Abstract
Neuroblastoma is a malignancy of the developing sympathetic nervous system that accounts for 12% of childhood cancer deaths. Like many childhood cancers, neuroblastoma shows a relative paucity of somatic single-nucleotide variants (SNVs) and small insertions and deletions (indels) compared to adult cancers. Here, we assessed the contribution of somatic structural variation (SV) in neuroblastoma using a combination of whole-genome sequencing (WGS) of tumor-normal pairs (n = 135) and single-nucleotide polymorphism (SNP) genotyping of primary tumors (n = 914). Our study design allowed for orthogonal validation and replication across platforms. SV frequency, type, and localization varied significantly among high-risk tumors. MYCN nonamplified high-risk tumors harbored an increased SV burden overall, including a significant excess of tandem duplication events across the genome. Genes disrupted by SV breakpoints were enriched in neuronal lineages and associated with phenotypes such as autism spectrum disorder (ASD). The postsynaptic adapter protein-coding gene, SHANK2, located on Chromosome 11q13, was disrupted by SVs in 14% of MYCN nonamplified high-risk tumors based on WGS and 10% in the SNP array cohort. Expression of SHANK2 was low across human-derived neuroblastoma cell lines and high-risk neuroblastoma tumors. Forced expression of SHANK2 in neuroblastoma cells resulted in significant growth inhibition (P = 2.6 × 10−2 to 3.4 × 10−5) and accelerated neuronal differentiation following treatment with all-trans retinoic acid (P = 3.1 × 10−13 to 2.4 × 10−30). These data further define the complex landscape of somatic structural variation in neuroblastoma and suggest that events leading to deregulation of neurodevelopmental processes, such as inactivation of SHANK2, are key mediators of tumorigenesis in this childhood cancer.
Neuroblastoma is a cancer of the developing sympathetic nervous system that most commonly affects children under 5 yr of age, with a median age at diagnosis of 17 mo (Maris 2010). Approximately 50% of cases present with disseminated disease at the time of diagnosis. Despite intense multimodal therapy, the survival rate for this high-risk subset remains <50% (Maris 2010). Recent whole-genome and exome sequencing studies of neuroblastoma have revealed relatively few recurrent protein-coding somatic mutations including single-nucleotide variations (SNVs) and small (<50 bp) insertion/deletions (indels) (Cheung et al. 2012; Molenaar et al. 2012; Pugh et al. 2013; Sausen et al. 2013). Large-scale structural variations (SVs) such as deletions, insertions, inversions, tandem duplications, and translocations can arise from mutational processes that alter chromosome structure and evade innate mechanisms of maintaining genomic stability. These diverse SVs are often acquired somatically in cancer and can act as driver mutations (Yang et al. 2013).
Multiple approaches to detect SVs in large array and sequencing data sets have been applied to cancer (Alkan et al. 2011; Yang et al. 2013; Tubio 2015; Macintyre et al. 2016). Methods to identify copy number variations (CNVs) from intensity data (log R ratios) have been applied to single-nucleotide polymorphism (SNP) genotyping and comparative genomic hybridization (CGH) arrays. More recently, these approaches were adapted and applied to read-depth measures from high-throughput sequencing. Numerous segmentation algorithms exist for both array (Carter 2007) and sequence-based (Zhao et al. 2013) approaches, with the resulting CNV calls ranging in size from a few hundred base pairs to whole chromosome alterations. Importantly, these calls are dosage sensitive, allowing for numerical quantification of amplifications and deletions.
Analysis of CNVs in neuroblastoma primary tumors and matched blood samples led to identification of recurrent somatically acquired CNVs. These include focal amplification of MYCN, gain of Chr 17q, and deletion of Chr 1p and Chr 11q. These events are associated with an undifferentiated phenotype, aggressive disease, and poor survival (Brodeur et al. 1984; Gilbert et al. 1984; Seeger et al. 1985; Gehring et al. 1995; Caron et al. 1996; Plantaz et al. 1997; Bown et al. 1999; Guo et al. 1999; Maris et al. 2001; Łastowska et al. 2002; Attiyeh et al. 2005; Michels et al. 2007; Deyell and Attiyeh 2011). In addition, focal deletions in the ATRX chromatin remodeler gene (ATRX) result in deleterious loss of function (Cheung et al. 2012; Kurihara et al. 2014). ATRX is implicated in the alternative lengthening of telomeres (ALT) phenotype. Focal CNVs involving other tumor suppressor genes, such as PTPRD (Stallings et al. 2006), ARID1A, and ARID1B (Sausen et al. 2013) have also been reported.
Although analysis of somatic CNVs has been productive, high-throughput sequencing approaches can profoundly expand our understanding of SVs in cancer (Macintyre et al. 2016). Alignment-based methods to identify SVs focus on reads and read pairs discordantly aligned to the reference genome. As such, these alignment-based approaches do not rely on dosage quantification and do not quantify numerical changes of deletions and tandem duplications. However, they provide essential information about inversions, translocations, and transposable elements, which are elusive to CNV callers. Furthermore, read coverage-based and alignment-based approaches have often been combined together to improve accuracy (Qi and Zhao 2011; Zhang and Wu 2011; Jiang et al. 2012); these and other available methods have been systematically reviewed (Tattini et al. 2015).
Recent studies using alignment-based detection of SVs from WGS of primary neuroblastomas revealed structural rearrangements as key oncogenic drivers. These SVs mediate enhancer hijacking or focal enhancer amplification, influencing telomere maintenance through activation of telomerase reverse transcriptase gene (TERT) (Peifer et al. 2015; Valentijn et al. 2015; Kawashima et al. 2016) or deregulating the MYC oncogene (Zimmerman et al. 2018). Despite the demonstrated importance of somatic CNVs and other SVs in neuroblastoma, studies systematically integrating CNV and alignment-based approaches are lacking. Therefore, the global landscape and mechanisms of pathogenicity for many of these events remain poorly understood.
Here, we studied the role of somatic SVs in a large neuroblastoma cohort comprising 997 distinct primary neuroblastoma tumors obtained at diagnosis. Specifically, we integrated whole-genome sequencing (WGS) from 135 tumor-normal pairs and single-nucleotide polymorphism (SNP) arrays from 914 primary tumors. Alternative approaches to SV detection were considered for both data sets, which overlapped in a subset of 52 cases. As such, this study allowed for cross-platform validation of SVs. We further explored the functional impact of SVs by integrating matched transcriptomic data from 153 RNA-sequencing samples and 247 HumanExon arrays comprising 361 distinct tumor samples. Finally, we performed in vitro studies to assess the functional relevance of SHANK2, a newly identified tumor suppressor gene disrupted by SVs. Taken together, the integration of multi-omic data sets with patient clinical profiles and biological experimentation serves to greatly expand the mutational landscape of neuroblastoma.
Results
Neuroblastoma patient characteristics and primary data sets for SV detection
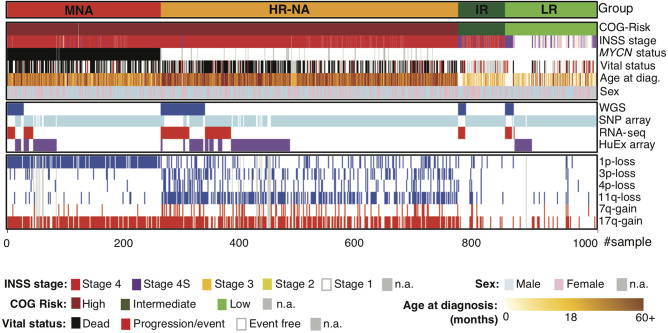
To establish the landscape of SVs in neuroblastoma, we sequenced the genomes of 135 primary diagnostic tumors and matched normal (blood leukocyte) DNA pairs through the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) initiative (https://ocg.cancer.gov/programs/target). Samples were obtained through the Children's Oncology Group (COG) and included 106 patients with high-risk tumors (29 MYCN amplified and 77 non-MYCN amplified), 14 with intermediate-risk tumors, and 15 with low-risk tumors (Fig. 1; Supplemental Tables S1, S2). Whole-genome sequencing (WGS) was performed by Complete Genomics (Drmanac et al. 2010) to a median average depth of 76×. To augment the WGS data, and to provide independent replication, we genotyped and analyzed 914 neuroblastoma patient tumors using Illumina SNP arrays (Fig. 1; Supplemental Tables S1, S2), including 52 cases overlapping the WGS data set. This cohort consisted of 696 high-risk (239 MYCN amplified and 457 non-MYCN amplified), 70 intermediate-risk, and 145 low-risk tumors. Throughout the study, we examined disease risk groups as defined by the COG and the International Neuroblastoma Risk Group (INRG) (Cohn et al. 2009). Specifically, the following subtypes were considered: low-risk neuroblastoma (LR); intermediate-risk neuroblastoma (IR); high-risk neuroblastomas with amplification of the MYCN oncogene (MNA); and high-risk neuroblastomas without MYCN amplification (HR-NA).
Figure 1.
Overview of samples, clinical information, and data types used. Survey of available samples, clinical information, and data types used throughout this study. Detailed patient and sample data are provided in Supplemental Tables S1 and S2.
Identification of novel regions of recurrent DNA copy number gain and loss
We first investigated DNA copy number profiles generated from WGS and SNP array data sets. Visualization using Integrative Genome Viewer (IGV) (Robinson et al. 2011) and analysis of segmentation profiles using GISTIC 2.0 (Mermel et al. 2011) confirmed well-established patterns of recurrent copy number alterations that differed across neuroblastoma clinical subtypes (Supplemental Fig. S1A–H; Wang et al. 2006; Michels et al. 2007). MNA and HR-NA subsets shared recurrent 17q gains and PTPRD deletions (9p23) and differed in 2p24 gain (MYCN locus) and prevalence of deletions at 1p, 3p, 4p, and 11q (Supplemental Fig. S1E–H). In the HR-NA group, we further observed deletions at 16q24.3 (Mosse et al. 2005), segmental gains of the q-arm of Chr 7 (Bosse et al. 2017), focal gains at Chr 5p15.33 (Q-value = 1.42 × 10−3) harboring the telomerase reverse transcriptase (TERT) gene, intragenic deletions of the ATRX chromatin remodeler gene at Xq21.1 (Q-value = 3.76 × 10−3), and a novel region of recurrent deletion at 10p15.3 (Q-value = 6.16 × 10−2) (Supplemental Fig. S1G,H).
