Abstract
Coronavirus disease 2019, known as COVID-19, is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The early, sensitive and specific detection of SARS-CoV-2 virus is widely recognized as the critical point in responding to the ongoing outbreak. Currently, the diagnosis is based on molecular real time RT-PCR techniques, although their implementation is being threatened due to the extraordinary demand for supplies worldwide. That is why the development of alternative and / or complementary tests becomes so relevant. Here, we exploit the potential of mass spectrometry technology combined with machine learning algorithms, for the detection of COVID-19 positive and negative protein profiles directly from nasopharyngeal swabs samples. According to the preliminary results obtained, accuracy = 67.66 %, sensitivity = 61.76 %, specificity = 71.72 %, and although these parameters still need to be improved to be used as a screening technique, mass spectrometry-based methods coupled with multivariate analysis showed that it is an interesting tool that deserves to be explored as a complementary diagnostic approach due to the low cost and fast performance. However, further steps, such as the analysis of a large number of samples, should be taken in consideration to determine the applicability of the method developed.
Keywords: Mass spectrometry, MALDI-TOF, Machine learning, COVID-19, SARS-CoV-2
1. Introduction
The novel coronavirus disease 2019, known as COVID-19, is caused by the SARS-CoV-2 virus and has been declared a pandemic by the World Health Organization on March 12th following its emergence in Wuhan, China (Yang et al., 2020). As of April 4th 2020, there were more than 1.2 million confirmed cases of COVID-19 in 175 countries, with more than 65,000 deaths (COVID-19 Map-Johns Hopkins Coronavirus Resource Center, 2020). SARS-CoV-2 is one of four new pathogenic viruses which have jumped from animal hosts to humans in the past 20 years, and the current pandemic alerts the entire scientific community about the need for research in different fields to help control it in a short time.
Clinical presentation of COVID-19 ranges from mild to severe illness with a high proportion of the population showing no symptoms but being equally infectious (Zhou et al., 2020). Together, these features have led to intensive lockdown measures in most countries with the aim to restrict the spread of the virus, limit the burden on healthcare systems and reduce mortality rate. In parallel, there has been an extraordinary response from the scientific community. These collective efforts aim to understand the pathogenesis of the disease, to evaluate treatment strategies and to develop a vaccine at unprecedented speeds in order to minimize its impact on individuals and on the global economy (Li, 2016; Wenzhong and Hualan, 2020).
The early, sensitive and specific detection of SARS-CoV-2 virus is currently widely recognized as the critical point in responding to the ongoing outbreak. Since the testing capacity of real time RT-PCR methodology is being challenged due to the extraordinary global demand of supplies such as RNA extraction kits and PCR reagents, alternative and/or complementary testing assays need to be deployed now in an effort to accelerate our understanding of COVID-19 disease (Chin et al., 2020; Antezack et al., 2020).
Recently, matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) has been used in many clinical laboratories with different applications. The peptide or protein MS fingerprint of a sample can be generated and stored in a library for further identification. In addition, MALDI-TOF MS also provides an alternative solution for molecular typing methods, thus enabling to detect an outbreak or a transmission route in time.
The aim of this work was to assess the potential of MALDI-TOF MS technology to create mass spectra directly from nasopharyngeal swabs in order to find specific discriminatory peaks by using machine learning algorithms, and whether those peaks were able to differentiate COVID-19 positive samples from COVID-19 negative samples, with the application of different approaches for the analysis.
2. Materials and methods
2.1. Sample preparation and MALDI-TOF data acquisition
2.1.1. Samples
All samples of nasopharyngeal swab (synthetic fiber swabs with plastic or wire shafts) in 2 ml of saline solution were submitted to the Reference Respiratory Virus Laboratory at INEI-ANLIS Dr Carlos G. Malbrán. The gold standard methodology consists in the detection of the specific genes (RdRp, E and N) of the virus by real-time RT-PCR (Corman et al., 2020). Then the samples are stored at −20 °C until their use in MALDI-TOF MS.
