Abstract
Solubility of recombinant proteins (i.e., the extent of soluble versus insoluble expression in heterogeneous hosts) is the first checkpoint criterion for determining recombinant protein quality. However, even soluble proteins often fail to represent functional activity because of the involvement of non-functional, misfolded, soluble aggregates, which compromise recombinant protein quality. Therefore, screening of solubility and folding competence is crucial for improving the quality of recombinant proteins, especially for therapeutic applications. The issue is often highlighted especially in bacterial recombinant hosts, since bacterial cytoplasm does not provide an optimal environment for the folding of target proteins of mammalian origin. Antibody fragments, such as single-chain variable fragment (scFv), single-chain antibody (scAb), and fragment antigen binding (Fab), have been utilized for numerous applications such as diagnostics, research reagents, or therapeutics. Antibody fragments can be efficiently expressed in microorganisms so that they offer several advantages for diagnostic applications such as low cost and high yield. However, scFv and scAb fragments have generally lower stability to thermal stress than full-length antibodies, necessitating a judicious combination of designer antibodies, and bacterial hosts harnessed with robust chaperone function. In this review, we discuss efforts on not only the production of antibodies or antibody fragments in microorganisms but also scFv stabilization via (i) directed evolution of variants with increased stability using display systems, (ii) stabilization of the interface between variable regions of heavy (VH) and light (VL) chains through the introduction of a non-native covalent bond between the two chains, (iii) rational engineering of VH-VL pair, based on the structure, and (iv) computational approaches. We also review recent advances in stability design, increase in avidity by multimerization, and maintaining the functional competence of chimeric proteins prompted by various types of chaperones.
Keywords: antibody fragments, solubility, stability, bacterial expression, scFv
Introduction
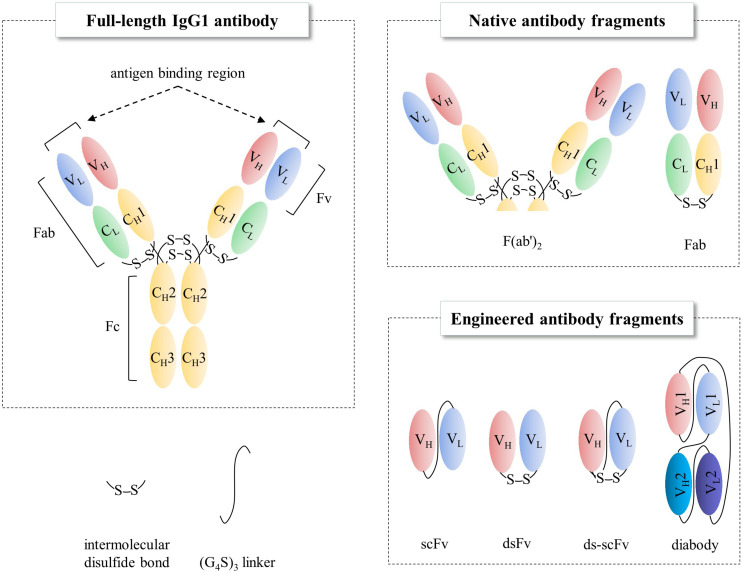
Antibodies are widely used for medical applications such as disease diagnosis and therapy (Grilo and Mantalaris, 2019). Valuable pharmaceutical properties of antibodies such as high affinity to their target molecules have led to them becoming constituted as key materials not only in antibody-based biosensors, which offer the promise of in-depth target detection capacity (Holliger and Hudson, 2005; Saerens et al., 2008; Conroy et al., 2009) but also in antibody-based proteomics, which provides insights into cancer disease states via cancer biomarker discovery (Brennan et al., 2010). In addition to the intrinsic property, industrial applications require high productivity and long shelf-life from thermal stress, organic solvents, and other stresses than physiological conditions. However, production of full-length antibodies cost extremely high, as they are typically expressed in mammalian cell lines such as Chinese hamster ovary (CHO) or NS0 murine myeloma cell lines (de la Cruz Edmonds et al., 2006; Huang et al., 2007; Liu et al., 2008; Grilo and Mantalaris, 2019) due to N-glycan heterogeneity among different species (Jefferis, 2009) and also the complex disulfide bond pattern; hence, the biopharmaceutics industry has devoted immense resources on its production processes (Chartrain and Chu, 2008). Instead, the single-chain variable fragment (scFv, Figure 1), a rational polypeptide design, consisting only of variable regions from heavy (VH) and light (VL) chains, joined together by a linker, not only maintains antigen binding capacity (Humphreys, 2003; Andersen and Reilly, 2004) but also can easily be produced in prokaryotes, such as Escherichia coli (E. coli) or Brevibacillus choshinensis (B. chosinensis) (Hu et al., 2017), along with high yield, which keeps the cost of production low (Power and Hudson, 2000; Terpe, 2006; Rosano and Ceccarelli, 2014; Gupta and Shukla, 2017). Despite the advantages of scFvs, they have a few drawbacks that limit their therapeutic potential, such as (i) deteriorated stability because of their propensity to readily aggregate under thermal stress (Jager and Pluckthun, 1999a; Demarest and Glaser, 2008); (ii) a short serum half-life of <1 day compared to 3 weeks for full-length immunoglobulin G (IgG)1, IgG2, and IgG4 antibodies (Kang and Jung, 2020); and (iii) reduced affinity compared to the full-length antibody counterpart. Therefore, scFv format is suitable for limited cases, such as macular degeneration or blood-related diseases (Table 1).
FIGURE 1.
Schematic representations of full-length IgG1 antibody and antibody fragments. Fab, fragment antigen binding; Fv, fragment variable; Fc, fragment crystallizable; scFv, single-chain Fv; dsFv, disulfide-stabilized Fv.
TABLE 1.
FDA approved therapeutic antibody fragments.
| Name1 | Brand name2 | Company3 | Target | Format | Indication4 | Year approved5 | Host6 |
| Abciximab | Reopro | Janssen Biotech, Inc. | GPIIb/IIIa | Chimeric IgG1 Fab | Prevention of blood clots in angioplasty | 1994 | Mammalian |
| Ranibizumab | Lucentis | Genentech, Inc. | VEGF | Humanized IgG1 Fab | Macular degeneration | 2006 | E. coli |
| Certolizumab pegol | Cimzia | UCB, Inc. | TNF | Humanized Fab, pegylated | Crohn disease | 2008 | E. coli |
| Blinatumomab | Blincyto | Amgen Inc. | CD19, CD3 | Murine bispecific tandem scFv | Acute lymphoblastic leukemia | 2014 | E. coli |
| Idarucizumab | Praxbind | Boehringer Ingelheim Pharmaceuticals, Inc. | Dabigatran | Humanized Fab | Reversal of dabigatran-induced anticoagulation | 2015 | CHO7 |
| Moxetumomab pasudotox | Lumoxiti | AstraZeneca Pharmaceuticals LP | CD22 | Murine IgG1 dsFv immunotoxin | Hairy cell leukemia | 2018 | E. coli |
| Caplacizumab | Cablivi | Ablynx N.V. | von Willebrand factor | Humanized Nanobody | Acquired thrombotic thrombocytopenic purpura | 2019 | E. coli |
| Brolucizumab | Beovu | Novartis Pharmaceuticals Corporation | VEGF-A | Humanized scFv | Neovascular age-related macular degeneration | 2019 | E. coli |
1Name: International non- proprietary names. 2Brand name: commercial names. 3Company: companies developed the antibody fragment drugs. 4Indication: indication first approved by the FDA. 5Year approved: the first year approved by the FDA. 6Host: expression cell-lines. 7CHO: Chinese Hamster Ovary cell-line. This table was classified using data from “The Antibody Society (2020)” (Antibody Society, 2020).
