Abstract
The overwhelming success of exome- and genome-wide association studies in discovering thousands of disease-associated genes necessitates developing novel high-throughput functional genomics approaches to elucidate the molecular mechanisms of these genes. Here, we have coupled multiplexed repression of neurodevelopmental disease–associated genes to single-cell transcriptional profiling in differentiating human neurons to rapidly assay the functions of multiple genes in a disease-relevant context, assess potentially convergent mechanisms, and prioritize genes for specific functional assays. For a set of 13 autism spectrum disorder (ASD)–associated genes, we show that this approach generated important mechanistic insights, revealing two functionally convergent modules of ASD genes: one that delays neuron differentiation and one that accelerates it. Five genes that delay neuron differentiation (ADNP, ARID1B, ASH1L, CHD2, and DYRK1A) mechanistically converge, as they all dysregulate genes involved in cell-cycle control and progenitor cell proliferation. Live-cell imaging after individual ASD-gene repression validated this functional module, confirming that these genes reduce neural progenitor cell proliferation and neurite growth. Finally, these functionally convergent ASD gene modules predicted shared clinical phenotypes among individuals with mutations in these genes. Altogether, these results show the utility of a novel and simple approach for the rapid functional elucidation of neurodevelopmental disease-associated genes.
The tremendous progress in identifying disease-associated genes and variants has far outpaced the discovery of the functions and pathological mechanisms of these genes. Exome- and genome-wide sequencing studies have identified approximately 5500 single-gene disorders and traits caused by mutations in over 3800 genes (Amberger et al. 2019). Over 1100 of these genes have been causally linked to neurodevelopmental disorders (Wright et al. 2015). In autism spectrum disorder (ASD) alone, recent exome sequencing studies have identified over 100 genes that cause ASD when a single copy is mutated to a loss-of-function allele (O'Roak et al. 2012; De Rubeis et al. 2014; Iossifov et al. 2014; Satterstrom et al. 2020). This genetic heterogeneity provides a substantial challenge to the development of broadly useful therapeutics. If, at an extreme, each disease-associated gene follows a separate mechanistic route, then each will require the development of an independent therapeutic. On the other hand, if subsets of these genes converge in their mechanisms, then these points of convergence would be logical targets for more broadly applicable therapeutics that apply to the entire subset. Identifying convergent mechanisms across diverse disease-associated genes first requires establishing a high-throughput and disease-relevant model system to both perturb numerous genes and systematically assess the functional consequences.
Although animal models and patient-derived induced pluripotent stem cell (iPSC) models are powerful tools for the study of disease mechanisms, these systems are generally low throughput and require long generation times, and results can vary across laboratories, strains, or individuals (Kilpinen et al. 2017; Zhao and Bhattacharyya 2018). A rapid, reproducible, and disease-relevant system in which multiple genes could be studied in parallel would fill an important gap in the functional genomics toolbox and enable direct comparison across genes to assess their mechanistic convergence. Recent technological advancements coupling CRISPR-Cas9 transcriptional repression to single-cell RNA sequencing (scRNA-seq) enable high-throughput perturbation of multiple genes in a single batch with a parallel functional readout of the transcriptional consequences (Gilbert et al. 2013; Adamson et al. 2016; Dixit et al. 2016; Datlinger et al. 2017). Such an approach holds great promise for efficiently defining the functional consequences of dominant loss-of-function mutations, as transcriptional repression can phenocopy haploinsufficiency. As the pathology of many neurodevelopmental diseases likely arise during neural development, especially when proliferating progenitors are differentiating into postmitotic neurons (Schafer et al. 2019), we set out to establish a scalable functional genomics approach in a simple human cellular model of neuron differentiation. Further, as a large fraction of causative genes in ASD are haploinsufficient transcriptional regulators (De Rubeis et al. 2014; Willsey et al. 2018; Satterstrom et al. 2020), we aimed to test this approach on a select subset of such genes to determine what insights into pathological mechanisms can be gleaned by measuring the transcriptional consequences of their perturbation.
Here, we used catalytically inactive Cas9-based transcriptional repression (dCas9-KRAB) to knockdown the expression of 13 ASD-related genes in a human cellular model of neuronal differentiation and captured the resulting transcriptional consequences using scRNA-seq. We sought to identify individual transcriptional signatures after repression for each candidate gene, compare these signatures to discover shared transcriptional changes, and examine the impact of knockdown on the trajectory of neuronal differentiation. We sought to generate mechanistic predictions from transcriptional profiling and test these by combining individual knockdown experiments with live-cell imaging. Finally, we aimed to compare our functional data to clinical phenotypes in individuals with mutations in these genes.
Results
Establishing a human model of neuronal differentiation for high-throughput disease gene perturbation
Human neuronal models are needed for studying neurodevelopmental disorders such as ASD (Zhao and Bhattacharyya 2018). Although human iPSC-derived neurons are a powerful cellular model system, the genetic heterogeneity, variability of neuronal differentiation, and technical difficulties achieving efficient transcriptional modulation in these cells complicate multiplexed transcriptional and phenotypic analyses (Ho et al. 2017; Hoffman et al. 2017). Therefore, we aimed to establish a tractable human neuronal model amenable to differentiation and transcriptional perturbation to enable high-throughput evaluation of the consequences of disease-associated gene repression. We selected the LUHMES neural progenitor cell line as such a model for their ease of use, capacity for rapid differentiation into postmitotic neurons, and suitability for high-content imaging (Scholz et al. 2011; Höllerhage et al. 2017; Tong et al. 2017; Pierce et al. 2018). Unlike many immortalized lines, LUHMES have a stable diploid karyotype and have been maintained by others in continuous culture for >5 yr without acquiring chromosomal abnormalities (Paul et al. 2007). Recent studies have used LUHMES to model neurodevelopmental disorders and their underlying pathways (Shah et al. 2016; Matelski et al. 2020).
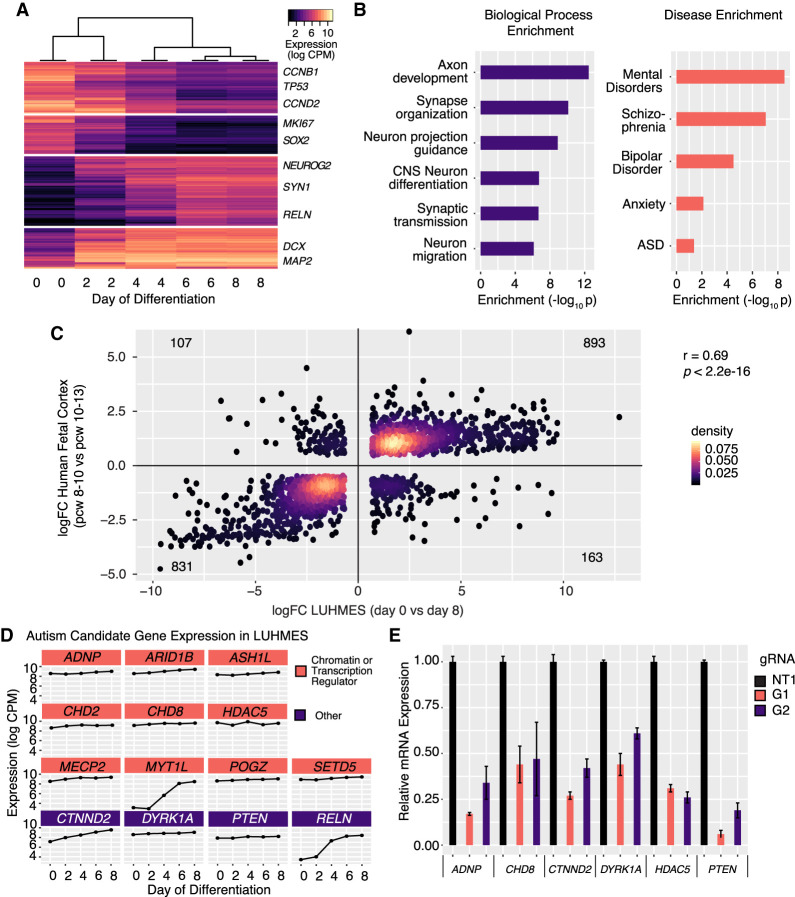
To further validate the relevance of these cells, we performed RNA sequencing (RNA-seq) analysis of LUHMES cells at multiple time points after inducing differentiation. Hierarchical clustering analysis of differentially expressed genes across the differentiation time course confirmed that differentiation of LUHMES was rapid and reproducible (Pearson's r2 between replicates >0.99), with biological replicates clustering together and samples arranged temporally by their day of differentiation (Fig. 1A; Supplemental Fig. S1A). Genes that were down-regulated during differentiation were enriched for cell-cycle markers such as CCND2, genes involved in proliferation (MKI67 and TP53), and the canonical neural stem cell marker gene SOX2. Genes that increased expression during differentiation included known neuronal markers MAP2 and DCX, and were heavily enriched for critical neurodevelopmental pathways including axon growth, synaptic development, and neuron migration (Fig. 1B, left). Genes expressed during differentiation were strongly enriched for genes implicated in a variety of neurological disorders, including schizophrenia, bipolar disorder, and ASD (Fig. 1C, right).
Figure 1.
LUHMES are a tractable, disease-relevant model of human neuronal differentiation amenable to perturbation. (A) Hierarchical clustering of bulk RNA-seq time course expression data indicates rapid and reproducible neuronal differentiation. Two replicates for each time point were performed. (B) Genes induced during LUHMES differentiation are enriched for relevant biological processes (left) and neurological disorders (right). (C) Differentially expressed genes during LUHMES differentiation are highly correlated with transcriptional changes that occur during early human fetal corticogenesis (Pearson's rho = 0.69, P = 2.2 × 1016). (logFC) log2 fold-change of differential expression between indicated time points; (pcw) postconception week. (D) High-confidence autism-causing genes, selected for perturbation experiments, are highly expressed at baseline or are increasingly expressed in LUHMES during differentiation and were selected for roles in transcriptional regulation. (E) Efficient dCas9-KRAB repression of individual target genes using the designated guide RNAs. n = 3 biological replicates for all qPCR experiments. Values represent mean ± SEM. (NT1) Nontargeting control gRNA; (G1) gRNA 1; (G2) gRNA 2.
