Abstract
Improved identification of structural variants (SVs) in cancer can lead to more targeted and effective treatment options as well as advance our basic understanding of the disease and its progression. We performed whole-genome sequencing of the SKBR3 breast cancer cell line and patient-derived tumor and normal organoids from two breast cancer patients using Illumina/10x Genomics, Pacific Biosciences (PacBio), and Oxford Nanopore Technologies (ONT) sequencing. We then inferred SVs and large-scale allele-specific copy number variants (CNVs) using an ensemble of methods. Our findings show that long-read sequencing allows for substantially more accurate and sensitive SV detection, with between 90% and 95% of variants supported by each long-read technology also supported by the other. We also report high accuracy for long reads even at relatively low coverage (25×–30×). Furthermore, we integrated SV and CNV data into a unifying karyotype-graph structure to present a more accurate representation of the mutated cancer genomes. We find hundreds of variants within known cancer-related genes detectable only through long-read sequencing. These findings highlight the need for long-read sequencing of cancer genomes for the precise analysis of their genetic instability.
Somatic mutations that drive cancer development range across all genomic scales, from single-nucleotide variants through large-scale genome rearrangements, and have been observed in nearly all types of cancer at every stage of the disease progression (Martincorena and Campbell 2015). Better detection, quantification, and reconciliation of mutation types in cancer samples can lead to a better understanding of disease progression and help improve existing and develop new, often patient-specific, therapeutic approaches for the disease (Baudino 2015). Furthermore, improvements in detecting germline genetic variants in healthy cells can allow for better risk assessment of both hereditary and de novo mutations of various cancer types, leading to a more proactive rather than reactive cancer treatment approach (Nielsen et al. 2016).
Our ability to detect genetic alterations has evolved over the last several decades. Prior to the completion of the Human Genome Project, only a small handful of oncogenes or tumor suppressors were known (Taparowsky et al. 1982; Li et al. 1997). Large-scale detection of cancer mutations began around the year 2000 after the initial sequencing of the human genome using either microarrays (Schena et al. 1995; Perou et al. 2000) or PCR amplification of known cancer-related genes sequenced on low-throughput ABI capillary instruments (Fearon and Vogelstein 1990). In the late-2000s, the advent of Solexa, which later became Illumina, second-generation sequencing instruments accelerated the pace of discovery so that whole cancer genomes could be sequenced for the first time (Ley et al. 2008; Pleasance et al. 2010). Since then, the improvements in the throughput and cost of whole-genome sequencing (WGS) and whole-exome sequencing (WES) (Hodges et al. 2007) have made these technologies increasingly important in cancer studies, opening the door to widespread sequencing of patients, and the advancement of precision and personalized medicine. Within The Cancer Genome Atlas project (TCGA) (The Cancer Genome Atlas Research Network et al. 2013), the International Cancer Genome Consortium (ICGC) (Zhang et al. 2011), and other large-scale efforts, several thousands of tumors have been sequenced using short-read Illumina sequencing across dozens of major cancer types. These studies have had a tremendous impact in cancer genomics, leading to the discovery, for example, of different signatures and mutation rates across cancer types and new insights into the clonal structural and evolution of tumors (Yates and Campbell 2012; Alexandrov et al. 2013; Schwartz and Schäffer 2017).
These results have substantially advanced our understanding of cancer susceptibility and progression, although the identification and understanding of the genetic alterations in cancer remain incomplete. A major contributor to our incomplete knowledge is that the known mutations have chiefly been detected using short-read Illumina sequencing (Goodwin et al. 2016). This technology is very effective for identifying single-nucleotide variants (SNVs) and large copy number variants (CNVs, especially those 100 kb or larger), however, several studies have found it has poor accuracy for structural variant (SV) detection (Sedlazeck et al. 2018b; Chaisson et al. 2019; Zook et al. 2020). SVs are larger mutations, 50 bp or larger, where sequence is added, removed, or rearranged in the genome. Because of the short-read lengths, Illumina sequencing is difficult to map across SV breakpoints, especially insertions that are not present in the reference genome. Furthermore, SVs are frequently flanked by repetitive sequences so that the short-read sequence data often cannot be unambiguously mapped back to its correct genomic position and de novo assembly techniques also fail to capture the novel sequences (Chaisson et al. 2019). Consequently, short-read analysis algorithms systematically fail to detect SVs, with false negative and false positive rates that can exceed 50% (Huddleston et al. 2017; Sedlazeck et al. 2018a; Chaisson et al. 2019). As a result, we are facing a major limitation with short-read sequencing studies of cancer where the field has systematically missed many important variants, potentially making it blind to entire classes of inherited genetic risk factors and blind to how SVs may mediate cancer progression and patient survival.
New long-read, single-molecule sequencing technologies from Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (ONT) have been shown to more reliably identify SVs with substantial improvements to both sensitivity and specificity. Reports by several groups have found a typical healthy human genome contains approximately 20,000 SVs, and that they can be detected with 95% or greater sensitivity and specificity with long reads (Sedlazeck et al. 2018b; Audano et al. 2019; De Coster et al. 2019). These variants are especially important to accurately identify for somatic mutations that are not in linkage disequilibrium with any nearby SNVs. A total of 748 genes have been identified that are inaccessible to short-read sequencing (Ebbert et al. 2019), including 193 medically relevant genes with at least one exon that cannot be sequenced with short reads, but are accessible to long reads (Mandelker et al. 2016; Wenger et al. 2019). Long reads also have improved power to resolve complex regions of the human genome, such as the highly variable major histocompatibility complex (MHC) or the lipoprotein(a) (LPA) gene sequence, and in some cases, identified causative SVs underlying genetic disease that had been missed by short reads (Merker et al. 2018; Miao et al. 2018).
Within cancer genetics, we previously published one of the first reports using PacBio long-read sequencing to study SVs in a cancer cell line genome and found that long reads could detect tens of thousands of variants that had been missed by short reads (Nattestad et al. 2018; Sedlazeck et al. 2018b). This included variants in known cancer genes such as ERBB2 (also known as HER2), APOBEC3B, and CDH1, as well as dozens of novel gene fusions and other complex rearrangements that had substantially altered the expression and regulation of genes in the cell. Since this work, the cost and quality of third-generation sequencing platforms make them more suitable than ever before in both academic and medical settings (De Coster et al. 2019; Sone et al. 2019) and thus require the improvement of existing and the development of new methods for mutation detection and analysis.
Here, we aim to provide a comprehensive analysis of the SKBR3 breast cancer cell line and patient-derived organoids representing tumor and matching normal cells from two breast cancer patients sequenced with ONT, PacBio, and Illumina/10x Genomics (10xG) third-generation sequencing technologies, building on the sequencing of SKBR3 with PacBio and Illumina we previously performed (Nattestad et al. 2018). We identify and reconcile different types of large-scale genomic mutations in observed samples with an ensemble of methods and highlight concordance and differences observed across different mutation inference methods and sequencing technologies, with the goal of identifying which technologies and methods are best able to reliably detect large-scale variation in cancer.
Results
Sample identification and sequencing
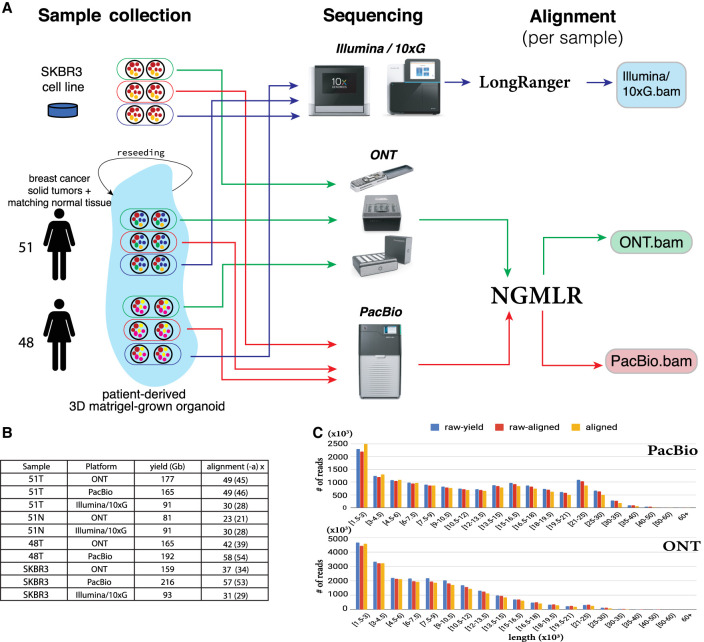
Building on our previous work, we first evaluated the widely studied SKBR3 breast cancer cell line. SKBR3 is one of the most widely studied ERBB2+ breast cancer cell lines, with applications ranging from basic to preclinical research (Neve et al. 2006; Lewis Phillips et al. 2008; Navin et al. 2011; Ichikawa et al. 2012). SKBR3 was chosen for this study because of its importance as a basic research model for cancer and because the SKBR3 genome contains many of the common features of cancer alterations, including a number of gene fusions, oncogene amplifications, and extensive rearrangements. We previously sequenced this cell line using short-read paired-end Illumina and long-read PacBio sequencing (Nattestad et al. 2018); here, we additionally examine 10x Genomics Linked Reads and Oxford Nanopore sequencing for this sample (Fig. 1A; Methods).
Figure 1.
Sample collection, sequencing, and alignment pipeline and statistics overview. (A) Biological data sample collection, sequencing, and alignment workflow for SKBR3 breast cancer cell line and 3D Matrigel-grown organoids for solid breast cancer tumor tissues obtained from two females, patient 51 and patient 48. (B) Yield and alignment coverage statistics for observed samples across WGS experiments various sequencing platforms. Suffixes T and N next to patient identifiers indicate tumor or matching normal tissue. Alignment values x (y) represent average read-depth x for aligned reads with (y) representing average read depth when all unresolved Ns in the reference are also taken into consideration. (C) Length distribution for reads of length 1.5+ kbp from PacBio and ONT sequencing runs for patient 51: (raw-yield) lengths of raw sequenced reads; (raw-aligned) lengths of raw reads that had any alignment inferred for them; (aligned) lengths of aligned parts of sequenced reads.
In addition, we sequenced tumor and normal patient-derived organoids from two breast cancer patients, here identified as patient 51 and patient 48, treated at Northwell Health and recruited in accordance with their Institutional Review Board protocol (Methods). Patient-derived organoids are three-dimensional cultures of normal and cancer cells, propagated inside a basement-membrane extract matrix, overlaid with a growth-factor-rich growth medium tailored toward the origin of the individual tissue (Sachs et al. 2018). The organoids were generated from cells harvested from surgical specimens from the patients and hence share the same genetic composition as the patient normal and tumor cells, with an estimated tumor purity of 0.98 (Methods). This allows for us to propagate the cells in a stable environment so that ample quantities of DNA were available for our three sequencing platforms (Fig. 1A). Furthermore, several studies have shown organoids are superior to standard 2D cell culture for recapitulating the molecular characteristics, physiology, and treatment response of patient tissues (Clevers 2016), allowing us to perform methylation analysis, RNA-seq, and other assays on the samples.
