Abstract
The 2019 SARS CoV-2 (COVID-19) pandemic has illustrated the need for rapid and accurate diagnostic tests. In this work, a multiplexed grating-coupled fluorescent plasmonics (GC-FP) biosensor platform was used to rapidly and accurately measure antibodies against COVID-19 in human blood serum and dried blood spot samples. The GC-FP platform measures antibody-antigen binding interactions for multiple targets in a single sample, and has 100% selectivity and sensitivity (n = 23) when measuring serum IgG levels against three COVID-19 antigens (spike S1, spike S1S2, and the nucleocapsid protein). The GC-FP platform yielded a quantitative, linear response for serum samples diluted to as low as 1:1600 dilution. Test results were highly correlated with two commercial COVID-19 antibody tests, including an enzyme linked immunosorbent assay (ELISA) and a Luminex-based microsphere immunoassay. To demonstrate test efficacy with other sample matrices, dried blood spot samples (n = 63) were obtained and evaluated with GC-FP, yielding 100% selectivity and 86.7% sensitivity for diagnosing prior COVID-19 infection. The test was also evaluated for detection of multiple immunoglobulin isotypes, with successful detection of IgM, IgG and IgA antibody-antigen interactions. Last, a machine learning approach was developed to accurately score patient samples for prior COVID-19 infection, using antibody binding data for all three COVID-19 antigens used in the test.
Keywords: COVID-19, Antibody, Biosensor, Plasmonic, Quantitative, Multiplex
Highlights
-
•
GC-FP biosensor yields rapid (30 min) detection of antibodies against COVID-19 infection.
-
•
Antibody detection is quantitative and can provide antibody titer information.
-
•
Diagnosis of prior COVID-19 infection using serum with 100% selectivity and sensitivity.
-
•
Multiplexed approach yields information on multiple antibody-antigen interactions.
-
•
Dried blood spot testing significantly simplifies sample collection and yields high sensitivity/specificity.
1. Introduction
The rapid spread of the 2019 SARS CoV-2 (COVID-19) virus has established an urgent need for accurate diagnostic technologies (Pascarella et al., 2020). Due to the wide range in severity of this disease, many individuals remain asymptomatic or have mild symptoms, defining a population that is not tested at the time of acute infection (Pascarella et al., 2020). For these patients, the immune response to past COVID-19 infection is the best measure of exposure. Immune response to COVID-19 infection is variable, and may be linked to disease symptom severity, length of infection, and multiple patient-specific factors (Sethuraman et al., 2020; To et al., 2020). Thus, quantitative detection of the antibody response to COVID-19 is critical to our response to this pandemic.
To measure antibody response to COVID-19 infection, a number of tests have been developed. Most tests detect binding of immunoglobulin G (IgG) and/or immunoglobulin M (IgM) to viral antigens. These tests are typically performed using whole blood, blood serum, or blood plasma. The most widely used approach is the enzyme linked immunosorbent assay (ELISA) (Amanat et al., 2020; Karp et al., 2020; Randad et al., 2020). ELISA-based testing enables high throughput (processing many samples in parallel), but is typically limited to a single antigen per well (Infantino et al., 2020; Younes et al., 2020). Alternatively, multiplexed testing enables detection of immunoglobulin binding to more than one antigen within a single tube, well, plate or slide. Multiplexed tests include, but are not limited to microsphere immunoassays (MIAs) (Ayouba et al., 2020; Randad et al., 2020), fluorescent protein microarrays (Hedde et al., 2020), and direct/label-free array technologies (Steiner et al., 2020).
Sample collection is a key challenge with implementing immunological/serological testing. Blood samples are typically obtained by venipuncture, followed by blood plasma or serum preparation. Alternatively, a simple finger stick and dried blood spotting allows self-collection, minimizing effort and likely increasing participation (Au - Grüner et al., 2015; Malsagova et al., 2020; Thevis et al., 2020; Vázquez-Morón et al., 2019). Samples collected in this manner may be maintained at ambient temperature and can be shipped using mail or courier service without the need for refrigeration. Dried blood spots (DBS) have been successfully utilized for immunological/serological testing for multiple viral diseases, including hepatitis C, HIV, and COVID-19 (Karp et al., 2020; Malsagova et al., 2020; Vázquez-Morón et al., 2019).
Previously, we reported an enhanced fluorescence biosensor for multiplexed detection of antibodies for Lyme disease diagnosis (Chou et al. 2018, 2019, 2020). In this approach, gold-coated nanoscale grating surfaces were modified with target antigens in a microarray format and then used to detect IgG or IgM binding from blood serum or plasma. Surface plasmons generated during illumination of the gold-coated biosensor chip actively enhance fluorescence emission intensity, yielding a high-sensitivity fluorescence detection platform. When measuring the fluorescence intensity of individual spots, the limit of detection of this approach was shown to be < 2 ng/spot. We have termed this approach “grating-coupled fluorescent plasmonics” (GC-FP). To achieve high sensitivity and an added measure of selectivity, fluorophore-tagged antibodies (against IgM or IgG) are applied during a labeling step. The entire detection process can be completed in less than 30 min with high sensitivity and specificity.
