Abstract
Purpose
Cataract, a clouding of the intraocular lens, is the leading cause of blindness. The lens-expressed long noncoding RNA OIP5-AS1 was upregulated in lens epithelial cells from patients with cataracts, suggesting its pathogenic role in cataracts. We investigated the regulatory role of OIP5-AS1 in the development of cataracts as well as potential RNA binding proteins, downstream target genes, and upstream transcription factors.
Methods
Clinical capsules and ex vivo and in vitro cataract models were used to test OIP5-AS1 expression. Cell apoptosis was detected using Western blots, JC-1 staining, and flow cytometry. Ribonucleoprotein immunoprecipitation-qPCR was performed to confirm the interaction of OIP5-AS1 and POLG. Chromatin immunoprecipitation-qPCR was used to determine the binding of TFAP2A and the OIP5-AS1 promoter region.
Results
OIP5-AS1 was upregulated in cataract lenses and B3 cells under oxidative stress. OIP5-AS1 knockdown protected B3 cells from H2O2-induced apoptosis and alleviated lens opacity in the ex vivo cataract model. HuR functioned as a scaffold carrying OIP5-AS1 and POLG mRNA and mediated the decay of POLG mRNA. POLG was downregulated in the cataract lens and oxidative-stressed B3 cells, and POLG depletion decreased the mtDNA copy number and MMP, increased reactive oxygen species production, and sensitized B3 cells to oxidative stress-induced apoptosis. POLG overexpression reversed these effects. TFAP2A bound the OIP5-AS1 promoter and contributed to OIP5-AS1 expression.
Conclusions
We demonstrated that OIP5-AS1, activated by TFAP2A, contributed to cataract formation by inhibiting POLG expression mediated by HuR, thus leading to increased apoptosis of lens epithelial cells and aggravated lens opacity, suggesting that OIP5-AS1 is a potential target for cataract treatment.
Keywords: OIP5-AS1, POLG, apoptosis, oxidative stress
Cataract is a clouding of the natural intraocular crystalline lens that leads to blindness, and it represents the most common eye disease globally, with age-related cataract, the most common form. The main causes of ARCs include excessive apoptosis of human lens epithelial cells (HLECs), reactive oxygen species (ROS) production, and dysfunction of the α-crystalline chaperone.1,2 The main ROS in the eye are superoxide anion (O2−), H2O2 and hydroxyl radical (OH−), which cause oxidative damage to DNA and proteins in the lens and lead to cell apoptosis.3,4 Excessive apoptosis of lens epithelial cells is the early common event of age-related cataract. Previous studies discovered that caspase 1-dependent pyroptosis of HLECs contributes to cataract formation5.5 Despite these findings, the molecular mechanisms underlying the pathogenesis of cataracts are still unclear.
Noncoding RNA (ncRNA) are functional molecules that regulate gene expression or modification through different mechanisms. Both short and long types of ncRNA could modulate gene expression and epigenetic processes, including methylation, histone modification, chromatin remodeling, and gene silencing6.6 Long ncRNAs (lncRNAs) are a class of noncoding transcripts longer than 200 nucleotides that play various regulatory roles in a wide array of cellular processes7.7 Many lncRNAs have been identified in the lens, and some have been shown to participate in the regulation of HLEC activities.8,9 For instance, MIAT acts as a competing endogenous RNA to form a feedback loop with Akt and miR-150-5p, thus regulating HLEC apoptosis and migration.10 MALAT1 can promote the apoptosis and oxidative stress of HLECs through the activation of p38 MAPK11.11 However, the majority of lens-expressed lncRNAs have not been functionally characterized.
The lncRNA OIP5-AS1 (known as Cyrano in zebrafish) is highly and ubiquitously expressed in various tissues and species12.12 Cyrano has been shown to promoting zebrafish brain development, and silencing of Cyrano by miR-7 without changing zygotic Cyrano altered the brain architecture at 24 hours post fertilization and 48 hours post fertilization13.13 In the lens, OIP5-AS1 is upregulated in the HLECs of patients with cataracts.10 This molecule has been reported as a tumor suppressor or oncogene depending on the tumor type.14,15 OIP5-AS1 is also involved in the self-renewal of embryonic stem cells by modulating the expression of miR-7.12,16 In human osteosarcoma U2OS cells, OIP5-AS1 depletion increases the expression of POLG, a mitochondrial DNA polymerase responsible for the synthesis of mitochondrial DNA (mtDNA), and RNA pulldown studies showed that OIP5-AS1 is associated with POLG mRNA.17 Mutations of POLG were reported to contribute to the formation of cataracts and progressive external ophthalmoplegia.18,19 However, whether OIP5-AS1 affects cataract formation by influencing POLG expression and the underlying regulatory mechanism have not been clarified.
In this study, we investigated the role of OIP5-AS1 in the development of cataracts in vitro and ex vivo and whether it interferes with POLG expression. We also attempted to identify the upstream regulatory factors of OIP5-AS1 in B3 cells.
Methods
Culture of the HLEC Line B3
The HLEC line B3 (CRL-11421, American Type Culture Collection, Manassas, VA) was used for all of the cell experiments. Cells were cultured in DMEM (CM10014, Macgene) supplemented with 10% fetal bovine serum (04-001-1ACS, Biological Industries, Cromwell, CT) and 1% penicillin-streptomycin (SV30010, HyClone, Logan, UT) at 37°C in a humidified atmosphere with 5% CO2. H2O2 was applied to induce oxidative stress in the medium. A total of 106 cells within 12 passages were seeded in culture flasks, and the medium was renewed every 2 days. All cells were free of mycoplasma.
Clinical Sample Collection
This study was approved by the Ethics Committee of the First Affiliated Hospital of Xi'an Jiaotong University and performed according to the Declaration of Helsinki. All of the lens capsules from the patients with cataracts were obtained at Xi'an Jiaotong University during cataract surgery. All of the patients provided informed consent, and the nature and possible consequences of the study were explained. All of the control lenses were from the Eye Bank of the First Affiliated Hospital of Xi'an Jiaotong University, and the capsules were acquired by continuous curvilinear capsulorhexis. All of the samples were preserved in TRIzol. Detailed information on the clinical participants is provided in Supplementary Table S1.
Establishment of the Rat Model of Cataract
Twenty healthy adult SD rats (provided by the Animal Experimentation Center Affiliated with Xi'an Jiaotong University, which adhered to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research) weighing 180 to 200 g with a gender ratio of 1:1 were chosen for this study. According to Lim,20 UVB is widely used to induce cataracts in rats. All rats were exposed to 9 kJ/m2 at 302 nm UVB (UVLM-26, UVP) within 7 days and then sacrificed 3 days after exposure. All of the rats were mydriatic before being anaesthetized by pentobarbital sodium, and all of the left eyes served as controls. A large incision was made using a 2.0-mm paracentesis knife, and then the intact lenses were isolated along the limbus and placed in warm PBS. Next, the zonular region was carefully cut away. Images were taken by a Canon M6 system.
Nuclear and Cytoplasmic OIP5-AS1 mRNA Quantification
Cell collection and fractionation were performed according as described by Wang et al21.21 B3 cells (approximately 3 × 106 cells) were digested using trypsin and collected in cold PBS. Then, the cells were centrifuged and resuspended in RSB buffer and lysed in RSBG40. The supernatant from high-speed centrifugation was collected as the cytoplasmic fraction, and the precipitated nuclear substances were released in RSBG40 containing one-tenth volume of detergent (3.3% [wt/wt] sodium deoxycholate and 6.6% [vol/vol] Tween-40). RNA was extracted using TRIzol, and RT-qPCR was used for quantification. Whole-cell RNA was used for normalization. The primers used are listed in Table 1.
Table 1.
