Abstract
Molecular evolution offers an insightful theory to interpret the genomic consequences of thermal adaptation to previous events of climate change beyond range shifts. However, disentangling often mixed footprints of selective and demographic processes from those due to lineage sorting, recombination rate variation, and genomic constrains is not trivial. Therefore, here we condense current and historical population genomic tools to study thermal adaptation and outline key developments (genomic prediction, machine learning) that might assist their utilization for improving forecasts of populations’ responses to thermal variation. We start by summarizing how recent thermal-driven selective and demographic responses can be inferred by coalescent methods and in turn how quantitative genetic theory offers suitable multi-trait predictions over a few generations via the breeder’s equation. We later assume that enough generations have passed as to display genomic signatures of divergent selection to thermal variation and describe how these footprints can be reconstructed using genome-wide association and selection scans or, alternatively, may be used for forward prediction over multiple generations under an infinitesimal genomic prediction model. Finally, we move deeper in time to comprehend the genomic consequences of thermal shifts at an evolutionary time scale by relying on phylogeographic approaches that allow for reticulate evolution and ecological parapatric speciation, and end by envisioning the potential of modern machine learning techniques to better inform long-term predictions. We conclude that foreseeing future thermal adaptive responses requires bridging the multiple spatial scales of historical and predictive environmental change research under modern cohesive approaches such as genomic prediction and machine learning frameworks.
Keywords: coalescent theory, genome-wide association studies, genome-wide selection scans, genome–environment associations, phylogeography, breeder’s equation, genomic prediction, machine learning
On the Challenges of Studying Genomic Thermal Adaptation
Warming is imposing an unprecedented climate emergency on nature, food, energy supply, and economy around the world (Ripple et al., 2020). While evolutionary genomics may improve prediction of populations’ responses to thermal change (Waldvogel et al., 2020a), geologic records of temperature and carbon dioxide (CO2) variations (Supplementary Figure S1) are also insightful into the coupling of biodiversity, climate, and the carbon cycle and hence may help predicting the consequences of future carbon emissions (Zachos et al., 2008). For instance, several reports of fire activity (Whitlock and Bartlein, 2003; Bush et al., 2008) and hydroclimate changes (Wang et al., 2017) as records of thermal changes during the Holocene have taught us that extinction is a slow process and that many species may already be functionally extinct (Cronk, 2016). A key modern advance has precisely been to couple the extinction risk with the migratory potential under an ecological niche conservatism scenario (Steinbauer et al., 2018), and predictions of population-level genomic and phenotypic responses to thermal change (Hoffmann and Sgro, 2011). Although atmospheric CO2 has been found to be better correlated with richness of (plant) species (Supplementary Figure S1C) than temperature itself throughout the Cenozoic up until 20 Mya (Jaramillo et al., 2006; Royer and Chernoff, 2013), we need to improve our understanding on how thermal change vulnerability impacts current and historical adaptive genetic variation in order to enhance populations response projections (Razgour et al., 2019).
Genomes are diverse in signatures of the populations’ evolutionary past across timescales (Wolf and Ellegren, 2017) and therefore are informative on historical adaptive responses to ancient and more recent events of climate change (Figure 1 and Table 1). By revealing the nature of these signatures and learning from previous reactions to environmental change, genomics can truly assist modern predictions aimed at incorporating responses beyond migration. Yet, disentangling often confused selective and demographic signatures from those due to genetic drift and genomic constrains is challenging (Ellegren and Galtier, 2016), consequently delaying the factual utilization of genomics for forecasting. Therefore, in this mini-review we envision summarizing modern tools from the genomic era that are enriching our comprehension of the genetic consequences of past and recent climate change, while offering a perspective on how to improve predictive models that incorporate thermal adaptation. Specifically, we aim prospecting how genomic prediction (GP) and machine learning (ML) approaches may offer cohesive frameworks to (1) integrate more traditional, but heterogeneous, genomic, and ecological datasets across temporal scales, by (2) maximizing prediction accuracies, while (3) understating the relative contribution of the underlying genomic processes. This is still a future avenue of research, and so we close by offering perspectives. Different drivers of the genomic landscape to thermal adaption (Gompert et al., 2014; Ravinet et al., 2017; Cortés and Blair, 2018; López-Hernández and Cortés, 2019), such as disruptive and background selection, gene flow (Miller et al., 2020), shared ancestral polymorphism, and mutation/recombination rate variation (Feder et al., 2012; Ellegren and Wolf, 2017; Cortés et al., 2018b), have been identified. In order to discern among them, a first necessary step toward the evaluation of the adaptive potential involves typifying the genomic landscape by using summary statistics like nucleotide diversity, π (Nei, 1987), and relative, FST (Weir and Cockerham, 1984), and absolute, DXY (Nei, 1987), divergence. FST vs. DXY contrasts inform population divergence in the presence of gene flow (co-occurrence of peaks in both profiles), recurrent selection across subpopulations (FST peaks match shallow DXY valleys), and selective sweeps predating the subpopulations’ split (FST peaks match deep DXY valleys) (Nachman and Payseur, 2012; Cruickshank and Hahn, 2014; Irwin et al., 2016). Inferences are more robust if carried out across replicated samplings of contrasting populations (e.g., in terms of thermal variation) within a hierarchically nested framework of divergence (Cortés et al., 2018b). A second step refers to the detection of selection signatures, if any – i.e., hard vs. soft selection sweeps (Pritchard et al., 2010; Zahn and Purnell, 2016), which must be followed by a third validation step across replicated demographics (Roesti et al., 2014; Lotterhos and Whitlock, 2015) and temporal levels (Nosil and Feder, 2011; Matos et al., 2015; Fragata et al., 2018).
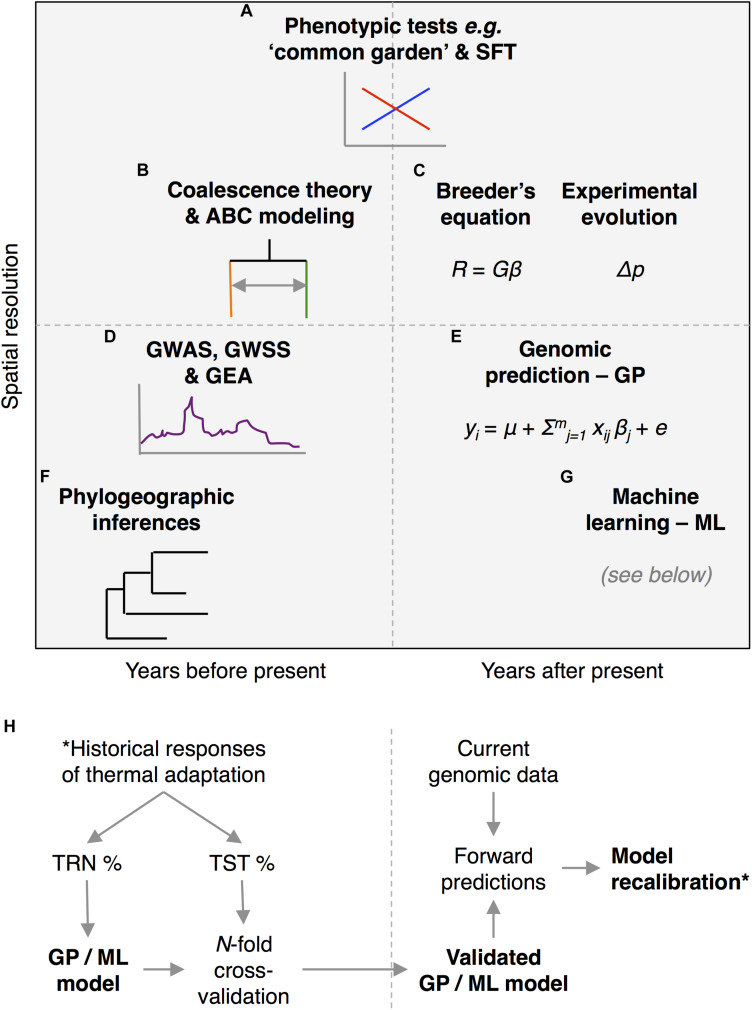
FIGURE 1.
Potential approaches to assess populations’ thermal adaptation by looking into their genomic past. Genomic analyses allow reconstructing populations’ adaptive responses to previous events of climate change across various temporal scales (A,B,D,F), as a tool to improve forecasting (C,E,G,H). (A) Empirical approaches such as replicated “common garden” (provenance) tests and space-for-time (SFT) substitution allow studying in situ ongoing genomic thermal adaptation. The inset plot exemplifies a significant genotype-by-environment (GxE) interaction, as can be quantified using reciprocal transplant experiments between habitat types that differ in their thermal stress. (B) Coalescent and approximate Bayesian computation (ABC) analyses help infer recent thermal-driven selective and demographic responses. The inset diagram shows a typical coalescent genealogy depicting divergence with gene flow. (C) The breeder’s equation predicts responses of genetically correlated traits over one generation (vector R) given standardized selection gradients to thermal stress (vector β) by means of the variance–covariance matrix (G) of additive genetic parameter estimates. Alternatively, experimental evolution traces real-time changes in allele frequencies (Δp) across generations. (D) When genomic signatures of thermal selection are under divergent selection after several generations, genome-wide association (GWAS), and selection (GWSS) scans, as well as genome–environment associations (GEA), allow characterizing the genomic architecture of thermal adaptation. The inset Manhattan plot schematizes a hypothetical genomic scan between populations that contrast in their thermal adaptation. (E) Modern high-throughput genotyping may facilitate predictions of the thermal adaptive potential over multiple generations using infinitesimal models under a genomic prediction (GP) framework. (F) Phylogeographic approaches offer an understanding of the genomic consequences of deep-time thermal shifts at an evolutionary time scale. The inset tree represents an imaginary phylogeny. Finally, (G) machine learning (ML) approaches (H) trained using heterogeneous past responses to thermal variation may enhance long-term predictions of the thermal adaptive potential. ML’s modus operandi, as GPs, requires partitioning the calibrating historical dataset between training (TRN) and testing (TST) subsets that are iteratively imputed into a N-fold cross-validation scheme.
TABLE 1.
Case studies that have addressed thermal adaptation at different temporal scales using diverse genetic analyses.