In the work presented here, we used the GC-FP biosensor platform to develop a rapid immunoassay for simultaneous detection of antibodies against three COVID-19 spike protein antigens (receptor binding domain, RBD; spike S1 fragment; spike S1S2 extracellular domain) and the COVID-19 nucleocapsid protein (Nuc). Using serum, we achieved 100% specificity and sensitivity for diagnosing prior COVID-19 infection, and for DBS we demonstrated 100% specificity and sensitivity as high as 86.9%. For serum samples, GC-FP results are highly correlated with established testing methods (ELISA and MIA). The assay also has a large linear dynamic range across multiple orders of concentration. Because antibody titer against COVID-19 antigens is positively correlated with viral neutralization capacity (To et al., 2020), our test has the potential to reveal the level of a subject's immune response.
2. Materials and methods
2.1. Materials
Nucleocapsid protein (Nuc), the S1 fragment of the spike protein (S1), the extracellular domain of the spike protein (S1S2), the receptor binding domain of the spike protein (RBD), human serum albumin (HSA), the S1 domain of the 2005 SARS coronavirus spike protein (WH20 isolate, abbreviated “SARS-S1”), and human Influenza B nucleoprotein (B/Florida/4/2006 isolate “Flu Nuc”) were all obtained from Sino Biological, Inc. Positive control protein, human IgG protein (Hum IgG), SuperBlock blocking buffer and phosphate buffered saline (PBS) were obtained from ThermoFisher Scientific. PBS-TWEEN (PBS-T) solution consisting of PBS + 0.05% v/v TWEEN-20 (Sigma-Aldrich) was prepared on a daily basis for all experiments. Alexa Fluor 647 labeled anti-human IgG (heavy and light chain) and anti-human IgM (heavy chain) were obtained from Invitrogen/ThermoFisher Scientific. Alexa Fluor 647 labeled anti-human IgA was obtained from Southern Biotech. ELISA testing was performed using COVID-19 human IgG testing kits from RayBiotech.
2.2. Grating-coupled fluorescent plasmonic (GC-FP) biosensor chip preparation
Gold coated grating-coupled fluorescent plasmonic (GC-FP) biosensor chips were fabricated as described previously (Chou et al. 2019, 2020). GC-FP chips were printed with an array of 400 μm diameter spots of target and control antigens/proteins using an ArrayIt SpotBot II microarray printer. Proteins/antigens were first diluted to 500 μg/μl in phosphate buffered saline (PBS) and then further diluted 1:1 just prior to printing with GBL protein array printing buffer (Grace Bio-Labs). For printing, a 180 μm diameter printing tip was used, at a relative humidity of 60–70%, at ambient temperature (~25 °C). After printing, chips were allowed to dry at ambient temperature (~25 °C) for 30 min, and were then transferred to a sealed container with desiccant for long-term storage (up to 4 weeks) before use.
2.3. Biological samples
Human blood samples were obtained from donors within New York state or from the Wadsworth Center, New York State Department of Health (see Supplementary Material for additional information and preparation details). Negative samples were collected prior to the 2019 SARS CoV-2 pandemic, and were obtained from the Lyme Disease Biobank. Additional blood samples were collected by finger stick. Lancet devices (27 ga.) and Whatman 903 protein saver collection cards were sent to volunteers with instructions. Blood sampling and testing was approved by the SUNY Polytechnic Institute Institutional Review Board (protocol #IRB-2020-10). Blood droplets were collected, allowed to dry, and then either hand delivered or mailed (via US Postal Service) to SUNY Polytechnic Institute. Following receipt of DBS samples, a sterile 6 mm diameter biopsy punch was used to remove samples from the collection cards. These disks were then soaked in 500 μl of PBS-T solution ~12 h at 4 °C with gentle rocking.
2.4. GC-FP detection assay
Prior to performing GC-FP detection assays, GC-FP chips were filled with SuperBlock blocking buffer, then incubated at room temperature for 15 min. Chips were then placed in a custom fluidic apparatus to provide sequential flow of sample and reagents using the following steps: 1) 500 μl of PBS-T at 100 μl/min, 2) 400 μl of diluted human blood serum or extracted dried blood spot sample at 50 μl/min, 3) 500 μl of PBS-T at 100 μl/min, 4) 400 μl of Alexa 647 anti-human IgG/IgM (diluted 1:400 in PBS-T) at 100 μl/min, and 5) 500 μl of PBS-T at 100 μl/min. GC-FP chips were then analyzed in a customized Ciencia, Inc. fluorescent plasmonic imaging instrument. For serum testing, a standard dilution of serum in PBS-T (1:25) was used. For dried blood spot testing, undiluted extract from the 6 mm diameter segment of the blood collection card was used in place of serum. Ciencia image analysis LabView software was used to define a region of interest (ROI) for each individual spot on the GC-FP biosensor chip and the fluorescence intensity of each spot was measured. The fluorescence intensity of all spots was normalized to the human IgG (Hum IgG) internal control spots on each chip, to account for variability between individual chips and individual experiments.