For the assay, samples of 311 patients were analyzed. At the moment of generating the spectra, the samples were thawed at room temperature and gently shaken. Without another prior enrichment, 1ul of the sample was spotted on each well of the steel plate (MSP 96 target ground steel; Bruker Daltonics), by triplicate, then allowed to dry for a few minutes at room temperature and covered with 1ul of commercial HCCA matrix (a solution containing α-cyano-4-hydroxycinnamic acid diluted into 500 μL of acetonitrile, 250 μL of 10 % trifluoroacetic acid and 250 μL of HPLC grade water). After the wells dried, the MALDI plate was introduced into the MicroFlex LT instrument version 3.4 (Bruker Daltonics, Bremen, Germany).
All manipulations were performed under certified class II biological safety cabinet TELSTARTM BIO IIA (Thermo Fischer Scientific, Villebon sur Yvette, France) and wearing all the appropriate personal protective equipment (PPE) required to comply with biosafety standards (World Health Organization, 2020).
2.1.2. Spectra acquisition
The mass spectra were acquired manually using FlexControl software v3.4 (Bruker Daltonics, Bremen, Germany) in the OFF mode by triplicate for each sample. Data were collected between 2000−20000 Da in linear positive-ionization mode. Each spectrum was a sum of 240 laser shots collected in increments of 40. Data acquisition was carried out at 40 % of the maximum laser energy. The platform was previously calibrated according to the manufacturer’s instructions using the Bruker Daltonics Bacterial Test Standard (Bruker Daltonics, Bremen, Germany).
All spectra collected were post processed using the Flex Analysis v3.4 software (Bruker Daltonics, Bremen, Germany), by the multiple spectrum display for spectra comparison and analysis (Flex Analysis 3.4 User Manual®).
2.2. MALDI-TOF MS spectra analysis
2.2.1. Database development. MSP library construction
Those spectra with high intensity and low noise peaks were selected to build an “in-house” database with Maldi Biotyper OC V3.1 software (Bruker Daltonics, Bremen, Germany): 20 Main Spectrum Profiles (MSPs), including 9 COVID-19 positive samples, 8 COVID-19 negative samples and 3 positive samples for other respiratory virus, were created from the proteomic data generated and according to manufacturer’s instructions. In order to incorporate the MSPs into the additional database, they had to meet the following criteria: to have at least 40 peaks conserved with a frequency of 100 % for each mass. This new database generated from local samples was named BE COVID-19.
Once the 20 MSPs were generated, an MSP dendrogram was constructed to assess the relatedness of these MSPs using default settings (Cipolla et al., 2018). See Fig. 1 .
Fig. 1.
Main spectra profiles (MSPs) based dendrogram of the 20 samples supplemented in the new in-house database. The horizontal axis of the dendrogram represents the calculated distance in the clustering analysis, displayed in relative units, corresponding to the similarity of MS spectra. The dendrogram was created using Biotyper v3.0 software.
2.3. Potential biomarkers detection
Mass spectrometry-based search for biomarker patterns is widely recognized (Pusch et al., 2003). Here, we use two different software to detect characteristic peaks of COVID-19 positive samples versus COVID-19 negativesamples. These potential biomarkers were searched based on the MSPs created.
2.3.1. Flex analysis v3.4 software analysis
Flex Analysis is the post processing software for spectra acquired with the Bruker time-of-flight mass spectrometers of the flex series, and proteomic data can be delivered in form of spectra and peak lists to further interactive data processing. In brief, spectra files from MSPs were exported as mzXML files using CompassXport CXP3.0.5. (Bruker Daltonics, Bremen, Germany). The flex programs support three different detection algorithms: Centroid, SNAP, and Sum peak finder. Centroid is often used in case of protein spectra and benefits from defining mass accuracy, which uses the first and second derivative to detect a peak. For the peak position, a specific cut off level above the baseline, a value around 80 %, was used. The result is stored in the peak list and the mass spectrum is displayed with the detected mass peak labels. Flex Analysis offers different parameters that can be displayed in the Mass List (Area, Background Peak, Chi Square, FWHM, Intensity, Relative intensity, Resolution and Signal to Noise), this peak list was exported to Excel (Microsoft, Redmond, WA) to analyze potential biomarkers. Also, a visual analysis of the spectra was performed, searching for significant differences between the two study groups.
