Abstract
The outbreak of novel coronavirus disease (COVID-19) caused by SARS-CoV-2 poses a serious threat to human health and world economic activity. There is no specific drug for the treatment of COVID-19 patients at this moment. Traditionally, people have been using spices like ginger, fenugreek and onion, etc. for the remedy of a common cold. This work identifies the potential inhibitors of the main protease (Mpro) and spike (S) receptor of SARS-CoV-2 from 10 readily available spices. These two proteins, S and Mpro, play an important role during the virus entry into the host cell, and replication and transcription processes of the virus, respectively. To identify potential molecules an in-house databank containing 1040 compounds was built-up from the selected spices. Structure-based virtual screening of this databank was performed with two important SARS-CoV-2 proteins using Glide. Top hits resulted from virtual screening (VS) were subjected to molecular docking using AutoDock 4.2 and AutoDock Vina to eliminate false positives. The top six hits against Mpro and top five hits against spike receptor subjected to 130 ns molecular dynamic simulation using GROMACS. Finally, the compound 1-, 2-, 3- and 5-Mpro complexes, and compound 17-, 18-, 19-, 20- and 21- spike receptor complexes showed stability throughout the simulation time. The ADME values also supported the drug-like nature of the selected hits. These nine compounds are available in onion, garlic, ginger, peppermint, chili and fenugreek. All the spices are edible and might be used as home remedies against COVID-19 after proper biological evaluation.
Keywords: Spices, SARS-CoV-2, main protease, spike receptor, virtual screening, molecular dynamics, ADME filtration
Graphical Abstract
Communicated by Ramaswamy H. Sarma
1. Introduction
The current pandemic eruption of coronavirus disease (COVID-19) caused by SARS-CoV-2 is an unprecedented global public health care threat (Huang et al., 2020; Israeli, 2020; Wu, Zhao, et al., 2020). This disease was first detected in Wuhan City, Hubei Province, China, which has now been detected worldwide and 18.9027 million confirmed infected cases and 0.7095 million deaths recorded as on 8th August 2020 (www.worldometers.info). Coronaviruses (CoVs) are enveloped, positive-sense, single-stranded RNA viruses, belonging to the Coronavirinae sub-family and Coronaviridae family of the order Nidovirales, which are divided into four genera (α, β, γ and δ) (Pillaiyar et al., 2020). In the last two decades, two highly pathogenic human coronaviruses such as Severe Acute Respiratory Syndrome (SARS) appeared in the year 2002 and Middle East Respiratory Syndrome (MERS) appeared in the year 2012, emerging from animal reservoirs, have led to global epidemics with high morbidity and mortality (http://www.who.int/emergencies/mers-cov/en/, https://www.cdc.gov/sars/about/fs-sars.html and Mohd et al., 2016). The fatal rate of SARS-CoV and MERS-CoV was 10% and 35%, respectively (Cheng et al., 2007; Lee et al., 2003). The new coronavirus, SARS-CoV-2 belongs to the β genus like MERS-CoV and SARS-CoV, and causes highly infectious pneumonia disease (Wang, Lan, et al., 2020; Zhu, Zhang, et al., 2020; Zhou, Yang, et al., 2020). The rapidly increasing number of infected persons and deaths worldwide prompted the World Health Organization to declare the SARS-CoV-2 outbreak as Global Public Health Emergency of International Concern (Jiang et al., 2020; Wang, Horby, et al., 2020). WHO estimated the crude mortality ratio of SARS-CoV-2 is in between 3 and 4% (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/). Several groups of researchers are working on the development of vaccines and drug molecules to prevent and treat the disease caused by SARS-CoV-2, but no successful treatment has yet been developed (Ton et al., 2020; Xu et al., 2020; Zhang, Saravanan, et al., 2020; Zumla et al., 2020). It is a well-known fact that drug discovery is a time demanding process. On an average, the discovery of a new drug from the initial findings to market takes months to years. While drug repurposing may be a short-term and non-specific solution to treat COVID-19 patients, development of more targeted inhibitors is highly desirable (Fischer et al., 2020; Li et al., 2020; Wu, Liu, et al., 2020; Li & De Clercq, 2020; Zhou, Hu, et al., 2020; Zhu, Wang, et al., 2020; Elfiky, 2020).
Human CoVs genome has several conserved structural proteins such as Spike (S) glycoprotein, envelope (E) protein, membrane (M) protein and nucleocapsid (N) protein and at least four non-structural proteins such as- Mpro, papain-like protease (PLpro), helicase and RNA-dependent RNA polymerase (RdRp). The replication of coronavirus begins with the binding of its spike protein (S) on the cell surface of the host. This receptor recognition is important for initiating virus entry into the host cells, thereby playing a pivotal role in mediating viral infection to the host receptor (Bosch et al., 2003; Li et al., 2003). A recent study showed that like SARS-CoV, SARS-CoV-2 is able to utilize angiotensin-converting enzyme 2 (ACE2) as an entry receptor in ACE2-expressing cells (Hoffmann et al., 2020; Zhou, Yang, et al., 2020), suggesting potential drug targets for therapeutic development. Among the non-structural proteins PLpro and Mpro are two important proteases, play a crucial role in the viral replication and transcription process through the extensive proteolysis of two replicase polyproteins, pp1a and pp1ab (Báez-Santos et al., 2015). The proteolytic cleavage of pp1a and pp1b produces 16 non-structural proteins (nsp1 to nsp16). The protease, PLpro is responsible for the cleavage of first three positions of its polyprotein to produce three non-structural proteins (nsP1 to nsP3), while 3CLpro (also known as Mpro) cleaves the remaining 11 locations, releasing non-structural proteins from nsP4 to nsP16. These nonstructural proteins (nsP1–nsP16) are assembled and form the replication-transcription complex which regulate the numerous functions of virus replication viz. replication of viral genome, sub-genomic RNA processing and packaging of new virion (Fehr et al., 2018). Interrupting any replication process would become a potential molecular target to develop therapeutics against coronavirus. Thus, inhibition of the function of Mpro may block the functional nsP-complex formation and may reduce the rapid multiplication of this virus, and can be a potentially effective target to combat COVID-19.
Most of the earlier efforts to target SARS-CoV-2 resulted in identification of several known drugs as inhibitors of SARS-CoV-2 proteins. A large number FDA approved drugs such as remdesivir, chloroquine phosphate, hydroxychloroquine, lopinavir, umifenovir, arbidol, ribavirin has been used as ad-hoc basis for the treatment of COVID-19 patients (Colson et al., 2020; Liu, Zhu, et al., 2020; Sanders et al., 2020). The clinical trials of some drugs are currently ongoing to determine the full efficacy spectrum of these drugs in patients (clinicaltrials.gov, NCT04280705, Ton et al., 2020). Recently, several efforts have been initiated to examine the potentiality of several plant secondary metabolites in inhibiting the SARS-CoV-2 proteases using molecular docking analysis. In this context, Khaerunnisa et al. (2020) reported that several secondary metabolites of plants such as kaempferol, quercetin, luteolin-7-glucoside, demethoxycurcumin, naringenin, apigenin-7-glucoside, oleuropein, curcumin, catechin and epicatechin-gallate have the potential to inhibit Mpro protease of SARS-CoV-2. Recently, Zhang, Lin, et al. (2020) reported α-ketoamide as a potential inhibitor of SARS-CoV-2 main protease. In a recent study, Sampangi-Ramaiah et al. (2020) also carried out the molecular docking analysis of some commonly occurring natural products against SARS-CoV-2 proteins 6LU7 and 6Y2E proteases. In another study, Zhang, Wu, et al. (2020) observed that 13 natural compounds collected from different Chinese herbs have the potential to inhibit SARS-CoV-2 virus. Aanouz et al. (2020) reported that crocin, digitoxigenin and β-eudesmol, available in different Moroccan medicinal and aromatic plants, also have inhibitory properties against SARS-CoV-2 main protease. Enmozhi et al. (2020) observed that Andrographolide available in Andrographis paniculata, acts as a main protease inhibitor of SARS-COV-2. In another study, Islam et al. (2020) found that the phytochemicals such as hypericin, cyanidin 3-glucoside, baicalin and glabridin have antiviral activities, are and also exhibits the inhibitory properties against SARS-CoV-2 main protease. In a similar study, Umesh et al. (2020) observed that the phytochemicals such as carnosol, rosmanol and arjunglucoside-I, available in Rosmarinus officinalis, Terminalia chebula showed potential binding affinity towards main protease.
From the above elegant contributions, we envisioned that compounds present in commonly used spices, fruits and vegetables may have the potentiality to inhibit the life cycle regulatory proteins of SARS-CoV-2. In the present study, 10 spices have been chosen, which are readily available and widely used in the cuisines worldwide. In order to identify promising inhibitors, we prepared an in-house database of 1040 compounds from the available literature. It is pertinent to mention that most of the previous studies related in-silico identification of SARS-CoV-2 proteins inhibitors have been carried out by utilizing natural phytochemical databank or medicinal plant sources. Only one report is available in the literature where the authors used spice compounds from PubChemand and Zinc database. Literature studies also revealed that some of the selected spices have diverse medicinal properties and traditionally used as a remedy for common cold. Moreover, the effectiveness of herbal treatment to control contagious viral disease was demonstrated during the SARS-CoV outbreak in the year 2003 (Chen & Nakamura, 2004).
In order to identify the inhibitors, the structure-based virtual screening using Glide and molecular docking using AutoDock 4.2 and AutoDock Vina were performed. In this study, three molecular docking softwares namely Glide, AutoDock 4.2 and AutoDock Vina were used to eliminate the false positive. During Virtual Screening (VS), the compounds were subjected to filtration through Lipinski rule of five, followed by ADME filtration and removal of reactive functional groups. The study predicts that a variety of natural compounds available in spices may inhibit SARS-CoV-2 and will provide the valuable information to the common peoples as well as researchers on compounds and spices that may be effective against COVID-19. Finally, based on binding affinity of the compounds with main protease and spike receptor, the spices are reported for in vitro activity studies. All of the spices are commercially available and edible. To the best of our knowledge such detailed study to identify the inhibitors of SARS-CoV-2 from the naturally occurring spices has not been reported earlier.
2. Materials and methods
2.1. Data collection
The X-ray crystal structures of two important SARS-CoV-2 proteins namely main protease in complexes with an inhibitor N3 (6LU7, 2.16 Å) (Liu, Zhang, et al., 2020; Jin et al. 2020) and spike receptor complexes with ACE2 (6M0J, 2.45 Å) (Wang, Lan, et al., 2020; Lan et al., 2020) were retrieved from RCSB protein data bank. The in-house databank was prepared from the available literature of the selected spices. This data bank has been utilized for virtual screening (VS) and identification of potential SARS-CoV-2 protein inhibitors. The compound numbers and their sources are listed in Table 1.
Table 1.
The common name, scientific name and reference of 10 selected spices.
| Sl. No | Common name | Scientific name and (Family) | Compound code (References) |
|---|---|---|---|
| 1 | Onion | Alium cepa L. (Liliaceae) | AC_01 –AC_70 (Bystricka et al., 2013; Teshika et al., 2019) |
| 2 | Garlic | Alium sativum L. (Liliaceae) | AS_01–AS_58 (Farag et al., 2017; Kamel & Saleh, 2000; Lanzotti 2012; Martins et al., 2016; Thomson & Ali, 2003) |
| 3 | Ginger | Zingiber officinale Roscoe. (Zingiberaceae) | ZO_01–ZO_99 (Feng et al., 2011; Mahboubi, 2019; Pancharoen et al., 2000) |
| 4 | Turmeric | Curcuma longa L. (Zingiberaceae) | CL_01-CL_161 (Meng et al., 2018) |
| 5 | Black piper | Piper nigrum L. (Piperaceae) | PN_01–PN_175 (Ee et al., 2009; Rasheed et al., 2005; Salehi et al., 2019; Takooree et al., 2019) |
| 6 | Red chilli | Capsicum annuum L. (Solanaceae) | CA_01–CA_120 (Antonio et al., 2018; Imran et al., 2018; Tundis et al., 2011) |
| 7 | Fenugreek (Methi) | Trigonella foenum-graecum L. (Fabaceae) | TF_01–TF_40 (Han et al., 2001; Omezzine et al., 2017; Yoshikawa et al., 1997) |
| 8 | Nigella/Kalonji/ Black cumin | Nigella sativa L. (Ranunculaceae) | NS_01–NS_120 (Ahmad et al., 2013; Islam et al., 2019; Kokoska et al., 2008; Mukhtar et al., 2019; Srinivasan, 2018) |
| 9 | Cumin (Zeera) | Cuminum cyminum L. (Apiaceae) | CC_01–CC_104 (Gachkar et al., Amin, 2012; 2007; Gohari & Saeidnia, 2011; Hajlaoui et al., 2010; Li & Jiang, 2004; Mandal & Mandal, 2016; Oroojalian et al., 2010; Singh et al., 2017; Topal et al., 2008) |
| 10 | Peppermint | Mentha piperita L. (Lamiaceae) | MP_01–MP_93 (Brahmi et al., 2017; Reddy et al., 2019; Saharkhiz et al., 2012; Soković et al., 2009) |
2.2. Ligand preparation
The structure of the compounds was drawn by using ChemDraw Professional 15 and was saved as a sdf file. The ligands were prepared using the LigPrep module of Schrodinger 2020 (LigPrep, Schrödinger, LLC, New York, NY, 2020) with default parameters and used for Virtual Screening (VS). The top 16 hits resulted against main protease and 14 hits resulted against spike receptor were exported as pdb format. These hits further converted to pdbqt format for docking using AutoDock 4.2 and AutoDock Vina with their respective proteins.
2.3. Protein preparation and receptor grid generation
The receptors for Glide docking were prepared by using ‘Protein Preparation Wizard’ (Protein Preparation Wizard; Epik, Schrödinger, LLC, New York, NY, 2016; Impact, Schrödinger, LLC, New York, NY, 2016; Prime, Schrödinger, LLC, New York, NY, 2020; Sastry et al., 2013) in Maestro 12.2 (Maestro, Schrödinger, LLC, New York, NY, 2020). In main protease (PDB ID: 6LU7) a peptide like co-ligand (N3) was covalently bonded with receptor CYS-145 residue. The receptor grid was generated by breaking the covalent bond between co-ligand and receptor and thereby selecting the co-ligand. In the spike receptor chain E was complexed with chain A of ACE2 (PDB ID: 6M0J). During preparation of spike protein, the chain A, water molecules and hetero atoms were removed. The active site of the spike receptor was determined by using CASTp 3.0 (Tian et al., 2018) and the receptor grid was generated by selecting the active site amino acid residues (residue number 338, 339, 342, 343, 367, 368, 371, 373, 374).
