Abstract
Recently, various studies have shown that angiotensin-converting enzyme 2 (ACE2) acts as the “doorknob” that can be bound by the spike protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which conduces to its entrance to the host cells, and plays an important role in corona virus disease 2019 (COVID-19). This paper aims to collect and sorts out the existing drugs, which exert the ability to block the binding of S protein and ACE2 so as to provide directions for the later drug development. By reviewing the existing literature, we expound the pathogenesis of SARS-CoV-2 from the perspective of S protein and ACE2 binding, and summarize the drugs and compounds that can interfere with the interaction of spike protein and ACE2 receptor from different ways. We summarized five kinds of substances, including peptide P6, griffithsin, hr2p analogs, EK1, vaccine, monoclonal antibody, cholesterol-depleting agents, and extracts from traditional Chinese medicine. They can fight SARS-CoV-2 by specifically binding to ACE2 receptor, S protein, or blocking membrane fusion between the host and virus. ACE2 is the key point for SARS-CoV-2 to enter the cells, and it is also the focus of drug intervention. Our drug summary on this pathomechanism is expected to provide ideas for the drug research on SARS-CoV-2 and help to develop anti-coronavirus drugs of broad spectrum for future epidemics.
Keywords: COVID-19, SARS-CoV-2, ACE2, Spike protein, Drug
Introduction
A novel coronavirus currently termed COVID-19 occurred in Wuhan city, Hubei province, China, on 31 December 2019. It was declared as a public health emergency of international concern on Jan 30, 2020, by WHO [1]. At present, more than 70,000 people have been diagnosed and thousands of people died which remains to be a great threat to human life in China and even the world. The initial symptoms of COVID-19 patients are mostly respiratory symptoms such as fever, cough, wheezing, and dyspnea. In more severe cases, patients may develop severe pneumonia, pulmonary edema, ARDS or multiple organ failure, or even death [2]. As the novel coronavirus and severe acute respiratory syndrome coronavirus (SARS-CoV) are genetically similar and belong to the same genus, the International Committee on Taxonomy of Viruses (ICTV) announced the official name of the new coronavirus: SARS-CoV-2 on February 11, 2020. Unfortunately, since the SARS-CoV-2 is an entirely new coronavirus in humans, there are no drugs or vaccines for treatment. Since 2013, the FDA has approved only 12 drugs to treat viral infections, 10 for hepatitis c virus (HCV) and HIV, one for cytomegalovirus (CMV), and one for influenza virus (IFV) (www.fda.gov). However, given the limitations of existing drugs in controlling other viral infections and the resistance of antiviral drugs, it is urgent to find effective drugs to control SARS-CoV-2 infections. Therefore, targeting host cell is important in the development of broad-spectrum drugs to overcome viral mutation and drug resistance. Several approaches to control or prevent new SARS-CoV-2 infections are envisaged, including vaccines, monoclonal antibodies, oligonucleotide-based therapies, peptides, interferon therapy, and small-molecule drugs [3–8]. New interventions, however, may take months to years to be developed. Considering the urgency of the SARS-CoV-2 outbreak, we should focus on the existing antiviral agents to block the virus from infecting the human. Studies of the SARS-CoV-2 genome sequence have shown that it is strongly homologous with the more thoroughly studied SARS-CoV, and some key drug targets including RdRp and 3CLpro show striking homology with SARS-CoV (> 95%) [9]. This makes the development of biologics and macrocyclic peptides feasible targets. Since S protein of SARS-CoV-2 shows a stronger binding to ACE2, it is expected to be a promising direction of drug development to find antibodies or compounds that can adsorb ACE2 and prevent SARS-CoV-2 from invading host cells. We propose potential drug candidates based on ACE2 so as to prevent and control the virus through summarizing extensive previous researches, which contributes to the development of broad-spectrum anti-coronavirus drugs for future epidemics.
Basic biological characteristics of SARS-CoV-2
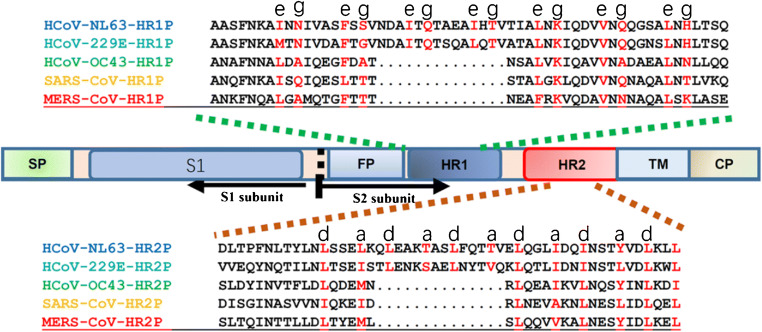
Four kinds of coronaviruses have been identified, including α, β, γ, and δ genera, according to its serological and genetic characteristics [10]. Although the host ranges of different coronaviruses are relatively specific, their genome structures are very similar, with a single-stranded plus strand with a capsule, which is the largest size among the known viral genome [11]. The coronavirus genome has a cap structure in the 5′ end and a poly(A) tail in the 3′ end. The 5′ end cap structure is followed by the pilot sequence (60–80 bp) and the 200–300-bp non-coding region, which may be related to coronavirus packaging [12]. The coronaviruses encode at least four major structural proteins, including spike glycoprotein, envelope protein, membrane protein (222aa), and nucleocapsid protein (419aa) [11]. Spike glycoprotein is an important structural protein of coronavirus. It is a homologous trimer protein on the surface of viral envelope involved in the process of receptor binding and membrane fusion. It is not only a key factor in determining host specificity but also an important target molecule for neutralizing antibodies [13]. The extracellular segment of the S protein can be cleaved by protease into the S1 region at the N-terminal and the C-terminal S2 region near to the viral envelope. The receptor-binding domain (RBD) in the S1 region is responsible for binding to virus receptors on host cells, and S2 is responsible for membrane fusion between the virus and the host cell. The S1 region contains two relatively independent domains: N-terminal domain (NTD) and C-terminal domain (CTD). The fusion state of the fusion center of coronavirus is a centrosymmetric six α helices. Among them, three long HR1 helices form the helix beam skeleton in the cooled coil mode and three short HR2 helices attach to the hydrophobic groove of HR1 skeleton through hydrophobic action (Fig. 1). Coronaviruses use S proteins to bind to specific receptors on the surface of target cells and then enter cells to replicate which results in infection ultimately. Thus, S proteins are very important in the process of coronaviruses infecting cells [15].
