Abstract
The COVID-19 pandemic has negatively affected human life globally. It has led to economic crises and health emergencies across the world, spreading rapidly among the human population and has caused many deaths. Currently, there are no treatments available for COVID-19 so there is an urgent need to develop therapeutic interventions that could be used against the novel coronavirus infection. In this research, we used computational drug design technologies to repurpose existing drugs as inhibitors of SARS-CoV-2 viral proteins. The Broad Institute’s Drug Repurposing Hub consists of in-development/approved drugs and was computationally screened to identify potential hits which could inhibit protein targets encoded by the SARS-CoV-2 genome. By virtually screening the Broad collection, using rationally designed pharmacophore features, we identified molecules which may be repurposed against viral nucleocapsid and non-structural proteins. The pharmacophore features were generated after careful visualisation of the interactions between co-crystalised ligands and the protein binding site. The ChEMBL database was used to determine the compound’s level of inhibition of SARS-CoV-2 and correlate the predicted viral protein target with whole virus in vitro data. The results from this study may help to accelerate drug development against COVID-19 and the hit compounds should be progressed through further in vitro and in vivo studies on SARS-CoV-2.
Keywords: SARS-CoV-2, COVID-19, drug repurposing, pharmacophore, virtual screening, nucleocapsid protein, non-structural proteins
Graphical Abstract
Communicated by Ramaswamy H. Sarma
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is the causative agent of the ongoing COVID-19 pandemic which has become a global public health emergency (WHO, 2020). The human-to-human transmission is rapid, making the virus highly contagious and at present, there are approximately 18.7 million cases reported worldwide (as of 5th August 2020, Worldometer, https://www.worldometers.info/coronavirus/). Currently, there are no drug treatments or vaccinations and the global research community has united in the search for a cure of this infectious disease.
1.1. Structural considerations of SARS-CoV-2
Phylogenetic relationships and genomic structures indicate that the virus causing COVID-19 disease (belonging to the Betacoronavirus genus) has a close sequence similarity to severe acute respiratory syndrome-related coronaviruses (SARS-CoV) (Mousavizadeh & Ghasemi, 2020). Due to these similarities, the coronavirus study experts of the International Committee on Taxonomy of Viruses (ICTV) named the new virus as SARS-CoV-2 (Cascella et al., 2020). Severe acute respiratory syndrome (SARS) and the Middle East respiratory syndrome (MERS) which also belong to the Betacoronavirus genus are associated mainly with nosocomial spread, but the SARS-CoV-2 is more widely transmitted in the community (Munster et al., 2020).
The SARS-CoV viruses have a positive-sense, single-stranded RNA genome (ranging from 27-32 kilobase pairs) in an enveloped structure with a 5’cap and 3′-poly-A-tail (Astuti & Ysrafil, 2020; Fehr & Perlman, 2015). The CoV genome has at least six open reading frames (ORFs), among these, there are two large overlapping ORFs, ORF 1a and ORF 1b, which guide the synthesis of polyproteins 1a/1ab (pp1a, pp1b). These polyproteins are then processed by the viral proteases, chymotrypsin-like protease (3CLpro) or main protease (Mpro) (Cascella et al., 2020). In addition to 3Clpro, the viral papain-like protease (PLpro) is responsible for producing 16 non-structural proteins (nsps) by cleaving polyproteins which are encoded by the large ORF1ab replicase gene (Cascella et al., 2020; Fehr & Perlman, 2015). The remaining ORFs encode for viral structural proteins, such as spike, membrane, envelope and nucleocapsid proteins (S, E, M, and N) (Perlman & Netland, 2009). In addition to these proteins, there are certain genes in the 3′ region between S-E-M-N which are known to encode for accessory proteins such as 3a, 3b, 6, 7a, 7b, 8a, 8b and 9b (Marra et al., 2003; Snijder et al., 2003). The genomic RNA is packed inside a bead-like structure which is known as a capsid and is formed by the N protein, while the remaining structural proteins form an envelope surrounding the capsid.
The remaining proteins such as several non-structural proteins contribute to viral RNA replication or transcription (Snijder et al., 2016) whereas, the accessory proteins interact with host cells to help the virus evade the host immune system thereby increasing their virulence (Menachery et al., 2017). The viral cycle begins with binding of the spike glycoprotein to the ACE2 host receptor (angiotensin-converting enzyme 2) with the help of the TMPRSS2 protease (Hoffmann et al., 2020). After viral entry into the host cells, the virus un-coats and releases the genome which is then transcribed and translated (Mousavizadeh & Ghasemi, 2020). The coronavirus replicase is reported to employ many RNA processing enzymes such as endoribonuclease, 3′-to-5′ exoribonuclease, ADP ribose 1′-phosphatase and 2′-O-ribose methyltransferase (Ziebuhr, 2005).
Scientists all over the world are in a race to find a vaccine for SARS-CoV-2 but until then there is an urgent need to discover potent anti-COVID-19 agents which can be used for treating the viral infection. Spike protein and viral proteases have been the major targets of investigation for researchers around the world but in this paper, we have focused on non-structural proteins and the nucleocapsid protein.
1.2. Repurposing of existing drugs against SARS-CoV-2
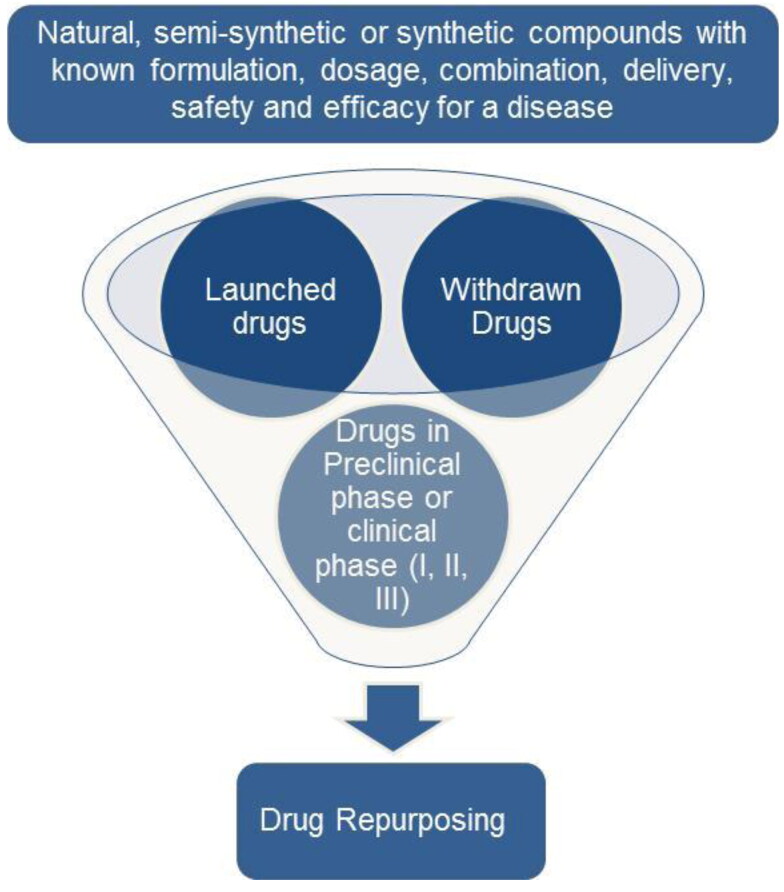
At present, there are no globally available drug treatments or a vaccination available for COVID-19. Favipiravir, which is an RNA polymerase inhibitor, is used as a second-line treatment of influenza outbreaks. It is marketed in Japan under the name Avigan and in China as Favilavir. Favipiravir under the brand name Avifavir was resynthesized in Russia and based on Phase II/III clinical trial results against SARS-CoV-2 infection, the Russian Ministry of Health granted a conditional marketing of Avifavir. This makes it the only approved oral drug for the treatment of moderate COVID-19 infection (Ivashchenko et al., 2020). The patients who develop severe symptoms with respiratory failure are given oxygen therapy and artificial ventilator support (Cascella et al., 2020). There is a need to find therapeutic interventions and to achieve this existing drugs can be repurposed (Cavalla et al., 2017; Schneider, 2018) for treating the SARS-CoV-2 infection (Pandey et al., 2020) (Figure 1). Approximately one-third of drug approvals involve repurposed drugs (Talevi & Bellera, 2020). There is a broad range of choices which can curb the effects of viral infection and also prevent it. These choices include the use of small molecule medicines, monoclonal antibodies, vaccines, interferon treatments and oligonucleotide-based drugs (Li & De Clercq, 2020) but the development process of a vaccine or a novel drug takes at least 10 years. The repurposing of existing vaccines has recently been proposed (O’Neill & Netea, 2020) but, considering the severity of the pandemic, we propose examining currently available drugs belonging to diverse therapeutic areas for possible treatment of COVID-19. Drug repurposing involves redeveloping a drug (can be an approved, shelved, discontinued or investigational drug) for a different use than originally intended (Ashburn & Thor, 2004). The main SARS-CoV-2 protein targets along with some proposed therapies are (Wu et al., 2020):
Figure 1.
The flow chart depicts the process of drug repurposing where a marketed drug or a drug which has already been tested safe in preclinical and clinical settings is repurposed for treating a new disease. These drugs can either be natural, semi-synthetic or synthetic compounds.
Therapies which act on viral enzymes or functional proteins, RNA synthesis and replication. (Targets – PLpro, RdRp (RNA-dependent RNA polymerase), 3CLpro, Helicase-nsp13).
Therapies which block the virus from binding to human cell receptors, acting on structural proteins. (Targets – Spike, nucleocapsid and envelope proteins).
Therapies which restore the host’s innate immunity, inhibiting virulence factor. (Targets – nsp1, nsp3c, ORF7a).
Therapies acting on hosts’ specific receptors thereby preventing viral entry. (Target – ACE2 receptor).
The combined efforts of various disciplines such as computational biology, chemistry, genetics, immunology and other related fields have contributed towards finding a cure for the coronavirus infection. Currently, there are ongoing clinical trials of primarily anti-viral (remdesivir, lopinavir/ritonavir, favipiravir) and anti-malarial (chloroquine and hydroxychloroquine) drugs which are showing mixed responses. In this paper, we describe an in-silico study which details compounds which are predicted to inhibit the viral enzymes or functional proteins, RNA synthesis and replication, thereby displaying the potential to act against SARS-CoV-2.
1.3. Computer-aided drug design and pharmacophore based virtual screening
Computational approaches have permeated throughout the field of medical sciences, augmenting the drug discovery and development process (Schneider, 2018). This has enabled researchers to find hit or lead compounds more reliably, exerting a therapeutic effect by binding to the 3D structure of the target receptor (primarily proteins). These 3D structures are obtained from X-ray crystallography or, to a lesser extent, nuclear magnetic resonance (NMR) spectroscopy and structures often have a co-resolved ligand. Hence, the structures can either be holo (having a co-crystallized moiety) or apo (no co-crystallized moiety). In silico studies involve selecting a biological target and then analysing/screening potentially active compounds against the chosen target (Walters et al., 1998).
