Abstract
Novel Coronavirus or SARS-CoV-2 outbreak has developed a pandemic condition all over the world. The virus is highly infectious and spreads by human to human local transmission mode. Till date, there is no vaccination or drugs been approved for the treatment by the World Health Organisation. Henceforth, the discovery of the potential drugs is an urgent and utmost requirement for the medical fraternity. Since, the side effects of plant-derived compounds will be lower compared to synthetic/chemical drugs. The Main protease (3CLpro or NSP5) and endoribonuclease (NSP15) proteins are necessity for viral replication and its survival in the host cell. In the present study, in-silico approach of drug development was used to search for potential antiviral plant-derived compounds as inhibitors against SARS-CoV-2 replication proteins. Eight plant-derived compounds of which the antiviral activity was known and available, and two reported drugs against SARS-CoV-2 selected for the molecular docking analysis. The docking results suggested that bisdemethoxycurcumin, demethoxycurcumin, scutellarin, quercetin and myricetin showed least binding energy, i.e., greater than −6.5 Kcal/mol against 3CLpro and endoribonuclease of SARS-CoV-2. Further studies of ADME-Tox and bioavailability of drugs were also performed that exhibited efficient parameters of drug likeness. Molecular dynamics simulation calculations were performed for the most negative binding affinity of the compound to evaluate the dynamic behavior,and stability of protein-ligand complex. Our findings suggest that these compounds could be potential inhibitors of SARS‐CoV-2 main protease and endoribonuclease. However, further in-vitro and pre-clinical experiments would validate the potential inhibitors of SARS‐CoV‐2 proteins.
Keywords: SARS-CoV-2, COVID-19, docking, plant-derived compounds, 3CLpro, endoribonuclease
Introduction
A mysterious breakdown of typical pneumonia by a novel coronavirus (COVID-19), created a pandemic situation all over the world by spreading the disease named as severe acute respiratory syndrome-related coronavirus-2 (SARS-CoV-2) by the World Health Organization (WHO). This disease is more contagious than previous outbreaks of severe acute respiratory syndrome (SARS) in the year 2002 and the Middle East respiratory syndrome (MERS) in 2012 (Dhama et al., 2020; Guo et al., 2020). By the end of the year 2019, an outbreak from COVID-19 occurred in Wuhan, Hubei Province of China. It was reported to infect over 3.3 million people and killed more than 0.23 million people in around 215 countries as of May 03, 2020 (https://www.who.int/emergencies/diseases/novel-coronavirus-2019). According to WHO situation report-71, India reported 1071 positive cases of SARS-CoV-2 and after one month, WHO situation report-99 reported 29435 positive cases of coronavirus affected patients. India’s death rate of COVID-19 is still lower than the global death rate of around 3.4 percent, according to data and statistics website worldometer (https://www.worldometers.info/coronavirus/).
SARS-CoV-2 is considered to be a seventh known human coronavirus (hCoV) from the same family after 229E, NL63, OC43, HKU1, MERS-CoV and SARS-CoV that belongs to the β-coronavirus family (coronaviridae). It contains a single-stranded positive-sense (+) RNA encapsulated within the lipid membrane. It has been reported that the genome of SARS-CoV-2 was 96.2%, 79%, and 50% nucleotide identical with bat-CoV RaTG13, SARS-CoV, and MERS genome, respectively (Dhama et al., 2020; Guo et al., 2020). Therefore, the bat was suspected to be a natural host of virus origin that infected human via pangolins (Manisjavanica) as an intermediate host.
The genome of SARS-CoV-2 is divided into two parts: open reading frames (ORFs) that encode accessory and structural proteins; and non-structural proteins (NSP). Structural protein involves spike (S) glycoprotein, small envelope (E) protein, matrix (M) protein, and nucleocapsid (N) protein (Boopathi et al., 2020). Sixteen NSP’s are known to encode various enzymes required for replication and transcription process such as main protease (NSP5 and NSP3), RNA dependent-RNA polymerase (NSP12), a helicase/triphosphatase (NSP13), an exoribonuclease (NSP14), an endonuclease (NSP15) (Chan et al., 2020). Studies have proved that protein NSP15 of SARS-CoV-2 has 88% and ∼50% sequence identity, 95% and 65% similarity with SARS-CoV and MERS, respectively (Kim et al., 2020). NSP15 protein, an endoribonuclease, one of the crucial NSP for viral replication and transcription. It is highly conserved in all known CoVs (Hackbart et al., 2020). It is a nidoviral RNA uridylate-specific (NendoU) endoribonuclease containing 346 amino acid and made up of 2 domains NSP-15A1 and NSP-15B. It is an interferon (IFN) antagonist and inhibits interferon-β production via an endoribonuclease activity-independent mechanism. (Liu et al., 2019)
There are two proteases encoded in the SARS-CoV-2 RNA strand: papain-like protease (PLP or NSP3) and 3-Chymotrypsin-like protease (3CLpro or NSP5) co-translationally cleave the two polypeptides into mature NSPs (Lim et al., 2000). The 3CLpro, also known as Mpro, has a dominant role in the post-translational processing of the replicase gene. Protease Mpro has been reported to be a conserved protein, in favour of new drug target in (+) strand coronaviruses (Chandel et al., 2020). It has two homodimer NSP5A and NSP5B. It belongs to the cysteine protease class of protein and responsible for the cleavage of viral peptides into functional units for replication and packaging within the host cells (Liu et al., 2020). It is produced by ORF pp1ab only. Targeting Mpro enzyme will inhibit the viral maturation and enhance the host innate immune response against COVID-19 (Elmezayen et al., 2020). Other studies suggested that the sequence similarity of NSP5 of SARS-CoV-2 reported nearly 95% with SARS-CoV (Chan et al., 2020). In the current study, two protein targets (3CLpro and endoribonuclease) of SARS-CoV-2 selected based on conserved sequence and their importance in virus replication according to the literature.
For the treatment of COVID-19, doctors are prescribing antimalarial, anti-HIV, anti-influenza drugs and their combinations, but these drugs are not permanent cure of coronavirus infection (Joshi et al., 2020). Drugs such as Hydroxychloroquine, Chloroquine, Nelfinavir, Rhein, Withaferin A, Withanolide D, Enoxacin, Mercaptopurine and Aloe-emodin are tested against various target proteins of viruses (Muralidharan et al., 2020). Conversely, various phytochemicals reported in the literature to possess potential anti-viral activity that could be an alternative to restrict the coronavirus replication (Elfiky, 2020). Natural compounds consist of high chemical diversity, lower cost of production than that of biotechnological products or compounds synthesised from combinatorial chemistry and also possess milder or inexistent side effects than chemical drugs (Kitazato et al., 2007). Therefore, in the present study attempt has been made to screen and evaluate the plant-derived compounds that have therapeutic applications against different genetically and functionally diverse viruses using molecular docking study.
Material and method
Retrieval of the target structure and their preparation
The protein structure of 3-chymotrypsin-like cysteine protease (3CLpro) enzyme (PDB ID: 6LU7) with 2.1Å resolution and endoribonuclease, non-structural protein 15 (NSP15) protein (PDB ID: 6VWW) with 2.2Å resolution of SARS-CoV- 2 were downloaded from RCSB protein databank (http://www.rcsb.org). Initially, different tasks were performed for protein preparation such as charge assignment, solvation parameters and fragmental volumes using SPDVB-4.10 version (Morris et al., 2009). The protein molecules were further optimized using AutoDock4 Tool for the molecular docking (Morris et al., 2009).
Secondary structure prediction
The secondary structure of proteins was speculated using the Self-Optimized Prediction Method with Alignment (SOPMA) server at https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html (Geourjon & Deléage, 1995). SOPMA predicts four conformational states including Helix, Beta sheets and bridges, Turns and Coils for each antigen.
Preparation of ligands
The natural compounds for the ligand dataset were selected from the previous literature (Table 2). Eight selected natural compounds and two drugs were obtained from the PubChem compound database (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4702940/) in InChI (Inch key) format. The two-dimensional (2D) ligand structures were generated using ACD/Chemsketch and save as mol file format. The three-dimensional (3D) structure of ligands were converted using OpenBabel tool (O’Boyle et al., 2011).
Table 2.
