Abstract
Background
Circular RNA (circRNA) is a noncoding RNA that forms a closed-loop structure, and its abnormal expression may cause disease. We aimed to find potential network for circRNA-related competitive endogenous RNA (ceRNA) in atrial fibrillation (AF).
Methods
The circRNA, miRNA, and mRNA expression profiles in the heart tissue from AF patients were retrieved from the Gene Expression Omnibus database and analyzed comprehensively. Differentially expressed circRNAs (DEcircRNAs), differentially expressed miRNAs (DEmiRNAs), and differentially expressed mRNAs (DEmRNAs) were identified, followed by the establishment of DEcircRNA-DEmiRNA-DEmRNA regulatory network. Functional annotation analysis of host gene of DEcircRNAs and DEmRNAs in ceRNA regulatory network was performed. In vitro experiment and electronic validation were used to validate the expression of DEcircRNAs, DEmiRNAs, and DEmRNAs.
Results
A total of 1611 DEcircRNAs, 51 DEmiRNAs, and 1250 DEmRNAs were identified in AF. The DEcircRNA-DEmiRNA-DEmRNA network contained 62 circRNAs, 14 miRNAs, and 728 mRNAs. Among which, two ceRNA regulatory pairs of hsa-circRNA-100053-hsa-miR-455-5p-TRPV1 and hsa-circRNA-005843-hsa-miR-188-5p-SPON1 were identified. In addition, six miRNA-mRNA regulatory pairs including hsa-miR-34c-5p-INMT, hsa-miR-1253-DDIT4L, hsa-miR-508-5p-SMOC2, hsa-miR-943-ACTA1, hsa-miR-338-3p-WIPI1, and hsa-miR-199a-3p-RAP1GAP2 were also obtained. MTOR was a significantly enriched signaling pathway of host gene of DEcircRNAs. In addition, arrhythmogenic right ventricular cardiomyopathy, dilated cardiomyopathy, and hypertrophic cardiomyopathy were remarkably enriched signaling pathways of DEmRNAs in DEcircRNA-DEmiRNA-DEmRNA regulatory network. The expression validation of hsa-circRNA-402565, hsa-miR-34c-5p, hsa-miR-188-5p, SPON1, DDIT4L, SMOC2, and WIPI1 was consistent with the integrated analysis.
Conclusion
We speculated that hsa-circRNA-100053-hsa-miR-455-5p-TRPV1 and hsa-circRNA-005843-hsa-miR-188-5p-SPON1 interaction pairs may be involved in AF.
1. Introduction
Atrial fibrillation (AF) is one of the most common arrhythmias and associated with heart failure [1–4]. Age, gender, obesity, and heart valve abnormalities are important factors of AF [4–6]. AF can also lead to heart failure hospitalization and death [7]. However, current treatment of AF may have adverse reactions [8, 9]. The pathogenesis of AF remains unclear. Further study of the underlying mechanisms of AF may provide new treatments for AF [10].
Circular RNAs (circRNAs) (with a covalent closed-loop structure) are considered to be the key to pathogenesis of heart disease, providing a new perspective for the pathogenesis of AF [11]. circRNA plays a crucial role in several pathophysiological processes [4, 12]. In heart disease, circRNAs function as the regulator of miRNA levels. circRNAs may be the potential biomarker. Moreover, bioinformatics analysis provides a novel perspective on circRNAs involved in AF and establishes the foundation for future research of the potential roles of circRNAs in AF [13]. miRNAs play a variety of roles in atrial fibrillation, including regulation of electrical remodeling and modulation of structural remodeling of cardiac tissue. Different miRNAs were confirmed to be up- or downregulated in AF patients [14]. Jiang et al. found the regulatory networks of has_circRNA_100612-has-miR-133b-KCNIP1/JPH2/ADRB1 and has_circRNA_405917/hsa_circRNA_008132/hsa_circRNA_104052/hsa_circRNA_101021/hsa_circRNA_101020/hsa_circRNA_102341-has-miR-892b-GJA1 in the heart tissue of AF patients [4]. However, the potential mechanism of AF remains to be studied. In this study, we performed integrated analysis based on GEO datasets to further identify dysregulated circRNAs in AF.
2. Methods
2.1. Data Collection
We obtained the expression profiles of circRNA, miRNA, and mRNA from GEO datasets by searching keywords (“Atrial fibrillation” [All Fields]) AND (“Homo sapiens” [porgn] AND “gse” [Filter]). We selected data according to the following criteria: (1) the selected dataset must be genome-wide circRNA/miRNA/mRNA transcriptome data; (2) these data were obtained from the heart tissues of the patients in the AF group and the normal control (NC) group (without drug stimulation or transfection); (3) standardized or original datasets were considered in this study. One circRNA expression dataset (GSE129409), two miRNA expression datasets (GSE68475 and GSE70887), and one mRNA expression dataset (GSE31821) were selected (Supplementary Table 1).
2.2. Identification of Differentially Expressed circRNAs, miRNAs, and mRNAs
Firstly, the probes corresponding to multiple circRNAs/miRNAs/mRNAs were removed. Only the single probe with the largest average expression was retained in multiple probes corresponding to circRNAs/miRNAs/mRNAs. After this treatment, qualified circRNAs/miRNAs/mRNAs were used for further analysis. Then, LIMMA package analysis was used to identify differentially expressed circRNAs and mRNAs. The metaMA package analysis was used to identify differentially expressed miRNAs. P value < 0.05 was the screening criteria for differentially expressed circRNAs, miRNAs, and mRNAs. Detailed data analysis process was performed as previously described [15].
2.3. Functional Annotation
To assess the functional annotations of host gene of DEcircRNAs and DEmRNAs in ceRNA regulatory network, Gene Ontology (GO) classification and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were conducted based on the online software GeneCodis3. Statistical significance was with the cutoff criteria of P value < 0.05.
2.4. Construction of ceRNA (DEcircRNA-DEmiRNA-DEmRNA) Regulatory Network
Firstly, starBase v3.0 was used to establish DEcircRNA-DEmiRNA regulatory network. Then, miRWalk 3.0 (http://mirwalk.umm.uni-heidelberg.de/) was utilized to find the target differentially expressed mRNAs of differentially expressed miRNAs. Finally, the DEcircRNA-DEmiRNA regulatory network was fused with the DEmiRNA-DEmRNA regulatory network to further construct the ceRNA (DEcircRNA-DEmiRNA-DEmRNA) regulatory through Cytoscape (version 3.6.1) software.
2.5. In Vitro Validation
The blood samples from 10 patients with AF and 10 healthy individuals were obtained for quantitative real-time polymerase chain reaction (qRT-PCR) validation. The inclusion criteria of AF patients were as follows: (1) patients were diagnosed according to the 2014 American Heart Association (AHA)/American College of Cardiology (ACC)/American Heart Rhythm Society (HRS) AF guidelines and confirmed by electrocardiogram (ECG) or Holter monitor, (2) the onset of AF in the patients was at least once a month, (3) patients were under the age of 85, (4) patients with normal liver and kidney function, and (5) patients visited the doctor at least 2 times and took medicine regularly. The exclusion criteria of AF patients were as follows: (1) patients with AF with hemodynamic disorders or malignant arrhythmia; (2) patients were 85 years old and above; (3) patients with AF with chronic cardiac insufficiency (grades II-IV); (4) patients with other severe systemic diseases, or liver and kidney failure; and (5) patients with ischemic heart disease, valvular disease, cardiomyopathy, rheumatic heart disease, primary pulmonary hypertension, and connective tissue disease. There were no statistically significant differences in age, sex, and body mass index (BMI) between AF patients and the normal individuals. We obtained the written informed consent and the approval from the ethics committee of the Second Hospital of Hebei Medical University.