Computer-aided drug design (CADD) technologies can be used for screening large databases of drug compounds, the results of which should be then validated with biological assay, providing insight into the underlying mechanism of action. This is the approach that we have taken in this study. The two leading strategies used for virtual screening (VS) are ligand-based drug design (LBDD) and structure-based drug design (SBDD). One of the main approaches for LBDD is pharmacophore-based virtual screening (PBVS) (McKay et al., 2012) and for SBDD is docking based virtual screening (DBVS) (Mawhinney et al., 2014). The 3D structure of the biological target is used for pharmacophore features generation and for docking of hit compounds. PBVS strategies can be used as a complementary approach to DBVS for filtering large drug libraries in order to remove compounds lacking the selected pharmacophore features (essential for binding affinity) which increases enrichment rates when compared with docking alone (McInnes, 2007; Muthas et al., 2008; Nevin et al., 2012).
A pharmacophore is a geometric representation of molecular interactions which enable the compound to trigger or block the biological target’s activity (Leach et al., 2010). The most common pharmacophore features are hydrogen-bond donors/acceptors, cations/anions, hydrophobic regions and the presence of aromatic rings. PBVS can further be classified into ligand-based pharmacophores (LBPs) and protein structure-based pharmacophores (SBPs). LBPs involve pharmacophore features that are derived from overlaid ligand structures whereas, SBPs involve pharmacophore feature selection from the 3D structures of protein targets (Leach et al., 2010; Sanders et al., 2012). In silico methods for drug design/development have been widely used in order to search existing drug databases for potential active compounds against the SARS-CoV-2 viral proteins.
1.4. Drug repurposing using in silico drug designing method for SARS-CoV-2
Many studies have focussed on repurposing of existing drugs for use against COVID-19 and VS of drug libraries has accelerated the search for a treatment of COVID-19. A global collaboration examining the SARS-CoV-2 protein interaction map, has identified 29 approved drugs that may have activity against SARS-CoV-2 (Gordon et al., 2020) opening up further treatment possibilities.
Mpro has been a popular inhibition target and diverse drugs have been reported to block its biological action (Jiménez-Alberto et al., 2020; Jin et al., 2020). A study by Pant et al. (2020) used a DBVS approach to calculate the binding affinity of FDA approved protease inhibitors against SARS-CoV-2 Mpro. The study screened a large drug database (ZINC/ChEMBL) and found that cobicistat, ritonavir, lopinavir and darunavir were among the top ranked molecules. The drugs reported to show inhibitory activity against Mpro belonged to various therapeutic domains, including an antidepressant drug named vortioxetine. However, the anti-viral properties of vortioxetine is not yet clear (Xiong et al., 2020). Apart from the well-known drugs (remdesivir, ritonavir, lopinavir) researchers are now also focusing on understudied drugs such as methisazone which is an antiviral drug known to inhibit mRNA and protein synthesis in POX viruses (Patskovsky et al., 1996). A PBVS study reported the inhibition activity of methisazone against SARS-CoV-2 proteins (Shah et al., 2020). A DBVS study by Kumar et al., 2020 showed the inhibition potential of tipranavir and raltegravir against SARS-CoV-2 Mpro. Phylogenetic analysis of the SARS-COV-2 genome shows no sign of diversification or mutation. Therefore the repurposed drugs can be used at the pan-community level against the COVID-19 (Kumar et al., 2020). Molecular docking and dynamics simulations indicated the potential use of a plant-based phenol derivative called theaflavin digallate against Mpro due to a good predicted binding affinity (Peele et al., 2020). Many plant-derived naturally occurring compounds are known to have anti-viral activity (Akram et al., 2018). Another interesting study screened dark chemical matter and food chemicals using DBVS to uncover Mpro inhibitors. The study indicated the potential of certain food chemicals such as folic acid, dihydrofolic acid and tetrahydrofolate to inhibit the Mpro active site (Santibáñez-Morán et al., 2020).
The SARS-CoV-2 nsps are now emerging as a new avenue of investigation for determining inhibitors that could help stop the viral replication process. A recent study using homology modelling and DBVS identified that toremifene inhibited SARS-CoV-2 nsp14 with a predicted binding affinity of −7.2 kcal/mol (Martin & Cheng, 2020). Another study focused on using DBVS to determine the inhibitory activity of FDA approved drugs against nsp16. The results indicated the potential of anti-viral drugs (maraviroc and raltegravir) and an anti-inflammatory drug (prednisolone) to be effective drug candidates against nsp16 (Tazikeh-Lemeski et al., 2020). Another such study integrated a data-driven repositioning framework to predict effective drug candidates with in silico screening followed by wet-lab validation. The result of this study indicated the potential of CVL218, which is a poly-ADP-ribose polymerase 1 (PARP1) inhibitor currently in a Phase 1 clinical trial, for the treatment of COVID-19 (Ge et al., 2020). The DBVS result highlighted the ability of CVL218 to bind to the N-terminal domain of the SARS-CoV-2 nucleocapsid protein, thereby providing insight into the anti-viral activity of the PARP1 inhibitor. To validate the in silico finding, an in vitro assay was performed which showed inhibitory activity of CVL218 against SARS-CoV-2 (Ge et al., 2020).
As data regarding SARS-CoV-2 studies are being released all around the world on numerous platforms, many agencies are working on providing a common source which can facilitate open data sharing and analysis to accelerate the development of COVID-19 treatments. The OpenData portal is an example of a platform which provides real-time datasets on assays that cover a wide spectrum of information on the novel coronavirus life cycle, host and viral targets, drug libraries and experimental therapeutics against the targets (Brimacombe et al., 2020).
This study used a rational PBVS CADD approach to repurpose existing drugs, detailed in the Broad Institute’s Drug Repurposing Hub, for utility against SARS-CoV-2 infection. The pharmacophore features were generated around the co-crystallized ligands in 3D crystal structures of SARS-CoV-2 proteins obtained from the PDB (currently 282 structures and increasing) followed by a computational search on the Broad database (5,836 molecules). The database contains drugs/compounds that are launched, pre-clinical, in clinical trials (phases I, II and III) and withdrawn drugs. Due to ongoing global drug design efforts against viral spike and protease proteins, we analysed alternative SARS-CoV-2 proteins: non-structural proteins and nucleocapsid protein. The hit compounds were then searched on the ChEMBL database for matching SARS-CoV-2 inhibition assay data.
We hypothesize that our predicted hit compounds demonstrating positive bioassay results against the SARS-CoV-2 in the ChEMBL database can indicate an underlying anti-viral mechanism of action. These hit compounds should be further tested in vitro or in vivo for their applicability against COVID-19 and to validate the viral target. We recommend that predicted hit compounds with negative in vitro results should be re-assayed against SARS-CoV-2 as confirmation.
2. Materials and methods
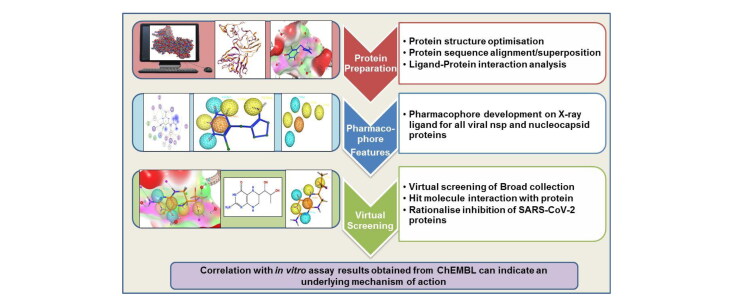
The research methodology conducted in this study involved generating pharmacophore features around the co-crystalized ligand in the 3D crystal structure of viral proteins followed by the VS for the hit compounds.
2.1. Websites, databases and software
The Broad Institute Drug Repurposing Hub was used for extracting the repurposing drug datasets. The database contained 5,836 molecules that were used for performing the VS step. (https://www.broadinstitute.org/drug-repurposing-hub). OMEGA 3.1.1.2 (OMEGA, 2018; Hawkins et al., 2010) was used to generate 50 conformers of each molecule. The ChEMBL website (https://www.ebi.ac.uk/chembl/) was accessed for compound inhibition data on SARS-CoV-2. The Protein Data Bank (PDB) (Berman et al., 2000) website was used for downloading 3D crystal structures of both holo or apo viral proteins. MOE 2019.0101 (Molecular Operating Environment (MOE),), 2020) was used to generate pharmacophore features from the visualised interactions, for VS of the Broad collection and analysing the results.
2.2. Protein preparation
The crystal structures of the SARS-CoV-2 proteins were downloaded from the PDB. In this study, the 3D crystal structures of non-structural proteins (nsp3, nsp7-nsp8-nsp12 complex, nsp16-nsp10 complex and nsp15) and the nucleocapsid protein of SARS-CoV-2 were studied as holo structures were available. Each protein was loaded in MOE and the Quick Preparation function was used with the default Amber10 force field in order to consider explicit hydrogen atoms, tautomeric states and possible breaks in protein structure prior to conducting restrained all atom molecular mechanics minimisation and electrostatics calculations.
2.3. 3D Protein structure alignment/superposition
The PDB had multiple crystal structures of a single protein with different co-crystallised ligands. Hence, it becomes necessary to analyse whether there were any structural differences among these protein structures. The protein alignment/superposition step was performed in MOE on different structures of the same protein (Table 1). First, the sequence alignment step was performed followed by 3D structural superposition using default parameters.
Table 1.
Proteins with their respective PDB IDs that were used for the superposition step with resulting RMSD values.
| S. No. | Protein | PDB IDs of the superposed proteins | RMSD values (Å) after protein superposition |
|---|---|---|---|
| 1. | Nsp 3 | 6YWL, 6YWK | 1.433 |
| 2 | Nsp 10-Nsp 16 complex | 6W4H, 7BQ7 | 0.478 |
| 3. | Nsp 7-Nsp 8- Nsp 12 complex | 7BV2, 7BW4 | 0.496 |
| 4. | Nsp 15 | 6WXC, 6WLC, 6X4I | 0.332 |
| 5. | Nucleocapsid protein | 6WKP, 6M3M | 1.092 |
After superposition of the proteins, the RMSD values were calculated. The holo protein structures were visually inspected to determine if there were any changes in the position of the binding site amino acids due to the ligand.
2.4. Pharmacophore generation
After studying the ligand-binding site interactions, pharmacophores were created in MOE using the unified scheme on the co-crystalised ligand. The pharmacophore features used were hydrogen bond acceptor (Acc – cyan colour), hydrogen bond donor (Don – purple colour), hydrophobic (Hyd – green colour), aromatic (Aro – orange colour) and charged groups such as cation (Cat – blue colour) or anion (Ani – red colour). The features were selected by carefully considering the ligand atoms responsible for binding interactions with the amino acid residues in the binding pocket and the default feature radius was used.
