Abstract
Background
The Great COVID-19 Shutdown aimed to eliminate or slow the spread of SARS-CoV-2, the virus that causes COVID-19. The United States has no national policy, leaving states to independently implement public health guidelines that are predicated on a sustained decline in COVID-19 cases. Operationalization of “sustained decline” varies by state and county. Existing models of COVID-19 transmission rely on parameters such as case estimates or R0 and are dependent on intensive data collection efforts. Static statistical models do not capture all of the relevant dynamics required to measure sustained declines. Moreover, existing COVID-19 models use data that are subject to significant measurement error and contamination.
Objective
This study will generate novel metrics of speed, acceleration, jerk, and 7-day lag in the speed of COVID-19 transmission using state government tallies of SARS-CoV-2 infections, including state-level dynamics of SARS-CoV-2 infections. This study provides the prototype for a global surveillance system to inform public health practice, including novel standardized metrics of COVID-19 transmission, for use in combination with traditional surveillance tools.
Methods
Dynamic panel data models were estimated with the Arellano-Bond estimator using the generalized method of moments. This statistical technique allows for the control of a variety of deficiencies in the existing data. Tests of the validity of the model and statistical techniques were applied.
Results
The statistical approach was validated based on the regression results, which determined recent changes in the pattern of infection. During the weeks of August 17-23 and August 24-30, 2020, there were substantial regional differences in the evolution of the US pandemic. Census regions 1 and 2 were relatively quiet with a small but significant persistence effect that remained relatively unchanged from the prior 2 weeks. Census region 3 was sensitive to the number of tests administered, with a high constant rate of cases. A weekly special analysis showed that these results were driven by states with a high number of positive test reports from universities. Census region 4 had a high constant number of cases and a significantly increased persistence effect during the week of August 24-30. This change represents an increase in the transmission model R value for that week and is consistent with a re-emergence of the pandemic.
Conclusions
Reopening the United States comes with three certainties: (1) the “social” end of the pandemic and reopening are going to occur before the “medical” end even while the pandemic is growing. We need improved standardized surveillance techniques to inform leaders when it is safe to open sections of the country; (2) varying public health policies and guidelines unnecessarily result in varying degrees of transmission and outbreaks; and (3) even those states most successful in containing the pandemic continue to see a small but constant stream of new cases daily.
Keywords: COVID-19, models, surveillance, reopening America, contagion, metrics, surveillance, health policy, public health
Introduction
Without question, SARS-CoV-2, the novel coronavirus that causes COVID-19 [1,2], has resulted in an unprecedented pandemic in modern history with significant morbidity and mortality [3-7]. Although some countries have had success in controlling COVID-19 [8-11], others have encountered much difficulty [12-17], resulting in significant adverse outcomes [18-21]. Beyond the overall implications related to infection and death [9,11,22-29], the COVID-19 pandemic has a deleterious impact on the global economy [27,30,31], violence [32-37], mental health [38-43], and food security [44-47], and disproportionately affects vulnerable populations such as the elderly [48-52], the poor [53,54], and racial and ethnic minorities [55-61]. We must establish COVID-19 control through good policy [10,12,62-68]; unfortunately, different states have implemented various and inconsistent COVID-19 policies [67,69-76] in the absence of a national plan [77,78]. Without a COVID-19 vaccine [79-81], we need systematic public health surveillance [7,82-89] to inform policies and guidelines for COVID-19 control and prevention such as quarantines, social distancing, face masks, crowd control, and hygiene to prevent viral spread [90-98]. Good surveillance can safely inform our leaders when, how, and where our country can reopen [76,99-103].
According to Teutsch and Churchill [85], public health surveillance is the “systematic, ongoing assessment of the health of a community, based on the collection, interpretation, and use of health data and information. Surveillance provides information necessary for public health decision making” (pg 1). Surveillance does not rely on a single indicator; it depends on a variety of metrics to identify high-priority COVID-19 health events such as incidence, prevalence, mortality, severity, cost, preventability, and communicability [104]. We need to meet these objectives of a surveillance system to prevent infectious diseases [104]. The United States must address several public health surveillance objectives, specifically to detect outbreaks (eg, the distribution and spread of COVID-19) and evaluate control strategies [104]. A surveillance system also includes “the functional capacity for data collection and analysis, as well as the timely dissemination of data” (pg 1) [87]. To this end, our study aims to create novel, validated metrics of speed, acceleration, and jerk in COVID-19 transmission in the United States.