2.5. Data analysis
Normalized spot intensity data was exported from the software and further analyzed using GraphPad Prism 8.0 software (for fitting, ROC analysis, correlation, and statistical analysis). To account for variation between chips and experiments, normalized intensity data for positive control and COVID-19 antigen spots (mean intensity, ) were divided by the average negative control spot intensity, plus three times the standard deviation (σ) of the negative control spot intensity ( + 3σ) to produce a detection metric as follows:
| (1) |
A support vector machine (SVM) based machine learning approach was used to analyze GC-FP detection data, and was implemented with freely available SVM software (LibSVM - http://www.csie.ntu.edu.tw/~cjlin/libsvm) (Chang and Lin, 2011). The nu-SVC package within LibSVM was utilized with sigmoid kernel, and a grid search for cost and gamma parameters was conducted to maximize the prediction accuracy of the SVM model.
3. Results and discussion
3.1. Detection assay development and characterization
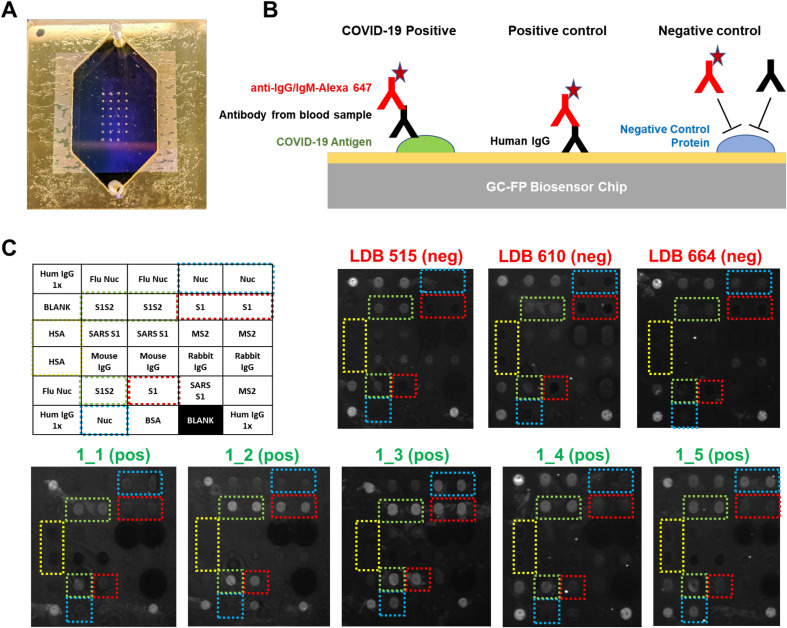
Rapid (less than 30 min), multiplexed detection of immunoglobulin binding to COVID-19 antigens was performed using our previously described GC-FP biosensing approach and a Ciencia, Inc. fluorescent plasmonic imaging instrument (Chou et al. 2018, 2019, 2020). An example GC-FP microchip for COVID-19 immunological analysis is shown in Fig. 1 A. COVID-19 specific antigens and control proteins were immobilized on the GC-FP biosensor chips in a variety of configurations (v1 – v4, Supplementary Figure S1). Testing utilized serum samples from subjects previously infected with COVID-19 who were expected to have an antibody response to COVID-19 antigens. All subjects had fully recovered from infection and were more than 2 weeks convalescent. Negative control samples showed no GC-FP response for IgG binding to COVID-19 Nuc and S1 antigens, and in some cases, very weak response for full-length spike S1S2 extracellular domain antigen (Fig. 1C).
Fig. 1.
A) GC-FP biosensor chip shown with gasket and fluidic cover attached. B) COVID-19 antigens or control proteins were spotted onto GC-FP biosensor chips, then assessed for antibody binding from human blood samples. Subsequent labeling with Alexa Fluor 647 tagged anti-human IgG was used for the enhanced fluorescence detection output. C) Enhanced fluorescence images of GC-FP biosensor chips (v1) processed with negative control serum (blood serum collected >2 years prior to the COVID-19 pandemic and serum from subjects who were >2 weeks convalescent from PCR-confirmed COVID-19 infection. Boxes with dotted outlines indicate paired spots of key COVID-19/SARS CoV-2 antigens, S1, S1S2, and Nuc.
3.2. GC-FP antibody detection in human serum and dried blood spot samples
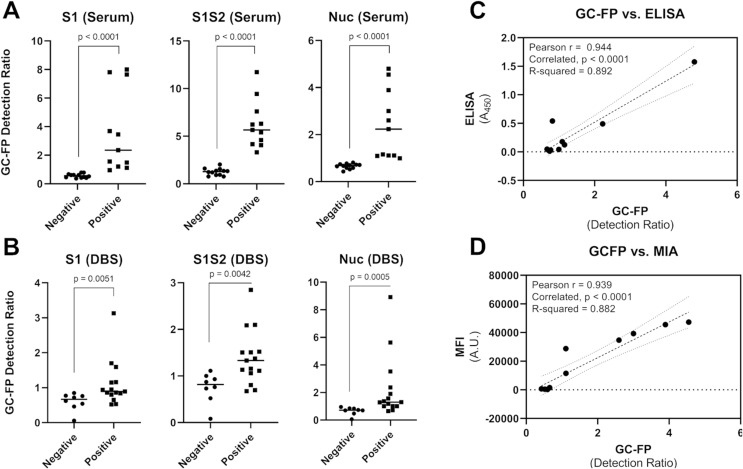
Using v2, v3 and v4 GC-FP chips, 23 different human blood serum samples, and 24 dried blood spot samples (with verified infection status) were tested. Samples were tested at a dilution rate of 1 part serum to 25 parts PBS-T (for serum) or undiluted (for dried blood spot extracts). Raw GC-FP fluorescence intensity data were normalized as described in the Materials and Methods section, and a “GC-FP detection ratio” was calculated to account for chip-to-chip differences and any variability in processing conditions. The GC-FP detection ratio provides a measure of signal above background, while also accounting for spot-to-spot variability. The results of these experiments are shown in Fig. 2 , Table 1 , and Supplementary Table T1. The average GC-FP detection ratio for COVID-19 positive samples vs. COVID-19 negative samples was significant for all antigens, for both serum and dried blood spot samples (Mann-Whitney U test, p < 0.05).
