Abstract
Last two decades have witnessed several global infectious outbreaks. Among these, coronavirus is identified as a prime culprit ranging from its involvement in severe acute respiratory syndrome (SARS), Middle East respiratory syndrome (MERS) to COVID-19. These infections involved in huge healthcare and economic cost incurred globally. Every time, coronavirus improved its infection ability and surprised the medical practitioners and researchers. Currently, COVID-19 is also causing numerous infections and stalled global activities. Global efforts are underway to identify potential viral targets for management of these outbreaks, but significant progress in prevention of these outbreaks is not yet achieved. We explored host–pathogen protein–protein interactions of MERS, SARS and COVID-19, and identified host targets common among all recent coronavirus outbreaks. Further, we tried to understand their potential for management of coronavirus. The common proteins involved in coronavirus host–pathogen interactions indicate their indispensable role in the pathogenesis and therefore targeting these proteins can give strategies to prevent current and future coronavirus outbreaks. Viral variability necessitates development of new therapeutic modalities for every outbreak, in contrast targeting necessary human proteins required by all coronaviruses can provide us a clue to prevent current and future coronavirus outbreaks. We found that targeting FURIN and TMPRSS2 can provide good results due to their common involvement in current and previous outbreaks. We also listed some known molecules against these two targets for their potential drug repurposing evaluation. Although, several recent studies undergoing with targeting these proteins for management of coronavirus, but safety evaluation and risk assessment must be given prime importance while targeting human proteins.
Keywords: coronavirus, host–pathogen interactions, FURIN, TMPRSS2, infection prevention
Introduction
During last two decades, Betacoronavirus genus gave us prominent pathogens leading to global public health emergency, which include Middle East respiratory syndrome (MERS), severe acute respiratory syndrome (SARS) and SARS-CoV-2 coronavirus [1]. Generally coronaviruses are involved in infections ranging from respiratory to enteric system, but recent outbreaks of SARS, MERS and current COVID-19 gave it a worldwide notoriety [2]. The cases of current coronavirus SARS-CoV-2 (causing COVID-19) overtook all previous coronavirus outbreaks in terms of incidence and mortalities. The symptoms of current outbreak range from mild illness, viral pneumonia with the power of affecting both lungs, acute respiratory distress syndrome to sepsis and septic shock, in addition to rapid infectivity and multiorgan involvement [3, 4]. Previous coronavirus outbreaks also created worldwide panic, but the current version of SARS-CoV-2 has affected all world activities in addition to worldwide panic and global public health emergency. Several researchers around the globe are working to contain the virus; unfortunately, the incidence and fatalities are out of control in the absence of proper treatment strategies. These all viruses are positive sense, single-stranded RNA virus with envelope and belongs to the Coronaviridae family. Coronaviruses generally emerge as zoonotic infections, where animal virus gains the ability to infect the human. The last three important coronaviruses are also suggested to be originated from the animal sources. Even the current SARS-CoV-2 is also considered to be a combination of bat and Malaysian pangolin coronavirus [5, 6].
Several factors are known to influence the ability of coronavirus severity, and it is considered that immune response subversion by viral or external factors is associated with its severity [7, 8]. Due to common involvement of three versions of betacoronavirus in recent outbreaks, several studies are going to understand the similarity between MERS, SARS and SARS2 coronavirus [9]. Some studies are finding common viral target to manage these infections, whereas some studies suggested strategies to develop broad-spectrum antivirals to manage all coronaviruses [10], but the virus is surprising researchers due to its remarkable variability and creating problems with development of antivirals [11].
Another strategy involved targeting of host as they are less susceptible to variation. During this study we performed comparative host–pathogen interaction (HPI) analysis of MERS, SARS and SARS-CoV-2 in order to identify common host targets necessary for current and previous outbreaks. We tried to understand about human proteins targeting which is likely to provide better response for the management of coronavirus outbreaks including the chances of their utilization in future coronavirus outbreaks due to their indispensable participation in all major coronavirus pathogenesis.
Materials and methods
Database
The Biological General Repository for Interaction Dataset (BioGrid) version 3.5.185 was used to download SARS-CoV-2 and other coronavirus-related interactions. The dataset was last modified up to 30 April 2020 at the time of analysis. All viral–viral, host–host and HPI derived from organisms other than human were filtered out. Only protein–protein interactions involving virus–human were selected for further analysis.