Total RNA was isolated with the TRIzol reagent following the manufacturer's protocol. Based on SuperReal PreMix Plus in ABI 7500 Real-Time PCR Detection System, the qRT-PCR reactions were performed. In the 2-ΔΔCt method, relative circRNA/miRNA/mRNA expression was determined. Human ACTB and GAPDH were used as endogenous controls for mRNA. In addition, human GAPDH and U6 were used as endogenous controls for circRNA and miRNA expression, respectively.
2.6. Electronic Validation
The GSE135445 dataset (involving 15 AF patients and 15 normal controls) was used to validate the expression of identified differentially expressed mRNAs. The result was presented as box plots. Statistical significance was ascribed to P value < 0.05.
3. Results
3.1. Differentially Expressed circRNAs, miRNAs, and mRNAs
Compared to normal heart tissues, a total of 1250 DEmRNAs (636 upregulated and 614 downregulated mRNAs) and 51 DEmiRNAs (14 upregulated and 38 downregulated miRNAs) were identified in AF (Figures 1(a) and 1(b)). Among them, RAP1GAP2 and ACTA1 were the most upregulated and downregulated mRNAs, respectively (Table 1); hsa-miR-508-5p and hsa-miR-99a were the most upregulated and downregulated miRNAs, respectively (Table 2). A total of 1611 DEcircRNAs (592 upregulated and 1019 downregulated circRNAs) were obtained in AF (Figure 1(c)). Among them, hsa_circRNA_405811 and hsa_circRNA_103752 were the most upregulated and downregulated circRNAs, respectively (Table 3).
Figure 1.
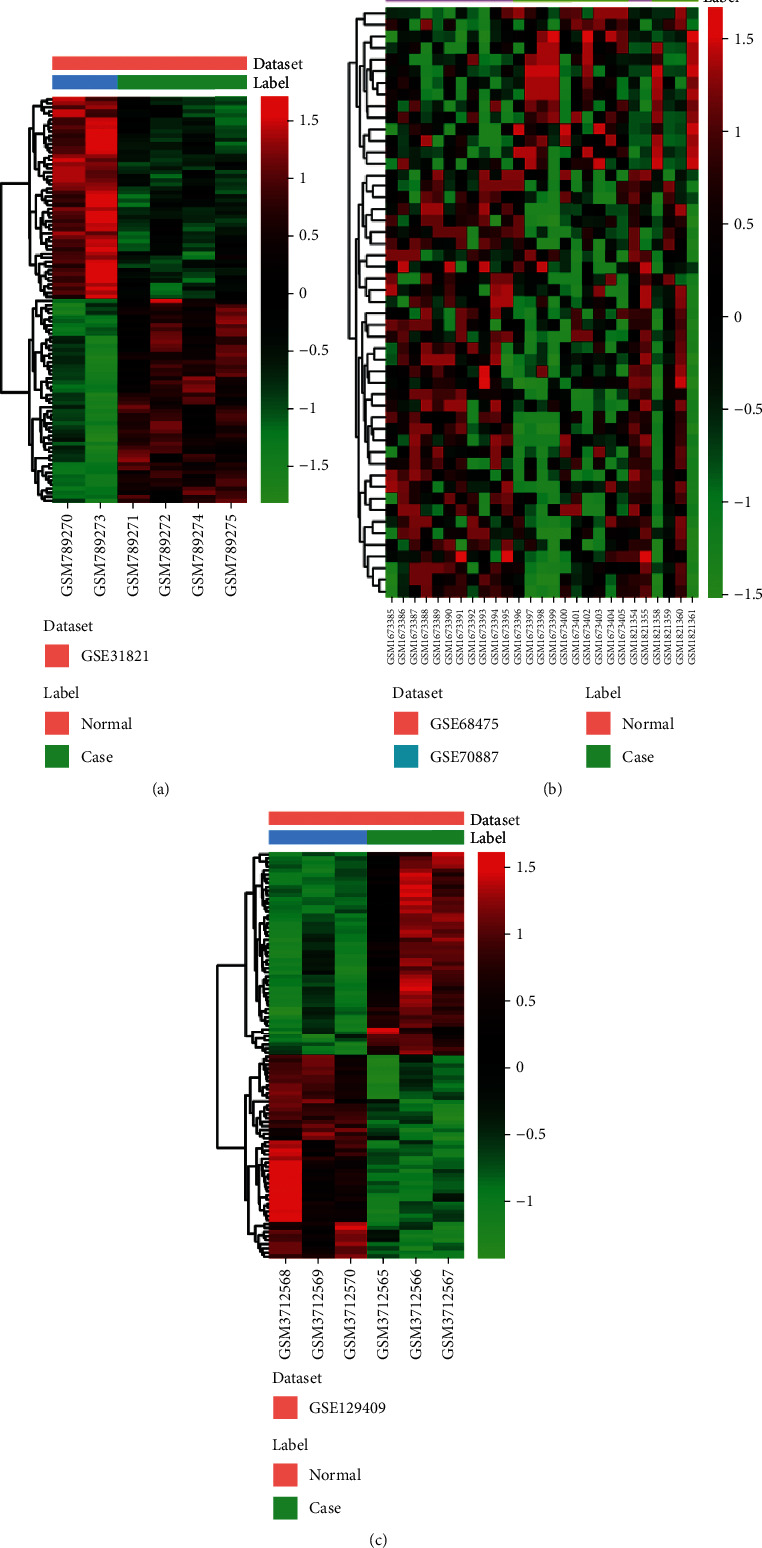
The heat map of top 100 upregulated and downregulated differentially expressed mRNAs (a), all differentially expressed miRNAs (b), and differentially expressed circRNAs (c) in AF.
Table 1.
Top 10 upregulated and downregulated mRNAs in AF.
| Symbol | Log FC | P value | Up/down |
|---|---|---|---|
| RAP1GAP2 | 0.564583 | 5.81E − 05 | Up |
| LOC388588 | 0.735415 | 0.000103 | Up |
| DDIT4L | 0.724298 | 0.000187 | Up |
| PCDHB5 | 0.282318 | 0.00063 | Up |
| TRPV1 | 0.332022 | 0.000843 | Up |
| LOC100509886 | 0.603618 | 0.000911 | Up |
| C1orf93 | 0.370358 | 0.000979 | Up |
| CCDC30 | 0.279824 | 0.001004 | Up |
| CCL18 | 0.579282 | 0.001226 | Up |
| MPHOSPH9 | 0.295236 | 0.001321 | Up |
| ACTA1 | -1.40916 | 8.30E − 06 | Down |
| COLEC11 | -0.68464 | 2.85E − 05 | Down |
| SMOC2 | -0.50736 | 8.63E − 05 | Down |
| ITGB1BP3 | -0.48115 | 0.000102 | Down |
| INMT | -0.43338 | 0.00019 | Down |
| WIPI1 | -0.43174 | 0.000304 | Down |
| B3GNT2 | -0.6447 | 0.000379 | Down |
| OSTF1 | -0.32814 | 0.000585 | Down |
| HEY1 | -0.50453 | 0.0006 | Down |
| SPON1 | -0.58698 | 0.000735 | Down |
FC: fold change.
Table 2.
