Abstract
Background and Aim:
Trimethoprim-sulfamethoxazole (TMP-SMX) is an important cause of idiosyncratic drug induced liver injury (DILI), but its genetic risk factors are not well understood. We investigated the relationship between variants in the HLA Class I and II genes and well characterized cases of TMP-SMX DILI.
Methods:
European American and African American persons with TMP-SMX DILI were compared to respective population controls. HLA sequencing was performed by Illumina MiSeq for cases. HLA genotype imputation with attribute bagging (HIBAG) program was used to impute HLA alleles for controls. Allele frequency difference between cases and controls was tested by Fisher exact tests per ethnic group. For European Americans, multivariable logistic regression with Firth penalization was used to test HLA allelic effect after adjusting for age and the top two principal components. Molecular docking was performed to assess the HLA binding with TMP and SMX.
Results:
The European American subset had 51 cases and 12,156 controls, while the African American subset had 10 cases and 5,439 controls. Four HLA alleles were significantly associated in the European American subset, with HLA-B*14:01 ranking at the top (OR: 9.20, 95% CI: 3.16–22.35, p=0.0003) after covariate adjustment. All HLA-B*14:01 carriers with TMP-SMX DILI possessed HLA-C*08:02, another significant allele (p=0.0026). This pattern was supported by HLA-B*14:01-HLA-C*08:02 haplotype association (p=1.33×10−5). For the African Americans, HLA-B*35:01 had 2.8-fold higher frequency in cases than in controls, with five of 10 patients carrying this allele. Molecular docking showed Cys67 in HLA-B*14:01 and Phe67 in HLA-B*35:01 to be the predictive binding sites to SMX metabolites.
Conclusion:
HLA-B*14:01 is associated with TMP-SMX DILI in European Americans, and HLA-B*35:01 may be a potential genetic risk factor for African Americans.
Keywords: DILI, HLA, Haplotype, amino acid, Hepatotoxicity
Introduction
Trimethoprim-sulfamethoxazole (TMP-SMX) is a fixed combination synthetic antimicrobial that is commonly used to treat various bacterial infections and as a prophylaxis against opportunistic infections(1). TMP-SMX use has been associated with life-threatening and presumed immune-mediated idiosyncratic adverse drug reactions such as Stevens-Johnson syndrome (SJS), toxic epidermal necrolysis (TEN), drug-reaction with eosinophilia and systemic symptoms (DRESS) , severe drug induced liver injury (DILI), and blood dyscrasias(2). TMP-SMX is among the top 5 leading causes of DILI in the United States(3). The risk factors for liver injury associated with TMP-SMX are largely unknown, other than that they are more common among African-Americans and those infected with human immunodeficiency virus(1, 4). The pattern of liver injury associated with TMP-SMX is typically cholestatic or mixed, although severe hepatocellular injury and acute liver failure have been reported as well(5)
Immunogenetic factors have been increasingly shown to play an important role in the pathogenesis of severe immune mediated adverse drug reactions including DILI(6). Previous studies have identified several genetic variants, especially in the HLA region associated with any-cause DILI or liver injury due to specific drugs(7–10). Variants in the HLA region have also been linked to a few TMP-SMX induced adverse reactions. A Turkish study found a significantly higher frequency of the HLA-A*30, HLA-A*30-HLA-B*13-Cw6 haplotype, and HLA-B*55 among 42 patients with TMP-SMX induced fixed drug eruption as compared to 2,378 healthy blood donors(11). In another study consisting of 43 Thai patients with TMP-SMX induced SJS and TEN and 91 TMP-SMX tolerant controls, HLA-B*15:02, HLA-C*06:02, and HLA-C*08:01 were found to be significantly associated with severe cutaneous adverse reactions(12). To our knowledge, there have been no studies to date which attempted to elucidate the genetic basis underlying liver injury caused by TMP-SMX using GWAS or targeted HLA allelic association analysis.
The main objectives of this study were to investigate the HLA allele association with TMP-SMX induced liver injury in a well-defined cohort of European Americans and African Americans enrolled into the Drug Induced Liver Injury Network (DILIN) Prospective and Retrospective studies.
Materials and Methods
Study Participants:
Individuals aged ≥ 2 years of age with suspected DILI and meeting predefined eligibility criteria were enrolled into the DILIN Prospective (NCT00345930) and Retrospective (NCT00360646) studies. The designs of these two studies have been previously published (13, 14). In brief, written informed consent were obtained from individuals with acute liver injury suspected due to prescription, over-the-counter, or herbal and dietary supplements prior to enroll at multiple centers throughout the United States. Our study followed the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the Institutional Review Board of each study site. Upon recruitment, participants underwent systematic evaluation for competing etiologies and all cases were adjudicated for causality by structured expert consensus. The causal relationship between suspected medication(s) and the liver injury episode was categorized into definite (>95% likelihood), highly likely (76–95% likelihood), probable (51–75% likelihood), possible (26–50% likelihood), and unlikely (≤25% likelihood) (14). Cases included in this study were adjudicated to have definite, highly likely or probable DILI due to TMP-SMX, referred to as high confidence DILI cases. For this study’s purposes, instead of self-reported race and ethnicity, we utilized the genetic ancestry inferred by genome-wide single nucleotide polymorphism (SNP) data from DILIN and 1000 genomes project(15) (Supplementary Materials). Of 1,916 DILI patients, 86 (4.5%) were suspected to have DILI due to TMP-SMX with 72 patients classified as high confidence cases. Based on genetic inferred ethnicity, these patients were then grouped to 51 European Americans, 10 African Americans, 6 Hispanics, 4 Asians, and 1 other (Supplementary materials Section I, Table S2). In this study, the 51 European American and 10 African American DILI cases were analyzed. Participants in this paper have been included in other publications from the DILIN (16).
Study design and HLA data:
We utilized a case-control study design to identify HLA alleles that may play a role in the risk of developing TMP-SMX DILI in European Americans and African Americans, respectively. Due to the lack of control data in DILIN, considering the low incidence rate of DILI in the population, we used large population cohorts as controls to overcome the potential bias of including potential subjects with DILI events. With the approval from NCBI dbGaP, we obtained the GWAS data from eMERGE-I: Genome Wide Association Studies of Network Phenotypes (phs000360.v3.p1) and PAGE: The Charles Bronfman Institute for Personalized Medicine BioMe BioBank (phs000925.v1.p1). The eMERGE-1 GWAS dataset consisted of 17,045 subjects who were predominantly Americans with European ancestry, while the PAGE GWAS dataset consisted of 12,932 subjects with diverse racial backgrounds. Genotyping platforms were Illumina Human 660W Quad for eMERGE-1 and MEGA for PAGE. Similarly, we used genome-wide SNP data to infer ethnicity for each dataset. This led to 12,156 European Americans from eMERGE-I and 5,439 African Americans from PAGE to serve as controls.
