Abstract
The spread of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) caused a worldwide outbreak of coronavirus disease 19 (COVID-19), which rapidly evolved as a global concern. The efforts of the scientific community are pointed towards the identification of promptly available therapeutic options. RNA-dependent RNA polymerase (RdRp) is a promising target for developing small molecules to contrast SARS-CoV-2 replication. Modern computational tools can boost identification and repurposing of known drugs targeting RdRp. We here report the results regarding the screening of a database containing more than 8800 molecules, including approved, experimental, nutraceutical, illicit, withdrawn and investigational compounds. The molecules were docked against the cryo-electron microscopy structure of SARS-CoV-2 RdRp, optimized by means of molecular dynamics (MD) simulations. The adopted three-stage ensemble docking study underline that compounds formerly developed as kinase inhibitors may interact with RdRp.
Communicated by Ramaswamy H. Sarma
Keywords: SARS-CoV-2, RdRp, remdesivir, molecular modelling, repurposing
Introduction
In December 2019, an outbreak of a novel coronavirus-related pneumonia was reported. The pandemic agent leading to this disease, named severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), spreads from human to human causing life-threatening coronavirus disease 19 (COVID-19) (Park et al., 2020; Spellberg et al., 2020). Epidemiological studies on the escalation of this pneumonia highlighted that human sneeze droplets are primarily involved in virus transmission. Most patients, lacking other risk factors, showed mild influenza-like disease symptoms and their social activity during infection lead to a high rate of community spread, which was proved to be especially threatening for weak elderly. In particular, severe pneumonia, secondary infections and cardiovascular events were the major reason for death due to SARS-CoV-2 (Chen et al., 2020; Li et al., 2020).
Global concern raised after COVID-19 and scientists worldwide focused their efforts in identifying new tools for accurate and timely diagnosis of infection and in promptly providing valid therapeutic options (Huang et al., 2020; Wu et al., 2020). Sequencing of SARS-CoV-2 genome showed that the virus exhibits 80% sequence identity to SARS-CoV and 96% to a bat coronavirus (Lu et al., 2020; Zhou et al., 2020). On the side of possible treatments, early reports highlighted that chloroquine and remdesivir inhibit viral replication (Fantini et al., 2020; Wang et al., 2020). Vaccines are also being studied, but mutations that may affect efficacy, immune complications and unavoidable development time cannot be ruled out (Tai et al., 2020).
During the past weeks, literature has been very prolific and several contributions reporting the identification of novel pharmacological targets to contrast SARS-CoV-2 spread appeared (Huang et al., 2020). One of the early studied targets is represented by the spike protein (S protein) of the coronavirus, which interacts with the alveolar epithelial cells expressing angiotensin-converting enzyme 2 (ACE2) (Xia et al., 2020). Main protease and papain-like protease inhibitors are also being studied in silico, in vitro and in vivo (Bhardwaj, Singh, Sharma, Rajendran, et al., 2020; Calligari et al., 2020; Huang et al., 2020; Li & De Clercq, 2020; Zhang et al., 2020).
In this work, we will focus our attention on RNA-dependent RNA polymerase (RdRp). This crucial enzyme is a replicase that operates the synthesis of a complementary RNA strand using viral RNA (Hillen et al., 2020; Yin et al., 2020). This machinery is targeted by remdesivir (GS-5734, Gilead), a compound that was formerly developed to contrast Ebola virus (EBOV) replication. Remdesivir acts as a non-functioning ATP mimic within RdRp, thus stopping genome transcription (Tchesnokov et al., 2019). Very recently, Gordon and colleagues investigated the mechanism of action, at the molecular level, of remdesivir against SARS-CoV-2 RdRp. The results of this study confirmed that RdRp efficiently incorporates remdesivir triphosphate, the active form of the drug, into RNA and, consequently, RNA synthesis is terminated (Gordon et al., 2020). Besides its potent antiviral activity in vitro, remdesivir efficiently contrasted SARS-CoV-2 replication in animal models. Ongoing clinical studies suggest that this compound improves recovery time, although further results from placebo-controlled trials are needed (Beigel et al., 2020; Goldman et al., 2020). Thus, SARS-CoV-2 RdRp represent an attractive and promising therapeutic target (Lung et al., 2020).
With the current study, we aim at identifying a set of compounds, by means of computational tools, that bear the potential of interacting with SARS-CoV-2 RdRp. In our comprehensive investigation, we screened the molecules present in the DrugBank database, containing more than 8800 molecular structures of drugs (Wishart et al., 2006). The database includes the 3D coordinates of compounds, which were docked against the cryo-electron microscopy structure of SARS-CoV-2 RdRp, optimized by means of molecular dynamics (MD) simulations (Gao et al., 2020). This approach is particularly attractive for medicinal chemists in the context of “drug repurposing”, which consists in the identification of novel therapeutic applications for known and promptly available compounds against COVID-19 (Pushpakom et al., 2019).
Material and methods
Database collection and refinement
The database of molecules used for this study was obtained from DrugBank (Wishart et al., 2006) and it is composed by 8815 drugs, comprehending approved, experimental, nutraceutical, illicit, withdrawn and investigational compounds. The database was downloaded in SDF format with 3D coordinates and was directly loaded on Schrödinger Maestro (Schrödinger, 2020d). All the structures were analysed with the QikProp tool (Schrödinger, 2020g) present in Schrödinger, obtaining an estimation of several pharmacokinetic properties. The molecules having one or more violations to the Lipinski’s rules (Lipinski et al., 1997) were discarded. Thus, the initial number of structures decreased to 6756 and it was further reduced to 6601 after the elimination of the molecules with a molecular weight lower than 100 g/mol.