The proteins were separately imported in AutoDock Tools1.5.6 for both AutoDock 4.2 and AutoDock Vina. Subsequently, water molecules and hetero atoms are removed, and then added polar hydrogen’s followed by computing Gasteiger and adding Kollman charge. Finally, proteins were saved as pdbqt format. The grid dimension of main protease was fixed by selecting the active site amino acid residue information generated from CASTp 3.0 (residue number 24, 26, 41, 49, 140–145, 163–168, 172, 187–192). The grid center dimension of Mpro is x = ‒10.883, y = 13.934, z = 68.209 and grid size is x = 58, y = 68, z = 70. Similarly, the grid dimension of the spike receptor was fixed by selecting the active site amino acid residue information from CASTp 3.0. The grid of spike receptor was generated at grid dimension center x = ‒32.00, y = 11.00, z = 28.00 and grid size x = 52, size y = 52, size z = 54.
2.4. Virtual screening of database
The virtual screening of in-house databank containing 1040 number of molecules was performed with receptor grid using Glide (Glide, Schrödinger, LLC, New York, NY, 2020, Friesner et al., 2004, 2006; Halgren et al., 2004). All compounds of the data bank were docked to each of the two receptor grid files. In Glide a rational workflow for virtual screening of database is provided. In the workflow initially the input molecules were filtered by Lipinski rule followed by removal of ligands with reactive functional groups by using QikProp (QikProp, Schrödinger, LLC, New York, NY, 2020). Next the molecules were enriched from HTVS to SP to XP, at every level such that only fewer numbers of molecules needed to be studied at the next higher accuracy level. The filtered molecules were docked with a receptor grid using default High Throughput Virtual Screening (HTVS) mode to filter out completely unreasonable molecules. Then 50% of the output molecules again docked with the receptor grid using Standard Precision (SP) mode of VS workflow. Finally, 50% of the output molecules from SP mode forwarded to dock using Glide Extra Precision (XP). The best hits resulted from each receptor being subjected to docking using AutoDock 4.2 (Morris et al., 2009) and AutoDock Vina (Trott & Olson, 2010). The best six hits against main protease and best five hits against spike receptor were selected and subjected to130 ns molecular dynamic simulation.
2.5. Validation of docking
The validation of the docking protocol of a protein having co-ligand in the active site is measured by calculating the root mean square deviation (RMSD). The RMSD is predicted by superimposing the docked co-ligand on its original crystallographic bound conformation. The RMSD of Mpro (6LU7) was 1.47, which is in the acceptable range (Figure S1). Ten SARS-CoV Mpro inhibitors and two known SARS-CoV-2 Mpro inhibitor namely Remdesivir and Favipiravir were retrieved from the literature (Ramajayam et al., 2010; Tu et al., 2020) and compared the binding affinity of hits of main protease inhibitors of SARS-CoV-2. The SARS-CoV-2 known spike receptor inhibitors (Arbidol and Nafamostat) were collected from literature (Chen et al., 2020; Li & De Clercq, 2020; Sanders et al., 2020) and compared the selected hits with these known inhibitors.
Figure 1.
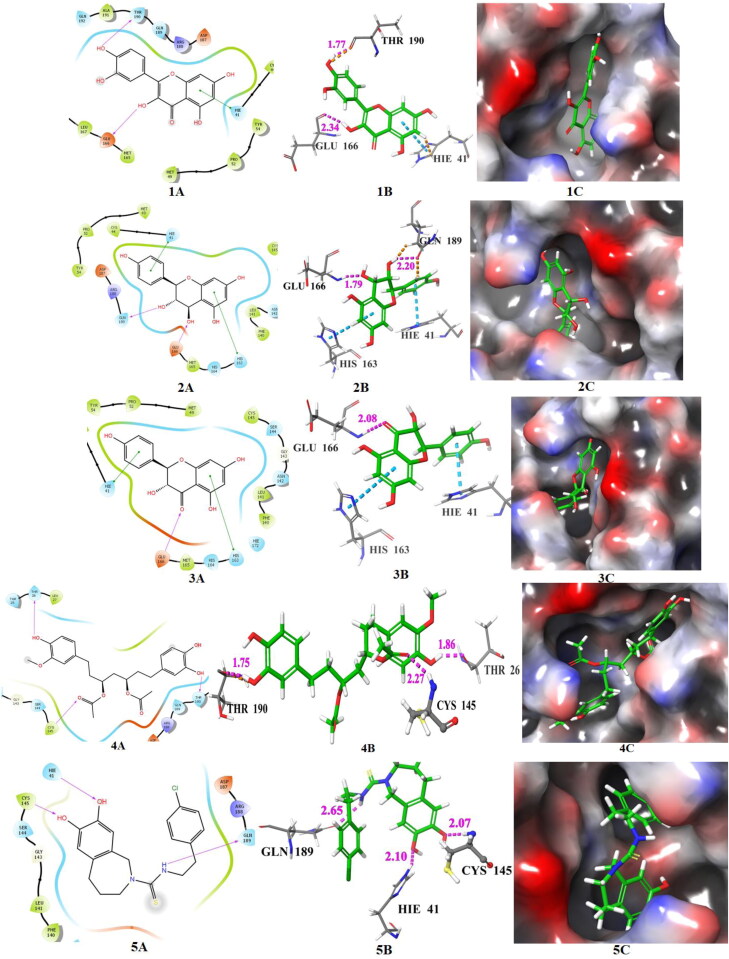
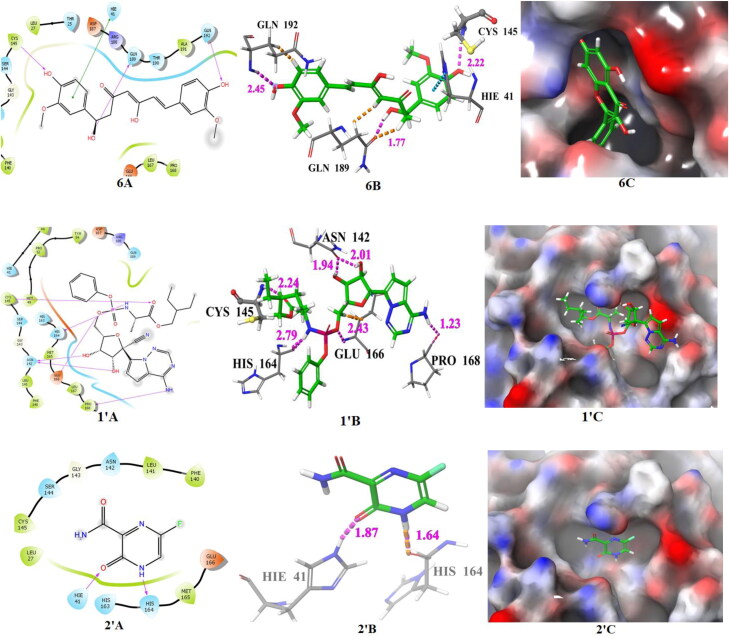
The docking poses of hits 1‒6 and known main protease inhibitors 1′ and 2′ (A: 2 D ligand interaction diagram like hydrogen bond donor, hydrogen bond acceptor, π-π stacking, B:3D ligand interactions like hydrogen bond donor, hydrogen bond acceptor, π-π stacking, C: Docking pose of hits in the active site), docked with main protease. The 2 D ligand interactions are depicted with different colors: pi-pi (green line), hydrogen bond (violet arrow line). The 3 D ligand interactions depicted hydrogen bond (purple dotted line), pi-pi (doted skyline).
2.6. Molecular dynamic (MD) stimulation study
All atom 130 nano second (ns) molecular dynamics (MD) simulation of selected protein-ligand complex as well as for the apo-proteins were conducted. In the present study the NVIDIA RTX 2070ti GPU accelerated GROMACS 2018.1 software (Abraham et al., 2015), running over Linux Mint 2019.2 operating system supported by Intel i7 9700k processor was used. The VMD software (Humphrey et al., 1996) was used to visualize the trajectory and Grace Software (https://plasma-gate.weizmann.ac.il/Grace/) was used as a plotting program. The Charmm36 force field was used to generate protein topology. The ligand topology and parameters required for MD simulation were generated by using Swissparam server (Zoete et al., 2011). The TIP3P (Jorgensen et al., 1983) water model was used for solvating each systems followed by neutralization with appropriate numbers of Na+ and Cl‒. Then energy of each system was minimized by using the steepest descent minimization algorithm with maximum 1,00,000 steps and < 10.0 kJ/mol force. The 2 ns NVT and 10 ns NPT ensemble equilibration process were conducted for each system. A positional restraint of 100 ps for backbone and C-α atom was subjected for both NVT and NPT ensemble equilibration process. However, free movements were allowed for the solvent molecules in order to establish solvent equilibrium in the system. Particle mesh Eshwald (PME) was considered for recording of long range electrostatic interactions by setting 1.2 nm cut-off and 1.2 nm Fourier spacing. In order to constrained covalent bonds, a linear constraint solver algorithm was used (Bowers et al., 2006). Each equilibrated system was subjected for 130 ns MD simulation with a time step of 2 femto-seconds (fs). V-rescale weak coupling method was used to regulate the temperature (315.10 K) of the systems. Using Parrinello − Rahman method the pressure of each system was set at 1 atm (Martyna et al., 1994). The structural coordinates were saved for every 2 pico-second intervals. After PBC (periodic boundary condition) correction and removing solvents, ions etc., from the 130 ns atomistic molecular dynamics trajectory various parameters like root mean square deviation (RMSD), root mean square fluctuation (RMSF), radius of gyration (Rg), number of hydrogen bonds occurred between protein and ligand (HB), were calculated (Chatterjee et al., 2020) using in-built functions of GROMACS 2018.1 software.
2.7. Mm-PBSA calculation
The binding free energy (ΔGbinding) of protein-ligand complexes was calculated by combining the Molecular Mechanic Poisson-Boltzmann Surface Area (MM-PBSA) with MD trajectory (Miller et al., 2012). The MM-PBSA was measured from the entire 130 ns trajectory as well as extracting snapshots of 10 ns of MD simulation. The ΔGbinding of protein-ligand complex provides an overview of the bio-molecular interactions between protein and ligand. The binding energy reflects the potential energy, polar and non-polar solvation energies. The MM-PBSA binding free energies were calculated by utilizing the ‘g_mmpbsa’ (Kumari et al., 2014) script programme. In this method the binding energy of the protein-ligand systems were calculated by using the following equation: ΔGbinding = Gcomplex − (Greceptor + Gligand), where ΔGbinding is the total binding energy of the complex, Greceptor is the binding energy of free receptor and Gligand is the energy of unbounded ligand.
3. Results and discussion
The surface area and volume of main protease (6LU7) were 351.125 Å2 and 319.370 Å3, respectively, predicted by CASTp 3.0. The VS of 1040 molecules with Mpro resulted 60 hits having XP glide score ranges from ‒6.510 to ‒9.523. Among the 60 hits, the XP glide score of sixteen hits are greater than 7.2 and these are 1(AC_01, AS_43, MP_71, TF_38), 2 (AC_19), 3 (AC_12), 4 (ZO_53), 5 (CA_59), 6 (CL_16), 7 (ZO_39), 8 (ZO_58), 9 (MP_67), 10 (MP_65, AC_15, CL_119), 11 (ZR_55), 12 (AC_28, CA_23), 13 (AS_44), 14 (AC_11), 15 (ZO_38), 16 (CL_121, AC_16, MP_64, TF_39). The range of binding energy values of these hits are ‒4.89 to ‒8.77 kcal/mol and the range of Ki values are 258.39 to 0.37 μM predicted by AutoDock 4.2. The ranges of binding affinities of these selected hits predicted by AutoDock Vina are ‒5.8 to ‒7.9 kcal/mol. The XP glide scores were predicted by Glide, binding energy and Ki values were predicted by AutoDock 4.2, and binding affinity were predicted by AutoDock Vina of these sixteen hits are listed in Table 2. The 2 D and 3 D ligand interaction diagram and binding pose of ligands (1–6) in the active site of Mpro are shown in Figure 1. The 2 D and 3 D ligand interaction diagram and binding pose of ligands (7–16) in the active site of Mpro are shown in Figure S2. Among SARS-CoV known inhibitors and SARS-CoV-2 known inhibitors, the best binding inhibitor was remdesivir. The XP glide score, binding energy predicted by AutoDock 4.2, Ki value and binding affinity predicted by AutoDock Vina of remdesivir are −8.061, −9.05, 0.2334 and −8.0, respectively (Figure 1). The structures, and 2 D ligand interactions and XP glide scores of 10 known SARS-CoV main protease inhibitors are shown in Figure S3 and Figure S4, respectively.
Table 2.
Potential Mpro(PDB ID: 6LU7) inhibitors, spice source number, standard inhibitors, active site interacting amino acid residues of main protease, extra precision glide score (XPGS), predicted binding energy in kcal/mol (BE) and Ki (AutoDock 4.2) and binding affinity (AutoDock Vina).
| Compound number | Compound ID | Active site interacting amino acid residues | XPGS | BE | Ki (μM) | BA |
|---|---|---|---|---|---|---|
| 1 | AC_01, AS_43, MP_71, TF_38 | HIE-41 (π-π), GLU-166 (2.34), THR-190 (1.77) | ‒8.333 | ‒8.2 | 0.9758 | ‒7.5 |
| 2 | AC_19 | HIE-41 (π-π), HIS-163 (π-π), GLU-166 (1.79), GLN-189 (2.20) | ‒9.523 | ‒7.8 | 1.8600 | ‒6.7 |
| 3 | AC_12 | HIE-41 (π-π), HIS-163 (π-π), GLU-166 (2.08) | ‒8.694 | ‒8.73 | 0.3956 | ‒7.9 |
| 4 | ZO_53 | THR-26 (1.86), CYS-145 (2.27), THR-190 (1.75) | ‒8.701 | ‒7.12 | 6.0300 | ‒7.2 |
| 5 | CA_59 | HIE-41 (2.10), CYS-145 (2.07), GLN-189 (2.65) | ‒7.333 | ‒8.77 | 0.3700 | ‒7.0 |
| 6 | CL_16 | HIE-41 (π-π), CYS-145 (2.22), GLN-189 (1.7), GLN-192 (2.45) | −8.027 | −6.64 | 13.64 | −6.7 |
| 7 | ZO_39 | GLY-143 (1.94), GLU-166 (1.76, 2.02), HIS-164 (1.95) | ‒8.461 | ‒6.72 | 11.7800 | ‒6.5 |
| 8 | ZO_58 | HIE-41 (2.00), HIS-164 (2,04), GLU-166 (1,91), THR-190 (2.12), | ‒7.913 | ‒4.89 | 258.3900 | ‒5.8 |
| 9 | MP_67 | HIE-41 (π-π), ASN-142 (2.63), HIS-163 (2.16), GLU-166 (1.91), GLN-189 (1.84) | ‒8.043 | ‒6.31 | 23.5900 | ‒7.3 |
| 10 | MP_65, AC_15, CL_119 | HIE-41 (π-π), GLU-166 (2.39), HIS-163 (π-π) | 7.775 | ‒7.55 | 2.9100 | ‒7.9 |
| 11 | ZO_55 | HIE-41 (π-π), CYS-145 (2.05) | ‒7.762 | ‒7.3 | 4.4600 | ‒6.3 |
| 12 | AC_28, CA_23 | LEU-141 (2.07), GLU-166 (2.34), ARG-188 (2.26), GLN-189 (1.96), THR-190 (2.12) | ‒7.724 | ‒7.37 | 3.9400 | ‒7.3 |
| 13 | AS_44 | HIE-41 (π-π), GLU-166 (2.02), GLN-192 (2.12) | ‒7.723 | ‒7.86 | 1.7500 | ‒7.0 |
| 14 | AC_11 | HIE-41 (π-π), HIS-164 (1,89), THR-190 (1.79) | ‒7.667 | ‒7.63 | 2.5500 | ‒7.3 |
| 15 | ZO_38 | HIE-41 (2.25), LEU-141 (1.81), GLU-166 (2.35), GLN-189 (1.93) | ‒7.321 | ‒6.13 | 32.0900 | ‒6.4 |
| 16 | CL_121, AC_16, MP_64, TF_39 | LEU-141 (2,09), GLU-166 (2.48), THR-190 (2.11, 1.91) | −7.201 | ‒7.55 | 2.9100 | ‒7.5 |
| 1′ | Remdesivir | CYS-145 (2.24), HIS-164 (2.79), GLU-166 (2.43), PRO-168 (1.23), ASN-142 (2.01, 1.94) | −8.061 | −9.05 | 0.2334 | −8.0 |
| 2′ | Favipiravir | HIE-41 (1.87), HIS-164 (1.64) | −4.373 | −4.82 | 295.28 | −4.9 |
Figure 2.
