Abstract
A biocompatible natural nanoparticle drug delivery system that has specific cancer-targeting function holds vast promise for cancer therapy. Here, a fucoidan/poly-lysine-functionalized layer-by-layer ginger-derived lipid vector (LbL-GDLV) was designed to target P-selectin (overexpressed by endothelial cells) and deliver a loaded drug into vascularized colon cancer. In vitro, LbL-GDLVs selectively bound to P-selectin, and the degradation of the fucoidan outer layer in a milieu similar to the cancer microenvironment resulted in rapid attachment of the cancer cell and internalization of the remaining positively charged poly-lysine coated-GDLVs. Upon enzymolysis of the poly-lysine layer inside the cancer cell, the GDLV core released loaded doxorubicin (Dox) which had the expected effects. In vivo bio-distribution studies showed that intravenously injected LbL-GDLVs exhibited enhanced accumulation at the vascularized tumor site (~ 4.4-fold higher than control vesicles), presumably due to P-selectin-mediated targeting plus the enhanced permeability and retention effect (EPR). In two animal models used to screen anti-cancer efficacy (Luc-HT-29 and HCT-116 xenografts), Dox-loaded LbL-GDLVs (LbL-GDLVs/Dox) significantly inhibited tumor growth and demonstrated much better therapeutic efficiency than free Dox. More importantly, LbL-GDLVs/Dox exhibited excellent biocompatibility, and LbL-GDLVs encapsulation largely reduced the cardiotoxicity of free Dox and avoided the notorious drug resistance of colon cells against free Dox. Together, these findings demonstrate the potential of our newly designed and highly biocompatible plant-derived LbL nanoparticles and their precise colon cancer drug delivery function.
Keywords: high-biocompatible nanoparticles, layer-by-layer, P-selectin targeted delivery, drug resistance, colon cancer therapy
1. Introduction
Colorectal cancer (CRC) is the second most common cause of cancer death among people in Europe and the third most common cancer in the United States.[1] Chemotherapy, either by itself or in combination with surgery and radiotherapy, remains the most common and effective strategy for treating CRC.[2] However, the majority of current chemotherapeutic CRC drugs cannot distinguish between malignant and normal cells, leading to severe systemic side effects.[3] To make things worse, the rich distribution of transporters on the colon cell membrane means that drug resistance develops in nearly all colon cancer patients, decreasing the therapeutic efficacies of anticancer agents.[3–4] Thus, we critically need to develop and employ drug delivery systems that can load chemotherapeutics into the cancer cells (to spare normal cells) and, ideally, avoid drug resistance. As pharmaceutical carriers, nanoparticles, including polymeric,[4a, 5] liposomal,[6] dendrimer-like,[7] inorganic,[8] and protein nanoparticles,[9] have been widely tested. These carriers tend to site-specifically deliver a higher payload of chemotherapeutic agents than obtained by systemic administration of conventional drugs, reduce untargeted bio-distribution, and yield lower systemic toxicity.[10]
The current efforts to develop target-specific delivery systems are based mainly on synthetic nanoparticles. Unfortunately, synthetic nanoparticles suffer from many limitations, such as their ability to induce off-target cytotoxicity, chronic inflammation, and host immune responses, as well as a high cost of large-scale production.[4b, 11] The use of natural nanoparticles may overcome these limitations. Our group and others have demonstrated that natural lipid nano vectors from edible plants (e.g., ginger, grapefruit, broccoli, and lemon) could serve as highly biocompatible drug delivery platforms [3, 4b, 11a, 12] that may solve the safety problems associated with synthetic nanoparticles. Interestingly, animal studies have shown that different plant-generated nanoparticles can target different tissues. For example, ginger-derived lipid vectors (GDLVs) mainly target the colon, while grapefruit-derived nanoparticles mostly accumulate in the liver.[4b] However, although plant-derived edible nanoparticles (PDNPs) can have tissue-specific targeting abilities, they do not specifically interact with or target cancer cells. Thus, further functionalization of these PDNPs with cell-specific targeting ability would be highly desirable for advanced drug delivery purposes.
P-selectin (CD62P) is a cancer-associated cell adhesion molecule that is found on the membranes of tumor-adjacent endothelial cells and the secretory α-granules of platelets.[13] It contributes to early leukocyte recruitment during inflammation and in the tumor microenvironment.[14] Importantly, elevated P-selectin expression had been reported in the vasculature of many types of cancer, including colon cancer,[15] suggesting that P-selectin could potentially serve as a cancer-specific delivery target for colon cancer therapy. In a pilot study, we evaluated the feasibility of using P-selectin as a colon cancer drug delivery target.[13–14] P-selectin was found to be overexpressed in human and murine colon cancer tissues, but not healthy tissues, strengthening the idea that P-selectin might be a potent colon cancer drug delivery target.
To design a P-selectin-targeting nanoparticle for delivering a drug to colon cancer cells, we constructed functionalized layer-by-layer multilayer biocompatible nanoparticles (LbL; Figure 1) based on the following design choices. As the outer coating of the LbL, we chose the polysaccharide, fucoidan, which is a natural seaweed-derived polymer that exhibits a high affinity for P-selectin.[13a] The theory is that P-selectin guides the accumulation of LbL in the tumor area, whereupon locally high concentrations of hydrogen peroxide (H2O2) and free radicals in the cancer microenvironment will trigger the degradation of fucoidan and expose the middle layer.[16] As a cell-attaching functionalized middle layer, we chose the bio-compatible cationic polymer, ε-poly-lysine (ε-PLL), which can adhere to the negatively charged cancer cell membrane and undergo internalization.[17] Thereafter, decomposition of the middle layer by cytosolic enzymes (inside the cancer cells) will release the drug-loaded nanoparticle core. This will free the loaded anti-cancer drug to exit the core and destroy the cancer cells. All of the components used to construct this novel LbL multilayer platform are highly biocompatible and functional nanomaterials that are derived from edible natural products. We hypothesized that this drug delivery system would be exceptionally safe, have a high cancer-targeting specificity, exhibit good stability outside the cancer microenvironment, and enhance the efficacy of the loaded drug against cancer cells. As the LbL structure boasts three different functional layers, we expected that our system could escort a loaded drug into cancer cells without triggering the upregulation of drug transporters, which are the root of drug resistance.
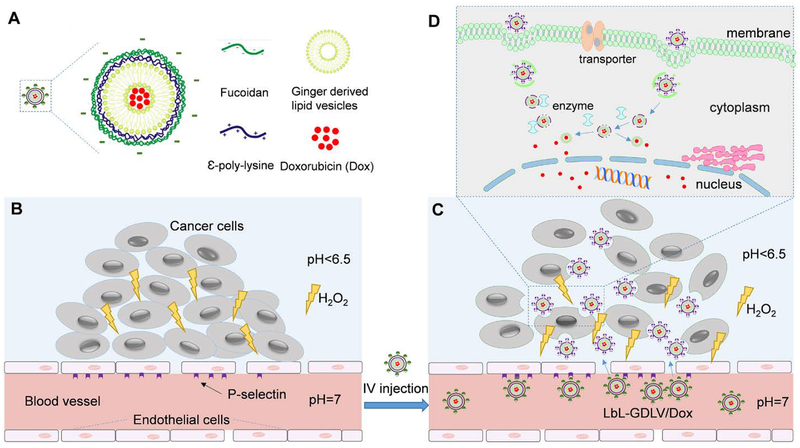
Figure 1. Schematic representing the P-selectin-mediated colon tumor tissue penetration and therapeutic drug delivery of LbL-GDLVs/Dox.
A. The biocompatible layer-by-layer nanoparticle (LbL) was composed of negatively charged fucoidan (outer layer), positively charged ε-poly-lysine (middle layer), ginger nano lipid vesicles (core), and doxorubicin (cargo). B. The cancer microenvironment is characterized by overexpression of P-selectin on adjacent endothelial cells, an acidic pH, and a high concentration of hydrogen peroxide (H2O2). C. Through P-selectin-mediated delivery, LbL-GDLVs/Dox accumulate and penetrate into tumors, undergo outer layer degradation in the tumor microenvironment, attach, and undergo internalization. D. Decomposition of the poly-lysine coating enables GDLVs to release Dox inside the cancer cells.
Our in vitro biocompatibility test demonstrated that the newly constructed LbL is very safe when applied to cultured cells. The target specificity test indicated that fucoidan-functionalized LbL has high affinity for recombinant P-selectin, but not L- or E-selectin. A microenvironment-dependent stability assay showed that the LbL nanoparticle is degraded in an H2O2-accumulated milieu at acidic pH and that the remaining ε-PLL-coated GDLVs are prone to being taken up into and digested by cancer cells. In vitro, fucoidan functionalization increased the transmigration ability of nanoparticles through an endothelial cell monolayer, in a manner that relied on the TNF-α-dependent activation of P-selectin. More importantly, fucoidan-LbL/Dox, but not free Dox, failed to up-regulate drug transporters on HT-29 cells (a cancer cell line), indicating that our novel LbL nanomedicine should not induce the notorious drug resistance of colon cancer against Dox.
Given these exciting in vitro data, we next looked at the in vivo bio-distribution of LbL-GDLVs. We found that LbL-GDLVs selectively accumulated at a vascularized tumor site after intravenous (i.v.) injection, showing P-selectin-guided cancer-specific enrichment compared to control nanoparticles (without fucoidan coating). In two animal models used to examine anti-tumor efficacy (Luc-HT-29 and HCT-116 xenografts), LbL-GDLVs/Dox treatment significantly inhibited tumor growth and demonstrated higher therapeutic efficiency than free Dox. More importantly, the in vivo biocompatibilities of LbL-GDLVs and LbL-GDLVs/DOX were excellent, with negligible toxicity to major organs (heart, liver, spleen, lung, and kidney), whereas free Dox induced severe cardiotoxicity in treated mice. To our knowledge, this is the first report of a functionalized layer-by-layer multilayer plant-derived nanodrug delivery system that is highly biocompatible and has cancer-specific targeting ability. Our findings could be exploited to develop new targeted drug delivery approaches for expanding the use of colon cancer-ineffective anti-cancer drugs to the chemotherapy of colon cancer.
