ABSTRACT
Coronavirus disease (COVID-19) is challenging many health, economic, and social systems. RT-PCR assays are diagnosis gold standard; however, they can lead to false-negative results. Therefore, anti-SARS-CoV-2 IgG, IgM, and IgA investigation can play a complementary role in assessing the individuals immune status. Majority of serological tests focus on IgM and IgG although IgA are the main immunoglobulins involved in mucosal immunity. It has been reported that digestive symptoms may occur in the absence of any typical respiratory symptom. Thus, a complete screening, comprising IgA, IgM, and IgG detection could be more consistent and useful in patients with atypical symptoms or in paucisymptomatic cases. Current literature describes over 200 immunoassays available worldwide, pointing out a great results variability, depending on methodology or antigens’ nature. In our study we evaluated anti-SARS-CoV-2 IgA, IgM, and IgG trend on a control group and on two COVID-19 patient groups (early and late infection time) with a lateral-flow combined immunoassay (LFIA) and an enzyme-linked immunosorbent assay (ELISA). Dissimilar antibodies time kinetics have been described in COVID-19 (decreasing IgM concentration with IgA/IgG persistence for a longer time; as well as persistent IgA, IgG, and IgM concentration); our results confirmed both of them depending on the methodology; therefore, it is difficult to compare different studies outcomes, suggesting the importance of a serological tests international standardization. Nevertheless, we propose a flowchart with combined anti-SARS-CoV-2 IgG/IgM/IgA detection as a screening on general population, where serological positivity should be considered as an “alert,” to avoid and contain possible new outbreaks.
KEYWORDS: SARS-CoV-2, COVID-19 serological test, lateral flow immunoassay (LFIA), ELISA assay
Introduction
The novel coronavirus named SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2), emerged in the region of Wuhan (China) on late December 2019, has rapidly spread all over the world originating coronavirus disease (COVID-19) and the subsequent COVID-19 pandemic (Wells et al. 2020; Zhu et al. 2020).
SARS-CoV-2, as other coronaviruses, causes a variety of possible symptoms ranging from mild rhinitis, fever, cough or diarrhea, to pneumonia and acute respiratory distress syndrome (ARDS). Number of hospitalizations, need of intensive care, and deaths are still rising; at time of writing (July 17, 2020) there are 13.616 million worldwide confirmed cases with 585.727 deaths (World Health Organization 2020), producing major impacts not only on public health systems but on state economies and social living.
COVID-19 is currently diagnosed through SARS-CoV-2 RNA detection in upper and lower respiratory specimens by molecular tests, such as real-time reverse-transcription polymerase chain reaction (RT-PCR) (Wells et al. 2020; Zhu et al. 2020). However, it has been reported several times that RT-PCR can give false negative results, depending on sampling and extraction methods or on the presence of a low viral load, thus originating severe consequences by facilitating contagious individuals circulation (Ai et al. 2020; West et al. 2020).
Knowledge of SARS-CoV-2 diagnostic tests is still evolving and a clear understanding of tests nature and interpretation of their findings is important. Although generally the timing of immunoglobulin production (from 4 days after symptoms onset, to 10–14 days) limits its applicability in the acute phase diagnosis (Padoan et al. 2020a; Xiang et al. 2020), anti-SARS-CoV-2 antibodies, by detecting IgM and IgA which are swiftly formed in response to infection, may represent a tool that can both help to close the RT-PCR negative gap, as well as significantly increase COVID-19 patients diagnostic sensitivity when combining serological tests with molecular tests. As it is clear that asymptomatic cases exist, the real percentage and how long they carry the virus is not known, therefore, screening for virus-specific IgM, IgG, and IgA antibodies will be an informative and decisive factor in controlling the pandemic as it is the main indicator of population immunity development (Azkur et al. 2020).
Moreover, serological data will enable the gathering of important epidemiological information, providing more realistic data on the epidemic spreading and on morbidity and mortality. In addition, anti-SARS-CoV-2 antibodies detection will play a key role in determining appropriate lockdown exit strategies, in vaccine or therapies development and to eventually manage and control further disease outbreaks (Nuccetelli et al. 2020).
COVID-19 incubation time is relatively long and has been reported to be 2–14 days. Specific IgM and IgA are the early antibody response that starts and peaks within 7 days; specific IgG antibodies develop few days after (ranging from 10 to 18 days), do not decrease to undetectable levels and are assumed to continue lifelong as protective antibodies (Guo et al. 2020). However, three types of seroconversion have been described: IgG and IgM synchronous seroconversion; IgM seroconversion earlier than IgG and IgM seroconversion later than IgG (Lee et al. 2020; Long et al. 2020).
Several serological assays have been developed since the beginning of COVID-19 pandemic, including enzyme-linked immunosorbent assays (ELISA), lateral flow immunoassays (LFIA), point-of-care test (POCT)-fluorescence assays, and chemiluminescence immunoassays (CLIA) (Okba et al. 2020), generating great variability among different serological kits results, thus demanding international standardization on methodology and SARS-CoV-2 antigens.
