Abstract
Background
Small cell lung cancer (SCLC) has the strongest association with smoking among lung cancers. The characteristics of never smokers with SCLC is not known.
Research Question
Are the clinical characteristics, prognostic factors, survival, genomic alterations, and tumor mutational burdens of SCLC in patients who have never smoked different from those who have smoked?
Study Design and Methods
A retrospective multicenter cohort study of patients with clinician-confirmed SCLC was performed with the use of a longitudinal and nationally representative electronic medical records database. Smoking history was assessed through technology-enabled abstraction and confirmed for never smokers via chart review. Genomic characteristics of never smoker patients with SCLC were examined with the use of a next-generation sequencing-based gene panel and whole exome sequencing.
Results
One hundred of 5,632 patients (1.8%) with SCLC were never smokers. Relative to smokers, never smokers were more likely to be female (66.0% vs 52.4%; P = .009) and present with extensive stage (70.0% vs 62.2%; P = .028). Never smokers had a higher proportion of patients in age groups 35 to 49 years (7.0% vs 3.0%; P = .006) and ≥80 years (17.0% vs 8.2%; P = .006). Known risk factors for lung cancer were found in <20% of never smokers. There were no overall survival differences between never smokers and smokers. Among patients with available genomic data (n = 9), never smoker SCLC were characterized by lower tumor mutational burden, a lower frequency of TP53 mutations, and an absence of mutational signatures related to tobacco exposure.
Interpretation
The sex- and age-specific distribution of SCLC among never smokers, along with differences that were identified by genomic analyses, suggests a distinct biology of SCLC in never smokers compared with smokers.
Key Words: COSMIC signature 4, never smoker, small cell lung cancer, tumor mutation burden
Abbreviations: EGFR, epidermal growth factor receptor; EHR, electric health record; ICD, International Classification of Diseases; NCI, National Cancer Institute; OS, overall survival; SCLC, small cell lung cancer; TMB, tumor mutational burden
Lung cancer is the leading cause of cancer death worldwide, with approximately 1.7 million deaths every year.1 Although most lung cancers are caused by long-term exposure to tobacco smoke, a significant fraction (up to 20% of all lung cancer in the United States1) occurs in the absence of known tobacco exposure. Lung adenocarcinoma in never smokers has markedly different epidemiologic, clinical, and molecular characteristics that are recognized as separate disease entities compared with lung adenocarcinoma in smokers2,3; therapies tailored towards molecular characteristics more prevalent among never smokers have resulted in substantial benefits for patients. With the decline in smoking prevalence in several countries, including the United States, the relative proportion of lung cancers in never smokers is expected to increase.4 There is growing interest in lung cancer in never smokers among patients, physicians, and policy makers, all of whom traditionally have conceptualized lung cancer as a “smokers” disease.
Take-home Points.
Study Question: Are the clinical characteristics, prognostic factors, survival, genomic alterations, and tumor mutational burdens of small cell lung cancer (SCLC) in patients who have never smoked different from those who have smoked?
Results: One hundred of 5632 patients (1.8%) with SCLC were never smokers, and clinical characteristics such as sex, age, and stage distributions were significantly different from those who had smoked. There were no overall survival differences among patients who were never smokers and patients who were smokers. Tumor mutational burden and frequency of TP53 mutations were lower in patients with SCLC who were never smokers.
Interpretation: The sex and age-specific distribution of SCLC among never smokers, along with differences identified by genomic analyses suggest a distinct biology of SCLC in patients who were never smokers compared with patients who were smokers.
Among the major lung cancer subtypes, small cell lung cancer (SCLC) has the strongest association with smoking.5 SCLC also has the most aggressive clinical course. Unlike the increasingly personalized approach to care of patients with lung adenocarcinoma, SCLC is still approached clinically as a single disease entity. The typical life expectancy for a patient who is diagnosed with SCLC has not changed much over the past three decades, which has caused the National Cancer Institute (NCI) to categorize it as a “recalcitrant” cancer.6
We hypothesized that the characteristics of SCLC in patients who have never smoked is different from those who have smoked. However, SCLC in patients who have never smoked is rare, and there are limited data regarding de novo SCLC in never smokers. An epidemiologic study reported that 3% of patients with SCLCs are never smokers.7 Published data on SCLC in never smokers have originated from single institution, retrospective studies.8, 9, 10, 11, 12
Given the rarity of cases, a large, population-based study was required to examine SCLC in never smokers. We used the Flatiron Health Inc (New York, NY) electronic health record (EHR)-derived database,13 a nationwide deidentified database to examine the clinical characteristics of never smoker patients with SCLC. We then used the deidentified Flatiron Health-Foundation Medicine Clinico-Genomic Database14 to examine genomic characteristics of a subset of never smoker patients with SCLC with next-generation sequencing. Additionally, we characterized the exomes of never smoker patients with SCLC who enrolled in a natural history study of lung cancer. Similar to what happened with lung adenocarcinoma, defining SCLC subgroups based on smoking may lead to treatment advances in this historically recalcitrant cancer.