2.3.2. ClinPro tools software
Spectra files from MSPs were loaded into the ClinPro Tools software (version 3.0, Bruker Daltonik GmbH, Bremen, Germany). The data preprocessing steps, including baseline subtraction, smoothing, and recalibration, were set as default for all analyses (Bruker Daltonik GmbH, 2011; Camoez et al., 2016; Zhang et al., 2015). Characteristic peaks among different profiles (Class 1= COVID-19 Positive Samples; Class 2= COVID-19 Negative Samples) were selected and sorted through several statistical tests, including the t-test, analysis of variance (ANOVA), the Wilcoxon or Kruskal–Wallis (W/KW) test, and the Anderson–Darling (AD) test. A P-value of 0.05 was set as the cutoff (Wang et al., 2018):
If P was <0.05 in the AD test, a characteristic peak was selected if the corresponding value of P in the W/KW test was also <0.05. When P was 0.05 in the AD test, then a characteristic peak was selected if the corresponding value of P in ANOVA was also <0.05 (Stephens, 1974). (Supplementary Table S1).
The two-dimensional distribution plot of characteristic peaks of each class can be seen in Fig. 2 .
Fig. 2.
2D Peak Distribution Plot of 2-class model (MSP database). This plot displays the distribution of two selected peaks in the non-excluded spectra on the loaded model generation classes. The data is shown on a two-dimensional plane. By default, the first two (best separating) peaks of the current statistic sort order are displayed. The ellipses represent the standard deviation of the class average of the peak area/intensities. The x-axis shows the peak area/intensity values with respect to the most important peak in accordance to the p-value, and the y-axis the peak area/intensity values for the second most important peak, respectively. The axis measures are given in arbitrary units which are chosen automatically to fit the plot optimal in the plane. Plot obtained by ClinPro Tools v3.0.
Biomarker peaks were identified by pairwise comparison of classes using the “Peak Statistic Table” function in ClinPro Tools followed by manual confirmation that the same peaks were distinguishable using Flex Analysis (Khot and Fisher, 2013); the discriminatory power for each putative biomarker was further described via analysis of area under the receiver operating characteristic (ROC) curve (AUC). The ROC curve gives a graphical overview about the specificity and the sensitivity of a test, and in this case an evaluation of the discrimination quality of a peak. An AUC value of 0 indicates that the considered peak is not discriminating, while an AUC of 1 indicates that the considered peak is discriminating.
2.4. Classifier models based on machine learning
The aim of model generation is to determine a common signature among spectra of each of the model classes in such a way that spectra of test isolates can be classified. This approach was performed using ClinPro Tools functions: data preparation, model generation, and spectra classification. Data preparation involved baseline subtraction (top hat; 10 % minimal baseline width), normalization (total ion current), recalibration (1,000 ppm maximal peak shift and 30 % match to calibrant peaks, with exclusion of spectra that could not be recalibrated), average spectrum calculation (resolution 800), average peak list calculation (signal-to-noise threshold 5), peak calculation in the individual spectra, and normalization of peak lists.
Classification models were generated using GA/ k-nearest neighbor algorithm.
2.4.1. Establishment of the training set
432 Spectra (corresponding to: 55 positive samples, 57 negative samples, 24 samples positive for influenza virus, named from now as FLU and 8 other respiratory virus samples) constituted the training group which was used to build several classification models. To determine the accuracy of the class prediction model, the software offers cross-validation and recognition capability. Cross-validation is a measure of the reliability of a calculated model and can be used to predict how a model will behave in the future. This method is used to evaluate the performance of a classifier for a given data set under a given parameterization. Recognition capability describes the performance of an algorithm, i.e., the proper classification of a given data set.
In this study, multiclass models for classifying an isolate were designed:
1- A two-class model named A (Class 1= COVID-19 positive samples; Class 2= COVID-19 Negative samples).
2- A three-class model named B (Class 1 = COVID-19 positive samples; Class 2 = COVID-19 negative samples and Class 3 = positive samples for other respiratory virus).
3- A two-class model named C (Class 1= COVID-19 positive samples; Class 2= Influenza positive samples). The design of this model was decided because many FLU samples were classified as COVID-19 positive when the 3 class model was used, so in order to optimize the results, a specific COVID-19 versus FLU model was created and applied only when a sample was positive either for COVID-19 or other respiratory virus in the 3 class model.