Comparative PPI map preparation and identification of common proteins
The Cytoscape v3.8.0 was used to create comparative protein–protein interaction network from known coronavirus interactions. The common human proteins known to interact with coronavirus were identified and filtered out for further analysis. These common interactors were ranked and categorized for their ability to interact with all or any two coronaviruses out of MERS, SARS-CoV and SARS-CoV-2.
Functional identification of common interactors
The Entrez gene IDs of common interactors were taken from BioGrid dataset and used to detect these proteins in Uniprot. The functional details of these common human proteins were collected from Uniprot and included for categorization of suitable host protein with therapeutic potential.
Identification of known inhibitors of selected common targets involved in coronavirus infection
The common human proteins detected in earlier steps with positive involvement in viral pathogenesis were screened at DrugBank, IUPHAR/BPS guide to Pharmacology and PubChem databases for finding known molecules targeting two proteins interacting with all three recent coronaviruses identified in earlier steps. In addition, literature was also searched for natural compounds involved in the inhibition of these two proteins activity.
Identification of metabolic pathways and biological processes affected by these common targets
The three common human proteins involved in all recent coronavirus outbreaks (identified in previous steps) were detected for their involvement in different pathways. Kyoto Encyclopedia of Genes and Genomes (KEGG) was considered for evaluation of their role in different pathways, whereas BioCyc database collection was used to detect gene ontology (biological process) to assess their role in different biological processes.
Results
We performed comparative HPI analysis of recent three coronavirus outbreaks and identified important human targets and their inhibitors in different databases. Figure 1 represents scheme for identification of important human proteins and their inhibitors. We used BioGrid version 3.5.185 for collecting HPIs of recent coronavirus outbreaks. BioGrid is a curated database and involve 72 164 literature references for 28 093 chemical association, 1814 182 protein and genetic interactions, and 874 796 posttranslational modification information at the time of data collection. The searching of database for SARS-CoV-2 and other related coronavirus-mediated interactions involved total 743 interactions including virus–human, human–human, virus–virus and interactions with host other than human. Filtering of all these interactions gave us 518 exclusive virus–human-mediated protein–protein interactions. These 518 interactions involved 63 redundant interaction derived from detection of same interactions by multiple approaches leaving 455 unique coronavirus HPIs. Table 1 represents detail about coronavirus-related HPI in BioGrid database. The details of interaction obtained from BioGrid version 3.5.185 is presented as Supplementary Table S1.
Figure 1. Work scheme representing identification of important human targets from comparative coronavirus HPI map.
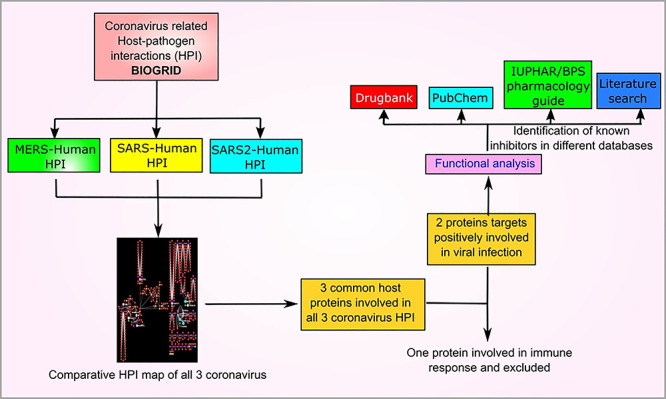
Table 1. Details about coronavirus-related interactions in BioGrid.
| Interacting partners | Number of interactions |
|---|---|
| Total interactions | 743 |
| Human–Human | 41 |
| Mus musculus–SARS | 01 |
| Cricetulus griseus–SARS | 01 |
| MERS–MERS | 03 |
| SARS–SARS | 165 |
| SARS CoV2–SARS CoV2 | 14 |
| MERS–human/human–MERS | 02/04 |
| SARS–human/human–SARS | 142/17 |
| SARS CoV2–human/human–SARS CoV2 | 340/13 |
| Total HPIs (human specific) | 518 |
For details please refer Table S1.
