Abstract
Eukaryotic genes often generate a variety of RNA isoforms that can lead to functionally distinct protein variants. The synthesis and stability of RNA isoforms is poorly characterized because current methods to quantify RNA metabolism use short-read sequencing and cannot detect RNA isoforms. Here we present nanopore sequencing–based isoform dynamics (nano-ID), a method that detects newly synthesized RNA isoforms and monitors isoform metabolism. Nano-ID combines metabolic RNA labeling, long-read nanopore sequencing of native RNA molecules, and machine learning. Nano-ID derives RNA stability estimates and evaluates stability determining factors such as RNA sequence, poly(A)-tail length, secondary structure, translation efficiency, and RNA-binding proteins. Application of nano-ID to the heat shock response in human cells reveals that many RNA isoforms change their stability. Nano-ID also shows that the metabolism of individual RNA isoforms differs strongly from that estimated for the combined RNA signal at a specific gene locus. Nano-ID enables studies of RNA metabolism at the level of single RNA molecules and isoforms in different cell states and conditions.
In metazoan cells, a single gene locus can give rise to a variety of different RNA molecules that are generally referred to as isoforms. These RNA isoforms can differ in their 5′- and 3′-ends that arise from the use of different transcription start sites and polyadenylation sites, respectively (Pelechano et al. 2013; Core et al. 2014; Chen et al. 2016; Turner et al. 2018). In addition, alternative splicing results in RNA isoforms that differ in the composition of their RNA body (Tilgner et al. 2015; Garalde et al. 2018). Different mRNA isoforms can result in functionally different proteins. Vulnerabilities in splicing can lead to nonfunctional protein products. Diseases have been linked to alternative splicing, which can generate aberrant RNA isoforms (Li et al. 2016). Duchenne muscular dystrophy (DMD), for example, can be pinpointed to a single gene encoding the protein dystrophin. The underlying aberrant RNA isoform shows a different splicing pattern and leads to a nonfunctional protein, which disrupts muscular cell integrity (Long et al. 2018). Likewise, the three most common types of breast tumors are linked to exon skipping and intron retention (Eswaran et al. 2013).
RNA isoforms can also differ in their stability. The untranslated region of an RNA molecule, which contains regulatory elements, can differ in length between isoforms and influence stability (Mayr 2017). The length of the poly(A)-tail at the 3′-end of RNA molecules can also vary between isoforms, affecting RNA stability (Houseley and Tollervey 2009; Falcone and Mazzoni 2018) and, in some cases, resulting in disease (Yamaguchi et al. 2018).
Not much is known, however, about the synthesis and stability of single RNA isoforms in cells because the systematic characterization of RNA isoforms and their metabolism is technically difficult. In particular, the detection, quantification, and estimation of the stability of RNA isoforms is essentially impossible with short-read RNA sequencing methods because those reads generally cannot be assigned to RNA isoforms. Also, alternative splicing patterns can be manifold and are difficult to identify using short-read sequencing approaches (Steijger et al. 2013). Finally, although the length of poly(A)-tails of RNAs can be measured on a genome-wide basis (Chang et al. 2014; Subtelny et al. 2014), they cannot currently be obtained at the level of individual RNA isoforms.
The architecture of RNA isoforms has been addressed so far by short-read RNA sequencing approaches such as DARTS (Zhang et al. 2019), VastDB (Tapial et al. 2017), and MPE-seq (Xu et al. 2019) to study alternative splicing or TIF-seq (Pelechano et al. 2013; Chen et al. 2016) to elucidate combinations of paired 5′- and 3′-ends of individual RNAs. More recent approaches include long-read sequencing on the Pacific Biosciences (PacBio) SMRT sequencing platform (Tilgner et al. 2015) or Oxford Nanopore Technologies (ONT) nanopore sequencing platform (Garalde et al. 2018; Clark et al. 2020; Tang et al. 2020). With these methods, however, it is not possible to study the metabolism of individual RNA isoforms as they lack the ability to assign age to single reads.
Methods to measure the synthesis and stability of RNA in a combined manner for entire gene loci are available (Dolken et al. 2008; Miller et al. 2011; Rabani et al. 2011). Transient transcriptome sequencing (TT-seq) is a protocol used to distinguish newly synthesized from pre-existing RNA in human cells (Schwalb et al. 2016). TT-seq involves a brief exposure of cells to the nucleoside analog 4-thiouridine (4SU). 4SU is incorporated into RNA during transcription, and the resulting 4SU-labeled RNA can be purified and sequenced to provide a snapshot of immediate transcription activity. RNA synthesis and stability at the level of the combined RNA signal can then be computationally inferred for a given gene locus.
Recent methods to assess RNA stability include SLAM seq (Herzog et al. 2017) and TimeLapse-seq (Schofield et al. 2018). Like TT-seq, SLAM seq and TimeLapse-seq involve an exposure of cells to 4SU for labeling of newly synthesized RNA. A chemical modification of the incorporated 4SU then facilitates the identification of labeled RNA in silico without the need for purification. All of these methods, however, have limitations. First, sequencing reads can usually be assigned to only entire gene loci and not to RNA isoforms and thus only allow for a combined RNA stability assessment. Second, they require template amplification, which can lead to an imbalance in measured sequences and information loss, for example, modified RNA bases (Shendure et al. 2017). Finally, labeled RNA purification (TT-seq) and cDNA library preparation (TT-seq, SLAM seq, and TimeLapse-seq) can also introduce biases, such as cross-contamination with unlabeled RNA and read duplication.
Therefore, monitoring RNA metabolism at the level of RNA isoforms requires a method that can detect individual RNA molecules. Recent advances in long-read nanopore sequencing indeed enable the sequencing of single, full-length RNA molecules (Garalde et al. 2018). Nanopore technology can directly sequence the original native RNA molecule with its modifications, either natural or acquired by metabolic RNA labeling. Moreover, with the availability of the entire RNA and coding sequence (CDS) within a single read, one can unambiguously and directly determine exon usage (Clark et al. 2020). Direct RNA long-read nanopore sequencing also has the potential to detect the position and length of the poly(A)-tail along with each individual isoform.
Here we aim at developing nanopore sequencing–based isoform dynamics (nano-ID), a method that combines metabolic RNA labeling, native RNA long-read nanopore sequencing, machine learning, and computational modeling to fully characterize RNA isoform dynamics.
Results
Experimental design
To monitor the RNA metabolism at the level of single isoforms, we sought to combine metabolic RNA labeling with direct, single-molecule RNA nanopore sequencing (Fig. 1A). By culturing cells in the presence of a nucleoside analog, cells will take up and incorporate the analog in nascent RNA during transcription, making it possible to distinguish newly synthesized RNA isoforms from pre-existing RNA isoforms in silico based on the quantification of analog-containing subpopulations. From this, the synthesis rate and stability of single RNA isoforms can be inferred. To dynamically characterize functional and fully processed RNA transcripts, we decided to measure polyadenylated RNA species. The library preparation kit offered by Oxford Nanopore Technologies for direct RNA sequencing is specifically optimized for this purpose. A 3′ poly(A)-tail–specific adapter is first ligated to the RNA transcript. This is followed by ligation of a second sequencing adapter equipped with a motor protein to the transcript-specific adapter. The preparation of RNA libraries from biological samples for direct RNA nanopore sequencing is established and can be performed within 2 h (Garalde et al. 2018). However, we faced significant challenges both in finding a suitable nucleoside analog for RNA labeling and in detection of labeled RNA isoforms. Challenges in detection are rooted in a low labeling efficiency, which is known to be limited to ∼2%–3%, that is, only two or three out of 100 natural nucleosides are replaced by the analog (Jao and Salic 2008). Furthermore, the nucleoside context, namely, sequences flanking the analog, poses additional difficulties for detection.
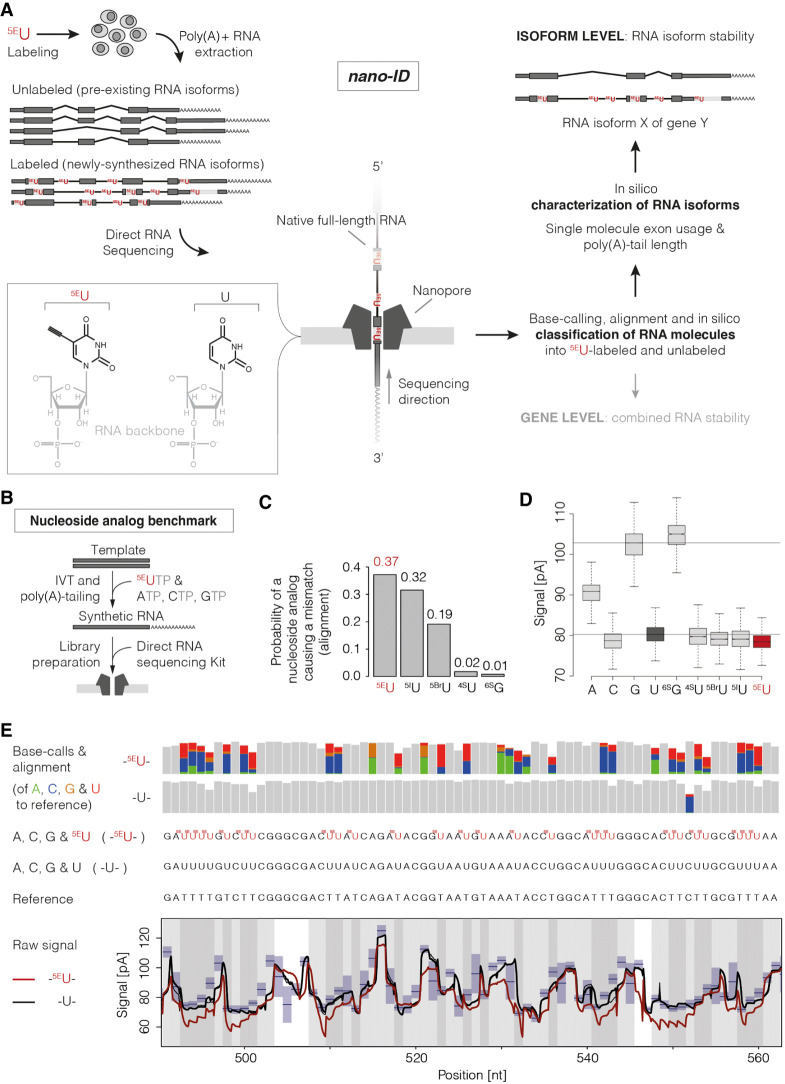
Figure 1.