Macromolecules preparation
A 10 µs MD trajectory (DESRES-ANTON-10917618) was obtained from DESHAW research website (https://www.deshawresearch.com). This model represents the SARS-CoV-2 nsp7-nsp8-nsp12 RNA polymerase complex determined in the absence of reducing agent (PDB id: 6M71) (D. E. Shaw Research, 2020). The simulation performed by DESRES research team with the Anton 2 supercomputer (Shaw et al., 2014) was based on the Amber ff99SB-ILDN force field (Lindorff-Larsen et al., 2010) for proteins and on the TIP3P model for water. The C- and N-peptide termini were capped with amide and acetyl groups respectively. The missing loops in the available structural models were manually built as extended peptide conformation. The missing part of chain D was built through homology modelling using the structure of SARS-CoV polymerase complex (PDB id: 6NUR). The system was neutralized and a 0.15 M concentration of NaCl was set. The simulation was conducted at 310 K in the NPT ensemble. The long trajectory was analysed with VMD (Humphrey et al., 1996) plotting the protein backbone RMSD (Root Mean Square Deviation) over the simulation time, as the system reached the stability after 5 µs (see Figure S1 in the Supplemental material file). One frame every 5 ns was extracted from 6.65 to 10.00 µs. The frames were then clustered basing on their RMSD in 10 different clusters, for each of which a representative frame was saved. Prior their use as targets for the ensemble docking, the obtained 10 frames were prepared with the Protein Preparation Wizard (Gupta et al., 2019; Schrödinger, 2020f) included in the Schrödinger suite using default settings, i.e. adding hydrogens, assigning disulphide bonds, removing surrounding waters, adjusting charges, capping termini, adding missing side-chains using Prime (Schrödinger, 2020e), optimizing hydrogen bonds and a with a final minimization under the OPLS3e force field (Roos et al., 2019).
Ligands preparation
Every ligand was prepared for docking with LigPrep application (Schrödinger, 2020c) included in the Schrödinger suite under the OPLS3e force field. Using Epik ioinizer (Schrödinger, 2020a), all the ionization states were generated in the 7 ± 2 pH range, and a final number of 10244 molecular structures to be used in the docking stage was obtained.
Molecular docking
The docking protocol consisted in a multiple-frame ensemble docking performed with Glide (Schrödinger, 2020b), under the OPLS3e force field, using in sequence the high throughput virtual screening, the standard and the extra precision docking protocols (respectively HTVS, SP and XP) with default settings. Starting from the 10244 structures initially prepared, after each docking step the 10% best scoring molecules were identified and used for the next higher precision screening (1020 for SP and 102 for XP docking). The dockings were performed manually to each different frame and then the results were merged considering the best scoring pose for each ligand. All the frames were obtained and prepared as previously described, and were analysed with SiteMap tool (Schrödinger, 2020h) included in the Schrödinger suite identifying the best putative receptor binding site that often corresponded to extended areas of the macromolecule. For each considered frame, a docking grid was prepared with Receptor Grid Generation tool included in the Schrödinger suite with custom settings. A cubic grid was generated setting the centre in the centroid of the potential receptor binding sites previously generated with SiteMap. The dimensions of the grids is related to the dimensions of the putative receptor binding sites, while the ligand diameter midpoint box was set at the maximum size of 40 × 40 × 40 Å.
Pose rescoring with MM-GBSA
The poses obtained with the last XP docking stage were re-scored with Prime Molecular Mechanical/Generalized Born Surface Area (MM-GBSA) tool included in the Schrödinger suite. The complexes were refined with Prime under the OPLS3e force field with the VSGB continuum solvation model (Variable Dielectric Surface Generalized Born model) (J. Li et al., 2011), the protein flexibility was enabled for the residues in the 12 Å range from the ligand position. The energies obtained from the complexes were automatically calculated with the five following energy terms and the equation systems reported in the following.
Optimized free receptor = Receptor, optimized free ligand = Ligand, optimized complex = Complex, receptor from minimized/optimized complex, ligand from minimized/optimized complex
Receptor Strain = Receptor (from optimized complex) – Receptor
Lig Strain = Ligand (from optimized complex) – Ligand
MM-GBSA dG Bind (NS) = Complex – Receptor (from optimized complex) – Ligand (from optimized complex) = MM-GBSA dG Bind – Rec Strain – Lig Strain
Results and discussion
Experimental design
Previous computational and experimental contribution in the literature were focused on the investigation of the interaction of drug-like small molecules with RdRp models (Elfiky, 2020b). Nevertheless, to the best of our knowledge, available in silico studies are based on homology models (Elfiky, 2020b, 2020a) and/or have been carried out using very limited pools of compound, such as know antivirals like ribavirin, sofosbuvir, favipiravir, galidesivir and tenofovir, hydroxychloroquine (Elfiky, 2020c) or natural compounds from folk medicine (Lung et al., 2020). Moreover, computational studies that follow the in silico-aided repurposing approach reported to date focus on small, pre-selected libraries and generally consist in preliminary, multi-target docking studies that lack of MD optimization and MM-GBSA stage (Shah et al., 2020). With this contribution, we aimed at providing a comprehensive screening of a database of more than 8800 drugs against the optimized structure of SARS-CoV-2 RdRp solved by cryo-electron microscopy.
