Abstract
The pine mushroom (Tricholoma matsutake; Agaricales, Tricholomataceae) is an ectomycorrhizal fungus that produces a commercially valuable, edible mushrooms. Attempts to artificially cultivate T. matsutake has so far been unsuccessful. One method used to induce T. matsutake to produce fruiting bodies of in the wild is shiro (mycelial aggregations of T. matsutake) transplantation. In vitro ectomycorrhization of T. matsutake with seedlings of Pinus densiflora has been successful, but field trials showed limited production of fruiting bodies. Few studies have been done to test what happens after transplantation in the wild, whether T. matsutake persists on the pine seedling roots or gets replaced by other fungi. Here, we investigated the composition and the interaction of the root fungal microbiome of P. densiflora seedlings inoculated with T. matsutake over a 3 year period after field transplantation, using high-throughput sequencing. We found a decline of T. matsutake colonization on pine roots and succession of mycorrhizal fungi as P. densiflora seedlings grew. Early on, roots were colonized by fast-growing, saprotrophic Ascomycota, then later replaced by early stage ectomycorrhiza such as Wilcoxina. At the end, more competitive Suillus species dominated the host roots. Most of the major OTUs had negative or neutral correlation with T. matsutake, but several saprotrophic/plant pathogenic/mycoparasitic species in genera Fusarium, Oidiodendron, and Trichoderma had positive correlation with T. matsutake. Four keystone species were identified during succession; two species (Fusarium oxysporum, and F. trincintum) had a positive correlation with T. matsutake, while the other two had a negative correlation (Suillus granulatus, Cylindrocarpon pauciseptatum). These findings have important implications for further studies on the artificial cultivation of T. matsutake.
Keywords: fungal diversity, microbiome, network analysis, Pinus densiflora, Tricholoma matsutake, pine mushroom, ectomycorrhizal fungi
Introduction
Ectomycorrhizal fungi are one of the most common forms of plant-fungal root symbioses in woody plants (Brundrett, 2009; Van Der Heijden et al., 2015), and improve nutrition and stress resistance of the host plant (Smith and Read, 2010; Berendsen et al., 2012; Van Der Heijden et al., 2015). Ectomycorrhizal fungi compete with each other to colonize root tips (Koide et al., 2005; Kennedy et al., 2009; Bakker et al., 2014) or co-exist (Perry et al., 1989; Yamamoto et al., 2014). Succession of the mycorrhizal community was reported in several host plants (Twieg et al., 2007). This phenomenon not only occurs in mature trees, but also in seedlings, where the dominant ectomycorrhizal taxa can change (Matsuda et al., 2009; Obase et al., 2009). Early stage ectomycorrhizal fungi (e.g., members of Inocybe, Rhizopogon, or Suillus) require small amount of carbon from hosts and are usually found in pine seedling in disturbed area (Colpaert et al., 1996; Sim and Eom, 2009). Arrival sequence of ectomycorrhizal fungi often influences colonization at early stages, with negative consequences for later colonizers (Alford and Wilbur, 1985; Shorrocks and Bingley, 1994). This phenomenon is called the priority effect, and has been reported in the early stage of interaction between ectomycorrhizal fungi and pine seedlings (Kennedy and Bruns, 2005; Fukumi, 2015).
The pine mushroom (Tricholoma matsutake; Agaricales, Tricholomataceae) produces edible fruiting bodies during symbiosis with members of Pinaceae, especially Pinus densiflora (Yamada et al., 2010). Due to its commercial value, artificial cultivation of T. matsutake has been attempted, but thus far been unsuccessful. Three unsuccessful methods to induce fruiting bodies of T. matsutake in the wild are inoculating cultured T. matsutake hyphae in soil (Lee et al., 2007), spraying of T. matsutake spores from fruiting body (Eto and Taniguchi, 2000), and transplanting shiro (aggregate of T. matsutake mycorrhiza) to uninfected pine trees (Kareki and Kawakami, 1985). The last approach of transplanting shiro to uninfected pine trees has been tried extensively in Korea (Park et al., 2007). In vitro ectomycorrhization of T. matsutake has been successful (Yamada et al., 1999, 2006; Saito et al., 2018), but field trials showed limited production of fruiting bodies (Ka et al., 2018). In order for this method to be efficient, T. matsutake must persist the pine seedling roots. Currently, it is unclear after pine seedlings are transplanted to the wild, T. matsutake persists on the pine seedling roots or gets replaced by other fungi.
Advances in high-throughput sequencing have greatly contributed to our understanding the diversity and function of fungi in various environments (Nilsson et al., 2019), and have been used to study the succession of fungal communities (Dickie et al., 2013, 2017; Voříšková et al., 2014; Hannula et al., 2017). In this study, we used high-throughput sequencing to examine the change in the root microbiome of T. matsutake inoculated pine seedlings after transplantation, focusing on the succession of mycorrhiza and interaction between root associated fungi. We hypothesize that (i) there is a significant change in root fungal communities during transplantation and seedling growth, and (ii) there are some fungi with positive or negative correlation with T. matsutake that affect the survival of T. matsutake on pine seedling roots.
Materials and Methods
Study Design and Sample Collection
This experiment was conducted at Gyeongsangbuk-do Forest Environment Research Institute in Gyeongju, South Korea. Tricholoma matsutake strain KBFERI 20T05 (GenBank accession no. AF367417) was cultured in K-liquid media (Park et al., 2007), and transferred to autoclaved culture vessels filled with mixed soil (perlite: peat moss = 80:1) as described by Park et al. (2007). For surface sterilization, P. densiflora seeds were placed in 70% ethanol for 60 s, and transferred to 2% NaClO solution for 4 min. Cleaned seeds were washed 3 times with sterile water then germinated in nutrient broth agar plates (Scharlau). Uncontaminated seedlings were transferred to culture vessels inoculated with T. matsutake in a sterilized culture room, then co-cultured for 3 months in a clean room illuminated with a fluorescent lamp (20°C; 25,000 lux; 24 h). Then, P. densiflora seedlings were moved to a greenhouse filled with autoclaved soil from a nearby pine forest. Sixteen pine seedlings were sampled at 6 different post T. matsutake-inoculation periods: 3 months (M03; in a sterilized culture room), 10, 17, 24, 31, and 38 months (M10, M17, M24, M31, M38; in a greenhouse). In total, 96 seedling roots were harvested.
DNA Extraction
Harvested seedlings were placed on ice, transported to the laboratory at Seoul National University (Seoul, South Korea), and stored at−80°C prior to DNA extraction. We did a preliminary morphological examination of roots to confirm the presence of T. matsutake (Gill et al., 2000; Yamada et al., 2010). Seedling roots were gently washed with running water to remove debris and sterilized with 3% sodium hypochlorite for 2 min. Samples were then washed with distilled water for 5 min. Surface-sterilized roots were cut into 5 cm fragments and air-dried. For each sample, three root fragments were wet with 500 μl of cetyltrimethylammonium bromide buffer (Biosesang, Seongnam, South Korea) and ground with a mortar and pestle. For each sample, genomic DNA was extracted from seedling root using modified CTAB methods (Rogers and Bendich, 1994). We confirmed the presence of T. matsutake in M03 samples with T. matsutake-specific primers (Kim and Han, 2009).
PCR Amplification and High Throughput Sequencing
The fungal internal transcribed spacer 2 (ITS2) region was amplified with primers ITS3 and ITS4 (White et al., 1990) with Illumina sequencing adaptors attached. PCR was conducted 3 times for each samples using AccuPower PCR PreMix kit (Bioneer, Daejeon, South Korea). PCR conditions were as follows: 94°C for 5 min, 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 40 s, and 72°C for 10 min as final extension. PCR products were confirmed on 1% agarose gel (BIOFACT, Daejeon, South Korea) with gel electrophoresis. After purification using the ExpinTM PCR SV kit (GeneAll Biotechnology, Seoul, South Korea), a unique identifier sequence was attached to each PCR products with a second round PCR following the Nextera XT index kit protocol (Illumina, San Diego, CA, United States). Second PCR products were purified as above. Concentration of each amplicon library were measured using a NanoDrop2000 (Thermo Fisher Scientific, Waltham, MA, United States). Amplicon libraries were pooled in equimolar quantities and sequenced using Illumina MiSeq platform at Macrogen (Seoul, South Korea).