2.5. Virtual screening
The Broad Institute’s Drug Repurposing Hub Database was downloaded, the molecules were washed in MOE and the MMFF94x force field was used to generate 3D structures of each molecule. For each molecule, fifty conformers were generated using OMEGA (Hawkins et al., 2010, OMEGA, 2019) software. This database was used for all pharmacophore searches. In MOE, hit compounds were viewed in the binding site to observe the interactions that these molecules made with the binding pocket of the target protein. The hits obtained from VS were then combined with the washed Broad database to obtain other relevant information such as clinical phase, disease area, indication, mechanism of action and target for these hit molecules.
2.6. Chembl database SARS-CoV-2 activity data
The ChEMBL database was used to determine if the in silico hit molecules were active or inactive against SARS-CoV-2 by referring to inhibition assay data. The ChEMBL website had a separate link to only SARS-CoV-2 related data and search filters such as type of bioassay result. The data used was generated by Ellinger et al., 2020): Antiviral activity was determined as inhibition of SARS-CoV-2 induced cytotoxicity of Caco-2 cells at 10 µM after 48 h by high content imaging. The database file was downloaded to aid interrogation. These assay results were used to differentiate between active and inactive in silico hits against SARS-CoV-2.
3. Results
The PBVS CADD approach in our study resulted in multiple hit molecules predicted to bind to a variety of viral proteins. A number of the predicted active compounds have been previously validated in vitro to inhibit the activity of SARS-CoV-2 so our results indicate a possible mechanism of action of these compounds. The next step should be to conduct direct assay against these viral proteins to confirm on-target activity. For confirmation of inactivity, in silico hit compounds which displayed no response following in vitro studies should be re-assayed.
3.1. Non-structural protein 3
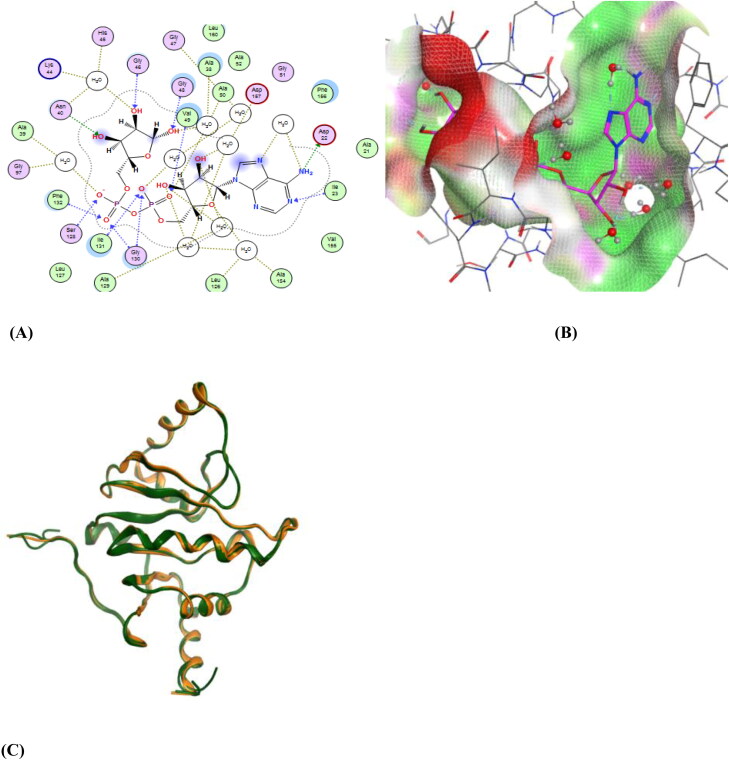
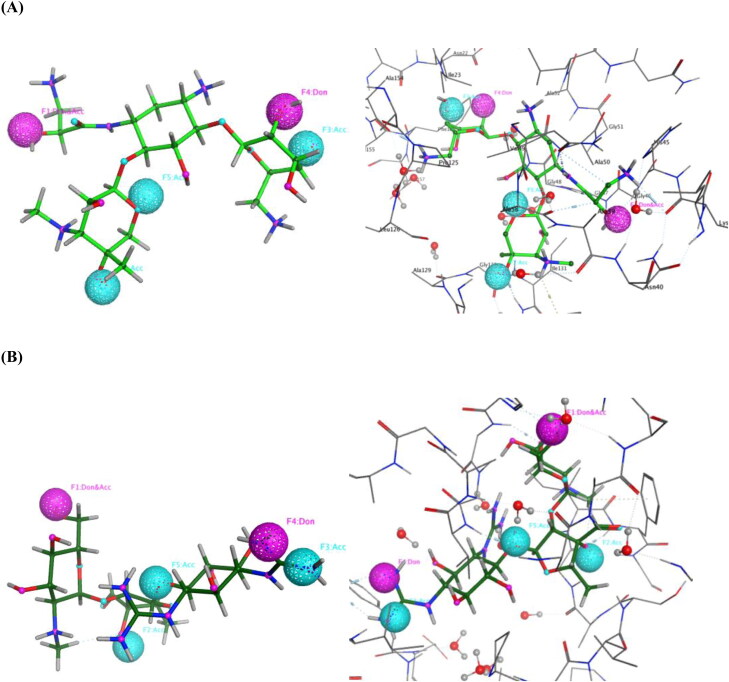
The SARS-CoV-2 non-structural protein 3 (nsp3) is known to cleave polypeptides, promote cytokine expression and block host innate immune response (Chen et al., 2020; Lei et al., 2018). The crystal structures of nsp3 (PDB IDs: 6YWL, 6YWK) were aligned and superposed which resulted in an RMSD of 1.433 Å after protein preparation. The two structures were similar and were properly superposed on each other. The co-crystallised ligands in the binding pocket of the two proteins were different. The 6YWL had adenosine-5-diphosphoribose (APR) as the main ligand in the binding site and 6YWK had 4-(2-hydroxyethyl)-1-piperazine ethanesulfonic acid (HEPES). The ligand present in the binding site caused slight changes in the amino acid position. The pharmacophore was generated using the 6YWL crystal structure as HEPES is a buffering agent and made fewer interactions in the binding site. Figure 2 shows the ligand interactions (2D) and site view (3D) of the ligand APR which is surrounded by hydrophilic and hydrophobic amino acid residues. Among them Asp22, Asp157 are polar acidic residues that interact with the ligand. There is also the presence of water molecules which form bridges between the ligand and protein. Other residues such as Gly46, Asn40, Gly48, Ser128, Gly130 are polar in nature whereas, Ile23, Val49, Phe132 are hydrophobic residues which contribute to the ligand interactions.
Figure 2.
(A) A 2D representation of the ligand-binding pocket showing attractive interactions between APR and amino acid residues. (B) A 3D representation of ligand APR (pink colour) interactions with amino acid residues with a surface representation. (C) Superposition of 6YWL and 6YWK showing structural similarity (Green colour – 6YWL, Orange colour – 6YWK).
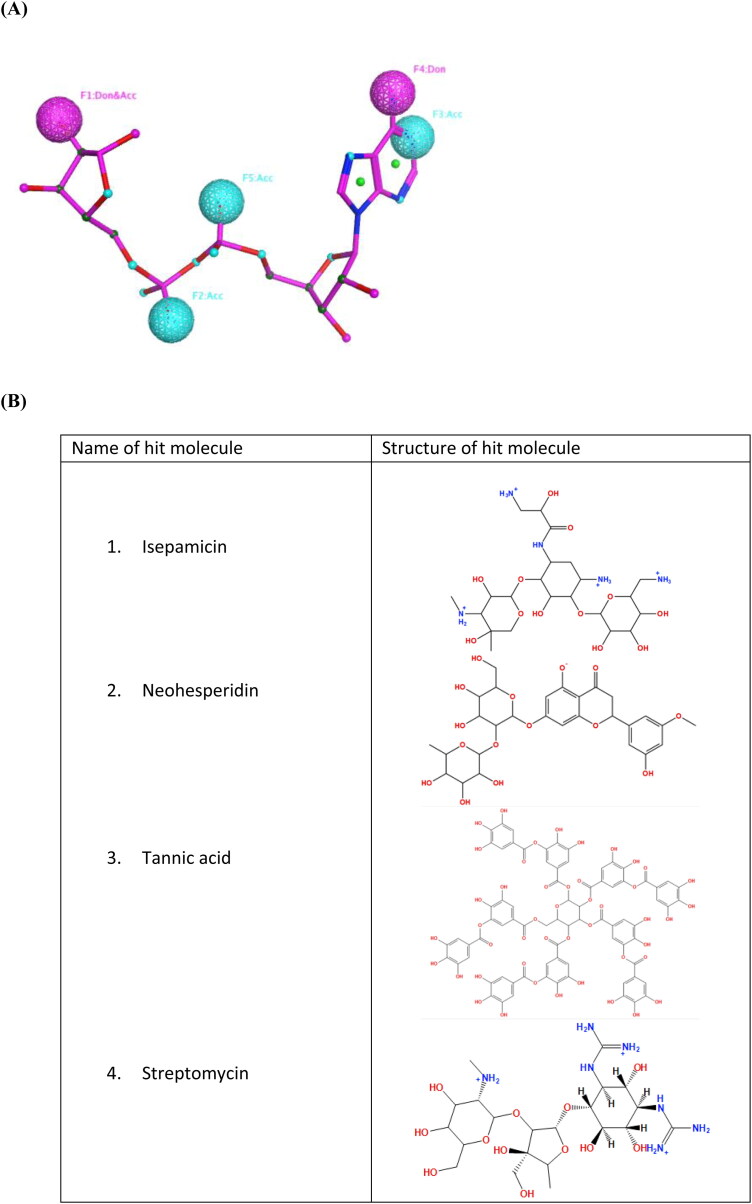
The pharmacophore features (5 in total) on the ligand were three acceptor, one donor and one dual donor & acceptor features. After completing the pharmacophore search, six hit molecules were identified. The six hits, with results for ChEMBL reported in vitro SARS-CoV-2 inhibition in brackets, were dihydrostreptomycin (-8.94%), hesperidin (-3.03%), isepamicin (20.73%), neohesperidin (5.55%), streptomycin (-5.98%) and tannic acid (28.53%) (Figure 3). These hit molecules had pharmacophore rmsdx values (rmsdx is the root‐mean‐square deviation between query features and their matching ligand annotation points including a penalty for each missing feature) ranging from 0.5444 to 0.7203 Å.
Figure 3.
(A) Pharmacophore features on APR (pink colour) from the 6YWL protein structure. Don: Hydrogen bond donor; Acc: Hydrogen bond acceptor. (B) Structures of selected hit molecules (isepamicin, neohesperidin, tannic acid and streptomycin) obtained after VS.
By visually analysing the interactions it was observed that dihydrostreptomycin, streptomycin neohesperidin and isepamicin fitted well inside the binding pocket of the protein. In addition to mapping to the pharmacophore, these molecules made extra hydrogen bonding interactions with the protein. Isepamicin and tannic acid are launched drugs and were active against SAR-CoV-2. Isepamicin fitted better inside the binding pocket than tannic acid as it made extra interactions with the Asn37 and Ala154 residues. Whereas, tannic acid was observed to clash with amino acid residues of the binding pocket due to its large size. In addition to isepamicin, streptomycin also made good interactions with the binding pocket (Figure 4) but in vitro assay showed no inhibition of SARS-CoV-2.