Selected plant-derived compounds based on their known pharmacological properties.
| S.no. | Plant-derived compound | Reported activity | Disease | References |
|---|---|---|---|---|
| 1. | Curcumin | Anti-viral | Herpes virus, Ebola | (Kutluay et al.,2008) |
| 2. | Demethoxycurcumin | Anti-viral | Ebola | (Baikerikar,2017) |
| 3. | Bisdemethoxycurcumin | Anti-viral | Ebola | (Baikerikar,2017) |
| 4. | Scutellarin | Anti-viral | Covid-19 (Nsp-13) | (Yu et al.,2012) |
| 5. | Myricetin | Anti-inflammatory | Diabetes mellitus | (Dormán et al., 2016) |
| 6. | Quercetin | Anti-viral | Mingo virus, Poliovirus, Herpes small virus-2 | (Veckenstedt et al.,1987) |
| 7. | Bergapten | Anti-viral | Coxsackievirus B3, Herpes small virus-1 | (Rajtar et al.,2017) |
| 8. | Isoflavone | Anti-viral | Herpes small Virus-1 | (Akula et al.,2002) |
Molecular docking
The docking analysis was performed by molecular docking program AutoDock4 Tool (Morris et al., 2009) to explore the poses of potential inhibitors within the targets active site. For protein-ligand interactions, the Lamarckian genetic algorithm was utilized to perform the docking with the preset parameters. The total number of poses was set to 50. Poses were further clustered using all atom RMSD cutoff of 0.3 Å to remove redundancy and on average 20 cluster representatives were kept. All other parameters for docking and scoring were used as default sets. The protein structure was kept rigid in all steps. All the docking poses and interaction analysis has been performed using the UCSF Chimera (Pettersen et al., 2004) and Discovery Studio Visualizer programs (http://accelrys.com/products/collaborative-science/biovia-discovery-studio/visualization-download.php). The amino acids residues that displayed interactions with the ligand were documented in Table 3 and Figure S1 and S2 (Supplementary material).
Table 3.
Details of molecular docking analysis of Plant-based compounds against SARS-CoV-2 endoribonuclease and 3CLpro proteins.
| Compound Name | PubChem CID | Structure | 3CLPro |
Endoribonuclease |
||
|---|---|---|---|---|---|---|
| Binding affinity | Amino acid residues | Binding affinity | Amino acid residues | |||
| Demethoxycurcumin | 5469424 | 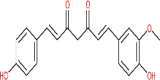 |
−7.02 | His 163 | −7.51 | Lys 290; Gly 248; His 235; Thr 341; Glu 340 |
| Curcumin | 969516 | 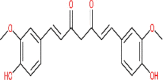 |
−6.04 | Asn 119; His 163 | −6.48 | Lys 290 |
| Bisdemethoxycurcumin | 5315472 | 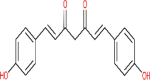 |
−7.3 | Cys 145;His 41; His 41; Phe 140; His 163 | −6.56 | Lys 290 |
| Quercetin | 5280343 | 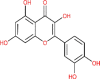 |
−6.58 | Asn 142; Gly 143; Ser 144; Leu 141; Cys 145; His 41; Thr 26; Thr 26 | −6.49 | Lys 290; His 235;His 250; Glu 340 |
| Myricetin | 5281672 | 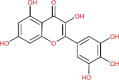 |
−6.15 | Gly 143; Leu 141; His 41; Thr 26 | −6.52 | Lys 290; His 235;His 250; Glu 340 |
| Bergapten | 2355 | 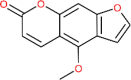 |
−5.98 | His 163; His 163; Phe 140 | −5.92 | Lys 290; His 235;Ser 294; Thr 341 |
| Scutellarin | 5281697 | 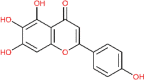 |
−7.13 | Gly 143; His 163 | −6.97 | Lys 290; His 250;Gly 248; Glu 340 |
| Isoflavone | 72304 | 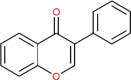 |
−5.69 | Gly 143, Cys 145 | −5.47 | His 235; Thr 341 |
| Remdesivir | 121304016 | 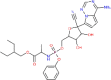 |
196 | Phe 140; His 164 | −7.7 | Glu 340 |
| Ribavirin | 37542 | 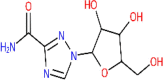 |
−5.43 | Ser 144; Ser 144; Gly 143; Thr 26; Thr 26; Thr 26; His 41; Cys 145 | −5.84 | Ser 294; Ser 294; Pro 344; Lys 290; His 235; His 235; Tyr 343; His 250; Thr 341 |
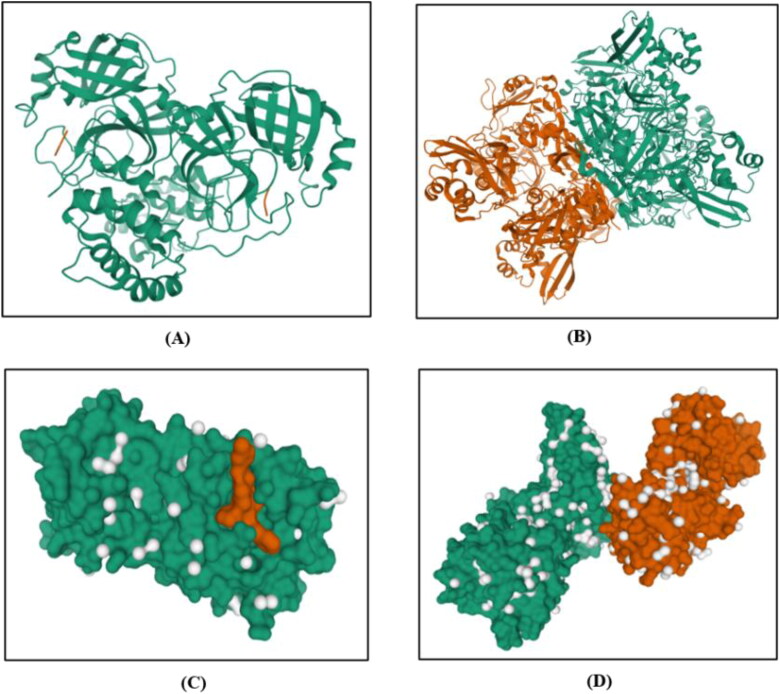
Figure 1.
Ribbon structure of (A) 3CLpro and (B) NSP15 protein. Crystal structure of (C) 3CLpro (PDB-ID:6LU7) and (D) NSP15 endoribonuclease protein (PDB-ID:6VWW). Source- PDB databank.
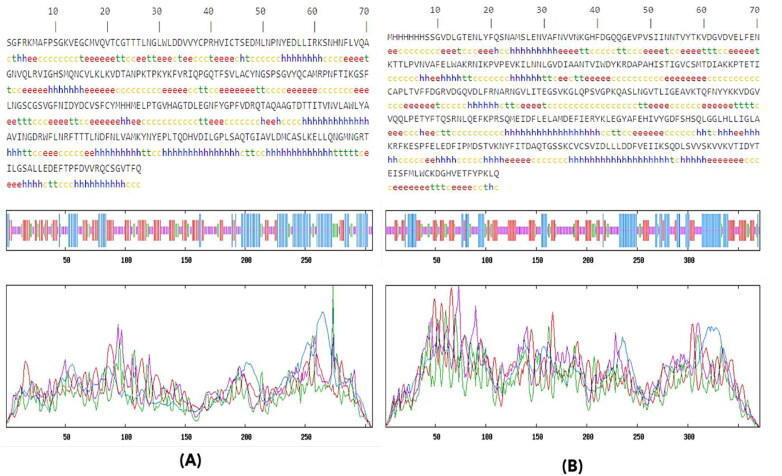
Figure 2.
The predicted secondary structure of the (A) 3CLpro and (B) NSP15 endoribonuclease using SOPMA software, whereas h: Alpha helix, e: Extended strand, t: Beta-turn and c: Random coil. The graphical representation of secondary elements in 3CLpro and NSP15 endoribonuclease protein (blue: alpha helix, red: extended, green: beta turn, yellow: random coil).
ADME-Tox prediction
The qualitative measurement of absorption, distribution, metabolism, excretion, and toxicity profile of selected compounds were predicted by using SwissADME program (http://www.swissadme.ch/index.php) (Hoda et al., 2019). The major ADME-Tox properties parameters include Molecular weight, H-bond acceptor, H-bond donors, Predicted octanol/water partition coefficient (MLogP), Total Polar Surface Area (TPSA), Lipinski (drug likeness), GI absorption, BBB permeant, Bioavailability score.
Molecular dynamics simulation setup and analysis
To analyze the conformational dynamics of the unbound and bound state of 3CLpro and endoribonuclease enzyme, molecular dynamics simulation has been performed using the Amber16 package (Gajula et al., 2016). After the preparation of the docked ligand molecules using the antechamber (Wang et al., 2006), tleap module is used to prepare the cubic simulation box of size 10 Å considering the protein at the center, the box was filled with TIP3P water molecules (Jorgensen et al., 1983) for solvation under periodic boundary condition. Some water molecules were replaced by the counterions to neutralize the systems. The Generalized Amber Force Field (GAFF) for ligand (Wang et al., 2004) and FF14SB FF for protein (Maier et al., 2015) was used during the simulation setup. For all electrostatic interactions calculation, Particle Mesh Ewald (PME) method (Essmann et al., 1995) was used and 2 fs time step is used with the SHAKE algorithm (Ryckaert et al., 1977) to constrain the hydrogen bonds.