The most used viral proteins as antigens in the available serological assays are: nucleocapsid protein (NP), transmembrane spike protein (SP), and spike protein subunits S1 and S2. S1 contains the receptor binding domain (RBD) for the host angiotensin-converting enzyme (ACE2) receptor; S2 contains elements needed for membrane fusion (Espejo et al. 2020; Tian et al. 2020).
Whatever the method used, nature of the antigen is important, considering that detection of antibodies directed against spike protein or its subunits are more likely to have a neutralizing activity and would better describe the immunization state.
Majority of serological tests focus on IgM and IgG antibodies although IgA antibodies have an important role in mucosal immunity, being the most important immunoglobulin to counteract infectious pathogens in respiratory and digestive systems (Chao et al. 2020).
The development of mucosal immunity via IgA may be important in preventing SARS-CoV-2 infections (Fox et al. 2020) given that the virus enters and attacks the respiratory epithelial cell by docking to the angiotensin-converting enzyme-2 (ACE2) protein on the surface of type-2 alveolar cells (Mahmoodpoor and Nader 2020).
Moreover, besides typical respiratory symptoms, digestive symptoms have been frequently reported including nausea, vomiting, diarrhea, and anorexia; in some cases digestive symptoms may occur in the absence of any respiratory symptom. In patients with COVID-19, diarrhea is also a common digestive symptom, with the incidence ranging from 1.3% to 29.3% (Agarwal et al. 2020).
In addition, SARS-CoV-2 induced diarrhea could be the onset symptom in patients with COVID-19 (Song et al. 2020). Nevertheless, the incidence of diarrhea varied widely among different reports, suggesting that clinicians might underestimate the value of digestive symptom in clinical practice, affecting the preliminary diagnostic accuracy (Liang et al. 2020).
Furthermore, COVID-19 disease in a patient with positive fecal but negative pharyngeal and sputum viral tests has been reported (Chen et al. 2020).
In this perspective, IgA assays could be useful, along with IgG and IgM, either in patients presenting with atypical symptoms and in paucisymptomatic cases (e.g. with transient mild conjunctivitis and low fever) or when naso-pharyngeal swab RT-PCR repeatedly remains negative in suspected subjects. To this end, anti-SARS-CoV-2 humoral response time kinetics can aid in COVID-19 diagnosis, including subclinical cases.
It has been reported that anti-SARS-CoV-2 IgM and IgA levels increased from days 0 to 14 and did not increase further between days 15 and 21 or after day 21; anti-SARS-CoV-2 IgG were detected on days 0–7, increased on days 8–14, continued to increase until days 15–21, and reached a plateau by day 21 (Guo et al. 2020).
Another study described the anti-SARS-CoV-2 IgA kinetics peculiar characteristics in comparison to anti-SARS-CoV-2 IgM: IgA and IgM levels increased since days 6–8 from symptoms onset, than IgA showed persistently higher levels over 38 days, with a peak level at days 20–22, whereas IgM levels peaked at days 10–12 and significantly declined at day 18 (Padoan et al. 2020b).
Taking into account all the above considerations, the aim of our study was to evaluate the serological IgA, IgM, and IgG anti-SARS-CoV-2 antibodies time kinetics trend, as well as kits sensitivities and specificities, with two different assays: a lateral-flow combined immunoassay (LFIA) and an enzyme-linked immunosorbent assay (ELISA). The assays were performed on a control group (healthcare workers with negative swabs) and on two different COVID-19 patient groups: early infection time patients (ranging from 1 to 9 days from first access to Emergency Department and from first positive nasopharyngeal swab); late infection time patients (ranging from 19 to 41 days from first access to Emergency Department and from first positive nasopharyngeal swab).
Materials and methods
Patients and serum specimens
Serum samples were recovered, in accordance with local ethical approvals (R.S.44.20), from “Tor Vergata” University Covid-Hospital of Rome hospitalized patients as follows: 44 positive RT-PCR-diagnosed SARS-CoV-2 patients (mean age 67.3 years ± 16.6 years; 25 males and 19 females), collected on days 1 to 9 from first access to Emergency Department and from first positive nasopharyngeal swab (GROUP 1); 48 positive RT-PCR-diagnosed SARS-CoV-2 patients (mean age 69.7 years ± 13.3 years; 27 males and 21 females), collected on days 19–41 from first access to Emergency Department and from first positive nasopharyngeal swab (GROUP 2) and 44 negative RT-PCR-diagnosed SARS-CoV-2 subjects (mean age 41.7 years ± 11.1 years; 23 males and 21 females) collected from Tor Vergata Hospital physicians and healthcare workers screened for internal surveillance (CONTROL GROUP) (Figure 1). Sera were separated by centrifugation at 2500 g for 10 min, within 1 h from collection.
Figure 1.