Primers Used for RT-qPCR
| Gene | Forward | Reverse |
| h-OIP5-AS1 | TTTCGTGGATGCCACAGGAC | TAGTTCCTCTCCTCTGGCCG |
| h-GAPDH | GTCAAGGCTGAGAACGGGAA | AAATGAGCCCCAGCCTTCTC |
| h-ACTIN | TACCTCATGAAGATCCTCACC | TTTCGTGGATGCCACAGGAC |
| h-POLG | TAGTTCCTCTCCTCTGGCCG | GAGGCAGCTTGAAAAACCAG |
| h-HuR | GTCATGATGGCGGAGTAAT | AGGACCCGCGAGTTGATGATCCG |
| h-7SL | ACTGAGCAGTCCACGTAGAAA | GGCTGGAGTGCAGTGGCTAT |
| h-MALAT1 | GATTGAGGAGGCTGTGCTGT | CAGCTGCCTGCTGTTTTCTG |
| r-OIP5-AS1 | AGGACCCGCGAGTTGATGATCCG | ACTGAGCAGTCCACGTAGAAA |
| r-ACTIN | CCATGCTCGTTCAGTGACTACCTC | AAGGGTGTAAAACGCAGCTC |
| h-TFAP2A | GTTCACGCCGATCCATGAAAA | AGATTGACCTACAGTGCCCAG |
| mt-ACTIN | ATCATGTTTGAGACCTTCAAC | ATCTCTTGCTCGAAGTCCA |
| mt-ND1 | TCTAGCCACCTCTAGCCTAG | GTCATGATGGCGGAG TAAT |
| ChIP-region1 | GGAAACATTCATCGAGGGGC | AACCAGAGTATGTCACCGCT |
| ChIP-region2 | AGAATCCACCCAGATGACAGC | GCAGGGAGGGTATACGTGGG |
| ChIP-region3 | GGGCCTTCAGAAAAGTGACG | GCGCCCAGGCTTTTAATCCA |
| ChIP-region4 | CCTCGGGCCTGAGAATGAAA | TTTCGATCCGGATTGGGTCC |
| ChIP-region3’ | ATGCAGAAGAAAGGATGCCCT | CCCTCCTGCATAGAGGAAACC |
Western Blot
Whole cells were lysed using Laemmli sample buffer (1610737, Bio‑Rad, Hercules, CA). Proteins were separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane. For Western blot analysis, the membrane was blocked in 1% BSA for 1 hour and then incubated in primary antibodies that recognize POLG (ab128899, Abcam, Cambridge, UK), Bax (ab32503, Abcam), Bcl-2 (ab32124, Abcam), and caspase3 (#9664, CST). After incubation with appropriate secondary antibodies, the protein signals were detected using an enhanced chemiluminescence Western blotting detection kit (1705060, Bio-Rad).
Small Interfering RNA (siRNA) Knockdown and Plasmid Transfection
Transfection of siRNA was performed as described by the data sheet. Briefly, 30 pM of siRNA and 9 µL of RNAiMAX reagent were diluted in Opti-MEM and then the diluted siRNA was added to the diluted lipofectamine RNAiMAX Reagent (1:1 ratio). After 5 minutes of incubation at room temperature, the siRNA-lipid complex was added to the cells. The sequences were as follows: siRNA for human HuR: TTACCAGTTTCAATGGTCA; siRNA for human OIP5-AS1: CACCAAACAGGCUUUGUGUUCCUTA; siRNA for human POLG: 1: GGUGCACAGACUUUAUGUA, 2: GGAUGGUAAUAGCUGUAAUTT; siRNA for rat OIP5-AS1: 1: GGTTTGTTACAGTAGTGAA, 2: GGTTAGTCAGATTGGACAA, 3: GGAAGTACAAGATAAACAA; siRNA for TFAP2A: AACAUCCCAGAUCAAACUGUA.
All sequences were from Guangzhou RiboBio Co., Ltd. Two POLG siRNAs were mixed and transfected into cells together. The POLG overexpression plasmid was purchased from Shanghai Genechem Co., Ltd (Shanghai, China). The POLG plasmid or negative control vector was transfected using jetPRIME (Polyplus) according to the manufacturer's instructions. Briefly, 2 µg of vector and 4 µL of jetPRIME transfection reagent were added to 200 µL of jetPRIME buffer and incubated for 15 minutes at room temperature. Then, the mix was added to the medium.
Mitochondrial Membrane Potential (MMP) Assay With JC-1
After appropriate treatment, the cells were washed in PBS twice, and 2 µM JC-1 (C2006, Beyotime, Shanghai, China) was loaded. Then, the cells were incubated at 37°C for 20 minutes in the dark. Cell visualization was performed by a fluorescence microscope (Eclipse Ti, Tokyo, Japan), and the red/green fluorescence intensity was determined by a microplate reader (SpectraMax i3, Molecular Devices, Eugene, OR).
Quantitative RT-qPCR
Total RNA samples from the rat lens or cultured cells were extracted using TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The iScript gDNA Clear cDNA Synthesis Kit (1725035, Bio-Rad) was used for cDNA synthesis, and SsoFast EvaGreen (1725201AP, Bio-Rad) was used for real-time PCR. The primers are listed in Table 1.
Analysis of mtDNA Copy Number
Whole-cell DNA was extracted using an assay kit from Tiangen Biotech Co., Ltd. (Beijing, China), and 50 ng DNA was used as the template for RT-qPCR. Actin was used as a nuclear single copy control and analyzed as described by Foote et al.22 The primers used in this assay are listed in Table 1.
ROS Detection
After induction of H2O2 for 24 hours, 10 µM DCFH-DA (D6883, Sigma-Aldrich, St Louis, MO) was added to the medium and the cells were incubated at 37°C for 30 minutes in the dark. Then, the cells were washed with PBS and resuspended. The ROS mean intensity was measured using a CytoFLEX system (Beckman Coulter, Brea, CA) in the FITC channel. The excitation wavelength was 488 nm, and the emission wavelength was 525 nm.
Ribonucleoprotein Immunoprecipitation (RIP)-qPCR
Ribonucleoprotein is composed of RNA and RNA-binding proteins and can regulate post-transcriptional gene expression. For the RIP assay, Protein A/G PLUS-Agarose beads (sc-2003, Santa Cruz Biotechnology, Santa Cruz, CA) were precoated with 5 µg HuR antibody (ab200342, Abcam) or rabbit normal IgG overnight at 4°C. A total of 107 cells were prepared in NT-2 buffer with 1 mM DTT and recombinant RNasin ribonuclease inhibitor. Then, the lysates were incubated with precoated agarose beads overnight at 4°C. After six washes in NT-2 buffer, RNA was extracted using TRIzol. RT-qPCR was used to detect enrichment. Primers for OIP5-AS1 and POLG are shown in Table 1.
Establishment of a Rat Ex Vivo Model of Cataract
Rat selection and animal care were performed as described elsewhere in this article. For the analysis of ex vivo opacified lens, the intact lenses were isolated along the limbus and the zonular region was carefully cut away. Whole lenses were cultured in DMEM containing 5% fetal bovine serum and 1% penicillin-streptomycin. Sixty picomoles of siRNA were supplied to silence rat OIP5-AS1. The lenses were visualized using a Canon M6 system.
Annexin V/PI Apoptosis Assay
Cell apoptosis was detected using an APC-PE-PI Apoptosis Assay kit (Biolegend, San Diego, CA) on a CytoFLEX system. All of the cells in the plate along with floating cells after treatment with 200 µM H2O2 for 24 hours were collected and washed in PBS. Then, the cells were resuspended in 100 µL of annexin V binding buffer. Next, 5 µL of annexin V together with 10 µL of propidium iodide was added to the suspension. After 15 minutes of incubation in the dark at room temperature, 400 µL of binding buffer was added to terminate the reaction and the cells were counted by flow cytometry.
Motif Prediction
We first selected the differentially expressed genes between control and patient lens epithelial cells from Wu et al23 and prioritized genes upregulated in HLECs from the cataract group. Then, motif prediction on OIP5-AS1 was performed using FIMO from the MEME Suite toolkit (v4.11.0)24 with default parameters and motifs available from five public motif databases: JASPAR (2018 version)25,25 HOCOMOCO (v11)26,26 SwissRegulon,27 Transfac, and Jolma201328.28 We selected the OIP5-AS1 promoter sequence (–2000 bp to +200 bp) for the motif analysis.
Chromatin Immunoprecipitation (ChIP)-qPCR
ChIP assays were performed for the B3 cells. All of the B3 cells in the culture dish (approximately 1 × 107 cells) were first crosslinked using methanol and quenched by glycine. Then, the cells were rinsed and scraped off into conical tubes. The pellets were resuspended in cell lysis buffer and SDS lysis buffer in turn, and a VirTis Virsonic 100 Ultrasonic Homogenizer/Sonicate (VCX500, Socics, Connecticut, USA) was used to shear chromatin. The sonicated lysates were diluted using DNA dilution buffer and precleared with protein A/G PLUS-agarose beads for 2 hours. Two percent of the total supernatant was taken as the input. Two micrograms of rabbit normal IgG or TFAP2A antibody (ab52222, Abcam) and 40 µL of beads were added together for immunoprecipitation. The DNA–protein complex was eluted, reverse crosslinked, and purified for RT-PCR. The PCR primers were designed according to the four clusters to evaluate the IP efficacy of the four regions as shown in Table 1. Because ultrasound can break the chromatin fragments into independent nucleosomes, the 3' primer pair was used to amplify short regions (<180 bp) in region 3 for ChIP-qPCR.