Pretreatment, normalization, baseline subtraction, peak defining (range 1960−20000 m/z), recalibration; and then, the automatic comparison of multiple spectra were performed. Values of m/z from the average spectra of each class and informative peaks were identified according to their statistical significance, as determined by the different statistical tests supported by ClinProTools: Anderson–Darling test, t-/ analysis of variance (ANOVA) test and Wilcoxon/Kruskal–Wallis tests. Informative peaks were those showing a significant difference between the classes as mentioned above. The best TOP TEN peaks are summarized in Table 1 (Full table can be found in Supplementary material, Table S2).
Table 1.
Characteristic MALDI-TOF MS peaks (best top ten) obtained by ClinPro Tools software for each model.
| 2 Class model A |
3 Class model B |
2 Class model C |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mass | DAve | PTTA | PWKW | PAD | Mass | DAve | PTTA | PWKW | PAD | Mass | DAve | PTTA | PWKW | PAD |
| 3372,19 | 7,85 | 0,000171 | 0,0000138 | < 0.000001 | 3443,31 | 13,35 | 0,00149 | 0,00000568 | < 0.000001 | 3443,19 | 13,35 | 0,00332 | 0,000319 | < 0.000001 |
| 3443,14 | 7,79 | 0,00834 | 0,000042 | < 0.000001 | 3372,29 | 11,52 | 0,000023 | 0,00000427 | < 0.000001 | 3372,15 | 11,52 | 0,00108 | 0,00059 | < 0.000001 |
| 4966,6 | 4,79 | 0,000277 | 0,000093 | < 0.000001 | 3487,4 | 11,42 | 0,00969 | 0,0359 | < 0.000001 | 4965,83 | 6,69 | 0,0000165 | 0,00000382 | < 0.000001 |
| 5236,08 | 3,88 | 0,00584 | 0,000764 | < 0.000001 | 5236,12 | 8,63 | < 0.000001 | < 0.000001 | < 0.000001 | 5235,02 | 4,62 | 0,00000929 | 0,000128 | < 0.000001 |
| 4078,24 | 3,49 | 0,000011 | < 0.000001 | 0 | 4966,4 | 6,74 | 0,0000157 | < 0.000001 | < 0.000001 | 4985,26 | 4,2 | 0,00000651 | 0,00000106 | < 0.000001 |
| 3487,1 | 3,24 | 0,0737 | 0,0191 | < 0.000001 | 4985,38 | 4,12 | 0,00000296 | < 0.000001 | < 0.000001 | 3465,33 | 3,61 | 0,0384 | 0,0414 | < 0.000001 |
| 4985,73 | 3,07 | 0,000033 | 0,0000101 | < 0.000001 | 5137,86 | 3,89 | < 0.000001 | < 0.000001 | < 0.000001 | 3394,17 | 3,3 | 0,00856 | 0,000829 | < 0.000001 |
| 3359,22 | 2,66 | 0,000032 | 0,000015 | < 0.000001 | 3465,33 | 3,6 | 0,0183 | 0,00758 | < 0.000001 | 4939,73 | 3,06 | < 0.000001 | 0,0000776 | < 0.000001 |
| 3393,78 | 2,54 | 0,00182 | 0,00418 | < 0.000001 | 5382,94 | 3,58 | < 0.000001 | 0,00000425 | < 0.000001 | 5381,66 | 2,93 | 0,0000266 | 0,0117 | < 0.000001 |
| 3475,85 | 2,48 | 0,000608 | 0,000148 | < 0.000001 | 5157,12 | 3,52 | < 0.000001 | < 0.000001 | < 0.000001 | 4077,47 | 2,5 | 0,00881 | 0,00794 | < 0.000001 |
DAve: Difference between the maximal and the minimal average peak intensity of all classes.
PTTA: P-value obtained through t-test. PWKW: P-value obtained through Wilcoxon/Kruskal-Wallis test. PAD: P-value obtained through Anderson-Darling test.
2.4.2. External validation test of the classification models
To show the efficacy and accuracy of the algorithm developed, an external validation was performed with a classification set of 501 spectra (n:167), different from the samples used to create the training set, using the “Classify” function in ClinPro Tools.
