Abstract
Microphysiological systems, also known as organ-on-a-chip platforms, show promise for the development of new testing methods that can be more accurate than both conventional two-dimensional cultures and costly animal studies. The development of more intricate microphysiological systems can help to better mimic the human physiology and highlight the systemic effects of different drugs and materials. Nanomaterials are among a technologically important class of materials used for diagnostic, therapeutic, and monitoring purposes; all of which and can be tested using new organ-on-a-chip systems. In addition, the toxicity of nanomaterials which have entered the body from ambient air or diet can have deleterious effects on various body systems. This in turn can be studied in newly developed microphysiological systems. While organ-on-a-chip models can be useful, they cannot pick up secondary and systemic toxicity. Thus, the utilization of multi-organ-on-a-chip systems for advancing nanotechnology will largely be reflected in the future of drug development, toxicology studies and precision medicine. Various aspects of related studies, current challenges, and future perspectives are discussed in this paper.
Keywords: organ-on-a-chips, multi-organ-on-a-chips, nanomaterials, toxicity, personalized medicine
1. INTRODUCTION
Nanomaterials (NMs) such as polymeric nanoparticles (NPs), vesicles, and dendrimers have revolutionized the biomedical field by enabling superior drug delivery, imaging, and sensing modalities [1]‥ The field of NMs is expected to grow tremendously as the nanotechnology market is projected to be worth $125 billion by 2024 [2]. In addition, the increasing number of NMs used for diagnostic and therapeutic [3] purposes impose new restrictions on their intended effects and unwanted complications. We are continuously exposed to environmental pollutants through inhalation [4], ingestion [5], and dermal contact [6]. Many of these materials are ultrafine in size (>100 nm in size), which enables their facile entry and interaction between different body compartments [7] (Figure 1). They may continue to have irreversible effects on deoxyribonucleic acid (DNA), cells, tissues, organs, or systems, which are not so well understood. Due to the increased risk of exposure, the field of nanotoxicology aims to accurately characterize the mechanisms of toxicity in nanoparticles induced in different organ systems. The most common targets include major organs associated with routes of exposure such as the skin, lungs, and gut, as well as downstream systems such as the brain and circulatory system. The small nature of NMs enables them to penetrate the skin and vessels to reach circulation [8]. As the field of nanotoxicology was emerging, Oberdörster et al. stated that an interdisciplinary approach involving the fields of “toxicology, materials science, medicine, molecular biology, and bioinformatics” would be necessary to fully capture the toxicity of NMs for appropriate risk assessment [8]. The major challenge facing nanotoxicologists is the development of a model system that can accurately mimic the physiology of human.
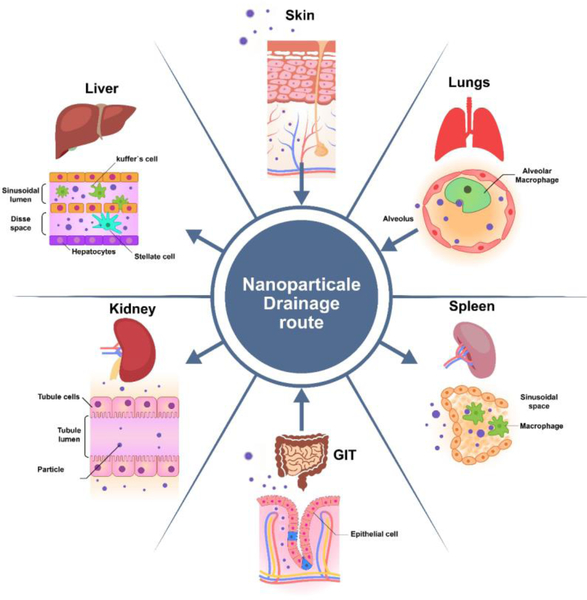
Figure 1:
Nanomaterial points of entry and drainage into the body. Nanomaterials can enter the body through the lungs, skin, and GI tract. Once inside, the circulatory system readily transports them throughout the body. Their small size enables facile translocation into a variety of tissue structures found in the kidney, spleen, liver, and lungs.
In vitro assessment is the primary means of evaluating the safety and behavior of NMs [9]. Generally, new drugs and chemicals administered to the body undergo testing with two-dimensional (2D) cell culture. Employing 2D cultures cannot reproduce events observed in the three-dimensional (3D) environment of native tissues in the body and thus, cannot be a reliable model of living tissues [10]. Animal studies offer more physiologically relevant environments to investigate various NMs; however, they suffer from limited predictability [11]. Since, in many instances, compounds that are safe in one species may cause different outcomes in another [10a, 12]. There were cases where therapeutics led to serious complications even after going through cell culture and animal testing, only to be discovered as toxic when they were in clinical trials [13]. The other disadvantage is longer period time animal experiments may take and the lack of high-throughput screening. In addition, new regulations in the US Toxicology in the 21st Century (Tox21) program asks to avoid using animal tests [14]. New EU regulations mandate toxicity testing for tens of thousands of chemicals [15], which would amount to billions of dollars in animal testing [16]. These changes will consequently be reflected as more expensive products, thus making medicines less available, particularly for the economically disadvantaged. The search for alternative biomimetic toxicity models has been suggested by researchers [17], encouraged by funding agencies, and required by industry [17].
With recent advances in microfluidics [18] and their integration with organ-on-a-chip (OoC) systems [19], it is becoming possible to more closely mimic key organ functions in vitro in 3D environments. The use of OoC platforms enables more control over the local cellular microenvironments including the ability to provide chemical [18], mechanical [20], and electrical cues similar to the native environments [21]. It has already been demonstrated that cells in OoCs perform differently than those in a 2D culture or in a static environment without dynamic flow [22]. Cell polarization, cytoskeletal arrangement and function in OoCs were also found to be more comparable to those seen in vivo [23]. Hence, it was suggested that OoC systems can be viable alternatives to 2D testing methods [24].
The platform of microphysiological systems represents an advanced in vitro cell culture model that utilizes physiologically accurate tissue and organ modeling for toxicology and pharmacology studies. For example, multi-cellular spheroids in 3D, with their increased physiological relevance, are emerging as a possible solution to test the efficacy and/or toxicity of compounds. Since OoC platforms have evolved from tissue engineering and microfluidics and associated microfabrication techniques, other in vitro 3D models including spheroid culture systems could be applied into the OoC platforms. With the developed tissue engineering models, the microengineering techniques allow the use of flow-associated shear stress or mechanical strain and integration of multiple sensing units for monitoring of pH, O2, temperature, or molecules secreted by cells and automatic systems for controlling dosing and dilution of multiple drugs in circulating media. These integrated systems possess several advantages over conventional cell culture systems; (1) an OoC platform can be manufactured for a low cost and allow to examine a wide range of concentration in the dose of medicine, which accelerate scientific research, (2) compared to cell culture in petri dishes or well plates, 3D dynamic culture of human cells including spheroid models using flow-associated shear stress or mechanical strain inside a chip leads to a better microenvironment to recapitulate important features in vivo, (3) since the system is based on miniaturized organoids and microfluidic-based technology, it lowers fluid volume consumption in terms of reagent costs and waste amounts, increases portability of the chips, and reduces fabrication costs. This system is a promising technology as the next generation toxicity testing model can produce critical functions of different human organs systems. Furthermore, the use of OoC systems allows us to make new observations which were otherwise impossible using 2D models, animal studies, or even clinical trials. An example includes being able to closely monitor cancer invasion and the influence of local mechanical factors [25].
Moreover, the use of cells that are directly derived from patients in OoC systems [26] enables the development of tools for personalized medicine, whereby medicinal dosages incorporating NMs can be tested and customized for individual patients [27]. Ideally, both the lack of toxicity and effectiveness of a drug could be ensured in preclinical studies [17]. This would avoid unnecessarily high doses and increased risk of complications [28], especially for applications pertaining to cancer [17, 29]. In cancer treatment, one major problem is the use of relatively toxic drugs in high doses which detrimentally influence the patients’ life quality. Another problem arises from the non-specificity of many drugs. Development of precision medicine tools will enable the use of effective drugs in appropriate dosage.
Recent studies using OoC system for nanotoxicology research have shown differential toxicities when using the chip technology versus mono or co-culture models. Huh et al. utilized an alveolar-capillary interface organ on chips with the ability to expand and contract, or breath, to model the cellular response to 12-nm silica nanoparticles [30]. They found that the mechanical strain enhanced nanoparticle uptake in the epithelial and endothelial cells and further into the microvascular space that is seen in in vivo models. The mechanical stress also heightened the toxic and inflammatory response as compared to monocultures [31]. A recent study used a liver-on-a-chip model to determine hepatotoxicity of NMs. Li et al. used superparamagnetic iron oxide nanoparticles (SPION) to demonstrate the metabolic capacity of the liver [32]. It was found that the OoC sustained higher liver function that monocultures over the course of a week of testing and produced the necessary cellular responses to SPION exposure that proved metabolism was occurring within the OoC [32]. This ability of microphysiological system to provide an in vitro platform with attributes of an in vivo system are highly valuable in the field of nanotoxicology to be able to generate more accurate conclusions about the potential toxicity of nanoparticles exposed to the humans to guide future human health risk assessments.
Despite the usefulness of using OoC models, the use of single organ model cannot pick up secondary toxicity that may result from drug metabolites or systemic toxicity that follow drug circulation to other organs. Thus, the utilization of multi-organ-on-a-chip (MoC) system is required to determine the effect and side effects of target drugs or chemical. In this system, more than one organ chip is linked together to form the MoC or body-on-a-chip. These systems allow for not only the study of systemic drug effects on different organs [33], but also of their metabolites and consequent secondary toxicity [34]. The use of MoCs can result in more representative models of normal clinical parameters or the whole body [20b, 26b]. In addition, these systems offer the possibility to study the cellular response from specific therapies in a high throughput fashion [35].
There have been limited reports on MoC systems [26b, 36] developed for studying drugs and chemicals, with even fewer on NMs [37]. By enabling the testing of NMs in biomimetic dynamic environments, MoC devices offer new opportunities to develop NMs for personalized medicine and nanotechnology. They are well poised to facilitate novel nanomedicine approaches and get translated to clinical applications more rapidly [38]. In this paper, the potential of OoC and MoC platforms for testing NMs are explained, and challenges are highlighted.
2. ORGAN-ON-A-CHIP MODELS
2.1. Organ-on-chip platform
OoC platforms are essentially 3D cell cultures with a dynamic microfluidic flow. The system allows one to compartmentalize cells [20b, 26a, 39] for the identification of biomarkers and for drug testing. Depending on the parameters studied, these platforms enable data collection on a variety of physical and biochemical processes. Thus, they are being increasingly used in physiological, pathophysiological, and drug development studies. Occasionally in this review, the term ‘organoid’ is used when referencing the inclusion of stem cells in on-chip devices. For a review article dedicated specifically to organoid-on-chip developments we direct the reader to the following article [40]. In addition to existing OoC devices [41], in the last few years new ones to replicate cervix [42], amnion [43], and placenta [44] functions have been developed. Furthermore, pathological models on-a-chip have been developed to focus on studies for both prevalent diseases such as cancer [45], inflammation [46], and thrombosis [47], as well as rare diseases such as Barth syndrome [48].
OoC devices can provide alternative nanomaterial testing platforms by evaluating multiple assays at the same time. Developed NMs can be tested on OoCs to perform dose assessment and when coupled with personalized therapies can create the most effective treatment for the individual patient. Induced pluripotent stem cells (iPSC) in OoC systems offers the possibility to develop personalized therapies by differentiating patient’s own donated cells to the cell type of interest [49]. So far, human iPSCs have been utilized in progeria-on-a-chip [50], liver-on-a-chip [51], glomerulus on a chip [26a], and cardiac tissue-on-a- chip [52] models with encouraging results. Heart-on-a-chip models in particular are vital for nanomaterial safety assessment, as all blood will be pumped through the heart before being transported to other organs. Lind et al. develop an instrumented cardiac-on-chip model by 3D printing six materials and subsequently seeding human iPSC derived cardiomyocytes [52b]. These devices were printed with embedded strain sensors capable of quantifying cellular contractile development as well as contractile stress during dose-response studies. The fabrication of such increasingly intricate devices with real-time data acquisition systems will lead to new insights in cardiac tissue morphogenesis, drug effects, and nanomaterial assessment. For an in-depth review of heart-on-a-chip models we direct the reader to the following review [53].
Despite their unprecedented potential to recapitulate in vivo responses, OoCs are limited to interactions within a single organ. Thus, OoCs need to be equipped with additional organ types to more accurately mimic human physiology and study secondary and systemic reactions to toxins and drugs [54].
