Abstract
The higher-order structural organization and dynamics of the chromosomes play a central role in gene regulation. To explore this structure–function relationship, it is necessary to directly visualize genomic elements in living cells. Genome imaging based on the CRISPR system is a powerful approach but has limited applicability due to background signals and nonspecific aggregation of fluorophores within nuclei. To address this issue, we developed a novel visualization scheme combining tripartite fluorescent proteins with the SunTag system and demonstrated that it strongly suppressed background fluorescence and amplified locus-specific signals, allowing long-term tracking of genomic loci. We integrated the multicomponent CRISPR system into stable cell lines to allow quantitative and reliable analysis of dynamic behaviors of genomic loci. Due to the greatly elevated signal-to-background ratio, target loci with only small numbers of sequence repeats could be successfully tracked, even under a conventional fluorescence microscope. This feature enables the application of CRISPR-based imaging to loci throughout the genome and opens up new possibilities for the study of nuclear processes in living cells.
Eukaryotic chromosomes are compacted inside the cell nucleus, within which they are organized in nonrandom positions (Bickmore 2013). The spatiotemporal organization of chromosomes results in a hierarchy of structural features, such as chromosome territories, topologically associated domains, and chromatin loops, which were suggested to play essential roles in regulating gene expression; however, it remains to be elucidated how the structure and dynamics of chromosomes regulate nuclear processes such as transcription, replication, and DNA repair (Misteli 2007; Gilbert et al. 2010; Bickmore 2013; Gibcus and Dekker 2013; Dixon et al. 2016). Visualization of the dynamics of chromosomes in living cells should yield major advances in understanding how chromosomes dynamically control genomic functions. However, currently available techniques do not enable us to track arbitrary genomic loci of interest.
Previously, genomic loci in living cells were visualized by integrating repeats of protein-binding DNA sequences such as lacO or tetO array into target sites (Robinett et al. 1996; Roukos et al. 2013) by targeting unaltered genomic loci with programmable DNA-binding systems such as transcription activator-like effectors (Ma et al. 2013; Miyanari et al. 2013) or, more recently, by the use of the CRISPR system (Chen et al. 2013, 2016; Anton et al. 2014; Deng et al. 2015; Ma et al. 2015, 2016, 2018a; Fu et al. 2016; Qin et al. 2017; Duan et al. 2018; Hong et al. 2018; Maass et al. 2018a). CRISPR-based genome imaging techniques utilize the sequence-specific target binding ability of a Cas9 variant with disabled restriction activity (dCas9) (Chen et al. 2013). This approach was extended to multilocus chromosome imaging by taking advantage of orthogonal dCas9 variants (Ma et al. 2015; Chen et al. 2016) and engineered sgRNA scaffolds (Cheng et al. 2016; Fu et al. 2016; Ma et al. 2016; Shao et al. 2016; Wang et al. 2016b). In addition, simultaneous tracking of genomic loci and transcription activity revealed the correlation between gene activation and chromosome mobility in embryonic stem cells (Ochiai et al. 2015).
Although CRISPR-based approaches show promising capabilities, they are limited in their practical application to a broad range of genomic targets. First, they require target loci to contain a large number of sequence repeats to obtain a signal-to-background ratio (S/B) high enough to rule out signals from unassociated fluorophores. This limits their applicability to arbitrary loci unless we first insert large repeat sequences at sites of interest. Techniques were developed to target nonrepeat genomic loci by packaging an array of multiple sgRNAs (Gu et al. 2018); nonetheless, the requirement for large numbers of CRISPR targets for reliable locus detection makes it difficult to perform live visualization of the genome without interfering with its dynamics. Second, protein-based fluorophores commonly exhibit nonuniform background and nonspecific speckles due to heterogeneous nuclear architecture and aggregation-prone proteins (Chen et al. 2013; Ma et al. 2015, 2016). Utilizing an sgRNA scaffold for fluorescent labeling decreases the amount of aggregation relative to direct fusion of dCas9 to a fluorescent protein (Ma et al. 2016) but, like dCas9, RNA-binding proteins also preferentially localize in nucleoli or Cajal bodies (Chen et al. 2013, 2016; Fu et al. 2016; Ma et al. 2016). Such behavior hampers the identification of target-specific foci, especially when we do not know the number of foci to expect, e.g., in studies of copy number variation, chromosomal replication, or temporary association between loci.
We envisioned that a repeating array of fluorophores could increase site-specific signals and reduce the required number of repeat sequences. Similar approaches were used to detect single protein or mRNA molecules in living cells (Bertrand et al. 1998; Tanenbaum et al. 2014; Morisaki et al. 2016; Wang et al. 2016a; Wu et al. 2016; Yan et al. 2016). However, this would also result in an elevated background level, as the unassociated dCas9 or sgRNA would emit equally strong signals. Tetracycline-inducible expression systems were used to control the expression levels of proteins, followed by selection of clonal cells expressing the optimal levels of fluorescent proteins for subsequent genomic imaging (Chen et al. 2013, 2016; Fu et al. 2016; Shao et al. 2016). This method suppresses the background signal by depleting unbound fluorophores, but it is still not ideal because the expression level may not be homogeneous among the population of cells. In addition, the foci photobleach quickly due to the lack of replacement fluorophores, and the method is not applicable to cases in which the number of target-specific foci varies over time. Thus, we pursued reliable detection and long-term tracking of target loci by suppressing background signals through engineering self-associating split superfolder green fluorescent protein (sfGFP) into the CRISPR-dCas9 system (Cabantous et al. 2005) and by integrating the SunTag system to amplify the signal. From this development, we aimed at tracking loci with small numbers of sequence repeats and ultimately tracking loci with nonrepeat sequences, in order to extend CRISPR-based genome imaging to loci throughout the genome.
Results
Design and optimization of the split fluorophore CRISPR system for background-suppressed genome imaging
As a reference for comparison, we tested the original CRISPR-dCas9 imaging system, in which nuclease-deactivated SpCas9 (dCas9) is directly fused to EGFP, targeting a pericentromeric region on Chromosome 9 (C9-1) (Grady et al. 1992; Sauter et al. 1995; Ma et al. 2015) in AD-293 cells. This approach showed sgRNA-dependent foci but also had a nonuniform background (Supplemental Fig. 1A). Even in the absence of sgRNA, speckles from the nonspecific aggregation of dCas9-EGFP were observed. This is a known issue with the CRISPR-dCas9-EGFP imaging system, which requires protein expression level to be precisely controlled by establishing clonal cell lines (Chen et al. 2013, 2016; Fu et al. 2016; Shao et al. 2016), limiting the applicability of CRISPR-based genome imaging. To suppress the background, we split sfGFP into two fragments, GFP1–10 and GFP11 (bipartite sfGFP), and fused GFP11 to either dCas9 or MS2 coat protein (MCP) (Fusco et al. 2003); the latter was to be used in combination with the RNA recognition motif, MS2, linked to the tail of the sgRNA (Supplemental Fig. 1B,C). A small solubility-enhancing tag, GB1, was also fused to each construct to suppress aggregation (Gronenborn et al. 1991). These bipartite sfGFP CRISPR systems exhibited fluorescence intensity comparable with that of dCas9-EGFP due to the high binding affinity between the GFP fragments (Cabantous et al. 2005). Consequently, however, they exhibited nonuniform residual background and aggregation in the absence of sgRNA.
We expected that the use of tripartite sfGFP could efficiently suppress background signal (Cabantous et al. 2013). Tripartite sfGFP consists of two 20-amino-acid peptides, GFP10 (between the ninth and 10th β-strands) and GFP11 (between the 10th and 11th β-strands), which are fused as tags to an interacting pair of proteins, and a large peptide, GFP1–9 (the first through to ninth β-strands), which serves as a detector of the interaction by reconstituting the full sfGFP chromophore. We reasoned that by conjugating GFP10 and GFP11 to dCas9 and sgRNA-binding proteins, respectively, it would be possible to acquire fluorescence signals exclusively from CRISPR complexes assembled at target loci, where the proximity between GFP10 and GFP11 would allow them to efficiently associate with GFP1–9.
To verify that tripartite sfGFP suppresses background signal, we first made a CRISPR complex containing a single set of tripartite sfGFP fragments by fusing GFP10 with dCas9, and GFP11 with MCP, which binds two MS2 sequences fused to the tail of the sgRNA (Supplemental Fig. 2A). GFP11 was also fused to another RNA-binding protein, PP7 coat protein (PCP), to measure the signal from the sgRNA-independent sfGFP assembly. GFP1–9 was cloned into a separate plasmid. All protein constructs contained nuclear localization signals (NLSs) to ensure proper transport to the nucleus and solubilizing tags to increase solubility. Expression of all proteins was driven by the CMV promoter. Transient expression of this set of constructs in AD-293 cells yielded a very low and uniform level of background fluorescence in the GFP channel in the absence of sgRNA, despite highly nonuniform localization of MCP-GFP11, as determined by the fluorescence from the mCherry fluorophore (Supplemental Fig. 2B).