Analysis of SV breakpoints using orthogonal approaches: SJ-BP, RD-BP, and CN-BP
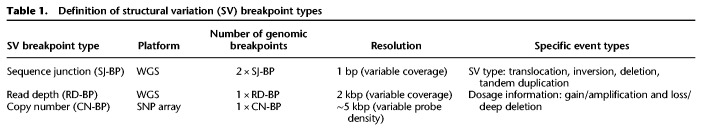
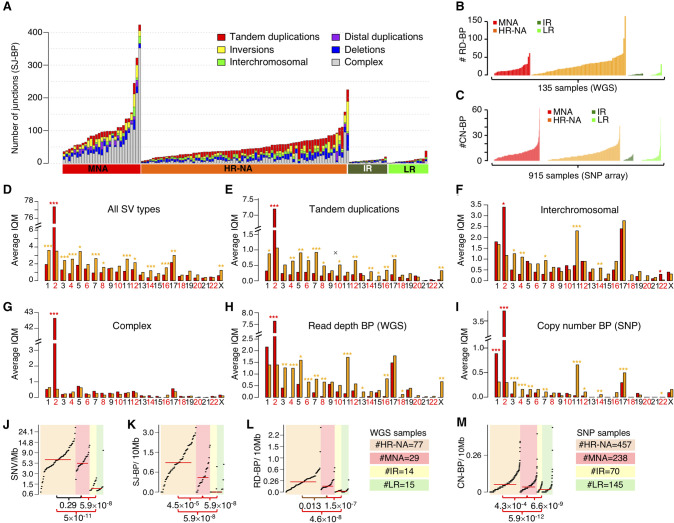
To expand and strengthen our study of SVs, we considered three approaches to SV breakpoint identification: sequence junction (SJ-BP), read-depth (RD-BP), and copy number breakpoint (CN-BP) (Table 1). First, we obtained alignment-based SVs reported by the Complete Genomics, Inc. (CGI) somatic pipeline, restricted to variants supported by at least three read pairs, and applied additional quality control filters (Methods). These SVs were defined as SJ-BP and were delimited by two breakpoints in the genome. SV types included deletions (>500 bp), tandem duplications (>200 bp), inversions (>200 bp), translocations, and complex events (Supplemental Fig. S2B–D). We observed a total of 7366 SJ-BP SV calls (Supplemental Table S3) distributed heterogeneously across neuroblastoma subtypes (Fig. 2A). We then mapped copy number dosage breakpoints derived from WGS read-depth segmentation profiles, hereafter referred to as read-depth breakpoints (RD-BP) (Methods). A total of 2836 RD-BPs SVs were identified (μ = 21) and were unevenly distributed across samples (Fig. 2B). Finally, analogous to the RD-BPs, we mapped copy number breakpoints from segmentation profiles derived from the larger SNP array cohort, referred to as copy number breakpoints (CN-BP) (Methods). A total of 6241 CN-BPs SVs were identified across 914 samples (μ = 6.8) (Fig. 2C). Overall, SV breakpoints from orthogonal approaches showed high concordance, in agreement with benchmarks from other available methods (Supplemental Fig. S3; Methods; Qi and Zhao 2011; Zhang and Wu 2011; Jiang et al. 2012). As expected (Wang et al. 2006; Michels et al. 2007; Schleiermacher et al. 2012), we observed substantially more SV events in high-risk compared to intermediate- and low-risk tumors.
Table 1.
Definition of structural variation (SV) breakpoint types
Figure 2.
Somatic structural variation burden differs among neuroblastoma subtypes by quantity, type, and genomic location. (A) Stacked bar chart of alignment-based SV calls by type and neuroblastoma subtype in WGS data set. (B) Bar plot representing the number of read-depth breakpoints (RD-BP) per sample across subtypes in the WGS data set. (C) Bar plot representing the number of copy number breakpoints (CN-BP) per sample across subtypes in the SNP data set. (D–I) By-chromosome comparison between MNA and HR-NA of the interquantile average number of SVs including all SJ-BP variant types (D), tandem duplications (E), inter-chromosomal translocations (F), complex events (G), as well as CNV breakpoints as defined by RD-BP (H) and CN-BP (I). A Wilcoxon test is obtained for every chromosome, and the P-value significance level is represented as follows: (***) P < 0.001; (**) P < 0.01; (*) P < 0.05. Asterisk color indicates the group with higher IQM: (red) MNA; (orange) HR-NA. Mutation burden analysis plot across neuroblastoma subtypes representing the burden of SNVs (J), SJ-BPs (K), RD-BPs (L), and CN-BP (M).
Chromothripsis associates with TERT and MYCN in neuroblastoma
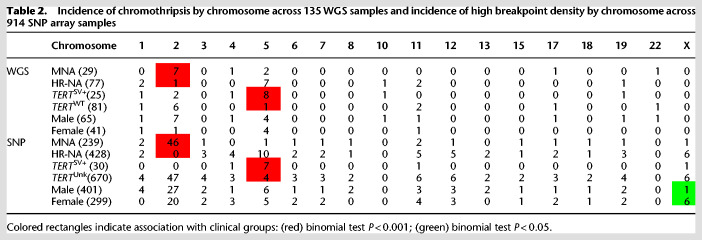
We next sought to leverage our large data set to evaluate chromothripsis. Previous studies have reported chromothripsis in up to 18% of high-risk neuroblastomas (Molenaar et al. 2012), and colocalization with key neuroblastoma oncogenes TERT and MYCN (Peifer et al. 2015; Valentijn et al. 2015). We observed rearrangements near the TERT locus in 24 HR-NA samples and 2 MNA from the WGS data set as well as 15 cases (14 HR-NA and 1 MNA) from the SNP array data set; one sample (PAPUTN) was present in both data sets (Supplemental Fig. S4). SVs near TERT were validated by Sanger sequencing in 11 cases from the WGS set with available DNA (Supplemental Fig. S5). MYCN amplification was determined diagnostically by fluorescence in situ hybridization (FISH) in 29 samples from the WGS data set (Supplemental Table S2) and confirmed by segmentation data in this study (Supplemental Fig. S6A). We also identified four SVs involving ALK (Supplemental Fig. S6B); these events were validated via Sanger sequencing (Supplemental Fig. S6C).
Chromothripsis was characterized based on clustered somatic rearrangements and alternating copy number states in defined chromosome regions (Maher and Wilson 2012). Chromothripsis events were identified at 27 regions (Supplemental Table S4) involving 20 distinct high-risk tumors (19%). These regions represented chromosome arms with high breakpoint densities (>2σ above the average of each sample's breakpoint burden distribution) and a minimum of six breakpoints (both SJ-BPs and RD-BPs). Chromothripsis was observed on Chr 2 in eight samples (Table 2; Supplemental Fig. S7) and showed enrichment in tumors harboring MYCN amplification (MNA) (7/8 samples, binomial test P = 7.4 × 10−4). Among them, two samples (PARETE and PATESI) involved coamplification of ALK with MYCN (Supplemental Fig. S7). Nine tumors harbored shattered Chr 5p and were enriched in samples with SVs near TERT (8/9, binomial test P = 7.3 × 10−5) (Table 2; Supplemental Fig. S8). Two samples (PAPSRJ and PAPUTN) (Supplemental Fig. S7) included inter-chromosomal events involving the MYCN and TERT gene loci and coamplification of both oncogenes. Other chromosomes involved in chromothripsis events included Chrs 1, 10, 11, and X in a female sample (Supplemental Fig. S9). Chromothripsis in most cases (15/20) was localized to a single chromosome.
Table 2.
Incidence of chromothripsis by chromosome across 135 WGS samples and incidence of high breakpoint density by chromosome across 914 SNP array samples
Finally, we sought to validate these results in the larger SNP array data set. In the absence of sequence junction information, we focused on unusual high density (>2σ above average) of CN-BPs (Table 2). We observed high breakpoint density on Chr 2 enriched in MNA samples (46/46, P ∼ 0). We also detected enrichment of high breakpoint density on Chr 5 involving cases harboring rearrangements or CN-BPs near TERT (7/11, P = 3.01 × 10−8). In addition, high breakpoint density on Chr X was enriched in female patients (6/7, P = 4.7 × 10−2), although no specific oncogenic associations were determined. Overall, SNP array analysis of high CN-BP density supported and replicated observations from the WGS analysis.
Patterns of SV mutational burden differ across neuroblastoma high-risk subtypes
We applied our SV filtering and assessed differences in SV type frequencies across other pediatric cancer types sequenced using CGI in the larger TARGET project (Methods). We observed that neuroblastomas harbored a higher frequency of tandem duplications (22%) compared with other tumor types (ALL = 7.9%, AML = 12.7%, OS = 16.2%, and WT = 12.0%) (Supplemental Fig. S10A). When comparing SVs across neuroblastoma subtypes, we observed this enrichment of tandem duplications was accentuated in HR-NA tumors (30.1%) (Supplemental Fig. S10B). Comparison of SJ-BPs in MNA and HR-NA tumors revealed these high-risk subsets also differed in SV genomic location (Fig. 2D–G; Supplemental Fig. S10C–F). MNA tumors harbored more SVs on Chr 2 (Wilcoxon test P = 1.6 × 10−14) (Fig. 2D), driven by complex junctions at the MYCN amplicon at Chr 2p24 (Supplemental Fig. S10C). Nearly all chromosomes displayed a higher frequency of SVs in HR-NA than MNA neuroblastoma (Fig. 2D). Specifically, HR-NA tumors harbored more tandem duplications in all chromosomes except Chr 2 (P = 4.0 × 10−12), in particular, Chr 7 (P = 3.91 × 10−5), Chr 5 (P = 1.2 × 10−3), and Chr 4 (P = 1.3 × 10−3) (Fig. 2E). Inter-chromosomal events were also more frequent in HR-NA tumors and overlapped with regions of known segmental copy number alterations including Chr 3p (P = 1.8 × 10−3), Chr 4p (P = 9.1 × 10−6), and Chr 11q (P = 1.9 × 10−8), but not Chr 1p and Chr 17q (Fig. 2F). In contrast, complex events showed no overall differences between high-risk groups with the exception of the aforementioned Chr 2 (Fig. 2G). Finally, RD-BP and CN-BP frequencies followed a similar pattern across chromosomes as that of SJ-BPs. MNA tumors harbored increased number of breakpoints in Chr 2 (PRD-BP = 2.4 × 10−9 and PCN-BP = 4.2 × 10−83) (Fig. 2H,I) while HR-NA harbored increased frequencies in most other chromosomes and in particular, Chr 11 (PRD-BP = 2.0 × 10−8 and PCN-BP = 4.0 × 10−25) (Fig. 2H,I).