Prepared simulation box was initially minimized for 500,000 steps (400,000SD +100,000CG) to relax the system. The minimized system was gradually heated from the 0 to 300 K over the period of 6000ps. A 1 ns simulation run was further used to equilibrate the system under NVT ensemble. The final MD production run of 50 ns was performed with an integration time step of 2.0 fs. Simulation conformations were collected after every 50ps and all trajectories were analyzed for the RMSD, RMSF, SASA and hydrogen bond dynamics using the CPPTRAJ module (Roe & Cheatham, 2013) of the Amber software.
Results
To perform the target protein analysis, FASTA sequences and 3 D structure of 3CLpro and endoribonuclease were retrieved from PDB databank (PDB ID: 6LU7 and 6VWW) (Figure 1). The secondary structure prediction indicated that protein 3CLpro consisted 29.08% α-helix (h), 27.12% extended strand (e), 11.44% beta-turn (t), 32.35% random coil (c) elements and NSP15 endoribonuclease comprised of 24.26% α-helix (h), 26.95% extended strand (e), 9.70% beta-turn (t), 39.08% random coil (c). There are two graphs shown in the result- 1) to visualize the prediction; 2) score curves for all predicted states (Figure 2). It also shows the parameters such as window width, number of states etc. that are used for the prediction.
Molecular docking was performed to evaluate the interaction of plant-derived compounds, having reported pharmacological activities against two non-structural proteins of SARS-CoV-2. Non-structural proteins, 3CLpro and endoribonuclease were targeted and the docking pocket was formed at the catalytic sites of the respective proteins as mentioned in Table 1. Also, the binding affinity of interacted compounds at the active site were assessed. Known anti-viral drugs such as remdesivir and ribavirin were docked against 3CLpro as well as endoribonuclease proteins. Plant-derived active compounds, reported with their pharmacological properties were listed in Table 2.
Table 1.
Active site residues of 3CLpro and NSP15 endoribonuclease protein.
| Protein Site | Residues | Amino Acids | Co-ordinates of amino acid residues |
|---|---|---|---|
| 3CLpro | Cys | 145 | x-axis: 3.032 y-axis: −4.728 z-axis: 39.052 |
| Endoribonuclease (NSP15) | Thr | 341 | x-axis: −93.206 y-axis: 14.072 z-axis: −5.47 |
Interaction of plant-derived compounds with active site of protein 3CLpro
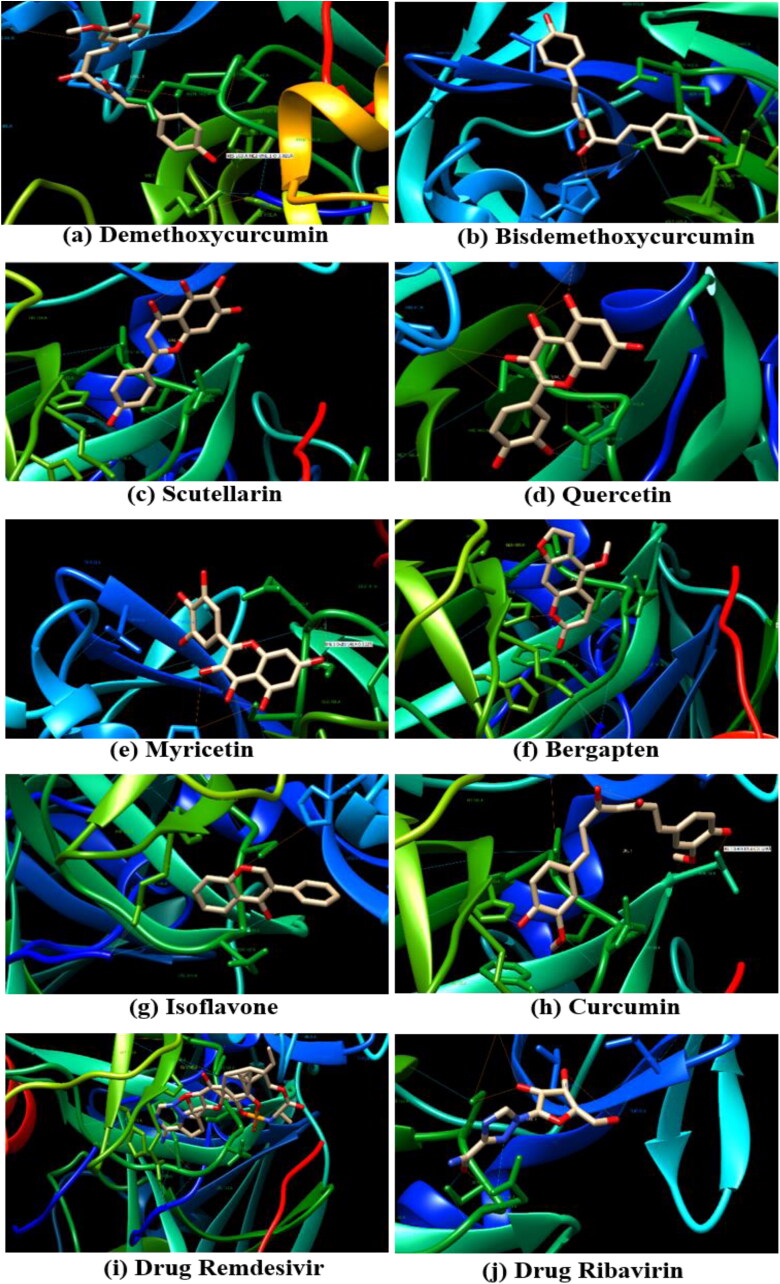
Bisdemethoxycurcumin and Demethoxycurcumin the analogues of curcumin, exhibited the docked score of −7.3Kcal/mol and −7.02Kcal/mol, and formed five hydrogen bonds with amino acid residues Lys-290, Gly-248, His-235, Thr-341, and Glu-340 and one hydrogen bond with His-163 amino acid residue, respectively (Figure 3). Scutellarin represented the binding affinity of −7.13 Kcal/mol was found to form two hydrogen bonds with amino acid residues Gly-143 and His-163, followed by flavonoid quercetin with binding energy of −6.58Kcal/mol and hydrogen bonds formed with amino acids Asn-142, Gly-143, Ser-144, Leu-141, Cys-145, His-41, and Thr-26. Among all, four plant-derived compounds with binding affinity more than −6.5Kcal/mol were myricetin (-6.15Kcal/mol), curcumin (-6.04Kcal/mol), bergapten (-5.98Kcal/mol) and isoflavone (-5.69Kcal/mol) with amino acids present in the docking pocket of 3CLpro shown in Table 3. Furthermore, the anti-viral drugs remedesivir and ribavirin showed +196 Kcal/mol and −5.43 Kcal/mol binding affinity, respectively.
Figure 3.
Binding interactions of eight plant-derived compounds (a-j) with the active site of SARS-CoV-2 3CLpro protein target site (PDB ID: 6LU7).