Improved sensitivity and high concordance in structural variation inference with ONT and PacBio long reads
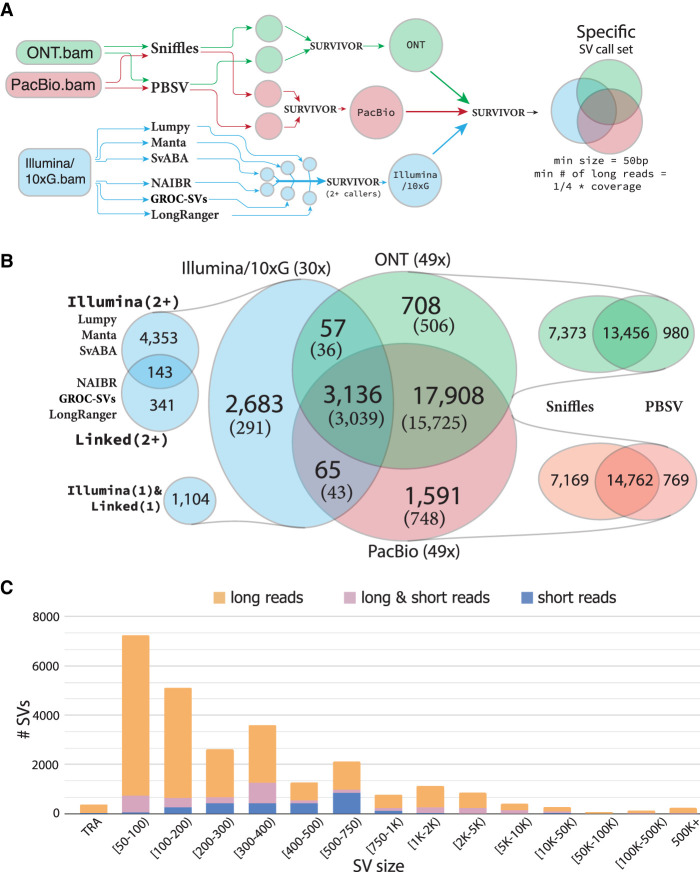
Using these data (Fig. 1B,C; Supplemental Fig. 1), we then used an ensemble of methods to infer all types of SVs at least 50 bp in size, including insertions, deletions, inversions, translocations, and duplications. For both the ONT and PacBio data sets, we used two state-of-the-art methods, Sniffles (Sedlazeck et al. 2018b) and PBSV (https://github.com/PacificBiosciences/pbsv), and for the Illumina/10xG data set, we used six SV inference methods, with three—LUMPY (Layer et al. 2014), Manta (Chen et al. 2016), and SvABA (Wala et al. 2018)—designed for regular paired-end short Illumina reads, and three—NAIBR (Elyanow et al. 2018), GROC-SVs (Spies et al. 2017), and LongRanger (Zheng et al. 2016)—which also use the single-molecule 10xG barcode information. We then iteratively merged SVs using the SURVIVOR (Jeffares et al. 2017) package, first merging calls from all SV detection methods for each sequencing technology separately and then merging across sequencing technologies to obtain sample-specific SV call sets (Fig. 2A).
Figure 2.
Structural variation inference across Illumina/10xG, ONT, and PacBio sequencing platforms for sample 51. (A) Ensemble workflow for SV inference, with multiple methods and technologies used to infer SVs, subsequent merging of, first method-specific results, and then technology-specific results, with size and support restrictions applied. (B) SV inference comparison across SVs inferred from platform (x) sequencing experiments, in which “platform” corresponds to sequencing technology, and (x) determines the average alignment read-depth coverage in the tumor sample. Methods-specific breakdown is provided for every sequencing technology. SVs detected in the normal sample are in parentheses. (C) Size distribution for SVs in sample 51T with SVs being either exclusively inferred from either long reads (either ONT, or PacBio, or both), or exclusively from Illumina/10xG short reads, or supported by both long and short reads.
Because SVs inferred from paired-end short reads have higher rates of false positives (Sudmant et al. 2015; Huddleston et al. 2017; Sedlazeck et al. 2018b), for the Illumina/10xG data set we only considered SVs supported by at least two methods. To mitigate false positives in the long-read SV calls, we only report SVs that were supported by at least one-quarter of the average alignment read depth in either ONT or PacBio data sets (coverage requirements are discussed below). During the merging, we optimize parameters to minimize the effects of small thresholding issues, such as a variant present in 10 reads in one sample, and hence called as a variant, but only nine reads in the other, and hence not called (for details, see Methods). Our results indicate a very strong concordance between SVs inferred with ONT and PacBio. Between 90% and 95% of variants called in at least one of the long-read data types were supported by both, with slightly lower concordance between PacBio-only calls (Fig. 2B; Supplemental Fig. 2A,C). Although >50% of SVs inferred from short-read data were also identified by long reads, the total quantity of SVs inferred from short reads, when support from two callers is required, is approximately four times less than for either of the long-read-based inferences.
We further examined the concordance between SVs inferred by individual short-read methods and long-read SVs, and find variable concordance, with some sample/short-read caller combinations having <10% of their SV calls also present in long-read calls (Supplemental Methods; Supplemental Table 1), highlighting the necessity of requiring calls from two short-read callers. We also note that short-read exclusive SVs are often located in areas of abnormally high coverage of Illumina/10xG reads compared to essentially uniform coverage for the ONT and PacBio data. These regions account for >60% of short-read exclusive SVs in SKBR3, and >70% in patient 51 (Supplemental Fig. 9). To examine these coverage abnormalities, we also considered independently generated Illumina/10xG, ONT, and regular paired-end Illumina reads for two additional noncancerous samples, NA12878 and HG002 (Supplemental Methods). From these data, we identified 7228 genomic regions shared across all five samples with abnormally high coverage of Illumina/10xG reads (Supplemental Table 2; Supplemental Figs. 13–16). We find that regular paired-end (non-10xG) Illumina reads do not show coverage abnormalities to the same degree (Supplemental Figs. 17, 18), suggesting a 10xG-specific amplification artifact. We additionally note that short-read exclusive SVs often have a nearby long-read SV of a different type, a previously described caveat of short-read SV calls (Sedlazeck et al. 2018b) and/or overlap tandem repeats, which are known to be difficult regions particularly for short-read alignment (Methods; Supplemental Figs. 9–12). In Supplemental Table 3, we quantify SVs that are supported by either long or long and short reads and overlap regulatory and functional genomic regions (Dreos et al. 2013; Fishilevich et al. 2017). Overall, these results show that across SVs sizes and types, long-read-based SV inference outperforms that of short reads (Fig. 2C; Supplemental Fig. 2B).
To quantify the level of patient-specific and common SVs in both the observed patients and the SKBR3 cancer cell line, we compared SVs inferred with multiple sequencing technologies in the presented study to SVs identified in 15 healthy human genomes sequenced with PacBio long reads presented in the recent study by Audano et al. (2019) and SVs called using the same analysis pipeline as our patient samples. We find a high level of agreement between the SVs themselves (20,116/26,148) and the distributions of their breakends locations identified in the cancer samples and the healthy samples (Supplemental Fig. 3), and in addition, we examine type and size distributions (Supplemental Figs. 4–8). We observe that in SVs identified exclusively with long reads, insertions and deletions dominate, comprising ∼50% and 36%, respectively. Duplications compose only 6% of the long-read exclusive SVs call sets, whereas the inclusion of SVs inferred with 2+ short-read SV callers increases that value several fold to 13%–16% in the multitechnology SV call set. Duplications constitute 71%–93% of the short-read exclusive SVs. Inversion and translocation SVs constitute similar fractions in both cancer and healthy SV sets in either short, long, or multitechnology SV sets.
For patient 51, both tumor and the matching normal cells were sequenced, allowing us to perform additional analyses of somatic variation. We observed that 77% (20,388/26,148) of the SVs identified in the tumor sample were also identified in the matching normal sample (Fig. 2B). A high fraction of SVs present both in the cancer and in the normal cells is expected because the cancer cells originate from normal tissue. Cancer cells, however, will generally acquire new mutations, although large deletions and loss of heterozygosity can potentially decrease the count of inherited SVs (Cavenee et al. 1983). We also observed that for SVs called exclusively by short reads in 51T, only ∼11% (291/2683) are also identified in the matching normal cells. This is several fold less than for SVs inferred both exclusively with long reads (88%) and with both long and short reads (97%), and we attribute this discrepancy to a high false positive rate in short-read SV inference.
We further examined the putatively somatic variants for patient 51 (present in 51T and not 51N SV calls) and found that of the 5760 SVs present in the tumor but not normal sample, 3368 are supported by long reads (Fig. 2B; Supplemental Fig. 4). We examined the size distribution and types of the somatic variants and found a similar size distribution to the overall SV set, but with somatic insertions more numerous than deletions relative to the full SV set (Supplemental Fig. 8). Of the long-read supported somatic variants, 28 and 470 overlapped promoter and enhancer regions, respectively, and 161 were exonic (Supplemental Table 3). Following best practices (Hiltemann et al. 2015; Franco et al. 2019), we further refined the somatic variant calls by filtering common germline variants identified in 15 healthy human PacBio sequenced genomes from Audano et al. (2019) (Supplemental Fig. 8). By considering variants present in this set as not likely somatic, this reduced the number of somatic, long-read supported variants from 3368 to 811, of which 17 and 144 overlapped promoter and enhancer regions and 61 were exonic (Supplemental Table 3). The most common somatic SVs are insertions, which may be in part attributable to reactivation of transposable elements in cancers, a phenomenon which has been reported previously (Burns 2017; Kong et al. 2019).
In addition to the above genomic analysis, we exploited a unique capability of Nanopore sequencing to identify cytosine methylation changes from DNA sequencing data without any additional library preparation (Simpson et al. 2017). Using this capability, we also examined methylation characteristics within the observed cancer and normal genomes (Methods). As previously reported (Hansen et al. 2011), we observed overall genome-wide hypomethylation in the tumor samples relative to normal (Supplemental Fig. 26A,B). Although this hypomethylation occurs genome-wide in the cancer genomes, promoter regions stand out as an exception to this trend (Supplemental Fig. 26C,D). Furthermore, promoter regions that had SVs in them showed a modest enrichment for hypomethylation when compared to promoter regions without SVs (Supplemental Fig. 26E). We also observed trends of similar averaged methylation frequencies around transcription start sites (TSS) both in cancer and normal samples (Supplemental Fig. 26F). We also identified several examples in which SVs located within promoter regions of known COSMIC genes coincide with methylation changes between normal and tumor cells in patient 51: (1) An insertion in PRDM1 coincides with hypomethylation of the respective promoter region in the tumor (Supplemental Fig. 26G); (2) a duplication in the promoter region of ZEB1 coincides with the increased methylation of the affected area in the tumor genome (Supplemental Fig. 26H); and (3) an insertion in the promoter region of USP6 coincides with the blocking of the TSS demethylation in tumor (Supplemental Fig. 26I).