We next studied overall differences in mutational burden and chromosomal instability across subtypes; we posit that the densities of breakpoints (SJ-BP, RD-BP, and CN-BP) throughout the genome represent a bona fide measure of chromosomal instability (CIN). We also obtained measures of somatic SNV density. To avoid skewing of results owing to the MYCN amplicon in MNA samples and regions showing chromothripsis, we implemented an SNV and SJ-BP tumor burden measure robust against outliers. To this end, the genome was divided into 41 sequence-mapped chromosome arms, and the density of SVs per megabase was measured. For each sample, the interquartile mean (IQM) was derived from the 41 arm measurements (Supplemental Fig. S10G,H). Similarly, we obtained IQM density measurements from RD-BP and CN-BP chromosomal burdens. As expected, LR and IR tumors carried very low mutational burden (Fig. 2J–M; Wang et al. 2006; Michels et al. 2007). We observed increased CIN (SJ-BP, RD-BP, and CN-BP) in HR-NA compared to MNA (Wilcoxon rank test: PSJ-BP = 4.5 × 10−5, PRD-BP = 1.3 × 10−2, PCN-BP = 4.6 × 10−8) (Fig. 2K–M, respectively), similar to previous reports (Caren et al. 2010). In contrast, no difference was observed in the average SNV burden (Wilcoxon rank test: P = 0.29) (Fig. 2J). These data support the notion that small SNVs and SVs arise from different mutational processes. These results also confirm the observation that HR-NA tumors show increased CIN (Wang et al. 2006; Caren et al. 2010) and expand this to include other classes of SVs, such as tandem duplication events identified by SJ-BP analysis.
Identification of genes recurrently altered by SVs in high-risk neuroblastomas
To identify genes affected by recurrent somatic SVs in neuroblastoma, SVs were assigned to different categories according to the inferred impact on the exonic sequence of known RefSeq genes (Supplemental Fig. S11). Using SJ-BP, we classified SVs into: (1) “coding” SVs that modify the exonic sequence of known genes including whole gene copy number alterations (duplications and deletions, size up to 2 Mb), or (2) “noncoding” SVs that do not modify the exonic sequences but might have an impact on regulatory regions proximal to known genes (100 kb upstream and 25 kb downstream) or intronic regions (Supplemental Fig. 11A). In contrast to SJ-BP junctions obtained from discordantly aligned mate read pairs, dosage-based breakpoints (RD-BP and CN-BP) cannot identify their counterpart location in the genome. Therefore, events such as translocations and inversions cannot be identified. With this in mind, for RD-BP and CN-BP, we assumed the impact as (1) “coding”: breakpoints within the transcription start and end positions of known genes; or (2) “noncoding”: breakpoints located on proximal upstream and downstream regions (Supplemental Fig. 11B). In addition, we localized CNVs involving amplification (CNWGS > 8; CNSNP > 4.5) and deep deletions (CNWGS < 0.5; CNSNP < 0.9) (Methods).
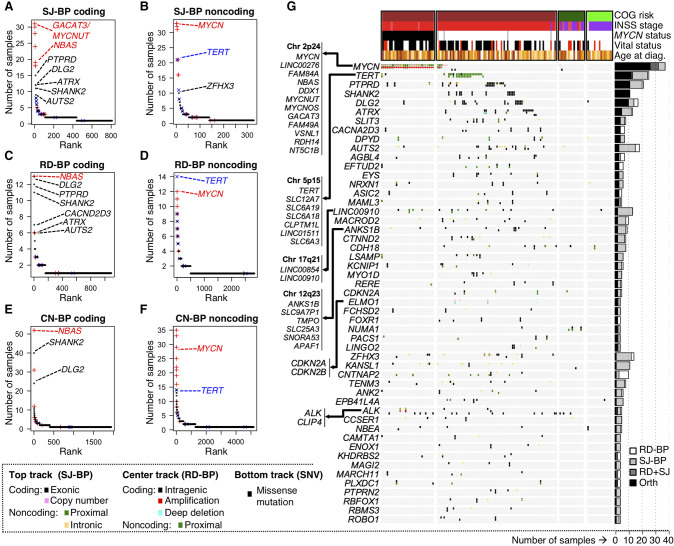
Based on the aforementioned definitions, we ranked genes according to the number of samples harboring “coding” and “noncoding” SVs for each of the three breakpoint analyses (SJ-BPs, RD-BPs, and CN-BPs) (Fig. 3A–F; Supplemental Table S5). We restricted CN-BPs analyses to SNP array samples that do not overlap with the WGS cohort. Therefore, SJ-BPs and RD-BPs are orthogonal approaches applied to the same discovery cohort (NCGI = 135), and CN-BPs are applied to an independent replication cohort (NSNP = 862). Overall, recurrently altered genes by “coding” and “noncoding” events return highly concordant results across the three approaches (Fig. 3A–F).
Figure 3.
Identification of recurrently altered genes in neuroblastoma by breakpoint analyses. (A–F) Recurrently altered genes ranked based on different breakpoint analyses and mode of impact: (A) gene coding sequences with recurrent SJ-BPs; (B) gene proximal and intronic sequences with recurrent SJ-BPs; (C) gene proximal sequences with recurrent RD-BPs; (D) gene coding sequences with recurrent RD-BPs; (E) gene coding sequences with recurrent CN-BPs; and (F) gene proximal sequences with recurrent CN-BPs. (G) OncoPrint based on the WGS data set recurrently altered genes by SVs detected through orthogonal approaches (SJ-BP and RD-BP) as depicted in bar plot (right). The oncoPrint aggregates three tracks per gene representing different BP analysis: (upper) SJ-BP; (center) RD-BP; and (lower) recurrent pathogenic SNVs.
To provide an integrated overview of the landscape of altered genes, we then combined WGS based methods (SJ-BP and RD-BP) into a ranking of recurrently altered genes with colocalizing breakpoints, hence, orthogonally validated. We shortlisted a total of 77 genes around 51 loci altered in four or more samples with at least one orthogonal (SJ-BP and RD-BP) breakpoint (Fig. 3G; Supplemental Table S6). In addition, we annotated likely pathogenic SNV calls within these genes (Supplemental Table S7). In addition to frequent alterations at the MYCN and TERT loci, the ALK gene harbored SVs in five samples (Supplemental Fig. S6B), and another 18 samples showed somatic SNVs in ALK, resulting in a combined set of 23 samples being affected (17% of all neuroblastomas). The PTPRD gene was altered in 20 samples, 11 of which were orthogonally validated (Supplemental Fig. S12A). Twelve ATRX intragenic deletions (five orthogonally validated) (Supplemental Fig. S12B) and one tandem duplication were observed in HR-NA tumors. Four ATRX SVs with available DNA were also validated by Sanger sequencing (Supplemental Fig. 13).
Genes lacking a well-established role in neuroblastoma were also disrupted by recurrent SVs in this study. Specifically, both SHANK2 and DLG2, located on Chr 11q at a region of prognostic significance in neuroblastoma, were frequently disrupted (Supplemental Fig. S14). The SHANK2 gene was disrupted in 11 HR-NA samples profiled by WGS and 40 HR-NA samples profiled by SNP array. The DLG2 gene was disrupted in 10 samples based on SJ-BP and 14 samples based on RD-BP analyses. In addition, 22 samples from the SNP array cohort harbored breakpoints involving DLG2. Across the larger TARGET cohort, we observed SHANK2 disruptions to be unique to neuroblastomas. In contrast, DLG2 disruptions were also observed frequently in osteosarcomas (Supplemental Table S5), where DLG2 has been described as a tumor suppressor (Smida et al. 2017; Shao et al. 2019). Alterations in SHANK2 and DLG2 occur predominantly in the HR-NA subset, where they are mutually exclusive (PFET = 0.042). Other novel altered genes included AUTS2 at Chr 7q (NWGS = 18, NSNP = 2) (Supplemental Fig. S15A), the calcium channel CACNA2D3 gene at Chr 3.p14.3 (NWGS = 11, NSNP = 10) (Supplemental Fig. S15B). Noncoding SVs affected a region proximal to the LINC00910 long noncoding RNA (lncRNA) on Chr 17 (Supplemental Fig. S16A), and focal deletions of CDKN2A and CDKN2B were identified in three tumors (Supplemental Fig. S16B). Sanger sequencing was used to validate 11 SHANK2 translocation events and 12 DLG2 variants (Supplemental Figs. S17, S18). Across this study, we validated 45 junctions of 49 variants tested by Sanger sequencing, representing a 92% success rate (Supplemental Table S8), consistent with previous validation efforts (Ma et al. 2015, 2018). Six of the SVs evaluated (13.3%, 3 ATRX and 3 DLG2) had low coverage (fewer than 10 reads) but were rescued by our pipeline and confirmed by Sanger sequencing.
Recurrent SVs have a regional transcriptional effect in neuroblastoma tumors
To gain further understanding of the functional relevance of SVs, we performed an expression quantitative trait loci (eQTL) analysis for each of the recurrent SV-associated genes (Supplemental Fig. S19A). The analysis, which was replicated in the two available transcriptional data sets (RNA-seq and HuEx array), reported consistent up-regulation of MYCN and TERT, including their neighbor genes in association with SVs. We also observed up-regulation of the lncRNA LINC00910 (PRNA = 7.0 × 10−3) at Chr 17.q21, a region with frequent inter-chromosomal translocations. In contrast, CDKN2A was down-regulated (PHuEx = 4.7 × 10−2; Pboth = 2.5 × 10−2) by focal deletions, and PLXDC1 at Chr 17.q12 was down-regulated (PHuEx = 4.7 × 10−2; Pboth = 2.5 × 10−2) in association with 17q gain breakpoints.