For each sample from the validation groups, a corresponding spectrum was presented to the selected classification model. Then, the software returned a result that was compared with the actual reference techniques.
2.5. Statistical analysis
For evaluating the performance, accuracy, sensitivity, specificity, positive prediction and negative prediction were calculated (ClinPro Tools 3.0: User Manual, 2011).
3. Results
3.1. Evaluation of the novel “in-house” database
The novel database was challenged with 223 (88 COVID-19 positive and 135 COVID-19 negatives) previously-characterized new samples different than those used to create it. They were processed by triplicate and the wells were read in the MALDI Biotyper OC v3.1 software for an offline classification.
MALDI-TOF MS identification was considered correct when the result obtained from BE-COVID-19 database presented a score value ≥ 2.0, according to the manufacturer's recommendations with the Bruker score system.
Only 22 % of the samples (50/223; 16 COVID-19 positive and 34 COVID-19 negative samples) passed the cut-off mentioned above.
According to the identification of the samples, we calculated the performance values of the complementary database that are presented in Table 2 .
Table 2.
Parameters of the “in-house” Database evaluated (n = 50).
| Parameters Evaluated | (%) | 95 % CI (%) |
|---|---|---|
| Accuracy | 38.0 | (24.65–52.83) |
| Specificity | 26.5 | (12.88–44.36) |
| Sensitivity | 62.5 | (35.43–84.80) |
| Positive Prediction Value | 28.6 | (20.65–38.07) |
| Negative Prediction Value | 60.0 | (39.19–77.74) |
3.2. Detection of potential biomarkers
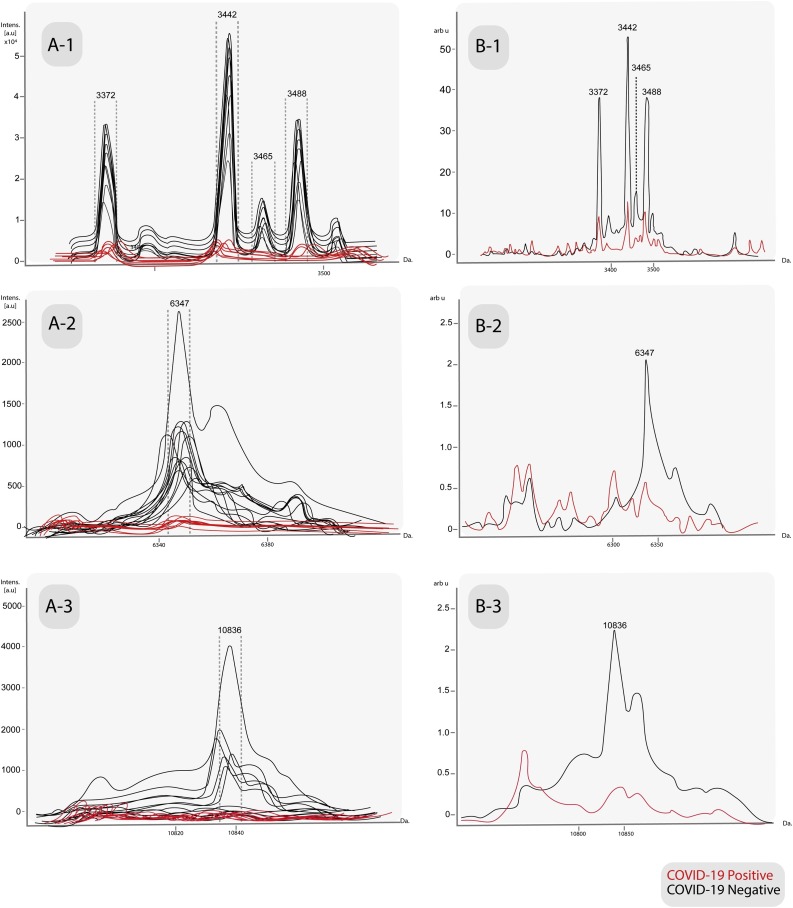
Flex Analysis v3.4 software revealed potential peaks of negativity which were not detected in the positive samples: 3372 Da, 3442 Da, 3465 Da, 3488 Da, 6347 Da and 10836 Da (Fig. 3 ). These peaks were statistically significant and also detected with reproducible intensity in the average spectrum of that same class in ClinPro Tools (Table 3 ); the peak at m/z 3488 was also found but it did not pass the cutoff mentioned above.