Upregulated and downregulated miRNAs in AF.
| ID | Combined ES | P value | Up/down |
|---|---|---|---|
| hsa-miR-99a | -1.948325744 | 1.58E − 05 | Down |
| hsa-miR-199a-5p | -1.477854197 | 0.000457589 | Down |
| hsa-miR-508-5p | 1.418488487 | 0.00088982 | Up |
| hsa-miR-199a-3p | -1.320142484 | 0.00135394 | Down |
| hsa-miR-365 | -1.280202941 | 0.002621664 | Down |
| hsa-miR-372 | 1.204514016 | 0.003633868 | Up |
| hsa-miR-26a-1∗ | -1.140485814 | 0.004661322 | Down |
| hsa-miR-622 | 1.119850033 | 0.005370597 | Up |
| hsa-miR-132 | 1.072803396 | 0.007384577 | Up |
| hsa-miR-302d | -1.047958964 | 0.008664093 | Down |
| hsa-let-7f | -1.033548505 | 0.009485724 | Down |
| hsa-miR-1181 | 1.006711572 | 0.011230985 | Up |
| hsa-miR-323-3p | -0.99327606 | 0.012491569 | Down |
| hsa-miR-514 | -0.982260644 | 0.013357392 | Down |
| hsa-miR-943 | -0.97025973 | 0.014154981 | Down |
| hsa-miR-328 | -1.027539395 | 0.014920866 | Down |
| hsa-miR-371-5p | 0.96345164 | 0.016747391 | Up |
| hsa-let-7i | -0.928557563 | 0.019493344 | Down |
| hsa-miR-34c-5p | 0.918481963 | 0.019551512 | Up |
| hsa-miR-298 | 0.914435147 | 0.020064925 | Up |
| hsa-miR-1224-5p | 0.921477354 | 0.020188607 | Up |
| hsa-miR-1253 | -0.910171471 | 0.020573062 | Down |
| hsa-let-7g | -0.914708625 | 0.020978097 | Down |
| hsa-miR-660 | -0.92439633 | 0.021108962 | Down |
| hsa-miR-455-5p | -1.065094646 | 0.021971426 | Down |
| hsa-miR-105 | -0.873885194 | 0.025653065 | Down |
| hsa-miR-7-2∗ | -0.870938842 | 0.026112318 | Down |
| hsa-miR-342-3p | -0.914017107 | 0.029399291 | Down |
| hsa-miR-412 | -0.848482991 | 0.029880717 | Down |
| hsa-miR-345 | 0.847634702 | 0.030183762 | Up |
| hsa-miR-188-5p | 0.84774053 | 0.03099467 | Up |
| hsa-miR-532-5p | -0.847445391 | 0.032511465 | Down |
| hsa-miR-1296 | -0.83188883 | 0.033097822 | Down |
| hsa-miR-150 | -0.92357357 | 0.034421637 | Down |
| hsa-miR-551b | -0.821543289 | 0.035474621 | Down |
| hsa-miR-181a-2∗ | -0.816558501 | 0.036074016 | Down |
| hsa-miR-338-3p | -0.822800891 | 0.03637736 | Down |
| hsa-miR-654-3p | -0.810794989 | 0.037860622 | Down |
| hsa-miR-498 | 0.806871169 | 0.038377453 | Up |
| hsa-miR-944 | -0.801311236 | 0.039480709 | Down |
| hsa-miR-208b | 0.79839323 | 0.040113362 | Up |
| hsa-miR-16-2∗ | -0.824929773 | 0.040444012 | Down |
| hsa-miR-143 | -0.791385474 | 0.042083126 | Down |
| hsa-miR-597 | -0.789967814 | 0.04214698 | Down |
| hsa-miR-433 | -0.788037698 | 0.042547929 | Down |
| hsa-miR-24-2∗ | -0.782812124 | 0.043926008 | Down |
| hsa-miR-450a | -0.781978746 | 0.044964884 | Down |
| hsa-miR-802 | -0.776547306 | 0.045503217 | Down |
| hsa-miR-1200 | -0.76825428 | 0.047587947 | Down |
| hsa-miR-513c | 0.767204868 | 0.04833756 | Up |
| hsa-miR-891a | -0.760813039 | 0.049622889 | Down |
ES: effect size. The bigger the absolute value of combined ES, the bigger the fold change.
Table 3.
Top 10 upregulated and downregulated circRNAs in AF.
| ID | Alias | Log FC | P value | Up/down |
|---|---|---|---|---|
| hsa_circRNA_405811 | 1.910908 | 5.86E − 05 | Up | |
| hsa_circRNA_058161 | hsa_circ_0058161 | 1.625918 | 0.000148 | Up |
| hsa_circRNA_100693 | hsa_circ_0020174 | 1.472556 | 0.00026 | Up |
| hsa_circRNA_404814 | 1.485693 | 0.000344 | Up | |
| hsa_circRNA_102950 | hsa_circ_0058794 | 1.575402 | 0.000409 | Up |
| hsa_circRNA_102949 | hsa_circ_0058792 | 1.48237 | 0.000424 | Up |
| hsa_circRNA_001873 | hsa_circ_0001873 | 1.394786 | 0.000522 | Up |
| hsa_circRNA_100372 | hsa_circ_0015004 | 1.350013 | 0.000533 | Up |
| hsa_circRNA_402565 | 1.299606 | 0.000622 | Up | |
| hsa_circRNA_406752 | 1.394682 | 0.00064 | Up | |
| hsa_circRNA_103752 | hsa_circ_0006867 | -1.27376 | 0.000302 | Down |
| hsa_circRNA_102831 | hsa_circ_0001074 | -1.41386 | 0.000534 | Down |
| hsa_circRNA_104315 | hsa_circ_0079480 | -1.49241 | 0.000718 | Down |
| hsa_circRNA_079477 | hsa_circ_0079477 | -1.28623 | 0.000724 | Down |
| hsa_circRNA_005791 | hsa_circ_0005791 | -1.7986 | 0.001027 | Down |
| hsa_circRNA_103416 | hsa_circ_0005299 | -1.87406 | 0.001123 | Down |
| hsa_circRNA_104750 | hsa_circ_0008678 | -1.07604 | 0.001253 | Down |
| hsa_circRNA_004825 | hsa_circ_0004825 | -1.08205 | 0.001293 | Down |
| hsa_circRNA_004491 | hsa_circ_0004491 | -1.17035 | 0.001311 | Down |
| hsa_circRNA_406303 | -1.30032 | 0.001433 | Down |
FC: fold change.
3.2. Functional Enrichment Analysis
A total of 342 host genes of DEcircRNAs were obtained. GO analysis indicated that these host genes were significantly enriched in biological processes of protein modification process (P = 1.99E − 09), RNA metabolic process (P = 2.57E − 08), and chromatin modification (P = 3.21E − 08) (Figure 2(a)). According to the KEGG pathway enrichment analysis, several pathways were identified, including ubiquitin-mediated proteolysis (P = 6.27E − 09), the mTOR signaling pathway (P = 0.000127701), and the MAPK signaling pathway (P = 0.000203168) (Figure 2(b)). In addition, significantly enriched cytological components and molecular functions of host genes are shown in Supplementary Figure 1.
Figure 2.
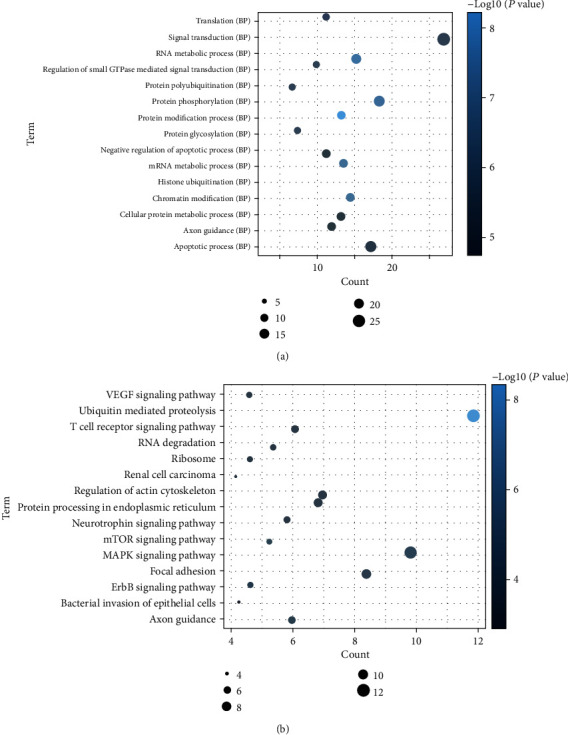
Significantly enriched biological processes and KEGG pathways of host genes of differentially expressed circRNAs. (a) BP: biological process. (b) KEGG pathways. The x-axis shows counts of host genes enriched in biological processes or KEGG pathways, and the y-axis shows biological processes or KEGG pathways.