2.2. Multi-organ-on-Chip
MoCs are the product of multiple OoC units linked together, enabling a physical interface and synergistic communication between multiple organ units [55]. Within MoCs, when distinct cell types are deposited in their corresponding microenvironments, they spontaneously express extracellular matrix (ECM) proteins, activate developmental plans, and organize to form differentiated tissue structures that closely resemble those of native organs. For drug testing, this means systemic toxicity resulting from the circulation of drugs or secondary toxicity resulting from its metabolites can be examined (Figure 2).
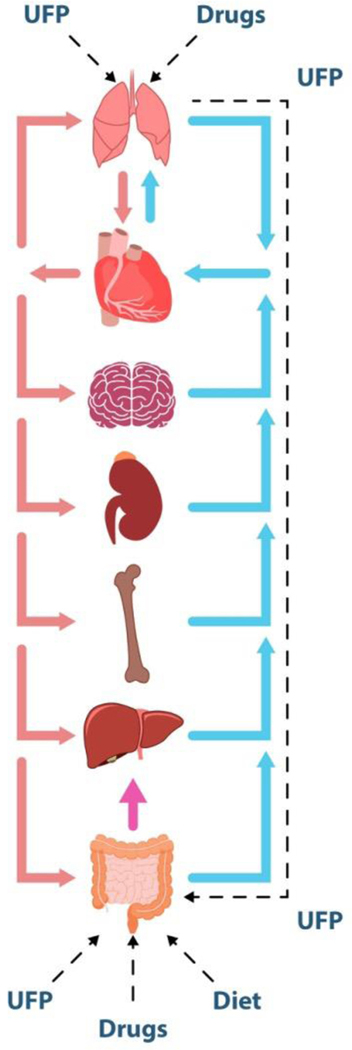
Figure 2.
Concept map for a human-on-a-chip device which can integrate multiple biomimetic microengineered organs interconnected with a physiologically relevant dynamic microfluidic circulatory system. The human-on-a-chip can be used to study the complex processes of drug distribution, absorption, excretion, and metabolism for a comprehensive evaluation of the drug efficiency, toxicity, and optimized dosage. As shown in this schematic, drug/ultrafine particle (UFP) absorption can be studied as they enter a microengineered lung model and pass into the circulation. From here, cardiotoxicity in a heart-mimetic microsystem, transport and clearance in the kidney-mimetic system, metabolic functions in the liver, and immune system of cells can all be studied. If the UFPs pass through relevant physiological barriers such as the BBB and intestinal metabolic pathways, organs such as the brain and gut can be included to study more systemic effects. Oral administration of drugs absorbed into the gut compartment can be modelled to study the interaction between drug and molecular transporters and metabolizing enzymes expressed in the various organs.
The ultimate success of OoC models will certainly depend on validating the extent of which these devices recapitulate in vivo responses. Many devices are traditionally validated in vitro with end-point molecular assays such as enzyme-linked immunosorbent assay (ELISA), RNA expression, and immunohistochemistry. More recently, non-invasive techniques such as electrochemical impedance spectroscopy (EIS) have been developed to monitor protein levels in real-time [56]. In the following sections, popular on-chip models for entry organs, metabolism and clearance, and toxicity-susceptible tissues and organs are discussed. The importance, fabrication, and function of these various platforms are explained. In particular, the applications of these organ chips for investigating and developing various NMs are discussed.
2.3. Device materials and fabrication
In an ongoing effort to engineer increasingly complex devices with multiple tissue types, a variety of materials and fabrication techniques have been utilized to develop the microfluidic devices essential to OoCs and MoCs. Polydimethylsiloxane (PDMS) has emerged as a predominant material for the fabrication of microfluidic devices owing to properties such as transparency, biocompatibility, flexibility, gas permeability, and ease of achieving micron scale resolution [18]. In a typical procedure, PDMS will first be mixed with its base polymer and curing agent with a weight ratio of 10:1 and degassed under vacuum to remove air bubbles. This mixture is then poured onto a master mold containing the design of interest and cured in an 80° convection oven for a few hours depending on the PDMS thickness. The cured PDMS can then be peeled from the master mold and the appropriate inlet/outlet ports are made by puncturing the PDMS with a biopsy punch. Finally, the PDMS layer can be bonded to other silicon substrates (e.g. glass, Si wafer, or another PDMS layer), typically by using oxygen plasma to expose hydroxyl groups on both surfaces. Despite its popularity, several technical limitations of PDMS such as drug absorption and maintaining sterility have encouraged the use of alternative device materials, such as polymethylmethacrylate (PMMA), polycarbonate (PC), and polystyrene (PS) [57].
In addition to material choice, several fabrications techniques commonly used to fabricate master molds include soft lithography [58], injection molding [59], hot embossing [59a], and 3D printing [60]. However, each fabrication method has specific limitations which must be considered according to the properties of the material. For example, the success of injection molding and hot embossing is highly dependent on material properties such as glass transition temperature, melting temperature, and thermal expansion coefficient. Microfluidic devices can be further customized by tuning their surface properties with chemical modifications post-fabrication.
Once fabricated, there are several cell sources which can be used to populate the MoC, such as cell lines, stem cells, or primary cells [17]. Although primary cells are often ideal in terms of capturing the phenotypes of interest, several barriers such as limited quantities and difficulties in expanding the cells prevent their widespread use in MoCs. Cell lines represent the most typical cell source owing to their ease of acquisition and expansion. However, traditional cell lines often lack the same functional characteristics as primary sources. iPSC’s represent a promising personalized and potentially unlimited cell source which could decouple any differences associated with the genotype and phenotype of the cells [17]. Depending on the cell-substrate and cell-cell interactions desired, the fabricated microfluidic devices can be coated with a variety of natural polymers prior to cell loading. For example, collagen [61], gelatin [62], and gelatin methacryloyl [63] have all been used to tune cell attachment and migration.
3. APPLICATION OF MOC MODELS IN TESTING OF NMS
3.1. Models including nanomaterial entry organs
3.1.1. Models including skin-on-a-chip
Skin is a complex tissue comprised of different cell types which perform many tasks, including immune protection, physical barrier, and sensory functions. Skin serves as the primary barrier against the entry of many agents and facilitates exposure of the body’s immune system to many pathogens and immunizations. NMs, because of their size, may enter the skin more easily and cause problems. In many industries, skin exposure to chemicals involving NMs may lead to internalization and could adversely affect one’s health [64].
Thus, human skin equivalents are being increasingly used for toxicity evaluation [65]. They are employed instead of experimental animals for drug development, chemical testing, and the cosmetic industry. There are currently several epidermis (one cell type) substitutes commercially available such as SkinEthik™, EpiSkin™, EpiCS®, EpiDerm™, and LabCyte EPI-MODELS24. Full-thickness skin equivalents (containing dermal layer) are also available, such as EpiDermFT®, GraftSkin®, and Phenion® [66]. These equivalents are actively utilized for substance absorption tests [66]. Recently, there has been increased attention to engineer full-thickness skin substitutes that mimic physiological properties more closely by integrating multiple skin components, including melanocytes and vasculature. However, due to difficulties associated with sourcing different cell types, determining appropriate co-culture conditions, and recapitulating the 3D physiological of the skin, many challenges remain.
To address these challenges, skin OoC and MoC systems have been integrated with various cells, biomaterials, microfluidics, and biosensors. Alexander et al. [67] created an artificial epidermis skin OoC model with murine fibroblasts cultures for in vitro skin toxicity studies by designing polycarbonate membrane-bound fluidic systems for continuous and automated perfusion. Transepithelial electrical resistance and extracellular acidification measurements demonstrated that the designed platform could maintain on-chip cell cultures, monitor metabolic changes, and track tissue damage over the course of the study.
Recent breakthroughs in microfluidic platforms and microfabrication techniques have enabled more reliable and intricate in vitro skin constructs integrated physiologically relevant ECM structures. These organ chips could transform current early stage drug discovery approaches by facilitating reliable and patient-specific evaluations of pharmaceuticals [68]. To develop next generation nanomedicines, the effects of NP surface properties and size must be understood to characterize their passage through biological barriers. According to Arends et al. [69], the ECM of blood vessels plays an important role in controlling the movement of NPs between the blood vessels and the surrounding extravascular space due to its selective permeability properties. To investigate this effect, they designed a microfluidic system to evaluate the diffusion of molecules through the basal layer, with varying net charges and molecular weights. In the gel interface, a charge-dependent retention of molecules was determined to be consistent with the temporary binding of molecules to the gel constituents. This in vitro OoC showed good correlation in vivo with a similar charge-dependent accumulation of molecules found in a murine model.
Ataç et al. [70] developed a MoC to extend the measurement time of a skin substitute and enhance the cellular deposition and nutritional delivery in the subcutis. A skin OoC was coupled with a hair OoC to support hair follicle maintenance and facilitate the emergence of complete follicular units. In this respect, ex vivo single hair follicles and skin were grown in a bioreactor to increase the static cell maintenance periods. As compared to the standard Philpott assay, hair follicle cultures in the MoC stimulated hair fiber elongation from the epidermis in addition to tripling the culture period (Figure 3). When compared to conventional in vitro assays, the MoC bioreactor allowed enhanced spatiotemporal control of the cellular environment [70]. For the first time, Maschmeyer et al. [36a] reported a human four-organ (skin, liver, intestine, and kidney) chip co-culture to study the distribution, absorption, excretion, and metabolic profiles of drugs, along with repeated doses to assess systemic toxicity (Figure 4). After 28 days of co-culture, they evaluated the tissue composition of the different organ-like structures, namely intestine, skin, kidney, and liver. It was found that a) 3D villi structures formed from the small intestine epithelia and expressed an apical brush border, b) stratified stratum corneum formed from the skin biopsies, c) a polarized monolayer barrier remained in the human renal proximal tubule epithelia, and d) a constant glucose gradient was present between the blood circuit, the intestinal lumen, and the excretory circuit. In addition, gene and metabolic analysis showed the creation of a reproducible homeostasis among all four tissue types over the 28-day culture period [36a]. Skin-on -a-chip involving MOC models will be useful in studying nanomaterial-based drug delivery and in vaccination, or in allergy and problems related to immune system disorders.
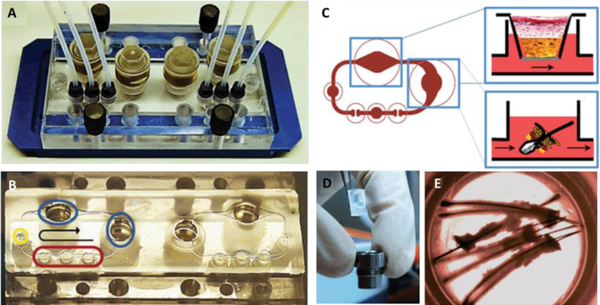
Figure 3.
Skin and hair on-a-chip: (A) A two-circuit skin and hair MoC device used to study the culture response under dynamic perfusion and static conditions simultaneously. (B) Breakdown of the fluidic circuit elements including integrated micropump (red), injection port (yellow), and Transwell® compatible tissue inserts (blue). (C) Schematic of two submerged tissue inserts culturing in vitro skin equivalents and ex vivo skin biopsies under dynamic perfusion. (D) Example of skin equivalent in a Transwell® insert before being placed into the MoC. (E) Image of the follicular unit extracts inside the MoC. Reproduced from [70] with permission from the Royal Society of Chemistry.
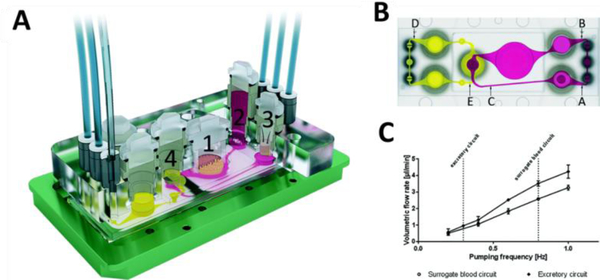
Figure 4.
(A) Four organ MoC device used to study the distribution, absorption, metabolism, and excretion profiles of drug candidates with repeated doses to assess systemic toxicity. The numbers correspond to the culture organs including (1) intestine, (2) liver, (3) skin, and (4) kidney. B) Schematic of locations in which microparticle image velocimetry was employed for flow analysis, where A, B and C are sample points for the surrogate blood circuit and D and E are for the excretory circuit. (C) The average volumetric flow rates versus pumping frequency for both the surrogate blood and excretory circuit. Pumping was driven by two on-chip peristaltic micropumps capable of circulating the media for weeks. Reproduced from [36a] with permission from the Royal Society of Chemistry.
Native skin is highly heterogenic by nature, comprised of layers forming complex vascular networks, sensory organs, and secretory glands [70]. A current shortcoming in artificial skin models is the lack of the immune system that is normally present in native tissue. Researchers have focused on studying immune responses of 3D skin models, but these fail to mimic physiological conditions due to the lack of immune cells. Moreover, artificial in vitro 3D skin models do not incorporate the dynamic transport of growth factors and nutrients. The incorporation of vascular structure within the artificial tissue represents a crucial role in diffusing nutrients, growth factors and signaling molecules [71], which must be considered in future MoC skin models.
