Abstract
Human C5a (hC5a), one of the pro-inflammatory glycoproteins of the complement system is known to undergo production hyperdrive in response to stress and infection. hC5a has been associated with the pathogenesis of many chronic and acute diseases, due to its proven ability in triggering the ‘cytokine storm’, by binding to its cognate receptor C5aR, expressed in myriad of tissues. Given the pleiotropic downstream function of hC5a, it is logical to consider the hC5a or its precursors as potential drug targets, and thus, we have been rationally pursuing the idea of neutralizing the harmful effect of excessive hC5a, by implementing the repurposing strategies for FDA-approved drugs. Indeed, the proof of principle biophysical studies published recently is encouraging, which strongly supports the potential of this strategy. Considering BSA-carprofen as a reference model system, the current study further explores the inherent conformational plasticity of hC5a and its effect in accommodating more than one drug molecule cooperatively at multiple sites. The data generated by recruiting a battery of experimental and computational biology techniques strongly suggest that hC5a can sequentially accommodate more than one raloxifene molecule with an estimated Ki ∼ 0.5 µM and Ki ∼ 3.58 µM on its surface at non-analogous sites. The study hints at exploration of polypharmacology approach, as a new avenue for discovering synergistic drug molecule pairs, or drug molecules with ‘broad-range’ binding affinity for targeting the different ‘hot spots’ on hC5a, as an alternative combination therapy for possible management of the ‘cytokine storm’-related inflammatory diseases, like COVID19.
Communicated by Ramaswamy H. Sarma
Keywords: COVID19, cytokine storm, C5a, carprofen, raloxifene, C5aR, circular dichroism, molecular dynamics
1. Introduction
Humans have evolved over time with an in-built line of defense system, described as complement (Atkinson et al., 2019) to coexist among plethora of microorganisms, including viruses in the natural environment (Stoermer & Morrison, 2011). Complement system that acts as a bridge between both adaptive and innate immunity plays a very important role in clearing the invading pathogens from the host body and gradually helps in restoring the normal physiology (Tomlinson, 1993). However, depending on the nature of the infection or type of the trigger, such as tissue stress, the complement can be often confused or impaired, resulting uncontrolled activation of the cascade, promoting inflammatory processes through production of several molecules of diverse biological functions, leading to a potentially lethal ‘cytokine storm’ (Tisoncik et al., 2012). The hyperinflammatory signal produced by the ‘cytokine storm’ (Hu et al., 2020; Mahmudpour et al., 2020; Mehta et al., 2020; Ye et al., 2020) is the prime contributor in the pathogenesis of many immunological and non-immunological diseases, including the current pandemic faced by the world in the name of COVID19 (Cevik et al., 2020; Pearce et al., 2020). While the system biology of the ‘cytokine storm’ remains to be clearly understood at the molecular level, it is evidenced that hC5a (Guo & Ward, 2005) can induce the secretion (Figure 1) of several pro-inflammatory cytokines (Zhang & An, 2007), including TNF-α (Okusawa et al., 1988), by binding to C5aR, widely expressed in both myeloid (neutrophils, macrophages, basophils, platelets) and non-myeloid (lung, liver, kidney, CNS) tissues (The GTEx Consortium, 2015). Generally, hC5a activates the generation of reactive oxygen species (ROS) through oxidative burst (Mollnes et al., 2002) post binding to C5aR expressed in a variety of cells, which subsequently triggers the downstream signaling cascades leading to the surge of synergistic actions of cytokines. Thus, it is reasonable to hypothesize that prolonged exposure to high concentration of ROS can be potentially damaging to the tissues and subsequently to the organs (Wood et al., 2018), which appears to be an important basis to the onset and progression of several diseases (Carroll & Sim, 2011; Holers, 2014; Ricklin & Lambris, 2007). Indeed, high concentration of hC5a has been linked to the acute lung injury (ALI), triggered by the exposure to highly contagious pathogens like influenza viruses, Middle East respiratory syndrome (MERS) and severe acute respiratory syndrome (SARS) coronaviruses (Wang et al., 2015). It is noteworthy that respiratory distress (Li & Ma, 2020) is one of the prime reasons reported in the ongoing COVID19-related mortality (Carsana et al., 2020) across the globe. Indeed, data available in preprint servers (Diao et al., 2020; Gao et al., 2020) indicates observation of high concentrations of hC5a in patients with severe COVID19 and treatment with anti-C5a or anti-C5 antibody appears to be therapeutically beneficial, which suggests the important role of complement system in COVID19 (Cugno et al., 2020; Gralinski et al., 2018). Thus, under the current scenario, it is quite tempting to link that therapeutic targeting of overtly produced hC5a by the pathogen assaulted complement system (Ricklin et al., 2019; Risitano et al., 2020) can be actually rewarding, as it can potentially downregulate the ‘cytokine storm’ (D’Elia et al., 2013; Huang et al., 2020), which can not only reduce tissue damage, but also can improve therapeutic responses of the known drugs by appreciably increasing the treatment window available to the healthcare system.
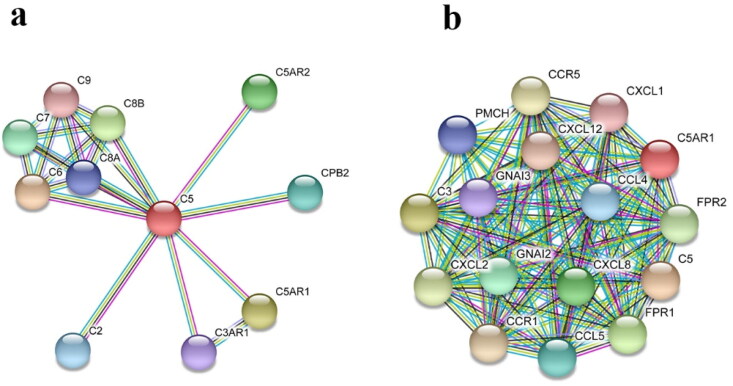
Figure 1.
Protein–protein interaction network of (a) C5 and (b) C5aR indicating the influence of hC5a–C5aR signaling axes on the various chemotactic and pro-inflammatory cytokines.
Given the fact that the hC5a is one of the important upstream glycoproteins of the complement cascade, we have been trying to understand (Rana et al., 2016a, 2016b; Rana & Sahoo, 2015; Sahoo et al., 2018) whether targeted antibodies, including small molecule drugs (Mishra & Rana, 2019) or drug-like compounds (Mishra et al., 2020) can be repurposed through design to effectively neutralize the adverse functionality of excessive hC5a. In fact, we have been reasonably successful in identifying a pool of potential ‘neutraligands’ (Mishra & Rana, 2019) targeting the ‘hotspots’ on hC5a (Figure 2) and our proof of principle study involving few nonsteroidal anti-inflammatory drugs (NSAIDs) (carprofen, oxaprozin, sulindac and raloxifene) strongly suggest that there is a scope for further exploration. The observation-based hypothesis is that small molecule drugs can bind and alter the conformation of hC5a significantly, which can alternatively modulate its biologically effective interaction with C5aR, and thus can control the ‘cytokine storm’. Interestingly, molecular dynamics (MD) studies, including spectroscopic studies on hC5a suggest that in response to stimuli, hC5a can behave like an intrinsically disordered protein (Rana et al., 2016a) and may exist in more than one major conformational ensembles (Mishra et al., 2020) with rearrangement of the ‘hotspots’, which may facilitate sequential binding of homologous or heterologous neutraligands.
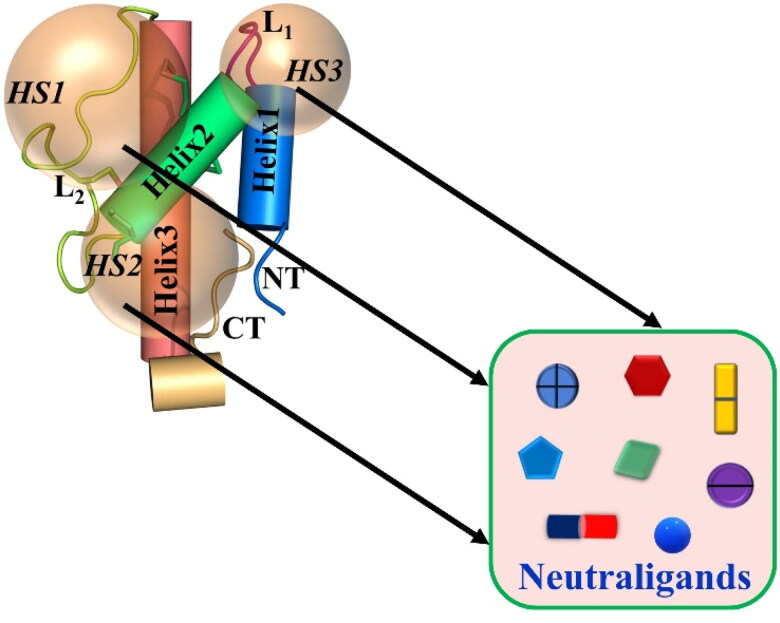
Figure 2.
Schematic illustration of the FDA-approved drugs or drug-like molecules as ‘neutraligands’ having diverse shape, size and molecular structure, which can be potentially repurposed for targeting the three ‘hotspots’ (HS) on hC5a.