Bioinformatics and Statistical Analysis
After sequencing, the raw data were processed using the Quantitative Insights Into Microbial Ecology v.1.8.0. (QIIME) pipeline (Caporaso et al., 2010). Fastq-join was used for merging paired-end sequences. After filtering low-quality sequences (Q < 20, length < 200 bp), 9,513,644 reads were retained for later analyses. Clustering of operational taxonomic units (OTUs) was performed with the open-source sequence search tool Vsearch v. 2.6.2 (Rognes et al., 2016) with 97% similarity level. For taxonomic identification, the most abundant sequence was selected as an OTU’s representative sequence. The UNITE v. 8.0 (Unite Community, 2019) database was used to determine OTU’s taxonomic identity with NCBI BLAST, following the criteria of Tedersoo et al. (2014). We removed chimeric sequences based on the reference database of UCHIME (Edgar et al., 2011). Singleton OTUs and non-fungal sequences were removed, and all samples were rarefied to a minimum number of sequences before further analysis. Taxonomic identity of major OTUs (OTUs with total relative abundance >0.5%) were checked manually with NCBI and UNITE databases (access date: August 26 2020). FUNGuild was used as a database for fungal trophic mode assignment (Nguyen et al., 2016).
Alpha diversity indices (Chao1 richness, Shannon’s diversity, equitability, and Good’s coverage) were calculated in QIIME. Statistical analysis was performed in R software (version 3.6.1, R Core Team, 2019). Kruskal-Wallis tests were performed to compare the diversity indices between sampling times with Dunn’s test as a post hoc test adjusted using the Benjamini and Hochberg method (Benjamini and Hochberg, 1995). Ordination analysis was performed by non-metric multidimensional scaling (NMDS) based on Bray-Curtis dissimilarity index using the phyloseq package (McMurdie and Holmes, 2013). Difference of community compositions among sampling times were tested with permutational multivariate analysis of variance (PERMANOVA) with 999 permutations, using the “adonis” function in the vegan package (Oksanen et al., 2018), and pairwise post hoc tests were done using the pairwiseAdonis package with Bonferroni correction of the Bray-Curtis dissimilarity matrix (Martinez Arbizu, 2017).
To test for correlations between species, Sparse Correlations for Compositional data (SparCC) (Friedman and Alm, 2012) network analysis was performed at the OTU level (OTUs with total relative abundance >0.5%) with the Galaxy-based analysis pipeline (Inter-Domain ecological network analysis pipeline, IDENAP, Feng et al., 2019). The significance of correlation was calculated by comparing the shuffled data from 100 permutations. Following previous studies, correlations with SparCC >0.3 and p < 0.05 were included (Kurtz et al., 2015). The network was visualized with Cytoscape version 3.7.2 (Shannon et al., 2003). Clusters were detected with Markov clustering algorithms (Van Dongen and Abreu-Goodger, 2012). For the overall network, species with high degree, betweenness centrality, and closeness centrality were selected as the keystone taxa. NMDS ordination and network analyses were performed without M03 samples as they were distinctly different from other samples due to high abundance of T. matsutake (>94% in average). Sequencing data were deposited in NCBI Sequence Read Archive (SRA) under Project ID PRJNA638021.
Results
Sequencing Results and Alpha Diversity Indices
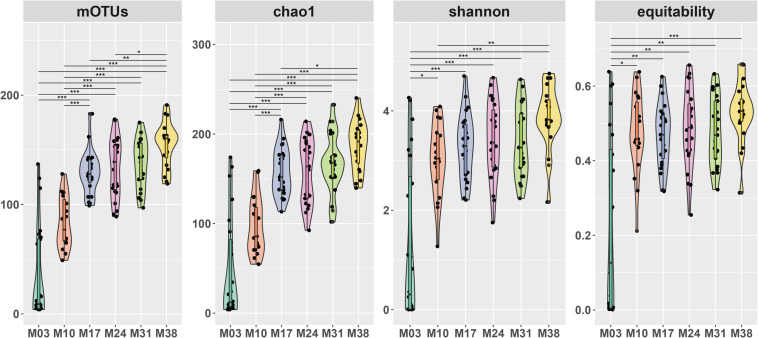
A total of 7,697,559 sequence reads were obtained from 96 samples through Illumina MiSeq sequencing with 25,244–228,456 sequence reads per sample. After rarefaction to 25,000 reads, 826 OTUs (range: 4–191) remained with a Good’s coverage of 0.998–0.999. Based on taxonomic level, the OTUs represented 8 phyla, 28 classes, 89 orders, 188 families, and 327 genera. The number of OTUs significantly increased with the age of P. densiflora seedlings, from 63 OTUs found in M03 (mean = 7.88 OTUs per sample) to 487 OTUs in M38 (mean = 155.88 OTUs per sample) (Figure 1). Chao1 richness, Shannon’s diversity, and equitability also showed significant increase following the growth of P. densiflora seedlings, especially between M03 and other sampling periods (Figure 1).
FIGURE 1.
Alpha diversity of fungal communities of Pinus densiflora seedlings. OTUs: the number of OTUs, Chao1: Chao1 index, Diversity: Shannon’s diversity, equitability: Shannon’s equitability. The number of OTUs were calculated after rarefaction (22,000 reads). Statistically significant differences between sampling times were determined by multiple Kruskal-Wallis tests with Dunn’s test as a post hoc test (*p < 0.05, **p < 0.01, ***p < 0.001).
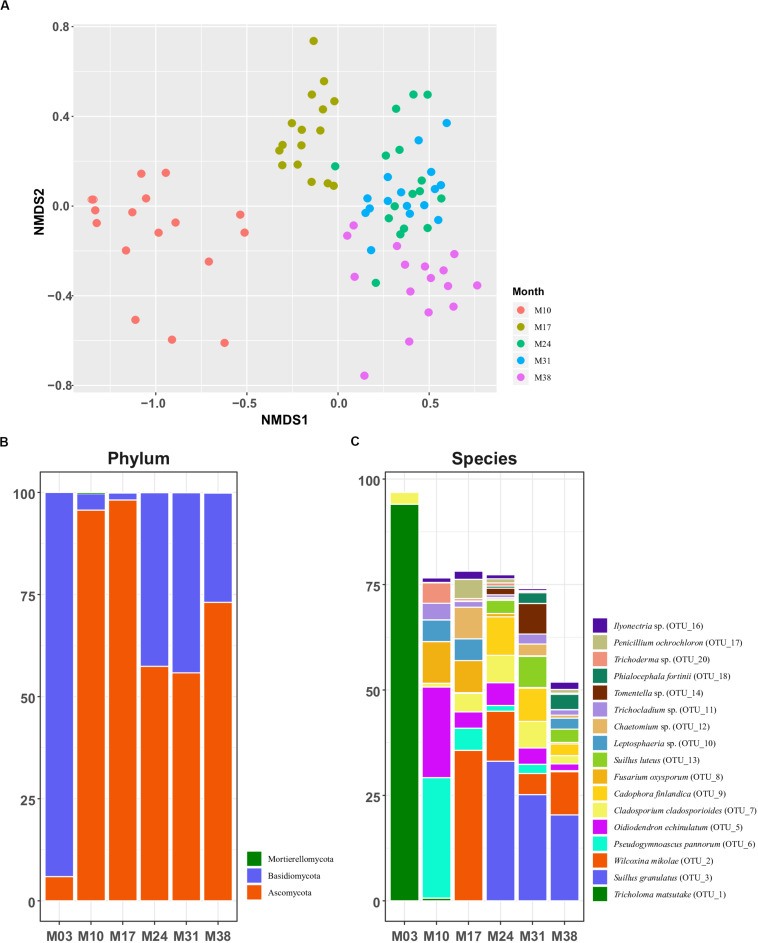
The NMDS ordination of Bray-Curtis dissimilarity based on OTU-level abundance revealed clear separation of fungal communities between most groups, except M24 and M31 (Figure 2A). This result was supported by pairwise adonis tests, where all but the M24-M31 comparison were statistically significant (Supplementary Table 1). We observed a significant shift of the overall fungal community in P. densiflora seedlings over time, based on the adonis analysis (R2 = 39.4%, p = 0.001; Figure 2A and Supplementary Table 1). The relative abundance of T. matsutake drastically decreased after the transplantation to greenhouse (M03 to M10; Figure 2C and Table 1), but T. matsutake was still detected in some samples (15/16 in M10; 8/16 in M17; 4/16 in M24 and M31; 2/16 in M38; Table 1).