In addition to eQTL-based changes in overall gene expression, translocations may lead to the expression of fusion transcripts
Three fusion transcript methods applied returned a wide range of events (NSTAR-fusion = 24,837, NfusionCATCHER = 6898, NDeFUSE = 22,837), and overlapped in only 68 events (0.1%) (Supplemental Fig. S19B; Supplemental Table S9). The subset of fusion events with matching DNA translocation events from WGS comprised 66 events (NSTAR-fusion = 45, NfusionCATCHER = 36, NDeFUSE = 44), with the three methods overlapping in 26 events (40%) (Supplemental Fig. S19C). We observed an in-frame fusion transcript and translocation involving FOXR1:DDX6, where oncogenic fusion events have previously been described in neuroblastoma (Santo et al. 2012). The most frequent gene fusion event with both RNA and DNA evidence involved SHANK2. However, none of the SHANK2 fusion transcripts appeared to be in-frame, suggesting the fusion transcripts may not be biologically relevant and that these are more likely loss of function events.
Neurodevelopmental genes are recurrently disrupted by SVs in neuroblastoma
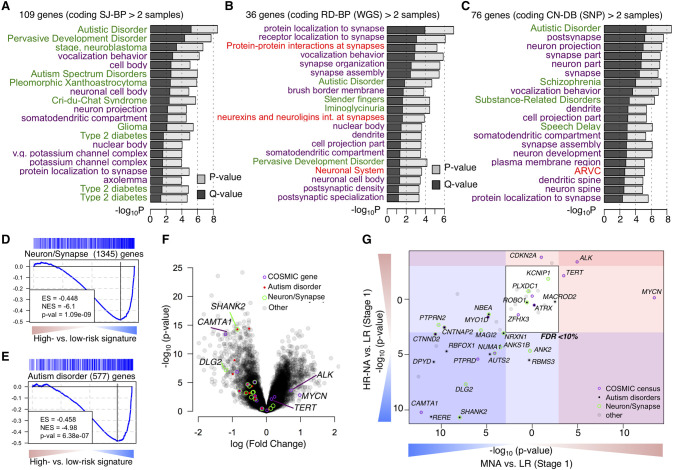
To identify pathways targeted by SVs, we considered recurrently altered genes from each of the coding (N > 2) and noncoding (N > 3) altered gene lists (number of genes: SJ-BPcoding = 109, SJ-BPnoncoding = 36, RD-BPcoding = 76, RD-BPnoncoding = 27, CN-BPcoding = 77, and CN-BPnoncoding = 88) (Fig. 3A–F). We tested each gene list for enrichment across Gene Ontology, pathway, and disease gene classes using ToppGene (Supplemental Table S10; Chen et al. 2009). Genes with coding sequences altered showed consistent results across the three breakpoint mappings and revealed strong enrichment in genes involved in autism spectrum disorder (ASD) susceptibility (PSJ-BP = 2.8 × 10−9; PRD-BP = 2.9 × 10−5; PCN-BP = 2.7 × 10−9) and other neurodevelopmental disorders (NDD) as well as protein localization to synapse (PSJ-BP = 1.2 × 10−5; PRD-BP = 1.1 × 10−7; PCN-BP = 2.4 × 10−6) and other neuronal related classes (Fig. 4A–C; Supplemental Table 10). These enrichments were not observed when the analysis was performed in other histotypes from the TARGET project, suggesting these findings are unique to neuroblastoma. Results in other histotypes showed enrichment in leukemia-related disease gene sets (Supplemental Fig. S20A; Supplemental Table S10). The neuroblastoma gene sets with “noncoding” alterations were more variable across the alternative breakpoint analyses, but were dominated by events involving MYCN and TERT in association with the disease class “stage, neuroblastoma” (PSJ-BP = 1.9 × 10−6; PRD-BP = 2.5 × 10−5; PCN-BP = 9.2 × 10−5) (Supplemental Fig. S20B–D; Supplemental Table S10).
Figure 4.
Neurodevelopmental genes are recurrently targeted by structural variations in neuroblastoma. (A–C) Function enrichment analysis bar plots for genes recurrently altered based on breakpoint analyses: (A) SJ-BPs; (B) RD-BPs; (C) CN-BPs. Analysis includes gene sets associated with diseases (green), Gene Ontology (purple), and Pathways (red). (D,E) Gene Set Enrichment Analysis across the signature of high- versus low-risk tumors from the HumanExon array show enrichment of neuronal and synapse part (D) and autism disorder predisposition genes (E). (F) Volcano plot showing differential expression between high- and low-risk highlighting genes with recurrent SVs and their functional classification. (G) Subtype-specific high- versus low-risk differential expression analysis of 77 recurrently altered genes from Figure 3I shown as scatter plot: (MNA) x-axis; (HR-NA) y-axis). (D–G) Analysis was replicated in two data sets: HuEx arrays (here) and RNA-seq (Supplemental Fig. S21A–D).
Neurodevelopmental genes disrupted by SVs are down-regulated in high-risk neuroblastoma
To further characterize the clinical relevance of recurrently altered genes in neuroblastoma, we studied their differential expression between high- and low-risk disease using the Affymetrix HuEx array data set (Fig. 4D,E). Using gene set enrichment analysis (GSEA) (Subramanian et al. 2005), we observed down-regulation of neuronal and synaptic genes (PHuEx = 1.09 × 10−9) and autism disorder susceptibility genes (PHuEx = 6.38 × 10−7) in high-risk tumors when compared to stage 1 low-risk tumors (Fig. 4D–F). We then focused on differential expression of genes with recurrent SVs in high-risk subtypes (Fig. 4G). Known oncogenes including TERT and ALK were up-regulated in both MNA and HR-NA, but MYCN was up-regulated only in MNA tumors. Known neuroblastoma tumor suppressor genes including CAMTA1 and RERE from the Chr 1p region and PTPRD were down-regulated in both subtypes. Most genes with a role in ASD, and those involved in neuron and synapse formation, were down-regulated in both high-risk subtypes. In particular, expression was significantly reduced for SHANK2 (PMNA = 2.15 × 10−11; PHR-NA = 1.05 × 10−8) and DLG2 (PMNA = 2.1 × 10−8; PHR-NA = 4.86 × 10−8) in high-risk compared with stage 1 low-risk tumors and compared to stage 4S low-risk tumors (PMNA = 1.41 × 10−3; PHR-NA = 1.82 × 10−5 and PMNA = 1.09 × 10−4; PHR-NA = 2.72 × 10−4, respectively). These results were replicated using the RNA-seq data set (Supplemental Fig. S21A–D).
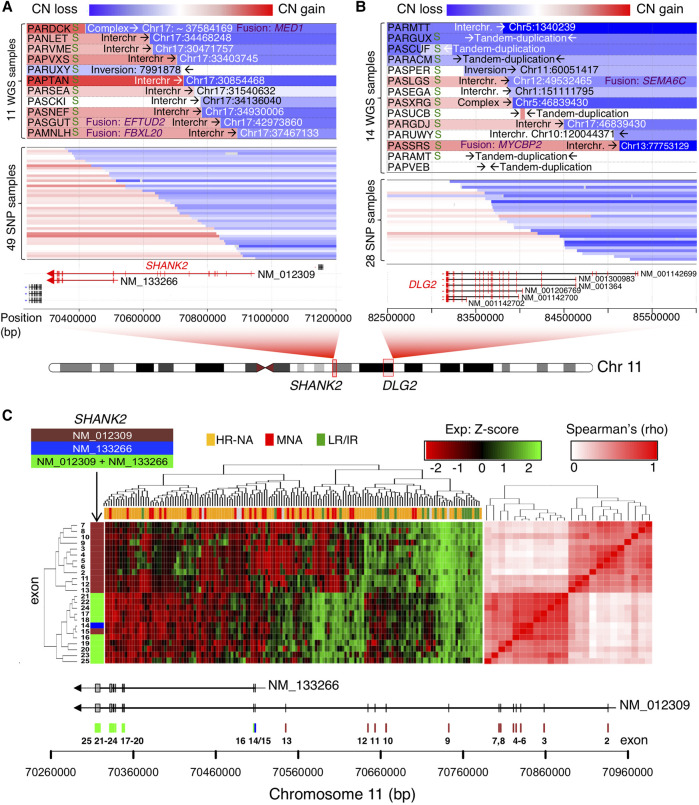
Neurodevelopmental genes SHANK2 and DLG2 are frequently disrupted by Chr 11 translocation events
High-risk neuroblastomas without MYCN amplification frequently show deletion of Chr 11q and this event is associated with a poor outcome (Guo et al. 1999; Attiyeh et al. 2005; Caren et al. 2010). The most frequent breakpoints observed in this study were located at Chr 11q.13 and 11q.14 and disrupted the SHANK2 and DLG2 gene loci, respectively (Fig. 5A,B; Supplemental Fig. S14). SHANK2 translocation partners involved Chr 17q in 10/11 WGS cases; in addition, we identified 49 samples from the SNP data set (10.7%) with breakpoints in SHANK2 (Fig. 5A). DLG2 translocation partners included multiple chromosomes. DLG2 breakpoints were also identified in 28 samples from the SNP array data set (Fig. 5B).
Figure 5.
Neuronal genes SHANK2 and DLG2 are frequently disrupted by translocation events involving Chr 11. (A,B) Copy number, junction location, and opposite break end destination location and types of SVs at genomic regions harboring rearrangements that span SHANK2 (A) and DLG2 loci (B); “S” at the left of the panel indicates positive validation by Sanger sequencing for SHANK2 (Supplemental Fig. S17) and DLG2 (Supplemental Fig. S18). Associated gene fusion events obtained from RNA-seq are also indicated in purple text. The top panel contains information derived from WGS, whereas the lower panel derives from SNP arrays and only represents CNV information. (C) Clustering analysis of SHANK2 exon level FPKM from RNA-seq data. The heatmap (left) shows higher exon expression level in low/intermediate risk compared to MNA and HR-NA samples. The correlation matrix (right) shows two well-defined clusters associated with the two known coding isoforms of the gene. Exons are color coded according to their isoform involvement.