Cotransfection of this tripartite sfGFP system with sgRNA targeting the C9-1 loci and containing two MCP binding sites (MBS) revealed distinct foci in the GFP channel (Supplemental Fig. 2C,D). BFP fused with GFP11-PCP did not colocalize with the GFP foci, confirming that the foci observed in the GFP and mCherry channels originated from CRISPR complexes assembled on the sgRNA and that association of free protein fragments did not generate notable signals. Cells with strong background signal in the mCherry channel exhibited a suppressed background in the GFP channel and revealed target-specific foci (Supplemental Fig. 2C, yellow circles), demonstrating the power of tripartite sfGFP to distinguish genomic loci over a broad range of fluorophore expression levels. The transient transfection was efficient, with ∼31% cells showing GFP fluorescence, and among those, ∼42% cells exhibited foci detected with our unbiased analysis procedure (Supplemental Fig. 3; Methods). However, the tripartite sfGFP system exhibited a weak fluorescence signal that diminished rapidly, limiting its application to live-cell imaging.
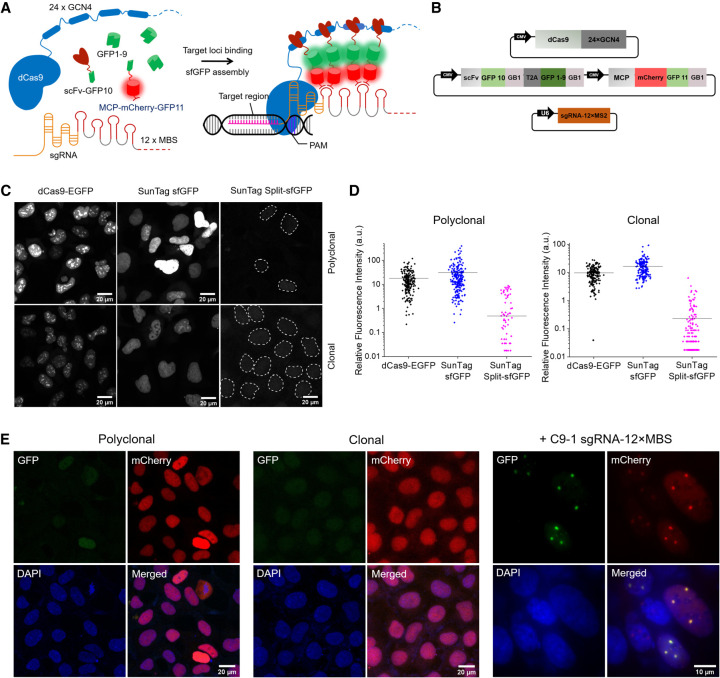
Because the stochastic assembly of the GFP fragments resulted in weak fluorescence, we integrated the SunTag system into our labeling scheme to amplify the signal (Tanenbaum et al. 2014). Specifically, we fused a 24-mer array of the GCN4 epitope to dCas9 and fused its minimal antibody, single-chain variable fragment (scFv), to GFP10 to recruit it to the GCN4 scaffold. This SunTag split-sfGFP system comprises dCas9-24 × GCN4, scFv-GFP10, GFP1–9, and MCP-mCherry-GFP11; mCherry was added to check the protein expression level (Fig. 1A,B; Supplemental Methods). We also constructed a SunTag labeling system with full sfGFP for comparison, which comprises dCas9-24 × GCN4 and scFv-GFP. For reliable comparison of the performance, we made stable cell lines expressing the dCas9-EGFP, SunTag sfGFP, and SunTag split-sfGFP systems using lentiviral constructs encoding the corresponding protein components. Transducing AD-293 for stable expression of the labeling components revealed nuclear aggregation with uneven background for the dCas9-EGFP or SunTag sfGFP systems, whereas the SunTag split-sfGFP system exhibited minimal background and no noticeable aggregation (Fig. 1C, upper panels).
Figure 1.
SunTag split-sfGFP CRISPR system enables background suppression and signal amplification for genome imaging. (A) Schematic design of the CRISPR-dCas9 system, integrated with tripartite sfGFP and the SunTag. dCas9 was fused to 24 × GCN4 peptides to recruit multiple scFv-GFP10 proteins. 12 × MBS motifs were added to the tail of sgRNA to recruit MCP-mCherry-GFP11 proteins. GFP1–9 was expressed separately. (B) Schematic representation of plasmid constructs for the components of the SunTag split-sfGFP CRISPR-dCas9 system. All protein components were expressed under the control of the CMV promoter. sgRNA-12 × MBS was expressed under the control of the U6 promoter. (GB1l) Solubility-enhancing tag, (T2A) self-cleaving peptide. (C) Representative images (GFP channel) of stable AD-293 cell lines transfected with the dCas9-EGFP, SunTag sfGFP, or SunTag split-sfGFP systems (top row) and of homogeneous stable cell lines obtained by colony picking (bottom row), in the absence of sgRNA. Dotted lines indicate nuclear boundaries detected in DAPI channel. (D) Nuclear fluorescence intensity compared between the fluorophore designs in C before and after colony picking (n > 200 cells). (E) Fluorescence images of fixed stable cell lines with the SunTag split-sfGFP CRISPR-dCas9 system in polyclonal and clonal after transient transfection with sgRNA-12 × MBS targeting a pericentromeric region of Chromosome 9, C9-1. mCherry images were acquired to check the expression level of sfGFP fragments.
Quantitative analysis of the imaging performance of engineered CRISPR systems
To perform quantitative comparisons between different CRISPR designs, we obtained single-clone stable cell lines by picking colonies (Methods; Supplemental Fig. 4), providing a uniform population of cells for each design (Fig. 1C, lower panels). Stable cell lines expressing the SunTag split-sfGFP system exhibited ninefold and 13-fold reductions in median background level relative to stable cell lines expressing the dCas9-EGFP and SunTag sfGFP systems, respectively (Fig. 1D). Fluorescence images in the mCherry channel revealed a high and uniform expression level of MCP-mCherry-GFP11; the other sfGFP fragments were expected to have similar expression levels because they were encoded by the same plasmid and driven by the same promoter (Fig. 1E). Thus, the reduction in the background was not due to a decrease in the expression of sfGFP fragments.
To increase the signal intensity of targeted foci, we added 12 repeats of MBS motifs to the tail of sgRNA. Because addition of more than two repeating MS2 or PP7 recruitment sequences at the tail of sgRNA may lead to degradation of the sgRNA (Zalatan et al. 2015), we modified the MBS motifs to enable stable expression of sgRNA with a long tail of MS2 repeats. We constructed different sgRNA designs to facilitate stable expression of a long sgRNA scaffold (Supplemental Fig. 5; Supplemental Methods). It was previously shown that such sgRNAs with long tails enable the targeted localization of various kinds of RNA constructs at specific genomic loci, named CRISPR-Display (Shechner et al. 2015). In all designs, the loop region (AUCA) of the MBS motif was kept unchanged, whereas the sequence in the stem region was varied to avoid formation of alternative RNA structures by minimizing the repetitive sequences (Shechner et al. 2015). The MBS motifs were separated by a 3-nt (UCU) linker. In design 4, the UUU sequence of tracrRNA region in design 3 was replaced with a UUG sequence to avoid interruption of transcription by a poly(T)-like sequence, which may act as a termination sequence as previously reported (Zaychikov et al. 1996; Chen et al. 2013; Ma et al. 2016).
With all designs, the stable AD-293 cell line containing the SunTag split-sfGFP system did not exhibit nonspecifically labeled foci in the absence of sgRNA or in the presence of negative control sgRNA that does not target the genome (Supplemental Fig. 5). However, upon transfection with the negative control sgRNA, the background fluorescence level increased 1.6-fold, possibly indicating the assembly of GFP fragments on freely diffusing sgRNAs (Supplemental Fig. 6). When the cells were transfected with each sgRNA construct targeting the C9-1 region, distinct foci were observed in all cases, but sgRNA design 4 yielded superior results in terms of the signal intensity and S/B; thus, it was used for the rest of the study.
