevention as part of their laboratory routine in combination with rRT-PCR for more sensitive and accurate detection of SARS-CoV-2 infection. Therefore, SENA is a useful technique that meets the urgent needs of combating COVID-19 pandemic.
Data sharing
All data is available in the main text or the Supplementary materials.
Funding sources
J.W. (Jin Wang) is supported by the National Natural Science Foundation of China (31922046); W.H. is supported by the National Key R&D Program of China (2019YFA0906000), NSFC (81772737), and Shenzhen High-level Hospital Construction Fund, Shenzhen Sanming Project of Medicine in Shenzhen (SZSM201412018, SZSM201512037); D.W. (Donghua Wen) is supported by NSFC (81972830) and Shanghai “Rising Stars of Medical Talent” Youth Development Program-Clinical Laboratory Practitioners Program; F.S. is supported by NSFC (31670757); G.Z. is supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB19040200). The funders had no role in the study design, data collection, data analysis, data interpretation and writing of the report.
Declarations of Interests
G.Z. and J.W. are cofounders of Tolo Biotechnology Co., Ltd. W.H., G.Z. and J.W. have filed patent applications relating to the work in this manuscript. All other authors declare no competing interests.
Acknowledgments
We thank Yucai Wang and Linxian Li for discussions and support.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.103036.
Contributor Information
Fei Song, Email: lst121@outlook.com.
Guoping Zhao, Email: gpzhao@sibs.ac.cn.
Jin Wang, Email: wangj01@hotmail.com.
Appendix. Supplementary materials
References
- 1.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Organization WH. 2020. Cornavirus disease 2019 (COVID-19) Situation Report-131.https://wwwwhoint/emergencies/diseases/novel-coronavirus-2019/situation-reports/ Available at: [Google Scholar]
- 3.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020 Jan;25(3) doi: 10.2807/1560-7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng H, Liu Y, Lv M, Zhong J. A case report of COVID-19 with false negative RT-PCR test: necessity of chest CT. Japan J Radiol. 2020 May;38(5):409–410. doi: 10.1007/s11604-020-00967-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y, Yao L, Li J, Chen L, Song Y, Cai Z. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J Med Virol. 2020 Mar 26 doi: 10.1002/jmv.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan Y, Long L, Zhang D, Yuan T, Cui S, Yang P. Potential false-negative nucleic acid testing results for severe acute respiratory syndrome coronavirus 2 from thermal inactivation of samples with low viral loads. Clin Chem. 2020 Jun 1;66(6):794–801. doi: 10.1093/clinchem/hvaa091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu J, Liu J, Li S, Peng Z, Xiao Z, Wang X. Detection and analysis of nucleic acid in various biological samples of COVID-19 patients. Travel Med Infect Dis. 2020 Apr 18 doi: 10.1016/j.tmaid.2020.101673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao AT, Tong YX, Zhang S. False-negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: Rather than recurrence. J Med Virol. 2020 Apr 9 doi: 10.1002/jmv.25855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017 Apr 28;356(6336):438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018 Apr 27;360(6387):439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li SY, Cheng QX, Liu JK, Nie XQ, Zhao GP, Wang J. CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res. 2018 Apr;28(4):491–493. doi: 10.1038/s41422-018-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018 Apr 27;360(6387):436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrington LB, Burstein D, Chen JS, Paez-Espino D, Ma E, Witte IP. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018 Nov 16;362(6416):839–842. doi: 10.1126/science.aav4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L, Li S, Wu N, Wu J, Wang G, Zhao G. HOLMESv2: A CRISPR-Cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation. ACS Synth Biol. 2019 Oct 18;8(10):2228–2237. doi: 10.1021/acssynbio.9b00209. [DOI] [PubMed] [Google Scholar]
- 15.Chertow DS. Next-generation diagnostics with CRISPR. Science. 2018 Apr 27;360(6387):381–382. doi: 10.1126/science.aat4982. [DOI] [PubMed] [Google Scholar]
- 16.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012 Sep 25;109(39):E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012 Aug 17;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li SY, Cheng QX, Wang JM, Li XY, Zhang ZL, Gao S. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018;4:20. doi: 10.1038/s41421-018-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012 Mar 410.1038/nmeth.1923;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.W.Z. Tieying Hou MY, Wenjing Chen, Lili Ren, Jingwen Ai, Ji Wu, Yalong Liao, Xuejing Gou, Yongjun Li, Xiaorui Wang, Hang Su, Bing Gu, Jianwei Wang, Teng Xu. Development and evaluation of A CRISPR-based diagnostic For 2019-novel coronavirus. medRxiv preprint2020; February 25, 2020. 10.1101/2020.02.22.20025460. [DOI] [PMC free article] [PubMed]
- 21.Forootan A, Sjoback R, Bjorkman J, Sjogreen B, Linz L, Kubista M. Methods to determine limit of detection and limit of quantification in quantitative real-time PCR (qPCR) Biomol Detect Quantif. 2017 Jun;12:1–6. doi: 10.1016/j.bdq.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bustin SA, Nolan T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J Biomol Techn JBT. 2004 Sep;15(3):155–166. http://www.ncbi.nlm.nih.gov/pubmed/15331581 [PMC free article] [PubMed] [Google Scholar]
- 23.Burns M, Valdivia H. Modelling the limit of detection in real-time quantitative PCR. Eur Food Res Technol. 2007;226(6):1513–1524. doi: 10.1007/s00217-007-0683-z. [DOI] [Google Scholar]
- 24.Fu W, Chen Q, Wang T. Letter to the editor: three cases of re-detectable positive SARS-CoV-2 RNA in recovered COVID-19 patients with antibodies. J Med Virol. 2020 May 5 doi: 10.1002/jmv.25968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Shuguang, Wang Tianhua, Yang Min. The evaluation of statistical methods for estimating the lower limit of detection. ASSAY and Drug Dev Technol. 2013;11(1):35–43. doi: 10.1089/adt.2011.438. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt AF, Finan C. Linear regression and the normality assumption. J Clin Epidemiol. 2018;98:146–151. doi: 10.1016/j.jclinepi.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 27.Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonák J, Lind K. The real-time polymerase chain reaction. Mol Asp Med. 2006;27(2):95–125. doi: 10.1016/j.mam.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 28.Ruijter JM, Ruiz Villalba A, Hellemans J, Untergasser A, van den Hoff MJB. Removal of between-run variation in a multi-plate qPCR experiment. Biomol Detect Quantif. 2015;5:10–14. doi: 10.1016/j.bdq.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.