FIGURE 2.
Fungal community structures of Pinus densiflora seedlings. (A) NMDS plots based on Bray-Curtis dissimilarity. Relative abundance of (B) major phyla and (C) major species. Taxa with total relative abundance higher than 1% were chosen as major taxa in (C).
TABLE 1.
Average relative abundance and frequency (the number of samples with T. matsutake) of Tricholoma matsutake (OTU 1) in Pinus densiflora seedling roots.
| Sampling groups | M03 | M10 | M17 | M24 | M31 | M38 |
| Sequence reads | 376,056 | 2,625 | 44 | 11 | 5 | 2 |
| Relative abundance (%) | 94.014 | 0.65625 | 0.011 | 0.00275 | 0.00125 | 0.0005 |
| Number of samples with T. matsutake | 16/16 | 15/16 | 8/16 | 4/16 | 4/16 | 2/16 |
Fungal Community Composition in P. densiflora Seedlings
The total abundances of major fungal phyla were relatively high: Ascomycota (64.353%) and Basidiomycota (35.516%). Abundance of the next most abundant phylum, Mortierellomycota, was low at less than 0.1% (Figure 2B). The abundance of Basidiomycota was high during the inoculation stage (M03, 94.1%), but drastically decreased after transplantation (4.0% in M10; 1.7% in M17), being replaced by Ascomycota. The abundance of Basidiomycota increased in M24 (42.5%) and M31 (44.1%), but decreased again in M38 (26.7%). The pattern of relative abundance at the species level was similar to that at the genus level. The most abundant OTUs of each sampling period were T. matsutake (OTU 1, 94.0%) and Cladosporium sp. (OTU 7, 2.81%) in M03, Pseudogymnoascus pannorum (OTU 6, 28.5%) and Oidiodendron echinulatum (OTU 5, 21.5%) in M10, and Wilcoxina mikolae (OTU 2, 35.7%) in M17. After M24, the most abundant OTU was Suillus granulatus (OTU 3, 33.11% in M24; 25.19% in M31; 20.39% in M38) followed by W. mikolae (OTU 2, 11.84%) in M24, S. luteus in M31 (OTU 13, 7.37%), and W. mikolae (10.22%) in M38 (Figure 2C).
Network Features and Correlation Within Fungal Community P. densiflora Seedlings
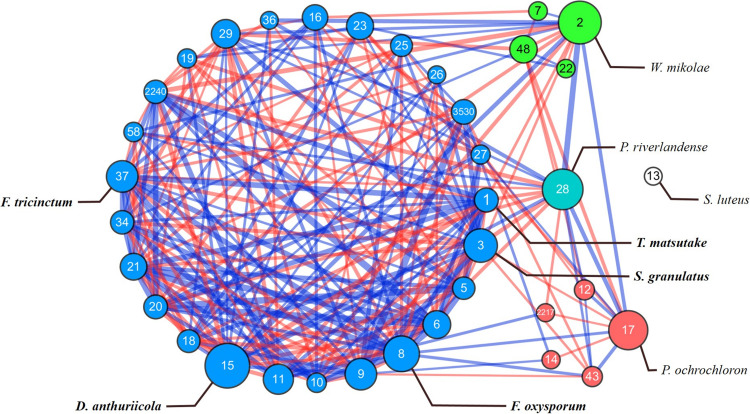
To identify the potential interactions among fungal species in P. densiflora seedlings, SparCC analysis was performed. Tricholoma matsutake (OTU 1) and 35 major OTUs with relative abundances >0.5%, accounting for 83.4% of total sequence reads, were clustered into four groups and one isolated OTU (Table 2). The network had a clustering coefficient of 0.589 and network centralization of 0.308 (Figure 3). Ten fungal OTUs showed significant positive correlations with T. matsutake (Supplementary Tables 1, 2). Most of these OTUs were saprotrophs or plant pathogens, with the exception of O. echinulatum (ericoid mycorrhiza). Based on our selection criteria, four OTUs were identified as keystone species during fungal succession of pine seedling roots: Cylindrocarpon pauciseptatum (OTU 15), Suillus granulatus (OTU 3), Fusarium oxysporum (OTU 8), and Fusarium sp. (OTU 37). All of the keystone taxa belonged to the same cluster. Among these keystone taxa, Cylindrocarpon pauciseptatum (OTU 15) and S. granulatus (OTU 3) showed significant negative correlation with T. matsutake, while Fusarium oxysporum (OTU 8) and Fusarium sp. (OTU 37) showed a positive correlation with T. matsutake (Supplementary Table 2).
TABLE 2.
Major OTUs (relative abundance >0.5%) identity and node properties of the Pinus densiflora root seedling microbiome network.
| ID | Species | Correlation with T. matsutake | Cluster | Betweenness centrality | Closeness centrality | Clustering coefficient | Degree | NCBI blast result | Accession No. | Identity | E-value |
| OTU 1 | Tricholoma matsutake | – | 1 | 0.017 | 0.667 | 0.669 | 17 | Tricholoma matsutake | JF908729 | 100.00% | 0.0 |
| OTU 2 | Wilcoxina mikolae | Negative | 3 | 0.068 | 0.63 | 0.438 | 15 | Wilcoxina mikolae | JQ310817.1 | 99.71% | 2E-179 |
| OTU 3 | Suillus granulatus | Negative | 1 | 0.043 | 0.694 | 0.544 | 19 | Suillus granulatus | AY898617.1 | 98.59% | 0.0 |
| OTU 5 | Oidiodendron echinulatum | Positive | 1 | 0.013 | 0.618 | 0.692 | 14 | Oidiodendron echinulatum | AF062791.1 | 100.00% | 1E-166 |
| OTU 6 | Pseudogymnoascus pannorum | Positive | 1 | 0.027 | 0.63 | 0.603 | 17 | Geomyces pannorum (= Pseudogymnoascus pannorum) | JX131373.1 | 100.00% | 9E-173 |
| OTU 7 | Cladosporium sp. | Neutral | 3 | 0.002 | 0.459 | 0.333 | 3 | Cladosporium cladosporioides | MK268136.1 | 100.00% | 7E-174 |
| OTU 8 | Fusarium oxysporum | Positive | 1 | 0.049 | 0.708 | 0.543 | 21 | Fusarium oxysporum | MT453296.1 | 100.00% | 2E-174 |
| OTU 9 | Cadophora finlandica | Negative | 1 | 0.038 | 0.642 | 0.581 | 17 | Cadophora finlandica | KT182905.1 | 97.82% | 3E-153 |
| OTU 10 | Leptosphaeria sp. | Positive | 1 | 0.004 | 0.586 | 0.821 | 13 | Leptosphaeria sp. | JX238777 | 100.00% | 4E-176 |
| OTU 11 | Trichocladium sp. | Positive | 1 | 0.034 | 0.642 | 0.617 | 16 | Trichocladium sp./ Humicola grisea (= Trichocladium griseum) |
MT348608.1/ MH860993.1 |
100.00%/ 100.00% |
4E-176/ 4E-176 |
| OTU 12 | Chaetomium sp. | Neutral | 2 | 0.006 | 0.507 | 0.5 | 5 | Chaetomium angustispirale/Humicola grisea |
MT453288.1/ MH860993.1 |
100.00%/ 100.00% |
4E-176/ 4E-176 |
| OTU 13 | Suillus luteus | Neutral | 0 | 0 | 0 | 0 | Suillus luteus | KX213740.1 | 100.00% | 0.0 | |
| OTU 14 | Tomentella sp. | Neutral | 2 | 0.001 | 0.459 | 0.333 | 3 | Tomentella tedersooi | NR121359.1 | 95.09% | 7E-180 |
| OTU 15 | Cylindrocarpon pauciseptatum | Negative | 1 | 0.073 | 0.739 | 0.515 | 22 | Dactylonectria pauciseptata (= Cylindrocarpon pauciseptatum) | MK602783.1 | 100.00% | 0.0 |
| OTU 16 | Ilyonectria sp. | Neutral | 1 | 0.021 | 0.531 | 0.778 | 10 | Ilyonectria liriodendri/Ilyonectria destructans | MK602788.1 | 100.00% | 0.0 |
| OTU 17 | Penicillium ochrochloron | Neutral | 2 | 0.058 | 0.596 | 0.236 | 11 | Penicillium ochrochloron | MK450704.1 | 100.00% | 0.0 |
| OTU 18 | Phialocephala fortinii | Negative | 1 | 0.013 | 0.618 | 0.714 | 15 | Phialocephala fortinii | KF313097.1 | 100.00% | 3E-167 |
| OTU 19 | Dactylonectria sp. | Neutral | 1 | 0.004 | 0.531 | 0.778 | 9 | Dactylonectria torresensis/Dactylonectria alcacerensis |