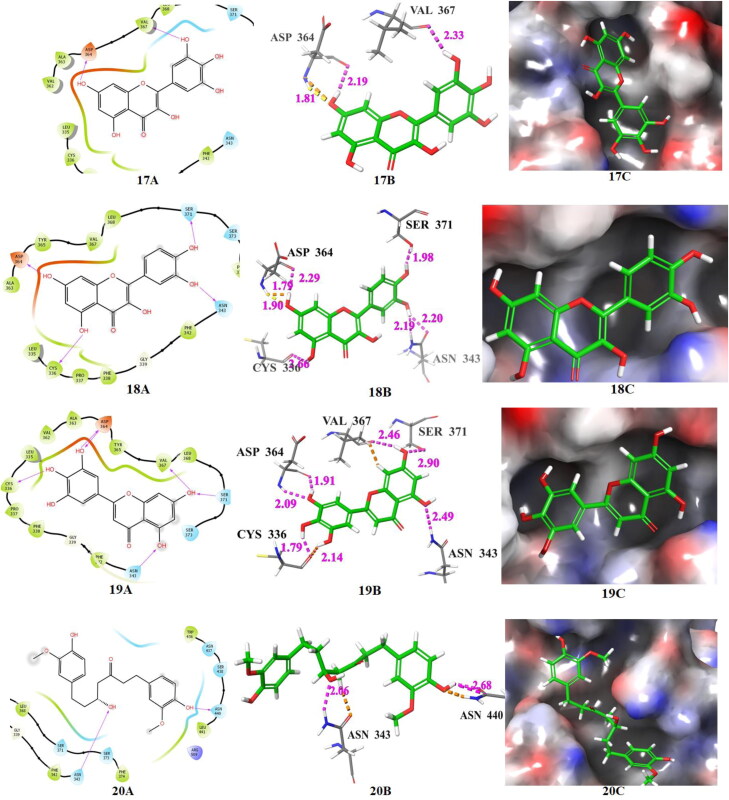
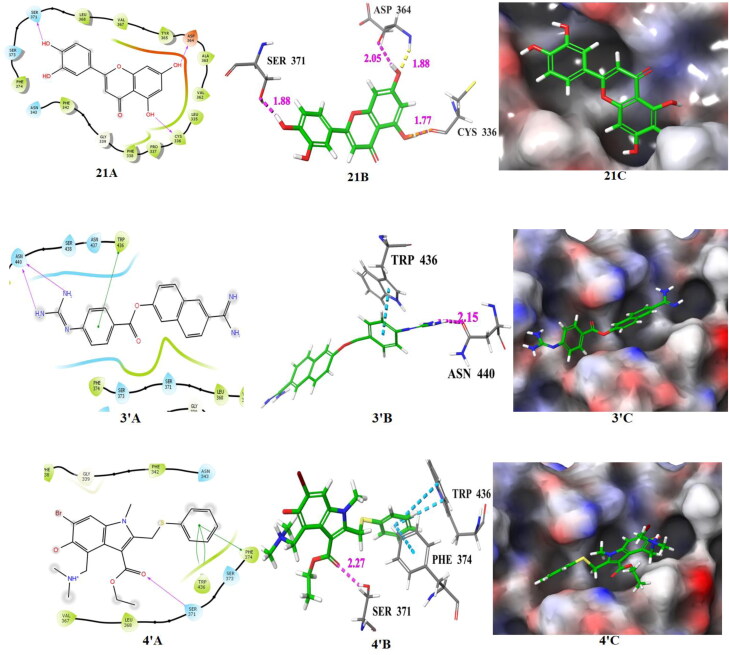
The docking poses of hits 17‒21 and two known spike receptor inhibitors 3′, 4′ (A: 2 D ligand interaction diagram like hydrogen bond donor, hydrogen bond acceptor, π-π stacking, B:3D ligand interactions like hydrogen bond donor, hydrogen bond acceptor, π-π stacking, C: Docking pose of hits in the active site), docked with spike receptor. The 2 D ligand interactions are depicted with different colors: pi-pi (green line), hydrogen bond (violet arrow line). The 3 D ligand interactions depicted hydrogen bond (purple dotted line), pi-pi (doted skyline).
Figure 3.
RMSD of apo-protein backbone and ligand (1‒6)-Mpro complexes vs time plot.
Figure 4.
RMSF profile of apo-protein and Mpro-ligand (1–6) complexes.
The important interacting amino acid residues of remdesivir with main protease are CYS-145 (2.24), HIS-164 (2.79), GLU-166 (2.43), PRO-168 (1.23), ASN-142 (2.01, 1.94). The XP glide score, binding energy and Ki value predicted by AutoDock 4.2, and binding affinity predicted by AutoDock Vina of compound 1 are ‒8.33, −8.20, 0.9758 and −7.5, respectively, and the interacting amino acid residues are HIE-41 (π-π), GLU-166 (2.34 Å), THR-190 (1.77 Å). The XP glide score, binding energy, Ki value (by AutoDock 4.2) and binding affinity (by AutoDock Vina) of compound 2 are −9.523, −7.8, 1.860 and −6.7, respectively, and the interacting amino acid residues are HIE-41 (π-π), HIS-163 (π-π), GLU-166 (1.79 Å), GLN-189 (2.20 Å). The XP glide score, binding energy, Ki value (by AutoDock 4.2) and binding affinity (by AutoDock Vina) of hit 3 are −8.694, −8.73, 0.3956 and −7.9, respectively, and interacting amino acid residues are HIE-41 (π-π), HIS-163 (π-π), GLU-166 (2.08 Å). The XP glide score, binding energy and Ki value and binding affinity of 4 are −8.701, −7.12, 6.030 and −7.2, respectively, and interacting amino acid residues are THR-26 (1.86 Å), CYS-145 (2.27 Å), THR-190 (1.75 Å). Compound 4 exhibits close docking score in all the three methods. The XP glide score, binding energy, Ki value and binding affinity of 5 are −7.33, −8.77, 0.3700 and −7.0, respectively, and interacting amino acid residues are HIE-41 (2.10 Å), CYS-145 (2.07 Å), GLN-189 (2.65 Å). The XP glide score, binding energy, Ki value and binding affinity (AutoDock Vina) of 6 are −8.027, −6.64, 13.64 and −6.7, respectively, and interacting amino acid residues are HIE-41 (π-π), CYS-145 (2.22 Å), GLN-192 (2.45 Å).
The docking score, interacting amino acid residues and binding pose in the active site of Mpro for compounds 1, 2 and 3 are compatible with remdesivir. These compounds also released similar binding energy in three docking methods. Therefore, the compounds 1, 2 and 3 may be considered as potential inhibitors of the main protease of SARS-CoV-2. There is evidences of flavonoid, baicalein showed good in vitro inhibitory activity (IC50: 0.50 μM against SARS-CoV-2 Mpro (Su et al., 2020). The compounds 1, 2 and 3 are also flavonoid types and are available in A. cepa, A. sativum, M. piperita T. foenum-graecum and C. annuum. Yu et al. (2012) also observed that two natural flavones-myricetin and scutellarein have potentiality to inhibit SARS-CoV helicase protein. The binding pose of 4 in the active site of Mpro is also good and it interacts with crucial amino acid residue CYS-145 from a close distance of 2.27 Å. The amino acid CYS-145 was covalently bonded with co-ligand (N3) in X-ray crystallography complex and also has close interaction with remedisivir (2.24 Å). This phenolic compound (4) is available in Z. officinale. The predicted Ki value of compound 5 was lowest (0.3700 μM) and it has close interaction with key amino acid residues HIE-41 (2.10 Å) and CYS-145 (2.07 Å) of main protease. It is an alkaloid, available in C. annuum. Kim et al. (2019) reported that three natural alkaloids bis-benzylisoquinoline, tetrandrine, cepharanthine exhibit antiviral activity against HCoV-OC43.The compound 6 shows π-π interactions with key amino acid HIE-41 and close hydrogen bonding interaction with CYS-145 (2.22 Å). It is also shown a comparable docking score in three methods and exhibits good binding pose in the active site of Mpro. The hits 4, 5 and 6 have common active site amino acid interacting residue CYS-145 with remedesivir. This compound (6) is also a phenolic compound and available in C. longa. The binding energy predicted by three methods was similar and the molecule is deeply inserted into the active site of Mpro. The XP glide score, binding energy, Ki values and binding affinity predicted by three methods and detailed interactions predicted by Glide of compounds 7–16 are shown in Table 2. Most of the compounds show comparable binding affinity with remdesivir.
The surface area and volume of the spike receptor were 73.389 Å2 and 60.837 Å3, respectively, as predicted by CASTp 3.0. The VS of 1040 molecules with spike receptor using Glide resulted 14 hits with XP glide score greater than ‒6.6 and these are 17 (AC_27, CA_23), 18 (AC_01), 19 (AC_17), 20 (ZO_48), 21 (TF_39), 22 (AC_10), 23 (ZO_53), 24 (CL_119, AC_15, MP_65), 25 (AC_12), 26 (ZO_55), 27 (TF_40, AC_09, MP_63), 28 (AC_13), 29 (MP_68), 30 (MP_67). The binding energies and Ki values of these fourteen hits were further predicted by AutoDock 4.2 and the values ranged from ‒4.79 to ‒6.57 kcal/mol, and 309.35 ‒15.29 μM, respectively. Further docking of these hits by AutoDock Vina gives the binding energy ranges from −5.7to −7.4 kcal/mol. The XP glide scores, binding energy, Ki values and binding affinity of top fourteen hits and two known inhibitors of spike receptor are listed in Table 3. The 2 D and 3 D ligand interaction diagram and binding pose of hits (17–21) in the active site of the spike receptor and known spike inhibitors are shown in Figure 2. The 2 D and 3 D ligand interaction diagram and binding pose of hits (22–30) in the active site are shown in Figure S5.
Table 3.
The potential spike receptor (PDB ID: 3M0J) inhibitors compound number, spice source compound number, standard inhibitor, active site interacting amino acid residues of spike, extra precision glide score (XPGS), predicted binding energy in kcal/mol(BE) and Ki values (AutoDock 4.2) and binding affinity (AutoDock Vina).
| Compound | Compound ID | XPGS | Interacting amino acid residues | BE | Ki μM | BA |
|---|---|---|---|---|---|---|
| 17 | AC_28, CA_23 | −8.729 | ASP-364 (1.81, 2.19), VAL-367(2.33) | −6.01 | 40.27 | −6.7 |
| 18 | AC_01 | −8.707 | ASP-364 (1.97, 2.02), SER-371 (1.96), CYS-336 (1.78), ASN-343 (2.14, 3.12, 3.25) | −5.88 | 48.76 | −6.4 |
| 19 | AC_17 | −8.222 | ASN-343 (2.49, 2.63), SER-371 (2.15), CYS-336 (2.14), 2.69, 2.71), ASP-364 (2.09, 2.87) | −6.34 | 22.62 | −7.2 |
| 20 | ZO_48 | −8.187 | ASN-343 (2.06), ASN-440 (2.20, 2.66) | 5.82 | 54.3 | −6.8 |
| 21 | TF_39 | −8.036 | CYS-336 (1.77), ASP-364 (1.88, 2.05), SER-371 (1.88) | −6.34 | 22.51 | −7.0 |
| 22 | AC_10 | −7.629 | CYS-336 (1.80), SER-371 (1.79), ASP-364 (1.80) | −6.57 | 15.29 | −7.4 |
| 23 | ZO_53 | −7.479 | CYS-336 (1.62), ASP-364 (1.78), SER-371 (2.46), TRP-436 (π-π), ASP-364 (π-π) | −4.79 | 309.35 | −6.5 |
| 24 | CL_119, AC_15, MP_65 | −7.061 | CYS-336 (1.79), ASP-364 (1.78), SER-371 (1.87) | −6.53 | 16.31 | −7.0 |
| 25 | AC_12 | −7.182 | CYS-336 (1.79), ASP-364 (1.80), SER-371 (1.80) | −5.84 | 52.31 | −6.4 |
| 26 | ZO_55 | −6.726 | ASN-440 (2.16, 2.09) | −6.37 | 21.3 | −6.9 |
| 27 | TF_40, AC_09, MP_63 | −6.687 | CYS-336 (1.79), ASP-364 (1.80), SER-371 (1.81) | −6.18 | 29.52 | −6.9 |
| 28 | AC_13 | −6.626 | CYS-336 (1.89, 1.61), GLY-339 (2.23), ASN-343 (1.73) | −5.9 | 68.03 | −6.4 |
| 29 | MP_68 | −6.619 | CYS-336 (1.77), ASP-364 (1.90, 2.03), SER-371 (1.87) | −6.44 | 18.88 | −6.7 |
| 30 | MP_67 | CYS-336 (1.68), ASP-364 (1.89, 2.00) | −6.01 | 39.38 | −6.8 | |
| 3′ | Nafamostat | −7.40 | TRP-436 ( π-π), ASN-440 (2.08, 2.15) | −6.21 | 28.63 | −8.7 |
| 4′ | Arbidol | −4.04 | CYS-336 (3.45), SER-371 (2.27), PHE-338 (3.03), TRP-436 (two, π-π) | −5.95 | 43.5 | −5.7 |
The XP glide score, binding energy, Ki value and binding affinity of known spike receptor inhibitor 3′ (Nafamostat) are ‒7.40, ‒6.21, 28.63 and ‒8.7, respectively, and the interacting amino acid residues are TRP-436 (π-π), ASN-440 (2.08, 2.15). The XP glide score, binding energy, Ki value and binding affinity of known spike receptor inhibitor 4′ (arbidol) are −4.04, −5.95, 43.5 and −5.7, respectively. The interacting amino acid residues of this inhibitor (4′) are CYS-336 (3.45 Å), SER-371 (2.27 Å), PHE-338 (3.03 Å), TRP-436 (two, π-π). The XP glide score predicted by Glide, binding energy and Ki value predicted by AutoDock 4.2 and docking score predicted by AutoDock Vina of hit 17 are −8.729, −6.01, 40.27 and −6.7, respectively. The interacting amino acid residues of 17 are ASP-364 (1.81 Å, 2.19 Å), VAL-367(2.33 Å).The XP glide score, binding energy, Ki value and binding affinity of compound 18 are −8.707, −5.88, 48.76 and −6.4, respectively, and the interacting amino acid residues of this molecule (18) are ASP-364 (1.97 Å, 2.02 Å), SER-371 (1.96 Å), ASN-343 (2.14 Å, 3.12 Å, 3.25 Å). The XP glide score, binding energy, Ki value and binding affinity of compound 19 are −8.222, −6.34, 22.62 and −7.2, respectively, and it interacts with the following amino acid residues ASN-343 (2.49 Å, 2.63 Å), SER-371 (2.15 Å), CYS-336 (2.14 Å, 2.69 Å, 2.71 Å), ASP-364 (2.09 Å, 2.87 Å). The XP glide score, binding energy, Ki value and binding affinity of hit 20 are −8.187, −6.02, 38.94 and −6.8, respectively, and the interacting amino acid residues are ASN-343 (2.06 Å), ASN-440 (2.20 Å, 2.66 Å). The XP glide score, binding energy, Ki value and binding affinity of compound 21 are ‒8.036, ‒6.34, 22.51 and ‒7.0, respectively, and its interacting amino acid residues are CYS-336 (1.77 Å), ASP-364 (1.88 Å, 2.05 Å), SER-371 (1.88 Å). The XP glide scores, binding energy, Ki value, binding affinity and interacting amino acid residues of compounds 22–30 are listed in Table 3.
