Abstract
The health sector has been on the race to find a potent therapy for coronavirus disease (COVID)-19, a diseases caused by severe acute respiratory syndrome coronavirus (SARS-CoV)-2. Repurposed anti-viral drugs have played a huge role in combating the virus, and most recently, dexamethasone (Dex) have shown its therapeutic activity in severe cases of COVID-19 patients. The study sought to provide insights on the anti-COVID-19 mechanism of Dex at both atomic and molecular level against SARS-CoV-2 targets. Computational methods were employed to predict the binding affinity of Dex to SARS-CoV-2 using the Schrodinger suite (v2020-2). The target molecules and ligand (Dex) were retrieved from PDB and PubChem, respectively. The selected targets were SARS-CoV-2 main protease (Mpro), and host secreted molecules glucocorticoid receptor, and Interleukin-6 (IL-6). Critical analyses such as Protein and ligand preparation, molecular docking, molecular dynamic (MD) simulations, and absorption, distribution, metabolism, excretion (ADME), and toxicity analyses were performed using the targets and the ligand as inputs. Dex showed stronger affinity to its theoretical (glucocorticoid) receptor with a superior docking score of −14.7 and a good binding energy value of −147.48 kcal/mol; while short hydrogen bond distances were observed in both Mpro and IL-6 when compared to glucocorticoid receptor. Based on these findings, Dex-target complexes were used to perform MD simulations to analyze Dex stability at 50 ns. This study demonstrates that Dex could bind to both the viral and host receptors as a potential drug candidate for COVID-19. To ascertain the biological fitness of this study, other SARS-CoV-2 targets should be explored. Also, the in vitro studies of dexamethasone against several SARS-CoV-2 targets warrant further investigation.
Communicated by Ramaswamy H. Sarma
Keywords: Dexamethasone, COVID-19, interleukins, glucocorticoid, SARS-CoV-2, main protease
1. Introduction
To date, the most effective practice against the transmission of COVID-19 is physical distancing and the use of personal protective equipment. As the world is gradually easing the lockdown measures to reopen the economy, the search for potent drugs and possible vaccines against this virus is pivotal. The binding of SARS-CoV-2 spike protein to the human angiotensin converting enzyme (hACE-2) causes pneumocytes damage. In response to this process, the pneumocytes release specific pro-inflammatory mediators such as IL-1, IL-6, and tumor necrotic factor-alpha (TNF-α). The persistence of this process leads to acute respiratory distress syndrome (ARDS) (Schett et al., 2020). So far, the antiviral drug remdesivir was shown to benefit patients with COVID-19 in a large randomized, controlled clinical trial. This drug was reported to shorten the amount of time that patients might need to spend in hospital but did not have a statistically significant effect on mortality rate (Beigel et al., 2020; Horby et al., 2020). Dex, an anti-inflammatory drug is recently considered for treatment of severe cases of COVID-19 (Horby et al., 2020). Similar to a natural hormone produced by the adrenal gland, it is classified as glucocorticosteroid. Also known as Decadron, Dex was considered a major breakthrough based on a recent randomized control trial in the United Kingdom. Based on the mode of action, cortisol binds to the glucocorticoid receptor to stimulate the production of anti-inflammatory proteins thereby suppressing pro-inflammatory cytokines causing lung damage. In a press release for the Randomized Evaluation of COVID-19 Therapy (RECOVERY) trial on 16 June 2020, Dex was recommended for use in COVID-19 patients with severe respiratory symptoms. Dex reduced deaths by approximately 33% in patients requiring ventilation and by 20% in those requiring oxygen (Healthcare, 2020). Studies show that Dex has been used to reduce inflammation in a wide range of conditions, including inflammatory disorders and certain cancers (Hollander, 1960; Stoll, 1960). The WHO has added this compound on the Model List of Essential Medicines since 1977 in multiple formulations, and is currently off-patent and commonly available in most countries (Organization WH, 1977; Villar et al., 2020). In the attempt to develop novel antiviral drugs against this infection, different strategies have been combined. Both experimental and computational approaches have been recently reported with the aim of identifying potent therapeutic agents (Elfiky, 2020; Elmezayen et al., 2020; Enayatkhani et al., 2020; Enmozhi et al., 2020; Mittal et al., 2020). Possible drug targets for the treatment of COVID-19 are currently under active investigation. These targets include; the Human coronavirus (SARS-CoV-2; COVID-19) proteins, membrane glycoprotein, Nucleocapsid, tumor necrosis factor receptor type 6, toll-like receptor 3, surface glycoprotein (spike glycoprotein), Interleukin-6 receptor subunit α (IL-6RA), and RNA-directed RNA polymerase (Sorbera et al., 2020).
Interleukin 6 receptor is a class I cytokine receptor for IL-6. The complex of this receptor with signal transducer subunit is essential for proper function. The complex is involved in immune response regulation, acute-phase reaction and hematopoiesis (Tanaka et al., 2014). The respiratory tract infection by SARS-CoV-2 results in mild or highly acute respiratory syndrome with the release of proinflammatory cytokines, including IL-6. Suppression of inflammation in inflammatory diseases can be achieved by inhibiting IL-6 activity. Additionally, based on the biological and the pathological roles of IL-6 in various diseases, it was anticipated that IL-6 targeting would constitute a novel treatment strategy for various immune-mediated diseases (Liuzzi et al., 2005; Nemeth et al., 2004; Nishimoto et al., 2005). Molecules capable of binding to the receptor subunit of IL-6 may render the receptor inactive and may potentially attenuate pulmonary inflammation in patients with COVID-19.
Coronaviruses are enveloped, positive-sense, single-stranded RNA viruses. The open reading frame ORF 1ab encodes the overlapping polyproteins (pp1a, pp1ab) and are cleaved into 16 non-structural proteins (nsp1-16) by Mpro and the papain-like protease (Chen et al., 2020). Due to the crucial role of Mpro in processing the polyproteins that are translated from the viral RNA, it is considered as an essential drug target (Boopathi et al., 2020). Currently, research efforts focus on the identification of potential inhibitors of this target (Zhavoronkov et al., 2020). Specifically, Mittal et al. (2020) identified potential molecules against COVID-19 main protease through structure-guided virtual screening approach using anti-protease molecules for drug repurposing purposes. Jin et al. (2020) also studied the crystal structure of the Mpro in relation to specific inhibitors.
Dexamethasone was initially developed as a glucocorticoid receptor-specific agonist (CGTP Collaborators, 2013) and was used to determine the first glucocorticoid receptor-ligand binding domain structure (Bledsoe et al., 2002). However, dexamethasone was later shown to also be a potent mineralocorticoid receptor ligand in a functional reporter gene assay (Rupprecht et al., 1993). The X-ray structure of mineralocorticoid receptor in complex with dexamethasone is similar to the corresponding glucocorticoid receptor-Dex structure. Also, the relationship between Dex and glucocorticoid were previously studied in neuroprogenitor cells in the hippocampus of rat pups (Sze et al., 2013), human monocyte cell line THP-1 (Bo et al., 2006), human lens epithelial cells (Gupta & Wagner, 2005), and in human ovarian carcinoma cell line 3AO (Xu et al., 2003). These studies suggest that Dex effects are mediated through glucocorticoid receptors. Additionally, the ligand binding mechanism in steroid receptors was studied by Edman et al. (2015) using glucocorticoid and solved the crystal structure using Dex as its inhibitor. This interaction was therefore considered as the reference complex and the co-crystalized ligand was regarded as control in the optimization of molecular docking protocol.