3.3. ceRNA (DEcircRNA-DEmiRNA-DEmRNA) Regulatory Network
We predicted 94 DEmiRNA-DEcircRNA interactions by starBase v3.0 with a strict mode. The network was constructed by Cytoscape 3.6.1, which included 79 nodes and 94 edges (Figure 3). We also collected 32932 experimentally validated DEmiRNA-DEmRNA interactions from miRWalk 3.0 and established network by Cytoscape 3.6.1, which included 785 nodes and 2099 edges (Supplementary Figure 2). The ceRNA network contained 62 circRNAs, 14 miRNAs, and 728 mRNAs (Supplementary Figure 3). Among which, two ceRNA regulatory pairs of hsa-circRNA-100053-hsa-miR-455-5p-TRPV1 and hsa-circRNA-005843-hsa-miR-188-5p-SPON1 were identified. In addition, GO enrichment analysis revealed that these DEmRNAs were significantly enriched in biological processes of signal transduction (P = 7.69E − 14), multicellular organismal development (P = 3.49E − 09), and regulation of transcription from RNA polymerase II promoter (P = 1.12E − 07) (Figure 4(a)). According to the KEGG pathway enrichment analysis, several pathways were significantly enriched, such as arrhythmogenic right ventricular cardiomyopathy (P = 2.28E − 05), dilated cardiomyopathy (P = 0.000590151), and hypertrophic cardiomyopathy (P = 0.00173912) (Figure 4(b)). In addition, significantly enriched cytological components and molecular functions of these DEmRNAs are shown in Supplementary Figure 2.
Figure 3.
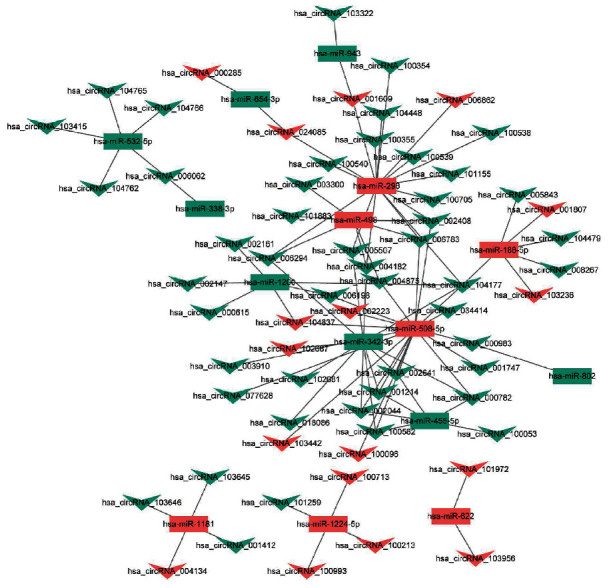
The cirRNA-miRNA network in AF. Rectangle and triangle represent circRNAs and miRNAs, respectively.
Figure 4.
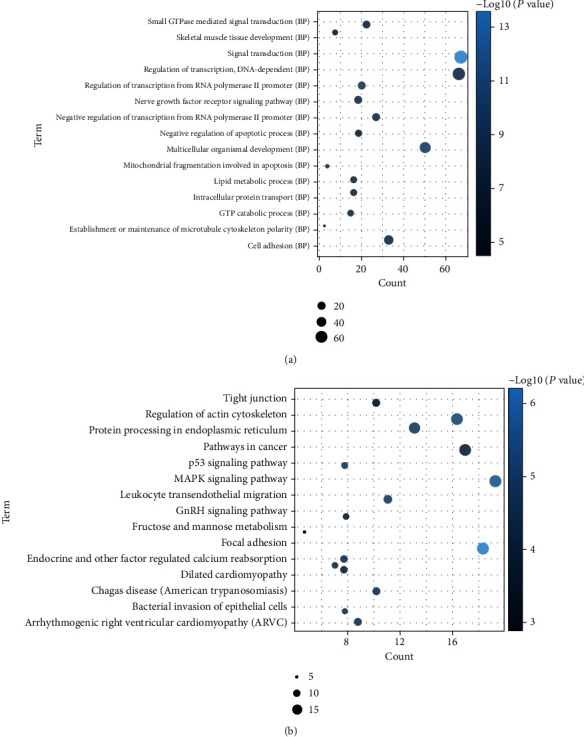
Significantly enriched biological processes and KEGG pathways of differentially expressed mRNAs in ceRNA regulatory network. (a) BP: biological process. (b) KEGG pathways. The x-axis shows counts of host genes enriched in biological processes or KEGG pathways, and the y-axis shows biological processes or KEGG pathways.
3.4. qRT-PCR Validation of Selected DEmRNAs, DEmiRNAs, and DEcircRNAs
In this study, 10 patients with AF and 10 normal individuals were enrolled. Clinical information of these patients is shown in Supplementary Table 2. Two DEmRNAs (including SPON1 and DDIT4L), two DEmiRNAs (including hsa-miR-34c-5p and hsa-miR-188-5p), and one DEcircRNAs hsa-circRNA-402565 were selected randomly for qRT-PCR validation (Figure 5). DDIT4L, hsa-miR-34c-5p, hsa-miR-188-5p, and hsa-circRNA-402565 were upregulated, while SPON1 was downregulated in AF. The qRT-PCR results were in line with our integrated analysis.
Figure 5.
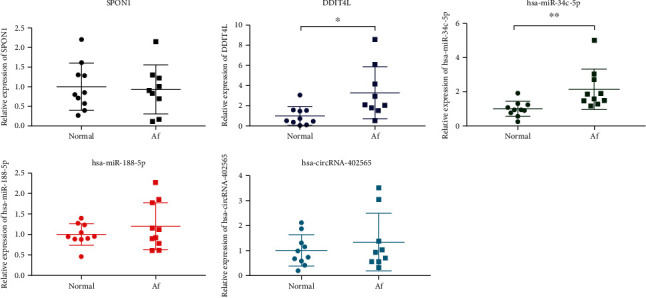
The qRT-PCR results of the DEmRNAs, DEmiRNAs, and DEcircRNAs in AF. The x-axis and the y-axis represent the group and relative expression of DEmRNAs/DEmiRNAs/DEcircRNAs, respectively. ∗P < 0.05 and ∗∗P < 0.01.
3.5. Electronic Validation of Selected mRNAs
In this study, two differentially expressed mRNAs (SMOC2 and WIPI1) and three host genes of circRNAs (MFN2, ZNF880, and LRBA) were randomly selected for validation (Figure 6). The result showed that SMOC2 and WIPI1 were downregulated in AF, which was consisted with our integrated analysis. The expression of MFN2, ZNF880, and LRBA was upregulated in AF. However, MFN2, ZNF880, and LRBA were not differentially expressed mRNAs in this study. Further study of MFN2, ZNF880, and LRBA in AF is needed.
Figure 6.