Interaction of plant-derived compounds with active site of protein endoribonuclease
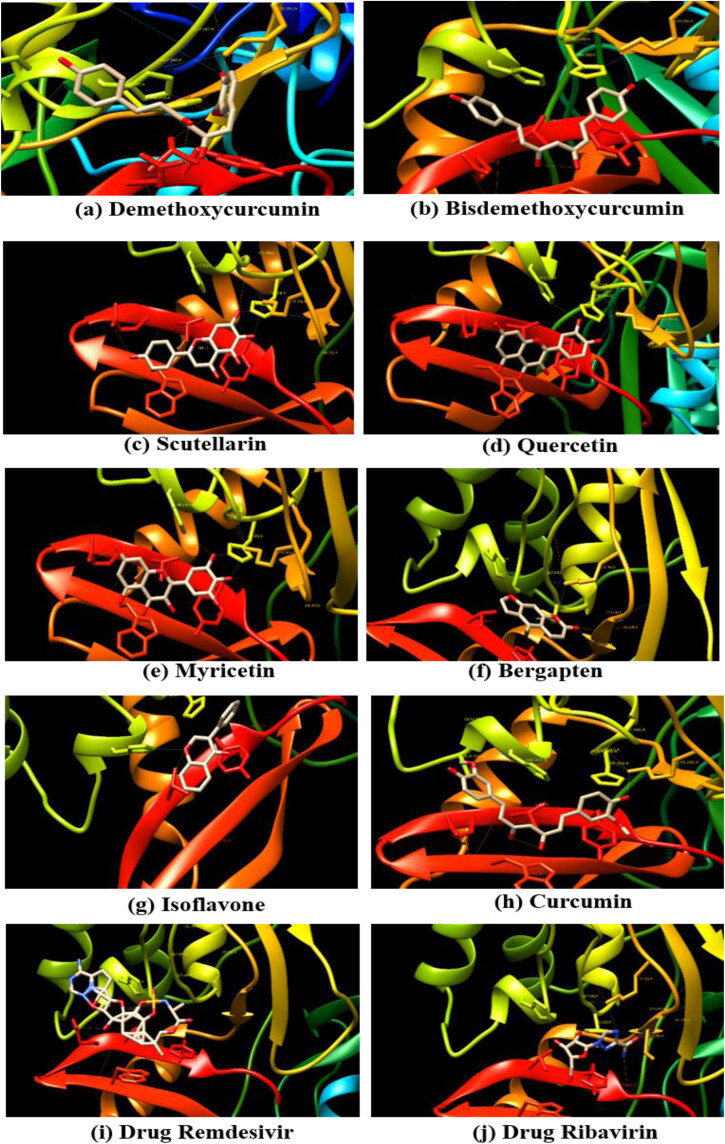
Compounds represented a similar kind of affinity with endoribonuclease as observed with 3CLpro. Based on binding affinities, three compounds were in common with both the protein targeted sites. Demethoxycurcumin with binding affinity of −7.51Kcal/mol and formed five hydrogen bonds with Lys-290, Gly-248, His-235, Thr-341, and Glu-340 amino acid residue. Another compound scutellarin, with binding affinity of −6.97Kcal/mol with Lys-290, His-250, Gly-248, and Glu-340 amino acids formed four hydrogen bonds within docking pocket followed by bisdemethoxycurcumin, binding affinity of −6.56Kcal/mol and one hydrogen bond formation with Lys-290. The flavonoid group of myricetin and quercetin exhibited negligible difference in their binding energies −6.52 and −6.49 Kcal/mol, but hydrogen bond formed with same amino acids residues in the pocket- Lys-290, His-235, His-250, and Glu-340. Bergapten (-5.89Kcal/mol) and isoflavone (-5.69Kcal/mol) also showed the minimum binding energy and formed hydrogen bonds. The anti-viral drugs remedesivir docked with −7.7Kcal/mol with one hydrogen bond, whereas ribavirin binding energy (-5.84Kcal/mol) was found close to that of 3CLpro target site (Table 3; Figure 4).
Figure 4.
Binding interactions of eight plant-derived compounds (a-j) with the active site of SARS-CoV-2 NSP15 endoribonuclease (PDB ID: 6VWW).
All the plant-derived compounds studied were analysed for their physicochemical properties and ADME to screen bioavailability via oral route. The eight compounds listed in Table 4 showed the five key criteria to qualify a compound to possess drug-like properties. The results explained that the plant-derived compounds studied would be more potent to be used as bioactive compounds as compared to the existing anti-viral drug remedesivir. All the compounds have molecular weight less than 500 g/mol, lipophilicity (MlogP) value less than 5, hydrogen bond donor and acceptor are less than 5 and 10, respectively which corresponds to no violations of Lipinski’s rule of five. Thus, it suggested that all of these studied compounds have the potential ability to work effectively as novel inhibitors.
Table 4.
Physicochemical analysis of potential inhibitors of 3CLpro and NSP15 endoribonuclease protein of SARS-CoV-2.
| Compounds | Molecular formula | ADME properties (Lipinki’s rule of five) | Values | |
|---|---|---|---|---|
| Demethoxycurcumin | C20H18O5 | Molecular weight (<500g/mol) LogP (<5) H-bond donor (<5) H-bond acceptor (<10) Violation |
338.4 g/mol 2.78 2 5 0 |
|
| Curcumin | C21H20O6 | Molecular weight (<500g/mol) LogP (<5) H-bond donor (<5) H-bond acceptor (<10) Violation |
368.4 g/mol 3.27 2 6 0 |
|
| Bisdemethoxycurcumin | C19H16O4 | Molecular weight (<500g/mol) LogP (<5) H-bond donor (<5) H-bond acceptor (<10) Violation |
308.3 g/mol 1.75 2 4 0 |
|
| Quercetin | C15H10O7 | Molecular weight (<500g/mol) LogP (<5) H-bond donor (<5) H-bond acceptor (<10) Violation |
302.23 g/mol 1.63 5 7 0 |
|
| Myricetin | C15H10O8 | Molecular weight (<500g/mol) LogP (<5) H-bond donor (<5) H-bond acceptor (<10) Violation |
318.23 g/mol 1.08 6 8 1 |
|
| Bergapten | C12H8O4 | Molecular weight (<500g/mol) LogP (<5) H-bond donor (<5) H-bond acceptor (<10) Violation |
216.19 g/mol 2.29 0 4 0 |
|
| Scutellarin | C15H10O6 | Molecular weight (<500g/mol) LogP (<5) H-bond donor (<5) H-bond acceptor (<10) Violation |
286.24 g/mol 2.08 4 6 0 |
|
| Isoflavone | C15H10O2 | Molecular weight (<500g/mol) LogP (<5) H-bond donor (<5) H-bond acceptor (<10) Violation |
222.24 g/mol 2.51 0 2 0 |
|
| Remdesivir | C27H35N6O8P | Molecular weight (<500g/mol) LogP (<5) H-bond donor (<5) H-bond acceptor (<10) Violation |
602.6 g/mol 3.24 4 13 2 |
|
| Ribavirin | C8H12N4O5 | Molecular weight (<500g/mol) LogP (<5) H-bond donor (<5) H-bond acceptor (<10) Violation |
244.2 g/mol 0.13 4 7 0 |
|
Oral bioavailability and gastrointestinal (GI) absorption of interacted plant-derived compounds
The potential inhibitors of enzymes involved in replication and transcription of SARS-CoV-2 studied were analysed for their oral bioavailability as well as GI tract absorption parameters. The potent compounds extracted from various natural sources were listed in the Table S1 (Supplementary material). Natural sources such as herbs, oils have many active compounds: Curcumin, Quercetin, Bergapten etc. which possess to have potential as anti-SARS-CoV-2 activity. The bioavailability of all the plant-based compounds is equal to 0.55 whereas remdesivir has lower bioavailability of 0.17. The absorption of the compounds by GI tract is high except for myricetin while the drugs have lower absorption rate.