Database collection and refinement
The first part of the work consisted in the preparation of a large library of molecules to be screened in silico for their ability to target SARS-CoV-2 RdRp and potentially inhibit this enzyme. With the intent of obtaining a set of hits that could ideally be easily repurposed against COVID-19 through the docking study, it was chosen to use the DrugBank database. This was imported in Schrödinger Maestro and analysed with the embedded QikProp application generating pharmacokinetic properties for all the molecules of the set. To assess the drug likeness of the considered compounds, the Lipinkski’s rule of five was adopted using the following criteria: molecular weight < 500 g/mol, logP < 5, number of hydrogen bond donors ≤ 5 and of hydrogen bond acceptors ≤ 10. As a result, 2059 molecules were not admitted to the following step, bringing the total number to 6756. This was further reduced to 6601 after the elimination of other 155 molecules having a molecular weight lower than 100 g/mol that, upon manual database inspection, corresponded to gases, solvents, salts and, in general, very low molecular weight compounds. These molecules, due to their simple chemical structure and to the lack of polar groups or aromatic rings, do not meet drug likeness criteria and are likely not able to establish the multiple interactions required for a site-specific binding motif.
Docking study
The docking study was designed as a three-stage ensemble docking using different frames of a long MD trajectory that are representative of several conformations of SARS-CoV-2 RdRp in. A 10 µs MD trajectory (DESRES-ANTON-10917618) was obtained from DESHAW research website (https://www.deshawresearch.com), representing the SARS-CoV-2 nsp7-nsp8-nsp12 RNA polymerase complex (PDB id: 6M71), particularly refined as described in the Material and Methods section. The simulation was conducted at 310 K in the NPT ensemble neutralizing the system and using a 0.15 M concentration of NaCl. The trajectory was analysed plotting its backbone RMSD over the simulation time and one frame every 5 ns was extracted from 6.65 to 10.00 µs. 10 representative frames were selected, which were finally prepared with Schrödinger as described previously to be used as targets for the docking stage. To focus the docking experiments towards the most promising binding sites, each one of the selected frames was inspected with SiteMap tool from Schrödinger suite obtaining extended areas that were then enclosed in grids with the Receptor Grid Generation tool. Lastly, the ligands were prepared as previously described obtaining 10244 molecular structures comprehending different protonation states. The three-stage ensemble docking was then performed starting from a low precision/high speed HTVS screening, followed by a medium precision/medium speed SP screening and, eventually, by the high precision/low speed XP screening. At the end of each docking stage, with the intent of gradually enriching of potential hits the compound library and at the same time of facilitating the following higher precision screening, the 10% of best scoring ligands was maintained in the set and carried to the subsequent stage. Consequently, starting from 10244 molecules, the number was reduced to 1020 after the first step and then to 102 molecules, that were analysed in the last XP docking stage. As a reference, even if the molecule is known to act through a covalent-binding inhibition mechanism, the active compound remdesivir triphosphate was docked using the described protocol showing a binding energy value of −9.244 kcal/mol.
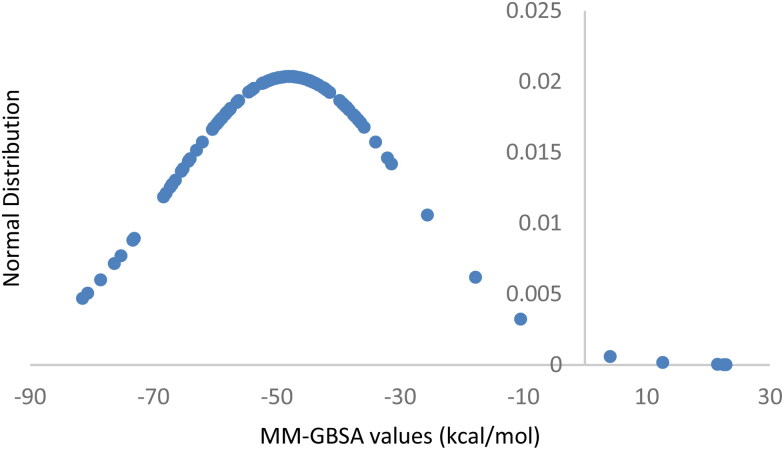
In order to increase simulation accuracy, the poses obtained from the final XP screening were also re-scored with the Prime MM-GBSA method from Schrödinger suite. The combined Molecular Mechanical/Generalized Born Surface Area (MM-GBSA) approach is a force-field based protocol that computes the free energy of binding from the difference between the free energies of the ligand, of the protein, and of the complex in solution. Moreover, the MM-GBSA method is based on the concept that the free energy of binding of a ligand to a receptor can be approximated by the combination of molecular mechanics (MM) energies, polar and nonpolar solvation terms, and an entropy term. The computed energies usually show good correlation with the experimental binding energy values, with correlation coefficients in the 0.4-0.7 range. In this work, the MM-GBSA calculations were performed allowing the protein flexibility for the residues in the 12 Å range from the ligand position and using VSGB as a continuum solvation model. The obtained values range from a minimum of +22.9 kcal/mol to a maximum of −84.6 kcal/mol, with an average value of −48.0 kcal/mol. More in detail, the gaussian plot reported in Figure 1 demonstrates that the scores are majorly concentrated in proximity of −50 kcal/mol value (Greenidge et al., 2013).
Figure 1.
Gaussian plot representing the distribution of the docking values obtained with the MM-GBSA protocol applied to the poses resulting from the XP docking stage.
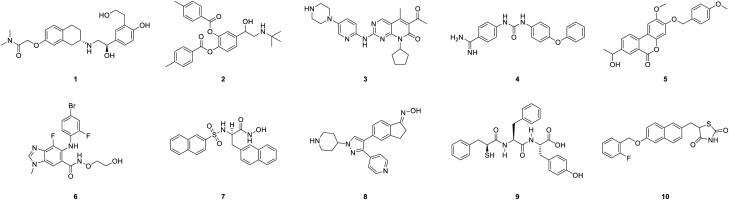
The 10 compounds showing the most promising energy values, based on the results of the MM-GBSA re-scoring, are reported in Figure 2. Remdesivir was not highlighted among this group of best scoring compounds, according to the MM-GBSA procedure.
Figure 2.
Best scoring compounds according to MM-GBSA screening.