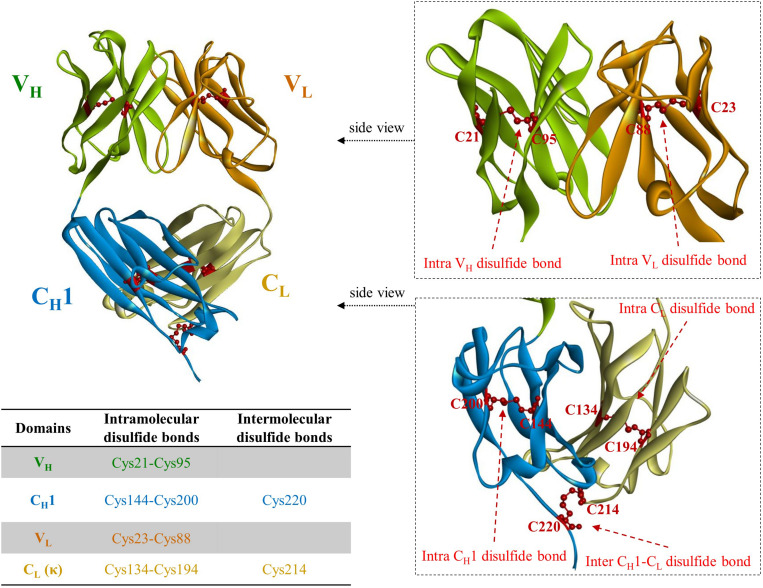
Immunoglobulin G, the most abundant monoclonal antibody (mAb) isotype in serum is composed of two antigen binding fragments (Fab) and one homodimeric fragment crystallizable (Fc) domain that contribute to the overall stability of the molecule (Figure 1; Kang and Jung, 2019; Saunders, 2019). Since Fab of an IgG becomes more sensitive to the heat denaturation when Fc region is removed (Tischenko et al., 1982; Vermeer and Norde, 2000; Ionescu et al., 2008), researchers have tried to engineer Fab to stabilize the interactions between constant heavy 1 (CH1) and constant light (CL) chains in order to obviate the need for using mammalian host cells for the expression of full-length antibodies because of N-glycan on the Fc region. This requires immense resources such as expensive media, facilities to maintain germfree conditions, and time. However, limited successes have been made (Demarest et al., 2006; Teerinen et al., 2006). Further elimination of CH1-CL pair in Fab, resulting in fragment variable (Fv), significantly discounts thermodynamic stability (Webber et al., 1995; Jager and Pluckthun, 1999b). This is presumably due to the unnatural exposure of the lower VL and VH regions, flanking CH1 and CL, where hydrophobic interaction used to contribute to the stability as a whole as well as the absence of the contribution of CH1, which controls the assembly of heavy and light chains of the whole IgG molecule (Feige et al., 2009). The only light-heavy intermolecular disulfide bond in native IgG antibodies on the residues Cys220 in CH1 and Cys214 in CL of Fab region (Figure 2 in canakinumab; PDB ID of 5BVJ) contributes to the thermodynamic stability of the whole Fab fragment. In addition, intramolecular disulfide bonds in both the VH and VL regions (Figure 2) is critical in the thermodynamic stability because elimination of them significantly enhanced the propensity of scFv aggregation (Montoliu-Gaya et al., 2017). In this article we review efforts on increasing expression yield as well as protein stability of antibody fragment and recent diverse designs of antibody fragments.
FIGURE 2.
A crystal structure of fragment antigen binding (Fab) region of canakinumab, a human monoclonal antibody neutralizing IL-1β (PDB ID: 5BVJ). VH regions are colored in light green; CH1 in light blue; VL in orange; CL in cyan. Cysteine residues involved in disulfide bonds are partially depicted in red ball and stick model. The only intermolecular disulfide bonds between heavy and light chain of IgG molecule is present on the cysteine residues in CH1 (Cys220) and CL (Cys214), respectively. The positions of the cysteine residues vary among different VH and VL due to the differences in CDR length.
Production of Antibody or Antibody Fragments in Bacteria
To reduce the cost of production of antibodies, researchers in both academia and industry put enormous efforts on elevating expression yield of IgG antibody or its fragment by (i) engineering expression plasmids, i.e., rhamnose-inducible expression system (Petrus et al., 2019) or comprehensive optimization via high-throughput screening (Makino et al., 2011), (ii) engineering global sigma factor RhoD, which regulates more than 1,000 gene expressions (McKenna et al., 2019), and (iii) devising bacterial strains capable of forming disulfide bonds in cytoplasm such as CyDisCo (Gaciarz and Ruddock, 2017) or SHuffle (Lobstein et al., 2012; Robinson et al., 2015; Yusakul et al., 2017). Despite the advantages, drawbacks limiting its potential are (i) the low stability of scFvs, known for their propensity to readily aggregate under thermal stress (Jager and Pluckthun, 1999a; Demarest and Glaser, 2008), (ii) absence of glycosylation machinery, (iii) lack of efficient secretory mechanism as compared to yeast or animal cells, functionally limited protein trafficking machinery from the cytoplasm to the periplasmic space or to the outside of the cells, and (iv) overproduction of acetic acid byproduct during fermentation (Holms, 1986; Wong et al., 2008).
Engineering Intrinsic Stability of scFvs: Directed Evolution, Rational Design, and Computational Approaches
Antibody fragments can be expressed in several compartments in E. coli: mostly as inclusion bodies in the cytoplasm, or as soluble forms displayed on (i) the inner membrane, (ii) in the periplasmic space, (iii) on the outer membrane, and/or (iv) outside the bacterium, facilitated by various signal sequences, such as outer membrane protein A (OmpA), pectate lyase B (PelB), or new lipoprotein A (NlpA) (Tseng et al., 2009; Frenzel et al., 2013; Khodabakhsh et al., 2013; Levy et al., 2013; DePalma, 2014; Mizukami et al., 2018). To overcome the drawbacks of scFvs, which comprise only VH and VL antigen-binding domains, to reduce the protein size in order to increase protein production but maintain high target molecule affinity, researchers have engineered scFvs with resistance to aggregation and enhanced intrinsic stability of antibody fragments. Table 2 summarizes the engineering efforts.
TABLE 2.