Despite being a mesencephalic-derived neuronal progenitor line best characterized for its ability to differentiate into dopaminergic neurons, cell type–specific expression analysis (CSEA) of differentiated LUHMES revealed that these neurons have transcriptional profiles that are highly similar to a range of neuronal subtypes relevant to neurological disorders (Supplemental Fig. S1B; Xu et al. 2014). Specifically, transcriptomes of differentiated cells resembled striatal dopaminergic neurons as expected but also matched some cortical, forebrain, and spinal cord neuron types. Differentiated LUHMES also expressed many markers of excitatory neurons (Supplemental Fig. S1C). Next, to assess the extent to which in vitro differentiation of LUHMES cells captures aspects of human brain development, we performed a transition-mapping approach comparing differentially expressed genes during LUHMES differentiation to the BrainSpan Atlas of Developing Human Brain (Stein et al. 2014; https://www.brainspan.org/). We found that changes in gene expression during in vitro differentiation closely mirror transcriptional differences that occur in the early developing human fetal neocortex (Pearson's r = 0.69) (Fig. 1C). This strong overlap indicates that LUHMES differentiation faithfully recapitulates many of the transcriptional pathways that are used during this critical neurodevelopmental window (Supplemental Fig. S1D–G). Because LUHMES in vitro differentiation produces only a single neuronal cell type, some important disease-associated phenomena such as shifts in neuronal cell fate decisions or aberrations in region-specific gene regulatory networks will not be captured by this system. However, as core transcriptional programs that control neuronal differentiation and maturation are largely conserved across neuronal subtypes (Li et al. 2018), we can model these critical disease-relevant processes using a simple in vitro system.
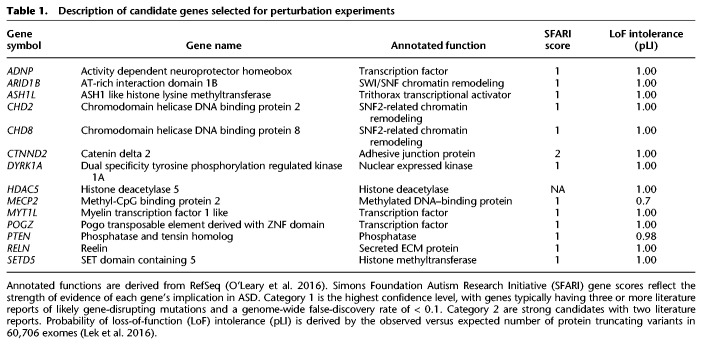
To establish that LUHMES cells are an appropriate model specifically for the study of ASD genes, we analyzed 25 high-confidence autism-causing genes in the SFARI database, a manually curated database of ASD-associated genes (Abrahams et al. 2013). We found that 22/25 (88%) were highly expressed in these cells across differentiation time points. We selected 13 of these genes for perturbation experiments (Table 1; Fig. 1D). HDAC5 was included as a nonassociated gene that is highly expressed in neuronal progenitors, where it may regulate stem cell proliferation (Sun et al. 2007). Genes were selected because of their roles in transcriptional regulation (10/14) (O'Leary et al. 2016) and because they are highly likely to act through haploinsufficiency (Table 1; Lek et al. 2016). Although many of these genes are coexpressed during neurodevelopment, module assignment of these genes by integrative bioinformatics approaches has not enabled specific mechanistic predictions about the potential convergence of their molecular targets (Parikshak et al. 2013; Li et al. 2018). We expect this set of genes to be broadly representative of transcriptional regulators implicated in neurodevelopmental disorders and well suited to show the feasibility of our approach.
Table 1.
Description of candidate genes selected for perturbation experiments
We next sought to determine whether the expression of candidate genes could be efficiently knocked down in LUHMES cells using CRISPR interference-mediated transcriptional repression (Gilbert et al. 2013), a prerequisite for perturbation assays. Three guide RNAs (gRNAs) per candidate gene were cloned into a CRISPR-repression optimized vector that also allows recovery of the gRNA from scRNA-seq (Hill et al. 2018; Sanson et al. 2018; Xie et al. 2018). We validated the efficacy of repression for two gRNAs targeting each of six candidate genes using quantitative real-time PCR (qRT-PCR) in LUHMES neural progenitor cells constitutively expressing dCas9-KRAB. All tested gRNAs induced significant down-regulation of their target gene, with 11/12 eliciting a knockdown >50% (Fig. 1E), a level that should phenocopy the autosomal-dominant loss-of-function modes of our candidate genes. Altogether, these data support LUHMES as a relevant and facile cellular model to evaluate the downstream consequences of transcriptional perturbation of neurodevelopmental genes.
Pooled repression of ASD genes and scRNA-seq
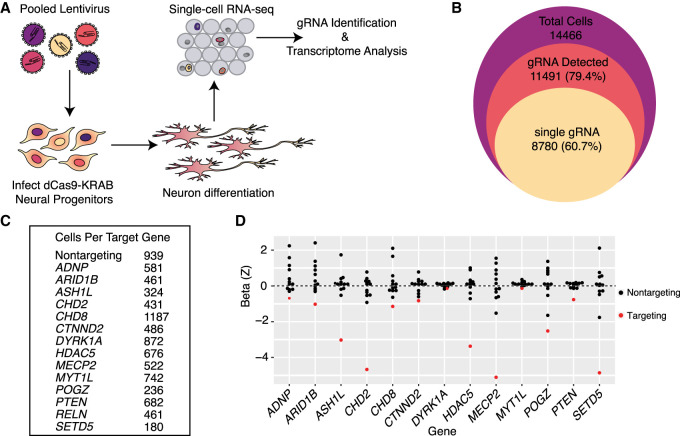
We produced a lentivirus pool that contained vectors expressing gRNAs targeting all 14 candidate genes (three gRNAs per gene), along with five nontargeting control gRNA sequences, for a total of 47 gRNAs. Because of the high success rate of gene knockdown in dCas9-KRAB LUHMES by all tested gRNAs and because we wanted to enable high-scale perturbation screening experiments, we did not validate the repression efficiency of all gRNAs individually. We infected dCas9-KRAB expressing LUHMES neuronal progenitors at a low multiplicity of infection such that most cells received zero or one gRNAs according to a Poisson distribution. Cells infected by a gRNA-expressing lentivirus were selected by growth in media containing puromycin for 4 d, and then the cells were induced to differentiate according to published protocols (Scholz et al. 2011). To allow sufficient time for CRISPR repression, we differentiated LUHMES for 7 d, a time point when differentiation appeared largely complete by RNA-seq. We then profiled the transcriptomes of more than 14,000 cells at this time point using droplet-based scRNA-seq across two replicate experiments (Fig. 2A,B; Zheng et al. 2017).
Figure 2.
Single-cell RNA-seq is an efficient readout of multiplexed gene repression in a human model of neuronal differentiation. (A) Schematic of pooled repression of ASD genes in LUHMES. (B) The numbers of total single cells that were collected (purple area), express at least one gRNA (red), and express a single unique gRNA (yellow) show the efficient recovery of gRNAs from single-cell RNA-seq data. (C) Hundreds of cells targeting all 14 genes were recovered, as well as 939 cells with nontargeting gRNAs. (D) Scaled MIMOSCA beta coefficients for targeting and nontargeting guides are shown for each targeted gene. Three targeting gRNAs for each gene are merged. Beta less than zero represents repression. In all cases, targeting gRNAs (red dots) have negative beta coefficients on target gene expression.
By using a specific gRNA enrichment PCR, we were able to detect gRNA expression for the vast majority (∼80%) of cells and restricted our analysis to the 8780 high-quality cells with only a single gRNA to ensure only one perturbation per cell (Fig. 2B). Cells with gRNAs targeting all candidate genes, and nontargeting controls, were represented in this data set (Fig. 2C). To evaluate the efficacy of ASD-gene repression in the pooled experiment, we grouped single cells by their detected gRNAs and visualized expression of all targeted genes across these groups using Seurat (Supplemental Fig. S2A; Satija et al. 2015; Stuart et al. 2019). This analysis revealed efficient on-target repression for 13/14 genes in our library. The detection of RELN was too low in single-cell data to evaluate efficiency of repression owing to its low expression level. We used the MIMOSCA pipeline to further evaluate the knockdown efficiency of gRNAs on their target genes, which confirmed strong on-target repression (Fig. 2D; Dixit et al. 2016). Almost all individual gRNAs elicited repression of their target genes (Supplemental Fig. S2B–E).