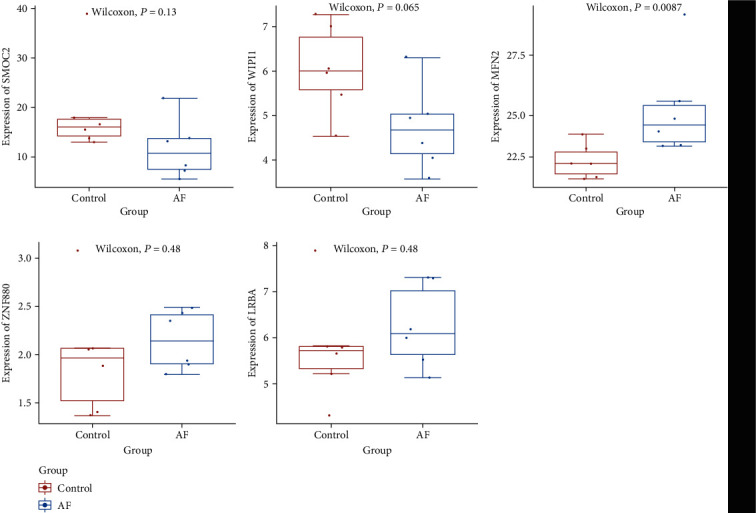
The box plots of SMOC2, WIPI1, MFN2, ZNF880, and LRBA in AF. The x-axis and the y-axis represent the group and expression of mRNAs, respectively.
4. Discussion
Mitofusin 2 (MFN2) is the host gene of hsa_circRNA_100053. MFN2 plays a key role in normal cardiac development [16]. MFN2 could regulate heart failure-related mitophagy by altering the mitochondrial membrane potential [17]. Chen et al. found that deletion of MFN2 leads to a spontaneous lethal dilated cardiomyopathy in mice [18]. In this study, we found that hsa_circRNA_100053 was downregulated in the heart tissue of AF patients. Moreover, downregulated hsa-miR-455-5p and target upregulated transient receptor potential vanilloid 1 (TRPV1) were under the regulation of hsa_circRNA_100053. Huang et al. reported that hsa-miR-455-5p was related to hypoxia-induced cardiomyocytes injury [19]. TRPV1 is a nonselective ion channel that preferentially obtains calcium from painful stimuli. In addition to traditional pain activation of TRPV1, TRPV1 can also be used as a universal sensor for cell damage including hypoxia. Direct activation of TRPV1 has been shown to produce cardioprotective effects on ischemia and reperfusion injury [20]. In addition, blocking TRPV1 limits the long-term preconditioning-induced cardioprotection of laparotomy [20]. An experimental study showed that TRPV1 inhibition blocked ischemic preconditioning- (IPC-) induced myocardial protection [21]. These reports suggested that hsa_circRNA_100053, hsa-miR-455-5p, and TRPV1 may play an important role in heart protection. Our study indicated that the interaction of hsa_circRNA_100053-hsa-miR-455-5p-TRPV1 may be involved in the process of AF.
Recently, there was no report about the association between hsa_circRNA_005843 and AF. Interestingly, we found that hsa_circRNA_005843 was downregulated in the heart tissue of patients with AF. Furthermore, upregulated hsa-miR-188-5p and target downregulated spondin 1 (SPON1) were regulated by hsa_circRNA_005843. hsa-miR-188-5p plays an important regulation role in the renin-angiotensin system [22]. It has been demonstrated that hsa-miR-188-5p is involved in murine cardiomyocyte biogenesis [23]. Decreased expression of hsa-miR-188-5p is found in hyperhomocysteinemia cardiomyocytes [24]. SPON1, a member of antiangiogenic family, is a sensitive plasma biomarker for early myocardial injury [25]. This indicated that hsa-miR-188-5p and SPON1 play roles in angiogenesis, which is associated with cardiomyocyte biogenesis. Our result suggested that the interaction between hsa_circRNA_005843, hsa-miR-188-5p, and SPON1 could be associated with AF.
We also found several DEmiRNA (upregulation)-DEmRNA (downregulation) regulatory pairs such as hsa-miR-34c-5p-indolethylamine N-methyltransferase (INMT), hsa-miR-1253-DNA damage-inducible transcript 4 like (DDIT4L), hsa-miR-508-5p-SPARC-related modular calcium binding 2 (SMOC2), hsa-miR-943-actin alpha 1, skeletal muscle (ACTA1), hsa-miR-338-3p-WD repeat domain, phosphoinositide-interacting 1 (WIPI1), and DEmiRNA (downregulation)-DEmRNA (upregulation) regulatory pair including hsa-miR-199a-3p-RAP1 GTPase-activating protein 2 (RAP1GAP2). Greco et al. found increased expression of hsa-miR-34c in the failing myocardium of diabetic patients [26]. INMT is differentially expressed in myocardial infarction [27]. No association between hsa-miR-1253 and AF has been reported. DDIT4L is expressed in cardiomyocytes and myocardial tissues of pathologically stressed mice, cultured neonatal rat ventricular myocyte (NRVM) models and patients with dilated cardiomyopathy. It is worth noting that pathological stress did not alter the abundance of the relevant DDIT4. DDIT4L, localizes to early endosomes, is a key regulator of NRVMs that inhibits stress-induced autophagy via mTORC1 [28]. Studies by Simonson et al. showed that cardiac pathological stress activates DDIT4L and induces autophagy by inhibiting the mTORC1 signaling pathway [28]. hsa-miR-508-5p is a potential diagnostic and prognostic marker for heart failure patients [29]. SMOC2 is involved in the inflammatory damage in the heart [30]. hsa-miR-943 is remarkably upregulated in acute ischemic stroke patients [31]. ACTA1, a contractile fiber gene, is associated with heart failure and cardiac hypertrophy [32, 33]. hsa-miR-338-3p is involved in heart failure and acute myocardial infarction [34, 35]. WIPI1 regulates mitochondrial oxidative signaling in cardiac myocytes [36]. hsa-miR-199a-3p is related to the pathophysiology of heart failure [37]. The expression of hsa-miR-199a-3p was downregulated in myocardial infarction [38]. RAP1GAP2 is associated with Chagas cardiomyopathy [39]. Thus, it can be seen that these miRNAs and target mRNAs play a key role in various cardiac pathologic processes, such as myocardial infarction, dilated cardiomyopathy, cardiac pathological stress, cardiac hypertrophy, and inflammatory damage. It is suggested that above DEmiRNAs and target DEmRNAs may be involved in the process of AF.
In addition, we found that hsa-circRNA-402565 was upregulated in the heart tissue of AF patients. Interestingly, qRT-PCR validated the expression of hsa-circRNA-402565. hsa-circRNA-402565 is downregulated in patients with ventricular septal defect [40]. It is noted that hsa-circRNA-405811 and hsa-circRNA-103752 were, respectively, the most upregulated and downregulated circRNAs in AF. hsa-miR-99a was the most downregulated miRNA in AF. Zinc finger protein 880 (ZNF880) is the host gene of hsa-circRNA-405811. The inactivating mutation of ZNF880 is found in isolated cardiac myxoma tissue samples [41]. LPS responsive beige-like anchor (LRBA) protein is the host gene of hsa-circRNA-103752. It is reported that LRBA is involved in signal transduction and vesicle trafficking in cardiogenesis [42]. The expression of LRBA is decreased 2 d after myocardial infarction [43]. It is suggested that hsa-miR-99a is involved in cardioprotective in postinfarction left ventricular remodelling [44]. Thus, it can be seen that these circRNAs involve in cardiogenesis and cardioprotection. Our result indicated that hsa-circRNA-402565, hsa-circRNA-405811, hsa-circRNA-103752, and hsa-miR-99a may be associated with the pathology of AF.
According to the functional annotation analysis, we found that mTOR was one of the most enriched signaling pathways of the host genes of DEcircRNAs. MTOR is a protein kinase that acts as an interface to a variety of metabolic pathways and is widely found in many species. MTOR plays a vital role in cellular metabolism [45]. Kinases are activated by extracellular growth factor signaling, enhancing cytoplasmic translation processes and protein synthesis. The mTOR pathway is involved in the steady-state process of the heart against stress [46]. The mTOR pathway contributes to the proliferation and survival of cardiomyocytes. In aged mice, knocking out and inhibiting mTOR can prolong survival and inhibit cardiac hypertrophy [47]. This indicated that the mTOR signaling pathway plays an important role in the heart against stress and cardiomyocyte survival, which may be associated with the development of AF.
