Abstract
In a developing nervous system, endogenous electric field (EF) influence embryonic growth. We reported the EF-directed migration of both rat Schwann cells (SCs) and oligodendrocyte precursor cells (OPCs) and explored the molecular mechanism using RNA-sequencing assay. However, previous studies revealed the differentially expressed genes (DEGs) associated with EF-guided migration of SCs or OPCs alone. In this study, we performed joint differential expression analysis on the RNA-sequencing data from both cell types. We report a number of significantly enriched gene ontology (GO) terms that are related to the cytoskeleton, cell adhesion, and cell migration. Of the DEGs associated with these terms, nine up-regulated DEGs and 32 down-regulated DEGs showed the same direction of effect in both SCs and OPCs stimulated with EFs, while the remaining DEGs responded differently. Thus, our study reveals the similarities and differences in gene expression and cell migration regulation of different glial cell types in response to EF stimulation.
Subject terms: Biophysics, Cell biology, Computational biology and bioinformatics, Genetics, Physiology
Introduction
In a developing nervous system, endogenous electric fields (EFs) affect embryonic growth1–4. Interestingly, EF vectors seem to be aligned with major embryonic axes5. Additionally, electrical activity regulates the path-finding of growing axons and the formation of initial connections in the developing nervous system. For example, studies have reported the influences of electrical activity on the projection of growing thalamic axons towards their appropriate cortical target area6 and the formation of layer-specific connections by axons of cortical pyramidal neurons 7.
Neural trauma causes neural cell necrosis, demyelination, and degeneration of axons, which lead to functional loss. Glial cell transplantation may enhance remyelination at the lesion. Oligodendrocytes and Schwann cells (SCs) are glial cells that myelinate axons in the central nervous system (CNS) and in the peripheral nervous system (PNS), respectively8–10. In post-spinal cord injury, the movement of oligodendrocyte precursor cells (OPCs) from gray and white matter to the lesion helps re-myelinate the axons11,12. Schwann cells can migrate to the injury lesion to myelinate spinal axons13,14. Both SCs and OPCs have been transplanted into wounded spinal cords, and the implantation of these cells enhanced spinal axonal regeneration15–17.
The effect of EFs on cell migration has been reported in a number of investigations. In previous studies, we found that both SCs18 and OPCs migrated toward the anodal pole of an applied EF19. The increase of directedness of anodal migration of these cells is in proportion to the strength of the applied EF (50 to 200 mV/mm). During directed cell migration, cells become polarized dynamically in response to guidance signals. The polarized cell shape is characterized by membrane ruffling and filopodia at the front edge20. The guided cell movement requires coordinated reorganization of the actin cytoskeleton and microtubules (MTs) in the leading and trailing processes and cell body. However, the molecular pathways regulating this critical process have not been elucidated.
To understand the signaling molecules that regulate the EF-guided migration of Schwann cells and OPCs, next-generation RNA sequencing (RNA-seq) was performed to identify the genes that play important roles in regulating directional cell migration18,19. In those studies, we determined the differentially expressed genes (DEGs) for SCs and OPCs subjected to EF stimulation. The RNA-seq assay of SCs identified 1045 up-regulated and 1636 down-regulated genes in cells with EF stimulation (100 mV/mm) versus control cells. A Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway assay showed that 21 pathways are down-regulated, while 10 pathways are up-regulated for EF-stimulated cells compared with control cells. Several cellular signaling pathways that are involved in the regulation of cell migration, including pathways regulating the actin cytoskeleton, focal adhesion, and PI3K-Akt were recognized. The RNA-seq assay of OPCs revealed that the mitogen-activated protein kinase (MAPK) pathway that signals cell migration was significantly up-regulated in EF-stimulated cells compared with control cells. Gene ontology (GO) enrichment analysis showed an enrichment of chemotaxis-related GO terms among down-regulated DEGs.
These studies revealed the differentially expressed genes that regulate the EF-guided migration of SCs or OPCs alone. However, the major similarities and differences in how EFs affect gene expression in different cell types have not been studied. In this study, we analyzed the DEGs in these two studies jointly and explored their function using GO terms, focusing on the cytoskeleton, cell adhesion, cell movement, and cell migration. Our results give insight into similarities and differences in DEGs and cell migration regulation of different glial cell types in response to EF stimulation.
Results
RSEM results
To better understand the similarities and differences in the response of Schwann cells and OPCs to an EF, we performed meta-analysis based on previously described RNA-seq data from two studies18,19. Four conditions were included in the analysis: EF-stimulated OPCs, control OPCs, EF-stimulated Schwann cells, and control Schwann cells. We identified 9364 genes that showed differential expression in at least one of these conditions. Of these DEGs, 1907 showed differential expression in at least one of the EF conditions and were sorted into eight categories based on the direction of expression change, if any, in each cell type (Supplemental Table 1), as follows: (1) EF-induced gene down-regulation in both OPCs and SCs (Both Down); (2) EF-induced gene up-regulation in both OPCs and SCs (Both Up); (3) EF-induced gene down-regulation in SCs and no change in OPCs (SC-Down/OPC-No Change); (4) EF-induced gene up-regulation in SCs and no change in OPCs (SC-Up/OPC-No Change); (5) EF-induced gene down-regulation in OPCs and no change in SCs (OPC-Down/SC-No Change); (6) EF-induced gene up-regulation in OPCs and no change in SCs (OPC-Up/SC-No Change); (7) EF-induced gene up-regulation in SCs and gene down-regulation in OPCs (SC-Up/OPC-Down); and (8) EF-induced gene up-regulation in OPCs and gene down-regulation in SCs (OPC-Up/SC-Down).
Table 1.
DEGs associated with GO terms related to cytoskeleton, cell adhesion, cell movement, and cell migration.