To probe the feasibility of such a hypothesis in hC5a, we first studied the interaction of carprofen with Bovine serum albumin (BSA) as a reference system and applied the knowledge to understand the interaction of raloxifene (Figure 3) with hC5a in a better way. It is worth mentioning that both carprofen and raloxifene have been identified as potential ‘neutraligands’ of hC5a in our previous study (Mishra & Rana, 2019). The logic behind selecting BSA as a model protein in the current study are primarily due to the following few parallels between hC5a (structurally helical with three disulfide bonds, including an unpaired cysteine) and BSA (Majorek et al., 2012): (i) BSA is a thoroughly studied protein with an all helical structure, which displays very good conformational sensitivity to both physical and chemical stimuli, (ii) BSA has several intra-chain disulfides, including an unpaired cysteine and is known to have multiple drug binding sites like Human serum albumin (HSA). So far as carprofen is concerned, it is an NSAID, generally prescribed to relieve pain and inflammation in animals (Malek et al., 2012), and thus it was logical to pair it with BSA in the current study. On the other hand, raloxifene is prescribed in humans (Jochems et al., 2008) to treat pain and inflammation caused by rheumatoid arthritis and also to reduce the risk of invasive breast cancer, in case of women. In addition, raloxifene’s beneficial role in maintaining cardiovascular health is also reported (Francucci et al., 2005; Saitta et al., 2001). It is worth mentioning that myocardial injury and cardiovascular dysfunction (Zheng et al., 2020) has been observed in ∼22% of critically ill patients (Clerkin et al., 2020) with COVID19. Interestingly, hC5a–C5aR interaction has been linked to have a profound effect on the progression of both cancer (Afshar-Kharghan, 2017; Ajona et al., 2019; Kleczko et al., 2019; Sayegh et al., 2014) and cardiovascular diseases (Fattahi & Ward, 2017; Niederbichler et al., 2006; Speidl et al., 2005) in humans. Further, in our previous studies (Mishra & Rana, 2019) we had observed that raloxifene provides better hydrophobic burial to the internal fluorophore probes of hC5a, and thus, it was natural to pair it with hC5a in the current study. Both the systems have been subjected to a battery of computational and experimental studies, which includes the circular dichroism (CD), fluorescence, ANS (8-anilino-1-naphthalenesulfonic acid) binding, isothermal calorimetry (ITC), automated docking, 1 µs MD and binding free energy calculation involving MM-PBSA (molecular mechanics Poisson–Boltzmann surface area) approach. The data documented in this report support the hypothesis that more than one molecule (homologous or heterologous type) can target hC5a in a sequential manner. This sequential or cooperative binding of homologous or heterologous small molecule drugs can be very important, as it can provide functional advantages of antibodies, but with a dose-dependent manner for managing inflammation-induced diseases.
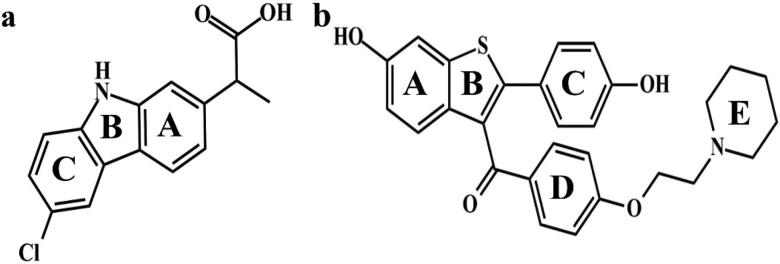
Figure 3.
The chemical structure of (a) carprofen and (b) raloxifene, respectively, paired against BSA and hC5a. The rings are labeled with alphabets for easy referencing to intermolecular interactions with the respective proteins.
2. Materials and methods
2.1. Generic computational methods and chemicals
PDB coordinates of the hC5a (1KJS) (Zhang et al., 1997) and BSA (3V03) (Majorek et al., 2012) were downloaded from www.rcsb.org. PyMOL (The PyMOL Molecular Graphics System, Version 1.1r1, Schrödinger, LLC) and Discovery studio (Accelrys) software were utilized for initial processing, visualization, analysis and presentation of the protein structures. The protein-protein network has been prepared using the STRING-v11 database (Szklarczyk et al., 2019) with highest confidence interaction score of 0.9. The maximum number of interactors has been limited to 10 only in the first shell. The topological parameter of both carprofen and raloxifene required for docking and MD simulation was generated by using the PRODRG server (Schuttelkopf & van Aalten, 2004), which were appropriately edited to suit the gromos-96 43a1 force field (Hess et al., 2008) built into GROMACS. Experimental and computational data were plotted in GraphPad Prism (version 6 for Windows, GraphPad Software, La Jolla, CA, USA, www.graphpad.com). The R&D Systems provided the recombinant hC5a protein. Sigma-Aldrich supplied the ligand molecules carprofen (racemic mixture) and raloxifene, including the dye ANS.
2.2. Automated docking studies
Both carprofen and raloxifene were, respectively, subjected to automated docking by recruiting the AutoDock (Morris et al., 2009) against BSA and hC5a. For probing sequential docking, the entire surface area of the most populated conformer of hC5a (Rana et al., 2016a) bound to one molecule of raloxifene (Supplementary Figure S1) at ‘HS2’ (Mishra & Rana, 2019) was further subjected to scanning by recruiting another molecule of raloxifene. Optimum grid dimension covering the entire protein surface along XYZ directions was achieved by using the AutoGrid program. The Lamarckian genetic approach (LGA) was applied for a population size of 250 with the maximum number of generations set to 27,000. Structurally distinct conformational clusters of the drugs were ranked in terms of increasing energy. Both the drug molecules were initially subjected to rigid dock, followed by flexi dock until the best-bound conformer was obtained (Supplementary Figure S1).
2.3. Circular dichroism studies
The CD studies were carried on a Chirascan CD spectropolarimeter system in far-UV region at 25 °C. Each sample was subjected to minimum of three scans with a time constant of 1 s and step size of 1 nm. The solvents and buffers used in the study were filtered and degassed by nitrogen bubbling method. The hC5a was solubilized only in 1× phosphate-buffered saline (PBS, pH ∼ 7.4) without the BSA and the carprofen and raloxifene stock solutions were prepared in appropriate solubility buffers maintaining the prescribed ratio of DMSO and 1× PBS. Further, the drug stocks were diluted in pure 1× PBS (pH ∼ 7.4) to the required concentrations prior to the spectroscopic studies. The BSA and hC5a concentrations were, respectively, maintained at 5 µM and 0.1 µM for far-UV region. The BSA and hC5a concentrations were, respectively, maintained at 15 µM and 1 µM for near-UV region. The drug concentrations were varied from 0 to 10 µM. Samples were incubated for minimum of 1 h at 4 °C prior to the CD studies. The molar ellipticity was converted to mean residue ellipticity (θMRE) after background subtraction, by using the methods described elsewhere (Greenfield, 2006).
2.4. Fluorescence studies
The fluorescence studies on hC5a and BSA were performed in pure 1× PBS (pH ∼ 7.4) by using the Horiba Fluoromax-4 spectrofluorometer system at 25 °C. Average of three scans was recorded and background spectra of the buffer were appropriately subtracted. The excitation and emission slit widths were adjusted between 2 and 5 nm with excitation wavelength set at 280 nm and emission range set between 290 and 500 nm for both BSA (5 µM) and hC5a (0.5 µM). The concentration of both the drugs was varied between 0 and 100 µM. ANS concentration was maintained at 50 µM (saturating concentration) both for BSA (Möller & Denicola, 2002) and hC5a, complexed to drugs with excitation wavelength set at 360 nm and emission range set between 370 and 600 nm. ANS concentration was varied between 0 and 100 µM for saturation binding studies with hC5a (0.1 µM) and the data were fitted to total binding equation defined in GraphPad Prism through non-linear regression to obtain the Kd. The obtained F/F0 data at the maximum emission wavelength were plotted against different concentrations of ANS and further fitted to a segmental linear regression to calculate the ‘x’ (abscissa value) and subsequently the binding stoichiometry (n) for the ANS–hC5a interaction, by recruiting the following assumption: x = nC + Kd, where C = concentration of hC5a, and Kd is the binding dissociation constant.
2.5. Isothermal calorimetry studies
The ITC experiments were performed over MicroCal PEAQ ITC (Malvern) with sample cell and injection volumes 200 µl and 40 µl, respectively. The experiment was conducted at 25 °C, with total injections set to 20 and a volume of 2 µl per injection. 100 µM of carprofen was titrated against 5 µM of BSA. Similarly, 80 µM of raloxifene was titrated against 5 µM of hC5a (data not shown). The drugs and proteins prepared in 1× PBS (pH ∼ 7.4) were thoroughly degassed prior to the titrations. The data obtained were corrected for buffer or ligand titrations, wherever possible by using the MicroCal PEAQ ITC software support and further processed through GraphPad Prism/Origin Pro for presentation.
2.6. MD studies of hC5a bound to two molecules of raloxifene
The hC5a complexed to two molecules of raloxifene, respectively, at ‘HS2’ and ‘HS3’ (Supplementary Figure S2) was subjected to further MD studies over 1 µs at 300 K in the presence of explicit water. Prior to MD simulation, the system was subjected to energy minimization in a cubic box with appropriate periodic boundary, by recruiting the gromos-96 43a1 united atom force field built into the GROMACS. Briefly, all the polar amino acids such as Lys and Arg were positively charged, whereas amino acids such as Asp and Glu were negatively charged on hC5a to roughly mimic the protonation state at the physiological pH. Further, the hC5a bound to two molecules of raloxifene was placed in the center of the periodic box, and energy minimized to 100 kJ mol−1 nm−1 tolerances with steepest descent, first in vacuum and then in the presence of simple point charge (SPC) water molecules, as provided in GROMACS. The system was solvated by using appropriate number of SPC water molecules with solvent density set to the value corresponding to 1 atm at 300 K and further, the net charge of the system was neutralized, by randomly placing requisite number of counterions to the box. Overall, the system contained 36,683 atoms, which included 11,948 water molecules and 7 chloride ions. The system was then equilibrated through NVT (0.5 ns) and NPT (1 ns) conditions prior to MD studies. Protein and solvents were coupled independently to V-rescale bath at 300 K, to the coupling time constant 0.1 ps. Bonds were constrained with LINCS with order 4. Non-bonded pair list cut-off was 1.2 nm with a grid function. Numerical integrations were performed in step size of 2 fs and the coordinates were updated every 5 ps. As a result, the 1 µs MD trajectory could sample 200,000 conformations for the system. The representative conformer of the major microstate, evolved over 1 µs of MD, is presented in Supplementary Figure S2. Solvent-accessible surface area (SASA) calculation for the representative conformers was performed by using the Discovery studio software with a solvent probe radius set to 1.4 Å. Conformational clustering across the trajectory was performed every 50 ps with RMSD cut-off ≤1.5 Å, by recruiting the gromos fitting method, as defined in GROMACS. The entire trajectory was thoroughly analyzed for understanding the intermolecular interactions between the raloxifene molecules and hC5a, by recruiting the utility modules available within the GROMACS. For example, the intermolecular hydrogen bonds between the raloxifene molecules, respectively, at the ‘HS2’ and ‘HS3’ of hC5a were computed, by recruiting the g_hbond module, which utilizes both the geometrical criterion (distance and angle) necessary for physical existence of a hydrogen bond between a given donor (D)–acceptor (A) pair. Generally, a distance cut-off of 0.35 nm between the heavy atoms of D and A and an angular cut-off of 30° between the HDA is set as default for computing a hydrogen bond in GROMACS. Further detailed information about any other utility module can be obtained from the GROMACS manual.