For such molecules, docking results obtained with the XP docking and the MM-GBSA re-scoring, alongside the pharmacokinetic properties calculated with the QikProp are reported in Table 1. Concerning the results of this procedure, it must be pointed out that some of the compounds highlighted by the screening correspond to experimental drugs. Two of them are in clinical use as anti-cancer drugs, namely binimetinib, a MEK inhibitor (Queirolo & Spagnolo, 2017), and palbociclib, a CDK4 and CDK6 inhibitor (Turner et al., 2015). MEK is a serine/tyrosine/threonine kinase while CDK4 and CDK6 are serine/threonine kinases. Palomid 529 is a tyrosine kinase inhibitor acting on the PI3K/Akt/mTOR pathway that was investigated as an antiproliferative agent (Xue et al., 2008), while netoglitazone is an antidiabetic drug interacting with peroxisome proliferator-activated receptor (PPAR) alpha (Khatik et al., 2018). Bedoradrine and bitolterol are beta-2-adrenergic receptor agonists, the first one remained investigational while the second reached the clinical use but was withdrawn from the market in 2001 (Cazzola et al., 2012; Tee et al., 2007). Moreover, according to the information retrieved from DrugBank (Wishart et al., 2006), experimental compound DB03337 is reported to target human trypsin-1, DB07861 N-acetylglucosamine deacetylase from Pseudomonas aeruginosa, DB08553 human serine/threonine-protein kinase B-raf, and DB03949 thermolysin from Geobacillus stearothermophilus.
Table 1.
MM-GBSA and docking values, pharmakocinetic properties, chemical names and database ids of the 10 best scoring compounds (according to MM-GBSA ΔG). The full table is reported in the Supplemental material file (Table S1).
| # | Name | Chemical name | DB id | ΔG MM-GBSA (kcal/mol) | Docking score (kcal/mol) | MW (g/mol) | pLogP | HB donor | HB acceptor |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Bedoradrine | 2-(((S)-7-(((R)-2-hydroxy-2-(4-hydroxy-3-(2-hydroxyethyl)phenyl)ethyl)amino)-5,6,7, 8-tetrahydronaphthalen-2-yl)oxy)-N,N-dimethylacetamide | DB05590 | −84.58 | −8.961 | 428.527 | 1.703 | 4.000 | 9.400 |
| 2 | Bitolterol | 4-(2-(tert-butylamino)-1-hydroxyethyl)-1,2-phenylene bis(4-methylbenzoate) | DB00901 | −81.49 | −9.024 | 461.557 | 4.852 | 2.000 | 7.700 |
| 3 | Palbociclib | 6-acetyl-8-cyclopentyl-5-methyl-2-((5-(piperazin-1-yl)pyridin-2-yl)amino)pyrido [2,3-d]pyrimidin-7(8H)-one | DB09073 | −80.65 | −5.516 | 447.539 | 2.151 | 2.000 | 11.000 |
| 4 | – | 4-(3-(4-phenoxyphenyl)ureido)benzimidamide | DB03337 | −78.56 | −9.493 | 346.388 | 2.545 | 5.000 | 4.000 |
| 5 | Palomid 529 | 8-(1-hydroxyethyl)-2-methoxy-3-((4-methoxybenzyl)oxy)-6H-benzo[c]chromen-6-one | DB12812 | −76.29 | −6.541 | 406.434 | 4.037 | 1.000 | 6.450 |
| 6 | Binimetinib | 5-((4-bromo-2-fluorophenyl)amino)-4-fluoro-N-(2-hydroxyethoxy)-1-methyl-1H- benzo[d]imidazole-6-carboxamide | DB11967 | −75.25 | −7.191 | 441.231 | 3.302 | 2.000 | 6.900 |
| 7 | – | (R)-N-hydroxy-3-(naphthalen-2-yl)-2-(naphthalene-2-sulfonamido)propanamide | DB07861 | −73.32 | −8.723 | 420.482 | 1.740 | 2.250 | 7.950 |
| 8 | – | (E)-5-(1-(piperidin-4-yl)-3-(pyridin-4-yl)-1H-pyrazol-4-yl)-2,3-dihydro-1H-inden-1-one oxime | DB08553 | −73.08 | −6.140 | 373.457 | 2.620 | 2.000 | 7.200 |
| 9 | – | ((S)-2-mercapto-3-phenylpropanoyl)-L-phenylalanyl-L-tyrosine | DB03949 | −68.32 | −10.145 | 492.589 | 4.194 | 3.300 | 6.750 |
| 10 | Netoglitazone | 5-((6-((2-fluorobenzyl)oxy)naphthalen-2-yl)methyl)thiazolidine-2,4-dione | DB09199 | −67.91 | −7.120 | 381.421 | 4.657 | 1.000 | 3.750 |
Considering the overall results of the study, and in particular the values reported in Table 1, it is not trivial to identify a direct correlation between docking and MM-GBSA scores. Nevertheless, if the results from MM-GBSA experiment are considered, it must be pointed out that 4 of the 10 highlighted compounds were previously developed as kinase inhibitors. This is an aspect of primary relevance, since such results shed new light on previous findings suggesting that tyrosine kinase inhibitors could play a role against SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV) replication, although multiple mechanisms may be involved (Coleman et al., 2016; Dong et al., 2020). Several previous contributions investigated kinase inhibitors in silico and (Bhardwaj, Singh, Sharma, Das, et al., 2020), in particular, as antiviral agents (Perwitasari et al., 2015; Schor & Einav, 2018). Moreover, growing pieces of evidence supporting the potential of tyrosine kinase (BTK and AXL) (Encinar & Menendez, 2020; Roschewski et al., 2020) and serine/threonine kinase (Rho) (Abedi et al., 2020) inhibitors against COVID-19 are recently emerging in the literature. Nevertheless, as in every repurposing study, main indications, contraindications, interactions, adverse effects and toxicity in general must be taken into account (Cha et al., 2018; Oprea et al., 2011). This especially holds true when the investigation focuses on a class of drugs for which the use of medication is often accompanied by intolerance and adverse effects (Bettiol et al., 2018; Fachi et al., 2018).