The Great COVID-19 Shutdown refers to the variety of “lockdown” [105] public health policies adopted by countries around the globe to prevent the further spread of COVID-19, ranging from strict and complete quarantines [106-108] to disorganized and piecemeal closures [105]. It worked in places when it was implemented properly and in a timely manner, such as China, South Korea, Singapore, and Vietnam [10,11,62,109]. Some countries eliminated COVID-19, defined as achieving zero new cases over 14 days, while others flattened the curve [64,110-113]. Governments that failed to effectively close down public movement and interactions resulted in increases in SARS-CoV-2 infections [50,114-120]. The United States had no national policy and was late in responding to the looming pandemic [105]. In fact, COVID-19 was technically classified as an epidemic by the Centers for Disease Control and Prevention when it accounted for >7.3% of all deaths in the United States. According to the National Center for Health Statistics, this was reached during the week of March 29-April 4 when COVID-19 accounted for 13.87% of all causes of death [121].
In response to the large death toll exacted by the epidemic, states independently implemented public health guidelines [14,19,70,71,122-125] regarding closures, social distancing, masks, and hand hygiene, which begs the question: when is it safe to reopen [126]? Reopening guidelines are predicated on a sustained decline in COVID-19 cases; however, operationalization of “sustained decline” varies by state [127]. Existing contagion models for COVID-19 rely on parameters such as case estimates or R0 and use intensive data collection efforts [128,129]. “Static” statistical models do not capture all of the relevant dynamics required to measure sustained declines [130-135]. Moreover, existing COVID-19 models use data that are subject to significant measurement error and other contaminants. Estimates of new SARS-CoV-2 infections suffer from undercounts due to asymptomatic carriers [136,137], access to testing [9,138-140], testing delays [141], testing sensitivity and specificity [142-145], and access to health care [60,146-150]. Surveillance systems and any enumeration of COVID-19 cases will err on the side of severity, meaning the most severe cases are more likely to be captured, the consequence of which is a significant undercount [71,104,130,151-156].
The conventional approach to modeling the spread of diseases such as COVID-19 is to posit an underlying contagion model and then to seek accurate direct measurement of the model parameters such as effective transmission rates or other parameters, often through labor-intensive methods relying on contact tracing to determine the spread of the virus in a sample population. For viral epidemics with an incubation period of up to 14 days, it takes weeks if not months to generate accurate parameter estimates even for simple contagion models [130]. For example, Li et al [157] provided early estimates of contagion parameters for COVID-19 using Wuhan data from contact tracing and methods developed by Lipsitch [158] but with weak statistical properties. It estimated the serial interval distribution and R0 from only six pairs of cases. These models also rely on underlying assumptions about immunity, common propensity for infection, well-mixed populations, etc [159]. Improvements in the models typically focus on relaxing these assumptions, for example, disaggregating the population by geography and modeling within-geography and cross-geographical personal interactions [160]. For example, Martcheva [161] provides an excellent dynamic analysis of a wide variety of contagion models and their possible dynamics. Unfortunately, the study had limited options for the statistical inference of parameter values from actual data.
In contrast, we take an empirical approach that focuses on statistical modeling of widely available empirical data such as the number of confirmed cases or the number of tests conducted that can inform estimates of the current value of critical parameters like the infection rate or reproduction rate. We explicitly recognize that the data generating process for the reported data contain an underlying contagion component, a political-economic component such as availability of accurate test kits, a social component such as how strongly people adhere to social-distancing and shelter-in-place policies, and a sometimes inaccurate data reporting process that may obscure the underlying contagion process. We therefore seek a statistical approach that can provide meaningful information despite the complex and sometimes obfuscating data generation process. Our approach is consistent with the principles of evidence-based medicine, including controlling for complex pathways that may include socioeconomic factors such as mediating variables, and policy recommendations “based on the best available knowledge, derived from diverse sources and methods” (pg S58) [162].