2.7. Estimation of the binding free energies of the raloxifene-hC5a complex
The 1 µs MD trajectory of hC5a complexed to raloxifene at ‘HS2’ and ‘HS3’ was used for calculating the binding free energies at the respective sites, by recruiting the MM-PBSA method, as described elsewhere (Kumari et al., 2014). Briefly, the following equation: ΔGbinding = Gcomplex − (Gprotein − Gligand), implemented in the g_mmpbsa program was recruited for calculating the binding free energies of the raloxifene complexed to hC5a at two independent sites. The free energy contribution of the raloxifene molecules was estimated by the following equation: G = EMM + Gsolv – TSsolute, where EMM (molecular mechanics energy) represents the summation of van der Waals and electrostatic, Gsolv represents the solvation energy contributed by both polar and non-polar solvation free energy, and TSsolute represents the temperature and entropy of the solute. Dielectric constant of solute and solvent were, respectively, fixed at 20 and 80, respectively, for calculation of polar solvation energy. Variation of solute dielectric in 2–20 range did not alter the relative difference in total binding energies between the two sites significantly. A value of 0.5 Å grid space was taken to calculate electrostatic energy. Probe radius was set to 1.4 Å to calculate the non-polar contribution to solvation free energy through SASA method. Finally, 1000 conformers collected at an interval of 1 ns, including 1000 conformers randomly selected from the first major cluster (Supplementary Figure S2) populated over 1 µs of the MD trajectory were, respectively, used for calculating the average binding free energy for the raloxifene molecules at the respective sites.
3. Results
3.1. Probing the potential binding sites of carprofen on BSA
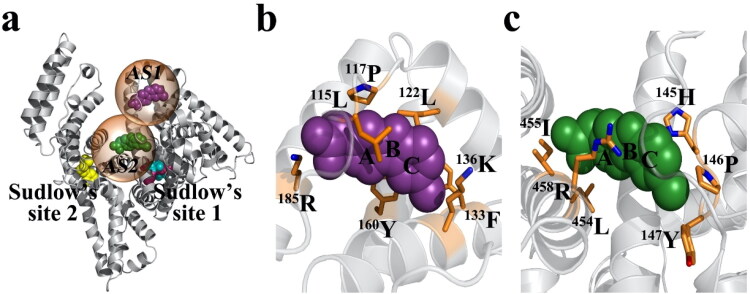
Given the ∼80% sequence homology between the HSA and BSA, it is assumed that BSA may have similar binding sites, as described (Sand et al., 2014) for HSA [active site 1 (AS1): warfarin site, active site 2 (AS2): diazepam site]. Thus, we screened the BSA (PDB: 3V03) to locate potential binding sites for carprofen, by recruiting the automated docking studies. The docking study revealed that carprofen can bind to BSA at three probable active sites [AS1 (L115, P117, L122, F133, K136, Y160, R185), Ki ∼ 12.58 µM, B.E. ∼ −6.69 kcal/mol; AS2 (H145, P146, Y147, A193, L454, I455, R458), Ki ∼ 20.34 µM, B.E. ∼ −6.4 kcal/mol; and AS3 (R208, A209, A212, L326, E353, S479, L480, V481), Ki ∼ 21.16 µM, B.E. ∼ −6.38 kcal/mol], not overlapping with either the warfarin or the diazepam binding sites described (Sand et al., 2014) for HSA (Figure 4). However, since the observed binding energies for AS2 and AS3 are almost similar, only two (AS1 > AS2 ≈ AS3) of the three probable binding sites of carprofen are presented on BSA (Figure 4). Further, this observation is in line with the prior titration study, which suggested the possible existence of two independent binding sites on BSA for carprofen (Kohita et al., 1994). It is worth mentioning that AS1 on BSA overlaps with the drug binding site, generally known to bind both endogenous (Hemin, Bilirubin) and exogenous (Fusidic acid, Lidocaine) ligands on HSA (Sand et al., 2014).
Figure 4.
(a) The probable binding sites identified for carprofen presented with reference to the warfarin and diazepam binding sites described for HSA. The interaction of carprofen, respectively, highlighted at AS1 (b) and at AS2 (c) on BSA.
3.2. Probing the effect of carprofen on the conformation of BSA
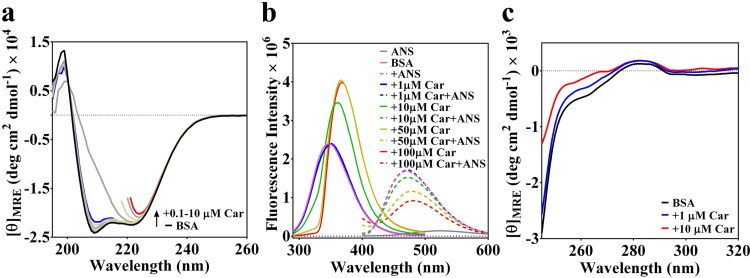
BSA like HSA is also known for its conformational sensitiveness toward binding of exogenous stimuli. In fact, allosteric modulation of the conformations due to the binding of ligands is well described in the literature (Fasano et al., 2005). Thus, we subjected BSA to a titration study against carprofen, by recruiting CD spectroscopy. The data presented in Figure 5 suggest that BSA undergoes a noticeable conformational change in the presence of 1–10 µM carprofen (Figure 5(a)), which may be most likely due to the binding of carprofen to BSA. Interestingly, the change in conformational signature observed for BSA, in response to 1–10 µM carprofen is broadly similar to the conformational signature observed for hC5a, in response to 1–10 µM carprofen. However, 1 µM carprofen produced significant conformational changes in case of hC5a (Mishra & Rana, 2019), compared to BSA. Though the CD signal of BSA (Figure 5(a)) got saturated in the presence of 10 µM carprofen, further conformational changes in BSA could be ascertained from the shift in fluorescence maxima, as noted, respectively, in the presence of 50–100 µM carprofen (Figure 5(b)).
Figure 5.
(a) Conformational perturbation observed for BSA in response to 0.1–10 µM carprofen (Car), suggesting the binding of carprofen to BSA. Few far-UV CD traces have been truncated beyond certain wavelengths to acknowledge the instrumental limitations. (b) The effect of 1–100 µM carprofen (Car) on the fluorescence spectra of both BSA and ANS. (c) The near-UV CD spectra highlighting the effect of 1–10 µM carprofen (Car) on the tertiary structure of BSA. The CD traces in the presence of 1 and 10 µM carprofen are, respectively, highlighted in blue and red lines.
Comparison of CD signatures observed for both BSA (data not shown) and hC5a in the presence of high concentrations (50–100 µM) of carprofen (Mishra & Rana, 2019) indicates that the high absorbing aromatic drugs can influence the dynode voltage of the CD instrument, beyond the acceptable cut-off range, resulting difficult to interpret CD spectra of proteins. However, in the presence of ≤10 µM drugs, the signature CD spectra of the proteins appear to be influenced by the presence of several inherently asymmetric disulfide linkages in the protein experiencing altered orientation of the peptide dipoles, due to the binding interaction with the drugs.
To probe the possible molecular interaction between carprofen and BSA further, we subjected BSA to fluorescence studies both in the presence and absence of carprofen, including ANS, the hydrophobic dye known for probing the conformational changes in proteins. The fluorescence data presented for BSA in Figure 5(b) strongly agree with the CD data (Figure 5(a)) and further confirms that 1–10 µM carprofen produces conformational changes in BSA affecting the microenvironment of the fluorophores, as evidenced from the observed red shift in fluorescence maxima of BSA. Further, in agreement with CD data, BSA incubated in the presence and absence of 1–10 µM carprofen altered the fluorescence intensity of 50 µM ANS appreciably, suggesting conformational perturbation induced by the carprofen. It is well known that ANS has ∼5 high-affinity binding sites (Kd ∼ 5 µM) on BSA (Möller & Denicola, 2002) and the 50 µM ANS used in the current study should be able to saturate all the binding sites on BSA. The significant drop in observed ANS fluorescence intensity in the presence of 50–100 µM carprofen could be due to the possible competition between ANS and carprofen in non-specific binding to BSA. In addition, it is observed that the fluorescence intensity of BSA sharply increases in the presence of 1–10 µM carprofen, subsequently saturating in the presence of 50–100 µM carprofen, which strongly suggests the specific binding of carprofen to BSA may be within 1–10 µM. In agreement to conformational changes observed in the secondary structure of BSA at the far-UV region (Figure 5(a)), the near-UV CD signature presented for BSA (Figure 5(c)) in the presence of 1–10 µM of carprofen indicates that there is also an appreciable change in the tertiary structure of BSA. Thus, it is clear from the CD and fluorescence data that carprofen actually binds to BSA and the binding interaction is most likely driven by the conformational selection mechanism rather than the induced fit interactions.