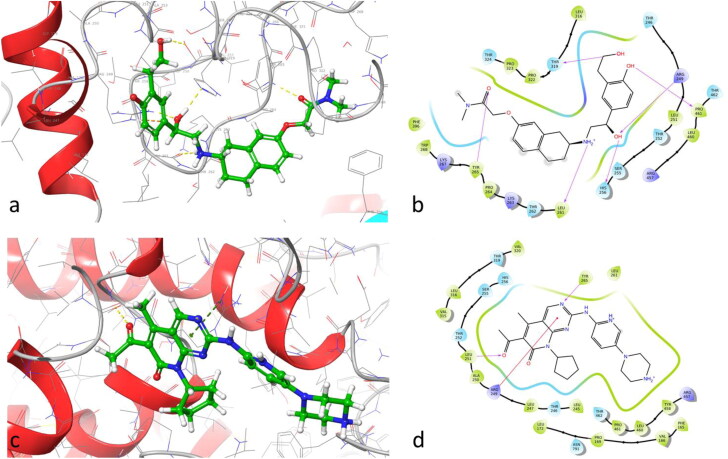
According to the results of the Prime MM-GBSA protocol, the docking poses corresponding to the best scoring compound bedoradrine and to the best scoring kinase inhibitor palbociclib are reported in Figure 3(a–d) as representative examples of the final screening results. Other depictive 2D and 3D interaction patterns for the selected compounds are shown in the Supplemental material file (Figures S2–S9).
Figure 3.
Docked pose of the ligands bedoradrine (a, b) and palbociclib (c, d) in RdRp showing the main interactions of the complex and two-dimensional interaction maps.
Conclusions
Computational tools can aid and boost the development or the repurposing of small molecules to contrast SARS-CoV-2 replication and spread. In this contribution, we screened a database of more than 8800 drugs against a MD-refined structure of RdRp using a three-stage ensemble docking approach. The results of the overall experimental workflow suggest that potential RdRp inhibitors should have very precise structural requirements, such as the presence of multiple rings and of highly polar moieties like secondary amines, alcohols and guanidine/urea groups. Very interestingly, the screening highlighted that some compounds belonging to the classes of kinase inhibitors are potential RdRp interactors. Thus, this study aims at prompting the scientific community to pursue the validation of this insight by in vitro and in vivo studies.
Authors contribution
Conceptualization, G.R. and A.G.; methodology, G.R. and A.O.; software, A.O. and E.O.; investigation, A.O., E.O. and G.R.; data curation, G.R. and A.G.; writing - original draft preparation, A.O., E.O. and G.R.; writing - review and editing, A.G., M.M. and G.Z.; supervision, G.R. and A.G.; funding acquisition, A.G.
Supplementary Material
Funding Statement
This work was granted by University of Brescia.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Abedi, F., Rezaee, R., & Karimi, G. (2020). Plausibility of therapeutic effects of Rho kinase inhibitors against severe acute respiratory syndrome Coronavirus 2 (COVID-19). Pharmacological Research, 156, 104808. 10.1016/j.phrs.2020.104808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel, J. H., Tomashek, K. M., Dodd, L. E., Mehta, A. K., Zingman, B. S., Kalil, A. C., Hohmann, E., Chu, H. Y., Luetkemeyer, A., Kline, S., Lopez de Castilla, D., Finberg, R. W., Dierberg, K., Tapson V., Hsieh, L., Patterson, T. F., Paredes, R., Sweeney, D. A., Short, W. R., … Lane, H. C. (2020). Remdesivir for the treatment of Covid-19—Preliminary Report. New England Journal of Medicine. 10.1056/NEJMoa2007764 [DOI] [Google Scholar]
- Bettiol, A., Marconi, E., Lombardi, N., Crescioli, G., Gherlinzoni, F., Walley, T., Vannacci, A., Chinellato, A., & Giusti, P. (2018). Pattern of use and long-term safety of tyrosine kinase inhibitors: A decade of real-world management of chronic myeloid leukemia. Clinical Drug Investigation, 38(9), 837–844. 10.1007/s40261-018-0676-7 [DOI] [PubMed] [Google Scholar]
- Bhardwaj, V. K., Singh, R., Sharma, J., Das, P., & Purohit, R. (2020). Structural based study to identify new potential inhibitors for dual specificity tyrosine-phosphorylation- regulated kinase. Computer Methods and Programs in Biomedicine, 194, 105494. 10.1016/j.cmpb.2020.105494 [DOI] [PubMed] [Google Scholar]
- Bhardwaj, V. K., Singh, R., Sharma, J., Rajendran, V., Purohit, R., & Kumar, S. (2020). Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1766572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calligari, P., Bobone, S., Ricci, G., & Bocedi, A. (2020). Molecular investigation of SARS–CoV-2 proteins and their interactions with antiviral drugs. Viruses, 12(4), 445. 10.3390/v12040445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzola, M., Page, C. P., Calzetta, L., & Matera, M. G. (2012). Pharmacology and therapeutics of bronchodilators. Pharmacological Reviews, 64(3), 450–504. 10.1124/pr.111.004580 [DOI] [PubMed] [Google Scholar]
- Cha, Y., Erez, T., Reynolds, I. J., Kumar, D., Ross, J., Koytiger, G., Kusko, R., Zeskind, B., Risso, S., Kagan, E., Papapetropoulos, S., Grossman, I., & Laifenfeld, D. (2018). Drug repurposing from the perspective of pharmaceutical companies: Drug repurposing in pharmaceutical companies. British Journal of Pharmacology, 175(2), 168–180. 10.1111/bph.13798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, N., Zhou, M., Dong, X., Qu, J., Gong, F., Han, Y., Qiu, Y., Wang, J., Liu, Y., Wei, Y., Xia, J., Yu, T., Zhang, X., & Zhang, L. (2020). Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. The Lancet, 395(10223), 507–513. 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman, C. M., Sisk, J. M., Mingo, R. M., Nelson, E. A., White, J. M., & Frieman, M. B. (2016). Abelson kinase inhibitors are potent inhibitors of severe acute respiratory syndrome coronavirus and Middle East respiratory syndrome coronavirus fusion. Journal of Virology, 90(19), 8924–8933. 10.1128/JVI.01429-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D. E. Shaw Research . (2020). Molecular dynamics simulations related to SARS-CoV-2, D. E. Shaw research technical data. http://www.deshawresearch.com/resources_sarscov2.html