Statistical Analysis
GraphPad Prism 6.0 statistical software was applied to perform the statistical analyses of the data. All of the data are presented as the mean ± SEM from at least three independent experiments. The differences between two groups were analyzed by unpaired Student t tests, comparisons between multiple groups were analyzed using one-way ANOVAs, and a P value of less than 0.05 was considered significant. The grade count data of the isolated turbid lens in Table 2 were analyzed using the Kruskal–Wallis test and the Mann–Whitney post hoc test with a Bonferroni correction in SPSS 21.0 (IBM Corp., Armonk, NY), with a P value of less than 0.05/6 considered statistically significant.
Table 2.
Morphologic Assessment of Lenticular Opacification of Lenses After 6 Days Incubation
| Grade 0 | Grade I | Grade II | Grade III | Grade IV | |
|---|---|---|---|---|---|
| siRNA-NC | 2 | 3 | 0 | 0 | 0 |
| siOIP5-AS1 | 2 | 3 | 0 | 0 | 0 |
| siRNA-NC+H2O2 | 0 | 0 | 0 | 0 | 5 |
| siOIP5-AS1+H2O2 | 0 | 1 | 3 | 1 | 0 |
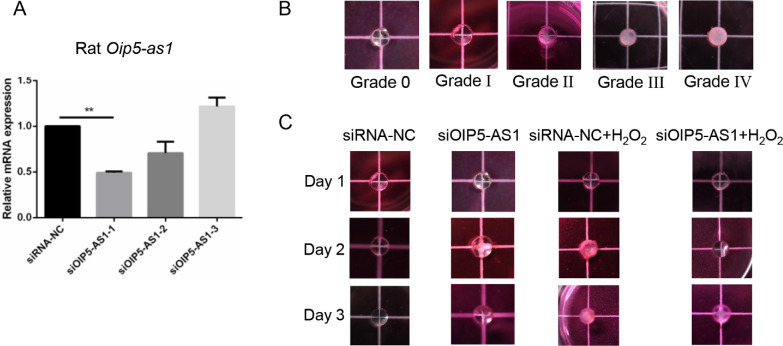
Incidence and grading of cataract formation of the lenses in each group. Grade 0: Absence of opacification (grid lines clearly visible); grade 1, minimal clouding of lens (grid lines easily visible); grade 2, presence of flaky opacification involving the part of lens (with gridlines visible), grade 3, Presence of opacification of the entire of lens (with grid lines faintly visible), and grade 4, presence of extensive opacification of the entire lens (grid lines not visible).
Results
OIP5-AS1 Was Upregulated in Clinical Cataract Samples and Cataract Models
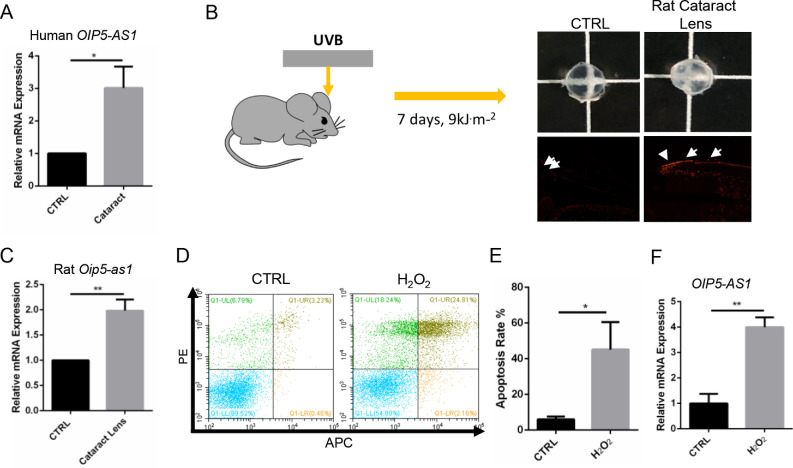
We first tested OIP5-AS1 expression in the clinical samples. RT-qPCR showed that the expression of OIP5-AS1 in the lens epithelial cells from the cataract group was much higher than that in the control group (Fig. 1A, P < 0.05). We then investigated the expression of Oip5-as1 in an in vivo rat model of UVB-induced cataract. Compared with the control lens, the UVB-radiated lens became turbid after 7 days and displayed an impaired tissue structure (Supplementary Fig. S1A). The opacification of the UVB-irradiated lens was associated with an increased apoptosis rate as shown by enhanced TUNEL staining (Fig. 1B) and increased expression of Oip5-as1 (P < 0.01, Fig. 1C), cleaved caspase 3 and Bax (Supplementary Fig. S1B). Moreover, we tested the expression of OIP5-AS1 in an H2O2-induced B3 cell model. The H2O2 treatment increased cell apoptosis from approximately 6% to 42% (P < 0.05, Figs. 1D, 1E) and concomitantly decreased the mtDNA copy number of B3 cells (P < 0.05, Supplementary Fig. S1C). Notably, OIP5-AS1 expression increased in response to the H2O2 treatment (P < 0.01, Fig. 1F). These results showed that OIP5-AS1 was upregulated in the lens epithelial cells in cataracts. Additionally, in ex vivo and in vitro cataract models, oxidative stress induced OIP5-AS1 expression.
Figure 1.
OIP5-AS1 expression was upregulated in clinical lens samples and cataract models. (A) RT-qPCR of OIP5-AS1 expression in lens capsules. (B) Rats underwent a total of 9 kJ.m2 UVB irradiation for 7 days and were then sacrificed. Their lenses were isolated, photographed, and stained using TUNEL dye for observation. The white arrows indicate the TUNEL-positive cells. (C) Oip5-as1 was upregulated in the lens of rats with cataract as shown by RT-qPCR. (D–E) H2O2 (200 µM) was chosen for the oxidative stress cell model. (F) OIP5-AS1 increased in the oxidative stress cell model, as shown by RT-qPCR. *P < 0.05, **P < 0.01.
OIP5-AS1 Knockdown Alleviated ROS Production and Cell Apoptosis
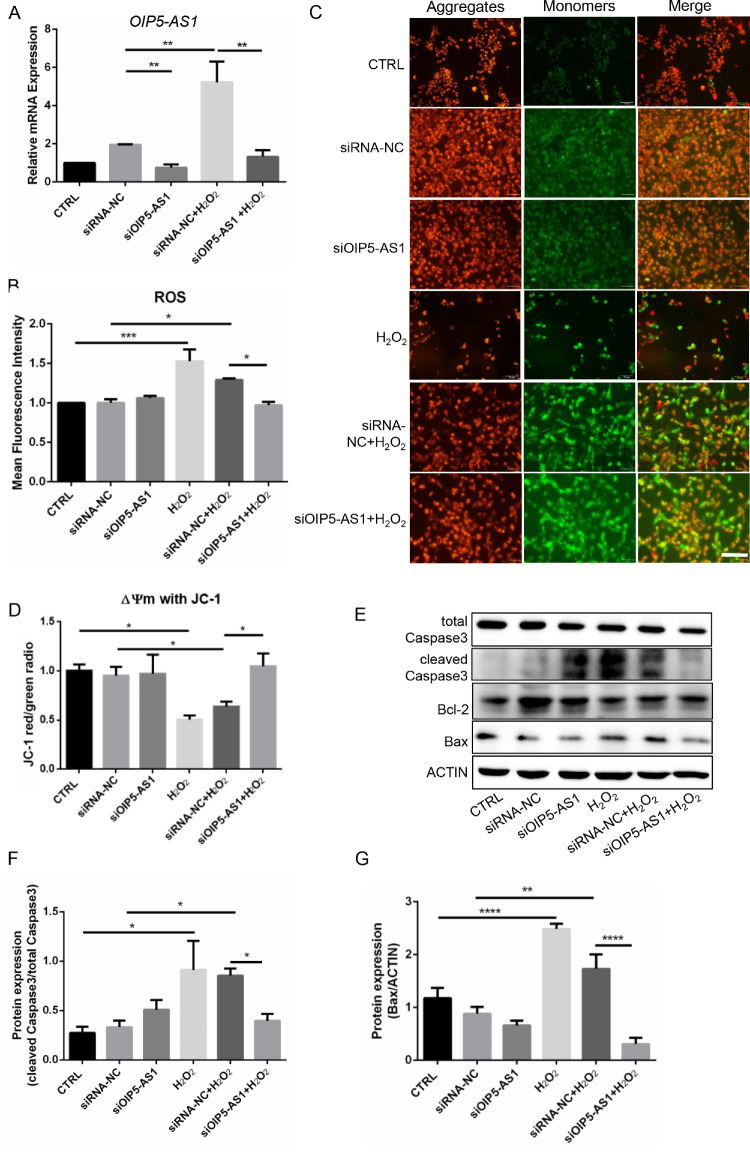
Oxidative stress-induced apoptosis of HLECs is one of the major contributing factors to cataract formation.4 To investigate the potential regulatory role of OIP5-AS1 in oxidative stress-induced apoptosis, we silenced OIP5-AS1 in the H2O2-treated B3 cells. H2O2 dramatically induced OIP5-AS1 upregulation, whereas siRNA silencing abolished the H2O2-enhanced OIP5-AS1 expression (P < 0.01, Fig. 2A). Under oxidative stress, the OIP5-AS1 knockdown group produced less ROS (P < 0.05, Fig. 2B) than the negative control group. We conducted JC-1 staining to measure MMP to demonstrate the early stage of cell death. As shown by fluorescence microscopy, H2O2 significantly reduced the MMP of B3 cells, but knockdown of OIP5-AS1 effectively blocked the decrease in MMP under oxidative stress (P < 0.05, Figs. 2C, 2D). Apoptosis is a common mechanism of generating cataracts.1 We further detected mitochondrial apoptosis-associated proteins, such as Bcl-2, Bax and cleaved caspase 3. The H2O2 treatment increased cleaved caspase 3 and Bax protein expression, although OIP5-AS1 knockdown decreased the levels under oxidative stress, but did not increase the protein expression (Figs. 2E-2G). These results confirmed that OIP5-AS1 contributed to oxidative stress-induced ROS production and the mitochondrial apoptosis pathway in B3 cells.