Nanopore sequencing–based isoform dynamics (nano-ID) combines metabolic RNA labeling with long-read nanopore sequencing of native RNA molecules. (A) Experimental schematic of 5EU-labeled RNA isoforms subjected to direct RNA long-read nanopore sequencing. Metabolic labeling of human K562 cells with the nucleoside analog 5-ethynyluridine (5EU) in vivo. Newly synthesized RNA isoforms will incorporate 5EU instead of standard uridine (U) residues. Newly synthesized RNA isoforms (labeled) can then be distinguished from pre-existing RNA isoforms (unlabeled) in silico after sequencing the native full-length molecules on an array of nanopores (Garalde et al. 2018). 5EU-containing RNA isoforms are computationally traceable and can thus be classified. Identification and quantification of single molecules subsequently enable assessment of exon usage, poly(A)-tail length, and RNA isoform stability. (B) Experimental schematic to derive synthetic RNAs for nucleoside analog benchmark. RNAs were in vitro transcribed: Either the standard bases A, U, C, and G were used as a control, or one of the natural bases was exchanged for a nucleoside analog (shown for 5EU). (C) Barplot showing the probability of a single-nucleoside analog to cause a mismatch in the alignment (compared with natural U or G, Methods) of all tested nucleoside analogs (5EU). (5BrU) 5-bromouridine; (5IU) 5-iodouridine; (4SU) 4-thiouridine; (6SG) 6-thioguanine. (D) Box plots showing the electric current readout (averaged per read) of the nanopore in pico-Amperes (pA; y-axis) associated with different nucleoside analogs 5EU, 5BrU, 5IU, 4SU, and 6SG (center position in 5-mer) in comparison to A, C, G, and U. Black horizontal lines indicate median raw electric current readout associated with G and U nucleosides. (E, top) Base miscalls (colored vertical bars) of the standard base-calling algorithm for synthetic RNAs containing 5EU instead of U (‘-5EU-,’ 7756 molecules) and synthetic control RNAs (‘-U-,’ 17,149 molecules) in comparison to the original sequence (reference) of an exemplary region on synthetic RNA “Spike-in 8” (Methods) (Supplemental Table S3). (Middle) Synthetic RNA sequences with (-5EU-) and without 5EU (-U-). (Bottom) Alignment of the raw signal readout of the nanopore in pico-Amperes (pA) to the reference sequence (nt). Synthetic control RNAs (-U-, 17,149 molecules) are shown in black. 5EU-containing synthetic RNAs are shown in red (-5EU-, 7756 molecules). Traces represent the average signal of all molecules. 5EU-containing synthetic RNAs show a clear deviation from the expected signal level in blue. Blue boxes indicate mean and standard deviation of 5-mers in the nanopore (provided by ONT).
5-Ethynyluridine can be detected in RNA by nanopore sequencing
To investigate if nucleoside analogs incorporated into RNA are detectable in the nanopore, we used synthetic RNAs derived from the ERCC RNA spike-in mix (Invitrogen). These synthetic RNAs of approximately 1000 nucleotides (nt) in length were chosen with similar U content (Supplemental Table S3). RNAs were transcribed in vitro either using the standard bases A, U, C, or G as a control or using one of the natural bases exchanged for a nucleoside analog (Fig. 1B; Methods). Subsequently, we subjected these synthetic RNAs to direct RNA nanopore sequencing (Supplemental Fig. S1A,B). We compared the nucleoside analogs 5-Ethynyluridine (5EU), 5-bromouridine (5BrU), 5-iodouridine (5IU), 4sU, and 6-thioguanine (6sG). To this end, we used the base-called and aligned direct RNA sequencing results to calculate how probable the identification would be at the level of single nucleotides. In particular, we assessed how likely a single-nucleoside analog was to cause a mismatch in the reference alignment in comparison to natural U or G (Fig. 1C, Methods).
The thiol-based analogs, 4sU and 6sG, showed lower incorporation efficiencies during in vitro transcription (IVT) and led to substantially shorter reads during nanopore sequencing. The original molecule was putatively full-length, but we observed that these reads did not span the entire molecule (Supplemental Fig. S1A). 5EU and 5IU could be detected to a similar extent by nanopore sequencing, whereas 5BrU was less easily recognized (Fig. 1C,D). Because 5EU has already been established to label endogenous RNAs in mammalian cells without toxic effects (Jao and Salic 2008; Bharmal et al. 2010; Abe et al. 2012), we used 5EU for a more detailed analysis. Approximately 37% of all U positions in 5EU-containing synthetic RNAs cause mismatches above background in reference sequence alignment after base-calling and can thus be discerned from U (Fig. 1E; Supplemental Fig. S1B,E). This observation is also supported by a shift in the electric current associated with 5EU in comparison to natural U (Fig. 1D). Aberrations caused by stretches of RNA containing 5EU are distinguishable from stretches of RNA containing the naturally occurring U in the nanopore (Fig. 1E). Further analysis showed that different 5-mers harboring the nucleoside analog show varying degrees of detectability (the probability of identification ranges from 0 to 0.6) with a tendency of better detectability toward higher U content (Supplemental Fig. S1F). Taken together, 5EU-based RNA labeling is well suited for nanopore sequencing.
Detection and sequencing of newly synthesized RNA isoforms
We next investigated whether it is possible to use metabolic RNA labeling with 5EU in human cells to detect single RNA molecules by nanopore sequencing. Identification probability assessment using the direct RNA nanopore sequencing of the 5EU-containing synthetic RNAs showed that RNAs are recognizable as 5EU containing with a probability of 0.75 once a minimum length of 500 nt is reached (Supplemental Fig. S1C). This covers the vast majority (93%) of all mature RNAs in the human organism (UCSC RefSeq GRCh38).
We then established direct RNA nanopore sequencing in the human myelogenous leukemia cell line K562. We cultured K562 cells in the presence of 5EU for 60 min (5EU 60 min) in six biological replicates (Methods) (Supplemental Tables S1, S2). The labeling duration was chosen to be in the range of the reported median mRNA half-life in the same cell type (∼50 min) (Supplemental Note 1; Supplemental Fig. S3D; Schwalb et al. 2016). For comparison, we created three biological replicates of cells exposed to 5EU labeling for 24 h (5EU 24 h) and three biological replicates of cells that were not labeled (control) (Methods; Supplemental Tables S1, S2). After base-calling, we could map reads to support 13,673 RefSeq annotated transcription units (RefSeq-TUs) (Methods), 8608 of these were supported in all conditions, and 2068 were supported in all samples (Supplemental Tables S1, S2).
All combined samples were then used to perform a full-length alternative RNA isoform analysis by means of the FLAIR algorithm (Methods; Tang et al. 2020). This makes it possible to define instances of unique exon–intron architecture with unique start and end sites in human K562 cells. Raw human direct RNA nanopore reads were corrected with the use of short-read sequencing data (RNA-seq) to increase splice site accuracy (Methods). We were able to detect 41,090 distinct RNA isoforms with an average of three isoforms per gene. Of which, 63% have not been annotated so far compared with RefSeq. This shows that direct RNA nanopore sequencing uncovers individual RNA isoforms in human K562 cells (Fig. 2D) with high reproducibility (Supplemental Fig. S2B,C).
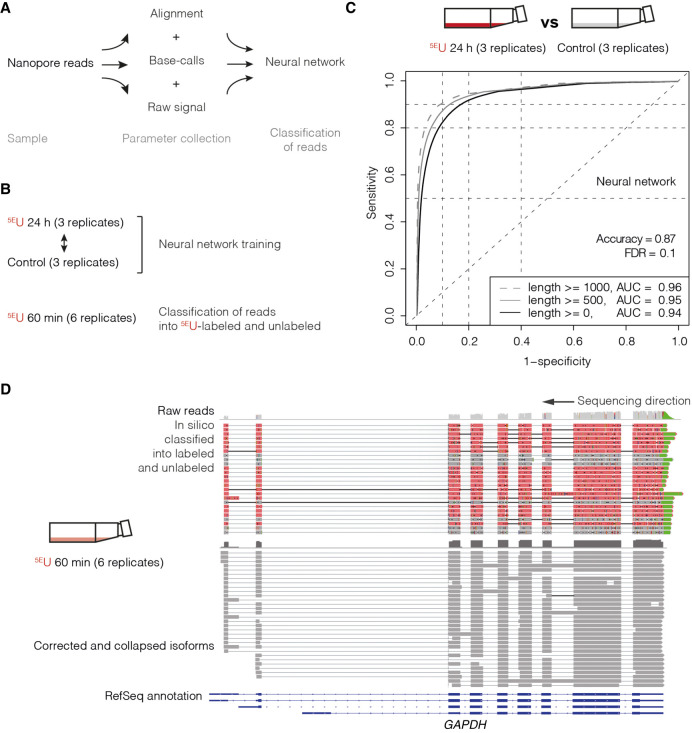
Figure 2.