Construction of comparative HPI maps in Cytoscape 3.8.0 is presented in Figure 2. The common human proteins involved in different HPI maps are presented with different node sizes and colors. The detail about common interacting human proteins and their involvement in different HPI is presented in Table 2.
Figure 2. Comparative host–pathogen protein–protein interactions of MERS, SARS and SARS2. Viral proteins are indicated with different node colors (MERS: fluorescent green, SARS: yellow, SARS2: blue). Human proteins are generally indicated with red colors but common human proteins are shown with large size nodes. In addition, the proteins common among all coronaviruses are shown with sky blue color, whereas human proteins common among other pathogens are also shown with different colors. Multiple edges indicate about detection of same interaction through multiple experimental evidences.
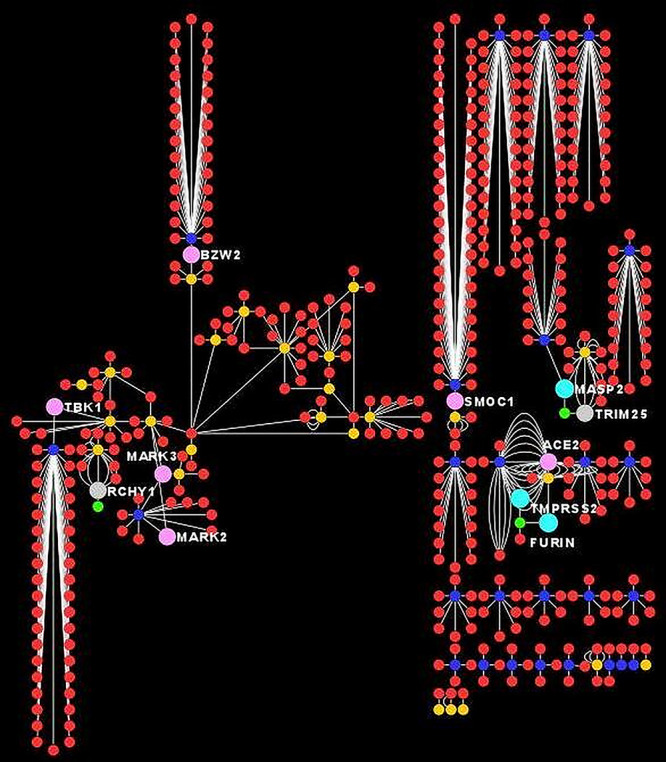
Table 2. Common human proteins found to interact with coronavirus of recent public health concern as per BioGrid database.
| Sr. No. | Virus | Common human interacting proteins |
|---|---|---|
| 1 | SARS, MERS and COVID | MASP2 |
| FURIN | ||
| TMPRSS2 | ||
| 2 | SARS and COVID | ACE2 |
| MASP2 | ||
| FURIN | ||
| TMPRSS2 | ||
| BZW2 | ||
| TBK1 | ||
| SMOC1 | ||
| MARK3 | ||
| MARK2 | ||
| 3 | MERS and COVID | MASP2 |
| FURIN | ||
| TMPRSS2 | ||
| 4 | SARS and MERS | MASP2 |
| TRIM25 | ||
| FURIN | ||
| TMPRSS2 | ||
| RCHY1 |
Human proteins commonly interacting with different coronaviruses and their functional role as per Uniprot is presented in Table 3. Proteins MASP2, FURIN and TMPRSS2 are involved in interaction with all three coronaviruses. Host protein MASP2 is known to interact with N protein, whereas FURIN and TMPRSS2 interact with S protein. These interactions are preserved in all three recent coronavirus outbreaks and indicate their importance in viral pathogenesis. MASP2 is involved in innate immune defense and therefore it was not considered for known inhibitors identification, in contrast FURIN and TMPRSS2 are involved in viral infection progression and therefore considered for further analysis. The results are presented in Table 3.