| Analytical approach | Diagram | Data sources | Main finding | References |
| Coalescence theory and ancestry distribution models | Figure 1B | 20 alpine plant species across the European Alps genotyped with AFLP markers and analyzed with ancestry distribution models | Ancestry distribution models open new perspectives to forecast population genetic changes within species | Jay et al., 2012 |
| Coalescence theory in a SFT framework | Figure 1B | 273 Salix genets in 12 SFT populations genotyped with 7 SSRs | There is asymmetric gene flow across a thermal gradient that may be affected under future climate conditions | Cortés et al., 2014 |
| Coalescence theory | Figure 1B | Exome re-sequencing of 48 Populus trichocarpa individuals | Effective population size has varied in concert with atmospheric temperature deviation from the past c. 120,000 years | Zhou et al., 2014 |
| Quantitative genetics | Figure 1C | Review of models on whether evolutionary changes within species can contribute to species adapting to global thermal change | Evolutionary processes and trait trade-offs (Q matrix) need to be incorporated into schemes that try to manage thermal impacts | Hoffmann and Sgro, 2011 |
| Quantitative genetics | Figure 1C | Review discussing thermal adaptation to climate change from an evolutionary physiological perspective | Species’ physiological, genetic and plastic (Nicotra et al., 2010) capacities can aid in forecasting their response to thermal change | Chown et al., 2010 |
| Quantitative genetics | Figure 1C | Physiological model that simulates thermal tolerance assays for multilocus quantitative traits in D. melanogaster | Realized heritabilities of knockdown temperature may underestimate the true heritability of the upper thermal limit | Rezende et al., 2010; Santos et al., 2012 |
| Breeder’s equation in 2-habitats SFT design | Figure 1C | 1,061 Salix herbacea genotypes, from 2 habitats in a SFT design, screened for 6 thermally influenced traits and 7 SSRs | Significant heritable variation in morphology and phenology might help S. herbacea adapt to thermal stress | Sedlacek et al., 2016 |
| Quantitative genetics and breeder’s equation | Figure 1C | 166 lines of D. melanogaster assessed for cold tolerance at 5 temperatures | Low thermal tolerance is environment specific and evolvability decreases with increasing developmental temperatures | Ørsted et al., 2019 |
| Quantitative genetics and breeder’s equation | Figure 1C | 4,267 25- to 35-year-old European larch trees growing in 21 reforestation installations across 4 distinct climatic regions in Austria | Genetic evaluation across broad thermal gradients permits delineation of suitable reforestation areas under future climates | Lstiburek et al., 2020 |
| GWAS | Figure 1D | Review on molecular-level regulation of the annual growth cycle in temperate and boreal regions | Merging genomic analyses with more quantitative approaches will aid studies on how species cope with thermal changes | Singh et al., 2017 |
| eGWAS | Figure 1D | Whole-genome transcriptional responses in D. subobscura subjected to threefold replicated laboratory thermal shocks | Many genes appear to be involved in thermal adaptation, as expected for the adaptive evolution of a complex trait | Laayouni et al., 2007 |
| GWAS across a SFT latitudinal gradient | Figure 1D | 446 Populus trichocarpa trees from a latitudinal gradient screened for bud-break in 2 provenance trials and with 2.2-M SNPs | Variation in bud-break reflects differential selection for thermal functions likely to be affected by climate warming | McKown et al., 2018 |
| GWSS across a SFT latitudinal gradient | Figure 1D | Two populations of D. subobscura from different latitudes introduced to a new common laboratory environment and WGS | Populations followed different genetic routes to reach predictable and similar adaptive phenotypic outcomes | Seabra et al., 2017 |
| GWSS given a modern heat wave | Figure 1D | Long-term time series of seasonal genetic data in D. subobscura | Genetic constitution of the populations transiently shifted to summer-like frequencies during the 2011 heat wave | Rodriguez-Trelles et al., 2013 |
| GWSS in 2 postglacial lineages | Figure 1D | 48 Populus alba ramets from 2 postglacial recolonization lineages genotyped with GWS for 1.7-M SNP markers | Selection from standing variation implies the potential for rapid evolution of P. alba populations in the face of thermal change | Stölting et al., 2015 |
| GEA at a continental scale | Figure 1D | 78 Andean and Mesoamerican wild bean accessions with 23,373 GBS-derived SNPs and 3 bioclimatic heat stress indices | 24 associated loci with contrasting habitat types flank 22 heat shock protein genes (Simões et al., 2003; Sørensen et al., 2003) | López-Hernández and Cortés, 2019 |
| GEA at a latitudinal gradient | Figure 1D | Four populations of D. subobscura from different latitudes screened for 4 candidate loci for thermal adaptation in inversions | Inversion frequency clines are being maintained by local thermal adaptation in face of gene flow | Simões and Pascual, 2018 |
| GEA at a regional scale | Figure 1D | 79 natural Fagus sylvatica populations, 144 SNPs out of 52 thermal candidate genes, and 87 environmental predictors | F. sylvatica exhibits local genetic adaptation to thermal heterogeneity at the regional scale (Swiss Alps) | Pluess et al., 2016 |
| GEA at a regional scale | Figure 1D | 140 wild tomato accessions, 6,830 SNPs, and redundancy analysis (RDA), structural equation modeling (SEM), and generalized dissimilarity modeling (GDM) | Regional differences in the abiotic environment contribute to genomic divergence within a wild tomato species | Gibson and Moyle, 2020 |
| Genomic prediction (GP) | Figure 1E | 48 cows genotypes with a BovineLD BeadChip and studied in climate-controlled chambers that simulate a heat wave event | GP for heat tolerance may increase resilience and welfare in animal breeding to increased incidence and duration of heat events | Garner et al., 2016 |
| Backward genomic prediction (GP) | Figure 1E | Re-sequencing of 15 1900-year-old maize cobs from Turkey Pen Shelter, and GBS data of 1,316 modern landraces for training | Thermal adaptation drove modern maize divergence and was selected in situ from ancient standing variation 2000 years ago | Swarts et al., 2017 |
| Genomic prediction (GP) | Figure 1E | 287 elite spring wheat lines assessed in a 90K Illumina array for traits as thermal time to flowering in 18 heat/drought environments | GP is capable to predict complex traits and find the best environments to adapt new crop lines to heat and drought stress events | Sukumaran et al., 2017 |
| Genomic prediction (GP) | Figure 1E | 3,485 wheat lines genotyped with 9,285 GBS-derived SNPs and phenotyped for grain yield in heat and drought environments | GP can be used to increase the size of plant nurseries by considering un-phenotyped lines for heat and drought stress-resilience | Juliana et al., 2019 |
| Fossil record | Figure 1F | Palynological neotropical plant diversity of 1,411 morpho-species and 287,736 occurrences (65–20 million years ago) | Low Paleocene flora diversity, more diverse early Eocene flora exceeding Holocene levels, and a decline at early Oligocene | Jaramillo et al., 2006 |
| Phylogenetics | Figure 1F | Thoreau’s dataset of the Concord (MA) flora that provides data on changes in species abundance and flowering time (150 years) | Thermal change has shaped the phylo-genetically biased pattern of species loss in species that do not respond to temperature | Willis et al., 2008 |
| Fossil record | Figure 1F | Pollen and macroscopic charcoal from the Erazo profile (Ecuador) | Global Pleistocene temperature change can radically alter vegetation communities on the Andean flank in western Amazonia | Cardenas et al., 2011 |
| Phylogeographic inferences – fossils | Figure 1F | Long-term ecological records and their relevance to climate change predictions for a warmer world | Range shifts, community turnover, genetic adaptation, and an increase in diversity are observed during warmer intervals | Willis and MacDonald, 2011 |
| Phylogeographic inferences | Figure 1F | 17 time-calibrated phylogenies of major tetrapod clades and climatic data from distributions of > 500 extant species | Rates of projected climate change dramatically exceed past rates of thermal niche evolution among vertebrate species | Quintero and Wiens, 2013 |
| Phylogeographic inferences | Figure 1F | Niche shifts among populations within 56 plant and animal species using time-calibrated phylogenetic trees | Rates of change in thermal niches in plant and animal populations have been much slower than projected climate change | Jezkova and Wiens, 2016 |
| Phylogenetic-assisted modeling | Figure 1F | 9,737 records for 1,312 plant species and phylogenetic correlation matrix as an additional random effect | Tropical plants do not have narrower heat tolerances, but are more at risk due to their upper thermal limits (Feeley et al., 2020) | Sentinella et al., 2020 |
| Dynamic eco-evolutionary modeling | Figure 1G | Four endemic Alpine plant species analyzed with niche modeling, and individual-based demographic and genetic simulations | Monitoring species’ local abundance instead of their range better informs on species’ extinction risks under thermal change | Cotto et al., 2017 |
| Machine learning (ML) | Figure 1G | Species geographic distributions modeling using maximum entropy (MaxEnt) | ML modeling can be used for discrimination of suitable vs. unsuitable areas for the species with presence-only datasets | Phillips et al., 2017 |
| Machine learning (ML) | Figure 1G | Temporal uncertainty framework to assess when and where cultivation of key crops in sub-Saharan Africa will become unviable | Incremental, preparatory and transformational adaptation phases enable projected crop transformational changes | Rippke et al., 2016 |
| Machine learning (ML) | Figure 1G | Random forest in Himalaya’s Betula for last inter-glaciation, present (1970–2000) and future (2061–2080) conditions | Biodiversity in high elevation ecosystems is sensitive to global environmental changes, especially temperature warming | Mohapatra et al., 2019 |
| Machine learning (ML) | Figure 1G | Modeling of the spatiotemporal distribution in the present and the future of pine in heat scenarios (RCP 4.5 y RCP 8.5) by MaxEnt | There were good predictions for both climate change scenarios, and two contrasted tendencies of progressive evolution | Garah and Bentouati, 2019 |
| Machine learning (ML) | Figure 1G | Association between gene expression and critical temperature in divergent trout populations was measured by random forest | The “gradient boosting” approach showed that evolution for higher upper thermal tolerance is possible | Chen et al., 2018 |
| Machine learning (ML) + phylogenetic diversity | Figure 1G | Predictive models of taxonomic and phylogenetic diversity using vascular plant database for the United States | Native phylogenetic diversity is likely to decrease over the next half century despite increases in species richness | Park et al., 2020 |
| The potential of big data | Figure 1G | Special issue inspired by the symposium “Fitness landscapes, big data, and the predictability of evolution” | Understanding evolutionary adaptive responses in the face of epistasis is a major need that could benefit from big data | Visser et al., 2018 |
| Genomic prediction (GP) + machine learning (ML) | Figures 1E,G | ca. 11,000 wheat landrace accessions assessed for 40,000 GBS-derived SNPs and traits possibly related with heat stress | Deep learning should be integrated with GBLUP for the study of complex traits and the GxE interaction | Montesinos-Lopez et al., 2018 |
| Genomic prediction (GP) + machine learning (ML) | Figures 1E,G | ca. 3,500 wheat landrace accessions examined for 2,038 GBS-derived SNPs in 4 environments of drought and 2 of heat stress | MLP and SVM were competitive in genomic prediction of complex traits possibly related to heat stress as days to heading | Montesinos-Lopez et al., 2019 |
Examples enlighten how analytical approaches that try to reconstruct populations’ past genetic adaptive responses to previous events of climate change could be proxies for better forecasting. This compilation is built for illustrative purposes and is not meant to be exhaustive. Examples are sorted as in Figure 1. SFT, space-for-time substitution, GWAS, genome-wide association study; eGWAS, expression GWAS; GWSS, genome-wide selection scans; GEA, genome–environment associations; SSRs, simple sequence repeats; SNP, single-nucleotide polymorphism; WGS, whole-genome sequencing; GBS, genotyping-by-sequencing; SVM, support vector machine; MLP, multilayer perceptron; GP, genomic prediction; ML, machine learning.
Exclusively phenotypic empirical methods (Figure 1A), such as in situ monitoring, growth chamber experiments, and “common garden” (provenance) tests (Miller et al., 2020), constitute baseline evidence of thermal adaptation and should therefore inform more advanced genomic approaches. Naturally available environmental gradients (e.g., elevation or latitudinal clines) can also be used as proxies for climate change (Wheeler et al., 2016; Cortés and Wheeler, 2018), which is known as space-for-time (SFT) substitution. Replicated “common garden” tests (a.k.a. reciprocal transplants) carried out in an SFT framework are in turn useful to test whether populations can cope with changes through local adaptation (standing variation) or via phenotypic plasticity, especially in long-living species (Bridle and Vines, 2007; Sedlacek et al., 2015). Within an SFT framework, restricted gene flow can lead to small-scale genetic structures (Stanton et al., 1997) or distorted source/sink-like patterns (e.g., Cortés et al., 2014) driven by environmental factors (Nathan and Muller-Landau, 2000). Asymmetric migratory potential in a local scale may provide suitable habitats within only a few meters of the current locations (Yamagishi et al., 2005; Scherrer and Körner, 2011) but may also lead to narrowly adapted populations, even in the face of gene flow (Fitzpatrick et al., 2015), that may respond poorly to future conditions (North et al., 2011; Miller et al., 2020).
From Recent Genetic Responses to Short-Term Predictions
Coalescence Informs on Contemporary Thermal-Driven Selective and Demographic Changes
In order to trace back thermal-driven selective and demographic changes at recent temporal scales (Figure 1B), coalescent theory (Wakeley, 2008) helps in discriminating among authentic signatures of selection and those related to demography (e.g., bottlenecks and among populations reduced gene flow), from spurious covariates (Yeaman and Otto, 2011) such as lineage sorting (Wolf and Ellegren, 2017; Becher et al., 2020) and inversions (Dolgova et al., 2010; Fragata et al., 2014). Recursive simulation-based tools to incorporate the mutation/selection balance (Bustamante et al., 2001) across various scenarios of divergence and gene flow are approximate Bayesian computation – ABC (Csilléry et al., 2010; Cornuet et al., 2014), and pairwise sequentially Markovian coalescent – PSMC (Nadachowska-Brzyska et al., 2016). These approaches can inform how isolated populations that usually occupy climates with scarce habitat complexity (Flantua et al., 2019) may favor thermal generalists, while intricate local-scale heterogeneity at larger scales could trigger (Hughes, 2006; Cortés et al., 2018a) thermal specialists with limited migration potential (Cuesta et al., 2019). They can also model population sizes (Beerli, 2006) in concert with thermal changes (Zhou et al., 2014; Lehnert et al., 2019). Yet, these approaches may be limited by computational burden as they rely on simulation-based rejection sampling, while much effort is gone into the design of multiple scenarios, dimensionality reduction, and feature selection (Schrider and Kern, 2018).