MK602787/ MK602786 |
100.00%/ 100.00% |
2E-180/ 2E-180 |
| OTU 20 | Trichoderma viride | Positive | 1 | 0.016 | 0.63 | 0.657 | 15 | Trichoderma viride | KU202217.1 | 100.00% | 0.0 |
| OTU 21 | Paraphaeosphaeria sporulosa | Positive | 1 | 0.024 | 0.68 | 0.643 | 19 | Paraphaeosphaeria sporulosa | MT576023.1 | 100.00% | 1E-176 |
| OTU 22 | Xenochalara juniperi | Neutral | 3 | 0.002 | 0.442 | 0.333 | 3 | Xenochalara juniperi | JX869564.1 | 100.00% | 3E-172 |
| OTU 23 | Hyaloscyphaceae sp. | Neutral | 1 | 0.025 | 0.567 | 0.472 | 9 | Hyaloscyphaceae sp. | AB986450.1 | 97.61% | 6E-160 |
| OTU 25 | Helotiales sp. | Neutral | 1 | 0.011 | 0.515 | 0.778 | 9 | Helotiales sp. | LC218319.1 | 100.00% | 4E-166 |
| OTU 26 | Trichoderma sp. | Neutral | 1 | 0 | 0.515 | 0.952 | 7 | Trichoderma sp. | MK871291.1 | 100.00% | 0.0 |
| OTU 27 | Oidiodendron rhodogenum | Neutral | 1 | 0.004 | 0.531 | 0.714 | 7 | Oidiodendron rhodogenum | AF062803.1 | 100.00% | 4E-166 |
| OTU 28 | Penicillium sp. | Negative | 4 | 0.062 | 0.654 | 0.442 | 16 | Penicillium sp. | MK450684.1 | 100.00% | 0.0 |
| OTU 29 | Oidiodendron sp. | Neutral | 1 | 0.03 | 0.654 | 0.6 | 16 | Oidiodendron tenuissimum/Oidiodendron griseum |
MH864345.1/ AF062797.1 |
99.69%/ 99.69% |
4E-166/ 6E-165 |
| OTU 34 | Knufia sp. | Positive | 1 | 0.015 | 0.618 | 0.725 | 14 | Knufia sp. | KX610444.1 | 98.79% | 8E-164 |
| OTU 36 | Entrophospora sp. | Neutral | 1 | 0 | 0.515 | 1 | 6 | Entrophospora sp. | AY035666.1 | 99.12% | 3E-172 |
| OTU 37 | Fusarium sp. | Positive | 1 | 0.038 | 0.708 | 0.59 | 21 | Fusarium acuminatum/Fusarium tricinctum |
MT294407.1/ MT453281.1 |
100.00%/ 100.00% |
0.0/ 0.0 |
| OTU 43 | Talaromyces sp. | Neutral | 2 | 0.007 | 0.531 | 0.524 | 7 | Talaromyces amestolkiae | MN511323.1 | 100.00% | 1E-177 |
| OTU 48 | Exophiala sp. | Neutral | 3 | 0.026 | 0.557 | 0.357 | 8 | Exophiala sp. | MF619956.1 | 100.00% | 0.0 |
| OTU 58 | Sebacina sp. | Negative | 1 | 0.005 | 0.586 | 0.711 | 10 | Sebacina sp. | KY271862.1 | 96.61% | 3E-178 |
| OTU 2217 | Corynascella inaequalis | Neutral | 2 | 0.001 | 0.453 | 0.667 | 3 | Corynascella inaequalis | MT453282.1 | 99.41% | 1E-172 |
| OTU 2240 | Pseudogymnoascus pannorum | Positive | 1 | 0.015 | 0.642 | 0.667 | 16 | Pseudogymnoascus pannorum | MH854616.1 | 99.70% | 4E-171 |
| OTU 3530 | Cadophora finlandica | Neutral | 1 | 0.019 | 0.596 | 0.679 | 13 | Cadophora finlandica | DQ069045.1 | 99.03% | 3E-153 |
OTUs in bold font indicate keystone taxa.
FIGURE 3.
SparCC network map in OTU level in microbiome of Pinus densiflora seedlings. Each point represents a fungal species; edges indicate the relationship between species. Each clusters are shown in different colors, with isolated species are marked with white. The size of the nodes follows betweenness centrality scores. The width of the edges follows SparCC correlation coefficients (blue: positive correlation; red: negative correlation). Species with total relative abundance higher than 0.5% were chosen. Only statistically significant edges corresponding to correlations with a magnitude higher than 0.3 (p < 0.05) were drawn. Identity of each OTU is described in Table 2.
Discussion
Change of Fungal Communities in Pine Seedlings After Transplantation
The root fungal communities significantly changed through seedling development. Our results showed that root colonization of T. matsutake dramatically decreased after being transplanted to the greenhouse, and they were replaced by other fungi. As expected, alpha diversity increased when seedlings were transplanted from a controlled environment to a more natural, open environment. After transplantation, fast-growing Ascomycota dominated, and then were replaced by early-stage ectomycorrhizal fungi. Previous studies looked at the mycorrhizal succession in pine seedlings (Peay et al., 2011; Herzog et al., 2019; Rudawska et al., 2019), and we found that the shift of root associated fungi in our study followed the general trend, despite being inoculated with T. matsutake.
After transplantation to the greenhouse, Ascomycota species became dominant in seedling roots. In M10, most of the dominant OTUs were saprotrophs or pathotrophs, such as Pseudogymnoascus and Fusarium, with one exception being Oidiodendron echinulatum, an ericoid mycorrhizal fungus. In M17, the abundance of symbiotrophs (e.g., Wilcoxina mikolae) increased, while pathotrophs decreased. The presence of saprotrophic fungi is commonly reported in roots (Vasiliauskas et al., 2007; Tedersoo and Smith, 2013; Lee et al., 2015; Rincón et al., 2015; Smith et al., 2017), so we believe their presence in our study is not a result of inappropriate sterilization of roots. Among the saprotroph species identified in our study, Pseudogymnoascus pannorum is widely distributed in the soil and adapted to nutrient poor environments (Minnis and Lindner, 2013; Chaturvedi et al., 2018). Previous studies of pine seedling roots also discovered the presence of Pseudogymnoascus species (Menkis and Vasaitis, 2011; Moler and Aho, 2018). Other taxa, like Oidiodendron and Wilcoxina, are well known species that are common in early successional or disturbed ecosystems (Berch et al., 2006; Lee et al., 2012; Lee and Eom, 2013; Rudawska et al., 2019). As these species were absent in samples from M03, and both taxa found in our study are expected to have been dispersed by wind (Horton, 2017). A noteworthy result is the high abundance of Fusarium in M10–M17 samples. Usually, Fusarium is considered a plant pathogen (Gordon, 2017), but Fusarium species have also been found as endophytes of a wide range of wild plants (Kuldau and Yates, 2000; Min et al., 2014). For example, growth-enhancement or pathogen-resistance conferred by non-pathogenic Fusarium species were widely reported (Forsyth et al., 2006; Waweru et al., 2014). Their role is uncertain in our study, and further study would be needed to understand Fusarium’s role in roots of pine seedlings.