Fig. 3.
A- Characteristic peaks in the individual spectra of the MSPs database among COVID-19 positive samples versus COVID-19 negative samples, obtained by manual analysis in Flex Analysis v3.4 software. B- Average spectra of same peaks, obtained by ClinProTools v3.0 software. A/B-1: 3372 Da, 3442 Da, 3465 Da and 3488 Da. A/B-2: 6347 Da. A/B-3:10836 Da.
Table 3.
Single-peak analysis for the discrimination of COVID-19 positive samples from the negative samples (n = 20), between Flex Analysis v3.4 and ClinPro Tools v3.0.
| ClinPro Tools |
FlexAnalysis |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Mass | PTTA | PWKW | PAD | AUC | AUC* | Sensitivity (%) | Specificity (%) | PPV (%) | PPN (%) |
| 3372,3 # | 0.0133 | 0.0199 | 0.05450 | 0.87 | 0.667 | 33.33 | 100.00 | 100.00 | 64,71 |
| 3443,28 # | 0.0133 | 0.0222 | 0.04480 | 0.86 | 0.667 | 33.33 | 100.00 | 100.00 | 64.71 |
| 3465,6 # | 0.0133 | 0.0244 | 0.05790 | 0.85 | 0.742 | 66.67 | 81.82 | 75.00 | 75.00 |
| 6347,57 # | 0.0146 | 0.0096 | 0.02110 | 0.92 | 0.636 | 100.00 | 27.27 | 52.94 | 100.00 |
| 10836,83 # | 0.0885 | 0,0236 | 0.00019 | 0.86 | 0.727 | 100.00 | 45.45 | 60.00 | 100.00 |
PTTA: P-value obtained through t-test. PWKW: P-value obtained through Wilcoxon/Kruskal-Wallis test. PAD: P-value obtained through Anderson-Darling test. *AUCs were obtained from a ROC curve constructed using Eng, J. ROC analysis: web-based calculator for ROC curves. from http://www.jrocfit.org. # the analysis include only these peaks, due to the other peaks of the table were not significant when the manual corroboration in Flex Analysis software was made.
All characteristic MALDI-TOF MS peaks from the MSPs, obtained with the ClinPro Tools software can be found in Supplementary material, Table S3.
None of the peaks found could be molecularly attributed to virus-specific proteins because they were conspicuously detected in most of the negative samples; this could be due to some type of host-virus interaction that requires further study in the future.
3.3. Machine learning models
A total of three GA algorithms (optimized by adjusting the number of neighbors for a k-nearest neighbor classification) were applied for a classifier model construction using spectral data from the training group; generated based on the comparison of the recognition capability and cross validation of all the models and exhibiting the best efficiency in the classification of test patients. The results of Recognition Capacity (RC) and Cross Validation (CV) values of all models used are summarized in Table 4 . The Integration Regions for the classification according to the different algorithms for each model are in Supplementary material, Table S4.
Table 4.
Complete results of Recognition capacity and Cross validation derived from the classification models calculated.
| Classifier Models | RC (%) | CV (%) |
|---|---|---|
| 2 Class model A | 100 | 92.98 |
| 3 Class model B | 100 | 87.16 |
| 2 Class model C | 100 | 92.87 |
RC: Recognition Capacity; CV: Cross Validation.
This novel approach based on Machine Learning algorithms with the combination of Potential Biomarkers was evaluated with 167 clinical samples and the results of the performance are summarized in Table 5 .
Table 5.
Parameters of the Machine Learning combined with Potential Biomarkers. (n = 167).
| Parameters Evaluated | (%) | 95 % CI (%) |
|---|---|---|
| Accuracy | 67.66 | (60.00–74.69) |
| Specificity | 71.72 | (61.78–80.31) |
| Sensitivity | 61.76 | (49.18–73.29) |
| Positive Prediction Value | 60.00 | (51.01–68.37) |
| Negative Prediction Value | 73.20 | (66.33–79.10) |
The table of the results obtained from clinical samples (n: 167) by Machine Learning and detection of Biomarkers compared to the current reference technique (RT-PCR) can be found in the Supplementary material, Table S5.