The Breeder’s Equation Assists Multi-Trait Predictions Over a Few Generations
In order for thermal adaptation to happen, there must be heritable trait variation upon which selection, enforced by climate change, acts (Darwin, 1874). A simple deterministic model that condenses this evolutionary paradigm, aiding in the forecast of adaptive trait responses across few generations, comes from the quantitative genetic discipline and is known as the breeder’s equation (Figure 1C). Its multivariate form (Walsh, 2008) allows predicting responses of genetically correlated traits (vector R) to standardized thermal selection gradients (vector β) over one generation, so that R = Gβ, where G is the variance–covariance matrix of additive genetic parameter estimates – a proxy for traits’ heritabilities and trade-offs (Falconer and Mackay, 1996). The potential evolutionary response can therefore be computed using selection-gradient estimates derived from fitness proxies (i.e., fitness values regressed as a function of standardized trait values) and marker-based heritabilities (Lynch and Ritland, 1999). This approach by itself is not novel, but what makes it powerful is that it can be coupled with SFT (Wheeler et al., 2014), among other trials, to predict thermal responses to thermal change (Sedlacek et al., 2016). Yet, a major drawback is that selection gradients heavily depend on the nature of the fitness proxies (Sedlacek et al., 2016). Alternatively, experimental evolution studies (Exposito-Alonso et al., 2019) could test more explicitly how rapidly growing populations may respond to different thermal scenarios (Kawecki et al., 2012) that, together with evolve and re-sequence analyses (Turner and Miller, 2012), may contribute to understand the genetic basis of short-term thermal adaptation.
From Deeper Genomic Signatures of Selection to Mid-Term Predictions
Genome-Wide Scans Reveal Signatures of Divergent Selection to Past Thermal Adaptation
Assuming that enough generations have passed as to exhibit divergent selection to thermal changes, genome-wide association (GWAS) (Hirschhorn and Daly, 2005) and selection (GWSS) (Sabeti et al., 2007) scans (Figure 1D) are essential analytical tools to reconstruct the genomic architecture of adaptive trait divergence to thermal stress (Lecheta et al., 2020; Zwoinska et al., 2020). These methods assume that some allele variants are in linkage disequilibrium (LD) (Slatkin, 2008) with causal variants that influence the adaptive phenotype (Morris and Borevitz, 2011; Tam et al., 2019), a.k.a. genetic “hitchhiking” (Maynard Smith and Haigh, 1974; Feder and Nosil, 2010). An interface between GWAS and GWSS studies where loci are directly correlated with niche’s thermal variables is named genome–environment association (GEA) (Forester et al., 2016) and is insightful to infer past thermal adaptation, too (Hancock et al., 2011; Pluess et al., 2016; López-Hernández and Cortés, 2019). Yet, these approaches partly disregard non-additive and highly polygenic architectures (Stephan, 2016; Csillery et al., 2018; Barghi et al., 2020) and may be misleading (Maher, 2008; Pennisi, 2014) if standardized data (Waldvogel et al., 2020b) and statistical covariates (Lambert and Black, 2012), such as population stratification (Barton et al., 2019) and genomic constrains (Wray et al., 2013; Huber et al., 2016), are incorrectly accounted for.
Genomic Prediction May Assist Forecasting of Adaptive Traits Over Multiple Generations
A cutting-edge development that materialized after bringing genomics into quantitative genetics theory is genomic prediction (GP) (Desta and Ortiz, 2014; Crossa et al., 2017; Grattapaglia et al., 2018). GP uses historical phenotypic data to adjust marker-based infinitesimal (Figure 1E) models (Meuwissen et al., 2001; Gianola et al., 2006; de los Campos et al., 2013) that may overcome some of the restraints described in the previous section. GP may offer a more thoughtful picture of complex traits (e.g., thermal adaptation), presumably regulated by many low-effect loci (Pritchard et al., 2010). GP has so far informed predictions of single adaptive traits in populations with known pedigrees (Saint Pierre et al., 2012; Cros et al., 2019) and hybrid origins (Technow et al., 2014; Tan et al., 2017), as well as multi-trait inferences across diverse unrelated populations (Crossa et al., 2007, 2016; Resende et al., 2012; Suontama et al., 2019) under genotype by environment interactions (GxE) (Montesinos-Lopez et al., 2018; Crossa et al., 2019) facing polygenic climate adaptation (Isabel et al., 2020). GP of thermal adaptive traits across multiple generations and populations may be incipient (Table 1), yet it harbors a promising potential, as was demonstrated by reversely predicting unobserved thermal phenology in 1900-year-old ancient corn (Swarts et al., 2017), and as we prospect in the last section of this mini-review.
From Deep-Time Genomic Consequences of Thermal Shifts to Long-Term Predictions
Phylogeography Offers Insights Into Past Responses at an Evolutionary Scale
Phylogeographic inferences (Figure 1F) offer insights into how species (1) diversify (Quintero and Wiens, 2013) and (2) face the effects of past thermal variation (Jezkova and Wiens, 2016; Richardson et al., 2019) by boosting complex interactions such as species facilitation (Wheeler et al., 2015), adaptive introgression, and hybrid speciation (Coyne and Orr, 2004; Abbott et al., 2013; Payseur and Rieseberg, 2016; Marques et al., 2019). For instance, interspecific hybrids with intermediate niche requirements may rescue population’s gene pools in the face of climate change, while they can also display signals of heterosis for thermal adaption due to dominance on recessive alleles or overdominance via novel allele combinations (Abdelmula et al., 1999; Leinonen et al., 2011). Modern phylogeographic inferences currently rely on abundant and unlinked genetic markers (Bryant et al., 2012) that are capable of bypassing traditional assumptions of single gene mutation models (Caliebe, 2008) while accounting for scenarios of reticulate evolution (Vargas et al., 2017). Marker-based inferences also offer higher resolution to validate cases where adaptive radiation (Madriñán et al., 2013), and ecological parapatric speciation resulted from local patterns of environmental variation (Cortés et al., 2018a) that may resemble those expected by thermal change. Mosaics of local-habitat heterogeneity can ultimately enforce thermal pre-adaptation (Cortés and Wheeler, 2018). Distance-based phylogenic reconstruction without proper out-groups (Baum et al., 2005; Cortés, 2013) is yet a major risk of these approaches.
Machine Learning May Bridge Historical Genomics and Long-Term Predictions
A promising way to simultaneously make sense of multiple sources of historical genomic data that can be utilized to predict populations’ adaptive responses is by merging them into a machine learning (ML) framework (Figures 1G,H). ML bypasses the “curse of dimensionality” and benefits from high-dimensional inputs of heterogeneous dependent variables (“features”) without a priori knowledge of their joint probability distribution (Schrider and Kern, 2018). This improves predictions’ “recall” (true positive) rate among a set of possible responses, especially when the classification is iteratively trained using “labeled” data (i.e., historical thermal responses may offer novel calibration datasets, Table 1) via N-fold cross-validation. ML has been routinely used to make ecological niche modeling (Phillips et al., 2017; Valencia et al., 2020) and functional predictions across genomes (Libbrecht and Noble, 2015). Yet, ML may likely displace other tools useful to characterize the genomic consequences of thermal adaptation, already introduced in this mini-review, such as ABC modeling (Liu et al., 2019) and GWSS (Schrider and Kern, 2018).
Concluding Remarks
Thermal adaptation is a complex polygenic trait well-described in terms of its genetic architecture and selection footprints across a wide range of phylogenetically diverse taxa (Way and Oren, 2010; Valladares et al., 2014; López-Hernández and Cortés, 2019). While genomics has enabled these achievements that rely on past events of thermal variation, forward predictions remain one step behind partly because (1) disentangling selective and demographic drivers of the genomic landscape from fortuitous genomic constrains (Logan and Cox, 2020) is puzzling (Ellegren and Galtier, 2016) and (2) merging these heterogeneous signatures and data sources into a cohesive predictive framework was unfeasible, until recently. In this mini-review, we advocated for novel approaches that may enhance our understanding of the genetic consequences of past climate change, while offering new avenues to calibrate more accurate predictive models of the thermal adaptive potential. For instance, ML advances are likely to now move beyond species distribution modeling (Phillips et al., 2017) and functional genomics (Libbrecht and Noble, 2015) to permeate the backward interpretation of recent genetic demographic responses and genomic signatures to historical thermal selection by updating popular but sometimes intractable methods such as ABC modeling and GWSS (Schrider and Kern, 2018). Meanwhile, GP and ML might boost forward predictions of the adaptive potential beyond a single generation by training multifactorial models that can try incorporating genomic heterogeneous evidence of historical thermal adaption across a wide spectrum of temporal scales. Ultimately, understanding how biotas formed in response to historical environmental change may improve our ability to predict and mitigate the threats to species posed by global warming (Ding et al., 2020).
Despite GP’s and ML’s being useful to comprehend and predict thermal adaptation, these new paradigms are not exempt of criticism. A reiterative misconception is that because these methodologies aim at strengthening predictions and classification boundaries, they do not offer a mechanistic understanding of the subjacent processes. However, even though GP and ML rely on algorithmically generated models, both are far from “black boxes” because they allow direct measurement of the contribution of each genetic marker (Resende et al., 2012; Spindel et al., 2016) and “feature” (Schrider and Kern, 2018), to the point that they can offer higher resolution than traditional genetic mapping (Hirschhorn and Daly, 2005) and deterministic model building (Otto and Day, 2007) techniques. A second misconception assumes computational burden. Although both GP and ML require a large number of simulations, they do not depend on rejection sampling, which means they may efficiently use all of the simulations to inform the mapping of historical thermal data to parameters (Schrider and Kern, 2018).
Future Directions
So far, GP and ML have been mostly utilized to address thermal adaptation individually (Table 1). For instance, GP has been used to project heat tolerance in diverse wheat lines (Sukumaran et al., 2017; Juliana et al., 2019), and bovine genotypes (Garner et al., 2016), in all cases more as a proof of concept. Similarly, ML approaches have not only deepened our understating on populations’ range shifts in the light of thermal variation (Rippke et al., 2016; Garah and Bentouati, 2019; Mohapatra et al., 2019) but also assisted eGWAS of critical temperature thresholds (Chen et al., 2018) and phylogenetic forecasting in plants (Park et al., 2020). However, since GP and ML are both cutting-edge tools, there is still room and need for new developments. For instance, merging more cohesively past adaptive responses to previous events of environmental change into cutting-edge analytical frameworks like GP and ML will ultimately allow predicting whether populations’ adaptive potential may keep up with the pace of current thermal increase (Franks and Hoffmann, 2012; Franks et al., 2014). Swarts et al. (2017) illustrates that across-temporal predictions may be useful not only to improve forecasting (Sweet et al., 2019) but also to better understand previous responses to thermal variation, since they used backward GP to demonstrate that thermal adaptation in maize was selected in situ from ancient standing variation 2000 years ago. By enlightening on the nature of these historical genetic signatures to past climate change, genomics can also enhance predictions that aim at incorporating adaptive responses beyond extirpation and range shifts (Chen et al., 2011).
Data sources incorporated into GP and ML can transcend those with a direct genomic connotation and involve others that can modulate or be informative of the thermal responses. For instance, from an abiotic point of view, nutrient availability (Little et al., 2016), absorption (Wu et al., 2020), and soil interactions (Sedlacek et al., 2014) could act as enhancers or limiting factors of the adaptive responses. From a biotic perspective, among-ecotype differentiation (Cortés et al., 2012a,b, 2013; Blair et al., 2016), intrapopulation divergence (Cortés et al., 2011; Blair et al., 2012, 2018; Kelleher et al., 2012), and within-family variation (Galeano et al., 2012; Blair et al., 2013) could encourage or coerce adaptation. Population’s functioning, abundance, distribution, and diversity, as predicted from controlled experiments (Way and Oren, 2010; Elmendorf et al., 2012; Wolkovich et al., 2012; Andresen et al., 2016; Becklin et al., 2017; Singh et al., 2017), experimental evolution (Tenaillon et al., 2012; Mallard et al., 2018; Pfenninger and Foucault, 2020), biological monitoring (Walther et al., 2002; Franks et al., 2013; Wipf et al., 2013; Reichstein et al., 2014; Hällfors et al., 2020), and shifts observed in the fossil record (Alsos et al., 2009; Willis and MacDonald, 2011; Lyons et al., 2016; Bruelheide et al., 2018), can feed back on climate change (Pearson et al., 2013) and so be considered as drivers themselves. Regardless of the exact nature and extent of the data type, both GP and ML may offer suitable scenarios to merge diverse, and even conflicting, data sources in order to pinpoint emergent properties (Street et al., 2011) out of a complex system, as is thermal genomic adaptation. Therefore, a key guideline for new developments concerns a better coupling of GP and ML approaches. Until now, only a few works have relied on both methodologies, in the context of thermal adaptation in wheat landraces (Montesinos-Lopez et al., 2018, 2019), but have not gone beyond technical comparisons/recommendations, nor have designed integrated pipelines. Also, reconciling modern genomics with last-generation predictive inferences of the thermal adaptive potential and stochastic demographic modeling (Jenouvrier et al., 2009) is necessary. Open-access resources and data sharing platforms are as crucial in this effort as new integrated analytical pipelines. We are looking forward to seeing more cohesive (Beyer et al., 2020) and systematic studies and predictions across the rich and informative temporal spectrum (Kristensen et al., 2018) of past and future environmental variation (Franks et al., 2013). These efforts should be carried out through a wide range of spatial scales (Parmesan and Hanley, 2015; Way et al., 2015; Gonzalez et al., 2020) spanning contrasting ecosystems (Lenoir et al., 2020), microhabitats (Zellweger et al., 2020), and unrelated taxa, which together may already be keeping heritable adaptive trait differentiation valuable for long-term thermal responses and informative for conservation prioritizations (Barnosky et al., 2017; Elsen et al., 2020).