Among other questions yet unanswered about the clinical use of Dex, the ideal dose and possible interactions against SARS-CoV-2 targets remain unclear. In order to investigate the mechanism of interactions between Dex and SARS-CoV-2 possible targets, this study examined the binding interaction of Dex in comparison with glucocorticoid in order to provide insight at both atomic and molecular level using computational approach.
2. Materials and methods
2.1. Modeling platform
The entire computational analysis was carried out using the Schrodinger suite (2020-2) using the Maestro v12.4 version packages including LigPrep, Protein preparation, Glide XP docking, grid generation, free energy calculations, ADME toxicity, and MD simulations. Linux was used as the operating system
2.2. Datasets
The receptors employed in this study include: (a) Glucocorticoid receptor in complex with Dex (Edman et al., 2015) (PDB ID: 4UDC); (b) SARS-CoV-2 Mpro bound to potent broad-spectrum non-covalent inhibitor X77 (A taxonomically-driven approach to development of potent, broad-spectrum inhibitors of coronavirus main protease including SARS-CoV-2 (COVID-19). Mesecar, A.D. (To be published)). (PDB ID: 6W63); (c) Human IL-6 (Somers et al., 1997) (PDB ID: 1ALU). Dex (decardron) structure was retrieved from PubChem at https://pubchem.ncbi.nlm.nih.gov/ as the ligand (compound CID: 5743).
2.3. Adme/tox analysis
The Schrodinger QikProp module (Release S. 4, 2017) and the AdmetSAR web-based tool (http://lmmd.ecust.edu.cn/admetsar1/predict) were used to analyze the absorption, distribution, metabolism, excretion, and toxicity of Dex, respectively. For toxicity, the Ames toxicity, carcinogenic properties, acute oral toxicity, and rat acute toxicity were predicted.
2.4. Preparation of Dex
LigPrep module in Schrodinger was used to prepare the 3D- structure of Dex. The ligand preparation process consists of a series of steps that perform conversions, apply corrections to the structures, generate variations on the structures, eliminate unwanted structures, and optimize the structures. The downloaded SD format from PubChem was converted to Maestro format using the command line ‘sdconvert’. Before the minimization of the 3D structure, hydrogen atoms were added using ‘applyhtreat’. Charged groups were neutralized before the generation of the ionization state by ‘neutralizer’ and ‘ionizer’. Prior to docking, low-energy ring conformation was generated (‘ring_conf’) and the geometries of the generated structure was optimized by ‘bmin’ (Release S. 2, 2017). The force field used in the minimization of Dex is the Optimized Potentials for Liquid Simulations known as OPLS_2005.
2.5. Preparation of Dex targets
Protein preparation module in Schrodinger was used to prepare the receptors for Dex: human IL-6, glucocorticoid receptor, and SARS-CoV-2 Mpro which were retrieved from protein data bank with the PDB IDs: 1ALU, 4UDC and 6W63, respectively. Prior to molecular modeling calculations, these proteins were subjected to protein preparation wizard in order to add hydrogen atoms, remove alternate conformation, correct missing or incorrectly specified residues, remove HetAtoms from the protein structure, correct missing or incorrectly specified residues amongst others.
2.6. Molecular docking study
Protein-ligand interactions study was performed between Dex and the aforementioned receptors. The binding pockets on the receptors were generated using the receptor grid generation module with either their co-crystalized ligands or specific coordinates. For 4UDC and 6W63, the co-crystalized ligands Dex and X77 (N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-1H-imidazole-4-carboxamide) were used to generate the grids by selecting the atoms of the ligands respectively. The docking grid of the active site of 1ALU was identified using the PDB file of the coordinates. This site provides information about the area around the active site (coordinates x, y and z). The receptor grid box resolution was positioned at coordinates −0.35 (x-axis), 74.13 (y-axis), and 350.16 (z-axis). Furthermore, the box length was maintained at 10 Å for x, y and z. Docking of Dex to the respective receptors was carried out using the Glide ligand docking tool in Maestro v12.4. (Release S. 4. , 2015). Docking and calculations were performed in the extra precision (XP) mode of Glide and XP visualizer.
2.7. Prime energy properties calculation
The prime MM-GBSA was used to generate the energy properties of the ligand, receptor, and complex structures as well as energy differences relating to strain. (Hayes & Archontis, 2012; Li et al., 2011). In this study, only the binding energies of ligand docked complexes were recorded.
2.8. Molecular dynamic (MD) simulations
Prior to simulation, the docked complexes were prepared using the system builder module in Maestro v12.4. MD simulations were carried out using the Desmond software. The optimized potentials for the liquid simulations OPLS-2005 force field were used in this system to determine the receptors interactions with Dex, which was solvated with the simple point charged (TIP-3P) water model (Blessy & Sharmila, 2015; Jorgensen et al., 1983). The orthorhombic water box was used to create a 10 Å buffer region between the atoms on the receptors and box sides. The volume of the box was minimized and the overall charge of the system was neutralized by adding Na+. The temperature and pressure were kept constant at 300 Kelvin and 1.01325 bar using Nose–Hoover thermostat (Hoover, 1985) and Martyna–Tobias–Klein barostat methods. The simulations were performed using NPT ensemble by considering number of atoms, pressure and timescale and the simulation time at 50 nanoseconds. The MD results were analyzed by simulation interactions diagram module and MS-MD trajectory analysis.
3. Results
Molecular interactions between Dex (Figure 1) and three targets involved in Covid-19; Glucocorticoid receptor, SARS-CoV-2 Mpro, and IL-6 receptors was investigated in silico through molecular docking analysis and simulation.
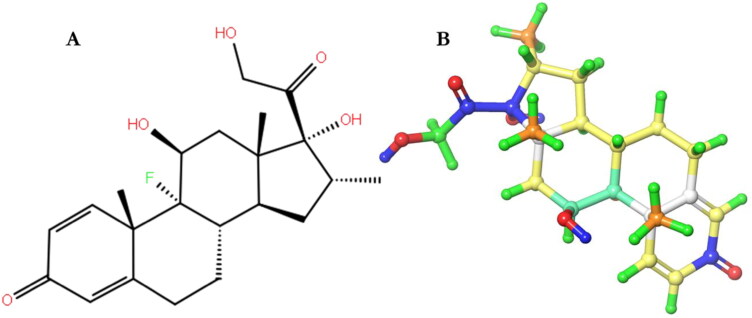
Figure 1.
The 2D (A) and 3D (B) structure of Dex colored by atomic partial charge.
3.1. Adme/tox analysis
The ADME and toxicological properties of Dex were analyzed by QikProp and AdmetSAR web-based tools, respectively. The physicochemical and biological properties of Dex (Tables 1 and 2) analyzed included the molecular weight, hydrogen bond acceptor, hydrogen bond donor, lipophilicity, number of rotatable bond, topological polar surface area, ames toxicity, carcinogens, acute oral toxicity, and rat acute toxicity. This analysis is crucial for analyzing the efficacy of molecules. All the parameters were within the ROF cut-off range for the test compound (Dex) and showed no violation of the Lipinski’s rule of five and Veber rules (Table 1) and presents no bystander toxicity effects (Table 2).