Molecular dynamics simulation
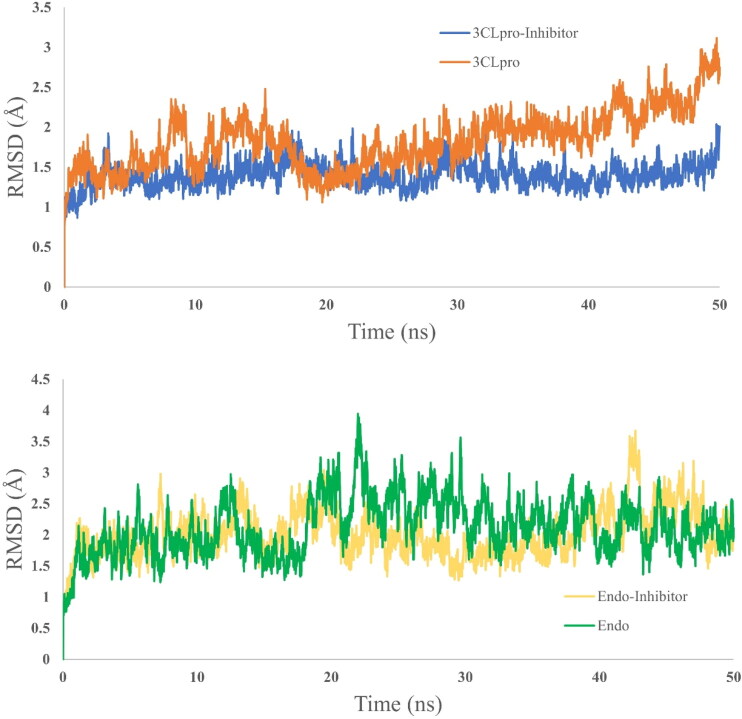
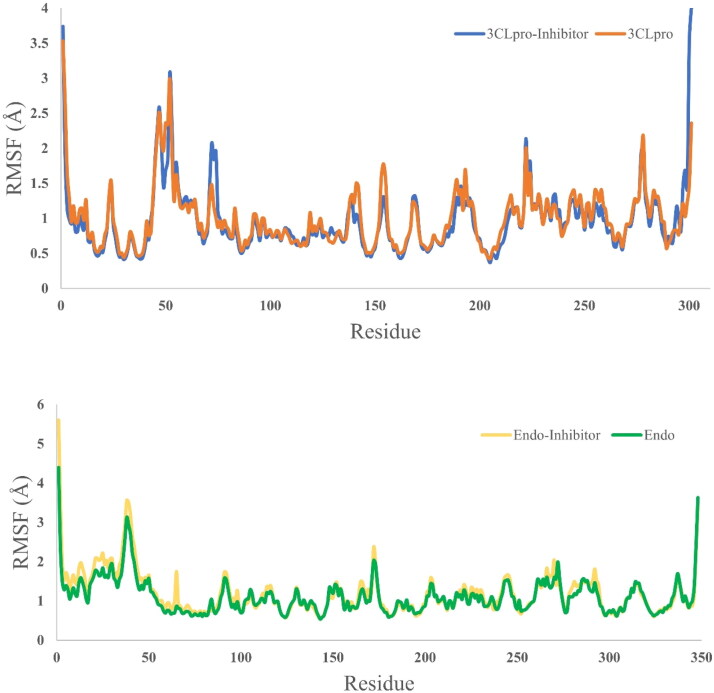
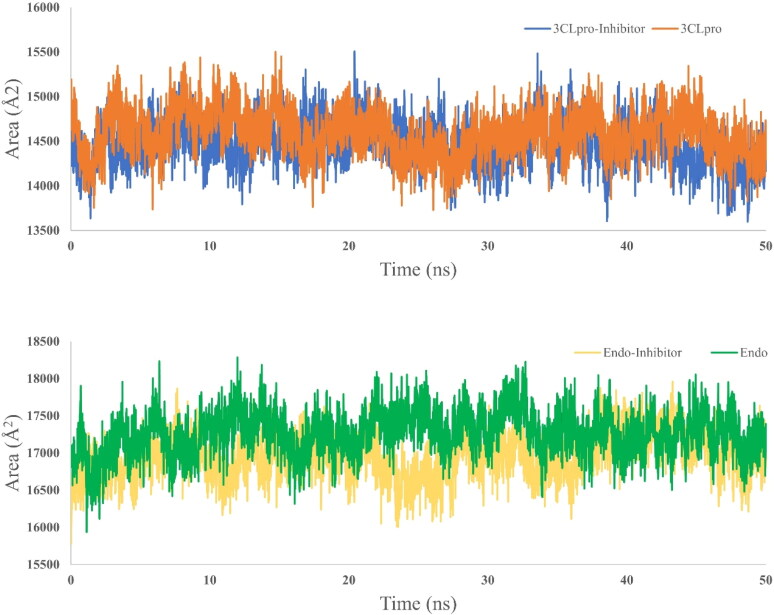
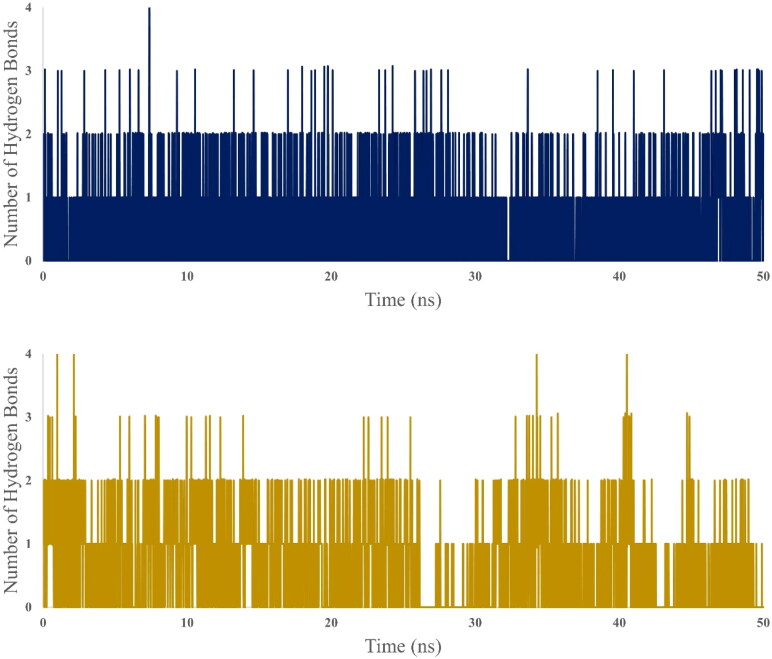
Molecular dynamics simulation has been preformed for 50 ns simulation time for four systems, two from 3CLpro (receptor only and with selected dock ligand complex) and two from endoribonuclease (receptor and docked ligand complex). For each case, receptor only dynamics is used as a reference to assess the stability dynamics of the ligand bound complex. RMSD (Figure 5), RMSF (Figure 6) and SASA (Figure 7) is calculated to assess the time dependent evolution of the systems and stability dynamics. At the same time, Hydrogen-bond analysis has been done to understand the engagement of the docked ligand with the corresponding receptor (Figure 8).
Figure 5.
RMSD plot of the 3CLpro system (above) and endoribonuclease system (below). For both cases, protein-ligand complex is showing better stability compared to the receptor alone.
Figure 6.
RMSF analysis plot for 3CLpro system (above) and endoribonuclease system (below). As expected, except the binding site residues, nearly similar fluctuations are recorded for the both unbound and bound system. Binding site residues are less fluctuating in ligand complex compared to receptor only dynamics.
Figure 7.
SASA plot for the 3CLpro system (above) and endoribonuclease system (below). For both systems, SASA are nearly consistent over time and reflects that after ligand binding receptor proteins do not undergo any large conformational change.
Figure 8.
Hydrogen bond dynamics between Bisdemethoxycurcumin-3CLpro complex (above) and Demethoxycurcumin-endoribonuclease complex (below). This analysis shows that selected ligands engage with receptors with nearly two hydrogen bonds and do not deviate much from the initial structure.
Discussion
The rapidly spreading coronavirus disease SARS-CoV-2, also known as COVID-19, is declared as a pandemic outbreak by the WHO. It has affected the public as well as social health of more than 200 countries around the globe (https://www.who.int/emergencies/diseases/novel-coronavirus-2019). Hence, it’s the need of an hour that has demanded the scientific community from all over the world to collaborate and make an extraordinary effort for the rapid detection of an effective anti-COVID-19 drug.
The NSPs, endoribonuclease (NSP15) and main protease enzyme encoded by 3CLpro have been sequenced and their X-ray crystallography 3-D structures are available in PDB databank (Jin et al., 2020; Kim et al., 2020) . Each monomeric unit of endoribonuclease protein is comprised of ∼345 amino acids which are folded into three domains: N-terminal, middle and NendoU C-terminal catalytic domain. The NendoU C-terminal catalytic domain reported to possess two β-sheets in antiparallel direction (Sinha et al., 2020). This protein is required by the virus as it takes part in various viral mechanisms including the formation of the replicase and transcriptase complex (RTC) (Deng et al., 2017). Also, in innate immune response, therefore NSP15 considered to be essential for biological progression of coronavirus. Study has reported that the main active sites of Nsp15 protein, conserved among SARS-CoV-2, SARS-CoV and MERS-CoV are located in a shallow groove between the two β-sheets that comprise six amino acids (His235, His250, Lys290, Thr341, Tyr343, and Ser294) (Sinha et al., 2020).
The main protease, 3CLpro protein is a homodimer with Cys-His catalytic dyad on the active site which is responsible for the protease activity and hence inhibiting the catalytic site present in enzyme would lead to the inhibition of activity of the protein(Islam et al., 2020; Lobo-Galo et al., 2020). The secondary structures of 3CLpro, has three domins: Domains I and II which have an antiparallel β-barrel structure and Domain III contains five α-helices arranged into a largely antiparallel globular cluster, and is connected to domain II by means of a long loop region. These features are similar to those of other Mpro reported previously (Ren et al., 2013; Wang et al., 2016; Xue et al., 2008).
At present, there are no approved medications from different drug administrations around the world for the treatment of novel coronavirus (SARS-CoV-2). Hence, the primary focus has been on clinical management which includes the prevention of infection, control measures and intensive supportive care. Recently, anti-malarial drugs like chloroquine, hydroxychloroquine have given positive results in the cell culture systems but has to proceed for the clinical trials to be established as an effective drug (Enmozhi et al., 2020). Several other anti-viral drugs such as remdesivir, ribavirin used in hepatitis C infection and Ebola, and many other vaccines are under rapid study to be used against coronavirus infection.