| Category | GO term | Gene number | Gene name |
|---|---|---|---|
| Both down | CC cytoskeleton | 32 | Apc2, Cdc42ep4, Dlgap5, Dsn1, Gtse1, Lasp1, Nckap5l, Sh2b2, Taok2, Tpx2, Triobp, Cdc7, Cep250, Cdk2, Ckap2, Dgkq, Espl1, Flnc, Fhod1, Klhl12, Klhl22, Kif20a, Kif2c, Mark4, Mcm2, Map3k11, Notch1, Nusap1, Plekhh3, Spag5, Timeless, Zw10 |
| Both up | MF focal adhesion | 9 | ADP-ribosylation factor 6(Arf6), Rho family GTPase 3(Rnd3), calponin 3(Cnn3), cortactin(Cttn), melanoma cell adhesion molecule(Mcam), plasminogen activator urokinase receptor(Plaur), poliovirus receptor(PVR), testin LIM domain protein(Tes), zyxin(Zyx) |
| OPC-down/SC-no change | CC cytoskeleton | 63 | Mtrr, Abl1, Bcar1, Clip2, Dennd2a, Dnaja3, Mdm1, Pms2, Pxk, Rimbp3, Racgap1, Src, Ssx2ip, Tbccd1, Xrcc2, Aif1l, Amot, Aurkb, Ambra1, Cenpe, Cenpf, Cenpj, Cep164, Cep55, Cep72, Ccnb1, Ccnf, Flna, Flnb, Gja1, Incenp, Ift80, Kif18b, Kif1c, Kif23, Kif4a, Kifap3, Knstrn, Lmnb1, Lmnb2, Lcp1, Mark2, Map4, Myo1c, Nphp4, Nf2, Nos2, Pik3r4, Plk1, Prc1, Ralbp1, Specc1l, Spdl1, Sybu, Ttc8, Top2a, Tmem214, Tsc2, LOC100909441, Tubd1, Tube1, Tubgcp2, Tubgcp5 |
| SC-up/OPC-no change | BP cell adhesion | 62 | Axl, Bcl2, Cd24, Cd44, Cited2, Ehd4, Ets1, L1cam, Lims2, Mb21d2, Pdlim1, Prdm1, Smad6, Actn4, Actn1, Ap3d1, Ajap1, Adgrg1, Bcan, Csnk1d, Cadm3, Cadm4, Col18a1, Ctgf, Cyp1b1, Dusp3, Epdr1, Efnb2, Ezr, Fermt2, Flrt2, Fbln2, Fstl3, Foxp1, Gcnt2, Hspa5, Hnrnpk, Has2, Il6st, Lama4, Lamc2, Lef1, Map2k1, Myh9l1, Nectin3, Nrg1, Parvb, Pxn, Postn, Pkp1, P2ry12, Ripk2, Runx2, Serpine1, Sdc4, Tspan5, Thbs1, Thbs2, Tinagl1, Tnfsf18, Vegfc, Vasn |
| BP cell migration | 53 | Arf4, Axl, Bcl2, Ccl22, Cd24, Cd44, Cited2, Ets1, Gata2, Sh3kbp1, Stard13, Abhd6, Actn4, Adgrg1, Clic4, Col18a1, Ctgf, Cyp1b1, Efnb2, Fgf2, Fgf7, Flrt2, Foxp1, Gdnf, Gcnt2, Hspa5, Hbegf, Has2, Igfbp3, Il6st, Lamc2, Lef1, Map2k1, Myh9l1, Nrg1, Nfe2l2, Ndel1, Postn, Plat, Pdgfb, Plxnd1, Pkn3, P2ry12, Serpine1, Shtn1, Sphk1, Sdc4, Thbs1, Tfap2a, Tnfsf18, Twist1, Tyk2, Vegfc | |
| CC focal adhesion | 29 | Akap12, Cd44, G3bp1, L1cam, Lims2, Pdlim1, Rab21, Arhgap22, Sh3kbp1, Actn4, Actn1, Efnb2, Ezr, Fermt2, Flrt2, Fzd2, Hspa5, Hnrnpk, Hmga1, Map2k1, Mprip, Myh9l1, Parvb, Pxn, Pabpc1, Pcbp2, Procr, LOC100360679, Sdc4 | |
| SC-down/OPC-no change | BP locomotion | 49 | Asap3, Cxcl6, Ephb4, Gli2, Ldb1, Six4, Slc9a3r1, Vangl2, Ank3, Aqp1, Celsr2, Col1a1, Col5a1, Csf1, Dst, Efnb1, Efnb3, Emp2, Erbb2, Epb41l4b, Fgf10, Foxo4, Hgf, Itga11, Itga4, Itgb4, Kif26b, Lama2, Lama5, Lamb1, Lpar1, Matn2, Mmp2, Msx2, Nefl, Nrp1, Nr2f2, Plekhg5, Pawr, Rreb1, Reck, Rffl, Sfrp2, Sema3b, Sema3g, Stat5a, Snai2, Tns1, Tgfbr3 |
| BP regulation of cell adhesion | 26 | Cd59, Ephb4, Lmo7, Ldb1, Ank3, Celsr2, Col1a1, Csf1, Efnb1, Efnb3, Emp2, Erbb2, Erbb3, Epb41l4b, Itga4, Kif26b, Lama2, Lama5, Lamb1, Mmp2, Nid1, Pawr, Rreb1, Stat5a, Snai2, Utrn |
Analysis of DEGs and GO analysis for OPCs and SCs subjected to EF stimulation
We performed GO term enrichment analysis on the DEGs in each of the eight categories. Gene ontology analysis provides information on the biological process (BP), molecular function (MF) and cellular component (CC) associated with particular genes. The significantly enriched GO terms of various DEGs in these categories were identified. GO terms that are related to the cytoskeleton, and adhesion and migration in the BP, MF, and CC classifications were of particular interest.
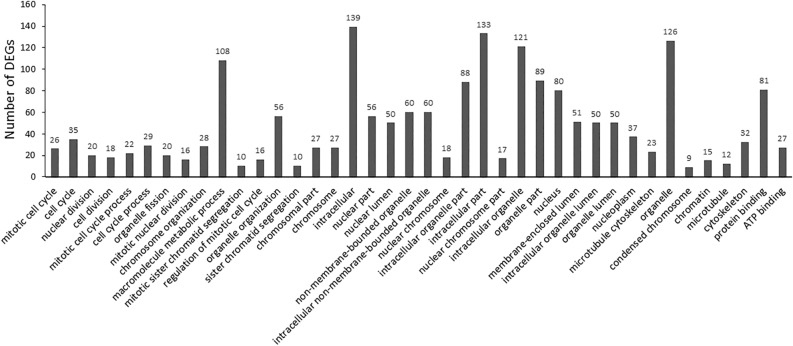
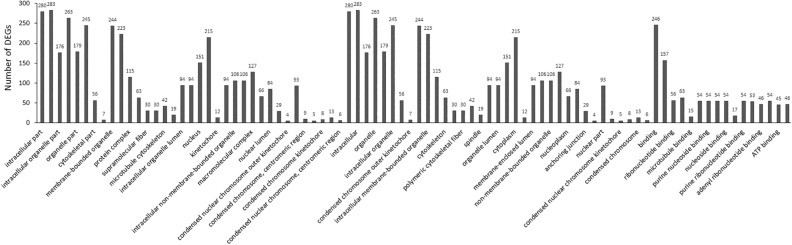
In the category of Both Down (Fig. 1), 193 DEGs of down-regulated gene were identified. In this category, the significantly enriched BP includes 14 terms, which are cell cycle and cell division-related terms, including mitotic cell cycle, cell division, and mitotic sister chromatid segregation. The significantly enriched CC terms (n = 24) are chromosome and intracellular-related terms as well as terms related to the cytoskeleton. The significantly enriched MF terms are protein binding and ATP binding.