Table 1.
Pharmacological properties of Dex.
| Compound | Lipinski rules |
Lipinski's | Veber rules |
||||
|---|---|---|---|---|---|---|---|
| MW | HBA | HBD | Log p | Violations | nRB | TPSA | |
| ROF cut-off | <500 | <10 | <5 | ≤5 | ≤1 | ≤10 | ≤140 |
| Dex | 392.46 | 6 | 3 | 2.15 | 0 | 2 | 94.83 Ų |
Note: ROF: Lipinski’s Rule of Five; MW: Molecular weight (g/mol); HBA: Hydrogen bond acceptor; HBD: Hydrogen bond donor; Log P: Lipophilicity; nRB: Number of rotatable bond; TPSA: Topological polar surface area.
Table 2.
Toxicological properties of Dex.
| Parameters | Compound name |
|---|---|
| Dexamethasone | |
| Ames toxicity | NAT |
| Carcinogens | NC |
| Acute oral toxicity | III |
| Rat acute toxicity | 2.1482 |
Note: NAT: Non Ames toxic; NC: Non-carcinogenic; Category-III means (500 mg/kg > LD50 < 5000 mg/kg).
3.2. Docking calculation
Molecular docking approach identifies suitable drug molecules for the target of interest (Roy et al., 2015; Sharma et al., 2011; Subhani et al., 2015; Vijayakumar et al., 2018). The docking result shows the potential of Dex to bind within the receptor pockets or active sites (Figure 2) and the hydrogen bond interactions within specified distances (Figures 3 and 4). The amino acid residues of the receptors crucial to binding Dex within specific distances (H-bond distance values) are depicted in Figure 4. Furthermore, the docking scores of the complexes were also generated (Table 3). To specify the nature of the receptor–Dex interactions, the Molecular Mechanics-Generalized Boltzmann surface area (MM-GBSA) binding energy calculations were carried out using the complexes as input in Prime module. The result of the binding energies of the complexes are given in Table 3. Glucocorticoid receptor was predicted to be notably more preferable for binding of Dex with corresponding binding free energy of −147.48 kcal/mol for Dex, being lower compared with the binding energies for Mpro and IL-6 which was −66.45 and −53.03 kcal/mol, respectively.
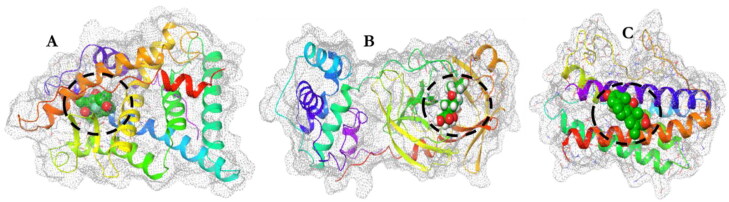
Figure 2.
The space filled model of the ligand (Dex) in the active sites of each receptor as depicted within the white broken circles. Glucocorticoid receptor (A); Mpro (B); and IL-6 (C) receptors.
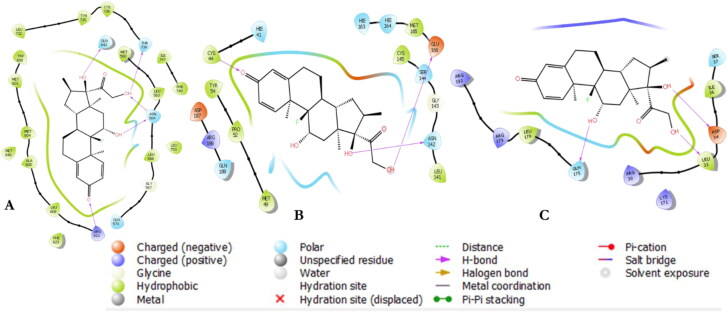
Figure 3.
2 D interactions of Dex within the receptors’ binding pockets. Glucocorticoid receptor (A); Mpro (B); and IL-6 (C).
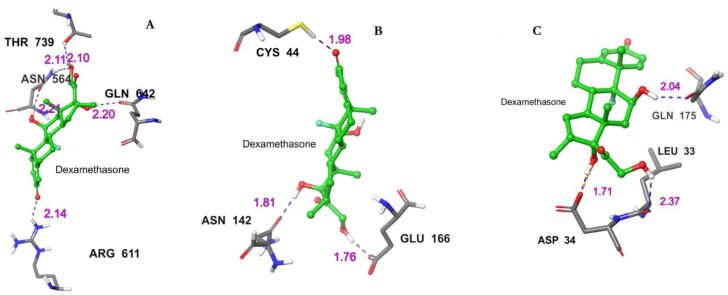
Figure 4.
Molecular interactions between Dex and selected receptors. Hydrogen bond contacts (blue dotted line) with their distance values (pink values) within the Dex-receptor complexes. (A) Interacting atoms of glucocorticoid receptor and Dex complex. (B) Interacting atoms of Mpro and Dex complex (C) Interacting atoms of IL-6 and Dex complex.
Table 3.
Docking results of Dex and the energy of interaction with the active site of the receptors.
| Receptor | Glide Gscore | Dock score | ΔGbind (kcal/mol) | (No) of H-bonds within 2.5 Å |
|---|---|---|---|---|
| Glucocorticoid | −14.3 | −14.3 | −147.48 | (5) ASN564 (2 bonds), THR739, GLN642, ARG611 |
| Mpro | −6.7 | −6.7 | −66.45 | (3) ASN142, GLU166, CYS44 |
| IL-6 | −3.6 | −3.6 | −53.03 | (3) GLN175, ASP34, LEU33 |
In this computational analysis, glucocorticoid receptor complexed with Dex had the highest docking score (−14.3) than other Mpro/IL-6-Dex complexes (−6.7 and −3.6 for Mpro and IL-6 respectively). Furthermore, based on specific interactions, ASN564, THR739, GLN642, and ARG611 in the glucocorticoid receptor were involved in hydrogen bonding with the atoms of Dex. Three amino acid residues (ASN142, GLU166, and CYS44) in the Mpro active site were involved in boding with Dex atoms. IL-6 also interacted with three residues (GLN175, ASP34, and LEU33) which were hydrogen bonded with Dex atoms in its binding pocket. However, Dex showed a better binding affinity for Mpro than the other receptors based on the distances of the interacting atoms in the active site. Overall, Dex showed good measurable binding affinities for the receptors residues. The binding affinities were indicative of the ligand's contribution to the flexibility for the targets. The present study also showed the H-bond distances and their contacts types for Dex.