3.1.2. Models including Lung-on-a-chip
The lung is an essential organ where many materials and agents from the environment may enter the body. The airways are lined with cells that have cilia and continuously clear out inhaled particulate matter. This cleared material may, however, get ingested and proceed to the gut. In any case, the lungs primary function is to facilitate oxygen exchange with carbon dioxide in the blood. The respiratory tract represents a vital port to the human body that is involved with the entry of toxins, pathogens, drugs, and other xenobiotic molecules. The lungs are also used as a common route for drug administration, such as in the case of inhalers for asthma treatment.
The respiratory system displays enormous diversity in its function, surrounding environment, and structure. The human lung is comprised of repeating basic units such as pulmonary alveoli. Each functional unit is in turn comprised of different cell types where NP uptake occurs easily. The lung epithelium cellular barrier is one of the first hindrances to NP entry of the body. The ability of NPs to enter and cross cellular barriers is a very important issue that warrants investigation. Conventional 3D engineered lung tissue models have been developed, but these are not capable of recapitulating dynamic microenvironments and intricate tissue−tissue interfaces. More recently, the advent of lung-on-a-chip mimics not only the structure, but some functional properties of in vivo tissue.
OoC platforms were developed to probe the complex functions associated with the lungs. Zhang et al., presented a novel 3D human lung-on-a-chip model by using three parallel channels which were seeded with lung alveolar epithelial cells, human vascular endothelial cells (ECs), and the ECM to determine NP toxicity [72]. These cells were used to determine any toxicity may be associated with TiO2 and ZnONPs. The NPs were scattered in the culture medium of the lung-on-a-chip platform at known concentrations. Permeability assay, immunofluorescence analysis, ROS assay and apoptosis analysis were used to characterize this multi-cell type OoC model.
Lung-on-a-chip systems commonly use lung epithelial cells and ECs to model the interactions between NMs and lung tissue. In an effort to develop biomimetic systems, one study used a poly(lactide-co-glycolide) (PLGA) electrospun nanofiber-based membrane as both the cell scaffold and chip substrate for a lung-on-a-chip platform. This design was developed based on the alveolar microenvironment and used to evaluate an anti-cancer drug on the lung-on-a-chip system for human fetal lung fibroblasts (HFL1) and human non-small cell lung cancer cells (A549) [45b].
It is known that lung cancer metastasis is a complex process which often metastasizes to the bone, liver and brain. MoCs have thus, been designed to better understand the processes behind cancer metastasis. For example, a study by Xu et al. described a new MoC platform for lung cancer metastasis which was very mimetic to the in vivo microenvironment [73]. The chip was prepared by combining an upstream lung organ made from silicone elastomer and three downstream organs made using osteoblasts, astrocytes, and hepatocyte cells. Different cell lines such as a human bronchial epithelial (16HBE) and human non-small cell lung cancer (A549) were used to replicate different tissue like brain, bone, and liver. This MoC system helped to better understand how lung cancer grows and metastasizes to distant organs and what the corresponding cell-cell interactions were. In this regard, a novel orthotopic model of nonsmall-cell lung cancer (NSCLC) was reported [25]. Here, the upper and lower tissue layers were composed of H1975 human NSCLC adenocarcinoma cells and small airway epithelial cells or primary lung alveolar. The cancer persister cells were analyzed in vitro based on these chip platforms. The results showed the in vitro human orthotopic lung cancer model could be developed by using microfluidic organ chip technology to study specific growth patterns. This study presents that only the combination of lung epithelium, endothelium, and breathing motions were enough to express NSCLC phenotypes. The model was innovative but had limitations because tumor stromal cells and immune cells were not integrated on the chip platforms. Two dimensional culture models and multi-well plate cultures of MCF-7 and A549 lung cancer cell lines have traditionally been used for testing NPs such as ZnONPs and silver NPs (AgNPs) to understand their impact on cell functions. However, the structure of the human lung and the alveoli environment is very complex. To more accurately mimic this complexity, 3D microfluidic systems should have precise control over fluid and solid mechanical forces representative of the human lung physiological environment [74]. These models can be sueful in testing nanomaterial drug delivery and its local and systemic effect, such as in cases of cancer chemotherapy.
3.1.3. Models including gut-on-a-chip
The gut is an important organ in which various materials and agents can access the body through ingestion, while also serving as a protective barrier [75]. In addition, the intestine is also responsible for the absorption of drugs [76], allowing their metabolism by organs such as the liver. In addition to the liver [77], the gut also communicates with the pancreas [78], cardiovascular system, and nervous system [79]. Particularly, gut function and health have been recently linked to intestinal health, immune modulation, and enteral and non-enteral diseases [80]. Therefore, understanding the multicellular environment of the gut in controlled systems can be highly advantageous to the understanding and development of treatments for various diseases.
The most common in vitro intestinal models for studying the gut’s barrier function and drug adsorption abilities have been done in Transwell insert culture devices. In these models, a monolayer of established human intestinal epithelial cells (such as Caco-2 [81] and HT-29 [82]) is cultured on a porous membrane with adsorbed ECM proteins. The use of 3D cultures has enabled both better structural (the formation of crypt and villi) and functional differences (production) as comparted to 2D systems. Because of these differences, traditional static cultures are unable to support the coculture of intestinal cells with microbiota, which are a critical part of the gut physiology. This is primarily due to the rapid overgrowth of bacterial contamination in the human cell culture, typically within 24 h. Multiple approaches have been considered to address the issues above by creating gut OoC models that better replicate the physiological environment.
Bein et al. have reviewed microfluidic approaches for modeling the human intestine [83]. Typical intestinal OoC models comprise two channels and a porous membrane separating them. Human intestinal Caco-2 cell line which via tight junctions can form a cell layer and selectively permeate certain molecules was used to culture on porous membrane to validate drug permeability of the integrated microfluidic device [84]. This configuration allowed for the probing of both the basal and apical sides of the epithelial layer, quantitative methods for measuring tight junction barrier function, and the adsorption of nutrients and drugs. However, membrane free methods have also been established [85]. While many gut OoC methods have used established cell lines, the integration of stem cells can allow for new applications in personalized medicine. Human enteroids or colonoids can be created from patient biopsies and be enzymatically fragmented to release intestinal stem cells. These cells have been cultured on Transwell inserts to provide primary intestinal monolayers, but still have the limitations associated with Transwell culture methods. Micro-bioreactors have been developed to overcome some of these limitations by providing fluid flow to organoid cultures, allowing for nutrient and oxygen delivery, as well as physiologically similar forces from fluid flow [56, 86].
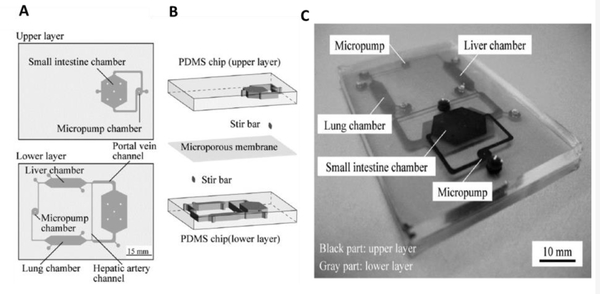
Due to the gut’s critical role in the administration and absorption of drugs, MoC systems have been developed for examining drug toxicity and metabolism, as well as pharmacokinetic and pharmacodynamic models. Kimura et al. constructed an intestine (Caco-2) – liver (HepG2) – lung (A549) MoC system for simulating the oral administration of anticancer drugs [87] (Figure 5). The device could sustain the cells for more than three days. For pharmacokinetic modeling, the developed system investigated three types of substrates, Epirubicin, irinotecan, and cyclophosphamide. The small intestine functionality was investigated using 10 μM Epirubicin injected into the luminal side of the unit. Liver function was evaluated with and without the intestinal chamber (as they also metabolize CPT-11)).
Figure 5.
Small intestine-liver on-a-chip: (A) Schematic displaying design of PDMS-based chip consisted of upper and lower layers. (B) Schematic representation of the device consisting of multiorgan in separated chambers (small intestine, liver, and lung) and stir-based micropumps linked to microchannels to mimic arteries and veins. (C) Photograph showing the microfluidic device. Black color represents microchannels on the upper layer and grey color displays microchannels on the lower layer of the device. Reproduced from [87] with permission from SAGE.
Esch et al. examined adsorption of 50 nm NPs (polystyrene) across Caco-2/HT29-MTX co-cultures [88]. Particles were flowed through the basal side of the intestinal barrier, where the basal side lead to liver, kidney, adipose, and bone marrow chambers. The use of an “other organ” (heart, skin, spleen, lung, muscle, brain, etc.) allowed for scaling the system’s volume. The adsorption of these particles led to the release of aspartate aminotransferase from downstream Hep-G2 cells, indicating cell injury. Mahler et al. also utilized this system for the adsorption and metabolism of acetaminophen [89]. Higher concentrations of acetaminophen in the apical chamber of the intestinal barrier showed decreased liver cell viability and glutathione production [89–90].
While the functionality and structure of the intestinal environment has been modeled using OoCs, the systems are still limited to studying intra-intestinal events. The integration of intestinal cells with MoCs is required to further drive these systems to physiological relevance. Particularly, smooth muscle and neuronal cells have been suggested owing to their roles in inflammatory diseases, blood flow, intestinal secretions, and motility [83].
3.2. Models including nanomaterial metabolism and clearance organs
3.2.1. Models including liver-on-a-chip
The liver is a vital organ with several important functions in the human body, especially for homeostasis and pathology. It has unique regeneration abilities and is the primary site of metabolism for several materials, such as medicine and absorbed food in the blood stream. The liver plays a crucial role not only in detoxification, but also in the production of other metabolites that can be useful or harmful to the body. For example, following the absorption of trimethylamine from the gut (produced from dietary choline [91] and L-carnitine [92]) into the liver, it is metabolized into trimethylamine N-oxide [93], which has adverse effects on the cardiovascular system [94]. For this reason, liver-on-a-chip platforms have emerged to improve our knowledge about liver function and to investigate drug metabolism. Particularly, the metabolism and hepatotoxicity of NPs have been studied with liver-on-a-chip systems which mimic the permeable sinusoidal ECs [95]. In addition, developing appropriate liver tissue models would be of great interest for testing and developing NMs.
The liver microenvironment is complex and dynamic, with multiple cell types, oxygen conditions, and vascularized systems. Many early efforts were made towards recapitulating liver function in vitro using 2D and 3D cultures to better understand underlying mechanisms behind liver regeneration, toxicity, and metabolism. Ho et al. presented a liver cell patterning technique based on an enhanced field-induced dielectrophoresis trap to reconstruct heterogenous lobule liver tissue [96]. Patterning of HepG2 and ECs in 2D culture revealed that liver function could be enhanced compared to non-patterned HepG2 cells. A functional 3D hepatic construct was fabricated by Tsang et al. using multilayer photopatterning of cellular hydrogels [97]. Photopolymerizable PEG diacrylate enabled the culture of encapsulated primary rat hepatocytes in a continuous flow bioreactor for 12 days. The success of these approaches encouraged the development of more complicated OoC and MoC devices.
Since the liver is a very important organ for drug metabolism and detoxification, a lot of work has been conducted to develop liver-on-a-chip models with high functionality. A study by Wang et al. [98] presented a new type of in vitro liver model that was comprised of liver organoids derived from iPSCs. These engineered liver organoids were combined with a liver-on-a-chip system to investigate organ development, disease mechanisms, and drug testing. The results showed that cell viability was improved under perfused culture conditions [98]. In a study by Nupura et al., bioprinted HepG2/C3A hepatic spheroid-laden hydrogels were incorporated in a perfusable bioreactor, which led to viable cultures for 30 days and enabled monitoring cell secretions for the duration of the culture [99]. Gori et al. reported a nonalcoholic fatty liver disease microfluidic model having a sinusoid-like organization which allows hepatic cell long-term culture with a permissive tissue like microenvironment [100]. Using this system, it was possible to recapitulate steatosis in vitro with controllable intracellular lipid accumulation.
Liver MoC models provide the ability to investigate a critical aspect of drug toxicity: metabolites produced in the liver may damage other interconnected organs. In the beginning of MoC models, 2D static models were mostly employed due to lack of fluidic device technologies. Lau et al. combined Caco-2 cells expanded on Transwell insert and hepatocytes placed in the basolateral receiver compartment representing liver [101]. This in vitro gut and liver hybrid model were created to test in vivo oral bioavailability of randomly chosen marketed drugs. Li et al. showed individual organs such as kidney, liver, central nervous system, lung, and blood vessels cultured in multi-well plates [102]. Cytotoxicity towards MCF-7 cells was evaluated, and the results indicated that multiple organ interactions are required to predict in vivo drug properties. In an effort toward understanding the effects of cosmetic substances on the liver, Wanger et al. reported a MoC capable of sustaining co-cultures of liver microtissue and skin biopsies. Fourteen days later, interaction between the tissues was evaluated and the platform was used to study the systemic toxicity effects the diabetic drug, troglitazone [103] (Figure 6A).