Additionally, we used RNA-seq gene expression data obtained from the tumor 51T and the matching normal 51N samples to investigate the impact of SVs on transcription. For this, we focused on differences in expression levels of those genes with SVs present in 51T but not present in 51N or 15 other healthy samples sequenced with long reads (for details, see Methods). Overall, we see that duplications and deletions generally increase and decrease gene expression, respectively, especially when >50% of the gene sequence is impacted by an SV (Supplemental Fig. 25A). Although in some examples (Supplemental Fig. 25B) we observe the SV link to gene expression change more clearly, there is a considerable range in the expression levels so that we cannot conclude that the magnitude of expression changes is solely explained by individual SVs. We note that SVs of different types that span genes often do not uniquely determine the copy number changes of the affected genomic regions owing to the rearranged nature of underlying cancer chromosomes. For example, localized deletions within larger highly amplified regions can still show an overall increase in genomic copy number and increase in expression.
Overall, our results show that single-molecule long-read sequencing is essential for comprehensive SV inference. We further highlight that ONT and PacBio produce highly concordant results, thus providing validation of the long-read variant calls. We also observe that a majority of SVs detected in tumor samples are also present in both matching normal cells as well as in the union set of SVs from healthy samples. These observations also underscore the need for a patient-specific reference genome approach in the analysis of structural variants and their role in cancer origination and progression. Finally, our RNA-seq analysis highlights an important, yet evidently nonexclusive, role that somatic SVs can play in tumor cells development and progression and thus the importance of SV detection in cancer studies.
Coverage requirements for accurate structural variation inference with long reads
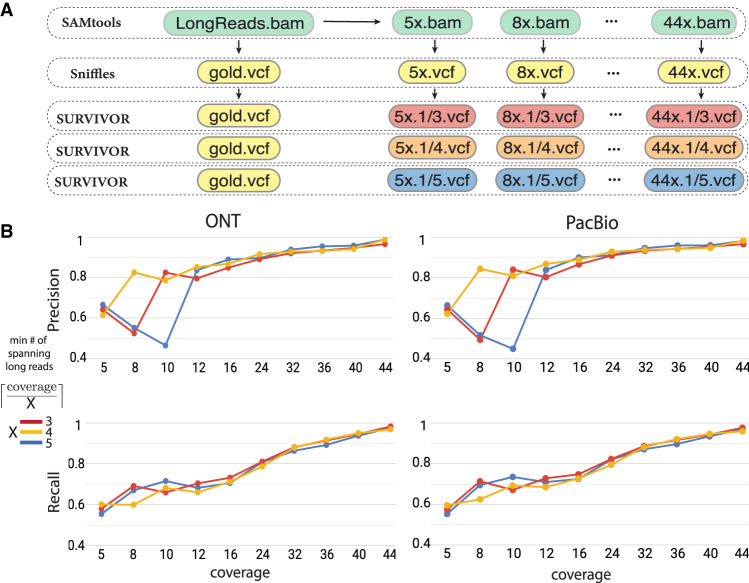
Because cost remains one of the final barriers for widespread long-read sequencing of patient genomes, we have measured how robust the SV inference with either ONT or PacBio reads is to lower sequencing coverage. For this, we randomly down-sampled our full coverage data sets to lower coverage levels and then compared SVs inferred on the down-sampled data sets to the ground truth SV call sets from the original full coverage data sets. As with all variant callers, long-read variant callers report variants supported by a certain minimum number of reads, although the higher error rate for long-read sequencing potentially makes this analysis more challenging. We measured this effect by adjusting the minimum number of long reads required to span (i.e., support) an SV for it to be classified as present from one-third to one-fifth of the average read-depth coverage (Fig. 3A). We found that for both ONT and PacBio reads, the recall reaches a robust value of >80% and the precision reaches >90% with 24× to 30× coverage available (Fig. 3B; Supplemental Fig. 19). Both ONT and PacBio data sets also showed generally high consistency for minimum read supports, except for very low coverage data sets (<10×). These observations allow us to conclude that robust SV detection with single-molecule long-read sequencing is possible even at relatively low coverage levels of 25×–30× average read depth, with very similar results achievable with either ONT or PacBio long reads.
Figure 3.
Structural variation inference on down-sampled long-read data sets. (A) Workflow for down-sampling full long-read data set and computing concordance between down-sampled and full coverage data sets with distinct minimum fractional x/y read support for an SV to be considered. (B) Precision and recall for SVs inferred on down-sampled ONT and PacBio data for sample 51T. SVs inferred on the full coverage data set at the matching read support threshold are used as the ground truth.
Integration and refinement of copy number and structural variations
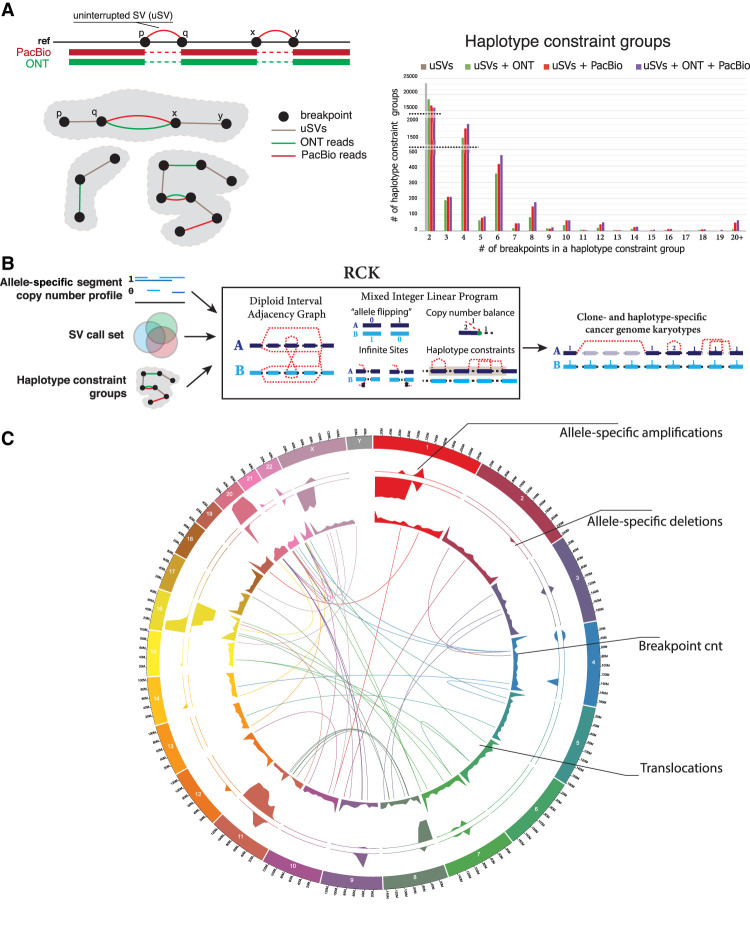
With the consensus SV call sets available, we then refined the rearranged structure of the cancer genome for patient 51 through an integrative analysis of SNVs, CNVs, and SVs. We first analyzed the short reads to infer large-scale allele-specific CNVs using two state-of-the-art methods, TitanCNA (Ha et al. 2014) and HATCHet (Zaccaria and Raphael 2018). As part of the automated copy number profile inference, both HATCHet and TitanCNA have identified a homogeneous composition of the tumor sample 51T with minimal admixture of normal cells. Tumor purity was estimated to be 0.982 and 0.979 by HACTCHet and TitanCNA, respectively. Although these methods provide a genome-wide view of large CNVs, the haplotype information is lost across both adjacent and distant fragments, and smaller (<50 kbp) CNVs are also often missed. To mitigate these limitations and to incorporate SV information, we extended our method RCK (Aganezov and Raphael 2020) to infer a haplotype-specific cancer karyotype-graph (Methods; Fig. 4B). This approach helps both to refine the boundaries of large CNVs, predicted by short-read CNV inference methods, as well as infer smaller CNVs based on the integration of SV information (Supplemental Fig. 20).
Figure 4.
Integration of SVs and CNVs for cancer genomes via karyotype-graph integration. (A) Haplotype constraint groups determined via uninterrupted SVs (uSVs) and long ONT and/or PacBio reads spanning multiple SVs. Distribution over the number of haplotype constraint groups inferred with only uSVs, and various combinations of uSVs and short/long reads in patient 51. (B) Workflow of the reconstruction of haplotype-specific cancer karyotype graphs (RCK) method with allele-specific copy number profiles on large fragments, resolved SV call set, and inferred haplotype constraint groups as inputs. (C) Circos plot of the CNVs and SVs from karyotype graph inferred by RCK for patient 51 with HATCHet segment copy number (CN) input. The top two tracks correspond to fractions x/y of the total length x of either amplified (CN ≥ 1) or deleted (CN = 0) fragments over the y = 5 × 106 long windows. Breakend track shows the total number (with 590 being the maximum value shown) of breakends inferred by RCK as being present.
In the new RCK v. 1.1 developed for this project, we added long-read-based haplotype constraints for SVs breakends (Methods). Both ONT and PacBio showed similar results in terms of haplotype grouping multiple SVs’ breakends. As expected, long reads most commonly introduced 2-breakend (i.e., single SV) haplotype constraint groups, but also identified several hundreds of 6+-breakend groups (i.e., 3+ SVs), as well as few 20+-breakend groups in which constraint information could be determined from multiple overlapping long reads (Fig. 4A).
Running RCK with our comprehensive SV call set along with either TitanCNA and HATCHet CNVs produced highly similar reconstructions of the rearranged cancer genome (Fig. 4C; Supplemental Fig. 21A). We also observed that even though the CNV profiles were refined by RCK to incorporate the input SVs, the resulting CNV profiles remained very similar to the input ones. (Supplemental Fig. 19B). Similarly, we further note that although up to 10% of input SVs were allowed to be dismissed by RCK as either erroneous or not concurring with the input CNVs during the karyotype-graph analysis, we see very similar SV breakend distribution across the input and refined SV call sets (Supplemental Fig. 22).