Table 3. Details of common interactors and their interaction with different coronaviruses.
| Common human protein interactor | Primary function according to Uniprot | Viral interactor proteins | ||
|---|---|---|---|---|
| MERS | SARS | SARS2 | ||
| MASP2 | Activation of complement by its serum protease activity via mannose-binding lectin. Cleaves and activates C2 and C4, resulting in formation of C3 convertase | N | N | N |
| FURIN | Endoprotease with the ability to cleave at the RX(K/R)R consensus motif. Required for H7N1 and H5N1 influenza virus infection by hemagglutinin cleavage [32] | S | S | S |
| TMPRSS2 | Promotes SARS-CoV and SARS-CoV-2 infections via proteolytic cleavage of Angiotensin-converting enzyme 2 (ACE2) receptor promoting viral uptake, and cleavage of spike glycoproteins of coronavirus and its activation for host cell entry [33, 34] | S | S | S |
| ACE2 | Receptor for entry of SARS-CoV and SARS-CoV-2, NL63/HCoV-NL63 | – | S | S |
| BZW2 | May be involved in neuronal differentiation | – | nsp8ab | M |
| TBK1 | Serine/threonine kinase playing an important role in inflammatory responses against foreign substances | – | nsp5ab | nsp13 |
| SMOC1 | Important roles in eye and limb development. Probably regulates differentiation of osteoblast | – | ORF7a | ORF8 |
| MARK3 | It is a serine/threonine-protein kinase [35], which is involved in the specific phosphorylation of microtubule-associated proteins for MAP2 and MAP4 and phosphorylate MAPT/TAU. It negatively regulates hippo signaling pathway | – | ORF9b,nsp13ab | ORF9b |
| MARK2 | It is a serine/threonine protein kinase [35], which is involved in cell polarity and microtubule dynamics regulation | – | nsp13ab | ORF9b |
| TRIM25 | Ubiquitin E3 ligase and ISG15 E3 ligase [36], which is implicated in innate immune response for viruses [37, 38]. Mediates signal transduction leading to the production of interferons against viral infection [37, 39] | N | N | – |
| RCHY1 | E3-dependent ubiquitination and proteasomal degradation of target proteins. Involved in the cell cycle progression regulation and enhance androgen receptor (AR) transcription factor activity | nsp3ab | nsp3ab | – |
The screening of FURIN and TMPRSS2 in different databases for known inhibitory molecules is presented in Table 4. The DrugBank database was found to have two molecules for FURIN and one molecule for TMPRSS2. Identification of targets in IUPHAR/BPS guide to Pharmacology and Pubchem database revealed several molecules for FURIN and TMPRSS2, which indicated about several RNAi molecules. Some natural compounds known for these proteins inhibitory activity are also presented in Table 4.
Table 4. Known molecule targeting of important human interactors and their possible reactions.
| Database | Human protein | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| FURIN | TMPRSS2 | ||||||||
| DrugBank | DRUGBANK ID | DB04951 | DB03600 | DB13729 | |||||
| Name | Pirfenidone (NCT04282902) | Capric acid | Camostat (NCT04353284, NCT04338906, NCT04321096, NCT04455815) | ||||||
| Drug group | Approved, investigational | Experimental | Experimental inhibitor | ||||||
| IUPHAR/BPS guide to Pharmacology | Ligand | MI-1148 [32] | Phenylacetyl-Arg-Val-Arg-4-amidinobenzylamide [40] | Peptide 18 [41] | Furin inhibitor peptide [42] | I-432 [43] | Compound 5 [44] | Nafamostat (NCT04418128, NCT04352400) [45] | Camostat (mentioned above) |
| Action | Inhibition | Inhibition | Inhibition | Inhibition | Inhibition | Inhibition | Inhibition | Inhibition | |
| PubChem | CID/SID | 16760442 | 44584947 | 57358476 | 85101385 | 85108608 | 70 siRNA with reagent vendors | ||
| Name | Furin inhibitor I | Furin inhibitor peptide | Hexa-D-arginine (furin inhibitor II) | RNAi of TMPRSS2 (R & D) | RNAi of TMPRSS2 (R & D) | ||||
| Literature search | Luteolin | Found in several food, inhibit viral replication through furin inhibition [46] | Geniposide (NPC306344) | A major iridoid glycosides of gardenia fruit predicted to inhibit TMPRSS2 during docking analysis [47] | |||||
| Baicalein, oroxylin A and chrysin | Isolated from stem bark of medicinally active plant Oroxylum indicum shoes modest to strong furin inhibitory activity [48] | Nafamostat (mentioned above) | Known to inhibit MERS [45] and proposed for COVID-2019 inhibition | ||||||
The few clinical trials (identifier) of certain molecule for COVID-19 are also mentioned in brackets.