Figure 1.
EF-induced gene down-regulation in both OPCs and SCs (both down).
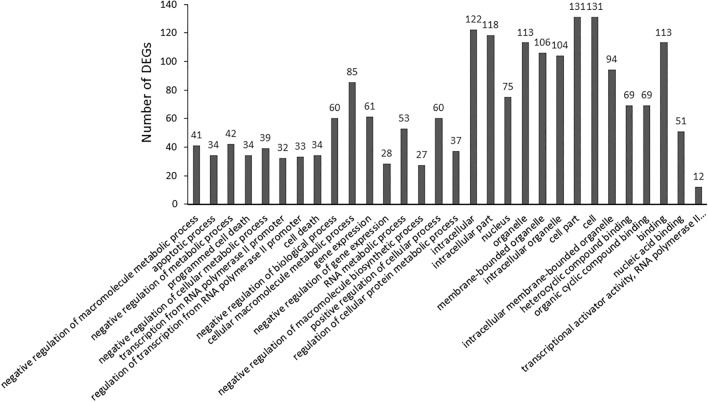
In the category of Both Up (Fig. 2), 154 DEGs were identified. The significantly enriched BP terms (n = 16) are negative regulation of macromolecule metabolic process, apoptotic process, and transcription from RNA polymerase II promoter-related terms. The significantly enriched CCs are intracellular, nucleus, and organelle-related terms (n = 9). Nine significantly up-regulated genes (Table 1) associated with focal adhesion (p = 0.9) were recognized, although this it is not a significantly enriched term. The significantly enriched MF terms are heterocyclic compound binding, organic cyclic compound binding, binding, nucleic acid binding transcriptional activator activity, and RNA polymerase II transcription regulatory region sequence-specific binding.
Figure 2.
EF-induced gene up-regulation in both OPCs and SCs (both up).
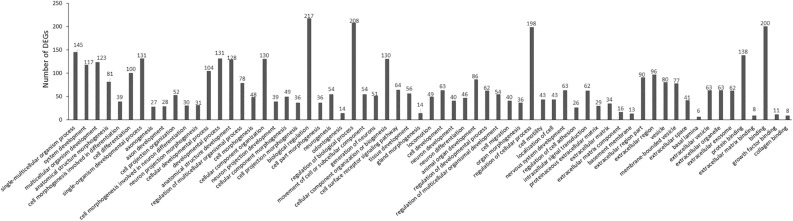
In the category of SC-Down/OPC-No Change (Fig. 3), 313 DEGs were identified. The significantly enriched BP terms (n = 47) include the single-multicellular organism process, system development, multicellular organism development, cell differentiation, and related terms. The significantly enriched CC terms (n = 13) include extracellular matrix, extracellular vesicle, and related terms. The significantly enriched MF terms (n = 5) include protein binding, extracellular matrix binding, binding, growth factor binding, and collagen binding.
Figure 3.
EF-induced gene down-regulation in SCs and no change in OPCs (SC-down/OPC-NO change).
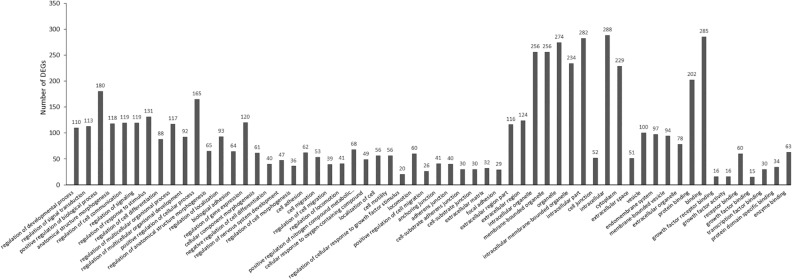
In the category of SC-Up/OPC-No Change (Fig. 4), 410 DEGs were identified. The significantly enriched BP terms (n = 256) include regulation of developmental process, regulation of signal transduction, positive regulation of biological process, anatomical structure morphogenesis, and related terms. The significantly enriched CC terms (n = 21) include adherens junction, cell-substrate adherens junction, cell-substrate junction, extracellular matrix, focal adhesion, and related terms. The significantly enriched MF terms (n = 9) include protein binding, growth factor, receptor binding, growth factor activity, receptor binding, growth factor binding, transcription factor binding, protein domain specific binding, and enzyme binding.
Figure 4.
EF-induced gene up-regulation in SCs and no change in OPCs (SC-Up/OPC-no change).
In the category of OPC-Down/SC-No Change (Fig. 5), 402 DEGs were identified. The significantly enriched BP terms (n = 20) include cell cycle, sister chromatid segregation, positive regulation of cellular process, and related terms. The microtubule-based process and microtubule cytoskeleton organization are also enriched terms. The significantly enriched CC terms (n = 35) include intracellular, organelle, nucleus, and related terms. The cytoskeletal part, cytoskeleton, and polymeric cytoskeletal fiber are also significantly enriched terms in this category. The significantly enriched MF terms (n = 16) include protein binding, ribonucleotide binding, carbohydrate derivative binding, and microtubule binding.
Figure 5.
EF-induced gene down-regulation in OPCs and no change in SCs (OPC-down/SC-no change).
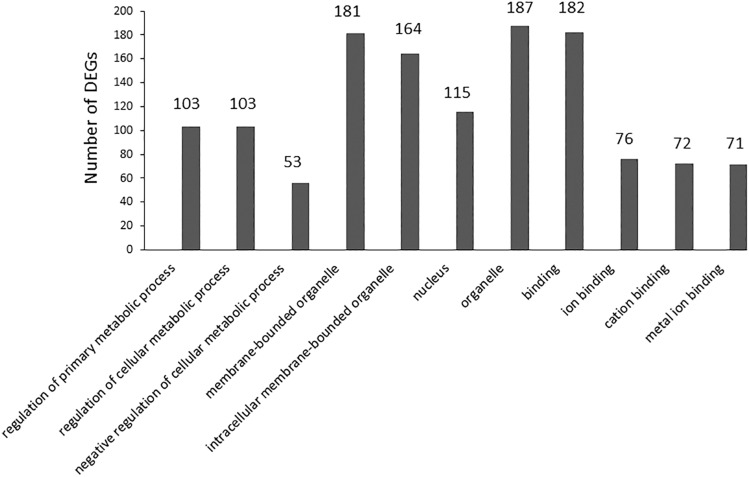
In the category of OPC-Up/SC-No Change (Fig. 6), 341 DEGs were identified. The significantly enriched BP terms (n = 3) include regulation of the primary metabolic process, regulation of the cellular metabolic process, and negative regulation of the cellular metabolic process. The significantly enriched CC terms (n = 4) include membrane-bounded organelle, intracellular membrane-bounded organelle, nucleus, organelle. The significantly enriched MF terms (n = 4) include binding, ion binding, cation binding, and metal ion binding.