Figure 6.
(A) MoC device which could sustain the co-culture of skin biopsies and liver microtissues. The chip contained two fluidic circuits to simultaneously study the culture’s response to direct fluid flow and shielded flow [103]. (B) throughput MoC device that evaluated the effect of anti-cancer drugs on two organs (colon and liver) and four organs system (cancer, liver, intestine, and connective tissue). Reproduced from [105] with permission from the Royal Society of Chemistry.
With the advancement of fluidic device technologies, single pass or recirculating systems have been combined with on-chip platforms incorporating multiple tissues to represent different organs in the body. For an example of a single pass system, gravity-driven unidirectional flow was used in one MoC model interconnected with single gut and liver cultures [104]. Another example utilized the manual transfer of media supernatant in isolated intestine, liver, kidney, and blood brain barrier (BBB) cultures for drug studies [26b]. However, microfluidic connections within a chip system enable both continuous perfusion to mimic blood circulation, and the recirculation of media for organ-organ communication both downstream and upstream. Satoh et al. recently developed a novel multi-throughput MoC system formed on a pneumatic pressure-driven medium circulation platform. This high-throughput MoC device was capable of simultaneously sustaining two-organ (liver and cancer) and four-organ/tissue (liver, intestine, cancer, and connective tissue) models. The platform could operate under one pressure source and was developed to examine the impacts of anticancer drugs [105] (Figure 6B). Towards improving on-chip sensing data acquisition, Zhang et al. created a MoC system integrated with sensors for monitoring various parameters in real-time. Namely, ECM status (by physical sensors), drug effects (by electrochemical sensors), and morphology changes (by mini-microscope) of multiple organs (Liver-heart or liver-cancer-heart) was monitored with a microfluidic-controlled breadboard system [56] (Figure 7).
Figure 7.
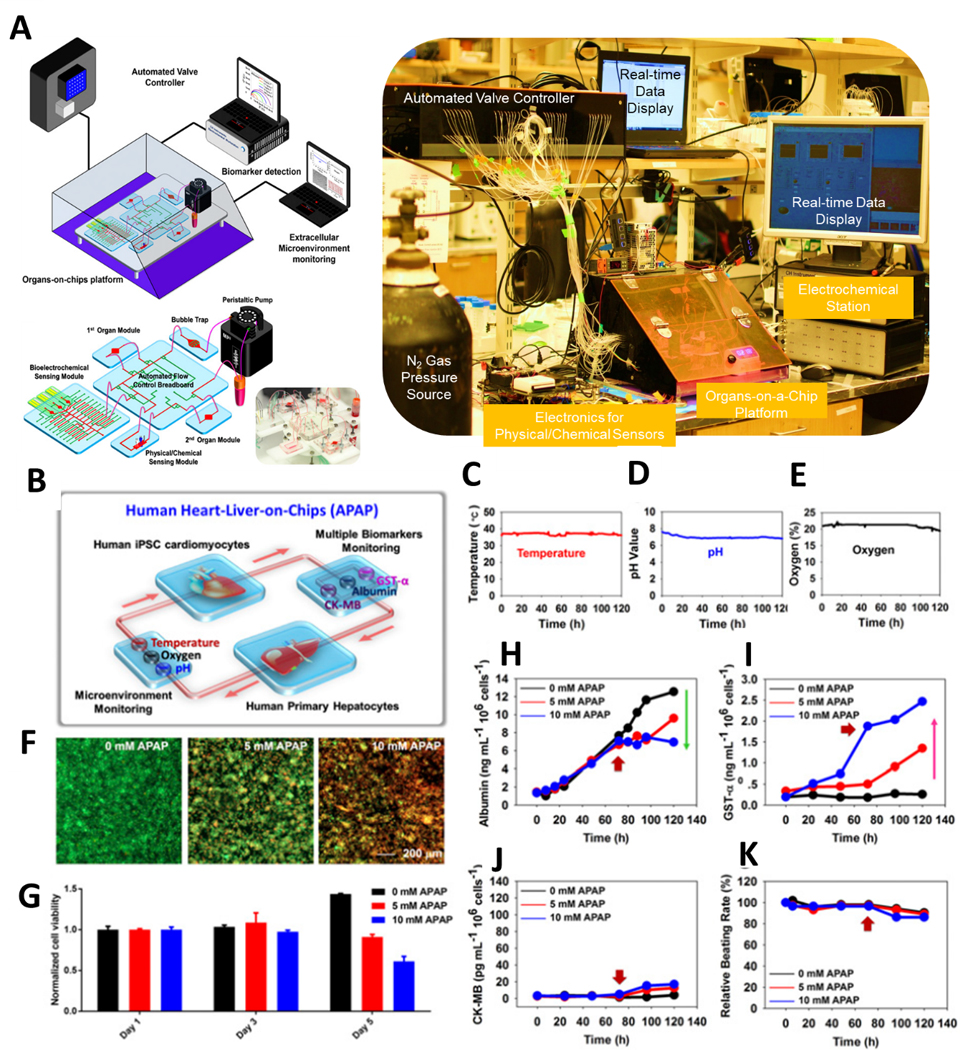
(A) Schematic and MoC microfluidic device layout with external device controls and monitors. (B) Liver-and-heart MoC and liver-cancer-heart MoC devices with integrated sensors (pH, O2, temperature, and electrochemical immunobiosensors) and a miniature microscope to study drug effects. Continual measurements of (C) Temperature, (D) pH and (E) oxygen with on-chip sensors. F) Live/dead staining of the liver organoids after drug administration and (G) normalized cell viability in response to acetaminophen. Continual electrochemical detection of (H) albumin, (I) GST-α, and (J) CK-MB with on-chip immunobiosensors. (K) Beating rate over time of cardiac tissue. (F-K) data monitored the cellular responses over 5 days, where the red arrows indicate the addition time of acetaminophen (72hr). Reproduced from [56] with permission from the National Academy of Sciences.
In another study, a new microfluidic two-organ-chip system was demonstrated by Bauer et al. with an on-chip micropump for recirculating the flow [106]. To understand insulin and glucose consumption and accumulation human pancreatic islet tissues and liver spheroids were cultured in this device. The spheroids were based on a carcinoma cell line and HepG2 cells were used for drug metabolism and toxicity. A liver, lung, and heart-tissue-organ-on-chip system was fabricated by Skardal et al. to investigate inter-organ responses to administered drugs [107]. Bioengineered tissue organoids were developed and integrated by bioprinting into microfluidic device that recirculated flow with a micro-peristaltic pump. The liver organoids were generated with multiple cell types which exist in vivo as 10% stellate cells, 10% Kupffer cells and 80% hepatocytes, ECM-derived bioink based on a hyaluronic acid and gelatin hydrogel enabled the formation of liver tissue constructs using cell aggregates in hanging drop culture. This study further demonstrates that the integration of multiple tissues/organ-on-chips may have a crucial work in assessing the effectiveness of candidate medicaments.
The interactions between NPs and hepatic cells have attracted recent attention due to targeted therapeutic approaches regarding the liver [108]. Exploring the design and application of NMs as promising vehicles to diagnose and treat hepatic diseases holds great potential in biomedical research. Therefore, it is required to combine 3D liver constructs with the same cell types that exist in natural liver tissue. Furthermore, microfluidic-based bioreactors should be used to enable the integration of multiple organ/tissue-on-a-chip modules to realize long-term monitoring of liver functionality and drug toxicity in MoC systems.
3.2.2. Models including kidney-on-a-chip
The kidney is an important organ for helping the body regulate homeostasis and several other functions including excretory, secretory, endocrine, etc. Many agents circulating in the blood are metabolized, excreted, and regulated in the kidneys. Drugs and chemicals have usually been tested in animals, and their accumulation in vital organs including the kidneys has been examined [109]. However, nephrotoxicity is often only seen at clinical trial stages, with many cases occurring in phase III and phase IV trials [13, 110]. For these reasons, understanding the underlying mechanism behind nephrotoxicity can help develop safer drugs. Nevertheless, a better predictive human in vitro model representing relevant kidney functions is still required for personalized medicine.
It remains difficult to use stem cell-based technologies to recreate complex structures, such as the kidneys. Yokoo et al. demonstrated a way to build complicated kidney tissues from autologous human mesenchymal stem cells [111]. These stem cells were reprogrammed into endogenous renal cell phenotypes and could contribute to functional complex kidney structures. Recent microengineering techniques allow the use of flow-associated shear stress or mechanical strain to study cellular behaviors. For instance, a microfluidic system with multilayered structures was developed by Jang et al. to evaluate the mechanical parameter such as shear stress on renal tubular cells cytoskeletal arrangement and cell polarization [112]. Since the kidney is a complex organ, model systems based on simple 2D and static culture or limited cell types are not suitable for drug-induced nephrotoxicity screening [113].
There has been a large effort in fabricating kidney-on-a-chip models [114]. There are several components of kidney tissue-on-a-chip that have been developed thus far, such as convoluted proximal tubule (PT) [113a], glomerulus [26a], and distal tubule [115]. Many studies have focused on the PT because of its higher susceptibility to drug toxicity due to its enzymes for gluconeogenesis [10c, 116]. Thus, many PT models focused on developing PT cell-based systems that could focus on how renal gluconeogenesis differs from the liver in terms of its affinity for substances and its response to pathological and physiological stimuli. In addition, there have been suggested designs to build a multicomponent kidney-on-a-chip [117]. A kidney-on-a-chip system was developed by Li et al. using a compartmentalized microfluidic device with primary rat glomerular ECs to study cadmium induced nephrotoxicity [118]. The system allowed the real-time assessment of cell viability, tight junction protein expression in glomerulus cells, and lactate dehydrogenase leakage under cadmium exposure. A study by Kim et al. presented kidney epithelial cells based microfluidic platform to examine pharmacokinetic responses [119]. The device used fluid perfusion to induce shear stress and promote tubular function to study injection and continuous infusion drug profiles.
MoC models could be used to mimic the metabolism and distribution of drugs in the human body. One study by Chang et al. integrated a kidney and liver-on-a-chip to investigate the mechanism responsible for the activation and distribution of aristolochic acid I [120]. Human primary cells (human epithelial cells from proximal tubule (hPTECs) from human kidney and hepatocytes from human liver tissues) were used, and the results identified how liver-kidney interactions occur in response to chemical toxicants. A similar study by Li et al. assessed the nephrotoxicity dependent on hepatic metabolism by using an integrated liver (HepG2 cells)-kidney (glomerular ECs) chip [121]. Liver and kidney cells were expanded on the bottom and upper layers, respectively in 2D, allowing for the evaluation of toxicity via filtration of kidney. The introduced drug was metabolized in the liver and found to be toxic towards kidney cells.
A MoC model integrating as many organ functions as possible on a microfluidic system has been suggested to predict organ communications. Miller et al. demonstrated a model containing 13 different organs-on-a-chip with barrier (gastrointestinal tract, skin, and lung) and without barrier (bone marrow, liver, heart, fat, kidney, brain, muscle, adrenal glands, spleen, and pancreas) types of cell culture [122]. The system mimicked the process of chemical or biological agent entry in the body by introducing them through the barrier tissues and then exposing the non-barrier tissues in 3D hydrogels. The MoC system used gravity to drive microfluidic circulation and measured cellular functions for a sustained period.
One of the major issues in recapitulating kidney function is the use of immortalized cell lines such as epithelial cells from proximal tubules [23b], tubular epithelial cells from porcine [123], canine (Madin-Darby canine kidney [MDCK]) tubular epithelial cells [119], and human (human kidney-2) proximal tubular epithelial cells [14] instead of employing primary human cells. The culture of these cell lines would cause blockages in the hollow fibers [123]), and the non-human cell lines may lead to limitations when evaluating therapeutic function of drugs and developing disease platform. To this end, hPTECs, embryonic stem cells [124], or iPSC-derived hPTEC-like cells [125] have been explored to recapitulate more functional kidney tissues. However, culturing primary cells has not been fast or readily achievable without using RNA interference or antisense nucleotides targeting p53 or p16INK4a which is known as cyclin dependent kinase inhibitor. Moreover, although it is promising to obtain hPTEC phenotypes from embryonic stem cells or iPSCs for realizing personalized medicine, these cells have not been sufficiently characterized to date.
Many cells such as glomerular vascular ECs and podocytes exist in the kidney and should also be integrated to express cellular metabolic and endocrine functions. Cell-cell interactions are highly required to generate an efficient kidney-on-a-chip platform. In addition, mechanical cues such as fluid flow dynamics, the renal tubular segments organizations, transcellular electrochemical, and osmotic pressure gradients that influence cellular functions are required. Topological cues are also required to better mimic the complex kidney structure due to their importance in cellular fate and function. Three dimensional bioprinting, which combines may help to develop these complicated structures [126]. These biomaterials can then be applied into MoCs and integrated with advanced microfluidic technologies, biosensors, and stem cell biology. Nanotechnological application of kidney related disease therapies, is in the progress, but will have transformative effects in medicine.