Summary of stability and avidity engineering of scFvs in bacteria.
| Classification | Group | Institute | Methodology | References |
| Directed evolution | Gregory Winter | University of Cambridge | Phage display | Jespers et al., 2004 |
| Daniel Christ | Garvan Institute of Medical Research | Phage display | Dudgeon et al., 2012 | |
| Dane Wittrup | MIT | Yeast display | Graff et al., 2004; Chao et al., 2006 | |
| Scott Glaser | Biogen Idec | Bacterial screening | Miller et al., 2010 | |
| Rational design | Ira Pastan | NIH | Non-native disulfide bond | Brinkmann et al., 1993 |
| Andreas Plückthun | University of Zurich | Disrupting the hydrophobic patches | Nieba et al., 1997 | |
| Wei Chen | Jiangnan University | Non-native disulfide bond | Zhao et al., 2010 | |
| Ning Deng | Jinan University | Non-native disulfide bond | Cai et al., 2016 | |
| Hiroshi Morioka | Kumamoto University | Cyclization of scFv | Yamauchi et al., 2019 | |
| Alexey Lugovskoy | Merrimack Pharmaceuticals | Tyrosine substitution | Zhang et al., 2015 | |
| An-Suei Yang | National Defense Medical Center in Taipei | Hydrophobic domains | Hsu et al., 2014 | |
| Robin Curtis | University of Manchester | Substitution of Arginine | Austerberry et al., 2019 | |
| Boris Steipe | University of Toronto | Consensus-based | Steipe, 2004 | |
| Alexey Lugovskoy | Biogen Idec | Consensus-based | Jordan et al., 2009 | |
| Computation | Andreas Plückthun | University of Zurich | Structure-based | Ewert et al., 2003, 2004 |
| David R. Liu | Harvard University | Supercharging | Lawrence et al., 2007 | |
| Andrew Ellington | University of Texas at Austin | Supercharging | Miklos et al., 2012 | |
| Peter Bowers | AnaptysBio | Integrative approach | McConnell et al., 2013; McConnell et al., 2014 | |
| George Georgiou and Andrew Ellington | University of Texas at Austin | Rosetta modeling | Lee et al., 2019 | |
| Fusion partner | Yechen Xiao | Jilin University | SUMO | Liu et al., 2015 |
| Junichi Takagi | Osaka University | Fv-clasp | Arimori et al., 2017 | |
| Chaperone coexpression | Shihua Wang | Fujian Agriculture and Forestry University | Skp | Wang et al., 2013 |
| Multimerization | Sergey Kipriyanov | Affimed Therapeutics AG | Diabody: bispecific | Le Gall et al., 2004 |
| C Ronald Geyer | University of Saskatchewan | SpyCatcher: trispecific | Alam et al., 2018 |
Directed Evolution
Greg Winter et al., utilized phage display directed evolution methodology to isolate VH variants that are more resistant to heat denaturation (Jespers et al., 2004). They further engineered VH and identified a key residue, Arg28 in VH that renders resistance to heat and acid aggregation (Famm et al., 2008). Daniel Christ’s group at the Garvan Institute of Medical Research selected critical residues for antigen binding in both VH and VL and constructed a phage library introducing aspartate or glutamate in those residues to screen for heat resistance. The isolated variants resulted in not only enhanced biophysical property but also structural conservation (Dudgeon et al., 2012). Dane Wittrup’s group at MIT devised a yeast surface display system to isolate scFv variants with high affinity to antigen and increased stability by constructing yeast mutant libraries, expressing scFv on the cell surface, followed by successive rounds of flow cytometry sorting (Graff et al., 2004; Chao et al., 2006). Brian Miller et al., at Biogen Idec, Inc. used sequence- and structure-based analyses to devise a high-throughput screening methodology that measure scFv extracellularly expressed by E. coli. This screening methodology resulted in enhanced melting temperature (Tm) by 14°C and additional Tm improvement by 12°C through combination of the resulting variants (Miller et al., 2010).
Rational Design
Unlike the presence of intermolecular CH1-CL disulfide bond, there is not one in the native VH-VL (Figures 1, 2). Instead of placing a linker between VH and VL (scFv in Figure 1) creating non-native disulfide bond between VH and VL via substituting amino acid residues in both framework 2 (FR2) in VH and FR4 in VL (dsFv; disulfide-stabilized Fv in Figure 1) led to indistinguishable specificity to antigen and similar cytotoxic activity when fused with exotoxin but exhibited superior protein stability at 37°C, compared to scFv counterpart (Brinkmann et al., 1993). Similarly, substitution of Val84 in VH to aspartic acid led to not only improved periplasmic production by 25-fold but also decreased the rate of thermally induced aggregation reaction (Nieba et al., 1997). In another study, introduction of Cys44 in VH and Cys100 in VL in anti-aflatoxin B(1) scFv resulted in improved stability and resistance to protein aggregation (Zhao et al., 2010). Introduction of the disulfide bond to anti-FGF2 diabody (ds-diabody, see section “multimerization” for diabody) also improved biological activity (Cai et al., 2016). This is presumably due to its lower propensity to the open state of VH-VL pair, in contrast to the presence of both assembled and disassembled state in case of scFv where VH and VL domains are simply connected by a G4S linker. These results indicate that bridging VH and VL by establishing intermolecular disulfide bond formation via cysteine residue incorporation can be a decent strategy for Fv stabilization (Trivedi et al., 2009). Another recent approach incorporating closed state of VH-VL pair is a cyclization of scFv using an enzyme sortase A, which ligate the pair, leading to cyclic scFv: this methodology markedly suppressed aggregation tendency without affecting affinity to antigen (Yamauchi et al., 2019).
Alexey Lugovskoy et al., at Merrimack Pharmaceuticals, Inc. showed that both essential and non-essential tyrosine residues for antigen binding in either CDR or FR can improve the biophysical property of scFv (Zhang et al., 2015). An-Suei Yang et al., at the National Defense Medical Center, Taipei, elucidated the nature of intra- and inter-hydrophobic domains of scFv: the former is flexible and indirectly affects antigen binding, as opposed to the latter affecting antigen binding directly (Hsu et al., 2014). Robin Curtis’s group at the University of Manchester investigated the aggregation propensity of arginine-rich scFv under denaturing condition: substitution of arginine residues in scFv with lysine significantly reduced aggregation (Austerberry et al., 2019). This diverse knowledge of protein nature in scFv may assist engineers with consensus-based design of antibody fragment for generating stabilizing mutations to pre-existing scFvs (Steipe, 2004) or bispecific antibody fragments (Jordan et al., 2009).
Computational Approaches
Andreas Plückthun’s group at University of Zurich designed a stabilized scFv from human VH germline sequences by analyzing hydrophobic core, pairing of hydrogen bonds, clusters of charge, and packing of β-sheets, leading to reduction of ΔG = 20.9 kJ/mol as well as improvement of scFv expression yield by 4-fold (Ewert et al., 2003). Furthermore, they could stabilize scFv by CDR grafting to more stable framework, using a structure-based analysis (Ewert et al., 2004).
Computational web servers, such as Prediction of Immunoglobulin Structure (PIGS) (Marcatili et al., 2008) or Web Antibody Modeling (WAM) (Whitelegg and Rees, 2000), made computational modeling of antibody variable regions possible. Importantly, recent advances in structural knowledge and computational protein modeling such as RosettaDesign accelerated antibody design toward improved antigen affinity as well as physicochemical properties (Borgo and Havranek, 2012; Buck et al., 2012). The homology modeling provides with guidance on not only prediction on the conformations of CDR loops but also VH-VL orientations via energy calculations such as antibody-antigen docking, comparing with known crystal structures (Kuroda et al., 2012). For example, a computational homology modeling significantly improved resistance of scFvs to heat inactivation by supercharging the protein through energy calculations (Lawrence et al., 2007; Miklos et al., 2012). In addition, combinatorial engineering approach, including CDR grafting onto stable frameworks, VH-VL interface stabilization, and in vitro somatic hypermutation significantly increased thermal stability of full-length antibody by 10°C, compared to the native IgG1 antibody (McConnell et al., 2013, 2014).