Materials and Methods
We performed a multicenter analysis using the longitudinal and nationwide Flatiron Health database, which is comprised of deidentified patient-level structured and unstructured data from an EHR data source, curated via technology-enabled abstraction. The Flatiron Health EHR-derived database includes deidentified data from >280 cancer clinics that represents >2.2 million US patients with cancer. The cohort for clinical analyses included patients with International Classification of Diseases (ICD-9) codes 162.x or ICD-10 codes C34.x or C39.9 who subsequently were confirmed as having pathologic findings that were consistent with SCLC, based on review of unstructured documents. Patients with both limited and extensive stage were included. Patients were diagnosed between January 1, 2013, and September 30, 2018, and had at least two documented clinical encounters. Patients with a history of lung cancer before the SCLC diagnosis were excluded. Given the retrospective nature of the study, independent pathologic review was not possible, and the possibility of misdiagnoses exists.
A personal history of tobacco smoking was assessed through abstraction across the entire cohort of patients with SCLC. Smoking status was reconfirmed by a second round of chart review across the cohort of never smokers (defined as having smoked >100 cigarettes in their lifetime) at which time patients who had tumors of mixed histologic findings were identified and excluded. Patients with second-hand smoke exposure who were identified during chart review remained in the never smoker cohort for the primary analysis because second-hand smoke exposure is difficult to quantify. Sensitivity analyses were performed by recategorizing such patients to the smoker cohort. Demographic information and clinical characteristics were obtained for both the smoker and the never smoker patients. The presence of other cancers was assessed via structured ICD codes and confirmed by chart review.
Survival curves were estimated with the use of the Kaplan-Meier method. Survival analyses were indexed to the diagnosis date and were censored at the last structured visit if no date of death was available. Association of patient characteristics with overall survival (OS), specifically stage, serum sodium, age, and race was assessed within never smokers with the use of a univariate Cox proportional hazards model. A multivariable Cox proportional hazards model was also used to assess the association of smoking status after adjustment for these covariates. Although stage and smoking were obtained from review of unstructured documents, serum sodium was obtained from structured laboratory data and evaluated within a time window of 60 days before or after the diagnosis date. Institutional Review Board approval of the study protocol for Flatiron Health data was obtained before the study was conducted (Copernicus Group Independent Review Board: approval number RWE-001) and included a waiver of informed consent. Data that were provided to third parties were deidentified, and provisions were in place to prevent reidentification to protect patient confidentiality.
Hybrid capture next-generation sequencing with the use of the FoundationOne platform was used for genomic analyses via Flatiron Health-Foundation Medicine Clinico-Genomic Database.15 Whole exome sequencing was performed on tumors of never smokers with SCLC who enrolled in an NCI natural history study (Clinicaltrials.gov identifier: NCT02146170). Genomic DNA isolated from peripheral blood and tumor was used for next-generation sequencing, bioinformatics, and variant interpretation as previously described.16 Human subjects committees approved the studies. Additional methodologic details are in e-Appendix 1.
Results
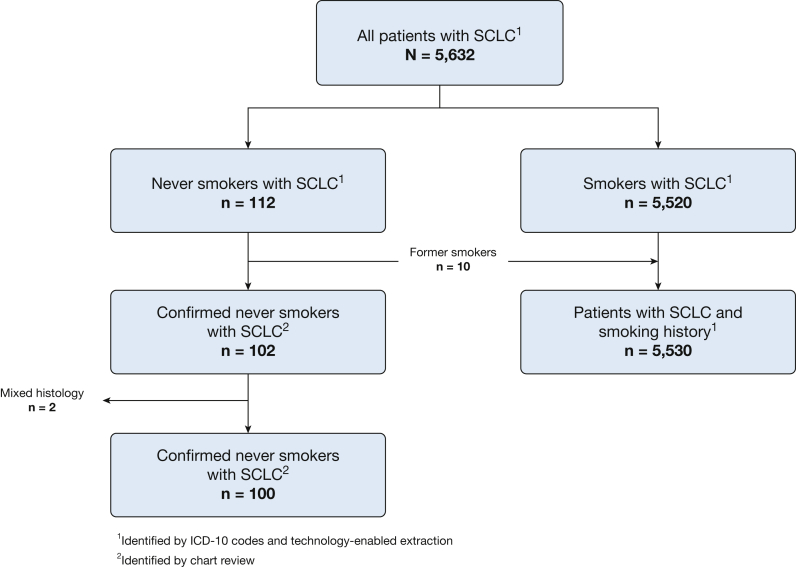
Of 5,632 patients with SCLC, 112 initially were identified as never smokers (Fig 1). Ten patients who were former smokers and two patients with mixed histologic findings were excluded after a second round of chart review. The final cohort consisted of 100 patients (1.8%) with SCLC with no smoking history and 5,530 patients with SCLC with smoking history.
Figure 1.
Never smoker SCLC cohort selection. ICD = International Classification of Diseases; SCLC = small cell lung cancer.
Clinical characteristics of never smokers with SCLC are summarized in Table 1. Compared with smokers with SCLC, never smokers with SCLC were more likely to be female (66.0% vs 52.4%; P = .009) and present with extensive-stage disease (70.0% vs 62.2%; P = .028). Compared with smokers, the age distribution of never smokers with SCLC was remarkable for higher frequencies in the following age groups: 35 to 49 years (7.0% vs 3.0%) and ≥80 years (17.0% vs 8.2%). No difference was observed between never smokers and smokers in terms of the region of residence or performance status at diagnosis.
Table 1.