ASD-gene repression alters trajectory of neuronal differentiation
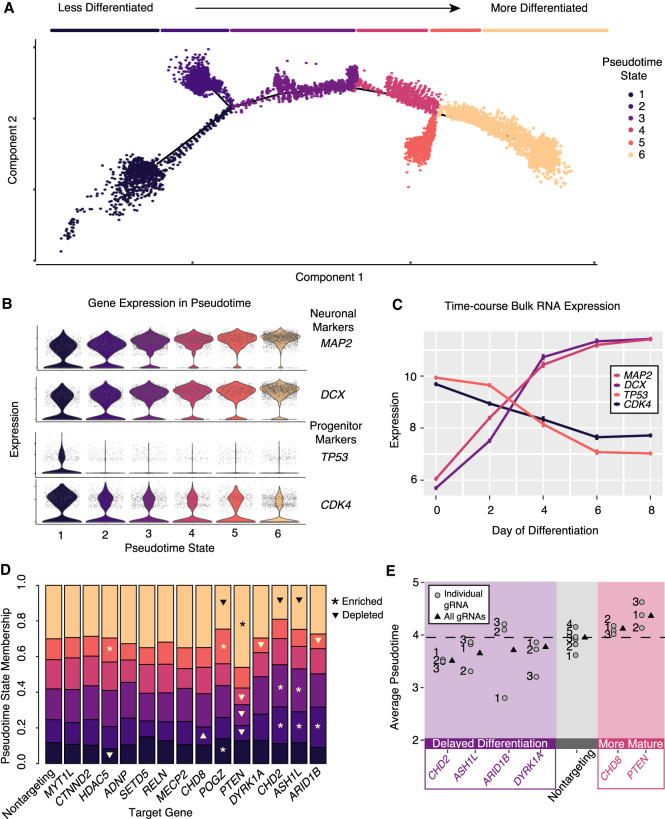
The efficient repression of targeted genes in the pooled experiment shown above led us to assess the unique and shared downstream consequences of ASD-gene repression in human neurons. Specifically, we wanted to directly test the hypothesis that some of the ASD genes might alter the dynamics of neuronal differentiation. To this end, we used Monocle to reconstruct a pseudotemporal trajectory reflecting gene expression changes in our data set and projected cells onto this pseudotime path (Fig. 3A; Trapnell et al. 2014). Recent single-cell CRISPR experiments have shown the advantages of trajectory analysis over global clustering-based approaches, which can be insensitive to detecting more subtle phenotypes in pooled experiments (Supplemental Fig. S3; Duan et al. 2019; McFaline-Figueroa et al. 2019; Yang et al. 2020). As all cells were differentiated for 7 d, >99% of single cells were postmitotic as assessed by the absence of proliferation markers MKI67 and TOP2A (Supplemental Fig. S3B). However, pseudotime and global clustering analysis indicated heterogeneity in the progression of differentiation at the single-cell level (Supplemental Fig. S3C–E). Two neuronal marker genes (MAP2 and DCX) showed a gradual increase in expression across pseudotime (Fig. 3B). In contrast, two genes known to be important for neural progenitor cell proliferation (TP53 and CDK4) showed a rapid drop in expression across pseudotime (Fig. 3B). These observations suggested the axis of pseudotime corresponds to the progression of neuronal differentiation. Consistent with this notion, these four genes show a similar pattern of expression over a time course of LUHMES differentiation (Fig. 3C). To further examine the relationship between pseudotime and neuronal differentiation, we identified the marker genes for each pseudotime state and plotted their expression across the differentiation time course RNA-seq data set. This analysis showed that marker genes of early pseudotime (states 1–3) are highly expressed in early neuron differentiation (differentiation days 0–4) (Supplemental Fig. S4A–C). Marker genes of late pseudotime (states 4–6) are highly expressed during later neuron differentiation (days 4–8) (Supplemental Fig. S4D–F). Altogether, these data support the interpretation of pseudotime as an axis corresponding to the progression of neuronal differentiation after cells have completed their final division.
Figure 3.
Pseudotime analysis reveals ASD-gene repression–induced alterations in differentiation trajectory and modules of ASD genes delaying or accelerating neuronal differentiation. (A) Pseudotime ordering of all single cells reveals a continuous trajectory of cell states corresponding to neuronal differentiation. Line segments along the trajectory are called “pseudotime states,” and cells are colored by these states. (B) Neuronal markers (MAP2 and DCX) increase along the pseudotime trajectory, whereas progenitor markers (TP53 and CDK4) decrease. (C) These marker genes show correlated patterns in time course bulk RNA expression. n = 2 replicates for each time point. Expression values, mean ± SEM. (D) Repression of some genes alter pseudotime state membership proportions. Significant enrichments and depletions are marked with asterisks and triangles, respectively (χ-squared test, P < 0.05). (E) Significantly decreased or increased average pseudotime scores relative to cells with nontargeting gRNAs (t-test, P < 0.01) indicate delayed or accelerated neuronal maturation. Boxed in purple is a set of ASD genes that delay neuronal differentiation by this metric. Boxed in red is a pair of genes, CHD8 and PTEN, that promote neuronal maturation. Overall (all gRNAs) and individual gRNA estimates of pseudotime are shown. Individual gRNAs numbers for each gene are indicated.
To assess whether any candidate gene knockdown shifted the developmental trajectory of the differentiating neurons, we compared the proportions of cells in each pseudotime state across each knockdown condition. Indeed, we found that several perturbations significantly altered the proportions of cells in specific pseudotime states (Fig. 3D). As an indicator of differentiation status, we next computed the average value of pseudotime across all cells in each knockdown condition. By this metric, we found that four of the ASD genes (CHD2, ASH1L, ARID1B, and DYRK1A) delayed neuronal differentiation when repressed, whereas two genes (PTEN and CHD8) accelerated neuronal differentiation (Fig. 3E). Estimated knockdown efficiencies and effects on pseudotime were highly reproducible between replicate experiments and concordant across individual gRNAs. (Supplemental Fig. S5A–E). These results show the utility of pseudotime analysis to investigate whether subsets of disease-associated genes alter the progression of neuronal differentiation in a pooled perturbation experiment.
Recurrently dysregulated genes highlight convergent mechanisms of ASD genes
We noticed that specific pseudotime state enrichment or depletion was not perfectly shared among the sets of genes that accelerated or delayed differentiation (Fig. 3D). This raises the possibility that although groups of genes may act similarly to promote (or delay) neuron differentiation, they may do so through different molecular mechanisms. We therefore sought to further dissect the transcriptional networks affected by gene perturbation using differential gene expression analysis to learn whether these networks converge across sets of ASD genes. Because of the low number of cells available for analysis of SETD5 and POGZ (Fig. 2C; Vieth et al. 2017), we excluded these cells from differential expression analyses. For each of the remaining genes, we found dozens to hundreds of differentially expressed genes for each knockdown (Supplemental Table S1).
To identify potential transcriptional convergence of diverse ASD-causing genes, we grouped cells by targeted gene and then clustered these aggregate transcriptional profiles using only genes that were found to be differentially expressed across three or more ASD-gene knockdowns. The grouping of ASD genes via hierarchical clustering largely recapitulated the results of pseudotime analysis (Supplemental Fig. S6A). Gene Ontology analysis of the set of dysregulated genes showed an enrichment of neuronal differentiation terms in the perturbed transcriptomes, supporting the interpretation that the misregulation of these ASD-associated genes alters neuronal differentiation (Supplemental Fig. S6B). This analysis identified ADNP as another gene delaying neuronal differentiation.
Heterogeneity in the progression of neuronal differentiation dominated the global differential gene expression analysis, and this may mask the discovery of distinct transcriptional changes that occurred early or later in neuronal differentiation. To address this possibility, we leveraged the power of single-cell data to explicitly account for differences in neuronal maturity across cells by stratifying samples based on pseudotime (Supplemental Fig. S6C). To retain enough cells per group required for differential gene expression analysis, we dichotomized pseudotime into two stages: “early” (states 1–3) and “late” (states 4–6). This stratification allowed us to evaluate gene expression differences across conditions within the same broad stage of differentiation without confounding by genes that are highly variable across the entire differentiation time course. To do so, we then recomputed differentially expressed genes for sets of cells within each pseudotime stage for each ASD-gene knockdown condition (Supplemental Tables S2, S3).
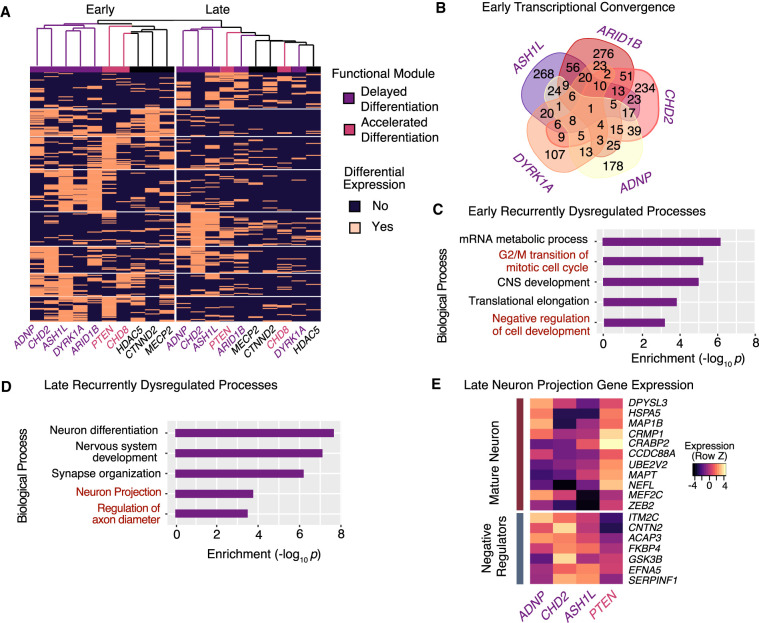
To investigate stage-specific convergence of ASD genes, we then clustered pseudotime-stratified samples based on recurrently dysregulated genes (i.e., genes that were differentially expressed in three or more ASD-gene knockdown samples) and found shared and distinct patterns of transcriptional dysregulation (Fig. 4A). This analysis grouped samples first by early and late pseudotime stage and then into stage-specific subsets of ASD genes. Because genes that acted to delay differentiation clustered together at the early stage, we can infer that the delayed neuronal maturation detected in our pseudotime analysis is a consequence of early-stage transcriptional dysregulation. Furthermore, the sets of transcriptional targets of these genes shared significant overlap (all pair-wise hypergeometric P-values < 10−22) (Fig. 4B), implying that these genes act through a convergent regulatory pathway that functions in early neuronal differentiation. This analysis also provided information about the regulatory hierarchy of these genes, with CHD2 down-regulated by ADNP or ARID1B repression and ASH1L down-regulated by ARID1B repression. Despite the cells being postmitotic, Gene Ontology enrichment of the recurrently dysregulated genes in the early-stage samples highlighted specifically disrupted processes, namely, the G2/M transition of cell cycle and negative regulation of cell development (Fig. 4C). The disrupted genes themselves are not core cell-cycle regulators so this signature may rather reflect cell-cycle disruptions that occurred earlier in the differentiation protocol (Supplemental Table S4). Together these results suggest that CHD2, ASH1L, ARID1B, DYRK1A, and ADNP comprise a convergent functional module of ASD genes acting on a shared gene regulatory pathway active in early neurodevelopment and that their haploinsufficiency impedes neuronal differentiation. Furthermore, our single-cell transcriptional analyses allowed us to make several explicit predictions about the consequences of ASD-gene repression on cellular phenotypes. Specifically, we predicted that if ADNP, ARID1B, ASH1L, CHD2, or DYRK1A are repressed, then we would observe a reduction in neural progenitor cell proliferation. In contrast, we expected PTEN repression to promote proliferation.