- Dong, W., Xie, W., Liu, Y., Sui, B., Zhang, H., Liu, L., Tan, Y., Tong, X., Fu, Z. F., Yin, P., Fang, L., & Peng, G. (2020). Receptor tyrosine kinase inhibitors block proliferation of TGEV mainly through p38 mitogen-activated protein kinase pathways. Antiviral Research, 173, 104651. 10.1016/j.antiviral.2019.104651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky, A. A. (2020. a). Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sciences, 248, 117477. 10.1016/j.lfs.2020.117477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky, A. A. (2020. b). Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sciences, 253, 117592. 10.1016/j.lfs.2020.117592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky, A. A. (2020. c). SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: An in silico perspective. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1761882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encinar, J. A., & Menendez, J. A. (2020). Potential drugs targeting early innate immune evasion of SARS-Coronavirus 2 via 2’-O-methylation of viral RNA. Viruses, 12(5), 525. 10.3390/v12050525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachi, M. M., Tonin, F. S., Leonart, L. P., Aguiar, K. S., Lenzi, L., Figueiredo, B. C., Fernandez-Llimos, F., & Pontarolo, R. (2018). Comparative efficacy and safety of tyrosine kinase inhibitors for chronic myeloid leukaemia: A systematic review and network meta-analysis. European Journal of Cancer, 104, 9–20. 10.1016/j.ejca.2018.08.016 [DOI] [PubMed] [Google Scholar]
- Fantini, J., Di Scala, C., Chahinian, H., & Yahi, N. (2020). Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. International Journal of Antimicrobial Agents, 55(5), 105960. 10.1016/j.ijantimicag.2020.105960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Y., Yan, L., Huang, Y., Liu, F., Zhao, Y., Cao, L., Wang, T., Sun, Q., Ming, Z., Zhang, L., Ge, J., Zheng, L., Zhang, Y., Wang, H., Zhu, Y., Zhu, C., Hu, T., Hua, T., Zhang, B., … Rao, Z. (2020). Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science (New York, N.Y.), 368(6492), 779–782. 10.1126/science.abb7498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman, J. D., Lye, D. C. B., Hui, D. S., Marks, K. M., Bruno, R., Montejano, R., Spinner C. D., Galli, M., Ahn, M.-Y., Nahass, R. G., Chen, Y.-S., SenGupta, D., Hyland, R. H., Osinusi, A. O., Cao, H., Blair, C., Wei, X., Gaggar, A., Brainard, D. M., … Subramanian, A. (2020). Remdesivir for 5 or 10 days in patients with severe Covid-19. New England Journal of Medicine. 10.1056/NEJMoa2015301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, C. J., Tchesnokov, E. P., Woolner, E., Perry, J. K., Feng, J. Y., Porter, D. P., & Götte, M. (2020). Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. Journal of Biological Chemistry, 295(20), 6785–6797. 10.1074/jbc.RA120.013679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenidge, P. A., Kramer, C., Mozziconacci, J.-C., & Wolf, R. M. (2013). MM/GBSA binding energy prediction on the PDBbind data set: Successes, failures, and directions for further improvement. Journal of Chemical Information and Modeling, 53(1), 201–209. 10.1021/ci300425v [DOI] [PubMed] [Google Scholar]
- Gupta, A. K., Wang, X., Pagba, C. V., Prakash, P., Sarkar‐Banerjee, S., Putkey, J., & Gorfe, A. A. (2019). Multi‐target, ensemble‐based virtual screening yields novel allosteric KRAS inhibitors at high success rate. Chemical Biology & Drug Design, 94(2), 1441–1456. 10.1111/cbdd.13519 [DOI] [PubMed] [Google Scholar]
- Hillen, H. S., Kokic, G., Farnung, L., Dienemann, C., Tegunov, D., & Cramer, P. (2020). Structure of replicating SARS-CoV-2 polymerase. Nature, 584(7819), 154–156. 10.1038/s41586-020-2368-8 [DOI] [PubMed] [Google Scholar]
- Huang, J., Song, W., Huang, H., & Sun, Q. (2020). Pharmacological therapeutics targeting RNA-dependent RNA polymerase, proteinase and spike protein: From mechanistic studies to clinical trials for COVID-19. Journal of Clinical Medicine, 9(4), 1131. 10.3390/jcm9041131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey, W., Dalke, A., & Schulten, K. (1996). VMD: Visual molecular dynamics. Journal of Molecular Graphics, 14(1), 33–38. 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Khatik, G. L., Datusalia, A. K., Ahsan, W., Kaur, P., Vyas, M., Mittal, A., & Nayak, S. K. (2018). A retrospect study on thiazole derivatives as the potential antidiabetic agents in drug discovery and developments. Current Drug Discovery Technologies, 15(3), 163–177. 10.2174/1570163814666170915134018 [DOI] [PubMed] [Google Scholar]
- Li, G., & De Clercq, E. (2020). Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nature Reviews. Drug Discovery, 19(3), 149–150. 10.1038/d41573-020-00016-0 [DOI] [PubMed] [Google Scholar]