Fig. 1.
Schematic representation of HCoV S protein [14]
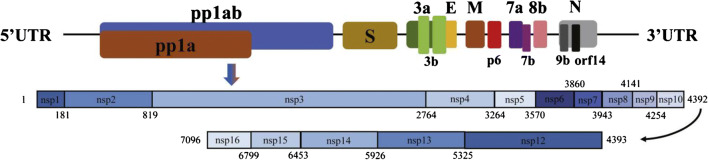
SARS-CoV-2 shows common characteristics of the above CoVs. Currently, the SARS-COV-2 genome has been found to contain 14 open reading frames (ORFs), encoding 27 proteins. The longest ORF located in 5’ terminus coding 15 nonstructural protein, jointly participate in viral replication and immune escape. 3’ terminus of the genome encoding protein structure and accessories [16] (Fig. 2). Roujian Lu et al. [17] found that SARS-CoV-2 is genetically closer to two bat-derived SARS-like coronaviruses such as bat-SL-CoVZC45 and bat-SL-CoVZXC21 (with about 88% genome sequence identity), than to SARS-CoV-1 (about 79% identity) and MERS-CoV (about 50% identity). Homology modeling has revealed that SARS-CoV-2 shows a similar receptor-binding domain structure to that of SARS-CoV-1, which suggests that COVID-19 infection might have a similar pathogenesis to SARS-CoV-1 infection. Phylogenetic analysis showed that SARS-CoV-2 belongs to the subgenus Sarbecovirus of the genus Betacoronavirus, with a relatively long branch length to its closest relatives bat-SL-CoVZC45 and bat-SL-CoVZXC21, which was genetically different from SARS-CoV. Homologous modeling shows that SARS-CoV-2 and SARS-CoV have similar receptor-binding domain structure. Studies show that the SARS-CoV-2 enters by binding its spike protein to a receptor on the surface of the host cell. It is clear that this receptor is ACE2 [18]. Currently, treatments for spike and ACE2 may be used for the prevention and treatment of SARS-CoV-2.
Fig. 2.
Genomic organization of SARS-CoV-2, indicating the coding regions for proteins that are potential drug targets [16]
ACE2 is the doorknob that SARS-CoV-2 enters the cell
ACE2 is a key regulator of blood pressure angiotensin-converting enzyme (ACE) [19]. Fifty years after ACE was identified, ACE2 was presented in the human genome through two separate genomic approaches [20]. ACE2 (GenBank No. 291820) has only one catalytic site domain (amino acid 147-555), which shows a similar exon/intron structure to the catalytic domain 2 in ACE1, indicating that they share a common ancestral gene [21]. Although ACE2 and ACE1 have homologous catalytic domains, their biochemical and pharmacological properties are completely different. ACE2 can convert the Ang of decapeptide into the Ang1-9 of the nine peptides by cutting off one amino acid residue from the C-end of the substrate. Ang1-9 could produce heptapeptide Ang1-7 and pentapeptide Ang1-5 after co-incubation with ACE1, but no transformation of Ang1-9 into Ang1-8 (Ang II) was found. ACE2 also acts on Ang II, which produces heptapeptide Ang1-7 [22]. Therefore, ACE2 cannot convert angiotensin I (Ang I) into Ang II and inactivate bradykinin with no vasoconstricting properties. However, it can convert substances that contend the pressurized proliferation and pro-fibrotic effects of Ang II and act through its own g protein–coupled receptors [23], which suggests that ACE and ACE2 may play a balancing role in the renin–angiotensin system (RAS) [24].