There are two primary advantages to this empirical approach. First, we can apply the empirical model relatively quickly to a short data set. This advantage stems from the panel nature of the model. We used US states as the cross-sectional variable, so that a week’s data from all US states provides a reasonable sample size. In addition to enabling parameter estimation early in a pandemic, using this property we tested to see if there has been a shift in the transmission or reproductive rates of the transmission process in the past week, that is, whether there is statistical evidence that the US pandemic is peaking.
The second advantage is that the approach directly measures and informs policy-relevant variables. For example, the White House issued guidance on reopening the US economy that depends on a decrease in the documented number of cases and in the proportion of positive test results over a 14-day period, among other criteria and considerations [163]. As noted above, the number and proportion of positive test results are the outcome of a data generating process that includes not just the underlying transmission process but a multitude of mediating factors as well as idiosyncrasies of the data collection and reporting process. We specifically modeled the number of positive test results in our empirical model, which provides evidence of direct use in policy dialog.
This study has two objectives: (1) to create a proof-of-concept COVID-19 surveillance system using the United States as a prototype for a global system; and (2) to validate novel surveillance metrics/techniques including speed, acceleration, and jerk to better inform public health leaders how the pandemic is spreading or changing course.
Methods
Overview of Methodology
First, we will provide standard surveillance metrics including new counts of SARS-CoV-2 infections, moving 7-day averages of SARS-CoV-2 infections, rates of SARS-CoV-2 infections per 100,000 population, new numbers of COVID-19 deaths, moving 7-day averages of COVID-19 deaths, and rates of COVID-19 deaths per 100,000 population plus testing and positive testing ratios. Standard surveillance metrics are useful and allow us to compare data even though standard techniques are limited to more severe cases and suffer from data contamination.
Second, to address these data limitations we will validate novel surveillance metrics of (1) speed, (2) acceleration, and (3) jerk (change in acceleration). The basic question we are trying to inform is: how are we doing this week relative to previous weeks? From a public health perspective, in the midst of a pandemic, we would like (at least) three affirmative responses: (1) there are fewer new cases per day this week than last week, (2) the number of new cases is declining from day to day, and (3) the day-to-day decline in the number of cases is even bigger this week than last week. Additionally, we would like some indicative information about significant shifts in how the pandemic is progressing — positive shifts could be the first indicators of the emergence of a new or recurrent hotspot, and positive shifts could be first indicators of successful public health policy.
This study derives indicators to inform the three questions specified in the study objective above. Next, we provide a regression-based decomposition of the indicators. While it is beyond the scope of this study to determine the underlying causes of the pandemic and its trajectory over time, we provide a decomposition into proximate contributory factors such as whether an acceleration is due to a “natural” progression of the pandemic (eg, due to an increasing infectious population) or to a shift in an underlying model parameter (eg, a parameter shift that could be associated with reopening, other health policy changes, a viral mutation, the end of summer vacation for K-12 schools, or other underlying causes). Other factors can affect acceleration by “shifting” the underlying parameters (eg, the virus can mutate to become more or less infectious, states can impose lockdowns, social pressures can encourage or discourage people from wearing masks and social distancing, etc). Therefore, we use the regression analysis to provide a decomposition of speed, acceleration, and jerk into proximate contributory factors. Finally, this study is an innovation over traditional agnostic surveillancesystems in that we go beyond presenting metrics of the transmission of COVID-19 by providing probable scenarios regarding the context in which the disease is spreading.
The COVID Tracking Project [164] compiles data from multiple state sources on the web [165]; data for the most recent 36 days were accessed from the GitHub repository [166]. After accounting for lagged and differenced regressors, this resulted in a panel of 50 states plus the District of Columbia with 29 days in each panel (n=1352). Following Oehmke et al [167], an empirical difference equation was specified in which the number of positive cases in each state at each day is a function of the prior number of cases, the level of testing, and weekly shift variables that measure whether the contagion was growing faster, at the same rate, or slower compared to the previous weeks. This resulted in a dynamic panel model that was estimated using the generalized method of moments approach by implementing the Arellano-Bond estimator in STATA/MP, version 16.1 (StataCorp LLC).