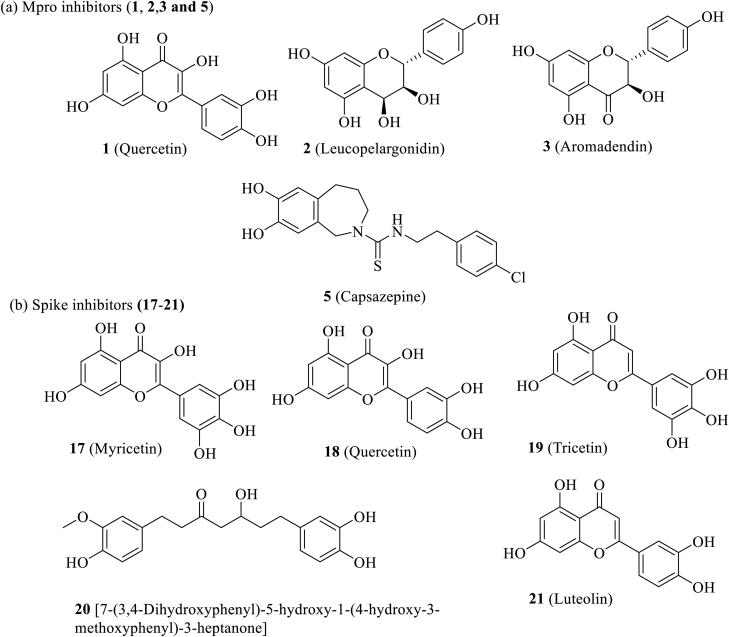
The compounds 17, 18 and 19 released fair binding energy in three docking methods and good XP binding pose in the active site of the spike receptor. The docking score of these hits is comparable with known SARS-CoV-2 spike receptor inhibitor 3′ and greater than from the inhibitor 4′. Compound 17 shows close hydrogen bond donor-acceptor interactions with ASP-364 and VAL-367. It is a flavonoid compound and is available in A. cepa and C. annuum. Compound 18 has a common binding interaction with SER-371 of known inhibitor 4′, two close hydrogen bonding interactions with ASP-364, and one close hydrogen bonding interaction with ASN-343. Compound 19 and known spike inhibitor 4′ show common interactions with amino acids CYS-336 and SER-371. This compound has one hydrogen bonding interaction with ASN-343 and two hydrogen boning interactions with ASP-364. The flavonoid compounds 18 and 19 are available in A. cepa. The compound 20 shows two close interactions with ASN-440 which is common with the known inhibitor 3′ and has one close interaction with ASN-343. It is observed that some portion of compound 20 resides outside the binding pocket. The compound 21 and 4′ have common interactions with CYS-336 and SER-371. This compound also shows two hydrogen bonding interactions with ASP-364. It shows a comparable docking score with nafamostat (3′) and better docking score than arbidol (4′). This flavonoid compound is available in T. foenum-graecum. The compounds 22‒30 also show superiority over the arbidol (4′) in terms of binding energy and binding interactions. The sources of these compounds are available in Table 1. Among these 22, 24, 25, 26 and 29 are flavone and flavonoids; show a similar type of interactions with active site amino acid residues like other flavones and flavonoids. The top scored five compounds 17‒21 were selected for MD simulation study for further screening.
Molecular dynamics simulation provides a significant insight about the stability of protein-ligand complex (Schreiner et al., 2012). In order to analyze the binding affinities of each identified hits from household spices, a 130 ns atomistic molecular dynamics simulation was conducted. To draw a conspicuous surmise about the stability of each protein-ligand system, the important parameters like RMSD, RMSF, Rg, intermolecular hydrogen bond as well as binding energy were calculated from the molecular dynamics trajectory. The RMSD, RMSF and Rg of each protein-ligand system were compared with the respective RMSD, RMSF and Rg of ligand free protein (apo-protein). The average values of each parameter with standard deviation were depicted in Table 4.
Table 4.
Results of 130 ns MD simulation of apo-proteins and individual protein-ligand complexes.
| System ID | Avg RMSD (Å) of backbone | Avg RMSF (Å) | Avg Rg (Å) | Avg number of HB | BE (total) kJ/mol | BE (last 10ns) kJ/mol |
|---|---|---|---|---|---|---|
| Main protease (PDB ID: 6LU7) | ||||||
| Apo-Protein | 2.382 ± 0.48 | 1.366 ± 1.05 | 22.51 ± 0.158 | NA | NA | NA |
| 1 | 2.314 ± 0.30 | 1.087 ± 0.613 | 22.36 ± 0.17 | 4.00 ± 0.2 | −211.2 ± 38.30 | −242.27 ± 25.20 |
| 2 | 2.226 ± 0.44 | 1.058 ± 0.54 | 22.5 ± 0.12 | 1.7 ± 0.2 | −106.6 ± 68.13 | −141.7 ± 29 |
| 3 | 2.477 ± 0.44 | 1.089 ± 0.61 | 22.37 ± 0.142 | 2.8 ± 0.17 | −222.3 ± 30.73 | −221.83 ± 31.7 |
| 4 | 3.34 ± 0.8 | 1.5 ± 0.96 | 22.4 ± 0.154 | 2.34 ± 0.23 | −180 ± 64.81 | −137 ± 39.37 |
| 5 | 2.112 ± 0.35 | 1.008 ± 0.517 | 22.50 ± 0.12 | 2.2 ± 0.97 | −139.014 ± 54.189 | −151.92 ± 20.72 |
| 6 | 4.605 ± 0.23 | 2.54 ± 0.915 | 22.64 ± 0.355 | 2.24 ± 0.22 | −158.41 ± 34.32 | −139.08 ± 22.86 |
| Spike receptor (PDB ID: 6M0J) | ||||||
| Apo-Protein | 1.7 ± 0.48 | 1.06 ± 0.865 | 18.62 ± 2.4 | NA | NA | NA |
| 17 | 1.362 ± 0.16 | 0.873 ± 0.42 | 18.42 ± 1.4 | 4.3 ± 2.20 | −177.43 ± 30.58 | −176.41 ± 31.37 |
| 18 | 1.402 ± 0.18 | 0.880 ± 0.52 | 18.37 ± 1.6 | 3.0 ± 2.24 | −167.15 ± 48.63 | −175.02 ± 36.77 |
| 19 | 1.356 ± 0.18 | 0.774 ± 0.33 | 18.25 ± 1.32 | 2.35 ± 1.6 | −172.4 ± 29 | −168.711 ± 27.6 |
| 20 | 1.5 ± 0.184 | 0.843 ± 0.37 | 18.5 ± 1.07 | 1.1 ± 0.83 | −163.1 ± 40.6 | −200.02 ± 27 |
| 21 | 1.36 ± 0.222 | 0.854 ± 0.5 | 18.21 ± 1.02 | 1.41 ± 0.85 | −168.50 ± 34.29 | −182.13 ± 30.13 |
HB = Hydrogen Bond, NA = Not Applicable
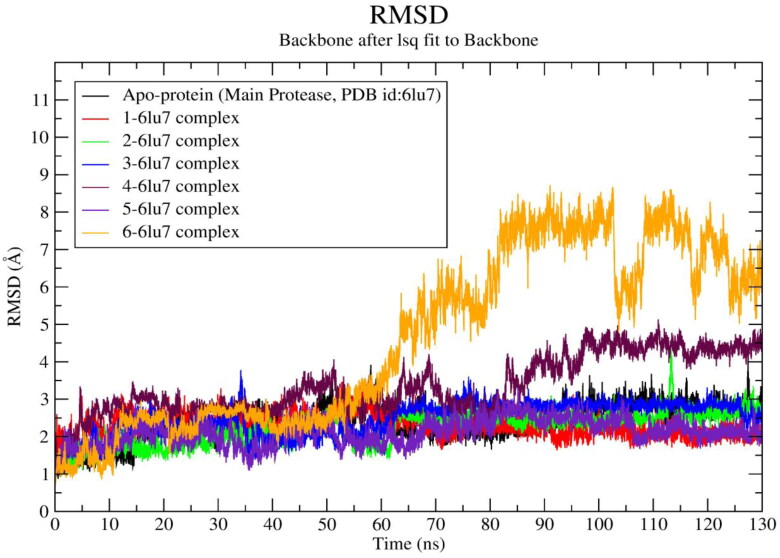
The molecular docking results of hits 1‒6 showed admirable docking score with the Mpro. All the ligand-protein systems exhibit lower average RMSD value compared to the RMSD value of apo-protein RMSD (∼2.38 Å), except ligand-protein complex of 4 and 6. The RMSD values of compound 4 and 6 were found to be ∼3.34 Å and ∼4.6 Å, respectively. Large changes in atomistic positions of residues like VAL-42 to TYR-54, ALA-94‒THR-93, PRO-168 to GLY-170, PRO-184 to THR-199, were observed in 4-6lu7 system after 85 ns to 130 ns of the simulation. All of these residues present in the loop region of the protein (Figure S6). Among them VAL-42 to THR-190 were reside in the binding site. This may be the probable cause of sudden jump of RMSD depicted by this system. After ∼60 ns sudden jump of RMSD towards higher values were found in 6-6lu7 system. Residues like ASP-48 to LEU-50, ALA-70 to ARG-76, ASP-92 to PRO-96, and ASP-263 to ALA-266 are present in the loop region of the protein (Figure S6). The residues like ASN-231 to ASP-229 present in alpha helix. All these residues exhibit large changes in atomistic position. Moreover, ligand 6 after ∼60 ns flipped inside the binding pocket. These events may responsible for the sudden jump in RMSD after ∼60 ns. Same higher RMSD profile was observed up to end of the simulation for this system. The ligand 5 shows lowest average RMSD value ∼2.11 Å. To evaluate the convergence of the trajectory, the RMSD of protein-ligand complexes (1‒6) and apo-protein were calculated and plotted against time in nanosecond (Figure 3).
Figure 6.
Number of average hydrogen bonding interactions between main protease–ligand (1–6) systems during 130 ns simulation time.
The RMSD parameter provides critical information about the structural conformation of protein-ligand complexes. In the present study distance-based RMSD values were calculated for protein-ligand complex backbone. A lower RMSD value indicates superior stability of the system. The acceptable range of RMSD value is < 3.0 Å (Kufareva & Abagyan, 2012) for a globular protein. It was observed that the hit 6 exhibited sudden rises in RMSD values after 60 ns and continued with higher value (∼6.5 Å) throughout the rest simulation time. The RMSD value of ligand 4 rises after 85 ns and fluctuates up to 130 ns with at highest RMSD value ∼3.8 Å. The ligand 1-, 2-, 3-, 5 -protein systems including native protein attained stable conformation after 35 ns of the simulation time. However, after 110 ns the RMSD values of ligand 1 and 5 shifted towards the lower range (∼1.3 Å). Analysis of RMSD results revealed that Mpro-ligand (1, 2, 3, 5) complexes showed lower RMSD value in comparison to apo-protein except the RMSD value exhibited by 4-, and 6-protein systems. This observation indicates that the compounds 1, 2, 3 and 5 form stable protein-ligand complexes and does not make any considerable conformational change in the protein structure during simulation.
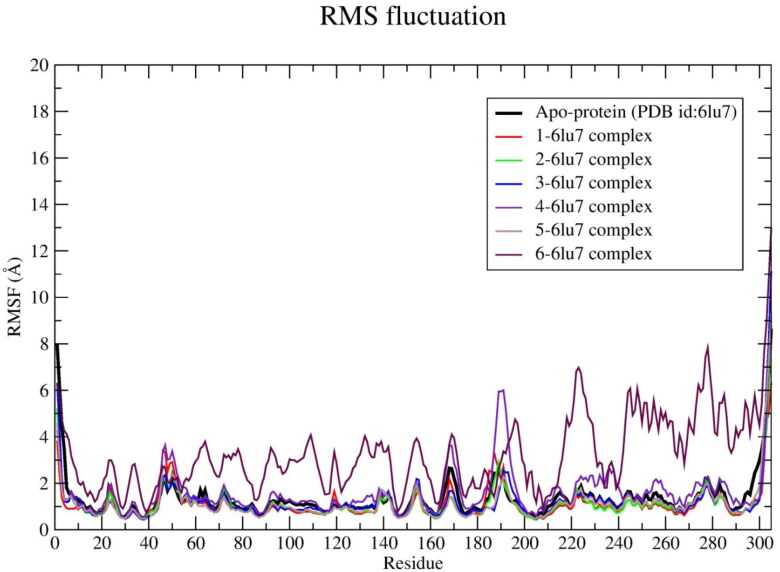
In order to explore the flexibility of individual amino acid residues of SARS-CoV-2 main protease RMSF was calculated from 130 ns MD trajectory as shown in Figure 4. The protein-ligand complexes of 1, 2, 3 and 5 exhibit lower RMSF values in comparison to apo-protein (Table 4). The lowest RMSF value (∼1.008 Å) was observed in case of ligand 5-protein system.