With the development of medical science and technology, ACE2 is not only recognized as an angiotensin-related peptide but also considered to be the receptor for SARS-CoV S protein by Michael Farzan’s team at Harvard University [25]. Li et al. [26] used the purified SARS-CoV protein to immunoprecipitate the susceptible cells as well as compared the target protein, and they finally isolated and identified ACE2 protein from Vero E 6 cells by mass spectrometry and protein database. The proteins could be infected by SARS-CoVs, which can efficiently bind to the S1 domain of SARS-CoV S protein. Besides, the ability to mediate cell–cell fusion of S protein in SARS-CoV is dependent on the presence of ACE2. And the 293T cells transfected with CD4 and CCR5 formed many syncytes with cells expressing HIV-1 g p160, but not with cells expressing the SARS-CoV S protein. In contrast, 293T cells expressing ACE2 did not form syncytes with cells expressing g p160, but formed many large syncytes with cells expressing S protein [27, 28]. Thus, ACE2’s facilitating effect on virus-mediated cell fusion was verified. In addition, ACE2 mediated viral replication. Li et al. incubated ACE2 with a control-transfected 293T cells in the presence or absence of SARS-CoV for 2 days and then measured viral genomic RNA in cell supernatant by RT-PCR. They found that viral replication increased by more than 100,000 times in the first 48 h in the ACE2-transformed cells. In contrast, in the control-transfected cells, the replication of viral genes increased only 10 fold over the same period [29]. Overall, these results suggest that ACE2 is significant to substantially promote SARS-CoV replication. ACE2 was also associated with adaptive immune response [30]. Through GSEA analysis, He et al. [31] found that the high expression of ACE2 was related to innate immune response, acquired immune response, B cell regulatory immunity, and cytokine secretion. It also enhanced the inflammatory response induced by cytokines such as IL-1, IL-10, IL-6, and IL-8, from which we can infer that the immune system dysfunction involved in the high expression of ACE2 was related to cytokine storm. The clinical study in Wuhan found that the levels of IL-1B, IL-10, and IL-8 were significantly increased in critical patients with new coronavirus infection, suggesting that there was cytokine-mediated inflammatory response in patients with coronavirus infection [32], which was consistent with the study of He.
Genome sequencing of the SARS-CoV-2 revealed strong homology with its more closely studied cousin, SARS-CoV. The results showed that the bat SARS–like coronavirus (GenBank NO. MG772933) was the closest to the SARS-CoV-2, with 88% nucleotide similarity [33]. Thus, we conformed that an ideal vaccine and antiviral drug might be developed through blocking the binding of S protein to the receptor ACE2. In the study of the SARS-CoV-2, researchers from the US National Institutes of Health (NIH) and the University of Texas determined a 3.5-Å resolution cryo-EM structure of the SARS-CoV-2 S trimer in the perfusion conformation. The predominant state of the trimer has one of the three RBDs rotated up in a receptor-accessible conformation. They also show biophysical and structural evidence that the SARS-CoV-2 S binds ACE2 with higher affinity than SARS-CoV S [34]. Qiang and his team [35] from China obtained a high-quality and stable complex of ACE2 with the intestinal amino acid transporter B0AT1 through co-expression, which was highly likely to stabilize ACE2 and thus obtain the full-length protein of ACE2 protein. Moreover, they successfully analyzed its three-dimensional structure using the cryo-electron microscope from Westlake University, with a resolution of 2.9 Å. The resolution of the extracellular domain, which was critical for virus recognition, was 2.7 Å. By analyzing the full-length protein structure of ACE2, Qiang et al. found that ACE2 existed as a dimer with open and closed conformation changes. The two conformations contained a mutual recognition interface with coronavirus. This discovery provided a strong basis for further analyzing the three-dimensional structure of full-length ACE2 and S protein complexes of neocoronavirus. It will be helpful to understand the structure basis and function characteristics of coronaviruses entering target cells and playing an important role in the optimization of inhibitors blocking the infection of coronaviruses with cells (Fig. 3).
Fig. 3.
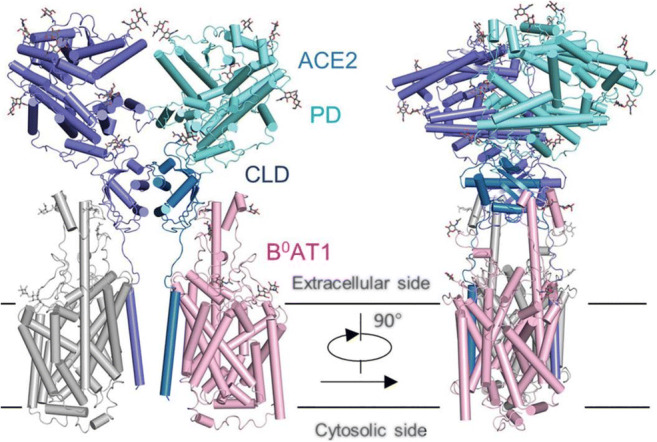
Structure diagram of ACE2-B0AT1 complex [35]
Clinical evidence shows that the multi-organ infection caused by SARS-CoV-2 is consistent with the tissue distribution of ACE2, which is mainly expressed in the vascular endothelial cells of the human lungs, heart, stomach, intestines, kidney, bone marrow, spleen, liver, brain, etc. These pieces of evidence suggest that ACE2 may be a key target in the pathogenesis of SARS-CoV-2. Pharmacological researches need to be studied to search for antibodies or compounds which can adsorb ACE2 to prevent the infection of SARS-CoV-2 with host cells.
Drugs that may act on SARS-CoV-2 through spike protein or ACE2
The important role of SARS-CoV-2 s protein makes it an important therapeutic target. Many researches have explored potential therapeutic methods. First of all, the drugs blocking the binding of RBD-ACE2 can be used as a new therapy for SARS-CoV-2 infection. Secondly, drugs binding to S protein interfere with the division of S1 and S2. This step inhibited the production of functional S1 and S2 subunits and the fusion of virus envelope and host cell membrane. Thirdly, the anti-SARS coronavirus peptide, which blocks the interaction of HR1–HR2 by forming fusion active core, has virus fusion inhibitory activity at the micro molar level [36–38].