For TMP-SMX DILI cases, HLA alleles were determined from the deep sequencing of HLA Class I and II genes using Illumina MiSeq (details in Supplementary Materials). For population controls, we utilized the GWAS SNPs to impute HLA alleles in HLA Class I and II genes for European Americans and African Americans, respectively. Specifically, four-digit HLA alleles were imputed based on the SNPs in the MHC region in chromosome 6 using the HLA genotype imputation with attribute bagging (HIBAG) program(17). All analyses were conducted for European Americans (51 cases vs. 12,156 controls) and African Americans (10 cases vs. 5,439 controls), respectively.
Statistical Analysis:
Descriptive statistics for demographic and patient characteristics were computed for 51 European American and 10 African American TMP-SMX DILI cases, respectively. We presented frequency (percentage) for categorical variables and median (interquartile range) for continuous variables. Allele frequencies (AF) and carriage frequencies (CF, proportion of patients carrying the allele) of all four-digit HLA alleles were computed for cases and controls, respectively, for each ethnic group. To evaluate the accuracy of imputed HLA alleles in controls, we compared the AF of imputed HLA alleles to population AF available from BeTheMatch(18) and AFND(19) for European Americans and African Americans, respectively. We set an arbitrary threshold to exclude imputed HLA alleles that have an absolute AF difference from population AF greater than 0.05. Since HLA alleles were determined by all variants in the coding sequence of the gene, this reduced variants to one marker with multiple alleles for each HLA gene (e.g., HLA-A*, HLA-B*, etc). Therefore, association tests were performed for a single allele at a time. Considering rare alleles will have low statistical power in single-allele association test, we excluded ultra-rare HLA alleles that have either allele count < 2 in TMP-SMX DILI cases or < 10 in controls.
For allelic association tests, each four-digit HLA allele was coded as 0, 1, or 2 based on the number of the allele an individual carried. The Fisher’s exact test was used to test AF differences between cases and controls for each HLA allele. For the European American subset, multivariable logistic regression with Firth penalization for rare events (Firth logistic regression) was used to test the association between each HLA allele and DILI with covariate adjustment. For covariate selection, univariate Firth logistic regression was performed for age, sex, and PC1 to PC10 individually first. We selected the top three variables meeting p<0.05 as covariates. Furthermore, since eMERGE-1 released age data in decades, we categorized age to three levels (1: < 40 years; 2: 40–69 years; 3: >69 years). False discovery rate (FDR) by Benjamini-Hochberg (20) was computed to correct multiple testing. Significant HLA alleles were determined based on FDR < 0.15 from the multivariable Firth logistic regression analysis. To delineate the relationship among the significant HLA alleles, we conducted conditional analyses by adjusting the most significant HLA allele along with other covariates. For the African American dataset, due to 10 cases only, we did not pursue multivariable regression analysis. Instead, we conducted Fisher exact test only and highlighted alleles meeting p<0.05.
Several secondary analyses were performed to understand the top HLA alleles further. First, haplotype association was tested for two and three gene combinations using the score statistics in the haplo.stats program (21). Second, using the NetMHCpan-2.8 algorithm in MHCcluster (22), HLA alleles in the gene of interest were clustered based on their predicted peptide-binding specificity. We then tested the association between each allelic cluster and TMP-SMX DILI. That is, HLA alleles in the same cluster were coded as the same marker and then the same association analysis as described above were followed. Third, we checked if the HLA allele of interest is present in persons with high confidence DILI due to non-antibacterial sulfonamides enrolled into the DILIN studies. Fourth, to evaluate if the HLA allele of interest is specific to TMP-SMX subset, we compared CF of the HLA allele of interest among DILIN drug groups that had at least 10 high confidence DILI cases. Finally, we tested if the HLA allele of interest is associated with the any-cause DILI for the corresponding ethnicity.
Replication of top HLA alleles:
We were able to identify five European Americans with TMP-SMX and three patients with TMP induced liver injury enrolled in the iDILIC consortium to evaluate our findings in DILIN. Causality assessment was performed as previously reported (23). The cases were previously analyzed as a part of the broad GWAS on susceptibility to DILI (24).
Amino Acid association analysis:
We first obtained the AA sequence alignment of all available HLA alleles in the same HLA gene from the Immuno Polymorphism Database(25). We then identified a set of polymorphic AA positions within the binding site. Using the HLA allele of interest as the reference allele, we converted the genotypes of the HLA allele to AA residue pairs of all polymorphic AA positions based on if the reference AA residue is present. In other words, assuming K polymorphic AA positions identified for a HLA gene, HLA genotype data of each subject is recoded as a vector of K markers with 0, 1, or 2 based on the number of reference AA residues present at the AA position. The same association tests, as described above for HLA alleles, were performed for each polymorphic AA position for European Americans and African Americans, respectively.
Molecular Docking:
Due to the lack of crystal structure of HLA-B*14:01, an atomic model of HLA-B*14:01 was generated based on the crystal structure of HLA-B*14:02, 99.6% identical, PDB code 3BVN (26) using SWISS-MODELLER (27). The HLA-B*14:01 model and HLA-B*35:01 crystal structure, PDB 6BJ8 (28), were used for molecular docking. AutoDock Tools was used for molecular docking of trimethoprim and sulfamethoxazole with AutoDock Vina(29) (e.g., add H, generate charges for each atom to be used in scoring). Scoring grids were 20 × 20 × 20 Å, and centered on 3 sites within the antigen binding clefts of HLA-B*14:01 and HLA-B*35:01, including sites corresponding to the Cα of the first peptide position (P1), Cα of the fifth peptide position (P5, the middle of the antigen binding cleft), and Cα of the terminal peptide position (P9). TMP and SMX were docked with exhaustiveness set to 10. The top 8 scoring orientations were output and compared. Sulfonamide was manually positioned in the modeled antigen binding cleft of HLA-B*14:01 with PyMol, which was used to generate molecular graphics (The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC.).
Results
From September 2004 through January 2019, the DILIN has enrolled a total of 2,256 participants with suspected DILI and 1,916 of them had undergone causality adjudication and had their HLA genes sequenced (Figure 1). There were 86 participants with suspected DILI in whom TMP-SMX was the primary implicated agent. Upon adjudication, 72 were deemed to have high-confidence DILI events (causality scores of definite, highly likely, or probable) with 51 European Americans and 10 African Americans. Table 1 describes the summary statistics for selected clinical characteristics of European Americans and African Americans, respectively. The median latency was longer in European Americans (25 days) than in African Americans (14.5 days). While the pattern of liver injury was evenly distributed among hepatocellular, cholestatic, and mixed categories in European Americans, it was predominantly hepatocellular in African Americans. Eleven patients (22%) of European Americans had peripheral eosinophilia but none appeared in African Americans. While a cutaneous rash was commonly reported (European Americans 47%, African Americans 40%), there were no reported instances of Stevens-Johnson syndrome in our TMP-SMX DILI patients.