Structural and copy number variants in COSMIC census genes
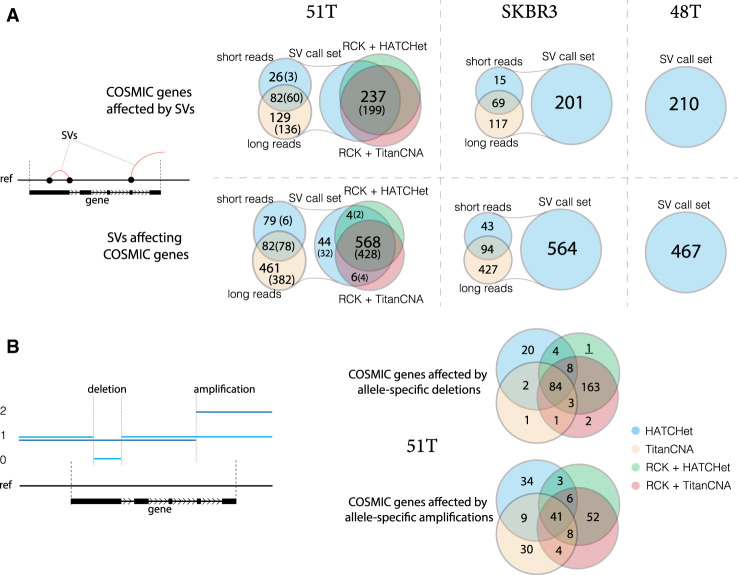
We then considered how many of the SVs in the three cancer samples are within known cancer-related genes cataloged in the COSMIC census gene set. We found 237 COSMIC census genes that intersect at least one SV in 51T, with 622 total SVs present in these genes (Fig. 5A). The majority (199/237) of the SV-intersecting COSMIC (Tate et al. 2019) census genes in patient 51 had intersecting SVs both the tumor and matching normal cells, and the individual SV calls were mostly in both as well (466/622 in the initial SV call set and 428/568 in inferred karyotype graphs). Long-read-based SV inference identified five times as many COSMIC census genes with SVs than were identified by short reads. Furthermore, the poor concordance between SVs inferred exclusively with short reads between the tumor and normal samples (6/79) provides additional evidence that the short-read SV calling is error-prone. In both patient 48 and the SKBR3 cell line, we observed similar results (Fig. 5A) with long-read SV inference outperforming short-read SV inference in both the number of COSMIC census genes with SVs, as well as the number of SVs intersecting them. We also observed strong concordance across COSMIC genes with allele-specific CNVs as determined by inferred karyotype graphs inferred with either TitanCNA or HATCHet input large-scale copy number profiles in sample 51T (Fig. 5B).
Figure 5.
Structural and copy number variants in COSMIC census genes. (A) Comparison of the number of COSMIC census genes containing SVs, as well as the number of SVs within COSMIC census genes, across inferred SV call set in 51T and N (parenthetical), SKBR3, and 48T, and SVs reported by RCK as being present in the karyotype graphs reconstructed with either HATCHet or TitanCNA copy number profiles in 51T. (B) Comparison of the number of COSMIC census genes with either allele-specific deletions or amplifications between copy number profiles from HATCHet, RCK + HATCHet, TitanCNA, and RCK + TitanCNA in 51T.
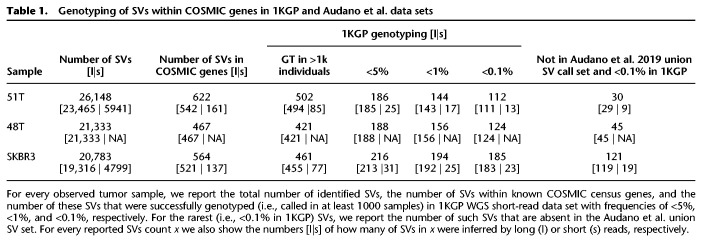
To assess the population frequency of SVs within COSMIC census genes, we genotyped the SVs from the three samples with Paragraph (Chen et al. 2019) across 2504 short-read WGS samples from the recent resequencing of the 1000 Genomes Project (1KGP) samples (Sudmant et al. 2015). Briefly, Paragraph genotypes SVs by constructing localized sequence graphs containing the reference allele and the candidate SV allele and performs a localized realignment of paired-end short reads to the graph. The genotype is then determined based on the coverage of reads uniquely supporting the reference or variant allele breakpoints. Not all variants can be genotyped by Paragraph, resulting in no genotype call when support is ambiguous, so we considered only SVs that Paragraph was capable of genotyping in at least 1000 samples. We then summarized rare variants identified in <5%, <1%, and <0.1% of the overall observed samples (Table 1). Because Paragraph v2.1 only genotypes insertions and deletions, we excluded inversions, translocations, and duplications from the genotyping analysis. As an additional approach to filtering common variants, we examined the presence of these variants in 15 healthy long-read-sequenced genomes from Audano et al. (2019). The approaches are largely complementary, although we do find some discordance with a subset of variants genotyped at low frequency (<5%) in the 1KGP individuals, but high frequency (≥20%) in the Audano data set (Audano et al. 2019; Supplemental Table 4). Of 275 COSMIC gene variants with discordant frequencies, nearly all (257) overlap tandem repeats (UCSC TRF track); Paragraph is known to have a higher false negative rate in such tandem repeat regions (Chen et al. 2019).
Table 1.
Genotyping of SVs within COSMIC genes in 1KGP and Audano et al. data sets
We combined these filtering approaches to identify a small set of variants found at very low frequency (<0.1%) in the 1KGP individuals and fully absent from the 15 healthy long-read genomes. These variants found at low-frequency in a healthy population are thus the most likely candidates for cancer risk-factor mutations (for full details, see Supplemental Table 5). These variants of interest are identified almost exclusively with long reads, and the ability of 15 long-read samples to additionally narrow the variants of interest further underscores the need for long-read sequenced genomes, both with healthy and disease phenotypes. Short-read genotype-based filtering of variants, although more conservative than filtering via comparison to long-read variants, remains a powerful tool to examine frequencies in larger cohorts than long-read data currently provide. Short-read genotyping may also prove especially valuable for examining frequencies in cohorts with phenotypes for which there is not long-read data available (e.g., TCGA, ICGC) and where this analysis could be used to select for high frequency variants within affected patients rather than against low frequency variants.
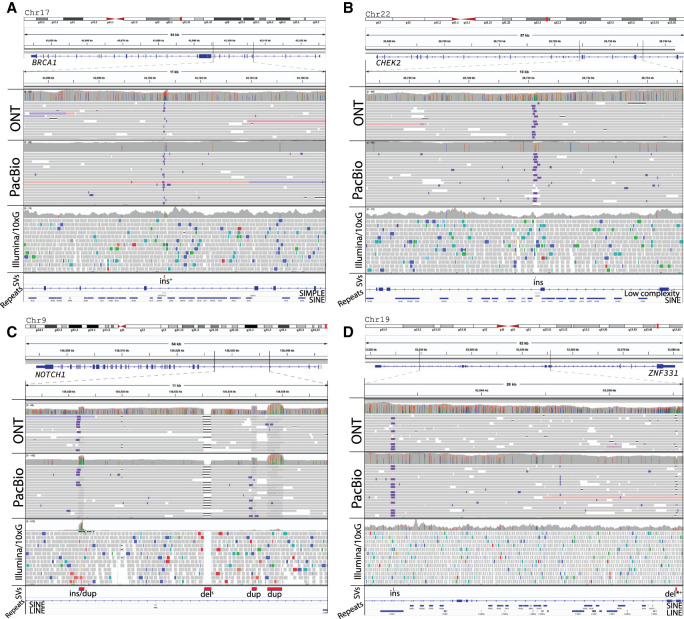
Four examples of genome rearrangements within COSMIC census genes in patient 51 are shown in Figure 6. We identified two insertions, which were missed by short-read SV inference, but were identified in both the ONT and PacBio data sets, in BRCA1 and CHEK2 breast cancer genes (Fig. 6A,B). Both insertions were also found exclusively with long reads in the matching normal tissue, with the insertion in the BRCA1 gene genotyped in <1% of 1KGP samples and present only in a single sample in the Audano et al. (2019) data set. In another example, we found multiple SVs present in NOTCH1 and ZNF331 (Fig. 6C,D). Both of these genes have been previously observed to play a significant role in tumor development (Yu et al. 2013; Nowell and Radtke 2017). Only one deletion (in NOTCH1) of the six SVs in NOTCH1 and ZNF331, was identified from short-read data, whereas all six of the considered SVs were identified in both ONT and PacBio long-read data sets. The exon-spanning deletion (Supplemental Fig. 23) in ZNF331 present in both 51T and 51N samples was found in <1% of 1KGP samples but was identified in 3/15 samples in the Audano et al. (2019) data set. Furthermore, by using long reads that span multiple SVs at the same time, we were able to phase SVs in NOTCH1 and ZNF331 and found these variants occurred on the same haplotype. The assignment of multiple SVs to either the same or different haplotypes helps to better understand relationships between allele-specific genetic alterations, which has been observed (Pastinen 2010) to provide important information in determining the possible effects on the functional activity of the genes.
Figure 6.
SVs identified in cancer-related COSMIC census genes in patient 51. All presented SVs are identified with both ONT and PacBio reads. Superscripts indicate the following: (*) marked SVs within known exons; (+) found as rare in 1KGP samples; and (s) identified by short-read SV inference methods, respectively. (A) An insertion in the BRCA1 gene identified in <1% of samples in 1KGP samples. (B) An insertion in the CHEK2 gene. (C) An insertion/duplication, deletion, and two duplications in the NOTCH1 gene, with deletion also found with short reads. All four SVs belong to the same haplotype as indicated by multiple long (both ONT and PacBio) reads spanning all of them at the same time. (D) An insertion and a deletion in the ZNF331 gene, with the later deletion within an exon in the NM_001317121 transcript and genotyped in <1% of 1KGP project samples. Both SVs belong to the same haplotype as indicated by long reads spanning all of them at the same time.
Finally, beyond the presence of individual SVs in particular genes, the somatic evolution of cancer genomes is also known to be propagated by various complex rearrangements that may require 3+ breakages happening simultaneously, some of which have been observed to have strong influence on the recovery prognosis of the patient (Hirsch et al. 2013; Fontana et al. 2018). We considered breakend groups in which pairs of breakends were either connected via SVs or located within 50 bp of each other. We identified several hundreds of breakend groups, mostly consisting of just three breakends (Supplemental Fig. 24), nearly all of which were only detectable with long reads. We note that not all complex k-breaks (k ≥ 3) produce breakend signatures, nor is there always an unambiguous way to reconstruct complex rearrangements from the breakend groups. However without observing or reconstructing the full somatic evolutionary history, such breakend grouping can be useful for emerging methods (Cortés-Ciriano et al. 2020) that infer and analyze genomic regions that are affected by a complex rearrangement history.