It was clearly observed that higher fluctuations were shown by each residue of the protein when bound with ligand 4 and 6, respectively (Figure 4). Residues like SER-46 and GLU-47 show slightly higher RMSF values in comparison with apo-protein’s SER-46, GLU-47 residues by 0.13 Å and 0.12 Å, respectively. The residue number PHE-185 shows slightly higher RMSF (∼0.12 Å) in 1-, 2-protein systems, respectively. However, these residues are present in the loop region of the protein, therefore, these values may disturb the binding interactions. The higher value of RMSF infers that the greater flexibility of the protein and vice versa (Chatterjee et al., 2020). Each protein-ligand system shows lower RMSF values than apo-protein. Only exceptions occur in case of 4- and 6-protein systems. These facts infer that the fluctuation of protein was minimized after binding with ligands 1, 2, 3, 5 and stable systems were achieved. In addition, high RMSF values were found for terminal residues. This may be due to the presence of terminal residues in the loop region. A similar observation in the RMSF plot was also reported by Muralidharan et al. (2020).
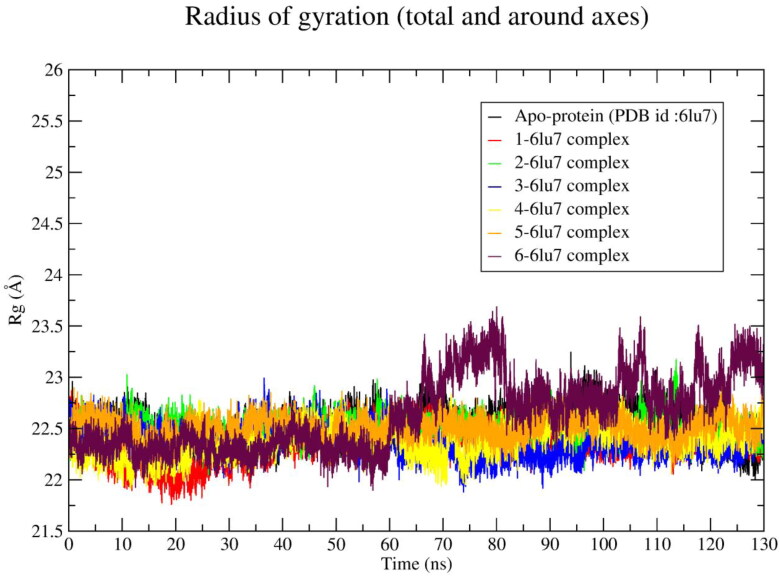
In order to inspect the compactness of the ligand bound protein Rg plot was constructed (Figure 5) and compared with the Rg value of apo-protein. The compactness of a protein was indicated by Rg value. Higher the Rg value indicates a distorted protein structure (Chatterjee et al., 2020).
Figure 5.
Radius of gyration of apo-protein and Mpro-ligand (1–6) complexes during 130 ns simulation time.
It was found that ligand 1–5 showed an average Rg value less than apo-protein Rg value (Table 4) except ligand 6. The highest Rg value (22.64 Å) was shown by ligand 6 and lowest Rg value (22.36 Å) was shown by ligand 1.
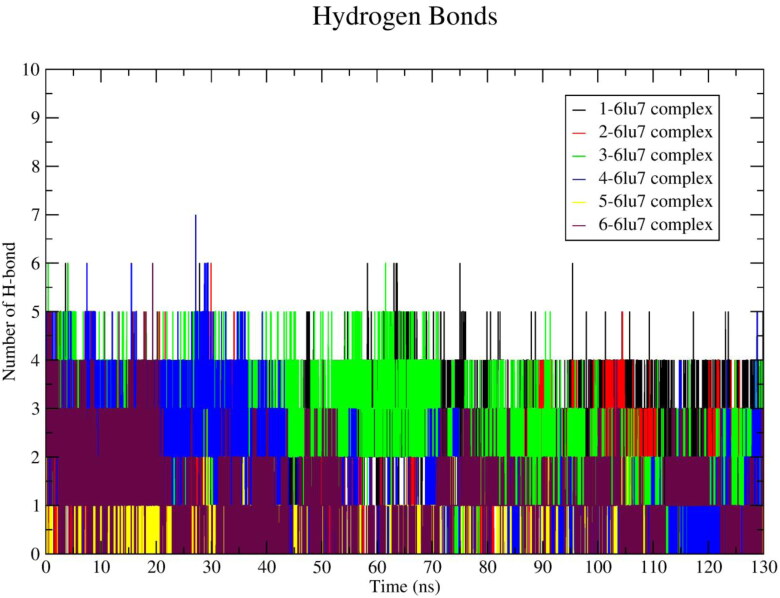
After 60 ns the Rg value of the 6-protein system was raised to ∼23.125 Å and rest of the simulation time it shows comparatively higher Rg value (∼23Å) than the apo-protein. The numbers of hydrogen bond interactions occurring in between protein and ligand was calculated and plotted against time in nanoseconds, shown in Figure 6. The average number of hydrogen bonds occurring in between protein and ligand is illustrated in Table 4.
Average ∼4, ∼1.7, ∼2.8, ∼2.34, ∼2.2, ∼2.24 numbers of hydrogen bonds were found in ligand 1, 2, 3, 4, 5, 6-protein complex, respectively. Ligand 3 exhibits the highest number of hydrogen bonds (∼3) at last 10 ns of MD simulation. The ligand 1 exhibited the highest average number of hydrogen bonds with lowest Rg values. The ligand 5 exhibits lowest RMSD, RMSF values and forms average ∼2.2 numbers of hydrogen bonds. Moreover, it depicts 0.01 Å less Rg value than apo-protein Rg value.
The binding affinity in terms of g_mmpbsa based binding energy of each ligands for the main protease enzyme were calculated from all frames of 130 ns MD trajectory as well as for the last 10 ns (that is 120‒130 ns) of molecular dynamics trajectory and are given in Table 4. The high negative binding energy infers greater stability of the protein-ligand complex system. Ligand 3 exhibits the highest binding energy (∼ ‒222.3 kJ/mol) when calculated from 130 ns molecular dynamics trajectory and shows second highest value (∼ ‒221.83 kJ/mol) when calculated from last 10 ns molecular dynamics trajectory with nominal (0.5 kJ/mol) difference. However, ligand 1 shows an average binding energy of ∼ ‒211.2 kJ/mol when calculated from 130 ns MD trajectory, which increases (high negative) at last, 10 ns and found to be ∼ ‒242.27 kJ/mol. Similar binding energy profiles were found for ligands 2 and 5. Whereas, the binding energy of ligand 4 (∼ ‒180 kJ/mol) and 6 (‒158.41 kJ/mol) were decreased to ∼ ‒137 kJ/mol and ∼ ‒139.08 kJ/mol, respectively, at last 10 ns.
The ligand 2-, 3-protein system exhibits acceptable RMSD, RMSF, Rg values. However, ligand 2 forms only ∼1.7 number of hydrogen bonds. In agreement with above information, it can be concluded that 1-, 2-, 3-, 5- ligand–protein systems show greater stability throughout 130 ns molecular dynamics simulation. Moreover, the binding energy analysis depicts that the ligand 1, 2, 5 shows high negative binding energy at last 10 ns. This finding is also evident that identified hits 1, 2, 3, 5 are present in the household spices which possess the superior affinity towards SARS-CoV-2 main protease enzyme.
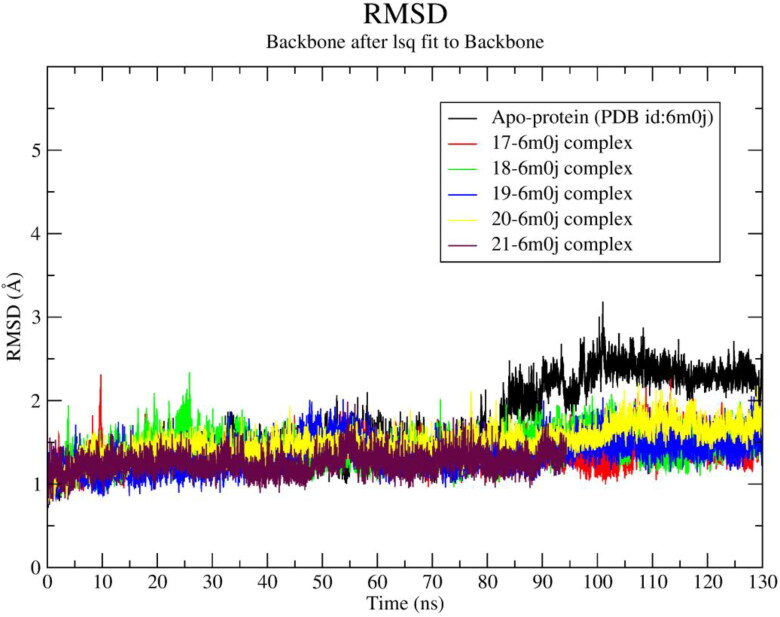
In order to analyze the stability of identified hit-spike protein complexes 130 ns MD simulation was conducted and various MD parameters were calculated from MD trajectory (Table 4). To inspect atomistic positions, the protein backbone RMSD of each ligand-spike protein complex was calculated and compared with native spike protein RMSD profile (Figure 7). It was clearly observed that ligand-protein systems of 17‒21 exhibit average ∼0.3 Å less RMSD value than apo-protein RMSD value (Table 4). Among all ligand-spike protein complexes the 20-spike protein system exhibits maximum average RMSD ∼1.5 Å, although it is also 0.20 Å less than apo-protein RMSD. The 18-spike protein system exhibits ∼0.16 Å higher RMSD than its average RMSD at 25 ns.
Figure 7.
RMSD of apo-protein backbone and ligand (17‒21)-spike protein complexes vs time plot.
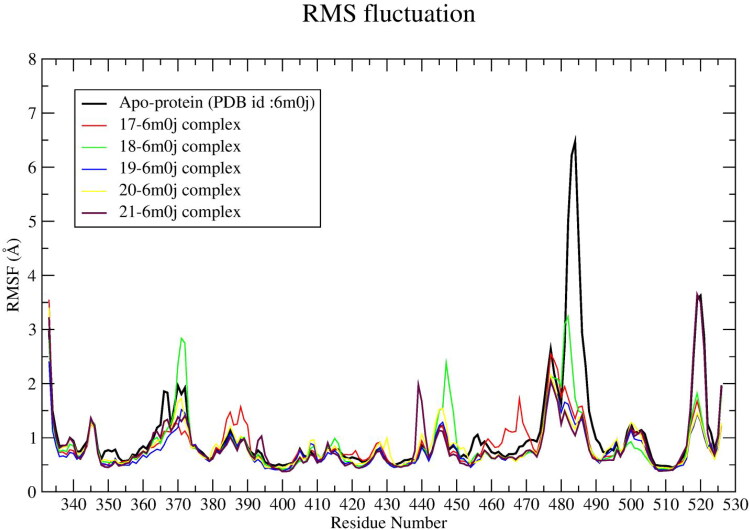
The RMSF parameters of ligand 17-, 18-, 19-, 20-, 21-spike protein systems were calculated from 130 ns MD trajectory, which is plotted against residue number (Figure 8). The average RMSF values of each system are given in Table 4. It was found that all the ligand–spike protein systems showed ∼ 0.218 Å lower RMSF value than apo-protein RMSF value.
Figure 8.
RMSF profile of apo-protein and ligand (17‒21)-spike protein complexes.
In comparison with apo-protein residue fluctuation, the SER-375, THR-376, PHE-377 residues in 18-spike protein complex exhibits 0.152, 0.206, 0.202 Å higher fluctuation, respectively. An average ∼0.0320 Å higher fluctuation depicted by SER-383, PRO-384, THR-385, LYS-386, LEU-387, ASN-388 and LEU-390 residues of 17-spike protein system than respective residues of apo-protein. The ASN-440 residue in the 21-spike protein system depicts 0.80 Å higher RMSF than apo-protein’s ASN-440. The LYS-444, VAL-445, GLY-446 residues of the 18-spike protein system exhibit ∼0.26 Å higher RMSF in comparison to the respective residues of apo-protein. The GLY-447 and ASN-448 shows 1.34 Å and 1.12 Å higher RMSF in comparison to the respective apo-protein residues. Moreover, in this system the amino acid residues lying in between ASN-460 to ALA-475 show 0.8 Å higher fluctuations than the respective residues of apo-protein. The ligand–spike protein system exhibits lower RMSD and RMSF values in comparison to apo-protein RMSD, RMSF values. The residues LYS-444, VAL-445 and GLY-446 show higher RMSF values although these residues do not reside in the receptor binding site. Moreover, LYS-444, VAL-445 and GLY-446 residues reside at the rear part of the binding site. Therefore, this high fluctuation may not interfere with the receptor binding affinity of the hits. The above observations are also supported by visual inspection of the trajectory.
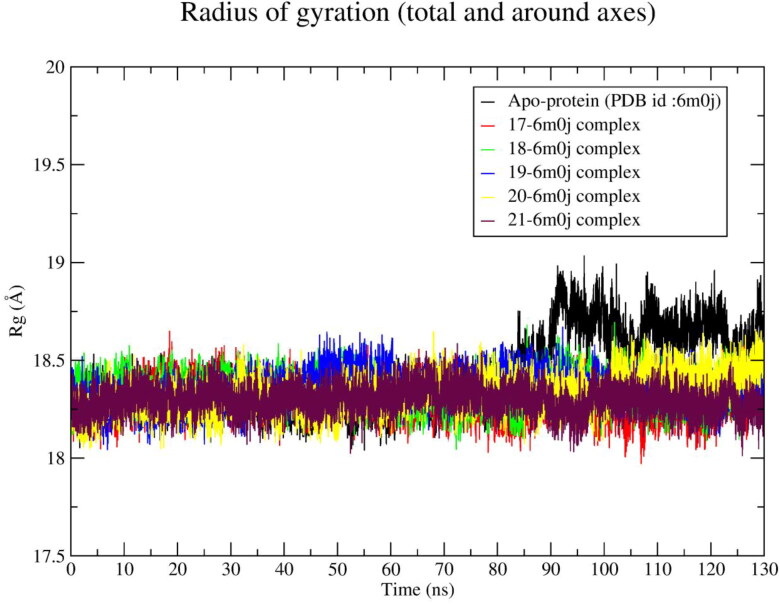
The radius of gyration (Rg) calculated from 130 ns MD trajectory and is plotted against time in nanosecond (Figure 9). The average values of Rg for each ligand-spike protein system along with apo-protein are given in Table 4. All the ligand-spike protein systems exhibit ∼0.24 Å lower Rg value than native protein Rg value (18.62 Å). It was observed that apo-protein system exhibits an average Rg of ∼18.76 Å after 90 ns (Figure 9). The Rg value of all ligand-spike protein systems was lower than the Rg value of apo-protein.
Figure 9.