Figure 4.
Single-cell differential gene expression analysis identifies early- and late-stage convergent modules of ASD genes. (A) Hierarchical clustering of ASD knockdown profiles using genes differentially expressed in three or more ASD-candidate perturbations reveals a convergence of the transcriptional pathways dysregulated at early and late stages. At the early stage (left column), ADNP, CHD2, ASH1L, DYRK1A, and ARID1B form a transcriptionally convergent module of “delayed differentiation” (purple). At the late stage (right column), ADNP, CHD2, and ASH1L continue to converge. (B) Venn diagram shows significant overlap of differentially expressed genes across five ASD genes at the early stage. All pairwise overlaps have P-values <10−22 by hypergeometric testing. (C) Gene Ontology enrichment analysis of early-stage recurrently dysregulated genes highlights relevant biological processes disrupted and predicts disrupted G2/M transition and cellular maturation for “delayed differentiation” genes. Genes driving this enrichment are provided in Supplemental Table S4. (D) Enrichment analysis of late-stage recurrently dysregulated genes highlights relevant biological processes disrupted. (E) Expression of neuron projection genes (Gene Ontology 0010975) in the late-stage samples predicts disrupted neurite extension for PTEN but an enhanced phenotype for ADNP, CHD2, and ASH1L.
In late-stage cells, ADNP, CHD2, and ASH1L transcriptional profiles clustered together, implying these genes continue to share downstream molecular targets in maturing neurons. In contrast, hierarchical clustering did not support a convergence of the genes that accelerate differentiation, PTEN and CHD8, at either the early or late stages. We found that the recurrently dysregulated genes among late-stage samples were enriched for processes specific to neuron maturation such as synapse organization, neuron projection, and regulation of axon diameter (Fig. 4D). The specificity of these terms highlights the added molecular resolution we achieved by accounting for differences in pseudotime in our analysis but still indicates substantial heterogeneity in the progression of neuronal differentiation even within late-stage samples.
Differential expression analysis allowed us to generate specific predictions regarding the effects of ASD-gene repression on neuronal projections (axons and dendrites). We clustered samples by the expression of the dysregulated neuron projection genes and found that the clustering of ADNP, CHD2, and ASH1L was driven by the decreased expression of neuron maturation markers such as MAPT, NEFL, and MAP1B with concomitant up-regulation of annotated negative regulators of neuron projection and differentiation (Fig. 4E). These genes were driven in the opposite direction by PTEN. From these results, we predicted that knockdown of ADNP, CHD2, and ASH1L would decrease the outgrowth of neuronal projections, whereas knockdown of PTEN would enhance this process.
Live-cell imaging reveals abnormalities in proliferation and neurite extension and confirms transcriptome-based predictions
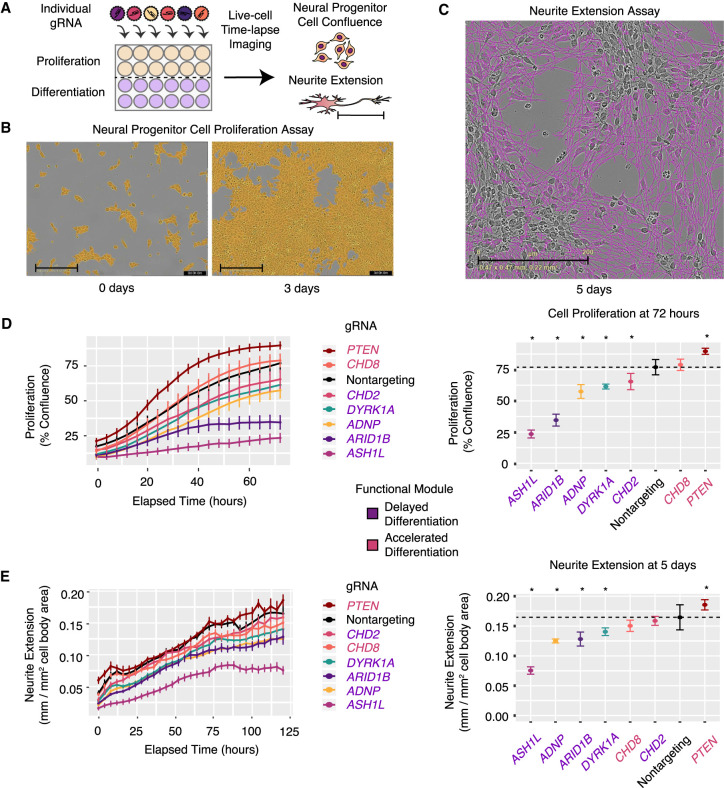
To test our transcriptome-based predictions, we implemented live-cell imaging to measure cellular proliferation and neurite extension after individual knockdown of ASD genes (Fig. 5A). We produced lentivirus expressing gRNAs that target candidate ASD genes and used them to infect dCas9-KRAB neuronal progenitor cells in an arrayed format. We imaged cells under both proliferative and differentiative conditions every 4 h for 3 or 5 d, respectively, using the IncuCyte live-cell imaging system. Representative images for proliferation and neurite extension are shown (Fig. 5B,C; Supplemental Fig. S7A,B). In these live-cell imaging experiments, we observed decreased proliferation after repression of each of the five proposed “delayed differentiation” genes (Fig. 5D). In contrast, PTEN repression caused a major increase in proliferation, consistent with our prediction and with its known function as an inhibitor of neural stem cell proliferation (Groszer et al. 2006). For the neurite extension assay, most of our predictions were also confirmed (Fig. 5E), as repression of four of the five “delayed differentiation” genes—ASH1L, ADNP, ARID1B, and DYRK1A—caused modest to severe reductions in neurite outgrowth. In agreement with our transcriptome-based prediction, PTEN repression increased neurite extension in this assay, a result consistent with an earlier observation that PTEN enhances the length of regenerating axons in vivo (Park et al. 2008). Together, proliferation and neurite extension assays confirmed the consequences of ASD-gene repression predicted by scRNA-seq analyses, showing the utility of our approach for high-throughput functional elucidation of neurodevelopmental disease–associated genes. Live-cell imaging further supports the functional convergence of some ASD genes acting at an early stage to delay neuron differentiation and decrease proliferation.
Figure 5.
Live-cell imaging after repression of individual ASD genes confirms defects in cellular proliferation and neurite extension. (A) Schematic overview of arrayed gRNA screening. Cells infected with a single gRNA are assayed by time course imaging for confluence and neurite extension. (B) Representative images of neural progenitor cell proliferation assay at the start- and end-points (day 0 and day 3). Neural progenitor cell proliferation is measured by creating a cell mask (orange) and computing the area of confluence at each time point. (C) Representative image of neurite extension assay at 5 d postdifferentiation. Neurite extension is measured with the NeuroTrack assay in the IncuCyte software. Neurite masks are shown (purple). Neurite extension lengths are normalized by cell cluster area to account for any differences in cell number. Scale bars, 200 µm. (D) Time-lapse imaging of cellular proliferation (left), assessed by the percentage of confluence, reveals significant decreases or increases (right). (*) P < 0.01, t-test, dotted horizontal line indicates average in control cells. (E) Time-lapse imaging of neurite extension (left) and quantification (right). (*) P < 0.01, t-test, dotted horizontal line indicates average in control cells). All values in D and E represent mean ± SEM. Cells with each individual gRNA were plated in duplicate or triplicate wells for each experiment. Images were captured from nine fields per well at each time point. Experiments were repeated two to three times for all gRNAs.