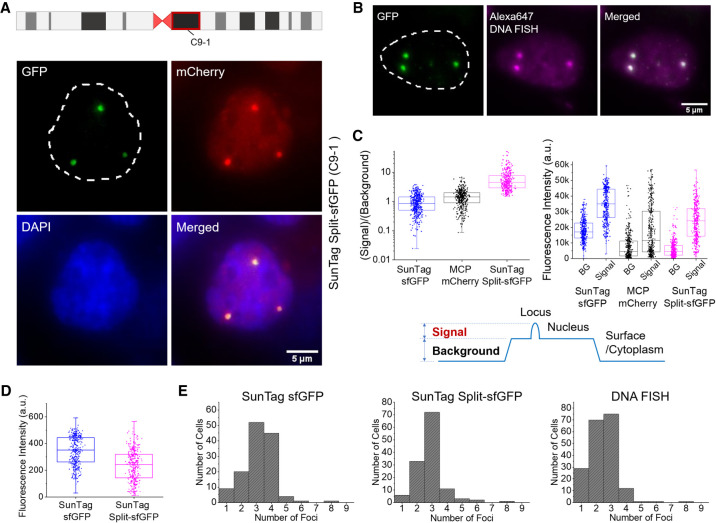
The stable cell line with the optimized sgRNA construct targeting the C9-1 region exhibited clear foci in 40%–60% of cells even without any further selection (Fig. 1E). The same foci were also distinguishable in the mCherry channel, but the background level was high and nonuniform relative to the GFP channel (Figs. 1E, 2A). To determine whether the observed foci are specific to the target loci, we combined CRISPR imaging with DNA fluorescence in situ hybridization (FISH) imaging (Methods). Foci observed by DNA FISH with a set of Alexa Fluor 647-labeled DNA probes targeting the C9-1 loci precisely colocalized with the sfGFP foci, confirming the target specificity of the CRISPR system with SunTag split-sfGFP (Fig. 2B). To assess applicability of this approach to other cell lines, we transiently transfected IMR-90 and RPE-1 cells with SunTag split-sfGFP constructs and sgRNA constructs targeting the C9-1 region or telomeres. The expected numbers of target-specific foci were observed in all cell lines, suggesting that our design is generally applicable to various cell types (Supplemental Fig. 7). In case of telomeres, detecting the foci from 2D images maximum-intensity-projected from z-stacked 3D images resulted in ∼35 foci per cell, but detecting the foci directly from 3D images gave ∼61 foci per cell, which is closer to the expected number of telomere foci in a diploid RPE1 cell line (Supplemental Fig. 7; Methods).
Figure 2.
SunTag split-sfGFP CRISPR system reveals target-specific foci with high S/B. (A) C9-1 region observed in a fixed stable AD-293 cell line expressing the SunTag split-sfGFP CRISPR system. Schematic shows the location of the C9-1 loci on Chromosome 9. (B) Simultaneous CRISPR and DNA FISH imaging of the C9-1 loci to verify the colocalization of GFP foci and DNA FISH foci. (C) Comparison of signal, background, and S/B in C9-1 region observed with SunTag sfGFP, MCP-mCherry, and SunTag split-sfGFP systems. BG stands for background. Schematic shows how S/B was defined from the images. (D) Distribution of fluorescence signal intensity of the detected C9-1 foci in fixed stable AD-293 cell lines expressing the SunTag sfGFP or SunTag split-sfGFP systems. (E) Histograms of the number of C9-1 foci detected with different labeling schemes.
We performed a quantitative analysis of S/B using the selected stable cell lines. The background level was defined as the average fluorescence level of the surrounding area of each peak subtracting that of the cytoplasmic region, which was nearly identical to that of bare surface, and the signal level was defined as the average fluorescence level within each peak area subtracting the background level as observed by confocal microscopy (Fig. 2C). Transfection conditions, laser excitation, and conditions for image acquisition were kept constant among all measurements. The median S/B for SunTag split-sfGFP was 5, which was sixfold higher than the value for SunTag sfGFP. It was also threefold higher than the median S/B for mCherry tagged on GFP11 for the same set of cells. This was threefold higher than the S/B from transient transfection of the tripartite sfGFP CRISPR system without SunTag (Supplemental Fig. 3). Although the SunTag split-sfGFP system had a lower signal than the SunTag sfGFP system (Fig. 2D), it achieved a superior S/B due to the strong suppression of background signal. The number of foci per nucleus detected by the SunTag split-sfGFP system exhibited a narrow distribution, with more than 50% of nuclei containing three foci, in contrast to those detected by the SunTag sfGFP system or DNA FISH (Fig. 2E). To obtain a reliable comparison, we performed unbiased detection of foci excluding any manual adjustment of parameters for different cells (Methods). Karyotyping on our AD-293 cell lines exhibited an average of 2.8–2.9 copies of Chromosome 9, supporting the target specificity of our SunTag split-sfGFP system (Supplemental Fig. 8). The elevated S/B and more consistent detection of chromosomal loci indicated that the SunTag split-sfGFP system can work with a smaller number of target sequence repeats or lower excitation conditions and still achieve equally reliable identification of target loci.
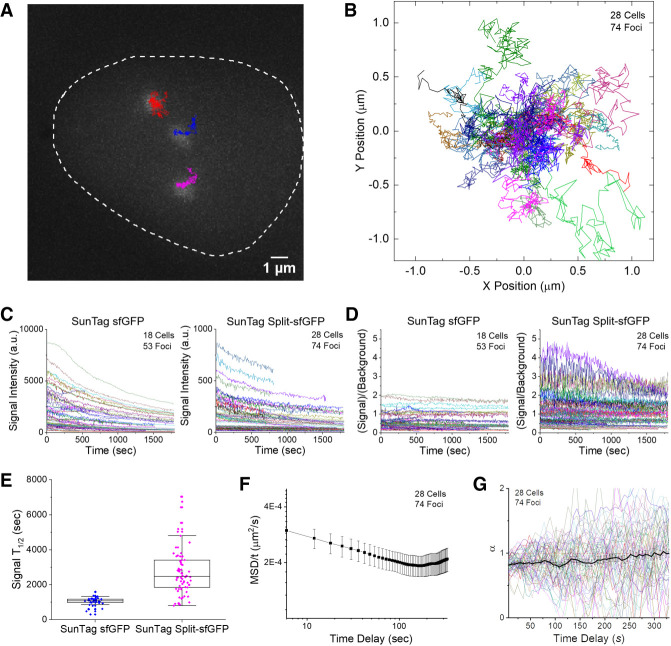
Using the engineered CRISPR system, we measured the diffusion dynamics of the C9-1 loci in live cells by taking z-stacked 3D images with a conventional fluorescence microscope (Fig. 3A,B; Supplemental Movies 1, 2). To exclude the motion of the cells and stage drift, we subtracted the average trajectory of multiple foci in a single nucleus from each trajectory. Under the same imaging conditions, the SunTag split-sfGFP system exhibited 10-fold lower signal but threefold higher S/B than the SunTag sfGFP system (Fig. 3C,D), as determined from the median behaviors from groups of traces. Reversible association between split sfGFP fragments would be responsible for the different fluorescence level, but the difference in the occupancy level of scFv between these SunTag systems might also be responsible for the difference. Though the SunTag sfGFP system exhibited stronger signal, its poorer S/B makes it inappropriate for identifying and tracking smaller loci. The SunTag split-sfGFP system also exhibited threefold slower decay of signal, presumably due to the fast exchange of sfGFP fragments to recover fluorescence during measurements (Fig. 3E). In order to assess the signal recovery of the tripartite sfGFP system, we performed fluorescence recovery after photobleaching measurements, and the tripartite sfGFP system showed fluorescence recovery within 10–20 min, nearly back to the original level, while the bipartite sfGFP system hardly recovered any signal within the same time (Supplemental Fig. 9), consistent with a previous report that bipartite sfGFP is virtually irreversible (Romei and Boxer 2019).
Figure 3.
Long-term tracking of the C9-1 loci reveals diffusion behavior dependent on the observation time scale. (A) Representative trajectories of three C9-1 foci in a single nucleus, shown along with the first-frame image of GFP channel, traced from a 2D-projected z-stack movie of live AD-293 cells integrated with the SunTag split-sfGFP CRISPR system. The trajectories represent the loci movement for 30 min with 6-sec frame intervals. (B) Collected trajectories of C9-1 foci (n = 74). All trajectories start at the origin. (C) Traces of signal intensity of C9-1 foci shown in B (n = 74) compared with those observed in another stable AD-293 cell line expressing the SunTag sfGFP CRISPR system (n = 53). Note the 10-fold difference in scale between two graphs. (D) Corresponding traces of S/B of C9-1 foci shown in C for the SunTag sfGFP and SunTag split-sfGFP systems, with the same color codes. (E) Decay half-life of fluorescence signal of C9-1 foci with the SunTag sfGFP and SunTag split-sfGFP systems, measured by fitting the traces in C longer than 600 sec to exponential decay functions with zero offset. (F) Mean-square displacement (MSD) divided by time, calculated over varying time delays. Error bars represent SEM between observed foci. (G) Diffusion exponent α found by fitting MSD curves of individual trajectories to locally at varying time delays. Average and SEM are shown as a black curve.