Figure 2.
OIP5-AS1 knockdown alleviated cell apoptosis and ROS production. (A) mRNA level of OIP5-AS1 quantified by RT-qPCR. SiRNA silencing abolished H2O2-enhanced OIP5-AS1 expression. (B) Flow cytometric detection of the mean fluorescence intensity of the cell ROS. (C–D) Analysis of MMP staining by JC-1with a fluorescence microscope and microplate spectrophotometer. The JC-1 monomer was green, indicating a low MMP, and JC aggregates were red, indicating a high MMP. Scale bar: 500 µm. (E) Western blot of apoptosis-associated proteins. (F–G) Quantification of the Western blot results. *P < 0.05, **P < 0.01, ***P < 0.0001, ****P < 0.00001.
OIP5-AS1 Knockdown Prevented Lens Opacity Ex Vivo
OIP5-AS1 is highly abundant in humans, rats, zebrafish, and chickens, indicating its importance. To verify the effects of OIP5-AS1, we established an ex vivo cataract model as described by Xiang et al 2018.29 After analyzing the efficacy of various siRNAs, an effective siRNA (siOIP5-AS1-1) (P < 0.01, Fig. 3A) was used in the following experiments. The clouding of the lens was evaluated by a doctor blinded to the study groups based on grade assessment8,30 on a scale of 0 to 4 as shown in Figure 3B. The counts of lens stages in every group are shown in Table 2. We observed that Oip5-as1 knockdown did not influence the opacity of the lens compared with the that of the negative control siRNA group (P > 0.008, Fig. 3C). With the H2O2 treatment, the lens in the negative siRNA transfection group became more turbid (P < 0.008, Fig. 3C) than that in the Oip5-as1 knockdown group, indicating the success of our H2O2-induced ex vivo cataract model. Notably, under oxidative stress, the lens in the Oip5-as1 silencing group was not turbid compared with that in the negative siRNA transfection group (P < 0.008, Fig. 3C). In conclusion, Oip5-as1 knockdown might prevent lens opacity under oxidative stress. OIP5-AS1 could be a new target for the treatment of cataracts.
Figure 3.
OIP5-AS1 knockdown might be effective in preventing lens opacity ex vivo. (A) OIP5-AS1 knockdown efficiency of siRNA in the rat lens. (B) Grades of isolated lenses on a scale of 0 to 4. (C) The lenses were kept in DMEM and photographed by a Canon M6 system. **P < 0.01.
POLG Was Negatively Regulated by OIP5-AS1 With HuR
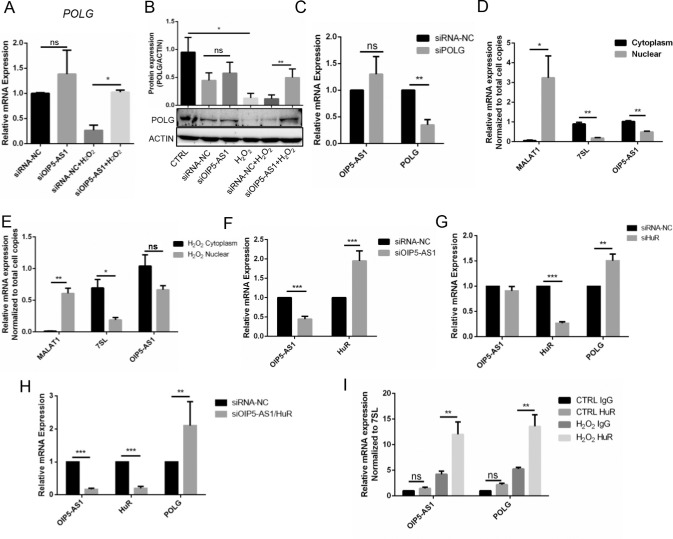
Previous research has suggested that OIP5-AS1 could negatively regulate POLG in the U2OS cell line17.17 Consistently, knockdown of OIP5-AS1 significantly increased POLG expression at both the mRNA and protein levels in the H2O2-induced B3 cells (P < 0.05, P < 0.01, Figs. 4A–4B). Additionally, we did not observe a significant change in the OIP5-AS1 expression after POLG knockdown, indicating that OIP5-AS1 was the upstream regulator of POLG (Fig. 4C). We further performed mRNA quantification on the cell fractions of the OIP5-AS1 cells. Compared with MALAT1 (in the nucleus) and 7SL (in the cytoplasm), OIP5-AS1 was found more frequently in the cytoplasm than the nucleus in both normal and H2O2-induced B3 cells (Figs. 4D, 4E). Generally, diverse lncRNA functions require interactions with one or more RNA binding proteins,31 thus inspiring our investigation of potential key RNA binding proteins in mediating the OIP5-AS1-POLG interaction. Previous research showed that OIP5-AS1 interacted with HuR at 4 sites.32 HuR has 3 target sites on OIP5-AS1 and 39 target sites on POLG in starBase (http://starbase.sysu.edu.cn/starbase2/browseRbpLncRNA.php), suggesting the important role of HuR in the OIP5-AS1-POLG interaction. In our study, OIP5-AS1 knockdown increased HuR mRNA expression (P < 0.001, Fig. 4F). HuR knockdown did not change the expression of OIP5-AS1, but upregulated POLG mRNA (P < 0.01, Fig. 4G). Simultaneous OIP5-AS1 and HuR knockdown enhanced POLG mRNA expression (P < 0.01, Fig. 4H). Although HuR has a well-known role in mRNA stabilization,33 this molecule was also found to destabilize p16 mRNA together with AUF1,34 which was consistent with our finding that HuR could suppress POLG expression. To further demonstrate the binding of HuR to both OIP5-AS1 and POLG mRNA in vitro, we performed RIP assays of B3 cells. The results showed that the specific HuR antibody bound to both OIP5-AS1 mRNA and POLG mRNA in normal HLECs at low affinity. However, we observed apparently higher binding of HuR on OIP5-AS1 mRNA and POLG mRNA than that of the IgG group in B3 cells under oxidative pressure (P < 0.01, Fig. 4I). Together, these results suggested that HuR might function as a scaffold carrying OIP5-AS1 and POLG mRNA and mediate the decay of POLG mRNA when lens oxidative stress is excessive.
Figure 4.