Comparison of Clinical Characteristics Between Patients Who Were Never Smokers and Patients Who Were Smokers
| Variable | Never Smokers (n = 100), No. (%) | Smokers (n = 5,530), No. (%) | P Value |
|---|---|---|---|
| Age at diagnosis, y | .006 | ||
| <35 | 0 | 6 (0.1) | |
| 35-49 | 7 (7.0) | 165 (3.0) | |
| 50-64 | 31 (31.0) | 2,013 (36.4) | |
| 65-79 | 45 (45.0) | 2,894 (52.3) | |
| ≥80 | 17 (17.0) | 452 (8.2) | |
| Sex | .009 | ||
| Female | 66 (66.0) | 2,898 (52.4) | |
| Male | 34 (34.0) | 2,632 (47.6) | |
| Race | < .001 | ||
| Asian | 4 (4.0) | 43 (0.8) | |
| Black | 7 (7.0) | 311 (5.6) | |
| Not known | 7 (7.0) | 560 (10.1) | |
| Other races | 21 (21.0) | 474 (8.6) | |
| White | 61 (61.0) | 4,142 (74.9) | |
| Region | .516 | ||
| Midwest | 15 (15.0) | 1,065 (19.3) | |
| Northeast | 22 (22.0) | 1,038 (18.8) | |
| Other/unknown | 6 (6.0) | 397 (7.2) | |
| South | 41 (41.0) | 2,380 (43.0) | |
| West | 16 (16.0) | 650 (11.8) | |
| Stage | .028 | ||
| Limited disease | 20 (20.0) | 1,761 (31.8) | |
| Extensive disease | 70 (70.0) | 3,440 (62.2) | |
| Unknown/not documented | 10 (10.0) | 329 (6.0) | |
| Group stage | .258 | ||
| IA/B | 3 (3.0) | 190 (3.4) | |
| IIA/B | 4 (4.0) | 184 (3.3) | |
| IIIA/B | 8 (8.0) | 871 (15.8) | |
| IV | 61 (61.0) | 3,079 (55.7) | |
| Unknown | 24 (24.0) | 1,206 (21.8) | |
| Eastern Cooperative Oncology Group Performance Status at diagnosis | .288 | ||
| 0 | 17 (17.0) | 648 (11.7) | |
| 1 | 13 (13.0) | 1,065 (19.3) | |
| 2 | 9 (9.0) | 539 (9.8) | |
| 3 | 4 (4.0) | 150 (2.7) | |
| 4 | 0 | 3 (0.1) | |
| Not known | 57 (57.0) | 3,125 (56.5) | |
| Serum sodium | .970 | ||
| >140 mEq/L | 28 (28.0) | 1,490 (26.9) | |
| 120-139 mEq/L | 55 (55.0) | 3,072 (55.6) | |
| Unknown | 17 (17.0) | 968 (17.5) |
Twenty-eight of the never smoker patients (28.0%) had structured ICD codes for other primary malignancies. The most common malignancies were breast (N = 6; 6.0%) and prostate (N = 4; 4%) cancers. Other malignancies that were identified in less than four patients each were anal, colon, bladder, meningeal, rectal, thyroid, and uterine cancers and Hodgkin’s lymphoma, basal cell carcinoma, and Merkel cell carcinoma. Five of the never smoker patients (5.0%) had organ transplants (lung, liver, or kidney) or pulmonary fibrosis.
Treatments that were received by never smokers with SCLC, as captured within the network of the EHR-derived database, are summarized in e-Table 1. Most patients received first-line therapy that consisted of cisplatin or carboplatin and etoposide. Commonly used second-line treatments were topotecan, paclitaxel, and immune checkpoint inhibitors.
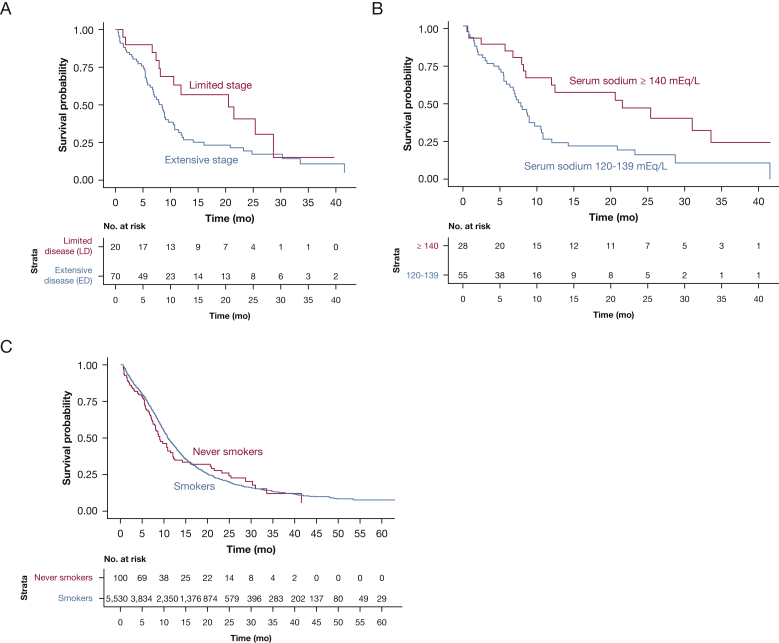
Based on univariate analyses, extensive-stage disease and low serum sodium at diagnosis were identified as poor prognostic factors among never smokers. Patients with limited-stage disease trended towards longer survival compared with patients with extensive-stage disease (median OS, 20.6 months [95% CI, 10.6 to not reached] vs 8.5 months [95% CI, 6.8-10.8 months]; hazard ratio, 1.82 [95% CI, 0.97-3.41]; P = .062) (Fig 2A). Median OS for patients with serum sodium ≥140 mEq/L at diagnosis was 21.5 months (95% CI, 8.5 to not reached) compared with 7.5 months (95% CI, 6.4-10.6) for patients with serum sodium 120 to 139 mEq/L (hazard ratio, 2.24 [95% CI, 1.23-4.08]; P = .009) (Fig 2B). The estimates yielded by the univariate and multivariable Cox proportional hazards models are shown in e-Table 2.