After M24, we witnessed an increase in proportion of ectomycorrhizal (Suillus and Tomentella) and endophytic fungi (Cadophora and Phialocephala), which are considered common fungi in an early successional stage (Colpaert et al., 1996; Berch et al., 2006; Sim and Eom, 2009; Lee et al., 2012; Lee and Eom, 2013; Lee and Koo, 2016). In particular, Suillus species are known to be important in the establishment of pine seedlings (Hayward et al., 2015). Suillus species might be more competitive than other mycorrhizal fungi found in first year, such as Wilcoxina. Wilcoxina is known as a weak competitor ectomycorrhizal fungus that prospers only in absence of competitor ectomycorrhizal fungi (Danielson and Prudel, 1990). Suillus species are known to form ectomycorrhiza with pine trees that span a large area, thanks to long distance dispersal of spores combined with large sporocarps and a high volume of spore production and (Peay et al., 2012; Horton, 2017). Other species, such as Cadophora and Tomentella, are considered common fungi of pine seedlings in an early successional stage or disturbed areas (Colpaert et al., 1996; Berch et al., 2006; Sim and Eom, 2009; Lee et al., 2012; Lee and Eom, 2013; Lee and Koo, 2016).
While T. matsutake was still found in several P. densiflora seedlings, its frequency and abundance steadily decreased over time after transplantation. Although the priority effect in ectomycorrhiza was reported in previous studies (Kennedy and Bruns, 2005; Kennedy et al., 2009; Fukumi, 2015), it did not apply to T. matsutake in our study. As T. matsutake is usually known to form symbiotic relationship with mature pine trees in the field (Wang et al., 2017), our results suggest that the symbiosis between T. matsutake and young seedlings is not sustainable outside of sterile environment without proper support. We suggest that this is due to a slow growth rate and higher carbon demand of T. matsutake as a late-stage ectomycorrhizal fungus (Smith and Read, 2010).
Network Analysis and Keystone Taxa
Microbial network analysis has been used to visualize taxa with a strong effect on network structure, or highly connected taxa in various environments (Barberán et al., 2012; Gilbert et al., 2012; Agler et al., 2016). We constructed a network of 35 fungal OTUs that were abundant during pine seedling growth with SparCC correlations. Interaction and network formation between functionally diverse fungi were previously reported (Toju et al., 2016), and our results were similar; a combination of functionally different OTUs were observed in each cluster in our network (Supplementary Table 1).
Among the 35 major OTUs, 10 OTUs were found to have positive correlations with T. matsutake, despite the abundance of T. matsutake decreasing after transplantation (Table 1, Supplementary Tables 1, 2). Among these positively correlated OTUs, Fusarium, Trichoderma, and/or Oidiodendron might improve survival of T. matsutake in our environment. While competition between ectomycorrhiza and other microfungi are common in soil (Leake et al., 2003), several studies reported growth promotion of ectomycorrhizal fungi by microfungi isolated from soil (Ogawa, 1976; Oh et al., 2018). For instance, Trichoderma and Oidiodendron species were exclusively isolated from the T. matsutake fruiting zone of P. densiflora forests (Ogawa, 1977; Oh et al., 2018, 2019), and high abundance of F. oxysporum was reported in Tuber magnatum-productive areas (Mello et al., 2010). Likewise, we found that ectomycorrhizal fungi, such as Suillus were less abundant in root samples with T. matsutake than those without T. matsutake. Trichoderma might help survival of T. matsutake by promoting plant growth, root branching, and development (Guzmán-Guzmán et al., 2019), or by inhibiting the growth of other ectomycorrhiza as reported between Trichoderma viride and Suillus bovinus in soil environment (Mucha et al., 2008; Sabella et al., 2015; Oh et al., 2018, 2019).
Four OTUs were identified as keystone taxa: S. granulatus, C. pauciseptatum, Fusarium sp. (OTU 37), and Fusarium oxysporum (Table 2). Keystone taxa are taxa highly connected to other network members and play important roles in the microbiome (Banerjee et al., 2018), and they are required to understand an ecosystem’s response to disturbance (Stinson et al., 2006). Of the four keystone taxa, S. granulatus and C. pauciseptatum showed a significantly negative correlation with T. matsutake, while F. oxysporum and Fusarium sp. (OTU 37) showed a significantly positive correlation with T. matsutake. As Suillus granulatus is a strong competitor and crucial in the establishment of pine seedlings (Dickie et al., 2010; Kohout et al., 2011; Hayward et al., 2015; Urcelay et al., 2017; Policelli et al., 2019), its negative relationship with T. matsutake is as expected. However, the significant correlation between T. matsutake and C. pauciseptatum or Fusarium species was interesting, as C. pauciseptatum and Fusarium species are known as soil saprotrophs or plant pathogens. The presence of C. pauciseptatum was reported in Pinus sylvestris (Menkis and Vasaitis, 2011), the relationship between C. pauciseptatum and P. densiflora is still unknown. It is possible that C. pauciseptatum indirectly influenced microbiome by affecting quality of pine seedlings (Agler et al., 2016). Fusarium oxysporum and F. trincintum are known as plant pathogens or mutualistic endophytes (Kuldau and Yates, 2000; Forsyth et al., 2006; Vu et al., 2006; Michielse and Rep, 2009; Min et al., 2014; Waweru et al., 2014; Vasundhara et al., 2016). While we do not understand their exact function in this study, both endophyte and plant pathogen might influence on root microbiome by positive or negative effects (Van Der Heijden et al., 2008).
Conclusion
We have documented the change in fungal community composition in pine seedlings after the T. matsutake inoculation, and introduced a SparCC analysis to predict the cross-fungi associations from NGS data. The root microbiome drastically changed at alpha- and beta-diversity levels after transplantation. Temporal succession of the mycorrhizal community suggests a weak priority effect as T. matsutake was rapidly replaced by W. mikolae, S. granulatus, and other fungi. While most of the major fungal OTUs showed negative or neutral correlation with T. matsutake, some of them showed a positive relationship. Fungi that had a positive correlation with T. matsutake were mostly known as saprotrophs or plant pathogens. In addition, we found four keystone species during microbiome succession that might play an important role in microbiome composition in pine seedlings. A further study is needed to verify the effect of fungi that have positive correlations with T. matsutake in an artificial cultivation of ectomycorrhizal fungi.
Data Availability Statement
The datasets generated for this study can be found in the NCBI Sequence Read Archive (SRA) under BioProject ID PRJNA638021 (https://www.ncbi.nlm.nih.gov/bioproject/PRJN A638021/) and also available on request to the corresponding author.
Author Contributions
S-YO, MP, and YL contributed to conceiving and designing the experiments. KP and S-YO performed the experiments and analyzed the data. KP, S-YO, and SY wrote the manuscript with revisions from JF, MP, and YL. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Gyeongsangbuk-do Forest Environment Research Institute (Gyeongju, South Korea) for their help in experiment and sample collection. We would also like to thank Yoonhee Cho for the English revision of the manuscript.