Radius of gyration of apo-protein and spike receptor-ligand (17‒21) complexes during 130 ns simulation time.
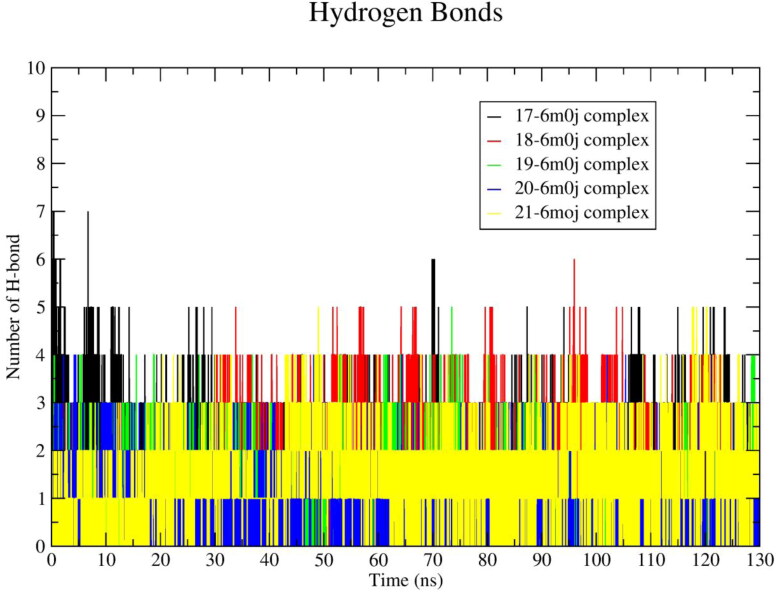
The hydrogen bonding interactions plays an important role in protein-ligand stability. In order to study the interactions, the average number of hydrogen bonds calculated from 130 ns MD trajectory and plotted against time (ns) are shown in Figure 10. The average ∼4.30, ∼3.00, ∼2.35, ∼1.10 and ∼1.41 numbers of hydrogen bonding interactions were found for ligand 17, 18, 19, 20 and 21, respectively. The results of the average number of hydrogen bonds formed in-between ligand and protein are shown in Table 4.
Figure 10.
Number of average hydrogen bonding interactions between spike receptor–ligand (17‒21) systems during 130 ns simulation time.
In order to analyze the affinity of the ligands towards SARS-CoV-2 spike protein, the binding energy of each ligand-spike protein complexes were calculated from all the frames of 130 ns MD trajectory. To calculate the binding energy of each system all snapshots from 120 to 130 ns were saved and used. The average binding energies with standard deviation for each system are illustrated in Table 4. The ligand 20 showed an average binding energy of ∼ ‒163.10 kJ/mol when calculated from 130 ns trajectory, which was increased by ∼ 36.92 kJ/mol in the last 10 ns (‒200.02 kJ/mol). Similarly, ligand 18 showed an average binding energy of ∼ ‒167.15 kJ/mol from 130 ns and ∼ ‒175.02 kJ/mol for last the 10 ns. The ligand 21 also showed a similar binding energy profile with ∼ ‒168.5 kJ/mol from 130 ns and ∼ ‒182.13 kJ/mol from last 10 ns. The ligand 17 showed an average binding energy of ∼ ‒177.43 kJ/mol calculated from130 ns MD trajectory and ‒176.41 kJ/mol for the last 10 ns. The drop of energy was found to be ‒1.02 kJ/mol for ligand 17. The ligand 19 exhibits binding energy ∼ ‒172.4 kJ/mol from 130 ns which was dropped around ‒3.7 kJ/mol in the last 10 ns and found to be ‒168.71 kJ/mol. All the ligands except ligand 19, exhibit high negative binding energy at the last 10 ns of simulation time. All the ligands showed lower Rg values than apo-protein. The Rg value of ligand 19 was lowest (18.25 Å). It was clearly identified that the Rg value of the 20-spike protein complex was slightly higher (0.008 Å), although it seems to be in acceptable range. The ligand 20 shows high negative binding energy at last 10 ns of simulation time. In agreement with the above findings, it can be concluded that the hits 17, 18, 19, 20, 21 showed stable binding affinities with SARS-CoV-2 spike protein.
One of the important parameters in drug discovery is ADME. The QikProp predicts the properties and descriptors of organic molecules by comparing with 95% of known drugs. According to the Lipinski’s rule, a molecule will be a drug like when molecular weight is < 500, octanol-water partition coefficient (QplogPo/w) should be < 5.0, hydrogen bond donor groups (donorHB) should be < 5.0, and hydrogen bond acceptor groups (accptHB) should be ≤ 10. All the identified potential hits follow Lipinski’s rule of five. The other important ADME properties like CNS, QplogS, QPPCaco, QplogBB, QPPMDCK, HOA, PHOA, ROF, ROT, etc. are presented in Table 5. The ADME properties of all the selected hits lie in the acceptable range and, therefore, all the selected hits are drug like. The structures of the selected potential hits are shown in Figure 11.
Table 5.
Predicted important ADMET properties of the eleven hits with their recommended values (in the table abbreviations are, RV: Recommended values; PHOA: Percent Human Oral Absorption; ROF: Rule of Five; ROT: Rule of Three; HOA: Human Oral Absorption; HB: hydrogen bond).
| Hits | #stars | CNS | mol_MW | donorHB | accptHB | QPlogPo/w | QPlogS |
|---|---|---|---|---|---|---|---|
| 1/18 | 0 | −2 | 302.240 | 4 | 5.25 | 0.358 | −2.893 |
| 2 | 0 | −2 | 290.272 | 5 | 6.40 | 0.300 | −2.510 |
| 3 | 0 | −2 | 288.256 | 3 | 5.70 | 0.748 | −2.917 |
| 4 | 0 | −2 | 446.496 | 3 | 7 | 3.444 | −4.218 |
| 5 | 0 | −1 | 376.900 | 3 | 4 | 4.315 | −5.977 |
| 6 | 0 | −2 | 386.401 | 2 | 5.45 | 2.938 | −3.668 |
| 17 | 1 | −2 | 318.239 | 5 | 6 | −0.307 | −2.656 |
| 19 | 0 | −2 | 302.240 | 4 | 5.25 | 0.244 | −2.780 |
| 20 | 1 | −2 | 374.433 | 2 | 5.70 | 3.600 | −5.101 |
| 21 | 0 | −2 | 286.240 | 3 | 4.50 | 0.913 | −3.030 |
| AR | 0–5 | −2(inactive) +2(active) |
130.0–725.0 | 0.0–6.0 | 2.0–20.0 | −2.0–6.5 | −6.5–0.5 |
| Hits | QPPCaco | QPlogBB | QPPMDCK | HOA | PHOA | ROF | ROT |
| 1/18 | 18.193 | −2.414 | 6.509 | 2 | 51.591 | 0 | 1 |
| 2 | 67.663 | −1.777 | 26.921 | 3 | 61.464 | 0 | 0 |
| 3 | 61.445 | −1.768 | 25.257 | 3 | 63.338 | 0 | 0 |
| 4 | 106.853 | −2.23 | 44.115 | 2 | 83.423 | 0 | 0 |
| 5 | 825.397 | −0.602 | 1642.128 | 3 | 100.00 | 0 | 1 |
| 6 | 76.911 | −2.395 | 30.92 | 2 | 77.903 | 0 | 0 |
| 17 | 6.562 | −2.939 | 2.162 | 2 | 26.815 | 1 | 1 |
| 19 | 14.773 | −2.449 | 5.197 | 2 | 49.306 | 0 | 1 |
| 20 | 182.04 | −2.218 | 78.465 | 3 | 88.476 | 0 | 1 |
| 21 | 40.797 | −1.942 | 15.581 | 3 | 61.115 | 0 | 0 |
| AR | <25 poor >500 great |
−3.0–1.2 | <25 poor >500 great |
1-low 2-medium 3-high |
>80% is high <25% is poor |
max 4 | max 3 |
Description of terms, CNS: Predicted central nervous system permeability; QPlogPo/w: Predicted octanol/water partition coefficient; QPlogS: Predicted aqueous solubility; QPPCaco: Predicted Caco-2 cell permeability in nm/s; QPlogBB: Predicted brain/blood partition coefficient; and QPPMDCK: Predicted apparent MDCK cell permeability in nm/s.
Figure 11.
Potential inhibitors of Mpro and Spike receptors of SARS-CoV-2.
4. Conclusion
The virtual screenings of in-house databank containing 1040 number of natural compounds retrieved from literature of readily available spices using Glide was performed. The best hits resulted from VS were subjected to molecular docking with AutoDock 4.2 and AutoDock Vina to eliminate false positive. The molecular docking study resulted six potential compounds (1–6) having good binding affinity towards active site amino acid residues of main protease (Mpro) and five compounds (17‒21) with good binding affinity towards active site amino acid residues of spike receptor of SARS-CoV-2. Finally, the 130 ns atomistic molecular dynamic simulation of two sets of selected hits with main protease and spike receptor were performed. Molecular dynamic simulation unveils that amongst the six hits, hits 1, 2, 3 and 5 formed stable complexes with Mpro throughout the simulation time. Similarly, the compounds 17, 18, 19, 20 and 21 formed stable complexes with spike receptor of SARS-CoV-2 and shows superior binding affinity during MD. The potential hits against main protease are 1 (Quercetin, present in Onion, Garlic, Peppermint, Fenugreek), 2 (Leucopelargonidin, present in Onion), 3 (Aronadendin, present in Onion) and 5 (Capsazepine, present in Chilli), identified on the basis of docking score predicted by three different softwares, MD simulation study, binding energy calculation and ADME study. The present studies have also identified five potential hits 17 (Myricetin, available in Onion, Chilli), 18 (Quercetin, available in Onion), 19 (Tricetin, available in Onion), 20 (5S-5-Hydroxy-1,7-bis(4-hydroxy-3-methoxyphenyl)-3-heptanone, available in Ginger) and 21 (Luteolin, available in Fenugreek) against SARS-CoV-2 spike protein. The predicted ADME properties of the selected hits are also within the acceptable range akin to 95% known drugs and supported the drug like nature of these molecules. All the selected spices are readily available and edible. Based on the above findings, it is hypothesized that under this pandemic situation the spices Onion, Garlic, Peppermint, Fenugreek, Chilli and Ginger might be used as precautionary home remedy against COVID-19 after appropriate biological screening.
Supplementary Material
Acknowledgments
The authors are thankful to Schrodinger, Bangalore, India for providing Software support for this research work. The authors are also thankful to Mrs. Sarbari Nath, Associate Professor, Department of English, Women’s College, Agartala, Tripura, India for English corrections.
Glossary
Abbreviations
- 3CLpro
3-Chymotrypsin-like Cysteine Protease
- ACE2
Angiotensin-Converting Enzyme II
- ADME
absorption, distribution, metabolism, and excretion
- CASTp
Computed Atlas of Surface Topography of proteins
- COVID-19
Novel Coronavirus Disease 2019
- CoVs
Coronaviruses
- FDA
Food and Drug Administration
- GROMACS
GROningen MAchine for Chemical Simulations
- HCoV-OC43
Human Coronavirus Strains OC43
- HIE
histidine with hydrogen on the epsilon nitrogen
- HTVS
high throughput virtual screening
- MD
molecular dynamics
- MERS
Middle East Respiratory Syndrome
- MM-PBSA
molecular mechanics Poisson-Boltzmann surface area
- Mpro
main protease
- nsP
non-structural proteins
- PBC
periodic boundary condition
- PLpro
papain-like protease
- PME
particle mesh Eshwald
- pp1a
polyproteins 1a
- pp1ab
polyproteins 1ab
- RdRp
RNA-dependent RNA polymerase
- Rg
radius of gyration
- RMSD
root mean square deviation
- RMSF
root mean square fluctuation
- S
spike
- SARS-CoV-2
Severe Acute Respiratory Syndrome Coronavirus 2
- SP
standard precision
- VMD
visual molecular dynamics
- WHO
World Health Organization
- XP
extra precision.
Disclosure statement
The authors declare that they have no competing interests.