2. Results and Discussion
2.1. P-selectin expression is increased in human and murine colon cancer tissues
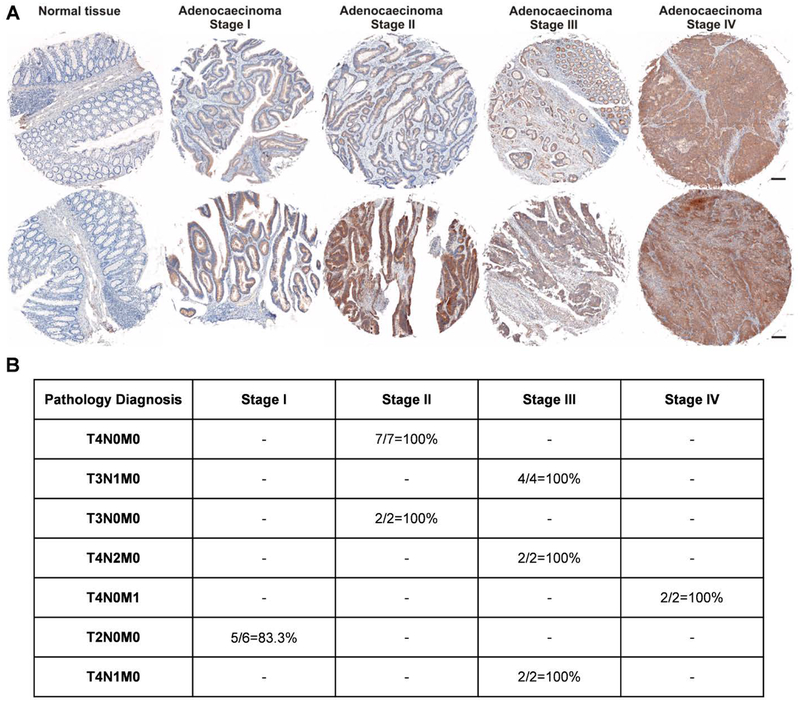
To assess P-selectin expression in colon cancer tissues, we obtained clinical samples from colon cancer patients and examined P-selectin expression and its association with cancer progression using immunohistochemistry (IHC) on a human tissue microarray (Figure 2A). We found that P-selectin expression was deficient in normal tissues but very high in tumor tissues, and that the expression levels appeared to be positively correlated with the clinical stages represented in the tissue microarray (Figure 2B). To confirm this finding in mouse tumor tissues, we generated a mouse colon tumor model induced by azoxymethane (AOM) and dextran sulfate sodium (DSS). As expected, colon tumor tissues of AOM/DSS-treated mice exhibited significantly higher mRNA expression of P-selectin (Figure S1) compared with normal colon tissues. These findings indicate that P-selectin is highly expressed in colon cancer tissues but not in the corresponding normal tissues and might thus be useful as a delivery target in colon cancer therapy.
Figure 2. P-selectin expression is increased in colon cancer tissues.
A. A human P-selectin antibody was used to observe P-selectin levels in a human tissue microarray counterstained with hematoxylin. The tissue microarray included samples from healthy individuals and cancer patients with colon adenocarcinoma of various stages. Scale bar: 20 μm. B. P-selectin antibody-stained slides were analyzed for tumor invasion and metastasis. The abbreviations are as follows: T refers to the primary tumor; T1, the tumor has invaded the submucosa; T2, the tumor has invaded the muscularis propria; T3, the tumor has invaded through the muscular propria into the subserosa or non-peritonealized pericolic or perirectal tissues; T4: the tumor has directly invaded other organs or structures and/or perforated the visceral peritoneum; N refers to the regional lymph nodes; N0, regional lymph node metastasis; N1, metastasis in one to three regional lymph nodes; N2, metastasis in four or more regional lymph nodes; M refers to distant metastasis; M0, no distant metastasis; and M1, distant metastasis.
2.2. Design, construction, and characterization of LbL-GDLVs
Given the high prevalence of P-selectin in the microenvironment of human colon cancer tissue, which was confirmed by our IHC staining results, we surmised that targeting of P-selectin would improve the ability of a nano-system to deliver a drug to colon cancer cells. We chose to target P-selectin with the polysaccharide, fucoidan, which was used as the exterior layer of our LbL-GDLVs. Fucoidan is expected to exhibit high and specific affinity for P-selectin by mimicking its principal ligand, P-selectin glycoprotein ligand 1 (PSGL-1).[18] As anticipated, fucoidan-functionalized nanoparticles were found to target recombinant P-selectin, but not L-selectin, E-selectin, or BSA (see section 2.5).
It is important to select an appropriate middle layer when designing an LbL nanoparticle. The middle layer should stabilize the LbL structure, bind to the cell membrane, guard the core, and release the drug-loaded core inside the cells. For these reasons, we chose the cationic amino acid derived polymer ε-poly-lysine (ε-PLL). The positively charged property of ε-PLL can fortify the LbL nanoparticle by electrostatically holding both the negatively charged core (GDLVs) and the outer layer (fucoidan). This electrostatic force is also expected to help the LbL particle attach to the cell surface after P-selectin-guided targeting and the tumor microenvironment-dependent decomposition of the outer layer. We expected that ε-PLL would be quickly biodegraded, as a multi-layered poly-lysine film was reported to undergo enzymatic degradation in PBS at 37 °C [17a]. Notably, poly-lysine is widely used in the food industry and is a generally recognized as safe (GRAS) agent, as confirmed by the US Food and Drug Administration (FDA); thus, this is a highly biocompatible nanomaterial.[19]
The core material of the LbL nano-structure is key for the particle’s biocompatibility. Many applications have employed non-degradable inorganic nanoparticles (i.e., gold, silver, iron, silica) or hard-to-decompose synthetic polymers that have the potential to trigger unexpected immune responses.[20] However, the recently described edible PDNPs, such as ginger-derived lipid vectors (GDLVs), can overcome these limitations. GDLVs offer many advantages, such as a lack of toxicity, negligible hazardous environmental effects, and the ability to undergo larger-scale production than synthetic nanoparticles. Therefore, GDLVs represent one of the safest existing nano therapy-based therapeutic delivery platforms.[12]
Before constructing the LbL-GDLV-based drug delivery system, we first validated the preparation of the LbL structure in a proof-of-concept study by employing negatively charged carboxyl-modified polystyrene latex nanoparticles (CMLs) as the core. CMLs are uniformly sized and resemble other drug delivery nanocarriers (e.g., liposomes) in terms of their stability and size.[21] As shown in Figure S2, the utilized CMLs had a hydrodynamic size of 98.9±1.7 nm and a zeta potential of −23.8±0.4 mV. The CMLs were modified with ε-PLL; this increased the particle size to 127.6±1.4 nm and changed the zeta potential to 33.8±0.3 mV, indicating that the CMLs had been successfully surface-coated with ε-PLL via electrostatic interaction to generate ε-PLL-CMLs. The ε-PLL-CMLs were further surface-coated with a layer of fucoidan to enable P-selectin targeting.[14] Our data showed that fucoidan-coated CML particles (LbL-CMLs) were successfully generated; they had a size of 160±2.1 nm and a zeta potential of −37.5±0.5 mV.
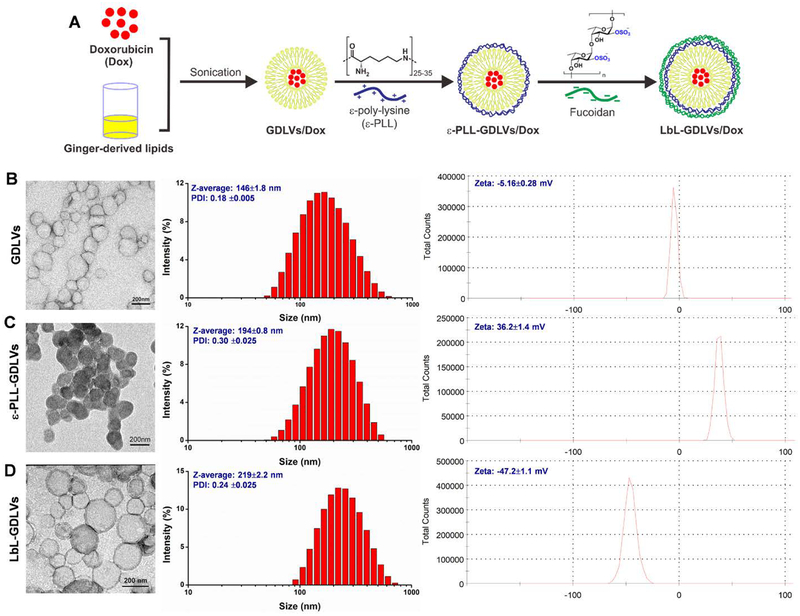
Next, we tested whether we could construct similar LbL nanoparticles with GDLVs generated from ginger lipids as the core (the composition was shown in Figure S3 and Table S1). As presented in Figure 3, native GDLVs had a hydrodynamic size of 146±1.8 nm and a zeta potential of −5.16±0.28 mV. Coating these particles with a layer of ε-PLL increased the size to 194±0.8 nm and reversed the zeta potential to 36.2±1.4 mV. Coating the resulting particles with an outer layer of fucoidan further increased the size to 219±2.2 nm and altered the zeta potential to −47.2±1.1 mV. These findings indicate that we successfully used GDLVs as the core for the layer-by-layer generation of LbL-GDLVs, which should represent a P-selectin-targeting drug delivery system that is highly biocompatible.
Figure 3. Construction and characterization of GDLVs, ε-PLL-GDLVs, and LbL-GDLVs.