3.3. Molecular dynamic simulation result
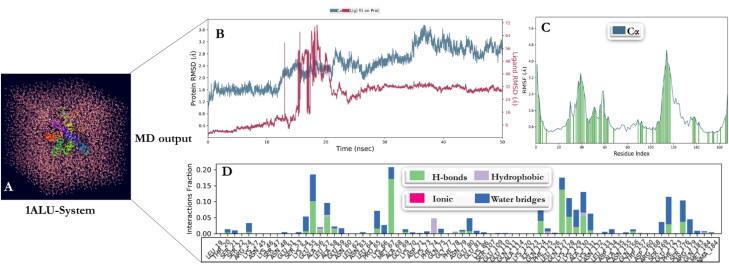
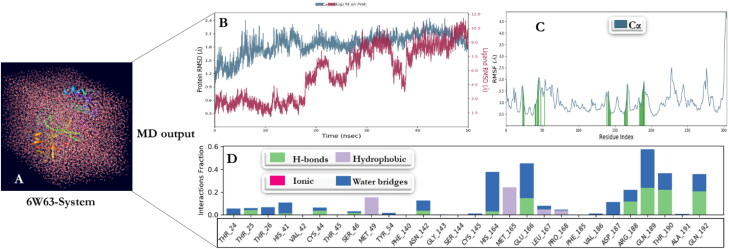
The MD simulation was performed for the three receptors (glucocorticoid receptor, Mpro, and IL-6) and Dex complexes to evaluate the structural stability and constancy by Desmond in Maestro v12.4 software. The simulation process for these complexes were computed for 50 ns. For human IL-6 (1ALU), Glucocorticoid receptor complex (4UDC), and Mpro complex (6W63) with Dex, the simulation processes were built with solvent model predefined as TIP-3P. The boundary conditions were adjusted with a box shape ‘orthorhombic’ and the box size calculation method ‘buffer’ with a minimized box volume. Ion replacement was recalculated by adding Na+ to neutralize the environment. The system builder was concluded by adding 0.15 M salt concentration (salt positive ion: Na+ and salt negative ion: Cl-) (Figures 5(A), 6(A), and 7(A)). The RMSD plot of human Interleukin-6 complex was stable within the first 12 ns of simulation and deviated for a certain period of time (14–25 ns) and attained equilibrium at 25 ns (Figure 5(B)). The RMSF was computed to include ligand contact as presented in Figure 5(C). The simulation of glucocorticoid receptor complex with Dex was at equilibrium from the beginning and throughout the simulation period of 50 ns (Figure 6(B), (C)). Initially, the RMSD plot of Mpro complex showed that the complex deviated for a certain period and attained equilibrium at 40 ns. Subsequently, it remained stable throughout the simulation time for up to 50 ns (Figure 7(B), (C)). Protein interactions with Dex were monitored throughout the simulation. These interactions were categorized by type and summarized, as shown in the plot above (Figures 5(D), 6(D), and 7(D)). Protein-ligand interactions (or 'contacts') are categorized into four types: Hydrogen Bonds, Hydrophobic, Ionic and Water Bridges. The stacked bar charts were normalized over the course of the trajectory. Notably, for glucocorticoid receptor-Dex complex, THR739 and GLN642 present interaction fragments above 0.75 suggesting that over 75% of the simulation time hydrogen bond is maintained with Dex. ASN564 residue showed multiple contacts of same subtype with Dex (above 1.75).
Figure 5.
MD simulations: (A) The system for Human IL-6 complex with Dex. (B) Root mean standard deviation (RMSD) of Dex with IL-6 complex as a function of simulation time. Protein residues that interact with the ligand are marked with green-colored vertical bars. (C) Root mean square fluctuation (RMSF) values of complex IL-6 residues with Dex. (D) The IL-6-Dex complex contacts.
Figure 6.
MD simulations: (A) The system for Glucocorticoid Receptor complex with Dex. (B) RMSD of Dex with glucocorticoid receptor complex as a function of simulation time. (C) RMSF values of complex glucocorticoid Receptor residues with Dex. Protein residues that interact with the ligand are marked with green-colored vertical bars. (D) The glucocorticoid receptor-Dex complex contacts.
Figure 7.
MD simulations: (A) The system for Mpro complex with Dex. (B) RMSD of Dex with Mpro complex as a function of simulation time. Protein residues that interact with the ligand are marked with green-colored vertical bars. (C) RMSF values of complex Mpro residues with dexamethasone. (D) The Mpro-Dex complex contacts.
4. Discussion
There are currently approximately 14 million global cases of COVID-19 and more than 500, 000 mortality rates (as at 23rd July 2020), yet there is no treatment that is specific for the disease. To keep the economy afloat, lockdown restrictions are being reduced, and this might cause another wave of COVID-19 infections and transmissions. Treatment so far, has been through repurposed anti-viral drugs such as chloroquine and its analogues, remdesivir, and currently an anti-inflammatory agent Dex. Although, they show promise in fighting against the disease, some are over-shadowed by their bystander effects (Healthcare, 2020). Dex have a long history in prevention of chronic lung infections in premature babies, and have been shown to bind to glucocorticoid receptor to induce its functions. Given its immunosuppressant properties, surprisingly Dex reduced of mortality of severe COVID-19 patients who requires oxygen and those on ventilators (Healthcare, 2020; Isidori et al., 2020; Sze et al., 2013). Khan and Htar (2020) reported the computational study of Dex against the SARS-CoV-2 Mpro in relation to remdesivir and reported that Dex interacted with a high affinity to the same sites of the SARS-COV-2 Mpro than remdesivir. Nayeem and Reddy (2020) also evaluated the binding capacity of Dex and Umifenovir to SARS-CoV-2 Mpro via molecular dynamic studies using Gromacs with OPLS-AA force field and concluded that Dex is more effective when compared to Umifenovir in binding SARS-CoV-2 Mpro. In this study, the effect of Dex was investigated against three targets associated with Covid-19 infections to determine the drug’s mode of action at both atomic and molecular levels and to further provide insight into its use as anti-COVID-19. The three targets namely glucocorticoid receptor, Mpro, and IL-6 are composed of 186, 280, and 336 amino acids with resolutions of 1.90, 2.50, and 2.10 Å, respectively. Dex and the three targets were prepared using LigPrep and protein preparation wizard, respectively. The outputs were further minimized using the force field OPLS_2005 and were used for molecular docking calculations.
Currently, there are no known drug that is safe and effective against COVID-19. However, the possible drug target in this virus is the Mpro, an enzyme responsible for gene expression and replication of the virus. This enzyme has been labeled an attractive target for drug discovery (Xue et al., 2008). The Mpro has over 11 cleavage sites on the replicase 1ab polyprotein that is approximately 780 kDa, inhibition of the protease activity would prevent viral replication. Also, because there are no known human protease with similar cleavage specificity, the inhibitor of the Mpro is less likely to be toxic (Zhang et al., 2020). The molecular docking study showed the potential of Dex to bind the receptor cavity of Mpro with specific interactions. The Mpro-Dex complex produced ligand docking scores with three H-bonds and their distance values and the consequent glide energy. Dex was also docked into the active site of glucocorticoid receptor in order to evaluate the binding interaction in relation to other two receptors. In glucocorticoid receptor-Dex complex, four hydrogen atoms were observed between the interacting atoms of Dex and ASN564, THR739, GLN642, and ARG611 amino acid residues in the binding cavity of the receptor. Additionally, the complex produced a dock score and binding energy of −14.3 and −147.48 kcal/mol, respectively. Previous study has reported the mediation of glucocorticoid receptor by Dex in pancreatic cancer (Liu et al., 2017). Several experimental studies have demonstrated the induction of resistance to chemotherapy by glucocorticoids in solid tumors (Herr & Pfitzenmaier, 2006; Volden & Conzen, 2013; Zhang et al., 2006). The third receptor (IL-6), is a pleiotropic cytokine involved in the regulation of cellular processes (Boulanger et al., 2003), and modulation of immune responses and acute immune reaction (Dowton et al., 1991; Eaves et al., 1991; Matsuda et al., 1989). Dysregulation of IL-6 or its receptor IL-6R correlates closely with cancer, inflammation diseases or autoimmune diseases (Hideshima et al., 2004; Nishimoto et al., 2005). SARS-CoV-2 infection in the lungs causes acute respiratory syndrome which in turn activates the release of proinflammatory cytokines. Suppression of inflammation can be achieved by inhibiting the activities of IL-6. Molecules that inhibit the IL-6RA would inactivate IL-6R and further decrease the inflammation of the lung tissue in individuals with COVID-19 (Conti et al., 2020; Mahmud-Al-Rafat et al., 2019; Zhang et al., 2020).