HuR bound OIP5-AS1 and POLG mRNA and mediated the decay of POLG mRNA. (A) POLG mRNA expression after OIP5-AS1 knockdown with or without H2O2 shown by RT-qPCR. (B) POLG protein expression after OIP5-AS1 knockdown with or without H2O2 shown by Western blots and quantification with ImageJ. (C) RT-qPCR detection of OIP5-AS1 expression after POLG knockdown. (D) RT-qPCR determination of OIP5-AS1 cell distribution in fractions of B3 cells. MALAT1 localized in the nucleus, and 7SL was expressed in the cytoplasm. (E) RT-qPCR determination of OIP5-AS1 cell distribution in cell fractions of B3 cells treated with H2O2. MALAT1 localized in the nucleus, and 7SL was expressed in the cytoplasm. (F) HuR mRNA expression after OIP5-AS1 knockdown shown by RT-qPCR. (G) OIP5-AS1 and POLG mRNA expression after HuR silencing shown by RT-qPCR. (H) POLG mRNA expression was upregulated after both OIP5-AS1 and HuR knockdown. (I) RIP detection of HuR binding of the mRNA of OIP5-AS1 and POLG with or without H2O2 stress. *P < 0.05, **P < 0.01, ***P < 0.001, ns: P > 0.05.
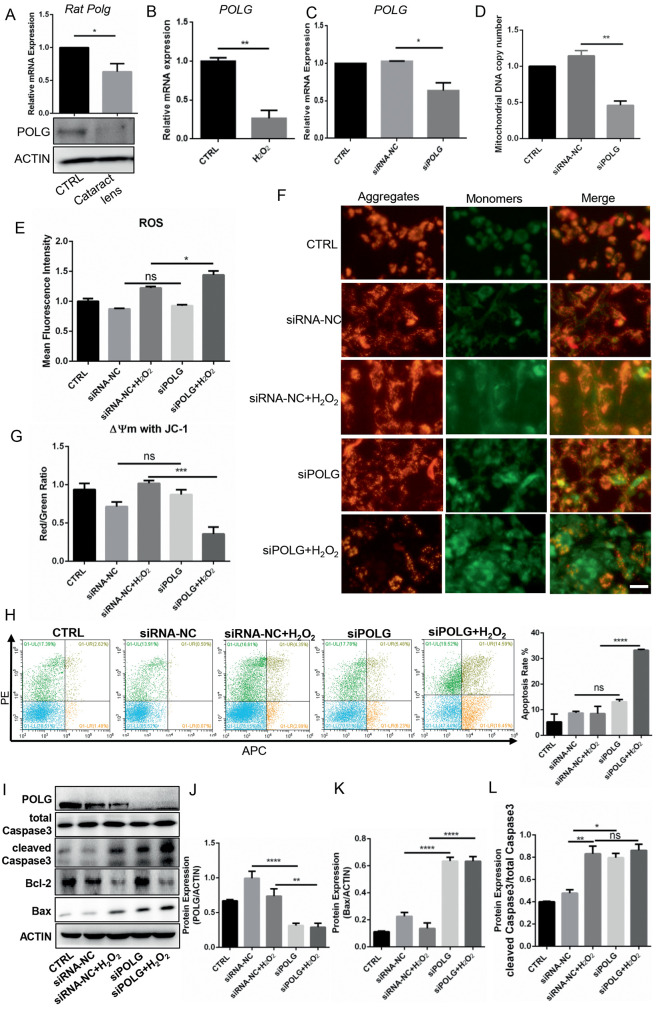
Depletion of POLG Increased B3 Cell Apoptosis
To explore the precise mechanism of POLG in cataracts, we first measured POLG expression in rat and cell models and detected significantly decreased POLG expression in both lens samples from a rat cataract model (P < 0.05, Fig. 5A) and a cell model (P < 0.01, Fig. 5B). We further knocked down POLG expression in B3 cells (P < 0.05, Fig. 5C) and detected a significant reduction in mtDNA copy number (P < 0.01, Fig. 5D), indicating the important role of PLOG in mitochondrial function. The mitochondrial electron transfer chain was reported to be one of the sources of ROS, suggesting the potential role of POLG in ROS. Indeed, we observed that POLG depletion increased the ROS level (P < 0.05, Fig. 5E) under oxidative stress. We also observed a decrease in MMP after POLG depletion under oxidative stress, indicating early apoptosis (P < 0.01, Figs. 5F, 5G). Finally, POLG knockdown contributed to an increase in both apoptotic cells (Fig. 5H) and proapoptotic protein expression (Bax and cleaved caspase 3) (Figs. 5I, 5L). Moreover, when H2O2 was added, POLG knockdown further increased the number of apoptotic cells (Fig. 5H) and Bax protein expression (Figs. 5I, 5K). Together, these results indicated the role of PLOG in mtDNA replication, ROS production and cell apoptosis, indicating that POLG depletion in B3 cells increased ROS production and induced cell apoptosis under oxidative stress.
Figure 5.
POLG knockdown increased the sensitivity of B3 cells to ROS. (A) Detection of Polg mRNA expression in the lenses of rats with cataracts by RT-qPCR. (B) POLG mRNA expression in the oxidative stress cell model shown by RT-qPCR. (C) Efficacy of POLG siRNA knockdown shown by RT-qPCR. (D) MtDNA copy number after POLG silencing shown by RT-qPCR. (E) Flow cytometric detection of the mean fluorescence intensity of ROS. (F–G) Microplate spectrophotometer analysis of MMP by JC-1 staining. The JC-1 monomer was green, representing a low MMP, and JC aggregates were red, representing a high MMP. (H) Flow cytometry was used to test apoptosis of the POLG-silenced cells with or without H2O2 loading and apoptotic cell rate quantification. (I) Western blot characterization of apoptosis-associated protein and POLG expression. (J–L) Quantification of the Western blot results. Scale bar: 100 µm. *P < 0.05, **P < 0.01, ***P < 0.0001, ****P < 0.00001, ns: P > 0.05.
Overexpression of POLG Maintained B3 Cell Viability
We demonstrated that POLG silencing could promote ROS production and cell apoptosis. Then, the following experiments were conducted to confirm whether POLG overexpression could reverse these effects. Our results showed that the mtDNA copy number was higher in the POLG vector transfection group than the null-vector group in B3 cells treated with H2O2 (P < 0.01, Fig. 6A). Moreover, transfection of the POLG overexpression vector decreased ROS generation compared to that of the control group in the H2O2-induced B3 cells (P < 0.05, Fig. 6B), although vector transfection caused excessive ROS production (fold change > 2, P < 0.0001, Fig. 6B). With H2O2 treatment, POLG overexpression rescued the decrease in MMP observed in the JC-1 analysis (P < 0.05, Figs. 6C, 6D) and the cell apoptosis of B3 cells detected by flow cytometry (P < 0.05, Figs. 6E, 6F). Finally, we detected significantly decreased expression of two proapoptotic proteins (cleaved caspase 3 and bax) in the POLG overexpression groups compared with the null-vector group (Figs. 6G–6J) under oxidative stress. Thus, POLG overexpression could maintain cell viability, avoid excessive ROS production, restrain mtDNA depletion and suppress apoptotic pathway activation, indicating that POLG was responsible for ROS production and cell survival under oxidative stress.
Figure 6.