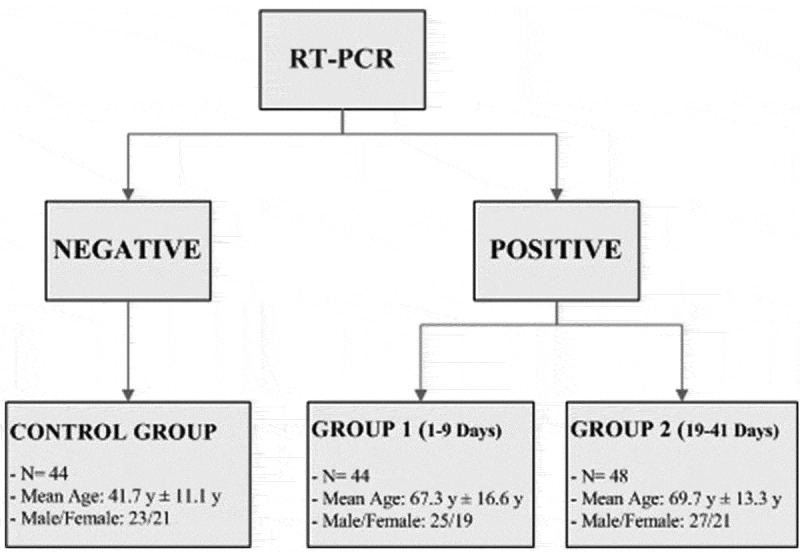
Characteristics of the groups involved in the study. All subjects have been tested for SARS-CoV-2 RNA detection in naso-pharyngeal swab by real-time polymerase chain reaction. Control group consists of serum samples from physicians and healthcare workers screened for internal surveillance, with negative RT-PCR results; Group 1 and Group 2 consist of serum samples from COVID-19 patients (collected on days 1 to 9 and 19 to 41 from first access to Emergency Department and from first positive nasopharyngeal swab, respectively), with positive RT-PCR results.
Real time polymerase chain reaction (RT-PCR)
Nasopharyngeal swabs were tested for SARS-CoV-2 infection with Seegene AllplexTM2019-nCoV Assay (Seegene, Seoul, South Korea), according to the manufacturer’s protocols.
Automated RNA extraction and PCR setup were carried out using Seegene NIMBUS, a liquid handling workstation. RT-PCR was run on a CFX96TMDx platform (Bio-Rad Laboratories, Inc., CA, USA) and subsequently interpreted by Seegene’s Viewer Software. The Seegene AllplexTM2019-nCoV Assay identifies the virus by multiplex real-time PCR targeting three viral genes (E, RdRP and N), thus complying with international validated testing protocols.
Lateral flow immunoassay (LFIA)
Lateral flow chromatographic immunoassay for combined qualitative detection of IgG and IgA/IgM antibodies to 2019-nCoV in human whole blood, serum, or plasma specimens (Beijing Zhongjian Antai Diagnostic Technology Co. Ltd., Beijing, China; distributed in Italy by Tregena srl, Roma, Italy). During testing, sample reacts with 2019-nCoV antigen-coated particles (recombinant nucleocapsid protein (NP), as declared by manufacturer) in the test cassette: a colored line will appear in IgG (T2) or combined IgM/IgA (T1) test line regions as a result, and in the control region (C) as an internal procedural control. T1 shows anti-SARS-CoV-2 IgA and/or IgM detection and gives feedback on the presence of infection. T2 shows anti-SARS-CoV-2 IgG detection, suggesting that patient may be currently infected or has had a previous infection. The results must be read strictly within 15–20 minutes. This test is CE approved.
Enzyme-linked immunosorbent assay (ELISA)
Immunoenzymatic assays for COVID-19 IgG, IgA, and IgM antibodies determination in human plasma and serum (DIA.PRO, Diagnostic Bioprobes srl, Milano, Italy; distributed by Alifax Srl, Padova, Italy), performed on the fully automated Immunomat Virion/Serion Analyzer (Serion Immundiagnostica GmbH Würzburg, Germany; distributed in Italy by Alifax Srl, Padova, Italy). The microplates are coated with COVID-19 recombinant antigens (nucleocapsid protein (NP), Spike glycoprotein subunits S1 and S2, as declared by manufacturer). The specific anti-SARS-CoV-2 immunoglobulins are bound to the antigens through incubation with diluted human serum (1:20), at 37°C. After washing to remove all not reacting proteins, the antibodies are detected by adding a conjugate solution containing polyclonal anti-human IgG, IgM, or IgA, labeled with horseradish peroxidase (HRP). Finally, a chromogenic solution with the HRP substrate (tetramethilbenzidine; TMB) is added, developing a blue color. The reaction is then stopped by adding 0.3 M sulphuric acid and the absorbances are read spectrophotometrically at 450 nm The optical density (OD) is proportional to the quantity of the specific anti-SARS-CoV-2 antibodies present in the samples. Results are calculated semi-quantitatively by a ratio between samples OD values and a cut-off value determined with the following formula:
Cut-Off (CO) = Negative Control Absorbance + 0.25
Ratio is considered negative for all the values < 0.9 COI (Cut Off Index); equivocal for all the values between 0.9 and 1.1 COI; positive for all the values >1.1 COI. This test is CE approved.
Total serum IgA assay was performed on the fully automated Abbott Architect c16000 Clinical Chemistry Analyzer (Abbott Diagnostics, Chicago, USA), by an immunoturbidimetric reaction measuring sample turbidity caused by insoluble immune complexes formation.