Direct RNA long-read nanopore sequencing of 5EU-labeled RNA isoforms in human K562 cells. (A) Multilayered data collection scheme. Parameter collection of samples was realized on three different layers: raw signal (electric current), base-call trace values, and alignment-derived mismatch properties (Methods). (B) In this study, data were collected in human K562 cells: control (three replicates) and 5EU 24 h (three replicates), as well as 5EU 60 min (six replicates) (Supplemental Tables S1, S2). The neural network was trained on the 5EU 24 h versus control samples and used to classify reads of the 5EU 60 min samples into 5EU labeled and unlabeled. (C) ROC analysis of fivefold cross-validated neural network training with an accuracy of 0.87 and a false-discovery rate (FDR) of 0.1. Plot shows ROC curves (1 – specificity versus sensitivity) for all reads of the test set (black; alignment length ≥0 nt, AUC = 0.94) (Methods; Supplemental Table S5), for reads with an alignment length >500 nt (gray; alignment length ≥500 nt, AUC = 0.95), and for reads with an alignment length >1000 nt (dashed gray; alignment length ≥1000 nt, AUC = 0.96). (D) Genome browser view of classified direct RNA long-read nanopore sequencing reads of the human GAPDH gene locus on Chromosome 12 (∼8 kbp; Chr12: 6532405–6540375) visualized with the Integrative Genomics Viewer (IGV; version 2.4.10; human hg38) (Robinson et al. 2011). From top to bottom, raw nanopore sequencing reads (unlabeled reads are shown in gray, 5EU-labeled reads are shown in red, and poly(A)-tail is shown in green; shown are typical aligned raw reads below the accumulated coverage of all measured reads), and corrected and collapsed isoforms (dark gray) determined with the FLAIR algorithm (Tang et al. 2020) based on raw reads and RefSeq GRCh38 annotation (blue).
A neural network identifies newly synthesized RNA isoforms
The next step was to derive a computational method that could classify each sequenced RNA molecule into one of two groups, newly synthesized (5EU-labeled) or pre-existing (unlabeled) RNA. To this end, the nucleoside analog 5EU had to be detected in RNA molecules, allowing the quantification of RNA isoforms generated during the 5EU-labeling pulse. Because of the high error rate of nanopore sequencing, the random nature of nucleoside analog incorporation, and the context dependence of detectability (Supplemental Fig. 1F), a single 5EU base-call is inappropriate as an indicator. We instead used the raw signal of the entire RNA nanopore read, including the base-calls and the alignment (Supplemental Fig. 1C,D), to discriminate 5EU-labeled from unlabeled RNAs. This discrimination was implemented as a classifying neural network trained on human K562 direct RNA nanopore sequencing data (Supplemental Tables S1, S2, S5, samples 10–15). We developed a custom multilayered data collection scheme to train a neural network for the classification of human RNA isoforms under the assumption that after 5EU 24 h 5EU-labeling (Supplemental Tables S1, S2, samples 13–15) samples exclusively contain labeled reads and that the control samples (Methods; Supplemental Tables S1, S2, samples 10–12) solely contain unlabeled reads (Fig. 2A,B; Methods). Support for this assumption (24 h 5EU-labeling) is given by the observation that upon 24 h of 5EU labeling, stable RNA species are strongly labeled as detected by fluorescence read-out (Jao and Salic 2008). Furthermore, human RNA half-life distributions of previous studies (Rabani et al. 2011; Schwalb et al. 2016) suggest that there is only a minor fraction of RNA species that lives >24 h. Thus, the majority of RNA molecules in the 5EU 24 h samples (Supplemental Tables S1, S2, samples 13–15) are putatively 5EU containing.
We then trained a neural network (Methods) on the 5EU 24 h versus control samples with an accuracy of 0.87 and a false-discovery rate (FDR) of 0.1 (fivefold cross-validated) (Fig. 2C). To mitigate the risk of overtraining the neural network, we introduced several drop-out layers in the network structure (Methods) (Supplemental Table S5). A ROC analysis (1 – specificity versus sensitivity) for all reads of the test set showed an area under the curve (AUC) of 0.94. For reads with an alignment length >500 nt and >1000 nt, the AUC improved to 0.95 and 0.96 (Fig. 2C; Supplemental Fig. S2B). Each single layer alone and all possible pairwise combinations were only able to yield an AUC between 0.86 and 0.92 (Supplemental Fig. S3C). All three different layers therefore contain additional unique information for training the neural network and classification of reads (Methods). Given that our carefully designed neural network mitigates the risk of overtraining and that we apply computational measures to validate this, we consider the degree of redundancy between layers harmless. Subsequently, we used the trained neural network to classify reads of the 5EU 60 min samples into 5EU labeled and unlabeled. Taken together, 5EU-containing RNA isoforms are computationally detectable with high accuracy (Fig. 2C). Thus, we were able to determine for each single RNA molecule with a low FDR if it had been produced during 5EU labeling or before (Fig. 2C).
Nano-ID estimates the stability of RNA isoforms
The ability to distinguish newly synthesized and pre-existing RNA molecules allowed us to derive estimates for the stability of RNA isoforms. For each single direct RNA nanopore read, we were able to assign the RNA isoform it represents. Additionally, we were able to assess the stability of RNA for single RNA isoforms by applying a first-order kinetic model (Methods) to derive estimates for RNA isoform–specific synthesis and stability. This can be performed based on the number of reads classified as 5EU labeled and unlabeled by the neural network. For a half-life estimate to be considered for further analysis, the corresponding RefSeq-TU had to be supported by at least five reads in each of the six biological replicates (5EU 60 min) (Methods; Supplemental Tables S1, S2). Comparison of our half-life estimates with estimates derived from the short-read approach 4sU-seq (Schwalb et al. 2016) yielded only a moderate correlation of 0.25, which is likely owing to technical differences such as the purification step of premature RNAs not present in nano-ID (Supplemental Fig. S3A). Taken together, nano-ID has the capability to infer synthesis rates and stability estimates of individual RNA isoforms in different cell states and conditions and to thus monitor their dynamic metabolism.
Determining factors of RNA isoform stability
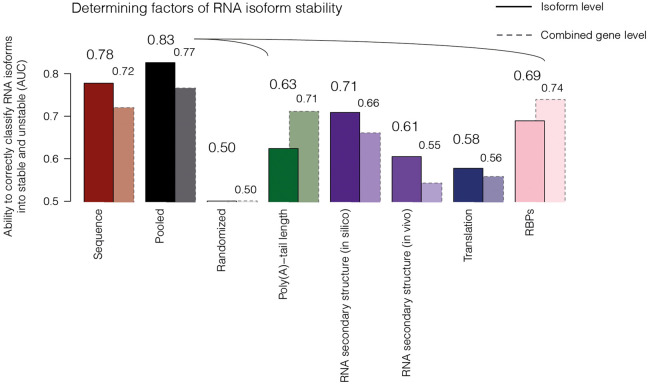
To show the strength of nano-ID derived stability estimates, we wanted to know to what extent determining factors such as sequence, poly(A)-tail length (Methods; Supplemental Fig. S3B, nanopolish) (Workman et al. 2019), RNA secondary structure in silico or in vivo (as measured by DMS-seq) (Rouskin et al. 2014), translation efficiency (as measured by Ribo-seq) (Ingolia et al. 2014), and RNA-binding proteins (RBPs, as measured by eCLIP) (Van Nostrand et al. 2020) influence RNA stability on the combined RNA (i.e., RNA that originates from the entire gene loci regardless of isoform assignment, gene level) as well as at the level of RNA isoforms. To address this, we asked whether we can predict if an RNA isoform is stable (above median half-life) or unstable (below median half-life) using a classifying neural network (Fig. 3) compared with the combined RNA (gene level). This analysis suggested that all of these factors are indeed associated with RNA stability. For features such as sequence, RNA secondary structure, and translation, the ability to predict RNA stability improved further at the level of RNA isoforms compared with the combined RNA (gene level). This suggests that these features have stronger variability among different isoforms even if they arise from the same genomic locus and, thus, molecular environment. In contrast, poly(A)-tail length and RBP association performed better on the combined gene level, suggesting a regulatory nature of these features on the gene level rather than on the RNA level. Taken together, nano-ID–derived RNA stability estimates can be validated by independent methods and, in principle, allow for further studying factors determining RNA stability.
Figure 3.
Determining factors of RNA isoform stability. Bar plot showing RNA stability related features (x-axis), namely, sequence (red), poly(A)-tail length (green), RNA secondary structure in silico or in vivo (violet), translation efficiency (dark blue), and RNA-binding proteins (RBPs; pink), and their abilities to correctly classify RNA isoforms into stable (above median half-life) or unstable (below median half-life) species (AUC; y-axis) (Methods; Supplemental Tables S4, S6). In black, all pooled features (listed above, excluding sequence and randomization) are represented (pooled). Bars with solid lines depict the features on RNA isoform level. Bars with dashed borders represent (AUC) of features calculated on combined RNA signal (gene level; all RNAs encoded by the entire gene loci regardless of isoform assignment).