Discussion
In this study, we presented a comprehensive analysis of three cancer genomes that we sequenced with Illumina/10xG, ONT, and PacBio sequencing technologies and subsequently analyzed for large-scale (≥50 bp) structural genomic mutations with an ensemble of methods. We observed how various SV and CNV inference methods compare to one another, and how SV inference results differ across sequencing technologies. We first showed that SVs called with PacBio versus ONT data show high concordance, with >90% of SVs called with one platform also called with the other platform. Owing to their highly different methods of sequencing, this cross-validation indicates a high true positive rate of SV inference from either long-read technology. We also showed how SV and CNV mutations can be used together, reconciled, and integrated to infer a haplotype-specific cancer genome karyotype graph, which provides a refined view into the rearranged structure of observed cancer genomes.
Our findings show the need for long-read sequencing technologies in clinical settings to improve genome-informed cancer risk assessment, analysis, and treatment. Using long reads we detect a large number of novel SVs that are missed by whole-genome short-read sequencing in genes in which other known variants have been shown to substantially increase cancer risk. Although the full functional impact of these variants is currently unknown, the detection of additional variants in cancer-relevant genes indicates that current analysis pipelines may underestimate the role of variants in hereditary cancer risk or the mutational burden in a tumor sample. We observed that although long-reads provide previously unprecedented resolution for SV detection, the sample preparation, sequencing, and analysis is on par with short-read genome sequencing assays in terms of complexity, time, and computational requirements. Although costs are a major consideration for a technology to become widely applicable for patient care, we show that robust SV detection is possible at relatively low ∼30× average read-depth coverage with either ONT or PacBio long-read sequencing platforms. When applied at scale, costs for this amount of coverage is below US$1000 per sample for ONT PromethION and below US$2000 for PacBio Sequel II, which is highly comparable to ∼US$800/$1000 (Illumina/10xG) for short-read sequencing. Even at the high end of this cost, it is very cost efficient for the information it provides. It will however exacerbate the computational and data storage issues that are brought about by large-scale clinical DNA sequencing.
In the presented study we showed the complementary nature of both short- and long-read sequencing technologies by integrating both SV (mostly inferred with from long-reads) and large-scale allele-specific CNVs (determined by short-read coverage alterations over the heterozygous germline SNP locations) into a unifying karyotype-graph structure, which better describes the structural alterations in the observed mutated cancer genomes. As both SV and CNV call sets describe complementary measurements of the true underlying rearranged chromosomes, their integration allows for refinement of both large-scale CNVs as well as identification of spurious SV calls. We noted that long-reads provide both unprecedented resolution in SVs inference as well as haplotype-of-origin constraints for groups of SVs breakends, which can be important when determining effects of multiple closely located SVs on the underlying functional sequences. Short-reads, although less suited to SV detection, remain essential for accurate detection of heterozygous germline SNPs and subsequent coverage analysis and large-scale CNV inference in an allele-specific fashion in tumor samples. Furthermore, although the cancer samples examined here were homogeneous owing to their cell-line/organoid nature, primary cancer patient samples are often heterogeneous and consist of multiple cancer clones with possibly distinct karyotypes (Gundem et al. 2015; Zaccaria and Raphael 2018). For such samples, long reads can provide valuable insight in assigning groups of SVs to particular clones, and future long-read powered cancer studies can illuminate previously unseen aspects of clonal evolution in cancer. As additional sequencing coverage may be required for a thorough reconstruction of cancer subclones, these types of analyses will become more feasible as sequencing costs continue to decrease. Further development of methods capable of incorporating SNPs, small indels, SVs, and large-scale CNVs, as well as examining somatic phylogenies and cancer clone trajectories, will require both short- and long-read sequencing, with long reads proving critical for sensitive and accurate inference of SVs. Integration of long-read SV analysis can also benefit methods (Aganezov et al. 2019; Cameron et al. 2019; Deshpande et al. 2019) focusing on recovering linear organization of rearranged cancer genomes.
We also noted that as long-read sequencing technologies become more and more advanced it becomes possible to move away from a generic haploid human genome reference into an era of patient-specific reference sequences. We believe that future extension of the presented methodology can benefit from incorporating patient-specific diploid healthy genome structure as a starting point for mutation inference. We further underscored the importance of extending existing and developing new methods for multisample, time-series, and multipatient integrative analysis of genetic instability that drives and propagates cancer development. It is only through a process of identifying these structural variants in a large number of individuals that we can begin to broadly understand their frequency in populations and their implications in human health and disease.
Methods
Patient-derived organoid culture
Tumor resections from breast cancer patients along with adjacent normal tissue were collected from Northwell Health in accordance with Institutional Review Board protocol IRB-03-012 (TAP16-08). The collection of genomic and phenotypic data for this project was consistent with 45 CFR Part 46 (Protection of Human Subjects) and the NIH Genomic Data Sharing (GDS) Policy. Patient-derived tumor and normal organoids were developed in accordance with a previously published protocol (Sachs et al. 2018). Briefly, the resected samples were manually cut into smaller pieces and treated with Collagenase IV at 37°C. The samples were then manually broken down by pipetting into smaller fragments and seeded in a dome of matrigel. The organoids were grown in organoid culture media that contained 10% R-Spondin 1 conditioned media, 5 nM Neuregulin 1 (Peprotech 100-03), 5 ng/mL FGF7 (Peprotech 100-19), 20 ng/mL FGF10 (Peprotech 100-26), 5 ng/mL EGF (Peprotech AF-100-15), 100 ng/mL Noggin (Peprotech 120-10C), 500 nM A83-01 (Tocris 2939), 5 uM Y-27632 (Abmole Y-27632), 1.2 uM SB202190 (Sigma-Aldrich S7067), 1× B27 supplement (Gibco 17504-44), 1.25 mM N-Acetylcysteine (Sigma-Aldrich A9165), 5 mM Nicotinamide (Sigma-Aldrich N0636), 1× GlutaMAX (Thermo Fisher Scientific 12634-034), 10 mM HEPES (Thermo Fisher Scientific 15630-056), 100 units/mL Pen-Strep (Thermo Fisher Scientific 15140-122), and 50 ug/mL Primocin (Thermo Fisher Scientific ant-pm-1) in 1× Advanced DMEM-F12 (Thermo Fisher Scientific 12634-034) (Sachs et al. 2018). Organoids were passaged every 2–4 wk using TrypLE (Thermo Fisher Scientific 12605028) to break down the organoids into smaller clusters of cells and replating them.
Organoid DNA and RNA extraction
RNA was extracted using TRIzol (Thermo Fisher Scientific 15596018) RNA extraction protocol. DNA was extracted by removing matrigel from organoids using ice cold PBS or TrypLE following by DNA extraction using Qiagen DNeasy Blood and Tissue kit (Qiagen 69504).
SKBR3 growth
SKBR3 cells were purchased from ATCC (ATCC HTB 30). Cells were grown in 5 mL of McCoy 5A Medium (ATCC 30-2007) with 10% Fetal Bovine Serum ATCC 30-2020) and 1% penicillin/streptomycin (Sigma-Aldrich 11074440001). Cells were grown at 37°C with 5% CO2. To harvest cells, the media was removed and 2 mL of Trypsin-EDTA 0.25% (Sigma-Aldrich 25200056) was added. Cells were allowed to sit at 37°C for 10 min. The harvested cells were washed in PBS and either reseeded or used for DNA extraction.
Sample sequencing
10x Genomics Linked Read sequencing followed standard protocols. For long-read sequencing, DNA was sheared to >20 kb via Covaris G-tube (Covaris 520079). Oxford Nanopore DNA sequencing was carried out on a MinION or GridION device. Sheared DNA was prepared for sequencing using standard Oxford Nanopore methods. Briefly, sheared DNA was repaired with the NEB FFPE repair module (NEB M6630L), ligated to Oxford Nanopore adapters (Oxford Nanopore SQK-LSK108) via the NEB blunt/TA master mix (NEB M0367L), and cleaned up with Ampure beads (Beckman Coulter A63881). The full volume of the prepared libraries was loaded on to a MinION R9.1 flow cell and run for 48 h.
PacBio sequencing was carried out on a Pacific Biosciences Sequel instrument using standard PacBio methods. Briefly, sheared DNA was prepared for sequencing via the SMRTbell template prep kit 1.0 (Pacific Biosciences 100-991-900). The prepared libraries were size selected on a Blue Pippin overnight with a 10–50 kb range (Sage BUF7510). Libraries were loaded for sequencing on a 1 M SMRTcell (Pacific Biosciences 101-531-000) with a concentration of 4–10 pM with diffusion loading and 10 h movies.
Read alignment
All read alignments were performed against the latest human genome reference GRCh38 (Schneider et al. 2017). ONT and PacBio long-reads were aligned with NGMLR (Sedlazeck et al. 2018b) v0.2.7. Illumina/10xG short reads were aligned with LongRanger (Zheng et al. 2016) v2.1.6 pipeline. Only major Chromosomes 1–22 and X were considered for the alignment and subsequent structural analysis. Alignment coverage was computed with SAMtools (Li et al. 2009) v 1.9 depth command both with and without -a flag and computes an average of the per-base coverage values. For long reads, the distribution of raw-yield lengths considers sequenced lengths of all reads. Distribution of raw-aligned lengths considers sequenced read lengths that have at least some part(s) of them aligned to the reference. The distribution of aligned lengths considers lengths of aligned portion(s) of sequenced reads.
SV inference workflow
For both ONT and PacBio long reads, we used Sniffles v1.0.11 and PBSV v2.2.0 for SV inference. For Sniffles the minimum number of reads supporting SV was set to 2, and the minimum SV size was set to 30 bp, although the final variant calls were a more stringent subset of these requiring higher read support and larger sizes. PBSV was run with default settings. For Illumina/10xG reads, we used SvABA v FH134, LUMPY v 0.2.13, Manta v 1.5.0, GROC-SVs v 0.2.5, NAIBR (version determined by 15eba96 commit GitHub master branch), and LongRanger v 2.1.6. All short-read SV callers were run with recommended settings. Some SV inference methods produced more than a single SV call set (usually with SVs segregated by size), which we subsequently concatenated into method-specific SV call sets. For example, for SvABA we concatenated indel and sv SV call sets, and for LongRanger we concatenated the dels and large_svs SV call sets.
For every sequencing technology, the SVs produced by all callers were merged together with the SURVIVOR (Jeffares et al. 2017) v1.0.6 software package into a ONT, PacBio, and Illumina/10xG technology-specific SV call sets. SURVIVOR merge was run with maximum distance between SVs set to 1000 and minimum SV size set to 30. SV types were not taken into account during the SURVIVOR merging as different methods may assign different types, especially insertions versus duplications, to the same inferred SV based on the respective method's terminology, but strand/orientation was required to match.
For SVs inferred on short reads, we removed any method-exclusive SVs (i.e., supported by only one out of six methods) and retained any SVs that had at least two methods inferring them. This was done to mitigate false positives as we previously showed in our SKBR3 analysis.