Figure 1:
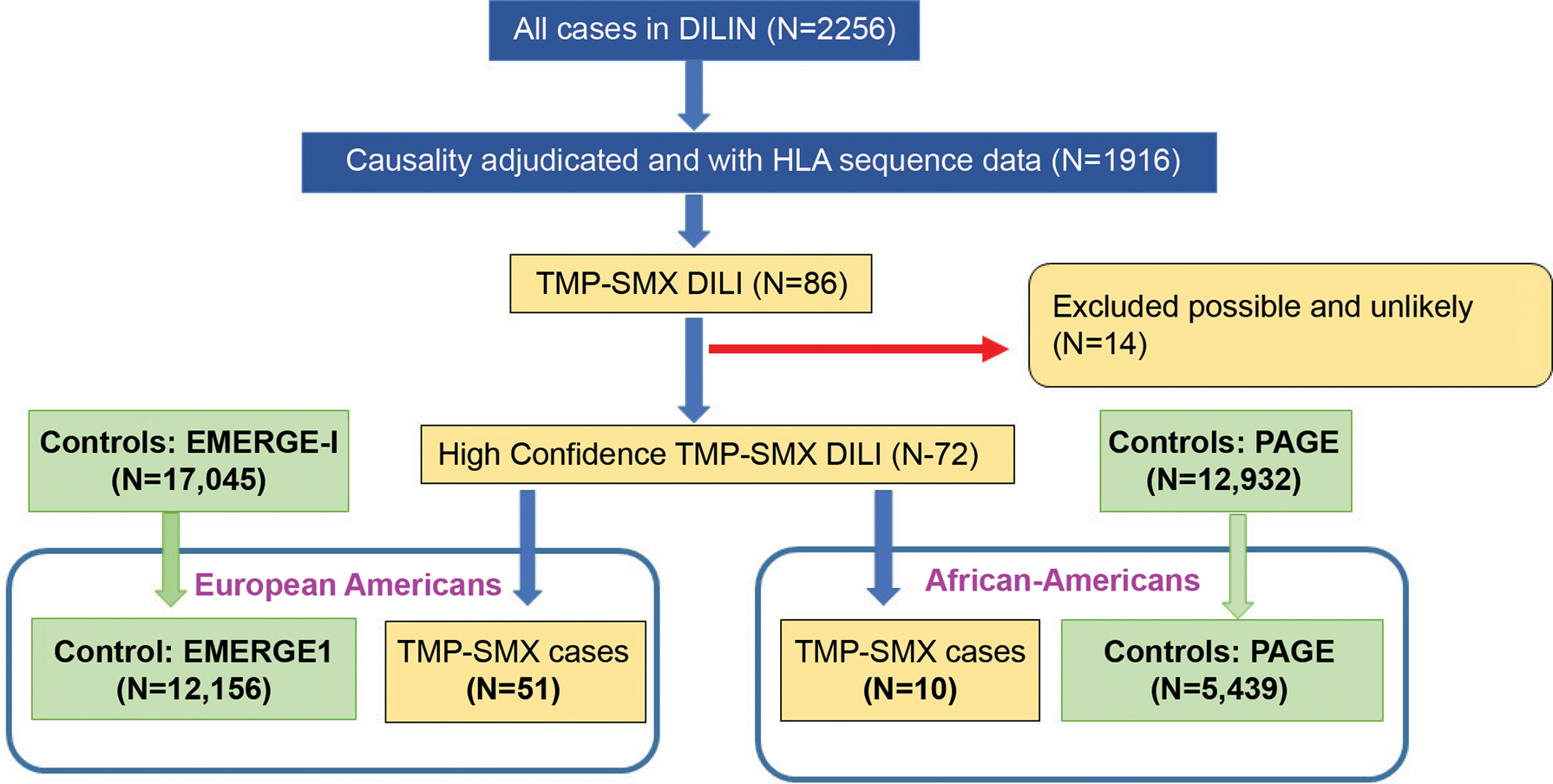
Consort diagram to outline the sample sizes used in this study
Table 1:
Selected clinical and laboratory characteristics for TMP-SMX DILI patients by ethnicity
| European Americans (N=51) | African-Americans (N=10) | |
|---|---|---|
| Age (years), mean (SD)) | 48 (20) | 45 (18) |
| Females, n (%) | 29 (57%) | 4 (40%) |
| BMI (kg/m2), mean (33)) | 25.8 (6.5) | 34.0 (13.5) |
| Diabetes mellitus, n (%) | 7 (14%) | 5 (50%) |
| Latency (days in median, IQR) | 25 (10, 34) | 14.5 (9, 20) |
| Jaundice (%) | 31 (61%) | 4 (40%) |
| INR (mean (SD)) | 1.4 (0.8) | 1.2 (0.25) |
| Peripheral eosinophilia (>500/μL), n (%) | 11 (22%) | 0 (0%) |
| Stevens Johnson syndrome, n (%) | 0 (0%) | 0 (0%) |
| Mixed | 20 (39%) | 1 (10%) |
| Fatal | 2 (3.9%) | 0 (0%) |
| Death, n (%) | 2 (3.9%) | 0 (0%) |
| Liver Transplantation, n (%) | 0 (0%) | 0 (0%) |
| Chronic DILI, n (%) | 8 (15.7%) | 2 (20%) |
Abbreviations:ALT, serum alanine aminotransferase; Alk P, serum alkaline phosphatase; BMI, body mass index; DILI, drug-induced liver injury; INR, international normalized ratio; IQR, interquartile range (25–75%)
We first examined the quality of imputed HLA alleles in controls: the eMERGE-1 and PAGE data. We identified four class II alleles in European Americans and seven class II alleles in African Americans with AF deviating from population AF more than 0.05 (Supplementary Table S3). These alleles were excluded from the association analysis. In addition, 114 ultra-rare HLA alleles in European Americans and 130 in African Americans were excluded. This led to 85 alleles in European Americans and 26 alleles in African Americans remained for interrogation in this study.
HLA allelic association:
Table 2 summarizes the top HLA alleles associated with TMP-SMX DILI for European Americans and African Americans, respectively. In European Americans, four Class I HLA alleles were considered significant with HLA-B*14:01 ranking at the top after adjusting for covariates and multiple testing. The AF of HLA-B*14:01 in cases was about 5.5-fold higher than in controls (AF=0.049 vs. 0.009, Fisher p=0.002). The effect size of HLA-B*14:01 on TMP-SMX DILI was estimated with an odds ratio (OR) of 9.20 (95% confidence interval (CI): 3.16–22.35, adjusted p=0.0003, FDR=0.024 based on 85 alleles tested). The other three top HLA alleles were HLA-A*34:02, HLA-C*08:02, and HLA-B*27:02. The OR estimates were unstable with large confidence intervals for both HLA-A*34:02 (95% CI = 5.17–320.43) and HLA-B*27:02 (95% CI= 2.50–48.04), which is likely due to low AF in both cases and controls. On the other hand, HLA-C*08:02, a relatively common allele (AF=0.03 in controls), had the most stable effect size estimate (narrower CI) among the four top alleles (OR (95% CI) = 3.78 (1.66–7.62), adjusted p=0.0026). However, once we adjusted for the effect of HLA-B*14:01, HLA-C*08:02 was no longer significant (p=0.230). This was due to the fact that all five patients carrying HLA-B*14:01 also carry HLA-C*08:02 (Supplementary Table S4).