Arellano-Bond estimation of difference equations has several statistical advantages: (1) it allows for statistical examination of the model’s predictive ability and the validity of the model specification; (2) it corrects for autocorrelation and heteroscedasticity; (3) it has good properties for data with a small number of time periods and large number of states; (4) it corrects for omitted variables issues and provides a statistical test of correction validity. With these advantages, the method is applicable to ascertaining and statistically validating changes in the evolution of the pandemic within a period of one week or less, such as changes in the reproduction rate [167-174].
Speed: New Cases Per Day
The basic indicator of the pandemic’s status on a given day is the number of new cases on that day. Since new cases per day is a rate (value per unit of time), we will adopt physics nomenclature and refer to this as the speed of the pandemic. This is consistent with heuristic descriptions of the pandemic as spreading rapidly (ie, a large number of new cases per day) or slowly (ie, a small number of new cases per day). The public health ideal is to bring the speed of the pandemic to zero.
We report the number of new cases for each state both as a number per day and as a number per 100,000 population per day (table and column references).
For mathematical formality, we write:
![]()
where we have suppressed the i subscript of the previous section. We will be reporting surveillance numbers for each state and for the District of Columbia.
Acceleration
We are also interested in whether the number of cases per day is increasing, peaking, or decreasing, and why. Again, we will adopt physics nomenclature and refer to this datum as the acceleration. Since acceleration is difficult to ascertain on a daily basis, and there are weekend effects, etc, in the data, we report the weekly average for the acceleration as:
![]()
where D. is the difference operator. A positive acceleration indicates an increasing number of cases per day, and a negative acceleration (deceleration) indicates a decreasing number of cases per day. An acceleration of 0 is indicative of a peak, valley, or inflection point depending in part on whether the previous acceleration was positive or negative. For example, acceleration in Illinois changed from positive to zero in mid-May, indicating a peak, and from negative to zero toward the end of June, indicating a valley (Figure 1) [69,175].
Figure 1.
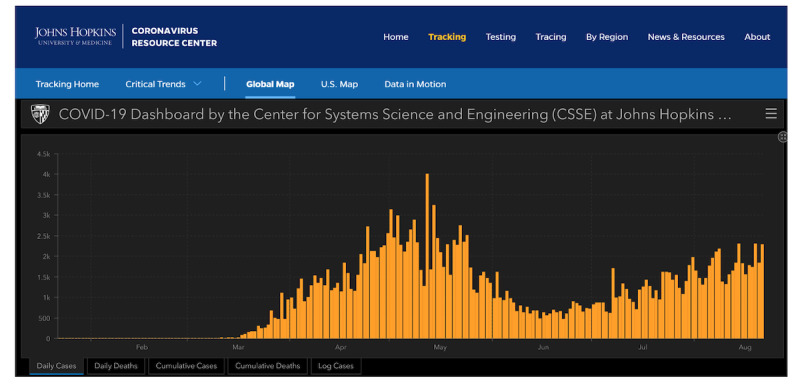
The number of positives per day in Illinois, according to the COVID-19 Dashboard of the Center for Systems Science and Engineering [175].
We provide a regression-based decomposition of accelerations into proximate components. That is, this is the systematic component of changes in acceleration, where t denotes the end date for the most recent week. Subtracting Oehmke et al’s [167] equation (3) at time t–1 from the same equation at time t results in:
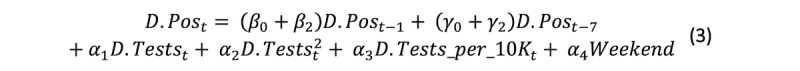
where we have suppressed the error terms and added a term for the “weekend effect.” We refer to the term containing Post–1 as the 1-day persistence effect. This, in turn, comprises a natural progression effect measured by β0D.Post–1 that represents the effect of a change over time in the number of new positive results where the magnitude of the effect is calibrated at the prior week’s parameter β0, and a shift effect β2D.Post–1 that measures the effect of the week’s shift in the parameter from β0 to β0 + β2. The second term in this equation is the 7-day persistence effect and is analogous to the 1-day persistence effect, including its decomposition into a natural progression effect y0D.Post–7 and a shift effect y2D.Post–7. The next part of equation 2 represents the portion of acceleration that is composed of changes in the contemporaneous component of the model.