3.3. Models including toxicity-susceptible tissues and organs
3.3.1. Models including bone marrow-on-a-chip
Bone marrow (BM) is a spongy tissue located inside the cancellous part of long bones and in flat bones. In early childhood, BM is of predominantly hematopoietic/osteogenic type. Increased accumulation of marrow adipose tissue (MAT) begins after birth, with the marrow of distal bones is affected more rapidly than the that of the proximal bones. In clinical studies of healthy and metabolic disordered populations, MAT has been reported as an important regulator of bone turnover which has been inversely related to bone mineral density and integrity [127]. BM also has important roles in the regulation of inflammation. Interestingly, gut microbiota can affect BM hematopoietic stem cells. In obesity, gut microbiota stimulated hematopoietic stem cell differentiation by damaging BM niche function [128].
BM is known to contain hematopoietic and stromal stem cells which are used to produce blood, bone, fat, and cartilage cells. Recently, interest in developing in vitro BM models has been increasing. However, it is difficult to reproduce the BM local microenvironment due to the many factors that affect the engineering of such BM models. Additionally, conventional blood stem cells lose their capacity to proliferate and differentiate while under in vitro conditions. It seems that reciprocal interactions among stromal, hematopoietic, and bone compartments are critical for hematopoiesis. Therefore, these issues must be considered when designing BM in vitro models that can provide appropriate marrow niches.
The BM niche and composition are important for the development of leukemia as well as leukemia’s resistance to drugs. Two-dimensional culture conditions that mimic leukemic BM niche failed to capture enough complexity. To study in vivo relevance, Bruce et al. engineered a multi-cell type OoC comprised of primary human BM stromal cells, leukemic cells, and osteoblasts encapsulated in a 3D collagen matrix and cultured on a microfluidic platform [129]. Tumor cell viability in the presence of the cytarabine (chemotherapeutic agent), was compared in tumor cells alone and tri-culture models either as 2D static, 3D static or 3D microfluidic cultures. According to this study, the 3D tri-culture model was associated with leukemic cells decreased drug sensitivity as compared to the 2D models. The protective role of the BM microenvironment for tumor cell survival during the treatment with drug was elucidated. Another microfluidic platform was developed by Houshmand et al. [130] to mimic the BM environment. TF-1 cells were co-cultured with BM stromal cells in 2D and 3D microfluidic systems. Drug screening was evaluated by using azacytidine and cytarabine. The number of leukemic cells in the 3D microfluidic culture were found to increase in comparison to 2D culture conditions. In 3D culture conditions, drug resistance of cells was found by cytotoxicity assay and higher BCL2 expression in the 3D microfluidic device was reported by qPCR. Once again, this research shows the importance of mimicking BM niche for the development of targeted therapies.
Taking another approach, Bourgine et al. designed an BM OoC model that utilized porous bone-resembling hydroxyapatite scaffolds. The scaffolds were seeded with human mesenchymal stromal cells and deposited osteogenic cell-derived ECM. A perfusion flow was employed to simulate the flow of interstitial fluid and related shear stress. Perfusion of Human purified cord blood-derived CD34+ cells were perfused into the system to add the blood compartment to the stromal tissue. It was found that exposure to bleomycin impairs the capacity of mesenchymal stromal cells to maintain hematopoietic stem cells quiescent [131].
A novel 3D MoC model was presented by Sieber et al., which was composed of a scaffold (hydroxyapatite coated zirconium oxide), containing human stromal cells and cord hematopoietic stem cells [132]. Based on their results, the HSPCs were found to maintain their stem cell state and capability of differentiating to erythrocyte, granulocyte, megakaryocyte, and macrophage colonies. This was the first human in vitro 3D BM system that demonstrated enhanced hematopoietic progenitor the long-term culture (28 days) in a microfluidic environment. Developing such long-term culture periods is critical for future MoC toxicity testing where systemic effects must be monitored over time.
3.3.2. Models including blood vessel-on-a-chip
It is known that blood vessels are liable to be affected by substances circulating in the blood such as NMs. NMs can move out of the vasculature into interstitial tissues or to other tissues from the blood. Fluid shear stress, vascular transport, and the toxicity on ECs are very important parameters regarding the interactions of NPs with the vasculature. Reported models have already reproduced several features of vasculature based on these perspectives [74]. For instance, Ahn and co-workers fabricated a 3D microvessel-on-a-chip model to understand how cationic polymer NPs effect vascular cytotoxicity and extravascular transport [133]. The extravascular transport mechanism for polyethylenimine (PEI), which was used as gene carrier for cationic NPs, was obtained in microvessel chip platform that was not possible for 2D platforms. In microvessel, the transcytotic effect of NPs was nearly the same as in vivo conditions.
In many early studies, ECs were cultured on a Transwell insert, petri dish, or hydrogel to make blood vessel walls and better understand vascular biology. However, 2D culture, Matrigel embedded culture, and microbead based models cannot mimic organ-specific microenvironments, making these models not suitable for gas exchange and nutrient transportation. More biomimetic structures considering shape and polarized surfaces are needed to investigate organ viability. In this respect, an in vitro model blood vessel-on-a-chip was reported from Pauty et al. that mimics effective endothelial angiogenesis which allowed the study of anti-angiogenic drugs and simultaneous testing of angiogenesis and permeability [134]. A vascular endothelial growth factor-A (VEGF-A) was selected as target for the development of drugs.
Sub-50 nm diameter mesoporous silica NPs were placed in blood vessel on a chip platform by Kim et al. [135] to investigate the effect of NPs on platelet functions such as adhesion. In another study from Kim et al. [136], blood circulation nanotoxicity was formalized and the cytotoxic effect of NPs on human umbilical vein ECs (HUVECs) was studied under microfluidics. This study showed that depending on NP modification, shear stress is a critical factor for toxicity to ECs. Recently, a microfluidic system was created by Fede et al. [137] to understand how human EC viability changes with different sizes (24 and 13 nm) and doses of AuNPs in a continuous flow of culture medium. The toxicity of AuNPs in flow conditions was found to be lower than those in static conditions. Additionally, the size of the particles was not critical for HUVEC viability whereas the HUVEC viability decreases as NP surface area increases.
To include organ-organ interactions, MoC systems employing engineered blood vessels can provide an opportunity to combine different numbers and types of organs together. The organization of a vascular network is a critical part of 3D culture formation in microfluidic chip devices. One study made by Zhang et al. consisted of the development of vascular module on a chip system to combine MoCs [138]. The group fabricated PDMS hollow tubes which showed very good gas exchange, optical visualization, and elasticity for vascular module and presented two different templating strategies. These templates were separated as the inner templates were prepared based on metal rods or airflow and the outer templates were fabricated with plastic tubes. HUVECs were used to functionalize the PDMS tubes to serve as elastomeric biomimetic blood vessels. These PDMS hollow tubes were easily integrated into a microfluidic platform. Similarly, Hasenberg et al. [139], reported a MoC system created from a combination of engineered vessels and neo-vascularization. Fibrin was used as a scaffold material because its structural and mechanical properties are close to those found in vivo ECMs. HUVECs were seeded in the fibrin gels to promote the formation of 3D capillary-like networks. The seeded cells showed good viability with the dynamic environment, and the network remained stable in serum.
Three-dimensional vascularized tissue constructs require ECs and ECM combination to achieve natural tissues and organs biomimetics. The blood circulatory system acts as a barrier for NPs injected intravenously as they reach the target organ. Advanced MoC platforms integrated with 3D microvascular networks can help explain how NMs behave in the human body. For future studies, MoCs will be applied to solve these problems owing to their superior biomimetic properties.
3.3.3. Models including spleen-on-a-chip
The spleen functions to filter aged, damaged and infected red blood cells via the splenic red pulp, while also serving to activate the adaptive immune system via splenic white pulp. An open-slow microcirculation system eliminates unhealthy red blood cells by two main mechanisms. Specialized macrophages target damaged cells and a physical constraint provided by unidirectional passage through inter-endothelial slits removes cells which lack of deformability [140].
The spleen is particularly susceptible to NP translocation via intestinal lymphatic tissue, potentially because of its blood cleansing and immune system functions [141]. As such, the development of models to study potential nanomaterial toxicity effects on the spleen have been suggested. Spleen-on-a-chip models offer a promising blood-cleansing solution to treat bloodstream infections by removing pathogens. A study was developed to mimic the hydrodynamic behavior, physical barriers properties, and filtering functions of the spleen [142]. This human splenon-on-a-chip includes two parts with different flow dynamics and two physical barriers with a pillar matrix to increase the hematocrit value of the blood. Unidirectional micro constrictions were used to mimic the filtration bed and inter-endothelial slits. This device can be used to improve our understanding of spleen pathophysiology in malaria and other hematological disorders, as well as for screening new drugs for hematological disorders. Another initiative was made to devise a spleen-on-a-chip for cleansing blood for sepsis therapy. In this effort, human opsonin (mannose-binding lectin) coated magnetic nanobeads were developed to bind different pathogens and toxins without leading to the activation of complement and coagulation systems. Infected blood can be mixed with these functionalized magnetic nanobeads to collect the opsonin-bound toxins and pathogens. The purified blood can then be returned to the patients. An in vivo study on infected rats showed that this spleen-on-a-chip can cleanse more than 90% of bacteria leading to decreased inflammatory cytokine levels [143].
One of main cause of failure in drug delivery and therapeutic NPs is the toxicity imposed to the immune system [144]. Because the spleen is an important immune organ which facilitates blood filtering and contains macrophages, it may be particularly suitable to such toxicity. Accordingly, further developments in spleen-on-a-chip models is of high relevance to elucidate immune physiology in response to NMs. Following examinations of developed models on large animals and clinical trials with longer observation periods can provide more blueprints to improve their functionality. However, less priority has been given to develop spleen mimicking models, due to the possibility of human survival without this organ. Spleen-on-a-chip devices for blood cleansing in combination with other multi-organ-on-a-chip units can not only help solicit more biomimetic immune models for developing NPs, but also serve as platform for cleansing pathogens after treatments.
4. CHALLENGES AND FUTURE OUTLOOK
Despite the fact that research on OoC models started about a decade ago and profound foundations have been established, robust clinical applications of OoCs have yet to be accomplished. Challenges such as reproducibility and feasibility of engineered OoC devices along with their validation and effectiveness as compared to biological systems should be overcome. More in depth studies are needed to incorporate functional characteristics of complex organs such as kidney, heart, and liver-on-a-chip. Some studies have reported the interaction between four organ-on-a-chip units [36a]; however, a comprehensive dynamic MoC has not been achieved yet. Recently, ‘fish-on-a-chip’ microfluidic systems have been developed, which allows monitoring of the small organism with fluorescent labeled nanoparticles and trafficking nanoparticle inside the cells in a high throughput manner [145]. These may have the potential to accelerate the “body-on-a-chip” system to assess new nanoparticle-based drugs and find out possible side effects.
To mimic the interconnectedness of organs within the human body, MOC systems need to maintain a variety of cell types. This creates another problem encountered in recirculating common medium for different cell types, transport of nutrient and soluble factors at the physiological level. Vascularization is one of the great challenges that tissue engineering faces in order to achieve sizeable tissue and organ substitutes that contain living cells. Since MOC systems are formed by linking several organs on a chip structures, it is important to ensure sterility and to remote air bubbles from the environment. In in vivo systems, cells obtain oxygen and nutrients via blood flow, as well as receive chemical and physical stimulation, such as stretching and shear stress, from the surrounding environment. This creates flow rate differences between platforms, ideal oxygenation and nutrient levels for different organs emerge as a problem for technical engineering. Complex microfluidic circulation allows automation and integration of organ models with biosensors, which could evaluate organoid responses to various stimuli. Challenges include reproducing the architectural complexity of the human tissues and organs in vitro with interconnected miniaturized biosensors and electrodes is not easy. Another technical and scientific problem is created by PDMS that has traditionally been the material of choice due to its favorable properties with regard to fabrication and permeability. However, for drug testing this material creates a problem due to the adsorption of drug [50, 146].
Additionally, OoC devices with routine production, reproducible quality, and robust functionality have yet to be developed. Latent opportunities must be identified, harnessed, and integrated to continue driving progress in the field [114]. Steps have been taken by researchers with experience in microfabrication to increase the use microfluidic devices as developers seek to make modular, plug-and-play platforms [147].