MSD was calculated from the 2D-projected trajectories. At short time intervals, MSD exhibited sublinear dependence on time delay, implying subdiffusive motion of the loci at short time scales, possibly due to their nuclear confinement or topological constraints imposed by chromosome structure (Fig. 3F). In contrast, at longer time scales, the diffusive motions exhibited a superlinear dependence on time delay, suggesting actively driven motions of the loci. Accordingly, the diffusion exponent, α, averaged over the traces, was around 0.8 at short time scales and gradually increased above 1 at longer time scales (Fig. 3G).
Such variation in diffusion behavior over different time scales is similar to what has been observed for telomeres (Bronstein et al. 2009). Diffusion behavior does not depend on the expression level of the CRISPR system because neither the diffusion coefficient (D) nor α correlated with the intensity of the foci (Supplemental Fig. 10A,B). We also tested whether the anomalous behavior was caused by photo-damage to the cells, e.g., the rounding up of dying cells. When we measured D or α from divided intervals of movies, parameters measured at later intervals did not show significant deviation from those measured at earlier intervals, suggesting that the effect of photo-damage under our imaging conditions had a negligible effect on the diffusion behavior (Supplemental Fig. 10C,D). Furthermore, C9-1 dynamics measured using the SunTag sfGFP system exhibited similar time scale-dependent diffusion behavior (Supplemental Fig. 11).
Tracking smaller repeat loci
We then took advantage of the improved imaging performance of the SunTag split-sfGFP system to track smaller repeat loci. To this end, we selected several genomic loci containing a small number of repeating CRISPR target sequences. As target loci of intermediate size, we selected a genomic region in Chromosome X containing 92 copies of target sequence (Chr X: 76,421–94,674) and another in Chromosome 1 containing 81 copies of target sequence (Chr 1:121,360,024–121,482,629) (Supplemental Fig. 12). Both regions exhibited clear foci with suppressed background. The foci were highly variable in size, with detected perimeters ranging from 0.5 to 4 μm. S/B was 4–6 times higher from split-sfGFP foci than from mCherry foci, demonstrating the background-suppressing capability of the split fluorophore design (Supplemental Fig. 12D,H).
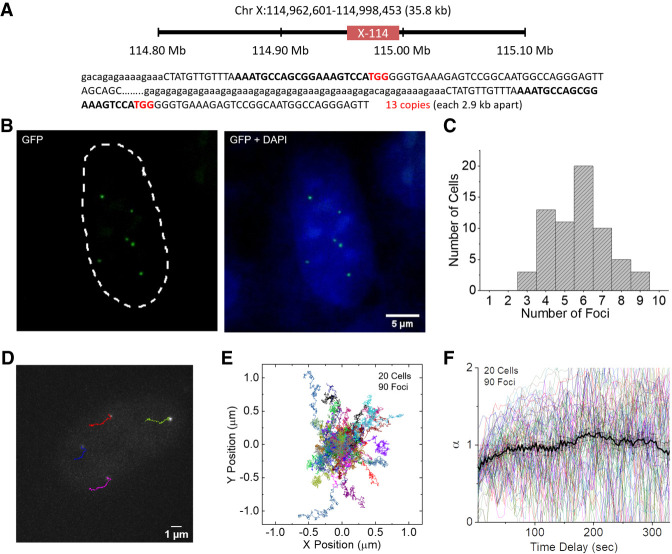
We further tested a small domain in Chromosome X containing 13 repeats within a 36-kb region (Chr X: 114,962,601–114,998,453; named X-114) (Fig. 4A). Targeting this region with the SunTag split-sfGFP system revealed clear foci (Fig. 4B). Automated detection of X-114 foci in the AD-293 cell line yielded a wide distribution, with a peak at six (Fig. 4C). In addition, we targeted a domain in Chromosome 9 containing 20 repeats of target sequence (Chr 9: 78,919,447–78,922,105; named C9-78), the FAM20C gene on Chromosome 7 (Chr 7: 215,244–216,445), and the HTT gene on Chromosome 4 (Chr 4: 3,228,971–3,229,570), which contain 20, 17, and 17 repeats of CRISPR target sequence, respectively (Supplemental Fig. 13). For these small target regions, the automatically detected number of foci yielded wider distributions than the C9-1 region, presumably due to the low S/B for these foci or possible translocation and insertion of these regions. The four sgRNA scaffold designs described above were tested on the HTT gene; only design 4 successfully detected the HTT loci, supporting the superiority of this optimized sgRNA design (Supplemental Fig. 5B). No foci were detected with the negative control sgRNA, confirming the specificity of the sgRNA design for targeting small-repeat loci.
Figure 4.
SunTag split-sfGFP CRISPR system allows imaging and tracking small-repeat loci. (A) Schematic diagram of the location of the X-114 loci in human Chromosome X shown with CRISPR target sequence (bold) and PAM (red). (B) A representative image of X-114 foci detected by the SunTag split-sfGFP system in fixed AD-293 cells, shown with the DAPI channel image. (C) Histogram of the number of X-114 foci automatically detected in each cell (n = 65 cells). (D) Exemplary trajectories of X-114 foci followed every 3 sec for 330 sec in a live AD-293 cell. (E) Collected trajectories of X-114 foci over 330 sec (n = 90). (F) Diffusion exponent α from MSD curves of the trajectories in E, calculated for varying time delays. Trajectories longer than 450 sec were used for this analysis. Average and SEM are shown as a black curve.
From continuous z-stacked imaging of living cells, live motions of the small-repeat loci could be successfully tracked for an extended period of time (Fig. 4D,E; Supplemental Movies 3, 4). The diffusion behavior of X-114 loci revealed subdiffusive motion at short time scales and superdiffusive motion around the 200-sec time scale, similar to what was observed for the C9-1 loci (Fig. 4F). We further tried to visualize a nonrepeat region using this CRISPR system. A set of 20 nonrepeat CRISPR target sites in a region downstream of the 9q12 domain proximal to the C9-1 region was designed and validated for off-target binding by the E-CRISP tool (Supplemental Table 2; Heigwer et al. 2014). Transfecting AD-293 cells with mixtures of five or 10 sgRNAs did not reveal clear foci under a conventional epifluorescence microscope, but transfecting with 20 sgRNAs revealed well-distinguished foci (Supplemental Fig. 14). S/B of the detected foci was lower than those for the small-repeat loci, possibly because the total expression level of sgRNA was higher in the cells revealing the nonrepeat loci, which would have raised the background level from sgRNAs not bound to the target (Supplemental Figs. 13, 14). These results show that the novel CRISPR design integrating tripartite sfGFP with the SunTag system enabled reliable detection and tracking of small genomic regions with fewer than 20 target sequence repeats, even using a conventional fluorescence microscope. Either by targeting sequence repeats near the region of interest (ROI) or by packaging a reasonable number of distinct sgRNAs in transfection plasmids, the CRISPR design developed here could be plausibly applied to the detection of loci throughout the genome.
Discussion
We developed a novel CRISPR-based genome labeling scheme for reliable visualization of genomic loci in live cells for an extended period of time. To achieve this, we integrated tripartite split-sfGFP and the SunTag system into the design of a CRISPR complex. This labeling scheme allowed us to detect genomic loci with a greatly reduced background level and high target specificity, thereby enabling successful tracking of genomic loci with a small number of sequence repeats in live cells. A critical problem with conventional approaches was nonspecific aggregation of the fluorophores, which was observed even in the absence of sgRNAs, which required the optimal expression of sgRNAs to deplete free fluorophores after assembling them at the target loci (Chen et al. 2013; Ma et al. 2015). Signal amplification using the SunTag system or RNA scaffolds still yielded high background level and nonspecific foci, but incorporation of split Venus fluorophore reduced false-positive signals (Hong et al. 2018; Wu et al. 2019). In another study, weak fluorescence signals and low S/B were improved by the use of mNeonGreen tetramer (Ye et al. 2017). Our design, incorporating tripartite sfGFP and the SunTag system, is unique in that it yielded lower signal levels but much higher S/B due to strong suppression of background signals. In addition, it allowed extended observation of the loci due to the fast exchange of photobleached sfGFP fragments. Thus, our design could be reliably applied to studies of dynamic chromosomal aberrations such as translocation, insertion, and deletion. A recent work demonstrated a method to label nonrepeat genomic loci with a few sgRNAs by inserting a cassette in the target loci for efficient CRISPR labeling (Chen et al. 2018). Another study reported that the use of lattice light-sheet microscopy decreases background signals by minimizing light illumination on off-focus volume, allowing the detection of genomic loci with a small number of sequence repeats (Qin et al. 2017). Combining our design with such advanced microscopy techniques should further decrease the required target size and lengthen the observation time.