The role of common human proteins in metabolic pathways according to KEGG is presented in Supplementary Figures S1–S3, whereas the role of these common human proteins in biological processes as determined by BioCyc database is presented in Table S2.
Discussion
The HPI plays an important role in the pathogenesis of any infectious organisms, but their role is more prominent in viral pathogenesis due to their reliance on effective utilization of host machinery for establishing the infection. The role of HPIs in coronavirus infections is also discussed earlier [12], but computational study of host–pathogen protein–protein interactions is adding more value for understanding the pathogenesis of coronavirus [13]. BioGrid is a well-known database of protein, genetic and chemical interactions between various organisms and host. We downloaded interactions from BioGrid with published experimental evidences and this provides reliability of high-quality interaction data [14]. Among these, few interactions are verified by multiple experimental approaches as evidenced in Figure 2 and Supplementary Table S1 and indicate about additional reliability of source data.
During identification of common human interactors in SARS, MERS and SARS-2 protein–protein HPI, three proteins were found to be involved in HPI with all three coronaviruses (Table 2). In addition, their host–pathogen interacting partners were also common among all three coronavirus HPI indicating indispensable nature of these interactions in coronavirus infection (Table 3). These three proteins, namely MASP2, FURIN and TMPRSS2 were detected for their function in human using Uniprot. Uniprot is a universal protein knowledgebase providing comprehensive information about major aspects of proteins [15]. We found that MASP2 is involved in complement activation and innate immune response, which is important for protection against pathogen. In contrast, FURIN and TMPRSS2 are positively involved in viral infection (Tables 3 and S2). This activity is also indicated while analyzing their role in metabolic pathways and biological processes through KEGG [16] and BioCyc [17] database (Figures S1–S3, Table S2). Therefore, we selected FURIN and TMPRSS2 for further analysis.
We detected the relevant databases for identification of known inhibitors of these two proteins. It included DrugBank v 5.1.6 [18], IUPHAR/BPS guide to Pharmacology [19] and PubChem [20] databases. These databases are renowned and provide extensive coverage of known and experimental molecules targeting particular proteins. Under certain situations, the references providing detail about these activities are also mentioned (Table 4). In addition, literature search for natural molecules inhibiting these two proteins also revealed some important findings. This cataloging is supposed to provide some hits about potential of these molecules in prevention of coronavirus outbreaks.
In addition to our results, some studies have also indicated the use of these proteins separately for management of these infections. For example, it has been proposed that targeting TMPRSS2 can help in the management of influenza and coronavirus infections [21]. In addition, TMPRSS2 inhibitor camostat is efficient to prevent entry and growth of SARS-CoV during in vitro experiments [22]. However, ACE2 proteins is known to interact with both SARS and SARS2 spike (S) proteins, and therefore several studies are undergoing to find S protein inhibitory molecules in addition to ACE2 inhibitors. It has been observed that emodin and promazine have ability to prevent interaction of S proteins with ACE2 and thereby inhibit coronavirus infection. These two molecules are also suggested as alternative choice for management of COVID-19 [23]. As per the previous information, the coronavirus entry into human cells depends on proteolytic cleavage of S protein through FURIN and TMPRSS2. The viral spike (S) protein is cleaved to S1 and S2 subunits through FURIN where S1 subunit binds to ACE2 receptor present on host cells, whereas S2 protein is further cleaved by TMPRSS2 resulting in membrane fusion and subsequent entry of virus into host cell, and therefore inhibition of FURIN and TMPRSS2 has been also suggested to control SARS-CoV-2 infection [24–27]. Indeed, several known FURIN and TMPRSS2 inhibitors are undergoing through clinical trials for COVID-19, including FURIN inhibitor pirfenidone (NCT04282902) and TMPRSS2 inhibitors camostat (NCT04353284, NCT04338906, NCT04321096, NCT04455815) and nafamostat (NCT04352400) (Table 4). While some other molecules need further investigation for their potential to control coronavirus infection. For example, luteolin is a flavonoid present in several edible plants; it is also proposed to have ability to manage COVID-19 and need detailed clinical investigations [28]. In addition, natural compound baicalein is also found to inhibit SARS-CoV-2 protease during in vitro study and draw special attention for development of antivirals against SARS-CoV-2 [29]. Our results validate these findings and indicate about preferential assessment of these two common human proteins due to their indispensable nature in all recently important coronavirus pathogenesis. The targeting of FURIN and TMPRSS2 is also suggested in several other studies and therefore supports our findings [25].