Fig. 2.
GC-FP results for serum (A) and dried blood spot (B) samples from subjects with verified positive COVID-19 infection status. The difference between the average positive and negative GC-FP detection ratios was confirmed using the Mann-Whitney U test, with corresponding p values shown in each plot. ELISA (450 nm absorbance values) for the COVID-19 nucleocapsid protein were compared to GC-FP detection ratio results for a subset of serum samples (C), while Luminex MIA results (MFI values) were compared to GC-FP detection ratio results for a second subset of samples (D). Pearson correlation coefficients (r) and R-squared values from correlation are shown, with associated p-values. Dashed lines indicate linear regression fit to the data, while dotted lines indicate the 95% confidence intervals for correlation analysis.
Table 1.
GC-FP detection ratio data for serum and dried blood spot samples with verified infection status. ELISA and Luminex MIA data are shown for serum samples tested with these assays. Samples scored positive (GC-FP detection ratio above the ROC threshold) are denoted by bold text. The Nuc diagnostic score and machine learning (ML) score are shown for dried blood spot samples.
| Serum Samples | ||||||
| Sample ID | S1 | S1S2 | Nuc | Nuc-ELISA (score/A450) | Nuc-MIA (score/MFI) | Notes |
| 2_1 | 0.67 | 1.55 | 0.81 | neg/0.054 | n/a | no known exposure |
| 2_2 | 0.61 | 0.94 | 0.72 | neg/0.027 | n/a | no known exposure |
| 2_3 | 0.79 | 0.77 | 0.74 | neg/0.013 | n/a | no known exposure |
| 2_4 | 0.79 | 0.77 | 0.67 | neg/0.046 | n/a | no known exposure |
| 2_5 | 0.52 | 1.33 | 0.77 | neg/0.032 | n/a | no known exposure |
| 2_6 | 0.61 | 1.60 | 0.70 | neg/0.031 | n/a | no known exposure |
| 2_7 | 0.62 | 1.37 | 0.71 | neg/0.037 | n/a | no known exposure |
| DIL0526-26 | 0.42 | 0.92 | 0.59 | n/a | neg/259 | 2009 pre-COVID |
| DIL0526-27 | 0.41 | 1.20 | 0.52 | n/a | neg/421 | 2009 pre-COVID |
| DIL0526-28 | 0.45 | 1.23 | 0.65 | n/a | neg/1563 | 2009 pre-COVID |
| DIL0526-29 | 0.48 | 2.03 | 0.59 | n/a | neg/238 | 2009 pre-COVID |
| DIL0526-30 | 0.37 | 1.36 | 0.43 | n/a | neg/719 | 2009 pre-COVID |
| 1_1 | 1.20 | 3.31 | 1.16 | pos/0.123 | n/a | Positive by RT-PCR |
| 1_2 | 3.69 | 5.65 | 1.09 | pos/0.178 | n/a | Positive by RT-PCR |
| 1_3 | 7.65 | 11.73 | 4.80 | pos/1.577 | n/a | Positive by RT-PCR |
| 1_4 | 1.49 | 5.41 | 0.99 | neg/0.043 | n/a | Positive by RT-PCR |
| 1_5 | 1.56 | 4.59 | 2.23 | pos/0.491 | n/a | Positive by RT-PCR |
| DIL0526-3 | 7.81 | 9.44 | 3.89 | n/a | pos/45,543 | Positive by RT-PCR |
| DIL0526-5 | 8.00 | 7.61 | 4.55 | n/a | pos/47,301 | Positive by RT-PCR |
| DIL0526-8 | 3.45 | 6.23 | 3.00 | n/a | pos/39,255 | Positive by RT-PCR |
| DIL0526-13 | 2.35 | 4.06 | 1.11 | n/a | pos/11,577 | Positive by RT-PCR |
| DIL0526-18 | 1.12 | 6.31 | 2.60 | n/a | pos/34,715 | Positive by RT-PCR |
| DIL0526-21 | 0.95 | 4.16 | 1.11 | n/a | pos/28,806 | Positive by RT-PCR |
| Dried Blood Spot Samples (Verified Infection Status) | ||||||
| Sample ID | S1 | S1S2 | Nuc | Nuc Score | ML Score | Notes |
| COV_5 | 0.54 | 0.73 | 0.70 | neg | neg | Negative by RT-PCR |
| COV_6 | 0.72 | 1.11 | 0.70 | neg | neg | Negative by RT-PCR |
| COV_7 | 0.06 | 0.08 | 0.06 | neg | neg | Negative by RT-PCR |
| COV_9 | 0.85 | 1.00 | 0.95 | neg | neg | Negative by RT-PCR |
| COV_14 | 0.46 | 0.52 | 0.48 | neg | neg | Negative by RT-PCR |
| COV_16 | 0.77 | 0.93 | 0.84 | neg | neg | Negative by RT-PCR |
| COV_18 | 0.62 | 0.76 | 0.74 | neg | neg | Negative by RT-PCR |