Peptide P6
Dong P. Han et al. [39] identified the determinants of SARS-CoV infection with ACE2 through alanine scanning mutagenesis analysis. They further tested the anti-SARS activity of six chemically synthesized peptides derived from ACE2. The most antiviral P6 peptide, which is composed of two noncontinuous ACE2 peptides 22-44 and 351-357 linked by glycine, exhibited a potent antiviral activity with IC50 of about 0.1 μM. Peptide P6 had only 8 amino acids longer than P4. However, its activity is 500 times higher. The effective antiviral activity of the P6 peptide might be attributed to its structure which is similar to that of ACE2 and can competitively bind to the viral S protein. Studies show that the interaction of S glycoprotein with ACE2 plays an important role in the pathogenesis of SARS [40]. The binding of S protein to ACE2 results in downregulation of the receptor, which leads to relaxation regulation of the renin–angiotensin system, and ultimately causes lung damage. Therefore, P6 peptide may be a promising therapeutic agent which can not only inhibit SARS-CoV infection but also prevent severe lung failure.
It could be seen that we developed a new chemically synthesized peptide similar to the P6 peptide based on the crystal structure of ACE2 to find a therapeutic agent for SARS-CoV-2.
Griffithsin
Griffithsin (GRFT) was originally isolated from the red alga Griffithsia sp. The unique protein with molecular weight of 12.7 kda has been shown to specifically bind to N-linked glycan end mannose residues on the surface of human immunodeficiency virus type 1 (HIV-1), HIV-2, and other envelope viruses (including hepatitis) and can be used for the treatment of HCV, SARS-CoV, and Ebola virus. At present, GRFT cannot promote the mitosis of human T cells and induce the production of proinflammatory cytokines in human cell lines. GRFT possessed three largely identical carbohydrate-binding domains orientated as an equatorial triangle, affording multivalent binding and thereby increasing potency.
SARS-CoV causes significant pathological changes in pulmonary tissues in animal models. O’Keefe et al. [41] assessed pulmonary histopathology at 2, 4, and 10 days post-infection in untreated, sham-treated (no virus), and GRFT-treated animals. Mice treated with GRFT and SARS-CoV exhibited robust perivascular infiltrates at levels greater than those with SARS-CoV alone, possibly due to the increasing immunogenicity of GRFT-aggregated viral particles; the GRFT-treated mice had reduced levels of pulmonary edema at both 2 and 4 days post-infection and reduced severity of necrotizing bronchiolitis at 2 days post-infection. It proved that GRFT treatment decreased pulmonary pathology during SARS-CoV infection.
To determine whether the activity of GRFT against SARS-CoV was due to specific interactions with the spike glycoprotein, enzyme-linked immunosorbent assay (ELISA) studies were performed using recombinant SARS-CoV spike glycoprotein which showed that GRFT binds to this protein in a concentration-dependent manner [41]. ELISA studies were performed to determine whether the binding of GRFT to SARS-CoV S glycoprotein was sufficient to inhibit the subsequent binding of the host cell receptor human ACE2 to S. These studies indicated that GRFT was unable to significantly inhibit the binding of the SARS-CoV S glycoprotein to ACE2 (data not shown). They speculated that the interaction between GRFT and S produced a complex that, although still able to bind to ACE2, might prevent the virus from entering the required next steps.
HR2P analogs
Lu et al. [42] characterized the six-helix bundle fusion core structure of MERS-CoV spike protein S2 subunit by X-ray crystallography and biophysical analysis. They found that two peptides, HR1P and HR2P, spanning residues 998–1039 in HR1 and 1251–1286 in HR2 domains respectively, can form a stable six-helix bundle fusion core structure, suggesting that MERS-CoV enters into the host cell mainly through the membrane fusion mechanism. They developed a MERS-CoV S protein–mediated cell–cell fusion assay to explore the effect of HR2P peptides on MERS-CoV S protein. At the same time, they compared the inhibitory effects of peptides HR1P, HR2P, T20 (anti-HIV-1 peptide) [43], and SC-1 (peptide derived from the HR2 region of SARS-CoV S protein) [44] on the cell fusion mediated by MERS-CoV S protein. The results showed that HR1P, T20, and SC-1 showed no significant inhibitory activity with the concentration of 40 mM, while HR2P showed strong inhibitory effect with IC50 of ~ 0.8 mM. These results indicated that HR2P can inhibit membrane fusion mediated by MERS-CoV S protein. In addition, they found that the introduction of Glu, Lys, or Arg residues into HR2P can improve its stability, solubility, and anti-MERS-CoV activity. So, we can draw the conclusion that in the drug development of SARS-CoV-2, we can make membrane fusion inhibitors according to the different structural characteristics of HR1P and HR2P.
A modified OC43-HR2P peptide-EK1
HCoV-OC43 (OC43) belongs to group β coronavirus. Xia et al. [16] screened a peptide OC43-HR2P with broad-spectrum fusion inhibitory activity. In addition, they have further improved the sequence of OC43-HR2P by introducing Glu or Lys at the appropriate site of the peptide to increase its solubility, thereby increasing the antiviral activity of the peptide. In addition, based on the structure of MERS-CoV S 6-HB, we also introduced mutations at some sites that are not expected to participate in HR1 binding, such as 4Q, 14Y, 32D, and 36L, to further enhance the fusion inhibitory activity of the peptide. Among a series of optimized peptides, peptide EK1 showed the most effective pan-CoV antiviral fusion activity, with IC50 values between 0.19 and 0.62 μM. SL coronaviruses, including WIV1, Rs3367, and SHC014 coronaviruses, all manifest potential for human infection [12, 45, 46]. To further evaluate the intensity of EK1 inhibition of fusion, Xia et al. [16] performed a cell–cell fusion assay mediated by the S protein of these SL-CoV. The results showed that EK1 showed a higher inhibitory activity than SARS-HR2P, and it had an inhibitory effect on the S protein–mediated cell fusion of the three SL-CoVs tested, while the EK1-disrupted peptide had no inhibitory effect. For intercellular fusion activity, the results show that EK1 peptide can effectively inhibit the fusion of multiple CoV cells and block various CoV infections.