Figure 6.
EF-induced gene up-regulation in OPCs and no change in SCs (OPC-up/SC-no change).
In the category of SC-Up/OPC-Down (Fig. 7), 52 DEGs were identified. The significantly enriched BP terms (n = 3) include regulation of developmental process, regulation of multicellular organismal development, and negative regulation of response to stimulus. We did not recognize any significantly enriched terms in CC and MF.
Figure 7.
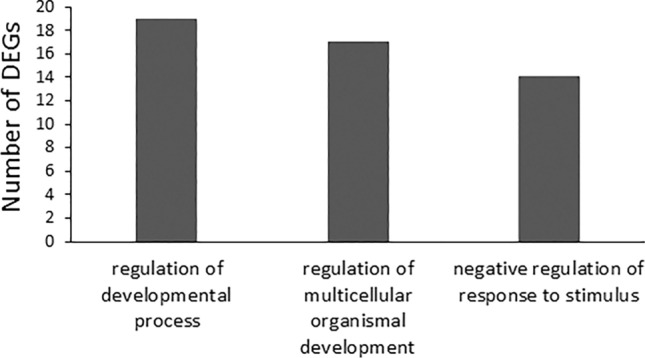
EF-induced gene up-regulation in SCs and down regulation in OPCs (SC-up/OPC-down).
In the category of OPC-Up/SC-Down, 42 DEGs were identified. We did not recognize any significantly enriched terms in BP, CC, and MF.
DEGs regulating cytoskeleton, adhesion, and migration
The cytoskeleton of Schwann cells and OPCs are organized by genes that regulate cell adhesion and migration. The DEGs associated with significantly enriched GO terms involving cytoskeleton, cell adhesion, or cell movement and migration are summarized in Table 1. In the category of Both Down, the significantly enriched CC-type GO term—cytoskeleton—includes 32 DEGs. In the category of OPC-Down/SC-No Change, the significantly enriched CC-type GO term—cytoskeleton—includes 63 DEGs. In the category of SC-Down/OPC-No Change, the significantly enriched BP-type GO terms—locomotion and cell motility—include 49 and 26 DEGs, respectively. In the category of SC-Up/OPC-No Change, the significantly enriched BP-type GO terms—cell adhesion and cell migration—include 62 and 53 DEGs, respectively. In this category, the significantly enriched CC-type GO term—focal adhesion—includes 29 DEGs.
Discussion
During directed cell migration, cells become polarized dynamically in response to guidance signals. The polarized cell shape is characterized by membrane ruffling and filopodia at the front edge, reorientation of the microtubule-organizing center (MTOC) and the Golgi apparatus in the direction of migration, and coordinated reorganization of the actin cytoskeleton and microtubules in the leading process21,22. In focal adhesions, F-actin and other associated molecules form a structure that enables cells to adhere to the extracellular matrix21,22. Signal transduction is involved in the assembly and disassembly of the protein complex thereby controlling cell motility and migration direction. In previous publications, we have reported that the migration of SCs and OPCs can be directed by an applied electric field, and we performed RNA-seq to explore the pathways that regulated the cell migration for each type18,19.
In this study, we jointly analyzed the RNA-seq data for OPCs and SCs subjected to EFs and then identified significantly enriched GO terms associated with DEGs. We were particularly interested in the significantly enriched GO terms related to cytoskeleton, cell adhesion, cell movement, and cell migration. Below we discuss some of the DEGs associated with these terms with respect to the direction of regulation in each glial cell type. Effective directional migration of OPCs and Schwann cells to a neural lesion is crucial in the neural remyelination process. Potentially an applied EF can guide the cells to migrate into the lesion to enhance the remyelination of axons. Our studies provide clues to understand the gene regulation of cell migration. To explore the gene expression for the neural cells subjected to EF stimulation may also lead to the improvement of strategies to regulate neural cell migration.
Both down: cytoskeleton (CC)
The organization of cytoskeleton controls cell migration. Previous studies showed that a number of DEGs that were down-regulated in both SC and OPCs subjected to EF stimulation regulate cytoskeleton function and cell migration. The TRIOBP gene is a member of the Rho guanine nucleotide exchange factor. It regulates cell spreading and cell contraction by organizing actin formation; therefore, it is involved in cell motility23,24. LIM and SH3 protein 1 (LASP1) binds to the actin cytoskeleton at the cell membrane extension and regulates the dynamic actin-based cell adhesion and motility. It was reported that overexpression of LASP1 is associated with proliferation, migration, and invasion in esophageal squamous cell carcinoma25. The FORMIN homology 2 domain containing protein 1 (FHOD1) is required for the assembly of F-actin structures, such as stress fibers. The Rho-ROCK signaling pathway regulates its function. One study of mouse fibroblast migration showed that FHOD1 is recruited to integrin clusters that lead to actin assembly26. That study suggested that FHOD1 is needed for directed forces and cell adhesion during cell spreading and migration. The CDC42 effector protein 4 (CDC42EP4) is a CDC42-binding protein that interacts with CDC42 to induce actin filament assembly, which regulates cell shape and movement27. A previous study showed that overexpression of PAK4, a CDC42-activated kinase that decreases adhesion, enhanced galvanotaxis speed of fibroblast28. However, we observed the decreased expression of CDC42EP4. Further investigation will help to explore the regulation of CDC42 and its effector proteins. The APC regulator of WNT signaling pathway 2 (APC2) regulates actin assembly and microtubule network formation, and therefore regulate cell adhesion and motility. The down-regulation of these genes in both SCs and OPCs subjected to EF stimulation suggests a similar function of these genes for the regulation of both cell types.