Recent advances in computational methodologies for both antibody sequencing and backbones (Goldenzweig and Fleishman, 2018) and for multistage processing of antibody engineering, capitalizing on computational design and experimental validation cycles (Baran et al., 2017), have enabled successful de novo antibody engineering (Chevalier et al., 2017), such as anti-influenza antibodies (Strauch et al., 2017; Sevy et al., 2019). Of note, Georgiou and Ellington at the University of Texas at Austin used the Rosetta modeling program (Sircar et al., 2009; Adolf-Bryfogle et al., 2018) to predict amino acid substitutions for anti-HA33 scFv stabilization and confirmed a melting temperature increase of 4.5°C by antigen-binding enzyme-linked immunosorbent assay (ELISA) after thermal stress for 2 h at 70°C (Lee et al., 2019).
A Fusion Partner
Tagging of anti-FGFR3 scFv with a solubilizing partner small ubiquitin-related modifier (Sumo), followed by removal of the Sumo protein using Sumo protease, enabled over 95% purity with the yield of 4 mg/L bacterial culture. The resulting anti-FGFR3 scFv has exhibited complete biological activity (Liu et al., 2015). Another example is an “Fv-clasp,” where scFv was fused with anti-parallel coiled coil structure, SRAH domain of human Mst1 with scFv. In addition, introduction of disulfide bond to the Fv greatly enhanced thermal stability and tendency of crystallization. This is presumably due to the shielding of hydrophobic residues exposed in Fv, according to the X-ray crystallography (Arimori et al., 2017). Alternatively, approaches based on the chaperone function of RNAs could also be considered (Choi et al., 2008). Fusion with an RNA-interaction domain (RID) greatly enhances the solubility (i.e., the ratio of soluble versus insoluble expression in heterogeneous hosts) and the overall yield of soluble proteins, harnessed with unique properties of RNAs as chaperone (chaperna; chaperone + RNA) (Kim et al., 2018; Yang et al., 2018), although this approach has not yet been documented for recombinant antibody fragments.
Engineering the Host Cell: Chaperone Coexpression or Genome-Level Screening
The folding of nascent polypeptides is often assisted by molecular chaperones (Hendrick and Hartl, 1995; Bukau and Horwich, 1998; Hartl and Hayer-Hartl, 2002), although their utility in recombinant expression has been documented only in limited cases (Structural Genomics Consortium et al., 2008). The stability problem associated with VH-VL pair in the scFv molecule can be circumvented through assistance with the chaperone of a pairing vehicle. Coexpression of the chaperone Skp enhanced binding activity of anti-TLH scFv by 3–4 fold, relative to the native counterpart, expressed in E. coli (Wang et al., 2013).
Multimerization
Diabodies not only render bivalency but also enhance stability of Fv by linker design (Le Gall et al., 2004). Introducing covalent bonds between VH1 and VL2 as well as VH2 and VL1 enables bispecific binding capacity of the two distinct scFv in one molecule (diabody in Figure 1). One example in clinic is blinatumomab (BLINCYTO,® 2014), a bispecific scFv for CD19 and CD3 (Table 1), which functions as a T lymphocyte engager to cancerous B lymphocytes for the treatment of acute lymphoblastc leukemia (Holliger et al., 1993; Mack et al., 1995; Suresh et al., 2014; Wu et al., 2015). In addition, constructing anti-HER3 trivalent scFv using SpyCatcher ligase system enhanced affinity by 12-fold as compared to a monomeric anti-HER3 counterpart (Alam et al., 2018). Another general approach of antibody fragments is the utilization of targeting ligands on nanoparticles in nanomedicine (Richards et al., 2017). Several antibody fragment-based nanoparticles are under clinical trials, including Erbitux-EDVSpac, which is a bacteria-derived mini-cell nanoparticle targeting EGFR currently under clinical phase II (Richards et al., 2017). Multimerization of scFvs as nanoparticles, using self-assembling scaffolds via chaperna approach (Kim et al., 2018; Yang et al., 2018) holds promise for further enhancing the avidity and thermostability of recombinant antibody fragments.
Conclusion
Up to March 2020, the US Food and Drug Administration (FDA) approved eight antibody fragments as drugs, six of which are produced from E. coli (75%, Table 1). The six therapeutic antibody fragments, produced by bacteria include ranibizumab (LUCENTIS®, 2006), certolizumab pegol (CIMZIA®, 2008), blinatumomab (BLINCYTO®, 2014), moxetumomab pasudotox (Kreitman and Pastan, 2011; LUMOXITITM, 2018), caplacizumab (CABLIVI®, 2019), and brolucizumab (BEOVU®, 2019), while those produced by mammalian hosts include abciximab (ReoPro®, 1994) and idarucizumab (PRAXBIND®, 2015), according to the data from “The Antibody Society” (Antibody Society, 2020).
Microorganisms are favorable expression hosts for antibody fragments, such as scFvs or Fab, in therapeutic applications (Table 1), because of the low production cost and lack of a carbohydrate chain. However, despite these advantages, scFvs expressed in bacteria have neither comparable stability relative to native full-length antibodies nor a comparable production yield of ∼1 g/L in bioreactors (Petrus et al., 2019) relative to mammalian hosts, that is, >10 g/L in CHO cells (Kunert and Reinhart, 2016). Therefore, scientists and engineers in both academia and industry put extensive efforts on increasing production yield as well as protein stability of scFv expressed in bacteria. To obtain improved yield various bacterial expression systems have been developed in terms of vector systems or engineered strains with engineered chaperone molecules.
The low intrinsic solubility and stability of native scFv protein with a relatively shorter shelf-life is a bottleneck for industrial application. To overcome the disadvantages there have been enormous research attempts on stability design via site-directed mutagenesis, generation of non-natural covalent bonds between the heavy and light variable chains, rational design, and recently computer-based engineering or chimeric approaches. Engineering of scFv with respect to increasing the stability lowers both the kinetic complexity in folding process and subsequently the propensity to aggregate into non-functional form. Folding into soluble, functional form with the desired level of avidity is often aided by exploiting the chaperone function of naturally existing molecular chaperones or artificial solubilizing tags. Besides thermodynamic aspects on overall stability, due consideration should be given to the kinetic aspects in de novo folding pathway for designer antibody fragments toward improved solubility, thermal stability, and productivity.
Author Contributions
TK and BS designed and wrote the manuscript. Both authors agreed to be accountable for the content of the work.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by grants from the Korean government (NRF-2020R1F1A1072124, NRF-2018M3A9H4079358, and HI20C0144).