Long-term 3D tracking of genomic loci enabled us to reveal the diffusion behavior of various genomic loci, which has implications for their physical environments, as well as for dynamic nuclear processes such as intra- and inter-chromosomal associations and structural rearrangement. The diffusion behavior of genomic loci changed over different time scales, similar to a previous observation that telomeric loci exhibit subdiffusive motion at shorter time scales and superdiffusive motion at longer time scales (Bronstein et al. 2009). Although telomeric loci were reported to have α values as low as 0.32 at time scales below 1 sec, and 0.51 at 1 − 200 sec, the centromeric and gene loci observed in this study had α values of 0.7–0.8 at short time scales. Such contrast may reflect the more confined motions of telomeres or may instead arise from systematic differences between the two labeling schemes. The increasing α value at longer time scales indicates the existence of actively driven motions, possibly at both the chromatin and chromosome levels, which provides insights for future studies, for instance, by controlling cell cycle, inducing localized replication stress, or stimulating localized gene expression.
Improvement in the design of the long sgRNA scaffold was critical for efficient genome labeling. Our final design, consisting of 12 × MBS with varying stem sequence with the UUU motif replaced by UUG, uniquely enabled the detection of loci with fewer sequence repeats. Avoidance of the UUU motif was proposed to be critical for successful CRISPR imaging (Chen et al. 2013; Ma et al. 2015). Because it requires fewer sequence repeats, our method can be applied to a broad range of genomic regions. We were able to discover potential CRISPR targets with more than a dozen local sequence repeats, including PAM, roughly every few megabases. Considering that target binding is only sensitive to 11 bases including PAM (Ma et al. 2016; Singh et al. 2016), the density of potential targets may be much higher. However, detection efficiency for loci with a small number of sequence repeats was not consistently high among target sites and cell lines, presumably due to varying transfection efficiency or the expression level of sgRNA, as previously suggested (Doench et al. 2014; Moreno-Mateos et al. 2015; Chen et al. 2016). We used AD-293 cells in the optimization of the SunTag split-sfGFP system and sgRNA design because of its high transfection efficiency and ease of imaging.
Other than the advantage of minimizing background fluorescence and enabling long-term tracking of genomic loci, our approach has several limitations as well. First, our improved design still does not yield sufficiently high labeling efficiency for small repeat or nonrepeat loci to reliably image such loci without careful cell selection or constructing stable cell lines. Thus, it was difficult to label nonrepeat genomic loci in cell lines with relatively lower transfection efficiency, like RPE1 tested here. Second, CRISPR labeling can be done with high efficiency by transient transfection of sgRNAs using exogenous plasmids but not at the same level by endogenous sgRNA expression through lentiviral incorporation. This is because a high level of sgRNA expression is required to match the levels of the fluorescent protein components, which was possible only with transient transfection. Third, our design requires the expression of four proteins to achieve a single-color genome imaging. We improved the efficiency of simultaneous transfection of all protein components by integrating multiple components in one plasmid, but this potentially limits the extension of the technique toward multicolor CRISPR imaging. It is also difficult to implement the optimal ratios between the proteins. This is exemplified by the signal amplification by SunTag being less than expected, which is possibly due to nonoptimal expression ratios. Fourth, the long sgRNA design with MS2 repeats is a drawback in packaging a large number of sgRNAs to target nonrepeat loci, while CRISPR designs in previous genome imaging studies adopted shorter sgRNA constructs (Chen et al. 2013; Shao et al. 2016; Qin et al. 2017; Gu et al. 2018; Maass et al. 2018a,b). It is possible to combine shorter sgRNAs having fewer protein binding motifs with the split fluorophore and SunTag system tested in this study to find an optimal design for labeling nonrepeat loci with multiple sgRNAs.
Technical breakthroughs in sgRNA transfection, especially for the reliable delivery of multiple sgRNAs, hold the key to extending the application of CRISPR labeling to targets throughout the genome not necessarily containing any sequence repeats. Cpf1 cleaves pre-crRNAs upstream of a hairpin structure, thereby generating multiple mature crRNAs, making it an ideal choice for packaging a large number of sgRNAs in plasmids (Fonfara et al. 2016). It is not directly applicable to our design because the sgRNA has a long tail of MBS motifs, which would not be properly processed by Cpf1. A recently developed strategy for assembling a large number of sgRNAs in a chimeric array might provide a plausible solution to this problem (Gu et al. 2018), as might engineering Csy4 to precisely cleave a tandem array of sgRNAs (Tsai et al. 2014). Our labeling schemes with split-sfGFP could be further extended to multicolor imaging using orthogonal RNA-binding motifs such as PP7 and Com, as well as orthogonal amplification tags such as the recently developed MoonTag (Boersma et al. 2019). Integrating additional colors of fluorophores would enable simultaneous tracking of multiple genomic loci, allowing visualization of dynamic nuclear processes such as DNA damage repair, recombination, and replication in real time.
Off-target binding of CRISPR complexes is a major technical challenge in CRISPR-based imaging, as well as in genome editing. The presence of off-target sites at high density may lead to false-positive locus detection, especially as we attempt to decrease the size of the CRISPR array (Kuscu et al. 2014; Zhang et al. 2015). As in this study, a DNA FISH assay is typically used to confirm the target specificity of CRISPR labeling. However, this method requires strongly denaturing conditions, making it incompletely compatible with protein-based CRISPR labeling; in addition, it may alter chromatin structure (Markaki et al. 2012; Williamson et al. 2014; Giorgetti and Heard 2016; Ma et al. 2018b). This limits the feasibility of FISH-based target validation, especially for confirmation of weak signals from small target regions. Because the target binding efficiency and off-target effect of CRISPR complexes depend on the stability of sgRNA and the accessibility of target loci (Doench et al. 2014; Kuscu et al. 2014; Wang et al. 2014; Wu et al. 2014), a systematic protocol for target selection will be needed to extend our method to small, nonrepeat target regions.
Methods
Plasmid construction
Mammalian expression and reporter plasmids were constructed by standard restriction digestion and ligation methods. The backbone for the lentiviral vector was derived from pHAGE-TO-DEST (Addgene 64108) containing the cytomegalovirus (CMV) promoter. Plasmid pHR-scFv-GCN4-sfGFP-GB1-NLS, encoding an antibody that binds to the GCN4 peptide from the SunTag system, was obtained from Addgene (60906) (Tanenbaum et al. 2014). GFP1–9, GFP10, and GFP11 fragments were synthesized as gBlocks (Integrated DNA Technologies). GFP11 was fused between MCP-mCherry (Addgene 27122) (Fusco et al. 2003) and GB1-NLS, yielding the MCP-mCherry-GFP11-GB1-NLS construct. The GCN4 peptide-binding module scFv-GCN4 was fused with GFP10-GB1-NLS, yielding the scFv-GCN4-GFP10-GB1-NLS construct. The human codon-optimized genes NLS-dCas9-NLS-EGFP (Addgene 51023) (Chen et al. 2013), NLS-dCas9-24 × GCN4-NLS (Addgene 60910) (Tanenbaum et al. 2014), NLS-dCas9-GFP10-GB1-NLS, GFP(1-9)-GB1-NLS, MCP-mCherry-GFP11-GB1-NLS, and scFv-GCN4-GFP10-GB1-NLS were constructed by PCR using Phusion High-Fidelity PCR Master Mix with HF buffer (New England BioLabs) under the following conditions: initial denaturation for 30 sec at 98°C, 25 thermal cycles (10 sec at 98°C, 30 sec at 60°C, and 2 min at 72°C), and final extension for 10 min at 72°C. The amplicons were cloned into the lentiviral backbone pHAGE-TO-DEST. Later, to facilitate transfection, scFv-GCN4-GFP10-GB1-NLS was fused with GFP(1–9)-GB1-NLS separated by the self-cleaving peptide T2A, yielding the scFv-GCN4-GFP10-GB1-NLS-T2A-GFP(1-9)-GB1-NLS construct, and then this was fused with another construct, Puro-T2A-MCP-mCherry-GFP11-GB1-NLS, both expressed by CMV promoters in the lentiviral backbone pHAGE-TO-DEST (all-in-one plasmid).