In order to evaluate the potential prevention and treatment of EK1 against MERS-CoV infection, Xia et al. [16] used well-defined transgenic (Tg) mice globally expressing human dipeptidyl peptidase IV (DPP4) viral receptor. The mice were then treated with 200 μg of EK1 or with PBS 30 min before or after challenge with MERS-CoV at 104 TCID50. Compared with the control group, the weight of EK1-treated Tg mice did not decrease significantly. The survival rate improved and no virus appeared in the lungs. The results indicated that the peptide EK1 displayed extensive and powerful potential antivirus. They further used a crystal structure to prove that EK1 would preclude binding of HR2 onto their corresponding 3HR1 core, thereby blocking formation of the 3HR1–3HR2 6-HB, which is an indispensable step during host–viral membrane fusion. Hence, administration of the EK1 peptide would block cellular entry of those HCoVs.
Neutralizing antibodies that target the S protein
During disease outbreaks, approaches to isolate neutralizing antibodies (nAbs) from recovered patients have proven successful. Up to now, many effective monoclonal antibodies against SARS-CoV have been found [47, 48]. The 193 amino acid length (n318-v510) RBD of spike protein(s) of SARS-CoV and SL-CoVs is a key target for neutralizing antibody [25]. Due to the relatively high identity (73%) of RBD in COVID-19 and SARS-CoV, the SARS-CoV-neutralizing antibodies are accordingly expected to effectively bind with RBD in COVID-19. However, several representative SARS-CoV-specific antibodies such as m396, CR3014, and CR3022 did not exhibit superior combining capacity with COVID-19 RBD, which may be attributed to the difference of C-terminus residues of RBD between SARS-CoV and COVID-19 [49]. Surprisingly, Tian et al. [49] found that CR3022, a SARS-CoV-specific antibody isolated from blood of a convalescent SARS patient, was capable of binding with COVID-19 RBD powerfully determined by ELISA and BLI. And they found that CR3022 recognized different epitopes on RBD when compared with other antibodies [50], which displayed that CR3022 would be a hopeful therapy for COVID-19 infections, alone or in combination with other neutralizing antibodies.
Besides, various attempts have been made to improve the monovalent affinities of neutralizing antibodies toward the S1 protein of COVID-19. Miao et al. [51] designed a biparatopic construct, in which the nAb (89C8) binding to the NTD of S1 was fused to recombinant ACE2 (89C8-ACE2). 89C8-ACE2 shows potent binding property to viral S1 protein. The highlight of this design is to avoid binding loss to a great extent due to the mutation of RBD, which ensures the neutralization ability of different coronavirus strains, thus providing a general treatment design idea for the treatment of other infectious diseases. To sum up, neutralizing antibody is a promising treatment method in this epidemic and even in the future.
Cholesterol depletion with methyl-β-cyclodextrin
Cholesterol depletion is widely used to study the role of cholesterol in cell processes. Cholesterol present in the plasma membrane of target cells has been shown to be important for the infection by SARS-CoV. Glende et al. [52] first compared the infective SARS-CoV with the viral stomatitis virus–based pseudotypes (VSV) containing SARS-CoV S protein; the results indicated that cholesterol was necessary for the virus S protein to bind to ACE2 cells. Then, the author used the acute cholesterol-depleting agent methyl-β-cyclodextrin (MBCD) to deplete cholesterol. The results showed that cholesterol depletion not only inhibited SARS-CoV infection but also inhibited virus S protein–mediated VSV pseudotype infection. A direct explanation for the effect of cholesterol depletion on S protein binding is that ACE2 is located in DRMS, and the concentration of ACE2 receptor in the microdomain increases the binding efficiency. The depletion of cholesterol in the target cell expressing ACE2 may lead to the dispersion of ACE2 molecules on the cell surface and the decrease of the spatial concentration of ACE2 in the membrane microdomain. It has been reported that cholesterol consumption reduces the expression of ACE2 on the cell surface to a certain extent [53]. Glende [52] believes that cholesterol consumption does not reduce the amount of ACE2 protein on the surface of Vero cells. He explained that cholesterol in the plasma membrane affects the conformation of ACE2 and thus the expression of epitopes.
Interestingly, MBCD, an acute cholesterol-depleting agent, had a certain effect on lipid rafts of aging T lymphocytes. Larbi et al. [54] found that MBCD did not affect the cholesterol content and fluidity of T cell membrane of young donors, but could reduce the cholesterol content and increase the fluidity of T cell membrane of old donors, so as to improve the activation of T cells. Whether MBCD has therapeutic effect on SARS-CoV-2 by affecting T lymphocytes is still unknown, which needs further exploration. In a word, cholesterol consumption can be used as a treatment strategy for SARS-CoV-2 infection in the future
Traditional Chinese medicine and its ingredients
Traditional Chinese medicine (TCM) is the precious wealth accumulated by Chinese people in their long-term struggle against diseases. TCM has played an active role in the treatment of pestilence in the history and in the prevention and treatment of new sudden infectious diseases such as SARS and H1N1 influenza virus. Nowadays, the diagnosis and treatment plan (trial version fifth) for the outbreak of COVID-19 were issued by the National Health Commission and the office of the State Administration of TCM. The plan details the relevant contents of the treatment plans by TCM. In addition, researchers have interpreted the mechanism of action of TCM against SARS-CoV or SARS-CoV-2 from the micro level by means of pharmacology, biochemistry, computer network, and other disciplines.