Statistical analysis
Qualitative rapid test kits specificity and sensitivity were calculated according to the following formulas:
Specificity (%) = 100 × [True negative/(True Negative + False Positive)].
Sensitivity (%) = 100 × [True Positive/(True Positive + False Negative)]
Specificity and sensitivity for ELISA tests were calculated by Receiver Operating Characteristic Curves (ROC Curve). All data were analyzed using Med Calc Ver.18.2.18 (MedCalc Software Ltd, Ostend, Belgium). The investigators were blinded to the group allocation during the experiment.
Ethical statement
The study was performed according to “Tor Vergata” University Covid-Hospital of Rome local ethical approvals (protocols no. R.S.44.20). Informed consent was obtained from all subjects enrolled in the study. The study was in accordance with the Helsinki Declaration, as revised in 2013.
Results
Sensitivities and specificities were calculated using receiver operating characteristic (ROC) curves, for the immunoenzymatic anti-SARS-CoV-2 IgA, IgG, IgM semi-quantitative assays (ELISA), and with formulas reported under “Materials and Methods” for the combined anti-SARS-CoV-2 IgA/IgM, IgG qualitative lateral-flow immunoassay (LFIA). Results with the analytical parameters for each test (area under curve (AUC), sensitivity and specificity) are summarized in Table 1.
Table 1.
Area under curve (AUC), sensitivity, and specificity of SARS-CoV-2 IgA-IgM-IgG serological tests.
| LFIA GROUP 1
(1–9 days) N = 44 |
LFIA GROUP 2
(19–41 days) N = 48 |
ELISA GROUP 1
(1–9 days) N = 44 |
ELISA GROUP 2
(19–41 days) N = 48 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CONTROL NEGATIVE GROUP N = 44 | IgA/IgM | IgG | IgA/IgM | IgG | IgA | IgM | IgG | IgA | IgM | IgG |
| Sensitivity (%) | 80 | 71 | 71 | 84 | 82 | 80 | 94 | 90 | 92 | 98 |
| Specificity (%) | 96 | 94 | 96 | 94 | 93 | 100 | 93 | 94 | 100 | 94 |
| Kit Cut-off | Qualitative Test | Qualitative Test | >1,1 COI | >1,1 COI | >1,1 COI | >1,1 COI | >1,1 COI | >1,1 COI | ||
| Area under the ROC curve (AUC); 95% Confidence interval | 0,955 | 0,932 | 0,962 | 0,984 | 0,978 | 0,997 | ||||
| Sensitivity (%) | 96 | 82 | 95 | 96 | 92 | 96 | ||||
| Specificity (%) | 91 | 98 | 93 | 93 | 100 | 100 | ||||
| Laboratory Cut-off | >0,61 COI | >0,94 COI | >0,91 COI | >0,68 COI | >1,1 COI | >1,97 COI | ||||
They have been correlated to the manufacturer’s cut-off and recalculated on a best fit cut-off that emerged from our data analysis.
The ROC curves have good AUC values in group 1, with an IgM moderately lower result (0.955 for IgA, 0.932 for IgM and 0.962 for IgG); the AUC values display better performances on group 2, where they increase in all the specific antibodies classes, reaching an optimal value close to 1 for IgG (0.984, 0.978, 0.997; IgA, IgM, and IgG respectively) (Figure 2). Interestingly, with our recalculated best-fit cut-off (0.61/0.68 compared to 1.1), sensitivities considerably increased especially for IgA in both groups (from 82% to 96% in group 1 and from 90% to 96% in group 2) (Table 1), suggesting to revise the cut-off values reported on datasheets by companies, because sometimes they are evaluated on a small number of subjects belonging to a specific ethnicity or region.
Figure 2.
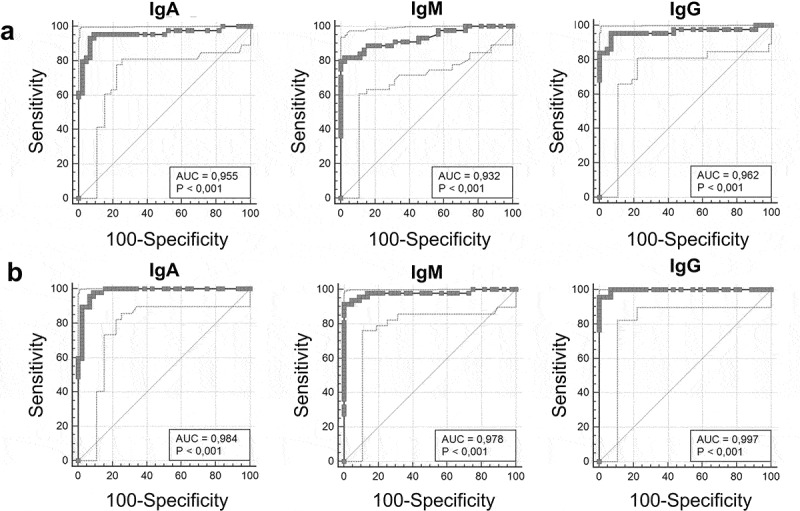
Anti-SARS-CoV-2 serological tests ROC curves. ELISA IgA, IgM and IgG results for group 1 are shown in Panel A (AUC 0.955, 0.932 and 0.962 respectively); ELISA IgA, IgM and IgG results for group 2 are shown in Panel B (AUC 0.984, 0.978, 0.997, respectively).