Figure 2.
A-C, Overall survival of patients with small cell lung cancer who were never smokers and smokers: A, stratified by stage at diagnosis; B, stratified by serum sodium at diagnosis; and C, based on smoking status.
Next, we compared OS between smokers and never smokers. In an unadjusted analysis, there was no statistically significant difference in OS between never smokers and smokers. Median OS of never smokers with SCLC was 9.0 months (95% CI, 7.8-12.0) and 11.0 months (95% CI, 10.7-11.3) for smokers (Fig 2C). OS was not statistically different between smokers and never smokers after adjustment for additional covariates (adjusted hazard ratio, 1.24; 95% CI, 0.94-1.64; P = .127) (e-Table 3). Clinical characteristics, prognostic factors, and survival were similar when the analyses recategorized 16 patients with a history of second-hand smoke exposure as smokers (e-Fig 1).
We then examined the genomic characteristics of never smokers with SCLC (Table 2; e-Fig 2) using the FoundationOne targeted sequencing platform. The genomic cohort consisted of 329 patients with SCLC that included 320 smokers and 9 never smokers. In this subgroup, never smokers (n = 9; 2.7%) were less likely to have an alteration in TP53 (33.3% vs 94.1%; P < .001), RB1 (22.2% vs 74.1%; P = .002), and more likely to have an epidermal growth factor receptor (EGFR) alteration (11.1% vs 0.94%). Never smokers had a lower tumor mutation burden (TMB), with a median of 1.74 mutations/Mb (interquartile range, 0.87-3.04) compared with the smokers (median, 8.70 mutations/Mb; interquartile range, 5.22-13.0).
Table 2.
Comparison of Genomic Characteristics Between Patients Who Were Never Smokers and Patients Who Were Smokers
| Variable | Never Smokers (n = 9), No. (%) | Smokers (n = 320), No. (%) | P Value |
|---|---|---|---|
| Age at diagnosis, y | .624 | ||
| 0-34 | 0 | 1 (0.3) | |
| 35-49 | 1 (11.1) | 14 (4.4) | |
| 50-64 | 4 (44.4) | 154 (48.1) | |
| 65-79 | 4 (44.4) | 135 (42.2) | |
| ≥80 | 0 | 16 (5.0) | |
| Sex | 1 | ||
| Female | 4 (44.4) | 150 (46.9) | |
| Male | 5 (55.6) | 170 (53.1) | |
| Race | .012 | ||
| Asian | 0 | 4 (1.4) | |
| Black | 3 (33.3) | 9 (3.1) | |
| Other races | 0 | 27 (9.2) | |
| White | 6 (66.7) | 255 (86.4) | |
| Stage | .226 | ||
| Extensive disease | 9 (100) | 225 (70.3) | |
| Limited disease | 0 | 88 (27.5) | |
| Unknown/not documented | 0 | 7 (2.19) | |
| Eastern Cooperative Oncology Group Performance Status at diagnosis | .71 | ||
| 0 | 1 (11.1) | 23 (7.2) | |
| 1 | 0 | 31 (9.7) | |
| >1 | 0 | 9 (2.8) | |
| Missing | 8 (88.9) | 257 (80.3) | |
| Clinical care setting | 1 | ||
| Academic | 0 | 14 (4.4) | |
| Community | 9 (100) | 306 (95.6) | |
| TP53 known/likely functional status alteration | < .001 | ||
| No TP53 alteration | 6 (66.7) | 19 (5.9) | |
| TP53 alteration | 3 (33.3) | 301 (94.1) | |
| TP53 alteration type where altered | 1 | ||
| Deletion | 0 | 3 (1.0) | |
| Short variant | 3 (100) | 298 (99.0) | |
| RB1 known/likely functional status alteration | .002 | ||
| No RB1 alteration | 7 (77.8) | 83 (25.9) | |
| RB1 alteration | 2 (22.2) | 237 (74.1) | |
| RB1 alteration type where altered | .258 | ||
| Deletion | 1 (50.0) | 28 (11.8) | |
| Rearrangement | 0 | 4 (1.7) | |
| Short variant | 1 (50.0) | 205 (86.5) | |
| EGFR known/likely functional status alteration | .105 | ||
| EGFR alteration | 1 (11.1) | 3 (0.9) | |
| No EGFR alteration | 8 (88.9) | 317 (99.1) | |
| EGFR alteration type where altered | .25 | ||
| Rearrangement | 1 (100) | 0 | |
| Short variant | 0 | 3 (100) | |
| EGFR coalterations | .25 | ||
| EGFR + TP53 | 0 | 3 (100) | |
| EGFR only | 1 (100) | 0 | |
| Tumor mutation burden scorea | 1.74 [0.87;3.04] | 8.70 [5.22;13.0] | .003 |
| Tumor mutation burden status | .005 | ||
| High (≥20 mutations/mB) | 1 (12.5) | 16 (5.2) | |
| Medium (<20, ≥5.5 mutations/mB) | 1 (12.5) | 209 (67.4) | |
| Low (<5.5 mutations/mB) | 6 (75.0) | 85 (27.4) | |
| TP53, RB1, or EGFR alteration present | < .001 | ||
| No TP53, RB1, or EGFR alteration | 5 (55.6) | 14 (4.4) | |
| TP53, RB1, or EGFR alteration | 4 (44.4) | 306 (95.6) |
EGFR = epidermal growth factor receptor.