Ho et al. [55] established an ELISA to evaluate the inhibitory effect of TCM on the interaction between S protein and ACE2. By screening 312 controlled Chinese medicinal herbs supervised by Committee on Chinese Medicine and Pharmacy at Taiwan, they found that emodin can inhibit the interaction of SARS-CoV S protein and ACE2. S protein–pseudotyped retrovirus infectivity was used to evaluate the inhibitory potential of emodin. Vero E6 cells transfected with the plasmid encoding ACE2 were infected with S protein–pseudotyped retrovirus in the presence or absence of compounds. Emodin inhibited the S protein–pseudotyped retrovirus infectivity in a dose-dependent manner. The percent inhibition of emodin at 50 μM was 94.12 ± 5.90%. The results indicated that emodin was a novel anti-SARS-CoV compound that was able to block the SARS-CoV S protein binding to Vero E6 cells.
Considering that SARS-CoV-2 and the SARS virus share great similarity in gene sequences, pathogenic mechanism, and clinical manifestations, Ming et al. [56] took the prescription of TCM for SARS and SARS-CoV-2 as clues to screen out effective drugs and core prescriptions against SARS-CoV-2 based on molecular docking technology and other virtual screening technologies. Finally, 46 active ingredients of novel coronavirus S protein and ACE2 binding region with high binding energy were found, which were mainly attributable to 7 traditional Chinese medicines, such as mulberry leaves, atractylodes, and Fritillaria thunbergii. To sum up, in addition to the clear competitive activity of ACE2 binding, TCM also shows the advantage of multi-target in antivirus, which can be used in combination with modern drugs to improve the effect of antiviral treatment. Last but not least, TCM contains huge resources for the development of antiviral drugs, which deserves our in-depth exploration to deal with more infectious diseases in the future.
Conclusions
At present, at least 120 COVID-19 vaccines have been studied, but it is still unclear whether safe and effective vaccines can be developed. And so far, there is no vaccine against coronavirus of any strain. Even if the development of an effective vaccine turns out to be possible, it may still take years to produce and manage the product. Moreover, studies have shown that patients tend to be afflicted again after initial exposure to SARS-CoV. Therefore, it is urgent to identify non-vaccine treatments, especially during new outbreaks. This article reviews the virological characteristics of SARS-CoV-2 and summarizes some therapy options from the aspect of inhibiting the binding of S protein to ACE2 receptor. Seven therapeutic substances were proposed, including peptide P6, griffithsin, HR2P analogs, EK1, vaccine, monoclonal antibody, cholesterol-depleting agents, and extracts from TCM, which are derived from the previous studies on SARS treatment. These drugs are expected to treat SARS-CoV-2 in consideration of extremely close homology between SARS-CoV-2 and SARS-CoV-1 by means of specifically binding to S protein and ACE2 receptor or blocking the membrane fusion between the host and virus. However, some of the above drugs have not been tested in animal or clinical trials at present, and their therapeutic effects need to be further verified. As there is little literature on non-vaccine drug development targeting SARS-CoV-2 currently, our study is indeed up-to-date. This summary is expected to provide direction for drug research and development and ideas for clinical drug selection. In addition, in-depth research in this field also provides valuable experience for future antiviral treatment, which deserves our attention.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
This article is about drug screening and development from the affinity of S protein of new coronavirus with ACE2. Because it does not involve clinical trials and collecting patients, there is no need for informed consent.
Consent for publication
The manuscript is approved by all authors for publication.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hui-Qing Lv, Email: hzlhq@126.com.
Cheng-Ping Wen, Email: wengcp@163.com.
References
- 1.Organization. WH (2005) Statement on the second meeting of the International Health Regulations (2005) Emergency Committee regarding the outbreak of novel coronavirus (2019-nCoV)
- 2.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J, Yu T, Zhang X, Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arunachalam R, Arumugaswami V (2020) Insights into cross-species evolution of novel human coronavirus 2019-nCoV and defining immune determinants for vaccine development. bioRxiv
- 4.Lu H (2020a) Drug treatment options for the 2019-new coronavirus (2019-nCoV). Biosci Trends [DOI] [PubMed]
- 5.Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, Shi Z, Hu Z, Zhong W, Xiao G (2020) Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res [DOI] [PMC free article] [PubMed]
- 6.Xin L, Wang X (2020) Potential inhibitors for 2019-nCoV coronavirus M protease from clinically approved medicines. bioRxiv [DOI] [PMC free article] [PubMed]
- 7.Xu Z (2020) Nelfinavir was predicted to be a potential inhibitor of 2019-nCov main protease by an integrative approach combining homology modelling, molecular docking and binding free energy calculation. bioRxiv
- 8.Yan L (2020) Therapeutic drugs targeting 2019-nCoV main protease by high-throughput screening. bioRxiv
- 9.Liu W, Morse JS, Lalonde T, Xu S (2020) Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. Chembiochem [DOI] [PMC free article] [PubMed]