The analogous expression for 1 week prior and 2 weeks prior are:
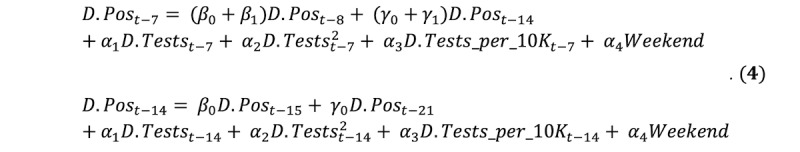
The expression for 2 weeks prior, D.Post–14, represents the baseline and does not contain any shift parameters. The shift parameters β1, β2, y1, and y2 represent shifts in the most recent 2 weeks relative to the week ending at time t–14.
The expressions for D.Post from equations 3 or 4 are easily adapted from time t to time t–j for each week and averaged over the week to provide a decomposition of acceleration as defined by equation 2.
Jerk: The Change in Acceleration
We now address the question of whether the day-to-day increase (or decrease) in new cases the current week is bigger or smaller than the day-to-day increase (or decrease) in new cases of the past week.
Formally, for the current week we are interested in is:
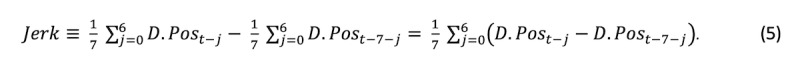
The first term to the right of the definitional equality is the average growth in the number of daily positive cases for the current week ending at time t, and the second term is the average increase in the number of daily positive cases for the prior week. Using physics nomenclature, the difference between these two acceleration rates is the “jerk.” A positive jerk indicates that the acceleration in the number of daily cases this week is greater than the average growth last week. Such a finding would be consistent with a scenario in which the pandemic was experiencing explosive growth; where a policy shift such as reopening had augmented the acceleration of the pandemic, possibly including a shift from deceleration to acceleration; or where a megaevent had “jerked” the acceleration upward, among other scenarios.
Using equation four, for the most recent week ending at time t, we can write:
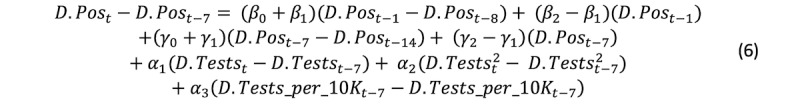
The top row contains the 1-day persistence effect’s contribution to the jerk. The first term on the right side of the equation represents the natural progression of the 1-day persistence effect on acceleration due to changes across weeks in the daily change in the number of new cases per day. The last term in the first row represents structural shifts in the 1-day persistence effect. The second row is analogous to the first row, except that it represents the 7-day persistence effect’s contribution to the jerk. The third row represents the contribution of contemporaneous effects to the jerk.
The analogous equation for the prior week is:
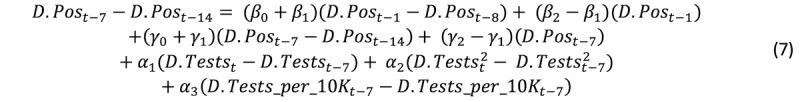
Equations 6 and 7 are easily averaged over the week to provide a decomposition of jerk as defined by equation 5.
Results
Regional Regression
Findings
We group the states according to Census region and present regression results for each region below. The biweekly surveillance products will be based on these regressions.
For Region 1 (Northeast), the regression Wald statistic shows that the model was statistically significant (χ210=132, P<.001), and the Sargan test fails to reject the validity of the overidentifying restrictions. (χ2252=258, P=.38) (Table 1).
Table 1.
