Abstract
Paenibacillus polymyxa is an important member of the plant growth-promoting rhizobacteria. P. polymyxa YC0136 inoculation had beneficial effect on growth promotion and biological control of tobacco (Nicotiana tabacum L.) under field conditions. This study aimed to reveal the growth-promoting mechanisms of strain YC0136. In growth-promotion assays, tobacco plant height was increased by 8.42% and 8.25% at 60 and 90 days, respectively, after inoculation with strain YC0136. Strain YC0136 also promoted the accumulation of tobacco biomass in varying degrees. Following inoculation with strain YC0136, 3,525 and 4,368 tobacco genes were up-regulated and down-regulated, respectively. Strain YC0136 induced the expression of plant hormone-related genes in tobacco, including auxin, cytokinin, and gibberellin, as well as transcription factors related to stress resistance such as WRKY and MYB. In addition, strain YC0136 induced the up-regulation of genes in the phenylpropanoid biosynthesis pathway by 1.51–4.59 times. Interaction with tobacco also induced gene expression changes in strain YC0136, with 286 and 223 genes up-regulated and down-regulated, respectively. Tobacco interaction induced up-regulation of the ilvB gene related to auxin biosynthesis in strain YC0136 by 1.72 times and induced expression of some nutrient transport genes. This study contributes to our understanding of the growth-promoting mechanisms of strain YC0136 on tobacco and provides a theoretical basis for the application of P. polymyxa YC0136 as a biological fertilizer.
Keywords: Paenibacillus polymyxa, tobacco, PGPR, growth-promoting mechanisms, RNA-seq
Introduction
The numerous microorganisms in the rhizosphere of plants can be divided into positive, negative, and neutral microorganisms. Plant growth-promoting rhizobacteria (PGPR) are a group of rhizobacteria colonizing the plant rhizosphere that can promote the growth of many plants (Salvo et al., 2018). In modern agriculture, the use of pesticides and chemical fertilizers is becoming more limited, and PGPR are attracting more and more attention. PGPR can not only be used as plant growth regulators but also as biological control agents (Recep et al., 2009; Bal et al., 2013a; Zhang and Kong, 2014; Djaya et al., 2019). PGPR’s associated with diverse plant hosts including tobacco variety K326 (Zhang and Kong, 2014), sudan grass var Sudanensis (Basak and Biswas, 2009), rice (cv. Naveen) (Bal et al., 2013a), potato cultivars Agria and Granola (Recep et al., 2009), cucumber (Han and Lee, 2006), common bean (Mohamed et al., 2019), Helianthus annuus (Khan et al., 2018), wheat (Mohite, 2013), and Arabidopsis thaliana (Jiang et al., 2012). PGPR promote growth through direct and indirect mechanisms (Santoro et al., 2015). Direct mechanisms include secretion of auxin (Lata et al., 2006) or cytokinin (Timmusk et al., 1999), phosphate solubilization (Son et al., 2006), dissolving potassium (Basak and Biswas, 2009), nitrogen fixation (Anand et al., 2013), siderophore production (Sarode et al., 2007), and 1-aminocyclopropane-1-carboxylate (ACC) deaminase production (Bal et al., 2013b). PGPR can not only promote plant growth directly, but can also be used as biocontrol agents to control plant pathogens, such as Aspergillus niger (Yuttavanichakul et al., 2012), Candida albicans (Ajilogba et al., 2016), Fusarium. sp. (Cao et al., 2018), Armillaria sp. (Vasconcellos and Cardoso, 2009), and Cercospora arachidicola (Sayyed and Patel, 2011). Inhibiting the growth of pathogens by biosynthesizing secondary metabolites is a major indirect growth-promoting mechanism of PGPR. The secondary metabolites are various, such as phenazines (Burkhead et al., 1994), iturin A (Ahimou et al., 2000), bacillomycin (Volpon et al., 1999), polymyxin (Kagan et al., 1951; Cruz et al., 2007; Niu et al., 2013), and fusaricidin (Vater et al., 2016). Inducing resistance in plants is another important indirect growth-promoting mechanism of PGPR (Tahir et al., 2017).
Paenibacillus polymyxa, formerly known as Bacillus polymyxa, was distinguished from other members of the genus Bacillus using a highly specific gene probe based on 16S rRNA (Ash et al., 1993). P. polymyxa is an important member of the PGPR that can promote the growth of many hosts through direct or indirect mechanisms (Michael et al., 1997; Ali, 2010; Muthukumar and Udaiyan, 2010; Lee et al., 2012; Anand et al., 2013; Kim et al., 2016; Abdallah et al., 2019). Research by Puri et al. (2016) suggested that P. polymyxa P2b-2R could increase the biomass and seedling height of canola through nitrogen fixation. P. polymyxa BRF-1 increased soybean dry weight and mineral absorption and has biocontrol ability against Phialophora gregata (Zhou et al., 2010). P. polymyxa ICA B01, a strain with phosphate solubilizing characteristic in vitro, promoted the growth of Zea mays under abiotic stress conditions (Mohd Din et al., 2020). Phi et al. (2010) reported that P. polymyxa KNUC265 improves the yield of pepper and antagonizes the pathogen Erwinia carotovora subsp. carotovora in tobacco. P. polymyxa ATCC 842T promoted the growth of tomato by producing indole-3-acetic acid (IAA) and inducing systemic resistance (Mei et al., 2014). P. polymyxa is a valuable species for agricultural application and has important research value.
Beneficial plant-microbe interactions that promote plant health and development have been the subject of considerable study. Azospirillum brasilense REC3, a PGPR strain, promoted the growth of strawberry plants (Elías et al., 2018), and the up-regulation of genes associated with ethylene (ET) signaling and IAA biosynthesis was detected by fluorescence quantitative PCR. Lee et al. (2012) demonstrated that P. polymyxa E681 elicited the induced systemic resistance (ISR) and plant growth promotion of Arabidopsis by producing C13 volatiles. Molecular aspects of the Gluconacetobacter diazotrophicus-sugarcane interaction have been investigated by quantitative mass spectrometry-based proteomics (Lery et al., 2010). Inoculation with PGPR strains could affect the transcriptomic profiles of tomato (Ibort et al., 2018). In Ibort’s research, Enterobacter sp. strain C7 alleviated the stress level of water through reducing the expression of ethylene biosynthesis gene in wild-type tomato. B. megaterium inoculation induced the expression of ethylene biosynthesis, signaling, and response genes in never ripe plants. In addition, B. megaterium could enhance the antioxidant capacity of wild-type tomato through affecting the expression of flavonoid biosynthesis genes. Plants also exert influence on microorganisms. Transcriptome profiling of P. aeruginosa PA01 response to sugar beet root exudates revealed that root exudates affected the expression of genes related to metabolism, chemotaxis, and other processes (Mark et al., 2005).
P. polymyxa YC0136 is a PGPR strain isolated from tobacco rhizosphere soil in Zunyi, Guizhou Province, China. It promotes the growth of tobacco and inhibits the growth of some tobacco pathogens, such as Ralstonia solanacearum, Phytophthora parasitica var. nicotianae (data not shown). In this study, we carried out RNA-seq of tobacco roots co-cultured with YC0136 cells under sterile conditions to analyze the promotion mechanism of strain YC0136 on improving the agronomic traits of tobacco and enhancing the resistance of tobacco. The effect of tobacco on gene expressing of strain YC0136 was also discussed.
Materials and Methods
Strains, Plant Material, and Growth Conditions
P. polymyxa YC0136 was previously isolated using Luria-Bertani (LB) agar medium from the rhizosphere soil of tobacco in Zunyi and stored in the PGPR laboratory, Shandong, China. Strain YC0136 was grown on Luria-Bertani (LB) agar medium (Ma et al., 2009) and cultured overnight at 37°C. A pure culture of strain YC0136 was grown in 5 mL of LB liquid medium for 12 hours (h) at 37°C and 180 rpm. Then 5 mL of the culture was transferred into 50 mL of fresh LB liquid medium and shaken for another 12 h at 37°C. The cell suspensions (diluted to ∼108 cells per milliliter) were used for growth promotion assays in pots. In addition, YC0136 cells cultured under the same conditions were harvested, washed with sterile water, and diluted to optical density OD600 = 0.9. These cells were used for transcript profiling of strain YC0136 and tobacco under sterile conditions.
Ralstonia solanacearum and Fusarium moniliforme were used for antagonistic tests. A pure culture of R. solanacearum was incubated in 5 mL of LB liquid medium for 10 h at 37°C with shaking (180 rpm). F. moniliforme was activated on Potato Dextrose Agar (PDA) medium (Cardwell et al., 2000) and cultured for 4 days, at 28°C.
Tobacco cultivar Nicotiana tabacum L. ‘K326’ was selected as plant material. It is a type of flue-cured tobacco widely planted in China.
Determination of Plant Growth-Promoting Traits
Preparation of cultures for detection of auxin secretion followed the method of Shao et al. (2015). Strain YC0136 was inoculated into Landy liquid medium with L-tryptophan and cultured at 25°C and 90 rpm with shaking for 4 days. Salkowski colorimetric solution was used to detect IAA secretion ability, and the absorbance of the mixture at 530 nm was measured (Thakuria et al., 2004), with 50 mg/L IAA used as a positive control. The negative control was bacteria-free Landy medium with L-tryptophan. IAA production was determined by the standard curve method (Yu et al., 2016).
Organophosphorus culture medium (Qingdao Hope Bio-technology Co., Ltd.) was used to test the phosphate solubilization ability of strain YC0136. Strain YC0136 was inoculated on organophosphorus medium and cultured at 30°C for 7 days until a transparent circle formed around the colonies. The ratio of transparent circle diameter (D)/colony diameter (d) indicated the capacity for phosphate solubilization (Kanse et al., 2015).
The ability of strain YC0136 to antagonize bacteria and fungi was also tested. The plant pathogens (R. solanacearum and F. moniliforme) were selected as indicators in the antagonistic test of this study. One hundred microliters of R. solanacearum culture were coated on LB agar medium, then strain YC0136 was inoculated on the same plate. The strains were co-cultured at 37°C for 24 h. Agar containing F. moniliforme was placed at the center of PDA agar medium plates and cultured for 2 days at 28°C. Strain YC0136 was then inoculated on the plate around the F. moniliforme (Príncipe et al., 2007). They were co-cultured at 28°C for another 2 days. The appearance of inhibition zones on the plates indicated that strain YC0136 had antagonistic abilities.
Growth-Promotion Assay in Pots
To evaluate the plant growth-promoting effects exerted by strain YC0136 on tobacco, plant height, leaf number, and biomass of tobacco were determined. The assay was carried out in the greenhouse of Shandong Agricultural University (36.200796°N, 117.125149°E). For pot experiments, tobacco seeds were sown in nutrition soil (total nutrient content ≥3%, organic matter content ≥30%) and cultured at room temperature. Tobacco seedlings with 5–6 leaves were planted in pots (diameter 22 cm, depth 15 cm) with 3 kg of soil per pot. Two hundred milliliters of water containing 5 mL of strain YC0136 cell suspension was poured into the rhizosphere of tobacco plants. There were 10 biological replicates in each group. At the same time, the control group was treated with 200 mL of water with 5 mL of LB medium. Plant height and leaf number were investigated every 30 days. For the investigation of plant height and leaf numbers, 10 plants were investigated at 30 days and 60 days after inoculation, and five plants were investigated at 90 days. At 60 days after strain inoculation, three plants in each group were collected for biomass analyses. These plants were dried at 105°C for 30 min, then dried at 80°C to constant weight.