In addition, comparative HPI analysis also revealed that SARS-CoV-2 is having more known HPI than SARS and MERS. It may be due to extensive global efforts to understand SARS-CoV-2 pathogenesis because of its more intensity and global coverage. Otherwise the results indicate that SARS-CoV-2 is able to affect host in better way and these viruses are increasing their overall infection ability without altering common HPIs. It can be assumed that targeting these common interactors may be more superior and must be given preferential importance for management of these outbreaks.
Although, our study indicates targeting of FURIN and TMPRSS2 for management of coronavirus outbreaks, but this study has certain limitations. For example, our study is based on interactions known up to 30 April 2020 in BioGrid Database, but future studies may uncover some other common interactions with coronavirus management potential. In addition, sometimes the proteins can interact without physical contact, like transcription factors can affect expression of several proteins without interacting with them. Therefore, much wider picture of HPIs may arise while considering viral influence on host cell without any physical HPIs. In addition, the targeting of human proteins can have widespread clinical implications in addition to their effects on viral pathogenesis. Although the role of protease inhibitors are widely discussed in the management of COVID-19, but using protease inhibitors can lead to several associated adverse events including their effects on cardiovascular, renal system etc. [30, 31]. These all associated adverse events can undermine the importance of these two proteins in management of coronavirus. However, we are optimistic with the global efforts that we will be able to find certain molecules targeting FURIN and/or TMPRSS2 without causing anticipated adverse events. Considering present situation, where virus is causing significant mortality with global economic impact, and improving its infection ability, we must consider these options not only for present situation but also as our preparation for future similar outbreaks.
Conclusion
Identification of FURIN and TMPRSS2 as common host proteins positively involved in all recent coronavirus outbreaks indicates toward therapeutic potential of these proteins in the management of these viral infections. Screening of several databases for identification of potential inhibitors of these proteins and subsequent identification of clinical studies going toward coronavirus management potential of few inhibitors supports our findings. Natural compounds known to inhibit these proteins may also be an important alternative and needs clinical investigations for management of coronavirus outbreaks. Targeting FURIN and TMPRSS2 can give us valuable insights about management of coronavirus outbreaks provided that these protease inhibitors must satisfy safety standards.
Key Points
Recently, the world has witnessed several outbreaks caused due to coronavirus. It includes SARS, MERS and COVID-19 that are posing challenges to healthcare workers and researchers.
We performed comparative host–pathogen interaction analysis of coronavirus involved in MERS, SARS and COVID-19, and identified common interactions among all outbreaks.
The common interactions among all recent coronavirus outbreaks indicate their indispensable role in pathogenesis.
Targeting common interactors can give clue to prevent recent and future coronavirus outbreaks, but viral variability is another challenge and targeting of host proteins can provide better response.
Several studies are undergoing that target human proteins interacting with coronavirus, but preferential assessment of common human interactors FURIN and TMPRSS2 can give important molecules with the potential to manage current and future outbreaks.
Funding
None.
Conflict of Interest
The authors declare no conflict of interest.
Supplementary Material
Abdul Arif Khan is currently associated with ICMR-National AIDS Research Institute, India. He is working to identify host–pathogen interactions and their role in microbial pathogenesis since last 17 years. He is using several computational approaches to decipher role of host–microbe interactions in microbial pathogenesis.
Zakir Khan is associated with Department of Biomedical Sciences, Department of Pathology and Laboratory Medicine, Cedars-Sinai Medical Center, Los Angeles, USA. He has major research interest in understanding of molecular mechanisms for identifying novel targets/strategies in different diseases. He is also involved in using computational approaches to understand molecular microbial pathogenesis.