A. LbL-GDLVs/Dox were constructed using a layer-by-layer technique. Ginger-derived lipids loaded with the therapeutic drug, doxorubicin (Dox), were first modified with ε-poly-lysine (ε-PLL) and then surface-coated with fucoidan via electrostatic interaction. B-D. Left: Transmission electron microscopic (TEM) images. Scale bar = 200 nm. Middle: Size distribution of nanoparticles, as assessed using dynamic light scattering (DLS). Right: Zeta potential (surface charge) of nanoparticles, as assessed using DLS.
2.3. Microenvironment-dependent layer stability and cell uptake test of LbL nanoparticles
According to Dr. Stephen Paget’s “seed and soil” hypothesis, the tumor microenvironment (the soil) plays a critical role in the progression and metastasis of a tumor (the seed).[22] Recent studies have shown that an abnormally high level of H2O2, which is produced by both cancer cells and cancer-associated fibroblasts, may provide the “fertilizer” in the soil (tumor microenvironment) that is needed for subsequent seed (tumor) development.[23] Hence, when designing our microenvironment-dependent drug delivery system, we took advantage of the locally high concentration of H2O2.
Using solutions containing 50 and 100 μM of H2O2 to mimic the tumor microenvironment,[16] we tested the degradation behavior of fucoidan-coated and ε-PLL-coated nanoparticles. As shown in Figure S4, the surface zeta potential of fucoidan-coated LbL GDLVs tended to time- and concentration-dependently decrease in absolute value upon exposure to the H2O2-containing environment, indicating that the fucoidan coating may be degraded under this condition, while the ε-PLL-coated particles were relatively stable even in the presence of 100 μM H2O2.
Having confirmed that our fucoidan-coated LbL nanoparticles targeted P-selectin and the outer layer could be decomposed by the tumor microenvironment, we next investigated whether the remaining stable ε-PLL-coated core could be internalized by cancer cells. We stained ε-PLL-coated GDLVs with the lipid dye, Dil-C18(3), and incubated stained ε-PLL-GDLVs with HT-29 cells. The obtained fluorescence images (Figure S5, middle column) showed that HT-29 cells started to take up ε-PLL/GDLVs after as little as 15 min of incubation. After just 1 h, most of the nanoparticles were engulfed by the cells (Figure S5 right column). Together, these results show that our fucoidan-coated LbL can operationalize the tumor microenvironment to deliver therapeutics into the cancer cells.
2.4. Comparing the drug resistance, cellular uptake and cytotoxicity of HT-29 to LbL-GDLVs/Dox and Free Dox
Multiple drug resistance (MDR), wherein cancer cells manage to block the effects of chemotherapeutic agents, is a huge obstacle in the successful treatment of cancer.[24] MDR may occur through various mechanisms, but it often correlates with the overexpression of membrane transporters, including ATP-binding cassette (ABC) transporters, multi-drug resistance protein, MDR-associated proteins (MRPs), glutathione transferase. These substances exhibit various functions in the induction of tumor cell MDR. Cellular MDR is divided into the ATP-dependent efflux pumps and non-ATP-dependent efflux pumps types. The ‘pumps’ type uses the energy obtained from ATP hydrolysis to efflux drugs via ATP-dependent transport proteins, such as P-Gp and MRPs, which decreases the intracellular drug concentration and subsequently results in drug resistance. The ‘non pumps’ type does not depend on the energy from ATP hydrolysis while activating the anti-apoptotic proteins, such as Bcl-2, and efflux drugs directly.[25] Pgp and MDR proteins are often unregulated on the colon cancer cell membrane upon Dox treatment, causing this commonly used cancer drug to gradually lose its efficacy against colon cancer.[26] Therefore, a delivery platform that can help avoid the up-regulation of drug transporters would be extremely useful for colon cancer therapy.
To this end, we studied whether our newly designed LbL-GDLVs/Dox, in which free Dox is covered with nano-layers, could circumvent the upregulation of MDR proteins. Real-time polymerase chain reaction (RT-PCR) showed that the expression level of MDR1 did not increase in HT-29 cells after LbL/Dox treatment for 10 passages, whereas the free Dox-treated cells showed significant upregulation of the mRNA encoding MDR1 (Figure S6). The reason might seem that LbL-GDLV/Dox altered Dox uptake in HT-29 cells. The system delivered Dox directly to HT-29 cells, which functioned to reverse MDR. LbL-GDLV/Dox affected the pharmacokinetic properties of Dox; free Dox crossed the cell membrane passively and were easily identified by the efflux pumps located on the cell membrane or captured by ABC transporter proteins, whereas LbL-GDLV/Dox avoided recognition by the efflux pumps and endocytosis by the transporters, leading to increased intracellular accumulation of Dox. Taken together, these data indicate that LbL-GDLVs could avoid inducing drug resistance.
In vitro drug release profile, cellular uptake and cytotoxicity were evaluated followed. LbL-GDLVs/Dox could achieve around 65% cumulative drug release within 24 h in the mimic tumor microenvironment (Figure S7). For the cellular uptake, obtained fluorescence signal of Dox indicated that Dox released from LbL-GDLVs/Dox yet at 24 h (Figure S8), combined with our abovementioned results, the possibly intracellular drug delivery route was described in Figure S9. In vitro cytotoxicity caused by Free Dox and LbL-GDLVs/Dox was also evaluated and compared by MTT assays (Figure S10), LbL-GDLVs/Dox induced more cell apoptosis at all tested concentrations (0.5, 1, 2 and 5 μg/ml Dox) in both HT-29 cells and HCT-116 cells. These results confirmed that LbL-GDLVs/Dox has a higher therapeutic effect than free Dox and can be served as a good drug delivery system.
2.5. Endothelial activation increases the penetration of P-selectin-targeted LbL nanoparticles across the endothelial barrier
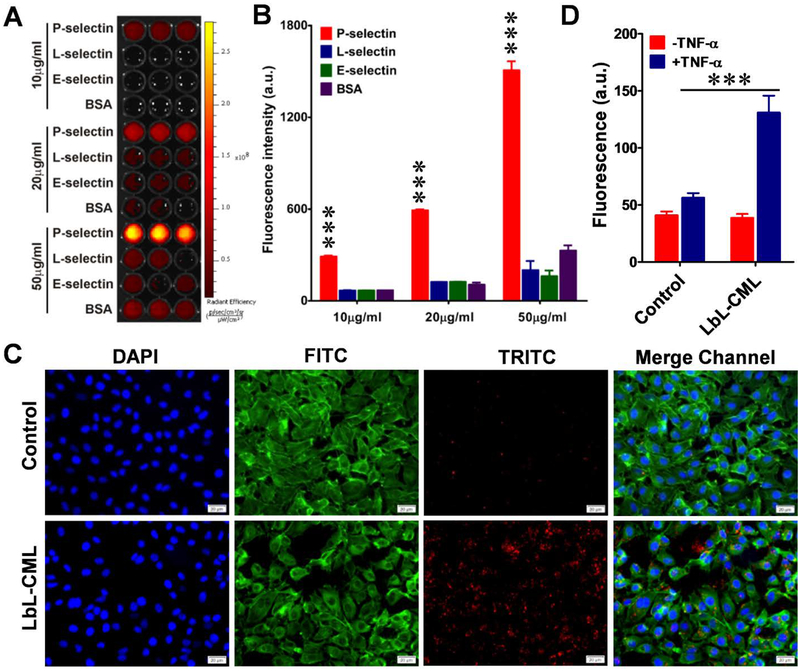
To assess the targeting selectivity of our fucoidan-coated LbL nanoparticles for P-selectin, we tested the binding of LbL-CMLs to immobilized human recombinant P-selectin, L-selectin, E-selectin, and bovine serum albumin (BSA). Indeed, P-selectin-coated wells exhibited stronger fluorescence signals than those coated with the other proteins, and LbL-CMLs demonstrated selective dose-dependent binding of their target (Figure 4A and B). We then measured the binding of LbL-CMLs to a monolayer of endothelial cells (ECs) subjected to an inflammatory stimulation known to induce P-selectin expression.[27] Upon activation with tumor necrosis factor-α (TNF-α), ECs-EA.hy926 cells expressed P-selectin and showed a large increase in the LbL-CML-derived fluorescence signal. In contrast, cells that did not receive TNF-α treatment expressed a low level of P-selectin and showed little or no fluorescent signal (Figure 4C). These findings validated the P-selectin-mediated targeting properties of our LbL-CML nanoparticles.
Figure 4. Fucoidan-coated LbL nanoparticles selectively bind to P-selectin and exhibit increased penetration across the endothelial barrier.
A. The binding capacity of fucoidan-functionalized dye (red)-labeled CML beads (LbL-CMLs) to immobilize recombinant P-selectin. Recombinant P-selectin, L-selectin, E-selectin, and BSA at three different concentrations (10, 20 and 50 μg/ml) were immobilized on ELISA plates, which were thoroughly washed and then incubated with dye (red)-labeled LbL-CMLs. Fluorescence was captured with an in vivo imaging system. B. The fluorescence intensity of each well in A was recorded and compared between groups (n=5), ***<0.001. C. Fluorescence images of human endothelial monolayers (EA.hy926 cells) treated with TNF-α to induce the expression of P-selectin, which increased the binding of fucoidan-functionalized LbL-CMLs. Control: cells without TNF-α treatment. Blue channel: DAPI staining of nuclei. FITC channel: fluorescein isothiocyanate-labeled phalloidin staining of actin. TRITC channel: dye (red)-labeled LbL-CMLs. Scale bar: 20 μm. D. The LbL-CMLs show high-level penetration through an activated endothelial monolayer barrier, as assessed by transwell assay (n=5), ***<0.001.