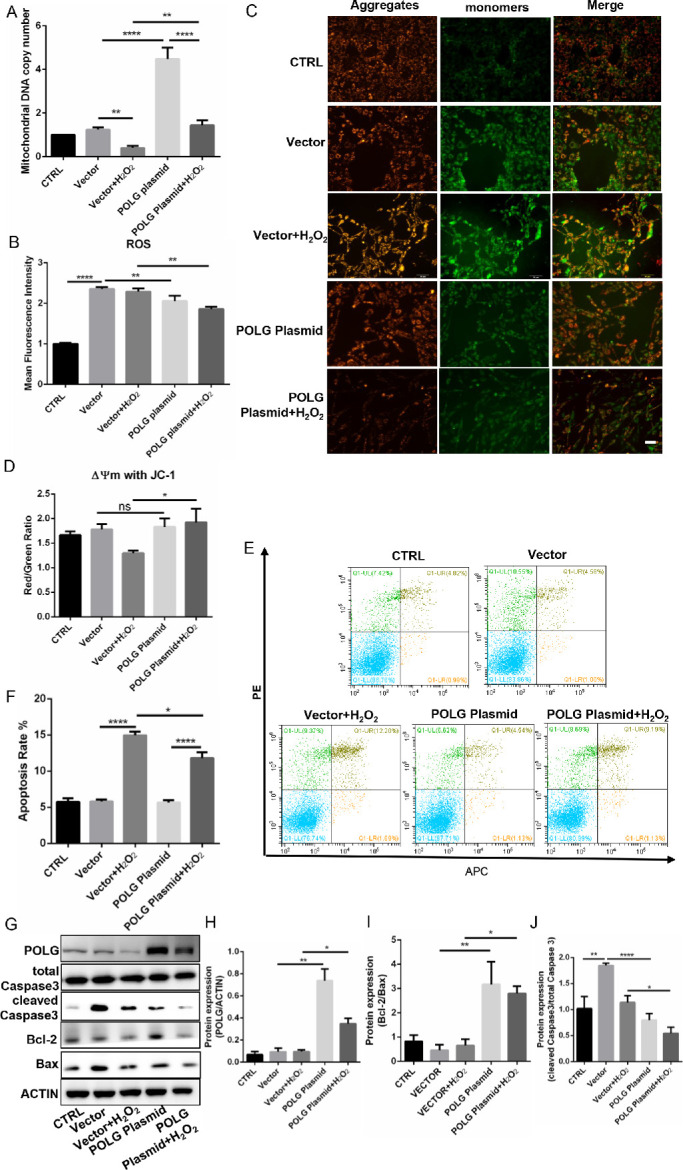
POLG maintained cell viability. (A) MtDNA copy number examination by RT-qPCR. (B) Flow cytometric detection of the mean fluorescence intensity of ROS in the cells transfected with the null vector or the POLG plasmid under oxidative stress. (C, D) Fluorescence microscopy and microplate spectrophotometry analysis of MMP by JC-1 staining. The JC-1 monomer was green, representing a low MMP, and JC aggregates were red, representing a high MMP. (E) Cell apoptosis detected by flow cytometry. (F) Apoptosis rate quantification of E. (G) Western blot analysis of apoptosis-associated proteins and POLG expression. (H–J) Quantification of the Western blot results. Scale bar: 100 µM. *P < 0.05, **P < 0.01, ****P < 0.00001, ns: P > 0.05.
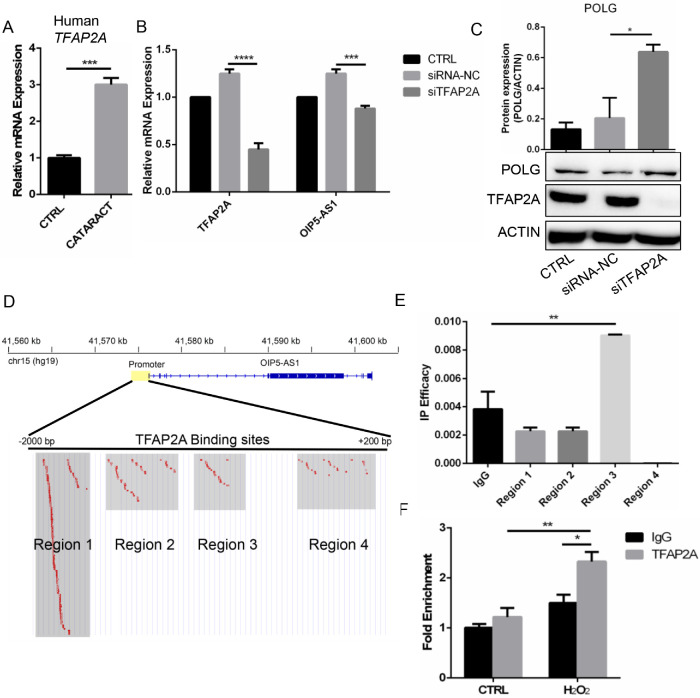
TFAP2A Was Responsible for OIP5-AS1 Transcriptional Activation
We demonstrated the important roles of OIP5-AS1 in cataract formation. To further explore potential upstream transcription factors of OIP5-AS1, we conducted a motif analysis in the region surrounding the promoter of OIP5-AS1. We found that the transcription factor TFAP2A had the most predicted binding sites (Supplementary Table S2). TFAP2A expression of the anterior capsule of patients with cataracts was significantly higher than that in healthy subjects (P < 0.001, Fig. 7A) in data from Wu et al,23 indicating the potential regulatory role of TFAP2A in cataracts. To validate the regulatory effect of TFAP2A on OIP5-AS1, we silenced TFAP2A in B3 cells and detected significantly decreased expression of OIP5-AS1 (P < 0.001, Fig. 7B) and elevated POLG expression (P < 0.05, Fig. 7C). According to our prediction, the TFAP2A binding sites in the OIP5-AS1 promoter could be divided into four clustered regions (Fig. 7D). qPCR revealed that region 3 had the highest binding affinity compared with that of IgG (P < 0.01, Fig. 7E). Region 3 was the nearest region to the transcriptional start site of OIP5-AS1, further supporting the potential key role of TFAP2A in regulating OIP5-AS1 transcription. We further performed ChIP-qPCR in the H2O2-induced B3 cells and observed significant enrichment in the TFAP2A treatment group compared with the negative control IgG group (P < 0.05, Fig. 7F). Altogether, these results indicated that TFAP2A might be involved in the upstream transcriptional regulation of OIP5-AS1 in B3 cells under oxidative stress.
Figure 7.
Prediction of OIP5-AS1 transcriptional activation. (A) TFAP2A expression in the anterior capsule of patients with cataracts was higher than that of healthy individuals. (B) OIP5-AS1 expression after TFAP2A knockdown. (C) POLG protein expression after OIP5-AS1 knockdown. (D) Prediction of OIP5-AS1–binding transcription factors identified TFAP2A. (E) IP efficacy of four regions around the OIP5-AS1 transcription start site manually divided according to the distribution. (F) ChIP-qPCR of TFAP2A after 6 hours of 50 µM H2O2 induction. Primers were located in region 3. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Discussion
Apoptosis is a common cell feature of the pathogenesis of age-related cataract, and multiple factors and genes have been confirmed to be involved in the occurrence or development of this process.35 Herein, we proposed (Supplementary Fig. S3) that lncRNA OIP5-AS1 was strongly transcriptionally activated in B3 cells from senile cataracts. Highly expressed OIP5-AS1 and POLG mRNA were bound and delivered by the HuR protein to stress granules, resulting in POLG mRNA decay and ROS production. These changes led to endogenous apoptotic pathway activation and finally cell death. Moreover, we found that TFAP2A might be the upstream transcription factor regulating OIP5-AS1 expression. We elucidated the specific functions of OIP5-AS1 and POLG, investigated the upstream regulators, and expanded our knowledge of the lncRNAs involved in ARCs. Thus, we demonstrated a potential mechanistic basis for the association between OIP5-AS1 and cataracts, which highlights the regulatory effects of lncRNAs on the pathogenesis of age-related ophthalmologic disease. More functional lncRNAs need to be identified and investigated. Despite the limitations, interfering with lncRNAs might be a novel strategy to prevent cataracts in the future.
Mutations in mtDNA have been found in many aging cells or tissues.36 Both depletion and deletion of mtDNA resulting from POLG alterations were associated with a group of heterogeneous diseases.37 The accumulation of ROS generated from oxidative phosphorylation damaged the mitochondrial membrane, causing the MMP to decrease and leading to the initiation of apoptosis.38 Lymphocytes with POLG mutations are more sensitive to oxidative stress-induced apoptosis than wild-type cells.39 In our study, POLG depletion led to a decrease in the mtDNA copy number and a decrease in MMP. With H2O2 stress, ROS production and proapoptotic protein expression increased, whereas POLG overexpression reversed these effects. In POLG knockdown experiments, H2O2 did not induce significantly more ROS production, apoptotic cells, or proapoptotic protein expression after POLG silencing, which was perhaps due to the severe oxidative damage induced by POLG depletion. In the POLG overexpression analyses, vector transfection created excessive oxidative damage; thus, H2O2 could not induce more oxidative stress. Additionally, we performed OIP5-AS1 and POLG double knockdown and analyzed the ROS, MMP, mtDNA copy number, and apoptotic proteins to study the effects of the knockdown. The results indicated that the double silencing of OIP5-AS1 and POLG had no significant effects on the cell fate and metabolic balance because POLG siRNA prevented the increases induced by OIP5-AS1 silencing (Supplementary Fig. S2). Thus, POLG regulated mitochondrial function and cell fate.