Data are given as mutations/mB.
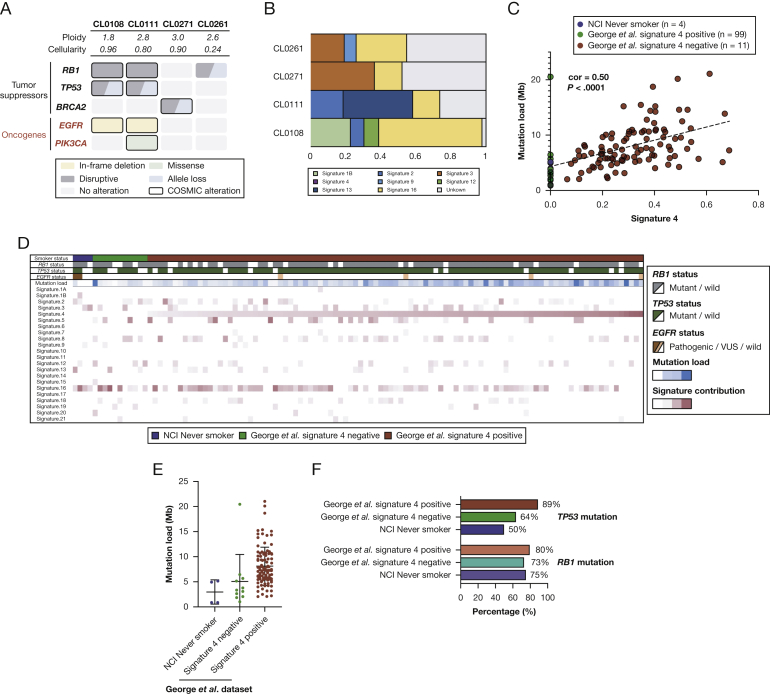
To better understand the genomic characteristics of never smoker SCLC, we performed whole exome sequencing of four never smoker SCLC tumors (e-Table 4) from patients who enrolled on a NCI natural history study. The diagnoses were confirmed by experienced thoracic pathologists who were blinded to the smoking status. All four patients were diagnosed with extensive-stage disease and received combination chemotherapy. Their median age was 61 years (range, 41-66 years). Two of four tumors had mutations in both TP53 and RB1; one tumor had an RB1 mutation, but no TP53 mutation; and one tumor had neither TP53 nor RB1 mutated (Fig 3A). TMB of these tumors ranged from 0.86 to 5.16 (0.86, 3.94, 4.96, and 5.16), with a mean of 3.73 mutations/Mb. Two patients, neither of whom had prior lung adenocarcinoma, had EGFR sensitizing mutations, in both cases exon 19 deletions. One patient had a somatic truncating mutation of BRCA2 (c.6645C>G, p.Tyr2215Ter) that was confirmed to be present at the germline level. This mutation is known to be associated with hereditary breast and ovarian cancers (ClinVar RCV000113621.3). Pathogenic germline mutations were not identified in the other three cases.
Figure 3.
A-E, Genomic characterization and signatures in never smokers. A, Genomic characteristics and B, mutational signatures of NCI never smoker small cell lung cancer. C, Correlation between signature 4 and tumor mutation burden; D, heat map shows smoking status, key genetic alterations, and mutational signatures; E, distribution of tumor mutation burden; and F, RB1 and TP53 mutations among the dataset of George et al19 and the NCI never smoker SCLC. EGFR = epidermal growth factor receptor; NCI = National Cancer Institute. See Figure 1 legend for expansion of other abbreviation.
Recent studies suggest that mutational signatures hint at the causative origins of cancer, especially when the environmental exposures have large effect sizes (eg, UV light with melanoma and smoking with lung cancer).17 Signature 4 characterized by C>A mutations is a direct mutational consequence of tobacco-induced misreplication of DNA. Signature 4 is found only in tobacco-related cancers that originate in tissues that are exposed directly to tobacco smoke.18 To characterize the mutational processes in never smoker SCLC, we investigated their mutational signatures and relative contributions of each signature (Fig 3B). Reassuringly, signature 4 was absent from all four NCI never smoker SCLC. Signature 3 associated with failure of DNA double-strand break-repair by homologous recombination was observed preferentially (36.5% of mutations) in the mutational spectrum of the tumor with mutated BRCA2. Other prevalent mutational signatures were the APOBEC-associated mutational signature (signature 13; 39.3%) and a signature associated with transcription coupled damage (signature 16; 28.5% and 58.7%).