Table 2:
HLA alleles associated with TMP-SMX DILI by ethnic group
| Fisher Exact Test | *Firth Logistic Regression | *Conditional analysis on B*14:01 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Race | Allele | Case AF | Control AF | Fisher P | OR (95% CI) | Adjusted P | **FDR | OR (95% CI) | Adjusted P |
| European American | B*14:01 | 0.049 | 0.009 | 0.002 | 9.20 (3.16, 22.35) | 0.0003 | 0.024 | --- | --- |
| A*34:02 | 0.020 | 0.001 | 0.002 | 47.52 (5.17, 320.43) | 0.0012 | 0.050 | 27.25 (3.85,150.33) | 0.002 | |
| C*08:02 | 0.078 | 0.032 | 0.016 | 3.78 (1.66, 7.62) | 0.0026 | 0.074 | 2.11(0.57, 5.56) | 0.230 | |
| B*27:02 | 0.020 | 0.002 | 0.024 | 13.53 (2.50, 48.04) | 0.0056 | 0.119 | 14.94 (2.74,53.53) | 0.004 | |
| African American | B*35:01 | 0.25 | 0.09 | 0.026 | |||||
| DPB1*04:01 | 0.20 | 0.06 | 0.034 | ||||||
| A*68:02 | 0.20 | 0.05 | 0.038 | ||||||
Firth logistic regression adjusted for age, PC2, and PC3; Conditional analysis: Firth logistic regression adjusted for age, PC2, PC3, and HLA-B*14:01
FDR: False discovery rate by Benjamini-Hochberg method to correct multiple testing of 85 alleles.
Given the rarity of the HLA-B*14:01 allele in the European American general population (population AF=0.008), we did not expect any carrier in the five TMP-SMX DILI cases from iDILI. However, instead, we found that one TMP-SMX DILI case carried the HLA-B*14:01 allele, a higher AF than expected (AF=0.1). Interestingly, this case with HLA-B*14:01 also carried HLA-C*08:02. None of the five TMP-SMX iDILI cases carried HLA-A*34:02 or HLA-B*27:02. Further, among three TMP cases in iDILI, they did not carry HLA-B*14:01 or HLA-B*14:02.
For African American subset, three HLA alleles showed nominally significant AF difference (Fisher p < 0.05) (Table 2). Among them, HLA-B*35:01 ranked at the top with 2.8 fold higher AF in cases than controls (AF: 0.25 vs. 0.087; p=0.026). More specifically, five out of 10 TMP-SMX DILI cases carried HLA-B*35:01. Both HLA-A*68:02 and HLA-DPB1*04:01 had four allele counts from 10 African American patients. HLA-A*68:02 was present in three TMP-SMX patients with one patient carrying homozygous HLA-A*68:02, while HLA-DPB1*04:01 was present in four patients with heterozygous HLA-DPB1*04:01.
HLA-B*14:01 haplotype association:
Haplotypes of HLA-B*14:01 was assessed for the following combinations of HLA Class I genes: A-B, B-C, and A-C-B in the European American subset. The combinations of HLA-A*32:01, HLA-C*08:02, and HLA-B*14:01 were the most promising HLA-B*14:01 haplotypes shown significant association with DILI (Supplementary Figure S3). The HLA-C*08:02-HLA-B*14:01 haplotype (frequency: 0.05, p=2.79×10−6) is more common than the other two haplotypes (A-C and A-C-B combinations; frequency: 0.02) in TMP-SMX DILI cases, which can be explained by all five HLA-B*14:01 carriers carrying HLA-C*08:02 but only two HLA-B*14:01 carriers having HLA-A*32:01 (Supplementary Table S4).
HLA allelic cluster association:
Figures 2A and 2B depict the HLA allelic clusters by peptide binding for European Americans and African Americans, respectively. For European Americans, the HLA-B*14:01 and HLA-B*14:02 cluster was the only cluster that showed significant association with TMP-SMX DILI (adjusted p=0.0028). In African Americans, the cluster of HLA-B*35:01, HLA-B*53:01, and HLA-B*51:01 had a significant higher cluster frequency in cases than controls (frequency: 0.55 vs. 0.23, Fisher p=0.002) (Table 3).
Figure 2:

HLA-B allele cluster from predicted peptide-binding specificity for European American (panel A) and African Americans (panel B). The color key was based on the distance between allele binding specificity. Red color (near 0) indicates high similarity on binding specificity between alleles, and white (near 1) indicates completely different binding specificity. Allele clusters tested for association with TMP-SMX DILI are marked by rectangles.
Table 3:
Association results between HLA-B allelic cluster and TMP-SMX DILI by ethnic group
| Race | Alleles in the cluster | Cluster Freq: Cases | Cluster Freq: Controls | Fisher P* | Adjusted P* |
|---|---|---|---|---|---|
| European American | B*14:01, B*14:02 | 0.078 | 0.032 | 0.018 | 0.0028 |
| B*57:01, B*58:01, B*15:17 | 0.049 | 0.046 | 0.810 | 0.799 | |
| B*35:02, B*35:12, B*51:01, B*35:01, B*53:01 | 0.115 | 0.115 | 1 | 0.604 | |
| B*07:02, B*42:01, B*81:01, B*55:01 | 0.137 | 0.158 | 0.683 | 0.891 | |
| B*37:01, B*49:01, B*47:01, B*13:01, B*44:02, B*44:03, B*18:01 | 0.245 | 0.217 | 0.472 | 0.537 | |
| B*13:02, B*52:01 | 0.020 | 0.029 | 1 | 0.874 | |
| African American | B*35:01, B*53:01, B*51:01 | 0.55 | 0.23 | 0.002 | - - - |
| B*07:02, B*81:01 | 0.1 | 0.1 | 1 | - - - | |
| B*44:03, B*18:01 | 0.15 | 0.08 | 0.224 | - - - |
Fisher P: p values from Fisher exact tests; Adjusted P: p values from Firth logistic regression with covariate adjustment of age, PC2, and PC3.
Note: For African American group, only Fisher exact test was performed.
Amino Acid association:
In the European American subset, a total of 45 AA positions in the binding sites of HLA-B were aligned, where 28 AA positions were polymorphic. HLA-B*14:01 and HLA-B*14:02 had identical AA residues in all sites except position 11 which is located at the carboxy terminal end of the first β strand on the floor of the antigen binding cleft, forming contact with the α1 helix (not oriented towards the peptide). Among all polymorphic AA residues in HLA-B*14:01, AA residues at positions 67 (Cys67: Cysteine), 97 (Trp97: Tryptophan), and 163 (Thr163: Threonine) showed significant association with DILI (Table 4). Trp97 was present only in HLA-B*14:01 and HLA-B*14:02 with estimated effect size of OR of 3.8 (95% CI: 1.7–7.6) (adjusted p=0.003). Cys67 was present in HLA-B*14:01, HLA-B*14:02 and nine other HLA-B* alleles with OR (95% CI) = 2.2 (1.2–3.7) (adjusted p=0.008), while Thr163 was present in HLA-B*14:01, HLA-B*14:02, and 15 other B* alleles with OR (95% CI) =1.9 (1.2–3.1) (adjusted p=0.007).