3.3. Determining the binding stoichiometry of carprofen to BSA
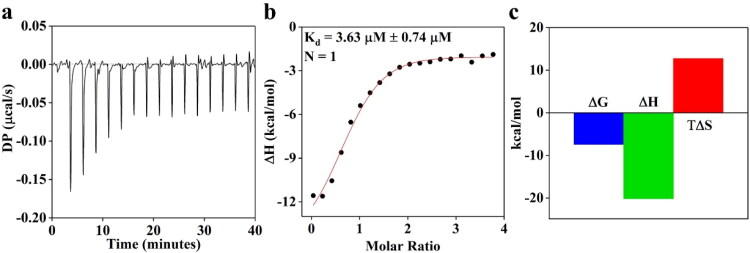
Both CD and fluorescence data presented in Figure 5 synergistically suggest that carprofen binds to BSA. To probe the binding stoichiometry of BSA–carprofen interaction further, we subjected BSA to ITC studies.
The data presented in Figure 6 further confirm that carprofen most likely binds to BSA at the hypothesized site AS1 (Figure 4) with 1:1 stoichiometry having Kd ∼ 3.63 ± 0.74 µM through favorable enthalpy. Interestingly, carprofen binds to hC5a with Kd ∼ 0.58 µM, as estimated from the CD titration studies reported earlier, suggesting an appreciable selectivity of carprofen toward hC5a over BSA. The unfavorable entropy observed for BSA-carprofen binding (Figure 6(c)) clearly indicates that the binding of carprofen triggers conformational changes in BSA, which strongly correlate with the observations made both in the CD and fluorescence studies (Figure 5).
Figure 6.
(a) Monitoring the binding of carprofen (100 µM) to BSA (5 µM) by recruiting ITC. (b) The binding isotherm of BSA–carprofen interaction suggesting 1:1 stoichiometry. (c) Thermodynamic profile (free energy, binding enthalpy and entropy factor) of BSA–carprofen interaction derived from the ITC studies.
3.4. Probing the effect of raloxifene on the conformation of hC5a
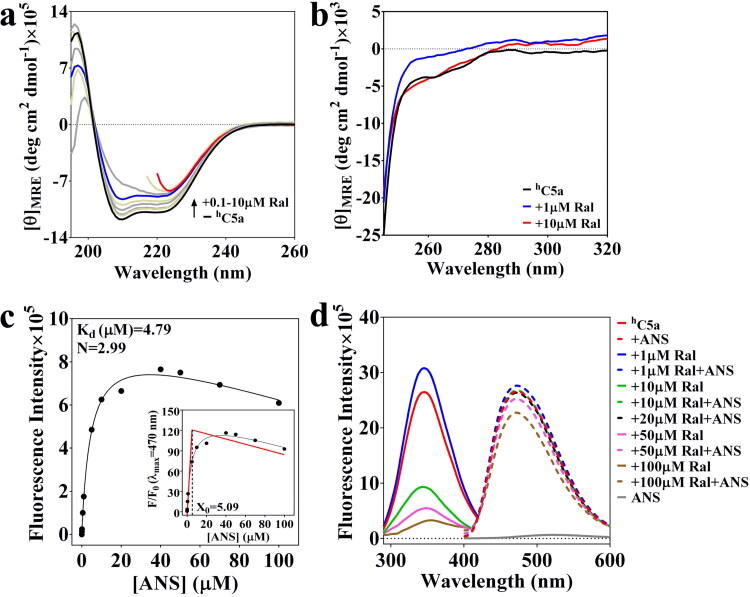
The far-UV and near-UV CD data presented in Figure 7(a,b) mutually agree with each other and clearly indicates that 1–10 µM raloxifene noticeably alters the conformation of hC5a, similar to the observation made for BSA-carprofen system (Figure 5). To probe the number of probable binding sites on hC5a, a saturation binding assay involving 0–100 µM ANS was conducted by recruiting fluorescence spectroscopy. The data presented in Figure 7(c) indicate that ANS can bind to hC5a with Kd ∼ 4.8 µM compared to the Kd ∼ 5 µM noted for BSA in earlier studies (Möller & Denicola, 2002). Further analysis of the data indicates that ∼3 molecules of ANS can bind to hC5a, matching to our earlier hypothesis that hC5a can have three possible ‘hotspots’ (Figure 2: HS1, HS2 and HS3), which can be targeted by the drugs acting as ‘neutraligands’. Thus, to probe whether more than one raloxifene can bind to hC5a, we subjected hC5a to competitive ANS binding studies both in the presence and absence of increasing concentration of raloxifene (Figure 7(d)), similar to the BSA-carprofen pair (Figure 5). The combined CD and fluorescence data presented in Figure 7 clearly suggest that raloxifene indeed binds to hC5a specifically between 0 and 1 µM. The binding is evidenced from the strong conformational changes, including the sharp increase in fluorescence intensity of 0.1 µM hC5a in the presence of 1 µM raloxifene, which substantially quenches as the concentration of raloxifene goes beyond 10 µM. On the other hand, the appreciable increase in the ANS fluorescence intensity in the presence of hC5a complexed to 1 µM raloxifene (Figure 7(c)), in comparison to free hC5a further indicates that the strong conformational changes observed in hC5a (Figure 7(a)) is due to the binding of raloxifene. In fact, the earlier CD titration studies (Mishra & Rana, 2019) performed in the presence of 1–10 µM raloxifene suggested that raloxifene can bind to hC5a with a Kd ∼ 0.71 µM. Further, this is in agreement with the near-UV CD spectra of hC5a (Figure 7(b)), which indicates appreciable change in the tertiary structure of hC5a in the presence of 1 µM raloxifene.
Figure 7.
(a) Conformational perturbation observed for hC5a in response to 0.1–10 µM raloxifene (Ral), suggesting the binding (Kd ∼ 0.71 µM) of raloxifene to hC5a (Mishra & Rana, 2019). Few far-UV CD traces have been truncated beyond certain wavelengths to acknowledge the instrumental limitations. (b) The near-UV CD spectra highlighting the effect of 1–10 µM raloxifene (Ral) on the tertiary structure of hC5a. (c) Determination of the ANS-hC5a binding stoichiometry through ANS fluorescence saturation study was performed in the presence of 0.1 µM hC5a. (d) The effect of 1–100 µM raloxifene (Ral) on the fluorescence spectra of both hC5a and ANS.
However, given the structure of raloxifene (Figure 3), the possibility of specific or non-specific binding cannot be ruled out, as the concentration of raloxifene goes beyond 10 µM. It is interesting to note that the fluorescence intensity of 50 µM ANS decreases substantially in the presence of hC5a incubated with 10 µM raloxifene, saturating at 20 µM raloxifene (Figure 7(c)), compared to hC5a incubated with 1 µM raloxifene, which could be due to the one more binding event at hC5a triggered in the presence of 1–20 µM of raloxifene. Further decrease in 50 µM ANS fluorescence in the presence of 50–100 µM raloxifene may be attributed to the conformational alteration in hC5a, triggered by competitive non-specific binding of raloxifene. It is worth mentioning that in our studies, neither carprofen nor raloxifene was found to influence the ANS fluorescence directly through possible cross-interactions. In light of these data (Figure 7), it appears that hC5a perhaps bind to at least two molecules of raloxifene between 0 and 20 µM, and it can be hypothesized that raloxifene binding to hC5a may be cooperative in nature. To probe this possibility, we revisited the ITC titration of raloxifene (80 µM) to hC5a (5 µM) again. The obtained titration data (not shown) were subsequently fitted to a sequential binding model, by considering a two-site binding scenario. Though the fitted data indicate the possible existence of 1:2 stoichiometry between hC5a-Raloxifene, the binding isotherm (data not shown) was too shaky to be reliable, which could be due to the possible formation of soluble oligomers of hC5a or masking of the binding heat changes due to the ligand dilutions affecting the protein-ligand binding isotherms observed in ITC.
3.5. Probing the possible sequential binding of raloxifene to hC5a by molecular dynamics
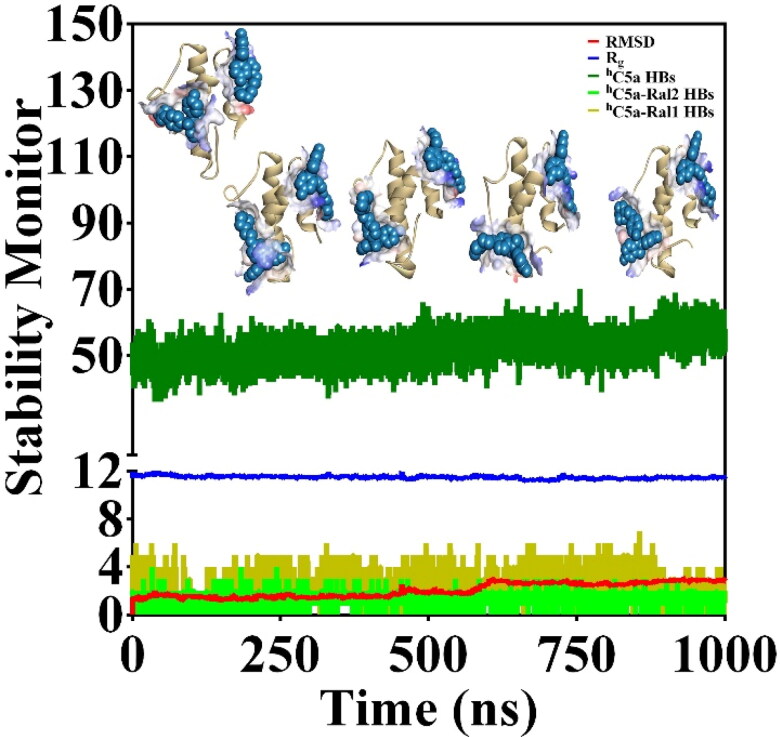
The CD and ANS binding titration studies suggest, there could be binding of more than one raloxifene molecules to hC5a. Though ITC indicates the possibility of 1:2 stoichiometry binding, the data (not shown) are not conclusive in nature. It is worth mentioning that our earlier docking studies have indicated that raloxifene can possibly bind at both HS2 (Ki ∼ 0.617 µM, B.E. ∼ −8.47 kcal/mol) and HS3 (Ki ∼ 132.50 µM, B.E. ∼ −5.29 kcal/mol) on hC5a (Figure 2) independently. The same idea was further extended to probe the possible existence of sequential binding of raloxifene to hC5a, by recruiting automated docking. To begin with, the hC5a complexed to one molecule of raloxifene at HS2 (Mishra & Rana, 2019) was subjected to further automated docking studies against another molecule of raloxifene. Interestingly, the second molecule of raloxifene could bind at HS3 of hC5a with much lower affinity than before. Subsequently, the hC5a bound to two molecules of raloxifene, respectively, at HS2 and HS3 of hC5a (Supplementary Figure S2) was subjected to 1 µs of MD at 300 K in the presence of explicit water to further probe the physical viability of the complex (Figure 8).