Interaction Between Strain YC0136 and Tobacco Under Sterile Conditions
For transcriptome tests, seeds of ‘K326’ were surface-sterilized and sown on 1/2 Murashige-Skoog (MS) medium for germination (Timmusk et al., 2005). Tobacco was grown in a chamber at 25°C with 10 h light period (13200 lux) and 14 h dark period. Seven-day-old seedlings were used for subsequent growth experiments. Tobacco seedlings were transplanted into tissue culture bottles with 90 mL of liquid MS medium. The tissue culture bottles contained a net to support the seedlings and a tube to support the net. Tobacco was grown in a chamber at 25°C with 10 h light period (13200 lux) and 14 h dark period for 30 days. One milliliter of YC0136 cells was then inoculated into each tissue culture bottle for co-culturing with the tobacco. The tobacco control group was inoculated with 1 mL of sterile water. Meanwhile, 1 mL of YC0136 cell suspension was inoculated in a bottle with MS medium but without tobacco as the control group for strain YC0136. Each treatment contained three biological replicates. Strain YC0136 and tobacco ‘K326’ were co-cultured for 20 h. Co-cultured YC0136 cells were then centrifuged at 8000 rpm for 2 min and rinsed three times with sterile PBS buffer to completely remove impurities. YC0136 cells in the YC0136-tobacco co-cultured group were labeled YB. YC0136 cells in the control group were labeled YB-CK. Tobacco roots were harvested and washed with sterile water three times. Tobacco roots from the co-cultured group were labeled YP, and tobacco roots from the control group were labeled YP-CK. Harvested bacteria and tobacco roots were placed in liquid nitrogen for 3 h, then stored in a freezer at −80°C for future tests.
Total RNA Extraction and Transcriptomic Analysis
Total RNAs of strain YC0136 and tobacco roots were extracted and purified using TRIzol Reagent (Invitrogen). The quality and quantity of RNAs were investigated using an Agilent 2100 (Agilent Technologies, Palo Alto, CA, United States) and NanoDrop (Thermo Fisher Scientific Inc.), and RNAs with RNA Integrity Number (RIN) > 7 were selected to construct the library. Next-generation sequencing library preparations were constructed according to the manufacturer’s protocol (NEBNext® UltraTM RNA Library Prep Kit for Illumina®). Libraries with different indices were multiplexed and loaded on an Illumina HiSeq instrument according to the manufacturer’s instructions (Illumina, San Diego, CA, United States). Sequencing was carried out using a 2 × 150 bp paired-end configuration. Image analysis and base calling were performed using the HiSeq Control Software (HCS) + OLB + GAPipeline-1.6 (Illumina) on the HiSeq instrument.
Raw data were stored in fastq file format. Low-quality data and adaptor sequences were filtered out to ensure the accuracy of data. The complete genome of strain YC0136 (GenBank accession number: CP017967.3) was selected as the reference genome. Although a draft genome of tobacco ‘K326’ is available, many genes have no annotation information. The genome of Nicotiana tabacum L. cultivar ‘TN90’ was therefore selected as the reference genome1. Clean data were aligned to the reference genome using the software Hisat2 (v2.0.1) (Kim et al., 2018). HTSeq (v0.6.1) was used to estimate gene and isoform expression levels from the paired-end clean data (Anders et al., 2015). Differentially expressed genes (DEGs) were analyzed using DESeq2 (V1.6.3) in the Bioconductor software package (Love et al., 2014). Kyoto Encyclopedia of Genes and Genomes (KEGG2) pathway enrichment analysis of DEGs was performed using KOBAS (Song et al., 2015).
RT-qPCR Assay for DEGs in RNA-Seq
One microgram of purified total RNA was used as template for first-strand cDNA synthesis using an Evo M-MLV RT Kit with gDNA Clean for qPCR (Accurate Biotechnology (Hunan) Co., Ltd.). Several genes identified through RNA-seq were selected for amplification using SYBR green qPCR. Primers were designed using Beacon Designer 7 and were listed in Supplementary Table S1. The tobacco actin gene was selected as a reference gene. In strain YC0136, the GAPDH gene (encoding glyceraldehyde-3-phosphate dehydrogenase) was used as the reference gene. Relative expression levels were calculated using the 2–(delta – delta Ct) method (Livak and Schmittgen, 2001). Three biological replicates were performed.
Statistical Analysis
The statistical analyses of agronomic characters used t-tests in GraphPad Prism 7 (GraphPad Software, San Diego, CA, United States), and P < 0.05 means that there was a significant difference. Due to the presence of plant sampling in the tests, there were no fewer than three replicates per statistical analysis. Columns were drawn using GraphPad Prism 7.
Results
General Characteristics of Strain YC0136
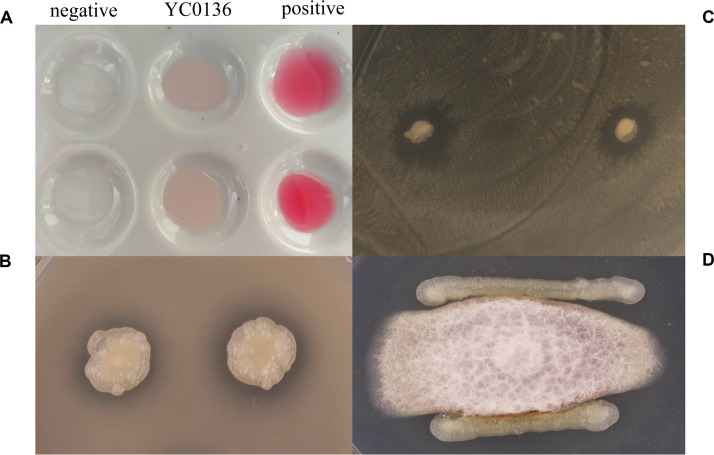
We cultured strain YC0136 in Landy medium for 4 days before mixing with Salkowski colorimetric solution and incubating in the dark for 30 min. Figure 1A shows that the culture of strain YC0136 turned red. These indicated that strain YC0136 could secrete IAA. At an OD600 of 1.0, the IAA yield of strain YC0136 was 24 μg/mL. For analysis of phosphate solubilization, transparent circles (as shown in Figure 1B) generated around colonies of strain YC0136 indicated that strain YC0136 could dissolve organic phosphorus. The ratio D/d was 1.32 ± 0.04.
FIGURE 1.
Growth-promoting characteristics of strain YC0136. (A) Detection of IAA secretion. A single clone of strain YC0136 was inoculated in Landy liquid medium and cultured for 4 days at 25°C, 90 rpm. Salkowski solution was then used to detect IAA secretion. A standard solution with 50 mg/L IAA was used as a positive control. Landy medium without inoculation was used as a negative control. (B) Qualitative analysis of phosphate solubilization. A single clone of strain YC0136 was cultivated on LB medium at 37°C. The pure bacterial culture was inoculated onto organophosphorus culture medium for qualitative analysis of phosphate solubilization. (C) Strain YC0136 inhibits the growth of R. solanacearum. R. solanacearum was coated on LB ager medium, and then strain YC0136 was inoculated onto the coated plate and cultured for 24 h at 37°C. (D) Strain YC0136 inhibits the growth of F. moniliforme. Hyphae of F. moniliforme were placed in the center of PDA medium and cultured for 2 days at 28°C. Strain YC0136 was then streaked around the fungi and cultured at 28°C for another 2 days.
The results of the antagonistic assay are shown in Figures 1C,D. Strain YC0136 formed antagonistic circles on plates coated with R. solanacearum. This indicated that strain YC0136 can secrete antibacterial substances. There were obvious inhibitory bands on the edge of the F. moniliforme culture. This indicated that strain YC0136 can also inhibit the growth of fungi by secreting antifungal substances.
Strain YC0136 Improves the Growth of Tobacco
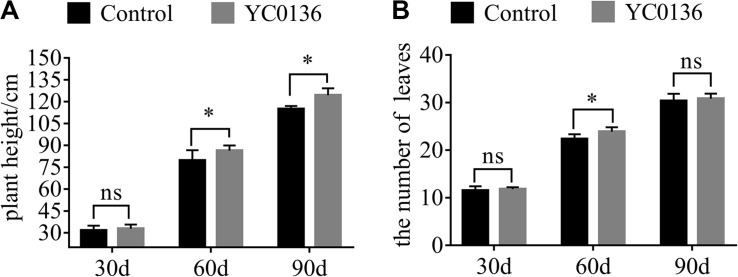
Agronomic traits of tobacco at different growth stages were promoted after inoculation with strain YC0136 (Figure 2). Strain YC0136 increased the plant height of tobacco at different growth stages (Figure 2A). At 30 days after inoculation, the plant height of tobacco in the group treated with strain YC0136 was slightly higher (3.79%) than that in the control group. There was a significant difference (P < 0.05) in the plant height of tobacco between the control group and the group treated with strain YC0136 at 60 days after inoculation. Compared with the control group, plant height in the group treated with strain YC0136 was increased by 8.42%. Moreover, the effects of strain YC0136 on tobacco plant height were also significant (P < 0.05) at 90 days after inoculation, with treated plants being 8.25% taller than controls. Strain YC0136 also increased the number of leaves on tobacco plants at different harvest intervals (Figure 2B). Plants treated with strain YC0136 had 6.97% more leaves than controls after 60 days of inoculation, and there was a significant difference at the P < 0.05 level.
FIGURE 2.
Effects of strain YC0136 on agronomic characteristics of tobacco at different growth stages. Two hundred milliliters of water with 5 mL of YC0136 cell suspension was poured into the rhizosphere of tobacco. In the control group, the cell suspension was replaced with 5 mL of LB medium. Plant height and leaf number were recorded every 30 days. (A) Statistical results for tobacco plant height. Plant height is the distance from the surface stem base to the first capsule base. (B) Statistical results for tobacco leaf number. Number of leaves is the number of leaves from the stem bottom to the top of the first flower branch. “∗” indicates a significant difference at the p < 0.05 level in t-tests. “ns” means that there is no significant difference at the p < 0.05 level. A total of two treatments were set up in the pot experiments with 10 replicates for each treatment at the beginning of the experiment. At 90 days after inoculation, there were five replicates. Error bars indicate the SD from the mean.
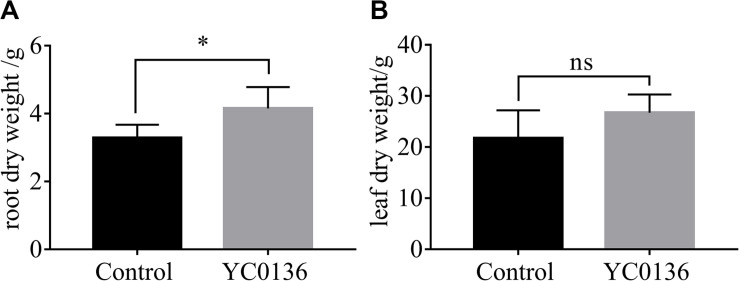
At 60 days after inoculation, we sampled the tobacco plants and analyzed the dry biomass of the plants (Figure 3). Strain YC0136 had different effects on the biomass of different tobacco tissues. After 60 days of inoculation, root dry weight of tobacco plants in the group treated with strain YC0136 was increased by 26.5% compared to that in the control group, and the difference was significant at the P < 0.05 level (Figure 3A). Leaf dry weight was increased by 23% in the group treated with strain YC0136 compared with that in the control group, but this difference was not significant (Figure 3B).
FIGURE 3.
Effects of YC0136 on tobacco dry biomass at 60 days after inoculation. Plants were collected at 60 days after inoculation, dried at 105°C for 30 min, and then dried at 80°C to maintain constant weight. The data were analyzed by t-tests. The error bars were standard deviation of the group. (A) Statistical results for root dry weight at 60 days after inoculation. (B) Statistical results for leaf dry weight at 60 days after inoculation. Three replicates were used for statistical analysis. Error bars indicate the SD from the mean. “*” Indicates a significant difference at the p < 0.05 level in t-tests. “ns” Means that there is no significant difference at the p < 0.05 level.
Transcriptome Profiling of the YC0136-Tobacco Interaction
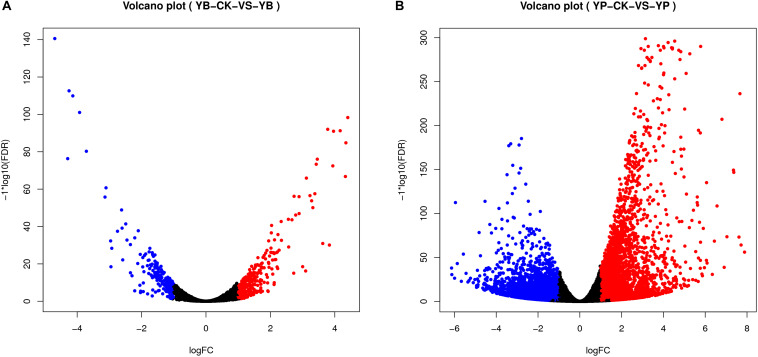
We analyzed the transcriptomes of strain YC0136 and tobacco ‘K326’ during their interaction. A total of 286 genes were up-regulated and 223 genes were down-regulated in strain YC0136 under interaction conditions compared with control conditions (Figure 4A). Among these DEGs, 109 genes were significantly up-regulated (logFC > 1.5) and 78 genes were significantly down-regulated (| logFC| > 1.5) in strain YC0136. After interacting with strain YC0136, 3,525 genes in tobacco roots were up-regulated while 4,368 genes were down-regulated compared with those in uninoculated roots. The number of DEGs showing significant up-regulation (logFC > 1.5) was 2,041, and the number showing significant down-regulation (| logFC| > 1.5) was 2,423 (Figure 4B).