Nano-ID monitors RNA isoform dynamics during heat shock
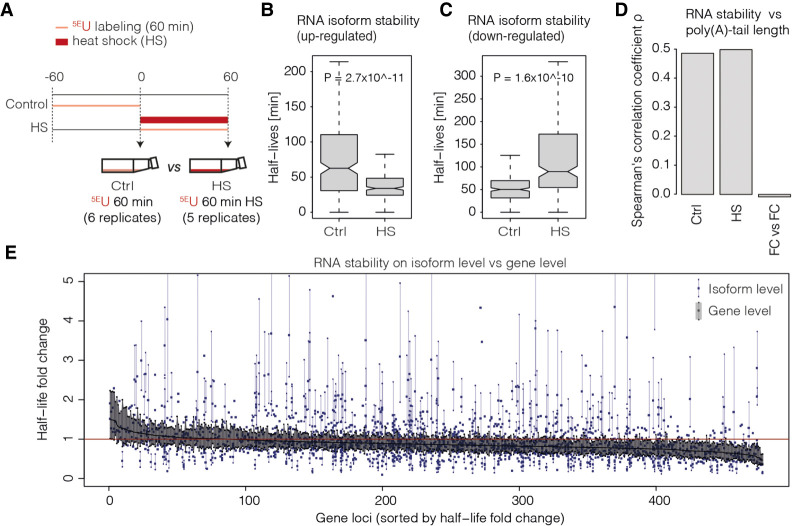
To show the advantages of nano-ID, we subjected human K562 cells to heat shock (42°C) for 60 min in the presence of 5EU (5EU 60 min HS) (Fig. 4A). The heat shock response provides a well-established model system (Supplemental Fig. S4; Theodorakis et al. 1989; Sistonen et al. 1992; Mathew et al. 1998; Vihervaara et al. 2013, 2017; Niskanen et al. 2015). We then asked if the RNA isoform synthesis rate is altered by heat shock and observed significant differential RNA isoform synthesis rates for 272 isoforms (fold change >1.25 and P-value < 0.1). One hundred fifty-four RNA isoforms were significantly up-regulated, whereas 118 were down-regulated (Supplemental Fig. S4B). RNA isoforms that changed their synthesis rate during heat shock were also observed to alter their stability (Fig. 4B,C). In particular, RNA isoforms that were up-regulated in their synthesis during heat shock also showed a lower stability and vice versa, resembling typical stress response behavior (Miller et al. 2011). The destabilization of up-regulated RNA isoforms is likely to ensure their rapid removal toward the end of the stress response.
Figure 4.
Nano-ID monitors RNA isoform dynamics during heat shock (HS). (A) Experimental set-up of the HS treatment (60 min at 42°C) in human K562 cells. 5EU labeling was performed for 60 min. (B) Box plot showing half-lives (min) of significantly up-regulated RNA isoforms in HS (5EU 60 min HS) against control samples (5EU 60 min). (C) As in B for significantly down-regulated RNA isoforms. (D) Bar plot showing correlation (Spearman's rank correlation coefficient ρ) of RNA half-lives and poly(A)-tail lengths before and after HS (1310 loci). The third bar shows the correlation of RNA half-life fold change upon HS and poly(A)-tail length fold change upon HS. (E) Half-life fold changes upon HS (y-axis) depicted for individual RNA isoforms alongside the half-life fold change derived from combined RNA (gene level; all RNAs encoded by the entire gene loci regardless of isoform assignment). Shown are 478 high-confident loci (x-axis). All estimates are supported across biological replicates (n ≥ 5) and conditions (HS; control). Half-life estimates for combined RNA (gene level) are depicted as a black line (sorted in decreasing order). Blue dots represent individual RNA isoform half-life estimates (1988 isoforms in total). All RNA isoform half-life estimates as well as one respective combined RNA half-life estimate (in black) sharing a common x-axis coordinate belong to the same gene loci. Vertical blue and black lines represent standard deviations of individual estimates. For individual RNA isoform half-life estimates, standard deviations are only shown if not overlapping with the standard deviation of the respective combined half-life estimates (black).
The metabolism of individual RNA isoforms differs from combined RNA estimates
To show the importance of individual RNA isoform assessment, we first derived estimates for the half-lives of combined RNAs (gene level) regardless of isoform assignment under steady-state conditions (Methods). We found a robust correlation of combined RNA stability with poly(A)-tail length (Spearman's rank correlation coefficient 0.48) (Fig. 4D). We then asked whether changes in RNA stability would also be reflected in changes in poly(A)-tail length upon heat shock, and this was not the case (Fig. 4D). Following this, we asked if there is differential behavior in RNA stability of individual RNA isoforms genome-wide or if, instead, RNA isoforms generally reflect the changes in RNA stability of the combined RNA from their respective gene loci. To this end, we compared changes in RNA stability estimates of individual RNA isoforms to those from combined RNAs and found that the dynamics of individual RNA isoforms during heat shock varies globally. This indicates that the observed RNA stability fold changes of individual RNA isoforms upon heat shock can differ significantly from RNA stability fold changes of the combined RNA level (gene level) that stems from entire gene loci (Fig. 4E). This analysis includes the uncertainty of individual estimates over replicate measurements and clearly indicates the need for detailed individual RNA isoform assessment as individual RNA isoforms can lead to functionally distinct protein variants. Thus, it is crucial to also study the behavior of individual RNA isoforms and not just an averaged and combined view of the entire gene locus. Taken together, this shows that conclusions made using only combined RNAs can be misleading and that much can be learned at the level of single RNA isoforms by using nano-ID.
Discussion
Here we develop nano-ID, a method that can resolve the dynamic metabolism of functional and fully processed RNA isoforms at the level of single native RNA molecules. Nano-ID combines metabolic RNA labeling with native RNA nanopore sequencing to enable RNA isoform identification. In combination with machine learning and computational modeling, nano-ID is able to fully characterize RNA isoform dynamics by means of age assignment and a measurement of the poly(A)-tail length of a single RNA molecule. Nano-ID visualizes changes in RNA isoform synthesis and stability, revealing a so-far-hidden layer of gene regulation. Nano-ID will, in principle, enable further study into the extent to which determining factors such as sequence, poly(A)-tail length, RNA secondary structure, translation efficiency, and RBPs influence RNA stability on the combined gene level as well as at the level of RNA isoforms. Nano-ID thus allows the study of transcriptional regulation in unprecedented detail and can prevent misleading conclusions that would be obtained when only combined RNA from an entire gene locus can be considered, as is the case with cDNA short-read sequencing approaches such as RNA-seq, 4sU-seq, or TT-seq.
Nano-ID has many advantages over other sequencing-based transcriptomic strategies because the original native RNA molecule is sequenced. In particular, there is no need for fragmentation of RNA before sequencing. Hence, in the present study, reads can be assigned to RNA isoforms with high confidence, leaving only residual ambiguity. Furthermore, nano-ID does not require template amplification and thus omits copying errors, sequence-dependent biases, and read duplication events. It therefore also prevents the loss of information on epigenetic modifications and artificially introduced RNA base analogs. Taken together, nano-ID overcomes significant drawbacks and limitations of state-of-the-art approaches, allowing study of RNA metabolism at the level of individual RNA isoforms.
Current limitations to nanopore sequencing are a lower throughput and lower accuracy compared with short-read cDNA sequencing approaches. In the specific context of nano-ID, the random nature of nucleoside analog incorporation and the sequence context (5-mer) dependence of nucleoside analog detectability may be limiting as well. Furthermore, we train the neural network with an approach that bears the risk of learning transcriptome-specific characteristics (i.e., human myelogenous leukemia cells) that might prevent immediate applicability to other model organisms. It is, however, possible to retrain the neural network for a different model organism once such data become available. Our neural network is carefully designed to mitigate the risk of overtraining, and we take strict computational measures to validate this.
These drawbacks, however, are outweighed by the novel information obtained about the age of the RNA sequencing substrates. The longer the sequenced molecules are, the less problematic the lack in accuracy in identifying their origin or classifying them into newly synthesized or pre-existing molecules. In the present study, we show that our algorithmic strategies are sufficient to address metabolic rate estimation in a reliable manner for abundant transcripts in human cells. The requirements for reliable RNA stability estimation that we implemented (threshold on the number of reads; Methods) as well as the depth to which we sequenced control and treated samples in biological replicates (Supplemental Tables S1, S2) are sufficient to detect rate changes in a significant manner for highly abundant RNA isoforms (Fig. 4E). Technical improvements in nanopore sequencing and its computational processing will further improve the sequencing depth and the accuracy of individual read sequences and thus the detectability of nucleoside analogs such as 5EU. This will have a direct impact on the robustness of RNA stability estimation at the level of combined RNA as well as RNA isoforms toward larger subpopulations that can be analyzed. Moreover, there are strategies to enrich for subpopulations of RNAs with transcript-specific adapters (ONT), which would allow an in-depth analysis of a certain loci of interest at a much higher sequencing depth.
A future challenge will involve the development of a novel signal-level base-calling algorithm for direct RNA nanopore sequencing with an extended base alphabet (A, C, G, U, and 5EU). This requires a large amount of artificial sequences to train on. These should ideally contain not only cases of non-5EU-containing 5-mer instances but also all possible combinations of 5EU-containing instances, especially those 5-mers carrying both 5EU and U. Furthermore, increased throughput of the nanopore sequencing platform will further improve the statistical precision of metabolic rate estimation to elucidate low abundance transcripts or transient processes.
Nanopore-based transcriptomic studies will allow us to monitor the formation of transcripts, post-transcriptional processing, export, and translation at the level of single RNA isoforms. Nano-ID is in principle also transferable to single-cell methodologies to catch heterogeneity of the RNA population in any state of the cell. This, however, requires sequencing library preparation with lower input amounts. The use of 5EU is widely established for in vivo applications in the field, such as for fluorescence microscopy. We thus envision that nano-ID is in principle applicable to a large range of organisms, cells, and conditions.