4. Discussion
This study aimed to evaluate the application of MALDI-TOF MS to COVID-19 identification, and to the detection of specific biomarkers, differentiating between virus-infected and uninfected patients, thus extending the use of this technology to a novel application.
At the moment of writing this manuscript, the only diagnostic methodology available in our laboratory was real-time reverse transcription-PCR (RT-PCR), which, as is known in developing countries, can be an expensive technique, and reagents are often difficult to obtain, even more in the current situation of demand. According to the values of performance achieved, we consider mass spectrometry as a potential alternative tool that can complement the molecular reference techniques.
We are aware that this outbreak has had a major impact on clinical microbiology laboratories, so new diagnosis methodologies are developed every day, some of which involves new genes RT-PCR targets to improve the molecular diagnosis (Chan et al., 2020), and others involving serological assays, such as rapid lateral flow assays (Li et al., 2020). However, MALDI-TOF MS is a simple, inexpensive and fast technique that analyses protein profiles with a high reliability rate, and could be used as a rapid screening method in a large population (Croxatto et al., 2012). These preliminary results suggest that MALDI-TOF MS coupled with ClinPro Tools software could represents an interesting alternative for diagnosis of SARS-CoV-2 virus as long as we could enhance the values of performance obtained in this first approach. To achieve that goal, we are already working on increasing the numbers of samples, evaluating different extraction methods, and making improvements of the machine learning algorithms.
Nevertheless, this study using MALDI-TOF MS combined with machine learning algorithms could be, as far as we know, a revolutionary alternative as a screening assay that deserves further development because it could greatly improve upon currently available methods.
5. Conclusions
The identification of specific biomarkers for each peak or group of peaks represents a difficult and demanding task that requires further studies from a large number of high-quality spectra.
Based on the promising preliminary results, this work constitutes the basis and encourages researchers to explore the potential of MALDI-TOF MS in order to assess the feasibility of this technology, widely available in clinical microbiology laboratories around the world, as a fast and inexpensive SARS-CoV-2 diagnostic tool.
In conclusion, in order to prevent the risks of a shortage of screening means, we propose to develop an innovative alternative strategy, PCR-free, based on the detection of specific protein signatures in human nasopharyngeal swabs by MALDI-TOF MS profiling to detect individuals infected with SARS-CoV2.
The development of this test is based on machine learning techniques, which involve "training" a mathematical model, in which the results of the mass spectrometry analysis of samples was used to determine the information needed to distinguish a sample from a SARS-CoV2-infected patients from no infected.
We could obtain peptide patterns from clinical samples with MALDI-TOF MS, and constructed classification models with moderate sensitivity and specificity. These results, though they need to be further refined, support the potential of MALDI-TOF MS technology, and provide a promising basis to extend its application to many other agents of medical interest and it can be a valuable tool for early routine screening approaches.
Author statement
Dr Maria Florencia Rocca is the corresponding author of this manuscript.
She is the responsable of the MALDITOF platform in the National Reference Institute in Argentina, she coordinates the National Network of Mass Spectrometry ReNaEM and she has recently been named as the Latin America coordinator of Microbenet by the CDC, Atlanta.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We gratefully thank the collaboration of Dr. Jorge Pidone, Dra. Natalia Carrion and group of work from Naval Hospital (Buenos Aires, Argentina).