Both up: focal adhesion (CC)
Although focal adhesion is not a significantly enriched term for the Both Up category, we summarized the function of nine significantly changed genes associated with this term. A number of studies have reported the role of small GTPase family members such as Rho29 and Rock30 in the regulation of EF-directed cell migration. In general, inhibition of these molecules abolished the EF-directed cell migration29,30. This study indicated the function of an additional GTPase member in EF-directed cell migration. The Rho family GTPase 3 (RND3) is a member of a small GTPase family. The protein acts as a negative regulator of fibroblast cytoskeletal organization that can lead to loss of adhesion31. Cortactin (CTTN) regulates the organization of the actin cytoskeleton and cell shape in processes such as lamellipodia formation. The cortactin N-terminal half binds and activates the ARP2/3 complex and therefore regulates actin dynamics. It was reported that overexpression of either full-length cortactin or the cortactin C-terminus can enhance the migration of mammary epithelial cells32. In a previous study, we reported the cathodal migration of neural stem cells–derived OPCs subjected to EF stimulation. The ARPC2−/− OPCs lost the directional migration in EF33. This study indicated that the function of ARPC2/3 complex and the activation of ARPC2/3 complex are required for EF-directed cell migration. The plasminogen activator, urokinase receptor (PLAUR), mediates the proteolysis-independent signal transduction activation effects of U-PA. The inhibition of PLAUR decreases the growth, invasion, angiogenesis, and metastasis of various cancers34–36. The ZYXIN (ZYX) protein concentrates at sites of focal adhesions and along the actin cytoskeleton in a cell. It was reported that in migrating epithelial cells, ZYXIN accumulates at force-bearing sites at the leading edge but not at the trailing edge. Rho-kinase and myosin II activation also regulated ZYXIN recruitment37. The result indicated that these up-regulated genes may be involved in the regulation of focal adhesion and migration of both SCs and OPCs subjected to EF stimulation.
SC-down/OPC-no change: locomotion (BP)
The significantly enriched GO terms are the BP terms locomotion and regulation of cell adhesion. There are 49 DEGs associated with the term locomotion, ARFGAP with SH3 domain, Ankyrin Repeat, and PH domain 3 (ASAP3) is a focal adhesion-associated ARFGAP and regulates cell migration and focal adhesion. It was reported that the reduced expression of ASPS3 decreased cell motility of glioblastoma cells and cell invasion of mammary carcinoma cells38. The cadherin EGF LAG seven-pass G-type receptor 2 (CELSR2) is a protein that is involved in the cell matrix or cell-to-cell communication in cell adhesion. Based on the phenotypes of knockout mice, one study showed that the CELSR2 and 3 genes controlled the migration ability of facial branchiomotor neurons. Ephrin B1 is a type I membrane protein and a ligand of Eph-related receptor tyrosine kinases, which are crucial for migration, repulsion, and adhesion. The interaction of Eph receptors and ephrins can lead either to cell repulsion or cell adhesion and invasion39. The epithelial membrane protein 2 (EMP2) functions as a key regulator of cell membrane composition and promotes the recruitment of integrins to lipid rafts. A previous study showed that Emp2 regulates cell migration and cell adhesion. EMP2 governs transepithelial migration of neutrophils into the airspace40. MATRILIN 2 (MATN2) is a multiadhesion adaptor protein that interacts with other ECM proteins and integrins. MATN2 promotes neurite outgrowth and Schwann cell migration41,42. NEUROPILIN 1 (NRP1) regulates actin network organization through RhoA and ROCK. It was reported that SEMA3D controls endothelial cell migration by molecular mechanisms via NRP1 and PLXND143.
SC-down/OPC-no change: cell adhesion (BP)
The integrin subunit alpha 4 (ITGA4) associates with a beta 1 or beta 7 subunit to form an integrin that is involved in cell motility and migration. An in vitro study showed that ITGA4 overexpression on mesenchymal stem cells (MSCs) enhances transendothelial migration44. The results indicated the involvement of down-regulated genes in the regulation of SC migration in an applied EF but not in OPCs.
SC-up/OPC-no change: cell adhesion (BP)
The AXL receptor tyrosine kinase (AXL) regulates the extracellular matrix protein expression and demonstrates that it is essential for invasion and metastasis of endometrial cancer. Studies have shown that AXL gene silencing inhibited the migration and invasion of endometrial cancer cells45. The L1 cell adhesion molecule (L1CAM) is an immunoglobulin superfamily protein that regulates neuronal migration and differentiation in nervous system development46. The adherens junctions associated protein 1 (AJAP1) is a type-I transmembrane protein that localizes and interacts with the E-cadherin-catenin complex. The endogenous AJAP1 is associated with the microtubule cytoskeleton and can attenuate sprouting angiogenesis by reducing endothelial migration and invasion capacities47.
SC-up/OPC-no change: cell migration (BP)
Actinin alpha 4 (ACTN4) is an actin cross-linking protein that regulates cell protrusion I cell movement. ACTN4 promotes migration and metastasis of osteosarcoma through the NF-κB Pathway48. Inhibition of ACTN4 decreased the formation of cell filopodia and therefore suppressed tumor cell migration49. The connective tissue growth factor, also known as CCN2, mediates cell adhesion, aggregation, and migration in a few cell types, such as vascular endothelial cells, fibroblasts, and epithelial cells50. The NudE neurodevelopment protein 1-like 1 (NDEL1) regulates microtubules and intermediate filaments. NDEL1 regulates cell movement by interacting with TRIO-associated repeat on actin (TARA), which is an actin-bundling protein51.
SC-up/OPC-no change: focal adhesion (BP)
PDZ and LIM domain 1 (PDLIM1) contains a C-terminal PDZ and one or more N-terminal LIM domain. It acts as an adapter that brings signal proteins to the cytoskeleton. It has been shown that the PDLIM protein CLP36 is critical for stress fiber formation and the assembly of focal adhesions in BeWo cells52. PDLIM1 overexpression attenuated the epithelial-mesenchymal transition of colorectal cancer cells53. Rho GTPase activating protein 22 (ARHGAP22) converts RAC1 to an inactive GDP-bound state that inhibits RAC1-dependent lamellipodium formation54. Forced expression of ARHGAP22 suppressed lamellae formation and cell spreading55. EZRIN (EZR) serves as an intermediate between the plasma membrane and the actin cytoskeleton, and regulates cell adhesion and migration. EZR overexpression causes the increased invasion of cancer cells56,57. The studies suggested that these up-regulated genes regulated Schwann cell migration in an EF. However, further studies will be required to determine how the EF regulates focal adhesion and therefore controls cell migration.
OPC-down/SC-no change: cytoskeleton (CC)
Integrin is cell surface receptor regulates the function of the small GTPase Rac1 and therefore control the cell membrane protrusion and cytoskeletal reorganization events in directional migration. The Rac GTPase-activating protein 1 (GAP1) links β1 integrin to Rac1. A study showed that siRNA-mediated knockdown of either filamin-A or IQ-motif-containing GTPase-activating protein 1 (IQGAP1) induced high, dysregulated Rac1 activity during cell spreading on fibronectin58. Tyrosine kinase SRC and its downstream signaling molecules, including the small GTPase Rac1 and Arp2/3 complex, regulate the polymerization of actin network that initiates membrane protrusion in cell migration59. It was reported that the cell front of a migrating fibroblast was defined by integrin activation. The subsequent FAK, Src, and p190RhoGAP induces reorientation of the nucleus and the establishment of front–rear polarity60. Microtubule-associated protein 4 (MAP4) protein stabilizes microtubules by modulating microtubule dynamics. A previous study showed that MAP4 regulates invasion and migration of esophageal squamous cancer cells. MAP4 promotes cell invasion and migration by activating the ERK-c-Jun-vascular endothelial growth factor A signaling pathway61.