Methods
Labeling and direct RNA nanopore sequencing of synthetic RNAs
Labeled synthetic RNAs and synthetic control RNAs are derived from selected RNAs of the ERCC RNA spike-in mix (Invitrogen) as previously described (Schwalb et al. 2016). Characteristics of selected RNAs of the ERCC RNA spike-in mix are listed in Supplemental Table S3. Briefly, selected spike-in sequences were cloned into a pUC19 cloning vector and verified by Sanger sequencing. For IVT template generation, the plasmid (3 µg) was linearized using a EcoRV-HF (blunt end cut) digestion mix containing a CutSmart buffer and EcoRV-HF enzyme. The digestion mix was incubated for 1 h at 37°C, and the reaction was terminated adding 1/20 volume of 0.5 M EDTA. Subsequently, DNA was precipitated in 1/10 volume of 3 M sodium acetate (pH 5.2) and 2 volumes of 100% ethanol for 15 min at −20°C. DNA was collected by centrifugation at 4°C and 16,000g for 15 min. The pellet was washed twice using 75% ethanol. DNA was air-dried and resuspended in 5 µL of H2O at a concentration of 0.1–1.0 µg/µL (quantified by NanoDrop). Synthetic RNAs were in vitro transcribed using the MEGAscript T7 kit (Invitrogen). IVT of synthetic control RNAs was performed following the manufacturer's instruction. For IVT of labeled synthetic RNAs, 100% of UTP (resp. GTP) was substituted with either 5-ethynyl-UTP (5EU; Jena Bioscience), 5-bromo-UTP (5BrU; Sigma-Aldrich), 5-iodo-UTP (5IU; TriLink BioTechnologies), 4-thio-UTP (4SU; Jena Bioscience), or 6-thio-GTP (6SG; Sigma-Aldrich). Note that, for performing a successful IVT with 4-thio-UTP and 6-thio-GTP, only a reduction to 80% substitution gave a successful yield. IVT reactions were incubated at 37°C. After 4 h, reaction volume was filled up with H2O to 40 µL, and then 2 µL of TURBO DNase was added and incubated for additional 15 min at 37°C. Synthetic RNAs were purified with RNAClean XP beads (Beckman Coulter) following the manufacturer's instructions. The final synthetic RNA pool contained an equal mass of all respective synthetic RNAs in a given library (Supplemental Tables S1, S2). RNA was quantified using Qubit (Invitrogen). RNA quality was assessed with the TapeStation system (Agilent). Synthetic RNA pools were poly(A)-tailed using the Escherichia coli Poly(A) Polymerase (NEB). The reaction was incubated for 5 min and stopped with 0.1 M EDTA. Poly(A)-tailed synthetic RNA pools were then purified with phenol:chloroform:isoamyl alcohol and precipitated. Purified poly(A)-tailed synthetic RNA pools were subsequently subjected to direct RNA nanopore sequencing library preparation (SQK-RNA001, Oxford Nanopore Technologies) following the manufacturer's protocol. All libraries were sequenced on a MinION Mk1B (MIN-101B) for 20 h, unless reads sequenced per second stagnated.
Culturing of human K562 cells
Human K562 erythroleukemia cells were obtained from DSMZ (ACC-10). K562 cells were cultured antibiotic-free in accordance with the DSMZ cell culture standards in RPMI 1640 medium (Thermo Fisher Scientific) containing 10% heat inactivated fetal bovine serum (FBS; Thermo Fisher Scientific) and 1× GlutaMAX supplement (Thermo Fisher Scientific) at 37°C in a humidified 5% CO2 incubator. Cells used in this study display the phenotypic properties, including morphology and proliferation rate, that have been described in literature. Cells were verified to be free of mycoplasma contamination using a Plasmo test mycoplasma detection kit (InvivoGen). Biological replicates were cultured independently.
5EU labeling and direct RNA nanopore sequencing of human K562 cells
K562 cells were kept at low passage numbers (fewer than six) and at optimal densities (3 × 105–8 × 105) during all experimental setups. Per biological replicate, K562 cells were diluted 24 h before the experiment was performed (Supplemental Tables S1, S2). Per 5EU 60 min sample (six replicates), cells were incubated in 5% CO2 for 1 h at 37°C after a final concentration of 500 µM 5EU (Jena Bioscience) was added. Per 5EU 24 h sample (three replicates), cells were incubated in 5% CO2 for 24 h at 37°C. 5EU was added three times during the 24-h incubation, namely, every 8 h (0 h, 8 h, 16 h) at a final concentration of 500 µM. Control samples were not labeled (three replicates). Per 5EU 60 min HS (heat shock) sample (five replicates), cells were incubated for 5 min at 42°C (until cell suspension reached 42°C), and then 5EU was added at a final concentration of 500 µM. Further, heat shock treatments were performed in a water bath (LAUDA, Aqualine AL12) for 1 h at 42°C. Temperature was monitored by thermometer. To avoid transcriptional changes by freshly added growth medium, fresh growth medium was added ∼24 h before heat shock treatments (Mahat and Lis 2017). Exactly after the labeling duration, cells were centrifuged at 1500g for 2 min at 37°C. Total RNA was extracted from K562 cells using QIAzol (Qiagen) according to the manufacturer's instructions. Poly(A) RNA was purified from 1 mg of total RNA using the µMACS mRNA isolation kit (Milteny Biotec) following the manufacturer's protocol. The quality of poly(A) RNA selection was assessed using the TapeStation system (Agilent). Poly(A)-selected RNAs were subsequently subjected to direct RNA nanopore sequencing library preparation (SQK-RNA001, SQK-RNA002, Oxford Nanopore Technologies) following the manufacturer's protocol with 1000 ng input. All libraries were sequenced on a MinION Mk1B (MIN-101B) for 48 h, unless reads sequenced per second stagnated.
RNA-seq
Two biological replicates of K562 cells were diluted 24 h before the experiment was performed. Per replicate, 3.6 × 107 cells in growth medium were labeled at a final concentration of 500 µM 4-thio-uracil (4sU; Sigma-Aldrich) and incubated in 5% CO2 for 5 min at 37°C. Exactly after 5 min of labeling, cells were harvested at 1500g for 2 min at 37°C. Total RNA was extracted from K562 cells using QIAzol according to the manufacturer's instructions except for the addition of 150 ng RNA spike-in mix (Schwalb et al. 2016) together with QIAzol. To isolate poly(A) RNA from 75 µg of total RNA, two subsequent rounds of purification by Dynabeads Oligo (dT)25 (Invitrogen) were performed. Purification based on the manufacturer's instructions was performed twice, using 1 mg of Dynabeads Oligo (dT)25 beads for the first round and 0.5 mg for the second round of purification. The quality of polyadenylated RNA selection was assessed using RNA ScreenTape on a TapeStation (Agilent). Sequencing libraries were prepared using the NuGEN ovation universal RNA-seq kit according to the manufacturer's instructions. Fragments were amplified by 10 cycles of PCR and sequenced on an Illumina NextSeq 550 in paired-end mode with a 75-bp read length.
Direct RNA nanopore sequencing data preprocessing of synthetic RNAs
Direct RNA nanopore sequencing reads were obtained for each of the samples (Supplemental Tables S1, S2). FAST5 files were base-called using Guppy 3.1.5 (Oxford Nanopore Technologies) with the following parameters: guppy_basecaller -i fast5 -s basecalled ‐‐num_callers 1 ‐‐cpu_threads_per_caller 12 -c rna_r9.4.1_70bps_hac.cfg -r ‐‐fast5_out ‐‐calib_detect ‐‐u_substitution on -q 0. Direct RNA nanopore sequencing reads were mapped with GraphMap 0.5.2 (Sovic et al. 2016) to the synthetic RNA reference sequence with the following parameters: graphmap align ‐‐evalue 10−10. Further data processing was performed using the R/Bioconductor environment. Note that for the IVT of our synthetic RNA pools, 100% of UTP was substituted with 5-ethynyl-UTP (among other analogs). This creates a sequence dissimilarity of >20% in the alignment necessary for mapping. Note also that these synthetic RNA pools just serve the purpose of benchmarking nucleosides to be combined with nanopore sequencing. These samples are thus not involved in training and validation of the neural network.
Direct RNA nanopore sequencing data preprocessing of human K562 cells
Direct RNA nanopore sequencing reads were obtained for each of the samples (Supplemental Tables S1, S2). FAST5 files were base-called using Guppy 3.1.5 (Oxford Nanopore Technologies) with the following parameters: guppy_basecaller -i fast5 -s basecalled ‐‐num_callers 1 ‐‐cpu_threads_per_caller 12 -c rna_r9.4.1_70bps_hac.cfg -r ‐‐fast5_out ‐‐calib_detect ‐‐u_substitution on -q 0. Direct RNA nanopore sequencing reads were mapped with minimap2 2.10 (Li 2018) to the GRCh38/hg38 genome assembly (Human Genome Reference Consortium) with the following parameters: minimap2 -ax splice -k14 ‐‐secondary = no. SAMtools 1.3.1 (Li et al. 2009) was used to quality filter SAM files, whereby alignments with MAPQ smaller than 20 (-q 20) were skipped. Further data processing was performed using the R/Bioconductor environment (v3.3.3; R Core Team 2017) and Python (see Software availability).
Probability of 5EU-labeled RNA isoform identification based on synthetic RNAs
The following parameters were collected on the direct RNA nanopore sequencing data of synthetic RNAs and used to calculate the probability of identification of a 5EU-labeled RNA isoform as labeled. Detectability d is the number of 5EU caused mismatches in the 5EU-labeled sample. Background b is the number of U caused mismatches in the unlabeled control sample. Given these parameters, the probability of identification p can be calculated as the probability of a U-based mismatch being caused by a 5EU in the transcript as
with 0.25 as the empirical probability of U content, and labeling efficiency of 0.028 as the empirical probability of a U being replaced by a 5EU in the labeling process (Jao and Salic 2008). This then allows to calculate the probability of labeled RNA identification pRNA as
the probability that an RNA contains at least one detectable 5EU.
Definition of transcription units based on the UCSC RefSeq genome assembly GRCh38 (RefSeq-TUs)
For each annotated gene, transcription units were defined as the interval from the first start site to the last poly(A)-site of all existing inherent transcript isoforms (UCSC RefSeq GRCh38). It thus defines the area of transcription of the entire gene locus. Transcription units were considered expressed with a minimum of two nanopore reads assigned in each 5EU 60 min samples.