| COV_12 | 0.76 | 0.87 | 0.85 | neg | neg | IgG and IgM negative |
| COV_2 | 0.65 | 0.80 | 0.72 | neg | neg | Positive by RT-PCR |
| COV_3 | 1.59 | 1.13 | 5.63 | pos | pos | Positive by RT-PCR |
| COV_13 | 0.53 | 0.67 | 0.65 | neg | neg | Positive by RT-PCR |
| COV_17 | 0.94 | 1.11 | 1.01 | pos | pos | Positive by RT-PCR |
| COV_28 | 0.80 | 1.37 | 3.53 | pos | pos | Positive by RT-PCR |
| COV_29 | 1.70 | 1.51 | 1.57 | pos | pos | Positive by RT-PCR |
| COV_40 | 1.15 | 1.50 | 2.37 | pos | pos | Positive by RT-PCR |
| COV_41 | 0.86 | 1.06 | 1.02 | pos | pos | Positive by RT-PCR |
| COV_43 | 0.52 | 0.70 | 1.19 | pos | neg | Positive by RT-PCR |
| COV_44 | 3.13 | 2.85 | 8.92 | pos | pos | Positive by RT-PCR |
| NYC_1 | 1.16 | 2.09 | 1.29 | pos | pos | Positive by RT-PCR |
| NYC_5 | 0.95 | 2.09 | 1.90 | pos | pos | Positive by RT-PCR |
| COV_19 | 0.89 | 1.52 | 1.37 | pos | pos | IgG pos., no PCR test |
| COV_38 | 0.86 | 1.33 | 1.30 | pos | pos | IgG pos., no PCR test |
| COV_24 | 0.84 | 1.16 | 1.01 | pos | pos | IgM pos., no PCR test |
GC-FP detection ratios for serum samples were compared to ELISA and a Luminex-based MIA (Yang et al., 2020). As shown in Fig. 2C and D, the measured GC-FP detection ratios for serum samples were highly correlated with both ELISA absorbance values (Pearson r = 0.944, R-squared = 0.892) and MIA fluorescence intensity (MFI) values (Pearson r = 0.939, R-squared = 0.882). These results demonstrate that the GC-FP detection approach provides comparable immunodetection results to established, gold-standard methods.
To determine if individual IgG/antigen responses could be used for diagnostic purposes, receiver operator characteristic (ROC) analysis was performed (Supplementary Figure S2). For serum samples, 100% sensitivity and specificity could be achieved when the following GC-FP detection ratio thresholds were met: S1 = 0.87, S1S2 = 2.67, Nuc = 0.9. For dried blood spot samples, ROC analysis yielded 100% specificity and variable sensitivity when the following GC-FP detection ratio thresholds were met: S1 = 0.855, S1S2 = 1.12, and Nuc = 0.98. When these GC-FP detection ratio thresholds were exceeded (to maintain 100% specificity) assay sensitivity was relatively low for S1 and S1S2 antigens (66.7% for both) but increased to 86.7% for the nucleocapsid antigen (Nuc).
The observed reduction in sensitivity for DBS vs. serum samples could be due to the sample format, especially since sample stability and extraction are more variable for dried blood spots vs. blood serum. Other reasons for the reduction in sensitivity could be variability in individual immune responses and/or reporting consistency for the research subjects who provided samples. Other studies have shown that individual antibody responses are variable (Amanat et al., 2020), which would also affect assay sensitivity. Importantly, no false positives were observed within the limited number of samples that were tested.
In addition to testing samples with known COVID-19 infection history, 39 additional dried blood spot samples were received and tested. For these samples, information was provided about exposure to infected individuals, potential disease symptoms, or complete lack of exposure, but none of the subjects had been tested with a COVID-19 RT-PCR test. The results from GC-FP testing for these samples are shown in Supplementary Table T1. Due to the fact that COVID-19 infection status was unverified for these samples, sensitivity and specificity was not determined.