To ensure consistency when comparing against 15 healthy genomes, sequenced with PacBio and reported in Audano et al. (2019), we performed alignment and variant calling on the 15 samples with the same pipeline described above. Raw reads from all 15 genomes were downloaded and aligned with NGMLR to the main chromosomes of GRCh38 with the -x pacbio setting. Structural variants were then called on each sample with Sniffles. Because all samples were reported in Audano et al. (2019) as above 40× coverage, we set Sniffles to require a minimum read support of 10 reads. A minimum SV size of 30 bp was used. Comparisons against the SVs in these 15 genomes were performed with SURVIVOR merge with a maximum distance of 1000, type and strand considered, and a minimum size of 30, with thresholding performed post merging to examine only variants of at least 50 bp, as described below.
Comparison of SVs inferred with different sequencing technologies
ONT, PacBio, and Illumina/10xG technology-specific SV call sets were subsequently merged together with the SURVIVOR package into a sample-specific sensitive SV call set. SV types were not taken into account for the same purpose as was described for the technology-specific merging procedure, minimum size for SVs to be considered was set to 30, maximum distance between SVs was set to 1000.
We further removed from the sensitive SV call set any of the SVs shorter than 50 bp to focus only on large-scale rearrangements. This filtration was done after the merging of technology-specific SV call sets, rather than before, to mitigate thresholding issues that may have arisen in cases when the same underlying SV had its length reported with slight imprecision around the size threshold value. For example, a variant called as 49 bp in one call set, and 51 bp in another, would have the 49 bp instance removed before it could have been merged with the 51 bp instance, producing a 50-bp-long merged SV, which would be retained.
To mitigate the relatively high per-base-pair error rate in long reads and its possible effect on false positive calls in long-read-exclusive (either from ONT, or PacBio, or both) SVs, we removed long-read exclusive SVs for which the number of long reads supporting them was less than a quarter of average read-depth in both ONT and PacBio data sets. After length and long-read-support filtration, we obtained the specific SV call set on which the agreement and discordance in SVs inference between sequencing technologies and methods was analyzed.
Methylation analysis
For calling methylation on ONT sequencing data, we used the nanopolish (Simpson et al. 2017) call-methylation module. A threshold of 2.5 was used to filter out ambiguous methylation calls. After aggregating methylation calls at each site, we smoothed the raw methylation frequencies using BSmooth function from R (R Core Team 2019) Bioconductor (Gentleman et al. 2004) package bsseq (Hansen et al. 2012). Briefly, we first choose a window such that at least 50 CpG sites and 500 bps of region are covered for each locus lj. Assuming that (1) the methylation frequency f(lj) follows a binomial distribution; and (2) is approximated by a second-degree polynomial, we fit a weighted generalized linear model inside each window. The weights are inversely proportional to the standard errors of the per-site measurements, and a tricube kernel is used in relation to the distance from the locus lj . For all subsequent analysis, we applied a coverage filter, removing data points on loci where the total number of calls was less than 5 in any of the samples.
For global comparisons of genomic context methylation, we used genomic contexts as determined by Ensembl (Cunningham et al. 2019) gene annotations and regulatory feature sets. For each region in the set, average methylation was calculated by dividing the sum of methylated calls by total calls.
RNA-seq expression analysis
RNA-seq libraries were prepared using Illumina TruSeq RNA Library prep kit v2 (RS-122-2001) and sequenced as 75-bp paired end. The reads were aligned using STAR (Dobin et al. 2013) v 2.5.3a. The built-in gene counts option was used to count raw reads using the GENCODE (The ENCODE Project Consortium 2012; Davis et al. 2018) v27 GTF reference file. The counts files were exported into R v3.5.1 and normalized using DESeq2 (Love et al. 2014) v1.22.2. Normalized counts were used to calculate log2 fold change of tumor versus normal samples.
SVs that were supported by long-read sequencing were used for this analysis. Structural variants overlapping genes were determined using the BEDTools (Quinlan and Hall 2010) intersect –wo command with GENCODE v27 GTF file as a reference. The graphs were plotted using ggplot2 (Wickham 2016) v3.2.1. Percent overlap was calculated by dividing the number of overlapping base pairs with the total length of the gene.
Down-sampling and SV inference
We designed and implemented the down-sampling workflow to analyze the robustness of long-read SV inference at various read depth coverages. Every full coverage long-read ONT or PacBio alignment data set reads.bam was down-sampled with SAMtools v1.9 command view -s x.y reads.bam, where x determines the seed for randomize alignments selection to be included in the produced down-sampled alignment, and y determines the fraction of the read alignments from the initial data set reads.bam to be selected.
For sample 51T, the down-sampled coverage levels were set to C = [5, 8, 10, 12, 16, 20, 24, 32, 38, 44]× for both ONT and PacBio, for sample 48T coverage levels were set at C = [5, 8, 10, 12, 16, 24, 32, 36, 40]× for both ONT and PacBio, and for sample SKBR3 down-sampled coverage levels were set at C = [5, 10, 16, 20, 24, 28, 32]× for ONT, and additional coverage levels at C = [40, 48, 52]× were set for PacBio, because the PacBio data set for SKBR3 had higher coverage available than the ONT one.
For both ONT and PacBio, for every down-sampled target coverage level we generated three distinct down-sampled read alignments data sets with different random seed values. SV inference on the down-sampled alignment data sets was computed using Sniffles v1.0.11 with the minimum number of reads required to support an SV set to 2, and the minimum SV size set to 30. As previously described, to mitigate a relatively high per-base-pair error rate in long reads, several reads are required to span an SV for it to be considered true. We observed how SV inference was affected by this parameter by considering various fractions of an average down-sampled data set–specific read depth coverage as a threshold for the minimum number of reads required to span an SV. We then generated distinct f-SVs call sets by removing all SVs that were supported by less than a fraction f of reads.
We then compared the technology-specific f-SVs call sets for every down-sampled coverage level c ∈ C with the gold standard (i.e., SVs call set on full coverage data set, using the matching read support threshold) with SURVIVOR and averaged the precision and recall results over three randomly created down-sampled data sets for every coverage target level c.
Integration and refinement of copy-number and structural variations
To measure large-scale CNVs we used Illumina/10xG short-read sequencing data sets from both the tumor and the matching normal cells. We used TitanCNA and HATCHet CNV-inference methods, which produce clone- and allele-specific segment copy number profiles. Both methods were run with recommended settings.
When considering possible errors in the measured copy number values, we take into account that both methods infer CNV profiles on rather large (≥50 kbp) segments, which will miss any smaller copy number variations (e.g., small deletions or duplications), and the specific boundaries of large CNVs. In order to combine the SV and CNV mutation inference, we used our RCK method, which integrates both SV and CNV mutations together and infers the underlying clone- and haplotype-specific karyotype graph or simply karyotype.
In the new RCK v. 1.1 developed for this project, we added long-read based haplotype constraints for SVs breakends, which helps to resolve ambiguities arising from equally plausible solutions in haplotype assignment. As single molecule platforms, both ONT and PacBio long reads that span multiple SVs introduce reference-haplotype-of-origin constraints, that is, ensuring that grouped SVs breakends are assigned to the same haplotype (Supplemental Methods).
We ran RCK v 1.1 on the inferred sample-specific SV call set and both HATCHet and TitanCNA CNV profiles separately with the required fraction P of input SVs to be used being set at 0.9. We also significantly improved the performance of the original version of RCK by introducing the per-chromosomal preprocessing step. In this step RCK first solves the karyotype-graph inference problem on a per chromosome basis, such that the union of solutions would equal to the whole-genome problem solution excluding the inter-chromosomal SVs. Per-chromosomal solutions are then used as starting vector for the whole-genome (with inter-chromosomal SVs) MILP problem solution search. Implementation of this pre-solve approach allowed us to improve performance by reducing the running time from 48 to 6 h and from 32 to 8 h wall clock time for TitanCNA and HATCHet CNV input, respectively. RCK was run on a 24 core (with ‐‐run-threads 24 flags) machine with 512GB RAM (with a peak usage of ∼200 GB of RAM). We note that time and high RAM usage is caused by tens of thousands of input SVs and haplotype constraint groups; on simpler cancer samples (with ∼1000 SVs), RCK can infer cancer karyotypes in several minutes.
Circos plots (Krzywinski et al. 2009) shown in Figure 4C and Supplemental Figures 3, 21, and 22 were constructed with Circa v 1.2.0 software (http://omgenomics.com/circa).
Analysis of COSMIC census genes intersecting SVs
For the analysis of SVs and COSMIC census gene interactions, we considered genes from the COSMIC Cancer Gene Census v88. We considered a COSMIC census gene g = A:[a, b], with start coordinate a and end coordinate b on Chromosome A, being intersected by SV s = {X:p, Y:q} with breakends p on Chromosome X and q on Chromosome Y if either X = A and a ≤ p ≤ b or Y = A and a ≤ q ≤ b, or both. We note that SV breakend strand orientations are not important in considering whether a gene is intersected by a SV, as in either case the breakend disrupts the genomic region that contains the gene. We say that a COSMIC census gene g is intersected by SVs if it is intersected by at least one SV, and we say that SV s intersects COSMIC census genes if at least one COSMIC census gene g is intersected by s.
We further analyzed the SVs interactions with the COSMIC census genes with their flanking sequences (i.e., for every gene g = A:[a, b], we considered , where Δ ∈ [500, 1000, 5000]). We did not discover any additional COSMIC census genes with flanking SVs nor have we observed any additional SVs flanking COSMIC census genes with any of the considered values of Δ.
Analysis of COSMIC census genes intersected by CNVs
For the analysis of CNVs and COSMIC census gene interactions, we considered genes from the COSMIC Cancer Gene Census v 88.
We considered a COSMIC census gene g = A:[a, b], with start coordinate a and end coordinate b on Chromosome A, being intersected by an allele-specific deletion (amplification) if there exists a segment j = A:[c, d] that overlaps g, or more formally, either c ≤ a ≤ d, or c ≤ b ≤ d, or both, and j has the respective allele-specific CNV, that is, either aj > 1( < 1) or bj > 1( < 1). Identification of the COSMIC census genes intersected by allele-specific CNVs was performed with a RCK-based utility script. We note that the same COSMIC census gene g (on either the same or different haplotypes) may be simultaneously intersected by both allele-specific deletion(s) and amplification(s).
Grouping of structural variation breakends
Complex rearrangements reflect an underlying double-stranded breakage event of k ≥ 3 double-stranded breaks. Segments resulting from the breakage are then often amplified or lost, and the subsequent ligation of involved segments are then detected as SVs. In general, by simple observation of a group U of SVs, we cannot determine whether all of the SVs in U we produced by a single complex rearrangement or by several sequential rearrangements.