Figure 4.
(A) The pharmacophore features around the hit molecule Isepamicinn (light green colour) and its interactions with binding pocket amino acids. (B) The pharmacophore features around the hit molecule streptomycin (dark green colour) and its interactions with binding pocket amino acids. Don: Hydrogen bond donor; Acc: Hydrogen bond acceptor.
3.2. Non-structural protein 7, non-structural protein 8, non-structural protein 12 complex
The non-structural protein 7 (nsp7) and non-structural protein 8 (nsp8) complex along with co-factors is known to stimulate RNA-dependent RNA polymerase (RdRp) activity together with non-structural protein 12 (nsp12). Nsp12 alone also possesses minimal RdRp activity (Subissi et al., 2014). The crystal structures of nsp7-nsp8-nsp12 complexes (PDB IDs: 7BV2, 7BW4) were aligned and superposed which resulted in an RMSD of 0.496 Å. The two structures were similar and were properly superposed on each other. The 7BV2 structure had remdesivir as the main ligand in the binding site and 7BW4 had only a zinc metal ion. The ligand present in the binding site caused slight changes in the amino acid position. The pharmacophore was generated using the 7BV2 crystal structure. Figure S1 (Supplementary material) shows the ligand interactions (2D) and site view (3D) of the ligand remdesivir which is surrounded by hydrophilic amino acid residues. Among them, AspA760, AspA761 and AspA623 are polar acidic residues in the binding pocket. There is also the presence of magnesium ion which forms a bridge between the ligand and protein. Other residues such as SerA759, AsnA691 and SecB20 are polar in nature which contributes to the ligand interactions.
The pharmacophore features (5 in total) around the ligand were two anionic acceptor, one donor, one acceptor and one dual donor & acceptor features. Six hit molecules were identified to have the assigned pharmacophore features. The six hits, with results for ChEMBL reported in vitro SARS-CoV-2 inhibition in brackets, were NADPH (-9.07%), adenosine triphosphate (2.34%), Coenzyme-I (29.03%), Coenzyme-A (6.97%), nadide (-1.83%) and Hydroxysafflor Yellow A (-7.05%) (Supplementary material Figure S2). These hit molecules had rmsdx value ranging from 0.4004 to 0.6174 Å.
By visually analysing the interactions we observed that NADPH, adenosine-triphosphate, Coenzyme-I and nadide fitted well inside the binding pocket of the protein. These molecules also had moieties that made hydrogen bond interactions in addition to the pharmacophore query features. Coenzyme-I is in phase 2 clinical trials and was active against SAR-CoV-2. Overall, these hit molecules were large in size and were observed to clash with other amino acid residues of the binding pocket.
Therefore, a second pharmacophore query was generated having five features (three acceptors and two donors) with modified positions and types (Supplementary material Figure S3). The search resulted in 44 hit molecules. Among these molecules, epigallocatechin-gallate-(-) (6.28%), kuromanin (3.66%), procyanidin-b-2 (3.96%) and rutin (40.82%) were the top four hits which fitted well to the specified pharmacophore query (Supplementary material Figure S4). Rutin is a launched drug and demonstrated the largest inhibition of SARS-CoV-2 whereas epigallocatechin-gallate-(-) and procyanidin-b-2 are in clinical trials while kuromanin is in the preclinical phase.
3.3. Non-structural protein 10 – non-structural protein 16 complex
The SARS-CoV-2 nsp10 protein interacts with nsp14 and nsp16 (Pan et al., 2008; von Brunn et al., 2007). The nsp16 in complex with nsp10 acts as a 2′-O-methyltransferase (2′-O-Mtase) to selectively 2’O-methylate the cap-0 structure to cap-1 structure (m7GpppAm-RNA) (Bouvet et al., 2010; Chen et al., 2011). The Mtase activity is crucial for the life-cycle of the coronavirus and its inhibition could suppress viral replication (Ke et al., 2012). The crystal structures of the nsp10-nsp16 complexes (PDB IDs: 6W4H, 7BQ7) were aligned and superposed which resulted in an RMSD of 0.478 Å after protein preparation. The two structures were similar and were properly superposed on each other. The co-crystallised ligand in the binding pocket of both proteins was the endogenous metabolite S-adenosyl methionine (SAM). The ligand present in the binding site caused slight changes in the amino acid position. The pharmacophore was generated using the 6W4H crystal structure. Figure S5 (Supplementary material) shows the ligand interactions (2D) and site view (3D) of the ligand SAM which is surrounded by hydrophilic and hydrophobic amino acids. Among them, Asp6897, Asp6928 and Asp6912 are polar acidic residues that interact with the ligand. There is also the presence of water molecules which form bridges between the ligand and the protein. Other residues such as Gly6869, Asn6841, Gly6879, Cys6913 and Gly6871 are polar in nature whereas, Leu6898 and Ala6870 are hydrophobic residues which contribute to the ligand interactions.
The pharmacophore features (5 in total) around the ligand were three donor and two acceptor features. Fifteen hit molecules were identified to have the assigned pharmacophore features. Key among the fifteen hits were adaptavir, dihydrostreptomycin, ATN-161, alarelin, gramicidin and exenatide (Supplementary material Figure S6). These hit molecules had rmsdx values ranging from 0.3747 to 0.66 Å. By visually analysing the interactions it was found that dihydrostreptomycin (-8.94%), ATN-161 (2.44%) and adaptavir (-1.32%) (in vitro SARS-CoV-2 inhibition in brackets) fitted well inside the binding pocket of the protein. In addition to mapping to the pharmacophore, these molecules made extra hydrogen bonding interactions with the protein.
All molecules had minimal in vitro activity which may be due to their large size and they were observed to clash with amino acid residues in the binding pocket. Therefore, a second pharmacophore query was generated having five features (four donors and one acceptor) with different positons and types (Supplementary material Figure S7). The second pharmacophore search resulted in eighteen hit molecules. Among these molecules, TMC-353121 (3.28%) and paromomycin (4.7%) were the top hit molecules (in vitro SARS-CoV-2 inhibition in brackets) which fitted well to the specified pharmacophore query (Supplementary material Figure S8). These are still very low activity values which may be due to using the endogenous metabolite SAM for creating the pharmacophore. Paromomycin is a launched drug whereas TMC-353121 is in phase 2 clinical trials.
3.4. Non-structural protein 15
The SARS-CoV-2 non-structural protein 15 (nsp15) is characterised as an endoribonuclease that cleaves RNA at uridylates at the 3′-position (Bhardwaj et al., 2006). Nsp15 targets and degrades the viral polyuridine sequences and double-stranded RNA (dsRNA) to prevent the host immune system from detecting the virus (Deng & Baker, 2018; Hackbart et al., 2020). The crystal structures of nsp15 (PDB IDs: 6WXC, 6WLC) were aligned and superposed which resulted in an RMSD of 0.466 Å after protein preparation. The two structures were similar and were properly superposed on each other. The A and the B chains of these proteins were aligned and superposed separately. The co-crystallised ligands in the binding pocket of the two proteins were different; 6WXC had tipiracil (CMU) as the main ligand in the binding site and 6WLC had uridine-5′-monophosphate (U5P). The ligand present in the binding site caused slight changes in the amino acid position. The pharmacophores were generated using both crystal structures since they had different ligands with different interactions. Figures S9 and S10 (Supplementary material) illustrates ligand interactions (2D) and site view (3D) representations which shows the ligands tipiracil and U5P are surrounded by hydrophilic and hydrophobic amino acid residues. U5P interacts with Lys290 and Lys345 which are polar basic residues. There is also the presence of water molecules which form bridges between the ligand and protein. Other residues such as Tyr343 and Ser294 are polar in nature whereas, Leu346 and Val292 are hydrophobic residues which contribute to ligand interactions. Tipiracil interacts with Lys290, Lys345 and His250 which are polar basic residues. There is also the presence of a phosphate ion and water molecules which form bridges between the ligand and protein. Other residues such as Tyr343, Ser294, Gly247 and Gly248 are polar in nature whereas, Leu346 is a hydrophobic residue which contributes to ligand interactions.
The first pharmacophore (6 features) on the ligand tipiracil contained three donor, two acceptor and one aromatic features. Twelve hit molecules were identified to have the assigned pharmacophore features. Key among the twelve hits, with results for ChEMBL reported in vitro SARS-CoV-2 inhibition in brackets, were AT-9283 (2.8%), acadesine (-4.27%), olomoucine (4.05%), sapropterin (14.33%), gossypol (66.34%) and tetrahydrofolic acid (28.84%) (Supplementary material Figure S11). These hit molecules had rmsdx values ranging from 0.6109 to 0.7910 Å.
By visually analysing the interactions it was observed that acadesine, olomoucine, sapropterin and tetrahydrofolic acid fitted well inside the binding pocket of the protein. These molecules also made hydrogen bond interactions in addition to the pharmacophore query features. Of the active compounds, olomoucine is in preclinical stages while sapropterin and tetrahydrofolic acid are launched drugs.
A second pharmacophore query was generated on the U5P ligand of 6WLC having one donor, three acceptor, one anionic acceptor and one aromatic features (Supplementary material Figure S12). The pharmacophore search resulted in six hit molecules: uridine-5-triphosphate (1.07%), baicalin (-2.99%), cyclic-AMP (1.15%), INS316 (25.9%), adenosine phosphate (43.81%) and diadenosine tetraphosphate (2.33%). INS316 is in phase 2 clinical trials and adenosine phosphate is launched on the market. The pharmacophore query search around the two ligands resulted in different hits due to the co-crystallised ligands displaying different interaction patterns (Supplementary material Figure S13).
Another crystal structure (PDB ID: 6X4I) was aligned with 6WLC which resulted in an RMSD of 0.332 Å and was also compared with 6WXC. The three structures were similar and were properly superposed on each other. The ligand present in the binding site of 6X4I was 3′-uridinemonophosphate (U3P). The site view (3D) showed that the three ligands had different interactions within the binding pocket (Supplementary material Figure S14). VS with the pharmacophore generated around U3P found two hit molecules (uridine-5-triphosphate and adenosine phosphate) that were also found as hits in 6WLC pharmacophore search.
3.5. Nucleocapsid protein
The SARS-CoV-2 nucleocapsid protein packages the single-stranded positive sense viral genome into a ribonucleoprotein (RNP) complex called the capsid. It has also been shown to modulate the host cellular machinery and may have regulatory roles in the viral life cycle (Hsieh et al., 2005; Surjit et al., 2006). The crystal structures of nucleocapsid proteins (PDB IDs: 6WKP, 6M3M) were aligned and superposed which resulted in an RMSD of 1.092 Å. 6WKP had 2-N-morpholino-ethanesulfonic acid (MES) as the main ligand in the binding site and 6M3M had no ligand so the pharmacophore was generated using the 6WKP structure. The ligand interactions (2D) and site view (3D) images show that the ligand MES is surrounded by hydrophilic and hydrophobic amino acid residues (Supplementary material Figure S15). Residues such as TyrA109, AsnB154, AsnB75 and ArgA107 are polar in nature whereas, AlaA55 is a hydrophobic residue which contributes to the ligand interactions. There is also the presence of water molecules which form bridges between the ligand and protein.