References
- 1. Chen Y, Liu Q, Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol 2020;92:418–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Habibzadeh P, Stoneman EK. The novel coronavirus: a bird's eye view. Int J Occup Environ Med 2020;11:65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhou L, Liu HG. Early detection and disease assessment of patients with novel coronavirus pneumonia. Zhonghua Jie He He Hu Xi Za Zhi 2020;43:E003. [DOI] [PubMed] [Google Scholar]
- 4. Zaim S, Chong JH, Sankaranarayanan V, et al. COVID-19 and multiorgan response. Curr Probl Cardiol 2020;45:100618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020;579:270–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hassanin A. Coronavirus origins: genome analysis suggests two viruses may have combined In: The Conversation. Australia: The Conversation Media Group Ltd, 2020. https://theconversation.com/coronavirus-origins-genome-analysis-suggests-two-viruses-may-have-combined-134059. . [Google Scholar]
- 7. Fehr AR, Channappanavar R, Jankevicius G, et al. The conserved coronavirus macrodomain promotes virulence and suppresses the innate immune response during severe acute respiratory syndrome coronavirus infection. MBio 2016;7(6):e01721–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 2020;395:507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Petrosillo N, Viceconte G, Ergonul O, et al. COVID-19, SARS and MERS: are they closely related? Clin Microbiol Infect 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Khan S, Siddique R, Shereen MA, et al. Emergence of a novel coronavirus, severe acute respiratory syndrome coronavirus 2: biology and therapeutic options. J Clin Microbiol 2020;58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Prajapat M, Sarma P, Shekhar N, et al. Drug targets for corona virus: a systematic review. Indian J Pharm 2020;52:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Miura TA, Holmes KV. Host-pathogen interactions during coronavirus infection of primary alveolar epithelial cells. J Leukoc Biol 2009;86:1145–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Khan AA, Khan Z. COVID-2019-associated overexpressed Prevotella proteins mediated host-pathogen interactions and their role in coronavirus outbreak. Bioinformatics 2020;36:4065–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oughtred R, Stark C, Breitkreutz BJ, et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res 2019;47:D529–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. UniProt Consortium T. UniProt: the universal protein knowledgebase. Nucleic Acids Res 2018;46:2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kanehisa M, Furumichi M, Tanabe M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45:D353–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Karp PD, Billington R, Caspi R, et al. The BioCyc collection of microbial genomes and metabolic pathways. Brief Bioinform 2019;20:1085–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wishart DS, Feunang YD, Guo AC, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 2018;46:D1074–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harding SD, Sharman JL, Faccenda E, et al. The IUPHAR/BPS guide to Pharmacology in 2018: updates and expansion to encompass the new guide to immunopharmacology. Nucleic Acids Res 2018;46:D1091–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim S. Getting the most out of PubChem for virtual screening. Expert Opin Drug Discov 2016;11:843–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shen LW, Mao HJ, Wu YL, et al. TMPRSS2: a potential target for treatment of influenza virus and coronavirus infections. Biochimie 2017;142:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kawase M, Shirato K, Hoek L, et al. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J Virol 2012;86:6537–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang L, Liu Y. Potential interventions for novel coronavirus in China: a systematic review. J Med Virol 2020;92:479–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020;181:271–280.e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bestle D, Heindl MR, Limburg H, et al. TMPRSS2 and furin are both essential for proteolytic activation and spread of SARS-CoV-2 in human airway epithelial cells and provide promising drug targets bioRxiv 2020:2020.2004.2015.042085.
- 26. Bergeron E, Vincent MJ, Wickham L, et al. Implication of proprotein convertases in the processing and spread of severe acute respiratory syndrome coronavirus. Biochem Biophys Res Commun 2005;326:554–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hoffmann M, Kleine-Weber H, Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell 2020;78(4):779–784.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ansari WA, Ahamad T, Khan MA, et al. Luteolin: a dietary molecule as potential anti-COVID-19 agent. Preprint available at Research Square 2020;(version 1. doi: 10.21203/rs.3.rs-35368/v1. [DOI] [Google Scholar]
- 29. Su H, Yao S, Zhao W, et al. Discovery of baicalin and baicalein as novel, natural product inhibitors of SARS-CoV-2 3CL protease in vitro. bioRxiv 2020:2020.2004.2013.038687.