Definition of isoform-independent exonic regions (constitutive exons)
Isoform-independent exonic regions were determined using a model for constitutive exons based on UCSC RefSeq annotation (GRCh38). It defines the set of consecutive exonic bases (i.e., portion of or entire exon) that belong to each isoform of the gene (Bullard et al. 2010).
Isoform determination and quantification for human K562 cells
The full-length alternative isoform analysis of RNA (FLAIR) algorithm (Tang et al. 2020) was used following the developer's instructions for the correction and isoform definition of raw human K562 direct RNA nanopore reads. Corrected and collapsed isoforms were obtained by adding short-read data (RNA-seq) to help increase splice site accuracy of the nanopore read splice junctions (https://github.com/BrooksLabUCSC/FLAIR). First, “flair align” was used to align all reads to the GRCh38/hg38 genome assembly (Human Genome Reference Consortium) using minimap2 2.10 (Li 2018) and SAMtools 1.3.1 (Li et al. 2009) with the following parameters: align -m ./minimap2 -sam ./SAMtools -c chromsizes.tsv -n –p. Second, “flair correct” was used to correct misaligned splice sites using genome annotation (UCSC RefSeq GRCh38) and short-read splice junctions (SJ.out.tab, extracted from assay 21 and 22 in Supplemental Table S1 via STAR 2.3.0; Dobin and Gingeras 2015) with the following parameters: flair correct -f RefSeq.gtf –c chromsizes.tsv -j SJ.out.tab -n. Third, “flair collapse” was used to define high-confidence isoforms from corrected reads using minimap2 2.10 (Li 2018) and SAMtools 1.3.1 (Li et al. 2009) with the following parameters: flair collapse -f RefSeq.gtf -m ./minimap2 -sam ./SAMtools. For nanopore read to isoform assignment and quantification of isoforms, reads were mapped with minimap2 2.10 (Li 2018) to the resulting high-confidence isoforms generated with flair collapse using the following parameters: minimap2 -ax splice -k14 ‐‐secondary = no. Further data processing was performed using the R/Bioconductor environment and Python (see Software availability).
Differential expression analysis
Differential expression analysis was performed using the R/Bioconductor package DESeq2 (Love et al. 2014). Isoforms with a fold change of at least 1.25 and a P-value lower than 0.05 were considered differentially induced or repressed dependent on the observed direction.
Parameter collection for neural network training and classification
For each read in each human K562 sample (5EU 60 min, control, 5EU 24 h, and 5EU 60 min HS), we obtained approximately 4700 parameters from three different layers: The raw signal layer consisting of electric current readout in [pA], the base-calls layer consisting of trace and move values (base-caller confidence and movement of substrate through the nanopore), and the alignment layer consisting of mismatch and indel properties derived from the reference sequence alignment. All three layers are described in more detail in the following.
Raw signal layer
Raw signal values are assigned to the (base-called) read sequence by means of the “move,” “stride,” and “num_events” values supplied by the base-caller. In brief, the raw electric current values are divided into “num_events” intervals with a length given by the “stride” parameter. Subsequently, the “move” parameter is used to combine intervals belonging to the same 5-mer. This enables to assign mean raw signal values to 5-mers (see Software availability). The raw signal layer consists of 2048 parameters in total. This entails raw electric current signal averaged for each possible 5-mer of nucleotides as well as the raw electric current signal averaged for each possible 3-mer centered in a 5-mer. The latter parameters were collected for U-containing and non-U-containing instances. In addition to that, raw electric current signal was gathered for 5-mers with all possible nucleotides in their center position also for U-containing and non-U-containing instances, as well as 5-mers exclusively leading or lagging U content. All collected raw signal parameters were standardized on all non-U-containing instances given the mean values of the pore model (mean electric current signal of 5-mers in the nanopore) provided by Oxford Nanopore Technologies. In brief, the raw signal is calibrated by means of a standardization that calibrates the entire signal to a unified signal level and magnitude (ONT pore model), but only considering raw signal assigned to non-U-containing 5-mers for the calculation of the standardization parameters. This approach unifies the raw signal on all non-U-containing 5-mers but keeps the relative signal changes on U-containing 5-mers unaltered. Note that it is necessary to calibrate global differences in the electric current readout between different reads. These global differences are likely owned to fluctuations in the voltage applied and different sensitivities of individual nanopores. Given the incompleteness of 5-mers (especially non-U-containing) in typical reads, we implemented the strategy of looking at the centered 3-mer contained in a 5-mer. Given that the centered 3-mer contained in a 5-mer is more influential on the electric current readout of the nanopore (Supplemental Fig. S3E), it makes the necessary standardization more robust.
The base-calls layer
The trace values (flip- and flop-bases) supplied by the base-caller (confidence in its output at each position of the signal considered) are assigned to the (base-called) read sequence and can thus be differentiated among different types of nucleotides. The base-calls layer consists of 2330 parameters derived from the trace table summarized for instances showing translocation and those that did not (“move” parameter) as mean, median, and centiles for all flip- and flop-bases and all different possible nucleotides in the called sequence. Note that although base-calls are performed on the raw signal, base identification adds on additional information that is not contained in the raw signal layer (please see Supplemental Fig. S3C). The base-call layer contains information produced by the flip-flop algorithm, which informs on confidence and alternatives of the called bases in the sequence. In some cases, for example, the base-caller will call an instance of 5EU as a native uridine. In such a case however, it will do so with a much lower probability. The base-call layer also contains the number of moves of the measured RNA molecule through the nanopore. This is largely determined by the motor protein and will likely be influenced by instances of 5EU in the measured molecule.
The alignment layer
The alignment is additional information that is added to the other two previously described layers of raw signal and base-calls, given that it informs on inserts, deletions, and homopolymer length with respect to the reference sequence (please see Supplemental Fig. S3C). The alignment layer consists of 147 parameters, including the length of the reads, nucleotide occurrences, number of nucleotide transitions (mismatch statistics, please also see Supplemental Fig. S1E), number of inserts, and deletions on a single-nucleotide basis, as well as on a 5-mer basis for U-containing and non-U-containing instances. Note that the model thus incorporates background frequencies of sequencing errors for the definitive classification of molecules into newly synthesized and pre-existing based on each entire read. In other words, reads with low sequencing quality are allowed to have more miscalls before they are classified as newly synthesized. For more detailed information on parameter collection, please see R code (see Software availability).
Neural network training, validation, and classification of human RNA isoforms into 5EU-labeled and unlabeled
Neural network was trained on the 5EU 24 h (Supplemental Tables S1, S2, samples 13–15) versus control (Supplemental Tables S1, S2, samples 10–12) sample parameters under the assumption that the 5EU 24 h sample solely contains labeled reads and the fact that the control sample solely contains unlabeled reads. Human RNA half-life distributions of previous studies (Rabani et al. 2011; Schwalb et al. 2016) suggest that there is only a minor fraction of RNA species that live >24 h. Thus, the majority of RNA molecules in the 5EU 24 h samples (Supplemental Tables S1, S2, samples 13–15) are putatively 5EU-containing. Further, 24 h of labeling with 5EU has been shown to strongly label stable RNA species (Jao and Salic 2008). The trained neural network consists of eight dense layers with decreasing output shape (dense; units = 512, 265, 128, 64, 32, 16, 8, and 1; activation = “relu” and “sigmoid”; see below) with preceding batch normalization layers. Seven dropout layers (with 25% dropout) in between regularize the attempted classification. Training was conducted on 294,467 reads; validation was performed on 126,130 reads in 40 epochs with the R interface to Keras on a TensorFlow backend (https://github.com/rstudio/keras). For more detailed information on neural network design, please also see Supplemental Table S5 and the R code (see Software availability). The neural network was fivefold cross-validated with an accuracy of 0.87 and a FDR of 0.1 and was used to classify reads of the 5EU 60 min (Supplemental Tables S1, S2, samples 4–9) and 5EU 60 min heat shock (Supplemental Tables S1, S2, samples 16–20) samples into 5EU-labeled and unlabeled. A ROC analysis (1 – specificity vs. sensitivity) for all reads of the test set showed an AUC of 0.94. For reads with an alignment length >500 nt and >1000 nt, the AUC improved to 0.95 and 0.96. This suggests a small potential length bias. This is, however, uncritical to RNA stability assessment, especially on the RNA isoform level, as non-full-length reads are not considered. Note that different network architectures, such as additional dense layers, batch normalization, and dropout percentage, did not change the performance of the classification. The accuracy of the neural network is likely delimited by the lower accuracy of the direct RNA platform rather than the number of reads in the training set. This will very likely improve in the future owing to technical improvements driven by ONT. Note that the implemented multilayered data collection scheme (raw signal, base-calls, and alignment) might cause a high level of redundancy of the underlying data/information. Our neural network, however, is carefully designed, and we take a lot of computational measures to validate this (fivefold cross-validation) and thereby mitigate the risk of overtraining.
Poly(A)-tail length determination
Poly(A)-tail length estimates were calculated with Nanopolishs’ poly(A)-tail feature (nanopolish polya) (Workman et al. 2019) according to the developers’ instructions (http://gensoft.pasteur.fr/docs/nanopolish/0.11.1/quickstart_polya.html).
RNA stability (degradation rate λij, half-life hlij) and synthesis rate μij estimation of human RNA isoforms
Each neural network classified direct RNA nanopore sequencing read of the 5EU 60 min and 5EU 60 min heat shock samples was assigned to a FLAIR-defined human isoform (or RefSeq-TU) either as 5EU-labeled Lij and unlabeled Tij − Lij. The resulting counts were subsequently converted into synthesis rates μij and degradation rates λij for isoform i in sample j assuming first-order kinetics as previously described (Miller et al. 2011) using the following equations:
where t is the labeling duration in minutes, and α is the growth rate (dilution rate, i.e., the reduction of concentration owing to the increase of cell volume during growth) defined as
with cell cycle length CCLj [min]. The half-life hlij for isoform i in sample j can thus be calculated as
in minutes [min].