CRISPR repression in iPSC neural progenitor cells confirms functional gene modules and transcriptional convergence at cell-cycle dysregulation
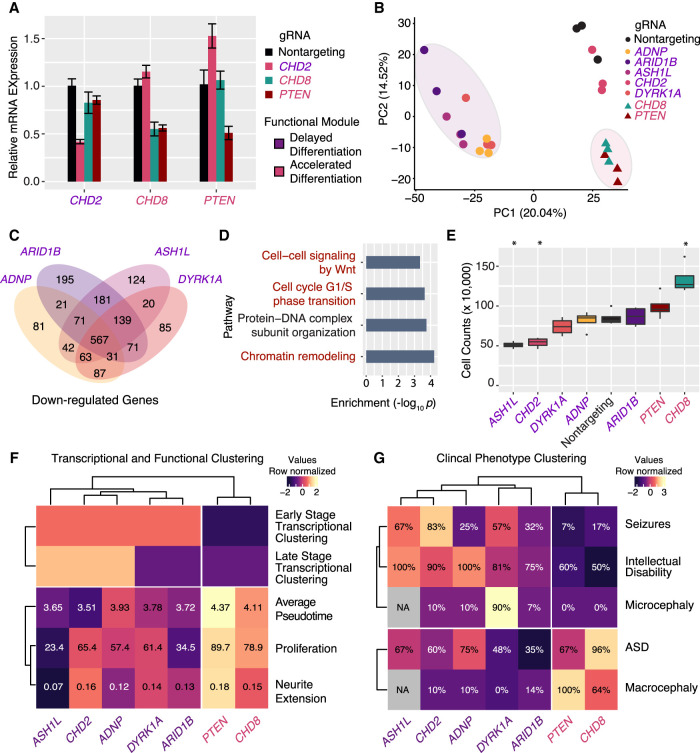
To validate the early transcriptional convergence and effects on cellular proliferation of the ASD gene modules in an orthogonal cellular model system, we performed CRISPR repression of individual genes in human iPSC-derived neural progenitor cells. First, we confirmed that dCas9-KRAB repression was efficient in these cells for a subset of genes using qPCR (Fig. 6A). Next, we performed RNA-seq after knockdown of seven individual ASD genes and a nontargeting control. RNA-seq confirmed efficient knockdown for 5/7 target genes (Supplemental Fig. S8A). Clustering transcriptomes using principal component analysis (PCA) closely reproduced the gene modules discovered in LUHMES, namely, the clustering of four members of the “delayed differentiation” gene set (ADNP, ARID1B, ASH1L, and DYRK1A) and the clustering of “accelerated differentiation” genes CHD8 and PTEN (Fig. 6B). The same clusters were also observed by unsupervised hierarchical clustering of transcriptomes using highly variable genes (Supplemental Fig. S8B). As in LUHMES, the “delayed differentiation” genes strongly converged at the level of transcriptional regulation (Fig. 6C), affecting genes enriched for roles in chromatin remodeling, Wnt signaling, and cell-cycle regulation (Fig. 6D). In these cells, the two “accelerated differentiation” genes had strongly overlapping transcriptional consequences on both down- and up-regulated genes (Supplemental Fig. S8C), leading to misregulation of cell-cycle genes and increased cell division pathways (Supplemental Fig. S8D). This transcriptional convergence could be explained by the observation that CHD8 repression also decreased the expression of PTEN (Fig. 6A), implying that these genes are in the same pathway. A proliferation assay in these cells after individual gene repression functionally confirmed that ASH1L and CHD2 decreased proliferation and CHD8 repression enhanced it (Fig. 6E). These results broadly confirm both the membership and functional interpretation of ASD-gene modules in a second human neural progenitor system, further validating the LUHMES as a relevant model for first-pass high-throughput functional genomics screening.
Figure 6.
CRISPR repression in iPSC neural progenitor cells confirms modules of ASD genes and transcriptional convergence at cell-cycle dysregulation. Experimental clustering of functional ASD gene modules matches clustering by clinical phenotypes. (A) Efficient dCas9-KRAB repression of individual target genes using the designated gRNAs in iPSC-NPCs. n = 3 biological replicates for all qPCR experiments. Values, mean ± SEM. (B) Clustering of RNA-seq profiles by principal component analysis reveals clustering of “delayed differentiation” and “accelerated differentiation” module ASD genes. (C) “Delayed differentiation” module gene repression elicits strongly overlapping transcriptional consequences. (D) Gene Ontology analysis of down-regulated genes shows enrichment for chromatin remodeling and cell-cycle genes. (E) Cellular proliferation measured by cell number after individual gene repression reveals significant decreases or increases. (*) P < 0.01, n = 4). (F) Integrating transcriptional and functional assays reveals and refines two functionally convergent modules of ASD genes. (G) Clinical phenotype data reveal the same two modules of ASD genes. (%) Prevalence of phenotype (percentage) in individuals from Stessman et al. (2017).
Functionally convergent ASD gene modules predict shared clinical phenotypes
Linking genotype to phenotype is the ultimate goal of functional genomics. To this end, we sought to determine if the functional convergence of ASD genes observed in our cellular model could predict a convergence of clinical phenotypes for these genes. To do so, we first integrated the results of our pseudotime analysis, transcriptional clustering, and functional profiling by hierarchical clustering (Fig. 6F). As expected, this integrated model clearly separated the “delayed differentiation” module genes from those in the “accelerated differentiation” module. Next, we performed hierarchical clustering on the prevalence of clinical phenotypes from one study on individuals with dominant loss-of-function mutations in these genes (Stessman et al. 2017). Clustering by clinical phenotypes fully recapitulated our proposed convergent modules and supports mechanistic links of convergent pathways to shared clinical outcomes (Fig. 6G). For example, individuals with mutations in the “delayed differentiation” module genes were highly likely to have intellectual disability, consistent with increased severity owing to early neurodevelopmental dysregulation. Comparing across the two clusters, these individuals have a higher incidence of microcephaly that is mechanistically consistent with the neural progenitor cell proliferation defects we observed. Likewise, individuals with PTEN and CHD8 mutations have a comparatively reduced prevalence of intellectual disability but a high prevalence of macrocephaly, consistent with the observed functional convergence of these genes on promoting neuronal differentiation, proliferation, and neurite outgrowth. Recent studies have confirmed that disruptive mutations in ADNP, ARID1B, CHD2, and DYRK1A are associated with a higher prevalence of severe neurodevelopmental delay. Conversely, CHD8 and PTEN mutations are associated with ASD without neurodevelopmental delay (Satterstrom et al. 2020).
Discussion
The genetic and phenotypic heterogeneity of neurodevelopmental disorders challenge the understanding and treatment of these conditions. Over 1100 genes have been discovered to cause neurodevelopmental disorders when mutated, and this number will continue to increase. As a result, it is imperative to develop better methods to cost-effectively unravel the functional contributions of these genes to both normal development and disease. Furthermore, the identification of convergent pathogenic mechanisms across diverse causative genes would facilitate the development of therapeutic interventions, but this requires a rapid, scalable, and disease-relevant model system in which tens to hundreds of genes can be modulated in parallel and their effects measured in a robust manner. Here, we have taken an important step toward establishing such a system by coupling pooled dCas9-based transcriptional repression to single-cell RNA-seq in a simple and highly tractable human model of neuron differentiation. By using this approach, we perturbed the expression of 13 diverse autism-associated genes and uncovered unique and overlapping consequences on transcriptional networks and pathways. This led to specific predictions about functional roles of these genes in growth and neurite extension, which we then validated through imaging. In addition to showing the utility of a high-throughput functional genomics approach to dissect gene function, our results suggest that many ASD genes might act on common pathways, namely, by modifying neural progenitor cell proliferation and cell-cycle.
By integrating pseudotime analysis, transcriptional clustering, and the cellular phenotyping of individual neurodevelopmental genes, we uncovered two consistent modules of genes with opposing functionality in altering the course of neuron differentiation. We identified a “delayed differentiation” module composed of five ASD genes and predicted that the knockdown of these genes would decrease neural progenitor cell proliferation. We further predicted, for a subset of this “delayed differentiation” module, that individual gene knockdown would decrease neuron projection. Finally, we also predicted that PTEN repression would increase both proliferation and neuron projection. Neural progenitor cell proliferation, the decision to exit the cell-cycle, and neuron differentiation are complex and interrelated processes (Hardwick and Philpott 2014). Disruptions to the progression of neuronal differentiation may also disrupt cell-cycle and vice versa. We have shown that repression of many of the ASD genes dysregulates the cell-cycle, which likely contributes to the altered progression of neuronal differentiation. However, we have not ruled out additional mechanisms by which these genes may alter differentiation. Increased proliferation could either drive an expansion of a progenitor pool or increase neurogenic cell divisions, leading to precocious differentiation. A parsimonious explanation of our data in LUHMES is that the progression of neuron differentiation is preceded by a few requisite rounds of neurogenic cell division. Delaying these divisions by disrupting cell cycle, or enhancing them by increasing proliferation rate, will consequently delay or accelerate neuronal differentiation progression, respectively. We confirmed these predictions by performing live-cell imaging of cell proliferation and neurite extension after gene knockdown, providing experimental functional validation for these ASD genes. Convergence of ASD genes at the regulation of cell-cycle genes was supported in an orthogonal model using human iPSC-derived NPCs. Heterochronicity of neurodevelopmental gene expression networks and consequent dysregulation of neuron differentiation is a plausible mechanism underlying ASD pathology and has been observed in other cellular models of ASD (Marchetto et al. 2017; Schafer et al. 2019).
We have shown that LUHMES cells enable the rapid and robust generation of postmitotic human neurons with transcriptional profiles that correspond closely to early human cortical development, a critical period of neurodevelopment that has been implicated in the etiology of ASD and other neurodevelopmental disorders. Confirmation of our results in human iPSC-derived NPCs further validates the utility of LUHMES for discovering potential mechanisms of a subset of neurodevelopmental disease-associated genes. Because transcriptional regulators are particularly enriched for high early-fetal expression (Satterstrom et al. 2020), LUHMES may be especially suited to studying this class of genes. However, for some other classes of neurodevelopmental genes, LUHMES cells may not be the most well-suited model system. For example, synaptic genes are more postnatally expressed across a broad range of neuronal types. iPSC-derived induced excitatory and inhibitory neurons, cocultures of these cells, or 3D organoids are likely to be more suitable than LUHMES for studying these genes and their roles in establishing synaptic phenotypes. These alternative human neurodevelopmental models may also be complementary for investigating genes and pathways involved in cell-fate specification, neuronal migration, and neuronal activity. However, the experimental complexity and heterogeneity of these models compared with the rapid and reproducible differentiation of LUHMES make them less suited for high-throughput analyses of sets of genes that are likely to be involved in neuronal differentiation or maturation. Overexpression of neurogenic transcription factors in iPSCs can reduce heterogeneity in differentiation to enable single-cell CRISPR screens in this model (Tian et al. 2019). Such models may bypass the neural progenitor cell stage, precluding the investigation of certain neurodevelopmental pathways (Schafer et al. 2019).