- Li, J., Abel, R., Zhu, K., Cao, Y., Zhao, S., & Friesner, R. A. (2011). The VSGB 2.0 model: A next generation energy model for high resolution protein structure modeling. Proteins, 79(10), 2794–2812. 10.1002/prot.23106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q., Guan, X., Wu, P., Wang, X., Zhou, L., Tong, Y., Ren, R., Leung, K. S. M., Lau, E. H. Y., Wong, J. Y., Xing, X., Xiang, N., Wu, Y., Li, C., Chen, Q., Li, D., Liu, T., Zhao, J., Liu, M., … Feng, Z. (2020). Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. The New England Journal of Medicine, 382(13), 1199–1207. 10.1056/NEJMoa2001316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindorff-Larsen, K., Piana, S., Palmo, K., Maragakis, P., Klepeis, J. L., Dror, R. O., & Shaw, D. E. (2010). Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins, 78(8), 1950–1958. 10.1002/prot.22711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski, C. A., Lombardo, F., Dominy, B. W., & Feeney, P. J. (1997). Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced Drug Delivery Reviews, 23(1–3), 3–26. 10.1016/S0169-409X(00)00129-0 [DOI] [PubMed] [Google Scholar]
- Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., Wang, W., Song, H., Huang, B., Zhu, N., Bi, Y., Ma, X., Zhan, F., Wang, L., Hu, T., Zhou, H., Hu, Z., Zhou, W., Zhao, L., … Tan, W. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. The Lancet, 395(10224), 565–574. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung, J., Lin, Y.-S., Yang, Y.-H., Chou, Y.-L., Shu, L.-H., Cheng, Y.-C., Liu, H. T., & Wu, C.-Y. (2020). The potential chemical structure of anti-SARS-CoV-2 RNA-dependent RNA polymerase. Journal of Medical Virology, 92(6), 693–697. 10.1002/jmv.25761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oprea, T. I., Bauman, J. E., Bologa, C. G., Buranda, T., Chigaev, A., Edwards, B. S., Jarvik, J. W., Gresham, H. D., Haynes, M. K., Hjelle, B., Hromas, R., Hudson, L., Mackenzie, D. A., Muller, C. Y., Reed, J. C., Simons, P. C., Smagley, Y., Strouse, J., Surviladze, Z., … Sklar, L. A. (2011). Drug repurposing from an academic perspective. Drug Discovery Today. Therapeutic Strategies, 8(3–4), 61–69. 10.1016/j.ddstr.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, M., Cook, A. R., Lim, J. T., Sun, Y., & Dickens, B. L. (2020). A systematic review of COVID-19 epidemiology based on current evidence. Journal of Clinical Medicine, 9(4), 967. 10.3390/jcm9040967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perwitasari, O., Yan, X., O'Donnell, J., Johnson, S., & Tripp, R. A. (2015). Repurposing kinase inhibitors as antiviral agents to control Influenza A virus replication. Assay and Drug Development Technologies, 13(10), 638–649. 10.1089/adt.2015.0003.drrr [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushpakom, S., Iorio, F., Eyers, P. A., Escott, K. J., Hopper, S., Wells, A., Doig, A., Guilliams, T., Latimer, J., McNamee, C., Norris, A., Sanseau, P., Cavalla, D., & Pirmohamed, M. (2019). Drug repurposing: Progress, challenges and recommendations. Nature Reviews. Drug Discovery, 18(1), 41–58. 10.1038/nrd.2018.168 [DOI] [PubMed] [Google Scholar]
- Queirolo, P., & Spagnolo, F. (2017). Binimetinib for the treatment of NRAS-mutant melanoma. Expert Review of Anticancer Therapy, 17(11), 985–990. 10.1080/14737140.2017.1374177 [DOI] [PubMed] [Google Scholar]
- Roos, K., Wu, C., Damm, W., Reboul, M., Stevenson, J. M., Lu, C., Dahlgren, M. K., Mondal, S., Chen, W., Wang, L., Abel, R., Friesner, R. A., & Harder, E. D. (2019). OPLS3e: Extending force field coverage for drug-like small molecules. Journal of Chemical Theory and Computation, 15(3), 1863–1874. 10.1021/acs.jctc.8b01026 [DOI] [PubMed] [Google Scholar]
- Roschewski, M., Lionakis, M. S., Sharman, J. P., Roswarski, J., Goy, A., Monticelli, M. A., Roshon, M., Wrzesinski, S. H., Desai, J. V., Zarakas, M. A., Collen, J., Rose, K., Hamdy, A., Izumi, R., Wright, G. W., Chung, K. K., Baselga, J., Staudt, L. M., & Wilson, W. H. (2020). Inhibition of Bruton tyrosine kinase in patients with severe COVID-19. Science Immunology, 5(48), eabd0110. 10.1126/sciimmunol.abd0110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schor, S., & Einav, S. (2018). Repurposing of kinase inhibitors as broad-spectrum antiviral drugs. DNA and Cell Biology, 37(2), 63–69. 10.1089/dna.2017.4033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger . (2020. a). Schrödinger Release 2020-1: Epik. Schrödinger, LLC. [Google Scholar]
- Schrödinger . (2020. b). Schrödinger Release 2020-1: Glide. Schrödinger, LLC. [Google Scholar]
- Schrödinger . (2020. c). Schrödinger Release 2020-1: LigPrep. Schrödinger, LLC. [Google Scholar]
- Schrödinger . (2020. d). Schrödinger Release 2020-1: Maestro. Schrödinger, LLC. [Google Scholar]
- Schrödinger . (2020. e). Schrödinger Release 2020-1: Prime. Schrödinger, LLC. [Google Scholar]