- 30. George PM, Wells AU, Jenkins RG. Pulmonary fibrosis and COVID-19: the potential role for antifibrotic therapy. Lancet Respir Med 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Olyaei AJ, deMattos AM, Bennett WM. Renal toxicity of protease inhibitors. Curr Opin Nephrol Hypertens 2000;9:473–6. [DOI] [PubMed] [Google Scholar]
- 32. Hardes K, Becker GL, Lu Y, et al. Novel Furin inhibitors with potent anti-infectious activity. ChemMedChem 2015;10:1218–31. [DOI] [PubMed] [Google Scholar]
- 33. Heurich A, Hofmann-Winkler H, Gierer S, et al. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J Virol 2014;88:1293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020;181:271–280 e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gu GJ, Lund H, Wu D, et al. Role of individual MARK isoforms in phosphorylation of tau at Ser(2)(6)(2) in Alzheimer's disease. Neuromolecular Med 2013;15:458–69. [DOI] [PubMed] [Google Scholar]
- 36. Zou W, Zhang DE. The interferon-inducible ubiquitin-protein isopeptide ligase (E3) EFP also functions as an ISG15 E3 ligase. J Biol Chem 2006;281:3989–94. [DOI] [PubMed] [Google Scholar]
- 37. Gack MU, Shin YC, Joo CH, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 2007;446:916–20. [DOI] [PubMed] [Google Scholar]
- 38. Lian H, Zang R, Wei J, et al. The zinc-finger protein ZCCHC3 binds RNA and facilitates viral RNA sensing and activation of the RIG-I-like receptors. Immunity 2018;49:438–448 e435. [DOI] [PubMed] [Google Scholar]
- 39. Oshiumi H, Miyashita M, Matsumoto M, et al. A distinct role of Riplet-mediated K63-linked polyubiquitination of the RIG-I repressor domain in human antiviral innate immune responses. PLoS Pathog 2013;9:e1003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Becker GL, Sielaff F, Than ME, et al. Potent inhibitors of furin and furin-like proprotein convertases containing decarboxylated P1 arginine mimetics. J Med Chem 2010;53:1067–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kwiatkowska A, Couture F, Levesque C, et al. Design, synthesis, and structure-activity relationship studies of a potent PACE4 inhibitor. J Med Chem 2014;57:98–109. [DOI] [PubMed] [Google Scholar]
- 42. Shiryaev SA, Remacle AG, Ratnikov BI, et al. Targeting host cell furin proprotein convertases as a therapeutic strategy against bacterial toxins and viral pathogens. J Biol Chem 2007;282:20847–53. [DOI] [PubMed] [Google Scholar]
- 43. Paszti-Gere E, Czimmermann E, Ujhelyi G, et al. In vitro characterization of TMPRSS2 inhibition in IPEC-J2 cells. J Enzyme Inhib Med Chem 2016;31:123–9. [DOI] [PubMed] [Google Scholar]
- 44. Sielaff F, Bottcher-Friebertshauser E, Meyer D, et al. Development of substrate analogue inhibitors for the human airway trypsin-like protease HAT. Bioorg Med Chem Lett 2011;21:4860–4. [DOI] [PubMed] [Google Scholar]
- 45. Yamamoto M, Matsuyama S, Li X, et al. Identification of Nafamostat as a potent inhibitor of Middle East respiratory syndrome coronavirus S protein-mediated membrane fusion using the split-protein-based cell-cell fusion assay. Antimicrob Agents Chemother 2016;60:6532–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Peng M, Watanabe S, Chan KWK, et al. Luteolin restricts dengue virus replication through inhibition of the proprotein convertase furin. Antiviral Res 2017;143:176–85. [DOI] [PubMed] [Google Scholar]
- 47. Rahman N, Basharat Z, Yousuf M, et al. Virtual screening of natural products against type II transmembrane serine protease (TMPRSS2), the priming agent of coronavirus 2 (SARS-CoV-2). Molecules 2020;25:2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Majumdar S, Mohanta BC, Chowdhury DR, et al. Proprotein convertase inhibitory activities of flavonoids isolated from Oroxylum indicum. Curr Med Chem 2010;17:2049–58. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