- 10.Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH, Bai R, Teng JL, Tsang CC, Wang M, Zheng BJ, Chan KH, Yuen KY. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 2012;86(7):3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su S, Wong G, Shi W, Liu J, Lai A, Zhou J, Liu W, Bi Y, Gao GF. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menachery VD, Yount BJ, Sims AC, Debbink K, Agnihothram SS, Gralinski LE, Graham RL, Scobey T, Plante JA, Royal SR, Swanstrom J, Sheahan TP, Pickles RJ, Corti D, Randell SH, Lanzavecchia A, Marasco WA, Baric RS. SARS-like WIV1-CoV poised for human emergence. Proc Natl Acad Sci U S A. 2016;113(11):3048–3053. doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(5742):1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 14.Xia S, Yan L, Xu W, Agrawal AS, Algaissi A, Tseng CK, Wang Q, Du L, Tan W, Wilson IA, Jiang S, Yang B, Lu L(2019 )A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci Adv 5(4):eaav4580 [DOI] [PMC free article] [PubMed]
- 15.Jiang S (2012) Receptor-binding domains of spike proteins of emerging or re-emerging viruses as targets for development of antiviral vaccines. Emerg Microbes Infect [DOI] [PMC free article] [PubMed]
- 16.Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, Meng J, Zhu Z, Zhang Z, Wang J, Sheng J, Quan L, Xia Z, Tan W, Cheng G, Jiang T (2020) Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 27(3):325–328 [DOI] [PMC free article] [PubMed]
- 17.Lu R (2020b) Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet [DOI] [PMC free article] [PubMed]
- 18.Hoffmann M (2020) The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells
- 19.Turner AJ, Hooper NM. The angiotensin-converting enzyme gene family: genomics and pharmacology. Trends Pharmacol Sci. 2002;23(4):177–183. doi: 10.1016/S0165-6147(00)01994-5. [DOI] [PubMed] [Google Scholar]
- 20.Acharya KR, Sturrock ED, Riordan JF, Ehlers MR. Ace revisited: a new target for structure-based drug design. Nat Rev Drug Discov. 2003;2(11):891–902. doi: 10.1038/nrd1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tipnis SR, Hooper NM, Hyde R, Karran E, Christie G, Turner AJ. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J Biol Chem. 2000;275(43):33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- 22.Donoghue M, Hsieh F, Baronas E, Godbout K, Gosselin M, Stagliano N, Donovan M, Woolf B, Robison K, Jeyaseelan R, Breitbart RE, Acton S. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ Res. 2000;87(5):E1–E9. doi: 10.1161/01.RES.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 23.Zisman LS, Keller RS, Weaver B, Lin Q, Speth R, Bristow MR, Canver CC. Increased angiotensin-(1-7)-forming activity in failing human heart ventricles: evidence for upregulation of the angiotensin-converting enzyme homologue ACE2. Circulation. 2003;108(14):1707–1712. doi: 10.1161/01.CIR.0000094734.67990.99. [DOI] [PubMed] [Google Scholar]
- 24.Crackower MA, Sarao R, Oudit GY, Yagil C, Kozieradzki I, Scanga SE, Oliveira-dos-Santos AJ, Da CJ, Zhang L, Pei Y, Scholey J, Ferrario CM, Manoukian AS, Chappell MC, Backx PH, Yagil Y, Penninger JM. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature. 2002;417(6891):822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- 25.Wong SK, Li W, Moore MJ, Choe H, Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J Biol Chem. 2004;279(5):3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling AE, Humphrey CD, Shieh WJ, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang JY, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 28.Kuiken T, Fouchier RA, Schutten M, Rimmelzwaan GF, van Amerongen G, van Riel D, Laman JD, de Jong T, van Doornum G, Lim W, Ling AE, Chan PK, Tam JS, Zambon MC, Gopal R, Drosten C, van der Werf S, Escriou N, Manuguerra JC, Stohr K, Peiris JS, Osterhaus AD. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362(9380):263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W (2003) Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature [DOI] [PMC free article] [PubMed]
- 30.Fischer D (2017) Protein malnutrition alters tryptophan and angiotensin-converting enzyme 2 homeostasis and adaptive immune responses in human rotavirus-infected gnotobiotic pigs with human infant fecal microbiota transplant. Clin Vaccine Immunol [DOI] [PMC free article] [PubMed]
- 31.He X (2020) Integrative bioinformatics analysis provides insight into the molecular mechanisms of 2019-nCoV. medRxiv [DOI] [PubMed]
- 32.C H . Clinical features of patients infected with 2019 coronavirus in Wuhan. Lancet: China; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phan T. Novel coronavirus: from discovery to clinical diagnostics. Infect Genet Evol. 2020;79:104211. doi: 10.1016/j.meegid.2020.104211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wrapp D (2020) Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. bioRxiv [DOI] [PMC free article] [PubMed]
- 35.Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bosch BJ, Martina BE, Van Der Zee R, Lepault J, Haijema BJ, Versluis C, Heck AJ, De Groot R, Osterhaus AD, Rottier PJ. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc Natl Acad Sci U S A. 2004;101(22):8455–8460. doi: 10.1073/pnas.0400576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan K, Yi L, Chen J, Qu X, Qing T, Rao X, Jiang P, Hu J, Xiong Z, Nie Y, Shi X, Wang W, Ling C, Yin X, Fan K, Lai L, Ding M, Deng H. Suppression of SARS-CoV entry by peptides corresponding to heptad regions on spike glycoprotein. Biochem Biophys Res Commun. 2004;319(3):746–752. doi: 10.1016/j.bbrc.2004.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng BJ, Guan Y, Hez ML, Sun H, Du L, Zheng Y, Wong KL, Chen H, Chen Y, Lu L, Tanner JA, Watt RM, Niccolai N, Bernini A, Spiga O, Woo PC, Kung HF, Yuen KY, Huang JD. Synthetic peptides outside the spike protein heptad repeat regions as potent inhibitors of SARS-associated coronavirus. Antivir Ther. 2005;10(3):393–403. [PubMed] [Google Scholar]