The sgRNA expression vectors were constructed in the pLKO.1 lentiviral expression plasmid (Addgene #64114) (Ma et al. 2015). To generate the sgRNA backbone plasmid containing the target insertion site and sgRNA scaffolds, two or 12 MS2 binding sites were chemically synthesized as a gBlock (IDT) and inserted into pLKO.1 lentiviral expression plasmid by AgeI and EcoRI digestion, followed by ligation. The tandem MS2 binding motifs were designed to minimize the sequence similarities by variations in stem sequences, separated by a 3-nt linker and then evaluated in NUPACK (Zadeh et al. 2011) for optimal folding. All target sites were selected by searching for NGG motifs in repeats identified by Tandem Repeat Finder (Benson 1999) from the genomic regions of the human chromosome downloaded from the UCSC Genome Browser (http://genome.ucsc.edu/; hg19). The specificity of sgRNAs was verified using the BLAT alignment tool (Kent 2002). Nonrepetitive sgRNAs downstream of 9q12 (C9-1) were designed and evaluated by using the E-CRISP tool (Heigwer et al. 2014). Target sequences were cloned into BbsI-digested sgRNA2 (Addgene #64114), sgRNA-2xMS2, and sgRNA-12xMS2 by ligation of annealed oligos. Sequences of sgRNA targets are provided in Supplemental Tables 1 and 2. The sgRNA target locations noted in this work were based on GRCh37, but they were also found in GRCh38 with the same copy numbers at the corresponding locations. Sequence information about the DNA constructs is provided in Supplemental Sequences.
Cell culture and lipofection
AD-293 (Stratagene 240085) and IMR-90 (ATCC CCL-186) cells were cultured in DMEM/high glucose (Gibco) supplemented with 10% fetal bovine serum (FBS; Gibco) and 1× penicillin/streptomycin (Gibco). RPE-1 ([ATCC CRL4000) cells were cultured in DMEM/F-12 (Gibco) supplemented with 10% FBS and 1× penicillin/streptomycin. Cells were maintained at 37°C and 5% CO2 in a humidified incubator. For transient transfection of AD-293 cells, 70%–80% confluent cells were transfected with 4 µg plasmid mix using 6 µL Lipofectamine 2000 (Invitrogen) in 2 mL medium. Cells were grown on a 6-well plate for 1 d before transfection, cotransfected with plasmids (1:3 ratio of transgene expression plasmids and sgRNA expression plasmids), and then incubated for another 24 h before paraformaldehyde fixation (4%) and imaging. In experiments with colony-picked stable SunTag sfGFP and SunTag split-sfGFP AD-293 cells, cells were transfected with 3 µg sgRNA plasmids and 6 µL Lipofectamine 2000. For RPE-1 and IMR-90 cells, 10 µL Lipofectamine 2000 was used for transfection.
Lentivirus production and transduction
Lentiviral particles were generated in HEK293T cells using second-generation packaging plasmids. HEK293T cells (3.8 × 106) were seeded in 10-cm plates in complete DMEM medium. After 24 h, complete growth medium was replaced with serum-free DMEM. Cells were cotransfected with 10 µg of the transgene, 5 µg of the viral envelope plasmids (pMD2G; Addgene #12259), and 5 µg of the viral packaging plasmids (psPAX2; Addgene #12260) in 60 µL of 1 mg/mL polyethylenimine (Polysciences). Twelve hours after transfection, media was replaced with complete DMEM growth medium. At 48 and 72 h post-transfection, lentivirus was harvested by filtering the medium through a sterile 0.45-µm syringe filter (Millipore #SLHV033RS). Lentivirus was concentrated using poly-(ethylene glycol) (PEG-8000; Sigma-Aldrich) as follows: 3× volume of medium containing lentivirus was mixed with 1× volume of PEG concentrator, incubated overnight at 4°C, and concentrated by centrifugation at 1600g for 60 min at 4°C. The viral pellet resuspended in serum-free DMEM was used for transduction (Kutner et al. 2009). AD-293 cells in six-well plates were transduced with dCas9-EGFP, SunTag sfGFP (pHR-scFv-GCN4-sfGFP-GB1-NLS, NLS-dCas9-24 × GCN4-NLS), or SunTag split-sfGFP (NLS-dCas9-24 × GCN4-NLS, all-in-one-plasmid containing the constructs of the split-sfGFP system) virus cocktail. For SunTag split-sfGFP, transduced cells were selected by culture for 5 d with 1 µg/mL puromycin, and clones with medium mCherry and high GFP fluorescence signals were selected. For dCas9-EGFP and SunTag sfGFP cells, clones with medium GFP fluorescence signals were selected.
Fluorescence microscopy
For fixed-cell imaging, cells were cultured on coverslips in six-well plates for 24 h, fixed with 4% paraformaldehyde, washed twice with PBS, and mounted onto glass slides using ProLong Diamond Antifade reagent (Molecular Probes). Z-stack images were acquired on a Zeiss LSM 880 confocal laser microscope with an oil-immersion 63× 1.4 NA objective lens. For live-cell imaging, cells were transfected in 35-mm glass-bottom confocal dishes (SPL Biosciences) and incubated overnight. Live-cell imaging was performed on an IX81-ZDC inverted wide-field fluorescence microscope with an oil-immersion 100× 1.4 NA objective lens equipped with a motorized stage and incubator in the UNIST-Olympus Biomed Imaging Center. The incubation chamber on the microscope was maintained at 37°C and 5% CO2 with humidity. To compare long-term continuous imaging between SunTag sfGFP and SunTag split-sfGFP designs, z-stacks of six layers were recorded for 30 min at intervals of 6 sec. Maximum z-projection of images at each time frame was used for the production of 2D-projected images.
For the fluorescence recovery after photobleaching (FRAP) experiments, cells were transfected as above before imaging. The FRAP measurements was performed on a Zeiss LSM 780 confocal laser microscope with an oil-immersion 63× 1.4 NA objective lens equipped with a motorized stage and incubator in the UNIST-Olympus Biomed Imaging Center. Round circular regions covering C9-1 foci were chosen for photobleaching. Selected regions of interest were photobleached with a 405-nm laser set to 100% transmission. Movies taken after photobleaching were analyzed to track the photobleached foci and measure their fluorescence signals.
Fluorescence in situ hybridization
Stable cells were transfected with sgRNAs on a 2-well chambered cover glass for 24 h. Cells were fixed with 4% paraformaldehyde for 10 min, washed with PBS twice, permeabilized with 0.7% Triton X-100 in PBS for 30 min at room temperature, washed twice again with PBS, treated with RNase A at 37°C for 1 h, washed twice again with PBS, and incubated with 0.05% (w/v) pepsin in 10 mM HCl for 10 min at 37°C. The cells were again washed twice in PBS and dehydrated by consecutive incubation for 5 min in 70%, 85%, and 100% ethanol. After drying for 10 min, cells were denatured at 80°C for 5 min in denaturation buffer (70% formamide/2× SSC) and washed in an ethanol series (70%, 85%, and 100%) for 5 min each. After air-drying for 10 min, 15 µL of Alexa Fluor 647-labeled probes in hybridization buffer (500 ng/mL of human Cot-1 DNA, 500 ng/mL of salmon sperm DNA, 50% formamide, 10% dextran sulphate in 20× SSC buffer; denatured at 80°C for 5 min; incubated at 37°C for 30 min) at a final concentration of 2 ng/µL was added to the sample and incubated at 37°C for 18 h in a moist chamber. After hybridization, cells were twice washed with 2× SSC buffer, stained with DAPI, and mounted on slides in ProLong Diamond Antifade reagent. Probe labeling was performed by nick translation. The C9-1 DNA FISH probe was generated from pHuR98 (ATCC 61078TM).
Image analysis
Fluorescence image analysis was performed on the maximum-intensity-projected images, except for telomere 3D counting. For foci detection, the projected images were first processed using the ImageJ Spot Detection plug-in from the MOSAIC group to select candidate foci (Sbalzarini and Koumoutsakos 2005). Within a region of interest around each focus, half of the pixels with weaker signal were flattened to remove the background gradient and noise. The flattened ROI was fitted by a 2D-Gaussian function (https://www.mathworks.com/matlabcentral/fileexchange/41938-fit-2d-gaussian-with-optimization-toolbox). Valid foci were selected based on whether the center coordinate is within the ROI and whether the ROI surrounds the boundary defined as the 95% area of the Gaussian curve. The background level was defined as the median value from the surrounding of each focus, which was defined by dilating the boundary by a factor of 2, and subtracting the level of bare surface. The signal value was defined as the average over the peak area, subtracting the background level. For telomere 3D counting, foci were detected from each image plane instead of the projected image and assembled by eliminating overlapping ones. For nucleus detection, simple thresholding in DAPI or GFP images did not give accurate results. Thus, images were Wiener-filtered and Gaussian-blurred. Then, nuclear boundaries were detected by adaptive Gaussian thresholding. Holes inside the boundaries were removed by a watershed filter. Then, the valid nuclei were further selected based on size and roughness.