Recent multidisciplinary technologies can be converted to achieve robust, functional, and physiologically relevant OoC systems to meet increasing demands for organ and tissue transplants [148]. For instance, the converging of bioelectronics with OoC systems and other modalities will enhance our technologies for diagnostics, prognosis, and therapies [149]. A variety of sensor types have been integrated into OoC systems to assess the viability and activity of cells, including O2, glucose, lactate, pyruvate, and glutamate [150]. Providing controls for multiple metrics of cell activity in real-time for each component of a MoC system will be of critical importance. This is particularly the case when distinguishing between healthy and diseased states for diagnoses or preliminary in vitro comparisons [107].
Engineering challenges also remain when scaling and sizing each organ system in MoC devices, particularly as they become more complex. These challenges have been discussed in detail by Wiskwo et al. [151]. Different approaches, each with their own advantages/disadvantages have been proposed, including 1) allometric scaling [152], interconnected “histological sections”, and functional scaling. Allometric scaling was applied in early pharmacokinetic models and could make a promising approach for the integration/validation of PK-PD modeling with the experimental results of MoCs. “Histological sections” aim to replicate a small portion of the tissue environment in vivo, and therefore the flow rates or external stimuli are scaled to the device. However, if the culture medium is recirculated, determining the optimal circuit volume is difficult and can lead to delays in the detection of various metabolites. The detections of highly reactive species with limited lifetimes is particularly difficult. Lastly, functional scaling sizes the OoC based on the functional component of the organ (i.e. lung and gas exchange). However, this may lead to over-simplifications of each organ and their interactions with other systems.
Another challenge in the clinical translation of OoC and MoC devices is cell sourcing. While culturing of these cells in individual devices has been done, the differentiation, maturation, formation, and equilibration of these cells often requires specialized organ-specific protocols/media, making it difficult to link multiorgan systems. iPSC cells have been used in OoC devices for experiments, however, they impose risks such as developing teratomas [50–52]. Accordingly, these risks must be taken in consideration when it comes to achieving clinical applications. This issue has encouraged researchers to look for cell sourcing alternatives. Additionally, nanotechnology developments can be tested in OoC and MoC systems to provide precision diagnostics, therapies, and specific cell-level analyses.
5. CONCLUSIONS
Although the development of NMs is on the rise, limited means to test their efficacy and has led to delayed translation in clinical and industrial applications. With the recent advent of OoC and MoC systems, it is becoming possible to leverage the advantages offered by these platforms to examine the interactions of NMs with representative target tissues and organs. Their potential is huge, yet to be exploited properly the data must be validated carefully using animal studies and clinical trials. With continued advancements, the use of MoC platforms should eventually lead to moving away from animal-testing.
Acknowledgement
The authors also acknowledge funding from the National Institutes of Health (AR057837, CA214411, HL137193, EB022403 and R01EB021857), Air Force Office of Sponsored Research under award # FA9550–15-1–0273, Department of Defense: BiofabUSA Quick Start Project, and the American Heart Association Transformational Project Award (18TPA34230036). R.T. would like to acknowledge the financial support from the Scientific and Technological Research Council of Turkey (TUBITAK) 2214-A International Doctorate Research Fellowship Program (App. No: 1059B141700084) and from Istanbul University Cerrahpasa, Engineering Faculty, Chemistry Department.
Footnotes
Disclosure
The authors declare no competing financial interest.
References
- [1].Lin L, Zhao H, Li J, Tang J, Duan M, Jiang L, Biochemical and biophysical research communications 2000, 274, 817. [DOI] [PubMed] [Google Scholar]
- [2].Anonymous, 2018, 191. [Google Scholar]
- [3].a) Youns M, Hoheisel JD, Efferth T, Current drug targets 2011, 12, 357; [DOI] [PubMed] [Google Scholar]; b) Menon JU, Jadeja P, Tambe P, Vu K, Yuan B, Nguyen KT, Theranostics 2013, 3, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].a) Miller MR, Raftis JB, Langrish JP, McLean SG, Samutrtai P, Connell SP, Wilson S, Vesey AT, Fokkens PHB, Boere AJF, Krystek P, Campbell CJ, Hadoke PWF, Donaldson K, Cassee FR, Newby DE, Duffin R, Mills NL, ACS Nano 2017, 11, 4542; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Järup L, British medical bulletin 2003, 68, 167. [DOI] [PubMed] [Google Scholar]
- [5].Rochman CM, Hoh E, Kurobe T, Teh SJ, Scientific reports 2013, 3, 3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Baudouin C, Charveron M, Tarroux R, Gall Y, Cell biology and toxicology 2002, 18, 341. [DOI] [PubMed] [Google Scholar]
- [7].Choi HS, Ashitate Y, Lee JH, Kim SH, Matsui A, Insin N, Bawendi MG, Semmler-Behnke M, Frangioni JV, Tsuda A, Nature Biotechnology 2010, 28, 1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Oberdorster G, Oberdorster E, Oberdorster J, Environ Health Perspect 2005, 113, 823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Arora S, Rajwade JM, Paknikar KM, Toxicol Appl Pharmacol 2012, 258, 151. [DOI] [PubMed] [Google Scholar]
- [10].a) Hartung T, Nature 2009, 460, 208; [DOI] [PubMed] [Google Scholar]; b) Jenkinson SE, Chung GW, van Loon E, Bakar NS, Dalzell AM, Brown CD, Pflugers Archiv : European journal of physiology 2012, 464, 601; [DOI] [PubMed] [Google Scholar]; c) Jang KJ, Mehr AP, Hamilton GA, McPartlin LA, Chung S, Suh KY, Ingber DE, Integrative biology : quantitative biosciences from nano to macro 2013, 5, 1119. [DOI] [PubMed] [Google Scholar]
- [11].Gintant G, Sager PT, Stockbridge N, Nature reviews. Drug discovery 2016, 15, 457. [DOI] [PubMed] [Google Scholar]
- [12].Wang B, Gray G, Risk analysis : an official publication of the Society for Risk Analysis 2015, 35, 1154. [DOI] [PubMed] [Google Scholar]
- [13].Naughton CA, American family physician 2008, 78, 743. [PubMed] [Google Scholar]
- [14].Adler M, Ramm S, Hafner M, Muhlich JL, Gottwald EM, Weber E, Jaklic A, Ajay AK, Svoboda D, Auerbach S, Kelly EJ, Himmelfarb J, Vaidya VS, Journal of the American Society of Nephrology : JASN 2016, 27, 1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hartung T, Rovida C, Nature 2009, 460, 1080. [DOI] [PubMed] [Google Scholar]
- [16].a) Blaauboer BJ, Andersen ME, Archives of toxicology 2007, 81, 385; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Greim H, Arand M, Autrup H, Bolt HM, Bridges J, Dybing E, Glomot R, Foa V, Schulte-Hermann R, Springer, 2006. [DOI] [PubMed] [Google Scholar]
- [17].Ronaldson-Bouchard K, Vunjak-Novakovic G, Cell Stem Cell 2018, 22, 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Whitesides GM, Nature 2006, 442, 368. [DOI] [PubMed] [Google Scholar]
- [19].a) João R, Hossein S, Amir M, Jeroen L, Katelyn B, Shrike ZY, Lino F, Ali K, Applied In Vitro Toxicology 2016, 2, 82;28971113 [Google Scholar]; b) Kim HJ, Huh D, Hamilton G, Ingber DE, Lab Chip 2012, 12, 2165. [DOI] [PubMed] [Google Scholar]
- [20].a) Ribas J, Zhang YS, Pitrez PR, Leijten J, Miscuglio M, Rouwkema J, Dokmeci MR, Nissan X, Ferreira L, Khademhosseini A, small 2017, 13, 1603737; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE, Science 2010, 328, 1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Pavesi A, Adriani G, Rasponi M, Zervantonakis IK, Fiore GB, Kamm RD, Scientific Reports 2015, 5, 11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Huh D, Hamilton GA, Ingber DE, Trends in Cell Biology 2011, 21, 745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].a) Weber EJ, Chapron A, Chapron BD, Voellinger JL, Lidberg KA, Yeung CK, Wang Z, Yamaura Y, Hailey DW, Neumann T, Kidney international 2016, 90, 627; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ferrell N, Ricci KB, Groszek J, Marmerstein JT, Fissell WH, Biotechnology and bioengineering 2012, 109, 797; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Shen C, Zhang G, Wang Q, Meng Q, ACS applied materials & interfaces 2015, 7, 19789; [DOI] [PubMed] [Google Scholar]; d) Banaeiyan AA, Theobald J, Paukštyte J, Wölfl S, Adiels CB, Goksör M, Biofabrication 2017, 9, 015014. [DOI] [PubMed] [Google Scholar]
- [24].Bhatia SN, Ingber DE, Nature Biotechnology 2014, 32, 760. [DOI] [PubMed] [Google Scholar]
- [25].Hassell BA, Goyal G, Lee E, Sontheimer-Phelps A, Levy O, Chen CS, Ingber DE, Cell reports 2017, 21, 508. [DOI] [PubMed] [Google Scholar]
- [26].a) Musah S, Mammoto A, Ferrante TC, Jeanty SSF, Hirano-Kobayashi M, Mammoto T, Roberts K, Chung S, Novak R, Ingram M, Fatanat-Didar T, Koshy S, Weaver JC, Church GM, Ingber DE, Nat Biomed Eng 2017, 1; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Vernetti L, Gough A, Baetz N, Blutt S, Broughman JR, Brown JA, Foulke-Abel J, Hasan N, In J, Kelly E, Scientific reports 2017, 7, 42296. [Google Scholar]
- [27].T P A, M SS, Jose A, Chandran L, Zachariah SM, Journal of clinical medicine research 2009, 1, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Esch EW, Bahinski A, Huh D, Nature reviews. Drug discovery 2015, 14, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kashaninejad N, Nikmaneshi MR, Moghadas H, Kiyoumarsi Oskouei A, Rismanian M, Barisam M, Saidi MS, Firoozabadi B, Micromachines 2016, 7, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE, Science 2010, 328, 1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Li X, Tian T, Analytical Methods 2018, 10, 3122. [Google Scholar]
- [32].Li L, Gokduman K, Gokaltun A, Yarmush ML, Usta OB, Nanomedicine (Lond) 2019, 14, 2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhang C, Zhao Z, Abdul Rahim NA, van Noort D, Yu H, Lab on a chip 2009, 9, 3185. [DOI] [PubMed] [Google Scholar]
- [34].Webb SPD, Biotechniques 2017, 62, 93. [DOI] [PubMed] [Google Scholar]
- [35].Lanz HL, Saleh A, Kramer B, Cairns J, Ng CP, Yu J, Trietsch SJ, Hankemeier T, Joore J, Vulto P, Weinshilboum R, Wang L, BMC cancer 2017, 17, 709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].a) Maschmeyer I, Lorenz AK, Schimek K, Hasenberg T, Ramme AP, Hübner J, Lindner M, Drewell C, Bauer S, Thomas A, Lab on a Chip 2015, 15, 2688; [DOI] [PubMed] [Google Scholar]; b) Li Z, Su W, Zhu Y, Tao T, Li D, Peng X, Qin J, Biomicrofluidics 2017, 11, 034114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Waseem H, Williams MR, Stedtfeld RD, Stedtfeld TM, Shanker R, Hashsham SA, in Nanotoxicology: Experimental and Computational Perspectives, DOI: 10.1039/9781782623922-00203, The Royal Society of Chemistry; 2018, p. 203. [DOI] [Google Scholar]