In addition, arrhythmogenic right ventricular cardiomyopathy, dilated cardiomyopathy, and hypertrophic cardiomyopathy were three remarkably enriched signaling pathways of DEmRNAs in ceRNA regulatory network. Arrhythmogenic right ventricular cardiomyopathy is a rare inherited cardiomyopathy characterized by fibro-fatty replacement of cardiomyocytes [48, 49]. Sudden cardiac death and ventricular enlargement are the most common clinical manifestations [50]. Arrhythmogenic right ventricular cardiomyopathy mainly involves the left and right ventricle during disease progression. The enlarged left atrial is associated with the incidence of risk for death in dilated cardiomyopathy patients [51–60]. In addition, extensive atrial fibrosis is observed at autopsy in dilated cardiomyopathy patients [61]. AF is an arrhythmia often complicating the course of hypertrophic cardiomyopathy. It is found that patients with hypertrophic cardiomyopathy have a higher risk (20%) for AF [62, 63]. These reports suggested that arrhythmogenic right ventricular cardiomyopathy, dilated cardiomyopathy, and hypertrophic cardiomyopathy may be involved in the process of AF.
5. Conclusion
Our study found two ceRNA (DEcircRNA-DEmiRNA-DEmRNA) regulatory networks including hsa-circRNA-100053-hsa-miR-455-5p-TRPV1 and hsa-circRNA-005843-hsa-miR-188-5p-SPON1 in AF. In addition, several miRNA-mRNA regulatory pairs including hsa-miR-34c-5p-INMT, hsa-miR-1253-DDIT4L, hsa-miR-508-5p-SMOC2, hsa-miR-943-ACTA1, hsa-miR-338-3p-WIPI1, and hsa-miR-199a-3p-RAP1GAP2 and four signaling pathways such as mTOR, arrhythmogenic right ventricular cardiomyopathy, dilated cardiomyopathy, and hypertrophic cardiomyopathy were also identified. The results of the present study may provide a potential novel field into the molecular mechanisms of AF. However, there are limitations to our study. Firstly, a sample size in the qRT-PCR was small. Larger numbers of samples are further needed to validate the expression of ceRNA (DEcircRNA-DEmiRNA-DEmRNA) regulatory networks including hsa-circRNA-100053-hsa-miR-455-5p-TRPV1 and hsa-circRNA-005843-hsa-miR-188-5p-SPON1 and the most upregulated or downregulated mRNAs/miRNAs/circRNAs in AF. Secondly, he potential deeper mechanism of AF is not investigated. In vivo animal model or in vitro cell experiment is further needed to study the potential biological function of identified circRNAs, miRNAs, and mRNAs.
Data Availability
All data are available in the article.
Conflicts of Interest
The authors declare that there is no conflict of interest.
Supplementary Materials
Supplementary Figure 1:significantly enriched cytological components and molecular functions of host genes of differentially expressed circRNAs. (A) CC, cytological components; (B) MF, molecular functions. The x-axis shows counts of host genes enriched in cytological components or molecular functions and the y-axis shows biological processes or molecular functions.
Supplementary Figure 2: the miRNA-mRNA network in AF. Rectangle and ellipse represent miRNAs and mRNA, respectively. Red and green colors represent upregulation and downregulation, respectively.
Supplementary Figure 3: ceRNA (DEcircRNA-DEmiRNA-DEmRNA) regulatory network. The trigonal nodes, rectangle nodes, and elliptical nodes indicate DEcircRNAs, DEmiRNAs, and DEmRNAs, respectively. Red and green colors represent upregulation and downregulation, respectively. Nodes with the black border were DEcircRNA/DEmiRNA/DEmRNA derived from the top 10 upregulated and downregulated DEcircRNA/DEmiRNA/DEmRNA in AF.
Supplementary Figure 4: significantly enriched cytological components and molecular functions of differentially expressed mRNAs in ceRNA regulatory network. (A) CC, cytological components; (B) MF, molecular functions. The x-axis shows counts of host genes enriched in cytological components or molecular functions, and the y-axis shows biological processes or molecular functions.
Supplementary Table 2: the clinical information of AF patients and normal individuals in the qRT-PCR.
Supplementary Table 1: selected circRNA, miRNA, and mRNA datasets.
References
- 1.Graham D. J., Reichman M. E., Wernecke M., et al. Cardiovascular, bleeding, and mortality risks in elderly Medicare patients treated with dabigatran or warfarin for nonvalvular atrial fibrillation. Circulation. 2015;131(2):157–164. doi: 10.1161/CIRCULATIONAHA.114.012061. [DOI] [PubMed] [Google Scholar]
- 2.Marrouche N. F., Brachmann J., Andresen D., et al. Catheter ablation for atrial fibrillation with heart failure. New England Journal of Medicine. 2018;378(5):417–427. doi: 10.1056/NEJMoa1707855. [DOI] [PubMed] [Google Scholar]
- 3.Lang K., Bozkaya D., Patel A. A., et al. Anticoagulant use for the prevention of stroke in patients with atrial fibrillation: findings from a multi-payer analysis. Bmc Health Services Research. 2014;14(1):p. 329. doi: 10.1186/1472-6963-14-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang S., Guo C., Zhang W., et al. The integrative regulatory network of circRNA, microRNA, and mRNA in atrial fibrillation. Frontiers in Genetics. 2019;10:p. 526. doi: 10.3389/fgene.2019.00526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dagres N., Anastasiou-Nana M. Atrial fibrillation and obesity: an association of increasing importance. Journal of the American College of Cardiology. 2010;55(21):2328–2329. doi: 10.1016/j.jacc.2010.01.045. [DOI] [PubMed] [Google Scholar]
- 6.Soliman E. Z., Safford M. M., Muntner P., et al. Atrial fibrillation and the risk of myocardial infarction. Jama Internal Medicine. 2014;174(1):107–114. doi: 10.1001/jamainternmed.2013.11912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voukalis C., Lip G. Y., Shantsila E. Emerging tools for stroke prevention in atrial fibrillation. eBioMedicine. 2016;4:26–39. doi: 10.1016/j.ebiom.2016.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vallabhajosyula S., Skiba J. F., Jr., Hashmi F., Kashani K. B. Cardiovascular critical care: therapeutic hypothermia, atrial fibrillation, and cardiopulmonary resuscitation. American Journal of Respiratory and Critical Care Medicine. 2016;194(6):762–764. doi: 10.1164/rccm.201601-0165RR. [DOI] [PubMed] [Google Scholar]
- 9.Wan E., Abrams J., Weinberg R. L., et al. Aberrant sodium influx causes cardiomyopathy and atrial fibrillation in mice. The Journal of clinical investigation. 2016;126(1):112–122. doi: 10.1172/JCI84669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogawa H., Senoo K., An Y., et al. Clinical features and prognosis in patients with atrial fibrillation and prior stroke: comparing the Fushimi and Darlington AF registries. eBioMedicine. 2017;18:199–203. doi: 10.1016/j.ebiom.2017.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z., Yang T., Xiao J. Circular RNAs: promising biomarkers for human diseases. eBioMedicine. 2018;34:267–274. doi: 10.1016/j.ebiom.2018.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang K., Long B., Liu F., et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. European Heart Journal. 2016;37(33):2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 13.Hu X., Chen L., Wu S., et al. Integrative analysis reveals key circular RNA in atrial fibrillation. Frontiers in Genetics. 2019;10:p. 108. doi: 10.3389/fgene.2019.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak K. J., Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C (T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Sun X., Ge X., Xu Z., Chen D. Identification of circular RNA–micro RNA–messenger RNA regulatory network in hepatocellular carcinoma by integrated analysis. Journal of Gastroenterology and Hepatology. 2020;35(1):157–164. doi: 10.1111/jgh.14762. [DOI] [PubMed] [Google Scholar]