FIGURE 4.
Visualization of differentially expressed genes (DEGs) in the interaction transcriptome (volcano map). DEGs were analyzed using DESeq2 (V1.6.3) in the Bioconductor software package and mapped to volcanoes. Red dots indicate gene up-regulation. Blue dots indicate gene down-regulation. The abscissa represents the fold change in gene expression in different samples. The ordinate represents the statistical significance of the difference in gene expression. (A) Visualization map of DEGs in strain YC0136. YC0136 cells in the YC0136-tobacco co-cultured group were labeled YB. YC0136 cells in the control group were labeled YB-CK. (B) Visualization map of DEGs in tobacco. Tobacco roots from the co-cultured group were labeled YP, and tobacco roots from the control group were labeled YP-CK.
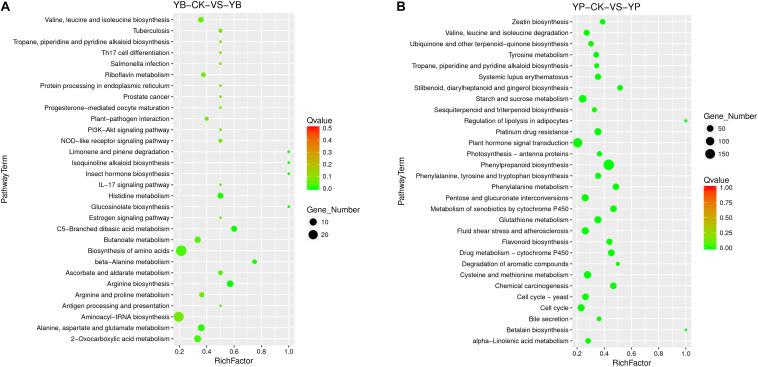
We performed enrichment analysis of the DEGs in strain YC0136 using the KEGG database (Figure 5A). A total of 306 genes were enriched in 73 categories of four major categories in the KEGG database. Among the DEGs of strain YC0136, 38 genes were enriched in the ABC transporters pathway. There were 28 genes enriched in the biosynthesis of amino acids pathway. In tobacco, 1,797 DEGs were enriched in 52 pathways in four major KEGG categories (as shown in Figure 5B). Through analysis, we found that 183 genes were enriched in the phenylpropanoid biosynthesis pathway. Up to 130 genes were enriched in the signal transduction pathway of plant hormones. The number of genes related to the starch and sucrose metabolism pathway was 75. About 39 genes were enriched in the flavonoid biosynthesis pathway. Thirty-one genes were enriched in the terpenoid backbone biosynthesis pathway. Furthermore, 26 genes were enriched in zeatin biosynthesis.
FIGURE 5.
KEGG enrichment analysis of differentially expressed genes (DEGs) in the interaction transcriptome. KEGG enrichment of different genes was performed using KOBAS software. The degree of KEGG enrichment is measured by the Rich factor, the Q-value, and the number of genes enriched in this pathway. Rich factor refers to the ratio of the number of DEGs in the pathway to the total number of all annotated genes in the pathway. The larger the Rich factor, the greater the degree of enrichment. Q-value is the p-value after correction for multiple hypothesis testing. The value range of Q-value is [0,1]; the closer to zero, the more significant the enrichment. (A) KEGG enrichment of DEGs in strain YC0136. YC0136 cells in the YC0136-tobacco co-cultured group were labeled YB. YC0136 cells in the control group were labeled YB-CK (B) KEGG enrichment of DEGs in tobacco. Tobacco roots from the co-cultured group were labeled YP, and tobacco roots from the control group were labeled YP-CK.
Verification of Partial DEGs Using RT-qPCR
Reverse-transcription quantitative PCR is an important method for verifying gene expression in RNA-seq. To verify the accuracy of RNA-seq data, we selected some DEGs in tobacco and strain YC0136 for RT-qPCR.
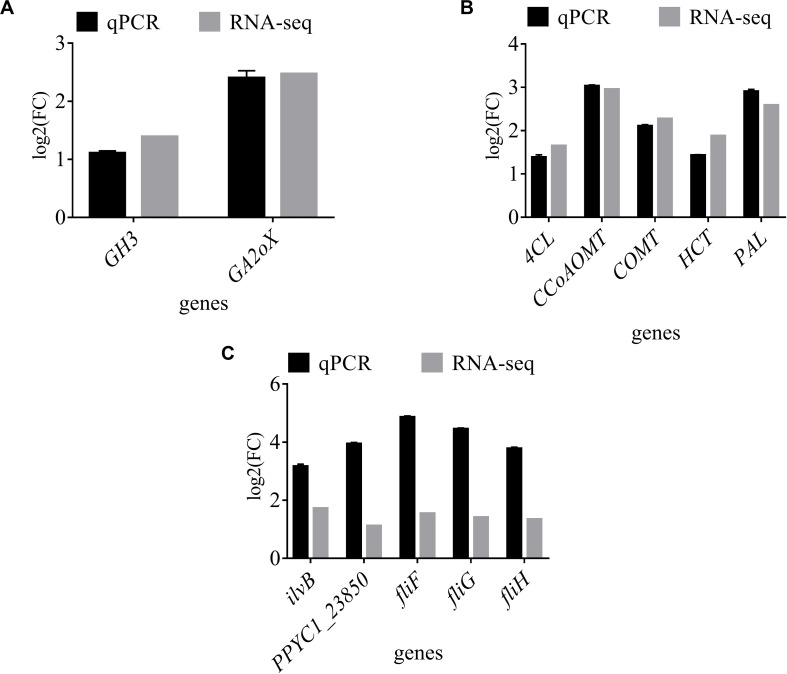
RNA-seq of tobacco co-cultured with strain YC0136 revealed that genes (GH3 and GA2ox) related to plant hormone transduction were up-regulated by 1.51–4.25 times and 1.57–4.98 times, respectively, compared with those in tobacco roots not cultured with strain YC0136. RT-qPCR showed that the expression of GH3 (gene56868) and GA2ox (gene14834) was up-regulated by 1.11 and 2.40 times, respectively (Figure 6A). The gene expression trends revealed by RT-qPCR were consistent with those determined by RNA-seq. In RNA-seq, five genes (4CL, CCoAOMT, COMT, HCT, and PAL) related to metabolism of phenylpropane compounds were up-regulated by 1.63–1.75 times, 1.63–2.61 times, 2.26–4.59 times, 1.5–1.96 times, and 1.51–3.26 times, respectively, compared with those in tobacco roots not cultured with strain YC0136. We used these five genes (gene65338, gene48930, gene51252, gene59104, and gene55404) encoding 4CL, CCoAOMT, COMT, HCT, and PAL, respectively, for RT-qPCR, and their expression was up-regulated by 1.39, 3.03, 2.11, 1.43, and 2.91 times, respectively (Figure 6B).
FIGURE 6.
RT-qPCR assay of genes in tobacco and YC0136. Fluorescence RT-qPCR to verify the accuracy of the transcriptome data. One microgram of purified total RNA was used as template for first-strand cDNA synthesis. Several genes from RNA-seq were selected for amplification by SYBR green RT-qPCR. The GAPDH gene of strain YC0136 and the actin gene of tobacco were selected as references. Relative expression levels were calculated using the delta-delta Ct method. (A) RT-qPCR results for genes related to plant hormone transduction in tobacco. (B) RT-qPCR results for genes related to phenylpropane compound metabolism in tobacco. (C) RT-qPCR results for genes in strain YC0136. Three biological replicates were carried out, and three technical replicates were performed. Error bars indicate the SD from the mean.
RT-qPCR results for strain YC0136 showed similar up-regulation of gene expression following interaction with tobacco as revealed by RNA-seq. Expression of ilvB, PPYC1_23850, fliF, fliG, and fliH was up-regulated by 3.17, 3.94, 4.85, 4.45, and 3.78 times, respectively, compared with that in strain YC0136 without tobacco roots (Figure 6C). The consistent trend in expression of these genes determined by RT-qPCR and RNA-seq indicated that the RNA-seq results were reliable.
Strain YC0136 Induces Plant Hormone Transduction in Tobacco
Early auxin response genes can be divided into three categories: Aux/IAAs, GH3, and SAURs (small auxin up RNAs) (Liu et al., 2005). After inoculating with strain YC0136, IAA, GH3, and SAUR genes in tobacco were up-regulated by varying degrees (Table 1). Five genes (gene28034, gene54165, gene2334, gene37942, and gene49814) encoding SAUR family proteins were up-regulated by 1.73–2.59 times compared with those in tobacco roots not cultured with strain YC0136. Following stimulation by strain YC0136, expression of the GH3 genes in tobacco increased by 1.51–4.25 times compared with that in tobacco not cultured with strain YC0136. The expression of auxin-responsive protein coding genes in tobacco roots was also up-regulated by 1.59–2.22 times.
TABLE 1.
Effects of strain YC0136 on the genes related to auxin and gibberellin in tobacco.
| Plant hormone | Gene ID | Log2(FC) | Gene | Product |
| IAA | gene28034 | 2.59 | SAUR | SAUR family protein |
| gene54165 | 2.20 | SAUR | SAUR family protein | |
| gene2334 | 1.89 | SAUR | SAUR family protein | |
| gene37942 | 1.84 | SAUR | SAUR family protein | |
| gene49814 | 1.73 | SAUR | SAUR family protein | |
| gene4468 | 4.25 | GH3 | Auxin-responsive GH3 gene family | |
| gene65249 | 1.69 | GH3 | Auxin-responsive GH3 gene family | |
| gene11692 | 1.63 | GH3 | Auxin-responsive GH3 gene family | |
| gene15243 | 1.51 | GH3 | Auxin-responsive GH3 gene family | |
| gene7537 | 2.22 | IAA | Auxin-responsive protein IAA | |
| gene32907 | 1.90 | IAA | Auxin-responsive protein IAA | |
| gene10414 | 1.74 | IAA | Auxin-responsive protein IAA | |
| gene1627 | 1.59 | IAA | Auxin-responsive protein IAA | |
| Gibberellin | gene45867 | 4.98 | GA2ox | Gibberellin 2-oxidase |
| gene22292 | 2.79 | GA2ox | Gibberellin 2-oxidase | |
| gene12319 | 2.68 | GA2ox | Gibberellin 2-oxidase | |
| gene14834 | 2.47 | GA2ox | Gibberellin 2-oxidase | |
| gene56942 | 1.91 | GA2ox | Gibberellin 2-oxidase | |
| gene55701 | 1.81 | GA2ox | Gibberellin 2-oxidase | |
| gene53350 | 1.60 | GA2ox | Gibberellin 2-oxidase | |
| gene11302 | 1.57 | GA2ox | Gibberellin 2-oxidase | |
| gene40263 | –1.69 | GA2ox | Gibberellin 2-oxidase | |
| gene1876 | –1.99 | GA2ox | Gibberellin 2-oxidase |
There are many enzymes involved in the biosynthesis of gibberellin in plants. Interaction with strain YC0136 increased the expression level of GA2ox genes coding gibberellin-2-oxidase (GA2ox) in tobacco by 1.57–4.98 times (Table 1).
Cis-zeatin O-glucosyltransferase converts cis-zeatin to zeatin glycoside. Interaction with strain YC0136 caused the expression of several genes encoding cis-zeatin O-glucosyltransferase in tobacco to change by varying degrees (Supplementary Table S2). Seven genes (gene51315, gene50, gene53518, gene68033, gene41639, gene48349, and gene24068) were down-regulated following culture with strain YC0136. Changes in the expression levels of genes encoding cis-zeatin O-glucosyltransferase showed that accumulation of zeatin is an effective response to interactions with strain YC0136.