The lateral flow immunochromatographic test shows the same specificities in groups 1 and 2 (94% and 96%, for IgG and combined IgA/IgM respectively), whereas sensitivities are different: 80% for IgA/IgM in group 1 and 71% for IgA/IgM in group 2; 71% and 84% for IgG, in groups 1 and 2 respectively (Table 1). The ELISA assays show the same specificities in groups 1 and 2 for IgM (100%) and a little increase in group 2 compared to group 1 for both IgG and IgA (94% versus 93% respectively); on the other hand, sensitivities show better performances on group 2 compared to group 1 (90% versus 82% for IgA; 92% versus 80% for IgM; 98% versus 94% for IgG). Since selective immunoglobulin A deficiency (SIGAD) is the most common inherited immunodeficiency disorder worldwide (with a prevalence ranging from 1 in 100 to 1 in 1000 people, depending by population), we detected total IgA in all anti-SARS-CoV-2 IgA negative samples to avoid false negative results. Only two samples displayed moderate IgA deficiency, with a concentration of 57 and 77 mg/dl (normal range: 101–645 mg/dl) that presumably did not interfere with our results.
Overall data show similar or identical IgG specificities in group 1 and 2 between the different assays (about 94%), but there is an excellent result on IgM detection when its assay is independent from IgA (100% in ELISA test; 96% in lateral flow test) and this value does not change between group 1 and 2. In contrast, ELISA assays show better performances on the analytical sensitivities, not only on IgG, IgM and IgA results but also on group 1 and group 2 differences; in fact, regarding IgG they were 94% and 98% for group 1 and 2 respectively, compared to the lateral flow results (71% for group 1 and 84% for group 2); regarding group 1 and 2 there was an increase from 82% to 90% for IgA and from 80% to 92% for IgM, whereas lateral flow results were completely opposite, showing an IgA/IgM decrease from 80% to 71%.
Dissimilar time kinetics have been described in COVID-19 specific antibodies (IgM decreasing concentration with IgA and IgG persistence for a longer time; as well as different times of early response for IgA and IgM with persistent IgA, IgG, and IgM concentration) and our results confirmed both of them, depending on the assay methodology.
Contrasting results are probably due to the antigen characteristics (NP in LFIA assay; NP and spike protein subunits S1 and S2 in ELISA assay), and to time and temperature used for sample incubation that could improve the antibody/antigen binding affinity (1-hour incubation/37°C versus 15 minutes/room temperature). Unfortunately, majority of in vitro diagnostic (IVD) companies do not report the antigen nature; therefore, it is difficult to compare different studies outcomes, suggesting the importance of an international standardization of serological tests.
Discussion
COVID-19 is probably one of the biggest health and economic burden of the last 100 years; as a zoonotic disease that has already spread globally to several millions of humans it will be practically impossible to eradicate it (Guo et al. 2020). From the first outbreak in China and a second large outbreak in Northern Italy, we are still dealing with an increase in the number of confirmed cases in different countries. European nations have been the first to be strongly affected by virus spreading that is now afflicting Russia, Brazil and the United States, where the epidemiological curve is not yet flattening, emphasizing the importance and need of a global strategy.
SARS-CoV-2 showed an unexpected high speed of transmission and spreadability related to different factors such as: a long infectious window period before symptoms onset, a large number of asymptomatic virus-carriers and super-spreaders, and the globalization extent with individual travelling. While there are estimates of disease severity and infection attack rates, a major knowledge gap still remains because mild or asymptomatic individuals are difficult to identify (Gao et al. 2020; Kronbichler et al. 2020).
Fast and reliable SARS-CoV-2 laboratory diagnostics are important to support implementation of appropriate public health interventions; in COVID-19 acute phase they relied on molecular methods, subsequently serological assays have been developed to allow epidemiological assessments through serosurveys, as well as retrospective diagnosis in targeted groups (Jaaskelainen et al. 2020). Nevertheless, it should be pointed out that molecular tests represent an “instantaneous” picture of possible virus presence, whereas serological tests display virus presence during a wider phase of the infectious process, whether or not it reaches a clinical relevance.
Immunoassays could provide identification of noncontagious and potentially protected individuals to support progressive deconfinement strategies in the process of gradually restoring safe economic and social activities (Gilbert et al. 2020; Montesinos et al. 2020; Yan et al. 2020). A complete serological screening, comprising IgA, IgM and IgG detection could be more consistent as a security strategy to prevent virus spreading (Jaaskelainen et al. 2020) and it could be applied to the massive testing challenge the world is currently facing, helping in containing the pandemic or to identify and promptly isolate little outbreaks that have become frequent after the lockdown exit.