Compared with traditional single-gene disease modeling experiments in mice or human cells (e.g., Cotney et al. 2015; Durak et al. 2016; Katayama et al. 2016; Bellmaine et al. 2017; Jung et al. 2017), our approach increases the number of genes that can be assayed in parallel while also overcoming many of the primary sources of variation in such models. Critically, this enables direct comparison of results across genes to discover convergent mechanisms. Current single-cell technology enables pathway-level inferences of transcriptional dysregulation and prioritization of candidate genes for further functional validation in a rapid and cost-effective manner for tens of disease-implicated genes. However, noise and sparsity in single-cell RNA-seq data limit its power to detect differentially expressed genes. Quantitative measurements of knockdown efficiency of individual gRNAs within single cells are difficult, and individually validating gRNAs is incompatible with the goal of high-throughput screening. Our qPCR measurements of gRNA efficiencies showed a range of 40%–90% knockdown for all tested target genes, which may better represent haploinsufficiency than a total knockout approach. Exquisite dosage sensitivity of certain genes raises the possibility of variable cellular phenotypes depending on the degree of knockdown. Including more gRNAs per gene, and enough cells to analyze effects on a per-gRNA instead of per-gene basis, would resolve these potential confounds. Improvements in the sensitivity and throughput of scRNA-seq, as well as declining costs, will enhance the utility of our approach in future experiments.
A major strength of our approach is its extensibility to different model systems for the rapid functional profiling of diverse gene sets, allowing for prioritization of candidate genes for low-throughput cellular phenotyping by imaging. Although we only measured proliferation and neurite extension with live-cell imaging, the integration of these data with pooled transcriptomes revealed consistent gene modules that were also convergent at the level of clinical phenotypes in individuals with mutations in these genes (Stessman et al. 2017), illustrating the potential of this approach for linking molecular pathways to clinical phenotypes. Furthermore, although not implemented in this study, this approach is amenable to screening of chemical libraries to discover effective pharmaceutical interventions for any observed defects and to determine whether convergent genetic modules will respond to common treatments. Pooled, rather than arrayed, optical phenotyping approaches would further accelerate these efforts (Feldman et al. 2019). Such high-content imaging and screening in future experiments will enable detailed characterization of perturbation-induced neuronal phenotypes and the discovery of convergent molecular endophenotypes of disease pathogenesis.
Methods
Cell culture
HEK293T cells were maintained in Dulbecco's Modified Eagle Media (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin, and passaged every 3–4 d after enzymatic dissociation using trypsin. LUHMES cells (ATCC CRL-2927) were cultured according to established protocols with minor modifications as described in the Supplemental Methods (Scholz et al. 2011). Polyclonal dCas9-KRAB-blast expressing LUHMES were generated by infecting cells with lentivirus and selecting using blasticidin (10 µg/mL). Lenti-dCas9-KRAB-blast was a gift from Gary Hon (Addgene 89567) (Xie et al. 2017). Human iPSC-derived neural progenitor cells (XCL4) were acquired from STEMCELL Technologies (catalog number 70902) and grown in neural progenitor medium 2. Although now discontinued by STEMCELL Technologies, these reagents are available from XCell Science. Tetracycline-inducible dCas9-KRAB NPCs were generated after neomycin selection (200 µg/mL). pHAGE TRE dCas9-KRAB was a gift from Rene Maehr and Scot Wolfe (Addgene 50917) (Kearns et al. 2014). For proliferation assays, TRE dCas9-KRAB XCL4 cells were infected in quadruplicate with individual gRNAs targeting seven ASD genes and one nontargeting gRNA. After puromycin selection and doxycycline induction, cells were plated at equal cell numbers and grown for 8 d, and total cells were counted using a hemocytometer. Cell counts were compared with cells infected with a nontargeting gRNA.
gRNA cloning
For each target gene, we selected three gRNAs optimized for repression from the Dolcetto library (Sanson et al. 2018) and cloned them into a CRISPR-repression optimized vector to enable pooled lentiviral preparation without guide-barcode swapping (Supplemental Methods; Hill et al. 2018; Sanson et al. 2018; Xie et al. 2018). CROP-seq-opti was a gift from Jay Shendure (Addgene 106280) (Hill et al. 2018).
Lentivirus production of individual gRNAs and pooled gRNA libraries
Lentivirus was produced according to established protocols. In brief, HEK293T cells were seeded at a density of 1 million cells per well of a six-well plate and transfected with 2 μg of DNA comprising 1 µg gRNA-transfer plasmid, 750 ng psPAX2, and 250 ng pMD2.G. psPAX2 and pMD2.G were gifts from Didier Trono (Addgene 12259 and 12260). Cells were transfected using the PEI method (Polysciences). Media was changed 12 h after transfection, and viral-containing supernatant was collected 24 and 48 h later. Lentivirus was concentrated using Lenti-X reagent and resuspended in 50-µL aliquots from each milliliter of original supernatant (20× concentration). Lentivirus was titrated on LUHMES cells by infecting cells with serial dilutions of virus, followed by antibiotic selection (puromycin for gRNAs, 1 µg/mL). For pooled gRNA libraries, equal amounts of DNA for each gRNA were mixed prior to transfection.
Lentiviral transduction of gRNAs
For individual or pooled gRNAs, LUHMES cells were infected with serial dilutions of virus. Virus-containing media were removed after 4–6 h of transduction. Antibiotic selection with puromycin (1 µg/mL) was applied 24 h after infection. Wells in which no more than 25% of cells survived, corresponding to multiplicity of infection <0.3, were used for experiments. Cells were expanded for 4 d before plating for differentiation. After replating, cells were differentiated for 6–8 d in the presence of tetracycline to allow efficient repression and differentiation. Puromycin and blasticidin were maintained for the duration of all experiments to ensure gRNA and dCas9-KRAB expression in all cells.
qRT-PCR
RNA was purified from 6–8 d differentiated LUHMES using TRIzol. One microgram of RNA was reverse-transcribed into cDNA using qScript cDNA SuperMix (Quantabio). qRT-PCR was performed on an ABI 7900HT using SYBR Green SuperMix (Quantabio). Relative expression levels were determined using the comparative threshold (ΔΔCT) method (Livak and Schmittgen 2001). Actin beta (ACTB) mRNA levels were used as a normalization control. qPCR primers sequences are provided in Supplemental Table S5.
Bulk RNA-seq
For time course differentiation analysis, two replicates of LUHMES cells were differentiated for each time point, and total mRNA was purified. RNA was sent for sequencing at the Genome Technology Access Center (GTAC) at the McDonnell Genome Institute (GTAG@MGI) at the Washington University School of Medicine. For dCas9-KRAB NPCs (XCL4), RNA-seq libraries were prepared after 8 d of repression by bulk RNA barcoding and sequencing (Supplemental Methods; Alpern et al. 2019). Custom barcoded oligo(dT) capture oligos are listed in Supplemental Table S6.
Single-cell transcriptome capture
Twelve thousand cells were loaded per lane of a 10x device using 10x V2 Single cell 3′ Solution reagents (10x Genomics). Two biological replicates of pooled single-cell experiments were performed independently. Each replicate was loaded across one or two lanes of a 10x Single Cell A Chip V2. Single-cell libraries were prepared following the Single Cell 3′ Reagent Kits v2 User Guide (rev B). Single-cell cDNA libraries were amplified for 12 initial cycles after reverse transcription. A fraction of the prefragmented cDNA libraries was reserved for gRNA-specific enrichment PCR.
gRNA-transcript enrichment PCR
Three gRNA-specific enrichment PCR replicates were performed for each single-cell library. Each reaction used 1 µL of the single-cell post-cDNA amplification product as a template to amplify captured gRNA sequences. A single-step PCR reaction was used to amplify gRNA from total captured cDNA libraries using custom primers in a 25-µL reaction volume using KAPA HiFi HotStart ReadyMix (2X, Roche) (Supplemental Table S7).
gRNA depletion analysis
Almost all of the gRNAs in our lentiviral pool (43/47) were well represented in perturbed cells at similar frequencies, yet four gRNAs (targeting ASH1L, POGZ gRNAs 1 and 2, and SETD5) were significantly depleted (χ-squared test, P < 0.01) (Supplemental Fig. S9A). We hypothesized that their depletion was the result of fitness defects caused by the repression of these genes (Shalem et al. 2014; Wang et al. 2014). To test this, we infected neural progenitor cells individually with three of the depleted gRNAs and monitored cell proliferation using live imaging. Compared with a nontargeting gRNA, all of the depleted gRNAs caused a significant reduction in cellular proliferation, explaining why few cells with these gRNAs were detected in our pooled experiment (Supplemental Fig. S9B).
Bioinformatic analyses
Sequencing data corresponding to single-cell transcriptomes were processed using the 10x Genomics software package Cell Ranger (v 2.1.0). Most analyses were performed using the Seurat (v3.0) (Satija et al. 2015; Stuart et al. 2019) and Monocle (v2.10.0) (Trapnell et al. 2014) packages in R (v3.5.1) (R Core Team 2018). Details of all analyses are provided in the Supplemental Methods. Individual cells were sequenced to an average depth of 50,208 ± 7310 mapped reads per cell (2145 ± 448 genes detected, 6625 ± 1766 unique molecular identifiers [UMIs]). Differences in total UMIs, experimental batch, and mitochondrial percentage were accounted for during data normalization. Normalized filtered data were used for the remaining analyses. For each perturbation, we grouped all cells with any of the three gRNAs targeting the same gene, as we have shown that all three gRNAs typically have strong on-target activity (Supplemental Fig. S2B,C).
Global clustering of single-cell transcriptomes
Dimensionality reduction was performed by running PCA and clustering cells by the first six PCAs using UMAP (Becht et al. 2019). To assess global variation in transcriptional states, we visualized all single-cell transcriptomes on the UMAP. This revealed that >99% of all cells formed a single cluster of postmitotic neurons as defined by the absence of proliferative marker expression (Supplemental Fig S3A,B). This is consistent with our experimental design capturing a single time point (day 7) in a rapid isogenic model of neuronal differentiation. Within the major cluster, however, the expression of markers of neuronal differentiation showed variable patterns across the UMAP (Supplemental Fig S3C–E). Moreover, the most variably expressed genes in single-cell transcriptomes were enriched for functional roles in neurogenesis and axon projection, suggesting heterogeneity of neuronal differentiation at the single-cell level. We did not observe batch effects on global clustering (Supplemental Fig. S3F).