Strain YC0136 Enhances Systemic Resistance in Tobacco
After interacting with strain YC0136, fls2 genes (gene32927, gene40590, and gene40591) of tobacco, coding flagellin-sensing 2 protein, were up-regulated by 1.87–3.35 times compared with those in tobacco roots not cultured with strain YC0136. This indicated that strain YC0136 may stimulate the early immune response in tobacco.
Strain YC0136 induces expression of transcription factors involved in plant stress tolerance
The WRKY transcriptional regulators belong to one of the largest transcriptional regulator families in plants. Interaction with strain YC0136 produced up-regulation of expression of some WRKY transcriptional regulator genes in tobacco (Table 2). Two genes (gene2945 and gene35472) encoding WRKY2 were up-regulated by 3.35 and 2.09 times, respectively, compared with those in tobacco roots not cultured with strain YC0136. Genes encoding WRKY33 (gene72767, gene6517, gene34934, and gene31508) were up-regulated by 1.67, 1.90, 1.84, and 1.52 times, respectively.
TABLE 2.
Effects of Strain YC0136 on the expression of transcription factors in tobacco.
| Type | Gene ID | Log2(FC) | Gene | Product |
| WRKY | gene2945 | 3.35 | WRKY2 | WRKY transcription factor 2 |
| gene35472 | 2.09 | WRKY2 | WRKY transcription factor 2 | |
| gene6517 | 1.90 | WRKY33 | WRKY transcription factor 33 | |
| gene34934 | 1.84 | WRKY33 | WRKY transcription factor 33 | |
| gene72767 | 1.67 | WRKY33 | WRKY transcription factor 33 | |
| gene31508 | 1.52 | WRKY33 | WRKY transcription factor 33 | |
| HSF | gene27131 | 3.47 | HSF | Heat shock transcription factor |
| gene62360 | 2.47 | HSF | Heat shock transcription factor | |
| gene38294 | 2.46 | HSF | Heat shock transcription factor | |
| gene23752 | 2.46 | HSF | Heat shock transcription factor | |
| gene33933 | 2.05 | HSF | Heat shock transcription factor | |
| gene38489 | 1.78 | HSF | Heat shock transcription factor | |
| gene3507 | 1.65 | HSF | Heat shock transcription factor | |
| gene9029 | −1.58 | HSF | Heat shock transcription factor | |
| MYB | gene16596 | 5.44 | MYB | Transcription factor MYB |
| gene62785 | 4.05 | MYB | Transcription factor MYB | |
| gene7813 | 3.68 | MYB | Transcription factor MYB | |
| gene19987 | 3.66 | MYB | Transcription factor MYB | |
| gene37246 | 3.37 | MYB | Transcription factor MYB | |
| gene41931 | 3.13 | MYB | Transcription factor MYB | |
| gene26405 | 3.03 | MYB | Transcription factor MYB | |
| gene41368 | 2.90 | MYB | Transcription factor MYB | |
| gene6921 | 2.73 | MYB | Transcription factor MYB | |
| gene37631 | 2.69 | MYB | Transcription factor MYB | |
| gene64315 | 2.65 | MYB | Transcription factor MYB | |
| gene32699 | 2.61 | MYB | Transcription factor MYB | |
| gene48183 | 2.49 | MYB | Transcription factor MYB | |
| gene41932 | 2.46 | MYB | Transcription factor MYB | |
| gene39829 | 2.40 | MYB | Transcription factor MYB | |
| gene70785 | 2.30 | MYB | Transcription factor MYB | |
| gene71054 | 2.25 | MYB | Transcription factor MYB | |
| gene7811 | 2.23 | MYB | Transcription factor MYB | |
| gene6521 | 2.22 | MYB | Transcription factor MYB | |
| gene39956 | 2.04 | MYB | Transcription factor MYB | |
| gene742 | 1.92 | MYB | Transcription factor MYB | |
| gene26446 | 1.90 | MYB | Transcription factor MYB | |
| gene18880 | 1.88 | MYB | Transcription factor MYB | |
| gene60170 | 1.55 | MYB | Transcription factor MYB | |
| gene15730 | 1.52 | MYB | Transcription factor MYB | |
| MYC2 | gene5340 | 1.73 | MYC2 | Transcription factor MYC2 |
Strain YC0136 caused changes in the expression of other transcription factors related to abiotic stress responses in plants, including heat shock transcription factors and MYB transcription factors. After interaction with strain YC0136, gene27131, gene62360, gene38294, gene23752, gene33933, gene38489, and gene3507 encoding HSFs in tobacco were up-regulated by 1.6–3.47 times compared with those in tobacco roots not cultured with strain YC0136. Strain YC0136 induced the expression of genes encoding MYB transcription regulators in tobacco. These genes were up-regulated by 1.52–5.44 times compared with those in tobacco roots not cultured with strain YC0136.
The MYC2 transcriptional regulatory factor is the core of the plant jasmonic acid signaling pathway. After interaction with strain YC0136, gene5340 encoding MYC2 in tobacco was up-regulated by 1.73 times.
Strain YC0136 induces expression of other genes related to systemic resistance in tobacco
Strain YC0136 can not only induce the expression of transcription factors related to stress tolerance, but also induce the systemic resistance of tobacco through the salicylic acid pathway and some secondary metabolites.
Salicylic acid is a widely recognized plant endogenous signaling molecule associated with a variety of plant resistances (Durner et al., 1997). In tobacco, we found that genes involved in the salicylic acid pathway were significantly up-regulated by strain YC0136 (Table 3). Expression of genes encoding SGT1 (pathogen-inducible salicylic acid glucosyltransferase) was significantly up-regulated compared with that in tobacco roots not cultured with strain YC0136. These nine genes (gene10335, gene32550, gene32965, gene34783, gene42409, gene42410, gene64161, gene64162, and gene64163) were up-regulated by 1.67–3.92 times.
TABLE 3.
Effects of strain YC136 on salicylic acid signaling pathway in tobacco.
| Gene ID | Log2 (FC) | Gene | Product |
| gene32550 | 3.92 | SGT1 | Pathogen-inducible salicylic acid glucosyltransferase |
| gene64162 | 3.91 | SGT1 | Pathogen-inducible salicylic acid glucosyltransferase |
| gene64163 | 3.85 | SGT1 | Pathogen-inducible salicylic acid glucosyltransferase |
| gene64161 | 3.5 | SGT1 | Pathogen-inducible salicylic acid glucosyltransferase |
| gene42409 | 3.14 | SGT1 | Pathogen-inducible salicylic acid glucosyltransferase |
| gene10335 | 3.05 | SGT1 | Pathogen-inducible salicylic acid glucosyltransferase |
| gene32965 | 2.18 | SGT1 | Pathogen-inducible salicylic acid glucosyltransferase |
| gene34783 | 1.71 | SGT1 | Pathogen-inducible salicylic acid glucosyltransferase |
| gene42410 | 1.67 | SGT1 | Pathogen-inducible salicylic acid glucosyltransferase |
Many secondary metabolites can enhance the resistance of plants to diseases. After inoculating with strain YC0136, some genes involved in the phenylpropanoid metabolic pathway in tobacco were up-regulated by varying degrees compared with those in tobacco roots not cultured with strain YC0136 (Table 4). Phenylalanine ammonia-lyase (PAL) is a key enzyme in the metabolic pathway of phenylpropanoids. The genes encoding PAL, gene23596, gene50105, gene55404, gene63885, gene13924, and gene2736, were up-regulated by 1.51–3.26 times following culture with strain YC0136. Four genes encoding cinnamic acid 4-hydroxylase (C4H) were up-regulated by 1.68–4.39 times, and the genes encoding shikimic acid O-hydroxycinnamoyl transferase, gene52491, gene59014, and gene68771, were up-regulated by 1.50–1.96 times. The expression of genes encoding caffeic acid 3-O-methyltransferase (COMT) (gene49647, gene51252, and gene57715) was up-regulated by 2.26–4.59 times, while nine genes related to caffeic acid CoA-O-methyl transferase (CCoAOMT) were up-regulated by 1.63–3.27 times. The up-regulation of genes encoding 4-coumarin-CoA ligase following culture with strain YC0136 was not significant.
TABLE 4.
Effects of Strain YC0136 on genes related to phenylpropane compound metabolism.
| Gene ID | Log2 (FC) | Gene | Product |
| gene48890 | 1.75 | 4CL | 4-Coumaroyl-CoA ligase |
| gene31505 | 1.66 | 4CL | 4-Coumaroyl-CoA ligase |
| gene65338 | 1.65 | 4CL | 4-Coumaroyl-CoA ligase |
| gene59318 | 1.63 | 4CL | 4-Coumaroyl-CoA ligase |
| gene14196 | 4.39 | C4H | Cinnamic acid 4-hydroxylase |
| gene45704 | 3.85 | C4H | Cinnamic acid 4-hydroxylase |
| gene16642 | 2.66 | C4H | Cinnamic acid 4-hydroxylase |
| gene73634 | 1.68 | C4H | Cinnamic acid 4-hydroxylase |
| gene49647 | 4.59 | COMT | Caffeic acid 3-O-methyltransferase |
| gene51252 | 2.27 | COMT | Caffeic acid 3-O-methyltransferase |
| gene57715 | 2.26 | COMT | Caffeic acid 3-O-methyltransferase |
| gene52406 | 2.61 | CCoAOMT | Caffeic acid CoA-O-methyl transferase |
| gene52407 | 3.27 | CCoAOMT | Caffeic acid CoA-O-methyl transferase |
| gene48930 | 2.95 | CCoAOMT | Caffeic acid CoA-O-methyl transferase |
| gene48931 | 2.74 | CCoAOMT | Caffeic acid CoA-O-methyl transferase |
| gene43754 | 2.2 | CCoAOMT | Caffeic acid CoA-O-methyl transferase |
| gene9422 | 1.86 | CCoAOMT | Caffeic acid CoA-O-methyl transferase |
| gene59406 | 1.74 | CCoAOMT | Caffeic acid CoA-O-methyl transferase |
| gene71741 | 1.73 | CCoAOMT | Caffeic acid CoA-O-methyl transferase |
| gene37383 | 1.63 | CCoAOMT | Caffeic acid CoA-O-methyl transferase |
| gene52491 | 1.96 | HCT | Shikimic acid O-hydroxycinnamoyl transferase |
| gene59014 | 1.88 | HCT | Shikimic acid O-hydroxycinnamoyl transferase |
| gene68771 | 1.50 | HCT | Shikimic acid O-hydroxycinnamoyl transferase |
| gene48554 | –2.57 | HCT | Shikimic acid O-hydroxycinnamoyl transferase |
| gene213 | –3.53 | HCT | Shikimic acid O-hydroxycinnamoyl transferase |
| gene23596 | 3.26 | PAL | Phenylalanine ammonia-lyase |
| gene50105 | 2.96 | PAL | Phenylalanine ammonia-lyase |
| gene55404 | 2.59 | PAL | Phenylalanine ammonia-lyase |
| gene63885 | 2.53 | PAL | Phenylalanine ammonia-lyase |
| gene13924 | 2.01 | PAL | Phenylalanine ammonia-lyase |
| gene2736 | 1.51 | PAL | Phenylalanine ammonia-lyase |
Laccase plays an important role in plant resistance to insects and fungi. The expression levels of genes encoding laccase were up-regulated by 1.85–4.81 times in tobacco roots cultured with strain YC0136 compared with those in tobacco roots not cultured with strain YC0136 (Supplementary Table S3). Up-regulation of laccase genes is helpful for enhancing the toughness of the tobacco rhizome, increasing the resistance of tobacco to pests and pathogens.