SHANK2 is a scaffold protein in the postsynaptic density (PSD) with two known coding isoforms (long isoform: NM_012309; short isoform: NM_133266). We therefore studied the expression pattern of SHANK2 at the exon level using both RNA-seq (Fig. 5C) and Affymetrix HumanExon arrays (Supplemental Fig. S22A,B) data. Clustering analysis of SHANK2 exon expression revealed two distinct clusters corresponding to the two known coding isoforms. Expression of the long isoform (NM_012309) was decreased in high-risk tumors compared to low- and intermediate-risk tumors as observed from RNA-seq (Fig. 5C) and array-based expression analysis (Supplemental Fig. S22A,B). Finally, in a large independent RNA-seq cohort (Wang et al. 2014a), reduced expression of the long isoform (NM_012309) was associated with increased tumor stage (P = 1.62 × 10−22) (Supplemental Fig. S22C) and poor overall survival (P = 7.21 × 10−13) (Supplemental Fig. S22D). The association with poor survival remained significant within the low- and intermediate-risk subsets of neuroblastoma that typically have favorable outcomes (P = 2.22 × 10−5) (Supplemental Fig. S22E). Consistent with the SHANK2 expression pattern, we observed decreased activation of PSD genes based on GSEA in high-risk compared to low-risk neuroblastomas in multiple prognostic signatures (Supplemental Fig. S23). We decided to further study the long isoform of SHANK2 (NM_012309), given that nearly all SVs uniquely disrupt this splice variant, leaving the short isoform (NM_133266) intact.
SHANK2 expression inhibits cell growth and viability of neuroblastoma cells
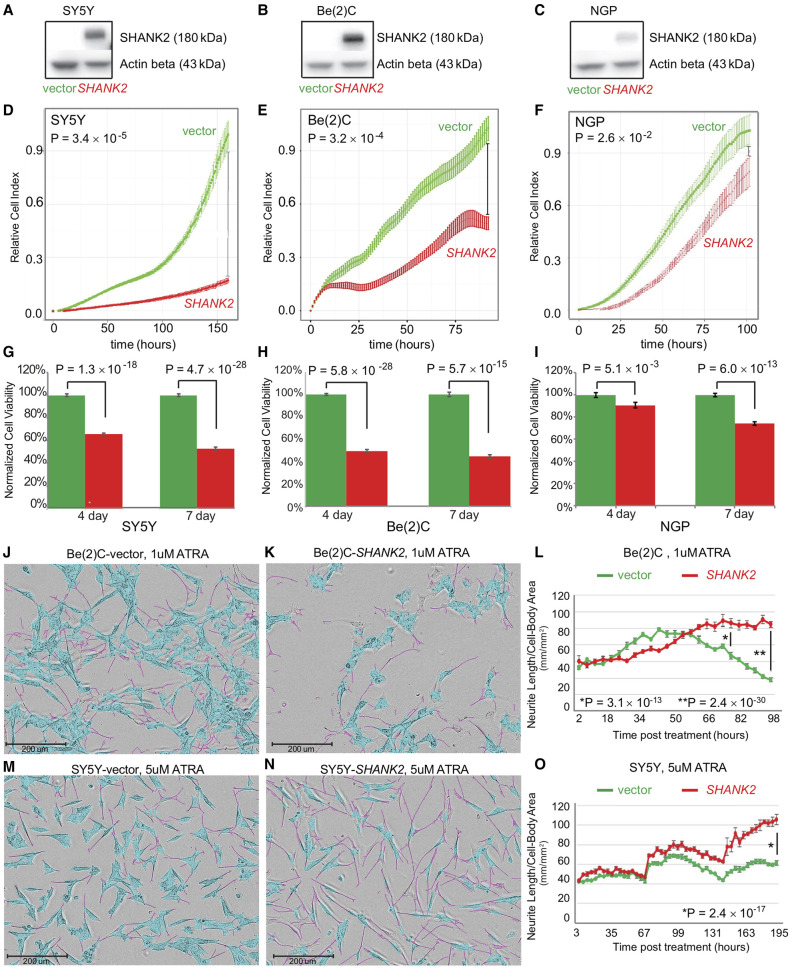
To further elucidate the role of SHANK2 in neuroblastoma, three neuroblastoma cell lines with low or no endogenous SHANK2 expression were selected. SHANK2 expression was low across 38 human-derived neuroblastoma cell models evaluated (Supplemental Fig. S24). The selected cell lines included SY5Y (MYCN nonamplified), Be(2)C (MYCN amplified), and NGP (MYCN amplified). Cells were stably transduced to constitutively overexpress SHANK2 long isoform or an empty vector control. SHANK2 expression was confirmed by western blot (Fig. 6A–C). When maintained in selection media and grown alongside empty vector controls, the SHANK2-overexpressing (SHANK2+) cells consistently showed decreased cell growth and viability as measured by RT-CES cell index (Fig. 6D–F) as well as CellTiter-Glo assay (Fig. 6G–I). For SY5Y, when control reached confluence, the comparable cell indexes of the SHANK2+ lines were reduced by 75% (P = 3.4 × 10−5) (Fig. 6D), Be(2)C cell index reduced by 62% (P = 3.16 × 10−4) (Fig. 6E), and NGP showed a 14% reduction (P = 2.62 × 10−2) (Fig. 6F). We also observed decreased cell viability in SHANK2+ cells at both 4- and 7-d endpoints using an ATP-dependent CellTiter-Glo assay. Specifically, viability of SY5Y-SHANK2+ cells was reduced to 65.51% (P = 1.34 × 10−18) and 52.64% (P = 4.72 × 10−26) of controls (Fig. 6G). This was reinforced in the similar results for Be(2)C-SHANK2+ cells (49.21% and 44.26%, P = 5.76 × 10-28 and 5.74 × 10−15) (Fig. 6H) and NGP-SHANK2+ (90.63% and 74.01%, P = 5.11 × 10−3 and 6.01 × 10−13) (Fig. 6I).
Figure 6.
SHANK2 reduces cell growth and promotes differentiation in neuroblastoma cell line models. (A–C) Western blots confirming overexpression of SHANK2 in all tested neuroblastoma cells: (A) SY5Y, (B) Be(2)C, (C) NGP. (D–F) Decreased proliferation in all three lines overexpressing SHANK2 (red) compared to controls (green), as measured by RT-CES. (G–I) Decreased viability in SHANK2 overexpressing cells (red) versus controls (green) as measured by ATP-dependent CellTiter-Glo Assay. (J,K) IncuCyte images of Be(2)C cells for vector control (J) and SHANK2-expressing cells (K) at 78 h post treatment with 1 µM ATRA. Neurite extensions masked in pink; cell bodies masked in blue. (L) Neurite length normalized to cell body area starting immediately after ATRA application corresponding to Be(2)C cells images at different time points. (M,N) SY5Y images from IncuCyte at day nine post ATRA treatment (5 µM). (O) Neurite outgrowth normalized to cell body area in corresponding to SY5Y cells images at different time points.
SHANK2 expression accelerates differentiation of neuroblastoma cells exposed to all-trans retinoic acid (ATRA)
We next investigated the role of SHANK2 in neuronal differentiation in Be(2)C and SY5Y cells exposed to ATRA. In the presence of ATRA, overexpression of SHANK2 accelerated differentiation as measured by presence and length of neurites compared to cell body (Fig. 6J–O; Supplemental Fig. S25A–F). Although decreases in growth can be measured even without drug application, once ATRA is applied, SHANK2+ cells developed neurites more quickly, and those neurites extended further than observed in empty vector controls (Supplemental Fig. S25C,D). In Be(2)C cells, a significant difference in neurite outgrowth normalized to cell body area was seen at 72 h post treatment with 1 µM ATRA (Fig. 6J–L), with SHANK2+ cells showing a 1.6-fold increase over controls (P = 3.09 × 10−13); the difference increased to 2.76-fold at 96 h (P = 2.37 × 10−30). Even with vehicle alone, SHANK2+ cells had more neurite outgrowth per cell body compared to their empty vector counterparts at both 72 and 96 h post treatment (P = 1.02 × 10−5 and P = 1.25 × 10−13, respectively). In SY5Y, although differentiation took longer and both SHANK2+ cells and controls eventually reach 100% confluence with vehicle alone, SHANK2+ cells still decreased confluence (P = 1.69 × 10−6) (Supplemental Fig. S25B). In analyzing total neurite outgrowth without normalization for cell body area, SY5Y ATRA-treated SHANK2+ cells outpaced controls starting at hour 144 post treatment and continued to lead until the experiment ended, with a total neurite measurement increased 1.55-fold over controls (P = 1.62 × 10−35) (Supplemental Fig. S25D). Once normalized, SHANK2+ cells have higher measured outgrowth starting at 75 h post treatment through hour 96, and maintain from there. At 195 h past treatment, SHANK2+ cells treated with 5 µM ATRA displayed neurites at 1.71-fold increase over their empty vector controls (P = 2.36 × 10−17) (Fig. 6M–O). Taken together, these data suggest SHANK2 is a newly identified haplo-insufficient tumor suppressor in high-risk neuroblastoma that is disrupted by recurrent somatic structural variation in the MYCN nonamplified subset of cases.
Discussion
Sequencing studies of neuroblastoma tumors have revealed a relatively low SNV burden and limited mutational landscape (Cheung et al. 2012; Molenaar et al. 2012; Pugh et al. 2013; Sausen et al. 2013), leaving aneuploidy and large segmental chromosomal alterations as the main candidate driver mutations in many tumors. Structural variations (insertions, deletions, duplications and translocations, and inversions) can also function as potent cancer drivers, as shown with the discovery of rearrangements near the TERT gene driving aberrant telomerase expression in many high-risk neuroblastomas (Peifer et al. 2015; Valentijn et al. 2015). Here, we expand the landscape and understanding of structural variation in neuroblastoma through an integrative genomic analysis of a large cohort of patient samples profiled by whole-genome sequencing and SNP arrays together with additional transcriptional data. To the best of our knowledge, this study represents the largest integrated genome-wide survey of structural variation in neuroblastoma including alignment-based (SJ-BP) and copy number–based (RD-BP and CN-BP) structural variation breakpoint analyses. This study uses human genome build hg19 based on CGI data analyses; however, use of GRCh38 would not significantly alter the results. Detection of smaller structural variants is limited for short-read sequencing and SNP array platforms used in this study. Complete Genomics short-read sequencing technology is restricted to SVs with sizes >200 bp for duplications and inversions and ∼500 bp for deletions. In addition, Complete Genomics technology does not provide information about the allelic fraction at which structural variants are identified. Future studies are warranted to address these issues, ideally using long-read sequencing technologies.