Author Contributions
AC conceived this mini-review. FL-H collected the literature and prepared diagrams. DO-R compiled the historical climate data. AC wrote the first draft of the mini-review with further contributions from FL-H and DO-R. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge thoughtful discussions with M. W. Blair regarding the genetic basis of thermal adaptation that took place with AC and DO-R during the Erasmus funded workshop “Molecular Breeding for Abiotic Constraints in Plants” held in Montpellier (France) during the summer of 2012. Some of the ideas discussed here were also framed into perspective, thanks to suggestions from A. A. Hoffmann to AC as part of the “Climate Change and Evolution” symposium during the XIV Congress of the European Society for Evolutionary Biology (ESEB) held at Lisbon (Portugal) in August 2013. The Evolutionary Biology Centre (EBC) Graduate School on Genomes and Phenotypes from Uppsala University is recognized for promoting AC participation in this meeting. AGROSAVIA’s Department for Research Capacity Building is credited for granting time to AC to carry out synergistic discussions and progress meetings during 2016 and 2017 in order to pursue this mini-review, as well as for sponsoring FL-H’s internship during 2018. We thank D. Royer for the Cenozoic temperature, CO2, and species richness dataset. Special thanks are given to M. J. Torres-Urrego for support while drafting and revising this mini-review. The topic editor and the two reviewers are recognized for their thoughtful suggestions to improve the scope of the mini-review, as well as for making possible the insightful special issue on “Coping with Climate Change: A Genomic Perspective on Thermal Adaptation.”
Footnotes
Funding. AC was supported by grants 4.1-2016-00418 and BS2017-0036 from Vetenskapsrådet (VR) and Kungliga Vetenskapsakademien (KVA), respectively. The National Science Foundation (NSF) and the SIMONS Collaboration on the Origins of Life support DO-R. The editorial fund from the Colombian Corporation for Agricultural Research (AGROSAVIA) was thanked for subsidizing the mini-review B-type processing charge.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.564515/full#supplementary-material
Past and future of thermal and CO2 variation, and their correlates with past biodiversity. (A) Temperature and richness of plant species (from pollen) for the Cenozoic Era (65 Mya – present). Temperature estimates (Supplementary Table S1) were computed by Hansen et al. (2013) using the original δ18O record from Zachos et al. (2008). Richness of plant species from pollen data (Supplementary Table S1) is based on 15 Neotropical stratigraphic sections inspected by Jaramillo et al. (2006). This profile goes from 65 to 20 Mya due to a lack of more recent suitable sampling records. (B) Projections of the near-surface temperature anomalies to 2,050 (Supplementary Table S2), which follow the CIMP5 RCP 8.5 scenario from the KNMI (http://climexp.knmi.nl/) repository averaged from an original 5-min resolution. Light gray shaded areas depict minimum and maximum estimates. (C) Atmospheric CO2 and richness of plant species (as in A) for the Cenozoic Era (65 Mya – present). CO2 records are an updated version (Supplementary Table S1) derived from Royer and Chernoff (2013), originally compiled by Beerling and Royer (2011). (D) Projected CO2 concentration (ppm) to 2,050 also follow the CIMP5 RCP 8.5 scenario, as in B (Supplementary Table S3).
Dataset of temperature, atmospheric CO2, and richness of plant species for the Cenozoic Era (65 Mya – present for temperature and CO2, and 65–20 Mya for richness of plant species). Temperature estimates were computed by Hansen et al. (2013) from five-point running means of the original temporal resolution of the δ18O record from Zachos et al. (2008), a profile of surface low-magnesium calcitic fossils (including planktonic foraminifera, belemintes, brachiopods, and bivalves) that was lower during periods with warmer seawater. Atmospheric CO2 corresponds to an updated version from Royer and Chernoff (2013), originally compiled by Beerling and Royer (2011). Richness of plant species is based on pollen data from Jaramillo et al. (2006), who analyzed 1,530 samples from 15 stratigraphic sections in Colombia and Venezuela (Neotropics).
Projections of thermal variation to 2,050. Simulation of Near-Surface Air Temperature Anomalies (°C) from 1,860 to 2,050 follow the CIMP5 RCP 8.5 scenario from the KNMI (http://climexp.knmi.nl/) database averaged from an original 5 min resolution. Minimum and maximum temperature estimates were generated by the coupled ACCESS v.1.0 model specifically designed for the CIMP5 project (Kowalczyk et al., 2013).
Projections of CO2 concentration (ppm) to 2,050. Simulations follow the CIMP5 RCP 8.5 scenario from 1,860 to 2,050 available at KNMI (http://climexp.knmi.nl/) database averaged from an original 5 min resolution.
References
- Abbott R., Albach D., Ansell S., Arntzen J. W., Baird S. J. E., Bierne N., et al. (2013). Hybridization and speciation. J. Evol. Biol. 26 229–246. [DOI] [PubMed] [Google Scholar]
- Abdelmula A. A., Link W., Von Kittlitz E., Stelling D. (1999). Heterosis and inheritance of drought tolerance in Faba Bean, Vicia Faba L. Plant Breed. 118 485–490. 10.1046/j.1439-0523.1999.00411.x [DOI] [Google Scholar]
- Alsos I. G., Alm T., Normand S., Brochmann C. (2009). Past and future range shifts and loss of diversity in Dwarf Willow (Salix Herbacea L.). Inferred from genetics, fossils and modelling. Glob. Ecol. Biogeogr. 18 223–239. 10.1111/j.1466-8238.2008.00439.x [DOI] [Google Scholar]
- Andresen L. C., Müller C., De Dato G., Dukes J. S., Emmett B. A., Estiarte M., et al. (2016). Shifting impacts of climate change. Adv. Ecol. Res. 55 437–473. [Google Scholar]
- Barghi N., Hermisson J., Schlötterer C. (2020). Polygenic adaptation: a unifying framework to understand positive selection. Nat. Rev. Genet. 10.1038/s41576-020-0250-z [DOI] [PubMed] [Google Scholar]
- Barnosky A. D., Hadly E. A., Gonzalez P., Head J., Polly P. D., Lawing A. M., et al. (2017). Merging paleobiology with conservation biology to guide the future of terrestrial ecosystems. Science 355: eaah4787. [DOI] [PubMed] [Google Scholar]
- Barton N., Hermisson J., Nordborg M. (2019). Why structure matters. eLife 8:e45380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum D. A., Smith S. D., Donovan S. S. (2005). The tree-thinking challenge. Science 310 979–970. 10.1126/science.1117727 [DOI] [PubMed] [Google Scholar]
- Becher H., Jackson B. C., Charlesworth B. (2020). Patterns of genetic variability in genomic regions with low rates of recombination. Curr. Biol. 30 94.e3–100.e3. [DOI] [PubMed] [Google Scholar]
- Becklin K. M., Walker S. M., II, Way D. A., Ward J. K. (2017). Co2 studies remain key to understanding a future world. New Phytol. 214 34–40. 10.1111/nph.14336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli P. (2006). Comparison of Bayesian and maximum-likelihood inference of population genetic parameters. Bioinformatics 22 341–345. 10.1093/bioinformatics/bti803 [DOI] [PubMed] [Google Scholar]
- Beerling D. J., Royer D. L. (2011). Convergent cenozoic Co2 history. Nat. Geosci. 4 418–420. 10.1038/ngeo1186 [DOI] [Google Scholar]
- Beyer R. M., Krapp M., Manica A. (2020). High-resolution terrestrial climate, bioclimate and vegetation for the last 120,000 years. Sci. Data 7:236. 10.1038/s41597-020-0552-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair M. W., Cortes A. J., Farmer A. D., Huang W., Ambachew D., Penmetsa R. V., et al. (2018). Uneven recombination rate and linkage disequilibrium across a reference Snp map for common Bean (Phaseolus Vulgaris L.). PLoS One 13:e0189597. 10.1371/journal.pone.0189597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair M. W., Cortés A. J., Penmetsa R. V., Farmer A., Carrasquilla-Garcia N., Cook D. R. (2013). A high-throughput snp marker system for parental polymorphism screening, and diversity analysis in common Bean (Phaseolus Vulgaris L.). Theoret. Appl. Genet. 126 535–548. 10.1007/s00122-012-1999-z [DOI] [PubMed] [Google Scholar]
- Blair M. W., Cortés A. J., This D. (2016). Identification of an Erecta gene and its drought adaptation associations with wild and cultivated common Bean. Plant Sci. 242 250–259. 10.1016/j.plantsci.2015.08.004 [DOI] [PubMed] [Google Scholar]
- Blair M. W., Soler A., Cortés A. J. (2012). Diversification and population structure in common Beans (Phaseolus Vulgaris L.). PLoS One 7:e49488. 10.1371/journal.pone.0049488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridle J. R., Vines T. H. (2007). Limits to evolution at range margins: when and why does adaptation fail? Trends Ecol. Evol. 22 140–147. 10.1016/j.tree.2006.11.002 [DOI] [PubMed] [Google Scholar]
- Bruelheide H., Dengler J., Purschke O., Lenoir J., Jiménez-Alfaro B., Hennekens S. M., et al. (2018). Global trait–environment relationships of plant communities. Nat. Ecol. Evol. 2 1906–1917. [DOI] [PubMed] [Google Scholar]
- Bryant D., Bouckaert R., Felsenstein J., Rosenberg N. A., Roychoudhury A. (2012). Inferring species trees directly from biallelic genetic markers: bypassing gene trees in a full coalescent analysis. Mol. Biol. Evol. 29 1917–1932. 10.1093/molbev/mss086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush M. B., Silman M. R., Mcmichael C., Saatchi S. (2008). Fire, climate change and biodiversity in amazonia: a late-holocene perspective. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363 1795–1702. 10.1098/rstb.2007.0014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante C. D., Wakeley J., Sawyer S., Hartl D. L. (2001). Directional selection and the site-frequency spectrum. Genetics 159 1779–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliebe A. (2008). Mathematical models in population genetics. Medizinische Genetik 20 282–287. [Google Scholar]
- Cardenas M. L., Gosling W. D., Sherlock S. C., Poole I., Pennington R. T., Mothes P. (2011). The response of vegetation on the Andean Flank in Western Amazonia to pleistocene climate change. Science 331 1055–1058. 10.1126/science.1197947 [DOI] [PubMed] [Google Scholar]
- Chen I. C., Hill J. K., Ohlemuller R., Roy D. B., Thomas C. D. (2011). Rapid range shifts of species associated with high levels of climate warming. Science 333 1024–1026. 10.1126/science.1206432 [DOI] [PubMed] [Google Scholar]
- Chen Z., Farrell A. P., Matala A., Narum S. R. (2018). Mechanisms of thermal adaptation and evolutionary potential of conspecific populations to changing environments. Mol. Ecol. 27 659–674. 10.1111/mec.14475 [DOI] [PubMed] [Google Scholar]
- Chown S. L., Hoffmann A. A., Kristensen T. N., Angilletta M. J., Stenseth N. C., Pertoldi C. (2010). Adapting to climate change: a perspective from evolutionary physiology. Clim. Res. 43 3–15. 10.3354/cr00879 [DOI] [Google Scholar]
- Cornuet J. M., Pudlo P., Veyssier J., Dehne-Garcia A., Gautier M., Leblois R., et al. (2014). Diyabc V2.0: a software to make approximate bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics 30 1187–1189. 10.1093/bioinformatics/btt763 [DOI] [PubMed] [Google Scholar]
- Cortés A. J. (2013). On the origin of the common Bean (Phaseolus Vulgaris L.). Am. J. Plant Sci. 4 1998–2000. [Google Scholar]
- Cortés A. J., Blair M. W. (2018). Genotyping by sequencing and genome – environment associations in wild common bean predict widespread divergent adaptation to drought. Front. Plant Sci. 9:128. 10.3389/fpls.2018.00128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés A. J., Chavarro M. C., Blair M. W. (2011). Snp marker diversity in common Bean (Phaseolus Vulgaris L.). Theoret. Appl. Genet. 123 827–845. 10.1007/s00122-011-1630-8 [DOI] [PubMed] [Google Scholar]