We also conducted in vitro experiments designed to mimic the biological barriers in the tumor microenvironment. A transwell assay was used to test the ability of LbL-CMLs to pass through an activated EC monolayer.[28] EA.hy926 cells were grown on the upper chambers, TNF-α treatment was used to induce P-selectin expression, LbL-CMLs were placed in the upper chambers, the system was incubated overnight, and the fluorescence of the medium in the bottom chamber was measured. As shown in Figure 4D, the TNF-α-treated group showed enhanced LbL-CML penetration compared to control cells (without TNF-α treatment), suggesting that endothelial activation increased the penetration of our P-selectin-targeting nanoparticles across the endothelial barrier. As TNF-α is known to induce EC activation similar to that seen in the tumor microenvironment, and is involved in inflammation-associated carcinogenesis,[29] our results suggest that the increased transmigration of fucoidan-functionalized nanoparticles across the endothelial barrier to the tumor site could be a specific advantage for employing fucoidan as the functional outer layer.
2.6. LbL-GDLVs show excellent in vitro and in vivo biocompatibility
Biocompatibility is a critical issue that should be investigated when evaluating a potential drug delivery system. Toward this end, we first used electric cell-substrate impedance sensing (ECIS) to perform real-time measurement of cell proliferation, as a means to assess the toxicity of LbL-GDLVs towards Caco2-bbe cells (Figure S11A). Cells were attached to the electrode surface and allowed to form a confluent monolayer, and then treated with LbL-GDLVs, DMEM (medium control), or Tween-100 (positive control). Caco2-bbe monolayers treated with LbL-GDLVs (up to the highest tested concentration of 200 μM) showed resistance curves similar to that of the DMEM control, whereas the resistance curve of the Tween-100 group showed an initial steep decrease and a subsequent decreasing tendency. These results indicated that LbL-GDLVs do not have cytotoxic effects in vitro. We further investigated the biocompatibility of LbL-GDLVs using a colony formation assay, which is an in vitro cell survival test based on the ability of a single cell to grow into a colony.[30] We observed that Luc-HT-29 cells exhibited similar colony formation in LbL-GDLV-treated and control experiments (Figure S11B), indicating LbL-GDLVs do not alter the proliferative potential of Luc-HT-29 cells. To confirm this, we assessed the luciferase activity of Luc-HT-29 cells treated with LbL-GDLVs. There was no significant change in luciferase activity, even among cells tested with the highest concentration (200 μM) of LbL-GDLVs (Figure S11C, D). Together, these results indicate that LbL-GDLVs exhibit excellent biocompatibility in vitro.
Next, we explored the potential in vivo cytotoxic effects of LbL-GDLVs. Blood was collected from mice that were i.v. injected with PBS, GDLVs, or LbL-GDLVs, and hemanalysis and biochemical analyses were performed. As shown in Table S2, there was no significant difference in the white blood cell count, red blood cell count, hemoglobin level or parameters of hepatic and renal function (i.e., blood urea nitrogen, total bilirubin, alanine aminotransferase [ALT] and total protein) between the PBS- and LbL-GDLV-treated groups. These results indicate that LbL-GDLVs did not have any appreciable effect on the blood and biochemical indexes of normal mice. Histological analysis of hematoxylin and eosin (H&E)-stained tissue sections did not find any clear evidence of organ damage in LbL-GDLV-treated mice compared with control mice. The liver hepatocytes appeared normal, no myocardial fibrillary loss or vacuolation was observed in the heart, no pulmonary fibrosis was detected in lung samples, and no necrosis was observed in any analyzed tissue (Figure S12). Collectively, our findings indicate that LbL-GDLVs exhibit excellent biocompatibility both in vitro and in vivo, and thus appear to be safe for use as a drug delivery vehicle.
2.7. LbL-GDLVs exhibit P-selectin-mediated tumor targeting and anti-tumor effects in mouse xenograft models
After confirming that no adverse side effect appeared following i.v. injection of mice with LbL-GDLVs, we next investigated the drug delivery capability of LbL-GDLVs. Our results revealed that LbL-GDLVs are capable of being loaded with chemo-drugs, such as doxorubicin (Dox), and showed a high loading capacity (Figure S13), the loading efficiency of Dox in LbL-GDLVs/Dox was up to 73.7±5.7%. As blood-transported nanoparticles may trigger hemolysis, severely limiting their in vivo application,[31] we tested the potential for LbL-GDLVs to trigger hemolysis. Triton X-100, which was used as a positive control, achieved 100% hemolysis. In contrast, LbL-GDLVs did not trigger hemolysis up to the highest tested concentration (200 μM) (Figure S14), suggesting that i.v. administrated LbL-GDLVs had a negligible effect on erythrocytes. Our stability analysis of LbL-GDLVs in blood indicated that circulating LbL-GDLVs were stable and detectable until 48 h post i.v. injection (Figure S15). This suggests that the particles would have sufficient time to reach and accumulate at the tumor site.
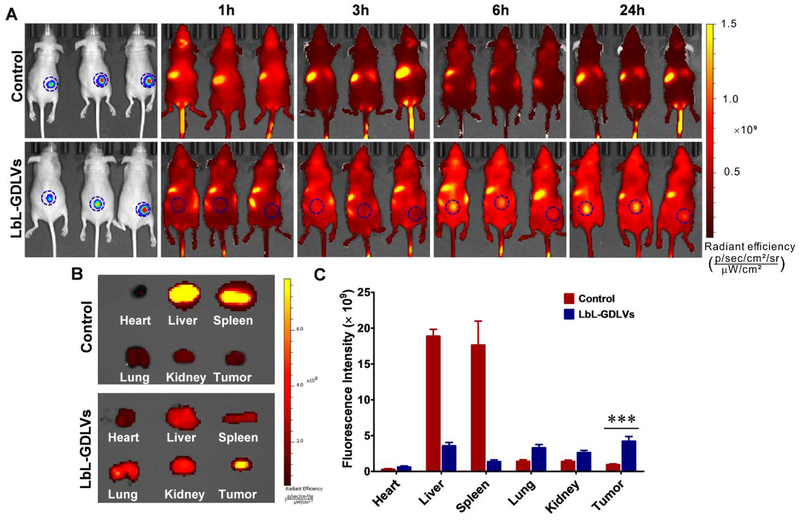
To estimate the targeting ability of LbL-GDLVs in vivo, we used a non-invasive near-infrared optical imaging technique to examine the bio-distribution of DiR-labeled LbL-GDLVs administrated intravenously into Luc-HT-29 tumor-bearing nude mice. As shown in Figure 5A, DiR-LbL-GDLVs exhibited a stronger DiR signal at the tumor site compared to normal tissues at 6 h post-injection. At 24 h post-injection, this DiR fluorescence signal was even higher at the tumor region compared with normal tissues, validating the significant tumor-targeting effect of LbL-GDLVs. In contrast, at 24 h after injection of control nanoparticles, we observed almost no signal at the tumor site or normal tissues, indicating that these nanoparticles underwent rapid clearance. To determine whether this held true throughout the animal, we sacrificed mice at 24 h post-injection, and harvested tumor samples and various organs (heart, liver, spleen, lung, and kidney) for ex vivo imaging of DiR. As shown in Figure 5B, a stronger DiR signal was observed in tumor tissues of mice injected with LbL-GDLVs compared to control nanoparticles. Additionally, fluorescence was found in the liver, spleen, and lung. The accumulation of nanoparticles in the liver and spleen may reflect their phagocytosis by the reticuloendothelial system (RES), while that in the lung may suggest that they are retained by lung capillaries.[32] Our quantitative region-of-interest (ROI) analysis determined that the fluorescence intensity at the tumor site was 4.4-fold higher in DiR-LbL-GDLV-treated mice compared to control-injected mice (Figure 5C). Together, our observations indicate that LbL-GDLVs have an excellent ability to target the tumor in our in vivo model. This may explain the superb anti-tumor capacity of LbL-GDLVs/Dox (see below).
Figure 5. LbL-GDLVs exhibit improved tumor delivery and penetration in Luc-HT-29 tumor-bearing mice in vivo.
A. In vivo imaging of mice at 1, 3, 6 and 24 h after intravenous injection (i.v.) of DiR-labeled LbL-GDLVs or control nanoparticles that had dextran sulfate sodium (DSS) as their outer layer (n=3). B. Ex vivo images of Luc-HT-29 tumors and other tissues (heart, liver, spleen, lung, and kidney) at 24 h after injection (n=3). C. Region of interest (ROI) data analysis of DiR fluorescent signals from tumors and normal tissues. Results are expressed as mean fluorescence intensity ± SD (n=3); ***p<0.001.
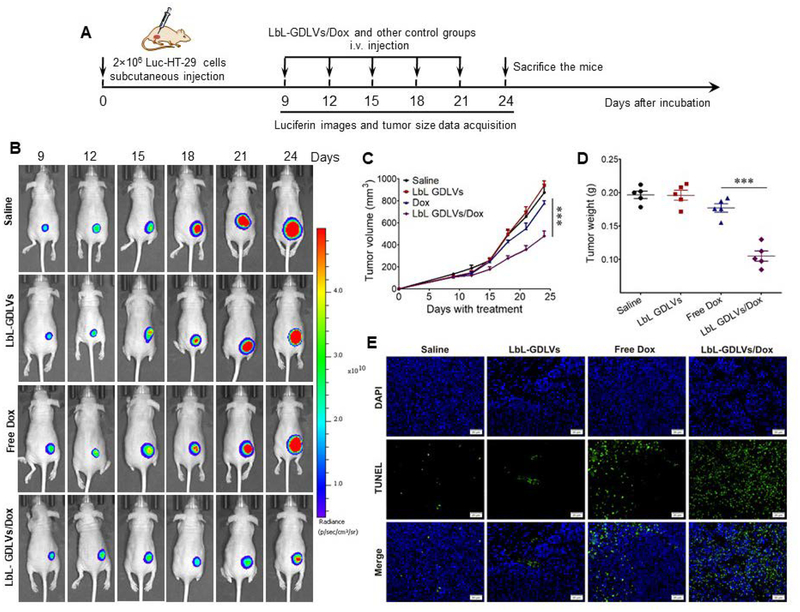
To determine the potential of LbL-GDLVs as a delivery system, we tested the ability of a drug loaded into LbL-GDLVs (doxorubicin; Dox) to exert tumoricidal effects in athymic BALB/c nu/nu mice bearing Luc-HT-29 tumors (Figure 6A). The tumor-bearing mice were randomly divided into four groups and intravenously injected with saline, LbL-GDLVs, free Dox, or LbL-GDLVs/Dox. Bioluminescence signal capture and tumor growth measurements revealed that tumor growth was significantly slower in the LbL-GDLVs/Dox-treated group compared to the other treatment groups (Figure 6B, C). When tumor weights were examined at the end of the experiment, the LbL-GDLVs/Dox-treated group showed the lowest tumor weight, and that this parameter was significantly lower than the tumor weights in the free Dox-, saline-, and LbL-GDLVs-treated control groups, which were similar (Figure 6D).