Strain YC0136 affects the expression of genes involved in tobacco metabolic pathways
After interacting with strain YC0136, some genes related to metabolism in tobacco were expressed differently. There were 51 DEGs related to amino acid metabolism (Supplementary Table S4). A total of 37 genes were up-regulated, while 14 genes were down-regulated following culture with strain YC0136. gene51112 encoding branched-chain amino acid transaminase was up-regulated by 5.76 times compared with that in tobacco roots not cultured with strain YC0136. In addition, gene57308 and gene9555, encoding asparagine synthetase involved in glutamine hydrolysis, were up-regulated by 5.0 and 3.96 times, respectively. Among the 14 down-regulated genes, gene42183 encoding glutamine synthetase was down-regulated by 1.96 times following culture with strain YC0136. gene60280 encoding threonine synthetase was down-regulated by 2.21 times. These results indicated that strain YC0136 can reduce glutamine content in tobacco by inducing glutamine degradation and inhibiting its biosynthesis.
There were 183 DEGs related to glycol metabolism, of which 71 genes were up-regulated and 112 genes were down-regulated following culture with strain YC0136 (Supplementary Table S5). Among the up-regulated genes, gene66000 and gene1858, which encode the reciprocal transformation of pentose and glucuronic acid, were up-regulated by 6.80 and 5.26 times, respectively, compared with those in tobacco roots not cultured with strain YC0136. Two genes, gene27593 and gene47222, involved in the metabolism of cyanidin-3-glucoside, were up-regulated by 5.14 and 4.54 times, respectively. Strain YC0136 also induced the expression of the gene (gene458809) encoding xylan 1,4-β-xylosidase, which was up-regulated by 1.56 times compared with that in tobacco roots not cultured with strain YC0136. In plants, β-fructofuranosidase is involved in the conversion of sucrose. The expression of genes (gene69223, gene6072, and gene53789) encoding β-fructofuranosidase was down-regulated by 4.44–4.54 times following culture with strain YC0136. These results indicated that strain YC0136 attenuates the sucrose conversion ability of tobacco.
Effects of Tobacco on Gene Expression in Strain YC0136
Tobacco stimulates the expression of auxin biosynthesis genes and motility genes in strain YC0136
Indole-3-pyruvate decarboxylase (IPDC) is a key enzyme in the IPyA pathway of auxin biosynthesis, which converts indole-3-pyruvic acid into indole-3-acetaldehyde (Malhotra and Srivastava, 2008). The similarity in protein sequence between ilvB (PPYC1_16985) of strain YC0136 and ipdC of P. polymyxa E681 was 100%. Gene PPYC1_23850 encodes aldehyde dehydrogenase, which converts indole-3-acetaldehyde into IAA. Under conditions of interaction with tobacco roots, expression of the ilvB gene in strain YC0136 was up-regulated by 1.72 times compared with that under conditions without tobacco roots, and expression of gene PPYC1_23850 was up-regulated by 1.12 times. This indicated that tobacco may promote auxin biosynthesis in strain YC0136. The flagellum motor switch is related to bacterial motility. Under interaction conditions, the expression of the flagellum motor switch gene fliG (PPYC1_14375) in strain YC0136 was up-regulated by 1.41 times compared with that in strain YC0136 without tobacco interaction. The transcription of genes encoding flagellum assembly proteins, fliH (PPYC1_14370) and fliF (PPYC1_14380), was up-regulated by 1.34 times and 1.54 times, respectively, following interaction with tobacco.
Tobacco affects the transporter genes of strain YC0136
After interacting with tobacco, genes related to transport in strain YC0136 were changed by varying degrees (Table 5). There were 29 genes up-regulated compared with their expression levels without tobacco interaction. Among them, 16 genes related to ABC transport were up-regulated by 1.59–4.32 times. The operon pstSABC, which is involved in inorganic phosphorus transport, was up-regulated 3.78–4.32 times. Three genes (PPYC1_06875, PPYC1_15095, and PPYC1_19620) belonging to the MFS transport family were up-regulated by 1.57, 4.34, and 2.20 times, respectively. The expression levels of partial genes in strain YC0136 were down-regulated following interaction with tobacco roots, compared with those in strain YC0136 without tobacco interaction. Four genes involved in ABC transport were down-regulated by 1.54–2.60 times. Two genes (PPYC1_19055 and PPYC1_22815) involved in MFS transfer were down-regulated by 1.67 times and 4.69 times, respectively. These benefit the growth of strain YC0136 by absorbing nutrients.
TABLE 5.
Effects of tobacco on the genes related to transport in strain YC0136.
| Gene ID | Log2 (FC) | Name | Product |
| PPYC1_17625 | 2.89 | Metal ABC transporter permease | |
| PPYC1_17630 | 2.78 | Manganese ABC transporter | |
| PPYC1_00910 | 1.6 | Nickel ABC transporter permease subunit NikC | |
| PPYC1_17640 | 1.94 | Manganese transporter | |
| PPYC1_03855 | 2.11 | phnD | Phosphate/phosphite/phosphonate ABC transporter substrate-binding protein |
| PPYC1_15975 | 3.78 | pstB | Phosphate ABC transporter ATP-binding protein |
| PPYC1_03845 | 1.75 | phnE | Phosphonate ABC transporter, permease protein PhnE |
| PPYC1_00905 | 1.72 | Nickel ABC transporter permease subunit NikB | |
| PPYC1_17635 | 2.56 | Metal ABC transporter ATP-binding protein | |
| PPYC1_15980 | 3.96 | pstA | Phosphate ABC transporter, permease protein PstA |
| PPYC1_15985 | 3.93 | pstC | Phosphate ABC transporter permease subunit PstC |
| PPYC1_15910 | 4.32 | pstB | Phosphate ABC transporter ATP-binding protein |
| PPYC1_15990 | 4.16 | pstS | Phosphate ABC transporter substrate-binding protein PstS |
| PPYC1_06875 | 1.57 | oxlT | MFS transporter |
| PPYC1_15095 | 4.34 | MFS transporter | |
| PPYC1_19620 | 2.2 | MFS transporter | |
| PPYC1_16575 | 1.67 | Lantibiotic ABC transporter permease | |
| PPYC1_20185 | 2.22 | Glutamine ABC transporter permease | |
| PPYC1_06535 | 1.5 | Efflux RND transporter periplasmic adaptor subunit | |
| PPYC1_03665 | 1.75 | arsB | Arsenic transporter |
| PPYC1_15275 | 3.31 | Ammonium transporter | |
| PPYC1_18230 | 1.91 | ABC transporter permease | |
| PPYC1_16580 | 1.59 | ABC transporter permease | |
| PPYC1_02025 | –1.54 | Amino acid ABC transporter substrate-binding protein | |
| PPYC1_01710 | –1.58 | ABC transporter permease | |
| PPYC1_19055 | –1.67 | araE | MFS transporter |
| PPYC1_00995 | –1.72 | PTS sugar transporter subunit IIB | |
| PPYC1_13110 | –2.04 | znuB | Metal ABC transporter permease |
| PPYC1_13115 | –2.6 | znuC | Metal ABC transporter ATP-binding protein |
| PPYC1_00515 | –3.13 | smr | QacE family quaternary ammonium compound efflux SMR transporter |
| PPYC1_22815 | –4.69 | MFS transporter |
To summarize, P. polymyxa YC0136 isolated from the rhizosphere of tobacco directly promoted the growth of tobacco by secreting auxin and solubilizing phosphate. Strain YC0136 also promoted the growth of tobacco by inducing the expression of genes related to plant hormone signal transduction in tobacco. Strain YC0136 antagonized pathogens by secreting secondary metabolites, and also induced systemic resistance in tobacco. This study contributes to our understanding of the growth-promoting mechanisms of strain YC0136 on tobacco and provides a theoretical basis for the application of P. polymyxa YC0136 as a biological fertilizer.
Discussion
This study was carried out to understand the promotion mechanism of strain YC0136 on improving the agronomic traits of tobacco and enhancing the resistance of tobacco. We found strain YC0136 can antagonize pathogenic bacteria and fungi. In our previous report (Liu et al., 2017), we used antiSMASH database to analyze the whole genome of strain YC0136 and found there were 14 gene clusters related to secondary metabolites controlling pathogenic bacteria and fungi. The existence of these gene clusters may cause the ability of strain YC0136 controlling pathogens. The results of pot experiments indicated that strain YC0136 can improve the agronomic characteristics of tobacco.
The transcriptome of tobacco roots during interaction with strain YC0136 revealed that strain YC0136 can stimulate auxin, gibberellin and cytokinin signal transduction in tobacco. Gibberellin-2-oxidase (GA2ox) is a key enzyme that plays a negative regulatory role in the gibberellin biosynthesis pathway (Wuddineh et al., 2015). Interaction with strain YC0136 increased the expression level of GA2ox genes in tobacco. Up-regulated expression of GA2ox can reduce gibberellin content and improve plant architecture (Wuddineh et al., 2015). High expression levels of genes encoding GA2ox could increase the response of tobacco to abiotic stress (Colebrook et al., 2014). In addition, gibberellin catabolism helps plants respond to adverse conditions, such as drought and high salinity (Zawaski and Busov, 2014; Zhou and Underhill, 2016). These results indicate that strain YC0136 might be beneficial to plant growth and promote plant stress tolerance. Zeatin is a natural cytokinin in plants that promotes cell division and delays plant senescence (Martin et al., 1999, 2001). Strain YC0136 attenuated the ability of zeatin conversion in tobacco by inhibiting the expression of cis-zeatin O-glucosyltransferase. This reduced the degradation of zeatin, allowing zeatin to play a role as a cytokinin (Kudo et al., 2012; Shang et al., 2016).
After interacting with strain YC0136, a large number of genes in tobacco were expressed differently. Flagellin-sensing 2 (FLS2) plays an important role in plant and microbial recognition (Chinchilla et al., 2007). FLS2 interacts with 22 N-terminal conserved amino acids in bacterial flagellin to induce plant defense responses. In Arabidopsis thaliana fls2 mutant, expression of VvFLS2 could restore flg22-induced H2O2 production, and VvFLS2 could induce grapevine immune response to Botrytis cinereal (Trdá et al., 2014). In tobacco interacted with strain YC0136, fls2 was up-regulated. We speculated that FLS2 protein stimulated the immune response of tobacco through recognizing with flg22 peptides in strain YC0136. Strain YC0136 may also overcome MAMP-triggered immunity to colonize plants as Burkholderia phytofirmans (Trdá et al., 2014). Transcriptional regulatory factors have extremely important effects on the growth and development of organisms (Yamaguchi-Shinozaki and Shinozaki, 2006; Pankaj et al., 2015; Lin et al., 2017). WRKY proteins are plant-specific transcriptional regulatory factors involved in plant growth and seed development (Ay et al., 2009; Jiang and Yu, 2009), plant disease resistance (Dang et al., 2013; Wang et al., 2013), plant signal transduction (Yang et al., 1999; Antoni et al., 2011; Peng et al., 2012), and abiotic stress (Zou et al., 2010; Sun et al., 2015). Mzid et al. (2007) reported that overexpression of VvWRKY2 in tobacco could enhance broad resistance to necrotrophic fungal pathogens. In this study, we found that gene2945 in tobacco, encoding transcription regulator WRKY2, was up-regulated by 3.35 times following culture with strain YC0136, compared with its expression level without strain YC0136. This is conducive to enhancing tobacco resistance to pathogens. Another transcription regulator in plants, WRKY33, also plays an important role in plant growth, development, and other life activities. WRKY33 can not only enhance the tolerance of plants to salt stress (Jiang and Deyholos, 2009; Jiang et al., 2014), but also plays an essential role in defense against necrotrophic fungi (Birkenbihl et al., 2012). After interacting with strain YC0136, the genes encoding WRKY33 were also up-regulated in tobacco. This indicates that strain YC0136 may induce tobacco plants to develop systemic resistance to better respond to abiotic and biological stresses.