3.3. Multiplexed data analysis
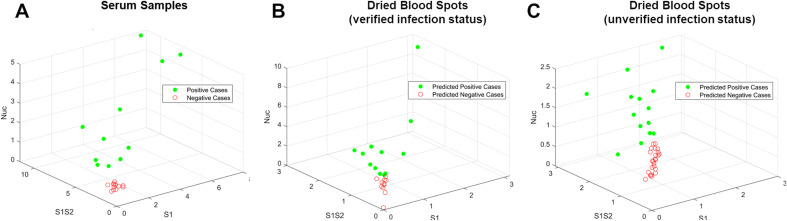
Scoring samples based on a composite antibody response (to all antigens) could provide increased diagnostic accuracy and a more complete understanding of a subject's antibody response to infection. To this end, a support vector machine (SVM) based machine learning (ML) approach was used to differentiate and classify samples based on their antibody response to three target antigens (S1, S1S2, and Nuc). ML approaches have been used extensively for classification and diagnosis when data from multiple biomarkers or targets is available (Sarkar and Saha, 2019; Uddin et al., 2019). SVM software (LibSVM) (Chang and Lin, 2011) was trained using GC-FP data from serum samples (Table 1). After training, the ML model was challenged with 10-fold cross-validation on unlabeled serum data, yielding 100% selectivity and sensitivity, which matched ROC analysis for individual antigens. After training and validation, DBS data were classified with the SVM model (Fig. 3 B & C, Table 1, Supplementary Table T1). For dried blood spots with verified prior infection status, the SVM model classified samples with 80% sensitivity and 100% selectivity. For dried blood spots with unverified prior infection status, SVM classification resulted in better correlation with presumed infection status than when scoring with individual antibody responses (S1, S1S2, or Nuc). While many additional samples will be needed to fully train the ML model, and to understand the true selectivity and sensitivity of the GC-FP assay, we have illustrated the potential for ML based scoring antibody responses to multiple antigens.
Fig. 3.
Visualization of testing data following machine learning-based analysis. (A) The GC-FP detection ratio data for all serum samples were plotted as a function of S1, S1S2, and Nuc. An SVM ML model was used to classify dried blood spot sample data from (B) subjects with verified COVID-19 infection status, and (C) subjects with unverified COVID-19 infection status.
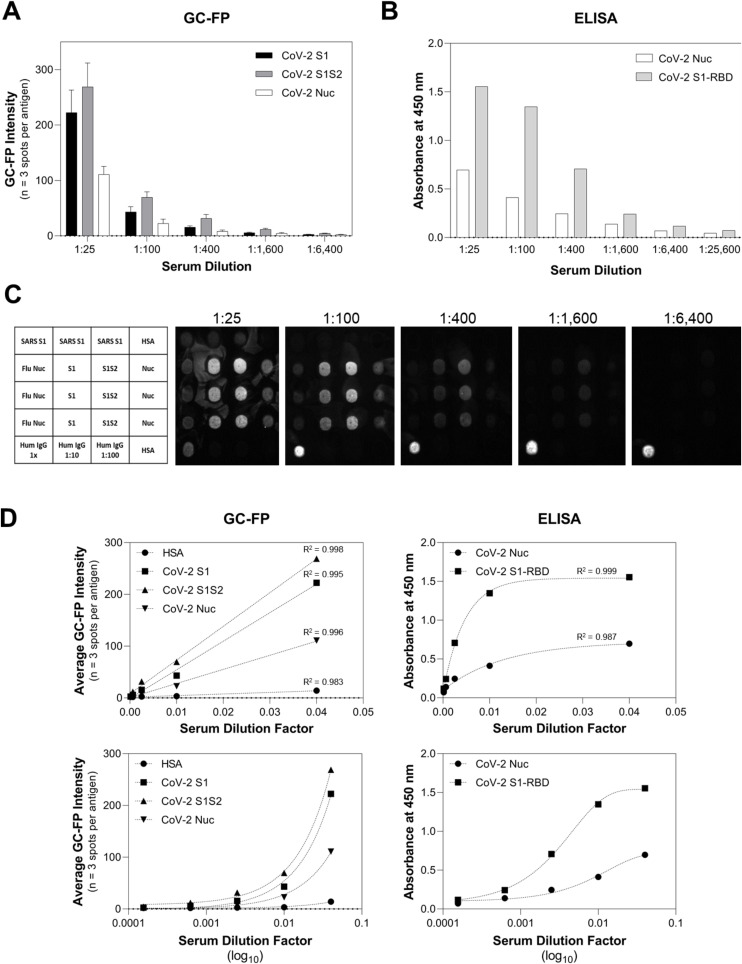
3.4. Quantification of antibody titer and comparison to ELISA
To assess GC-FP for quantitative determination of antibody concentration (titer), individual GC-FP chips were processed using dilutions of a COVID-19 positive serum sample received from the NYS Department of Health (sample DIL0526-3). Sample dilutions ranged from 1:25 to 1:25,600 and GC-FP testing results were compared to ELISA against S1 and Nuc, using commercial ELISA kits (Ray Biotech). The results of this experiment (Fig. 4 A–C), demonstrate that GC-FP could detect IgG at a minimum dilution of 1:1600 and that the commercial ELISA kit could detect IgG to a minimum dilution of 1:6,400. When GC-FP and ELISA data were plotted as a function of dilution factor (Fig. 4D) and fit with either linear regression (GC-FP) or partial least squares regression (ELISA), high goodness of fit (R-square > 0.98) was observed for all antigens.
Fig. 4.
Quantitative comparison of GC-FP and ELISA for detection of IgG against multiple COVID-19 antigens. Sample DIL0526-3 was used for both GC-FP and ELISA testing, at dilutions ranging from 1:25 to 1:26,600 in PBS-T.