Table 4:
Amino acids (AAs) showing significant association with TMP-SMX DILI by ethnic group
| Fisher Exact test | Firth logistic regression* | ||||||
|---|---|---|---|---|---|---|---|
| Race/Reference HLA allele | AA position (residue) | B* alleles shared the same residue | Case AA Freq | Control AA Freq | P | OR (95% CI) | Adjusted P |
| 67 (Cys) | B*14:01, B*14:02, B*15:10, B*15:18, B*27:02, B*27:05, B*38:01, B*39:01, B*39:06, B*39:24, B*73:01 | 0.18 | 0.11 | 0.057 | 2.2 (1.2,3.7) | 0.008 | |
| 97 (Trp) | B*14:01, B*14:02 | 0.08 | 0.03 | 0.02 | 3.8 (1.7,7.6) | 0.003 | |
| 163 (Thr) | B*14:01, B*14:02, B*08:01, B*18:01, B*18:02, B*18:13, B*18:18, B*37:01, B*38:01, B*39:01, B*39:06, B*39:24, B*41:01, B*41:02, B*42:01, B*42:02, B*55:01 | 0.41 | 0.27 | 0.002 | 1.9 (1.2,3.1) | 0.007 | |
| 63 (Asn) | B*14:01, B*14:02, B*07:02, B*08:01, B*15:10, B*18:01, B*38:01, B*39:01, B*39:03, B*39:06, B*39:10, B*42:01, B*42:02, B*51:01 | 0.85 | 0.58 | 0.02 | … | … | |
| 67 (Phe) | Phe: B*08:01, B*51:01, B*53:01, B*78:01 | 0.55 | 0.25 | 0.007 | … | … | |
Firth logistic regression included covariate adjustment of age, PC2, and PC3
In the African American subset, among the same 45 AA residues in the binding sites, there were 26 polymorphic AA residues in the available HLA-B alleles. The AA residues at positions 63 (Asn63: Asparagine) and 67 (Phe67: Phenylalanine) in HLA-B*35:01 reached the nominal significance threshold (Table 4). Phe67, present in HLA-B*35:01, HLA-B*08:01, and HLA-B*51:01, ranked at the top with a two-fold higher frequency in cases than in controls (frequency: 0.55 vs. 0.25, p=0.007).
Molecular docking of Sulfamethoxazole or Trimethoprim with B*14:01 and B*35:01:
For HLA-B*14:01, molecular docking predictions of non-covalent interactions (H bonds and van der Waal contacts) showed that neither SMX nor TMP were predicted to bind with high affinity to the antigen binding cleft of HLA-B*14:01. The estimated ΔG values were −6.7 kcal per mole for SMX and −6.4 kcal per mole for TMP (<−10 kcal per mole suggests high affinity binding, −6.5 to −9.0 moderate affinity, >−6.5 kcal per mole, low affinity). However, drug metabolites and peptides may form intermolecular contacts with HLA-B*14:01 by covalent interactions. It is notable that HLA-B*14:01 exhibited a chemically reactive side chain located at a site in the antigen binding cleft critical for peptide binding: a cysteine at position 67 (Cys67), the residue detected in our amino acid analysis. As the cysteine sulfhydryl group is often in a reactive thiolate form in cells, free unpaired cysteine residues have the potential to form covalent bonds with reactive groups (e.g., with peptides that contain cysteine, or other molecules with reactive groups). SMX is known to be metabolized into reactive forms, such as sulfonamide, that have the potential to bind Cys67 in the antigen binding cleft of HLA-B*14:01. These covalent bonds may partially explain the association between TMP-SMX and HLA-B*14:01, as metabolite binding at this site would result in altered peptide presentation to T cells. Based on the location of the reactive group in sulfonamide and the free sulfhydryl of Cys67 in the HLA-B*14:01 model, SMX was accommodated in a site comprised of residues associated with TMP-SMX DILI (Cys67, Trp97 and Thr163, Figures 3A and 3B). These data suggested functional interactions between HLA antigen binding clefts and reactive SMX metabolites.
Figure 3:

Results of molecular docking. Panels A and B: HLA-B*14:01 exhibited an unpaired cysteine at position 67 with the potential to bind reactive SMX metabolite sulfonamide. The molecular surface of a model of HLA-B*14:01 is shown in light blue. Sulfonamide is shown as sticks, white for carbon, blue for nitrogen, red for oxygen. Cys67, Trp97, and Thr163 are shown in red. Panels C and D: Molecular docking predicted interaction between SMX and the antigen binding cleft of HLA-B*35:01. The crystal structure of HLA-B*35:01 is shown (PDB code 6BJ8), colored based on sequence similarity to HLA-B*15:02. Blosum62 similarity values are: blue, 40–50, cyan, 50–60, green, 60–70, yellow, 70–80, orange 80–90, and red 90–100. Molecular docking (AutoDock Vina, ΔG=−7.6 kcal/mol) predicted SMX interaction with residues shared by HLA-B*35:01 and HLA-B*15:02 shown as sticks. Predicted H bond interactions are shown as black dashed lines. The position 67 (Phe67), which we detected in our AA analysis, was not predicted to form contact with SMX by molecular docking.
For HLA-B*35:01, molecular docking predicted SMX to bind better than TMP (estimated ΔG = −7.6 kcal/mol for SMX, versus ΔG = −6.9 kcal/mol for TMP) (Figures 3C and 3D). SMX was predicted to form hydrogen-bond interactions with residues in the antigen-binding clefts of HLA-B*35:01 that are identical in HLA-B*15:02, a previously identified HLA allele associated with TMP-SMX induced severe cutaneous adverse drug reactions such as SJS/TEN in a Thai population(12).
HLA-B*14:01 and HLA-B*35:01 in DILI patients due to other agents:
In the European American subset, among 20 drugs with at least 10 DILI cases, only six drug subsets, including TMP-SMX, have HLA-B*14:01 present (Supplementary Table S5). HLA-B*14:01 was the most prevalent in TMP-SMX with 5 of 51 patients carrying HLA-B*14:01 (CF=0.098). Isoniazid and minocycline rank second with two of 32 patients carrying HLA-B*14:01 (CF=0.063). We did not find HLA-B*14:01 in seven DILI patients due to non-antibacterial sulfonamides (3 celecoxib, 3 sulfasalazine, and 1 zonisamide DILI patients) (Supplementary Table S6). We further tested if HLA-B*14:01 is associated with DILI regardless of the causal drugs by performing multivariable logistic regression with age, gender, and PC1 through PC10 adjusted. Based on 903 high confidence DILI cases and 12,156 controls, the association between HLA-B*14:01 and DILI did not reach the significance level (OR (95% CI) = 1.58 (0.96–2.50), p=0.073).