In the transcriptome of tobacco interacted with stain YC0136, the genes of PAL, 4CL, COMT, CCoAOMT, shikimic acid O-hydroxycinnamoyl transferase, and C4H were up-regulated in varying degrees. The six enzymes mentioned above are all involved in the biosynthesis of phenylpropanoids (Ferrer et al., 2008). Phenylpropanoids are secondary metabolites biosynthesized from phenylalanine in plants and they can enhance the ability of plants to resist various biotic and abiotic stresses (Ye and Varner, 1995; Toquin et al., 2003; Korkina, 2007; Yu and Jez, 2008; Liu et al., 2016; Zhang et al., 2020). Chen et al. reported that overexpression of LjPAL could enhance the resistance to Pseudomonas syringae pv. phaseolicola NPS3121 infection in L. japonicus (Chen et al., 2017). Yamamoto et al. (1998) reported that the activity of PAL was increased in tobacco treated with phosphorus starvation and it protected tobacco cells from the cytotoxic lipid peroxidation caused by the combination of aluminum and iron. In wheat, overexpression of TaCOMT-3D can enhance the resistance of wheat to sharp eyespot disease (Wang et al., 2018). The overexpression of 4CL and STS in tobacco could not only enhance the antagonistic activity against the pathogen Monilinia fructicola, but also significantly enhance the tolerance to salt stress (He et al., 2018). In previous studies, pathogens can induce the up-regulation of PAL and other related enzymes genes in plants. In this study, although strain YC0136 was a beneficial bacteria, the PAL, COMT and other enzyme genes of tobacco interacted with strain YC0136 were also up-regulated. The results indicated that strain YC0136 could induce the systemic resistance of tobacco by stimulating phenylpropanoids metabolism in the absence of pathogenic bacteria and adversity. Besides, enzymes of PAL, COMT and CCoAOMT are also involved in different methylation pathways in lignin biosynthesis of plants (Ye and Varner, 1995; Chen et al., 2017). The up-regulation of PAL, COMT, and CCoAOMT might increase the content of lignin in tobacco. Lignin is an important part of secondary thickening of cell wall, it can enhance mechanical strength of plants, and protect plant against insects and pathogens (Dauwe et al., 2010). The accumulation of lignin helps to enhance the mechanical strength of tobacco, thus enhancing the resistance of tobacco to pests and pathogens.
Tobacco also affected the growth of strain YC0136 under conditions of their interaction. In strain YC0136, the expression of genes (ilvB and PPYC1_23850) involved in auxin biosynthesis was up-regulated in the presence of tobacco roots. This indicates that materials in root exudates may induce auxin biosynthesis in strain YC0136. Kwon et al. (2016) also reported that the interaction between P. polymyxa E681 and Arabidopsis thaliana results in the enhancement of auxin secretion in P. polymyxa E681. The expression of genes related to the phosphate-specific transport system (pstSCAB, phoU) was significantly up-regulated by interaction with tobacco roots. The pst operon is a high-affinity phosphate transport system (Qi et al., 1997; Aguena et al., 2002). The up-regulation of pst operon expression is beneficial to meet the phosphorus requirement and promote the growth of strain YC0136. The transcriptome revealed that 31 genes related to transport were expressed differently in strain YC0136 in the presence of tobacco roots. These genes are related to the transport of metal ions, phosphates, glutamine, arsenic, and ammonium. They are beneficial for enhancing the absorption of inorganic salt ions and the growth of strain YC0136.
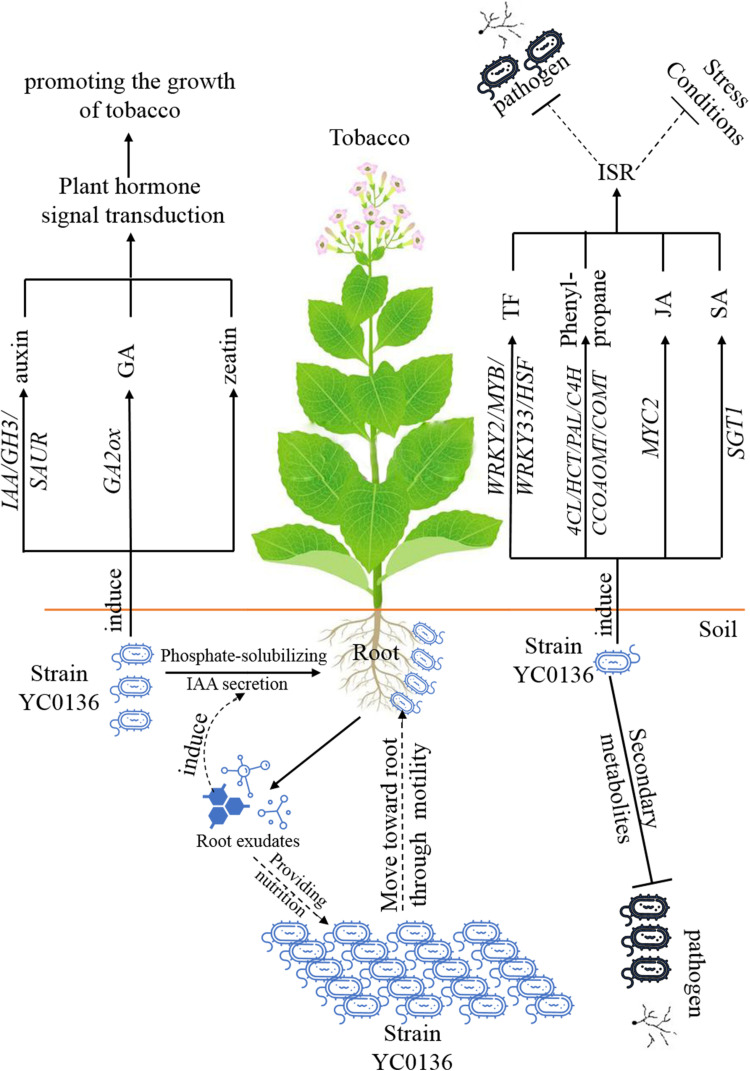
We summarize the relationship between strain YC0136 and tobacco in a pattern diagram, as shown in Figure 7. Tobacco root exudates may attract strain YC0136, then YC0136 cells move toward the tobacco root through flagella rotation. The tobacco root exudates may provide nutrients for the growth of strain YC0136. Strain YC0136 might promote the growth of tobacco by secreting auxin and solubilizing phosphate. At the same time, strain YC0136 promotes the growth of tobacco by inducing auxin, gibberellin, and zeatin signaling. Strain YC0136 biosynthesizes secondary metabolites to antagonize pathogens. Strain YC0136 also induces systemic resistance of tobacco by stimulating the expression of transcriptional regulatory factors (such as WRKY and MYB, etc.) and related genes of the phenylpropanoid biosynthesis pathway.
FIGURE 7.
Proposed model of the growth-promoting mechanisms of strain YC0136 on tobacco.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
CW, YD, and BD designed the study. HL, JW, and HS performed the laboratory work and analyzed the data. HL wrote the manuscript. KL, XH, YP, and JL advised the manuscript. YD, BD, and CW supported the study. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by the National Natural Science Foundation of China (31700094 and 31770115), the National Key Research and Development Program of China (2017YFD0200804), the funds of Shandong “Double Tops” Program (SYL2017XTTD03), the Science and Technology Project of Guizhou (ZYJ2017-8), and the Key Field Research and Development Program of Guangdong Province (2019B020218009).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.584174/full#supplementary-material
References
- Abdallah Y., Yang M., Zhang M., Masum M. M. I., Bin L. (2019). Plant growth promotion and suppression of bacterial leaf blight in rice by Paenibacillus polymyxa Sx3. Lett. Appl. Microbiol. 68 423–429. 10.1111/lam.13117 [DOI] [PubMed] [Google Scholar]
- Aguena M., Yagil E., Spira B. (2002). Transcriptional analysis of the pst operon of Escherichia coli. Mol. Genet. Genomics 268 518–524. 10.1007/s00438-002-0764-4 [DOI] [PubMed] [Google Scholar]
- Ahimou F. O., Jacques P., Deleu M. (2000). Surfactin and iturin A effects on Bacillus subtilis surface hydrophobicity. Enzyme Microb. Technol. 27 749–754. 10.1016/s0141-0229(00)00295-7 [DOI] [PubMed] [Google Scholar]
- Ajilogba C., Babalola O., Adebola P., Adeleke R. (2016). Evaluation of PGPR and biocontrol activities of bacteria isolated from Bambara groundnut rhizosphere. New Biotechnol. 33 S128–S129. [Google Scholar]
- Ali S. (2010). Inhibitory activity of Paenibacillus macerans and Paenibacillus polymyxa against Ralstonia solanacearum. Afr. J. Microbiol. Res. 4 2048–2054. [Google Scholar]
- Anand R., Grayston S., Chanway C. (2013). N2-fixation and seedling growth promotion of Lodgepole Pine by endophytic Paenibacillus polymyxa. Microb. Ecol. 66 369–374. 10.1007/s00248-013-0196-1 [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., Huber W. (2015). HTSeq-a python framework to work with high-throughput sequencing data. Bioinformatics 31 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoni R., Rodriguez L., Gonzalez-Guzman M., Pizzio G. A., Rodriguez P. L. (2011). News on ABA transport, protein degradation, and ABFs/WRKYs in ABA signaling. Curr. Opin. Plant Biol. 14 547–553. 10.1016/j.pbi.2011.06.004 [DOI] [PubMed] [Google Scholar]
- Ash C., Priest F. G., Collins M. D. (1993). Molecular identification of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test. Antonie Van Leeuwenhoek 64 253–260. 10.1007/bf00873085 [DOI] [PubMed] [Google Scholar]
- Ay N., Irmler K., Fischer A., Uhlemann R., Reuter G., Humbeck K. (2009). Epigenetic programming via histone methylation at WRKY53 controls leaf senescence in Arabidopsis thaliana. Plant J. 58 333–346. 10.1111/j.0960-7412.2009.03782.x [DOI] [PubMed] [Google Scholar]
- Bal H. B., Das S., Dangar T. K., Adhya T. K. (2013a). ACC deaminase and IAA producing growth promoting bacteria from the rhizosphere soil of tropical rice plants. J. Basic Microbiol. 53 972–984. 10.1002/jobm.201200445 [DOI] [PubMed] [Google Scholar]
- Bal H. B., Nayak L., Das S., Adhya T. K. (2013b). Isolation of ACC deaminase producing PGPR from rice rhizosphere and evaluating their plant growth promoting activity under salt stress. Plant Soil 366 93–105. 10.1007/s11104-012-1402-5 [DOI] [Google Scholar]
- Basak B. B., Biswas D. R. (2009). Influence of potassium solubilizing microorganism (Bacillus mucilaginosus) and waste mica on potassium uptake dynamics by sudan grass (Sorghum vulgare Pers.) grown under two Alfisols. Plant Soil 317 235–255. 10.1007/s11104-008-9805-z [DOI] [Google Scholar]
- Birkenbihl R. P., Diezel C., Somssich I. E. (2012). Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol. 159 266–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhead K. D., Schisler D. A., Slininger P. J. (1994). Pyrrolnitrin production by biological control agent Pseudomonas cepacia B37w in culture and in colonized wounds of potatoes. Appl. Environ. Microbiol. 60 2031–2039. 10.1128/aem.60.6.2031-2039.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Pi H., Chandrangsu P., Li Y., Wang Y., Zhou H., et al. (2018). Antagonism of two plant-growth promoting Bacillus velezensis isolates against Ralstonia solanacearum and Fusarium oxysporum. Sci. Rep. 8:4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardwell K. F., Kling J. G., Maziya-Dixon B., Bosque-Pérez N. A. (2000). Interactions between Fusarium verticillioides, Aspergillus flavus, and insect infestation in four maize genotypes in Lowland Africa. Phytopathology 90 276–284. 10.1094/phyto.2000.90.3.276 [DOI] [PubMed] [Google Scholar]
- Chen Y., Li F., Tian L., Huang M., Deng R., Li X., et al. (2017). The phenylalanine ammonia lyase gene LjPAL1 is involved in plant defense responses to pathogens and plays diverse roles in Lotus japonicus-rhizobium symbioses. Mol. Plant Microbe Interact. 30 739–753. 10.1094/mpmi-04-17-0080-r [DOI] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J. D., et al. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448 497–500. 10.1038/nature05999 [DOI] [PubMed] [Google Scholar]
- Colebrook E. H., Thomas S. G., Phillips A. L., Hedden P. (2014). The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 217 67–75. 10.1242/jeb.089938 [DOI] [PubMed] [Google Scholar]
- Cruz D. N., Perazella M. A., Bellomo R., Cal M. D. (2007). Effectiveness of polymyxin B-immobilized fiber column in sepsis: a systematic review. Crit. Care 11:R47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang F. F., Wang Y. N., Yu L., Eulgem T., Lai Y., Liu Z. Q., et al. (2013). CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 36 757–774. 10.1111/pce.12011 [DOI] [PubMed] [Google Scholar]
- Dauwe R., Morreel K., Goeminne G., Gielen B., Rohde A., Van Beeumen J., et al. (2010). Molecular phenotyping of lignin-modified tobacco reveals associated changes in cell-wall metabolism, primary metabolism, stress metabolism and photorespiration. Plant J. 52 263–285. 10.1111/j.1365-313x.2007.03233.x [DOI] [PubMed] [Google Scholar]