Drug discovery is a crucial aspect of research in which simulations can drive experiments (Borhani & Shaw, 2012; Durrant & McCammon, 2011). Recent advances in structural biology have led to structures for many key drug discovery targets. Fully exploiting the power of structure-based drug design requires taking the dynamic properties of proteins into account (Hollingsworth & Dror, 2018). Simulations have proven valuable in deciphering functional mechanisms of proteins and other biomolecules, in uncovering the structural basis for disease, and in the design and optimization of small molecules, peptides, and proteins. MD was used to capture the behavior of Dex and potential SARS-CoV-2 targets in full atomic detail and at very fine temporal resolution.
The RMSD of the backbone of the targets were used to evaluate the stability of the systems. This parameter is a measure of how much the protein targets changes with respect to the initial structure over the course of the simulation (structural distance between coordinates). The results of the RMSD of the Dex-targets were studied using 50 ns trajectories. The RMSD behavior of the complex in each system did not deviate drastically over the simulation testing period studied. The RMSD values of the system observed in this study was within the range of 2.25 − 3.6 Å. This range of value is a measure of how much the targets conformation changed with respect to Dex. RMSD around 1.0 − 3.0 Å is acceptable for small and globular proteins and changes much larger than 3.0 Å, however, shows that the target proteins underwent conformational changes during the simulation. It was also observed that the simulation converged and a stabilized RMSD values were seen at a fixed value (40 − 50 ns) in all the systems. This shows that the systems equilibrated and the simulation time was enough for rigorous analysis. The binding pose of Dex to glucocorticoid receptor seems to be stable throughout the MD simulation times (50 ns), maintaining complexation and hydrogen bonding for Dex as well as hydrophobic contacts with preferred partners on glucocorticoid receptor. The characterization of the local fluctuation of the targets was carried out using the RMSF in order to compare the time average representation per residue fluctuations representing the flexibility of each residue. The results show that the complexes maintained a reasonable stability in aqueous medium. For glucocorticoid-Dex, the residues around 25 and 250 fluctuated much more and these regions correspond to the terminal regions of glucocorticoid. In the Mpro-Dex system, only the residues at the N-terminal and C- terminal fluctuated the most. The system for IL-6-Dex fluctuated the most among the three systems. Additionally, the residues around 110 in the IL-6 (around 4.8 Å) were involved in larger fluctuation within the system and with respect to other systems. Secondary structure elements such as alpha helices and beta strands are usually more rigid than unstructured part of a protein and thus will fluctuate less. Hydrogen Bonds and their properties play a significant role in ligand binding specifically in drug design due to their strong influence on drug specificity, metabolization and adsorption. Hydrogen bonds between a protein and a ligand can be further broken down into four subtypes: backbone acceptor; backbone donor; side-chain acceptor; side-chain donor. These bonds stabilize the secondary, tertiary and quaternary structure of proteins which are formed by alpha helix, beta sheets, turns and loops. The current geometric criteria for protein-ligand H-bond in this study is a distance of 2.5 Å between the donor and acceptor atom. These contacts are one of the most important contributions to protein-ligand interactions governed by changes in entropy and enthalpy (Klebe & Böhm, 1997).Similarly, but less stable, Dex maintained complexation, hydrogen bonding, and hydrophobic contacts with Mpro and IL-6 specific residues. Other contacts observed in this study (SARS-CoV-2 potential targets and Dex interactions) include hydrophobic interaction, ionic contact, and salt bridges. Hydrophobic contacts include π-Cation; π-π; and other, non-specific interactions. These contacts are important for the folding of proteins keeping them stable, biologically active, and reduce the undesirable interactions with water. The importance of ionic contact was also observed due to their potent electrostatic attractions. In the hydrophobic interior of proteins, ionic bonds may approach the strength of covalent bonds making them crucial for stability (Pace et al., 2014). Water bridges are hydrogen-bonded protein-ligand interactions mediated by a water molecule. In the simulation study, the hydrogen-bond geometry of the water bridges is slightly relaxed from the standard H-bond interactions.
Based on the hydrogen bond contact and other interactions between the complexes during molecular docking study and simulation, five atoms of Dex were involved in the binding interactions with the receptors. These atoms may be significant and constant with other targets. Specifically, Dex interacted with the amino acid residues in the binding sites of the studied receptors through hydrogen bonding using three molecules of its hydroxyl groups at positions 12, 23, and 28 and additional oxygen atoms at position 10 and 25. During simulations, the hydrogen bonding observed in the docking analysis was stable and constantly in contact with glucocorticoid. GLN 642 was hydrogen bonded to the hydroxyl group of Dex at position 12 (hydrogen bond donor). This residue was in contact with Dex for about half the entire time of contact during simulation. ASN564 retained the hydrogen interaction in the binding domain and the hydroxyl group at position 21 and 28 of Dex throughout the entire simulation time. Although the hydrogen bond interaction between ARG611 and the oxygen atom at position 10 of Dex were highly unstable, water bridges were observed for about 50% of the entire simulation time. The interactions in the IL-6 system were totally unstable as the hydrogen interactions observed between the ASP34, GLN175, and LEU33 and the hydroxyl groups at position 12, 23, and 28, respectively, were replaced with residues GLU55, MET6, GLN12, and LYS 129 using both hydrogen bonding, hydrophobic interactions and water bridges throughout the simulation period. For Mpro system, all the target residues were stable and constantly in contact with Dex’s oxygen atoms at position 10 (CYS44), and two hydroxyl groups at position 12 (ASN142), and 28 (GLU166). The contact observation therefore shows that glucocorticoid and Mpro systems of Dex are more stable than the IL-6-Dex system.