- 39.Han DP, Penn-Nicholson A, Cho MW. Identification of critical determinants on ACE2 for SARS-CoV entry and development of a potent entry inhibitor. Virology. 2006;350(1):15–25. doi: 10.1016/j.virol.2006.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ksiazek TG (2003) A novel coronavirus associated with severe acute respiratory syndrome. New Engl J Med [DOI] [PubMed]
- 41.O’Keefe BR, Giomarelli B, Barnard DL, Shenoy SR, Chan PK, McMahon JB, Palmer KE, Barnett BW, Meyerholz DK, Wohlford-Lenane CL, McCray PJ. Broad-spectrum in vitro activity and in vivo efficacy of the antiviral protein griffithsin against emerging viruses of the family Coronaviridae. J Virol. 2010;84(5):2511–2521. doi: 10.1128/JVI.02322-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu L, Liu Q, Zhu Y, Chan KH, Qin L, Li Y, Wang Q, Chan JF, Du L, Yu F, Ma C, Ye S, Yuen KY, Zhang R, Jiang S. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat Commun. 2014;5:3067. doi: 10.1038/ncomms4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wild CT, Shugars DC, Greenwell TK, McDanal CB, Matthews TJ. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc Natl Acad Sci U S A. 1994;91(21):9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu S, Xiao G, Chen Y, He Y, Niu J, Escalante CR, Xiong H, Farmar J, Debnath AK, Tien P, Jiang S. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363(9413):938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, Epstein JH, Mazet JK, Hu B, Zhang W, Peng C, Zhang YJ, Luo CM, Tan B, Wang N, Zhu Y, Crameri G, Zhang SY, Wang LF, Daszak P, Shi ZL. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503(7477):535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Menachery VD, Yount BJ, Debbink K, Agnihothram S, Gralinski LE, Plante JA, Graham RL, Scobey T, Ge XY, Donaldson EF, Randell SH, Lanzavecchia A, Marasco WA, Shi ZL, Baric RS. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med. 2015;21(12):1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.ter Meulen J, Bakker AB, van den Brink EN, Weverling GJ, Martina BE, Haagmans BL, Kuiken T, de Kruif J, Preiser W, Spaan W, Gelderblom HR, Goudsmit J, Osterhaus AD. Human monoclonal antibody as prophylaxis for SARS coronavirus infection in ferrets. Lancet. 2004;363(9427):2139–2141. doi: 10.1016/S0140-6736(04)16506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu Z, Chakraborti S, He Y, Roberts A, Sheahan T, Xiao X, Hensley LE, Prabakaran P, Rockx B, Sidorov IA, Corti D, Vogel L, Feng Y, Kim JO, Wang LF, Baric R, Lanzavecchia A, Curtis KM, Nabel GJ, Subbarao K, Jiang S, Dimitrov DS. Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc Natl Acad Sci U S A. 2007;104(29):12123–12128. doi: 10.1073/pnas.0701000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tian X, Li C, Huang A, Xia S, Lu S, Shi Z, Lu L, Jiang S, Yang Z, Wu Y, Ying T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect. 2020;9(1):382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.ter Meulen J, van den Brink EN, Poon LL, Marissen WE, Leung CS, Cox F, Cheung CY, Bakker AQ, Bogaards JA, van Deventer E, Preiser W, Doerr HW, Chow VT, de Kruif J, Peiris JS, Goudsmit J. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3(7):e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miao X, Luo Y, Huang X, Lee S, Yuan Z, Tang Y, Chen L, Wang C, Wu F, Xu Y, Jiang W, Gao W, Song X, Yan Y, Pang T, Chen C, Zou Y, Fu W, Wan L, Gilbert-Jaramillo J, Knight M, Tan TK, Rijal P, Townsend A, Sun J, Liu X, James W, Tsun A, Xu Y. A novel biparatopic hybrid antibody-ACE2 fusion that blocks SARS-CoV-2 infection: implications for therapy. MABS-Austin. 2020;12(1):1804241. doi: 10.1080/19420862.2020.1804241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Glende J, Schwegmann-Wessels C, Al-Falah M, Pfefferle S, Qu X, Deng H, Drosten C, Naim HY, Herrler G. Importance of cholesterol-rich membrane microdomains in the interaction of the S protein of SARS-coronavirus with the cellular receptor angiotensin-converting enzyme 2. Virology. 2008;381(2):215–221. doi: 10.1016/j.virol.2008.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li GM, Li YG, Yamate M, Li SM, Ikuta K. Lipid rafts play an important role in the early stage of severe acute respiratory syndrome-coronavirus life cycle. Microbes Infect. 2007;9(1):96–102. doi: 10.1016/j.micinf.2006.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Larbi A, Douziech N, Khalil A, Dupuis G, Gherairi S, Guerard KP, Fulop TJ. Effects of methyl-beta-cyclodextrin on T lymphocytes lipid rafts with aging. Exp Gerontol. 2004;39(4):551–558. doi: 10.1016/j.exger.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 55.Ho TY, Wu SL, Chen JC, Li CC, Hsiang CY. Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction. Antivir Res. 2007;74(2):92–101. doi: 10.1016/j.antiviral.2006.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ming N (2020) Rapid screening model and application of anti-new coronavirus TCM prescription based on clinical experience and molecular docking technology