To identify potential signatures of complex rearrangements for a given set A of SVs, we constructed a complex rearrangements graph GC = (V, E), where a set of vertices is determined by reference adjacencies (within 50 bp threshold), and every edge ea = {u, v} ∈ E is determined by an SV a = {px, qy} such that px ∈ u and qy ∈ v, or more simply, if a connects extremities involved in u and v. Once the GC is constructed, complex rearrangements are determined as connected components with at least three vertices in them, as they correspond to groups of reciprocal SVs. We note that not every k-break produces reciprocal SVs. Grouping of SVs breakends for identifying genomic locations potentially intersected by complex rearrangements was implemented in RCK v1.1.
Software availability
The SV inference and comparison workflow is implemented with Snakemake (Köster and Rahmann 2012) v 5.5.4 and is available at GitHub (https://github.com/aganezov/EnsembleSV). RCK (Aganezov and Raphael 2020) v 1.1 used for cancer genome karyotype inference is available at GitHub (https://github.com/aganezov/RCK). A summary of the available data and workflows are also available at https://schatz-lab.org/publications/bcorganoid/.
Data access
All raw sequencing data generated in this study for SKBR3 have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA476239. All raw sequencing data and variant data in this study from the patient samples have been submitted to the NCBI database of Genotypes and Phenotypes (dbGaP; https://www.ncbi.nlm.nih.gov/gap/) under accession number phs038843.v1.
Competing interest statement
W.T. owns two patents currently licensed by Oxford Nanopore Technologies Limited. M.C.S. and W.T. have received travel funding from Oxford Nanopore Technologies Limited. W.R.M. has participated in Illumina-sponsored meetings over the last four years and received travel reimbursement and an honorarium for presenting at these events. W.R.M. has participated in Pacific Biosciences-sponsored meetings over the last four years and received travel reimbursement for presenting at these events. Oxford Nanopore, Illumina, and Pacific Biosciences had no role in decisions relating to the study/work to be published, data collection, analysis of the data, or decision to publish. W.R.M. is a founder and shareholder of Orion Genomics, which focuses on plant genomics and cancer genetics.
Supplementary Material
Acknowledgments
The authors thank Benjamin J. Raphael, Rebecca Elyanow, and Simonne Zaccaria for thoughtful and productive discussions and their help in running the NAIBR and HATCHet methods. We thank Gavin Ha for thoughtful and productive discussion and his help in running the TitanCNA method. This work was funded in part by the U.S. National Science Foundation (NSF) grant DBI-1350041 (M.C.S.); U.S. National Institutes of Health (NIH) grants R01-HG006677 (M.C.S.), R21-CA220411 (M.C.S.), 5P30CA045508 (W.R.M.), and 1R01-HG009190 (W.T.); the Bill and Melinda Gates Foundation (M.C.S.); and Cold Spring Harbor Laboratory/Northwell Health (W.R.M., M.C.S., and D.L.S.).
Author contributions: M.C.S. and W.R.M. designed the study. S.A. led the data analysis. S.G. led the sample preparation and sequencing. G.A. and R.W. prepared samples and assisted with sequencing. S.G., R.M.S., F.J.S., G.A., S.B., I.L., M. Kirsche, R.W., and M. Kramer performed analysis. K.K. collected the patient samples. D.L.S., W.T., W.R.M., and M.C.S. oversaw analysis. All authors reviewed and approved the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.260497.119.
References
- Aganezov S, Raphael BJ. 2020. Reconstruction of clone- and haplotype-specific cancer genome karyotypes from bulk tumor samples. Genome Res (this issue). 10.1101/gr.256701.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aganezov S, Zban I, Aksenov V, Alexeev N, Schatz MC. 2019. Recovering rearranged cancer chromosomes from karyotype graphs. BMC Bioinformatics 20: 641 10.1186/s12859-019-3208-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale AL, et al. 2013. Signatures of mutational processes in human cancer. Nature 500: 415–421. 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audano PA, Sulovari A, Graves-Lindsay TA, Cantsilieris S, Sorensen M, Welch AME, Dougherty ML, Nelson BJ, Shah A, Dutcher SK, et al. 2019. Characterizing the major structural variant alleles of the human genome. Cell 176: 663–675.e19. 10.1016/j.cell.2018.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudino TA. 2015. Targeted cancer therapy: the next generation of cancer treatment. Curr Drug Discov Technol 12: 3–20. 10.2174/1570163812666150602144310 [DOI] [PubMed] [Google Scholar]
- Burns KH. 2017. Transposable elements in cancer. Nat Rev Cancer 17: 415–424. 10.1038/nrc.2017.35 [DOI] [PubMed] [Google Scholar]
- Cameron DL, Baber J, Shale C, Papenfuss AT, Valle-Inclan JE, Besselink N, Cuppen E, Priestley P. 2019. GRIDSS, PURPLE, LINX: unscrambling the tumor genome via integrated analysis of structural variation and copy number. bioRxiv 10.1101/781013v1 [DOI]
- The Cancer Genome Atlas Research Network, Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM, et al. 2013. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 45: 1113–1120. 10.1038/ng.2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavenee WK, Dryja TP, Phillips RA, Benedict WF, Godbout R, Gallie BL, Murphree AL, Strong LC, White RL. 1983. Expression of recessive alleles by chromosomal mechanisms in retinoblastoma. Nature 305: 779–784. 10.1038/305779a0 [DOI] [PubMed] [Google Scholar]
- Chaisson MJP, Sanders AD, Zhao X, Malhotra A, Porubsky D, Rausch T, Gardner EJ, Rodriguez OL, Guo L, Collins RL, et al. 2019. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat Commun 10: 1784 10.1038/s41467-018-08148-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Källberg M, Cox AJ, Kruglyak S, Saunders CT. 2016. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 32: 1220–1222. 10.1093/bioinformatics/btv710 [DOI] [PubMed] [Google Scholar]
- Chen S, Krusche P, Dolzhenko E, Sherman RM, Petrovski R, Schlesinger F, Kirsche M, Bentley DR, Schatz MC, Sedlazeck FJ, et al. 2019. Paragraph: a graph-based structural variant genotyper for short-read sequence data. Genome Biol 20: 291 10.1186/s13059-019-1909-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. 2016. Modeling development and disease with organoids. Cell 165: 1586–1597. 10.1016/j.cell.2016.05.082 [DOI] [PubMed] [Google Scholar]
- Cortés-Ciriano I, Lee JJK, Xi R, Jain D, Jung YL, Yang L, Gordenin D, Klimczak LJ, Zhang CZ, Pellman DS, et al. 2020. Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. Nat Genet 52: 331–341. 10.1038/s41588-019-0576-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham F, Achuthan P, Akanni W, Allen J, Amode MR, Armean IM, Bennett R, Bhai J, Billis K, Boddu S, et al. 2019. Ensembl 2019. Nucleic Acids Res 47: D745–D751. 10.1093/nar/gky1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, Hilton JA, Jain K, Baymuradov UK, Narayanan AK, et al. 2018. The encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res 46: D794–D801. 10.1093/nar/gkx1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Coster W, De Rijk P, De Roeck A, De Pooter T, D'Hert S, Strazisar M, Sleegers K, Van Broeckhoven C. 2019. Structural variants identified by oxford nanopore PromethION sequencing of the human genome. Genome Res 29: 1178–1187. 10.1101/gr.244939.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande V, Luebeck J, Nguyen NPD, Bakhtiari M, Turner KM, Schwab R, Carter H, Mischel PS, Bafna V. 2019. Exploring the landscape of focal amplifications in cancer using AmpliconArchitect. Nat Commun 10: 392 10.1038/s41467-018-08200-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreos R, Ambrosini G, Cavin Périer R, Bucher P. 2013. EPD and EPDnew, high-quality promoter resources in the next-generation sequencing era. Nucleic Acids Res 41: D157–D164. 10.1093/nar/gks1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbert MTW, Jensen TD, Jansen-West K, Sens JP, Reddy JS, Ridge PG, Kauwe JSK, Belzil V, Pregent L, Carrasquillo MM, et al. 2019. Systematic analysis of dark and camouflaged genes reveals disease-relevant genes hiding in plain sight. Genome Biol 20: 97 10.1186/s13059-019-1707-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elyanow R, Wu HT, Raphael BJ. 2018. Identifying structural variants using linked-read sequencing data. Bioinformatics 34: 353–360. 10.1093/bioinformatics/btx712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon ER, Vogelstein B. 1990. A genetic model for colorectal tumorigenesis. Cell 61: 759–767. 10.1016/0092-8674(90)90186-I [DOI] [PubMed] [Google Scholar]
- Fishilevich S, Nudel R, Rappaport N, Hadar R, Plaschkes I, Iny Stein T, Rosen N, Kohn A, Twik M, Safran M, et al. 2017. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database 2017 10.1093/database/bax028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana MC, Marconi G, Feenstra JDM, Fonzi E, Papayannidis C, Ghelli Luserna Di Rorá A, Padella A, Solli V, Franchini E, Ottaviani E, et al. 2018. Chromothripsis in acute myeloid leukemia: biological features and impact on survival. Leukemia 32: 1609–1620. 10.1038/s41375-018-0035-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco I, Helgadottir HT, Moggio A, Larsson M, Vrtačnik P, Johansson A, Norgren N, Lundin P, Mas-Ponte D, Nordström J, et al. 2019. Whole genome DNA sequencing provides an atlas of somatic mutagenesis in healthy human cells and identifies a tumor-prone cell type. Genome Biol 20: 285 10.1186/s13059-019-1892-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin S, McPherson JD, McCombie WR. 2016. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet 17: 333–351. 10.1038/nrg.2016.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JMC, Papaemmanuil E, Brewer DS, Kallio HML, Högnäs G, Annala M, et al. 2015. The evolutionary history of lethal metastatic prostate cancer. Nature 520: 353–357. 10.1038/nature14347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha G, Roth A, Khattra J, Ho J, Yap D, Prentice LM, Melnyk N, McPherson A, Bashashati A, Laks E, et al. 2014. TITAN: inference of copy number architectures in clonal cell populations from tumor whole-genome sequence data. Genome Res 24: 1881–1893. 10.1101/gr.180281.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, Wen B, Wu H, Liu Y, Diep D, et al. 2011. Increased methylation variation in epigenetic domains across cancer types. Nat Genet 43: 768–775. 10.1038/ng.865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KD, Langmead B, Irizarry RA. 2012. BSmooth: from whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biol 13: R83 10.1186/gb-2012-13-10-r83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltemann S, Jenster G, Trapman J, van der Spek P, Stubbs A. 2015. Discriminating somatic and germline mutations in tumor DNA samples without matching normals. Genome Res 25: 1382–1390. 10.1101/gr.183053.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch D, Kemmerling R, Davis S, Camps J, Meltzer PS, Ried T, Gaiser T. 2013. Chromothripsis and focal copy number alterations determine poor outcome in malignant melanoma. Cancer Res 73: 1454–1460. 10.1158/0008-5472.CAN-12-0928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges E, Xuan Z, Balija V, Kramer M, Molla MN, Smith SW, Middle CM, Rodesch MJ, Albert TJ, Hannon GJ, et al. 2007. Genome-wide in situ exon capture for selective resequencing. Nat Genet 39: 1522–1527. 10.1038/ng.2007.42 [DOI] [PubMed] [Google Scholar]