Pseudotime analysis
We projected cells in pseudotime in Monocle by ordering cells by highly variable genes. Dimensionality reduction was performed using the “DDRTree” method. Trajectories based on different sets of highly variable genes were qualitatively similar, showing a single trajectory with only minor branching. The pseudotime trajectory is composed of individual line segments called pseudotime “states.” To ensure high correlation between pseudotime and neuron differentiation status, we computed the state-specific genes in the bulk RNA-seq data set for each day of differentiation and used these genes for pseudotime ordering. We then transferred pseudotime state labels into Seurat to discover marker genes for each pseudotime state. Transferring pseudotime labels onto the UMAP plot showed distinct banding patterns representing subtle transcriptional state differences within the main cluster (Supplemental Fig. S10A,B). Reclustering cells in the earliest and latest pseudotime states revealed two completely distinct cell states expressing either differentiation (NEUROD1) or maturation (STMN2) markers (Supplemental Fig. S10C–E; Dennis et al. 2019; Polioudakis et al. 2019). This confirms that pseudotime is more sensitive to detect biologically relevant transcriptional patterns than UMAP clustering in our data set. We tested for altered pseudotime state membership proportions for each gRNA using χ-squared tests, computed the distribution of pseudotime state scores for each gRNA, and compared their averages using t-tests (Supplemental Methods).
Transition mapping of LUHMES differentiation
Transition mapping allows the comparison of in vitro neuron differentiation to in vivo development by computing the overlap of differentially expressed genes at selected time points across data sets (Stein et al. 2014). We compared the in vitro LUHMES differentiation time points day 0 to day 8 to transcriptional changes across brain regions and developmental time points in the BrainSpan Atlas of the Developing Human Brain. LUHMES differentiation had the strongest overlap with transcriptional changes occurring in the cortex of postconception week-8 to week-10 embryos and week-10 to week-13 embryos.
Differential gene expression analysis
Differential gene expression testing was performed using the FindMarkerGenes function in Seurat using the Wilcoxon rank sum test and a relaxed log2 fold-change threshold of 0.1 to increase the number of differentially expressed genes. This cutoff was calibrated against a gold-standard data set comparing single-cell and bulk RNA-seq data to identify differentially expressed genes (Avey et al. 2018). To find marker genes of pseudotime state clusters, only positive markers were returned.
Pseudotime state was binarized with states 1–3 labeled as “early” and states 4–6 as “late.” We next created another label combining the targeted gene with binary pseudotime state (e.g., CHD8_early). Averaged transcriptional profiles were recomputed for each group based on these new labels, and differentially expressed genes were also recalculated. PCA and hierarchical clustering were performed on these samples. As expected, unsupervised clustering of the stratified profiles by PCA perfectly discriminated between “early” and “late” samples (Supplemental Fig. S6D). This analysis showed that the first principal component corresponds to pseudotime status and explains almost 20% of the total variance in the data set. Gene Ontology and pathway enrichment analyses were performed using WebGestalt (Wang et al. 2017).
Live-cell imaging
Cells were imaged using an IncuCyte S3 live imaging system (Essen BioScience) For each experiment, dCas9-KRAB LUHMES were infected in duplicate or triplicate with individual gRNAs and were plated in duplicate or triplicate in wells of a 24-well plate in either self-renewing or differentiation conditions. Nine fields per well were imaged every 4 h for either 3 or 5 d for proliferation or differentiation, respectively. These experiments were repeated two or three times for each individual gRNA. Images were analyzed using the IncuCyte Software. Specifically, we performed the proliferation analysis and NeuroTrack neurite tracing analyses with default parameters. Cell bodies and neurites were detected from phase contrast images. Representative images are shown in Supplemental Figure S7.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE142078. All in-house code/scripts used to perform the analyses are available as Supplemental Code.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
This work was supported by T32HL125241-05 (National Heart, Lung, and Blood Institute), U54HD087011-04 (National Institute of Child Health and Human Development), R21NS087230-01A1 (National Institute of Neurological Disorders and Stroke) (R.D.M. and J.M.), RF1MH117070 and U01MH109133 (National Institute of Mental Health) (R.D.M., J.D.D.), SFARI Explorer 500661 (Simons Foundation Autism Research Initiative) (R.D.M., J.D.D.), and T32CA009547-33 (National Cancer Institute) (D.A.). GTAC@MGI is partially supported by National Institutes of Health (NIH) grants P30 CA91842 and UL1 TR000448.
Author contributions: M.A.L. performed experiments, analyzed data, designed the study, and wrote the manuscript. D.A. performed experiments and wrote the manuscript. J.D.D., J.M., and R.D.M. analyzed data, designed the study, and wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.262295.120.
References
- Abrahams BS, Arking DE, Campbell DB, Mefford HC, Morrow EM, Weiss LA, Menashe I, Wadkins T, Banerjee-Basu S, Packer A. 2013. SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol Autism 4: 36 10.1186/2040-2392-4-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson B, Norman TM, Jost M, Cho MY, Nuñez JK, Chen Y, Villalta JE, Gilbert LA, Horlbeck MA, Hein MY, et al. 2016. A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell 167: 1867–1882.e21. 10.1016/j.cell.2016.11.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpern D, Gardeux V, Russeil J, Mangeat B, Meireles-Filho ACA, Breysse R, Hacker D, Deplancke B. 2019. BRB-seq: ultra-affordable high-throughput transcriptomics enabled by bulk RNA barcoding and sequencing. Genome Biol 20: 71 10.1186/s13059-019-1671-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberger JS, Bocchini CA, Scott AF, Hamosh A. 2019. OMIM.org: leveraging knowledge across phenotype–gene relationships. Nucleic Acids Res 47: D1038–D1043. 10.1093/nar/gky1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avey D, Sankararaman S, Yim AKY, Barve R, Milbrandt J, Mitra RD. 2018. Single-cell RNA-seq uncovers a robust transcriptional response to morphine by glia. Cell Rep 24: 3619–3629.e4. 10.1016/j.celrep.2018.08.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E, McInnes L, Healy J, Dutertre C-A, Kwok IWH, Ng LG, Ginhoux F, Newell EW. 2019. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol 37: 38–44. 10.1038/nbt.4314 [DOI] [PubMed] [Google Scholar]
- Bellmaine SF, Ovchinnikov DA, Manallack DT, Cuddy CE, Elefanty AG, Stanley EG, Wolvetang EJ, Williams SJ, Pera M. 2017. Inhibition of DYRK1A disrupts neural lineage specification in human pluripotent stem cells. eLife 6: e24502 10.7554/eLife.24502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotney J, Muhle RA, Sanders SJ, Liu L, Willsey AJ, Niu W, Liu W, Klei L, Lei J, Yin J, et al. 2015. The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat Commun 6: ncomms7404 10.1038/ncomms7404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datlinger P, Rendeiro AF, Schmidl C, Krausgruber T, Traxler P, Klughammer J, Schuster LC, Kuchler A, Alpar D, Bock C. 2017. Pooled CRISPR screening with single-cell transcriptome readout. Nat Methods 14: 297–301. 10.1038/nmeth.4177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis DJ, Han S, Schuurmans C. 2019. bHLH transcription factors in neural development, disease, and reprogramming. Brain Res 1705: 48–65. 10.1016/j.brainres.2018.03.013 [DOI] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, et al. 2014. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515: 209–215. 10.1038/nature13772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit A, Parnas O, Li B, Chen J, Fulco CP, Jerby-Arnon L, Marjanovic ND, Dionne D, Burks T, Raychowdhury R, et al. 2016. Perturb-Seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 167: 1853–1866.e17. 10.1016/j.cell.2016.11.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan B, Zhou C, Zhu C, Yu Y, Li G, Zhang S, Zhang C, Ye X, Ma H, Qu S, et al. 2019. Model-based understanding of single-cell CRISPR screening. Nat Commun 10: 2233 10.1038/s41467-019-10216-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durak O, Gao F, Kaeser-Woo YJ, Rueda R, Martorell AJ, Nott A, Liu CY, Watson LA, Tsai L-H. 2016. Chd8 mediates cortical neurogenesis via transcriptional regulation of cell cycle and Wnt signaling. Nat Neurosci 19: 1477–1488. 10.1038/nn.4400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman D, Singh A, Schmid-Burgk JL, Carlson RJ, Mezger A, Garrity AJ, Zhang F, Blainey PC. 2019. Optical pooled screens in human cells. Cell 179: 787–799.e17. 10.1016/j.cell.2019.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. 2013. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154: 442–451. 10.1016/j.cell.2013.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszer M, Erickson R, Scripture-Adams DD, Dougherty JD, Belle JL, Zack JA, Geschwind DH, Liu X, Kornblum HI, Wu H. 2006. PTEN negatively regulates neural stem cell self-renewal by modulating G0-G1 cell cycle entry. Proc Natl Acad Sci 103: 111–116. 10.1073/pnas.0509939103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick LJA, Philpott A. 2014. Nervous decision-making: to divide or differentiate. Trends Genet 30: 254–261. 10.1016/j.tig.2014.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill AJ, McFaline-Figueroa JL, Starita LM, Gasperini MJ, Matreyek KA, Packer J, Jackson D, Shendure J, Trapnell C. 2018. On the design of CRISPR-based single-cell molecular screens. Nat Methods 15: 271–274. 10.1038/nmeth.4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S-M, Hartley BJ, Flaherty E, Rajarajan P, Abdelaal R, Obiorah I, Barretto N, Muhammad H, Phatnani HP, Akbarian S, et al. 2017. Evaluating synthetic activation and repression of neuropsychiatric-related genes in hiPSC-derived NPCs, neurons, and astrocytes. Stem Cell Rep 9: 615–628. 10.1016/j.stemcr.2017.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman GE, Hartley BJ, Flaherty E, Ladran I, Gochman P, Ruderfer DM, Stahl EA, Rapoport J, Sklar P, Brennand KJ. 2017. Transcriptional signatures of schizophrenia in hiPSC-derived NPCs and neurons are concordant with post-mortem adult brains. Nat Commun 8: 2225 10.1038/s41467-017-02330-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höllerhage M, Moebius C, Melms J, Chiu W-H, Goebel JN, Chakroun T, Koeglsperger T, Oertel WH, Rösler TW, Bickle M, et al. 2017. Protective efficacy of phosphodiesterase-1 inhibition against α-synuclein toxicity revealed by compound screening in LUHMES cells. Sci Rep 7: 11469 10.1038/s41598-017-11664-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, O'Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, et al. 2014. The contribution of de novo coding mutations to autism spectrum disorder. Nature 515: 216–221. 10.1038/nature13908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung E-M, Moffat JJ, Liu J, Dravid SM, Gurumurthy CB, Kim W-Y. 2017. Arid1b haploinsufficiency disrupts cortical interneuron development and mouse behavior. Nat Neurosci 20: 1694–1707. 10.1038/s41593-017-0013-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S, Moriguchi T, Ohtsu N, Kondo T. 2016. A powerful CRISPR/Cas9-based method for targeted transcriptional activation. Angew Chem Int Ed 55: 6452–6456. 10.1002/anie.201601708 [DOI] [PubMed] [Google Scholar]