Given the strong interactions with Dex and its atoms in close contact with the amino acid residues in the binding pockets of the receptors, conformational changes could be induced which may affect binding interaction and lead to receptors inactivation. The role and expression of glucocorticoid receptor in COVID-19 is still elusive. Drugs that function as glucocorticoid mediators, either as agonists or antagonists, are used in many sections of therapeutic activities (Rathnayake & Weerasinghe, 2018). This data suggest that it is possible that Dex might interact with receptors other than glucocorticoid receptor to alleviate COVID-19. Taking note that the immune system plays an important role in the fight against COVID-19, the use of corticosteroids in the initial stages was previously discouraged. However, the current finding does suggest that this drug might be favorable in patients with severe COVID-19. The current clinical studies on Dex is expected to share insights on its mechanisms, and possibly identify the targets and validate if Dex use is only effective in severe cases and not the early stages. Currently, it is not clear whether Dex binds to host or the virus receptors but based on this study, the possibility of the drug binding to either of the two exists.
5. Conclusion
This study shows that the binding of Dex to the studied targets (Mpro, glucocorticoid, and IL-6 receptors) could prevent the synthesis and attenuate pulmonary inflammation in patients with severe COVID-19. The statistics indicates that the strength of binding energies of Dex to glucocorticoid and Mpro is stronger than IL-6. Furthermore, the elucidation of the detailed mechanism of interaction between Dex and these receptors will further enable rational design of highly effective inhibitory molecules for SARS-CoV-2 targets.
6. Recommendation
The mode of interaction with other molecules and effective dosage of Dex can further be investigated experimentally to ascertain its biological fitness. Other reported SARS-CoV-2 targets such as Protein Kinase Cα type which mediate lung endothelial injury and Basigin (CD147) which mediates viral entry into host cells can be explored for therapeutic development against COVID-19 using Dex.
Authors’ contributions
All authors have made significant contributions to the submission of the article. A.O.F, and M.M conceived the concept and the design of the study, N.R.S.S, A.M, and M.M supervised and provided the necessary supports and software required for analysis. The analysis and data interpretations were done collaboratively by all the authors while A.O.F and N.R.S.S prepared the initial draft and also substantially revised the manuscript. A.M, and M.M thoroughly revised the manuscript. Finally, all authors read and approved the submitted version of the manuscript for publication.
Acknowledgements
We would like to acknowledge the National Integrated Cyberinfrastructure system, Center for High Performance Computing (CHPC), Department of Science and Technology, Republic of South Africa for the license to the Lengau cluster and other modules under the Schrodinger suit.
Disclosure statement
The authors declare no competing interests.
References
- Alexander, S. P., Benson, H. E., Faccenda, E., Pawson, A. J., Sharman, J. L., Spedding, M., Peters, J. A., Harmar, A. J., Collaborators, C, & CGTP Collaborators (2013). The concise guide to PHARMACOLOGY 2013/14: Nuclear hormone receptors. British Journal of Pharmacology, 170(8), 1652–1675. 10.1111/bph.12448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel, J. H., Tomashek, K. M., Dodd, L. E., Mehta, A. K., Zingman, B. S., Kalil, A. C., Hohmann, E., Chu, H. Y., Luetkemeyer, A., & Kline, S. (2020). Remdesivir for the treatment of Covid-19—preliminary report. New England Journal of Medicine, 1–12. [DOI] [PubMed] [Google Scholar]
- Bledsoe, R. K., Montana, V. G., Stanley, T. B., Delves, C. J., Apolito, C. J., McKee, D. D., Consler, T. G., Parks, D. J., Stewart, E. L., Willson, T. M., Lambert, M. H., Moore, J. T., Pearce, K. H., & Xu, H. E. (2002). Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell, 110(1), 93–105. 10.1016/s0092-8674(02)00817-6 [DOI] [PubMed] [Google Scholar]
- Blessy, J. J., & Sharmila, D. J. S. (2015). Molecular simulation of N-acetylneuraminic acid analogs and molecular dynamics studies of cholera toxin-Neu5Gc complex. Journal of Biomolecular Structure & Dynamics, 33(5), 1126–1139. 10.1080/07391102.2014.931825 [DOI] [PubMed] [Google Scholar]
- Bo, L., Xiangjun, B., & Haiping, W. (2006). Effect of dexamethasone on expression of glucocorticoid receptor in human monocyte cell line THP-1. Journal of Huazhong University of Science and Technology. Medical Sciences = Hua Zhong ke ji da Xue Xue Bao. Yi Xue Ying De Wen Ban = Huazhong Keji Daxue Xuebao. Yixue Yingdewen Ban, 26(1), 25–27. 10.1007/BF02828029 [DOI] [PubMed] [Google Scholar]
- Boopathi, S., Poma, A. B., & Kolandaivel, P. (2020). Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1758788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borhani, D. W., & Shaw, D. E. (2012). The future of molecular dynamics simulations in drug discovery. Journal of Computer-Aided Molecular Design, 26(1), 15–26. 10.1007/s10822-011-9517-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulanger, M. J., Chow, D-c., Brevnova, E. E., & Garcia, K. C. (2003). Hexameric structure and assembly of the interleukin-6/IL-6 alpha-receptor/gp130 complex . Science (New York, N.Y.), 300(5628), 2101–2104. 10.1126/science.1083901 [DOI] [PubMed] [Google Scholar]
- Chen, Y., Liu, Q., & Guo, D. (2020). Emerging coronaviruses: Genome structure, replication, and pathogenesis. Journal of Medical Virology, 92(4), 418–423. 10.1002/jmv.25681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti, P., Ronconi, G., Caraffa, A., Gallenga, C., Ross, R., Frydas, I., & Kritas, S. (2020). Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): Anti-inflammatory strategies. J Biol Regul Homeost Agents, 34, 1. [DOI] [PubMed] [Google Scholar]
- Dowton, S. B., Waggoner, D. J., & Mandl, K. D. (1991). Developmental regulation of expression of C-reactive protein and serum amyloid A in Syrian hamsters. Pediatric Research, 30(5), 444–449. 10.1203/00006450-199111000-00010 [DOI] [PubMed] [Google Scholar]
- Durrant, J. D., & McCammon, J. A. (2011). Molecular dynamics simulations and drug discovery. BMC Biology, 9, 71–79. 10.1186/1741-7007-9-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaves, C. J., Cashman, J. D., Kay, R. J., Dougherty, G. J., Otsuka, T., Gaboury, L., Hogge, D., Lansdorp, P., Eaves, A., & Humphries, R. (1991). Mechanisms that regulate the cell cycle status of very primitive hematopoietic cells in long-term human marrow cultures. II. Analysis of positive and negative regulators produced by stromal cells within the adherent layer.Blood, 78, 110–117. [PubMed] [Google Scholar]
- Edman, K., Hosseini, A., Bjursell, M. K., Aagaard, A., Wissler, L., Gunnarsson, A., Kaminski, T., Köhler, C., Bäckström, S., Jensen, T. J., Cavallin, A., Karlsson, U., Nilsson, E., Lecina, D., Takahashi, R., Grebner, C., Geschwindner, S., Lepistö, M., Hogner, A. C., & Guallar, V. (2015). Ligand binding mechanism in steroid receptors: From conserved plasticity to differential evolutionary constraints. Structure (London, England: 1993), 23(12), 2280–2290. 10.1016/j.str.2015.09.012 [DOI] [PubMed] [Google Scholar]
- Elfiky, A. A. (2020). SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: An in silico perspective. Journal of Biomolecular Structure and Dynamics, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmezayen, A. D., Al-Obaidi, A., Şahin, A. T., & Yelekçi, K. (2020). Drug repurposing for coronavirus (COVID-19): In silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. Journal of Biomolecular Structure and Dynamics, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enayatkhani, M., Hasaniazad, M., Faezi, S., Guklani, H., Davoodian, P., Ahmadi, N., Einakian, M. A., Karmostaji, A., & Ahmadi, K. (2020). Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: An in silico study. Journal of Biomolecular Structure and Dynamics, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enmozhi, S. K., Raja, K., Sebastine, I., & Joseph, J. (2020). Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: An in silico approach. Journal of Biomolecular Structure and Dynamics, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, V., & Wagner, B. (2005). Dexamethasone induces changes in glucocorticoid receptor phosphorylation and gene expression in human lens epithelial cells. Investigative Ophthalmology & Visual Science, 46, 1890. [Google Scholar]
- Hayes, J. M., & Archontis, G. (2012). MM-GB (PB) SA calculations of protein-ligand binding free energies. Molecular Dynamics-Studies of Synthetic and Biological Macromolecules, 171–190. [Google Scholar]
- Healthcare, G. (2020). Dexamethasone, an unexpected drug, shows promise in treating Covid-19.