The trajectories of the detected foci from live-cell movies were obtained by successive 2D Gaussian fitting, followed by further analysis using the MSD Analyzer program (Tarantino et al. 2014). The calculated mean-squared displacement (MSD) versus time delay was plotted on a log–log scale and then locally fitted by a linear curve to find the diffusion coefficient (D) and the diffusion exponent α from the equation log (MSD) = αlog (t) + log(2D). The value of α at each time delay was determined by the instantaneous tangent in this log–log graph at each time delay for each MSD trajectory.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
We thank Dr. Sang-Hee Shim, Dr. Bo Huang, Dr. Hiroshi Ochiai, and Dr. Jae-Hyung Jeon for helpful discussions. This work was supported by the National Research Foundation of Korea (2017R1D1A1B03036239, 2017M3A9E2062181, 2018R1A5A1024340), the UNIST research fund (1.190091.01), and the Institute for Basic Science (IBS-R022-D1).
Author contributions: N.C. and H.K. conceived the project. N.C. designed protein and sgRNA constructs with the help of H.K. and H.C. N.C. constructed all plasmids and stable cell lines and performed all measurements with the help of N.G. N.C. and S.N. performed quantitative analysis of the data. J.R. and K.M. performed karyotyping analysis. N.C. and H.K. wrote the manuscript with input from all other authors.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.260018.119.
References
- Anton T, Bultmann S, Leonhardt H, Markaki Y. 2014. Visualization of specific DNA sequences in living mouse embryonic stem cells with a programmable fluorescent CRISPR/Cas system. Nucleus 5: 163–172. 10.4161/nucl.28488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27: 573–580. 10.1093/nar/27.2.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. 1998. Localization of ASH1 mRNA particles in living yeast. Mol Cell 2: 437–445. 10.1016/S1097-2765(00)80143-4 [DOI] [PubMed] [Google Scholar]
- Bickmore WA. 2013. The spatial organization of the human genome. Annu Rev Genomics Hum Genet 14: 67–84. 10.1146/annurev-genom-091212-153515 [DOI] [PubMed] [Google Scholar]
- Boersma S, Khuperkar D, Verhagen BMP, Sonneveld S, Grimm JB, Lavis LD, Tanenbaum ME. 2019. Multi-color single-molecule imaging uncovers extensive heterogeneity in mRNA decoding. Cell 178: 458–472.e19. 10.1016/j.cell.2019.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronstein I, Israel Y, Kepten E, Mai S, Shav-Tal Y, Barkai E, Garini Y. 2009. Transient anomalous diffusion of telomeres in the nucleus of mammalian cells. Phys Rev Lett 103: 018102 10.1103/PhysRevLett.103.018102 [DOI] [PubMed] [Google Scholar]
- Cabantous S, Terwilliger TC, Waldo GS. 2005. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat Biotechnol 23: 102–107. 10.1038/nbt1044 [DOI] [PubMed] [Google Scholar]
- Cabantous S, Nguyen HB, Pedelacq J-D, Koraïchi F, Chaudhary A, Ganguly K, Lockard MA, Favre G, Terwilliger TC, Waldo GS. 2013. A new protein-protein interaction sensor based on tripartite split-GFP association. Sci Rep 3: 2854 10.1038/srep02854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, et al. 2013. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155: 1479–1491. 10.1016/j.cell.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Hu J, Almeida R, Liu H, Balakrishnan S, Covill-Cooke C, Lim WA, Huang B. 2016. Expanding the CRISPR imaging toolset with Staphylococcus aureus Cas9 for simultaneous imaging of multiple genomic loci. Nucleic Acids Res 44: e75 10.1093/nar/gkv1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Zou W, Xu H, Liang Y, Huang B. 2018. Efficient labeling and imaging of protein-coding genes in living cells using CRISPR-Tag. Nat Commun 9: 5065 10.1038/s41467-018-07498-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AW, Jillette N, Lee P, Plaskon D, Fujiwara Y, Wang W, Taghbalout A, Wang H. 2016. Casilio: a versatile CRISPR–Cas9–Pumilio hybrid for gene regulation and genomic labeling. Cell Res 26: 254–257. 10.1038/cr.2016.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Shi X, Tjian R, Lionnet T, Singer RH. 2015. CASFISH: CRISPR/Cas9-mediated in situ labeling of genomic loci in fixed cells. Proc Natl Acad Sci 112: 11870–11875. 10.1073/pnas.1515692112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Gorkin DU, Ren B. 2016. Chromatin domains: the unit of chromosome organization. Mol Cell 62: 668–680. 10.1016/j.molcel.2016.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, Root DE. 2014. Rational design of highly active sgRNAs for CRISPR–Cas9–mediated gene inactivation. Nat Biotechnol 32: 1262–1267. 10.1038/nbt.3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Lu G, Hong Y, Hu Q, Mai X, Guo J, Si X, Wang F, Zhang Y. 2018. Live imaging and tracking of genome regions in CRISPR/dCas9 knock-in mice. Genome Biol 19: 192 10.1186/s13059-018-1530-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfara I, Richter H, Bratovič M, Le Rhun A, Charpentier E. 2016. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532: 517–521. 10.1038/nature17945 [DOI] [PubMed] [Google Scholar]
- Fu Y, Rocha PP, Luo VM, Raviram R, Deng Y, Mazzoni EO, Skok JA. 2016. CRISPR–dCas9 and sgRNA scaffolds enable dual-colour live imaging of satellite sequences and repeat-enriched individual loci. Nat Commun 7: 11707 10.1038/ncomms11707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, Singer RH, Bertrand E. 2003. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol 13: 161–167. 10.1016/S0960-9822(02)01436-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibcus JH, Dekker J. 2013. The hierarchy of the 3D genome. Mol Cell 49: 773–782. 10.1016/j.molcel.2013.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM, Takebayashi S-I, Ryba T, Lu J, Pope BD, Wilson KA, Hiratani I. 2010. Space and time in the nucleus: developmental control of replication timing and chromosome architecture. Cold Spring Harb Symp Quant Biol 75: 143–153. 10.1101/sqb.2010.75.011 [DOI] [PubMed] [Google Scholar]
- Giorgetti L, Heard E. 2016. Closing the loop: 3C versus DNA FISH. Genome Biol 17: 215 10.1186/s13059-016-1081-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady DL, Ratliff RL, Robinson DL, McCanlies EC, Meyne J, Moyzis RK. 1992. Highly conserved repetitive DNA sequences are present at human centromeres. Proc Natl Acad Sci 89: 1695–1699. 10.1073/pnas.89.5.1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronenborn AM, Filpula DR, Essig NZ, Achari A, Whitlow M, Wingfield PT, Clore GM. 1991. A novel, highly stable fold of the immunoglobulin binding domain of streptococcal protein G. Science 253: 657–661. 10.1126/science.1871600 [DOI] [PubMed] [Google Scholar]
- Gu B, Swigut T, Spencley A, Bauer MR, Chung M, Meyer T, Wysocka J. 2018. Transcription-coupled changes in nuclear mobility of mammalian cis-regulatory elements. Science 359: 1050–1055. 10.1126/science.aao3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heigwer F, Kerr G, Boutros M. 2014. E-CRISP: fast CRISPR target site identification. Nat Methods 11: 122–123. 10.1038/nmeth.2812 [DOI] [PubMed] [Google Scholar]
- Hong Y, Lu G, Duan J, Liu W, Zhang Y. 2018. Comparison and optimization of CRISPR/dCas9/gRNA genome-labeling systems for live cell imaging. Genome Biol 19: 39 10.1186/s13059-018-1413-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT—the BLAST-Like Alignment Tool. Genome Res 12: 656–664. 10.1101/gr.229202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. 2014. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol 32: 677–683. 10.1038/nbt.2916 [DOI] [PubMed] [Google Scholar]
- Kutner RH, Zhang XY, Reiser J. 2009. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat Protoc 4: 495–505. 10.1038/nprot.2009.22 [DOI] [PubMed] [Google Scholar]