Figure 6. LbL-GDLVs/Dox have in vivo therapeutic efficacy in Luc-HT-29 tumor-bearing mice.
A. Illustration of the experimental design. Luc-HT-29 tumor-bearing mice were established, randomly divided into four groups, and treated with the LbL-GDLVs/Dox or controls consisting of saline, LbL-GDLVs, and free Dox (n=5). B. Representative bioluminescence images of mice following the administration of luciferin substrate. On days (D)9, D12, D15, D18, D21 and D24 post-injection of Luc-HT-29, tumor cells were visualized using an in vivo imaging system (n=5). C. Tumor growth profiles in mice treated with LbL-GDLV/Dox or control formulations. Data are presented as the mean value ± SD (n=5); ***p<0.001. D. Tumor weights in each group were measured at the end of the experiment (n=5); ***p<0.001. E. Terminal deoxynucleotidyl transferase-mediated nick-end labeling (TUNEL) assay was used to evaluate the levels of apoptotic cells (green) in Luc-HT-29 tumors (n=5). Scale bar: 20 μm.
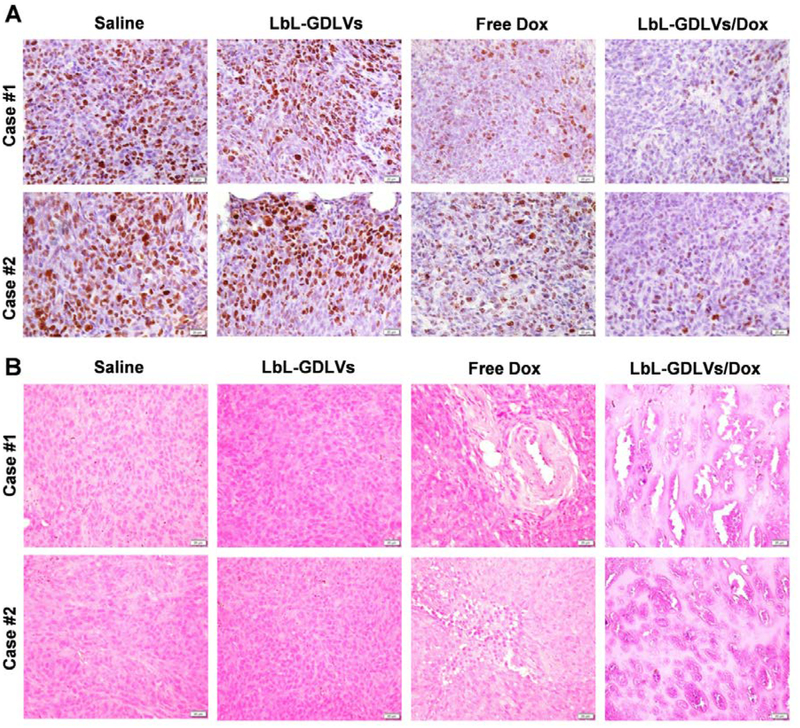
To explore the mechanism underlying the tumor growth inhibition conferred by LbL-GDLVs/Dox, we stained for apoptotic cells using terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) and performed immunohistochemical detection of the cellular proliferation marker, Ki67. Few TUNEL-positive apoptotic cells (green dots) were detected in tumor sections from the saline- and LbL-GDLVs-treated groups, providing additional evidence that LbL-GDLVs show good biocompatibility in vivo. In contrast, tumor sections from the LbL-GDLVs/Dox-treated group showed a high degree of apoptosis, much higher than that in the free Dox group (Figure 6E). This indicates that LbL-GDLVs/Dox inhibit tumor growth in a solid tumor model by inducing apoptosis. Immunohistochemical staining of the Ki67 antigen in tumor sections followed by visual inspection revealed that tumor cells in the LbL-GDLVs/Dox-treated group exhibited decreased Ki67 expression (Figure 7A), indicating that cell proliferation was markedly reduced in the tumors of these mice. To confirm the improved therapeutic efficacy of LbL-GDLVs/Dox, we performed histologic analysis. H&E staining of tumor sections suggested that there was considerably more necrosis in the LbL-GDLVs/Dox-treated group compared with the control groups (Figure 7B).
Figure 7. Therapeutic efficacy of LbL-GDLVs/Dox in Luc-HT-29 tumor-bearing mice, as evaluated by histological analysis.
A. Immunohistochemical staining of the cell proliferation marker, Ki67. Scale bar: 20 μm. B. H&E staining of tumor tissues. Scale bar: 20 μm.
To investigate the potential side effects of LbL-GDLVs/Dox, we performed histologic examinations of major organs (heart, liver, spleen, lung, and kidney) at the end of the experiment. As shown in Figure S16, mice of the saline, LbL-GDLVs, and LbL-GDLVs/Dox groups did not show any noticeable sign of tissue or cellular damage (e.g., myocardial fibrillar loss or vacuolation in the heart; edema, ballooning, and/or degeneration of hepatocytes; increased numbers of granulocytes in the spleen; tubular vacuolization or tubular dilation with hemorrhagic areas in the kidney; or increased alveolar wall thickness or cellular infiltration in the lung). In contrast, free Dox treatment induced typical myocardial damage with intensive vacuolization and myofibril loss (Figure S16, blue circles). Finally, we evaluated the anti-tumor capacity of LbL-GDLVs/Dox in an HCT-116 tumor-bearing model generated in athymic BALB/c nu/nu mice. Our results consistently demonstrated that LbL-GDLVs/Dox treatment significantly inhibited tumor growth and showed better therapeutic efficiency than free Dox (Figure S17–19).
3. Conclusion
The intrinsic limits of conventional cancer therapies, such as chemotherapy, have prompted the development and application of various nanotechnologies for more effective and safer cancer treatment. There have been considerable technological successes in the development of nanoparticle-based drug delivery systems.[5, 20a, 32b] However, the potential toxicity of many synthetic nanoparticles and issues surrounding economic and large-scale production have limited their application. We speculate that nanoparticles from natural sources, such as edible plants, might address these limitations if we use plants that are regularly consumed, have no known potential toxicity to humans, and can be produced in large quantities. Accordingly, we herein designed a highly biocompatible layer-by-layer drug delivery system by employing edible ginger-derived lipid vehicles (GDLVs) as the core and functional targeting natural polymers as the layers.
This newly designed LbL-GDLV system exhibited excellent biocompatibility both in vitro and in vivo, and thus appears to be safe for use as a drug delivery vehicle. Biodistribution studies in Luc-HT-29 tumor-bearing nude mice revealed that LbL-GDLVs selectively targeted the vascularized tumor site by 24 h after injection and that this accumulation was ~4.4-fold higher than that seen with control nanoparticles. LbL loaded with the chemotherapeutic drug, doxorubicin (Dox) (LbL-GDLVs/Dox), significantly inhibited the tumor growth and showed better therapeutic efficiency than free Dox in two xenograft colon cancer models. Notably, many potentially potent chemotherapeutic drugs have failed to treat colon cancer because they lack specificity and cause drug resistance. The drug delivery system described herein might prove extremely useful for the testing of such drugs, as the disease-specific over-expression of P-selectin in colon cancer makes it an excellent target molecule for site-specific therapy,[33] and the multi-layer functional structure camouflages the chemotherapeutic agents, enabling them to bypass the drug resistance of colon cancer.
In summary, our introduction of LbL-GDLVs made of an edible ginger-derived lipid core surrounded by functionalized layers could shift the current paradigm of drug delivery from artificially synthesized nanoparticles toward the use of natural nanovectors derived from edible plants. Because such nanovectors are non-toxic and can be produced on a large scale, nanovectors derived from edible plants could represent a very safe therapeutic delivery platform. We herein demonstrate that LbL-GDLVs loaded with Dox successfully inhibited tumor growth in two xenograft tumor models of colon cancer. Our novel findings expand current perspectives on drug delivery systems and could lay the foundation for a new targeted drug delivery approach for the treatment of colon cancer.
4. Experimental Section
Materials:
Doxorubicin (D-4000) was purchased from LC Laboratories (Woburn, MA, USA). ε-poly-lysine (ε-PLL) was obtained from Zhengzhou Bainafo Bioengineering Co., Ltd (Zhengzhou, Henan, China). Fucoidan from Macrocystis pyrifera, gentian violet, phalloidin-FITC, dextran sulfate sodium salt and in situ cell death detection kit were purchased from Sigma (St. Louis, MO, USA). The XenoLight D-Luciferin-K+ salt bioluminescent substrate was purchased from PerkinElmer (Shelton, CT, USA). Carboxyl Modified Latex (CML) Beads (4% w/v, 0.1 μm), FluoSpheres™ Carboxylate-Modified Latex (CML) Latex Beads (2% w/v, 0.1 μm, 580/605), and P-selectin rabbit monoclonal antibody (3H20L10) were obtained from Thermo Scientific (Eugene, OR, USA). Recombinant human P-selectin/CD62P protein, recombinant human E-Selectin/CD62E protein, recombinant human L-Selectin/CD62L protein, and recombinant human TNF-alpha protein were purchased from R&D Systems (Minneapolis, MN, USA). Hydrogen peroxide solution (3%) and paraformaldehyde (4%) was purchased from Sigma (St. Louis, MO, USA). Dil and DiR dyes were obtained from Thermo Fisher Scientifics (Eugene, OR, USA)
Staining of tissue microarrays:
Colon normal and tumor tissue microarrays (TMAs) (BC05002b) were obtained from US Biomax, Inc. (Derwood, MD, USA). The TMAs were deparaffinized and rehydrated with xylene and a graded series of ethanol. Antigen retrieval was performed with sodium citrate buffer (10 mM Sodium citrate, 0.05% Tween 20, pH 6.0), and sections were blocked with 10% normal goat serum in PBS. Slides were incubated with antibody against P-selectin (Thermo, 1:200 dilution) for 4 h, followed by biotinylated secondary antibody for 1 h.