References
- Aanouz, I., Belhassan, A., El-Khatabi, K., Lakhlifi, T., El-Ldrissi, M., & Bouachrine, M. (2020). Moroccan medicinal plants as inhibitors against SARS-CoV-2 main protease: Computational investigations. Journal of Biomolecular Structure & Dynamics, 1–9. 10.1080/07391102.2020.1758790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham, M. J., Murtola, T., Schulz, R., Páll, S., Smith, J. C., Hess, B., & Lindah, E. (2015). GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX, 1-2, 19–25. 10.1016/j.softx.2015.06.001 [DOI] [Google Scholar]
- Ahmad, A., Husain, A., Mujeeb, M., Khan, S. A., Najmi, A. K., Siddique, N. A., Damanhouri, Z. A., & Anwar, F. (2013). A review on therapeutic potential of Nigella sativa: A miracle herb. Asian Pacific Journal of Tropical Biomedicine, 3(5), 337–352. 10.1016/S2221-1691(13)60075-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin, G. (2012). Cumin. In Peter K. V. (Ed.), Handbook of herbs and spices (pp. 250–259). CRC Press. [Google Scholar]
- Antonio, A. D., Wiedemann, L. S., & Júnior, V. F. (2018). The genus Capsicum: A phytochemical review of bioactive secondary metabolites. RSC Advances, 8(45), 25767–25784. 10.1021/np300898z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Báez-Santos, Y. M., St John, S. E., & Mesecar, A. D. (2015). The SARS-coronavirus papain-like protease: Structure, function and inhibition by designed antiviral compounds. Antiviral Research, 115, 21–38. 10.1016/j.antiviral.2014.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch, B. J., van der Zee, R., de Haan, C. A., & Rottier, P. J. (2003). The coronavirus spike protein is a class I virus fusion protein: Structural and functional characterization of the fusion core complex. Journal of Virology, 77(16), 8801–8811. 10.1128/jvi.77.16.8801-8811.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers, K. J., Chow, D. E., Xu, H., Dror, R. O., Eastwood, M. P., Gregersen, B. A., Klepeis, J. L., Kolossvary, I., Moraes, M. A., Sacerdoti, F. D., & Salmon, J. K. (2006). Scalable algorithms for molecular dynamics simulations on commodity clusters [Paper presentation]. SC'06: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing ( pp. 43–43). 10.1109/SC.2006.54 [DOI] [Google Scholar]
- Brahmi, F., Khodir, M., Mohamed, C., & Pierre, D. (2017). Chemical composition and biological activities of Mentha species. In El-Shemy H. (Ed.), Aromatic and medicinal plants - Back to nature (pp. 47–79). London: IntechOpen. 10.5772/67291 [DOI] [Google Scholar]
- Bystricka, J., Musilova, J., Vollmannova, A., Timoracka, M., & Kavalcova, P. (2013). Bioactive components of onion (Allium cepa L.) – A review. Acta Alimentaria, 42(1), 11–22. [Google Scholar]
- Chatterjee, S., Maity, A., Chowdhury, S., Islam, M. A., Muttinini, R. K., & Sen, D. (2020). In silico analysis and identification of promising hits against 2019 novel coronavirus 3C-like main protease enzyme. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1787228 [DOI] [PubMed] [Google Scholar]
- Chen, C., Zhang, Y., Huang, J., Yin, P., Cheng, Z., Wu, J., Chen, S., Zhang, Y., Chen, B., Lu, M., Luo, Y., Ju, L., Zhang, J., & Wang, X. (2020). Favipiravir versus Arbidol for COVID-19: A randomized clinical trial. medRxiv Preprint, 10.1101/2020.03.17.20037432 [DOI] [Google Scholar]
- Chen, Z., & Nakamura, T. (2004). Statistical evidence for the usefulness of Chinese medicine in the treatment of SARS. Phytotherapy Research, 18(7), 592–594. 10.1002/ptr.1485 [DOI] [PubMed] [Google Scholar]
- Cheng, V. C., Lau, S. K., Woo, P. C., & Yuen, K. Y. (2007). Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clinical Microbiology Reviews, 20(4), 660–694. 10.1128/CMR.00023-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson, P., Rolain, J. M., Lagier, J. C., Brouqui, P., & Raoult, D. (2020). Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. International Journal of Antimicrobial Agents, 55(4), 105932. 10.1016/j.ijantimicag.2020.105932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ee, G. C., Lim, C. M., Lim, C. K., Rahmani, M., Shaari, K., & Bong, C. F. (2009). Alkaloids from Piper sarmentosum and Piper nigrum . Natural Product Research, 23(15), 1416–1423. 10.1080/14786410902757998 [DOI] [PubMed] [Google Scholar]
- Elfiky, A. A. (2020). SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: An in silico perspective. Journal of Biomolecular Structure & Dynamics, 1–9. 10.1080/07391102.2020.1761882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enmozhi, S. K., Raja, K., Sebastine, I., & Joseph, J. (2020). Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: An in silico approach. Journal of Biomolecular Structure & Dynamics, 1–7. 10.1080/07391102.2020.1760136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag, M. A., Ali, S. E., Hodaya, R. H., El-Seedi, H. R., Sultani, H. N., Laub, A., Eissa, T. F., Abou-Zaid, F., & Wessjohann, L. A. (2017). Phytochemical profiles and antimicrobial activities of Allium cepa Red cv. and A. sativum subjected to different drying methods: A comparative MS-based metabolomics. Molecules, 22(5), 761. 10.3390/molecules22050761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr, A. R., Jankevicius, G., Ahel, I., & Perlman, S. (2018). Viral macrodomains: Unique mediators of viral replication and pathogenesis. Trends in Microbiology, 26(7), 598–610. 10.1016/j.tim.2017.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, T., Su, J., Ding, Z. H., Zheng, Y. T., Li, Y., Leng, Y., & Liu, J. K. (2011). Chemical constituents and their bioactivities of “Tongling White Ginger” (Zingiber officinale). Journal of Agricultural and Food Chemistry, 59(21), 11690–11695. 10.1021/jf202544w [DOI] [PubMed] [Google Scholar]
- Fischer, A., Sellner, M., Neranjan, S., Lill, M. A., & Smieško, M. (2020). Inhibitors for novel coronavirus protease identified by virtual screening of 687 million compounds. ChemRxiv Preprint, 10.26434/chemrxiv.11923239.v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner, R. A., Banks, J. L., Murphy, R. B., Halgren, T. A., Klicic, J. J., Mainz, D. T., Repasky, M. P., Knoll, E. H., Shelley, M., Perry, J. K., Shaw, D. E., Francis, P., & Shenkin, P. S. (2004). Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. Journal of Medicinal Chemistry, 47(7), 1739–1749. 10.1021/jm0306430 [DOI] [PubMed] [Google Scholar]
- Friesner, R. A., Murphy, R. B., Repasky, M. P., Frye, L. L., Greenwood, J. R., Halgren, T. A., Sanschagrin, P. C., & Mainz, D. T. (2006). Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. Journal of Medicinal Chemistry, 49(21), 6177–6196. 10.1021/jm051256o [DOI] [PubMed] [Google Scholar]
- Gachkar, L., Yadegari, D., Rezaei, M. B., Taghizadeh, M., Astaneh, S. A., & Rasooli, I. (2007). Chemical and biological characteristics of Cuminum cyminum and Rosmarinus officinalis essential oils. Food Chemistry, 102(3), 898–904. 10.1016/j.foodchem.2006.06.035 [DOI] [Google Scholar]
- Gohari, A. R., & Saeidnia, S. (2011). A review on phytochemistry of Cuminum cyminum seeds and its standards from field to market. Pharmacognosy Journal, 3(25), 1–5. 10.5530/pj.2011.25.1 [DOI] [Google Scholar]
- Hajlaoui, H., Mighri, H., Noumi, E., Snoussi, M., Trabelsi, N., Ksouri, R., & Bakhrouf, A. (2010). Chemical composition and biological activities of Tunisian Cuminum cyminum L. essential oil: A high effectiveness against Vibrio spp. strains. Food and Chemical Toxicology : An International Journal Published for the British Industrial Biological Research Association, 48(8-9), 2186–2192. 10.1016/j.fct.2010.05.044 [DOI] [PubMed] [Google Scholar]
- Halgren, T. A., Murphy, R. B., Friesner, R. A., Beard, H. S., Frye, L. L., Pollard, W. T., & Banks, J. L. (2004). Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. Journal of Medicinal Chemistry, 47(7), 1750–1759. 10.1021/jm030644s [DOI] [PubMed] [Google Scholar]
- Han, Y., Nishibe, S., Noguchi, Y., & Jin, Z. (2001). Flavonol glycosides from the stems of Trigonella foenum-graecum. Phytochemistry, 58(4), 577–580. 10.1016/S0031-9422(01)00273-4 [DOI] [PubMed] [Google Scholar]
- Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., Schiergens, T. S., Herrler, G., Wu, N. H., Nitsche, A., Müller, M. A., Drosten, C., & Pöhlmann, S. (2020). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell, 181(2), 271–280. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., Zhang, L., Fan, G., Xu, J., Gu, X., Cheng, Z., Yu, T., Xia, J., Wei, Y., Wu, W., Xie, X., Yin, W., Li, H., Liu, M., … Cao, B. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet, 395(10223), 497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey, W., Dalke, A., & Schulten, K. (1996). VMD: Visual molecular dynamics. Journal of Molecular Graphics, 14(1), 33–38. 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Imran, M., Butt, M. S., & Suleria, H. A. R. (2018). Capsicum annuum Bioactive compounds: Health promotion perspectives. In Mérillon J. M., & Ramawat K. (Eds.), Bioactive molecules in food. Reference series in phytochemistry (pp 1–22). Springer. 10.1007/978-3-319-54528-8_47-1 [DOI] [Google Scholar]
- Islam, M. T., Khan, M. R., & Mishra, S. K. (2019). An updated literature-based review: Phytochemistry, pharmacology and therapeutic promises of Nigella sativa L. Oriental Pharmacy and Experimental Medicine, 19(2), 115–129. 10.1007/s13596-019-00363-3 [DOI] [Google Scholar]
- Islam, R., Parves, M. R., Paul, A. S., Uddin, N., Rahman, M. S., Mamun, A. A., Hossain, M. N., Ali, M. A., & Halim, M. A. (2020). A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. Journal of Biomolecular Structure & Dynamics, 1–12. 10.1080/07391102.2020.1761883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israeli, E. (2020). Novel coronavirus that recently emerged in China. Harefuah, 159(1), 70–71. PMID: 32048481 [PubMed] [Google Scholar]
- Jiang, F., Deng, L., Zhang, L., Cai, Y., Cheung, C. W., & Xia, Z. (2020). Review of the clinical characteristics of Coronavirus Disease 2019 (COVID-19). Journal of General Internal Medicine, 35(5), 1545–1549. 10.1007/s11606-020-05762-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., Zhang, B., Li, X., Zhang, L., Peng, C., Duan, Y., Yu, J., Wang, L., Yang, K., Liu, F., Jiang, R., Yang, X., You, T., Liu, X., … Yang, H. (2020). Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature, 582, 289–293. 10.1038/s41586-020-2223-y [DOI] [PubMed] [Google Scholar]
- Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W., & Klein, M. L. (1983). Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics, 79(2), 926–935. 10.1063/1.445869 [DOI] [Google Scholar]
- Kamel, A., & Saleh, M. (2000). Recent studies on the chemistry and biological activities of the organosulfur compounds of garlic (Allium sativum). In Atta-ur-Rahman (Ed.), Studies in natural products chemistry (Vol. 23, pp. 455–485). 10.1016/S1572-5995(00)80135-0 [DOI] [Google Scholar]
- Khaerunnisa, S., Kurniawan, H., Awaluddin, R., Suhartati, S., & Soetjipto, S. (2020). Potential inhibitor of COVID-19 main protease (Mpro) from several medicinal plant compounds by molecular docking study. Preprints, 2020030226. 10.20944/preprints202003.0226.v1 [DOI] [Google Scholar]
- Kim, D. E., Min, J. S., Jang, M. S., Lee, J. Y., Shin, Y. S., Park, C. M., Song, J. H., Kim, H. R., Kim, S., Jin, Y.-H., & Kwon, S. (2019). Natural bis-benzylisoquinoline alkaloids-tetrandrine, fangchinoline, and cepharanthine, inhibit human Coronavirus OC43 infection of MRC-5 human lung cells. Biomolecules, 9(11), 696–1012. https://doi:10.3390/biom9110696 10.3390/biom9110696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokoska, L., Havlik, J., Valterova, I., Sovova, H., Sajfrtova, M., & Jankovska, I. (2008). Comparison of chemical composition and antibacterial activity of Nigella sativa seed essential oils obtained by different extraction methods. Journal of Food Protection, 71(12), 2475–2480. 10.4315/0362-028x-71.12.2475 [DOI] [PubMed] [Google Scholar]
- Kufareva, I., & Abagyan, R. (2012). Methods of protein structure comparison. Methods in Molecular Biology (Clifton, N.J.), 857, 231–257. 10.1007/978-1-61779-588-6_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari, R., Kumar, R., & Lynn, A, Open Source Drug Discovery Consortium. (2014). g_mmpbsa-a GROMACS tool for high-throughput MM-PBSA calculations. Journal of Chemical Information and Modeling, 54(7), 1951–1962. 10.1021/ci500020m [DOI] [PubMed] [Google Scholar]
- Lan, J., Ge, J., Yu, J., Shan, S., Zhou, H., Fan, S., Zhang, Q., Shi, X., Wang, Q., Zhang, L., & Wang, X. (2020). Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature, 581(7807), 215–220. 10.1038/s41586-020-2180-5 [DOI] [PubMed] [Google Scholar]
- Lanzotti, V. (2012). Bioactive polar natural compounds from garlic and onions. Phytochemistry Reviews, 11(2-3), 179–196. 10.1007/s11101-012-9247-3 [DOI] [Google Scholar]
- Lee, N., Hui, D., Wu, A., Chan, P., Cameron, P., Joynt, G. M., Ahuja, A., Yung, M. Y., Leung, C. B., To, K. F., Lui, S. F., Szeto, C. C., Chung, S., & Sung, J. J. (2003). A major outbreak of severe acute respiratory syndrome in Hong Kong. The New England Journal of Medicine, 348(20), 1986–1994. 10.1056/NEJMoa030685 [DOI] [PubMed] [Google Scholar]
- Li, G., & De Clercq, E. (2020). Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nature Reviews. Drug Discovery, 19(3), 149–150. 10.1038/d41573-020-00016-0 [DOI] [PubMed] [Google Scholar]
- Li, R., & Jiang, Z. T. (2004). Chemical composition of the essential oil of Cuminum cyminum L. from China. Flavour and Fragrance Journal, 19(4), 311–313. 10.1002/ffj.1302 [DOI] [Google Scholar]
- Li, W., Moore, M. J., Vasilieva, N., Sui, J., Wong, S. K., Berne, M. A., Somasundaran, M., Sullivan, J. L., Luzuriaga, K., Greenough, T. C., Choe, H., & Farzan, M. (2003). Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature, 426(6965), 450–454. 10.1038/nature02145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Zhan, J., Wang, N., Li, H., Shi, Y., Guo, G., Liu, K., Zeng, H., & Zou, Q. (2020). Therapeutic drugs targeting 2019-nCoV main protease by high-throughput screening. bioRxiv, 10.1101/2020.01.28.922922 [DOI] [Google Scholar]
- Liu, W., Zhu, H. L., & Duan, Y. (2020). Effective chemicals against novel coronavirus (COVID-19) in China. Current Topics in Medicinal Chemistry, 20(8), 603–605. 10.2174/1568026620999200305145032 [DOI] [PubMed] [Google Scholar]
- Liu, X., Zhang, B., Jin, Z., Yang, H., Rao, Z. (2020). The crystal structure of COVID-19 main protease in complex with an inhibitor N3. RCSB Protein Data Bank. https://doi.org/10.2210/pdb6LU7/pdb
- Mahboubi, M. (2019). Zingiber officinale Rosc. Essential oil, a review on its composition and bioactivity. Clinical Phytoscience, 5(1), 1–12. 10.1186/s40816-018-0097-4 [DOI] [Google Scholar]