Footnotes
Funding. This project was supported by the research project for exploring potential fungal diversity in forest soil (KNA1-1-25, 19-2) from the Korea National Arboretum.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.574146/full#supplementary-material
References
- Agler M. T., Ruhe J., Kroll S., Morhenn C., Kim S. T., Weigel D., et al. (2016). Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 14:e1002352. 10.1371/journal.pbio.1002352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alford R. A., Wilbur H. M. (1985). Priority effects in experimental pond communities: competition between Bufo and Rana. Ecology 66 1097–1105. 10.2307/1939161 [DOI] [Google Scholar]
- Bakker M. G., Schlatter D. C., Otto-Hanson L., Kinkel L. L. (2014). Diffuse symbioses: roles of plant–plant, plant–microbe and microbe–microbe interactions in structuring the soil microbiome. Mol. Ecol. 23 1571–1583. 10.1111/mec.12571 [DOI] [PubMed] [Google Scholar]
- Banerjee S., Schlaeppi K., van der Heijden M. G. A. (2018). Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 16 567–576. 10.1038/s41579-018-0024-1 [DOI] [PubMed] [Google Scholar]
- Barberán A., Bates S. T., Casamayor E. O., Fierer N. (2012). Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 6 343–351. 10.1038/ismej.2011.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57 289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- Berch S. M., Brockley R. P., Battigelli J. P., Hagerman S., Holl B. (2006). Impacts of repeated fertilization on components of the soil biota under a young lodgepole pine stand in the interior of British Columbia. Can. J. For. Res. 36 1415–1426. 10.1139/x06-037 [DOI] [Google Scholar]
- Berendsen R. L., Pieterse C. M., Bakker P. A. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17 478–486. 10.1016/j.tplants.2012.04.001 [DOI] [PubMed] [Google Scholar]
- Brundrett M. C. (2009). Mycorrhizal associations and other means of nutrition of vascular plants: understanding the global diversity of host plants by resolving conflicting information and developing reliable means of diagnosis. Plant Soil 320 37–77. 10.1007/s11104-008-9877-9 [DOI] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi V., DeFiglio H., Chaturvedi S. (2018). Phenotype profiling of white-nose syndrome pathogen Pseudogymnoascus destructans and closely-related Pseudogymnoascus pannorum reveals metabolic differences underlying fungal lifestyles. F1000Res. 7:665. 10.12688/f1000research.15067.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colpaert J. V., Van Laere A., van Assche J. A. (1996). Carbon and nitrogen allocation in ectomycorrhizal and non-mycorrhizal Pinus sylvestris L. seedlings. Tree Physiol. 16 787–793. 10.1093/treephys/16.9.787 [DOI] [PubMed] [Google Scholar]
- Danielson R. M., Prudel M. (1990). “Ectomycorrhizae of spruce seedlings growing in disturbed soils and in undisturbed mature forests,” in Proceedings of the 8th North American Conference on Mycorrhizae, eds Allen M. F., Williams S. E. (Jackson, WY: ), 68. [Google Scholar]
- Dickie I. A., Bolstridge N., Cooper J. A., Peltzer D. A. (2010). Co-invasion by Pinus and its mycorrhizal fungi. New Phytol. 187 475–484. 10.1111/j.1469-8137.2010.03277.x [DOI] [PubMed] [Google Scholar]
- Dickie I. A., Bufford J. L., Cobb R. C., Desprez-Loustau M. L., Grelet G., Hulme P. E., et al. (2017). The emerging science of linked plant–fungal invasions. New Phytol. 215 1314–1332. 10.1111/nph.14657 [DOI] [PubMed] [Google Scholar]
- Dickie I. A., Martínez-García L. B., Koele N., Grelet G. A., Tylianakis J. M., Peltzer D. A., et al. (2013). Mycorrhizas and mycorrhizal fungal communities throughout ecosystem development. Plant Soil 367 11–39. 10.1007/s11104-013-1609-0 [DOI] [Google Scholar]
- Edgar R. C., Haas B. J., Clemente J. C., Quince C., Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27 2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eto S., Taniguchi M. (2000). Development of the spawns of Tricholoma matsutake using fungicides for the inoculation in forests. J. Mushroom Sci. Soc. Jpn. 8 197–202. [Google Scholar]
- Feng K., Zhang Y., He Z., Ning D., Deng Y. (2019). Interdomain ecological networks between plants and microbes. Mol. Ecol. Resour. 19 1565–1577. 10.1111/1755-0998.13081 [DOI] [PubMed] [Google Scholar]
- Forsyth L. M., Smith L. J., Aitken E. A. (2006). Identification and characterization of non-pathogenic Fusarium oxysporum capable of increasing and decreasing Fusarium wilt severity. Mycol. Res. 110 929–935. 10.1016/j.mycres.2006.03.008 [DOI] [PubMed] [Google Scholar]
- Friedman J., Alm E. J. (2012). Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 8:e1002687. 10.1371/journal.pcbi.1002687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumi T. (2015). Historical contingency in community assembly: integrating niches, species pools, and priority effects. Annu. Rev. Ecol. Evol. Syst. 46 1–23. 10.1146/annurev-ecolsys-110411-160340 [DOI] [Google Scholar]
- Gilbert J. A., Steele J. A., Caporaso J. G., Steinbrück L., Reeder J., Temperton B., et al. (2012). Defining seasonal marine microbial community dynamics. ISME J. 6 298–308. 10.1038/ismej.2011.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill W. M., Guerin-Laguette A., Lapeyrie F., Suzuki K. (2000). Matsutake–morphological evidence of ectomycorrhiza formation between Tricholoma matsutake and host roots in a pure Pinus densiflora forest stand. New Phytol. 147 381–388. 10.1046/j.1469-8137.2000.00707.x [DOI] [Google Scholar]
- Gordon T. R. (2017). Fusarium oxysporum and the Fusarium wilt syndrome. Annu. Rev. Phytopathol. 55 23–39. 10.1146/annurev-phyto-080615-095919 [DOI] [PubMed] [Google Scholar]
- Guzmán-Guzmán P., Porras-Troncoso M. D., Olmedo-Monfil V., Herrera-Estrella A. (2019). Trichoderma species: versatile plant symbionts. Phytopathology 109 6–16. 10.1094/PHYTO-07-18-0218-RVW [DOI] [PubMed] [Google Scholar]
- Hannula S. E., Morriën E., de Hollander M., Van Der Putten W. H., van Veen J. A., De Boer W. (2017). Shifts in rhizosphere fungal community during secondary succession following abandonment from agriculture. ISME J. 11 2294–2304. 10.1038/ismej.2017.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward J., Horton T. R., Nuñez M. A. (2015). Ectomycorrhizal fungal communities coinvading with Pinaceae host plants in Argentina: Gringos bajo el bosque. New Phytol. 208 497–506. 10.1111/nph.13453 [DOI] [PubMed] [Google Scholar]
- Herzog C., Hartmann M., Frey B., Stierli B., Rumpel C., Buchmann N., et al. (2019). Microbial succession on decomposing root litter in a drought-prone Scots pine forest. ISME J. 13 2346–2362. 10.1038/s41396-019-0436-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton T. R. (2017). “Spore dispersal in ectomycorrhizal fungi at fine and regional scales”,” in Biogeography of Mycorrhizal Symbiosis, ed. Tedersoo L. (New York, NY: Springer; ), 61–78. 10.1007/978-3-319-56363-3_3 [DOI] [Google Scholar]
- Ka K. H., Kim H. S., Hur T. C., Park H., Jeon S. M., Ryoo R., et al. (2018). Analysis of Environment and Production of Tricholoma matsutake in Matsutake-infected Pine Trees. Korean J. Med. Mycol. 46 34–42. 10.4489/KJM.20180005 [DOI] [Google Scholar]
- Kareki K., Kawakami Y. (1985). Artificial formation of shiro (fungus colony) by planting the pine saplings infected with Tricholoma matsutake (Ito et Imai) Sing. Bull. Hiroshima Prefect. For. Exp. Station 20 13–23. [Google Scholar]
- Kennedy P. G., Bruns T. D. (2005). Priority effects determine the outcome of ectomycorrhizal competition between two Rhizopogon species colonizing Pinus muricata seedlings. New Phytol. 166 631–638. 10.1111/j.1469-8137.2005.01355 [DOI] [PubMed] [Google Scholar]
- Kennedy P. G., Peay K. G., Bruns T. D. (2009). Root tip competition among ectomycorrhizal fungi: are priority effects a rule or an exception? Ecology 90 2098–2107. 10.1890/08-1291.1 [DOI] [PubMed] [Google Scholar]