To further characterize the genomic characteristics of never smokers, we integrated NCI never-smoker SCLC exomes and a published dataset of 110 SCLC whole genome sequences (George et al19 dataset). In the combined dataset, signature 4 showed a highly significant correlation with TMB (correlation, 0.50; P < .0001), which is consistent with the known link between smoking and increased mutational load (Fig 3C). Interestingly, in addition to the NCI never smoker SCLC, several cases in the dataset of George et al19 lacked signature 4 (Fig 3C, D). Specifically, 11 of 110 cases (10%) in the dataset were signature 4-negative, which suggests that these cases likely may not be smoking related.
NCI never smoker SCLC had lower TMB (3.7 vs 8.1 mutations/Mb) (Fig 3E) and a lower frequency of TP53 (50% vs 89%) (Fig 3F) mutations compared with signature 4-positive SCLC from George et al.19 This was not the case for RB1 (75% vs 80%) (Fig 3F). As with NCI never smoker SCLC, signature 4-negative SCLC also had lower TMB and a lower frequency of TP53 mutations than signature 4-positive tumors from the database of George et al19 (Fig 3E, F). Collectively, exome sequencing of the four NCI never smoker SCLC and the published dataset validates some of the key findings from the targeted sequencing that included lower TMB, a lower frequency of TP53 mutations, and the possible presence of EGFR mutations among never smoker SCLC.
Discussion
Patients with SCLC have one of the worst survival rates of all patients with cancer. Clinically relevant SCLC subsets that could inform prognosis, clinical course, or treatment have not been identified. The incidence, clinical and genomic characteristics, and outcomes of SCLC arising in patients who have never smoked has not been described previously. In our analyses of a nationally representative cohort of >5,600 patients, approximately 2% of patients with SCLC were never smokers. Compared with smokers with SCLC, a significantly higher proportion of never smokers were women and were more likely to present with extensive-stage disease. Serum sodium level and stage were statistically significant prognostic factors. There were no significant differences of risk of death between smokers and never smokers after adjustment for key covariates, specifically age, sex, race, stage, and sodium serum. Genomic analyses in a limited number of patients demonstrated a lower TMB and a lower frequency of TP53 mutations among never smoker SCLC. This analysis to our knowledge represents the largest report of SCLC in patients who have never smoked. The sex- and age-specific distribution of SCLC among never smokers, along with potential differences identified by genomic analyses, may be reflective of a distinct biologic condition of these tumors.
Although it is well-established that the majority of SCLCs are caused by tobacco smoking, a minority of patients have no known history of smoking, and the cause of cancer in these patients is not clear. Factors other than active tobacco use are thought to account for approximately 15% of cases of lung cancer in women and 10% in men.4 Major risk factors among never smokers include exposure to second-hand smoke, radiation, occupational exposures (such as asbestos),20 domestic radon,21 previous lung disease,22 organ transplantation,23 and genetic factors.24 Approximately 20% of the patients in this study had known risk factors such as second-hand smoking, pulmonary fibrosis, and organ transplants. Additionally, one patient had an inherited loss-of-function BRCA2 mutation with loss of heterozygosity of the wild-type allele that could have predisposed to SCLC. Although larger studies would be required to ascertain the cause, this study suggest that, in most cases, SCLC in never smokers cannot be explained by known risk factors.
One notable finding from this study that initially was observed in the Foundation Medicine cohort and confirmed in the NCI cohort and in the dataset of George et al19 is the lower TMB and TP53 mutations among never smokers. TP53 along with RB1 is inactivated almost universally in SCLC.19 In SCLC, these inactivation events are believed to be the essential, initiating molecular events, usually from normal neuroendocrine cells that are located predominantly in the larger airways.25 One possibility that might explain the lower rates of TP53 mutations in this cohort is an alternative mechanism of deregulation in never smokers (intronic or epigenetic changes or other mechanisms) that were not identified by our genomic analyses. Another possibility is that SCLC in these patients have a nonneuroendocrine cell of origin, such as the alveolar type 2 cells, which are prevalent in the distal lung alveoli that may transform to SCLC independent of TP53 mutations.25 A third possibility is that oncogenic events that include activating EGFR mutations and/or mutations in tumor suppressor genes other than TP53 may be driving SCLC in never smokers. Supporting the latter possibility, one never smoker patient with SCLC in this study had a pathogenic BRCA2 mutation, with no alterations detected in TP53 or RB1.
Previous studies have reported instances of EGFR mutations in de novo SCLC.8,26 In this study, 3 of 13 cases (23%) in the combined NCI/Foundation Medicine cohort had an EGFR alteration. Although patients with mixed histologic condition and previous lung cancers were excluded, these cases, nevertheless, could potentially represent sampling error in the setting of tumor heterogeneity. It is also possible that these cases represent SCLC transformation from EGFR-mutated adenocarcinoma where the SCLC clone emerged in the absence of known effective EGFR inhibition.27
In several cancers, TMB is elevated in smokers vs never smokers, and mutational signature 4 is correlated with pack years smoked.18 Consistent with these observations and with the notions of increased mutation load caused by tobacco smoke that contributes to an increased cancer risk, we found higher TMB in SCLC with more signature 4 mutations. Interestingly, we found a subset of SCLC in the dataset of George et al19 where signature 4 was absent, which suggests that tobacco smoke constituents may not have contributed substantially to tumorigenesis in these cases.18 These signature 4-negative tumors shared several features that included lower TMB and lower frequency of TP53 mutations with the four confirmed NCI never smokers, pointing possibly to a different SCLC pathogenesis in these cases. Despite this, we did not observe a significant difference in survival between never smokers and smokers, possibly because the outcomes are uniformly poor regardless of genomic background due to the lack of effective therapies, especially at relapse. Underscoring this notion, no clear genomically driven subsets of prognostic relevance have been identified to date in SCLC.