- Mandal, M., & Mandal, S. (2016). Fenugreek (Trigonellafoenum-graecum L.) oils. In Preedy V. R. (Ed.), Essential oils in food preservation, flavor and safety (pp 421–429). Academic Press. 10.1016/B978-0-12-416641-7.00047-X [DOI] [Google Scholar]
- Martins, N., Petropoulos, S., & Ferreira, I. C. (2016). Chemical composition and bioactive compounds of garlic (Allium sativum L.) as affected by pre- and post-harvest conditions: A review. Food Chemistry, 211, 41–50. 10.1016/j.foodchem.2016.05.029 [DOI] [PubMed] [Google Scholar]
- Martyna, G. J., Tobias, D. J., & Klein, M. L. (1994). Constant pressure molecular dynamics algorithms. The Journal of Chemical Physics, 101(5), 4177–4189. 10.1063/1.467468 [DOI] [Google Scholar]
- Meng, F. C., Zhou, Y. Q., Ren, D., Wang, R., Wang, C., Lin, L. G., Zhang, X. Q., Ye, W. C., & Zhang, Q. W. (2018). Turmeric: A review of its chemical composition, quality control, bioactivity, and pharmaceutical application. InGrumezescu A. M. & Holban A. M. (Eds.), Natural and artificial flavoring agents and food dyes (pp. 299–350). Academic Press. 10.1016/B978-0-12-811518-3.00010-7 [DOI] [Google Scholar]
- Miller, B. R., McGee, T. D., Swails, J. M., Homeyer, N., Gohlke, H., & Roitberg, A. E. (2012). MMPBSA.py: An efficient program for end-state free energy calculations. Journal of Chemical Theory and Computation, 8(9), 3314–3321. 10.1021/ct300418h [DOI] [PubMed] [Google Scholar]
- Mohd, H. A., Al-Tawfiq, J. A., & Memish, Z. A. (2016). Middle East Respiratory Syndrome Coronavirus (MERS-CoV) origin and animal reservoir. Virology Journal, 13, 87. 10.1186/s12985-016-0544-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., & Olson, A. J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30(16), 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtar, H., Qureshi, A. S., Anwar, F., Mumtaz, M. W., & Marcu, M. (2019). Nigella sativa L. seed and seed oil: Potential sources of high-value components for development of functional foods and nutraceuticals/pharmaceuticals. Journal of Essential Oil Research, 31(3), 171–183. 10.1080/10412905.2018.1562388 [DOI] [Google Scholar]
- Muralidharan, N., Sakthivel, R., Velmurugan, D., & Gromiha, M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. Journal of Biomolecular Structure and Dynamics, 10.1080/07391102.2020.1752802 [DOI] [PubMed] [Google Scholar]
- Omezzine, F., Bouaziz, M., Daami-Remadi, M., Simmonds, M. S. J., & Haouala, R. (2017). Chemical composition and antifungal activity of Trigonellafoenum-graecum L. varied with plant ploidy level and developmental stage. Arabian Journal of Chemistry, 10, S3622–S3631. 10.1016/j.arabjc.2014.03.013 [DOI] [Google Scholar]
- Oroojalian, F., Oroojalian, F., Kasra-Kermanshahi, R., Azizi, M., & Bassami, M. R. (2010). Phytochemical composition of the essential oils from three Apiaceae species and their antibacterial effects on food-borne pathogens. Food Chemistry, 120(3), 765–770. 10.1016/j.foodchem.2009.11.008 [DOI] [Google Scholar]
- Pancharoen, O., Prawat, U., & Tutiwachwuttikul, P. (2000). Phytochemistry of the zingiberaceae bioactive natural products (Part D). In Rahman A. (Ed.), Studies in natural products chemistry (Vol. 23, pp. 797–865). Amsterdam, Lausanne, New York: Elsevier. [Google Scholar]
- Pillaiyar, T., Meenakshisundaram, S., & Manickam, M. (2020). Recent discovery and development of inhibitors targeting coronaviruses. Drug Discovery Today, 25(4), 668–688. 10.1016/j.drudis.2020.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramajayam, R., Tan, K. P., Liu, H. G., & Liang, P. H. (2010). Synthesis and evaluation of pyrazolone compounds as SARS-coronavirus 3C-like protease inhibitors. Bioorganic and Medicinal Chemistry, 18(22), 7849–7854. 10.1016/j.bmc.2010.09.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasheed, M., Afshan, F., Tariq, R. M., Siddiqui, B. S., Gulzar, T., Mahmood, A., Begum, S., & Khan, B. (2005). Phytochemical studies on the seed extract of Piper nigrum Linn. Natural Product Research, 19(7), 703–712. 10.1080/14786410512331330657 [DOI] [PubMed] [Google Scholar]
- Reddy, D. N., Al-Rajab, A. J., Sharma, M., Moses, M. M., Reddy, G. R., & Albratty, M. (2019). Chemical constituents, in vitro antibacterial and antifungal activity of Mentha piperita L. (peppermint) essential oils. Journal of King Saud University - Science, 31(4), 528–533. 10.1016/j.jksus.2017.07.013 [DOI] [Google Scholar]
- Saharkhiz, M. J., Motamedi, M., Zomorodian, K., Pakshir, K., Miri, R., & Hemyari, K. (2012). Chemical composition, antifungal and antibiofilm activities of the essential oil of Mentha piperita L. ISRN Pharmaceutics, 2012, 718645. 10.5402/2012/718645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi, B., Zakaria, Z. A., Gyawali, R., Ibrahim, S. A., Rajkovic, J., Shinwari, Z. K., Khan, T., Sharifi-Rad, J., Ozleyen, A., Turkdonmez, E., Valussi, M., Tumer, T. B., Monzote Fidalgo, L., Martorell, M., & Setzer, W. N. (2019). Piper species: A comprehensive review on their phytochemistry, biological activities and applications. Molecules, 24(7), 1364. 10.3390/molecules24071364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampangi-Ramaiah, M. H., Vishwakarma, R., & Shaanker, R. U. (2020). Molecular docking analysis of selected natural products from plants for inhibition of SARS-CoV-2 main protease. Current Science, 118(7), 1087–1092. [Google Scholar]
- Sanders, J. M., Monogue, M. L., Jodlowski, T. Z., & Cutrell, J. B. (2020). Pharmacologic treatments for coronavirus disease 2019 (COVID-19): A review. JAMA - Journal of the American Medical Association, 323(18), 1824–1836. 10.1001/jama.2020.6019 [DOI] [PubMed] [Google Scholar]
- Sastry, G. M., Adzhigirey, M., Day, T., Annabhimoju, R., & Sherman, W. (2013). Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments . Journal of Computer Aided Molecular Design, 27(3), 221–234. 10.1007/s10822-013-9644-8 [DOI] [PubMed] [Google Scholar]
- Schreiner, W., Karch, R., Knapp, B., & Ilieva, N. (2012). Relaxation estimation of RMSD in molecular dynamics immunosimulations. Computational and Mathematical Methods in Medicine, 2012, 173521. 10.1155/2012/173521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R. P., Gangadharappa, H. V., & Mruthunjaya, K. (2017). Cuminum cyminum– A popular spice: An updated review. Pharmacognosy Journal, 9(3), 292–301. 10.5530/pj.2017.3.51 [DOI] [Google Scholar]
- Soković, M. D., Vukojević, J., Marin, P. D., Brkić, D. D., Vajs, V., & van Griensven, L. J. (2009). Chemical composition of essential oils of Thymus and Mentha species and their antifungal activities. Molecules (Basel, Switzerland), 14(1), 238–249. 10.3390/molecules14010238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan, K. (2018). Cumin (Cuminum cyminum) and black cumin (Nigella sativa) seeds: Traditional uses, chemical constituents, and nutraceutical effects. Food Quality and Safety, 2(1), 1–16. 10.1093/fqsafe/fyx031 [DOI] [Google Scholar]
- Su, H., Yao, S., Zhao, W., Li, M., Liu, J., Shang, W., Xie, H., Ke, C., Gao, M., Yu, K., Liu, H., Shen, J., Tang, W., Zhang, L., Zuo, J., Jiang, H., Bai, F., Wu, Y., Ye, Y., & Xu, Y. (2020). Discovery of baicalin and baicalein as novel, natural product inhibitors of SARS-CoV-2 3CL protease in vitro. Preprint bioRxiv, 10.1101/2020.04.13.038687 [DOI] [Google Scholar]
- Takooree, H., Aumeeruddy, M. Z., Rengasamy, K. R., Venugopala, K. N., Jeewon, R., Zengin, G., & Mahomoodally, M. F. (2019). A systematic review on black pepper (Piper nigrum L.): From folk uses to pharmacological applications. Critical Reviews in Food Science and Nutrition, 59, S210–S243. 10.1080/10408398.2019.1565489 [DOI] [PubMed] [Google Scholar]
- Teshika, J. D., Zakariyyah, A. M., Zaynab, T., Zengin, G., Rengasamy, K. R., Pandian, S. K., & Fawzi, M. M. (2019). Traditional and modern uses of onion bulb (Allium cepa L.): A systematic review. Critical Reviews in Food Science and Nutrition, 59(sup1), S39–S70. 10.1080/10408398.2018.1499074 [DOI] [PubMed] [Google Scholar]
- Thomson, M., & Ali, M. (2003). Garlic [Allium sativum]: A review of its potential use as an anti-cancer agent. Current Cancer Drug Targets, 3(1), 67–81. 10.2174/1568009033333736 [DOI] [PubMed] [Google Scholar]
- Tian, W., Chen, C., Lei, X., Zhao, J., & Liang, J. (2018). CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Research, 46(W1), W363–W367. 10.1093/nar/gky473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton, A. T., Gentile, F., Hsing, M., Ban, F., & Cherkasov, A. (2020). Rapid identification of potential inhibitors of SARS-CoV-2 Main Protease by deep docking of 1.3 Billion Compounds . Molecular Informatics, 39(8), e2000028. 10.1002/minf.202000028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topal, U., Sasaki, M., Goto, M., & Otles, S. (2008). Chemical compositions and antioxidant properties of essential oils from nine species of Turkish plants obtained by supercritical carbon dioxide extraction and steam distillation. International Journal of Food Sciences and Nutrition, 59(7-8), 619–634. 10.1080/09637480701553816 [DOI] [PubMed] [Google Scholar]
- Trott, O., & Olson, A. J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, Y. F., Chien, C. S., Yarmishyn, A. A., Lin, Y. Y., Luo, Y. H., Lin, Y. T., Lai, W. Y., Yang, D. M., Chou, S. J., Yang, Y. P., Wang, M. L., & Chiou, S. H. (2020). A review of SARS-CoV-2 and the ongoing clinical trials. International Journal of Molecular Sciences, 21(7), 2657. 10.3390/ijms21072657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tundis, R., Loizzo, M. R., Menichini, F., Bonesi, M., Conforti, F., Statti, G., De Luca, D., de Cindio, B., & Menichini, F. (2011). Comparative study on the chemical composition, antioxidant properties and hypoglycaemic activities of two Capsicum annuum L. cultivars (Acuminatum small and Cerasiferum). Plant Foods for Human Nutrition (Dordrecht, Netherlands), 66(3), 261–269. 10.1007/s11130-011-0248-y [DOI] [PubMed] [Google Scholar]
- Umesh, K. D., Selvaraj, C., Singh, S. K., & Dubey, V. K. (2020). Identification of new anti-nCoV drug chemical compounds from Indian spices exploiting SARS-CoV-2 main protease as target. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1763202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C., Horby, P. W., Hayden, F. G., & Gao, G. F. (2020). A novel coronavirus outbreak of global health concern. Lancet (London, England), 395(10223), 470–473. 10.1016/S0140-6736(20)30185-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Lan, J., Ge, J., Yu, J., Shan, S. (2020). Crystal structure of SARS-CoV-2 spike receptor-binding domain bound with ACE2 receptor. Nature, 581(7807), 215-220. 10.2210/pdb6M0J/pdb [DOI] [PubMed]
- Wu, C., Liu, Y., Yang, Y., Zhang, P., Zhong, W., Wang, Y., Wang, Q., Xu, Y., Li, M., Li, X., Zheng, M., Chen, L., & Li, H. (2020). Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica B, 10(5), 766–788. 10.1016/j.apsb.2020.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F., Zhao, S., Yu, B., Chen, Y. M., Wang, W., Song, Z. G., Hu, Y., Tao, Z. W., Tian, J. H., Pei, Y. Y., Yuan, M. L., Zhang, Y. L., Dai, F. H., Liu, Y., Wang, Q. M., Zheng, J. J., Xu, L., Holmes, E. C., & Zhang, Y. Z. (2020). A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Z., Peng, C., Shi, Y., Zhu, Z., Mu, K., Wang, X., & Zhu, W. (2020). Nelnavir was predicted to be a potential inhibitor of 2019-nCov main protease by an integrative approach combining homology modeling, molecular docking and binding free energy calculation. bioRxiv, 10.1101/2020.01.27.921627 [DOI] [Google Scholar]
- Yoshikawa, M., Murakami, T., Komatsu, H., Murakami, N., Yamahara, J., & Matsuda, H. (1997). Medicinal foodstuffs. IV. Fenugreek seed. (1): Structures of trigoneosides Ia, Ib, IIa, IIb, IIIa, and IIIb, new furostanol saponins from the seeds of Indian Trigonella foenum-graecum L. Chemical & Pharmaceutical Bulletin, 45(1), 81–87. 10.1248/cpb.45.81 [DOI] [PubMed] [Google Scholar]
- Yu, M. S., Lee, J., Lee, J. M., Kim, Y., Chin, Y. W., Jee, J. G., Keum, Y. S., & Jeong, Y. J. (2012). Identification of Myricetin and Scutellarein as novel chemical inhibitors of the SARS Coronavirus Helicase, nsP13. Bioorganic & Medicinal Chemistry Letters, 22(12), 4049–4054. 10.1016/j.bmcl.2012.04.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D. H., Wu, K. L., Zhang, X., Deng, S. Q., & Peng, B. (2020). In silico screening of Chinese herbal medicines with the potential to directly inhibit 2019 novel coronavirus. Journal of Integrative Medicine, 18(2), 152–158. 10.1016/j.joim.2020.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Saravanan, K. M., Yang, Y., Hossain, M. T., Li, J., Ren, X., Pan, Y., & Wei, Y. (2020). Deep learning based drug screening for novel Coronavirus 2019-nCov. Interdisciplinary Sciences, Computational Life Sciences, 12(3), 368–376. 10.1007/s12539-020-00376-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., Becker, S., Rox, K., & Hilgenfeld, R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science (New York, N.Y.), 368(6489), 409–412. 10.1007/s12539-020-00376-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P., Yang, X.-L., Wang, X.-G., Hu, B., Zhang, L., Zhang, W., Si, H.-R., Zhu, Y., Li, B., Huang, C.-L., Chen, H.-D., Chen, J., Luo, Y., Guo, H., Jiang, R.-D., Liu, M.-Q., Chen, Y., Shen, X.-R., Wang, X., … Shi, Z.-L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., Hu, Y., Shen, J., Huang, Y., Martin, W., & Cheng, F. (2020). Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discovery, 6(1), 14. 10.1038/s41421-020-0153-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., Zhao, X., Huang, B., Shi, W., Lu, R., Niu, P., Zhan, F., Ma, X., Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., Zhao, X., Huang, B., Shi, W., Lu, R., Niu, P., Zhan, F., Ma, X., Wang, D., Xu, W., Wu, G., Gao, G. F., & Tan, W., China Novel Coronavirus Investigating and Research Team. (2020). A novel coronavirus from patients with pneumonia in China, 2019 . The New England Journal of Medicine, 382(8), 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Z., Wang, X., Yang, Y., Zhang, Z., Mu, K., Shi, Y., Peng, C., Xu, Z., & Zhu, W. (2020). D3Similarity: A ligand-based approach for predicting drug targets and for virtual screening of active compounds against COVID-19. ChemRxiv, 10.26434/chemrxiv.11959323.v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoete, V., Cuendet, M. A., Grosdidier, A., & Michielin, O. (2011). SwissParam: A fast force field generation tool for small organic molecules. Journal of Computational Chemistry, 32(11), 2359–2368. 10.1002/jcc.21816 [DOI] [PubMed] [Google Scholar]
- Zumla, A., Hui, D. S., Azhar, E. I., Memish, Z. A., & Maeurer, M. (2020). Reducing mortality from 2019-nCoV: Host-directed therapies should be an option. Lancet (London, England), 395(10224), e35–e36. 10.1016/S0140-6736(20)30305-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.