Recent COVID-19 antibody testing studies have shown that antibody titers in the range of 1:320 or higher could be considered eligible for convalescent plasma donation (for convalescent plasma therapy) (Wajnberg et al., 2020). As shown in this work, GC-FP can detect antibodies down to 1:1,600 titer, and thus has the necessary sensitivity for determining clinically and therapeutically relevant seroconversion status. The fact that GC-FP has a linear response for all antigens tested makes it easier to quantify antibody concentrations across the full dynamic range, and to directly compare a subject's response to different viral antigens.
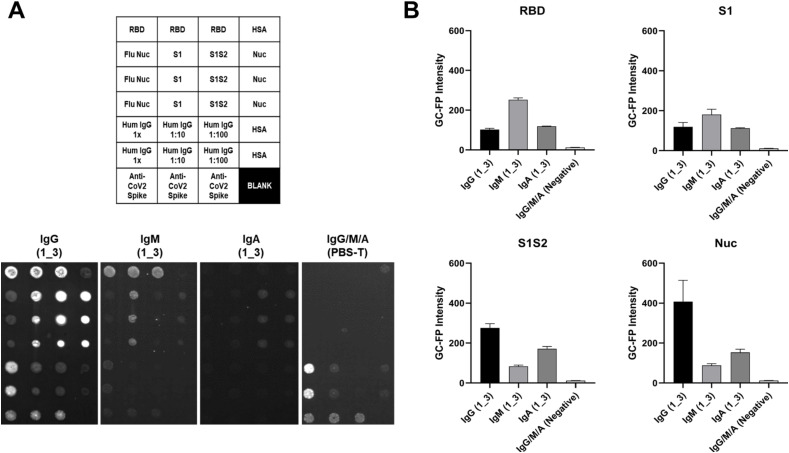
3.5. GC-FP detection of multiple immunoglobulin isotypes
As a final experiment, three individual v4 GC-FP chips were processed with a COVID-19 positive, convalescent serum sample (subject 1_3) and then assessed for binding by different antibody isotypes (IgG, IgM, and IgA) (Fig. 5 ). A negative control chip was processed with dilution buffer (PBS-T) instead of serum. Different antibody binding patterns were observed, based on the immunoglobulin isotype and the antigen being targeted (Fig. 5B). IgG binding was highest for the Nuc antigen, IgM showed the highest binding to RBD, and IgA showed similar binding for RBD, S1, S1S2, and Nuc.
Fig. 5.
Serum sample 1_3 was tested on three separate GC-FP biosensor chips that included RBD, S1, S1S2, and Nuc antigens. A fourth chip was processed using PBS-T as a negative control. The three chips tested with 1_3 serum were labeled with Alexa 647-tagged anti-human IgG, anti-human IgM, or anti-human IgA, while the negative control chip was labeled with a mixture of all three secondary antibodies. Mean GC-FP intensity (n = 3 spots per chip) is shown, for each COVID-19 antigen.
These results demonstrate the potential of the GC-FP approach to not only perform simultaneous, quantitative detection of antibody binding to multiple antigens, but also discrimination of antibody binding based on immunoglobulin isotype. Understanding antibody responses across various immunoglobulin classes may enable determining the stage of an individual's seroconversion response (Randad et al., 2020).
4. Conclusions
Rapid, accurate, and quantitative antibody tests are needed as part of the global response to the COVID-19 pandemic. In addition to epidemiological and seroconversion studies, such tests have the potential to confirm an individual's immunity status following prior infection or vaccination. In this work, we have demonstrated that GC-FP can be used to simultaneously measure antibody levels for multiple antigens in a single sample. The assay is quantitative across a large dynamic range (serum dilutions ranging from 1:25–1:1,600) and is highly correlated with gold-standard antibody detection tests, including ELISA and MIA (Fig. 2, Fig. 4). Although GC-FP was not able to match ELISA for detecting antibodies in the lowest dilution (1:1,600 vs. 1:6,400), this may be overcome by modifying the GC-FP imaging system, including addition of a more sensitive camera. Future iterations of the GC-FP system could also include a point-of-care platform for on-site clinical diagnostics.
The entire assay time for performing antibody detection is below 30 min (27 min for all fluidic processing steps, and less than 1 min required for GC-FP fluorescence imaging), which is significantly shorter than either ELISA or MIA, which take 2–3 h to complete. Alternative antibody testing approaches, such as lateral flow assays, may be completed in <10 min, but suffer from low accuracy and lack of multiplexing (Lisboa Bastos et al., 2020; Sethuraman et al., 2020; Younes et al., 2020). This study also demonstrates that dried blood spots are a viable sample matrix for COVID-19 antibody testing. Using dried blood spots reduces the complexity of sample collection, handling and storage versus venipuncture-based whole blood collection.