Ginger derived nanoparticles (GDNPs) isolation and purification:
For isolation of GDNPs, ginger was first ground in a blender to obtain juice. Then the juice was centrifuged at 3000×g for 20 min, 10 000×g for 40 min to remove the large fiber of ginger with Type 45Ti rotor. The supernatant was then ultra-centrifuged at 150 000×g for 2 h; the pellet was suspended in PBS under sonication. Followed, the suspension was transferred to a sucrose gradient (8 %, 30 %, 45 % and 60 % (g/v)) and ultra-centrifuged at 150 000×g for another 2 h. The band between 30/45% was harvested and noted as ginger derived nanoparticles (GDNPs) according to our previous study [3, 12]. To test the stability of GDNPs in serum, GDNPs nanoparticle were added in mice serum which isolated from whole blood. Size and zeta potential of GNDPs at different times (0, 3, 6, 12, 24, 48 h) were tested by dynamic light scattering (DLS) (Figure S20).
Lipid analysis of GDNPs:
Lipids from GDNPs were submitted to the Lipidomic Research Center, Kansas State University (Manhattan, KS, USA). Briefly, the lipid composition was determined using a triple quadrupole mass spectrometer (Applied Biosystems Q-TRAP; Applied Biosystems, Foster City, CA, USA) as described in the online protocol (http://www.k-state.edu/lipid/lipidomics/profiling.html). Data for each lipid molecular species were presented as mol % of the total lipids.
Preparation of ginger derived lipid vehicles (GDLVs):
Firstly, total lipids were extracted using the Bligh and Dyer method. Briefly, 6 ml of methyl alcohol/chloroform with a volume ratio of 2:1 (v/v) was added to 1.6 ml of GDNPs in PBS and mixed well. Chloroform (2 ml) and ddH2O (2 ml) were added sequentially and vortexed. The mixture was centrifuged at 2,000 r.p.m. for 10 min at room temperature in glass tubes to separate the mixture into two phases (aqueous phase and organic phase). For collection of the organic phase, a glass pipette was inserted through the aqueous phase with gently positive pressure and the bottom phase (organic phase) was aspirated and dispensed into new glass tubes. Then the samples were washed once with a small volume KCl (1 M, 0.5 ml) and once with a small amount of water (0.5 ml). Finally, the organic phase samples were dried by heating (60 °C) under nitrogen. For GDLVs fabrication, residual chloroform was removed using a vacuum pump for 15 min, and the dried lipids were immediately suspended in 500 μL HEPES buffer (20 mM, sigma, pH 7.4). After a bath sonication for 5 min, an equal volume of HEPES buffer was added and sonicated for another 5 min. Finally, the solution was passed through AVESTIN liposomes extruder 20 times (Ottawa, ON, Canada) with 200 nm polycarbonate membrane. For GDLVs/Dox fabrication, Doxorubicin (Dox) in aqueous solution was added in lipids film, then an identical protocol as described above was used to make GDLVs/Dox.
Phosphorus quantification:
Phosphate in ginger derived lipid vehicles (GDLVs) was quantified by using a standard phosphorus solution (0.65 mM, Sigma). First, different amounts of phosphate standard solutions (0, 6.5, 13, 26, 52, 104 nmol) and GDLVs samples were added to glass test tubes, then 30 μL 10% Mg(NO3)2·6H2O (prepared in 95% alcohol) was added, and the mixture was heated by shaking the tubes over the strong flame of burden burner until the brown fumes disappeared. After cooling, 300 μL HCl (prepared in 0.5 N) was added to each tube and allowed to heat in a boiling water bath for 15 min to hydrolyze any pyrophosphate to phosphate. After the tube has cooled, 0.1 mL 10 % ascorbic acid (prepared in ddH2O) and 0.6 mL 0.42 % ammonium molybdate.4H2O (prepared in 1 N H2SO4) were added in sequence. Finally, the mix solutions were incubated at 45 °C for 20 min and read the absorbance at OD820.
Preparation of layer-by-layer (LbL) nanoparticles:
Briefly, the negatively charged cores used were either carboxyl modified latex beads (CML) (with or without dye) or GDLVs. For LbL assembly, CML nanoparticles (2 mg/mL) or GDLVs (200 nmol/mL) were first mixed with ε-poly-lysine (ε-PLL) (1 mg) in H2O (adjust pH=4), the mixture solution was stirred for 30 min, followed by a washing step through centrifugation (30,000 ×g for 1 h). The purified ε-PLL-modified nanoparticles were further mixed with fucoidan (1 mg) or dextran sulfate sodium salt (1mg) in an aqueous solution (adjust pH=11), followed by a washing step through centrifugation (30,000 ×g for 1 h). Finally, the LbL nanoparticles were dispersed in PBS buffer for further use.
Characterization of nanoparticles:
Size and zeta potential of nanoparticles were measured by dynamic light scattering using a Zetasizer Nano ZS 90 (Malvern, Southborough, MA). For TEM imaging, a drop of samples was deposited onto the surface of formvar-coated copper grids, then add 1% uranyl acetate for 15s, the samples were left to dry at room temperature.
High-Performance Liquid Chromatography (HPLC) analysis:
Samples were analyzed using high-performance liquid chromatography (HPLC) method. The auto-sampler temperature was set at 20°C, and an injection volume of 5 μL was used for analysis. Separation was achieved using Agilent 1200 series LC (Agilent Technologies, Wilmington, DE) with an Agilent Zorbax reverse-phase column (SB-C18, 3.0×250 mm, 5.0 μm). LC system was equipped with a UV detector; the detection wavelength was set at 480 nm. An isobaric method was employed to separate the doxorubicin using 50 % of mobile phase A (0.1% formic acid in water) and 50% of mobile phase B (Acetonitrile). The total acquisition time was 5 min for each injection at a flow rate of 0.4 mL/min. The retention time of doxorubicin was 2.5 min.
Binding studies of nanoparticles to immobilized P-selectin:
Human recombinant P-selectin, L-selectin, E-selectin and bovine serum albumin (BSA) with different concentrations (10, 20 and 50 μg/mL) were added to 96-well enzyme-linked immunosorbent assay (ELISA) plates and incubated at 4 °C overnight. The wells were washed with PBS and incubated with dye-labeled LbL-CML in PBS solution for 4 h. The plate was gently washed with PBS 3 times. Fluorescence of the plate was captured by IVIS series pre-clinical in vivo imaging systems (PerkinElmer, Massachusetts).
Cell culture:
Luciferase-expressing HT-29 colon carcinoma cells (Luc-HT-29) were purchased from PerkinElmer (Shelton, CT, USA), Caco-2bbe enterocyte cells, HCT-116 colon carcinoma cells and EA.hy926 vascular endothelium cells were all purchased from ATCC. Luc-HT-29 and HCT-116 cells were cultured in McCoy’s Modified Medium (Life Technologies, NY, USA), supplemented with penicillin (100 U/ml), streptomycin (100 U/ml), and fetal bovine serum (10 %) (Atlanta Biologicals, GA, USA), puromycin at a final concentration of 2 μg/ml was added for Luc-HT-29 cells. Caco-2bbe enterocyte cells and EA.hy926 cells were cultured in Dulbecco’s Modified Eagle’s Medium supplemented with penicillin (100 U/ml), streptomycin (100 U/ml), and fetal bovine serum (10 %). All the cell lines were tested and authenticated via morphology and PCR to rule out interspecies and intraspecies contamination.
Binding of nanoparticles to a monolayer of endothelial cells (ECs):
EA.hy926 vascular endothelium cells were seeded on eight-chamber slides (Tissue-Tek, Sakura, USA) and cultured overnight at 37 °C. TNF-alpha (100 ng/mL) was pre-incubated with cells for 1 h and then with dye-labeled LbL-CML nanoparticles for 2 h. Subsequently, the cells were fixed with 4 % paraformaldehyde (PFA) for 10 min and extracted with acetone at −20°C for 5 min. After blocking with 1% BSA in PBS for 30 min, 100 μl of phalloidin-FITC (1:40 dilution) was added, and the mixture was incubated for an additional 30 min. Finally, cells were coverslip-mounted with mounting medium containing 4,6-diamidino-2-phenylindole (DAPI, H-1500; Vector Laboratories, Burlingame, CA, USA). Images were captured using an Olympus fluorescence microscopy equipped with a Hamamatsu Digital Camera ORCA-03G.
Evaluating the penetration of nanoparticles through endothelial cells (ECs):
We used a modified Transwell assay (Corning) to test the penetration of nanoparticles through a monolayer of ECs. EA.hy926 vascular endothelium cells with a concentration of 1×104 cells were seeded on transwell inserts in 12-well plates for 3 days. The medium was changed every other day. Experiments were conducted in the absence of gaps in cell junctions; the confluence of the monolayer was confirmed under a microscope. After activation with TNF-alpha (100 ng/mL) for 1 h, the cells were then incubated with nanoparticles overnight. Samples of the medium from the bottom chamber (50 μL) were collected, and fluorescence intensity was measured with a microplate reader.