Most cancer registries including SEER (Surveillance, Epidemiology, and End Results) do not collect information on smoking history. A singular strength of our analysis is the use of nationwide, high-quality population-based data that included patient-level data on smoking status with linked tumor genomic data. The data represent the general SCLC population, not a selected subgroup, drawn from regions throughout the United States. The demographic characteristics of smokers with SCLC that were included in this study mirrored that of the SEER SCLC patient population (2003-2012; n = 16,920; data not shown).
Limitations of this study include the lack of independent pathologic review of the Flatiron cohort. Thus, misclassification of other neuroendocrine tumors as SCLC cannot be ruled out entirely. For example, one patient had an ICD code for Merkel cell carcinoma, which could represent either miscoding or disease misclassification. Nevertheless, the pathology report, clinician confirmation of diagnosis, and treatment regimens were all consistent with SCLC. Patients who had been exposed to second-hand smoke (n = 16) were found during chart review. These patients were retained for the primary analysis as never smokers; a sensitivity analysis showed no difference in results when these patients were recategorized. Patients with metastatic small-cell neuroendocrine tumors that could have come from sites other than lung (ie, a lung origin could not be confirmed; n = 12) were also found during a chart review. These patients remained in the cohort because the treating clinicians had determined a diagnosis of SCLC and followed treatment protocols for the disease.
In conclusion, although SCLC in never smokers is uncommon, the distinctive clinical and genomic characteristics observed in never smokers (including age and sex; lower frequency of TMB, RB1 and TP53 mutations; and absence of mutational signature 4) point to a different disease biology from SCLC in smokers. Further exploration of genomic characteristics of never smoker SCLC that can be challenging due to effects of passive smoking, misreporting of smoking habits, or annotation errors may benefit from a mutational signature-based approach and could improve our understanding of SCLC tumorigenesis. It remains to be seen whether such knowledge could be leveraged to personalize therapy for never smokers with SCLC.
Acknowledgments
Author contributions: A. T. and K. R. C. had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. I. M., C. T., and L. P. contributed equally to this work. A. T, E. S., A. Z. T., and K. R. C. were responsible for the concept and design of the study. All of the authors contributed to the acquisition, analysis, or interpretation of data, to the drafting of the manuscript, and to critical revision of the manuscript for important intellectual content. A. T., A. Z. T., and K. R. C. obtained funding and participated in a supervisory capacity.
Financial/nonfinancial disclosures: The authors have reported to CHEST the following: A. Z. T., J. S., K. M., and K. R. C. report employment at Flatiron Health, Inc, which is an independent subsidiary of the Roche Group, along with stock ownership in Roche and equity ownership in Flatiron Health Inc (initiated before acquisition by Roche in April 2018). K. R. C. is also an employee of Washington University School of Medicine in St. Louis, MO, is an employee of the US Department of Veterans Affairs, and was formerly a consultant for the Roche Group. G. F. , G. L., and S. A. are employed by Foundation Medicine Inc and own stock in Foundation Medicine Inc. The remaining authors declare no conflict of interest. None declared (A. T., I. M., C. T., L. P., N. T., S. K., S. N., V. R., U. G., E. S., J. F., C. A. M., I. I. W., J. S. W., J. K., E. S.).
Other contributions: The authors gratefully acknowledge the patients who participated in the study as well as the efforts of Kellie Ciofalo and Nicholas Brown (Flatiron Health Inc). This work represents an academic and industry partnership. Both academic and industry authors were involved in study design, data collection, data analysis, data interpretation, and writing of the report.
Additional information: The e-Appendix, e-Figures, and e-Tables can be found in the Supplemental Materials section of the online article.
Footnotes
FUNDING/SUPPORT: This study was supported by the Center for Cancer Research, the Intramural Program of the National Cancer Institute [Grant ZIA BC 011793 to A. T.].