Our data show high analytical performances in detecting anti-SARS-CoV-2 IgA, IgG, and IgM, especially with the ELISA methodology and on the late infection patients (19–41 days). In this group specificities are 94% for IgA and IgG, and 100% for IgM; sensitivities are 90% for IgA, 92% for IgM and 98% for IgG, and increased from group 1 to group 2 for all the specific antibodies. Since the latter results denote a persistent positivity over the time, serological tools could be useful to understand the infection overall rate in the communities, including the rate of asymptomatic infections.
We previously proposed a flowchart in which serological tests were included to help social and work activities implementation after the pandemic acute phase, integrated with nasopharyngeal RT-PCR swab when necessary (Nuccetelli et al. 2020). The flowchart took into account anti-SARS-CoV-2 IgG and IgM detection, as a tool for a safe readmission at work, but with these new evidences, anti-SARS-CoV-2 IgA measurement can be added for a complete and more reliable screening on general population (Figure 3). IgA molecules are mostly produced by plasma cells in the lamina propria: they can be secreted as a dimer into mucosal surfaces and as a monomer in serum (Chao et al. 2020; Krammer 2019). A simultaneous rise of IgA levels both in serum and saliva has been reported in COVID-19 patients, thus hypothesizing that high circulating IgA levels could represent a mirror of mucosal infiltration (Randad et al. 2020). However, the role of serum anti-SARS-CoV-2 IgA is not fully understood and it cannot be excluded that patients could have high protective mucosal IgA levels without detectable circulating IgA levels.
Figure 3.
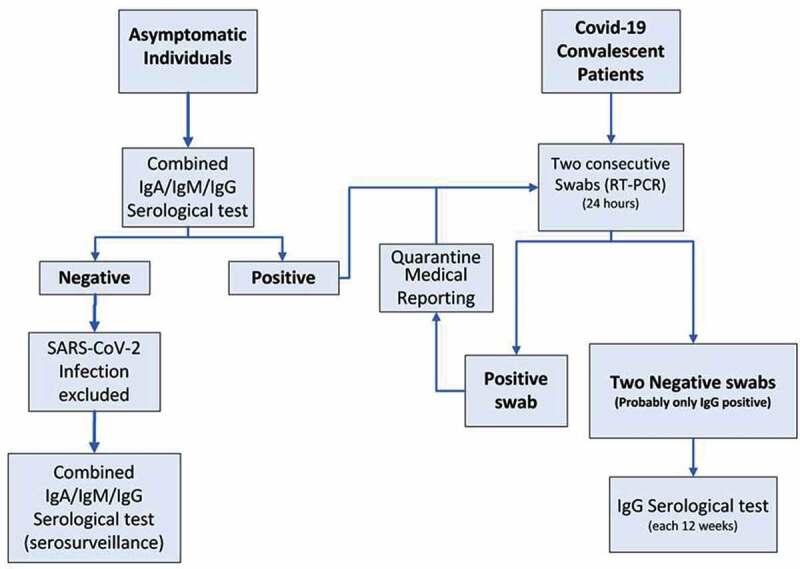
Flowchart proposal on general population. Path 1 describes combined anti-SARS-CoV-2 IgA/IgM/IgG serological test on asymptomatic general population: in case of negative results, SARS-CoV-2 infection is excluded; in case of positive results two consecutive nasopharyngeal swabs RT-PCR are mandatory. Further combined anti-SARS-CoV-2 IgA/IgM/IgG detection could be considered as serosurveillance. Path 2 describes serological anti-SARS-CoV-2 IgG assays on COVID-19 convalescent patients to detect natural immunization span.
Nevertheless, the impact on our screening flowchart is minimal because a greater number of positive individuals could be lost when anti-SARS-CoV-2 IgA are not tested.
In fact, serological positivity should be considered as an “alert,” to better investigate if the subjects are currently infectious or not, and to avoid and contain possible new outbreaks. At this regard, in the screening sustainability it should be also evaluated the cost/benefit ratio: with ELISA tests, each sample should be analyzed three times for each specific antibody, whereas in the immunochromatographic combined card it is analyzed in the same assay. From this point of view, a combined commercial chemiluminescent immunoassay (CLIA) able to detect anti-SARS-CoV-2 IgM, IgG and IgA simultaneously has been already developed, thus reducing considerably costs and time of analysis. It should be also pointed out that majority of ELISA assays (as well as CLIA assays) can be run in total automation laboratories (where traceability is complete), and are able to send results directly to Laboratory Information System, also limiting technologists exposition to blood samples; on the other hand, the immunochromatographic cards are fully manual and the reading could be subjective, but they have the advantages to be ready-to-use on capillary blood, time-saving and in some contexts easier to be analyzed by non healthcare workers (airports, prisons, religious communities, sports associations and centers for the elderly).
Finally, we acknowledge our study limitations related to the small number of patients analyzed (due to the limited kits availability) and to the lack of information on their clinical status.