- Herr, I., & Pfitzenmaier, J. (2006). Glucocorticoid use in prostate cancer and other solid tumours: Implications for effectiveness of cytotoxic treatment and metastases. The Lancet Oncology, 7(5), 425–430. 10.1016/S1470-2045(06)70694-5 [DOI] [PubMed] [Google Scholar]
- Hideshima, T., Bergsagel, P. L., Kuehl, W. M., & Anderson, K. C. (2004). Advances in biology of multiple myeloma: Clinical applications. Blood, 104(3), 607–618. 10.1182/blood-2004-01-0037 [DOI] [PubMed] [Google Scholar]
- Hollander, J. L. (1960). Clinical use of dexamethasone: Role in treatment of patients with arthritis. Journal of the American Medical Association, 172, 306–310. 10.1001/jama.1960.03020040004002 [DOI] [PubMed] [Google Scholar]
- Hollingsworth, S. A., & Dror, R. O. (2018). Molecular dynamics simulation for all. Neuron, 99(6), 1129–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover, W. G. (1985). Canonical dynamics: Equilibrium phase-space distributions. Physical Review A, General Physics, 31(3), 1695–1697. 10.1103/physreva.31.1695 [DOI] [PubMed] [Google Scholar]
- Horby, P., Lim, W. S., Emberson, J. R., Mafham, M., Bell, J. L., Linsell, L., Staplin, N., Brightling, C., Ustianowski, A., & Elmahi, E. (2020). Dexamethasone in hospitalized patients with COVID-19-Preliminary Report. The New England journal of medicine. [Google Scholar]
- Isidori, A. M., Arnaldi, G., Boscaro, M., Falorni, A., Giordano, C., Giordano, R., Pivonello, R., Pofi, R., Hasenmajer, V., Venneri, M. A., Sbardella, E., Simeoli, C., Scaroni, C., & Lenzi, A. (2020). COVID-19 infection and glucocorticoids: Update from the Italian Society of Endocrinology Expert Opinion on steroid replacement in adrenal insufficiency. Journal of Endocrinological Investigation, 43(8), 1141–1147. 10.1007/s40618-020-01266-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., Zhang, B., Li, X., Zhang, L., Peng, C., Duan, Y., Yu, J., Wang, L., Yang, K., Liu, F., Jiang, R., Yang, X., You, T., Liu, X., … Yang, H. (2020). Structure of M pro from SARS-CoV-2 and discovery of its inhibitors. Nature, 582(7811), 289–285. 10.1038/s41586-020-2223-y [DOI] [PubMed] [Google Scholar]
- Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W., & Klein, M. L. (1983). Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics, 79(2), 926–935. 10.1063/1.445869 [DOI] [Google Scholar]
- Khan, S. U., & Htar, T. (2020). Deciphering the binding mechanism of Dexamethasone against SARS-CoV-2 Main Protease: Computational molecular modelling approach. ChemRxiv, 1–27.
- Klebe, G., & Böhm, H. J. (1997). Energetic and entropic factors determining binding affinity in protein-ligand complexes. Journal of Receptor and Signal Transduction Research, 17(1–3), 459–473. [DOI] [PubMed] [Google Scholar]
- Li, J., Abel, R., Zhu, K., Cao, Y., Zhao, S., & Friesner, R. A. (2011). The VSGB 2.0 model: A next generation energy model for high resolution protein structure modeling. Proteins, 79(10), 2794–2812. 10.1002/prot.23106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L., Aleksandrowicz, E., Schönsiegel, F., Gröner, D., Bauer, N., Nwaeburu, C. C., Zhao, Z., Gladkich, J., Hoppe-Tichy, T., Yefenof, E., Hackert, T., Strobel, O., & Herr, I. (2017). Dexamethasone mediates pancreatic cancer progression by glucocorticoid receptor, TGFβ and JNK/AP-1 . Cell Death & Disease, 8(10), e3064. 10.1038/cddis.2017.455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liuzzi, J. P., Lichten, L. A., Rivera, S., Blanchard, R. K., Aydemir, T. B., Knutson, M. D., Ganz, T., & Cousins, R. J. (2005). Interleukin-6 regulates the zinc transporter Zip14 in liver and contributes to the hypozincemia of the acute-phase response. Proceedings of the National Academy of Sciences of the United States of America, 102(19), 6843–6848. 10.1073/pnas.0502257102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmud-Al-Rafat, A., Majumder, A., Rahman, K. T., Hasan, A. M., Islam, K. D., Taylor-Robinson, A. W., & Billah, M. M. (2019). Decoding the enigma of antiviral crisis: Does one target molecule regulate all?. Cytokine, 115, 13–23. 10.1016/j.cyto.2018.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda, T., Yamasaki, K., Taga, T., Hirano, T., & Kishimoto, T. (1989). Current concepts of B cell modulation. International Reviews of Immunology, 5(2), 97–109. 10.3109/08830188909061976 [DOI] [PubMed] [Google Scholar]
- Mittal, L., Kumari, A., Srivastava, M., Singh, M., & Asthana, S. (2020). Identification of potential molecules against COVID-19 main protease through structure-guided virtual screening approach. Journal of Biomolecular Structure and Dynamics, 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth, E., Rivera, S., Gabayan, V., Keller, C., Taudorf, S., Pedersen, B. K., & Ganz, T. (2004). IL-6 mediates hypoferremia of inflammation by inducing the synthesis of the iron regulatory hormone hepcidin. The Journal of Clinical Investigation, 113(9), 1271–1276. 10.1172/JCI20945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayeem, S. M., & Reddy, M. S. (2020). Target SARS-CoV-2: Computation of Binding energies with drugs of Dexamethasone/Umifenovir by Molecular Dynamics using OPLS-AA force field. Research square, 1–17.