- Ma H, Reyes-Gutierrez P, Pederson T. 2013. Visualization of repetitive DNA sequences in human chromosomes with transcription activator-like effectors. Proc Natl Acad Sci 110: 21048–21053. 10.1073/pnas.1319097110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Naseri A, Reyes-Gutierrez P, Wolfe SA, Zhang S, Pederson T. 2015. Multicolor CRISPR labeling of chromosomal loci in human cells. Proc Natl Acad Sci 112: 3002–3007. 10.1073/pnas.1420024112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Tu LC, Naseri A, Huisman M, Zhang S, Grunwald D, Pederson T. 2016. Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nat Biotechnol 34: 528–530. 10.1038/nbt.3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Tu L-C, Naseri A, Chung Y-C, Grunwald D, Zhang S, Pederson T. 2018a. CRISPR-Sirius: RNA scaffolds for signal amplification in genome imaging. Nat Methods 15: 928–931. 10.1038/s41592-018-0174-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Chen L, Shi M, Niu J, Zhang X, Yang X, Zhanghao K, Wang M, Xi P, Jin D, et al. 2018b. Developing novel methods to image and visualize 3D genomes. Cell Biol Toxicol 34: 367–380. 10.1007/s10565-018-9427-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass PG, Barutcu AR, Shechner DM, Weiner CL, Melé M, Rinn JL. 2018a. Spatiotemporal allele organization by allele-specific CRISPR live-cell imaging (SNP-CLING). Nat Struct Mol Biol 25: 176–184. 10.1038/s41594-017-0015-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass PG, Barutcu AR, Weiner CL, Rinn JL. 2018b. Inter-chromosomal contact properties in live-cell imaging and in Hi-C. Mol Cell 69: 1039–1045.e3. 10.1016/j.molcel.2018.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markaki Y, Smeets D, Fiedler S, Schmid VJ, Schermelleh L, Cremer T, Cremer M. 2012. The potential of 3D-FISH and super-resolution structured illumination microscopy for studies of 3D nuclear architecture. Bioessays 34: 412–426. 10.1002/bies.201100176 [DOI] [PubMed] [Google Scholar]
- Misteli T. 2007. Beyond the sequence: cellular organization of genome function. Cell 128: 787–800. 10.1016/j.cell.2007.01.028 [DOI] [PubMed] [Google Scholar]
- Miyanari Y, Ziegler-Birling C, Torres-Padilla M-E. 2013. Live visualization of chromatin dynamics with fluorescent TALEs. Nat Struct Mol Biol 20: 1321–1324. 10.1038/nsmb.2680 [DOI] [PubMed] [Google Scholar]
- Moreno-Mateos MA, Vejnar CE, Beaudoin JD, Fernandez JP, Mis EK, Khokha MK, Giraldez AJ. 2015. CRISPRscan: designing highly efficient sgRNAs for CRISPR–Cas9 targeting in vivo. Nat Methods 12: 982–988. 10.1038/nmeth.3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisaki T, Lyon K, DeLuca KF, DeLuca JG, English BP, Zhang Z, Lavis LD, Grimm JB, Viswanathan S, Looger LL, et al. 2016. Real-time quantification of single RNA translation dynamics in living cells. Science 352: 1425–1429. 10.1126/science.aaf0899 [DOI] [PubMed] [Google Scholar]
- Ochiai H, Sugawara T, Yamamoto T. 2015. Simultaneous live imaging of the transcription and nuclear position of specific genes. Nucleic Acids Res 43: e127 10.1093/nar/gkv624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin P, Parlak M, Kuscu C, Bandaria J, Mir M, Szlachta K, Singh R, Darzacq X, Yildiz A, Adli M. 2017. Live cell imaging of low- and non-repetitive chromosome loci using CRISPR–Cas9. Nat Commun 8: 14725 10.1038/ncomms14725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, Murray A, Belmont AS. 1996. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol 135: 1685–1700. 10.1083/jcb.135.6.1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romei MG, Boxer SG. 2019. Split green fluorescent proteins: scope, limitations, and outlook. Annu Rev Biophys 48: 19–44. 10.1146/annurev-biophys-051013-022846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roukos V, Voss TC, Schmidt CK, Lee S, Wangsa D, Misteli T. 2013. Spatial dynamics of chromosome translocations in living cells. Science 341: 660–664. 10.1126/science.1237150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter G, Moch H, Carroll P, Kerschmann R, Mihatsch MJ, Waldman FM. 1995. Chromosome-9 loss detected by fluorescencein situ hybridization in bladder cancer. Int J Cancer 64: 99–103. 10.1002/ijc.2910640205 [DOI] [PubMed] [Google Scholar]
- Sbalzarini IF, Koumoutsakos P. 2005. Feature point tracking and trajectory analysis for video imaging in cell biology. J Struct Biol 151: 182–195. 10.1016/j.jsb.2005.06.002 [DOI] [PubMed] [Google Scholar]
- Shao S, Zhang W, Hu H, Xue B, Qin J, Sun C, Sun Y, Wei W, Sun Y. 2016. Long-term dual-color tracking of genomic loci by modified sgRNAs of the CRISPR/Cas9 system. Nucleic Acids Res 44: e86 10.1093/nar/gkw066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechner DM, Hacisuleyman E, Younger ST, Rinn JL. 2015. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat Methods 12: 664–670. 10.1038/nmeth.3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D, Sternberg SH, Fei J, Doudna JA, Ha T. 2016. Real-time observation of DNA recognition and rejection by the RNA-guided endonuclease Cas9. Nat Commun 7: 12778 10.1038/ncomms12778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. 2014. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159: 635–646. 10.1016/j.cell.2014.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarantino N, Tinevez J-Y, Crowell EF, Boisson B, Henriques R, Mhlanga M, Agou F, Israël A, Laplantine E. 2014. TNF and IL-1 exhibit distinct ubiquitin requirements for inducing NEMO-IKK supramolecular structures. J Cell Biol 204: 231–245. 10.1083/jcb.201307172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. 2014. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol 32: 569–576. 10.1038/nbt.2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. 2014. Genetic screens in human cells using the CRISPR–Cas9 system. Science 343: 80–84. 10.1126/science.1246981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Han B, Zhou R, Zhuang X. 2016a. Real-time imaging of translation on single mRNA transcripts in live cells. Cell 165: 990–1001. 10.1016/j.cell.2016.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Su J-H, Zhang F, Zhuang X. 2016b. An RNA-aptamer-based two-color CRISPR labeling system. Sci Rep 6: 26857 10.1038/srep26857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson I, Berlivet S, Eskeland R, Boyle S, Illingworth RS, Paquette D, Dostie J, Bickmore WA. 2014. Spatial genome organization: contrasting views from chromosome conformation capture and fluorescence in situ hybridization. Genes Dev 28: 2778–2791. 10.1101/gad.251694.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Scott DA, Kriz AJ, Chiu AC, Hsu PD, Dadon DB, Cheng AW, Trevino AE, Konermann S, Chen S, et al. 2014. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol 32: 670–676. 10.1038/nbt.2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Eliscovich C, Yoon YJ, Singer RH. 2016. Translation dynamics of single mRNAs in live cells and neurons. Science 352: 1430–1435. 10.1126/science.aaf1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Mao S, Ying Y, Krueger CJ, Chen AK. 2019. Progress and challenges for live-cell imaging of genomic loci using CRISPR-based platforms. Genomics Proteomics Bioinformatics 17: 119–128. 10.1016/j.gpb.2018.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Hoek TA, Vale RD, Tanenbaum ME. 2016. Dynamics of translation of single mRNA molecules in vivo. Cell 165: 976–989. 10.1016/j.cell.2016.04.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Rong Z, Lin Y. 2017. Live cell imaging of genomic loci using dCas9-SunTag system and a bright fluorescent protein. Protein Cell 8: 853–855. 10.1007/s13238-017-0460-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadeh JN, Steenberg CD, Bois JS, Wolfe BR, Pierce MB, Khan AR, Dirks RM, Pierce NA. 2011. NUPACK: analysis and design of nucleic acid systems. J Comput Chem 32: 170–173. 10.1002/jcc.21596 [DOI] [PubMed] [Google Scholar]
- Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M, Tsai JC, Weissman JS, Dueber JE, Qi LS, et al. 2015. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 160: 339–350. 10.1016/j.cell.2014.11.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaychikov E, Martin E, Denissova L, Kozlov M, Markovtsov V, Kashlev M, Heumann H, Nikiforov V, Goldfarb A, Mustaev A. 1996. Mapping of catalytic residues in the RNA polymerase active center. Science 273: 107–109. 10.1126/science.273.5271.107 [DOI] [PubMed] [Google Scholar]
- Zhang X-H, Tee LY, Wang X-G, Huang Q-S, Yang S-H. 2015. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther Nucleic Acids 4: e264 10.1038/mtna.2015.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.