An anti-viral activity of curcumin and its analogues demethoxycurcumin and bisdemethoxycurcumin was reported against several different viruses including hepatitis viruses, influenza viruses and emerging arboviruses like the Zika virus (ZIKV) (Mounce et al., 2017) or chikungunya virus (CHIKV) (von Rhein et al., 2016). These analogues have described role as a metabolite, an anti-neoplastic agent and an anti-inflammatory agent in PubChem databank. They possess much greater chemical stability at physiological pH. Scutellarin is a glycosyloxyflavone, a glucosiduronic acid, a trihydroxyflavone and a monosaccharide derivative. (Cheng et al., 2013) observed that it could inactivate porcine reproductive and respiratory syndrome virus (PRRSV) and restrict the early stage of PRRSV replication under in vitro conditions. Similarly, Zhang et al.(2005) demonstrated that it possessed anti-HIV-1 activity by inhibiting HIV-1 reverse transcriptase activity, HIV-1 particle attachment and cell fusion (Cheng et al., 2013; Sun et al., 2015). Quercetin is a polyphenol, known as flavonoids, its various pharmacological activities described in the literature such as anti-inflammatory, anti-oxidant, anti-enzymatic activities. (Polansky & Lori, 2020) concluded that quercetin and its derivatives were reported to inhibit the SARS and MERS virus replication via protease inhibition (Cheng et al., 2013). Myricetin is structurally related to several well-known phenolic compounds namely quercetin, morin, kaempferol and fisetin. Myricetin demonstrated antiviral activity as a strong inhibitor of reverse transcriptase against Rauscher murine leukemia virus (RLV) and human immunodeficiency virus (HIV). Some studies reported that bergapten exhibited potent anti-HIV activity via inhibition of HIV-1 replication with therapeutic index value greater than 5.0 (Shikishima et al., 2001). According to literature, it has been reported that 5,7,3′,4′-tetrahydroxy-2′-(3,3-dimethylallyl) isoflavone used as an anti-leishmanial agent (Salem & Werbovetz, 2006), and it is also described in traditional Chinese medicine records (Zhou et al., 2011).
The preliminary screening of potential molecules having anti-viral activity via computational approach would help in providing the fast selection of promising compounds towards the development of therapeutics against SARS- CoV-2. To elucidate the binding affinity of plant-derived compounds with known properties of anti-viral, were docked against two NSPs of COVID-19 using AutoDock4 software (Table 2; Figure 1 and 2). The binding scores from the docking study revealed the affinity of a specific ligand and strength by which a compound interacts with and binds to the pocket of a target protein. Docking results were successful with least binding scores and presence of significant hydrogen bonding with demethoxycurcumin, bisdemethoxycurcumin and a flavone scutellarin. Compounds having binding affinity of −6.5 Kcal/mol or less are considered to be a good inhibitor of enzymatic activities (Shah et al., 2020). In the present study, five plant-derived compounds demonstrated binding affinity more negative to −6.5Kcal/mol with 3CLpro, whereas with NSP15 endoribonuclease, five different plant-derived compounds exhibited the same results. These compounds could be potent drugs for inhibition of two essential viral proteins and thus inhibiting the viral replication in the infected cells. Additionally, the in-silico experiment represented the inability of remdesivir to bind with 3CLpro, as binding affinity of remdesivir was +196 Kcal/mol, which showed instability. In contrast, Remdesivir contributes −6.58 kcal/mol to the binding free energy at the electrostatic S1 subsite of protease protein (Khan et al., 2020).
Further, the physicochemical properties of these compounds were studied by ADME-Tox following the Lipinski’s rule of five for evaluating drug likeness. Lipinski’s rule determines the molecular properties which are important for a drug’s pharmacokinetics in the human body such as absorption, distribution, metabolism, and excretion (ADME). Lipinski’s rule of five criteria for an ideal drug are (i) a molecular mass less than 500 Daltons (ii) no more than 5 hydrogen bond donors, (iii) no more than 10 hydrogen bond acceptors, (iv) an octanol–water partition coefficient log P not greater than 5. Three or more than 3 violations of Lipinski’s rule do not fit into the criteria of drug likeliness via oral route administration (Benet et al., 2016). ADME-Tox studies of docked eight plant-derived compounds showed that all the compounds had virtual hits and were successful at passing through these ADME-Tox test filters, stating that they could be potential drugs which can be easily administered orally (Table 4).
The other crucial experiments like bioavailability and GI absorptions are of detecting the low bioavailability is a major concern for drug design, as it may lead to failure of a novel drug in the clinical trials, in spite of the fact that the compounds have high efficacy in in-vitro and/or in-vivo tests (Gyebi et al., 2020). Therefore, oral bioavailability of a compound is essential to be taken into account when predicting the compound as a drug candidate. The compounds in the present study exhibited bioavailability of 0.55 with low as well as high GI absorption. The absorption into GI was found to be at a high rate in all compounds except that of Myricetin.
Protein flexibility plays an essential role in the inhibitor designing process (Teague, 2003) and as molecular docking has been performed using the single structure, molecular dynamic simulation can provide an in-depth understanding of the protein-ligand interaction information. 3CLpro and endoribonuclease proteins, along with selected compounds, were used for MDS analysis, which suggests that the ligand-bound receptor adopt stability over the receptor only dynamics (Figure 5 and 7). H-bond based interaction analysis suggests that both docked compounds with respective proteins utilize the available binding interactions and maintain these interactions throughout the simulation time (Figure 8). Apart from the receptor-based interaction, docked ligand also formed on average two H-bonds with the nearly water molecules which additionally provide the stability to the docked ligand (Supplementary material Figure S3).
Conclusion
In the current scenario, there is an urgent requirement of the potential treatment of SARS-CoV-2 infection. Compounds extracted from natural sources might be a source in the development of new potent drugs which can improve the novel coronavirus treatment. Various plant-derived compounds have been reported for their anti-viral activity, may found to be effective against SARS-CoV-2. The bioinformatics approach could be a very useful and efficient tool to identify potent inhibitors against the specific targets of novel coronavirus. The docking study concluded that plant-derived compounds (Demethoxycurcumin, Bisdemethoxycurcumin, Scutellarin, Quercetin and Myricetin) exhibited potential drug candidates against two enzymes, main protease (3CLpro) and endoribonuclease (NSP15) proteins of SARS-CoV-2. Also, molecular dynamics and simulations of two best docked protein-ligand structures revealed the dynamics information of their stability in the biological system. Instability of these two enzyme activities would lead to inhibition of viral replication, which in response would restrict the infection in the host cell.
Supplementary Material
Acknowledgements
The authors would like to thank Amity University Uttar Pradesh, India for providing facilities to carry out this work. The authors also would like to thank the researchers who generated and shared the sequencing and protein data of main protease (3CLpro) and endoribonuclease (NSP15) proteins of SARS-CoV-2 in Protein Data Bank website (https://www.rcsb.org/). The authors would like to acknowledge Ms. Sapna Ratnakar, Ph.D. Scholar, School of Life Sciences, JNU for providing her support to carry out MDS. P.K. would like to thank DBT Project for supporting the research work and also thank Dr. Debabsisa Mohanty for providing the NII computational facility to carry out the simulation analysis.
Glossary
Abbreviations
- 2D
Two-dimensional
- 3CLpro
3-Chymotrypsin-like protease
- 3D
Three-dimensionsal
- ADME
absorption, distribution, metabolism, and excretion
- BBB
Blood Brain Barrier
- COVID-19
Coronavirus Disease
- GI
Gastrointestinal
- hCoV
human coronavirus
- HIV
human immunodeficiency virus
- IFN
interferon
- InChI
Inch key
- MERS-CoV
Middle East respiratory syndrome- coronavirus
- MlogP
lipophilicity
- Mpro
Main protease domain
- NSP
Non-structural proteins
- NSP15
endoribonuclease
- ORF
Open reading frame
- PDB
Protein Data Bank
- PLP
papain-like protease
- RMSD
Root mean square deviation
- RMSF
Root mean square fluctuation
- SASA
Solvent accessible surface area
- RNA
Ribonucleic Acid
- SARS-CoV
Severe acute respiratory syndrome- coronavirus
- SARS-CoV-2
Severe acute respiratory syndrome-related coronavirus 2
- SOPMA
Self-Optimized Prediction Method with Alignment
- TPSA
Total Polar Surface Area
- WHO
World Health Organization
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Disclosure statement
The authors declare no potential conflicts of interest to disclose.
Authors contributions
All authors conceived, conducted and analyzed the results of experiment(s) equally. L.G. performed docking studies, manuscript editing; A.S. and S.G. drafted manuscript, performed ADMET studies; A.K.Y. manuscript editing, result analysis and P.K. performed molecular dynamics and simulations.