- Schrödinger . (2020. f). Schrödinger Release 2020-1: Protein Preparation Wizard; Epik, 2016; Impact, 2016; Prime, 2020. Schrödinger, LLC. [Google Scholar]
- Schrödinger . (2020. g). Schrödinger Release 2020-1: QikProp. Schrödinger, LLC. [Google Scholar]
- Schrödinger . (2020. h). Schrödinger Release 2020-1: SiteMap. Schrödinger, LLC. [Google Scholar]
- Shah, B., Modi, P., & Sagar, S. R. (2020). In silico studies on therapeutic agents for COVID-19: Drug repurposing approach. Life Sciences, 252, 117652. 10.1016/j.lfs.2020.117652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, D. E., Grossman, J. P., Bank, J. A., Batson, B., Butts, J. A., Chao, J. C., Deneroff, M. M., Dror, R. O., Even, A., Fenton, C. H., Forte, A., Gagliardo, J., Gill, G., Greskamp, B., Ho, C. R., Ierardi, D. J., Iserovich, L., Kuskin, J. S., Larson, R. H., … Young, C. (2014). Anton 2: Raising the bar for performance and programmability in a special-purpose molecular dynamics supercomputer [Paper presentation]. SC14: International Conference for High Performance Computing, Networking, Storage and Analysis (pp. 41–53), New Orleans, LA, USA. 10.1109/SC.2014.9 [DOI] [Google Scholar]
- Spellberg, B., Haddix, M., Lee, R., Butler-Wu, S., Holtom, P., Yee, H., & Gounder, P. (2020). Community prevalence of SARS-CoV-2 among patients with influenzalike illnesses presenting to a Los Angeles medical center in March 2020. JAMA, 323(19), 1966–1967. 10.1001/jama.2020.4958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai, W., He, L., Zhang, X., Pu, J., Voronin, D., Jiang, S., Zhou, Y., & Du, L. (2020). Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: Implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cellular & Molecular Immunology, 17(6), 613–620. 10.1038/s41423-020-0400-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchesnokov, E. P., Feng, J. Y., Porter, D. P., & Götte, M. (2019). Mechanism of inhibition of ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses, 11(4), 326. 10.3390/v11040326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee, A., Koh, M. S., Gibson, P. G., Lasserson, T. J., Wilson, A., & Irving, L. B. (2007). Long-acting beta2-agonists versus theophylline for maintenance treatment of asthma. Cochrane Database of Systematic Reviews. 10.1002/14651858.CD001281.pub2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, N. C., Ro, J., André, F., Loi, S., Verma, S., Iwata, H., Harbeck, N., Loibl, S., Huang Bartlett, C., Zhang, K., Giorgetti, C., Randolph, S., Koehler, M., & Cristofanilli, M. (2015). Palbociclib in hormone-receptor–positive advanced breast cancer. New England Journal of Medicine, 373(3), 209–219. 10.1056/NEJMoa1505270 [DOI] [PubMed] [Google Scholar]
- Wang, M., Cao, R., Zhang, L., Yang, X., Liu, J., Xu, M., Shi, Z., Hu, Z., Zhong, W., & Xiao, G. (2020). Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Research, 30(3), 269–271. 10.1038/s41422-020-0282-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart, D. S., Knox, C., Guo, A. C., Shrivastava, S., Hassanali, M., Stothard, P., Chang, Z., & Woolsey, J. (2006). DrugBank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Research, 34(Database issue), D668–D672. 10.1093/nar/gkj067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F., Zhao, S., Yu, B., Chen, Y.-M., Wang, W., Song, Z.-G., Hu, Y., Tao, Z.-W., Tian, J.-H., Pei, Y.-Y., Yuan, M.-L., Zhang, Y.-L., Dai, F.-H., Liu, Y., Wang, Q.-M., Zheng, J.-J., Xu, L., Holmes, E. C., & Zhang, Y.-Z. (2020). A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, S., Zhu, Y., Liu, M., Lan, Q., Xu, W., Wu, Y., Ying, T., Liu, S., Shi, Z., Jiang, S., & Lu, L. (2020). Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cellular & Molecular Immunology, 17(7), 765–763. 10.1038/s41423-020-0374-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, Q., Hopkins, B., Perruzzi, C., Udayakumar, D., Sherris, D., & Benjamin, L. E. (2008). Palomid 529, a novel small-molecule drug, is a TORC1/TORC2 inhibitor that reduces tumor growth, tumor angiogenesis, and vascular permeability. Cancer Research, 68(22), 9551–9557. 10.1158/0008-5472.CAN-08-2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, W., Mao, C., Luan, X., Shen, D.-D., Shen, Q., Su, H., Wang, X., Zhou, F., Zhao, W., Gao, M., Chang, S., Xie, Y.-C., Tian, G., Jiang, H.-W., Tao, S.-C., Shen, J., Jiang, Y., Jiang, H., Xu, Y., … Xu, H. E. (2020). Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science, 368(6498), eabc1560. 10.1126/science.abc1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., Becker, S., & Rox, K., Hilgenfeld, R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science, eabb3405. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P., Yang, X.-L., Wang, X.-G., Hu, B., Zhang, L., Zhang, W., Si, H.-R., Zhu, Y., Li, B., Huang, C.-L., Chen, H.-D., Chen, J., Luo, Y., Guo, H., Jiang, R.-D., Liu, M.-Q., Chen, Y., Shen, X.-R., Wang, X., … Shi, Z.-L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.