Figure 8.
(a) Monitoring the various structural parameters (deep green: total hydrogen bonds; blue: radius of gyration; red: backbone RMSD; fluorescent green and deep olive: intermolecular hydrogen bonds, respectively, between hC5a and Ral1, Ral2) for probing the physical viability of the hC5a-Raloxifene (1:2) complex at an interval of 0.25 µs over 1 µs of MD at 300 K. The two molecules of raloxifene (Ral1 and Ral2) are, respectively, complexed at HS2 and HS3 on hC5a.
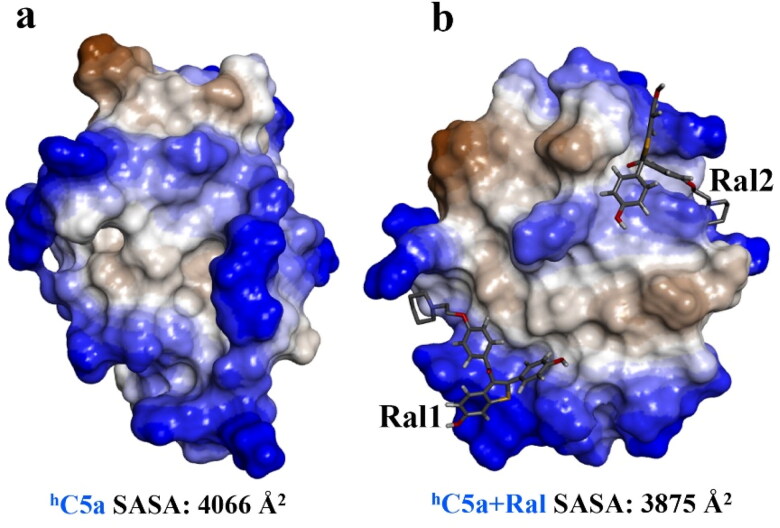
The structural snapshot of the central conformer of hC5a complexed to two molecules of raloxifene evolved over 1 µs of MD (Figure 9) strongly suggests that two molecules of raloxifene can be comfortably accommodated, respectively, at the HS2 and HS3 of hC5a. It is certain that the solution conformers of hC5a are altered in response to binding of raloxifene. Both far- and near-UV CD, including the fluorescence data presented in Figure 7 support the argument. In addition, the 1 µs MD data presented in Figure 8 seem to suggest that the binding of raloxifene induces moderate structural changes in the hC5a and the observed changes in ANS binding in the presence of the raloxifene also suggest the change in tertiary structure of hC5a. It is clear from the data (Figure 8) that 2 and 4 intermolecular hydrogen bonds are respectively sustained between hC5a and the raloxifene molecules over the course of the MD trajectory. In addition, the SASA calculation performed over the free (4066 Å2) and raloxifene bound conformers of hC5a (3875 Å2) also support the physical viability of the 1:2 complex (Figure 9).
Figure 9.
(a) Surface area representation of the free and the central conformer of hC5a-raloxifene (1:2) complex evolved over 1 µs of MD at 300 K, indicating the overall physical viability of the complex. The two raloxifene molecules, respectively, bound at the HS2 and HS3 of hC5a are represented as stick models.
Further, sequential redocking data of the raloxifene molecules to the above conformer of hC5a, respectively, at HS2 (Ki ∼ 0.5 µM, B.E. ∼ −8.59 kcal/mol) and HS3 (Ki ∼ 3.58 µM, B.E. ∼ −7.43 kcal/mol) strongly supports the hypothesis of the existence of cooperative/sequential binding of ligands in hC5a. It is noteworthy that titration studies also indicate that raloxifene bind to hC5a with a Kd ∼ 0.71 µM. The above observation is also synergistically supported by the estimated binding free energy calculated for raloxifene molecules by recruiting the MM-PBSA, respectively, at HS2 (−43.59 ± 0.21 kcal/mol) and HS3 (−35.02 ± 0.18 kcal/mol) over the entire 1 µs MD trajectory.
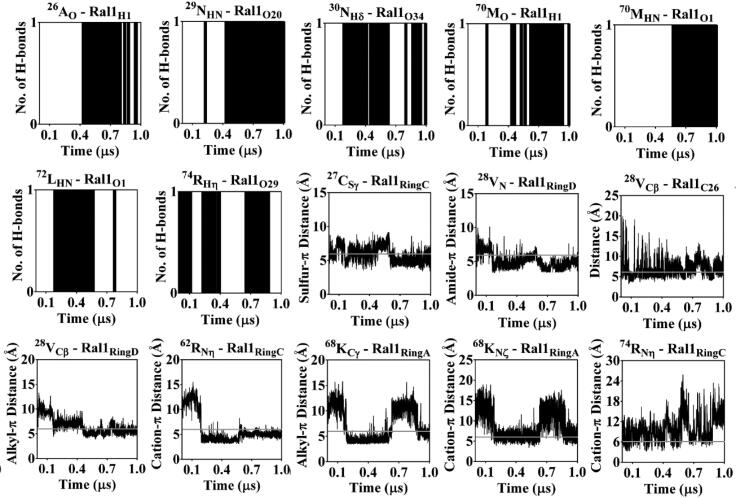
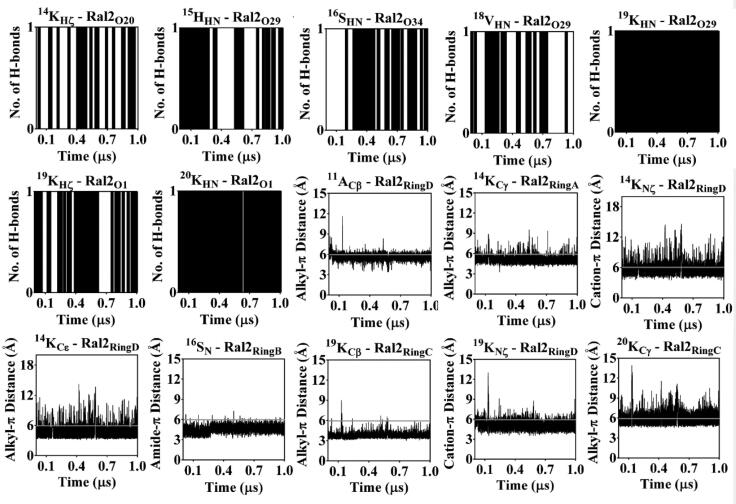
The intermolecular interactions sustained over 1 µs of MD are considered to be responsible for anchoring the raloxifene molecules, respectively, at the HS2 and HS3 on hC5a are further detailed in Figures 10 and 11. It is clear from the data that raloxifene is anchored to hC5a through multiple hydrogen bonds, including several other commonly observed noncovalent interactions, such as hydrophobic, ‘amide-π’ (Ferreira de Freitas & Schapira, 2017), ‘sulfur-π’ (Motherwell et al., 2018) and ‘cation-π’ (Dougherty, 2013) interactions. Overall, the energetic data and MD data presented here strongly support the possible formation of a 1:2 hC5a-raloxifene complex in solution.
Figure 10.
Monitoring the sustained intermolecular hydrogen bonding, hydrophobic and other (‘amide-π’, ‘sulfur-π’ and ‘cation-π’) interactions of raloxifene (Ral1) at HS2 of hC5a in explicit water over 1 µs of MD at 300 K. The hydrophobic and other interactions between the raloxifene and hC5a are, respectively, presented with cut-off distances (6 Å), highlighted in gray lines.
Figure 11.
Monitoring the sustained intermolecular hydrogen bonding, hydrophobic and other (‘amide-π’, ‘sulfur-π’ and ‘cation-π’) interactions of raloxifene (Ral2) at HS3 of hC5a in explicit water over 1 µs of MD at 300 K. The hydrophobic and other interactions between the raloxifene and hC5a are, respectively, presented with cut-off distances (6 Å), highlighted in gray lines.