Arellano-Bond dynamic panel data modeling of the number of daily infections reported by state, August 2-30, 2020.
| Variable | Region 1 | Region 2 | Region 3 | Region 4 | ||||||
|
|
Coefficient | P value | Coefficient | P value | Coefficient | P value | Coefficient | P value | ||
| L1Pos | 0.084 | .22 | –0.102 | .02 | –0.012 | .77 | 0.273 | <.001 | ||
| L1shiftAug17 | –0.129 | .16 | 0.069 | .24 | 0.221 | .02 | –0.213 | .03 | ||
| L1shiftAug24 | –0.112 | .19 | 0.093 | .10 | –0.021 | .84 | –0.737 | <.001 | ||
| L7Pos | 0.151 | .02 | 0.288 | <.001 | 0.269 | <.001 | 0.006 | .93 | ||
| L7shiftAug17 | –0.014 | .87 | –0.024 | .67 | –0.334 | <.001 | 0.018 | .79 | ||
| L7shiftAug24 | 0.004 | .96 | 0.046 | .49 | –0.265 | .003 | 0.397 | .02 | ||
| Tests | 0.003 | .12 | 0.047 | <.001 | 0.091 | <.001 | 0.017 | .048 | ||
| Tests_squared | 7.12E-09 | .61 | –5.05E-07 | <.001 | –4.89E-07 | <.001 | 2.74E-08 | .48 | ||
| Tests_per_10K | 1.072 | .04 | 8.023 | .002 | –15.986 | <.001 | —a | — | ||
| Weekend | –14.751 | .20 | 23.581 | .28 | –33.948 | .58 | 51.977 | .34 | ||
| Constant | 124.637 | <.001 | 46.461 | .89 | 429.167 | <.001 | 397.678 | <.001 | ||
| Wald statistic for regression | χ210=132 | <.001 | χ210=590 | <.001 | χ210=475 | <.001 | χ210=316 | <.001 | ||
| Sargan statistic for validity | χ2252=258 | .38 | χ2338=373 | .09 | χ2483=446 | .89 | χ2368=370 | .46 | ||
aRegion 4 did not include the Tests_per_100K variable due to collinearity.
The coefficient on the first lag of the dependent variable is not statistically significant, nor are the shift parameters for the weeks of August 17 and August 24 for this coefficient. The coefficient on the 7th lag is positive and statistically significant (0.151, P=.02). Neither of the shift parameters for the weeks of August 17 and August 24 are statistically significant. Of the variables representing the number of tests administered, the number per 100,000 population is significant (1.072, P=.04). The weekend variable is not significant. The constant is positive and significant (124.637, P<.001).
For Region 2 (Midwest), the regression Wald statistic shows that the model was statistically significant (χ210=590, P<.001), and the Sargan test fails to reject the validity of the overidentifying restrictions (χ2338=373, P=.09).
The coefficient on the first lag of the dependent variable is not statistically significant. The shift for the week of August 17 for this coefficient is positive and statistically significant (0.221, P=.02), but the shift for the week of August 24 is not significant. The coefficient on the 7th lag of the dependent variable is positive and significant (0.269, P<.001). Neither of the weekly shift variables for this coefficient are significant. The tests, tests squared, and tests per 10,000 population are all statistically significant (0.047, P<.001; –5.05E-07, P<.001; 8.023, P=.002). Neither the weekend variable nor the constant are significant.
For Region 3 (South), the regression Wald statistic shows that the model was statistically significant (χ210=475, P<.001), and the Sargan test fails to reject the validity of the overidentifying restrictions (χ2483=446, P=.89).
The coefficient on the first lag of the dependent variable is negative and statistically significant (–0.102, P=.02). Neither of the weekly shift variables for this coefficient are significant. The coefficient on the 7th lag of the dependent variable is positive and significant (0.288, P<.001). The shifts for the weeks of August 17 and August 24 are negative and significant (–0.334, P<.001; and –0.265, P=.003, respectively). The tests, tests squared, and tests per 10,000 population are all statistically significant (0.091, P<.001; –4.89E-07, P<.001; and –15.986, P<.001, respectively). The weekend variable is not significant. The constant is positive and significant (429.167, P<.001).