References
- Adolf-Bryfogle J., Kalyuzhniy O., Kubitz M., Weitzner B. D., Hu X., Adachi Y., et al. (2018). RosettaAntibodyDesign (RAbD): a general framework for computational antibody design. PLoS Comput. Biol. 14:e1006112. 10.1371/journal.pcbi.1006112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam M. K., Brabant M., Viswas R. S., Barreto K., Fonge H., Ronald Geyer C. (2018). A novel synthetic trivalent single chain variable fragment (tri-scFv) construction platform based on the SpyTag/SpyCatcher protein ligase system. BMC Biotechnol. 18:55. 10.1186/s12896-018-0466-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen D. C., Reilly D. E. (2004). Production technologies for monoclonal antibodies and their fragments. Curr. Opin. Biotechnol. 15 456–462. 10.1016/j.copbio.2004.08.002 [DOI] [PubMed] [Google Scholar]
- Antibody Society (2020). Available online at: https://www.antibodysociety.org/resources/approved-antibodies/ (accessed March 10, 2020). [Google Scholar]
- Arimori T., Kitago Y., Umitsu M., Fujii Y., Asaki R., Tamura-Kawakami K., et al. (2017). Fv-clasp: an artificially designed small antibody fragment with improved production compatibility, stability, and crystallizability. Structure 25 1611–1622. 10.1016/j.str.2017.08.011 [DOI] [PubMed] [Google Scholar]
- Austerberry J. I., Thistlethwaite A., Fisher K., Golovanov A. P., Pluen A., Esfandiary R., et al. (2019). Arginine to Lysine mutations increase the aggregation stability of a single-chain variable fragment through unfolded-state interactions. Biochemistry 58 3413–3421. 10.1021/acs.biochem.9b00367 [DOI] [PubMed] [Google Scholar]
- Baran D., Pszolla M. G., Lapidoth G. D., Norn C., Dym O., Unger T., et al. (2017). Principles for computational design of binding antibodies. Proc. Natl. Acad. Sci. U.S.A. 114 10900–10905. 10.1073/pnas.1707171114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- BEOVU® (2019). Prescribing Information Approved by the U.S. Food and Drug Administration. (U.S. License Number: 1244). East Hanover, NJ: Novartis Pharmaceuticals Corporation. [Google Scholar]
- BLINCYTO® (2014). Prescribing Information Approved by the U.S. Food and Drug Administration. (U.S. License No. 1080). Thousand Oaks, CA: Amgen Inc. [Google Scholar]
- Borgo B., Havranek J. J. (2012). Automated selection of stabilizing mutations in designed and natural proteins. Proc. Natl. Acad. Sci. U.S.A. 109 1494–1499. 10.1073/pnas.1115172109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan D. J., O’Connor D. P., Rexhepaj E., Ponten F., Gallagher W. M. (2010). Antibody-based proteomics: fast-tracking molecular diagnostics in oncology. Nat. Rev. Cancer 10 605–617. 10.1038/nrc2902 [DOI] [PubMed] [Google Scholar]
- Brinkmann U., Reiter Y., Jung S. H., Lee B., Pastan I. (1993). A recombinant immunotoxin containing a disulfide-stabilized Fv fragment. Proc. Natl. Acad. Sci. U.S.A. 90 7538–7542. 10.1073/pnas.90.16.7538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck P. M., Kumar S., Wang X., Agrawal N. J., Trout B. L., Singh S. K. (2012). Computational methods to predict therapeutic protein aggregation. Methods Mol. Biol. 899 425–451. 10.1007/978-1-61779-921-1_26 [DOI] [PubMed] [Google Scholar]
- Bukau B., Horwich A. L. (1998). The Hsp70 and Hsp60 chaperone machines. Cell 92 351–366. 10.1016/s0092-8674(00)80928-9 [DOI] [PubMed] [Google Scholar]
- CABLIVI® (2019). Prescribing Information Approved by the U.S. Food and Drug Administration. (U.S. License No. 2085). Ghent: Ablynx NV. [Google Scholar]
- Cai Y., Zhang J., Lao X., Jiang H., Yu Y., Deng Y., et al. (2016). Construction of a disulfide-stabilized diabody against fibroblast growth factor-2 and the inhibition activity in targeting breast cancer. Cancer Sci. 107 1141–1150. 10.1111/cas.12981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao G., Lau W. L., Hackel B. J., Sazinsky S. L., Lippow S. M., Wittrup K. D. (2006). Isolating and engineering human antibodies using yeast surface display. Nat. Protoc. 1 755–768. 10.1038/nprot.2006.94 [DOI] [PubMed] [Google Scholar]
- Chartrain M., Chu L. (2008). Development and production of commercial therapeutic monoclonal antibodies in Mammalian cell expression systems: an overview of the current upstream technologies. Curr. Pharm. Biotechnol. 9 447–467. 10.2174/138920108786786367 [DOI] [PubMed] [Google Scholar]
- Chevalier A., Silva D. A., Rocklin G. J., Hicks D. R., Vergara R., Murapa P., et al. (2017). Massively parallel de novo protein design for targeted therapeutics. Nature 550 74–79. 10.1038/nature23912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S. I., Han K. S., Kim C. W., Ryu K. S., Kim B. H., Kim K. H., et al. (2008). Protein solubility and folding enhancement by interaction with RNA. PLoS One 3:e2677. 10.1371/journal.pone.0002677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CIMZIA® (2008). Prescribing Information Approved by the U.S. Food and Drug Administration. (US License No. 1736). Smyrna, GE: UCB, Inc. [Google Scholar]
- Conroy P. J., Hearty S., Leonard P., O’Kennedy R. J. (2009). Antibody production, design and use for biosensor-based applications. Semin. Cell Dev. Biol. 20 10–26. 10.1016/j.semcdb.2009.01.010 [DOI] [PubMed] [Google Scholar]
- de la Cruz Edmonds M. C., Tellers M., Chan C., Salmon P., Robinson D. K., Markusen J. (2006). Development of transfection and high-producer screening protocols for the CHOK1SV cell system. Mol. Biotechnol. 34 179–190. 10.1385/mb:34:2:179 [DOI] [PubMed] [Google Scholar]
- Demarest S. J., Chen G., Kimmel B. E., Gustafson D., Wu J., Salbato J., et al. (2006). Engineering stability into Escherichia coli secreted Fabs leads to increased functional expression. Protein Eng. Design Select. 19 325–336. 10.1093/protein/gzl016 [DOI] [PubMed] [Google Scholar]
- Demarest S. J., Glaser S. M. (2008). Antibody therapeutics, antibody engineering, and the merits of protein stability. Curr. Opin. Drug Discov. Dev. 11 675–687. [PubMed] [Google Scholar]
- DePalma A. (2014). Advances in protein expression. Genet. Eng. Biotechnol. News 34 24–25. 10.1089/gen.34.01.14 [DOI] [Google Scholar]
- Dudgeon K., Rouet R., Kokmeijer I., Schofield P., Stolp J., Langley D., et al. (2012). General strategy for the generation of human antibody variable domains with increased aggregation resistance. Proc. Natl. Acad. Sci. U.S.A. 109 10879–10884. 10.1073/pnas.1202866109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewert S., Honegger A., Pluckthun A. (2003). Structure-based improvement of the biophysical properties of immunoglobulin VH domains with a generalizable approach. Biochemistry 42 1517–1528. 10.1021/bi026448p [DOI] [PubMed] [Google Scholar]