- Cortés A. J., Chavarro M. C., Madriñán S., This D., Blair M. W. (2012a). Molecular ecology and selection in the drought-related Asr gene polymorphisms in wild and cultivated common Bean (Phaseolus Vulgaris L.). BMC Genet. 13:58. 10.1186/1471-2156-13-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés A. J., This D., Chavarro C., Madriñán S., Blair M. W. (2012b). Nucleotide diversity patterns at the drought-related Dreb2 encoding genes in wild and cultivated common Bean (Phaseolus Vulgaris L.). Theoret. Appl. Genet. 125 1069–1085. 10.1007/s00122-012-1896-5 [DOI] [PubMed] [Google Scholar]
- Cortés A. J., Garzón L. N., Valencia J. B., Madriñán S. (2018a). On the causes of rapid diversification in the páramos: isolation by ecology and genomic divergence in Espeletia. Front. Plant Sci. 9:1700. 10.3389/fpls.2018.01700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés A. J., Skeen P., Blair M. W., Chacón-Sánchez M. I. (2018b). Does the genomic landscape of species divergence in Phaseolus Beans coerce parallel signatures of adaptation and domestication? Front. Plant Sci. 9:1816. 10.3389/fpls.2018.01816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés A. J., Monserrate F., Ramírez-Villegas J., Madriñán S., Blair M. W. (2013). Drought tolerance in wild plant populations: the case of common Beans (Phaseolus Vulgaris L.). PLoS One 8:e62898. 10.1371/journal.pone.0062898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés A. J., Waeber S., Lexer C., Sedlacek J., Wheeler J. A., Van Kleunen M., et al. (2014). Small-scale patterns in snowmelt timing affect gene flow and the distribution of genetic diversity in the Alpine Dwarf Shrub Salix Herbacea. Heredity 113 233–239. 10.1038/hdy.2014.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés A. J., Wheeler J. A. (2018). “The environmental heterogeneity of mountains at a fine scale in a changing world,” in Mountains, Climate, and Biodiversity, eds Hoorn C., Perrigo A., Antonelli A. (New York, NY: Wiley; ). [Google Scholar]
- Cotto O., Wessely J., Georges D., Klonner G., Schmid M., Dullinger S., et al. (2017). A dynamic eco-evolutionary model predicts slow response of alpine plants to climate warming. Nat. Commun. 8:15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne J. A., Orr H. A. (2004). Speciation. Sunderland, MA: Sinauer. [Google Scholar]
- Cronk Q. (2016). Plant extinctions take time. Science 353 446–447. 10.1126/science.aag1794 [DOI] [PubMed] [Google Scholar]
- Cros D., Mbo-Nkoulou L., Bell J. M., Oum J., Masson A., Soumahoro M., et al. (2019). Within-family genomic selection in rubber tree (Hevea Brasiliensis) increases genetic gain for rubber production. Ind. Crops Prod. 138:111464 10.1016/j.indcrop.2019.111464 [DOI] [Google Scholar]
- Crossa J., Burgueno J., Dreisigacker S., Vargas M., Herrera-Foessel S. A., Lillemo M., et al. (2007). Association analysis of historical bread wheat germplasm using additive genetic covariance of relatives and population structure. Genetics 177 1889–1913. 10.1534/genetics.107.078659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossa J., Jarquin D., Franco J., Perez-Rodriguez P., Burgueno J., Saint-Pierre C., et al. (2016). Genomic prediction of gene bank wheat landraces. G3 6 1819–1834. 10.1534/g3.116.029637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossa J., Martini J. W. R., Gianola D., Perez-Rodriguez P., Jarquin D., Juliana P., et al. (2019). Deep Kernel and deep learning for genome-based prediction of single traits in multienvironment breeding trials. Front. Genet. 10:1168. 10.3389/fgene.2019.01168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossa J., Perez-Rodriguez P., Cuevas J., Montesinos-Lopez O., Jarquin D., De Los Campos G., et al. (2017). Genomic selection in plant breeding: methods, models, and perspectives. Trends Plant Sci. 22 961–975. [DOI] [PubMed] [Google Scholar]
- Cruickshank T. E., Hahn M. W. (2014). Reanalysis suggests that genomic Islands of speciation are due to reduced diversity, not reduced gene flow. Mol. Ecol. 23 3133–3157. 10.1111/mec.12796 [DOI] [PubMed] [Google Scholar]
- Csilléry K., Blum M. G. B., Gaggiotti O. E., François O. (2010). Approximate Bayesian computation (Abc) in practice. Trends Ecol. Evol. 25 410–418. 10.1016/j.tree.2010.04.001 [DOI] [PubMed] [Google Scholar]
- Csillery K., Rodriguez-Verdugo A., Rellstab C., Guillaume F. (2018). Detecting the genomic signal of polygenic adaptation and the role of epistasis in evolution. Mol. Ecol. 27 606–612. 10.1111/mec.14499 [DOI] [PubMed] [Google Scholar]
- Cuesta F., Tovar C., Llambí L. D., Gosling W. D., Halloy S., Carilla J., et al. (2019). Thermal Niche traits of high alpine plant species and communities across the tropical andes and their vulnerability to global warming. J. Biogeogr. 47 408–420. 10.1111/jbi.13759 [DOI] [Google Scholar]
- Darwin C. R. (1874). The Descent of Man and Selection in Relation to Sex. New York, NY: Hurst and Company. [Google Scholar]
- de los Campos G., Hickey J. M., Pong-Wong R., Daetwyler H. D., Calus M. P. (2013). Whole-genome regression and prediction methods applied to plant and animal breeding. Genetics 193 327–345. 10.1534/genetics.112.143313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desta Z. A., Ortiz R. (2014). Genomic selection: genome-wide prediction in plant improvement. Trends Plant Sci. 19 592–601. 10.1016/j.tplants.2014.05.006 [DOI] [PubMed] [Google Scholar]
- Ding W. N., Ree R. H., Spicer R. A., Xing Y. W. (2020). Ancient orogenic and monsoon-driven assembly of the world’s richest temperate alpine flora. Science 369 578–581. 10.1126/science.abb4484 [DOI] [PubMed] [Google Scholar]
- Dolgova O., Rego C., Calabria G., Balanya J., Pascual M., Rezende E. L., et al. (2010). Genetic constraints for thermal coadaptation in Drosophila Subobscura. BMC Evol. Biol. 10:363. 10.1186/1471-2148-10-363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H., Galtier N. (2016). Determinants of genetic diversity. Nat. Rev. Genet. 17 422–433. [DOI] [PubMed] [Google Scholar]
- Ellegren H., Wolf J. B. W. (2017). Parallelism in genomic landscapes of differentiation, conserved genomic features and the role of linked selection. J. Evol. Biol. 30 1516–1518. 10.1111/jeb.13113 [DOI] [PubMed] [Google Scholar]
- Elmendorf S. C., Henry G. H. R., Hollister R. D., Björk R. G., Bjorkman A. D., Callaghan T. V., et al. (2012). Global assessment of experimental climate warming on tundra vegetation: heterogeneity over space and time. Ecol. Lett. 15 164–175. 10.1111/j.1461-0248.2011.01716.x [DOI] [PubMed] [Google Scholar]
- Elsen P. R., Monahan W. B., Dougherty E. R., Merenlender A. M. (2020). Keeping pace with climate change in global terrestrial protected areas. Sci. Adv. 6:eaay0814 10.1126/sciadv.aay0814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exposito-Alonso M., Burbano H. A., Bossdorf O., Nielsen R., Weigel D. (2019). Natural selection on the Arabidopsis thaliana genome in present and future climates. Nature 573 126–129. 10.1038/s41586-019-1520-9 [DOI] [PubMed] [Google Scholar]
- Falconer D. S., Mackay T. F. C. (1996). Introduction to Quantitative Genetics. Essex: Longman. [Google Scholar]
- Feder J. L., Gejji R., Yeaman S., Nosil P. (2012). Establishment of new mutations under divergence and genome hitchhiking. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367 461–474. 10.1098/rstb.2011.0256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder J. L., Nosil P. (2010). The efficacy of divergence hitchhiking in generating genomic islands during ecological speciation. Evolution 64 1729–1747. 10.1111/j.1558-5646.2009.00943.x [DOI] [PubMed] [Google Scholar]
- Feeley K., Martinez-Villa J., Perez T., Silva Duque A., Triviño Gonzalez D., Duque A. (2020). The thermal tolerances, distributions, and performances of tropical montane tree species. Front. For. Glob. Change 3:25 10.3389/ffgc.2020.00025 [DOI] [Google Scholar]
- Fitzpatrick S. W., Gerberich J. C., Kronenberger J. A., Angeloni L. M., Funk W. C. (2015). Locally adapted traits maintained in the face of high gene flow. Ecol. Lett. 18 37–47. 10.1111/ele.12388 [DOI] [PubMed] [Google Scholar]
- Flantua S. G. A., O’dea A., Onstein R. E., Giraldo C., Hooghiemstra H. (2019). The flickering connectivity system of the North Andean Páramos. J. Biogeogr. 46 1808–1825. 10.1111/jbi.13607 [DOI] [Google Scholar]
- Forester B. R., Jones M. R., Joost S., Landguth E. L., Lasky J. R. (2016). Detecting spatial genetic signatures of local adaptation in heterogeneous landscapes. Mol. Ecol. 25 104–120. 10.1111/mec.13476 [DOI] [PubMed] [Google Scholar]
- Fragata I., Lopes-Cunha M., Barbaro M., Kellen B., Lima M., Santos M. A., et al. (2014). How much can history constrain adaptive evolution? A real-time evolutionary approach of inversion polymorphisms in Drosophila Subobscura. J. Evol. Biol. 27 2727–2738. 10.1111/jeb.12533 [DOI] [PubMed] [Google Scholar]
- Fragata I., Simões P., Matos M., Szathmáry E., Santos M. (2018). playing evolution in the laboratory: from the first major evolutionary transition to global warming. Europhys. Lett. 122:38001 10.1209/0295-5075/122/38001 [DOI] [Google Scholar]
- Franks P. J., Adams M. A., Amthor J. S., Barbour M. M., Berry J. A., Ellsworth D. S., et al. (2013). Sensitivity of plants to changing atmospheric Co2 concentration: from the geological past to the next century. New Phytol. 197 1077–1094. 10.1111/nph.12104 [DOI] [PubMed] [Google Scholar]
- Franks S. J., Hoffmann A. A. (2012). Genetics of climate change adaptation. Annu. Rev. Genet. 46 185–208. [DOI] [PubMed] [Google Scholar]
- Franks S. J., Weber J. J., Aitken S. N. (2014). Evolutionary and plastic responses to climate change in terrestrial plant populations. Evol. Appl. 7 123–139. 10.1111/eva.12112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galeano C. H., Cortés A. J., Fernandez A. C., Soler A., Franco-Herrera N., Makunde G., et al. (2012). Gene-based single nucleotide polymorphism markers for genetic and association mapping in common Bean. BMC Genet. 13:48. 10.1186/1471-2156-13-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garah K., Bentouati A. (2019). Using the maxent model for assessing the impact of climate change on the Aurasian Aleppo pine distribution in algeria. Afr. J. Ecol. 57 500–511. 10.1111/aje.12630 [DOI] [Google Scholar]
- Garner J. B., Douglas M. L., Williams S. R., Wales W. J., Marett L. C., Nguyen T. T., et al. (2016). Genomic selection improves heat tolerance in dairy cattle. Sci. Rep. 6:34114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianola D., Fernando R. L., Stella A. (2006). Genomic-assisted prediction of genetic value with semiparametric procedures. Genetics 173 1761–1776. 10.1534/genetics.105.049510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson M. J. S., Moyle L. C. (2020). Regional differences in the abiotic environment contribute to genomic divergence within a wild tomato species. Mol. Ecol. 29 2204–2217. 10.1111/mec.15477 [DOI] [PubMed] [Google Scholar]
- Gompert Z., Comeault A. A., Farkas T. E., Feder J. L., Parchman T. L., Buerkle C. A., et al. (2014). Experimental evidence for ecological selection on genome variation in the wild. Ecol. Lett. 17 369–379. 10.1111/ele.12238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A., Germain R. M., Srivastava D. S., Filotas E., Dee L. E., Gravel D., et al. (2020). Scaling-up biodiversity-ecosystem functioning research. Ecol. Lett. 23 757–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grattapaglia D., Silva-Junior O. B., Resende R. T., Cappa E. P., Muller B. S. F., Tan B., et al. (2018). Quantitative genetics and genomics converge to accelerate forest tree breeding. Front. Plant Sci. 9:1693. 10.3389/fpls.2018.01693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hällfors M. H., Antao L. H., Itter M., Lehikoinen A., Lindholm T., Roslin T., et al. (2020). Shifts in timing and duration of breeding for 73 boreal bird species over four decades. PNAS 117 18557–18565. 10.1073/pnas.1913579117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock A. M., Brachi B., Faure N., Horton M. W., Jarymowycz L. B., Sperone F. G., et al. (2011). Adaptation to climate across the Arabidopsis Thaliana genome. Science 334 83–86. 10.1126/science.1209244 [DOI] [PubMed] [Google Scholar]