Determining factors of RNA isoform stability
For all features, for example, sequence, poly(A)-tail length, RNA secondary structure, translation (Ribo-seq) (Ingolia et al. 2014), and RBP peak occupancy (eCLIP) (Van Nostrand et al. 2020), a classifying neural network was trained to distinguish stable (above median half-life) and unstable (below median half-life) RNA (combined gene level) and RNA isoforms. For each feature, the area under the ROC curve was calculated as a predictor. Only expressed transcription units (see above) were used for gene level predictions. For isoform level predictions, only isoforms were used, which were overlapping with expressed transcription units. Quantification or sequence assessment was either performed on entire isoforms (isoform level) or on constitutive exons (see above) for the gene level.
Sequence
For each RNA isoform (or constitutive exon) not exceeding 5000 nt, the underlying sequence was split into all possible consecutive k-mers (k = 1,…,9, overlapping by k−1). For each k, a neural network was constructed containing an embedding layer (with input_dimension = 5k + 1, output_dimension = 16), a global average pooling layer (1d), and a fully connected layer (dense, units = 16, activation = “relu”). All layers were subsequently merged into a final dense layer with sigmoid activation. The neural network was trained against a binary classification of the respective half-life into stable (above median) and unstable (below median) isoforms. Note that embedding layers possess the ability to account for position of a k-mer.
Poly(A)-tail length: data (see above)
A neural network was constructed containing a batch normalization, followed by three fully connected layers (dense, units = 64, 8 and 1, activation = “relu,” “relu,” and “sigmoid”). Additional batch normalization and dropout (25%) layers were placed between the fully connected layer. The neural network was trained against a binary classification of the respective half-life into stable (above median) and unstable (below median) isoforms.
RNA secondary structure: in silico
For each isoform (or constitutive exon), the mean minimum free energy was calculated from subsequent minimum free energy estimates of 13-bp RNA sequence fragments tiling the entire length of the respective feature using RNAfold from the ViennaRNA package (Lorenz et al. 2011). Minimum free energy estimates were aggregated as mean, min, and centiles. In addition, the minimum free energy was calculated on the entire sequence. The neural network was constructed and trained as for poly(A)-tail length.
In vivo
For each isoform (or constitutive exon), the DMS-seq (Rouskin et al. 2014) coverage was calculated using the processed files of following samples: 300DMS, 400DMS, vitro, and denatured. The neural network was constructed and trained as for poly(A)-tail length.
Translation
For each isoform (or constitutive exon), the Ribo-seq (Ingolia et al. 2014) coverage was calculated using the following samples: GFPshCtrl and GFP0d (K562). Reads were mapped with STAR 2.3.0 (Dobin and Gingeras 2015) to the hg38 (GRCh38) genome assembly (Human Genome Reference Consortium). SAMtools 1.3.1 (Li et al. 2009) was used to quality filter SAM files, whereby alignments with MAPQ smaller than seven (-q 7) were skipped and only proper pairs (-f2) were selected. The neural network was constructed and trained as for poly(A)-tail length.
RBPs
For each isoform (or constitutive exon), the RBP peak occupancy (eCLIP) (Van Nostrand et al. 2020) was calculated using the processed files of all available K562 samples. The neural network was constructed and trained as for poly(A)-tail length.
RNA-seq data preprocessing and antisense bias correction
Paired-end 75-base reads with additional six-base reads of barcodes were obtained for each of the samples (Supplemental Table S1). Reads were demultiplexed and mapped with STAR 2.3.0 (Dobin and Gingeras 2015) to the hg38 (GRCh38) genome assembly (Human Genome Reference Consortium). SAMtools 1.3.1 (Li et al. 2009) was used to quality filter SAM files, whereby alignments with MAPQ smaller than seven (-q 7) were skipped and only proper pairs (-f2) were selected. Further data processing was performed using the R/Bioconductor environment. We used a spike-in (RNAs) normalization strategy essentially as previously described (Schwalb et al. 2016) to allow observation of antisense bias ratio cj (ratio of spurious reads originating from the opposite strand introduced by the reverse transcription reaction). Antisense bias ratios were calculated for each sample j according to
for all available spike-ins i. Read counts (kij) for spike-ins were calculated using HTSeq (Anders et al. 2015). The number of transcribed bases (tbj) for all samples was calculated as the sum of the coverage of evident (sequenced) fragment parts (read pairs only) for all fragments in addition to the sum of the coverage of nonevident fragment parts for fragments with an inner mate interval not entirely overlapping a RefSeq annotated intron (UCSC RefSeq GRCh38). The number of transcribed bases (tbj) or read counts (kj) for all features (RefSeq-TUs) were corrected for antisense bias cj as follows using the parameter calculated as described above. The real number of read counts or coverage sij for transcribed unit i in sample j was calculated as
where Sij and Aij are the observed numbers of read counts or coverage on the sense and antisense strand. RPKs were calculated upon antisense bias corrected read counts (kj) falling into the region of a RefSeq-TU divided by its length in kilobases. Coverages were calculated upon antisense bias-corrected number of transcribed bases (tbj) falling into the region of a RefSeq-TU divided by its length in bases.
Software availability
All information and detailed description regarding data collection and the open source and custom code used in this study is explained in detail in the Methods section, and an additional conceptual description of the data collection scheme and neural network design is provided in Figure 2A and Supplemental Table S5. Custom R and Python code was used to analyze the data and has been deposited in a GitHub repository (https://github.com/birdumbrella/nano-ID) and as Supplemental Code. R add-on software packages used in custom R code are listed and cited in the respective parts of the Methods section.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE127890 and to the Göttingen Research Online Data Archive (GRO) under the DOI https://doi.org/10.25625/XNSXV6. GEO contains *.fastq and *.bw files; GRO contains base-called *.fast5 files. Modified *.bam files for the 5EU 60 min and 5EU 60 min heat shock samples contain an additional tag “YC” to highlight 5EU-labeled reads in Integrative Genomics Viewer (IGV; human hg38), as well as the poly(A)-tail length estimate per read as elongated alignment, and are available at https://www3.mpibpc.mpg.de/downloads/cramer/illuMinatION/ or as a UCSC Genome Browser Track Hub at https://www3.mpibpc.mpg.de/downloads/cramer/nano-ID/. If reads are not highlighted in two different colors (red and gray for labeled and unlabeled, respectively), right click on the display to open the “configure 5EU 60 min combined” tab. Under “additional coloring modes,” you can activate the field “use R,G,B colors specified in user-defined tag” and enter “YC” in the adjacent box. After application, the reads should be colored accordingly.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
We thank Johannes Söding, Christian Roth, and Dmitry Tegunov (Max Planck Institute for Biophysical Chemistry) for advice on machine learning aspects. We also thank Nikolaos Papadopoulos and Noah Wulff Mottelson (Max Planck Institute for Biophysical Chemistry) for help with Python. In addition, we thank Michael Lidschreiber (Max Planck Institute for Biophysical Chemistry) and Brian Lee (UCSC Genomics Institute) for help with the UCSC Genome Browser Track Hub. Moreover, we thank Anna Sawicka and Kristina Žumer (Max Planck Institute for Biophysical Chemistry) for sharing the pUC19 spike-in plasmids. P.C. was supported by the Deutsche Forschungsgemeinschaft (SFB860, SPP1935), the European Research Council Advanced Investigator Grant TRANSREGULON (grant agreement no. 693023), and the Volkswagen Foundation.
Author contributions: K.M., B.S., and S.G. performed experiments. B.S. designed and performed all bioinformatics analysis. B.S. conceptualized, designed, and supervised research. B.S. and P.C. prepared the manuscript, with input from all authors.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.257857.119.