Cell attachment assays:
Cell attachment assays were performed to investigate the real-time cytotoxicity of nanoparticles using electrical impedance sensing (ECIS) technology (Applied BioPhysics, Troy, NY, USA), which is based on AC impedance measurements using weak and noninvasive AC signals. The attachment and spread of cells on the electrode surface change the impedance in such a way that morphological information about attached cells can be inferred. The measurement system consists of an 8-well culture dish (ECIS 8W1E plate) with the surface treated for cell culture. The bottom of each well contains a small, active electrode and a large counter electrode. A lock-in amplifier with an internal oscillator is used to switch among the different wells, and a personal computer controls the measurement and stores the data. For the experiment, Caco2-bbe cells at a density of 2×105/well were seeded in the plate. Once cells reached confluence, LbL-GDLVs nanoparticles with different concentrations (5, 10, 20, 50, 100, and 200 μM) were added to the wells. Basal resistance measurements were performed using the ideal frequency for Caco2-bbe cells, 500Hz, and a voltage of 1V.
Colony formation assay:
After treated with LbL-GDLVs (200 μM) for 24 h, Luc-HT-29 and HCT-116 cells were harvested, 500 cells of each cell line were seeded in 6-well plates and cultured for 2 weeks. Medium was changed every other day. At the end of the experiment, cells were rinsed carefully with PBS and added 2–3 mL of a mixture of 6% glutaraldehyde and 0.5% crystal violet for 1 h. Finally, colonies were counted using a marker pen.
Luciferase activity evaluation:
Luc-HT-29 cells were seeded in 96-well plates overnight. Then different concentrations (5, 10, 20, 50, 100, and 200 μM) of LbL-GDLVs nanoparticles were added in the well and incubated for 24 h. Cells were washed with PBS 3 times, and the bioluminescent substrate was then added. Bioluminescence of Luc-HT-29 cells was captured by IVIS series pre-clinical in vivo imaging systems (PerkinElmer, Massachusetts).
Stability of LbL nanoparticles in cancer microenvironment alike solutions:
Fucoidan coated LbL-GDLVs or ε-PLL coated GDLVs was incubated in pH 6.3 buffer solution with different concentrations of H2O2 (50 and 100 μM) at 37 °C. At different time points (0.5, 1, 2, 4, 6, 12 hrs), samples were collected and quenched by icy cold PBS (1:1) and submitted to Zetasizer Nano ZS 90 (Malvern, Southborough, MA) to characterize the surface zeta-potential.
qRT-PCR Analysis for drug transporter (MDR-1):
After cultured with LbL-GDLVs/Dox or free-Dox for 10 passages, HT-29 cells were lysed, total RNA was extracted using the RNeasy Plus Mini Kit (Qiagen, Valencia, CA, USA). The complementary DNA (cDNA) was generated from the total RNAs isolated above using the Maxima First Strand cDNA Synthesis Kit (Thermo Fisher, Waltham, MA, USA) according to the manufacturer’s instructions. Levels of MDR1 RNA expression were quantified by reverse-transcription polymerase chain reaction using Maxima SYBR Green/ROX qPCR Master Mix (Thermo Fisher, Waltham, MA, USA). The data were normalized to the internal control: 36B4. Relative gene expression levels were calculated using the 2−ΔΔCt method. Sequences of the primers are MDR1: 5’- AAAGCTGGAGCAGTTGCTGA-3’ (Forward) and 5’-TGCCTATCGAAATGCTGGCT-3’(Reverse), 36B4: 5’-TCCAGGCTTTGGGCATCA-3’ (Forward) and 5’-CTTTATCAGCTGCACATCACTCAGA-3’ (Reverse).
Cell uptake assay of ε-PLL coated GDLVs nanoparticles:
Dil-stained ε-PLL-GDLVs was used to analyze the cellular uptake by fluorescence microscopy imaging. HT-29 cells with a concentration of 1 × 105/well were seeded in eight-chamber tissue culture glass slides [TissueTek; Sakura, USA] for overnight. Dil-stained ε-PLL-GDLVs were incubated with cells for 0, 15 min, or 1 h. Then the cells were fixed with 4 % paraformaldehyde [PFA] for 15 min. After blocking with 1% bovine serum albumen [BSA] in PBS for 30 min, 100 μL of phalloidin-FITC [1:40 dilution in blocking buffer] was added and the mixture was incubated for an additional 30 min. Finally, cells were coverslip-mounted with mounting medium containing 4-,6-diamidino-2-phenylindole (DAPI). Images were observed using an Olympus fluorescence microscopy equipped with a Hamamatsu Digital Camera ORCA-03G.
Animal Studies:
Athymic BALB/c nu/nu mice and FVB/NJ mice (4–6 wk old) were purchased from Jackson Laboratories (Bar Harbor, ME, USA). Mice were housed under specific pathogen-free conditions. All the experiments involving mice were approved by the Institutional Animal Care and Use Committee (IACUC) of Georgia State University (Atlanta, GA, USA).
Circulation of LbL-GDLVs in vivo:
To evaluate the stability of circulating LbL-GDLVs in vivo, we injected 200 nmol of DiR dye-labeled LbL-GDLVs into mice via tail vein. Blood was drawn into BD Microtainer MAP tube with K2EDTA (1 mg) at various time points (1, 3, 6, 24, and 48 h) post-injection. The intensity of DiR signals from same volume blood samples was measured using IVIS series pre-clinical in vivo imaging system. To determine whether the circulating LbL-GDLVs in mice are free or not with blood cells, DiR dye-labeled LbL-GDLVs were injected into FVB mice, whole blood from mice was collected at various time points (1, 3, 6, 24, and 48 h) post-injection. Blood cells and plasma were separated by centrifuging at 8000×g for 8 min. Then the intensity of DiR signals from an equal volume of blood cells and plasma were measured using IVIS series pre-clinical in vivo imaging system.
Hemolysis assay:
Fresh mouse blood was collected in BD Microtainer MAP tube with K2EDTA (1 mg). Then red blood cells (RBC) were separated, purified and diluted to 1×108 for the hemolysis test according to our previous study. Different concentrations of LbL-GDLVs and were incubated with RBC at 37 °C for 2 h. The absorbance of the supernatants from each group was measured using a microplate reader at 540 nm. The group which treated by Triton-100 (1 %) was used as a positive control, and the release rate of hemoglobin for this group was set as 100 %. The percentage of hemolysis was calculated as follows:
Blood hemanalysis and biochemical analysis:
Blood samples were collected from PBS-, GDLVs- and LbL-GDLVs-treated groups. Blood cells and biochemical studies were performed using a hematology analyzer (VetScan HM5; Abaxis, CA) and a biochemical analyzer (VetScan VS2; Abaxis, CA), respectively.
In vivo bio-distribution of LbL-GDLVs:
For the biodistribution analysis of LbL-GDLVs in tumor-bearing mice, Luc-HT-29 xenograft tumor-bearing mice were i.v. administrated with DiR dye-labeled LbL-GDLVs (200 nmol) and DiR dye-GDLVs (Control). Whole body imaging was done at 1, 3, 6, and 24 h post-injection. DiR dye signal in major organs (heart, liver, spleen, lung, and kidney) and tumor tissue were also quantified at the end of the experiment using IVIS series pre-clinical in vivo imaging system (PerkinElmer, Massachusetts).
In vivo anti-tumor effects:
Xenograft tumor growth model was used to determine LbL-GDLVs mediated targeted delivery of therapeutic drug-doxorubicin (Dox) to tumors. To grow subcutaneous tumor xenografts, we implanted female athymic BLAB/c nu/nu mice with Luc-HT-29 cells (2×106) or HCT-116 (1×106) subcutaneously in the right flank. When tumors reached approximately 100 mm3 in volume, the mice were randomly assigned to four different treatment groups: 1) Saline; 2) LbL-GDLVs; 3) Free Dox (5mg/kg) and 4) LbL-GDLVs/Dox (equal to 5mg/kg free Dox). Mice were treated every 3 days for 5 times. Tumor growth in Luc-HT-29 cells was monitored for luciferase expression using PerkinElmer IVIS in vivo imaging system every 3 days for five consecutive times from the 9th day after the injection of tumor cells. The tumor volumes and body weights of the mice were measured and recorded at the end of the experiment. Tumor volume was calculated as follows: volume =1/2LW2, where L is the long diameter, and W is the short diameter of a tumor. Animals were euthanized when the signals of sickness, such as breathing problems, failure to eat and drink, lethargy or abnormal posture, were observed.
H&E staining and TUNEL examination:
To conduct histological analysis, we fixed tumors and the major organs (heart, liver, spleen, lung, and kidney) for 2 days in 10 % buffered formalin solution and embedded in paraffin. After deparaffinization, the tissue sections (5 μm) were stained with hematoxylin/eosin (H&E). Images were acquired using an Olympus equipped with a Hamamatsu Digital Camera DP-26. The apoptosis of tumor cells was determined using 4,6-diamidino-2–2-phenylindole (DAPI) and terminal deoxynucleotidyl transferase-mediated nick end labeling (TUNEL) assays, with a commercial apoptosis detection kit according to the manufacturer’s standard protocols. Images were acquired using an Olympus equipped with a Hamamatsu Digital Camera ORCA-03G.
Immunohistochemistry (IHC):
For the proliferation marker Ki67 staining, 6-μm-thick paraffin-embedded tissue sections were deparaffinized in xylene, incubated in 3% hydrogen peroxide in PBS for 30 min, then treated with 10 mM sodium citrate buffer (pH 6.0) containing 0.05% Tween 20 and heated in a pressure cooker for 10 min (antigen retrieval). Sections were blocked with goat serum for 45 min at 37 °C, followed by incubation with the anti-Ki67 antibody (1:50; Vector Laboratories) for 1 hour at 37 °C. After washing with PBS containing 0.01% Tween 20, sections were incubated first with the appropriate biotinylated secondary antibody for 30 min at 37 °C, and then with reagents from the Vectastain ABC kit (Vector Laboratories) to allow color development. Sections were counterstained with hematoxylin, dehydrated, and coverslip-mounted. Images were acquired using an Olympus microscope equipped with a DP-26 digital camera.
Statistical analysis:
One- and two-way analyses of variance (ANOVA) and t-tests were used to determine statistical significance (*p<0.05, **p<0.01, ***p<0.001), as indicated in the figure legends.