- Huddleston J, Chaisson MJP, Steinberg KM, Warren W, Hoekzema K, Gordon D, Graves-Lindsay TA, Munson KM, Kronenberg ZN, Vives L, et al. 2017. Discovery and genotyping of structural variation from long-read haploid genome sequence data. Genome Res 27: 677–685. 10.1101/gr.214007.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa T, Sato F, Terasawa K, Tsuchiya S, Toi M, Tsujimoto G, Shimizu K. 2012. Trastuzumab produces therapeutic actions by upregulating miR-26a and miR-30b in breast cancer cells. PLoS One 7: e31422 10.1371/journal.pone.0031422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffares DC, Jolly C, Hoti M, Speed D, Shaw L, Rallis C, Balloux F, Dessimoz C, Bähler J, Sedlazeck FJ. 2017. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat Commun 8: 14061 10.1038/ncomms14061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y, Rose CM, Cass AA, Williams AG, Darwish M, Lianoglou S, Haverty PM, Tong AJ, Blanchette C, Albert ML, et al. 2019. Transposable element expression in tumors is associated with immune infiltration and increased antigenicity. Nat Commun 10: 5228 10.1038/s41467-019-13035-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köster J., Rahmann S. 2012. Snakemake—a scalable bioinformatics workflow engine. Bioinformatics 28: 2520–2522. 10.1093/bioinformatics/bts480 [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer RM, Chiang C, Quinlan AR, Hall IM. 2014. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol 15: R84 10.1186/gb-2014-15-6-r84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis Phillips GD, Li G, Dugger DL, Crocker LM, Parsons KL, Mai E, Blättler WA, Lambert JM, Chari RVJ, Lutz RJ, et al. 2008. Targeting HER2-positive breast cancer with trastuzumab-DM1, an antibody–cytotoxic drug conjugate. Cancer Res 68: 9280–9290. 10.1158/0008-5472.CAN-08-1776 [DOI] [PubMed] [Google Scholar]
- Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, et al. 2008. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 456: 66–72. 10.1038/nature07485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, et al. 1997. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 275: 1943–1947. 10.1126/science.275.5308.1943 [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelker D, Schmidt RJ, Ankala A, McDonald Gibson K, Bowser M, Sharma H, Duffy E, Hegde M, Santani A, Lebo M, et al. 2016. Navigating highly homologous genes in a molecular diagnostic setting: a resource for clinical next-generation sequencing. Genet Med 18: 1282–1289. 10.1038/gim.2016.58 [DOI] [PubMed] [Google Scholar]
- Martincorena I, Campbell PJ. 2015. Somatic mutation in cancer and normal cells. Science 349: 1483–1489. 10.1126/science.aab4082 [DOI] [PubMed] [Google Scholar]
- Merker JD, Wenger AM, Sneddon T, Grove M, Zappala Z, Fresard L, Waggott D, Utiramerur S, Hou Y, Smith KS, et al. 2018. Long-read genome sequencing identifies causal structural variation in a Mendelian disease. Genet Med 20: 159–163. 10.1038/gim.2017.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao H, Zhou J, Yang Q, Liang F, Wang D, Ma N, Gao B, Du J, Lin G, Wang K, et al. 2018. Long-read sequencing identified a causal structural variant in an exome-negative case and enabled preimplantation genetic diagnosis. Hereditas 155: 32 10.1186/s41065-018-0069-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nattestad M, Goodwin S, Ng K, Baslan T, Sedlazeck FJ, Rescheneder P, Garvin T, Fang H, Gurtowski J, Hutton E, et al. 2018. Complex rearrangements and oncogene amplifications revealed by long-read DNA and RNA sequencing of a breast cancer cell line. Genome Res 28: 1126–1135. 10.1101/gr.231100.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, et al. 2011. Tumour evolution inferred by single-cell sequencing. Nature 472: 90–94. 10.1038/nature09807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, et al. 2006. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10: 515–527. 10.1016/j.ccr.2006.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen FC, Van Overeem Hansen T, Sørensen CS. 2016. Hereditary breast and ovarian cancer: new genes in confined pathways. Nat Rev Cancer 16: 599–612. 10.1038/nrc.2016.72 [DOI] [PubMed] [Google Scholar]
- Nowell CS, Radtke F. 2017. Notch as a tumour suppressor. Nat Rev Cancer 17: 145–159. 10.1038/nrc.2016.145 [DOI] [PubMed] [Google Scholar]
- Pastinen T. 2010. Genome-wide allele-specific analysis: insights into regulatory variation. Nat Rev Genet 11: 533–538. 10.1038/nrg2815 [DOI] [PubMed] [Google Scholar]
- Perou CM, Sørile T, Eisen MB, Van De Rijn M, Jeffrey SS, Ress CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. 2000. Molecular portraits of human breast tumours. Nature 406: 747–752. 10.1038/35021093 [DOI] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordóñez GR, Bignell GR, et al. 2010. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463: 191–196. 10.1038/nature08658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2019. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: http://www.R-project.org/. [Google Scholar]
- Sachs N, de Ligt J, Kopper O, Gogola E, Bounova G, Weeber F, Balgobind AV, Wind K, Gracanin A, Begthel H, et al. 2018. A living biobank of breast cancer organoids captures disease heterogeneity. Cell 172: 373–386.e10. 10.1016/j.cell.2017.11.010 [DOI] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270: 467–470. 10.1126/science.270.5235.467 [DOI] [PubMed] [Google Scholar]
- Schneider VA, Graves-Lindsay T, Howe K, Bouk N, Chen H-C, Kitts PA, Murphy TD, Pruitt KD, Thibaud-Nissen F, Albracht D, et al. 2017. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res 27: 849–864. 10.1101/gr.213611.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz R, Schäffer AA. 2017. The evolution of tumour phylogenetics: principles and practice. Nat Rev Genet 18: 213–229. 10.1038/nrg.2016.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlazeck FJ, Lee H, Darby CA, Schatz MC. 2018a. Piercing the dark matter: bioinformatics of long-range sequencing and mapping. Nat Rev Genet 19: 329–346. 10.1038/s41576-018-0003-4 [DOI] [PubMed] [Google Scholar]
- Sedlazeck FJ, Rescheneder P, Smolka M, Fang H, Nattestad M, von Haeseler A, Schatz MC. 2018b. Accurate detection of complex structural variations using single-molecule sequencing. Nat Methods 15: 461–468. 10.1038/s41592-018-0001-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JT, Workman RE, Zuzarte PC, David M, Dursi LJ, Timp W. 2017. Detecting DNA cytosine methylation using nanopore sequencing. Nat Methods 14: 407–410. 10.1038/nmeth.4184 [DOI] [PubMed] [Google Scholar]
- Sone J, Mitsuhashi S, Fujita A, Mizuguchi T, Hamanaka K, Mori K, Koike H, Hashiguchi A, Takashima H, Sugiyama H, et al. 2019. Long-read sequencing identifies GGC repeat expansions in NOTCH2NLC associated with neuronal intranuclear inclusion disease. Nat Genet 51: 1215–1221. 10.1038/s41588-019-0459-y [DOI] [PubMed] [Google Scholar]
- Spies N, Weng Z, Bishara A, McDaniel J, Catoe D, Zook JM, Salit M, West RB, Batzoglou S, Sidow A. 2017. Genome-wide reconstruction of complex structural variants using read clouds. Nat Methods 14: 915–920. 10.1038/nmeth.4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Hsi-Yang Fritz M, et al. 2015. An integrated map of structural variation in 2,504 human genomes. Nature 526: 75–81. 10.1038/nature15394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taparowsky E, Suard Y, Fasano O, Shimizu K, Goldfarb M, Wigler M. 1982. Activation of the T24 bladder carcinoma transforming gene is linked to a single amino acid change. Nature 300: 762–765. 10.1038/300762a0 [DOI] [PubMed] [Google Scholar]
- Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, Boutselakis H, Cole CG, Creatore C, Dawson E, et al. 2019. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res 47: D941–D947. 10.1093/nar/gky1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wala JA, Bandopadhayay P, Greenwald NF, O'Rourke R, Sharpe T, Stewart C, Schumacher S, Li Y, Weischenfeldt J, Yao X, et al. 2018. SvABA: genome-wide detection of structural variants and indels by local assembly. Genome Res 28: 581–591. 10.1101/gr.221028.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger AM, Peluso P, Rowell WJ, Chang PC, Hall RJ, Concepcion GT, Ebler J, Fungtammasan A, Kolesnikov A, Olson ND, et al. 2019. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat Biotechnol 37: 1155–1162. 10.1038/s41587-019-0217-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York. [Google Scholar]
- Yates LR, Campbell PJ. 2012. Evolution of the cancer genome. Nat Rev Genet 13: 795–806. 10.1038/nrg3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Liang QY, Wang J, Cheng Y, Wang S, Poon TCW, Go MYY, Tao Q, Chang Z, Sung JJY. 2013. Zinc-finger protein 331, a novel putative tumor suppressor, suppresses growth and invasiveness of gastric cancer. Oncogene 32: 307–317. 10.1038/onc.2012.54 [DOI] [PubMed] [Google Scholar]
- Zaccaria S, Raphael BJ. 2018. Accurate quantification of copy-number aberrations and whole-genome duplications in multi-sample tumor sequencing data. bioRxiv 10.1101/496174 [DOI] [PMC free article] [PubMed]
- Zhang J, Baran J, Cros A, Guberman JM, Haider S, Hsu J, Liang Y, Rivkin E, Wang J, Whitty B, et al. 2011. International cancer genome consortium data portal—a one-stop shop for cancer genomics data. Database 2011: bar026 10.1093/database/bar026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GXY, Lau BT, Schnall-Levin M, Jarosz M, Bell JM, Hindson CM, Kyriazopoulou-Panagiotopoulou S, Masquelier DA, Merrill L, Terry JM, et al. 2016. Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat Biotechnol 34: 303–311. 10.1038/nbt.3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zook JM, Hansen NF, Olson ND, Chapman L, Mullikin JC, Xiao C, Sherry S, Koren S, Phillippy AM, Boutros PC, et al. 2020. A robust benchmark for detection of germline large deletions and insertions. Nat Biotechnol 10.1038/s41587-020-0538-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.