For HLA-B*35:01 in the African American subset, three drug subsets, including TMP-SMX, have at least 10 DILI cases. HLA-B*35:01 was the most prevalent in the TMP-SMX cases, with 5 of 10 patients carrying HLA-B*35:01 (CF=0.5). The other two drugs were Isoniazid with 2 of 19 DILI patients carrying HLA-B*35:01 (CF=0.105), and amoxicillin with clavulanic acid with one of 10 patients carrying HLA-B*35:01 (CF=0.1). Among two African American patients with DILI caused by Darunavir, a non-antibacterial sulfonamide, one patient carried HLA-B*35:01 (Supplementary Table S6). We also tested if HLA-B*35:01 is associated with overall DILI, regardless of the causal drugs, on 124 cases and 5,439 controls by adjusting age and PC1, HLA-B*35:01 did not have a significant effect on overall DILI (OR (95% CI) = 1.06 (0.35–2.42), p >0.99).
Patient characteristics for HLA-B*14:01 and HLA-B*35:01 carriers and non-carriers:
We observed slightly longer latency of liver injury (median 29 vs. 21) and higher proportion of peripheral eosinophilia (40% vs. 16%) in TMP-SMX DILI patients with HLA-B*14:01 than those without the allele, but the differences were not statistically significant. No significant differences were found for liver biochemistries at onset and at peak, pattern of liver injury, recovery of liver biochemistries, severity and outcomes either (Supplementary Table S7). Similarly, the characteristics and the outcomes of African Americans with and without HLA-B*35:01 were not significantly different from each other.
Discussion
Our study found that HLA-B*14:01 was associated with TMP-SMX related DILI in European Americans with the risk being 9.2-fold higher among individuals carrying the allele than those without. The HLA-B*14:01 allele was about 5.5-fold more frequent in European American patients with TMP-SMX DILI than controls. This large frequency difference was also present in one of five TMP-SMX DILI validation cases carrying HLA-B*14:01 from the iDILIC cohort. For the African American subset, HLA-B*35:01 ranked at the top as a potential risk factor for TMP-SMX DILI. While only 10 African American TMP-SMX DILI patients are available in our dataset, half of them carried HLA-B*35:01, a much higher allele and carriage frequencies than in the control group. Although replication studies with a larger sample size are needed, our results are promising and consistent with previous genetic studies of drug-specific DILI that identified HLA Class I and II genes(8, 30, 31) as genetic risk factors for DILI.
In the European American subset, HLA-A*34:02, HLA-B*27:02, and HLA-C*08:02 were the other top alleles that showed a significant association with TMP-SMX DILI. However, we considered HLA-B*14:01 as the most promising risk factor for the following reasons. First, HLA-B*14:01 remained significant after correcting for multiple testing. Second, both HLA-A*34:02 and HLA-B*27:02 were present in only two TMP-SMX DILI patients. Together with their low frequency in controls, the estimated effect sizes (OR) were unstable, as shown by the large 95% CI. Therefore, further replication is definitely needed. Third, while HLA-C*08:02 is the most common allele among these four top alleles, HLA-C*08:02 is known to be in strong linkage disequilibrium with B*14:01/B*14:02 (32). In our data, all eight HLA-C*08:02 carriers carried either HLA-B*14:01 (five patients) or HLA-B*14:02 (three patients). Since HLA-B*14:02 did not show association with TMP-SMX DILI, the association signal of HLA-C*08:02 is more likely dependent on HLA-B*14:01. As we demonstrated in the conditional analysis, after adjusting for the effect of HLA-B*14:01, the significant effect of HLA-C*08:02 on TMP-SMX DILI was diminished.
HLA-B*14:01 and HLA-B*14:02 are highly correlated in terms of their predicted peptide-binding specificity. Despite this similarity, HLA-B*14:01 has much lower frequency than HLA-B*14:02 in the general population (e.g, AF= 0.008 vs. 0.03 in European Americans). Interestingly, within the TMP-SMX DILI subset, we observed an opposite phenomenon where HLA-B*14:01 had higher frequency than HLA-B*14:02 (AF=0.049 vs. 0.029). Further, the HLA-B*14:02 frequency was similar between cases and controls. This implied that the effect of HLA-B*14:01 was independent from HLA-B*14:02. To date, we have not found any reports linking HLA-B*14:01 to DILI in general or to drug-specific cases. Among all causal drugs in the DILIN cohort, HLA-B*14:01 was more prevalent in the TMP-SMX subset than in other drug-specific subsets or the subset caused by non-antibacterial sulfonamide like drugs. Therefore, if the effect HLA-B*14:01 on DILI risk we observed in this study is correct, it is specific to TMP-SMX only (and likely to sulfamethoxazole).
In the literature, the HLA-B*14:01 allele has been linked to protection against HIV progression and with lower plasma viral levels (pVL) in HIV patients (32, 33). Different AA residues at a single AA position can also a play role (protective or risk) in disease outcomes as illustrated in HIV related studies (33). In the current study, the main identified risk allele, HLA-B*14:01, has an unpaired cysteine at position 67. This provides clues not only to a shared epitope with other HLA alleles that could in theory be implicated in risk of DILI but also a clue to the structure of the reactive metabolites and specific groups of SMX such as sulfonamide RS(O)NR’2 that might be expected to covalently bind with host proteins.
In the smaller group of African American persons with TMP-SMX related DILI, HLA-B*35:01 appeared to be an important HLA risk allele. HLA-B*35:01 was previously reported to be associated with DILI caused by Polygonum multiflorum, a Chinese herbal medicine (34), in Chinese patients. However, the relationship between HLA-B*35:01 and DILI with other drugs has not been reported. HLA-B*35:01 is known to share peptide-binding specificities with HLA-B*51:01 and HLA-B*53:01. Our allelic cluster analysis also showed that the cluster of these three alleles had stronger association with TMP-SMX DILI than HLA-B*35:01 alone. The two significant AA residues in positions 63 (Asn63) and 67 (Phe67) identified are also present in HLA-B*51:01 and HLA-B*53:01. Therefore, it is likely that these three alleles are equally important, but this conclusion deserves further study in large cohorts of African Americans with DILI. On the other hand, molecular docking led us to HLA-B*15:01, which shares nearly identical peptide binding preference as HLA-B*35:01. Since HLA-B*15:01 was reported to associate with SJS/TEN due to TMP-SMX in Thai population, the similarity in the antigen-binding cleft between these two alleles offer potential clues to explain the role of HLA-B*35:01 in TMP-SMX DILI in African Americans.