For Region 4 (West), the regression Wald statistic shows that the model was statistically significant (χ210=316, P<.001), and the Sargan test fails to reject the validity of the overidentifying restrictions (χ2368=370, P=.46).
The coefficient on the first lag of the dependent variable is negative and statistically significant (0.273, P<.001). The shifts for the week of August 17 and August 24 for this coefficient are negative and statistically significant (–0.213, P=.03; and –0.737, P<.001, respectively). The coefficient on the 7th lag of the dependent variable is not significant. The shift for the week of August 24 for this coefficient is positive and significant (0.397, P=.02), but the shift for the week of August 17 is not significant. Of the test variables, only the coefficient on the number of tests administered is significant (0.017, P=.048). The weekend variable is not significant. The constant is positive and significant (397.678, P<.001).
Interpretation
Region 1 appears to be fairly calm, with the only statistically significant persistence effect being a small 7-day lag effect. Region 2 is slightly less calm, but with a larger and statistically significant persistence effect and a noticeable positive effect of both the number of tests and the number of tests per 10,000. Region 3 has the largest constant (average of state-specific effects) and the largest coefficient on tests, suggesting that the number of people newly tested for the virus is an important explanatory factor for the number of new cases. Region 4 has a high constant (average state-specific value) and significant shifts in both the 1-day and 7-day persistence values.
University Reopenings
Regression Results
A significant advantage of the panel data approach is that it can provide statistically valid quantifications of shifts in a fairly short period such as 1 week. Perhaps the biggest pandemic issue during the week of August 24 was the high number of cases reported on university campuses as they reopened. We address this with an additional regression analysis. Six states in Region 3 reported 500 or more cases; at least one other university in these states reported 200 or more cases (Alabama, Florida, Georgia, North Carolina, South Carolina, and Texas). To inform this university effect, we split Region 3 into two groups of states—one with a high prevalence of university COVID-19 positives (denoted as group 3a) and another comprising the remaining Region 3 states (denoted as group 3b)—and then ran the regression analysis on the two groups (Table 2).
Table 2.
Arellano-Bond dynamic panel data modeling of the number of daily infections reported by states in Region 3, grouped by the university effect, August 2-30, 2020.
| Variable | Group 3a (with university effect) | Group 3b (without university effect) | |||
|
|
Coefficient | P value | Coefficient | P value | |
| L1Pos | –0.023 | .76 | 0.037 | .34 | |
| L1shiftAug17 | 0.249 | .13 | –0.029 | .74 | |
| L1shiftAug24 | –0.075 | .69 | 0.005 | .95 | |
| L7Pos | 0.268 | <.001 | 0.213 | <.001 | |
| L7shiftAug17 | –0.364 | .007 | –0.100 | .22 | |
| L7shiftAug24 | –0.252 | .12 | 0.092 | .26 | |
| Tests | 0.121 | <.001 | 0.029 | <.001 | |
| Tests_squared | –6.59E-09 | <.001 | –5.61E-07 | <.001 | |
| Tests_per_10K | –39.704 | .005 | –4.402 | .06 | |
| Constant | 910.482 | .008 | 245.307 | <.001 | |
| Wald statistic for regression | χ29=169 | <.001 | χ29=491 | <.001 | |
| Sargan statistic for validity | χ2165=149 | .81 | χ2310=301 | .63 | |
For each group, the Wald statistic shows that the model was statistically significant (χ29=169, P<.001; and χ29=491, P<.001, respectively), and the Sargan test fails to reject the validity of the overidentifying restrictions (χ2165=149 P=.81; χ2310=301, P=.63).
Without belaboring the individual coefficients, there are two important differences between the two groups. First is the coefficient on Tests, which numerically is the most important of the three test coefficients; group 3a (0.121, P<.001) is more than four times the size of the coefficient for group 3b (0.029, P<.001). The second important difference is that the constant for group a is more than three times the size of the constant for group b.