- Hansen J., Sato M., Russell G., Kharecha P. (2013). Climate sensitivity, sea level and atmospheric carbon dioxide. Philos. Trans. A Math. Phys. Eng. Sci. 371:20120294. 10.1098/rsta.2012.0294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn J. N., Daly M. J. (2005). Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 6 95–108. 10.1038/nrg1521 [DOI] [PubMed] [Google Scholar]
- Hoffmann A. A., Sgro C. M. (2011). Climate change and evolutionary adaptation. Nature 470 479–485. [DOI] [PubMed] [Google Scholar]
- Huber C. D., Degiorgio M., Hellmann I., Nielsen R. (2016). Detecting recent selective sweeps while controlling for mutation rate and background selection. Mol. Ecol. 25 142–156. 10.1111/mec.13351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes C. (2006). From the cover: Island radiation on a continental scale: exceptional rates of plant diversification after uplift of the andes. Proc. Natl. Acad. Sci. U.S.A. 103 10334–10339. 10.1073/pnas.0601928103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin D. E., Alcaide M., Delmore K. E., Irwin J. H., Owens G. L. (2016). Recurrent selection explains parallel evolution of genomic regions of high relative but low absolute differentiation in a ring species. Mol. Ecol. 25 4488–4507. 10.1111/mec.13792 [DOI] [PubMed] [Google Scholar]
- Isabel N., Holliday J. A., Aitken S. N. (2020). Forest genomics: advancing climate adaptation, forest health, productivity, and conservation. Evol. Appl. 13 3–10. 10.1111/eva.12902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo C., Rueda M. J., Mora G. (2006). Cenozoic plant diversity in the neotropics. Science 311 1893–1896. 10.1126/science.1121380 [DOI] [PubMed] [Google Scholar]
- Jay F., Manel S., Alvarez N., Durand E. Y., Thuiller W., Holderegger R., et al. (2012). Forecasting changes in population genetic structure of alpine plants in response to global warming. Mol. Ecol. 21 2354–2368. 10.1111/j.1365-294x.2012.05541.x [DOI] [PubMed] [Google Scholar]
- Jenouvrier S., Caswell H., Barbraud C., Holland M., Stroeve J., Weimerskirch H. (2009). Demographic models and Ipcc climate projections predict the decline of an emperor penguin population. PNAS 106 1844–1847. 10.1073/pnas.0806638106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jezkova T., Wiens J. J. (2016). Rates of change in climatic niches in plant and animal populations are much slower than projected climate change. Proc. Biol. Sci. 283:20162104. 10.1098/rspb.2016.2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliana P., Montesinos-López O. A., Crossa J., Mondal S., González Pérez L., Poland J., et al. (2019). Integrating genomic-enabled prediction and high-throughput phenotyping in breeding for climate-resilient bread wheat. Theoret. Appl. Genet. 132 177–194. 10.1007/s00122-018-3206-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawecki T. J., Lenski R. E., Ebert D., Hollis B., Olivieri I., Whitlock M. C. (2012). Experimental evolution. Trends Ecol. Evol. 27 547–560. [DOI] [PubMed] [Google Scholar]
- Kelleher C. T., Wilkin J., Zhuang J., Cortés A. J., Quintero ÁL. P., Gallagher T. F., et al. (2012). Snp discovery, gene diversity, and linkage disequilibrium in wild populations of Populus Tremuloides. Tree Genet. Genomes 8 821–829. 10.1007/s11295-012-0467-x [DOI] [Google Scholar]
- Kowalczyk E. A., Stevens L., Law R. M., Dix M., Wang Y. P., Harman I. N., et al. (2013). The land surface model component of access: description and impact on the simulated surface climatology. Aust. Meteorol. Oceanogr. 63 65–82. 10.22499/2.6301.005 [DOI] [Google Scholar]
- Kristensen T. N., Ketola T., Kronholm I. (2018). Adaptation to environmental stress at different timescales. Ann. N. Y. Acad. Sci. [Epub ahead of print] 10.1111/nyas.13974 [DOI] [PubMed] [Google Scholar]
- Laayouni H., Garcia-Franco F., Chavez-Sandoval B. E., Trotta V., Beltran S., Corominas M., et al. (2007). Thermal evolution of gene expression profiles in Drosophila Subobscura. BMC Evol. Biol. 7:42. 10.1186/1471-2148-7-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert C. G., Black L. J. (2012). Learning from our gwas mistakes: from experimental design to scientific method. Biostatistics 13 195–203. 10.1093/biostatistics/kxr055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecheta M. C., Awde D. N., O’leary T. S., Unfried L. N., Jacobs N. A., Whitlock M. H., et al. (2020). Integrating Gwas and transcriptomics to identify the molecular underpinnings of thermal stress responses in Drosophila Melanogaster. Front. Genet. 11:658. 10.3389/fgene.2020.00658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnert S. J., Kess T., Bentzen P., Kent M. P., Lien S., Gilbey J., et al. (2019). Genomic signatures and correlates of widespread population declines in salmon. Nat. Commun. 10:2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen P. H., Remington D. L., Savolainen O. (2011). Local adaptation, phenotypic differentiation, and hybrid fitness in diverged natural populations of Arabidopsis Lyrata. Evolution 65 90–107. 10.1111/j.1558-5646.2010.01119.x [DOI] [PubMed] [Google Scholar]
- Lenoir J., Bertrand R., Comte L., Bourgeaud L., Hattab T., Murienne J., et al. (2020). Species better track climate warming in the oceans than on land. Nat. Ecol. Evol. 4 1044–1059. 10.1038/s41559-020-1198-2 [DOI] [PubMed] [Google Scholar]
- Libbrecht M. W., Noble W. S. (2015). Machine learning applications in genetics and genomics. Nat. Rev. Genet. 16 321–332. 10.1038/nrg3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little C. J., Wheeler J. A., Sedlacek J., Cortés A. J., Rixen C. (2016). Small-scale drivers: the importance of nutrient availability and snowmelt timing on performance of the Alpine Shrub Salix Herbacea. Oecologia 180 1015–1024. 10.1007/s00442-015-3394-3 [DOI] [PubMed] [Google Scholar]
- Liu S., Cornille A., Decroocq S., Tricon D., Chague A., Eyquard J. P., et al. (2019). The complex evolutionary history of apricots: species divergence, gene flow and multiple domestication events. Mol. Ecol. 28 5299–5314. 10.1111/mec.15296 [DOI] [PubMed] [Google Scholar]
- Logan M. L., Cox C. L. (2020). Genetic constraints, transcriptome plasticity, and the evolutionary response to climate change. Front. Genet. 10.3389/fgene.2020.538226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Hernández F., Cortés A. J. (2019). Last-generation genome–environment associations reveal the genetic basis of heat tolerance in common Bean (Phaseolus Vulgaris L.). Front. Genet. 10:22. 10.3389/fgene.2019.00954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotterhos K. E., Whitlock M. C. (2015). The relative power of genome scans to detect local adaptation depends on sampling design and statistical method. Mol. Ecol. 24 1031–1046. 10.1111/mec.13100 [DOI] [PubMed] [Google Scholar]
- Lstiburek M., Schueler S., El-Kassaby Y. A., Hodge G. R., Stejskal J., Korecky J., et al. (2020). In Situ genetic evaluation of european larch across climatic regions using marker-based pedigree reconstruction. Front. Genet. 11:28. 10.3389/fgene.2020.00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Ritland K. (1999). Estimation of pairwise relatedness with molecular markers. Genetics 152 1753–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons S. K., Amatangelo K. L., Behrensmeyer A. K., Bercovici A., Blois J. L., Davis M., et al. (2016). Holocene shifts in the assembly of plant and animal communities implicate human impacts. Nature 529 80–83. 10.1038/nature16447 [DOI] [PubMed] [Google Scholar]
- Madriñán S., Cortés A. J., Richardson J. E. (2013). Páramo is the world’s fastest evolving and coolest biodiversity hotspot. Front. Genet. 4:192. 10.3389/fgene.2013.00192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher B. (2008). Missing heritability. Nature 456 18–21. 10.1038/456018a [DOI] [PubMed] [Google Scholar]
- Mallard F., Nolte V., Tobler R., Kapun M., Schlötterer C. (2018). A simple geneticbasis of adaptation to a novel thermal environment results in complex metabolic rewiring in Drosophila. A simple genetic basis of adaptation to a novel thermal environment results in complex metabolic rewiring in Drosophila. Genome Biol. 19:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques D. A., Meier J. I., Seehausen O. (2019). A combinatorial view on speciation and adaptive radiation. Trends Ecol. Evol. 34 531–544. 10.1016/j.tree.2019.02.008 [DOI] [PubMed] [Google Scholar]
- Matos M., Simoes P., Santos M. A., Seabra S. G., Faria G. S., Vala F., et al. (2015). History, chance and selection during phenotypic and genomic experimental evolution: replaying the tape of life at different levels. Front. Genet. 6:71. 10.3389/fgene.2015.00071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J., Haigh J. (1974). The hitch-hiking effect of a favourable gene. Genet. Res. 23 23–35. 10.1017/s0016672300014634 [DOI] [PubMed] [Google Scholar]
- McKown A. D., Klapste J., Guy R. D., El-Kassaby Y. A., Mansfield S. D. (2018). Ecological genomics of variation in bud-break phenology and mechanisms of response to climate warming in Populus Trichocarpa. New Phytol. 220 300–316. 10.1111/nph.15273 [DOI] [PubMed] [Google Scholar]
- Meuwissen T. H. E., Hayes B. J., Goddard M. E. (2001). Prediction of total genetic value using genome-wide dense marker maps. Genetics 157 1819–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A. D., Coleman M. A., Clark J., Cook R., Naga Z., Doblin M. A., et al. (2020). Local thermal adaptation and limited gene flow constrain future climate responses of a marine ecosystem engineer. Evol. Appl. 13 918–934. 10.1111/eva.12909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra J., Singh C., Hamid M., Verma A., Semwal S. C., Gajmer B., et al. (2019). Modelling Betula Utilis distribution in response to climate-warming scenarios in Hindu-Kush Himalaya using random forest. Biodiv. Conserv. 28 2295–2317. 10.1007/s10531-019-01731-w [DOI] [Google Scholar]
- Montesinos-Lopez A., Montesinos-Lopez O. A., Gianola D., Crossa J., Hernandez-Suarez C. M. (2018). Multi-environment genomic prediction of plant traits using deep learners with dense architecture. G3 8 3813–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesinos-Lopez O. A., Martin-Vallejo J., Crossa J., Gianola D., Hernandez-Suarez C. M., Montesinos-Lopez A., et al. (2019). A benchmarking between deep learning, support vector machine and bayesian threshold best linear unbiased prediction for predicting ordinal traits in plant breeding. G3 9 601–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris B. B. G. P., Borevitz J. O. (2011). Genome-wide association studies in plants: the missing heritability is in the field. Genome Biol. 12:232. 10.1186/gb-2011-12-10-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman M. W., Payseur B. A. (2012). Recombination rate variation and speciation: theoretical predictions and empirical results from rabbits and mice. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367 409–421. 10.1098/rstb.2011.0249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadachowska-Brzyska K., Burri R., Smeds L., Ellegren H. (2016). Psmc analysis of effective population sizes in molecular ecology and its application to black-and-white Ficedula Flycatchers. Mol. Ecol. 25 1058–1072. 10.1111/mec.13540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan R., Muller-Landau H. C. (2000). Spatial patterns of seed dispersal, their determinants and consequences for recruitment. Trends Ecol. Evol. 15 278–285. 10.1016/s0169-5347(00)01874-7 [DOI] [PubMed] [Google Scholar]
- Nei M. (1987). Molecular Evolutionary Genetics. New York, NY: Columbia University Press. [Google Scholar]