In the current study, structural variation complexity is most evident in high-risk tumors without amplification of MYCN (HR-NA). This observation is consistent with other reports of increased chromosomal instability in this high-risk subset (Caren et al. 2010). The current study strengthens and extends previous reports of chromosomal instability by including additional structural variation types and breakpoint burden analyses. Specifically, we observed significantly more tandem duplications HR-NA tumors. These affected nearly every chromosome, except for Chr 2, which contains the MCYN amplicon. HR-NA tumors harbored more SVs in known cancer genes as well as novel genes. In contrast, the SNV burden was very similar between MNA and HR-NA groups. As suggested by pancancer studies, the underlying mechanisms potentiating chromosomal instability and somatic SNV burden may differ (Ciriello et al. 2013). The mechanism leading to the observed increase in chromosomal instability in HR-NA tumors remains unknown. Pancancer studies have reported TP53 mutations as a major driver of chromosomal instability; however, TP53 loss of function is rarely observed in neuroblastoma at diagnosis.
Chromothripsis has been reported in as many as 18% of high-stage neuroblastomas (Molenaar et al. 2012). Similarly, in the current study, 19% of high-risk tumors from the TARGET cohort showed chromothripsis (N = 20/105), involving a total of 27 chromosomal regions. These events largely overlap with amplification of MYCN (as well as some ALK cases) on Chr 2p and TERT on Chr 5p, suggesting an important role of chromothripsis followed by purifying selection as an underlying cause of those alterations. We also observed high breakpoint density in the Chr X of females based on the SNP array data, which could be explained by higher tolerance to chromothripsis in diploid regions. Future studies are required to determine whether the oncogenic role of chromothripsis represents an opportunity for therapeutic intervention.
Along this study, we report a shared repertoire of genes altered in neuroblastoma and neurodevelopmental disorders (NDD), including autism spectrum disorder. A link between cancer and autism has been previously established in PTEN-associated germline syndromes (Goffin et al. 2001), and multiple autism susceptibility genes also have a known role in cancer (Crawley et al. 2016). Moreover, certain germline deletions associated with NDD, such as 10p15 (DeScipio et al. 2012) and 16p24.3 (Willemsen et al. 2010), are reported here to occur somatically in neuroblastoma. In addition, we have recently discovered that individuals with germline deletions of 16p11.2 microdeletions, also associated with NDDs, have increased risk of developing neuroblastoma (Egolf et al. 2019). Transcriptomic analyses have shown that neural lineage pathways are commonly down-regulated in high-risk neuroblastomas compared to low-risk signatures (Fredlund et al. 2008). Here, we show that structural variation preferentially disrupts neurodevelopmental genes in neuroblastoma. We hypothesize that structural variations in SHANK2 and other coding proteins of the postsynaptic density (PSD) comprise novel neuroblastoma candidate tumor suppressors involved in neuronal differentiation. Additional candidate genes identified here with a proposed role in neurotransmission and synapsis and involvement in autism include DLG2, AUTS2, CNTNAP2, NRXN1, and CTNND2 (Gai et al. 2012). Structural variants affecting these genes are more prevalent in high-risk neuroblastomas without amplification of MYCN, which is itself a potent driver of dedifferentiation (Westermark et al. 2011).
We propose that disruption and deregulation of SHANK2 promotes the undifferentiated state of neuroblastoma cells, and that other synaptic genes may play a similar role in this childhood cancer. Synaptogenesis is a key process in neuronal differentiation and mutations in the SHANK family of proteins are frequently implicated in NDD (also termed shankopathies), offering potential therapeutic opportunities for these disorders (Wang et al. 2014b). In the current study, we show that SHANK2 is disrupted by recurrent somatic SVs in HR-NA tumors and that SHANK2 expression is low across high-risk tumors. The mechanism driving low SHANK2 levels in the MNA high-risk tumors remains to be identified. We further show that decreased SHANK2 expression is associated with poor survival in neuroblastoma, even within the low- and intermediate-risk subsets of patients that typically have good outcomes. This suggests that SHANK2 expression, or the undifferentiated cell state, may serve as a biomarker for non-high-risk tumors requiring more aggressive treatment. Our in vitro studies show that forced expression of SHANK2 reduces cell growth and increases neurite outgrowth (indicative of differentiation) in human-derived neuroblastoma cell lines exposed to ATRA. Given that retinoids are currently used as maintenance therapy in high-risk neuroblastoma standard of care (Matthay et al. 1999, 2009), the sensitizing effect of SHANK2 expression to ATRA treatment underscores the importance of understanding the mechanisms driving and maintaining the undifferentiated phenotype of high-risk neuroblastoma. Subsequent studies with larger cohorts should evaluate the role of mutated and deregulated neurodevelopmental genes in retinoic acid treatment response. Taken together, we depict a substantially expanded landscape of structural variation in neuroblastoma and provide mechanistic insight into the aberrant neuronal development hallmark of the high-risk form of this childhood cancer.
Methods
Sequencing and array data availability and processing
All TARGET DNA and RNA-sequencing data analyzed in this study are available in the NCBI database of Genotypes and Phenotypes (dbGaP; https://www.ncbi.nlm.nih.gov/gap/) under study ID phs000218/phs000467. In addition, WGS processed data files from the Cancer Pipeline 2.0 (Complete Genomics: http://www.completegenomics.com/documents/DataFileFormats_Cancer_Pipeline_2.0.pdf) including coverage info (“somaticCnvDetailsDiploidBeta-[ASM-ID]-N1.tsv”), structural variant calls (“somaticAllJunctionsBeta-[ASM-ID]-N1.tsv”), and small variants (“masterVarBeta-[ASM-ID]-T1.tsv”) can be downloaded from the TARGET data matrix (https://target-data.nci.nih.gov/Controlled/NBL/WGS/CGI/). The SEQC RNA-sequencing data set analyzed in this study is available through the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo) under accession GSE62564. The cell line RNA-sequencing and SNP genotyping array data analyzed in this study are available through GEO at accessions GSE89413 and GSE89968, respectively. We applied further processing of these files to obtain CNV segmentation data, filtered structural variants, and filtered SNVs. Custom R language scripts (v3.4.0; R Core Team 2017) were used to annotate recurrently altered genes and mutational burden (Supplemental Code); further details are provided in Supplemental Methods. We have organized all downstream analysis code and resulting processed data in a dedicated public GitHub repository (https://github.com/diskin-lab-chop/NB_structural_variants) (see Data access).
Generating DNA copy number segmentation from SNP arrays
We genotyped 914 patient tumor samples using Illumina SNP arrays (HumanHap550, Human610-Quad, and HumanOmniExpress). Intensities were analyzed using GenomeStudio to obtain Log R Ratio (LRR) and B-allele frequencies (BAF). GC content bias correction was applied for the common set of 316,210 SNPs. Segmentation was obtained using the SNPRank algorithm in Nexus Copy Number version 8.0 (BioDiscovery), which implements circular binary segmentation (CBS) version 3.0.
Gene fusion analysis
Gene fusion analysis from RNA-seq data was studied using three available tools: STAR-fusion (Dobin et al. 2013), fusionCATCHER (Nicorici et al. 2014), and DeFUSE (McPherson et al. 2011). We then collected fusion events that matched inter-chromosomal events from the CGI SV calls.
Statistical and survival analyses
Statistics performed on genomic data including Wilcoxon rank-sum test, Kruskal–Wallis test, and survival analyses were performed using R programming. For the survival analyses, we used the CRAN “survival” library (http://cran.r-project.org/web/packages/survival/). To assess the association between SHANK2 long isoform expression and survival, we obtained the optimal separation (lower log-rank test P-value) from all possible expression thresholds and then used Benjamini and Hochberg (false discovery rate) for multiple testing correction and Q-value estimation.
Sanger sequence validation
Primers were designed based on sequence junction breakpoints using Primer3. PCR reactions were then carried out on 25 ng of DNA using optimized conditions for each reaction. Products with multiple bands were run out using the remaining sample, bands of interest were excised, and the DNA extracted using MinElute Gel Extraction Kit from Qiagen. Products with single bands were cleaned and prepared for sequencing using the MinElute PCR Purification Kit (Qiagen). Samples were then sequenced with 2 pM of the same primer used to create amplicon.
Cell culture
Cells were grown in RPMI-1640 with HEPES, L-glutamine, and phenol red (catalog 22400-089), supplemented with 10% Fetal Bovine Serum, 1% antibiotic-antimycotic (catalog 15240-062), and 1% L-glutamine (catalog 25-005-CI) in 5% CO2 at 37°C in the dark. Transduced cells also had the appropriate concentration of puromycin in media for selection.
Lentivirus infection
Lentiviral vector plasmid for the long isoform of SHANK2 (NM_012309) was obtained commercially from GeneCopoeia (EX-H5274-Lv105). Empty vector control plasmid pLv105 was originally from GeneCopoeia. Creation of the virus media was accomplished using Lipofectamine 3000 applied to 293TN cells with packaging plasmid psPAX2, envelope plasmid pMD2.g, and the Lentiviral backbone plasmid containing the ORF for NM_012309 or empty vector. Infectious viral media was pooled over 2 d then filtered through 0.45 μm nitrocellulose and combined with polybrene at 8 μg/mL media and applied to cells. Following infection, transduced cells were selected with puromycin in line-dependent concentrations.
Growth and proliferation assay using RT-CES
Cells were plated in 96-well RTCES microelectronic sensor arrays (ACEA Biosciences). Density measurements were made every hour. Cell densities were normalized to 5 h post plating.
Cell viability assays
Cells were plated in clear-bottomed, 96-well plates in 200 μL media and allowed to grow under normal conditions for either 4 or 7 d. Before reading, 100 μL media was replaced with equal volume of CellTiter-Glo reagent and read on a GloMax Multidetection instrument (Promega). Arbitrary luminescence units were normalized to empty vector-transduced controls and results expressed as percentages of control levels from the same assay.
ATRA-induced differentiation
Cells were plated in normal media at optimized densities for each parental line in 96-well plates and allowed 24–48 h to firmly attach to plates. Media was then switched for low-serum media containing either 1% or 3% FBS and allowed 24 h to equilibrate, after which it was replaced with low-serum media supplemented with varying concentrations of ATRA (all-trans-retinoic-acid, Sigma-Aldrich R2625) or vehicle (DMSO) alone, in volume corresponding to the highest concentration of ATRA for each experiment. Plates were then left in normal growth conditions and protected from light. RA media was refreshed every 72 h to prevent oxidation. Plates were placed in an IncuCyte ZOOM instrument to use live cell imaging. Each well was imaged every 4 h, and the Incucyte Neurotrack software module was used to quantify neurite outgrowth.