- Djaya L., Istifadah N., Hartati S., Joni I. M. (2019). In vitro study of plant growth promoting rhizobacteria (PGPR) and endophytic bacteria antagonistic to Ralstonia solanacearum formulated with graphite and silica nano particles as a biocontrol delivery system (BDS). Biocatal. Agric. Biotechnol. 19:101153 10.1016/j.bcab.2019.101153 [DOI] [Google Scholar]
- Durner J., Shah J., Klessig D. F. (1997). Salicylic acid and disease resistance in plants. Trends Plant Sci. 2 266–274. [Google Scholar]
- Elías J. M., Guerrero-Molina M. F., Martínez-Zamora M. G., Díaz-Ricci J. C., Pedraza R. O. (2018). Role of ethylene and related gene expression in the interaction between strawberry plants and the plant growth-promoting bacterium Azospirillum brasilense. Plant Biol. 20 490–496. 10.1111/plb.12697 [DOI] [PubMed] [Google Scholar]
- Ferrer J. L., Austin M. B., Stewart C., Jr., Noel J. P. (2008). Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol. Biochem. 46 356–370. 10.1016/j.plaphy.2007.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H. S., Lee K. (2006). Effect of co-inoculation with phosphate and potassium solubilizing bacteria on mineral uptake and growth of pepper and cucumber. Plant Soil Environ. 52 130–136. 10.17221/3356-pse [DOI] [Google Scholar]
- He X. C., Xue F. Y., Zhang L. L., Guo H. L., Ma L. Q., Yang M. F. (2018). Overexpressing fusion proteins of 4-coumaroyl-CoA ligase (4CL) and stilbene synthase (STS) in tobacco plants leading to resveratrol accumulation and improved stress tolerance. Plant Biotechnol. Rep. 12 295–302. 10.1007/s11816-018-0494-7 [DOI] [Google Scholar]
- Ibort P., Molina S., Ruiz-Lozano J. M., Aroca R. (2018). Molecular insights into the involvement of a never Ripe Receptor in the interaction between two beneficial soil bacteria and tomato plants under well-watered and drought conditions. Mol. Plant Microbe Interact. 31 633–650. 10.1094/mpmi-12-17-0292-r [DOI] [PubMed] [Google Scholar]
- Jiang W., Yu D. (2009). Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol. 9:96. 10.1186/1471-2229-9-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Deyholos M. K. (2009). Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 69 91–105. 10.1007/s11103-008-9408-3 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Duan Y., Yin J., Ye S., Zhu J., Zhang F., et al. (2014). Genome-wide identification and characterization of the Populus WRKY transcription factor family and analysis of their expression in response to biotic and abiotic stresses. J. Exp. Bot. 65 6629–6644. 10.1093/jxb/eru381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Wu Y., Xu W. S., Cheng Y. H., Chen J. D., Xu L., et al. (2012). IAA-producing bacteria and bacterial-feeding nematodes promote Arabidopsis thaliana root growth in natural soil. Eur. J. Soil Biol. 52 20–26. 10.1016/j.ejsobi.2012.05.003 [DOI] [Google Scholar]
- Kagan B. M., Krevsky D., Milzer A., Locke M. (1951). Polymyxin B and polymyxin E; clinical and laboratory studies. J. Lab. Clin. Med. 37 402–414. [PubMed] [Google Scholar]
- Kanse O. S., Whitelawweckert M., Kadam T., Bhosale H. J. (2015). Phosphate solubilization by stress-tolerant soil fungus Talaromyces funiculosus SLS8 isolated from the Neem rhizosphere. Ann. Microbiol. 65 85–93. 10.1007/s13213-014-0839-6 [DOI] [Google Scholar]
- Khan N., Zandi P., Ali S., Mehmood A., Adnan Shahid M. (2018). Impact of salicylic acid and PGPR on the drought tolerance and phytoremediation potential of Helianthus annus. Front. Microbiol. 9:2507. 10.3389/fmicb.2018.02507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T., Seo H. D., Hennighausen L., Lee D., Kang K. (2018). Octopus-toolkit: a workflow to automate mining of public epigenomic and transcriptomic next-generation sequencing data. Nucleic Acids Res. 46:e53. 10.1093/nar/gky083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. S., Kotnala B., Kim Y. H., Jeon Y. (2016). Biological characteristics of Paenibacillus polymyxa GBR-1 involved in root rot of stored Korean ginseng. J. Ginseng Res. 40 453–461. 10.1016/j.jgr.2015.09.003 [DOI] [Google Scholar]
- Korkina L. G. (2007). Phenylpropanoids as naturally occurring antioxidants: from plant defense to human health. Cell. Mol. Biol. 53 15–25. [PubMed] [Google Scholar]
- Kudo T., Makita N., Kojima M., Tokunaga H., Sakakibara H. (2012). Cytokinin activity of cis-zeatin and phenotypic alterations induced by overexpression of putative cis-zeatin-O-glucosyltransferase in rice. Plant Physiol. 160 319–331. 10.1104/pp.112.196733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y. S., Lee D. Y., Rakwal R., Baek S. B., Lee J. H., Kwak Y. S., et al. (2016). Proteomic analyses of the interaction between the plant-growth promoting rhizobacterium Paenibacillus polymyxa E681 and Arabidopsis thaliana. Proteomics 16 122–135. 10.1002/pmic.201500196 [DOI] [PubMed] [Google Scholar]
- Lata H., Li X. C., Silva B., Moraes R. M., Halda-Alija L. (2006). Identification of IAA-producing endophytic bacteria from micropropagated Echinacea plants using 16S rRNA sequencing. Plant Cell Tissue Organ Cult. 85 353–359. 10.1007/s11240-006-9087-1 [DOI] [Google Scholar]
- Lee B., Farag M. A., Park H. B., Kloepper J. W., Lee S. H., Ryu C. M. (2012). Induced resistance by a long-chain bacterial volatile: elicitation of plant systemic defense by a C13 volatile produced by Paenibacillus polymyxa. PLoS One 7:e48744. 10.1371/journal.pone.0048744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lery L. M. S., Hemerly A. S., Nogueira E. M., Krüger W. M. A. V., Bisch P. M. (2010). Quantitative proteomic analysis of the interaction between the endophytic plant-growth-promoting bacterium. Mol. Plant Microbe Interact. 24 562–576. 10.1094/mpmi-08-10-0178 [DOI] [PubMed] [Google Scholar]
- Lin T., Yang W., Lu W., Wang Y., Qi X. (2017). Transcription factors PvERF15 and PvMTF-1 form a cadmium stress transcriptional pathway. Plant Physiol. 173 1565–1573. 10.1104/pp.16.01729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Liu K., Li Y. H., Wang C. Q., Hou Q. H., Xu W. F., et al. (2017). Complete genome sequence of Paenibacillus polymyxa YC0136, a plant growth–promoting rhizobacterium isolated from tobacco rhizosphere. Genome Announc. 5:e01635-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Kang B. C., Jiang H., Moore S. L., Li H., Watkins C. B., et al. (2005). A GH3-like gene, CcGH3, isolated from Capsicum chinense L. fruit is regulated by auxin and ethylene. Plant Mol. Biol. 58 447–464. 10.1007/s11103-005-6505-4 [DOI] [PubMed] [Google Scholar]
- Liu Z., Luan Y., Li J., Yin Y. (2016). Expression of a tomato MYB gene in transgenic tobacco increases resistance to Fusarium oxysporum and Botrytis cinerea. Eur. J. Plant Pathol. 144 607–617. 10.1007/s10658-015-0799-0 [DOI] [Google Scholar]
- Livak K., Schmittgen T. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Rajkumar M., Freitas H. (2009). Inoculation of plant growth promoting bacterium Achromobacter xylosoxidans strain Ax10 for the improvement of copper phytoextraction by Brassica juncea. J. Environ. Manage. 90 831–837. 10.1016/j.jenvman.2008.01.014 [DOI] [PubMed] [Google Scholar]
- Malhotra M., Srivastava S. (2008). An ipdC gene knock-out of Azospirillum brasilense strain SM and its implications on indole-3-acetic acid biosynthesis and plant growth promotion. Antonie Van Leeuwenhoek 93 425–433. 10.1007/s10482-007-9207-x [DOI] [PubMed] [Google Scholar]
- Mark G. L., Dow J. M., Kiely P. D., Higgins H., Haynes J., Baysse C., et al. (2005). Transcriptome profiling of bacterial responses to root exudates identifies genes involved in microbe-plant interactions. Proc. Natl. Acad. Sci. U.S.A. 102 17454–17459. 10.1073/pnas.0506407102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R. C., Mok M. C., Habben J. E., Mok D. W. S. (2001). A maize cytokinin gene encoding an O-glucosyltransferase specific to cis-zeatin. Proc. Natl. Acad. Sci. U.S.A. 98 5922–5926. 10.1073/pnas.101128798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R. C., Mok M. C., Mok D. W. S. (1999). Isolation of a cytokinin gene, ZOG1, encoding zeatin O-glucosyltransferase from Phaseolus lunatus. Proc. Natl. Acad. Sci. U.S.A. 96 284–289. 10.1073/pnas.96.1.284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei L., Liang Y., Zhang L., Wang Y., Guo Y. (2014). Induced systemic resistance and growth promotion in tomato by an indole-3-acetic acid-producing strain of Paenibacillus polymyxa. Ann. Appl. Biol. 165 270–279. 10.1111/aab.12135 [DOI] [Google Scholar]
- Michael L., Thierry H., Anton H. (1997). Production of auxin and other indolic and phenolic compounds by Paenibacillus polymyxa strains isolated from different proximity to plant roots. FEMS Microbiol. Ecol. 22 325–334. 10.1111/j.1574-6941.1997.tb00384.x [DOI] [Google Scholar]
- Mohamed I., Eid K. E., Abbas M. H. H., Salem A. A., Ahmed N., Ali M., et al. (2019). Use of plant growth promoting rhizobacteria (PGPR) and mycorrhizae to improve the growth and nutrient utilization of common bean in a soil infected with white rot fungi. Ecotoxicol. Environ. Saf. 171 539–548. 10.1016/j.ecoenv.2018.12.100 [DOI] [PubMed] [Google Scholar]
- Mohd Din A. R. J., Rosli M. A., Mohamad Azam Z., Othman N. Z., Sarmidi M. R. (2020). Paenibacillus polymyxa role involved in phosphate solubilization and growth promotion of Zea mays under abiotic stress condition. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 90 63–71. 10.1007/s40011-019-01081-1 [DOI] [Google Scholar]
- Mohite B. (2013). Isolation and characterization of indole acetic acid (IAA) producing bacteria from rhizospheric soil and its effect on plant growth. J. Soil Sci. Plant Nutr. 13 638–649. [Google Scholar]
- Muthukumar T., Udaiyan K. (2010). Growth response and nutrient utilization of Casuarina equisetifolia seedlings inoculated with bioinoculants under tropical nursery conditions. New For. 40 101–118. 10.1007/s11056-009-9186-z [DOI] [Google Scholar]
- Mzid R., Marchive C., Blancard D., Deluc L., Barrieu F., Corio-Costet M. F., et al. (2007). Overexpression of VvWRKY2 in tobacco enhances broad resistance to necrotrophic fungal pathogens. Physiol. Plant. 131 434–447. 10.1111/j.1399-3054.2007.00975.x [DOI] [PubMed] [Google Scholar]