- Kim J. H., Han Y. H. (2009). Development of specific primer for Tricholoma matsutake. Mycobiology 37 317–319. 10.4489/MYCO.2009.37.4.317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohout P., Sýkorová Z., Bahram M., Hadincová V., Albrechtová J., Tedersoo L., et al. (2011). Ericaceous dwarf shrubs affect ectomycorrhizal fungal community of the invasive Pinus strobus and native Pinus sylvestris in a pot experiment. Mycorrhiza 21 403–412. 10.1007/s00572-010-0350-2 [DOI] [PubMed] [Google Scholar]
- Koide R. T., Xu B., Sharda J., Lekberg Y., Ostiguy N. (2005). Evidence of species interactions within an ectomycorrhizal fungal community. New Phytol. 165 305–316. 10.1111/j.1469-8137.2004.01216 [DOI] [PubMed] [Google Scholar]
- Kuldau G. A., Yates I. E. (2000). “Evidence for Fusarium endophytes in cultivated and wild plants,” in Microbial Endophytes, eds Bacon C. W., White J. F. (New York, NY: Marcel Dekker; ), 85–117. [Google Scholar]
- Kurtz Z. D., Müller C. L., Miraldi E. R., Littman D. R., Blaser M. J., Bonneau R. A. (2015). Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput. Biol. 11:e1004226. 10.1371/journal.pcbi.1004226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leake J. R., Donnelly D. P., Boddy L. (2003). “Interactions between ecto-mycorrhizal and saprotrophic fungi,” in Mycorrhizal Ecology, eds Heijden M. G. A., Sanders I. R. (Berlin: Springer-Verlag; ), 345–371. 10.1007/978-3-540-38364-2_14 [DOI] [Google Scholar]
- Lee E. H., Eo J. K., Lee C. S., Eom A. H. (2012). Effect of soil ameliorators on ectomycorrhizal fungal communities that colonize seedlings of Pinus densiflora in abandoned coal mine spoils. Mycobiology 40 168–172. 10.5941/MYCO.2012.40.3.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. H., Eom A. H. (2013). Ectomycorrhizal fungal communities of red pine (Pinus densiflora) seedlings in disturbed sites and undisturbed old forest sites. Mycobiology 41 77–81. 10.5941/MYCO.2013.41.2.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. Y., Koo C. D. (2016). Genet variation of ectomycorrhizal Suillus granulatus fruiting bodies in Pinus strobus stands. Mycobiology 44 7–13. 10.5941/MYCO.2016.44.1.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. H., Han S. K., Kim B. S., Shrestha B., Lee S. Y., Ko C. S., et al. (2007). Proliferation of Tricholoma matsutake mycelial mats in pine forest using mass liquid inoculum. Mycobiology 35 54–61. 10.4489/MYCO.2007.35.2.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. I., Yang C. K., Gebauer G. (2015). The importance of associations with saprotrophic non-Rhizoctonia fungi among fully mycoheterotrophic orchids is currently under-estimated: novel evidence from sub-tropical Asia. Ann. Bot. 116 423–435. 10.1093/aob/mcv085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Arbizu P. (2017). Pairwise Multilevel Comparison Using Adonis. R Package Version 0.3. Available online at: https://github.com/pmartinezarbizu/pairwiseAdonis (accessed December 2, 2019). [Google Scholar]
- Matsuda Y., Noguchi Y., Ito S. I. (2009). Ectomycorrhizal fungal community of naturally regenerated Pinus thunbergii seedlings in a coastal pine forest. J. For. Res. 14 335–341. 10.1007/s10310-009-0140-x [DOI] [Google Scholar]
- McMurdie P. J., Holmes S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello A., Miozzi L., Vizzini A., Napoli C., Kowalchuk G., Bonfante P. (2010). Bacterial and fungal communities associated with Tuber magnatum-productive niches. Plant Biosyst. 144 323–332. 10.1080/11263500903374724 [DOI] [Google Scholar]
- Menkis A., Vasaitis R. (2011). Fungi in roots of nursery grown Pinus sylvestris: ectomycorrhizal colonisation, genetic diversity and spatial distribution. Microb. Ecol. 61 52–63. 10.1007/s00248-010-9676-8 [DOI] [PubMed] [Google Scholar]
- Michielse C. B., Rep M. (2009). Pathogen profile update: Fusarium oxysporum. Mol. Plant Pathol 10 311–324. 10.1111/j.1364-3703.2009.00538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min Y. J., Park M. S., Fong J. J., Quan Y., Jung S., Lim Y. W. (2014). Diversity and saline resistance of endophytic fungi associated with Pinus thunbergii in coastal shelterbelts of Korea. J Microbiol. Biotechnol. 24 324–333. 10.4014/jmb.1310.10041 [DOI] [PubMed] [Google Scholar]
- Minnis A. M., Lindner D. L. (2013). Phylogenetic evaluation of Geomyces and allies reveals no close relatives of Pseudogymnoascus destructans, comb. nov., in bat hibernacula of eastern North America. Fungal Biol. 117 638–649. 10.1016/j.funbio.2013.07.001 [DOI] [PubMed] [Google Scholar]
- Moler E. R., Aho K. (2018). Whitebark pine foliar fungal endophyte communities in the southern Cascade Range, USA: host mycobiomes and white pine blister rust. Fungal Ecol. 33 104–114. 10.1016/j.funeco.2018.02.003 [DOI] [Google Scholar]
- Mucha J., Zadworny M., Werner A., Napierała-Filipiak A., Łakomy P. (2008). Antagonistic activity of the ectomycorrhizal fungus Suillus bovinus challenged by saprotrophic fungi from different soils. Nova Hedwigia 87 373–385. 10.1127/0029-5035/2008/0087-0373 [DOI] [Google Scholar]
- Nguyen N. H., Song Z., Bates S. T., Branco S., Tedersoo L., Menke J., et al. (2016). FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20 241–248. 10.1016/j.funeco.2015.06.006 [DOI] [Google Scholar]
- Nilsson R. H., Anslan S., Bahram M., Wurzbacher C., Baldrian P., Tedersoo L. (2019). Mycobiome diversity: high-throughput sequencing and identification of fungi. Nat. Rev. Microbiol. 17 95–109. 10.1038/s41579-018-0116-y [DOI] [PubMed] [Google Scholar]
- Obase K., Cha J. Y., Lee J. K., Lee S. Y., Lee J. H., Chun K. W. (2009). Ectomycorrhizal fungal communities associated with Pinus thunbergii in the eastern coastal pine forests of Korea. Mycorrhiza 20 39–49. 10.1007/s00572-009-0262-1 [DOI] [PubMed] [Google Scholar]
- Ogawa M. (1976). Studies on the artificial reproduction of Tricholoma matsutake (S. Ito et Imai) Sing. III. Effects of growth promotion of natural products on the vegetative growth of T. matsutake. Trans. Mycol. Soc. Jpn. 17 492–498. [Google Scholar]
- Ogawa M. (1977). Microbial ecology of mycorrhizal fungus, Tricholoma matsutake (Ito et Imai) Sing. in pine forest. III: fungal florae in Shiro soil and on the mycorrhiza. Bull. Gov. For. Exp. Stn. Tokyo 293 105–170. [Google Scholar]
- Oh S. Y., Park M. S., Cho H. J., Lim Y. W. (2018). Diversity and effect of Trichoderma isolated from the roots of Pinus densiflora within the fairy ring of pine mushroom (Tricholoma matsutake). PLoS One 13:e0205900. 10.1371/journal.pone.0205900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S. Y., Park M. S., Lim Y. W. (2019). The Influence of Microfungi on the Mycelial Growth of Ectomycorrhizal Fungus Tricholoma matsutake. Microorganisms 7:169. 10.3390/microorganisms7060169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J., Blanchet F. G., Friendly M., Kindt R., Legendre P., McGlinn D., et al. (2018). vegan: Community Ecology Package. R package version 2.5–2. Available online at: https://CRAN.R-project.org/package=vegan (accessed December 2, 2019). [Google Scholar]
- Park M. C., Sim S. G., Cheon W. J. (2007). Method of preparing Tricholoma matsutake-infected young pine by coculturing aseptic pine seedlings and T. matsutake. U.S. Patent No 7,269,923. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Peay K. G., Kennedy P. G., Bruns T. D. (2011). Rethinking ectomycorrhizal succession: are root density and hyphal exploration types drivers of spatial and temporal zonation? Fungal Ecol. 4 233–240. 10.1016/j.funeco.2010.09.010 [DOI] [Google Scholar]
- Peay K. G., Schubert M. G., Nguyen N. H., Bruns T. D. (2012). Measuring ectomycorrhizal fungal dispersal: macroecological patterns driven by microscopic propagules. Mol. Ecol. 21 4122–4136. 10.1111/j.1365-294X.2012.05666 [DOI] [PubMed] [Google Scholar]
- Perry D. A., Margolis H., Choquette C., Molina R., Trappe J. M. (1989). Ectomycorrhizal mediation of competition between coniferous tree species. New Phytol. 112 501–511. 10.1111/j.1469-8137.1989.tb00344 [DOI] [PubMed] [Google Scholar]