- 16.Kasahara A., Cipolat S., Chen Y., Dorn G. W., Scorrano L. Mitochondrial fusion directs cardiomyocyte differentiation via calcineurin and Notch signaling. Science. 2013;342(6159):734–737. doi: 10.1126/science.1241359. [DOI] [PubMed] [Google Scholar]
- 17.Mori J., Zhang L., Oudit G. Y., Lopaschuk G. D. Impact of the renin-angiotensin system on cardiac energy metabolism in heart failure. Journal of Molecular & Cellular Cardiology. 2013;63:98–106. doi: 10.1016/j.yjmcc.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y., Liu Y., Dorn G. W., II Mitochondrial fusion is essential for organelle function and cardiac homeostasis. Circulation Research. 2011;109(12):1327–1331. doi: 10.1161/CIRCRESAHA.111.258723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang S., Tao W., Guo Z., Cao J., Huang X. Suppression of long noncoding RNA TTTY15 attenuates hypoxia-induced cardiomyocytes injury by targeting miR-455-5p. Gene. 2019;701:1–8. doi: 10.1016/j.gene.2019.02.098. [DOI] [PubMed] [Google Scholar]
- 20.Heymann H. M., Wu Y., Lu Y., Qvit N., Gross G. J., Gross E. R. Transient receptor potential vanilloid 1 inhibitors block laparotomy- and opioid-induced infarct size reduction in rats. British Journal of Pharmacology. 2017;174(24):4826–4835. doi: 10.1111/bph.14064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Randhawa P. K., Jaggi A. S. TRPV1 and TRPV4 channels: potential therapeutic targets for ischemic conditioning-induced cardioprotection. European Journal of Pharmacology. 2015;746:180–185. doi: 10.1016/j.ejphar.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 22.Kemp J. R., Unal H., Desnoyer R., Yue H., Bhatnagar A., Karnik S. S. Angiotensin II-regulated microRNA 483-3p directly targets multiple components of the renin–angiotensin system. Journal of Molecular & Cellular Cardiology. 2014;75:25–39. doi: 10.1016/j.yjmcc.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Humphreys D. T., Hynes C. J., Patel H. R., et al. Complexity of murine cardiomyocyte miRNA biogenesis, sequence variant expression and function. PLoS One. 2012;7(2, article e30933) doi: 10.1371/journal.pone.0030933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mishra P. K., Tyagi N., Kundu S., Tyagi S. C. MicroRNAs are involved in homocysteine-induced cardiac remodeling. Cell Biochemistry & Biophysics. 2009;55(3):153–162. doi: 10.1007/s12013-009-9063-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keshishian H., Burgess M. W., Gillette M. A., et al. Multiplexed, quantitative workflow for sensitive biomarker discovery in plasma yields novel candidates for early myocardial injury. Molecular & Cellular Proteomics. 2015;14(9):2375–2393. doi: 10.1074/mcp.M114.046813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greco S., Fasanaro P., Castelvecchio S., et al. MicroRNA dysregulation in diabetic ischemic heart failure patients. Diabetes. 2012;61(6):1633–1641. doi: 10.2337/db11-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao L., Gao Y., Li X., et al. Aquaporins mediate the chemoresistance of human melanoma cells to arsenite. Molecular oncology. 2012;6(1):81–87. doi: 10.1016/j.molonc.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simonson B., Subramanya V., Chan M. C., et al. DDiT4L promotes autophagy and inhibits pathological cardiac hypertrophy in response to stress. Science Signaling. 2017;10(468, article eaaf 5967) doi: 10.1126/scisignal.aaf5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiang L., Hong L., Ningfu W., Huaihong C., Jing W. Expression of miR-126 and miR-508-5p in endothelial progenitor cells is associated with the prognosis of chronic heart failure patients. International Journal of Cardiology. 2013;168(3):2082–2088. doi: 10.1016/j.ijcard.2013.01.160. [DOI] [PubMed] [Google Scholar]
- 30.Laugier L., Frade A. F., Ferreira F. M., et al. Whole-genome cardiac DNA methylation fingerprint and gene expression analysis provide new insights in the pathogenesis of chronic Chagas disease cardiomyopathy. Clinical Infectious Diseases. 2017;65(7):1103–1111. doi: 10.1093/cid/cix506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Modak J. M., Roy-O'Reilly M., Zhu L., Staff I., McCullough L. D. Differential microribonucleic acid expression in cardioembolic stroke. Journal of Stroke and Cerebrovascular Diseases. 2019;28(1):121–124. doi: 10.1016/j.jstrokecerebrovasdis.2018.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hulanicka M., Garncarz M., Parzeniecka-Jaworska M., Jank M. The transcriptomic profile of peripheral blood nuclear cells in dogs with heart failure. BMC Genomics. 2014;15(1):p. 509. doi: 10.1186/1471-2164-15-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gurung I. S., Medina-Gomez G., Kis A., et al. Deletion of the metabolic transcriptional coactivator PGC1β induces cardiac arrhythmia. Cardiovascular Research. 2011;92(1):29–38. doi: 10.1093/cvr/cvr155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barsanti C., Trivella M. G., D’Aurizio R., et al. Differential regulation of microRNAs in end-stage failing hearts is associated with left ventricular assist device unloading. BioMed Research International. 2015;2015:13. doi: 10.1155/2015/592512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maciejak A., Kostarska-Srokosz E., Gierlak W., et al. Circulating miR-30a-5p as a prognostic biomarker of left ventricular dysfunction after acute myocardial infarction. Scientific Reports. 2018;8(1):p. 9883. doi: 10.1038/s41598-018-28118-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tzimas C., Rau C. D., Buergisser P. E., et al. WIPI1 is a conserved mediator of right ventricular failure. JCI Insight. 2019;4(11) doi: 10.1172/jci.insight.122929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matkovich S. J., van Booven D. J., Youker K. A., et al. Reciprocal regulation of myocardial microRNAs and messenger RNA in human cardiomyopathy and reversal of the microRNA signature by biomechanical support. Circulation. 2009;119(9):1263–1271. doi: 10.1161/CIRCULATIONAHA.108.813576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eulalio A., Mano M., Ferro M. D., et al. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492(7429):376–381. doi: 10.1038/nature11739. [DOI] [PubMed] [Google Scholar]