Conclusions
Serological tests continue to hold promise in COVID-19 applications but there are still knowledge gaps that must be clarified to give meaningful recommendations for their use in different conditions. Almost all of the current literature focused on the serologic testing results in symptomatic patients; for screening and seroprevalence studies it will be essential to define specific antibody responses in individuals with subclinical and mild disease. In this perspective, our proposal for a combined anti-SARS-CoV-2 IgG/IgM/IgA testing on general population could provide useful information to correctly identify cases, predict clinical outcomes and develop treatment strategies to deal with the disease and to avoid major second infection waves.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Acknowledgments
The Authors would like to thank Dr. Fabio Cherubini, Dr. Paolo Casalino, Dr. Ilio Giambini, and all the Clinical Biochemistry Laboratory staff of Tor Vergata Hospital, for their support; Beijing Zhongjian Antai Diagnostic Technology Co. Ltd., Tregena srl, DIA.PRO, Diagnostic Bioprobes srl and Alifax Srl, for kindly having provided kits and instruments for this study.
Declaration of interest statement
The authors declare that they have no conflict of interest.
References
- Agarwal A, Chen A, Ravindran N, To C, Thuluvath PJ.. 2020. Gastrointestinal and liver manifestations of COVID-19. J Clin Exp Hepatol. 10(3):263–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, Tao Q, Sun Z, Xia L.. 2020. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: A report of 1014 cases. Radiology. 296(2): E32–E40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azkur AK, Akdis M, Azkur D, Sokolowska M, Veen W, Brüggen M-C, O’Mahony L, Gao Y, Nadeau K, Akdis CA. 2020. Immune response to SARS-CoV-2 and mechanisms of immunopathological changes in COVID-19. Allergy. 75(7):1564–81. doi: 10.1111/all.14364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao YX, Rotzschke O, Tan E-K. 2020. The role of IgA in COVID-19. Brain Behav Immun. 87:182–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Lou J, Bai Y, Wang M. 2020. COVID-19 disease with positive fecal and negative pharyngeal and sputum viral tests. Am J Gastroenterol. 115(5):790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espejo AP, Akgun Y, Al Mana AF, Tjendra Y, Millan NC, Gomez-Fernandez C, Cray C. 2020. Review of current advances in serologic testing for COVID-19. Am J Clin Pathol 5;154(3):293–304. doi: 10.1093/ajcp/aqaa112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox A, Marino J, Amanat F, Krammer F, Hahn-Holbrook J, Zolla-Pazner S, Powell RL. 2020. Evidence of a significant secretory-IgA-dominant SARS-CoV-2 immune response in human milk following recovery from COVID-19. medRxiv. 2020.2005.2004.20089995. https://ind01.safelinks.protection.outlook.com/?url=https%3A%2F%2Fdoi.org%2F10.1101%2F2020.05.04.20089995&data=02%7C01%7Csathyan.dhanasekaran%40integra.co.in%7Ce068a4872f9b4566ba1808d859687bdd%7C70e2bc386b4b43a19821a49c0a744f3d%7C0%7C1%7C637357652590048529&sdata=dvDJTfxQSDvSqzAzD04PuQdYR5r2I6kZqrhos2or%2BUg%3D&reserved=0. [Google Scholar]
- Gao Z, Xu Y, Sun C, Wang X, Guo Y, Qiu S, Ma K. 2020. A systematic review of asymptomatic infections with COVID-19. J Microbiol Immunol Infect. doi: 10.1016/j.jmii.2020.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M, Dewatripont M, Muraille E, Platteau J-P, Goldman M. 2020. Preparing for a responsible lockdown exit strategy. Nat Med. 26(5):643–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Ren L, Yang S, Xiao M, Chang D, Yang F, Dela Cruz CS, Wang Y, Wu C, Xiao Y. 2020. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin Infect Dis. 71(15):778–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaaskelainen AJ, Kekalainen E, Kallio-Kokko H, Mannonen L, Kortela E, Vapalahti O, Kurkela S, Lappalainen M. 2020. Evaluation of commercial and automated SARS-CoV-2 IgG and IgA ELISAs using coronavirus disease (COVID-19) patient samples. Euro Surveill. 25(18). doi: 10.2807/1560-7917.ES.2020.25.18.2000603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F. 2019. The human antibody response to influenza A virus infection and vaccination. Nat Rev Immunol. 19(6):383–97. [DOI] [PubMed] [Google Scholar]
- Kronbichler A, Kresse D, Yoon S, Lee KH, Effenberger M, Shin JI. 2020. Asymptomatic patients as a source of COVID-19 infections: A systematic review and meta-analysis. Int J Infect Dis. 98:180–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YL, Liao CH, Liu P-Y, Cheng C-Y, Chung M-Y, Liu C-E, Chang S-Y, Hsueh P-R. 2020. Dynamics of anti-SARS-Cov-2 IgM and IgG antibodies among COVID-19 patients. J Infect. 81(2):e55–e58. doi: 10.1016/j.jinf.2020.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W, Feng Z, Rao S, Xiao C, Xue X, Lin Z, Zhang Q, Qi W. 2020. Diarrhoea may be underestimated: a missing link in 2019 novel coronavirus. Gut. 69(6):1141–43. [DOI] [PubMed] [Google Scholar]