4. Discussion
Physiology of complement system is both intricate and delicate. Proper regulation of the complement is key to survival under stressful assaults of both known and unknown family of microbes populated or likely to populate in our habitat. Circumstantial deregulation of the complement can trigger the collision between several intertwined signaling pathways (Chauhan et al., 2020), resulting severe life-threatening pathophysiology. It is evidenced that infection due to SARS-CoV can activate complement system in the lungs (Wang et al., 2015), as early as 24 hrs post-infection (Gralinski et al., 2018). Similarly, the role of hC5a in endothelial damage (Marchetti, 2020; Narasaraju et al., 2011; Pons et al., 2020; Twaddell et al., 2019) and ALI due to the viral infection is well known. It is worth highlighting that about 15% of the patients (Cugno et al., 2020) who develops life-threatening complications in COVID19 (Guan et al. 2020) due to SARS-CoV2 infection have elevated levels of pro-inflammatory cytokines in the plasma and most of them are likely to develop severe acute respiratory distress syndrome (ARDS). It appears that post-infection, continued deterioration in lung function has no direct correlation with the viral load, which could be most likely attributed to the uncontrolled immune response of the body (Risitano et al., 2020). Though the precise contribution of complement proteins in progression of COVID19 is still unclear, recent pathologic studies (Chauhan et al., 2020) evidence the activation of the complement in COVID19. The data available in preprint servers demonstrate the colocalization of SARS-CoV2 spike proteins with mannan-binding lectin serine peptidase 2 (MASP2) in human lungs (Gao et al., 2020), which suggests the likely involvement of mannose-binding lectin (MBL) arm of complement in SARS-CoV2 infection, as noted earlier for SARS-CoV1 (Gralinski et al., 2018). Nevertheless, growing bodies of data seem to suggest the involvement of complement in the pathophysiological conditions triggered by COVID19 (Risitano et al., 2020). In fact, a recent clinical study performed over a small group of patients with moderate to severe COVID19 (Cugno et al., 2020), including data available in preprints strongly evidence elevated level of hC5a and other complement proteins in plasma. Thus, it is not surprising that several clinical trials targeting the complement proteins (Diurno et al. 2020; Giamarellos-Bourboulis et al., 2020; Mastellos et al., 2019) C3 (trial id: NCT04395456), C5 (trial id: NCT04288713) and C5a (trial id: NCT04333420) have been initiated recently with a hope to combat COVID19. Considering the gravity of the current healthcare conditions across the globe, novel combination therapy targeting the cross-talking signaling cascades have also been initiated for managing COVID19. In a recent control study, both coagulation and complement cascade have been targeted together, by recruiting a combination of ruxolitinib, a JAK1/2 inhibitor alongside eculizumab (Giudice et al., 2020), an anti-C5 antibody as an alternative therapy to manage the outcome of COVID19. In similar lines, some even hypothesize that a combination of heparin and hC5a antagonist (Shin, 2020) can be potentially successful in alleviating the symptoms related to COVID19. Thus, controlling undesired assault of complement is very important and hC5a, which has established itself, both as a chemoattractant and anaphylatoxin can be the perfect candidate.
Truly speaking, there is nothing currently in the market, which can directly neutralize the excessive concentration of hC5a in the plasma. MEDI7814 (Colley et al., 2018) and IFX-1 (Giamarellos-Bourboulis et al., 2020) are the few known monoclonal antibodies that are expected to block the biological activity of hC5a. It is noteworthy that small molecules targeting hC5a with an aim to control its biological function are not known in the literature, except the recent efforts made by our group to find out potential ‘neutraligands’ of hC5a through drug repurposing approach. While we continue to explore our version of hC5a-antibody, carprofen and raloxifene are few such candidate ‘neutraligands’ from the pool, which we have subjected to proof of principle biophysical studies earlier (Mishra & Rana, 2019). The current study pursues the idea further with an aim to understand whether raloxifene alone or combined with other suitable drugs can be effective in modulating the harmful function of hC5a. Raloxifene is a prescription drug that is known to alleviate the pain and inflammation in rheumatoid arthritis. Raloxifene belongs to the class of selective estrogen receptor modulators (SERM), generally prescribed 60 mg/day with low reported side effects. Very recently, hydroxychloroquine (EC50 ∼ 0.72 µM), a primary drug prescribed for malaria and rheumatoid arthritis was repurposed for possible treatment of COVID19 (Jean et al., 2020). Our studies suggest that raloxifene can comfortably bind to hC5a probably at two sites, respectively, with estimated Ki ∼ 0.5 µM and Ki ∼ 3.58 µM. Interestingly, the two sites (HS2 and HS3) on hC5a, at which raloxifene potentially binds contains amino acids that are known to be important for its downstream signaling (Sahoo et al., 2018). Even some residues at the HS3 to which raloxifene binds have been shown to interact with the MEDI7164 antibody (Colley et al., 2018).
Growing number of evidence suggests that ‘cytokine storm’ plays a major role in patients with moderate to severe COVID19. It is well known that hC5a, a pro-inflammatory glycoprotein has a significant role in triggering (Figure 1) severe inflammation through ‘cytokine storm’ in response to the virus assaults. In the current study, we provide a perspective that binding of drug molecules, such as raloxifene to hC5a can potentially damper (D’Elia et al., 2013) the virus-induced ‘cytokine storm’ triggered by the excessive hC5a produced in the body in general and thus, raloxifene can be explored either solo or in combination with other synergistic drugs (Hung et al. 2020), as an alternative therapy for possible management of COVID19.
5. Conclusion
The role of hC5a in triggering ‘cytokine storm’ and ARDS due to inflammation is well documented and history has records, which can be linked to how ‘cytokine storm’ could have played a disastrous role in mortality (Wang et al., 2015) caused by common to severe influenza-like pandemic, faced by the mankind over 100 years (Taubenberger et al., 2019), including the recent COVID19. Though it would be ideal to find an effective vaccine for COVID19, given the complex learning curve associated with SARS-CoV2, currently, it appears to be a long wide-open road ahead. While the race for developing the perfect cure is on, renewed focus on finding novel synergistic combination of drugs should be considered, which can not only reduce both the viral load and inflammation, but also the associated co-infections (Lai et al., 2020) manifested in patients with COVID19. Though the current study is quite primitive, it instills optimism that the synergistic action of combination of drugs, capable of both checking viral shedding and acting on complement proteins like hC5a may be reasonably effective against managing COVID19.
Supplementary Material
Acknowledgments
This research is supported by the SERB (EMR/2016/000681). Use of supercomputing facility at IIT Delhi is highly appreciated. Usage of the analytical instrumentation facility of IIT Bhubaneswar and ILS Bhubaneswar is strongly acknowledged.
Disclosure statement
The authors declare no competing interests.
Author contributions
R.M. conducted most of the biophysical experiments. L.M.B. conducted some fluorescence experiments. R.M., L.M.B. and S.R. prepared the figures. S.R. conducted the computational modeling and MD studies. R.M. and S.R. analyzed the data. S.R. wrote the manuscript and conceived the idea for the project.
References
- Afshar-Kharghan, V. (2017). The role of the complement system in cancer. The Journal of Clinical Investigation, 127(3), 780–789. 10.1172/JCI90962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajona, D., Ortiz-Espinosa, S., & Pio, R. (2019). Complement anaphylatoxins C3a and C5a: Emerging roles in cancer progression and treatment. Seminars in Cell & Developmental Biology, 85, 153–163. 10.1016/j.semcdb.2017.11.023 [DOI] [PubMed] [Google Scholar]
- Atkinson, J. P., Du Clos, T. W., Mold, C., Kulkarni, H., Hourcade, D., & Wu, X. (2019). The human complement system: Basic concepts and clinical relevance. In Rich R. R., Fleisher T. A., Shearer W. T., Schroeder H. W., Frew A. J., & Weyand C. M. (Eds.), Clinical immunology (5th ed., pp. 299–317). Elsevier. [Google Scholar]
- Carroll, M. V., & Sim, R. B. (2011). Complement in health and disease. Advanced Drug Delivery Reviews, 63(12), 965–975. 10.1016/j.addr.2011.06.005 [DOI] [PubMed] [Google Scholar]
- Carsana, L., Sonzogni, A., Nasr, A., Rossi, R. S., Pellegrinelli, A., Zerbi, P., Rech, R., Colombo, R., Antinori, S., Corbellino, M., Galli, M., Catena, E., Tosoni, A., Gianatti, A., & Nebuloni, M. (2020). Pulmonary post-mortem findings in a series of COVID-19 cases from northern Italy: A two-centre descriptive study. Lancet Infectious Diseases, S1473–3099(20), 30434–5. 10.1016/S1473-3099(20)30434-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevik, M., Bamford, C. G. G., & Ho, A. (2020). COVID-19 pandemic – A focused review for clinicians. Clinical Microbiology and Infection, 26(7), 842–847. 10.1016/j.cmi.2020.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan, A. J., Wiffen, L. J., & Brown, T. P. (2020). COVID-19: A collision of complement, coagulation and inflammatory pathways. Journal of Thrombosis and Haemostasis, 18(9), 2110–2117. 10.1111/jth.14981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerkin, K. J., Fried, J. A., Raikhelkar, J., Sayer, G., Griffin, J. M., Masoumi, A., Jain, S. S., Burkhoff, D., Kumaraiah, D., Rabbani, L., Schwartz, A., & Uriel, N. (2020). COVID-19 and cardiovascular disease. Circulation, 141(20), 1648–1655. 10.1161/CIRCULATIONAHA.120.046941 [DOI] [PubMed] [Google Scholar]
- Colley, C. S., Popovic, B., Sridharan, S., Debreczeni, J. E., Hargeaves, D., Fung, M., An, L. L., Edwards, B., Arnold, J., England, E., Eghobamien, L., Sivars, U., Flavell, L., Renshaw, J., Wickson, K., Warrener, P., Zha, J., Bonnell, J., Woods, R., … Vaughan, T. J. (2018). Structure and characterization of a high affinity C5a monoclonal antibody that blocks binding to C5aR1 and C5aR2 receptors. MAbs, 10(1), 104–117. 10.1080/19420862.2017.1384892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cugno, M., Meroni, P. L., Gualtierotti, R., Griffini, S., Grovetti, E., Torri, A., Panigada, M., Aliberti, S., Blasi, F., Tedesco, F., & Peyvandi, F. (2020). Complement activation in patients with COVID-19: A novel therapeutic target. The Journal of Allergy and Clinical Immunology, 146(1), 215–217. 10.1016/j.jaci.2020.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Elia, R. V., Harrison, K., Oyston, P. C., Lukaszewski, R. A., & Clark, G. C. (2013). Targeting the “cytokine storm” for therapeutic benefit. Clinical and Vaccine Immunology, 20(3), 319–327. 10.1128/CVI.00636-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao, B., Wang, C., Wang, R., Feng, Z., Tan, Y., Wang, H., Wang, C., Liu, L., Liu, Y., Liu, Y., Wang, G., Yuan, Z., Ren, L., Wu, Y., & Chen, Y. (2020). Human kidney is a target for novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. medRxiv: 2020.2003.2004.20031120.