Despite these comprehensive analyses suggesting HLA-B*14:01 and HLA-B*35:01 as risk factors for TMP-SMX induced DILI in European Americans and African Americans, respectively, the limitations in this study need to be considered. First, the numbers of cases were limited, a common problem in most drug-specific genetic association studies. Using our European American subset for a post-hoc power calculation, given 51 cases and 12,156 controls and assuming AF of the risk allele at 0.009 (same as HLA-B*14:01) and complete LD between the risk allele and the testing allele, we have 80% power to detect genetic relative risk at 5.9 for an allele at the same AF. This calculation implies that our sample size was sufficient to detect HLA-B*14:01 but will require more samples to detect other HLA risk alleles with smaller effect sizes. In addition, because of this sample size limitation, several approaches were used to enhance the analysis quality, including using large population control datasets for comparison, Fisher exact tests to accommodate the low counts, and logistic regression models with Firth penalization to reduce the potential bias of parameter estimates. With these strategies, we expected a reduced chance of biased findings. Second, while HLA genotype data in DILI patients were obtained from advanced sequencing techniques, the HLA data in population controls (eMERGE-1 and PAGE) were obtained fromimputation of SNPs in the MHC region. This may raise concerns if HLA calls are inaccurate in the control group. The imputation quality by various programs have been compared before. Karnes et al.(35) reported 97.6% concordance rate for European Americans and 92.9% concordance rate for African Americans using HIBAG. Particularly, alleles in Class I genes had over 98% concordance rates. Therefore, with the large sample size of the control groups used in this study, the impact of minor imputation errors on the result should be small, particularly for the Class I genes. Third, binding interactions between SMX and TMP were predicted, rather than being experimentally determined. Moreover, predictions of SMX and TMP contact with HLA-B*14:01 were based on an atomic model (as opposed to a solved crystal structure). However, HLA-B*14:01 is nearly identical to the crystal structure of HLA-B*14:02, with only one difference in α1 and α2 domains, residues 1–180, that form the antigen binding cleft: (Ser11 vs. Ala11). Since the side chain at this position is not oriented toward peptide ligands (in class I HLA molecules, the side chain at position 11 is oriented towards the α1 helix), this polymorphic difference between HLA-B*14:01 and –B*14:02 is unlikely to influence reduce confidence in modeling the antigen binding cleft of HLA-B*14:01. Overall, the results from our molecular docking may to help us understanding interactions with SMX and its metabolites.
Given the rarity of liver injury due to TMP-SMZ and the relatively small effect size for the associated HLA alleles, our observations are not necessarily immediately clinically actionable from the perspective of pre-prescription screening to avoid a clinical event. However, our observations may help to both inform the immunopathogenesis of DILI related to trimethoprim-sulfamethoxazole and also help in the risk stratification and early diagnosis of such patients. Indeed, in future state where integrated health care systems (e.g., Geisenger) undertake whole exome or whole genome sequencing of all their patients, our genetic association findings may assist in the thorough evaluation of risk - benefit ratios and help promote safe prescription practices.
Supplementary Material
Acknowledgements:
We are extremely grateful to all patients and study coordinators participated in the DILIN study. We also thank NCBI/dbGaP at http://www.ncbi.nlm.nih.gov/gap approved our application to obtain control datasets. The data of non-DILI control subjects were from eMERGE-I and PAGE GWAS studies from dbGaP. The eMERGE network is a consortium of five participating sites (Group Health Seattle, Marshfield Clinic, Mayo Clinic, Northwestern University, and Vanderbilt University) funded by the National Human Genome Research Institute (NHGRI). Funding supports for the cohort and genotyping of eMERGE samples are as the following: (1) Group Health Seattle: U01AG006781 and U01HG004438 with genotyping performed at Johns Hopkins University; (2) Mayo clinic: UOIHG004599 and U01HG004424 with genotyping performed at The Broad Institute; (3) Marshfield Clinic: U01HG004608 and U01HG004438 with genotyping performed at Johns Hopkins University; (4) Northwestern University: U01HG04603 and U01HG004424 with the genotyping performed at The Broad Institute; (5) Vanderbilt University: U01HG004603 and U01HG004424 with the genotyping performed at The Broad Institute. The eMERGE GWAS datasets were from dbGaP accession number phs000360.v3.p.
PAGE dataset was from the Charles Bronfman Institute for Personalized Medicine (IPM) BioMe BioBank at the Icahn School of Medicine at Mount Sinai (New York). Phenotype data collection was supported by The Andrea and Charles Bronfman Philanthropies. Funding support for genotyping, which was performed at The Center for Inherited Disease Research (CIDR), was provided by the NIH (U01HG007417). The GWAS data were from dbGaP accession number phs000925.v1.p1.
Financial Support: The DILIN (https://dilin.dcri.duke.edu/) is supported by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health (NIH) as a Cooperative Agreement (U01s) under the following grants: U01-DK065176 and U24-DK065176 (Duke), U01-DK065211 (Indiana), U01-DK065201 (UNC), U01-DK065184 (Michigan), U01-DK065193 (UConn), U01-DK065238 (UCSF/CPMC), U01-DK083023 (UTSW), U01-DK083020 (USC), U01-DK082992 (Mayo), U01-DK083027 (TJH/UPenn), and U01-DK100928 (Icahn).
Disclosures: Chalasani and Watkins have consulting agreements and research grants from several pharmaceutical companies. Nicoletti is also an employee of Sema4. They all declare that their engagement does not have direct or significant relationship to this paper.
Abbreviations:
- AA
amino acid
- AF
allele frequency
- CF
carriage frequency
- CI
confidence interval
- DILI
drug-induced liver injury
- DILIN
drug-induced liver injury network
- GWAS
genome wide association study
- HLA
human leukocyte antigen
- MHC
major histocompatibility Complex
- OR
odds ratio
- PC
principal component
- TMP-SMX
trimethoprim-sulfamethoxazole
- SNP
single nucleotide polymorphism
Footnotes
Website links:
1000 genomes: https://www.internationalgenome.org/
NCBI dbGap: https://www.ncbi.nlm.nih.gov/gap/
BeTheMatch: https://bioinformatics.bethematchclinical.org
AFND: http://www.allelefrequencies.net/
MHCcluster: http://www.cbs.dtu.dk/services/MHCcluster/)
Immuno Polymorphism Database: https://www.ebi.ac.uk/ipd/imgt/hla/align.html
References:
- 1.Sulfamethoxazole-Trimethoprim In: LiverTox: Clinical and Research Information on Drug-Induced Liver Injury (https://livertox.nlm.nih.gov/SulfamethoxazoleTrimethoprim.htm). Bethesda (MD), 2012. [PubMed] [Google Scholar]
- 2.US Food & Drug Admission. FDA approved drug: Bactrim. In; 2019. p. https://www.accessdata.fda.gov/drugsatfda_docs/label/2013/017377s017068s017073lbl.pdf.