- Nicotra A. B., Atkin O. K., Bonser S. P., Davidson A. M., Finnegan E. J., Mathesius U., et al. (2010). Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 15 684–692. 10.1016/j.tplants.2010.09.008 [DOI] [PubMed] [Google Scholar]
- North A., Pennanen J., Ovaskainen O., Laine A.-L. (2011). Local adaptation in a changing world: the roles of gene-flow, mutation, and sexual reproduction. Evolution 65 79–89. 10.1111/j.1558-5646.2010.01107.x [DOI] [PubMed] [Google Scholar]
- Nosil P., Feder J. L. (2011). Genomic divergence during speciation: causes and consequences. Philos. Trans. R. Soc. B Biol. Sci. 367 332–342. 10.1098/rstb.2011.0263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ørsted M., Hoffmann A. A., Rohde P. D., Sørensen P., Kristensen T. N. (2019). Strong impact of thermal environment on the quantitative genetic basis of a key stress tolerance trait. Heredity 122 315–325. 10.1038/s41437-018-0117-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto S. P., Day T. (2007). A Biologist’s Guide to Mathematical Modeling in Ecology and Evolution. Princeton, NJ: Princeton University Press. [Google Scholar]
- Park D. S., Willis C. G., Xi Z., Kartesz J. T., Davis C. C., Worthington S. (2020). Machine learning predicts large scale declines in native plant phylogenetic diversity. New Phytol. 227 1544–1556. 10.1111/nph.16621 [DOI] [PubMed] [Google Scholar]
- Parmesan C., Hanley M. E. (2015). Plants and climate change: complexities and surprises. Ann. Bot. 116 849–864. 10.1093/aob/mcv169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payseur B. A., Rieseberg L. H. (2016). A genomic perspective on hybridization and speciation. Mol. Ecol. 25 2337–2360. 10.1111/mec.13557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson R. G., Phillips S. J., Loranty M. M., Beck P. S. A., Damoulas T., Knight S. J., et al. (2013). Shifts in arctic vegetation and associated feedbacks under climate change. Nat. Clim. Change 3 673–677. 10.1038/nclimate1858 [DOI] [Google Scholar]
- Pennisi E. (2014). Disputed Islands. Science 345 611–613. 10.1126/science.345.6197.611 [DOI] [PubMed] [Google Scholar]
- Pfenninger M., Foucault Q. (2020). Genomic processes underlying rapid adaptation of a natural Chironomus Riparius population to unintendedly applied experimental selection pressures. Mol. Ecol. 29 536–548. 10.1111/mec.15347 [DOI] [PubMed] [Google Scholar]
- Phillips S. J., Anderson R. P., Dudiík M., Schapire R. E., Blair M. E. (2017). Opening the black box: An open-source release of maxent. Ecography 40 887–893. 10.1111/ecog.03049 [DOI] [Google Scholar]
- Pluess A. R., Frank A., Heiri C., Lalague H., Vendramin G. G., Oddou-Muratorio S. (2016). Genome-environment association study suggests local adaptation to climate at the regional scale in Fagus Sylvatica. New Phytol. 210 589–601. 10.1111/nph.13809 [DOI] [PubMed] [Google Scholar]
- Pritchard J. K., Pickrell J. K., Coop G. (2010). The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr. Biol. 20 R208–R215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero I., Wiens J. J. (2013). Rates of projected climate change dramatically exceed past rates of climatic niche evolution among vertebrate species. Ecol. Lett. 16 1095–1103. 10.1111/ele.12144 [DOI] [PubMed] [Google Scholar]
- Ravinet M., Faria R., Butlin R. K., Galindo J., Bierne N., Rafajlovic M., et al. (2017). Interpreting the genomic landscape of speciation: a road map for finding barriers to gene flow. J. Evol. Biol. 30 1450–1477. 10.1111/jeb.13047 [DOI] [PubMed] [Google Scholar]
- Razgour O., Forester B., Taggart J. B., Bekaert M., Juste J., Ibanez C., et al. (2019). Considering adaptive genetic variation in climate change vulnerability assessment reduces species range loss projections. PNAS 116 10418–10423. 10.1073/pnas.1820663116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichstein M., Bahn M., Mahecha M. D., Kattge J., Baldocchi D. D. (2014). Linking plant and ecosystem functional biogeography. Proc. Natl. Acad. Sci. U.S.A. 111 13697–13702. 10.1073/pnas.1216065111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende M. D. V., Jr., Resende M. F. R., Sansaloni C. P., Petroli C. D., Missiaggia A. A., Aguiar A. M., et al. (2012). Genomic selection for growth and wood quality in Eucalyptus: capturing the missing heritability and accelerating breeding for complex traits in forest trees. New Phytol. 194 116–128. 10.1111/j.1469-8137.2011.04038.x [DOI] [PubMed] [Google Scholar]
- Rezende E. L., Tejedo M., Santos M. (2010). Estimating the adaptive potential of critical thermal limits: methodological problems and evolutionary implications. Funct. Ecol. 25 111–121. 10.1111/j.1365-2435.2010.01778.x [DOI] [Google Scholar]
- Richardson J. E., Madriñán S., Gómez-Gutiérrez M. C., Valderrama E., Luna J., Banda-R K., et al. (2019). Using dated molecular phylogenies to help reconstruct geological, climatic, and biological history: Examples from Colombia. Geol. J. 53 2935–2943. 10.1002/gj.3133 [DOI] [Google Scholar]
- Rippke U., Ramirez-Villegas J., Jarvis A., Vermeulen S. J., Parker L., Mer F., et al. (2016). Timescales of transformational climate change adaptation in Sub-Saharan African agriculture. Nat. Clim. Change 6 605–609. 10.1038/nclimate2947 [DOI] [Google Scholar]
- Ripple W. J., Wolf C., Newsome T. M., Barnard P., Moomaw W. R. (2020). World Scientists’ warning of a climate emergency. BioScience 70 8–12. [Google Scholar]
- Rodriguez-Trelles F., Tarrio R., Santos M. (2013). Genome-wide evolutionary response to a heat wave in Drosophila. Biol. Lett. 9:20130228. 10.1098/rsbl.2013.0228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesti M., Gavrilets S., Hendry A. P., Salzburger W., Berner D. (2014). The genomic signature of parallel adaptation from shared genetic variation. Mol. Ecol. 23 3944–3956. 10.1111/mec.12720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer D. L., Chernoff B. (2013). Diversity in neotropical wet forests during the cenozoic is linked more to atmospheric Co2 than temperature. Proc. Biol. Sci. 280:20131024. 10.1098/rspb.2013.1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti P. C., Varilly P., Fry B., Lohmueller J., Hostetter E., Cotsapas C., et al. (2007). Genome-wide detection and characterization of positive selection in human populations. Nature 449 913–918. 10.1038/nature06250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint Pierre C., Crossa J. L., Bonnett D., Yamaguchi-Shinozaki K., Reynolds M. P. (2012). Phenotyping transgenic wheat for drought resistance. J. Exp. Bot. 63 1799–1808. 10.1093/jxb/err385 [DOI] [PubMed] [Google Scholar]
- Santos M., Castaneda L. E., Rezende E. L. (2012). Keeping pace with climate change: what is wrong with the evolutionary potential of upper thermal limits? Ecol. Evol. 2 2866–2880. 10.1002/ece3.385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherrer D., Körner C. (2011). Topogaphically controlled thermal-habitat differentiation buffers Alpine plant diversity against climate warming. J. Biogeogr. 38 406–416. 10.1111/j.1365-2699.2010.02407.x [DOI] [Google Scholar]
- Schrider D. R., Kern A. D. (2018). Supervised machine learning for population genetics: a new paradigm. Trends Genet. 34 301–312. 10.1016/j.tig.2017.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabra S. G., Fragata I., Antunes M. A., Faria G. S., Santos M. A., Sousa V. C., et al. (2017). Different genomic changes underlie adaptive evolution in populations of contrasting history. Mol. Biol. Evol. 35 549–563. 10.1093/molbev/msx247 [DOI] [PubMed] [Google Scholar]
- Sedlacek J., Bossdorf O., Cortés A. J., Wheeler J. A., Van-Kleunen M. (2014). What role do plant-soil interactions play in the habitat suitability and potential range expansion of the Alpine Dwarf Shrub Salix Herbacea? Basic Appl. Ecol. 15 305–315. 10.1016/j.baae.2014.05.006 [DOI] [Google Scholar]
- Sedlacek J., Cortés A. J., Wheeler J. A., Bossdorf O., Hoch G., Klapste J., et al. (2016). Evolutionary potential in the Alpine: trait heritabilities and performance variation of the Dwarf Willow Salix Herbacea from different elevations and microhabitats. Ecol. Evol. 6 3940–3952. 10.1002/ece3.2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlacek J., Wheeler J. A., Cortés A. J., Bossdorf O., Hoch G., Lexer C., et al. (2015). The response of the Alpine Dwarf Shrub Salix Herbacea to altered snowmelt timing: lessons from a multi-site transplant experiment. PLoS One 10:e0122395. 10.1371/journal.pone.0122395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentinella A. T., Warton D. I., Sherwin W. B., Offord C. A., Moles A. T., Wang Z. (2020). Tropical plants do not have narrower temperature tolerances, but are more at risk from warming because they are close to their upper thermal limits. Glob. Ecol. Biogeogr. 29 1387–1398. 10.1111/geb.13117 [DOI] [Google Scholar]
- Simões J. L., Gouvêa N., Margis M. (2003). Small heat shock proteins genes are differentially expressed in distinct varieties of common Bean. Braz. J. Plant Physiol. 15 33–41. 10.1590/s1677-04202003000100005 [DOI] [Google Scholar]
- Simões P., Pascual M. (2018). Patterns of geographic variation of thermal adapted candidate genes in Drosophila Subobscura sex chromosome arrangements. BMC Evol. Biol. 18:60. 10.1186/s12862-018-1178-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. K., Svystun T., Aldahmash B., Jonsson A. M., Bhalerao R. P. (2017). Photoperiod- and temperature-mediated control of phenology in trees - a molecular perspective. New Phytol. 213 511–524. 10.1111/nph.14346 [DOI] [PubMed] [Google Scholar]
- Slatkin M. (2008). Linkage disequilibrium — understanding the evolutionary past and mapping the medical future. Nat. Rev. Genet. 9 477–485. 10.1038/nrg2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen J. G., Kristensen T. N., Loeschcke V. (2003). The evolutionary and ecological role of heat shock proteins. Ecol. Lett. 6 1025–1037. 10.1046/j.1461-0248.2003.00528.x [DOI] [Google Scholar]
- Spindel J. E., Begum H., Akdemir D., Collard B., Redona E., Jannink J. L., et al. (2016). Genome-wide prediction models that incorporate de novo gwas are a powerful new tool for tropical rice improvement. Heredity 116 395–408. 10.1038/hdy.2015.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton M. L., Galen C., Shore J. (1997). Population structure along a steep environmental gradient: consequences of flowering time and habitat variation in the snow buttercup, Ranunculus Adoneus. Evolution 51 79–94. 10.2307/2410962 [DOI] [PubMed] [Google Scholar]
- Steinbauer M. J., Grytnes J. A., Jurasinski G., Kulonen A., Lenoir J., Pauli H., et al. (2018). Accelerated increase in plant species richness on mountain summits is linked to warming. Nature 556 231–234. [DOI] [PubMed] [Google Scholar]
- Stephan W. (2016). Signatures of positive selection: from selective sweeps at individual loci to subtle allele frequency changes in polygenic adaptation. Mol. Ecol. 25 79–88. 10.1111/mec.13288 [DOI] [PubMed] [Google Scholar]
- Stölting K. N., Paris M., Meier C., Heinze B., Castiglione S., Bartha D., et al. (2015). Genome-wide patterns of differentiation and spatially varying selection between postglacial recolonization lineages of Populus Alba (Salicaceae), a widespread forest tree. New Phytol. 207 723–734. 10.1111/nph.13392 [DOI] [PubMed] [Google Scholar]
- Street N. R., Jansson S., Hvidsten T. (2011). A systems biology model of the regulatory network in Populus leaves reveals interacting regulators and conserved regulation. BMC Plant Biol. 11:13. 10.1186/1471-2229-11-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran S., Crossa J., Jarquin D., Lopes M., Reynolds M. P. (2017). Genomic prediction with pedigree and genotype x environment interaction in spring wheat grown in South and West Asia, North Africa, and Mexico. G3 7 481–495. 10.1534/g3.116.036251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suontama M., KláPšTě J., Telfer E., Graham N., Stovold T., Low C., et al. (2019). Efficiency of genomic prediction across two Eucalyptus Nitens seed orchards with different selection histories. Heredity 122 370–379. 10.1038/s41437-018-0119-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts K., Gutaker R. M., Benz B., Blake M., Bukowski R., Holland J., et al. (2017). Genomic estimation of complex traits reveals ancient maize adaptation to temperate North America. Science 357 512–515. 10.1126/science.aam9425 [DOI] [PubMed] [Google Scholar]