Protein isolation and western blotting
Whole cell lysates were created by applying denaturing lysis buffer containing protease/phosphatase inhibitors (Cell Signaling Technology 5872) to cells on ice and allowing lysis for 30 min. The total sample was sonicated for 5 sec and spun at max speed in a microcentrifuge for 15 min at 4°C before collecting supernatant to clean tube. Quantification of protein was done using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific 23227). Protein was loaded on 4%–12% Tris-Glycine gels, transferred to PVDF membrane, and probed with antibodies in 5% milk in TBST. Antibody stripping used Restore Stripping buffer (Thermo Fisher Scientific 21059). Detection of HRP-conjugated secondary antibodies used SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific 34096). Primary and secondary antibodies used were the following: Actin Beta (Santa Cruz Biotechnology sc-47778), used at 1:2500; SHANK2 (Santa Cruz Biotechnology sc-271834), used at 1:1500; and Goat anti-mouse HRP (Santa Cruz Biotechnology sc-2005), used at 1:2500.
Data access
All raw and processed SNP genotyping array data generated in this study was submitted to NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE131189. In addition to these resources, custom scripts used in this study are available as Supplemental Code and at a dedicated public GitHub repository (https://github.com/diskin-lab-chop/NB_structural_variants).
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health (NIH) grants R01-CA124709 (S.J.D.), R35-CA220500 (J.M.M.), the Roberts Collaborative Forefront Award (G.L.), and the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. This project was also funded in part by a supplement to the Children's Oncology Group Chair's grant CA098543 and with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E to S.J.D. and Complete Genomics.
Author contributions: S.J.D designed the experiment. G.L. and S.J.D. drafted the manuscript. G.L. and S.J.D. performed analyses of SVs from WGS. G.L. performed RNA data analysis. G.L. and A.M. performed de novo transcript analyses. G.L. and K.S.R. performed fusion transcript analyses. K.L.C. and M.D. performed Sanger sequencing. K.L.C., M.D., L.M.F., and E.H. performed SHANK2 experiments. Z.V. assisted with sequence data analysis. J.S.W. and J.K. generated RNA-sequencing data. S.A. and R.C.S. generated array-based expression data. All authors commented on or contributed to the current manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.252106.119.
References
- Alkan C, Coe BP, Eichler EE. 2011. Genome structural variation discovery and genotyping. Nat Rev Genet 12: 363–376. 10.1038/nrg2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attiyeh EF, London WB, Mossé YP, Wang Q, Winter C, Khazi D, McGrady PW, Seeger RC, Look AT, Shimada H, et al. 2005. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N Engl J Med 353: 2243–2253. 10.1056/NEJMoa052399 [DOI] [PubMed] [Google Scholar]
- Bosse KR, Raman P, Zhu Z, Lane M, Martinez D, Heitzeneder S, Rathi KS, Kendsersky NM, Randall M, Donovan L, et al. 2017. Identification of GPC2 as an oncoprotein and candidate immunotherapeutic target in high-risk neuroblastoma. Cancer Cell 32: 295–309.e12. 10.1016/j.ccell.2017.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bown N, Cotterill S, Łastowska M, O'Neill S, Pearson AD, Plantaz D, Meddeb M, Danglot G, Brinkschmidt C, Christiansen H, et al. 1999. Gain of chromosome arm 17q and adverse outcome in patients with neuroblastoma. N Engl J Med 340: 1954–1961. 10.1056/NEJM199906243402504 [DOI] [PubMed] [Google Scholar]
- Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM. 1984. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science 224: 1121–1124. 10.1126/science.6719137 [DOI] [PubMed] [Google Scholar]
- Caren H, Kryh H, Nethander M, Sjoberg RM, Trager C, Nilsson S, Abrahamsson J, Kogner P, Martinsson T. 2010. High-risk neuroblastoma tumors with 11q-deletion display a poor prognostic, chromosome instability phenotype with later onset. Proc Natl Acad Sci 107: 4323–4328. 10.1073/pnas.0910684107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron H, van Sluis P, de Kraker J, Bökkerink J, Egeler M, Laureys G, Slater R, Westerveld A, Voûte PA, Versteeg R. 1996. Allelic loss of chromosome 1p as a predictor of unfavorable outcome in patients with neuroblastoma. N Engl J Med 334: 225–230. 10.1056/NEJM199601253340404 [DOI] [PubMed] [Google Scholar]
- Carter NP. 2007. Methods and strategies for analyzing copy number variation using DNA microarrays. Nat Genet 39: S16–S21. 10.1038/ng2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Bardes EE, Aronow BJ, Jegga AG. 2009. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37: W305–W311. 10.1093/nar/gkp427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung NK, Zhang J, Lu C, Parker M, Bahrami A, Tickoo SK, Heguy A, Pappo AS, Federico S, Dalton J, et al. 2012. Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA 307: 1062–1071. 10.1001/jama.2012.228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N, Sander C. 2013. Emerging landscape of oncogenic signatures across human cancers. Nat Genet 45: 1127–1133. 10.1038/ng.2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn SL, Pearson AD, London WB, Monclair T, Ambros PF, Brodeur GM, Faldum A, Hero B, Iehara T, Machin D, et al. 2009. The International Neuroblastoma Risk Group (INRG) classification system: an INRG Task Force report. J Clin Oncol 27: 289–297. 10.1200/JCO.2008.16.6785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley JN, Heyer WD, LaSalle JM. 2016. Autism and cancer share risk genes, pathways, and drug targets. Trends Genet 32: 139–146. 10.1016/j.tig.2016.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeScipio C, Conlin L, Rosenfeld J, Tepperberg J, Pasion R, Patel A, McDonald MT, Aradhya S, Ho D, Goldstein J, et al. 2012. Subtelomeric deletion of chromosome 10p15.3: clinical findings and molecular cytogenetic characterization. Am J Med Genet A 158A: 2152–2161. 10.1002/ajmg.a.35574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyell RJ, Attiyeh EF. 2011. Advances in the understanding of constitutional and somatic genomic alterations in neuroblastoma. Cancer Genet 204: 113–121. 10.1016/j.cancergen.2011.03.001 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG, Carnevali P, Nazarenko I, Nilsen GB, Yeung G, et al. 2010. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 327: 78–81. 10.1126/science.1181498 [DOI] [PubMed] [Google Scholar]
- Egolf LE, Vaksman Z, Lopez G, Rokita JL, Modi A, Basta PV, Hakonarson H, Olshan AF, Diskin SJ. 2019. Germline 16p11.2 microdeletion predisposes to neuroblastoma. Am J Hum Genet 105: 658–668. 10.1016/j.ajhg.2019.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredlund E, Ringnér M, Maris JM, Påhlman S. 2008. High Myc pathway activity and low stage of neuronal differentiation associate with poor outcome in neuroblastoma. Proc Natl Acad Sci 105: 14094–14099. 10.1073/pnas.0804455105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai X, Xie HM, Perin JC, Takahashi N, Murphy K, Wenocur AS, D'Arcy M, O'Hara RJ, Goldmuntz E, Grice DE, et al. 2012. Rare structural variation of synapse and neurotransmission genes in autism. Mol Psychiatry 17: 402–411. 10.1038/mp.2011.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Berthold F, Edler L, Schwab M, Amler LC. 1995. The 1p deletion is not a reliable marker for the prognosis of patients with neuroblastoma. Cancer Res 55: 5366–5369. [PubMed] [Google Scholar]
- Gilbert F, Feder M, Balaban G, Brangman D, Lurie DK, Podolsky R, Rinaldt V, Vinikoor N, Weisband J. 1984. Human neuroblastomas and abnormalities of chromosomes 1 and 17. Cancer Res 44: 5444–5449. [PubMed] [Google Scholar]
- Goffin A, Hoefsloot LH, Bosgoed E, Swillen A, Fryns JP. 2001. PTEN mutation in a family with Cowden syndrome and autism. Am J Med Genet 105: 521–524. 10.1002/ajmg.1477 [DOI] [PubMed] [Google Scholar]
- Guo C, White PS, Weiss MJ, Hogarty MD, Thompson PM, Stram DO, Gerbing R, Matthay KK, Seeger RC, Brodeur GM, et al. 1999. Allelic deletion at 11q23 is common in MYCN single copy neuroblastomas. Oncogene 18: 4948–4957. 10.1038/sj.onc.1202887 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Wang Y, Brudno M. 2012. PRISM: pair-read informed split-read mapping for base-pair level detection of insertion, deletion and structural variants. Bioinformatics 28: 2576–2583. 10.1093/bioinformatics/bts484 [DOI] [PubMed] [Google Scholar]
- Kawashima M, Kojima M, Ueda Y, Kurihara S, Hiyama E. 2016. Telomere biology including TERT rearrangements in neuroblastoma: a useful indicator for surgical treatments. J Pediatr Surg 51: 2080–2085. 10.1016/j.jpedsurg.2016.09.042 [DOI] [PubMed] [Google Scholar]
- Kurihara S, Hiyama E, Onitake Y, Yamaoka E, Hiyama K. 2014. Clinical features of ATRX or DAXX mutated neuroblastoma. J Pediatr Surg 49: 1835–1838. 10.1016/j.jpedsurg.2014.09.029 [DOI] [PubMed] [Google Scholar]
- Łastowska M, Cotterill S, Bown N, Cullinane C, Variend S, Lunec J, Strachan T, Pearson ADJ, Jackson MS. 2002. Breakpoint position on 17q identifies the most aggressive neuroblastoma tumors. Genes Chromosomes Cancer 34: 428–436. 10.1002/gcc.10089 [DOI] [PubMed] [Google Scholar]
- Ma X, Edmonson M, Yergeau D, Muzny DM, Hampton OA, Rusch M, Song G, Easton J, Harvey RC, Wheeler DA, et al. 2015. Rise and fall of subclones from diagnosis to relapse in pediatric B-acute lymphoblastic leukaemia. Nat Commun 6: 6604 10.1038/ncomms7604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Liu Y, Liu Y, Alexandrov LB, Edmonson MN, Gawad C, Zhou X, Li Y, Rusch MC, Easton J, et al. 2018. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature 555: 371–376. 10.1038/nature25795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macintyre G, Ylstra B, Brenton JD. 2016. Sequencing structural variants in cancer for precision therapeutics. Trends Genet 32: 530–542. 10.1016/j.tig.2016.07.002 [DOI] [PubMed] [Google Scholar]