- Policelli N., Bruns T. D., Vilgalys R., Nuñez M. A. (2019). Suilloid fungi as global drivers of pine invasions. New Phytol. 222 714–725. 10.1111/nph.15660 [DOI] [PubMed] [Google Scholar]
- R Core Team (2019). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Rincón A., Santamaría-Pérez B., Rabasa S. G., Coince A., Marçais B., Buée M. (2015). Compartmentalized and contrasted response of ectomycorrhizal and soil fungal communities of Scots pine forests along elevation gradients in France and Spain. Environ. Microbiol. 17 3009–3024. 10.1111/1462-2920.12894 [DOI] [PubMed] [Google Scholar]
- Rogers S. O., Bendich A. J. (1994). “Extraction of total cellular DNA from plants, algae and fungi,” in Plant Molecular Biology Manual, eds Gelvin S. B., Schilperoort R. A. (Dordrecht: Springer; ), 183–190. 10.1007/978-94-011-0511-8_12 [DOI] [Google Scholar]
- Rognes T., Flouri T., Nichols B., Quince C., Mahé F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. 10.7717/peerj.2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudawska M., Kujawska M., Leski T., Janowski D., Karliñski L., Wilgan R. (2019). Ectomycorrhizal community structure of the admixture tree species Betula pendula, Carpinus betulus, and Tilia cordata grown in bare-root forest nurseries. For. Ecol. Manag. 437 113–125. 10.1016/j.foreco.2019.01.009 [DOI] [Google Scholar]
- Sabella E., Nutricati E., Aprile A., Miceli A., Sorce C., Lorenzi R., et al. (2015). Arthrinium phaeospermum isolated from Tuber borchii ascomata: the first evidence for a “Mycorrhization Helper Fungus”? Mycol. Prog. 14:59 10.1007/s11557-015-1083-6 [DOI] [Google Scholar]
- Saito C., Ogawa W., Kobayashi H., Yamanaka T., Fukuda M., Yamada A. (2018). In vitro ectomycorrhization of Tricholoma matsutake strains is differentially affected by soil type. Mycoscience 59 89–97. 10.1016/j.myc.2017.09.002 [DOI] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13 2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorrocks B., Bingley M. (1994). Priority effects and species coexistence: experiments with fungal-breeding Drosophila. J Anim. Ecol. 63 799–806. 10.2307/5257 [DOI] [Google Scholar]
- Sim M. Y., Eom A. H. (2009). Diversity of Ectomycorrhizal Fungi of Pinus densiflora Siebold et Zucc. Seedlings in a Disturbed Forest on Mt. Songni. J Ecol. Environ. 32 13–18. 10.5141/JEFB.2009.32.1.013 [DOI] [Google Scholar]
- Smith G. R., Finlay R. D., Stenlid J., Vasaitis R., Menkis A. (2017). Growing evidence for facultative biotrophy in saprotrophic fungi: data from microcosm tests with 201 species of wood-decay basidiomycetes. New Phytol. 215 747–755. 10.1111/nph.14551 [DOI] [PubMed] [Google Scholar]
- Smith S. E., Read D. J. (2010). Mycorrhizal Symbiosis. New York, NY: Academic Press. [Google Scholar]
- Stinson K. A., Campbell S. A., Powell J. R., Wolfe B. E., Callaway R. M. (2006). Invasive plant suppresses the growth of native tree seedlings by disrupting belowground. PLoS Biol. 4:e140. 10.1371/journal.pbio.0040140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedersoo L., Bahram M., Põlme S., Kõljalg U., Yorou N. S., Wijesundera R., et al. (2014). Global diversity and geography of soil fungi. Science 346:1256688. 10.1126/science.1256688 [DOI] [PubMed] [Google Scholar]
- Tedersoo L., Smith M. E. (2013). Lineages of ectomycorrhizal fungi revisited: foraging strategies and novel lineages revealed by sequences from belowground. Fungal Biol. Rev. 27 83–99. 10.1016/j.fbr.2013.09.001 [DOI] [Google Scholar]
- Toju H., Kishida O., Katayama N., Takagi K. (2016). Networks depicting the fine-scale co-occurrences of fungi in soil horizons. PLoS One 11:e0165987. 10.1371/journal.pone.0165987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twieg B. D., Durall D. M., Simard S. W. (2007). Ectomycorrhizal fungal succession in mixed temperate forests. New Phytol. 176 437–447. 10.1111/j.1469-8137.2007.02173 [DOI] [PubMed] [Google Scholar]
- Unite Community (2019). UNITE QIIME Release for Fungi. Version 18.11.2018. Available online at: https://forum.qiime2.org/t/unite-v-8-0-2018-11-18-classifiers-for-qiime2-available-here/8750 (accessed December 2, 2019). [Google Scholar]
- Urcelay C., Longo S., Geml J., Tecco P. A., Nouhra E. (2017). Co-invasive exotic pines and their ectomycorrhizal symbionts show capabilities for wide distance and altitudinal range expansion. Fungal Ecol. 25 50–58. 10.1016/j.funeco.2016.11.002 [DOI] [Google Scholar]
- Van Der Heijden M. G., Bardgett R. D., Van Straalen N. M. (2008). The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 11 296–310. 10.1111/j.1461-0248.2007.01139 [DOI] [PubMed] [Google Scholar]
- Van Der Heijden M. G., Martin F. M., Selosse M. A., Sanders I. R. (2015). Mycorrhizal ecology and evolution: the past, the present, and the future. New Phytol. 205 1406–1423. 10.1111/nph.13288 [DOI] [PubMed] [Google Scholar]
- Van Dongen S., Abreu-Goodger C. (2012). Using MCL to extract clusters from networks. Methods Mol. Biol. 804 281–295. 10.1007/978-1-61779-361-5_15 [DOI] [PubMed] [Google Scholar]
- Vasiliauskas R., Menkis A., Finlay R. D., Stenlid J. (2007). Wood-decay fungi in fine living roots of conifer seedlings. New Phytol. 174 441–446. 10.1111/j.1469-8137.2007.02014.x [DOI] [PubMed] [Google Scholar]
- Vasundhara M., Baranwal M., Kumar A. (2016). Fusarium tricinctum, an endophytic fungus exhibits cell growth inhibition and antioxidant activity. Indian J. Microbiol. 56 433–438. 10.1007/s12088-016-0600-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voříšková J., Brabcová V., Cajthaml T., Baldrian P. (2014). Seasonal dynamics of fungal communities in a temperate oak forest soil. New Phytol. 201 269–278. 10.1111/nph.12481 [DOI] [PubMed] [Google Scholar]
- Vu T., Sikora R., Hauschild R. (2006). Fusarium oxysporum endophytes induced systemic resistance against Radopholus similis on banana. Nematology 8 847–852. 10.1163/156854106779799259 [DOI] [Google Scholar]
- Wang Y., Zhang C., Li S. (2017). Tricholoma matsutake: an edible mycorrhizal mushroom of high socioeconomic relevance in China. Rev. Mex. Mic. 46 55–61. [Google Scholar]
- Waweru B., Turoop L., Kahangi E., Coyne D., Dubois T. (2014). Non-pathogenic Fusarium oxysporum endophytes provide field control of nematodes, improving yield of banana (Musa sp.). Biol. Control 74 82–88. 10.1016/j.biocontrol.2014.04.002 [DOI] [Google Scholar]
- White T. J., Bruns T., Lee S. J. W. T., Taylor J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, eds Innis M. A., Gelfand D. H., Sninsky J. J., White T. J. (New York, NY: Academic Press, Inc; ), 315–322. 10.1016/b978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Yamada A., Kobayashi H., Murata H., Kalmiş E., Kalyoncu F., Fukuda M. (2010). In vitro ectomycorrhizal specificity between the Asian red pine Pinus densiflora and Tricholoma matsutake and allied species from worldwide Pinaceae and Fagaceae forests. Mycorrhiza 20 333–339. 10.1007/s00572-009-0286-6 [DOI] [PubMed] [Google Scholar]
- Yamada A., Maeda K., Kobayashi H., Murata H. (2006). Ectomycorrhizal symbiosis in vitro between Tricholoma matsutake and Pinus densiflora seedlings that resembles naturally occurring ‘shiro’. Mycorrhiza 16 111–116. 10.1007/s00572-005-0021-x [DOI] [PubMed] [Google Scholar]
- Yamada A., Maeda K., Ohmasa M. (1999). Ectomycorrhiza formation of Tricholoma matsutake isolates on seedlings of Pinus densiflora in vitro. Mycoscience 40 455–463. 10.1007/BF02461022 [DOI] [Google Scholar]
- Yamamoto S., Sato H., Tanabe A. S., Hidaka A., Kadowaki K., Toju H. (2014). Spatial segregation and aggregation of ectomycorrhizal and root-endophytic fungi in the seedlings of two Quercus species. PLoS One 9:e96363. 10.1371/journal.pone.0096363 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study can be found in the NCBI Sequence Read Archive (SRA) under BioProject ID PRJNA638021 (https://www.ncbi.nlm.nih.gov/bioproject/PRJN A638021/) and also available on request to the corresponding author.