References
- Akula, S. M., Hurley, D. J., Wixon, R. L., Wang, C., & Chase, C. C. L. (2002). Effect of genistein on replication of bovine herpesvirus type 1. American Journal of Veterinary Research, 63(8), 1124–1128. 10.2460/ajvr.2002.63.1124 [DOI] [PubMed] [Google Scholar]
- Baikerikar, S. (2017). Curcumin and natural derivatives inhibit Ebola viral proteins: An In silico approach. Pharmacognosy Research, 9(5), 15. 10.4103/pr.pr_30_17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benet, L. Z., Hosey, C. M., Ursu, O., & Oprea, T. I. (2016). BDDCS, the Rule of 5 and drugability. Advanced Drug Delivery Reviews, 101, 89–98. 10.1016/j.addr.2016.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boopathi, S., Poma, A. B., & Kolandaivel, P. (2020). Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure and Dynamics, 38,1–10. 10.1080/07391102.2020.1758788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, J. F. W., Kok, K. H., Zhu, Z., Chu, H., To, K. K. W., Yuan, S., & Yuen, K. Y. (2020). Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerging Microbes & Infections, 9(1), 221–236. 10.1080/22221751.2020.1719902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, J. F.-W., Yuan, S., Kok, K.-H., To, K. K.-W., Chu, H., Yang, J., Xing, F., Liu, J., Yip, C. C.-Y., Poon, R. W.-S., Tsoi, H.-W., Lo, S. K.-F., Chan, K.-H., Poon, V. K.-M., Chan, W.-M., Ip, J. D., Cai, J.-P., Cheng, V. C.-C., Chen, H., Hui, C. K.-M., … Yuen, K.-Y. (2020). A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. The Lancet, 395(10223), 514–523. 10.1016/S0140-6736(20)30154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel, V., Raj, S., Rathi, B., & Kumar, D. (2020). In silico identification of potent COVID-19 main protease inhibitors from FDA approved antiviral compounds and active phytochemicals through molecular docking: A drug repurposing approach. 7(3), 1-9. 10.20944/preprints202003.0349.v1 [DOI] [Google Scholar]
- Cheng, J., Sun, N., Zhao, X., Niu, L., Song, M., Sun, Y., Jiang, J., Guo, J., Bai, Y., He, J., & Li, H. (2013). In vitro screening for compounds derived from traditional Chinese medicines with antiviral activities against porcine reproductive and respiratory syndrome virus. Journal of Microbiology and Biotechnology, 23(8), 1076–1083. 10.4014/jmb.1303.03074 [DOI] [PubMed] [Google Scholar]
- Deng, X., Hackbart, M., Mettelman, R. C., O'Brien, A., Mielech, A. M., Yi, G., Kao, C. C., & Baker, S. C. (2017). Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proceedings of the National Academy of Sciences of the United States of America, 114(21), E4251–E4260. 10.1073/pnas.1618310114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhama, K., Khan, S., Tiwari, R., Sircar, S., Bhat, S., Malik, Y. S., Singh, K. P., Chaicumpa, W., Bonilla-Aldana, D. K., & Rodriguez-Morales, A. J. (2020). Coronavirus Disease 2019–COVID-19. Clinical Microbiology Reviews, 33(4), e00028–20. 10.1128/CMR.00028-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormán, G., Flachner, B., Hajdú, I., & András, C. D. (2016). Target Identification and Polypharmacology of Nutraceuticals. In Nutraceuticals (pp. 263–286). Elsevier. 10.1016/B978-0-12-802147-7.00021-8 [DOI] [Google Scholar]
- Elfiky, A. A. (2020). Natural products may interfere with SARS-CoV-2 attachment to the host cell. Journal of Biomolecular Structure and Dynamics, 38, 1–10. 10.1080/07391102.2020.1761881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmezayen, A. D., Al-Obaidi, A., Şahin, A. T., & Yelekçi, K. (2020). Drug repurposing for coronavirus (COVID-19): In silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. Journal of Biomolecular Structure and Dynamics, 38, 1–13. 10.1080/07391102.2020.1758791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enmozhi, S. K., Raja, K., Sebastine, I., & Joseph, J. (2020). Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: An in silico approach. Journal of Biomolecular Structure and Dynamics, 38, 1–7. 10.1080/07391102.2020.1760136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann, U., Perera, L., Berkowitz, M. L., Darden, T., Lee, H., & Pedersen, L. G. (1995). A smooth particle mesh Ewald method. The Journal of Chemical Physics, 103(19), 8577–8593. 10.1063/1.470117 [DOI] [Google Scholar]
- Gajula, M. P., Kumar, A., & Ijaq, J. (2016). Protocol for molecular dynamics simulations of proteins. Bio-Protocol, 6(23), e2051–e2051. 10.21769/BioProtoc.2051 [DOI] [Google Scholar]
- Geourjon, C., & Deléage, G. (1995). SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Computer Applications in the Biosciences: Cabios, 11(6), 681–684. 10.1093/bioinformatics/11.6.681 [DOI] [PubMed] [Google Scholar]
- Guo, Y. R., Cao, Q. D., Hong, Z. S., Tan, Y. Y., Chen, S. D., Jin, H. J., Tan, K. S., Wang, D. Y., & Yan, Y. (2020). The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Military Medical Research, 7(1), 11. 10.1186/s40779-020-00240-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyebi, G. A., Ogunro, O. B., Adegunloye, A. P., Ogunyemi, O. M., & Afolabi, S. O. (2020). Potential inhibitors of coronavirus 3-chymotrypsin-like protease (3CLpro): An in silico screening of Alkaloids and Terpenoids from African medicinal plants. Journal of Biomolecular Structure and Dynamics, 38, 1–19. 10.1080/07391102.2020.1764868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackbart, M., Deng, X., & Baker, S. C. (2020). Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proceedings of the National Academy of Sciences of the United States of America, 117(14), 8094–8103. 10.1073/pnas.1921485117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoda, S., Gupta, L., Agarwal, H., Raj, G., Vermani, M., & Vijayaraghavan, P. (2019). Inhibition of aspergillus fumigatus biofilm and cytotoxicity study of natural compound cis-9-hexadecenal. Journal of Pure and Applied Microbiology, 13(2), 1207–1216. 10.22207/JPAM.13.2.61 [DOI] [Google Scholar]
- Islam, R., Parves, R., Paul, A. S., Uddin, N., Rahman, M. S., Mamun, A. A., Hossain, M. N., Ali, M. A., & Halim, M. A. (2020). A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. Journal of Biomolecular Structure and Dynamics, 38, 1–20. 10.1080/07391102.2020.1761883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., Zhang, B., Li, X., Zhang, L., Peng, C., Duan, Y., Yu, J., Wang, L., Yang, K., Liu, F., Jiang, R., Yang, X., You, T., Liu, X., … Yang, H. (2020). Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature, 582(7811), 289–293. 10.1038/s41586-020-2223-y [DOI] [PubMed] [Google Scholar]
- Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W., & Klein, M. L. (1983). Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics, 79(2), 926–935. 10.1063/1.445869 [DOI] [Google Scholar]
- Joshi, R. S., Jagdale, S. S., Bansode, S. B., Shankar, S. S., Tellis, M. B., Pandya, V. K., Chugh, A., Giri, A. P., & Kulkarni, M. J. (2020). Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. Journal of Biomolecular Structure and Dynamics, 38, 1–16. 10.1080/07391102.2020.1760137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, S. A., Zia, K., Ashraf, S., Uddin, R., & Ul-Haq, Z. (2020). Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. Journal of Biomolecular Structure and Dynamics, 38, 1–10. 10.1080/07391102.2020.1751298 [DOI] [PubMed] [Google Scholar]
- Kim, Y., Jedrzejczak, R., Maltseva, N. I., Wilamowski, M., Endres, M., Godzik, A., Michalska, K., & Joachimiak, A. (2020). Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Science, 29(7), 1596–1605. 10.1002/pro.3873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y., Jedrzejczak, R., Maltseva, N. I., Wilamowski, M., Endres, M., Godzik, A., Michalska, K., & Joachimiak, A. (2020). Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2 . Protein Sci, 29(7), 1596–1605. 10.1002/pro.3873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazato, K., Wang, Y., & Kobayashi, N. (2007). Viral infectious disease and natural products with antiviral activity. Drug Discoveries & Therapeutics, 1(1), 14–22. [PubMed] [Google Scholar]