- [38].Zhang YS, Zhang YN, Zhang W, Drug discovery today 2017, 22, 1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].a) Brown JA, Pensabene V, Markov DA, Allwardt V, Neely MD, Shi M, Britt CM, Hoilett OS, Yang Q, Brewer BM, Samson PC, McCawley LJ, May JM, Webb DJ, Li D, Bowman AB, Reiserer RS, Wikswo JP, Biomicrofluidics 2015, 9, 054124; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Jang K-J, Mehr AP, Hamilton GA, McPartlin LA, Chung S, Suh K-Y, Ingber DE, Integrative Biology 2013, 5, 1119; [DOI] [PubMed] [Google Scholar]; c) Kim HJ, Ingber DE, Integr Biol (Camb) 2013, 5, 1130; [DOI] [PubMed] [Google Scholar]; d) Kim HJ, Huh D, Hamilton G, Ingber DE, Lab on a Chip 2012, 12, 2165; [DOI] [PubMed] [Google Scholar]; e) Booth R, Kim H, Lab on a chip 2012, 12, 1784; [DOI] [PubMed] [Google Scholar]; f) Achyuta AKH, Conway AJ, Crouse RB, Bannister EC, Lee RN, Katnik CP, Behensky AA, Cuevas J, Sundaram SS, Lab on a chip 2013, 13, 542; [DOI] [PubMed] [Google Scholar]; g) Shin SR, Kilic T, Zhang YS, Avci H, Hu N, Kim D, Branco C, Aleman J, Massa S, Silvestri A, Kang J, Desalvo A, Hussaini MA, Chae SK, Polini A, Bhise N, Hussain MA, Lee H, Dokmeci MR, Khademhosseini A, Advanced science (Weinheim, Baden-Wurttemberg, Germany) 2017, 4, 1600522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Park SE, Georgescu A, Huh D, Science 2019, 364, 960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].a) Ashammakhi N, Elkhammas E, Hasan A, Journal of Biomedical Materials Research Part B: Applied Biomaterials 2019, 107, 2006; [DOI] [PubMed] [Google Scholar]; b) Shuler ML, Lab on a Chip 2019, 19, 9. [DOI] [PubMed] [Google Scholar]
- [42].Zhao Y, Yao R, Ouyang L, Ding H, Zhang T, Zhang K, Cheng S, Sun W, Biofabrication 2014, 6, 035001. [DOI] [PubMed] [Google Scholar]
- [43].Gnecco JS, Anders AP, Cliffel D, Pensabene V, Rogers LM, Osteen K, Aronoff DM, Current Pharmaceutical Design 2017, 23, 6115. [DOI] [PubMed] [Google Scholar]
- [44].Lee JS, Romero R, Han YM, Kim HC, Kim CJ, Hong J-S, Huh D, The Journal of Maternal-Fetal & Neonatal Medicine 2016, 29, 1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].a) Fan Y, Nguyen DT, Akay Y, Xu F, Akay M, Scientific Reports 2016, 6, 25062; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Yang X, Li K, Zhang X, Liu C, Guo B, Wen W, Gao X, Lab on a chip 2018, 18, 486; [DOI] [PubMed] [Google Scholar]; c) Elmusrati M, Ashammakhi N, J Craniofac Surg 2018, 29, 1682. [DOI] [PubMed] [Google Scholar]
- [46].Benam KH, Mazur M, Choe Y, Ferrante TC, Novak R, Ingber DE, Methods in molecular biology (Clifton, N.J.) 2017, 1612, 345. [DOI] [PubMed] [Google Scholar]
- [47].Zhang YS, Davoudi F, Walch P, Manbachi A, Luo X, Dell’Erba V, Miri AK, Albadawi H, Arneri A, Li X, Wang X, Dokmeci MR, Khademhosseini A, Oklu R, Lab on a chip 2016, 16, 4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wang G, McCain ML, Yang L, He A, Pasqualini FS, Agarwal A, Yuan H, Jiang D, Zhang D, Zangi L, Geva J, Roberts AE, Ma Q, Ding J, Chen J, Wang D-Z, Li K, Wang J, Wanders RJA, Kulik W, Vaz FM, Laflamme MA, Murry CE, Chien KR, Kelley RI, Church GM, Parker KK, Pu WT, Nature medicine 2014, 20, 616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ashammakhi NA, Elzagheid A, J Craniofac Surg 2018, 29, 823. [DOI] [PubMed] [Google Scholar]
- [50].Ribas J, Zhang YS, Pitrez PR, Leijten J, Miscuglio M, Rouwkema J, Dokmeci MR, Nissan X, Ferreira L, Khademhosseini A, Small 2017, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hong S, Lee LP, Biophysical Journal 2014, 106, 245a. [Google Scholar]
- [52].a) Mathur A, Loskill P, Shao K, Huebsch N, Hong S, Marcus SG, Marks N, Mandegar M, Conklin BR, Lee LP, Healy KE, Sci Rep 2015, 5, 8883; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lind JU, Busbee TA, Valentine AD, Pasqualini FS, Yuan H, Yadid M, Park S-J, Kotikian A, Nesmith AP, Campbell PH, Vlassak JJ, Lewis JA, Parker KK, Nature materials 2017, 16, 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Conant G, Lai BFL, Lu RXZ, Korolj A, Wang EY, Radisic M, Stem Cell Reviews and Reports 2017, 13, 335. [DOI] [PubMed] [Google Scholar]
- [54].Skardal A, Shupe T, Atala A, Drug discovery today 2016, 21, 1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ingber DE, Development (Cambridge, England) 2018, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Zhang YS, Aleman J, Shin SR, Kilic T, Kim D, Shaegh SAM, Massa S, Riahi R, Chae S, Hu N, Proceedings of the National Academy of Sciences 2017, 201612906. [Google Scholar]
- [57].Sosa-Hernández JE, Villalba-Rodríguez AM, Romero-Castillo KD, Aguilar-Aguila-Isaías MA, García-Reyes IE, Hernández-Antonio A, Ahmed I, Sharma A, Parra-Saldívar R, Iqbal HMN, Micromachines 2018, 9, 536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Yang Q, Lian Q, Xu F, Biomicrofluidics 2017, 11, 031301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].a) Sosa-Hernández JE, Villalba-Rodríguez AM, Romero-Castillo KD, Aguilar-Aguila-Isaías MA, García-Reyes IE, Hernández-Antonio A, Ahmed I, Sharma A, Parra-Saldívar R, Iqbal HMN, Micromachines (Basel) 2018, 9; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Díaz Lantada A, Pfleging W, Besser H, Guttmann M, Wissmann M, Plewa K, Smyrek P, Piotter V, García-Ruíz JP, Polymers 2018, 10, 1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Lee H, Cho DW, Lab Chip 2016, 16, 2618. [DOI] [PubMed] [Google Scholar]
- [61].Kasendra M, Tovaglieri A, Sontheimer-Phelps A, Jalili-Firoozinezhad S, Bein A, Chalkiadaki A, Scholl W, Zhang C, Rickner H, Richmond CA, Li H, Breault DT, Ingber DE, Scientific reports 2018, 8, 2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Ahn S, Ardoña HAM, Lind JU, Eweje F, Kim SL, Gonzalez GM, Liu Q, Zimmerman JF, Pyrgiotakis G, Zhang Z, Beltran-Huarac J, Carpinone P, Moudgil BM, Demokritou P, Parker KK, Anal Bioanal Chem 2018, 410, 6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Chen MB, Srigunapalan S, Wheeler AR, Simmons CA, Lab on a Chip 2013, 13, 2591. [DOI] [PubMed] [Google Scholar]
- [64].Borm PJ, Robbins D, Haubold S, Kuhlbusch T, Fissan H, Donaldson K, Schins R, Stone V, Kreyling W, Lademann J, Krutmann J, Warheit D, Oberdorster E, Particle and fibre toxicology 2006, 3, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Zhang Z, Michniak-Kohn BB, Pharmaceutics 2012, 4, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Schimek K, Hsu HH, Boehme M, Kornet JJ, Marx U, Lauster R, Portner R, Lindner G, Bioengineering (Basel, Switzerland) 2018, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Alexander FA, Eggert S, 2018, 9. [Google Scholar]
- [68].[] Abaci HE, Guo Z, Doucet Y, Jackow J, Christiano A, Experimental biology and medicine (Maywood, N.J.) 2017, 242, 1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Arends F, Sellner S, Seifert P, Gerland U, Rehberg M, Lieleg O, Lab on a chip 2015, 15, 3326. [DOI] [PubMed] [Google Scholar]
- [70].Ataç B, Wagner I, Horland R, Lauster R, Marx U, Tonevitsky AG, Azar RP, Lindner G, Lab on a Chip 2013, 13, 3555. [DOI] [PubMed] [Google Scholar]
- [71].Lee S, Jin S-P, Kim YK, Sung GY, Chung JH, Sung JH, Biomedical Microdevices 2017, 19, 22. [DOI] [PubMed] [Google Scholar]
- [72].Zhang M, Xu C, Jiang L, Qin J, Toxicology Research 2018, 7, 1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Xu Z, Li E, Guo Z, Yu R, Hao H, Xu Y, Sun Z, Li X, Lyu J, Wang Q, ACS Applied Materials & Interfaces 2016, 8, 25840. [DOI] [PubMed] [Google Scholar]
- [74].Bhise NS, Ribas J, Manoharan V, Zhang YS, Polini A, Massa S, Dokmeci MR, Khademhosseini A, Journal of Controlled Release 2014, 190, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Silverthorn DU, Ober WC, Garrison CW, Silverthorn AC, Johnson BR, Human physiology: an integrated approach, Pearson/Benjamin Cummings San Francisco, CA:, 2004. [Google Scholar]
- [76].Benet LZ, Wu C-Y, Hebert MF, Wacher VJ, Journal of controlled release 1996, 39, 139. [Google Scholar]
- [77].a) Moore FA, Moore EE, Poggetti R, Mcanena OJ, Peterson VM, Abernathy CM, Parsons PE, The Journal of trauma 1991, 31, 629; [DOI] [PubMed] [Google Scholar]; b) Bloemen JG, Venema K, van de Poll MC, Damink SWO, Buurman WA, Dejong CH, Clinical nutrition 2009, 28, 657. [DOI] [PubMed] [Google Scholar]
- [78].Ahuja M, Schwartz DM, Tandon M, Son A, Zeng M, Swaim W, Eckhaus M, Hoffman V, Cui Y, Xiao B, Cell metabolism 2017, 25, 635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].a) Cryan JF, Dinan TG, Nature reviews neuroscience 2012, 13, 701; [DOI] [PubMed] [Google Scholar]; b) Mayer EA, Nature Reviews Neuroscience 2011, 12, 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Zhang Y-J, Li S, Gan R-Y, Zhou T, Xu D-P, Li H-B, Int J Mol Sci 2015, 16, 7493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].a) Hidalgo IJ, Raub TJ, Borchardt RT, Gastroenterology 1989, 96, 736; [PubMed] [Google Scholar]; b) Artursson P, Karlsson J, Biochemical and biophysical research communications 1991, 175, 880. [DOI] [PubMed] [Google Scholar]
- [82].a) Pinto MGV, Gómez MR, Seifert S, Watzl B, Holzapfel WH, Franz CM, International journal of food microbiology 2009, 133, 86; [DOI] [PubMed] [Google Scholar]; b) Eveillard M, Fourel V, Bare MC, Kernéis S, Coconnier MH, Karjalainen T, Bourlioux P, Servin AL, Molecular microbiology 1993, 7, 371. [DOI] [PubMed] [Google Scholar]
- [83].Bein A, Shin W, Jalili-Firoozinezhad S, Park MH, Sontheimer-Phelps A, Tovaglieri A, Chalkiadaki A, Kim HJ, Ingber DE, Cellular and molecular gastroenterology and hepatology 2018, 5, 659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Gao D, Liu H, Lin J-M, Wang Y, Jiang Y, Lab on a chip 2013, 13, 978. [DOI] [PubMed] [Google Scholar]
- [85].Trietsch SJ, Naumovska E, Kurek D, Setyawati MC, Vormann MK, Wilschut KJ, Lanz HL, Nicolas A, Ng CP, Joore J, Nature communications 2017, 8, 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, Yao B, Hamersky GR, Jacob F, Zhong C, Cell 2016, 165, 1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Kimura H, Ikeda T, Nakayama H, Sakai Y, Fujii T, Journal of Laboratory Automation 2015, 20, 265. [DOI] [PubMed] [Google Scholar]
- [88].Esch MB, Mahler GJ, Stokol T, Shuler ML, Lab on a chip 2014, 14, 3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Mahler GJ, Esch MB, Glahn RP, Shuler ML, Biotechnol Bioeng 2009, 104, 193. [DOI] [PubMed] [Google Scholar]