HuR (formally known as ELAVL1) is a well-known RNA binding protein with broad function in diverse cellular activities.40 Generally, HuR was found to stabilize mRNA, resulting in enhanced protein translation.41,42 HuR was also found to function with AUF1, a cofactor, to promote p16 mRNA decay.34 Additionally, HuR could repress translation of p27, an important cell growth inhibitor.43 The lncRNAs often act as molecular sponges or decoys for RNA-binding proteins.32,44,45 Notably, OIP5-AS1 is an AT-rich (including 29 continuous A) lncRNA, indicating that it could be bound by RNA binding protein.46 A previous study suggested that high level OIP5-AS1 could prohibit the interaction of HuR with other mRNA by being a sponge increasing OIP5-AS1-HuR complex interaction.32 In our study, HuR not only bound OIP5-AS1 mRNA, but also POLG mRNA, and POLG mRNA finally decreased. Therefore, we concluded that HuR might act as a scaffold delivering OIP5-AS1 and POLG mRNA to the stress granules, in which POLG mRNA was destabilized or partially decayed. Intriguingly, HuR mRNA expression was enhanced by OIP5-AS1 knockdown. Indeed, a positive feedback loop might be formed.47 In sum, our findings are consistent with HuR's role in controlling cell senescence.
Our study indicated that excess OIP5-AS1 was expressed in lens epithelial cells from individuals with cataract. However, the reason for OIP5-AS1 overexpression or downregulation in tumors or other tissues is unknown. To address this issue, we predicted transcription factors by bioinformatics methods. TFAP2A was identified, and further experiments verified that knockdown of TFAP2A decreased OIP5-AS1 expression and that TFAP2A could bind to the OIP5-AS1 promoter region near the transcription start site, which is consistent with previous findings.48 However, silencing TFAP2A only induced a slight downregulation of OIP5-AS1, suggesting that other transcription factors may be involved. Indeed, in the human genome, the majority of genes are regulated by multiple promoters and display tissue-specific regulation patterns.49 Further investigations are required to elucidate the precise mechanisms based on specific analyses, such as DNA pulldown-mass spectrometry.
TFAP2A was shown to regulate OIP5-AS1 expression according to our predictions and experiments. Usually, TFAP2A is only expressed in lens epithelial cells, but not fiber cells.50 TFAP2A mutations were found to be associated with lens morphologic defects50 or cataracts with branchio-oculo-facial syndrome,51 suggesting the essential function of this molecule in the development of cataracts. Specifically, TFAP2A has cell-autonomous regulatory effects on lens vesicle separation. Cell adhesion and epithelial cell phenotype maintenance associated genes, such as E-cadherin, were regulated by TFAP2A. Upregulated expression of TFAP2A in fiber cells resulted in microphthalmia and cataracts in transgenic mice but blocked fiber cell differentiation.52,53 Moreover, TFAP2A was involved in the original differentiation and development of the neural crest as a pioneer transcription factor, indicating that TFAP2A could constitutively bind nucleosomal target sites and reorganize local chromatin to increase its accessibility.54,55 TFAP2A should be further investigated in both congenital and age-related cataracts.
We also analyzed TFAP2A/OIP5-AS1/POLG expression in human normal corneas based on the work of Kabza et al56 and retinas (GSE102485). Specifically, OIP5-AS1 expression is higher than that of TFAP2A and POLG in the cornea and retina (Supplementary Fig. S4A, B). We found that OIP5-AS1 could negatively regulate POLG expression in human corneal epithelial (HCE) cells and TFAP2A could positively regulate POLG expression in RPE (retinal pigment epithelium) cells (Supplementary Fig. S4D, G). However, we did not observe similar transcription activation of TFAP2A on OIP5-AS1 in HCE (Supplementary Fig. S4C) and OIP5-AS1 does not seem to be involved in TFAP2A/POLG regulation in RPE (Supplementary Fig. S4G, H). In addition, we have not detected the apoptosis-inducible effects of OIP5-AS1 in HCE or RPE cells (Supplementary Fig. S4E, F, I, J), suggesting the lens-specific role of OIP5-AS1. Collectively, these experiments indicated that the whole TFAP2A/OIP5-AS1/POLG regulatory axis was only partially demonstrated in HCE/RPE cells, indicating the lens-specific regulatory mechanisms.
In summary, loss-of-function and gain-of-function experiments as well as interaction analyses elucidated a potential mechanistic basis for the relationship of OIP5-AS1 to its target gene, POLG. Herein, we used immortalized cell line B3 to investigate the specific molecular mechanisms underlying TFAP2A/OIP5-AS1/POLG axis in cataract pathopoeias in vitro. The immortalized cell line likely could not fully adequately represent the primary lens cell line or lens in vivo. Further confirmation in primary cells or OIP5-AS1 knockout animals is required.
Supplementary Material
Acknowledgments
The authors thank the School of Life Science and Technology for providing experimental assistance during the study.
The clinical sample collection, animal care, handling and surgical procedures in this study were approved by the Ethics Committee of the First Affiliated Hospital of Xi'an Jiaotong University.
Supported by the National Natural Science Foundation of China under grant (Nos. 81470614 and 81800812); the Xi'an Science and Technology Project Fund under grant (201905097YX5SF31) (1); the Postdoctoral Natural Science Foundation under grant (2018 M633528); the Natural Science Foundation of Shaanxi Province under grant (No. 2018JQ8021); the Fundamental Research Funds for the Central Universities under grant (xjj2018099); and the Xi'an Jiaotong University First Affiliated Hospital Fund under grant (2016QN-04).
Disclosure: R. Jing, None; B. Ma, None; T. Qi, None; C. Hu, None; C. Liao, None; C. Wen, None; Y. Shao, None; C. Pei, None
References
- 1. Li WC, Kuszak JR, Dunn K, et al.. Lens epithelial cell apoptosis appears to be a common cellular basis for non-congenital cataract development in humans and animals. J Cell Biol. 1995; 130: 169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lam D, Rao SK, Ratra V, et al.. Cataract. Nat Rev Dis Primers. 2015; 1: 15014. [DOI] [PubMed] [Google Scholar]
- 3. Thrimawithana TR, Rupenthal ID, Räsch SS, Lim JC, Morton JD, Bunt CR. Drug delivery to the lens for the management of cataracts. Adv Drug Deliv Rev. 2018; 126: 185–194. [DOI] [PubMed] [Google Scholar]
- 4. Rong X, Rao J, Li D, Jing Q, Lu Y, Ji Y. TRIM69 inhibits cataractogenesis by negatively regulating p53. Redox Biol. 2019; 22: 101157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jin X, Jin H, Shi Y, Guo Y, Zhang H. Pyroptosis, a novel mechanism implicated in cataracts. Mol Med Rep. 2018; 18: 2277–2285. [DOI] [PubMed] [Google Scholar]
- 6. Gerson KD, Haviland MJ, Neo D, et al.. Pregnancy-associated changes in cervical noncoding RNA. Epigenomics. 2020; 12: 1013–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nair L, Chung H, Basu U. Regulation of long non-coding RNAs and genome dynamics by the RNA surveillance machinery. Nat Rev Mol Cell Biol. 2020; 21: 123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xiang J, Chen Q, Kang L, et al.. LncRNA PLCD3-OT1 functions as a CeRNA to prevent age-related cataract by sponging miR-224-5p and regulating PLCD3 expression. Invest Ophthalmol Vis Sci. 2019; 60: 4670–4680. [DOI] [PubMed] [Google Scholar]
- 9. Zhang N, Zhang C, Wang X, Qi Y. High-throughput sequencing reveals novel lincRNA in age-related cataract. Int J Mol Med. 2017; 40: 1829–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shen Y, Dong LF, Zhou RM, et al.. Role of long non-coding RNA MIAT in proliferation, apoptosis and migration of lens epithelial cells: a clinical and in vitro study. J Cell Mol Med. 2016; 20: 537–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gong W, Zhu G, Li J, Yang X. LncRNA MALAT1 promotes the apoptosis and oxidative stress of human lens epithelial cells via p38MAPK pathway in diabetic cataract. Diabetes Res Clin Pract. 2018; 144: 314–321. [DOI] [PubMed] [Google Scholar]
- 12. Smith KN, Starmer J, Miller SC, Sethupathy P, Magnuson T. Long noncoding RNA moderates microRNA activity to maintain self-renewal in embryonic stem cells. Stem Cell Rep. 2017; 9: 108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sarangdhar MA, Chaubey D, Srikakulam N, Pillai B. Parentally inherited long non-coding RNA Cyrano is involved in zebrafish neurodevelopment. Nucleic Acids Res. 2018; 46: 9726–9735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hu GW, Wu L, Kuang W, et al.. Knockdown of linc-OIP5 inhibits proliferation and migration of glioma cells through down-regulation of YAP-NOTCH signaling pathway. Gene. 2017; 610: 24–31. [DOI] [PubMed] [Google Scholar]