Supplementary Material
Acknowledgments
Mingzhen Zhang and Chunhua Yang contributed equally to this manuscript. This work was supported by the National Institutes of Health of Diabetes and Digestive and Kidney (R01DK116306, R01DK107739 to D.M.), the Department of Veterans Affairs (Merit Award BX002526 to D.M.), the “Young Talent Support Plan” of Xi’an Jiaotong University (YX6J001) and the Fundamental Research Funds for the Central Universities (xzy012019070). D.M. is a recipient of a Senior Research Career Scientist Award (BX004476) from the Department of Veterans Affairs. MZ was recipient of the Career Development Award from the Crohn’s and Colitis Foundation (2016–2018).
Footnotes
Conflict of interest
The authors declare no competing financial interest.
References
- [1].a) Fortina P, Kricka LJ, Graves DJ, Park J, Hyslop T, Tam F, Halas N, Surrey S, Waldman SA, Trends in biotechnology 2007, 25, 145; [DOI] [PubMed] [Google Scholar]; b) Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RG, Barzi A, Jemal A, CA: a cancer journal for clinicians 2017, 67, 177. [DOI] [PubMed] [Google Scholar]
- [2].a) Gustavsson B, Carlsson G, Machover D, Petrelli N, Roth A, Schmoll HJ, Tveit KM, Gibson F, Clin Colorectal Cancer 2015, 14, 1; [DOI] [PubMed] [Google Scholar]; b) Murcia O, Jover R, Egoavil C, Perez-Carbonell L, Juarez M, Hernandez-Illan E, Rojas E, Alenda C, Balaguer F, Andreu M, Llor X, Castells A, Boland CR, Goel A, Clin Cancer Res 2018, 24, 2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Zhang M, Xiao B, Wang H, Han MK, Zhang Z, Viennois E, Xu C, Merlin D, Mol Ther 2016, 24, 1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].a) Masood F, Mater Sci Eng C Mater Biol Appl 2016, 60, 569; [DOI] [PubMed] [Google Scholar]; b) Wang Q, Zhuang X, Mu J, Deng ZB, Jiang H, Zhang L, Xiang X, Wang B, Yan J, Miller D, Zhang HG, Nat Commun 2013, 4, 1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ulbrich K, Hola K, Subr V, Bakandritsos A, Tucek J, Zboril R, Chem Rev 2016, 116, 5338. [DOI] [PubMed] [Google Scholar]
- [6].a) Torchilin VP, Nat Rev Drug Discov 2014, 13, 813; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sercombe L, Veerati T, Moheimani F, Wu SY, Sood AK, Hua S, Front Pharmacol 2015, 6, 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wang H, Huang Q, Chang H, Xiao J, Cheng Y, Biomater Sci 2016, 4, 375. [DOI] [PubMed] [Google Scholar]
- [8].Ma P. a., Xiao H, Li C, Dai Y, Cheng Z, Hou Z, Lin J, Materials Today 2015, 18, 554. [Google Scholar]
- [9].Molino NM, Wang SW, Curr Opin Biotechnol 2014, 28, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sun T, Zhang YS, Pang B, Hyun DC, Yang M, Xia Y, Angew Chem Int Ed Engl 2014, 53, 12320. [DOI] [PubMed] [Google Scholar]
- [11].a) Yang C, Zhang M, Merlin D, J Mater Chem B 2018, 6, 1312; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Xiaoying Zhuang YT, Samykutty Abhilash, Mu Jingyao, Lifeng Zhongbin Deng, Pengxiao Cao zhang, Rong Yuan, Yan Jun, Miller Donald, Zhang Huang-Ge, Mol Ther 2016, 24, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhang M, Viennois E, Prasad M, Zhang Y, Wang L, Zhang Z, Han MK, Xiao B, Xu C, Srinivasan S, Merlin D, Biomaterials 2016, 101, 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].a) Juenet M, Aid-Launais R, Li B, Berger A, Aerts J, Ollivier V, Nicoletti A, Letourneur D, Chauvierre C, Biomaterials 2018, 156, 204; [DOI] [PubMed] [Google Scholar]; b) Ferber S, Tiram G, Sousa-Herves A, Eldar-Boock A, Krivitsky A, Scomparin A, Yeini E, Ofek P, Ben-Shushan D, Vossen LI, Licha K, Grossman R, Ram Z, Henkin J, Ruppin E, Auslander N, Haag R, Calderon M, Satchi-Fainaro R, Elife 2017, 6; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Yosi Shamay ME, Li Hongyan, Shah Janki, Brook Samuel, Wang Feng, Adler Keren Baut Emily, Scaltriti Maurizio, Jena PrakritV, Gardner Eric E., Poirier John T., Rudin Charles M., Baselga José, Haimovitz-Friedman Adriana, Heller Daniel A., Science Translational Medicine 2016, 8, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Simion V, Constantinescu CA, Stan D, Deleanu M, Tucureanu MM, Butoi E, Manduteanu I, Simionescu M, Calin M, Mediators Inflamm 2016, 2016, 1625149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lisa ZW. Coussens M, Nature 2002, 420, 8.12422178 [Google Scholar]
- [16].a) Ted CFN Szatrowski P, Cancer Research 1991, 51, 5;1988106 [Google Scholar]; b) Hou Y, Wang J, Jin W, Zhang H, Zhang Q, Carbohydrate Polymers 2012, 87, 153. [DOI] [PubMed] [Google Scholar]
- [17].a) Ren K, Ji J, Shen J, Biomaterials 2006, 27, 1152; [DOI] [PubMed] [Google Scholar]; b) Li Z, Shuai C, Li X, Li X, Xiang J, Li G, J Biomed Mater Res A 2013, 101, 2846. [DOI] [PubMed] [Google Scholar]
- [18].Vishwanath Ramachandran MUN, Qiu Haiying, Liu Wen-Jun, Cummings Richard D., Zhu Cheng, and McEver Rodger P., PNAS 1999, 96, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hyldgaard M, Mygind T, Vad BS, Stenvang M, Otzen DE, Meyer RL, Appl Environ Microbiol 2014, 80, 7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].a) McClements DJ, Prog Lipid Res 2013, 52, 409; [DOI] [PubMed] [Google Scholar]; b) Yun JW, Kim SH, You JR, Kim WH, Jang JJ, Min SK, Kim HC, Chung DH, Jeong J, Kang BC, Che JH, J Appl Toxicol 2015, 35, 681. [DOI] [PubMed] [Google Scholar]
- [21].a) Tatsiana SSB Shutava G, Vangala Pranitha, Steffan Joshua J., Bigelow Rebecca L., Cardelli James A., Patrick O’Neal D, Lvov Yuri M., ACS Nano 2009, 3, 8; [DOI] [PubMed] [Google Scholar]; b) Zhou SWM Deng J, Ben-Akiva Elana, Dreaden Erik C., Shopsowitz Kevin E., Hammond Paula T., ACS NAno 2013, 7, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Fidler IJ, Poste G, The Lancet Oncology 2008, 9. [DOI] [PubMed] [Google Scholar]
- [23].Lisanti MP, Martinez-Outschoorn UE, Lin Z, Pavlides S, Whitaker-Menezes D, Pestell RG, Howell A, Sotgia F, Cell Cycle 2011, 10, 2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ai S, Jia T, Ai W, Duan J, Liu Y, Chen J, Liu X, Yang F, Tian Y, Huang Z, Br J Pharmacol 2013, 168, 1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hu T, Li Z, Gao CY, Cho CH, World J Gastroenterol 2016, 22, 6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Chiara Riganti EM, Viarisio Daniele, Costamagna Costanzo, Pescarmona Gianpiero, Ghigo Dario, Bosia Amalia, Cancer Research 2005, 65, 10. [PubMed] [Google Scholar]
- [27].Dore JSM, Vet. Pathol 1996, 33, 10. [DOI] [PubMed] [Google Scholar]
- [28].a) Josef Friedl MP, Bartlett David L., Libutti Steven K., Turner Ewa N., Gnant Michael F. X., and Richard Alexander H, Blood 2002, 100, 6; [PubMed] [Google Scholar]; b) Tay CY, Setyawati MI, Leong DT, ACS Nano 2017, 11, 2764. [DOI] [PubMed] [Google Scholar]
- [29].a) Zhang H, Park Y, Wu J, Chen X, Lee S, Yang J, Dellsperger KC, Zhang C, Clin Sci (Lond) 2009, 116, 219; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhang C, Basic Res Cardiol 2008, 103, 398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Franken NA, Rodermond HM, Stap J, Haveman J, Van Bree C, Nature protocols 2006, 1, 2315. [DOI] [PubMed] [Google Scholar]
- [31].a) Li F, Zhao X, Wang H, Zhao R, Ji T, Ren H, Anderson GJ, Nie G, Hao J, Advanced Functional Materials 2015, 25, 788; [Google Scholar]; b) Jiang T, Mo R, Bellotti A, Zhou J, Gu Z, Advanced Functional Materials 2014, 24, 2295. [Google Scholar]
- [32].a) Aaron VG Anselmo C, Zern Blaine J., Pan Daniel, Zakrewsky Michael, Muzykantov Vladimir, Mitragotri Samir, ACS Nano 2013, 7, 9; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Blanco E, Shen H, Ferrari M, Nat Biotechnol 2015, 33, 941; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Tsoi KM, MacParland SA, Ma XZ, Spetzler VN, Echeverri J, Ouyang B, Fadel SM, Sykes EA, Goldaracena N, Kaths JM, Conneely JB, Alman BA, Selzner M, Ostrowski MA, Adeyi OA, Zilman A, McGilvray ID, Chan WC, Nat Mater 2016, 15, 1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].a) Shi J, Kantoff PW, Wooster R, Farokhzad OC, Nat Rev Cancer 2017, 17, 20; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Schroeder A, Heller DA, Winslow MM, Dahlman JE, Pratt GW, Langer R, Jacks T, Anderson DG, Nat Rev Cancer 2011, 12, 39. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.