- Nishimoto, N., Kanakura, Y., Aozasa, K., Johkoh, T., Nakamura, M., Nakano, S., Nakano, N., Ikeda, Y., Sasaki, T., Nishioka, K., Hara, M., Taguchi, H., Kimura, Y., Kato, Y., Asaoku, H., Kumagai, S., Kodama, F., Nakahara, H., Hagihara, K., … Kishimoto, T. (2005). Humanized anti-interleukin-6 receptor antibody treatment of multicentric Castleman disease. Blood, 106(8), 2627–2632. 10.1182/blood-2004-12-4602 [DOI] [PubMed] [Google Scholar]
- Organization WH (1977). The selection of essential drugs: report of a WHO expert committee [meeting held in Geneva from 17 to 21 October 1977]. World Health Organization; [PubMed]
- Pace, C. N., Scholtz, J. M., & Grimsley, G. R. (2014). Forces stabilizing proteins. FEBS Letters, 588(14), 2177–2184. 10.1016/j.febslet.2014.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathnayake, S., & Weerasinghe, S. (2018). Exploring the binding properties of agonists interacting with glucocorticoid receptor: An in silico approach. Journal of Molecular Modeling, 24(12), 342 10.1007/s00894-018-3879-1 [DOI] [PubMed] [Google Scholar]
- Release S. 2 (2017). LigPrep. Schrödinger, LLC. [Google Scholar]
- Release S. 4 (2017). QikProp. Schrödinger, LLC. [Google Scholar]
- Release S. 4. (2015). Glide. Schrödinger, LLC; (Google Scholar There is no corresponding record for this reference. 2015). [Google Scholar]
- Roy, S., Kumar, A., Baig, M. H., Masařík, M., & Provazník, I. (2015). Virtual screening, ADMET profiling, molecular docking and dynamics approaches to search for potent selective natural molecules based inhibitors against metallothionein-III to study Alzheimer's disease. Methods (San Diego, California), 83, 105–110. 10.1016/j.ymeth.2015.04.021 [DOI] [PubMed] [Google Scholar]
- Rupprecht, R., Reul, J. M., van Steensel, B., Spengler, D., Söder, M., Berning, B., Holsboer, F., & Damm, K. (1993). Pharmacological and functional characterization of human mineralocorticoid and glucocorticoid receptor ligands. European Journal of Pharmacology: Molecular Pharmacology, 247(2), 145–154. 10.1016/0922-4106(93)90072-H [DOI] [PubMed] [Google Scholar]
- Schett, G., Manger, B., Simon, D., & Caporali, R. (2020). COVID-19 revisiting inflammatory pathways of arthritis. Nature Reviews. Rheumatology, 16(8), 465–466. 10.1038/s41584-020-0451-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, J., Ramanathan, K., & Sethumadhavan, R. (2011). Identification of potential inhibitors against acetylcholinesterase associated with Alzheimer's diseases: A molecular docking approach. Journal of Computational Methods in Molecular Design, 1, 44–51. [Google Scholar]
- Somers, W., Stahl, M., & Seehra, J. S. (1997). 1.9 A crystal structure of interleukin 6: Implications for a novel mode of receptor dimerization and signaling. The EMBO Journal, 16(5), 989–997. 10.1093/emboj/16.5.989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorbera, L., Graul, A., & Dulsat, C. (2020). Taking aim at a fast-moving target: Targets to watch for SARS-CoV-2 and COVID-19. Drugs of the Future, 45(4), 239–244. 10.1358/dof.2020.45.4.3150676 [DOI] [Google Scholar]
- Stoll, B. A. (1960). Dexamethasone in advanced breast cancer. Cancer, 13(5), 1074–1080. [DOI] [PubMed] [Google Scholar]
- Subhani, S., Jayaraman, A., & Jamil, K. (2015). Homology modelling and molecular docking of MDR1 with chemotherapeutic agents in non-small cell lung cancer. Biomedicine & Pharmacotherapy = Biomedecine & Pharmacotherapie, 71, 37–45. 10.1016/j.biopha.2015.02.009 [DOI] [PubMed] [Google Scholar]
- Sze, C.-I., Lin, Y.-C., Lin, Y.-J., Hsieh, T.-H., Kuo, Y. M., & Lin, C.-H. (2013). The role of glucocorticoid receptors in dexamethasone-induced apoptosis of neuroprogenitor cells in the hippocampus of rat pups. Mediators of Inflammation, 2013, 628094. 10.1155/2013/628094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, T., Narazaki, M., & Kishimoto, T. (2014). IL-6 in inflammation, immunity, and disease. Cold Spring Harbor Perspectives in Biology, 6(10), a016295. 10.1101/cshperspect.a016295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayakumar, S., Manogar, P., Prabhu, S., & Singh, R. A. S. (2018). Novel ligand-based docking; molecular dynamic simulations; and absorption, distribution, metabolism, and excretion approach to analyzing potential acetylcholinesterase inhibitors for Alzheimer's disease. Journal of Pharmaceutical Analysis, 8(6), 413–420. 10.1016/j.jpha.2017.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar, J., Confalonieri, M., Pastores, S. M., & Meduri, G. U. (2020). Rationale for prolonged corticosteroid treatment in the acute respiratory distress syndrome caused by coronavirus disease 2019. Critical Care Explorations, 2, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volden, P. A., & Conzen, S. D. (2013). The influence of glucocorticoid signaling on tumor progression. Brain, Behavior, and Immunity, 30, S26–S31. 10.1016/j.bbi.2012.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, M., Song, L., & Wang, Z. (2003). Effects of Dexamethasone on glucocorticoid receptor expression in a human ovarian carcinoma cell line 3AO. Chinese Medical Journal (England), 116, 392–395. [PubMed] [Google Scholar]
- Xue, X., Yu, H., Yang, H., Xue, F., Wu, Z., Shen, W., Li, J., Zhou, Z., Ding, Y., Zhao, Q., Zhang, X. C., Liao, M., Bartlam, M., & Rao, Z. (2008). Structures of two coronavirus main proteases: Implications for substrate binding and antiviral drug design. Journal of Virology, 82(5), 2515–2527. 10.1128/JVI.02114-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C., Kolb, A., Büchler, P., Cato, A. C. B., Mattern, J., Rittgen, W., Edler, L., Debatin, K.-M., Büchler, M. W., Friess, H., & Herr, I. (2006). Corticosteroid co-treatment induces resistance to chemotherapy in surgical resections, xenografts and established cell lines of pancreatic cancer. BMC Cancer, 6, 61. 10.1186/1471-2407-6-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., Becker, S., Rox, K., & Hilgenfeld, R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science (New York, N.Y.), 368(6489), 409–412. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Zhang, Z., Ju, M., Li, J., Jing, Y., Zhao, Y., Gu, C., Dong, M., Li, G., & Liu, Y. (2020). Pretreatment with interleukin 35-engineered mesenchymal stem cells protected against lipopolysaccharide-induced acute lung injury via pulmonary inflammation suppression. Inflammopharmacology, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhavoronkov, A., Zagribelnyy, B., Zhebrak, A., Aladinskiy, V., Terentiev, V., Vanhaelen, Q., Bezrukov, D. S., Polykovskiy, D., Shayakhmetov, R., & Filimonov, A. (2020). Potential non-covalent SARS-CoV-2 3C-like protease inhibitors designed using generative deep learning approaches and reviewed by human medicinal chemist in virtual reality. ChemRxiv, 1–18.