- Ewert S., Honegger A., Pluckthun A. (2004). Stability improvement of antibodies for extracellular and intracellular applications: CDR grafting to stable frameworks and structure-based framework engineering. Methods 34 184–199. 10.1016/j.ymeth.2004.04.007 [DOI] [PubMed] [Google Scholar]
- Famm K., Hansen L., Christ D., Winter G. (2008). Thermodynamically stable aggregation-resistant antibody domains through directed evolution. J. Mol. Biol. 376 926–931. 10.1016/j.jmb.2007.10.075 [DOI] [PubMed] [Google Scholar]
- Feige M. J., Groscurth S., Marcinowski M., Shimizu Y., Kessler H., Hendershot L. M., et al. (2009). An unfolded CH1 domain controls the assembly and secretion of IgG antibodies. Mol. Cell. 34 569–579. 10.1016/j.molcel.2009.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenzel A., Hust M., Schirrmann T. (2013). Expression of recombinant antibodies. Front. Immunol. 4:217. 10.3389/fimmu.2013.00217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaciarz A., Ruddock L. W. (2017). Complementarity determining regions and frameworks contribute to the disulfide bond independent folding of intrinsically stable scFv. PLoS One 12:e0189964. 10.1371/journal.pone.0189964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenzweig A., Fleishman S. J. (2018). Principles of protein stability and their application in computational design. Annu. Rev. Biochem. 87 105–129. 10.1146/annurev-biochem-062917-12102 [DOI] [PubMed] [Google Scholar]
- Graff C. P., Chester K., Begent R., Wittrup K. D. (2004). Directed evolution of an anti-carcinoembryonic antigen scFv with a 4-day monovalent dissociation half-time at 37 degrees C. Protein Eng. Design Select. 17 293–304. 10.1093/protein/gzh038 [DOI] [PubMed] [Google Scholar]
- Grilo A. L., Mantalaris A. (2019). The increasingly human and profitable monoclonal antibody market. Trends Biotechnol. 37 9–16. 10.1016/j.tibtech.2018.05.014 [DOI] [PubMed] [Google Scholar]
- Gupta S. K., Shukla P. (2017). Microbial platform technology for recombinant antibody fragment production: a review. Crit. Rev. Microbiol. 43 31–42. 10.3109/1040841X.2016.1150959 [DOI] [PubMed] [Google Scholar]
- Hartl F. U., Hayer-Hartl M. (2002). Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295 1852–1858. 10.1126/science.1068408 [DOI] [PubMed] [Google Scholar]
- Hendrick J. P., Hartl F. U. (1995). The role of molecular chaperones in protein folding. FASEB J. 9 1559–1569. 10.1096/fasebj.9.15.8529835 [DOI] [PubMed] [Google Scholar]
- Holliger P., Hudson P. J. (2005). Engineered antibody fragments and the rise of single domains. Nat. Biotechnol. 23 1126–1136. 10.1038/nbt1142 [DOI] [PubMed] [Google Scholar]
- Holliger P., Prospero T., Winter G. (1993). “Diabodies”: small bivalent and bispecific antibody fragments. Proc. Natl. Acad. Sci. U.S.A. 90 6444–6448. 10.1073/pnas.90.14.6444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holms W. H. (1986). The central metabolic pathways of Escherichia coli: relationship between flux and control at a branch point, efficiency of conversion to biomass, and excretion of acetate. Curr. Top. Cell Regul. 28 69–105. 10.1016/b978-0-12-152828-7.50004-4 [DOI] [PubMed] [Google Scholar]
- Hsu H. J., Lee K. H., Jian J. W., Chang H. J., Yu C. M., Lee Y. C., et al. (2014). Antibody variable domain interface and framework sequence requirements for stability and function by high-throughput experiments. Structure 22 22–34. 10.1016/j.str.2013.10.006 [DOI] [PubMed] [Google Scholar]
- Hu W., Xiang J. Y., Kong P., Liu L., Xie Q., Xiang H. (2017). Expression and characterization of a single-chain variable fragment against human LOX-1 in Escherichia coli and Brevibacillus choshinensis. J. Microbiol. Biotechnol. 27 965–974. 10.4014/jmb.1702.02007 [DOI] [PubMed] [Google Scholar]
- Huang Y., Li Y., Wang Y. G., Gu X., Wang Y., Shen B. F. (2007). An efficient and targeted gene integration system for high-level antibody expression. J. Immunol. Methods 322 28–39. 10.1016/j.jim.2007.01.022 [DOI] [PubMed] [Google Scholar]
- Humphreys D. P. (2003). Production of antibodies and antibody fragments in Escherichia coli and a comparison of their functions, uses and modification. Curr. Opin. Drug Discov. Dev. 6 188–196. [PubMed] [Google Scholar]
- Ionescu R. M., Vlasak J., Price C., Kirchmeier M. (2008). Contribution of variable domains to the stability of humanized IgG1 monoclonal antibodies. J. Pharm. Sci. 97 1414–1426. 10.1002/jps.21104 [DOI] [PubMed] [Google Scholar]
- Jager M., Pluckthun A. (1999a). Domain interactions in antibody Fv and scFv fragments: effects on unfolding kinetics and equilibria. FEBS Lett. 462 307–312. 10.1016/s0014-5793(99)01532-x [DOI] [PubMed] [Google Scholar]
- Jager M., Pluckthun A. (1999b). Folding and assembly of an antibody Fv fragment, a heterodimer stabilized by antigen. J. Mol. Biol. 285 2005–2019. 10.1006/jmbi.1998.2425 [DOI] [PubMed] [Google Scholar]
- Jefferis R. (2009). Glycosylation as a strategy to improve antibody-based therapeutics. Nat. Rev. Drug Discov. 8 226–234. 10.1038/nrd2804 [DOI] [PubMed] [Google Scholar]
- Jespers L., Schon O., Famm K., Winter G. (2004). Aggregation-resistant domain antibodies selected on phage by heat denaturation. Nat. Biotechnol. 22 1161–1165. 10.1038/nbt1000 [DOI] [PubMed] [Google Scholar]
- Jordan J. L., Arndt J. W., Hanf K., Li G., Hall J., Demarest S., et al. (2009). Structural understanding of stabilization patterns in engineered bispecific Ig-like antibody molecules. Proteins 77 832–841. 10.1002/prot.22502 [DOI] [PubMed] [Google Scholar]
- Kang T. H., Jung S. T. (2019). Boosting therapeutic potency of antibodies by taming Fc domain functions. Exp. Mol. Med. 51 1–9. 10.1038/s12276-019-0345-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang T. H., Jung S. T. (2020). Reprogramming the constant region of immunoglobulin G subclasses for enhanced therapeutic potency against cancer. Biomolecules 10:382. 10.3390/biom10030382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodabakhsh F., Zia M. F., Moazen F., Rabbani M., Sadeghi H. M. (2013). Comparison of the cytoplasmic and periplasmic production of reteplase in Escherichia coli. Prep. Biochem. Biotechnol. 43 613–623. 10.1080/10826068.2013.764896 [DOI] [PubMed] [Google Scholar]