The pharmacophore (5 features) around MES of the B chain were two acceptor, two anionic acceptor and one hydrophobic features. 122 hit molecules were identified to have the assigned pharmacophore features. Key hits, with results for ChEMBL reported in vitro SARS-CoV-2 inhibition in brackets, were varespladib (68.26%), hexonic acid (37.6%), citric acid (46.04%), OSI-027 (104.83%), MK-5108 (77.92%), stepronin (42.06%), calcium gluceptate (31.49%), sodium glucoheptonate (3.84%), CPP (33.36%), pirenoxine (13%), midafotel (19.88%), dexamethasone sodium phosphate (-1.11%) and maltobionic acid (13.12%) (Supplementary material Figure S16). These hit molecules had rmsdx values ranging from 0.3303 to 0.6789 Å.
By visually analysing the interactions it was found that citric acid, varespladib, stepronin and calcium gluceptate fitted well to the specified pharmacophore features and the binding site. These molecules also had moieties that made hydrogen bond interactions in addition to the pharmacophore query features. Stepronin, calcium gluceptate, pirenoxine and maltobionic acid are already launched drugs. Varespladib and midafotel are in phase 3 clinical trials. CPP is in phase 2 clinical trials. OSI-027 and MK-5108 are in phase 1 clinical trials. Hexonic acid and citric acid are in the preclinical stage.
4. Discussion
Computational drug design approaches can speed up the process to discover urgently needed treatments for SARS-CoV-2 infection. Currently, there is no specific antiviral treatment or vaccine available for COVID-19. There is an urgent need to find therapeutic interventions by repurposing existing drugs for possible use against SARS-CoV-2 by targeting the proteins encoded by the viral genome. Several studies are ongoing, including clinical trials, on repurposing of approved drugs (Pacios et al., 2020). The major focus of research interest has been on the spike glycoprotein and the proteases. So as to examine other therapeutic intervention options, in this paper, we have focussed on all X-ray structures published on the PDB (as of 9th July 2020) of non-structural proteins and the nucleocapsid protein. Non-structural proteins are known to participate in viral RNA replication or transcription (Snijder et al., 2020). We have also examined the nucleocapsid protein which is known to bind to the viral RNA genome and package them into helical nucleocapsid structures (Masters & Sturman, 1990; McBride et al., 2014). PBVS was used to create ligand interaction guided pharmacophores from the holo X-ray structures and VS the Broad Institute’s Drug Repurposing collection for known drugs mapping to viral pharmacophores.
The SARS-CoV-2 nsp3 crystal structure (PDB ID: 6YWL) had five features that were assigned on the co-crystallized large ligand APR. PBVS resulted in eight hits and among them, the top hits were antibiotics (dihydrostreptomycin, streptomycin, isepamicin) and a flavanone glycoside (neohesperidin) as they fitted well inside the binding pocket of the protein and made extra interactions with surrounding amino acids. These extra interactions may contribute to improved inhibition of the target protein. Isepamicin and streptomycin are launched drugs used against infectious diseases and are known to treat pneumonia and tuberculosis respectively. Isepamicin was active against SARS-CoV-2 in an inhibition bioassay (Ellinger et al., 2020) reported in the ChEMBL database. Streptomycin had better interactions within the active pocket and fitted well to the given pharmacophore query. Antibiotics such as Teicoplanin were reported to inhibit the growth of virus in human cells (Colson & Raoult, 2016), is used to treat staphylococci infections and was also efficacious in the first stage of MERS-CoV viral cycle (Colson & Raoult, 2016; Zhou et al., 2016). Azithromycin is another antibiotic that was found to be effective in treating viral infections caused by Zika and Ebola viruses by inhibiting growth in vitro (Bosseboeuf et al., 2018; Madrid et al., 2015). Combination use of hydroxychloroquine and azithromycin has been reported to treat COVID-19 patients (Gautret, 2020). These findings suggest that antibiotics found as VS hits can possibly inhibit SARS-CoV-2 and should be progressed to in vitro and in vivo studies.
Another SARS-CoV-2 target called the nsp7-nsp8-nsp12 complex, which is known to be involved with RNA-dependent-RNA polymerase activity (Subissi et al., 2014), was used for pharmacophore generation. The superposition/alignment step of the two nsp7-nsp8 complex proteins (7BV2, 7BW4) gave a low RMSD, indicating structural similarity. It was observed that whenever a ligand binds in a protein binding pocket there can be a wide range of structural changes, ranging from hinge movement of domains to small side-chain rearrangements within the binding pocket (Najmanovich et al., 2000). The ligand remdesivir was a large molecule and had many atoms making interactions with the protein but there was only a slight change within the binding pocket due to interaction with remdesivir. The identification of compounds capable of inhibiting proteins involved in viral replication has proven to be successful in the drive to discover potential anti-viral drugs (Ashraf et al., 2019; Pár & Pár, 2018; Qadir et al., 2018). The hits found from PBVS can possibly inhibit RdRp activity thereby affecting the viral replication of SAR-CoV-2. The top hits from the pharmacophore search were NADPH, adenosine-triphosphate (ATP), Coenzyme-I and nadide. Coenzyme-I, NADPH and ATP are all coenzymes necessary for biological activities. It is interesting, that these biological molecules could be used as therapeutic interventions. It has been reported that ATP can act as an immune system modulator in the treatment of HIV/AIDS (Wagner, 2011). The vitamin A derivative, retinazone, was shown to inhibit growth of a range of viruses (Kesel et al., 2014). But the potential of these molecules to inhibit viral proteins is not fully explored. However, Coenzyme-I demonstrated a 29% inhibiting of SARS-CoV-2 in vitro which indicates anti-viral activity.
During analysis of the VS results, we observed that the majority of the hit molecules clashed with the binding pocket amino acids due to their large size. Often molecules that are large in size tend to form the required interactions with amino acid residues, but a significant portion of the molecule is outside of the binding site or clashing with nearby amino acid residues. A second pharmacophore was generated with new features to search for compounds displaying fewer binding site clashes. PBVS found four top hit compounds: epigallocatechin-gallate-(-) (EGCg), kuromanin, procyanidin-b-2 and rutin. These hit molecules are naturally occurring compounds and displayed varying active against SARS-CoV-2 in vitro. Among these, EGCg (6.28% inhibition) and rutin (40.82% inhibition) possess known anti-viral properties. EGCg is a catechin component of green tea and was reported to have antiviral activity against human immunodeficiency virus (HIV) type 1 as it impinges on each step of the viral life cycle (Yamaguchi et al., 2002). Rutin is a flavonoid glycoside which not only has antiviral activity against various infections (hepatitis B, herpes virus and retrovirus) but also possesses anti-asthmatic activity (Ganeshpurkar & Saluja, 2017). The use of natural compounds like lycorine was reported to show potent antiviral activity against SARS-CoV (Li et al., 2005). These studies indicate the potential utility of natural compounds against viral infections and that they should be considered as possible therapeutic interventions.
The nsp10-nsp16 complex is known to have the methyltransferase activity. Two nsp10-nsp16 crystal structures were aligned, superposed and had no structural differences. The ligand SAM was co-crystallised in the binding pocket, made several interactions with the amino acids in addition to bridging water molecules which assisted with ligand binding. The presence of bridging water molecules can be used to increase ligand specificity and affinity so has proven useful in drug design (Ladbury, 1996; Schiebel et al., 2018). Due to the large size of the ligand, two pharmacophore queries were generated to examine the effect of different feature combinations. Large ligands are likely to have multiple protein interactions which makes the query search more precise but results in fewer hits. The first pharmacophore search found dihydrostreptomycin, ATN-161 and adaptavir to fit well inside the protein binding pocket but none had appreciable in vitro inhibition. Adaptavir is in phase 2 clinical trials for the treatment of HIV and hepatitis B virus (HBV) infections whereas, dihydrostreptomycin is an antibiotic used for treating tuberculosis. The second pharmacophore search found TMC-353121 and paromomycin as hits. TMC-353121 is a potent respiratory syncytial virus fusion inhibitor (Ispas et al., 2015) and in vitro had 3.28% inhibition of SARS-CoV-2. Paromomycin is an antimicrobial used to treat parasitic infections which had 4.7% inhibition of SARS-CoV-2. A recent study indicated the potential of an anti-parasitic drug called Ivermectin to have anti-viral activity in vitro against the SARS-CoV-2 (Caly et al., 2020). Repurposing of these drugs which belong to diverse therapeutic areas (anti-viral, anti-parasitic and anti-bacterial) against the novel coronavirus is an important approach to finding a quick therapeutic alternative.
The two structures (6WXC, 6WLC) of SARS-CoV-2 nsp15 contained different ligands (tipiracil and U5P), were involved in dissimilar interactions, so separate pharmacophore searches were undertaken. Tipiracil made good interactions within the binding pocket and a phosphate ion was also involved in the network of hydrogen bonds. A recent study indicated that tipiracil inhibits the nsp15 endoribonuclease (Kim et al., 2020) hence generating pharmacophore features from it can help to rapidly identify other possible drugs against SARS-CoV-2. The pharmacophore search generated hits such as AT-9283, acadesine, sapropterin, tetrahydrofolic acid, gossypol and olomoucine. Among them gossypol was found to be active in vitro against SARS-CoV-2 with inhibition of 66.34%. Gossypol is known to have various therapeutic effects, 433 distinct activities in biological assays as described by Bisson et al. (2016) and is often found as a hit molecule in computational drug design and follow-on in vitro assays so we do recommend caution if progressing this compound further. Sapropterin (14.33% inhibition), a phenylalanine-4-hydroxylase stimulant and tetrahydrofolic acid (28.84% inhibition) were also confirmed in vitro to be SARS-CoV-2 inhibitors. Folic acid and its derivatives (tetrahydrofolic acid and 5-methyl tetrahydrofolic acid) have been reported to have good binding affinity to viral proteins in various VS studies (Kumar & Jena, 2020). The second pharmacophore search based on the U5P ligand resulted in uridine-5-triphosphate, baicalin, cyclic-AMP, INS316, adenosine phosphate and diadenosine tetraphosphate. Baicalin was found to be inactive against SARS-CoV-2 in vitro, but we cannot fully ignore the potential of baicalin as, in conflict with this finding, it was previously reported to inhibit SARS-CoV-2 3CLpro in vitro (Su et al., 2020). INS316 (25.9% inhibition) and adenosine phosphate (43.81% inhibition) are promising candidates for further in vitro and in vivo studies. To analyse the binding site interaction with different ligands (U5P, tipiracil and U3P) and to observe the variation in the position of the amino acid residues we performed an alignment/superposition step with three different nsp15 crystal structures (6X4I, 6WLC and 6WXC). The three ligands made different interactions within the binding site, however, the pharmacophore generated around U3P had two common hit molecules (uridine-5-triphosphate, adenosine phosphate) with the U5P pharmacophore search which is unsurprising due to the close similarity between the co-crystallised ligands.