Effective directional migration of OPCs and Schwann cells to a neural lesion is crucial in the neural remyelination process. Potentially an applied EF can guide the cells to migrate into the lesion to enhance the remyelination of axons. Our studies provide clues to understand the gene regulation of cell migraiton. Exploring the gene expression in neural cells subjected to EF stimulation may also lead to the improvement of strategies to regulate neural cell migration.
Conclusion
In summary, in this study, we performed comparative differential expression analysis for RNA-seq reads from OPCs and SCs that were subjected to EF stimulation in our previous reports in Yao et al.18 and Li et al.19, respectively. GO term analysis of the DEGs revealed a number of significantly enriched GO terms related to cytoskeleton, cell adhesion, cell movement, and cell migration. Of the DEGs associated with these terms, nine up-regulated DEGs and 32 down-regulated DEGs showed the same direction of effect in both SCs and OPCs stimulated with EFs, while the remaining DEGs showed different effects in SCs and OPCs subjected to EF stimulation. The DEGs revealed in this study support the findings in previous reports regarding the function of genes that regulated EF-directed cell migration, for example the expression of cortactin, an activator of the ARP2/3 complex, increased in both SC and PCs. Additionally, this study revealed that the genes that generally regulate cell adhesion and migration in some cases follow the same transcription trend during the regulation of EF-guided migration of different cell types, but in other cases show different transcriptional effects. This outcome suggests that some processes involved in the migration of different glial cell types are similar, but there are likely processes specific to each cell type as well. Although the DEGs that regulate cell migration are recognized in the GO term analysis, no signaling pathways show significant enrichment of DEGs. This suggests that existing established signaling pathways may not be the key regulators of EF-guided cell migration.
Methods
The RNA-seq reads from OPCs and Schwann cells exposed to electrical fields were previously described in the work of Yao et al.18 and Li et al.19, respectively. The OPCs and Schwann cells that were used in those studies were isolated from cerebral cortexes of postnatal rats (days 1–2) and the sciatic nerves of postnatal rats (days 1–3) respectively. After culturing, OPCs or Schwann cells (100,000 cells) were then seeded in a cell culture chamber for EF stimulation. RNA was extracted for RNA sequencing studies after the cells were subjected to EF stimulation. The cells without EF stimulation were used as control. In this study, for our comparative analysis, we omitted the reads from OPCs exposed to 100 V, since only exposure to 200 mV showed a significant effect on cell migration. We also used only the R1 reads from the SCs, since the OPC reads were single-end reads. All reads were trimmed for adapter contamination and sequence quality with a custom pipeline using the adaptive trimming tools Scythe62, Trimmomatic-0.3363, and Sickle64. Differential expression results comparing four conditions (200 V OPCs, control OPCs, 100 mV SCs, and control SCs) were calculated using the RSEM-EBSeq pipeline65,66). First, reads were aligned to the Rnor_6.0 reference genome with Bowtie 2, and then gene-level counts from RSEM were used by EBSeq to calculate the probability of differential expression for each of 15 possible expression patterns (Supplemental Table 1). False discovery rate (FDR) cutoffs were applied to the results to generate lists of differentially expressed genes at less than 5 percent and 1 percent FDR (Supplemental Table 2).
The patterns that EBSeq uses to classify genes are based on the expression level in each condition. For example, genes with the same expression level in all conditions are classified as Pattern 1. Genes that show differential expression between OPCs and Schwann cells regardless of electrical stimulation but have the same level of expression in cells of the same type are classified as Pattern 4. Similarly, genes that are differentially expressed between EF-stimulated and control cells but have the same level regardless of cell type are classified as Pattern 6. However, there are several other patterns that include genes that show differential expression between EF-stimulated and control cells, but do not have the same expression level in both types of EF-stimulated or control cells (Patterns 10–13). The patterns reported by EBSeq also do not differentiate between the up-regulation and down-regulation of genes. Since we were interested in the direction of differential expression after the EF, regardless of the initial level of expression in each particular cell type, we manually combined genes differentially expressed genes (FDR < 0.05) from multiple RSEM patterns into a new set of more informative patterns. (Supplemental Table 1).
Gene ontology functional enrichment analysis and signaling pathway analysis of the significantly changed genes were performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID)67.
Supplementary information
Acknowledgements
This work was supported by the National Institute of General Medical Sciences (P20 GM103418) from the National Institutes of Health.
Author contributions
L.Y., T.S. and Y.L. conceived the project. T.S., L.Y. and Y.L. conducted the analysis of data. L.Y. and T.S. wrote the manuscript. The authors read and approved the final manuscript.
Data availability
All data generated or analysed during this study are included in this published article and its supplementary information files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-74085-x.