- 15. Dai J, Xu L, Hu X, et al.. Long noncoding RNA OIP5-AS1 accelerates CDK14 expression to promote osteosarcoma tumorigenesis via targeting miR-223. Biomed Pharmacother. 2018; 106: 1441–1447. [DOI] [PubMed] [Google Scholar]
- 16. Chiu HS, Somvanshi S, Patel E, et al.. Pan-cancer analysis of lncRNA regulation supports their targeting of cancer genes in each tumor context. Cell Rep. 2018; 23: 297–312.e212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim J, Noh JH, Lee SK, et al.. LncRNA OIP5-AS1/cyrano suppresses GAK expression to control mitosis. Oncotarget. 2017; 8: 49409–49420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bekheirnia MR, Zhang W, Eble T, et al.. POLG mutation in a patient with cataracts, early-onset distal muscle weakness and atrophy, ovarian dysgenesis and 3-methylglutaconic aciduria. Gene. 2012; 499: 209–212. [DOI] [PubMed] [Google Scholar]
- 19. Van Goethem G, Martin JJ, Van Broeckhoven C. Progressive external ophthalmoplegia and multiple mitochondrial DNA deletions. Acta Neurol Belg. 2002; 102: 39–42. [PubMed] [Google Scholar]
- 20. Lim JC, Umapathy A, Donaldson PJ. Tools to fight the cataract epidemic: a review of experimental animal models that mimic age related nuclear cataract. Exp Eye Res. 2016; 145: 432–443. [DOI] [PubMed] [Google Scholar]
- 21. Wang Y, Zhu W, Levy DE. Nuclear and cytoplasmic mRNA quantification by SYBR green based real-time RT-PCR. Methods (San Diego, Calif). 2006; 39: 356–362. [DOI] [PubMed] [Google Scholar]
- 22. Foote K, Reinhold J, Yu EPK, et al.. Restoring mitochondrial DNA copy number preserves mitochondrial function and delays vascular aging in mice. Aging Cell. 2018; 17: e12773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu C, Liu Z, Ma L, et al.. MiRNAs regulate oxidative stress related genes via binding to the 3' UTR and TATA-box regions: a new hypothesis for cataract pathogenesis. BMC Ophthalmol. 2017; 17: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bailey TL, Mikael B, Buske FA, et al.. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 2009; 37: W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aziz K, Oriol F, Arnaud S, et al.. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018; 46(D1): D1284, doi: 10.1093/nar/gkx1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kulakovskiy IV, Vorontsov IE, Yevshin IS, et al.. HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res. 2017; D1: D61–D67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pachkov M, Balwierz PJ, Arnold P, Ozonov E, van Nimwegen E. SwissRegulon, a database of genome-wide annotations of regulatory sites: recent updates. Nucleic Acids Res. 2012;41: D214–D220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jolma A, Yan J, Whitington T, et al.. DNA-binding specificities of human transcription factors. Cell. 2013; 152: 327–339. [DOI] [PubMed] [Google Scholar]
- 29. Xiang J, Kang L, Gao H, et al.. BLM can regulate cataract progression by influencing cell vitality and apoptosis. Exp Eye Res. 2018; 178: 99–107. [DOI] [PubMed] [Google Scholar]
- 30. Makri OE, Ferlemi AV, Lamari FN, Georgakopoulos CD. Saffron administration prevents selenite-induced cataractogenesis. Mol Vis. 2013; 19: 1188–1197. [PMC free article] [PubMed] [Google Scholar]
- 31. Ferrè F, Colantoni A, Helmer-Citterich M. Revealing protein-lncRNA interaction. Brief Bioinform. 2016; 17: 106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim J, Abdelmohsen K, Yang X, et al.. LncRNA OIP5-AS1/cyrano sponges RNA-binding protein HuR. Nucleic Acids Res. 2016; 44: 2378–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stellos K, Gatsiou A, Stamatelopoulos K, et al.. Adenosine-to-inosine RNA editing controls cathepsin S expression in atherosclerosis by enabling HuR-mediated post-transcriptional regulation. Nat Med. 2016; 22: 1140–1150. [DOI] [PubMed] [Google Scholar]
- 34. Chang N, Yi J, Guo G, et al.. HuR uses AUF1 as a cofactor to promote p16INK4 mRNA decay. Mol Cell Biol. 2010; 30: 3875–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu YC, Wilkins M, Kim T, Malyugin B, Mehta JS. Cataracts. Lancet. 2017; 390: 600–612. [DOI] [PubMed] [Google Scholar]
- 36. Zsurka G, Peeva V, Kotlyar A, Kunz WS. Is There still any role for oxidative stress in mitochondrial DNA-dependent aging? Genes. 2018; 9: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nogueira C, Almeida LS, Nesti C, et al.. Syndromes associated with mitochondrial DNA depletion. Ital J Pediatr. 2014; 40: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zorov DB, Juhaszova M, Sollott SJ. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol Rev. 2014; 94: 909–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Formichi P, Radi E, Branca C, et al.. Oxidative stress-induced apoptosis in peripheral blood lymphocytes from patients with POLG-related disorders. J Neurol Sci. 2016; 368: 359–368. [DOI] [PubMed] [Google Scholar]
- 40. Blackinton JG, Keene JD.. Functional coordination and HuR-mediated regulation of mRNA stability during T cell activation. Nucleic Acids Res. 2016; 44: 426–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ganguly K, Giddaluru J, August A, Khan N. Post-transcriptional regulation of immunological responses through Riboclustering. Front Immunol. 2016; 7: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Grammatikakis I, Abdelmohsen K, Gorospe M. Posttranslational control of HuR function. Wiley Interdiscip Rev RNA. 2017; 8: 10.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kullmann M, Gopfert U, Siewe B, Hengst L. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5'UTR. Genes Dev. 2002; 16: 3087–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013; 154: 26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Alvarez-Dominguez JR, Lodish HF.. Emerging mechanisms of long noncoding RNA function during normal and malignant hematopoiesis. Blood. 2017; 130: 1965–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dejardin J. Switching between epigenetic states at pericentromeric heterochromatin. Trends Genet. 2015; 31: 661–672. [DOI] [PubMed] [Google Scholar]
- 47. Panneerdoss S, Eedunuri VK, Yadav P, et al.. Cross-talk among writers, readers, and erasers of m(6)A regulates cancer growth and progression. Sci Adv. 2018; 4: eaar8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bansal M, Kumar A, Yella VR. Role of DNA sequence based structural features of promoters in transcription initiation and gene expression. Curr Opin Struct Biol. 2014; 25: 77–85. [DOI] [PubMed] [Google Scholar]
- 49. Demircioğlu D, Cukuroglu E, Kindermans M, et al.. A pan-cancer transcriptome analysis reveals pervasive regulation through alternative promoters. Cell. 2019; 178: 1465–1477.e1417. [DOI] [PubMed] [Google Scholar]
- 50. Kerr CL, Zaveri MA, Robinson ML, Williams T, West-Mays JA. AP-2α is required after lens vesicle formation to maintain lens integrity. Dev Dynam. 2014; 243: 1298–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dumitrescu AV, Milunsky JM, Longmuir SQ, Drack AV. A family with branchio-oculo-facial syndrome with primarily ocular involvement associated with mutation of the TFAP2A gene. Ophthalm Genet. 2012; 33: 100–106. [DOI] [PubMed] [Google Scholar]
- 52. Pontoriero GF, Deschamps P, Ashery-Padan R, et al.. Cell autonomous roles for AP-2alpha in lens vesicle separation and maintenance of the lens epithelial cell phenotype. Dev Dynam. 2008; 237: 602–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. West-Mays JA, Coyle BM, Piatigorsky J, Papagiotas S, Libby D. Ectopic expression of AP-2alpha transcription factor in the lens disrupts fiber cell differentiation. Dev Biol. 2002; 245: 13–27. [DOI] [PubMed] [Google Scholar]
- 54. Donaghey J, Thakurela S, Charlton J, et al.. Genetic determinants and epigenetic effects of pioneer-factor occupancy. Nat Genet. 2018; 50: 250–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rothstein M, Simoes-Costa M.. Heterodimerization of TFAP2 pioneer factors drives epigenomic remodeling during neural crest specification. Genome Res. 2020; 30: 35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kabza M, Karolak JA, Rydzanicz M, et al.. Collagen synthesis disruption and downregulation of core elements of TGF-β, Hippo, and Wnt pathways in keratoconus corneas. Eur J Human Genet. 2017; 25: 582–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.