The majority of the crystal structures of the nucleocapsid protein had no co-crystalized ligand. 6WKP had MES as the ligand which made a number of interactions with the amino acid residues of the protein. Buffers like MES are used for NMR and X-ray crystallography and it is generally believed that the interactions between the protein and buffer agents have no significant influence on protein structure. But it is important to understand the effects of buffer agents on protein dynamics. One such study indicated a change in protein dynamics due to the interactions between MES buffer and human liver fatty acid-binding protein (Long & Yang, 2009). Although MES is a buffer, it made well-defined interactions within a protein binding site so, we assumed that MES was located in the active site of the protein and that the interactions made were relevant for generation of pharmacophore features. The first pharmacophore resulted in multiple hits also displaying good in vitro inhibition of SARS-CoV-2. The hits belonged to diverse therapeutic areas: Varespladib has anti-inflammatory properties, stepronin is a mucolytic drug, calcium glucepatate is a calcium supplement and citric acid has antioxidant properties. Nearly all of the VS predicted inhibitors of the nucleocapsid protein also display promising in vitro inhibition of SARS-CoV-2 indicating that, if on-target binding is subsequently confirmed, this protein is a promising anti-viral target.
5. Conclusions
The COVID-19 pandemic has negatively affected normal human life. It has been estimated that due to underlying health conditions, about one in five individuals worldwide could be at greater risk of developing severe COVID-19 after they become infected (Clark et al., 2020). When time is a critical factor, repurposing of existing drugs can be a successful strategy leading to the discovery of therapeutic interventions which have already proven to be sufficiently safe in human trials.
To the best of our knowledge, this is the first comprehensive PBVS campaign against all publically available holo SARS-CoV-2 nsp and nucleocapsid proteins which has discovered multiple existing drugs that are predicted to inhibit SARS-CoV-2 proteins. The following compounds are proposed to inhibit the activity of viral proteins, all except streptomycin display in vitro inhibition of SARS-CoV-2 and are recommended for biological follow-up: isepamicin and streptomycin (nsp3); Coenzyme-I, rutin, epigallocatechin-gallate-(-) and procyanidin-b-2 (nsp7/nsp8/nsp12); paromomycin (nsp10/nsp16); olomoucine, sapropterin, tetrahydrofolic acid, INS316 and adenosine phosphate (nsp15); varespladib, hexonic acid, citric acid, OSI-027, MK-5108, stepronin, calcium gluceptate, CPP, pirenoxine, midafotel and maltobionic acid (Nucleocapsid).
By undertaking computational studies, we analysed key protein binding site amino acid residues, generated bespoke pharmacophores and obtained hits predicted to inhibit viral proteins. All excluding one of these hit molecules were previously validated in vitro to inhibit the activity of SARS-CoV-2. Our results indicate the underlying mechanism of action for the viral inhibitory activity of these molecules. We recommend conducting confirmatory SARS-CoV-2 biological assays of the in silico hit molecules which were reported inactive in the bioassay conducted by Ellinger et al. (2020). For hit compunds with in vitro activity, we recommend that confirmatory on target assays are undertaken, followed by detailed SARS-CoV-2 in vitro and in vivo studies. These hit compunds may have utility in fighting SARS-CoV-2 infection after further validation work, which will be particularly important if there are mutations of the spike or protease proteins which render future developed vaccine or small molecule treatments ineffective.
Supplementary Material
Acknowledgements
The Trinity Biomedical Sciences Institute (TBSI) is supported by a capital infrastructure investment from Cycle 5 of the Irish Higher Education Authority’s Programme for Research in Third Level Institutions (PRTLI). We thank the software vendors for their continuing support of academic research efforts, in particular the contributions of the Chemical Computing Group, Biovia and OpenEye Scientific. The support and provisions of Dell Ireland is also acknowledged. The Trinity College Dublin MSc in Molecular Medicine is also gratefully acknowledged for supporting this work.
Disclosure statement
The authors report no potential conflicts of interest.
References
- Akram, M., Tahir, I. M., Shah, S. M. A., Mahmood, Z., Altaf, A., Ahmad, K., Munir, N., Daniyal, M., Nasir, S. and Mehboob, H. (2018). Antiviral potential of medicinal plants against HIV, HSV, influenza, hepatitis, and coxsackievirus: A systematic review. Phytotherapy Research, 32(5), 811–822. 10.1002/ptr.6024 [DOI] [PubMed] [Google Scholar]
- Ashburn, T. T., & Thor, K. B. (2004). Drug repositioning: Identifying and developing new uses for existing drugs. Nature Reviews. Drug Discovery, 3(8), 673–683. https://doi.org/10. 1038/nrd1468 10.1038/nrd1468 [DOI] [PubMed] [Google Scholar]
- Ashraf, M. U., Iman, K., Khalid, M. F., Salman, H. M., Shafi, T., Rafi, M., Javaid, N., Hussain, R., Ahmad, F., Shahzad-Ul-Hussan, S., Mirza, S., Shafiq, M., Afzal, S., Hamera, S., Anwar, S., Qazi, R., Idrees, M., Qureshi, S. A., & Chaudhary, S. U. (2019). Evolution of efficacious pangenotypic hepatitis C virus therapies. Medicinal Research Reviews, 39(3), 1091–1136. 10.1002/med.21554 [DOI] [PubMed] [Google Scholar]
- Astuti, I. & Ysrafil. (2020). Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): An overview of viral structure and host response. Diabetology & Metabolic Syndrome, 14(4), 407–412. 10.1016/j.dsx.2020.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N., & Bourne, P. E. (2000). The Protein Data Bank. Nucleic Acids Research, 28(1), 235–242. 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj, K., Sun, J., Holzenburg, A., Guarino, L. A., & Kao, C. C. (2006). RNA recognition and cleavage by the SARS coronavirus endoribonuclease. Journal of Molecular Biology, 361(2), 243–256. 10.1016/j.jmb.2006.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson, J., McAlpine, J. B., Friesen, J. B., Chen, S. N., Graham, J., & Pauli, G. F. (2016). Can invalid bioactives undermine natural product-based drug discovery? Journal of Medicinal Chemistry, 59(5), 1671–1690. 10.1021/acs.jmedchem.5b01009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosseboeuf, E., Aubry, M., Nhan, T., de Pina, J. J., Rolain, J. M., & Raoult, D. (2018). Azithromycin inhibits the replication of Zika virus. J Antivirals Antiretrovirals, 10(1), 6–11. 10.4172/1948-5964.1000173 [DOI] [Google Scholar]
- Bouvet, M., Debarnot, C., Imbert, I., Selisko, B., Snijder, E. J., Canard, B., & Decroly, E. (2010). In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathogens, 6(4), e1000863. 10.1371/annotation/a0dde376-2eb1-4ce3-8887-d29f5ba6f162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brimacombe, K. R., et al. (2020). An OpenData portal to share COVID-19 drug repurposing data in real time. bioRxiv, p. 2020.06.04.135046. 10.1101/2020.06.04.135046 [DOI]
- Caly, L., Druce, J. D., Catton, M. G., Jans, D. A., & Wagstaff, K. M. (2020). The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Research, 178, 104787. 10.1016/j.antiviral.2020.104787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascella, M., Rajnik, M., Cuomo, A., Dulebohn, S. C., Di, N. R. (2020). Features, evaluation and treatment coronavirus (COVID-19). StatPearls, StatPearls Publishing. http://www.ncbi.nlm.nih.gov/books/NBK554776/ [PubMed]
- Cavalla, D., Oerton, E., & Bender, A. (2017). 1.02 – Drug repurposing review. In Chackalamannil, S., Rotella, D. P., Ward, S. E. (Eds.), Reference module in chemistry, molecular sciences and chemical engineering (pp. 11–47). Elsevier. 10.1016/B978-0-12-409547-2.12283-8 [DOI] [Google Scholar]
- Chen, Y., Liu, Q., & Guo, D. (2020). Emerging coronaviruses: Genome structure, replication, and pathogenesis. Journal of Medical Virology, 92(4), 418–423. 10.1002/jmv.25681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Su, C., Ke, M., Jin, X., Xu, L., Zhang, Z., Wu, A., Sun, Y., Yang, Z., Tien, P., Ahola, T., Liang, Y., Liu, X., & Guo, D. (2011). Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex. PLoS Pathogens, 7(10), e1002294. 10.1371/journal.ppat.1002294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, A., Jit, M., Warren-Gash, C., Guthrie, B., Wang, H. H. X., Mercer, S. W., Sanderson, C., McKee, M., Troeger, C., Ong, K. L., Checchi, F., Perel, P., Joseph, S., Gibbs, H. P., Banerjee, A., Eggo, R. M., Nightingale, E. S., O'Reilly, K., Jombart, T., … Jarvis, C. I. (2020). Global, regional, and national estimates of the population at increased risk of severe COVID-19 due to underlying health conditions in 2020: A modelling study. The Lancet Global Health, 8(8), e1003–e1017. 10.1016/S2214-109X(20)30264-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson, P., & Raoult, D. (2016). Fighting viruses with antibiotics: An overlooked path. International Journal of Antimicrobial Agents, 48(4), 349–352. 10.1016/j.ijantimicag.2016.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, X., & Baker, S. C. (2018). An “Old” protein with a new story: Coronavirus endoribonuclease is important for evading host antiviral defenses. Virology, 517, 157–163. 10.1016/j.virol.2017.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinger, B., et al. (2020). Identification of inhibitors of SARS-CoV-2 in-vitro cellular toxicity in human (Caco-2) cells using a large scale drug repurposing collection. Research Square. 10.21203/RS.3.RS-23951/V1 [DOI] [Google Scholar]