- Kutluay, S. B., Doroghazi, J., Roemer, M. E., & Triezenberg, S. J. (2008). Curcumin inhibits herpes simplex virus immediate-early gene expression by a mechanism independent of p300/CBP histone acetyltransferase activity. Virology, 373(2), 239–247. 10.1016/j.virol.2007.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, K. P., Ng, L. F. P., & Liu, D. X. (2000). Identification of a novel cleavage activity of the first papain-like proteinase domain encoded by open reading frame 1a of the coronavirus avian infectious bronchitis virus and characterization of the cleavage products. Journal of Virology, 74(4), 1674–1685. 10.1128/jvi.74.4.1674-1685.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C., Zhou, Q., Li, Y., Garner, L. V., Watkins, S. P., Carter, L. J., Smoot, J., Gregg, A. C., Daniels, A. D., Jervey, S., & Albaiu, D. (2020). Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Central Science, 6(3), 315–331. 10.1021/acscentsci.0c00272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X., Fang, P., Fang, L., Hong, Y., Zhu, X., Wang, D., Peng, G., & Xiao, S. (2019). Porcine deltacoronavirus nsp15 antagonizes interferon-β production independently of its endoribonuclease activity. Molecular Immunology, 114, 100–107. 10.1016/j.molimm.2019.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo-Galo, N., Terrazas-López, M., Martínez-Martínez, A., & Díaz-Sánchez, Á. G. (2020). FDA-approved thiol-reacting drugs that potentially bind into the SARS-CoV-2 main protease, essential for viral replication. Journal of Biomolecular Structure and Dynamics, 38, 1–12. 10.1080/07391102.2020.1764393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, J. A., Martinez, C., Kasavajhala, K., Wickstrom, L., Hauser, K. E., & Simmerling, C. (2015). ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. Journal of Chemical Theory and Computation, 11(8), 3696–3713. 10.1021/acs.jctc.5b00255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., & Olson, A. J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30(16), 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounce, B. C., Cesaro, T., Carrau, L., Vallet, T., & Vignuzzi, M. (2017). Curcumin inhibits Zika and chikungunya virus infection by inhibiting cell binding. Antiviral Research, 142, 148–157. 10.1016/j.antiviral.2017.03.014 [DOI] [PubMed] [Google Scholar]
- Muralidharan, N., Sakthivel, R., Velmurugan, D., & Gromiha, M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. Journal of Biomolecular Structure and Dynamics, 38, 1–6. 10.1080/07391102.2020.1752802 [DOI] [PubMed] [Google Scholar]
- O’Boyle, N. M., Banck, M., James, C. A., Morley, C., Vandermeersch, T., & Hutchison, G. R. (2011). Open Babel: An open chemical toolbox. Journal of Cheminformatics, 3, 33. 10.1186/1758-2946-3-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., & Ferrin, T. E. (2004). UCSF Chimera-a visualization system for exploratory research and analysis. J Comput Chem, 25(13), 1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Polansky, H., & Lori, G. (2020). Coronavirus disease 2019 (COVID-19): first indication of efficacy of Gene-Eden-VIR/Novirin in SARS-CoV-2 infection. International Journal of Antimicrobial Agents, 55(6), 105971. 10.1016/j.ijantimicag.2020.105971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajtar, B., Skalicka-Woźniak, K., Świątek, Ł., Stec, A., Boguszewska, A., & Polz-Dacewicz, M. (2017). Antiviral effect of compounds derived from Angelica archangelica L. on Herpes simplex virus-1 and Coxsackievirus B3 infections. Food and Chemical Toxicology: An International Journal Published for the British Industrial Biological Research Association, 109(Pt 2), 1026–1031. 10.1016/j.fct.2017.05.011 [DOI] [PubMed] [Google Scholar]
- Ren, Z., Yan, L., Zhang, N., Guo, Y., Yang, C., Lou, Z., & Rao, Z. (2013). The newly emerged SARS-Like coronavirus HCoV-EMC also has an “Achilles' heel”: current effective inhibitor targeting a 3C-like protease. Protein & Cell, 4(4), 248–250. 10.1007/s13238-013-2841-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe, D. R., & Cheatham, T. E. (2013). PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. Journal of Chemical Theory and Computation, 9(7), 3084–3095. 10.1021/ct400341p [DOI] [PubMed] [Google Scholar]
- Ryckaert, J. P., Ciccotti, G., & Berendsen, H. J. C. (1977). Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. Journal of Computational Physics, 23(3), 327–341. 10.1016/0021-9991(77)90098-5 [DOI] [Google Scholar]
- Salem, M. M., & Werbovetz, K. A. (2006). Isoflavonoids and other compounds from Psorothamnus arborescens with antiprotozoal activities. Journal of Natural Products, 69(1), 43–49. 10.1021/np0502600 [DOI] [PubMed] [Google Scholar]
- Shah, B., Modi, P., & Sagar, S. R. (2020). In silico studies on therapeutic agents for COVID-19: Drug repurposing approach. Life Sciences, 252, 117652. 10.1016/j.lfs.2020.117652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikishima, Y., Takaishi, Y., Honda, G., Ito, M., Takfda, Y., Kodzhimatov, O. K., Ashurmetov, O., & Lee, K. H. (2001). Chemical constituents of Prangos tschiniganica; structure elucidation and absolute configuration of coumarin and furanocoumarin derivatives with anti-HIV activity . Chem. Pharm. Bull, 49(7), 877–880. 10.1248/cpb.49.877 [DOI] [PubMed] [Google Scholar]
- Sinha, S. K., Shakya, A., Prasad, S. K., Singh, S., Gurav, N. S., Prasad, R. S., & Gurav, S. S. (2020). An in-silico evaluation of different Saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets. Journal of Biomolecular Structure and Dynamics, 38, 1–13. 10.1080/07391102.2020.1762741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, N., Yu, T., Zhao, J. X., Sun, Y. G., Jiang, J. B., Duan, Z. B., Wang, W. K., Hu, Y. L., Lei, H. M., & Li, H. Q. (2015). Antiviral activities of natural compounds derived from traditional chinese medicines against porcine circovirus type 2 (PCV2). Biotechnology and Bioprocess Engineering, 20(1), 180–187. 10.1007/s12257-014-0520-8 [DOI] [Google Scholar]
- Teague, S. J. (2003). Implications of protein flexibility for drug discovery. Nature Reviews. Drug Discovery, 2(7), 527–541. 10.1038/nrd1129 [DOI] [PubMed] [Google Scholar]
- Veckenstedt, A., Güttner, J., & Béládi, I. (1987). Synergistic action of quercetin and murine alpha/beta interferon in the treatment of Mengo virus infection in mice. Antiviral Research, 7(3), 169–178. 10.1016/0166-3542(87)90005-2 [DOI] [PubMed] [Google Scholar]
- von Rhein, C., Weidner, T., Henß, L., Martin, J., Weber, C., Sliva, K., & Schnierle, B. S. (2016). Curcumin and Boswellia serrata gum resin extract inhibit chikungunya and vesicular stomatitis virus infections in vitro. Antiviral Research, 125, 51–57. 10.1016/j.antiviral.2015.11.007 [DOI] [PubMed] [Google Scholar]
- Wang, F., Chen, C., Tan, W., Yang, K., & Yang, H. (2016). Structure of Main Protease from Human Coronavirus NL63: Insights for Wide Spectrum Anti-Coronavirus Drug Design. Scientific Reports, 6(1), 22677. 10.1038/srep22677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Wang, W., Kollman, P. A., & Case, D. A. (2006). Automatic atom type and bond type perception in molecular mechanical calculations. Journal of Molecular Graphics & Modelling, 25(2), 247–260. 10.1016/j.jmgm.2005.12.005 [DOI] [PubMed] [Google Scholar]
- Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A., & Case, D. A. (2004). Development and testing of a general amber force field. J Comput Chem, 25(9), 1157–1174. 10.1002/jcc.20035 [DOI] [PubMed] [Google Scholar]
- Xue, X., Yu, H., Yang, H., Xue, F., Wu, Z., Shen, W., Li, J., Zhou, Z., Ding, Y., Zhao, Q., Zhang, X. C., Liao, M., Bartlam, M., & Rao, Z. (2008). Structures of two coronavirus main proteases: Implications for substrate binding and antiviral drug design. Journal of Virology, 82(5), 2515–2527. 10.1128/JVI.02114-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, M. S., Lee, J., Lee, J. M., Kim, Y., Chin, Y. W., Jee, J. G., Keum, Y. S., & Jeong, Y. J. (2012). Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13. Bioorganic & Medicinal Chemistry Letters, 22(12), 4049–4054. 10.1016/j.bmcl.2012.04.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J., Xie, G., & Yan, X. (2011). Volume 6 indexes. In Zhou J., Xie G., & Yan X. (Eds.), Encyclopedia of traditional Chinese medicines - molecular structures, pharmacological activities, natural sources and applications (pp. 1–730). Springer. 10.1007/978-3-642-16744-7_1 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.