- Niu B., Vater J., Rueckert C., Blom J., Borriss R. (2013). Polymyxin P is the active principle in suppressing phytopathogenic Erwinia spp. by the biocontrol rhizobacterium Paenibacillus polymyxa M-1. BMC Microbiol. 13:137. 10.1186/1471-2180-13-137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankaj B., Naika B. N. M., Doni J. N., Ramanathan S., Khader S., Bones A. M. (2015). Transcriptional regulatory networks in Arabidopsis thaliana during single and combined stresses. Nucleic Acids Res. 44 3147–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X., Hu Y., Tang X., Zhou P., Deng X., Wang H., et al. (2012). Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta 236 1485–1498. 10.1007/s00425-012-1698-7 [DOI] [PubMed] [Google Scholar]
- Phi Q. T., Park Y. M., Seul K. J., Ryu C. M., Ghim S. Y. (2010). Assessment of root-associated Paenibacillus polymyxa groups on growth promotion and induced systemic resistance in pepper. J. Microbiol. Biotechnol. 20 1605–1613. [PubMed] [Google Scholar]
- Príncipe A., Alvarez F., Castro M. G., Zacchi L. F., Fischer S. E., Mori G. B., et al. (2007). Biocontrol and PGPR features in native strains isolated from saline soils of Argentina. Curr. Microbiol. 55 314–322. 10.1007/s00284-006-0654-9 [DOI] [PubMed] [Google Scholar]
- Puri A., Padda K. P., Chanway C. P. (2016). Evidence of nitrogen fixation and growth promotion in canola (Brassica napus L.) by an endophytic diazotroph Paenibacillus polymyxa P2b-2R. Biol. Fertil. Soils 52 119–125. 10.1007/s00374-015-1051-y [DOI] [Google Scholar]
- Qi Y., Kobayashi Y., Hulett F. M. (1997). The pst operon of Bacillus subtilis has a phosphate-regulated promoter and is involved in phosphate transport but not in regulation of the pho regulon. J. Bacteriol. 179 2534–2539. 10.1128/jb.179.8.2534-2539.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recep K., Fikrettin S., Erkol D., Cafer E. (2009). Biological control of the potato dry rot caused by Fusarium species using PGPR strains. Biol. Control 50 194–198. 10.1016/j.biocontrol.2009.04.004 [DOI] [Google Scholar]
- Salvo L. P. D., Cellucci G. C., Carlino M. E., De Salamone I. E. G. (2018). Plant growth-promoting rhizobacteria inoculation and nitrogen fertilization increase maize (Zea mays L.) grain yield and modified rhizosphere microbial communities. Appl. Soil Ecol. 126 113–120. 10.1016/j.apsoil.2018.02.010 [DOI] [Google Scholar]
- Santoro M. V., Cappellari L. R., Giordano W., Banchio E. (2015). Plant growth-promoting effects of native Pseudomonas strains on Mentha piperita (peppermint): an in vitro study. Plant Biol. 17 1218–1226. [DOI] [PubMed] [Google Scholar]
- Sarode P. D., Rane M. P., Chaudhari B. L., Chincholkar S. B. (2007). Screening for siderophore producing PGPR from black cotton soils of North Maharashtra. Curr. Trends Biotechnol. Pharm. 1 96–105. [Google Scholar]
- Sayyed R. Z., Patel P. R. (2011). Biocontrol potential of siderophore producing heavy metal resistant Alcaligenes sp. and Pseudomonas aeruginosa RZS3 vis-à-vis organophosphorus fungicide. Indian J. Microbiol. 51 266–272. 10.1007/s12088-011-0170-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang X. L., Xie R. R., Tian H., Wang Q. L., Guo F. Q. (2016). Putative zeatin O-glucosyltransferase OscZOG1 regulates root and shoot development and formation of agronomic traits in rice. J. Integr. Plant Biol. 58 627–641. 10.1111/jipb.12444 [DOI] [PubMed] [Google Scholar]
- Shao J. H., Li S. Q., Zhang N., Cui X. S., Zhou X., Zhang G. S., et al. (2015). Analysis and cloning of the synthetic pathway of the phytohormone indole-3-acetic acid in the plant-beneficial Bacillus amyloliquefaciens SQR9. Microb Cell Fact. 14:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son H. J., Park G. T., Cha M. S., Heo M. S. (2006). Solubilization of insoluble inorganic phosphates by a novel salt- and pH-tolerant Pantoea agglomerans R-42 isolated from soybean rhizosphere. Bioresour. Technol. 97 204–210. 10.1016/j.biortech.2005.02.021 [DOI] [PubMed] [Google Scholar]
- Song W. Q., Gu W. Q., Qian Y. B., Ma X., Liu W. J. (2015). Identification of long non-coding RNA involved in osteogenic differentiation from mesenchymal stem cells using RNA-Seq data. Genet. Mol. Res. 14 18268–18279. 10.4238/2015.december.23.14 [DOI] [PubMed] [Google Scholar]
- Sun X. C., Gao Y. F., Li H. R., Yang S. Z., Liu Y. S. (2015). Over-expression of SlWRKY39 leads to enhanced resistance to multiple stress factors in tomato. J. Plant Biol. 58 52–60. 10.1007/s12374-014-0407-4 [DOI] [Google Scholar]
- Tahir H., Gu Q., Wu H., Raza W., Safdar A., Huang Z., et al. (2017). Effect of volatile compounds produced by Ralstonia solanacearum on plant growth promoting and systemic resistance inducing potential of Bacillus volatiles. BMC Plant Biol. 17:133. 10.1186/s12870-017-1083-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakuria D., Talukdar N. C., Goswami C., Hazarika S., Boro R. C., Khan M. R. (2004). Characterization and screening of bacteria from rhizosphere of rice grown in acidic soils of Assam. Curr. Sci. 86 978–985. [Google Scholar]
- Timmusk S., Grantcharova N., Wagner E. G. H. (2005). Paenibacillus polymyxa invades plant roots and forms biofilms. Appl. Environ. Microbiol. 71 7292–7300. 10.1128/aem.71.11.7292-7300.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmusk S., Nicander B., Granhall U., Tillberg E. (1999). Cytokinin production by Paenibacillus polymyxa. Soil Biol. Biochem. 31 1847–1852. 10.1016/s0038-0717(99)00113-3 [DOI] [Google Scholar]
- Toquin V., Grausem B., Geoffroy P., Legrand M. (2003). Structure of the tobacco caffeic acid O-methyltransferase (COMT) II gene: identification of promoter sequences involved in gene inducibility by various stimuli. Plant Mol. Biol. 52 495–509. [DOI] [PubMed] [Google Scholar]
- Trdá L., Fernandez O., Boutrot F., Héloir M. C., Kelloniemi J., Daire X., et al. (2014). The grapevine flagellin receptor VvFLS2 differentially recognizes flagellin-derived epitopes from the endophytic growth-promoting bacterium Burkholderia phytofirmans and plant pathogenic bacteria. New Phytol. 201 1371–1384. 10.1111/nph.12592 [DOI] [PubMed] [Google Scholar]
- Vasconcellos R. L. F. D., Cardoso E. J. B. N. (2009). Rhizospheric streptomycetes as potential biocontrol agents of Fusarium and Armillaria pine rot and as PGPR for Pinus taeda. Biocontrol 54 807–816. 10.1007/s10526-009-9226-9 [DOI] [Google Scholar]
- Vater J., Herfort S., Doellinger J., Weydmann M., Lasch P. (2016). Fusaricidins from Paenibacillus polymyxa M-1, a family of lipohexapeptides of unusual complexity-a mass spectrometric study. J. Mass Spectrom. 52 7–15. 10.1002/jms.3891 [DOI] [PubMed] [Google Scholar]
- Volpon L., Besson F., Lancelin J. M. (1999). NMR structure of active and inactive forms of the sterol-dependent antifungal antibiotic bacillomycin L. Eur. J. Biochem. 264 200–210. 10.1046/j.1432-1327.1999.00605.x [DOI] [PubMed] [Google Scholar]
- Wang M., Zhu X., Wang K., Lu C., Luo M., Shan T., et al. (2018). A wheat caffeic acid 3-O-methyltransferase TaCOMT-3D positively contributes to both resistance to sharp eyespot disease and stem mechanical strength. Sci. Rep. 8:6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. N., Dang F. F., Liu Z. Q., Wang X., Eulgem T., Lai Y., et al. (2013). CaWRKY58, encoding a group I WRKY transcription factor of Capsicum annuum, negatively regulates resistance to Ralstonia solanacearum infection. Mol. Plant Pathol. 14 131–144. 10.1111/j.1364-3703.2012.00836.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuddineh W. A., Mazarei M., Zhang J., Poovaiah C. R., Mann D. G. J., Ziebell A., et al. (2015). Identification and overexpression of gibberellin 2-oxidase (GA2ox) in Switchgrass (Panicum virgatum L.) for improved plant architecture and reduced biomass recalcitrance. Plant Biotechnol. J. 13 636–647. 10.1111/pbi.12287 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57 781–803. 10.1146/annurev.arplant.57.032905.105444 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y., Hachiya A., Hamada H., Matsumoto H. (1998). Phenylpropanoids as a protectant of aluminum toxicity in cultured tobacco cells. Plant Cell Physiol. 39, 950–957. 10.1093/oxfordjournals.pcp.a029459 [DOI] [Google Scholar]
- Yang P., Chen C., Wang Z., Fan B., Chen Z. (1999). A pathogen-and salicylic acid-induced WRKY DNA-binding activity recognizes the elicitor response element of the tobacco class I chitinase gene promoter. Plant J. 18 141–149. 10.1046/j.1365-313x.1999.00437.x [DOI] [Google Scholar]
- Ye Z. H., Varner J. E. (1995). Differential expression of two O-methyltransferases in lignin biosynthesis in Zinnia elegans. Plant Physiol. 108, 459–467. 10.1104/pp.108.2.459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Yu Z. H., Fan G. Q., Wang G. H., Liu X. B. (2016). Isolation and characterization of indole acetic acid producing root endophytic bacteria and their potential for promoting crop growth. J. Agric. Sci. Technol. 18 1381–1391. [Google Scholar]
- Yu O., Jez J. M. (2008). Nature’s assembly line: biosynthesis of simple phenylpropanoids and polyketides. Plant J. 54 750–762. 10.1111/j.1365-313x.2008.03436.x [DOI] [PubMed] [Google Scholar]
- Yuttavanichakul W., Lawongsa P., Wongkaew S., Teaumroong N., Boonkerd N., Nomura N., et al. (2012). Improvement of peanut rhizobial inoculant by incorporation of plant growth promoting rhizobacteria (PGPR) as biocontrol against the seed borne fungus, Aspergillus niger. Biol. Control 63 87–97. 10.1016/j.biocontrol.2012.06.008 [DOI] [Google Scholar]
- Zawaski C., Busov V. B. (2014). Roles of gibberellin catabolism and signaling in growth and physiological response to drought and short-day photoperiods in Populus trees. PLoS One 9:e86217. 10.1371/journal.pone.0086217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Kong F. (2014). Isolation and identification of potassium-solubilizing bacteria from tobacco rhizospheric soil and their effect on tobacco plants. Appl. Soil Ecol. 82 18–25. 10.1016/j.apsoil.2014.05.002 [DOI] [Google Scholar]
- Zhang Y., Peng D., Song Y., Jin C., Guan C. (2020). Enhancement of methyl salicylate accumulation promotes early flowering in transgenic tobacco plants by overexpressing a carboxymethyl transferase (SAMT) gene from Lycium chinense. Mol. Breed. 40:52. [Google Scholar]
- Zhou K. Q., Yamagishi M., Osaki M. (2010). Paenibacillus BRF-1 has biocontrol ability against Phialophora gregata disease and promotes soybean growth. Soil Sci. Plant Nutr. 54 870–875. 10.1111/j.1747-0765.2008.00308.x [DOI] [Google Scholar]
- Zhou Y., Underhill S. J. R. (2016). Breadfruit (Artocarpus altilis) gibberellin 2-oxidase genes in stem elongation and abiotic stress response. Plant Physiol. Biochem. 98 81–88. 10.1016/j.plaphy.2015.11.012 [DOI] [PubMed] [Google Scholar]
- Zou C., Jiang W., Yu D. (2010). Male gametophyte-specific WRKY34 transcription factor mediates cold sensitivity of mature pollen in Arabidopsis. J. Exp. Bot. 61 3901–3914. 10.1093/jxb/erq204 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.