- Fehr, A. R., & Perlman, S. (2015). Coronaviruses: An overview of their replication and pathogenesis. Methods in Molecular Biology, 1282, 1–23. doi: 10.1007/978-1-4939-2438-7_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganeshpurkar, A., & Saluja, A. K. (2017). The pharmacological potential of rutin. Saudi Pharmaceutical Journal, 25(2), 149–164. 10.1016/j.jsps.2016.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautret, P., Lagier, J.-C., Parola, P., Hoang, V. T., Meddeb, L., Mailhe, M., Doudier, B., Courjon, J., Giordanengo, V., Vieira, V. E., Tissot Dupont, H., Honoré, S., Colson, P., Chabrière, E., La Scola, B., Rolain, J.-M., Brouqui, P., & Raoult, D. (2020). Hydroxychloroquine and azithromycin as a treatment of COVID-19: Results of an open-label non-randomized clinical trial. International Journal of Antimicrobial Agents, 56(1), 105949. doi: 10.1016/j.ijantimicag.105949 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ge, Y., et al. (2020). A data-driven drug repositioning framework discovered a potential therapeutic agent targeting COVID-19. bioRxiv, p. 2020.03.11.986836. 10.1101/2020.03.11.986836 [DOI] [PMC free article] [PubMed]
- Gordon, D. E., Jang, G. M., Bouhaddou, M., Xu, J., Obernier, K., White, K. M., O'Meara, M. J., Rezelj, V. V., Guo, J. Z., Swaney, D. L., Tummino, T. A., Hüttenhain, R., Kaake, R. M., Richards, A. L., Tutuncuoglu, B., Foussard, H., Batra, J., Haas, K., Modak, M., … Krogan, N. J. (2020). A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature, 583(7816), 459–468. 10.1038/s41586-020-2286-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackbart, M., Deng, X., & Baker, S. C. (2020). Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proceedings of the National Academy of Sciences of the United States of America, 117(14), 8094–8103. 10.1073/pnas.1921485117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins, P. C. D., Skillman, A. G., Warren, G. L., Ellingson, B. A., & Stahl, M. T. (2010). Conformer generation with OMEGA: Algorithm and validation using high quality structures from the protein databank and Cambridge structural database. Journal of Chemical Information and Modeling, 50(4), 572–584. 10.1021/ci100031x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., Schiergens, T. S., Herrler, G., Wu, N.-H., Nitsche, A., Müller, M. A., Drosten, C., & Pöhlmann, S. (2020). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell, 181(2), 271–280.e8. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh, P. K., Chang, S. C., Huang, C. C., Lee, T. T., Hsiao, C. W., Kou, Y. H., Chen, I. Y., Chang, C. K., Huang, T. H., & Chang, M. F. (2005). Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. Journal of Virology, 79(22), 13848–13855. 10.1128/JVI.79.22.13848-13855.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ispas, G., Koul, A., Verbeeck, J., Sheehan, J., Sanders-Beer, B., Roymans, D., Andries, K., Rouan, M. C., De Jonghe, S., Bonfanti, J. F., Vanstockem, M., Simmen, K., & Verloes, R. (2015). Antiviral activity of TMC353121, a respiratory syncytial virus (RSV) fusion inhibitor, in a non-human primate model. PLoS One, 10(5), e0126959. 10.1371/journal.pone.0126959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashchenko, A. A., Dmitriev, K. A., Vostokova, N. V., Azarova, V. N., Blinow, A. A., Egorova, A. N., Gordeev, I. G., Ilin, A. P., Karapetian, R. N., Kravchenko, D. V., Lomakin, N. V., Merkulova, E. A., Papazova, N. A., Pavlikova, E. P., Savchuk, N. P., Simakina, E. N., Sitdekov, T. A., Smolyarchuk, E. A., Tikhomolova, E. G., Yakubova, E. V., & Ivachtchenko, A. V. (2020). AVIFAVIR for treatment of patients with moderate COVID-19: Interim results of a phase II/III multicenter randomized clinical trial. Clinical Infectious Diseases. 10.1093/cid/ciaa1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Alberto, A., Ribas-Aparicio, R. M., Aparicio-Ozores, G., & Castelán-Vega, J. A. (2020). Virtual screening of approved drugs as potential SARS-CoV-2 main protease inhibitors. Computational Biology and Chemistry, 88, 107325. 10.1016/j.compbiolchem.2020.107325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., Zhang, B., Li, X., Zhang, L., Peng, C., Duan, Y., Yu, J., Wang, L., Yang, K., Liu, F., Jiang, R., Yang, X., You, T., Liu, X., … Yang, H. (2020). Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature, 582(7811), 289–229. 10.1038/s41586-020-2223-y [DOI] [PubMed] [Google Scholar]
- Ke, M., Chen, Y., Wu, A., Sun, Y., Su, C., Wu, H., Jin, X., Tao, J., Wang, Y., Ma, X., Pan, J.-A., & Guo, D. (2012). Short peptides derived from the interaction domain of SARS coronavirus nonstructural protein nsp10 can suppress the 2'-O-methyltransferase activity of nsp10/nsp16 complex. Virus Research, 167(2), 322–328. 10.1016/j.virusres.2012.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesel, A. J., Huang, Z., Murray, M. G., Prichard, M. N., Caboni, L., Nevin, D. K., Fayne, D., Lloyd, D. G., Detorio, M. A., & Schinazi, R. F. (2014). Retinazone inhibits certain blood-borne human viruses including Ebola virus Zaire. Antiviral Chemistry & Chemotherapy, 23(5), 197–215. 10.3851/IMP2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y., et al. (2020). Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2. bioRxiv, p. 2020.06.26.173872. 10.1101/2020.06.26.173872 [DOI] [PMC free article] [PubMed]
- Kumar, V., & Jena, M. (2020). In silico virtual screening-based study of nutraceuticals predicts the therapeutic potentials of folic acid and its derivatives against COVID-19. PREPRINT (Version 1) Available at Research Square. 10.21203/rs.3.rs-31775/v1 [DOI] [PMC free article] [PubMed]
- Kumar, Y., Singh, H., & Patel, C. N. (2020). In silico prediction of potential inhibitors for the Main protease of SARS-CoV-2 using molecular docking and dynamics simulation based drug-repurposing. Journal of Infection and Public Health, 13(9), 1210–1223. 10.1016/j.jiph.2020.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladbury, J. E. (1996). Just add water! The effect of water on the specificity of protein- ligand binding sites and its potential application to drug design. Chemistry & Biology, 3(12), 973–980. 10.1016/S1074-5521(96)90164-7 [DOI] [PubMed] [Google Scholar]
- Leach, A. R., Gillet, V. J., Lewis, R. A., & Taylor, R. (2010). Three-dimensional pharmacophore methods in drug discovery. Journal of Medicinal Chemistry, 53(2), 539–558. 10.1021/jm900817u [DOI] [PubMed] [Google Scholar]
- Lei, J., Kusov, Y., & Hilgenfeld, R. (2018). Nsp3 of coronaviruses: Structures and functions of a large multi-domain protein. Antiviral Research, 149, 58–74. 10.1016/j.antiviral.2017.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, G., & De Clercq, E. (2020). Therapeutic options for the 2019 novel coronavirus (2019-nCoV)). Nature Reviews. Drug Discovery, 19(3), 149–150. 10.1038/d41573-020-00016-0 [DOI] [PubMed] [Google Scholar]
- Li, S. Y., Chen, C., Zhang, H. Q., Guo, H. Y., Wang, H., Wang, L., Zhang, X., Hua, S. N., Yu, J., Xiao, P. G., Li, R. S., & Tan, X. (2005). Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antiviral Research, 67(1), 18–23. 10.1016/j.antiviral.2005.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, D., & Yang, D. (2009). Buffer interference with protein dynamics: A case study on human liver fatty acid binding protein. Biophysical Journal, 96(4), 1482–1488. 10.1016/j.bpj.2008.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid, P. B., Panchal, R. G., Warren, T. K., Shurtleff, A. C., Endsley, A. N., Green, C. E., Kolokoltsov, A., Davey, R., Manger, I. D., Gilfillan, L., Bavari, S., & Tanga, M. J. (2015). Evaluation of Ebola virus inhibitors for drug repurposing. ACS Infectious Diseases, 1(7), 317–326. 10.1021/acsinfecdis.5b00030 [DOI] [PubMed] [Google Scholar]
- Marra, M. A., Jones, S. J. M., Astell, C. R., Holt, R. A., Brooks-Wilson, A., Butterfield, Y. S. N., Khattra, J., Asano, J. K., Barber, S. A., Chan, S. Y., Cloutier, A., Coughlin, S. M., Freeman, D., Girn, N., Griffith, O. L., Leach, S. R., Mayo, M., McDonald, H., Montgomery, S. B., … Roper, R. L. (2003). The genome sequence of the SARS-associated coronavirus. Science, 300(5624), 1399–1404. 10.1126/science.1085953 [DOI] [PubMed] [Google Scholar]
- Martin, W. R., & Cheng, F. (2020). Repurposing of FDA-approved toremifene to treat COVID-19 by blocking the spike glycoprotein and NSP14 of SARS-CoV-2. chemRxiv. 10.26434/CHEMRXIV.12431966.V1 [DOI] [PMC free article] [PubMed]
- Masters, P. S., & Sturman, L. S. (1990). Background paper. Functions of the coronavirus nucleocapsid protein. Advances in Experimental Medicine and Biology, 276, 235–238. 10.1007/978-1-4684-5823-7_32 [DOI] [PubMed] [Google Scholar]
- Mawhinney, L., Armstrong, M. E., O’Reilly, C., Bucala, R., Leng, L., Fingerle-Rowson, G., Fayne, D., Keane, M. P., Tynan, A., Maher, L., Cooke, G., Lloyd, D., Conroy, H., & Donnelly, S. C. (2014). Macrophage migration inhibitory factor (MIF), enzymatic activity & lung cancer. Molecular Medicine, 20(1), 729–735. 10.2119/molmed.2014.00136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride, R., van Zyl, M., & Fielding, B. C. (2014). The coronavirus nucleocapsid is a multifunctional protein. Viruses, 6(8), 2991–3018. 10.3390/v6082991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes, C. (2007). Virtual screening strategies in drug discovery. Current Opinion in Chemical Biology, 11(5), 494–502. 10.1016/j.cbpa.2007.08.033 [DOI] [PubMed] [Google Scholar]
- McKay, P. B., Fayne, D., Horn, H. W., James, T., Peters, M. B., Carta, G., Caboni, L., Nevin, D. K., Price, T., Bradley, G., Williams, D. C., Rice, J. E., & Lloyd, D. G. (2012). Consensus computational ligand-based design for the identification of novel modulators of human Estrogen Receptor alpha. Molecular Informatics, 31(3-4), 246–258. 10.1002/minf.201100127 [DOI] [PubMed] [Google Scholar]
- Menachery, V. D., Mitchell, H. D., Cockrell, A. S., Gralinski, L. E., Yount, B. L., Graham, R. L., McAnarney, E. T., Douglas, M. G., Scobey, T., Beall, A., Dinnon, K., Kocher, J. F., Hale, A. E., Stratton, K. G., Waters, K. M., & Baric, R. S. (2017). MERS-CoV accessory ORFs play key role for infection and pathogenesis. mBio, 8(4), e0066517. 10.1128/mBio.00665-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molecular Operating Environment (MOE) . (2020). 2019.01; Chemical Computing Group ULC, 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7.