References
- Abe K, Ishigami T, Shyu AB, Ohno S, Umemura S, Yamashita A. 2012. Analysis of interferon-β mRNA stability control after poly(I:C) stimulation using RNA metabolic labeling by ethynyluridine. Biochem Biophys Res Commun 428: 44–49. 10.1016/j.bbrc.2012.09.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. 2015. HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharmal H, Clarke S, Lemire A, Agnew B, Kumar K. 2010. Capture and analysis of newly synthesized RNA a novel enabling technology to study high resolution gene expression. J Biomol Tech 21: S43. [Google Scholar]
- Bullard JH, Purdom E, Hansen KD, Dudoit S. 2010. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics 11: 94 10.1186/1471-2105-11-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H, Lim J, Ha M, Kim VN. 2014. TAIL-seq: genome-wide determination of poly(A) tail length and 3′ end modifications. Mol Cell 53: 1044–1052. 10.1016/j.molcel.2014.02.007 [DOI] [PubMed] [Google Scholar]
- Chen Y, Pai AA, Herudek J, Lubas M, Meola N, Järvelin AI, Andersson R, Pelechano V, Steinmetz LM, Jensen TH, et al. 2016. Principles for RNA metabolism and alternative transcription initiation within closely spaced promoters. Nat Genet 48: 984–994. 10.1038/ng.3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MB, Wrzesinski T, Garcia AB, Hall NAL, Kleinman JE, Hyde T, Weinberger DR, Harrison PJ, Haerty W, Tunbridge EM. 2020. Long-read sequencing reveals the complex splicing profile of the psychiatric risk gene CACNA1C in human brain. Mol Psychiatry 25: 37–47. 10.1038/s41380-019-0583-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Martins AL, Danko CG, Waters CT, Siepel A, Lis JT. 2014. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet 46: 1311–1320. 10.1038/ng.3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Gingeras TR. 2015. Mapping RNA-seq reads with STAR. Curr Protoc Bioinformatics 51: 11.14.1–11.14.19. 10.1002/0471250953.bi1114s51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolken L, Ruzsics Z, Radle B, Friedel CC, Zimmer R, Mages J, Hoffmann R, Dickinson P, Forster T, Ghazal P, et al. 2008. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA 14: 1959–1972. 10.1261/rna.1136108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswaran J, Horvath A, Godbole S, Reddy SD, Mudvari P, Ohshiro K, Cyanam D, Nair S, Fuqua SA, Polyak K, et al. 2013. RNA sequencing of cancer reveals novel splicing alterations. Sci Rep 3: 1689 10.1038/srep01689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone C, Mazzoni C. 2018. RNA stability and metabolism in regulated cell death, aging and diseases. FEMS Yeast Res 18 10.1093/femsyr/foy050 [DOI] [PubMed] [Google Scholar]
- Garalde DR, Snell EA, Jachimowicz D, Sipos B, Lloyd JH, Bruce M, Pantic N, Admassu T, James P, Warland A, et al. 2018. Highly parallel direct RNA sequencing on an array of nanopores. Nat Methods 15: 201–206. 10.1038/nmeth.4577 [DOI] [PubMed] [Google Scholar]
- Herzog VA, Reichholf B, Neumann T, Rescheneder P, Bhat P, Burkard TR, Wlotzka W, von Haeseler A, Zuber J, Ameres SL. 2017. Thiol-linked alkylation of RNA to assess expression dynamics. Nat Methods 14: 1198–1204. 10.1038/nmeth.4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Tollervey D. 2009. The many pathways of RNA degradation. Cell 136: 763–776. 10.1016/j.cell.2009.01.019 [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne GJ, Jackson SE, Wills MR, Weissman JS. 2014. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep 8: 1365–1379. 10.1016/j.celrep.2014.07.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao CY, Salic A. 2008. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc Natl Acad Sci 105: 15779–15784. 10.1073/pnas.0808480105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34: 3094–3100. 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YI, van de Geijn B, Raj A, Knowles DA, Petti AA, Golan D, Gilad Y, Pritchard JK. 2016. RNA splicing is a primary link between genetic variation and disease. Science 352: 600–604. 10.1126/science.aad9417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long C, Li H, Tiburcy M, Rodriguez-Caycedo C, Kyrychenko V, Zhou H, Zhang Y, Min YL, Shelton JM, Mammen PPA et al. 2018. Correction of diverse muscular dystrophy mutations in human engineered heart muscle by single-site genome editing. Sci Adv 4: eaap9004 10.1126/sciadv.aap9004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. 2011. ViennaRNA package 2.0. Algorithms Mol Biol 6: 26 10.1186/1748-7188-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahat DB, Lis JT. 2017. Use of conditioned media is critical for studies of regulation in response to rapid heat shock. Cell Stress Chaperones 22: 155–162. 10.1007/s12192-016-0737-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew A, Mathur SK, Morimoto RI. 1998. Heat shock response and protein degradation: regulation of HSF2 by the ubiquitin-proteasome pathway. Mol Cell Biol 18: 5091–5098. 10.1128/MCB.18.9.5091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C. 2017. Regulation by 3′-untranslated regions. Annu Rev Genet 51: 171–194. 10.1146/annurev-genet-120116-024704 [DOI] [PubMed] [Google Scholar]
- Miller C, Schwalb B, Maier K, Schulz D, Dümcke S, Zacher B, Mayer A, Sydow J, Marcinowski L, Dölken L, et al. 2011. Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol Syst Biol 7: 458 10.1038/msb.2010.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niskanen EA, Malinen M, Sutinen P, Toropainen S, Paakinaho V, Vihervaara A, Joutsen J, Kaikkonen MU, Sistonen L, Palvimo JJ. 2015. Global SUMOylation on active chromatin is an acute heat stress response restricting transcription. Genome Biol 16: 153 10.1186/s13059-015-0717-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, Wei W, Steinmetz LM. 2013. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature 497: 127–131. 10.1038/nature12121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabani M, Levin JZ, Fan L, Adiconis X, Raychowdhury R, Garber M, Gnirke A, Nusbaum C, Hacohen N, Friedman N, et al. 2011. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat Biotechnol 29: 436–442. 10.1038/nbt.1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2017. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: https://www.R-project.org/. [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. 2014. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 505: 701–705. 10.1038/nature12894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield JA, Duffy EE, Kiefer L, Sullivan MC, Simon MD. 2018. TimeLapse-seq: adding a temporal dimension to RNA sequencing through nucleoside recoding. Nat Methods 15: 221–225. 10.1038/nmeth.4582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwalb B, Michel M, Zacher B, Frühauf K, Demel C, Tresch A, Gagneur J, Cramer P. 2016. TT-seq maps the human transient transcriptome. Science 352: 1225–1228. 10.1126/science.aad9841 [DOI] [PubMed] [Google Scholar]
- Shendure J, Balasubramanian S, Church GM, Gilbert W, Rogers J, Schloss JA, Waterston RH. 2017. DNA sequencing at 40: past, present and future. Nature 550: 345–353. 10.1038/nature24286 [DOI] [PubMed] [Google Scholar]
- Sistonen L, Sarge KD, Phillips B, Abravaya K, Morimoto RI. 1992. Activation of heat shock factor 2 during hemin-induced differentiation of human erythroleukemia cells. Mol Cell Biol 12: 4104–4111. 10.1128/MCB.12.9.4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sović I, Šikić M, Wilm A, Fenlon SN, Chen S, Nagarajan N. 2016. Fast and sensitive mapping of nanopore sequencing reads with GraphMap. Nat Commun 7: 11307 10.1038/ncomms11307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steijger T, Abril JF, Engström PG, Kokocinski F, RGASP Consortium, Hubbard TJ, Guigó R, Harrow J, Bertone P. 2013. Assessment of transcript reconstruction methods for RNA-seq. Nat Methods 10: 1177–1184. 10.1038/nmeth.2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. 2014. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508: 66–71. 10.1038/nature13007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang AD, Soulette CM, van Baren MJ, Hart K, Hrabeta-Robinson E, Wu CJ, Brooks AN. 2020. Full-length transcript characterization of SF3B1 mutation in chronic lymphocytic leukemia reveals downregulation of retained introns. Nat Commun 11: 1438 10.1038/s41467-020-15171-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapial J, Ha KCH, Sterne-Weiler T, Gohr A, Braunschweig U, Hermoso-Pulido A, Quesnel-Vallieres M, Permanyer J, Sodaei R, Marquez Y, et al. 2017. An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms. Genome Res 27: 1759–1768. 10.1101/gr.220962.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodorakis NG, Zand DJ, Kotzbauer PT, Williams GT, Morimoto RI. 1989. Hemin-induced transcriptional activation of the HSP70 gene during erythroid maturation in K562 cells is due to a heat shock factor-mediated stress response. Mol Cell Biol 9: 3166–3173. 10.1128/MCB.9.8.3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Jahanbani F, Blauwkamp T, Moshrefi A, Jaeger E, Chen F, Harel I, Bustamante CD, Rasmussen M, Snyder MP. 2015. Comprehensive transcriptome analysis using synthetic long-read sequencing reveals molecular co-association of distant splicing events. Nat Biotechnol 33: 736–742. 10.1038/nbt.3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner RE, Pattison AD, Beilharz TH. 2018. Alternative polyadenylation in the regulation and dysregulation of gene expression. Semin Cell Dev Biol 75: 61–69. 10.1016/j.semcdb.2017.08.056 [DOI] [PubMed] [Google Scholar]
- Van Nostrand EL, Freese P, Pratt GA, Wang X, Wei X, Xiao R, Blue SM, Chen J-Y, Cody NAL, Dominguez D, et al. 2020. A large-scale binding and functional map of human RNA-binding proteins. Nature 583: 711–719. 10.1038/s41586-020-2077-3 [DOI] [PMC free article] [PubMed]
- Vihervaara A, Sergelius C, Vasara J, Blom MA, Elsing AN, Roos-Mattjus P, Sistonen L. 2013. Transcriptional response to stress in the dynamic chromatin environment of cycling and mitotic cells. Proc Natl Acad Sci 110: E3388–E3397. 10.1073/pnas.1305275110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihervaara A, Mahat DB, Guertin MJ, Chu T, Danko CG, Lis JT, Sistonen L. 2017. Transcriptional response to stress is pre-wired by promoter and enhancer architecture. Nat Commun 8: 255 10.1038/s41467-017-00151-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman RE, Tang AD, Tang PS, Jain M, Tyson JR, Razaghi R, Zuzarte PC, Gilpatrick T, Payne A, Quick J, et al. 2019. Nanopore native RNA sequencing of a human poly(A) transcriptome. Nat Methods 16: 1297–1305. 10.1038/s41592-019-0617-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Fair BJ, Dwyer ZW, Gildea M, Pleiss JA. 2019. Detection of splice isoforms and rare intermediates using multiplexed primer extension sequencing. Nat Methods 16: 55–58. 10.1038/s41592-018-0258-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Suzuki T, Sato T, Takahashi A, Watanabe H, Kadowaki A, Natsui M, Inagaki H, Arakawa S, Nakaoka S, et al. 2018. The CCR4-NOT deadenylase complex controls Atg7-dependent cell death and heart function. Sci Signal 11: eaan3638 10.1126/scisignal.aan3638 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Pan Z, Ying Y, Xie Z, Adhikari S, Phillips J, Carstens RP, Black DL, Wu Y, Xing Y. 2019. Deep-learning augmented RNA-seq analysis of transcript splicing. Nat Methods 16: 307–310. 10.1038/s41592-019-0351-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.