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jviromet.2020.113991.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Antezack A., Chaudet H., Tissot-Dupont H., Brouqui P., Monnet-Corti V. Rapid diagnosis of periodontitis, a feasibility study using MALDI-TOF mass spectrometry. PLoS One. 2020;15(3) doi: 10.1371/journal.pone.0230334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruker Daltonik GmbH . Bruker Daltonik GmbH; Bremen: 2011. ClinPro Tools User Manual Version 3.0. [Google Scholar]
- Camoez M., Sierra J.M., Dominguez M.A., Ferrer-Navarro M., Vila J., Roca I. Automated categorization of methicillin-resistant Staphylococcus aureus clinical isolates into different clonal complexes by MALDI-TOF mass spectrometry. Clin. Microbiol. Infect. 2016;22(2) doi: 10.1016/j.cmi.2015.10.009. 161.e1-161.e7. [DOI] [PubMed] [Google Scholar]
- Chan J.F., Yip C.C., To K.K., Tang T.H., Wong S.C., Leung K.H., Fung A.Y., Ng A.C., Zou Z., Tsoi H.W., Choi G.K., Tam A.R., Cheng V.C., Chan K.H., Tsang O.T., Yuen K.Y. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/hel real-time reverse Transcription-PCR assay validated in vitro and with clinical specimens. J. Clin. Microbiol. 2020;58(5):e00310–00320. doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin A., Chu J., Perera M., Hui K., Yen H.L., Chan M., Peiris M., Poon L. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe. 2020 doi: 10.1016/S2666-5247(20)30003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipolla L., Rocca M.F., Armitano R., Martinez C., Almuzara M., Faccone D., Vay C., Prieto M. Desarrollo y evaluación de una base de datos in house para la identificación rápida de Burkholderia contaminans por EM MALDI-TOF. Rev. Argent. Microbiol. 2018 doi: 10.1016/j.ram.2018.09.001. [DOI] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COVID-19 Map - Johns Hopkins Coronavirus Resource Center . 2020. COVID-19 Map - Johns Hopkins Coronavirus Resource Center.https://coronavirus.jhu.edu/map.html (Accessed 5 April 2020) [Google Scholar]
- Croxatto A., Prod’hom G., Greub G. Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol. Rev. 2012;36:380–407. doi: 10.1111/j.1574-6976.2011.00298.x. [DOI] [PubMed] [Google Scholar]
- Khot P.D., Fisher M.A. Novel approach for differentiating Shigella species and Escherichia coli by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2013;51(11):3711–3716. doi: 10.1128/JCM.01526-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Yi Y., Luo X., et al. Development and clinical application of a rapid IgM‐IgG combined antibody test for SARS‐CoV‐2 infection diagnosis. J. Med. Virol. 2020;92:1518–1524. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusch W., Flocco M.T., Leung S.M., Thiele H., Kostrzewa M. Mass spectrometry-based clinical proteomics. Pharmacogenomics. 2003;4(4):463–476. doi: 10.1517/phgs.4.4.463.22753. [DOI] [PubMed] [Google Scholar]
- Stephens M.A. EDF statistics for goodness of fit and some comparisons. JASA. 1974;69:730–737. [Google Scholar]
- Wang H.Y., Lien F., Liu T.P., Chen C.H., Chen C.J., Lu J.J. Application of a MALDI-TOF analysis platform (ClinProTools) for rapid and preliminary report of MRSA sequence types in Taiwan. PeerJ. 2018;6:e5784. doi: 10.7717/peerj.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzhong L., Hualan L. COVID-19: attacks the 1-beta chain of hemoglobin and captures the porphyrin to inhibit human heme metabolism. ChemRxiv Preprint. 2020 doi: 10.26434/chemrxiv.11938173.v8. [DOI] [Google Scholar]
- World Health Organization . World Health Organization; 2020. Rational Use of Personal Protective Equipment (PPE) for Coronavirus Disease (COVID-19): Interim Guidance, 19 March 2020.https://apps.who.int/iris/handle/10665/331498 (Accessed 5 April 2020) [Google Scholar]
- Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T., Wang Y., Pan S., Zou X., Yuan S., Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. Preprint. 2020 doi: 10.1016/S2213-2600(20)30079-5. https://www.thelancet.com/lancet/article/S2213-2600(20)30079-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Cao J., Li L., Liu Y., Zhao H., Li N., Li B., Zhang A., Huang H., Chen S., Dong M., Yu L., Zhang J., Chen L. Identification of urine protein biomarkers with the potential for early detection of lung cancer. Sci. Rep. 2015;5:11805. doi: 10.1038/srep11805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Zhang X., Qu J. Coronavirus disease 2019 (COVID-19): a clinical update. Front. Med. 2020:1–10. doi: 10.1007/s11684-020-0767-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.