Supplementary Data
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Sun S., Schiller J.H., Gazdar A.F. Lung cancer in never smokers: a different disease. Nat Rev Cancer. 2007;7(10):778–790. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 3.Subramanian J., Govindan R. Molecular genetics of lung cancer in people who have never smoked. Lancet Oncol. 2008;9(7):676–682. doi: 10.1016/S1470-2045(08)70174-8. [DOI] [PubMed] [Google Scholar]
- 4.Jemal A., Miller K.D., Ma J. Higher lung cancer incidence in young women than young men in the United States. N Engl J Med. 2018;378(21):1999–2009. doi: 10.1056/NEJMoa1715907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kenfield S.A., Wei E.K., Stampfer M.J., Rosner B.A., Colditz G.A. Comparison of aspects of smoking among the four histological types of lung cancer. Tob Control. 2008;17(3):198–204. doi: 10.1136/tc.2007.022582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwiatkowski N., Zhang T., Rahl P.B. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature. 2014;511(7511):616–620. doi: 10.1038/nature13393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ou S.H., Ziogas A., Zell J.A. Prognostic factors for survival in extensive stage small cell lung cancer (ED-SCLC): the importance of smoking history, socioeconomic and marital statuses, and ethnicity. J Thorac Oncol. 2009;4(1):37–43. doi: 10.1097/JTO.0b013e31819140fb. [DOI] [PubMed] [Google Scholar]
- 8.Varghese A.M., Zakowski M.F., Yu H.A. Small-cell lung cancers in patients who never smoked cigarettes. J Thorac Oncol. 2014;9(6):892–896. doi: 10.1097/JTO.0000000000000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torres-Duran M., Ruano-Ravina A., Kelsey K.T. Small cell lung cancer in never-smokers. Eur Respir J. 2016;47(3):947–953. doi: 10.1183/13993003.01524-2015. [DOI] [PubMed] [Google Scholar]
- 10.Tavares e Castro A., Clemente J., Carvalho L., Freitas S., Cemlyn-Jones J. Small-cell lung cancer in never-smokers: a case series. Lung Cancer. 2016;93:82–87. doi: 10.1016/j.lungcan.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 11.Sun J.M., Choi Y.L., Ji J.H. Small-cell lung cancer detection in never-smokers: clinical characteristics and multigene mutation profiling using targeted next-generation sequencing. Ann Oncol. 2015;26(1):161–166. doi: 10.1093/annonc/mdu504. [DOI] [PubMed] [Google Scholar]
- 12.Kurahara Y., Kawaguchi T., Tachibana K. Small-cell lung cancer in never-smokers: a case series with information on family history of cancer and environmental tobacco smoke. Clin Lung Cancer. 2012;13(1):75–79. doi: 10.1016/j.cllc.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Abernethy A.P., Gippetti J., Parulkar R., Revol C. Use of electronic health record data for quality reporting. J Oncol Pract. 2017;13(8):530–534. doi: 10.1200/JOP.2017.024224. [DOI] [PubMed] [Google Scholar]
- 14.Agarwala V., Khozin S., Singal G. Real-world evidence in support of precision medicine: clinico-genomic cancer data as a case study. Health Aff (Millwood) 2018;37(5):765–772. doi: 10.1377/hlthaff.2017.1579. [DOI] [PubMed] [Google Scholar]
- 15.Singal G., Miller P.G., Agarwala V. Association of patient characteristics and tumor genomics with clinical outcomes among patients with non-small cell lung cancer using a clinicogenomic database. JAMA. 2019;321(14):1391–1399. doi: 10.1001/jama.2019.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang W., Brohl A.S., Patidar R. Multidimensional clinomics for precision therapy of children and adolescent young adults with relapsed and refractory cancer: a report from the Center for Cancer Research. Clin Cancer Res. 2016;22(15):3810–3820. doi: 10.1158/1078-0432.CCR-15-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kucab J.E., Zou X., Morganella S. A compendium of mutational signatures of environmental agents. Cell. 2019;177(4):821–836.e16. doi: 10.1016/j.cell.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alexandrov L.B., Ju Y.S., Haase K. Mutational signatures associated with tobacco smoking in human cancer. Science. 2016;354(6312):618–622. doi: 10.1126/science.aag0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.George J., Lim J.S., Jang S.J. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524(7563):47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samet J.M., Avila-Tang E., Boffetta P. Lung cancer in never smokers: clinical epidemiology and environmental risk factors. Clin Cancer Res. 2009;15(18):5626–5645. doi: 10.1158/1078-0432.CCR-09-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darby S., Hill D., Auvinen A. Radon in homes and risk of lung cancer: collaborative analysis of individual data from 13 European case-control studies. BMJ. 2005;330(7485):223. doi: 10.1136/bmj.38308.477650.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Littman A.J., Thornquist M.D., White E., Jackson L.A., Goodman G.E., Vaughan T.L. Prior lung disease and risk of lung cancer in a large prospective study. Cancer Causes Control. 2004;15(8):819–827. doi: 10.1023/B:CACO.0000043432.71626.45. [DOI] [PubMed] [Google Scholar]
- 23.Engels E.A., Pfeiffer R.M., Fraumeni J.F., Jr. Spectrum of cancer risk among US solid organ transplant recipients. JAMA. 2011;306(17):1891–1901. doi: 10.1001/jama.2011.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y., McKay J.D., Rafnar T. Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nat Genet. 2014;46(7):736–741. doi: 10.1038/ng.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Semenova E.A., Nagel R., Berns A. Origins, genetic landscape, and emerging therapies of small cell lung cancer. Genes Dev. 2015;29(14):1447–1462. doi: 10.1101/gad.263145.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le X., Desai N.V., Majid A. De novo pulmonary small cell carcinomas and large cell neuroendocrine carcinomas harboring EGFR mutations: lack of response to EGFR inhibitors. Lung Cancer. 2015;88(1):70–73. doi: 10.1016/j.lungcan.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee J.K., Lee J., Kim S. Clonal history and genetic predictors of transformation into small-cell carcinomas from lung adenocarcinomas. J Clin Oncol. 2017;35(26):3065–3074. doi: 10.1200/JCO.2016.71.9096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.