- Long QX, Liu BZ, Deng H-J, Wu G-C, Deng K, Chen Y-K, Liao P, Qiu J-F, Lin Y, Cai X-F. 2020. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 26(6):845–48. [DOI] [PubMed] [Google Scholar]
- Mahmoodpoor A, Nader ND. 2020. Immune responses to the novel coronavirus-2: friend or Foe? Immunol Invest. 1–3. doi: 10.1080/08820139.2020.1795191 [DOI] [PubMed] [Google Scholar]
- Montesinos I, Gruson D, Kabamba B, Dahma H, Van den Wijngaert S, Reza S, Carbone V, Vandenberg O, Gulbis B, Wolff F, et al. 2020. Evaluation of two automated and three rapid lateral flow immunoassays for the detection of anti-SARS-CoV-2 antibodies. J Clin Virol. 128:104413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuccetelli M, Pieri M, Grelli S, Ciotti M, Miano R, Andreoni M, Bernardini S. 2020. SARS-CoV-2 infection serology: a useful tool to overcome lockdown? Cell Death Discov. 6(1):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okba NMA, Muller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, Lamers MM, Sikkema RS, de Bruin E, Chandler FD. 2020. Severe acute respiratory syndrome coronavirus 2-specific 2−specific antibody responses in coronavirus disease patients. Emerg Infect Dis. 26(7):1478–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padoan A, Cosma C, Sciacovelli L, Faggian D, Plebani M. 2020a. Analytical performances of a chemiluminescence immunoassay for SARS-CoV-2 IgM/IgG and antibody kinetics. Clin Chem Lab Med. 58(7):1081–88 doi: 10.1515/cclm-2020-0443 [DOI] [PubMed] [Google Scholar]
- Padoan A, Sciacovelli L, Basso D, Negrini D, Zuin S, Cosma C, Faggian D, Matricardi P, Plebani M. 2020b. IgA-Ab response to spike glycoprotein of SARS-CoV-2 in patients with COVID-19: A longitudinal study. Clin Chim Acta. 507:164–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randad PR, Pisanic N, Kruczynski K, Manabe YC, Thomas D, Pekosz A, Klein S, Betenbaugh MJ, Clarke WA, Laeyendecker O, et al. 2020. COVID-19 serology at population scale: SARS-CoV-2-specific antibody responses in saliva. medRxiv. https://ind01.safelinks.protection.outlook.com/?url=https%3A%2F%2Fdoi.org%2F10.1101%2F2020.05.24.20112300&data=02%7C01%7Csathyan.dhanasekaran%40integra.co.in%7Ce068a4872f9b4566ba1808d859687bdd%7C70e2bc386b4b43a19821a49c0a744f3d%7C0%7C1%7C637357652590048529&sdata=NOcL1p9ngq56CcmHKWHlDiurjJBLjV%2BjO6Lj2uKAQZ4%3D&reserved=0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Liu P, Shi XL, Chu YL, Zhang J, Xia J, Gao XZ, Qu T, Wang MY. 2020. SARS-CoV-2 induced diarrhoea as onset symptom in patient with COVID-19. Gut. 69(6):1143–44. [DOI] [PubMed] [Google Scholar]
- Tian X, Li C, Huang A, Xia S, Lu S, Shi Z, Lu L, Jiang S, Yang Z, Wu Y. 2020. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect. 9(1):382–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells CR, Sah P, Moghadas SM, Pandey A, Shoukat A, Wang Y, Wang Z, Meyers LA, Singer BH, Galvani AP, et al. 2020. Impact of international travel and border control measures on the global spread of the novel 2019 coronavirus outbreak. Proc Natl Acad Sci U S A. 117(13):7504–09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West C, Montori V, Sampathkumar P. 2020. COVID-19 testing: the threat of false-negative results. Mayo Clin Proc. 95(6):1127–29. doi: 10.1016/j.mayocp.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2020. Coronavirus disease 2019 (COVID-19) Situation Report – 179. [accessed 2020 July 17]. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200803-covid-19-sitrep-196-cleared.pdf?sfvrsn=8a8a3ca4_4.
- Xiang F, Wang X, He X, Peng Z, Yang B, Zhang J, Zhou Q, Ye H, Ma Y, Li H, et al. 2020. Antibody detection and dynamic characteristics in patients with COVID-19. Clin Infect Dis. https://ind01.safelinks.protection.outlook.com/?url=https%3A%2F%2Fdoi.org%2F10.1093%2Fcid%2Fciaa461&data=02%7C01%7Csathyan.dhanasekaran%40integra.co.in%7Ce068a4872f9b4566ba1808d859687bdd%7C70e2bc386b4b43a19821a49c0a744f3d%7C0%7C1%7C637357652590048529&sdata=P%2FKGNvdYZU6AcE4QENQZIZIztnvirLd3KAfZahlQgAU%3D&reserved=0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Chang L, Wang L. 2020. Laboratory testing of SARS-CoV, MERS-CoV, and SARS-CoV-2 (2019-nCoV): current status, challenges, and countermeasures. Rev Med Virol. 30(3):e2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 382(8):727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]