- Diurno, F., Numis, F. G., Porta, G., Cirillo, F., Maddaluno, S., Ragozzino, A., De Negri, P., Di Gennaro, C., Pagano, A., Allegorico, E., Bressy, L., Bosso, G., Ferrara, A., Serra, C., Montisci, A., D’Amico, M., Schiano Lo Morello, S., Di Costanzo, G., Tucci, A. G., … Facchini, G. (2020). Eculizumab treatment in patients with COVID-19: Preliminary results from real life ASL Napoli 2 Nord experience. European Review for Medical and Pharmacological Sciences, 24(7), 4040–4047. 10.26355/eurrev_202004_20875 [DOI] [PubMed] [Google Scholar]
- Dougherty, D. A. (2013). The cation-π interaction. Accounts of Chemical Research, 46(4), 885–893. 10.1021/ar300265y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano, M., Curry, S., Terreno, E., Galliano, M., Fanali, G., Narciso, P., Notari, S., & Ascenzi, P. (2005). The extraordinary ligand binding properties of human serum albumin. IUBMB Life, 57(12), 787–796. 10.1080/15216540500404093 [DOI] [PubMed] [Google Scholar]
- Fattahi, F., & Ward, P. A. (2017). Complement and sepsis-induced heart dysfunction. Molecular Immunology, 84, 57–64. 10.1016/j.molimm.2016.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira de Freitas, R., & Schapira, M. (2017). A systematic analysis of atomic protein-ligand interactions in the PDB. MedChemComm, 8(10), 1970–1981. 10.1039/C7MD00381A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francucci, C. M., Camilletti, A., & Boscaro, M. (2005). Raloxifene and cardiovascular health: Its relationship to lipid and glucose metabolism, hemostatic and inflammation factors and cardiovascular function in postmenopausal women. Current Pharmaceutical Design, 11(32), 4187–4206. 10.2174/138161205774913237 [DOI] [PubMed] [Google Scholar]
- Gao, T., Hu, M., Zhang, X., Li, H., Zhu, L., Liu, H., Dong, Q., Zhang, Z., Wang, Z., Hu, Y., Fu, Y., Jin, Y., Li, K., Zhao, S., Xiao, Y., Luo, S., Li, L., Zhao, L., Liu, J., … Cao, C. (2020). Highly pathogenic coronavirus N protein aggravates lung injury by MASP-2-mediated complement over-activation. medRxiv: 2020.2003.2029.20041962.
- Giamarellos-Bourboulis, E. J., Argyropoulou, M., Kanni, T., Spyridopoulos, T., Otto, I., Zenker, O., Guo, R., & Riedemann, N. C. (2020). Clinical efficacy of complement C5a inhibition by IFX-1 in hidradenitis suppurativa: An open-label single-arm trial in patients not eligible for adalimumab. The British Journal of Dermatology, 183(1), 176–178. 10.1111/bjd.18877 [DOI] [PubMed] [Google Scholar]
- Giudice, V., Pagliano, P., Vatrella, A., Masullo, A., Poto, S., Polverino, B. M., Gammaldi, R., Maglio, A., Sellitto, C., Vitale, C., Serio, B., Cuffa, B., Borrelli, A., Vecchione, C., Filippelli, A., & Selleri, C. (2020). Combination of ruxolitinib and eculizumab for treatment of severe SARS-CoV-2-related acute respiratory distress syndrome: A controlled study. Frontiers in Pharmacology, 11, 857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski, L. E., Sheahan, T. P., Morrison, T. E., Menachery, V. D., Jensen, K., Leist, S. R., Whitmore, A., Heise, M. T., & Baric, R. S. (2018). Complement activation contributes to severe acute respiratory syndrome coronavirus pathogenesis. mBio, 9(5), e01753–18. 10.1128/mBio.01753-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield, N. J. (2006). Using circular dichroism spectra to estimate protein secondary structure. Nature Protocols, 1(6), 2876–2890. 10.1038/nprot.2006.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, W. J., Ni, Z. Y., Hu, Y., Liang, W. H., Ou, C. Q., He, J. X., Liu, L., Shan, H., Lei, C. L., Hui, D. S. C., Du, B., Li, L. J., Zeng, G., Yuen, K. Y., Chen, R. C., Tang, C. L., Wang, T., Chen, P. Y., Xiang, J., … Zhong, N. S. (2020). Clinical characteristics of coronavirus disease 2019 in China. The New England Journal of Medicine, 382(18), 1708–1720. 10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, R. F., & Ward, P. A. (2005). Role of C5a in inflammatory responses. Annual Review of Immunology, 23, 821–852. 10.1146/annurev.immunol.23.021704.115835 [DOI] [PubMed] [Google Scholar]
- Hess, B., Kutzner, C., van der Spoel, D., & Lindahl, E. (2008). GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. Journal of Chemical Theory and Computation, 4(3), 435–447. 10.1021/ct700301q [DOI] [PubMed] [Google Scholar]
- Holers, V. M. (2014). Complement and its receptors: New insights into human disease. Annual Review of Immunology, 32, 433–459. 10.1146/annurev-immunol-032713-120154 [DOI] [PubMed] [Google Scholar]
- Hu, B., Huang, S., & Yin, L. (2020). The cytokine storm and COVID-19. Journal of Medical Virology. 10.002/jmv.26232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Q., Wu, X., Zheng, X., Luo, S., Xu, S., & Weng, J. (2020). Targeting inflammation and cytokine storm in COVID-19. Pharmacological Research, 159, 105051. 10.1016/j.phrs.2020.105051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung, I. F., Lung, K. C., Tso, E. Y., Liu, R., Chung, T. W., Chu, M. Y., Ng, Y. Y., Lo, J., Chan, J., Tam, A. R., Shum, H. P., Chan, V., Wu, A. K., Sin, K. M., Leung, W. S., Law, W. L., Lung, D. C., Sin, S., Yeung, P., … Yuen, K. Y. (2020). Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: An open-label, randomised, phase 2 trial. Lancet, 395(10238), 1695–1704. 10.1016/S0140-6736(20)31042-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean, S. S., Lee, P. I., & Hsueh, P. R. (2020). Treatment options for COVID-19: The reality and challenges. Journal of Microbiology, Immunology and Infection, 53(3), 436–443. 10.1016/j.jmii.2020.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jochems, C., Lagerquist, M., Håkansson, C., Ohlsson, C., & Carlsten, H. (2008). Long-term anti-arthritic and anti-osteoporotic effects of raloxifene in established experimental postmenopausal polyarthritis. Clinical and Experimental Immunology, 152(3), 593–597. 10.1111/j.1365-2249.2008.03660.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleczko, E. K., Kwak, J. W., Schenk, E. L., & Nemenoff, R. A. (2019). Targeting the complement pathway as a therapeutic strategy in lung cancer. Frontiers in Immunology, 10, 954. 10.3389/fimmu.2019.00954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohita, H., Matsushita, Y., & Moriguchi, I. (1994). Binding of carprofen to human and bovine serum albumins. Chemical & Pharmaceutical Bulletin, 42(4), 937–940. 10.1248/cpb.42.937 [DOI] [PubMed] [Google Scholar]
- Kumari, R., Kumar, R., Open Source Drug Discovery Consortium , & Lynn, A. (2014). g_mmpbsa – A GROMACS tool for high-throughput MM-PBSA calculations. Journal of Chemical Information and Modeling, 54(7), 1951–1962. 10.1021/ci500020m [DOI] [PubMed] [Google Scholar]
- Lai, C. C., Wang, C. Y., & Hsueh, P. R. (2020). Co-infections among patients with COVID-19: The need for combination therapy with non-anti-SARS-CoV-2 agents? Journal of Microbiology, Immunology and Infection, 53(4), 505-512. 10.1016/j.jmii.2020.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., & Ma, X. (2020). Acute respiratory failure in COVID-19: Is it “typical” ARDS? Critical Care, 24(1), 198. 10.1186/s13054-020-02911-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmudpour, M., Roozbeh, J., Keshavarz, M., Farrokhi, S., & Nabipour, I. (2020). COVID-19 cytokine storm: The anger of inflammation. Cytokine, 133, 155151. 10.1016/j.cyto.2020.155151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majorek, K. A., Porebski, P. J., Dayal, A., Zimmerman, M. D., Jablonska, K., Stewart, A. J., Chruszcz, M., & Minor, W. (2012). Structural and immunologic characterization of bovine, horse, and rabbit serum albumins. Molecular Immunology, 52(3–4), 174–182. 10.1016/j.molimm.2012.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek, S., Sample, S. J., Schwartz, Z., Nemke, B., Jacobson, P. B., Cozzi, E. M., Schaefer, S. L., Bleedorn, J. A., Holzman, G., & Muir, P. (2012). Effect of analgesic therapy on clinical outcome measures in a randomized controlled trial using client-owned dogs with hip osteoarthritis. BMC Veterinary Research, 8(1), 185. 10.1186/1746-6148-8-185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti, M. (2020). COVID-19-driven endothelial damage: Complement, HIF-1, and ABL2 are potential pathways of damage and targets for cure. Annals of Hematology, 99(8), 1701–1707. 10.1007/s00277-020-04138-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastellos, D. C., Ricklin, D., & Lambris, J. D. (2019). Clinical promise of next-generation complement therapeutics. Nature Reviews Drug Discovery, 18(9), 707–729. 10.1038/s41573-019-0031-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta, P., McAuley, D. F., Brown, M., Sanchez, E., Tattersall, R. S., Manson, J. J., & HLH Across Speciality Collaboration, UK . (2020). COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet, 395(10229), 1033–1034. 10.1016/S0140-6736(20)30628-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, R., Das, A., & Rana, S. (2020). Resveratrol binding to human complement fragment 5a (hC5a) may modulate the C5aR signaling axes. Journal of Biomolecular Structure and Dynamics, 1–15. 10.1080/07391102.2020.1738958 [DOI] [PubMed] [Google Scholar]