- Kim Y. S., Son A., Kim J., Kwon S. B., Kim M. H., Kim P., et al. (2018). Chaperna-mediated assembly of ferritin-based middle east respiratory syndrome-coronavirus nanoparticles. Front. Immunol. 9:1093. 10.3389/fimmu.2018.01093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitman R. J., Pastan I. (2011). Antibody fusion proteins: anti-CD22 recombinant immunotoxin moxetumomab pasudotox. Clin. Cancer Res. 17 6398–6405. 10.1158/1078-0432.CCR-11-0487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunert R., Reinhart D. (2016). Advances in recombinant antibody manufacturing. Appl. Microbiol. Biotechnol. 100 3451–3461. 10.1007/s00253-016-7388-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda D., Shirai H., Jacobson M. P., Nakamura H. (2012). Computer-aided antibody design. Protein Eng. Design Select. 25 507–521. 10.1093/protein/gzs024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M. S., Phillips K. J., Liu D. R. (2007). Supercharging proteins can impart unusual resilience. J. Am. Chem. Soc. 129 10110–10112. 10.1021/ja071641y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gall F., Reusch U., Little M., Kipriyanov S. M. (2004). Effect of linker sequences between the antibody variable domains on the formation, stability and biological activity of a bispecific tandem diabody. Protein Eng. Des. Sel. 17 357–366. 10.1093/protein/gzh039 [DOI] [PubMed] [Google Scholar]
- Lee J., Der B. S., Karamitros C. S., Li W., Marshall N. M., Lungu O. I., et al. (2019). Computer-based engineering of thermostabilized antibody fragments. AIChE J. 66:e16864. 10.1002/aic.16864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy R., Ahluwalia K., Bohmann D. J., Giang H. M., Schwimmer L. J., Issafras H., et al. (2013). Enhancement of antibody fragment secretion into the Escherichia coli periplasm by co-expression with the peptidyl prolyl isomerase, FkpA, in the cytoplasm. J. Immunol. Methods 394 10–21. 10.1016/j.jim.2013.04.010 [DOI] [PubMed] [Google Scholar]
- Liu C., Dalby B., Chen W., Kilzer J. M., Chiou H. C. (2008). Transient transfection factors for high-level recombinant protein production in suspension cultured mammalian cells. Mol. Biotechnol. 39 141–153. 10.1007/s12033-008-9051-x [DOI] [PubMed] [Google Scholar]
- Liu Z., Zhang J., Fan H., Yin R., Zheng Z., Xu Q., et al. (2015). Expression and purification of soluble single-chain Fv against human fibroblast growth factor receptor 3 fused with Sumo tag in Escherichia coli. Electron. J. Biotechnolo. 18 302–306. 10.1016/j.ejbt.2015.05.006 [DOI] [Google Scholar]
- Lobstein J., Emrich C. A., Jeans C., Faulkner M., Riggs P., Berkmen M. (2012). SHuffle, a novel Escherichia coli protein expression strain capable of correctly folding disulfide bonded proteins in its cytoplasm. Microb Cell Fact 11:56. 10.1186/1475-2859-11-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUCENTIS® (2006). Prescribing Information Approved by the U.S. Food and Drug Administration. South San Francisco, CA: Genentech Inc. [Google Scholar]
- LUMOXITITM (2018). PRESCRIBING INFORMATION Approved by the U.S. Food and Drug Administration (U.S. License No. 2059). Wilmington: AstraZeneca Pharmaceuticals LP. [Google Scholar]
- Mack M., Riethmuller G., Kufer P. (1995). A small bispecific antibody construct expressed as a functional single-chain molecule with high tumor cell cytotoxicity. Proc. Natl. Acad. Sci. U.S.A. 92 7021–7025. 10.1073/pnas.92.15.7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino T., Skretas G., Kang T. H., Georgiou G. (2011). Comprehensive engineering of Escherichia coli for enhanced expression of IgG antibodies. Metab. Eng. 13 241–251. 10.1016/j.ymben.2010.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcatili P., Rosi A., Tramontano A. (2008). PIGS: automatic prediction of antibody structures. Bioinformatics 24 1953–1954. 10.1093/bioinformatics/btn341 [DOI] [PubMed] [Google Scholar]
- McConnell A. D., Spasojevich V., Macomber J. L., Krapf I. P., Chen A., Sheffer J. C., et al. (2013). An integrated approach to extreme thermostabilization and affinity maturation of an antibody. Protein Eng. Design Select. 26 151–164. 10.1093/protein/gzs090 [DOI] [PubMed] [Google Scholar]
- McConnell A. D., Zhang X., Macomber J. L., Chau B., Sheffer J. C., Rahmanian S., et al. (2014). A general approach to antibody thermostabilization. mAbs 6 1274–1282. 10.4161/mabs.29680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna R., Lombana T. N., Yamada M., Mukhyala K., Veeravalli K. (2019). Engineered sigma factors increase full-length antibody expression in Escherichia coli. Metab. Eng. 52 315–323. 10.1016/j.ymben.2018.12.009 [DOI] [PubMed] [Google Scholar]
- Miklos A. E., Kluwe C., Der B. S., Pai S., Sircar A., Hughes R. A., et al. (2012). Structure-based design of supercharged, highly thermoresistant antibodies. Chem. Biol. 19 449–455. 10.1016/j.chembiol.2012.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller B. R., Demarest S. J., Lugovskoy A., Huang F., Wu X., Snyder W. B., et al. (2010). Stability engineering of scFvs for the development of bispecific and multivalent antibodies. Protein Eng. Design Select. 23 549–557. 10.1093/protein/gzq028 [DOI] [PubMed] [Google Scholar]
- Mizukami M., Onishi H., Hanagata H., Miyauchi A., Ito Y., Tokunaga H., et al. (2018). Efficient production of Trastuzumab Fab antibody fragments in Brevibacillus choshinensis expression system. Protein Expr. Purif. 150 109–118. 10.1016/j.pep.2018.05.013 [DOI] [PubMed] [Google Scholar]
- Montoliu-Gaya L., Martinez J. C., Villegas S. (2017). Understanding the contribution of disulfide bridges to the folding and misfolding of an anti-Abeta scFv. Protein Sci. 26 1138–1149. 10.1002/pro.3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieba L., Honegger A., Krebber C., Pluckthun A. (1997). Disrupting the hydrophobic patches at the antibody variable/constant domain interface: improved in vivo folding and physical characterization of an engineered scFv fragment. Protein Eng. 10 435–444. 10.1093/protein/10.4.435 [DOI] [PubMed] [Google Scholar]
- Petrus M. L. C., Kiefer L. A., Puri P., Heemskerk E., Seaman M. S., Barouch D. H., et al. (2019). A microbial expression system for high-level production of scFv HIV-neutralizing antibody fragments in Escherichia coli. Appl. Microbiol. Biotechnol. 103 8875–8888. 10.1007/s00253-019-10145-10141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power B. E., Hudson P. J. (2000). Synthesis of high avidity antibody fragments (scFv multimers) for cancer imaging. J. Immunol. Methods 242 193–204. 10.1016/s0022-1759(00)00201-5 [DOI] [PubMed] [Google Scholar]
- PRAXBIND® (2015). Prescribing Information Approved by the U.S. Food and Drug Administration. (US License No. 2006). Ingelheim am Rhein: Boehringer Ingelheim Pharmaceuticals Inc. [Google Scholar]
- ReoPro® (1994). Prescribing Information Approved by the U.S. Food and Drug Administration. (U.S. License Number: 1864). Horsham: Janssen Biotech, Inc. [Google Scholar]