- Mousavizadeh, L., & Ghasemi, S. (2020). Genotype and phenotype of COVID-19: Their roles in pathogenesis. Journal of Microbiology, Immunology and Infection. 10.1016/j.jmii.2020.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster, V. J., Koopmans, M., van Doremalen, N., van Riel, D., & de Wit, E. (2020). A novel coronavirus emerging in China – Key questions for impact assessment. The New England Journal of Medicine, 382(8), 692–694. 10.1056/NEJMp2000929 [DOI] [PubMed] [Google Scholar]
- Muthas, D., Sabnis, Y. A., Lundborg, M., & Karlen, A. (2008). Is it possible to increase hit rates in structure-based virtual screening by pharmacophore filtering? An investigation of the advantages and pitfalls of post-filtering. Journal of Molecular Graphics & Modelling, 26(8), 1237–1251. 10.1016/j.jmgm.2007.11.005 [DOI] [PubMed] [Google Scholar]
- Najmanovich, R., Kuttner, J., Sobolev, V., & Edelman, M. (2000). Side-chain flexibility in proteins upon ligand binding. Proteins: Structure, Function, and Genetics, 39(3), 261–268. [DOI] [PubMed] [Google Scholar]
- Nevin, D. K., Peters, M. B., Carta, G., Fayne, D., & Lloyd, D. G. (2012). Integrated virtual screening for the identification of novel and selective Peroxisome proliferator-activated receptor (PPAR) scaffolds. Journal of Medicinal Chemistry, 55(11), 4978–4989. 10.1021/jm300068n [DOI] [PubMed] [Google Scholar]
- OMEGA . (2019). OMEGA 3.1.1.2: OpenEye Scientific Software. http://www.eyesopen.com
- O’Neill, L. A. J., & Netea, M. G. (2020). BCG-induced trained immunity: Can it offer protection against COVID-19? Nature Reviews. Immunology, 20(6), 335–337. 10.1038/s41577-020-0337-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacios, O., Blasco, L., Bleriot, I., Fernandez-Garcia, L., González Bardanca, M., Ambroa, A., López, M., Bou, G., & Tomás, M. (2020). Strategies to combat multidrug-resistant and persistent infectious diseases. Antibiotics, 9(2), 65. 10.3390/antibiotics9020065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, J., Peng, X., Gao, Y., Li, Z., Lu, X., Chen, Y., Ishaq, M., Liu, D., Dediego, M. L., Enjuanes, L., & Guo, D. (2008). Genome-wide analysis of protein-protein interactions and involvement of viral proteins in SARS-CoV replication. PLoS One, 3(10), e3299. 10.1371/journal.pone.0003299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, A., Nikam, A. N., Shreya, A. B., Mutalik, S. P., Gopalan, D., Kulkarni, S., Padya, B. S., Fernandes, G., Mutalik, S., & Prassl, R. (2020). Potential therapeutic targets for combating SARS-CoV-2: Drug repurposing, clinical trials and recent advancements. Life Sciences, 256, 117883. 10.1016/j.lfs.2020.117883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant, S., Singh, M., Ravichandiran, V., Murty, U., & Srivastava, H. K. (2020). Peptide-like and small-molecule inhibitors against Covid-19. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1757510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pár, A., & Pár, G. (2018). [Three decades of the hepatitis C virus from the discovery to the potential global elimination: The success of translational researches]. Orvosi Hetilap, 159(12), 455–465. 10.1556/650.2018.30997 [DOI] [PubMed] [Google Scholar]
- Patskovsky, Y. V., Negrebetskaya, E. N., Chernomaz, A. A., Voloshchuk, T. P., Rubashevsky, E. L., Kitam, O. E., Tereshchenko, M. I., Nosach, L. N., & Potopalsky, A. I. (1996). Aromatic thiosemicarbazones, their antiviral action and interferon. 1. The decreasing of adenovirus type 1 resistance against interferon by methisazone in vitro. Biopolymers and Cell, 12(2), 74–83. 10.7124/bc.000425 [DOI] [Google Scholar]
- Peele, K. A., Chandrasai, P., Srihansa, T., Krupanidhi, S., Sai, A. V., Babu, D. J., Indira, M., Reddy, A. R., & Venkateswarulu, T. C. (2020). Molecular docking and dynamic simulations for antiviral compounds against SARS-CoV-2: A computational study. Informatics in Medicine Unlocked, 19, 100345. 10.1016/j.imu.2020.100345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman, S., & Netland, J. (2009). Coronaviruses post-SARS: Update on replication and pathogenesis. Nature Reviews Microbiology, 7(6), 439–450. 10.1038/nrmicro2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadir, A., Riaz, M., Saeed, M., & Shahzad-Ul-Hussan, S. (2018). Potential targets for therapeutic intervention and structure based vaccine design against Zika virus. European Journal of Medicinal Chemistry, 156, 444–460. 10.1016/j.ejmech.2018.07.014 [DOI] [PubMed] [Google Scholar]
- Sanders, M. P. A., McGuire, R., Roumen, L., de Esch, I. J. P., de Vlieg, J., Klomp, J. P. G., & de Graaf, C. (2012). From the protein’s perspective: The benefits and challenges of protein structure-based pharmacophore modelling. MedChemComm, 3(1), 28–38. 10.1039/C1MD00210D [DOI] [Google Scholar]
- Santibáñez-Morán, M. G., López-López, E., Prieto-Martínez, F. D., Sánchez-Cruz, N., & Medina-Franco, J. L. (2020). Consensus virtual screening of dark chemical matter and food chemicals uncover potential inhibitors of SARS-CoV-2 main protease. Version 1. ChemRxiv. 10.26434/chemrxiv.12420860.v1 [DOI] [PMC free article] [PubMed]
- Schiebel, J., Gaspari, R., Wulsdorf, T., Ngo, K., Sohn, C., Schrader, T. E., Cavalli, A., Ostermann, A., Heine, A., & Klebe, G. (2018). Intriguing role of water in protein-ligand binding studied by neutron crystallography on trypsin complexes. Nature Communications, 9(1), 3559. 10.1038/s41467-018-05769-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, G. (2018). Automating drug discovery. Nature Reviews Drug Discovery, 17(2), 97–113. 10.1038/nrd.2017.232 [DOI] [PubMed] [Google Scholar]
- Schneider, P., & Schneider, G. (2018). Polypharmacological drug-target inference for chemogenomics. Molecular Informatics, 37 (9-10), e1800050. 10.1002/minf.201800050 [DOI] [PubMed] [Google Scholar]
- Shah, B., Modi, P., & Sagar, S. R. (2020). In silico studies on therapeutic agents for COVID-19: Drug repurposing approach. Life Sciences, 252, 117652. 10.1016/j.lfs.2020.117652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder, E. J., Bredenbeek, P. J., Dobbe, J. C., Thiel, V., Ziebuhr, J., Poon, L. L. M., Guan, Y., Rozanov, M., Spaan, W. J. M., & Gorbalenya, A. E. (2003). Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-o from the coronavirus group 2 lineage. Journal of Molecular Biology, 331(5), 991–1004. 10.1016/S0022-2836(03)00865-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder, E. J., Decroly, E., & Ziebuhr, J. (2016). The nonstructural proteins directing coronavirus RNA synthesis and processing. Advances in Virus Research, 96, 59–126. 10.1016/bs.aivir.2016.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder, E. J., Limpens, R., de Wilde, A. H., de Jong, A., Zevenhoven-Dobbe, J. C., Maier, H. J., Faas, F., Koster, A. J., & Bárcena, M. (2020). A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis. PLoS biology, 18(6), e3000715. 10.1371/journal.pbio.3000715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, H., et al. (2020). Discovery of baicalin and baicalein as novel, natural product inhibitors of SARS-CoV-2 3CL protease in-vitro. bioRxiv, p. 2020.04.13.038687. 10.1101/2020.04.13.038687 [DOI]
- Subissi, L., Posthuma, C. C., Collet, A., Zevenhoven-Dobbe, J. C., Gorbalenya, A. E., Decroly, E., Snijder, E. J., Canard, B., & Imbert, I. (2014). One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proceedings of the National Academy of Sciences of the United States of America, 111(37), E3900. 10.1073/pnas.1323705111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit, M., Liu, B., Chow, V. T., & Lal, S. K. (2006). The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclin-dependent kinase complex and blocks S phase progression in mammalian cells. Journal of Biological Chemistry, 281(16), 10669–10681. 10.1074/jbc.M509233200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talevi, A., & Bellera, C. L. (2020). Challenges and opportunities with drug repurposing: Finding strategies to find alternative uses of therapeutics. Expert Opinion on Drug Discovery, 15(4), 397–401. 10.1080/17460441.2020.1704729 [DOI] [PubMed] [Google Scholar]
- Tazikeh-Lemeski, E., Moradi, S., Raoufi, R., Shahlaei, M., Janlou, M., & Zolghadri, S. (2020). Targeting SARS-COV-2 non-structural protein 16: A virtual drug repurposing study. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1779133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Brunn, A., Teepe, C., Simpson, J. C., Pepperkok, R., Friedel, C. C., Zimmer, R., Roberts, R., Baric, R., & Haas, J. (2007). Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PLoS One, 2(5), e459. 10.1371/journal.pone.0000459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, M. C. (2011). The therapeutic potential of adenosine triphosphate as an immune modulator in the treatment of HIV/AIDS: A combination approach with HAART. Current HIV Research, 9(4), 209–222. 10.2174/157016211796320289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters, W. P., Stahl, M. T., & Murcko, M. A. (1998). Virtual screening – An overview. Drug Discovery Today, 3(4), 160–178. 10.1016/S1359-6446(97)01163-X [DOI] [Google Scholar]
- WHO . (2020). WHO Director-General's statement on IHR Emergency Committee on Novel Coronavirus (2019-nCoV). Accessed 15 July 2020. https://www.who.int/dg/speeches/detail/who-director-general-s-statement-on-ihr-emergency-committee-on-novel-coronavirus-(2019-ncov)
- Wu, C., Liu, Y., Yang, Y., Zhang, P., Zhong, W., Wang, Y., Wang, Q., Xu, Y., Li, M., Li, X., Zheng, M., Chen, L., & Li, H. (2020). Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica B, 10(5), 766–788. 10.1016/j.apsb.2020.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, H.-L., et al. (2020). Several FDA-approved drugs effectively inhibit SARS-CoV-2 infection in vitro. bioRxiv. 10.1101/2020.06.05.135996 [DOI] [PMC free article] [PubMed]
- Yamaguchi, K., Honda, M., Ikigai, H., Hara, Y., & Shimamura, T. (2002). Inhibitory effects of (-)-epigallocatechin gallate on the life cycle of human immunodeficiency virus type 1 (HIV-1). Antiviral Research, 53(1), 19–34. 10.1016/S0166-3542(01)00189-9 [DOI] [PubMed] [Google Scholar]
- Zhou, N., Pan, T., Zhang, J., Li, Q., Zhang, X., Bai, C., Huang, F., Peng, T., Zhang, J., Liu, C., Tao, L., & Zhang, H. (2016). Glycopeptide antibiotics potently inhibit cathepsin L in the late endosome/lysosome and block the entry of Ebola virus, Middle East respiratory syndrome coronavirus (MERS-CoV), and severe acute respiratory syndrome coronavirus (SARS-CoV). The Journal of Biological Chemistry, 291(17), 9218–9232. 10.1074/jbc.M116.716100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr, J. (2005). The coronavirus replicase. Current Topics in Microbiology and Immunology, 287, 57–94. 10.1007/3-540-26765-4_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.