- 3.Chalasani N, Bonkovsky HL, Fontana R, Lee W, Stolz A, Talwalkar J, Reddy KR, et al. Features and Outcomes of 899 Patients With Drug-Induced Liver Injury: The DILIN Prospective Study. Gastroenterology 2015;148:1340–1352 e1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chalasani N, Reddy KRK, Fontana RJ, Barnhart H, Gu J, Hayashi PH, Ahmad J, et al. Idiosyncratic Drug Induced Liver Injury in African-Americans Is Associated With Greater Morbidity and Mortality Compared to Caucasians. Am J Gastroenterol 2017;112:1382–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bjornsson E, Jerlstad P, Bergqvist A, Olsson R. Fulminant drug-induced hepatic failure leading to death or liver transplantation in Sweden. Scand J Gastroenterol 2005;40:1095–1101. [DOI] [PubMed] [Google Scholar]
- 6.Pavlos R, Mallal S, Phillips E. HLA and pharmacogenetics of drug hypersensitivity. Pharmacogenomics 2012;13:1285–1306. [DOI] [PubMed] [Google Scholar]
- 7.Urban TJ, Daly AK, Aithal GP. Genetic basis of drug-induced liver injury: present and future. Semin Liver Dis 2014;34:123–133. [DOI] [PubMed] [Google Scholar]
- 8.Urban TJ, Nicoletti P, Chalasani N, Serrano J, Stolz A, Daly AK, Aithal GP, et al. Minocycline hepatotoxicity: Clinical characterization and identification of HLA-B *35:02 as a risk factor. J Hepatol 2017;67:137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicoletti P, Werk AN, Sawle A, Shen Y, Urban TJ, Coulthard SA, Bjornsson ES, et al. HLA-DRB1*16: 01-DQB1*05: 02 is a novel genetic risk factor for flupirtine-induced liver injury. Pharmacogenet Genomics 2016;26:218–224. [DOI] [PubMed] [Google Scholar]
- 10.Nicoletti P, Aithal GP, Bjornsson ES, Andrade RJ, Sawle A, Arrese M, Barnhart HX, et al. Association of Liver Injury From Specific Drugs, or Groups of Drugs, With Polymorphisms in HLA and Other Genes in a Genome-Wide Association Study. Gastroenterology 2017;152:1078–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ozkaya-Bayazit E, Akar U. Fixed drug eruption induced by trimethoprim-sulfamethoxazole: evidence for a link to HLA-A30 B13 Cw6 haplotype. J Am Acad Dermatol 2001;45:712–717. [DOI] [PubMed] [Google Scholar]
- 12.Kongpan T, Mahasirimongkol S, Konyoung P, Kanjanawart S, Chumworathayi P, Wichukchinda N, Kidkeukarun R, et al. Candidate HLA genes for prediction of co-trimoxazole-induced severe cutaneous reactions. Pharmacogenet Genomics 2015;25:402–411. [DOI] [PubMed] [Google Scholar]
- 13.Rochon J, Protiva P, Seeff LB, Fontana RJ, Liangpunsakul S, Watkins PB, Davern T, et al. Reliability of the Roussel Uclaf Causality Assessment Method for assessing causality in drug-induced liver injury. Hepatology 2008;48:1175–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontana RJ, Watkins PB, Bonkovsky HL, Chalasani N, Davern T, Serrano J, Rochon J, et al. Drug-Induced Liver Injury Network (DILIN) prospective study: rationale, design and conduct. Drug Saf 2009;32:55–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, et al. A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chalasani N, Fontana RJ, Bonkovsky HL, Watkins PB, Davern T, Serrano J, Yang H, et al. Causes, clinical features, and outcomes from a prospective study of drug-induced liver injury in the United States. Gastroenterology 2008;135:1924–1934, 1934 e1921–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng X, Shen J, Cox C, Wakefield JC, Ehm MG, Nelson MR, Weir BS. HIBAG--HLA genotype imputation with attribute bagging. Pharmacogenomics J 2014;14:192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maiers M, Gragert L, Klitz W. High-resolution HLA alleles and haplotypes in the United States population. Hum Immunol 2007;68:779–788. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez-Galarza FF, Takeshita LY, Santos EJ, Kempson F, Maia MH, da Silva AL, Teles e Silva AL, et al. Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res 2015;43:D784–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Statist. Soc 1995:289–300. [Google Scholar]
- 21.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet 2002;70:425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoof I, Peters B, Sidney J, Pedersen LE, Sette A, Lund O, Buus S, et al. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics 2009;61:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aithal GP, Watkins PB, Andrade RJ, Larrey D, Molokhia M, Takikawa H, Hunt CM, et al. Case definition and phenotype standardization in drug-induced liver injury. Clin Pharmacol Ther 2011;89:806–815. [DOI] [PubMed] [Google Scholar]
- 24.Cirulli ET, Nicoletti P, Abramson K, Andrade RJ, Bjornsson ES, Chalasani N, Fontana RJ, et al. A Missense Variant in PTPN22 is a Risk Factor for Drug-induced Liver Injury. Gastroenterology 2019;156:1707–1716 e1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SG. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res 2015;43:D423–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar P, Vahedi-Faridi A, Saenger W, Merino E, Lopez de Castro JA, Uchanska-Ziegler B, Ziegler A. Structural basis for T cell alloreactivity among three HLA-B14 and HLA-B27 antigens. J Biol Chem 2009;284:29784–29797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T. Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc 2009;4:1–13. [DOI] [PubMed] [Google Scholar]
- 28.Sibener LV, Fernandes RA, Kolawole EM, Carbone CB, Liu F, McAffee D, Birnbaum ME, et al. Isolation of a Structural Mechanism for Uncoupling T Cell Receptor Signaling from Peptide-MHC Binding. Cell 2018;174:672–687 e627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem 2009;30:2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lucena MI, Molokhia M, Shen Y, Urban TJ, Aithal GP, Andrade RJ, Day CP, et al. Susceptibility to amoxicillin-clavulanate-induced liver injury is influenced by multiple HLA class I and II alleles. Gastroenterology 2011;141:338–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singer JB, Lewitzky S, Leroy E, Yang F, Zhao X, Klickstein L, Wright TM, et al. A genome-wide study identifies HLA alleles associated with lumiracoxib-related liver injury. Nat Genet 2010;42:711–714. [DOI] [PubMed] [Google Scholar]
- 32.Valenzuela-Ponce H, Alva-Hernandez S, Garrido-Rodriguez D, Soto-Nava M, Garcia-Tellez T, Escamilla-Gomez T, Garcia-Morales C, et al. Novel HLA class I associations with HIV-1 control in a unique genetically admixed population. Sci Rep 2018;8:6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.International HIVCS, Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 2010;330:1551–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li C, Rao T, Chen X, Zou Z, Wei A, Tang J, Xiong P, et al. HLA-B*35:01 Allele Is a Potential Biomarker for Predicting Polygonum multiflorum-Induced Liver Injury in Humans. Hepatology 2019;70:346–357. [DOI] [PubMed] [Google Scholar]
- 35.Karnes JH, Shaffer CM, Bastarache L, Gaudieri S, Glazer AM, Steiner HE, Mosley JD, et al. Comparison of HLA allelic imputation programs. PLoS One 2017;12:e0172444. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