- Mishra, R., & Rana, S. (2019). A rational search for discovering potential neutraligands of human complement fragment 5a (hC5a). Bioorganic & Medicinal Chemistry, 27(19), 115052. 10.1016/j.bmc.2019.115052 [DOI] [PubMed] [Google Scholar]
- Möller, M., & Denicola, A. (2002). Study of protein-ligand binding by fluorescence. Biochemistry and Molecular Biology Education, 30(5), 309–312. 10.1002/bmb.2002.494030050089 [DOI] [Google Scholar]
- Mollnes, T. E., Brekke, O. L., Fung, M., Fure, H., Christiansen, D., Bergseth, G., Videm, V., Lappegård, K. T., Köhl, J., & Lambris, J. D. (2002). Essential role of the C5a receptor in E. coli-induced oxidative burst and phagocytosis revealed by a novel lepirudin-based human whole blood model of inflammation. Blood, 100, 1869–1877. [PubMed] [Google Scholar]
- Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., & Olson, A. J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30(16), 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motherwell, W. B., Moreno, R. B., Pavlakos, I., Arendorf, J. R. T., Arif, T., Tizzard, G. J., Coles, S. J., & Aliev, A. E. (2018). Noncovalent interactions of π systems with sulfur: The atomic chameleon of molecular recognition. Angewandte Chemie, 57(5), 1193–1198. 10.1002/anie.201708485 [DOI] [PubMed] [Google Scholar]
- Narasaraju, T., Yang, E., Samy, R. P., Ng, H. H., Poh, W. P., Liew, A. A., Phoon, M. C., van Rooijen, N., & Chow, V. T. (2011). Excessive neutrophils and neutrophil extracellular traps contribute to acute lung injury of influenza pneumonitis. The American Journal of Pathology, 179(1), 199–210. 10.1016/j.ajpath.2011.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederbichler, A. D., Hoesel, L. M., Westfall, M. V., Gao, H., Ipaktchi, K. R., Sun, L., Zetoune, F. S., Su, G. L., Arbabi, S., Sarma, J. V., Wang, S. C., Hemmila, M. R., & Ward, P. A. (2006). An essential role for complement C5a in the pathogenesis of septic cardiac dysfunction. The Journal of Experimental Medicine, 203(1), 53–61. 10.1084/jem.20051207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okusawa, S., Yancey, K. B., van der Meer, J. W., Endres, S., Lonnemann, G., Hefter, K., Frank, M. M., Burke, J. F., Dinarello, C. A., & Gelfand, J. A. (1988). C5a stimulates secretion of tumor necrosis factor from human mononuclear cells in vitro. Comparison with secretion of interleukin 1 beta and interleukin 1 alpha. The Journal of Experimental Medicine, 168(1), 443–448. 10.1084/jem.168.1.443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce, L., Davidson, S. M., & Yellon, D. M. (2020). The cytokine storm of COVID-19: A spotlight on prevention and protection. Expert Opinion on Therapeutic Targets, 1–8. 10.1080/14728222.2020.1783243 [DOI] [PubMed] [Google Scholar]
- Pons, S., Fodil, S., Azoulay, E., & Zafrani, L. (2020). The vascular endothelium: The cornerstone of organ dysfunction in severe SARS-CoV-2 infection. Critical Care, 24(1), 353. 10.1186/s13054-020-03062-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana, S., & Sahoo, A. R. (2015). Model structures of inactive and peptide agonist bound C5aR: Insights into agonist binding, selectivity and activation. Biochemistry and Biophysics Reports, 1, 85–96. 10.1016/j.bbrep.2015.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana, S., Sahoo, A. R., & Majhi, B. K. (2016. a). Allosterism in human complement component 5a (hC5a): A damper of C5a receptor (C5aR) signaling. Journal of Biomolecular Structure & Dynamics, 34(6), 1201–1213. 10.1080/07391102.2015.1073634 [DOI] [PubMed] [Google Scholar]
- Rana, S., Sahoo, A. R., & Majhi, B. K. (2016. b). Structural complexes of the agonist, inverse agonist and antagonist bound C5a receptor: Insights into pharmacology and signaling. Molecular Biosystems, 12(5), 1586–1599. 10.1039/c6mb00031b [DOI] [PubMed] [Google Scholar]
- Ricklin, D., & Lambris, J. D. (2007). Complement-targeted therapeutics. Nature Biotechnology, 25(11), 1265–1275. 10.1038/nbt1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklin, D., Mastellos, D. C., & Lambris, J. D. (2019). Therapeutic targeting of the complement system. Nature Reviews Drug Discovery. 10.1038/s41573-019-0055-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risitano, A. M., Mastellos, D. C., Huber-Lang, M., Yancopoulou, D., Garlanda, C., Ciceri, F., & Lambris, J. D. (2020). Complement as a target in COVID-19? Nature Reviews Immunology, 20(6), 343–344. 10.1038/s41577-020-0320-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo, A. R., Mishra, R., & Rana, S. (2018). The model structures of the complement component 5a receptor (C5aR) bound to the native and engineered hC5a. Scientific Reports, 8(1), 2955. 10.1038/s41598-018-21290-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitta, A., Morabito, N., Frisina, N., Cucinotte, D., Corrado, F., D’Anna, R., Altavilla, D., Squadrito, G., Minutoli, L., Arcoraci, V., Cancellieri, F., & Squadrito, F. (2001). Cardiovascular effects of raloxifene hydrochloride. Cardiovascular Drug Reviews, 19(1), 57–74. 10.1111/j.1527-3466.2001.tb00183.x [DOI] [PubMed] [Google Scholar]
- Sand, K. M., Bern, M., Nilsen, J., Noordzij, H. T., Sandlie, I., & Andersen, J. T. (2014). Unraveling the interaction between FcRn and albumin: Opportunities for design of albumin-based therapeutics. Frontiers in Immunology, 5, 682. 10.3389/fimmu.2014.00682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayegh, E. T., Bloch, O., & Parsa, A. T. (2014). Complement anaphylatoxins as immune regulators in cancer. Cancer Medicine, 3(4), 747–758. 10.1002/cam4.241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuttelkopf, A. W., & van Aalten, D. M. F. (2004). PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallographica. Section D, Biological Crystallography, 60(Pt 8), 1355–1363. 10.1107/S0907444904011679 [DOI] [PubMed] [Google Scholar]
- Shin, H. S. (2020). Empirical treatment and prevention of COVID-19. Infection & Chemotherapy, 52(2), 142–153. 10.3947/ic.2020.52.2.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speidl, W. S., Exner, M., Amighi, J., Kastl, S. P., Zorn, G., Maurer, G., Wagner, O., Huber, K., Minar, E., Wojta, J., & Schillinger, M. (2005). Complement component C5a predicts future cardiovascular events in patients with advanced atherosclerosis. European Heart Journal, 26(21), 2294–2299. 10.1093/eurheartj/ehi339 [DOI] [PubMed] [Google Scholar]
- Stoermer, K. A., & Morrison, T. E. (2011). Complement and viral pathogenesis. Virology, 411(2), 362–373. 10.1016/j.virol.2010.12.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., Simonovic, M., Doncheva, N. T., Morris, J. H., Bork, P., Jensen, L. J., & Mering, C. V. (2019). STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research, 47(D1), D607–D613. 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger, J. K., Kash, J. C., & Morens, D. M. (2019). The 1918 influenza pandemic: 100 years of questions answered and unanswered. Science Translational Medicine, 11(502), eaau5485. 10.1126/scitranslmed.aau5485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The GTEx Consortium . (2015). The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science, 348, 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisoncik, J. R., Korth, M. J., Simmons, C. P., Farrar, J., Martin, T. R., & Katze, M. G. (2012). Into the eye of the cytokine storm. Microbiology and Molecular Biology Reviews, 76(1), 16–32. 10.1128/MMBR.05015-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson, S. (1993). Complement defense mechanisms. Current Opinion in Immunology, 5(1), 83–89. 10.1016/0952-7915(93)90085-7 [DOI] [PubMed] [Google Scholar]
- Twaddell, S. H., Baines, K. J., Grainge, C., & Gibson, P. G. (2019). The emerging role of neutrophil extracellular traps in respiratory disease. Chest, 156(4), 774–782. 10.1016/j.chest.2019.06.012 [DOI] [PubMed] [Google Scholar]
- Wang, R., Xiao, H., Guo, R., Li, Y., & Shen, B. (2015). The role of C5a in acute lung injury induced by highly pathogenic viral infections. Emerging Microbes & Infections, 4(5), e28. 10.1038/emi.2015.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, A. J. T., Vassallo, A., Summers, C., Chilvers, E. R., & Conway-Morris, A. (2018). C5a anaphylatoxin and its role in critical illness-induced organ dysfunction. European Journal of Clinical Investigation, 48(12), e13028. 10.1111/eci.13028 [DOI] [PubMed] [Google Scholar]
- Ye, Q., Wang, B., & Mao, J. (2020). The pathogenesis and treatment of the ‘cytokine storm’ in COVID-19. The Journal of Infection, 80(6), 607–613. 10.1016/j.jinf.2020.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. M., & An, J. (2007). Cytokines, inflammation, and pain. International Anesthesiology Clinics, 45(2), 27–37. 10.1097/AIA.0b013e318034194e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Boyar, W., Toth, M. J., Wennogle, L., & Gonnella, N. C. (1997). Structural definition of the C5a C terminus by two-dimensional nuclear magnetic resonance spectroscopy. Proteins: Structure, Function, and Genetics, 28(2), 261–267. [DOI] [PubMed] [Google Scholar]
- Zheng, Y. Y., Ma, Y. T., Zhang, J. Y., & Xie, X. (2020). COVID-19 and the cardiovascular system. Nature Reviews Cardiology, 17(5), 259–260. 10.1038/s41569-020-0360-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.