- Sweet L. C., Green T., Heintz J. G. C., Frakes N., Graver N., Rangitsch J. S., et al. (2019). Congruence between future distribution models and empirical data for an iconic species at Joshua Tree National Park. Ecosphere 10:e0276 10.1002/ecs2.2763 [DOI] [Google Scholar]
- Tam V., Patel N., Turcotte M., Bosseì Y., Pareì G., Meyre D. (2019). Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 20 467–484. 10.1038/s41576-019-0127-1 [DOI] [PubMed] [Google Scholar]
- Tan B., Grattapaglia D., Martins G. S., Ferreira K. Z., Sundberg B. R., Ingvarsson P. R. K. (2017). Evaluating the accuracy of genomic prediction of growth and wood traits in two eucalyptus species and their F1 hybrids. BMC Plant Biol. 17:110. 10.1186/s12870-017-1059-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Technow F., Schrag T. A., Schipprack W., Bauer E., Simianer H., Melchinger A. E. (2014). Genome properties and prospects of genomic prediction of hybrid performance in a breeding program of maize. Genetics 197 1343–1355. 10.1534/genetics.114.165860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O., Rodriguez-Verdugo A., Gaut R. L., Mcdonald P., Bennett A. F., Long A. D., et al. (2012). The molecular diversity of adaptive convergence. Science 335 457–461. 10.1126/science.1212986 [DOI] [PubMed] [Google Scholar]
- Turner T. L., Miller P. M. (2012). Investigating natural variation in drosophila courtship song by the evolve and resequence approach. Genetics 191 633–642. 10.1534/genetics.112.139337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia J. B., Mesa J., León J. G., Madriñán S., Cortés A. J. (2020). Climate vulnerability assessment of the Espeletia complex in Páramo sky islands of the northern Andes. Front. Ecol. Evol. 10.3389/fevo.2020.565708 [DOI] [Google Scholar]
- Valladares F., Matesanz S., Guilhaumon F., Araujo M. B., Balaguer L., Benito-Garzon M., et al. (2014). The effects of phenotypic plasticity and local adaptation on forecasts of species range shifts under climate change. Ecol. Lett. 17 1351–1364. 10.1111/ele.12348 [DOI] [PubMed] [Google Scholar]
- Vargas O. M., Ortiz E. M., Simpson B. B. (2017). Conflicting phylogenomic signals reveal a pattern of reticulate evolution in a recent high-andean diversification (Asteraceae: Astereae: Diplostephium). New Phytol. 214 1736–1750. 10.1111/nph.14530 [DOI] [PubMed] [Google Scholar]
- Visser J. A. G. M. D., Elena S. F., Fragata I. S., Matuszewski S. (2018). The utility of fitness landscapes and big data for predicting evolution. Heredity 121 401–405. 10.1038/s41437-018-0128-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley J. (2008). Coalescent Theory: An Introduction. Cambridge: Harvard University. [Google Scholar]
- Waldvogel A. M., Feldmeyer B., Rolshausen G., Exposito-Alonso M., Rellstab C., Kofler R., et al. (2020a). Evolutionary genomics can improve prediction of species’ responses to climate change. Evol. Lett. 4 4–18. 10.1002/evl3.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldvogel A. M., Schreiber D., Pfenninger M., Feldmeyer B. (2020b). Climate change genomics calls for standardised data reporting. Front. Ecol. Evol. 8:242 10.3389/fevo.2020.00242 [DOI] [Google Scholar]
- Walsh B. (2008). Evolutionary quantitative genetics. Handb. Stat. Genet. 1 533–586. [Google Scholar]
- Walther G. R., Post E., Convey P., Menzel A., Parmesan C., Beebee T. J. C., et al. (2002). Ecological responses to recent climate change. Nature 416 389–395. [DOI] [PubMed] [Google Scholar]
- Wang X., Edwards R. L., Auler A. S., Cheng H., Kong X., Wang Y., et al. (2017). Hydroclimate changes across the amazon lowlands over the past 45,000 years. Nature 541 204–207. 10.1038/nature20787 [DOI] [PubMed] [Google Scholar]
- Way D. A., Oren R. (2010). Differential responses to changes in growth temperature between trees from different functional groups and biomes: a review and synthesis of data. Tree Physiol. 30 669–688. 10.1093/treephys/tpq015 [DOI] [PubMed] [Google Scholar]
- Way D. A., Oren R., Kroner Y. (2015). The space-time continuum: the effects of elevated Co2 and temperature on trees and the importance of scaling. Plant Cell Environ. 38 991–1007. 10.1111/pce.12527 [DOI] [PubMed] [Google Scholar]
- Weir B. S., Cockerham C. (1984). Estimating F-Statistics for the analysis of population structure. Evolution 38 1358–1370. 10.2307/2408641 [DOI] [PubMed] [Google Scholar]
- Wheeler J. A., Cortés A. J., Sedlacek J., Karrenberg S., Van Kleunen M., Wipf S., et al. (2016). The snow and the willows: accelerated spring snowmelt reduces performance in the low-lying Alpine Shrub Salix Herbacea. J. Ecol. 104 1041–1050. 10.1111/1365-2745.12579 [DOI] [Google Scholar]
- Wheeler J. A., Hoch G., Cortés A. J., Sedlacek J., Wipf S., Rixen C. (2014). Increased Spring freezing vulnerability for Alpine Shrubs under early snowmelt. Oecologia 175 219–229. 10.1007/s00442-013-2872-8 [DOI] [PubMed] [Google Scholar]
- Wheeler J. A., Schnider F., Sedlacek J., Cortés A. J., Wipf S., Hoch G., et al. (2015). With a little help from my friends: community facilitation increases performance in the Dwarf Shrub Salix Herbacea. Basic Appl. Ecol. 16 202–209. 10.1016/j.baae.2015.02.004 [DOI] [Google Scholar]
- Whitlock C., Bartlein P. J. (2003). holocene fire activity as a record of past environmental change. Dev. Quatern. Sci. 1 479–490. 10.1016/s1571-0866(03)01022-4 [DOI] [Google Scholar]
- Willis C. G., Ruhfel B., Primack R. B., Miller-Rushing A. J., Davis C. C. (2008). Phylogenetic patterns of species loss in Thoreau’s woods are driven by climate change. PNAS 105 17029–17033. 10.1073/pnas.0806446105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis K. J., MacDonald G. M. (2011). Long-term ecological records and their relevance to climate change predictions for a warmer world. Annu. Rev. Ecol. Evol. Syst. 42 267–287. 10.1146/annurev-ecolsys-102209-144704 [DOI] [Google Scholar]
- Wipf S., Stöckli V., Herz K., Rixen C. (2013). The oldest monitoring site of the alps revisited: accelerated increase in plant species Richness on Piz Linard summit since 1835. Plant Ecol. Divers. 6 447–455. 10.1080/17550874.2013.764943 [DOI] [Google Scholar]
- Wolf J. B., Ellegren H. (2017). Making sense of genomic islands of differentiation in light of speciation. Nat. Rev. Genet. 18 87–100. 10.1038/nrg.2016.133 [DOI] [PubMed] [Google Scholar]
- Wolkovich E. M., Cook B. I., Allen J. M., Crimmins T. M., Betancourt J. L., Travers S. E., et al. (2012). Warming experiments underpredict plant phenological responses to climate change. Nature 485 494–497. 10.1038/nature11014 [DOI] [PubMed] [Google Scholar]
- Wray N. R., Yang J., Hayes B. J., Price A. L., Goddard M. E., Visscher P. M. (2013). Pitfalls of predicting complex traits from Snps. Nat. Rev. Genet. 14 507–515. 10.1038/nrg3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Islam A. S. M. F., Limpot N., Mackasmiel L., Mierzwa J., Cortés A. J., et al. (2020). Genome-wide Snp identification and association mapping for seed mineral concentration in Mung Bean (Vigna Radiata L.). Front. Genet. 11:656. 10.3389/fgene.2020.00656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi H., Allison T. D., Ohara M. (2005). Effect of snowmelt timing on the genetic structure of an Erythronium Grandiflorum population in an Alpine environment. Ecol. Res. 20 199–204. 10.1007/s11284-004-0032-7 [DOI] [Google Scholar]
- Yeaman S., Otto S. P. (2011). Establishment and maintenance of adaptive genetic divergence under migration, selection, and drift. Evolution 65 2123–2129. 10.1111/j.1558-5646.2011.01277.x [DOI] [PubMed] [Google Scholar]
- Zachos J. C., Dickens G. R., Zeebe R. E. (2008). An early cenozoic perspective on greenhouse warming and carbon-cycle dynamics. Nature 451 279–283. 10.1038/nature06588 [DOI] [PubMed] [Google Scholar]
- Zahn L. M., Purnell B. A. (2016). Genes under pressure. Science 354:52. 10.1126/science.354.6308.52 [DOI] [PubMed] [Google Scholar]
- Zellweger F., De Frenne P., Lenoir J., Vangansbeke P., Verheyen K., Bernhardt-Römermann M., et al. (2020). Forest microclimate dynamics drive plant responses to warming. Science 368 772–775. [DOI] [PubMed] [Google Scholar]
- Zhou L., Bawa R., Holliday J. A. (2014). Exome resequencing reveals signatures of demographic and adaptive processes across the genome and range of Black Cottonwood (Populus Trichocarpa). Mol. Ecol. 23 2486–2499. 10.1111/mec.12752 [DOI] [PubMed] [Google Scholar]
- Zwoinska M. K., Rodrigues L. R., Slate J., Snook R. R. (2020). Phenotypic responses to and genetic architecture of sterility following exposure to sub-lethal temperature during development. Front. Genet. 11:573. 10.3389/fgene.2020.00573 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Past and future of thermal and CO2 variation, and their correlates with past biodiversity. (A) Temperature and richness of plant species (from pollen) for the Cenozoic Era (65 Mya – present). Temperature estimates (Supplementary Table S1) were computed by Hansen et al. (2013) using the original δ18O record from Zachos et al. (2008). Richness of plant species from pollen data (Supplementary Table S1) is based on 15 Neotropical stratigraphic sections inspected by Jaramillo et al. (2006). This profile goes from 65 to 20 Mya due to a lack of more recent suitable sampling records. (B) Projections of the near-surface temperature anomalies to 2,050 (Supplementary Table S2), which follow the CIMP5 RCP 8.5 scenario from the KNMI (http://climexp.knmi.nl/) repository averaged from an original 5-min resolution. Light gray shaded areas depict minimum and maximum estimates. (C) Atmospheric CO2 and richness of plant species (as in A) for the Cenozoic Era (65 Mya – present). CO2 records are an updated version (Supplementary Table S1) derived from Royer and Chernoff (2013), originally compiled by Beerling and Royer (2011). (D) Projected CO2 concentration (ppm) to 2,050 also follow the CIMP5 RCP 8.5 scenario, as in B (Supplementary Table S3).
Dataset of temperature, atmospheric CO2, and richness of plant species for the Cenozoic Era (65 Mya – present for temperature and CO2, and 65–20 Mya for richness of plant species). Temperature estimates were computed by Hansen et al. (2013) from five-point running means of the original temporal resolution of the δ18O record from Zachos et al. (2008), a profile of surface low-magnesium calcitic fossils (including planktonic foraminifera, belemintes, brachiopods, and bivalves) that was lower during periods with warmer seawater. Atmospheric CO2 corresponds to an updated version from Royer and Chernoff (2013), originally compiled by Beerling and Royer (2011). Richness of plant species is based on pollen data from Jaramillo et al. (2006), who analyzed 1,530 samples from 15 stratigraphic sections in Colombia and Venezuela (Neotropics).
Projections of thermal variation to 2,050. Simulation of Near-Surface Air Temperature Anomalies (°C) from 1,860 to 2,050 follow the CIMP5 RCP 8.5 scenario from the KNMI (http://climexp.knmi.nl/) database averaged from an original 5 min resolution. Minimum and maximum temperature estimates were generated by the coupled ACCESS v.1.0 model specifically designed for the CIMP5 project (Kowalczyk et al., 2013).
Projections of CO2 concentration (ppm) to 2,050. Simulations follow the CIMP5 RCP 8.5 scenario from 1,860 to 2,050 available at KNMI (http://climexp.knmi.nl/) database averaged from an original 5 min resolution.