- [90].Wang L, Nagesha DK, Selvarasah S, Dokmeci MR, Carrier RL, Journal of Nanobiotechnology 2008, 6, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL, The New England journal of medicine 2013, 368, 1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, DiDonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WHW, Bushman FD, Lusis AJ, Hazen SL, Nature Medicine 2013, 19, 576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Bennett BJ, de Aguiar Vallim TQ, Wang Z, Shih DM, Meng Y, Gregory J, Allayee H, Lee R, Graham M, Crooke R, Edwards PA, Hazen SL, Lusis AJ, Cell Metab 2013, 17, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung Y-M, Wu Y, Schauer P, Smith JD, Allayee H, Tang WHW, DiDonato JA, Lusis AJ, Hazen SL, Nature 2011, 472, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Beckwitt CH, Clark AM, Wheeler S, Taylor DL, Stolz DB, Griffith L, Wells A, Experimental cell research 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Ho C-T, Lin R-Z, Chen R-J, Chin C-K, Gong S-E, Chang H-Y, Peng H-L, Hsu L, Yew T-R, Chang S-F, Liu C-H, Lab on a chip 2013, 13, 3578. [DOI] [PubMed] [Google Scholar]
- [97].Tsang VL, Chen AA, Cho LM, Jadin KD, Sah RL, DeLong S, West JL, Bhatia SN, 2007, 21, 790. [DOI] [PubMed] [Google Scholar]
- [98].Y. Wang, H. Wang, P. Deng, W. Chen, Y. Guo, T. Tao, J. Qin, 2018, DOI: 10.1039/c8lc00869h. [DOI]
- [99].Nupura SB, Vijayan M, Solange M, Ali T, Masoumeh G, Mario M, Qi L, Yu Shrike Z, Su Ryon S, Giovanni C, Nasim A, Thomas DS, Colin EB, Anthony A, Mehmet RD, Ali K, Biofabrication 2016, 8, 014101. [Google Scholar]
- [100].Gori M, Simonelli MC, Giannitelli SM, Businaro L, Trombetta M, Rainer A, PLOS ONE 2016, 11, e0159729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Lau YY, Chen YH, Liu TT, Li C, Cui X, White RE, Cheng KC, Drug Metab Dispos 2004, 32, 937. [PubMed] [Google Scholar]
- [102].Li AP, Bode C, Sakai Y, Chemico-Biological Interactions 2004, 150, 129. [DOI] [PubMed] [Google Scholar]
- [103].Wagner I, Materne E-M, Brincker S, Süßbier U, Frädrich C, Busek M, Sonntag F, Sakharov DA, Trushkin EV, Tonevitsky AG, Lauster R, Marx U, Lab on a chip 2013, 13, 3538. [DOI] [PubMed] [Google Scholar]
- [104].Esch MB, Ueno H, Applegate DR, Shuler ML, Lab on a chip 2016, 16, 2719. [DOI] [PubMed] [Google Scholar]
- [105].Satoh T, Sugiura S, Shin K, Onuki-Nagasaki R, Ishida S, Kikuchi K, Kakiki M, Kanamori T. J. L. o. a. C., Lab on a chip 2018, 18, 115. [DOI] [PubMed] [Google Scholar]
- [106].Bauer S, Wennberg Huldt C, Kanebratt KP, Durieux I, Gunne D, Andersson S, Ewart L, Haynes WG, Maschmeyer I, Winter A, Ämmälä C, Marx U, Andersson TB, Scientific reports 2017, 7, 14620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Skardal A, Murphy SV, Devarasetty M, Mead I, Kang H-W, Seol Y-J, Shrike Zhang Y, Shin S-R, Zhao L, Aleman J, Hall AR, Shupe TD, Kleensang A, Dokmeci MR, Jin Lee S, Jackson JD, Yoo JJ, Hartung T, Khademhosseini A, Soker S, Bishop CE, Atala A, Scientific Reports 2017, 7, 8837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Li L, Wang H, Ong ZY, Xu K, Ee PLR, Zheng S, Hedrick JL, Yang Y-Y, Nano Today 2010, 5, 296. [Google Scholar]
- [109].Swindle MM, Makin A, Herron AJ, Clubb FJ, Frazier KS, 2012, 49, 344. [DOI] [PubMed] [Google Scholar]
- [110].Redfern WS, Bialecki R, Ewart L, Hammond TG, Kinter L, Lindgren S, Pollard CE, Rolf M, Valentin J-P, Journal of Pharmacological and Toxicological Methods 2010, 2, e29. [DOI] [PubMed] [Google Scholar]
- [111].Yokoo T, Ohashi T, Shen JS, Sakurai K, Miyazaki Y, Utsunomiya Y, Takahashi M, Terada Y, Eto Y, Kawamura T, Osumi N, Hosoya T, 2005, 102, 3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Jang KJ, Suh KY, Lab on a chip 2010, 10, 36. [DOI] [PubMed] [Google Scholar]
- [113].a) Homan KA, Kolesky DB, Skylar-Scott MA, Herrmann J, Obuobi H, Moisan A, Lewis JA, Scientific reports 2016, 6, 34845; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lin NYC, Homan KA, Robinson SS, Kolesky DB, Duarte N, Moisan A, Lewis JA, Proceedings of the National Academy of Sciences 2019, 116, 5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Ashammakhi N, Wesseling-Perry K, Hasan A, Elkhammas E, Zhang YS, Kidney Int 2018, 94, 1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Baudoin R, Griscom L, Monge M, Legallais C, Leclerc E, Biotechnology progress 2007, 23, 1245. [DOI] [PubMed] [Google Scholar]
- [116].a) Tiong HY, Huang P, Xiong S, Li Y, Vathsala A, Zink D, Mol Pharm 2014, 11, 1933; [DOI] [PubMed] [Google Scholar]; b) Perazella MA, Clinical Journal of the American Society of Nephrology 2009, 4, 1275. [DOI] [PubMed] [Google Scholar]
- [117].Weinberg E, Kaazempur-Mofrad M, Borenstein J, Int J Artif Organs 2008, 31, 508. [DOI] [PubMed] [Google Scholar]
- [118].Li Z, Jiang L, Tao T, Su W, Guo Y, Yu H, Qin J, Toxicology Research 2017, 6, 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Kim S, LesherPerez SC, Kim BCC, Yamanishi C, Labuz JM, Leung B, Takayama S, Biofabrication 2016, 8, 015021. [DOI] [PubMed] [Google Scholar]
- [120].Chang SY, Weber EJ, Sidorenko VS, Chapron A, Yeung CK, Gao C, Mao Q, Shen D, Wang J, Rosenquist TA, Dickman KG, Neumann T, Grollman AP, Kelly EJ, Himmelfarb J, Eaton DL, JCI Insight 2017, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Li Z, Jiang L, Zhu Y, Su W, Xu C, Tao T, Shi Y, Qin J, Toxicology in Vitro 2018, 46, 1. [DOI] [PubMed] [Google Scholar]
- [122].Miller PG, Shuler ML, Biotechnol Bioeng 2016, 113, 2213. [DOI] [PubMed] [Google Scholar]
- [123].Ozgen N, Terashima M, Aung T, Sato Y, Isoe C, Kakuta T, Saito A, Nephrology Dialysis Transplantation 2004, 19, 2198. [DOI] [PubMed] [Google Scholar]
- [124].Narayanan K, Schumacher KM, Tasnim F, Kandasamy K, Schumacher A, Ni M, Gao S, Gopalan B, Zink D, Ying JY, Kidney International 2013, 83, 593. [DOI] [PubMed] [Google Scholar]
- [125].Chang-Panesso M, Humphreys BD, Nature Reviews Nephrology 2016, 13, 39. [DOI] [PubMed] [Google Scholar]
- [126].Ashammakhi N, Ahadian S, Xu C, Montazerian H, Ko H, Nasiri R, Barros N, Khademhosseini A, Materials Today Bio 2019, 1, 100008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Scheller EL, Rosen CJ, Annals of the New York Academy of Sciences 2014, 1311, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Luo Y, Chen G-L, Hannemann N, Ipseiz N, Krönke G, Bäuerle T, Munos L, Wirtz S, Schett G, Bozec A, Cell Metabolism 2015, 22, 886. [DOI] [PubMed] [Google Scholar]
- [129].Bruce A, Evans R, Mezan R, Shi L, Moses BS, Martin KH, Gibson LF, Yang Y, PLoS ONE 2015, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Mohammad H, Masoud S, Amir A, Giuseppe S, Mohammad A, Mahin NZ, Tissue Engineering Part C: Methods 2017, 23, 72.28007011 [Google Scholar]
- [131].Bourgine PE, Klein T, Paczulla AM, Shimizu T, Kunz L, Kokkaliaris KD, Coutu DL, Lengerke C, Skoda R, Schroeder T, Martin I, Proceedings of the National Academy of Sciences 2018, 115, E5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Sieber S, Wirth L, Cavak N, Koenigsmark M, Marx U, Lauster R, Rosowski M, 2018, 12, 479. [DOI] [PubMed] [Google Scholar]
- [133].Ahn J, Cho C-S, Cho SW, Kang JH, Kim S-Y, Min D-H, Song JM, Park T-E, Jeon NL, Acta Biomaterialia 2018, 76, 154. [DOI] [PubMed] [Google Scholar]
- [134].Pauty J, Usuba R, Cheng IG, Hespel L, Takahashi H, Kato K, Kobayashi M, Nakajima H, Lee E, Yger F, Soncin F, Matsunaga YT, EBioMedicine 2018, 27, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Kim D, Finkenstaedt-Quinn S, Hurley KR, Buchman JT, Haynes CL, The Analyst 2014, 139, 906. [DOI] [PubMed] [Google Scholar]
- [136].Kim D, Lin YS, Haynes CL, Analytical chemistry 2011, 83, 8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Fede C, Fortunati I, Weber V, Rossetto N, Bertasi F, Petrelli L, Guidolin D, Signorini R, De Caro R, Albertin G, Ferrante C, Microvascular research 2015, 97, 147. [DOI] [PubMed] [Google Scholar]
- [138].Zhang W, Zhang YS, Bakht SM, Aleman J, Shin SR, Yue K, Sica M, Ribas J, Duchamp M, Ju J, Sadeghian RB, Kim D, Dokmeci MR, Atala A, Khademhosseini A, Lab on a Chip 2016, 16, 1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Hasenberg T,. T. Emulating the human vasculature in a Multi-Organ-Chip platform. Journal of Biotechnology 2018, 216, 1. [DOI] [PubMed] [Google Scholar]
- [140].Rigat-Brugarolas LG, Elizalde-Torrent A, Bernabeu M, De Niz M, Martin-Jaular L, Fernandez-Becerra C, Homs-Corbera A, Samitier J, del Portillo HA, Lab on a Chip 2014, 14, 1715. [DOI] [PubMed] [Google Scholar]
- [141].a) Chen Z, Meng H, Xing GM, Chen CY, Zhao YL, Int J Nanotechnol 2007, 4, 179; [Google Scholar]; b) Oberdörster G, Maynard A, Donaldson K, Castranova V, Fitzpatrick J, Ausman K, Carter J, Karn B, Kreyling W, Lai DJP, toxicology f., Particle and fibre toxicology 2005, 2, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Rigat-Brugarolas LG, Elizalde-Torrent A, Bernabeu M, De Niz M, Martin-Jaular L, Fernandez-Becerra C, Homs-Corbera A, Samitier J, del Portillo HA, Lab on a chip 2014, 14, 1715. [DOI] [PubMed] [Google Scholar]
- [143].Kang JH, Super M, Yung CW, Cooper RM, Domansky K, Graveline AR, Mammoto T, Berthet JB, Tobin H, Cartwright MJ, Watters AL, Rottman M, Waterhouse A, Mammoto A, Gamini N, Rodas MJ, Kole A, Jiang A, Valentin TM, Diaz A, Takahashi K, Ingber DE, Nature Medicine 2014, 20, 1211. [DOI] [PubMed] [Google Scholar]
- [144].Guengerich FP, Drug Metabolism and Pharmacokinetics 2011, 26, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].a) Yang F, Gao C, Wang P, Zhang G-J, Chen Z, Lab on a Chip 2016, 16, 1106; [DOI] [PubMed] [Google Scholar]; b) Baker M, Nature Methods 2010, 8, 47. [DOI] [PubMed] [Google Scholar]
- [146].a) Low LA, Tagle DA, Experimental Biology and Medicine 2017, 242, 1573; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kimura H, Sakai Y, Fujii T, Drug Metabolism and Pharmacokinetics 2018, 33, 43; [DOI] [PubMed] [Google Scholar]; c) Bovard D, Iskandar A, Luettich K, Hoeng J, Peitsch MC, Toxicology Research and Application 2017, 1, 2397847317726351. [Google Scholar]
- [147].a) Loskill P, Marcus SG, Mathur A, Reese WM, Healy KE, PLoS One 2015, 10, e0139587; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Fujii T, Sando Y, Higashino K, Fujii Y, Lab on a chip 2003, 3, 193; [DOI] [PubMed] [Google Scholar]; c) Yuen PK, Lab on a chip 2008, 8, 1374. [DOI] [PubMed] [Google Scholar]
- [148].Fetah K, Tebon P, Goudie MJ, Eichenbaum J, Ren L, Barros N, Nasiri R, Ahadian S, Ashammakhi N, Dokmeci MR, Khademhosseini A, Progress in Biomedical Engineering 2019, 1, 012001. [Google Scholar]
- [149].a) Ashammakhi N, Ahadian S, Darabi MA, El Tahchi M, Lee J, Suthiwanich K, Sheikhi A, Dokmeci MR, Oklu R, Khademhosseini A, Adv Mater 2019, 31, e1804041; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ashammakhi N, Darabi MA, Pountos I, J Craniofac Surg 2019, 30, 623. [DOI] [PubMed] [Google Scholar]
- [150].Kieninger J, Weltin A, Flamm H, Urban GA, Lab on a chip 2018, 18, 1274. [DOI] [PubMed] [Google Scholar]
- [151].Wikswo JP, Curtis EL, Eagleton ZE, Evans BC, Kole A, Hofmeister LH, Matloff WJ, Lab on a chip 2013, 13, 3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Wikswo JP, Block F, Cliffel DE, Goodwin CR, Marasco CC, Markov DA, McLean DL, McLean JA, McKenzie JR, Reiserer RS, IEEE Transactions on Biomedical Engineering 2013, 60, 682. [DOI] [PMC free article] [PubMed] [Google Scholar]