- Maher CA, Wilson RK. 2012. Chromothripsis and human disease: piecing together the shattering process. Cell 148: 29–32. 10.1016/j.cell.2012.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris JM. 2010. Recent advances in neuroblastoma. N Engl J Med 362: 2202–2211. 10.1056/NEJMra0804577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris JM, Guo C, Blake D, White PS, Hogarty MD, Thompson PM, Rajalingam V, Gerbing R, Stram DO, Matthay KK, et al. 2001. Comprehensive analysis of chromosome 1p deletions in neuroblastoma. Med Pediatr Oncol 36: 32–36. [DOI] [PubMed] [Google Scholar]
- Matthay KK, Villablanca JG, Seeger RC, Stram DO, Harris RE, Ramsay NK, Swift P, Shimada H, Black CT, Brodeur GM, et al. 1999. Treatment of high-risk neuroblastoma with intensive chemotherapy, radiotherapy, autologous bone marrow transplantation, and 13-cis-retinoic acid. N Engl J Med 341: 1165–1173. 10.1056/NEJM199910143411601 [DOI] [PubMed] [Google Scholar]
- Matthay KK, Reynolds CP, Seeger RC, Shimada H, Adkins ES, Haas-Kogan D, Gerbing RB, London WB, Villablanca JG. 2009. Long-term results for children with high-risk neuroblastoma treated on a randomized trial of myeloablative therapy followed by 13-cis-retinoic acid: a children's oncology group study. J Clin Oncol 27: 1007–1013. 10.1200/JCO.2007.13.8925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson A, Hormozdiari F, Zayed A, Giuliany R, Ha G, Sun MG, Griffith M, Heravi Moussavi A, Senz J, Melnyk N, et al. 2011. deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol 7: e1001138 10.1371/journal.pcbi.1001138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. 2011. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 12: R41 10.1186/gb-2011-12-4-r41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels E, Vandesompele J, De Preter K, Hoebeeck J, Vermeulen J, Schramm A, Molenaar JJ, Menten B, Marques B, Stallings RL, et al. 2007. ArrayCGH-based classification of neuroblastoma into genomic subgroups. Genes Chromosomes Cancer 46: 1098–1108. 10.1002/gcc.20496 [DOI] [PubMed] [Google Scholar]
- Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, Hamdi M, van Nes J, Westerman BA, van Arkel J, et al. 2012. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 483: 589–593. 10.1038/nature10910 [DOI] [PubMed] [Google Scholar]
- Mosse YP, Greshock J, Margolin A, Naylor T, Cole K, Khazi D, Hii G, Winter C, Shahzad S, Asziz MU, et al. 2005. High-resolution detection and mapping of genomic DNA alterations in neuroblastoma. Genes Chromosomes Cancer 43: 390–403. 10.1002/gcc.20198 [DOI] [PubMed] [Google Scholar]
- Nicorici D, Satalan M, Edgren H, Kangaspeska S, Murumägi A, Kallioniemi O, Virtanen S, Kilkku O. 2014. FusionCatcher—a tool for finding somatic fusion genes in paired-end RNA-sequencing data. bioRxiv 10.1101/011650. [DOI] [Google Scholar]
- Peifer M, Hertwig F, Roels F, Dreidax D, Gartlgruber M, Menon R, Krämer A, Roncaioli JL, Sand F, Heuckmann JM, et al. 2015. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature 526: 700–704. 10.1038/nature14980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plantaz D, Mohapatra G, Matthay KK, Pellarin M, Seeger RC, Feuerstein BG. 1997. Gain of chromosome 17 is the most frequent abnormality detected in neuroblastoma by comparative genomic hybridization. Am J Pathol 150: 81–89. [PMC free article] [PubMed] [Google Scholar]
- Pugh TJ, Morozova O, Attiyeh EF, Asgharzadeh S, Wei JS, Auclair D, Carter SL, Cibulskis K, Hanna M, Kiezun A, et al. 2013. The genetic landscape of high-risk neuroblastoma. Nat Genet 45: 279–284. 10.1038/ng.2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Zhao F. 2011. inGAP-sv: a novel scheme to identify and visualize structural variation from paired end mapping data. Nucleic Acids Res 39: W567–W575. 10.1093/nar/gkr506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2017. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: https://www.R-project.org/. [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santo EE, Ebus ME, Koster J, Schulte JH, Lakeman A, van Sluis P, Vermeulen J, Gisselsson D, Øra I, Lindner S, et al. 2012. Oncogenic activation of FOXR1 by 11q23 intrachromosomal deletion-fusions in neuroblastoma. Oncogene 31: 1571–1581. 10.1038/onc.2011.344 [DOI] [PubMed] [Google Scholar]
- Sausen M, Leary RJ, Jones S, Wu J, Reynolds CP, Liu X, Blackford A, Parmigiani G, Diaz LA, Papadopoulos N, et al. 2013. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat Genet 45: 12–17. 10.1038/ng.2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiermacher G, Mosseri V, London WB, Maris JM, Brodeur GM, Attiyeh E, Haber M, Khan J, Nakagawara A, Speleman F, et al. 2012. Segmental chromosomal alterations have prognostic impact in neuroblastoma: a report from the INRG project. Br J Cancer 107: 1418–1422. 10.1038/bjc.2012.375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger RC, Brodeur GM, Sather H, Dalton A, Siegel SE, Wong KY, Hammond D. 1985. Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. N Engl J Med 313: 1111–1116. 10.1056/NEJM198510313131802 [DOI] [PubMed] [Google Scholar]
- Shao YW, Wood GA, Lu J, Tang QL, Liu J, Molyneux S, Chen Y, Fang H, Adissu H, McKee T, et al. 2019. Cross-species genomics identifies DLG2 as a tumor suppressor in osteosarcoma. Oncogene 38: 291–298. 10.1038/s41388-018-0444-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smida J, Xu H, Zhang Y, Baumhoer D, Ribi S, Kovac M, von Luettichau I, Bielack S, O'Leary VB, Leib-Mosch C, et al. 2017. Genome-wide analysis of somatic copy number alterations and chromosomal breakages in osteosarcoma. Int J Cancer 141: 816–828. 10.1002/ijc.30778 [DOI] [PubMed] [Google Scholar]
- Stallings RL, Nair P, Maris JM, Catchpoole D, McDermott M, O'Meara A, Breatnach F. 2006. High-resolution analysis of chromosomal breakpoints and genomic instability identifies PTPRD as a candidate tumor suppressor gene in neuroblastoma. Cancer Res 66: 3673–3680. 10.1158/0008-5472.CAN-05-4154 [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattini L, D'Aurizio R, Magi A. 2015. Detection of genomic structural variants from next-generation sequencing data. Front Bioeng Biotechnol 3: 92 10.3389/fbioe.2015.00092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubio JM. 2015. Somatic structural variation and cancer. Brief Funct Genomics 14: 339–351. 10.1093/bfgp/elv016 [DOI] [PubMed] [Google Scholar]
- Valentijn LJ, Koster J, Zwijnenburg DA, Hasselt NE, van Sluis P, Volckmann R, van Noesel MM, George RE, Tytgat GA, Molenaar JJ, et al. 2015. TERT rearrangements are frequent in neuroblastoma and identify aggressive tumors. Nat Genet 47: 1411–1414. 10.1038/ng.3438 [DOI] [PubMed] [Google Scholar]
- Wang Q, Diskin S, Rappaport E, Attiyeh E, Mosse Y, Shue D, Seiser E, Jagannathan J, Shusterman S, Bansal M, et al. 2006. Integrative genomics identifies distinct molecular classes of neuroblastoma and shows that multiple genes are targeted by regional alterations in DNA copy number. Cancer Res 66: 6050–6062. 10.1158/0008-5472.CAN-05-4618 [DOI] [PubMed] [Google Scholar]
- Wang C, Gong B, Bushel PR, Thierry-Mieg J, Thierry-Mieg D, Xu J, Fang H, Hong H, Shen J, Su Z, et al. 2014a. The concordance between RNA-seq and microarray data depends on chemical treatment and transcript abundance. Nat Biotechnol 32: 926–932. 10.1038/nbt.3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Bey AL, Chung L, Krystal AD, Jiang YH. 2014b. Therapeutic approaches for shankopathies. Dev Neurobiol 74: 123–135. 10.1002/dneu.22084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermark UK, Wilhelm M, Frenzel A, Henriksson MA. 2011. The MYCN oncogene and differentiation in neuroblastoma. Semin Cancer Biol 21: 256–266. 10.1016/j.semcancer.2011.08.001 [DOI] [PubMed] [Google Scholar]
- Willemsen MH, Fernandez BA, Bacino CA, Gerkes E, de Brouwer AP, Pfundt R, Sikkema-Raddatz B, Scherer SW, Marshall CR, Potocki L, et al. 2010. Identification of ANKRD11 and ZNF778 as candidate genes for autism and variable cognitive impairment in the novel 16q24.3 microdeletion syndrome. Eur J Hum Genet 18: 429–435. 10.1038/ejhg.2009.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Luquette LJ, Gehlenborg N, Xi R, Haseley PS, Hsieh CH, Zhang C, Ren X, Protopopov A, Chin L, et al. 2013. Diverse mechanisms of somatic structural variations in human cancer genomes. Cell 153: 919–929. 10.1016/j.cell.2013.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wu Y. 2011. SVseq: an approach for detecting exact breakpoints of deletions with low-coverage sequence data. Bioinformatics 27: 3228–3234. 10.1093/bioinformatics/btr563 [DOI] [PubMed] [Google Scholar]
- Zhao M, Wang Q, Wang Q, Jia P, Zhao Z. 2013. Computational tools for copy number variation (CNV) detection using next-generation sequencing data: features and perspectives. BMC Bioinformatics 14 (Suppl 11): S1 10.1186/1471-2105-14-S11-S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman MW, Liu Y, He S, Durbin AD, Abraham BJ, Easton J, Shao Y, Xu B, Zhu S, Zhang X, et al. 2018. MYC drives a subset of high-risk pediatric neuroblastomas and is activated through mechanisms including enhancer hijacking and focal enhancer amplification. Cancer Discov 8: 320–335. 10.1158/2159-8290.CD-17-0993 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.