- Kearns NA, Genga RMJ, Enuameh MS, Garber M, Wolfe SA, Maehr R. 2014. Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells. Development 141: 219–223. 10.1242/dev.103341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpinen H, Goncalves A, Leha A, Afzal V, Alasoo K, Ashford S, Bala S, Bensaddek D, Casale FP, Culley OJ, et al. 2017. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature 546: 370–375. 10.1038/nature22403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536: 285–291. 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Santpere G, Kawasawa YI, Evgrafov OV, Gulden FO, Pochareddy S, Sunkin SM, Li Z, Shin Y, Zhu Y, et al. 2018. Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science 362: eaat7615 10.1126/science.aat7615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Marchetto MC, Belinson H, Tian Y, Freitas BC, Fu C, Vadodaria KC, Beltrao-Braga PC, Trujillo CA, Mendes APD, Padmanabhan K, et al. 2017. Altered proliferation and networks in neural cells derived from idiopathic autistic individuals. Mol Psychiatry 22: 820–835. 10.1038/mp.2016.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matelski L, Morgan RK, Grodzki AC, Van de Water J, Lein PJ. 2020. Effects of cytokines on nuclear factor-κB, cell viability, and synaptic connectivity in a human neuronal cell line. Mol Psychiatry 10.1038/s41380-020-0647-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFaline-Figueroa JL, Hill AJ, Qiu X, Jackson D, Shendure J, Trapnell C. 2019. A pooled single-cell genetic screen identifies regulatory checkpoints in the continuum of the epithelial-to-mesenchymal transition. Nat Genet 51: 1389–1398. 10.1038/s41588-019-0489-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, et al. 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44: D733–D745. 10.1093/nar/gkv1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, et al. 2012. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485: 246–250. 10.1038/nature10989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, Horvath S, Geschwind DH. 2013. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 155: 1008–1021. 10.1016/j.cell.2013.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KK, Liu K, Hu Y, Smith PD, Wang C, Cai B, Xu B, Connolly L, Kramvis I, Sahin M, et al. 2008. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science 322: 963–966. 10.1126/science.1161566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul G, Christophersen NS, Raymon H, Kiaer C, Smith R, Brundin P. 2007. Tyrosine hydroxylase expression is unstable in a human immortalized mesencephalic cell line: sudies in vitro and after intracerebral grafting in vivo. Mol Cell Neurosci 34: 390–399. 10.1016/j.mcn.2006.11.010 [DOI] [PubMed] [Google Scholar]
- Pierce SE, Tyson T, Booms A, Prahl J, Coetzee GA. 2018. Parkinson's disease genetic risk in a midbrain neuronal cell line. Neurobiol Dis 114: 53–64. 10.1016/j.nbd.2018.02.007 [DOI] [PubMed] [Google Scholar]
- Polioudakis D, de la Torre-Ubieta L, Langerman J, Elkins AG, Shi X, Stein JL, Vuong CK, Nichterwitz S, Gevorgian M, Opland CK, et al. 2019. A single-cell transcriptomic atlas of human neocortical development during mid-gestation. Neuron 103: 785–801.e8. 10.1016/j.neuron.2019.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2018. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: https://www.R-project.org/. [Google Scholar]
- Sanson KR, Hanna RE, Hegde M, Donovan KF, Strand C, Sullender ME, Vaimberg EW, Goodale A, Root DE, Piccioni F, et al. 2018. Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat Commun 9: 5416 10.1038/s41467-018-07901-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A. 2015. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 33: 495–502. 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterstrom FK, Kosmicki JA, Wang J, Breen MS, De Rubeis S, An J-Y, Peng M, Collins R, Grove J, Klei L, et al. 2020. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell 180: 568–584.e23. 10.1016/j.cell.2019.12.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer ST, Paquola ACM, Stern S, Gosselin D, Ku M, Pena M, Kuret TJM, Liyanage M, Mansour AA, Jaeger BN, et al. 2019. Pathological priming causes developmental gene network heterochronicity in autistic subject-derived neurons. Nat Neurosci 22: 243–255. 10.1038/s41593-018-0295-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz D, Pöltl D, Genewsky A, Weng M, Waldmann T, Schildknecht S, Leist M. 2011. Rapid, complete and large-scale generation of post-mitotic neurons from the human LUHMES cell line. J Neurochem 119: 957–971. 10.1111/j.1471-4159.2011.07255.x [DOI] [PubMed] [Google Scholar]
- Shah RR, Cholewa-Waclaw J, Davies FCJ, Paton KM, Chaligne R, Heard E, Abbott CM, Bird AP. 2016. Efficient and versatile CRISPR engineering of human neurons in culture to model neurological disorders. Wellcome Open Res 1: 13 10.12688/wellcomeopenres.10011.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343: 84–87. 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JL, de la Torre-Ubieta L, Tian Y, Parikshak NN, Hernández IA, Marchetto MC, Baker DK, Lu D, Hinman CR, Lowe JK, et al. 2014. A quantitative framework to evaluate modeling of cortical development by neural stem cells. Neuron 83: 69–86. 10.1016/j.neuron.2014.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stessman HAF, Xiong B, Coe BP, Wang T, Hoekzema K, Fenckova M, Kvarnung M, Gerdts J, Trinh S, Cosemans N, et al. 2017. Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat Genet 49: 515–526. 10.1038/ng.3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, Satija R. 2019. Comprehensive integration of single-cell data. Cell 177: 1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Yu RT, Evans RM, Shi Y. 2007. Orphan nuclear receptor TLX recruits histone deacetylases to repress transcription and regulate neural stem cell proliferation. Proc Natl Acad Sci 104: 15282–15287. 10.1073/pnas.0704089104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian R, Gachechiladze MA, Ludwig CH, Laurie MT, Hong JY, Nathaniel D, Prabhu AV, Fernandopulle MS, Patel R, Abshari M, et al. 2019. CRISPR interference-based platform for multimodal genetic screens in human iPSC-derived neurons. Neuron 104: 239–255.e12. 10.1016/j.neuron.2019.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Z-B, Hogberg H, Kuo D, Sakamuru S, Xia M, Smirnova L, Hartung T, Gerhold D. 2017. Characterization of three human cell line models for high-throughput neuronal cytotoxicity screening. J Appl Toxicol 37: 167–180. 10.1002/jat.3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. 2014. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32: 381–386. 10.1038/nbt.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieth B, Ziegenhain C, Parekh S, Enard W, Hellmann I. 2017. powsimR: power analysis for bulk and single cell RNA-seq experiments. Bioinformatics 33: 3486–3488. 10.1093/bioinformatics/btx435 [DOI] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. 2014. Genetic screens in human cells using the CRISPR-Cas9 system. Science 343: 80–84. 10.1126/science.1246981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Vasaikar S, Shi Z, Greer M, Zhang B. 2017. WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res 45: W130–W137. 10.1093/nar/gkx356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willsey AJ, Morris MT, Wang S, Willsey HR, Sun N, Teerikorpi N, Baum TB, Cagney G, Bender KJ, Desai TA, et al. 2018. The psychiatric cell map initiative: a convergent systems biological approach to illuminating key molecular pathways in neuropsychiatric disorders. Cell 174: 505–520. 10.1016/j.cell.2018.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CF, Fitzgerald TW, Jones WD, Clayton S, McRae JF, van Kogelenberg M, King DA, Ambridge K, Barrett DM, Bayzetinova T, et al. 2015. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. The Lancet 385: 1305–1314. 10.1016/S0140-6736(14)61705-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S, Duan J, Li B, Zhou P, Hon GC. 2017. Multiplexed engineering and analysis of combinatorial enhancer activity in single cells. Mol Cell 66: 285–299.e5. 10.1016/j.molcel.2017.03.007 [DOI] [PubMed] [Google Scholar]
- Xie S, Cooley A, Armendariz D, Zhou P, Hon GC. 2018. Frequent sgRNA-barcode recombination in single-cell perturbation assays. PLoS One 13: e0198635 10.1371/journal.pone.0198635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Wells AB, O'Brien DR, Nehorai A, Dougherty JD. 2014. Cell type–specific expression analysis to identify putative cellular mechanisms for neurogenetic disorders. J Neurosci 34: 1420–1431. 10.1523/JNEUROSCI.4488-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Zhu Y, Yu H, Cheng X, Chen S, Chu Y, Huang H, Zhang J, Li W. 2020. scMAGeCK links genotypes with multiple phenotypes in single-cell CRISPR screens. Genome Biol 21: 19 10.1186/s13059-020-1928-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Bhattacharyya A. 2018. Human models are needed for studying human neurodevelopmental disorders. Am J Hum Genet 103: 829–857. 10.1016/j.ajhg.2018.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. 2017. Massively parallel digital transcriptional profiling of single cells. Nat Commun 8: 14049 10.1038/ncomms14049 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.