- 39.Deng X., Sabino E. C., Cunha-Neto E., et al. Genome wide association study (GWAS) of Chagas cardiomyopathy in Trypanosoma cruzi seropositive subjects. PLoS One. 2013;8(11, article e79629) doi: 10.1371/journal.pone.0079629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu H., Hu Y., Zhuang B., et al. Differential expression of circRNAs in embryonic heart tissue associated with ventricular septal defect. International Journal of Medical Sciences. 2018;15(7):703–712. doi: 10.7150/ijms.21660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jian H., Sun M., Li E., Hou Y., Yang L. Recurrent somatic mutations of PRKAR1A in isolated cardiac myxoma. Oncotarget. 2017;8:103968–103974. doi: 10.18632/oncotarget.21916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chamberlain A. A., Lin M., Lister R. L., et al. DNA methylation is developmentally regulated for genes essential for cardiogenesis. Journal of the American Heart Association. 2014;3, article e000976 doi: 10.1161/JAHA.114.000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y., Huang Y., Zhang M., Zhang X., Tang X., Kang Y. Bioinformatic analysis of the possible regulative network of miR-30a/e in cardiomyocytes 2 days post myocardial infarction. Acta Cardiologica Sinica. 2018;34(2):175–188. doi: 10.6515/ACS.201803_34(2).20170926A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Q., Xie J., Li R., et al. Overexpression of microRNA-99a attenuates heart remodelling and improves cardiac performance after myocardial infarction. Journal of Cellular & Molecular Medicine. 2014;18(5):919–928. doi: 10.1111/jcmm.12242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saxton R. A., Sabatini D. M. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168(6):960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang D., Contu R., Latronico M. V. G., et al. MTORC1 regulates cardiac function and myocyte survival through 4E-BP1 inhibition in mice. Journal of Clinical Investigation. 2010;120(8):2805–2816. doi: 10.1172/JCI43008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sciarretta S., Forte M., Frati G., Sadoshima J. New insights into the role of mTOR signaling in the cardiovascular system. Circulation Research. 2018;122(3):489–505. doi: 10.1161/CIRCRESAHA.117.311147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pilichou K., Thiene G., Bauce B., et al. Arrhythmogenic cardiomyopathy. Orphanet Journal of Rare Diseases. 2016;11(1, article 33) doi: 10.1186/s13023-016-0407-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soni R., Oade Y. Arrhythmogenic right ventricular cardiomyopathy. Bmj Case Reports. 2011;2011:565–570. doi: 10.1136/bcr.05.2011.4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corrado D., Basso C., Judge D. P. Arrhythmogenic cardiomyopathy. Circulation Research. 2017;121(7):784–802. doi: 10.1161/CIRCRESAHA.117.309345. [DOI] [PubMed] [Google Scholar]
- 51.Dini F. L., Cortigiani L., Baldini U., et al. Prognostic value of left atrial enlargement in patients with idiopathic dilated cardiomyopathy and ischemic cardiomyopathy. American Journal of Cardiology. 2002;89(5):518–523. doi: 10.1016/S0002-9149(01)02290-1. [DOI] [PubMed] [Google Scholar]
- 52.Kim H., Cho Y. K., Jun D. H., et al. Prognostic implications of the NT-ProBNP level and left atrial size in non-ischemic dilated cardiomyopathy. Circulation Journal Official Journal of the Japanese Circulation Society. 2008;72(10):1658–1665. doi: 10.1253/circj.CJ-07-1087. [DOI] [PubMed] [Google Scholar]
- 53.Modena M. G., Muia N., Jr., Sgura F. A., Molinari R., Castelli A., Rossi R. Left atrial size is the major predictor of cardiac death and overall clinical outcome in patients with dilated cardiomyopathy: a long-term follow-up study. Clinical Cardiology. 1997;20(6):553–560. doi: 10.1002/clc.4960200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quiñones M. A., Greenberg B. H., Kopelen H. A., et al. Echocardiographic predictors of clinical outcome in patients with left ventricular dysfunction enrolled in the SOLVD registry and trials: significance of left ventricular hypertrophy. Journal of the American College of Cardiology. 2000;35(5):1237–1244. doi: 10.1016/S0735-1097(00)00511-8. [DOI] [PubMed] [Google Scholar]
- 55.Sabharwal N., Cemin R., Rajan K., Hickman M., Lahiri A., Senior R. Usefulness of left atrial volume as a predictor of mortality in patients with ischemic cardiomyopathy. American Journal of Cardiology. 2004;94(6):760–763. doi: 10.1016/j.amjcard.2004.05.060. [DOI] [PubMed] [Google Scholar]
- 56.Maddukuri P. V., Vieira M. L., DeCastro S., et al. What is the best approach for the assessment of left atrial size? Comparison of various unidimensional and two-dimensional parameters with three-dimensional echocardiographically determined left atrial volume. Journal of the American Society of Echocardiography. 2006;19(8):1026–1032. doi: 10.1016/j.echo.20c06.03.011. [DOI] [PubMed] [Google Scholar]
- 57.Wang Y., Gutman J. M., Heilbron D., Wahr D., Schiller N. B. Atrial volume in a normal adult population by two-dimensional echocardiography. Chest. 1984;86(4):595–601. doi: 10.1378/chest.86.4.595. [DOI] [PubMed] [Google Scholar]
- 58.Whitlock M., Garg A., Gelow J., Jacobson T., Broberg C. Comparison of left and right atrial volume by echocardiography versus cardiac magnetic resonance imaging using the area-length method. The American Journal of Cardiology. 2010;106(9):1345–1350. doi: 10.1016/j.amjcard.2010.06.065. [DOI] [PubMed] [Google Scholar]
- 59.Aune E., Baekkevar M., Roislien J., Rodevand O., Otterstad J. E. Normal reference ranges for left and right atrial volume indexes and ejection fractions obtained with real-time three-dimensional echocardiography. European Journal of Echocardiography. 2009;10(6):738–744. doi: 10.1093/ejechocard/jep054. [DOI] [PubMed] [Google Scholar]
- 60.Peluso D., Badano L. P., Muraru D., et al. Right atrial size and function assessed with three-dimensional and speckle-tracking echocardiography in 200 healthy volunteers. European Heart Journal Cardiovascular Imaging. 2013;14(11):1106–1114. doi: 10.1093/ehjci/jet024. [DOI] [PubMed] [Google Scholar]
- 61.Ohtani K., Yutani C., Nagata S., Koretsune Y., Hori M., Kamada T. High prevalence of atrial fibrosis in patients with dilated cardiomyopathy. Journal of the American College of Cardiology. 1995;25(5):1162–1169. doi: 10.1016/0735-1097(94)00529-Y. [DOI] [PubMed] [Google Scholar]
- 62.Kubo T., Kitaoka H., Okawa M., et al. Clinical impact of atrial fibrillation in patients with hypertrophic cardiomyopathy. Circulation Journal. 2009;73(9):1599–1605. doi: 10.1253/circj.CJ-09-0140. [DOI] [PubMed] [Google Scholar]
- 63.Siontis K. C., Geske J. B., Ong K., Nishimura R. A., Ommen S. R., Gersh B. J. Atrial fibrillation in hypertrophic cardiomyopathy: prevalence, clinical correlations, and mortality in a large high-risk population. Journal of the American Heart Association. 2014;3, article e001002 doi: 10.1161/JAHA.114.001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1:significantly enriched cytological components and molecular functions of host genes of differentially expressed circRNAs. (A) CC, cytological components; (B) MF, molecular functions. The x-axis shows counts of host genes enriched in cytological components or molecular functions and the y-axis shows biological processes or molecular functions.
Supplementary Figure 2: the miRNA-mRNA network in AF. Rectangle and ellipse represent miRNAs and mRNA, respectively. Red and green colors represent upregulation and downregulation, respectively.
Supplementary Figure 3: ceRNA (DEcircRNA-DEmiRNA-DEmRNA) regulatory network. The trigonal nodes, rectangle nodes, and elliptical nodes indicate DEcircRNAs, DEmiRNAs, and DEmRNAs, respectively. Red and green colors represent upregulation and downregulation, respectively. Nodes with the black border were DEcircRNA/DEmiRNA/DEmRNA derived from the top 10 upregulated and downregulated DEcircRNA/DEmiRNA/DEmRNA in AF.
Supplementary Figure 4: significantly enriched cytological components and molecular functions of differentially expressed mRNAs in ceRNA regulatory network. (A) CC, cytological components; (B) MF, molecular functions. The x-axis shows counts of host genes enriched in cytological components or molecular functions, and the y-axis shows biological processes or molecular functions.
Supplementary Table 2: the clinical information of AF patients and normal individuals in the qRT-PCR.
Supplementary Table 1: selected circRNA, miRNA, and mRNA datasets.
Data Availability Statement
All data are available in the article.