While this work establishes the potential of GC-FP for detecting antibodies against COVID-19 antigens in both serum and dried blood spots, it is still limited by the total number of samples tested (86 samples tested at the writing of this manuscript). As more samples are tested with the GC-FP platform, the major goal will be to retain 100% selectivity while maximizing sensitivity. One way to achieve this will be to compare antibody responses to additional COVID-19 antigens, as we have shown with the addition of the RBD antigen. Further, GC-FP results should be compared to other tests, such as antibody neutralization, which provides a measure of whether an individual's antibodies can neutralize the virus to prevent infection. It is possible that maximizing sensitivity and specificity, as well as correlating antibody levels with neutralization testing will require knowledge of antibody levels against multiple antigens. Understanding antibody responses across various immunoglobulin classes may also be useful for determining the stage of an individual's seroconversion response, and could be useful when analyzing other bodily fluids, such as saliva (Randad et al., 2020). Thus, the multiplexed microarray format of the GC-FP antibody detection assay will likely have great utility in the future.
CRediT authorship contribution statement
Nathaniel C. Cady: conducted experimental design, experimental work, data analysis, and was the primary author of this manuscript. Natalya Tokranova: assisted with experimental work. Armond Minor: assisted with experimental work. Nima Nikvand: performed data analysis and machine learning analysis. Klemen Strle: assisted with writing, editing and data analysis. William T. Lee: provided serum samples and Luminex MIA data. William Page: fabricated the GC-FP fluorescent imaging platform and wrote the analysis software used for testing. Ernest Guignon: fabricated the GC-FP fluorescent imaging platform and wrote the analysis software used for testing. Arturo Pilar: fabricated the GC-FP fluorescent imaging platform and wrote the analysis software used for testing. George N. Gibson: fabricated the GC-FP fluorescent imaging platform and wrote the analysis software used for testing.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: A.P., E.G., and W.P. are all employees of Ciencia, Inc. and G.G. is a consultant for Ciencia, Inc.
Acknowledgements:
The authors thank Dr. B. Winston and Capital Cardiology Associates (Albany, NY) for assistance with dried blood spot sampling, K. Howard, M. Marchewka, and K. Kulas for MIA specimen characterization, and the State University of New York COVID-19 seed funding program for funding.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2020.112679.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Amanat F., Stadlbauer D., Strohmeier S. Nat. Med. 2020;26(7):1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au - Grüner N., Au - Stambouli O., Au - Ross R.S. JoVE. 2015;(97) doi: 10.3791/52619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayouba A., Thaurignac G., Morquin D. J. Clin. Virol. 2020;129:104521. doi: 10.1016/j.jcv.2020.104521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.-C., Lin C.-J. ACM Transactions on Itelleigent Systems and Technology. 2011;2 Article 27. [Google Scholar]
- Chou E., Lasek-Nesselquist E., Taubner B. PloS One. 2020;15(2) doi: 10.1371/journal.pone.0228772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou E., Pilar A., Guignon E. SPIE BiOS. 2019 [Google Scholar]
- Chou E., Zenteno G., Taubner B. SPIE Defense + Security. 2018 [Google Scholar]
- Hedde P.N., Abram T.J., Jain A. Lab Chip. 2020;20(18):3302–3309. doi: 10.1039/d0lc00547a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infantino M., Damiani A., Gobbi F.L. Isr. Med. Assoc. J. 2020;22(4):203–210. [PubMed] [Google Scholar]
- Karp D.G., Danh K., Seftel D. medRxiv. 2020 2020.2005.2029.20116004. [Google Scholar]
- Lisboa Bastos M., Tavaziva G., Abidi S.K. BMJ. 2020;370:m2516. doi: 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malsagova K., Kopylov A., Stepanov A. Diagnostics. 2020;10(4):248. doi: 10.3390/diagnostics10040248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascarella G., Strumia A., Piliego C. J. Intern. Med. 2020;288(2):192–206. doi: 10.1111/joim.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randad P.R., Pisanic N., Kruczynski K. medRxiv. 2020 2020.2005.2024.20112300. [Google Scholar]
- Sarkar D., Saha S. J. Biosci. 2019;44(4):104. [PubMed] [Google Scholar]
- Sethuraman N., Jeremiah S.S., Ryo A. J. Am. Med. Assoc. 2020;323(22):2249–2251. doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- Steiner D.J., Cognetti J.S., Luta E.P. bioRxiv. 2020 2020.2006.2015.153064. [Google Scholar]
- Thevis M., Knoop A., Schaefer M.S. Drug Test. Anal. 2020;12(7):994–997. doi: 10.1002/dta.2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K.-W., Tsang O.T.-Y., Leung W.-S. Lancet Infect. Dis. 2020;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin S., Khan A., Hossain M.E. BMC Med. Inf. Decis. Making. 2019;19(1):281. doi: 10.1186/s12911-019-1004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Morón S., Ardizone Jiménez B., Jiménez-Sousa M.A. Sci. Rep. 2019;9(1):7316. doi: 10.1038/s41598-019-41139-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajnberg A., Amanat F., Firpo A. medRxiv. 2020 2020.2007.2014.20151126. [Google Scholar]
- Yang H.S., Racine-Brzostek S.E., Lee W.T. Clin. Chim. Acta. 2020;509:117–125. doi: 10.1016/j.cca.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younes N., Al-Sadeq D.W., Al-Jighefee H. Viruses. 2020;12(6):582. doi: 10.3390/v12060582. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.