- Richards D. A., Maruani A., Chudasama V. (2017). Antibody fragments as nanoparticle targeting ligands: a step in the right direction. Chem. Sci. 8 63–77. 10.1039/c6sc02403c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. P., Ke N., Lobstein J., Peterson C., Szkodny A., Mansell T. J., et al. (2015). Efficient expression of full-length antibodies in the cytoplasm of engineered bacteria. Nat. Commun. 6:8072. 10.1038/ncomms9072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosano G. L., Ceccarelli E. A. (2014). Recombinant protein expression in Escherichia coli: advances and challenges. Front. Microbiol. 5:172. 10.3389/fmicb.2014.00172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saerens D., Huang L., Bonroy K., Muyldermans S. (2008). Antibody fragments as probe in biosensor development. Sensors 8 4669–4686. 10.3390/s8084669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders K. O. (2019). Conceptual approaches to modulating antibody effector functions and circulation half-life. Front. Immunol. 10:1296. 10.3389/fimmu.2019.01296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevy A. M., Wu N. C., Gilchuk I. M., Parrish E. H., Burger S., Yousif D., et al. (2019). Multistate design of influenza antibodies improves affinity and breadth against seasonal viruses. Proc. Natl. Acad. Sci. U.S.A. 116 1597–1602. 10.1073/pnas.1806004116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sircar A., Kim E. T., Gray J. J. (2009). RosettaAntibody: antibody variable region homology modeling server. Nucleic Acids Res. 37 W474–W479. 10.1093/nar/gkp387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steipe B. (2004). Consensus-based engineering of protein stability: from intrabodies to thermostable enzymes. Methods Enzymol. 388 176–186. 10.1016/S0076-6879(04)88016-9 [DOI] [PubMed] [Google Scholar]
- Strauch E. M., Bernard S. M., La D., Bohn A. J., Lee P. S., Anderson C. E., et al. (2017). Computational design of trimeric influenza-neutralizing proteins targeting the hemagglutinin receptor binding site. Nat. Biotechnol. 35 667–671. 10.1038/nbt.3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Structural Genomics Consortium, China Structural Genomics Consortium, Northeast Structural Genomics, Consortium, Graslund S., Nordlund P., Weigelt J., et al. (2008). Protein production and purification. Nat. Methods 5 135–146. 10.1038/nmeth.f.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suresh T., Lee L. X., Joshi J., Barta S. K. (2014). New antibody approaches to lymphoma therapy. J Hematol. Oncol. 7:58. 10.1186/s13045-014-0058-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teerinen T., Valjakka J., Rouvinen J., Takkinen K. (2006). Structure-based stability engineering of the mouse IgG1 Fab fragment by modifying constant domains. J. Mol. Biol. 361 687–697. 10.1016/j.jmb.2006.06.073 [DOI] [PubMed] [Google Scholar]
- Terpe K. (2006). Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 72 211–222. 10.1007/s00253-006-0465-8 [DOI] [PubMed] [Google Scholar]
- Tischenko V. M., Zav’yalov V. P., Medgyesi G. A., Potekhin S. A., Privalov P. L. (1982). A thermodynamic study of cooperative structures in rabbit immunoglobulin G. Eur. J. Biochem. 126 517–521. 10.1111/j.1432-1033.1982.tb06811.x [DOI] [PubMed] [Google Scholar]
- Trivedi M. V., Laurence J. S., Siahaan T. J. (2009). The role of thiols and disulfides on protein stability. Curr. Protein Peptide Sci. 10 614–625. 10.2174/138920309789630534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng T. T., Tyler B. M., Setubal J. C. (2009). Protein secretion systems in bacterial-host associations, and their description in the gene ontology. BMC Microbiol. 9(Suppl. 1):S2. 10.1186/1471-2180-9-S1-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer A. W., Norde W. (2000). The thermal stability of immunoglobulin: unfolding and aggregation of a multi-domain protein. Biophys. J. 78 394–404. 10.1016/S0006-3495(00)76602-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Xiang S., Feng Y., Srinivas S., Zhang Y., Lin M., et al. (2013). Engineering production of functional scFv antibody in E. coli by co-expressing the molecule chaperone Skp. Front. Cell Infect. Microbiol. 3:72. 10.3389/fcimb.2013.00072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber K. O., Reiter Y., Brinkmann U., Kreitman R., Pastan I. (1995). Preparation and characterization of a disulfide-stabilized Fv fragment of the anti-Tac antibody: comparison with its single-chain analog. Mol. Immunol. 32 249–258. 10.1016/0161-5890(94)00150-y [DOI] [PubMed] [Google Scholar]
- Whitelegg N. R. J., Rees A. R. (2000). WAM: an improved algorithm for modelling antibodies on the WEB. Protein Eng. 13 819–824. 10.1093/protein/13.12.819 [DOI] [PubMed] [Google Scholar]
- Wong M. S., Wu S., Causey T. B., Bennett G. N., San K. Y. (2008). Reduction of acetate accumulation in Escherichia coli cultures for increased recombinant protein production. Metab. Eng. 10 97–108. 10.1016/j.ymben.2007.10.003 [DOI] [PubMed] [Google Scholar]
- Wu J., Fu J., Zhang M., Liu D. (2015). Blinatumomab: a bispecific T cell engager (BiTE) antibody against CD19/CD3 for refractory acute lymphoid leukemia. J. Hematol. Oncol. 8:104. 10.1186/s13045-015-0195-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi S., Kobashigawa Y., Fukuda N., Teramoto M., Toyota Y., Liu C., et al. (2019). Cyclization of Single-Chain Fv antibodies markedly suppressed their characteristic aggregation mediated by inter-chain VH-VL Interactions. Molecules 24:2620. 10.3390/molecules24142620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. W., Jang Y. H., Kwon S. B., Lee Y. J., Chae W., Byun Y. H., et al. (2018). Harnessing an RNA-mediated chaperone for the assembly of influenza hemagglutinin in an immunologically relevant conformation. FASEB J. 32 2658–2675. 10.1096/fj.201700747RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusakul G., Nuntawong P., Sakamoto S., Ratnatilaka Na Bhuket P., Kohno T., Kikkawa N., et al. (2017). Bacterial expression of a single-chain variable fragment (scFv) Antibody against ganoderic Acid A: a Cost-effective approach for quantitative analysis using the scFv-Based enzyme-linked immunosorbent assay. Biol. Pharm. Bull. 40 1767–1774. 10.1248/bpb.b17-00531 [DOI] [PubMed] [Google Scholar]
- Zhang K., Geddie M. L., Kohli N., Kornaga T., Kirpotin D. B., Jiao Y., et al. (2015). Comprehensive optimization of a single-chain variable domain antibody fragment as a targeting ligand for a cytotoxic nanoparticle. mAbs 7 42–52. 10.4161/19420862.2014.985933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J. X., Yang L., Gu Z. N., Chen H. Q., Tian F. W., Chen Y. Q., et al. (2010). Stabilization of the single-chain fragment variable by an interdomain disulfide bond and its effect on antibody affinity. Intern. J. Mol. Sci. 12 1–11. 10.3390/ijms12010001 [DOI] [PMC free article] [PubMed] [Google Scholar]