References
- 1.Robinson KR. Electrical currents through full-grown and maturing Xenopus oocytes. Proc. Natl. Acad. Sci. USA. 1979;76:837–841. doi: 10.1073/pnas.76.2.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaffe LF, Stern CD. Strong electrical currents leave the primitive streak of chick embryos. Science. 1979;206:569–571. doi: 10.1126/science.573921. [DOI] [PubMed] [Google Scholar]
- 3.Robinson KR. Endogenous electrical current leaves the limb and prelimb region of the Xenopus embryo. Dev. Biol. 1983;97:203–211. doi: 10.1016/0012-1606(83)90077-5. [DOI] [PubMed] [Google Scholar]
- 4.Hotary KB, Robinson KR. Endogenous electrical currents and the resultant voltage gradients in the chick embryo. Dev. Biol. 1990;140:149–160. doi: 10.1016/0012-1606(90)90062-N. [DOI] [PubMed] [Google Scholar]
- 5.McCaig CD, Rajnicek AM, Song B, Zhao M. Controlling cell behavior electrically: current views and future potential. Physiol. Rev. 2005;85:943–978. doi: 10.1152/physrev.00020.2004. [DOI] [PubMed] [Google Scholar]
- 6.Catalano SM, Shatz CJ. Activity-dependent cortical target selection by thalamic axons. Science. 1998;281:559–562. doi: 10.1126/science.281.5376.559. [DOI] [PubMed] [Google Scholar]
- 7.Dantzker JL, Callaway EM. The development of local, layer-specific visual cortical axons in the absence of extrinsic influences and intrinsic activity. J. Neurosci. 1998;18:4145–4154. doi: 10.1523/JNEUROSCI.18-11-04145.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanno H, Pearse DD, Ozawa H, Itoi E, Bunge MB. Schwann cell transplantation for spinal cord injury repair: its significant therapeutic potential and prospectus. Rev. Neurosci. 2015;26:121–128. doi: 10.1515/revneuro-2014-0068. [DOI] [PubMed] [Google Scholar]
- 9.Sun L, et al. Inhibition of TROY promotes OPC differentiation and increases therapeutic efficacy of OPC graft for spinal cord injury. Stem Cells Dev. 2014;23:2104–2118. doi: 10.1089/scd.2013.0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie XM, et al. Co-transplantation of MRF-overexpressing oligodendrocyte precursor cells and Schwann cells promotes recovery in rat after spinal cord injury. Neurobiol. Dis. 2016;94:196–204. doi: 10.1016/j.nbd.2016.06.016. [DOI] [PubMed] [Google Scholar]
- 11.Almad A, Sahinkaya FR, McTigue DM. Oligodendrocyte fate after spinal cord injury. Neurotherapeutics. 2011;8:262–273. doi: 10.1007/s13311-011-0033-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S, et al. Notch receptor activation inhibits oligodendrocyte differentiation. Neuron. 1998;21:63–75. doi: 10.1016/S0896-6273(00)80515-2. [DOI] [PubMed] [Google Scholar]
- 13.Guest JD, Hiester ED, Bunge RP. Demyelination and Schwann cell responses adjacent to injury epicenter cavities following chronic human spinal cord injury. Exp. Neurol. 2005;192:384–393. doi: 10.1016/j.expneurol.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 14.Thuret S, Moon LD, Gage FH. Therapeutic interventions after spinal cord injury. Nat. Rev. Neurosci. 2006;7:628–643. doi: 10.1038/nrn1955. [DOI] [PubMed] [Google Scholar]
- 15.Fu H, Hu D, Zhang L, Shen X, Tang P. Efficacy of oligodendrocyte progenitor cell transplantation in rat models with traumatic thoracic spinal cord injury: a systematic review and meta-analysis. J. Neurotrauma. 2018;35:2507–2518. doi: 10.1089/neu.2017.5606. [DOI] [PubMed] [Google Scholar]
- 16.Bastidas J, et al. Human Schwann cells exhibit long-term cell survival, are not tumorigenic and promote repair when transplanted into the contused spinal cord. Glia. 2017;65:1278–1301. doi: 10.1002/glia.23161. [DOI] [PubMed] [Google Scholar]
- 17.Bunge MB, Monje PV, Khan A, Wood PM. From transplanting Schwann cells in experimental rat spinal cord injury to their transplantation into human injured spinal cord in clinical trials. Prog. Brain Res. 2017;231:107–133. doi: 10.1016/bs.pbr.2016.12.012. [DOI] [PubMed] [Google Scholar]
- 18.Yao L, Li Y, Knapp J, Smith P. Exploration of molecular pathways mediating electric field-directed Schwann cell migration by RNA-seq. J. Cell. Physiol. 2015;230:1515–1524. doi: 10.1002/jcp.24897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Wang X, Yao L. Directional migration and transcriptional analysis of oligodendrocyte precursors subjected to stimulation of electrical signal. Am. J. Physiol. Cell Physiol. 2015;309:C532–540. doi: 10.1152/ajpcell.00175.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gov NS, Gopinathan A. Dynamics of membranes driven by actin polymerization. Biophys. J. 2006;90:454–469. doi: 10.1529/biophysj.105.062224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geiger B, Yamada KM. Molecular architecture and function of matrix adhesions. Cold Spring Harbor Perspect. Biol. 2011;3:005033. doi: 10.1101/cshperspect.a005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parsons JT, Horwitz AR, Schwartz MA. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell Biol. 2010;11:633–643. doi: 10.1038/nrm2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seipel K, O'Brien SP, Iannotti E, Medley QG, Streuli M. Tara, a novel F-actin binding protein, associates with the Trio guanine nucleotide exchange factor and regulates actin cytoskeletal organization. J. Cell Sci. 2001;114:389–399. doi: 10.1242/jcs.114.2.389. [DOI] [PubMed] [Google Scholar]
- 24.Bellanger JM, et al. The two guanine nucleotide exchange factor domains of Trio link the Rac1 and the RhoA pathways in vivo. Oncogene. 1998;16:147–152. doi: 10.1038/sj.onc.1201532. [DOI] [PubMed] [Google Scholar]
- 25.He B, et al. Overexpression of LASP1 is associated with proliferation, migration and invasion in esophageal squamous cell carcinoma. Oncol. Rep. 2013;29:1115–1123. doi: 10.3892/or.2012.2199. [DOI] [PubMed] [Google Scholar]
- 26.Iskratsch T, et al. FHOD1 is needed for directed forces and adhesion maturation during cell spreading and migration. Dev. Cell. 2013;27:545–559. doi: 10.1016/j.devcel.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farrugia AJ, Calvo F. The Borg family of Cdc42 effector proteins Cdc42EP1-5. Biochem. Soc. Trans. 2016;44:1709–1716. doi: 10.1042/BST20160219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finkelstein E, et al. Roles of microtubules, cell polarity and adhesion in electric-field-mediated motility of 3T3 fibroblasts. J. Cell Sci. 2004;117:1533–1545. doi: 10.1242/jcs.00986. [DOI] [PubMed] [Google Scholar]
- 29.Feng JF, et al. Guided migration of neural stem cells derived from human embryonic stem cells by an electric field. Stem Cells. 2012;30:349–355. doi: 10.1002/stem.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao L, McCaig CD, Zhao M. Electrical signals polarize neuronal organelles, direct neuron migration, and orient cell division. Hippocampus. 2009;19:855–868. doi: 10.1002/hipo.20569. [DOI] [PubMed] [Google Scholar]
- 31.Nobes CD, et al. A new member of the Rho family, Rnd1, promotes disassembly of actin filament structures and loss of cell adhesion. J. Cell Biol. 1998;141:187–197. doi: 10.1083/jcb.141.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kowalski JR, et al. Cortactin regulates cell migration through activation of N-WASP. J. Cell Sci. 2005;118:79–87. doi: 10.1242/jcs.01586. [DOI] [PubMed] [Google Scholar]
- 33.Li Y, Wang PS, Lucas G, Li R, Yao L. ARP2/3 complex is required for directional migration of neural stem cell-derived oligodendrocyte precursors in electric fields. Stem Cell Res. Ther. 2015;6:41. doi: 10.1186/s13287-015-0042-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lakka SS, et al. Specific interference of urokinase-type plasminogen activator receptor and matrix metalloproteinase-9 gene expression induced by double-stranded RNA results in decreased invasion, tumor growth, and angiogenesis in gliomas. J. Biol. Chem. 2005;280:21882–21892. doi: 10.1074/jbc.M408520200. [DOI] [PubMed] [Google Scholar]
- 35.Pulukuri SM, et al. RNA interference-directed knockdown of urokinase plasminogen activator and urokinase plasminogen activator receptor inhibits prostate cancer cell invasion, survival, and tumorigenicity in vivo. J. Biol. Chem. 2005;280:36529–36540. doi: 10.1074/jbc.M503111200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Rao JS, et al. Inhibition of invasion, angiogenesis, tumor growth, and metastasis by adenovirus-mediated transfer of antisense uPAR and MMP-9 in non-small cell lung cancer cells. Mol. Cancer Ther. 2005;4:1399–1408. doi: 10.1158/1535-7163.MCT-05-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uemura A, Nguyen TN, Steele AN, Yamada S. The LIM domain of zyxin is sufficient for force-induced accumulation of zyxin during cell migration. Biophys. J. 2011;101:1069–1075. doi: 10.1016/j.bpj.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ha VL, et al. ASAP3 is a focal adhesion-associated Arf GAP that functions in cell migration and invasion. J. Biol. Chem. 2008;283:14915–14926. doi: 10.1074/jbc.M709717200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poliakov A, Cotrina M, Wilkinson DG. Diverse roles of eph receptors and ephrins in the regulation of cell migration and tissue assembly. Dev. Cell. 2004;7:465–480. doi: 10.1016/j.devcel.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 40.Lin WC, et al. Epithelial membrane protein 2 governs transepithelial migration of neutrophils into the airspace. J. Clin. Investig. 2020;130:157–170. doi: 10.1172/JCI127144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Korpos E, Deak F, Kiss I. Matrilin-2, an extracellular adaptor protein, is needed for the regeneration of muscle, nerve and other tissues. Neural Regener. Res. 2015;10:866–869. doi: 10.4103/1673-5374.158332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malin D, et al. The extracellular-matrix protein matrilin 2 participates in peripheral nerve regeneration. J. Cell Sci. 2009;122:995–1004. doi: 10.1242/jcs.040378. [DOI] [PubMed] [Google Scholar]
- 43.Hamm MJ, Kirchmaier BC, Herzog W. Sema3d controls collective endothelial cell migration by distinct mechanisms via Nrp1 and PlxnD1. J. Cell Biol. 2016;215:415–430. doi: 10.1083/jcb.201603100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cui LL, et al. Integrin alpha4 overexpression on rat mesenchymal stem cells enhances transmigration and reduces cerebral embolism after intracarotid injection. Stroke. 2017;48:2895–2900. doi: 10.1161/STROKEAHA.117.017809. [DOI] [PubMed] [Google Scholar]
- 45.Divine LM, et al. AXL modulates extracellular matrix protein expression and is essential for invasion and metastasis in endometrial cancer. Oncotarget. 2016;7:77291–77305. doi: 10.18632/oncotarget.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pace KR, Dutt R, Galileo DS. Exosomal L1CAM stimulates glioblastoma cell motility, proliferation, and invasiveness. Int, J. Mol. Sci. 2019;20:1. doi: 10.3390/ijms20163982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hotte K, Smyrek I, Starzinski-Powitz A, Stelzer EHK. Endogenous AJAP1 associates with the cytoskeleton and attenuates angiogenesis in endothelial cells. Biol. Open. 2017;6:723–731. doi: 10.1242/bio.022335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang, Q., et al. ACTN4 promotes the proliferation, migration, metastasis of osteosarcoma and enhances its invasive ability through the NF-kappaB pathway. Pathol. Oncol. Res. (2019). [DOI] [PMC free article] [PubMed]
- 49.Agarwal N, et al. MTBP suppresses cell migration and filopodia formation by inhibiting ACTN4. Oncogene. 2013;32:462–470. doi: 10.1038/onc.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aguiar DP, et al. New strategy to control cell migration and metastasis regulated by CCN2/CTGF. Cancer Cell Int. 2014;14:61. doi: 10.1186/1475-2867-14-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hong JH, et al. Regulation of the actin cytoskeleton by the Ndel1-Tara complex is critical for cell migration. Sci. Rep. 2016;6:31827. doi: 10.1038/srep31827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tamura N, Ohno K, Katayama T, Kanayama N, Sato K. The PDZ-LIM protein CLP36 is required for actin stress fiber formation and focal adhesion assembly in BeWo cells. Biochem. Biophys. Res. Commun. 2007;364:589–594. doi: 10.1016/j.bbrc.2007.10.064. [DOI] [PubMed] [Google Scholar]
- 53.Chen HN, et al. PDLIM1 stabilizes the E-cadherin/beta-catenin complex to prevent epithelial-mesenchymal transition and metastatic potential of colorectal cancer cells. Can. Res. 2016;76:1122–1134. doi: 10.1158/0008-5472.CAN-15-1962. [DOI] [PubMed] [Google Scholar]
- 54.Sanz-Moreno V, et al. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135:510–523. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 55.Mori M, Saito K, Ohta Y. ARHGAP22 localizes at endosomes and regulates actin cytoskeleton. PLoS ONE. 2014;9:e100271. doi: 10.1371/journal.pone.0100271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Naba A, Reverdy C, Louvard D, Arpin M. Spatial recruitment and activation of the Fes kinase by ezrin promotes HGF-induced cell scattering. EMBO J. 2008;27:38–50. doi: 10.1038/sj.emboj.7601943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoskin V, et al. Ezrin regulates focal adhesion and invadopodia dynamics by altering calpain activity to promote breast cancer cell invasion. Mol. Biol. Cell. 2015;26:3464–3479. doi: 10.1091/mbc.E14-12-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jacquemet G, et al. Rac1 is deactivated at integrin activation sites through an IQGAP1-filamin-A-RacGAP1 pathway. J. Cell Sci. 2013;126:4121–4135. doi: 10.1242/jcs.121988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhuo Y, et al. Subcellular and dynamic coordination between src activity and cell protrusion in microenvironment. Sci. Rep. 2015;5:12963. doi: 10.1038/srep12963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maninova M, et al. The reorientation of cell nucleus promotes the establishment of front-rear polarity in migrating fibroblasts. J. Mol. Biol. 2013;425:2039–2055. doi: 10.1016/j.jmb.2013.02.034. [DOI] [PubMed] [Google Scholar]
- 61.Chen X, et al. Effect of microtubule-associated protein-4 on epidermal cell migration under different oxygen concentrations. J. Dermatol. 2016;43:674–681. doi: 10.1111/1346-8138.13192. [DOI] [PubMed] [Google Scholar]
- 62.Scythe, B. A Bayesian adapter trimmer. (2014).
- 63.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Joshi Na, F.J. Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files. (2011).
- 65.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leng N, et al. EBSeq-HMM: a Bayesian approach for identifying gene-expression changes in ordered RNA-seq experiments. Bioinformatics. 2015;31:2614–2622. doi: 10.1093/bioinformatics/btv193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.