Interpretation
The larger coefficient on Tests means that a higher percentage of tests are associated with positive results, possibly as large as 10% for group a (considering only the linear term). The larger value of the constant (which is an average of state-specific effects) means that there are larger state-specific risk factors, possibly related to the degree of “lockdown” and social compliance with recommendations such as social distancing or wearing masks. Coupling these two effects suggests that for the week of August 24, the university effect is mostly due to increases in the number of asymptomatic students who got tested for the first time as they returned to university. This is consistent with the comparison of regional results across regions. It also suggests that the following week may be much worse if a significant fraction of the students are infectious and fail to practice social distancing, etc, thereby infecting others, who will likely show up in that week’s numbers.
These results may also help to explain spikes in other states, such as Iowa, Kansas, North Dakota, and South Dakota (which is also potentially affected by the Sturgis Motorcycle Rally), which all had significant numbers of COVID-19 cases at universities.
Surveillance Results
Surveillance results are presented in Tables 3 and 4. The seven data elements in this proof-of-concept surveillance system are calculated as weekly averages and the speed, acceleration, and jerk are normalized per 100,000 population to compare the transmission of COVID-19 from week to week. These surveillance system data elements include (1) average weekly number of daily tests; (2) average weekly number of daily tests per 100,000 population; (3) average weekly number of daily positive tests; (4) average weekly number of daily positive tests per 100,000 population referred to as speed; (5) weekly average of day-to-day change in the number of positives per day per 100,000 population, referred to as acceleration; (6) change in acceleration, referred to as jerk, which is the acceleration in the current week minus the acceleration in the prior week; a sustained positive jerk is typically associated with explosive growth; and finally, (7) the 7-day lag, which is the number of new cases of COVID-19 reported today per 100,000 population (ie, today’s speed) that are associated with new cases reported 7 days ago (ie, last week’s speed), and measures how much the increase in speed from last week persists into this week. Data are presented according to US Census regions. Data element 1 is reported as a number while 2-7 are reported as a rate, which better allows for comparison between US states.
Table 3.
Surveillance metrics for the week of August 17-23, 2020.
| State | Tests per day, n (weekly average) | Daily tests per 100K people, n (daily average for the week) | Positives, n (reported number of new positive test results or confirmed cases per day per 100K people, weekly average) | Speed, n (daily positives per 100K people, weekly average) | Acceleration (day-to-day change in the number of positives per day, weekly average, per 100K people) | Jerk (week-over-week change in acceleration, per 100K people) | 7-day persistence effect on speed (number of new cases per day per 100K people) | |
| Region 1 |
|
|
|
|
||||
|
|
CT | 16,936 | 475 | 127 | 3.56 | 0.49 | –0.16 | 0.32 |
|
|
ME | 3068 | 228 | 24 | 1.78 | –0.06 | –0.15 | 0.18 |
|
|
MA | 14,815 | 215 | 309 | 4.48 | –0.63 | –0.57 | 0.61 |
|
|
NH | 1591 | 117 | 17 | 1.25 | 0.07 | 0.13 | 0.23 |
|
|
NJ | 22,687 | 255 | 291 | 3.28 | 0.39 | 0.93 | 0.59 |
|
|
NY | 78,995 | 406 | 604 | 3.11 | –0.03 | –0.09 | 0.47 |
|
|
PA | 13,737 | 107 | 655 | 5.12 | –0.05 | 0.07 | 0.86 |
|
|
RI | 5884 | 555 | 107 | 10.10 | 0.18 | 0.16 | 1.12 |
|
|
VT | 1273 | 204 | 6 | 0.96 | –0.05 | –0.07 | 0.18 |
| Region 2 |
|
|
|
|
||||
|
|
IL | 48,181 | 380 | 2026 | 15.99 | 0.37 | 0.17 | 3.58 |
|
|
IN | 10,136 | 151 | 788 | 11.71 | –0.26 | 0.38 | 3.41 |
|
|
IA | 4398 | 139 | 550 | 17.42 | –0.43 | –1.01 | 4.35 |
|
|
KS | 4654 | 160 | 594 | 20.40 | 6.63 | –2.55 | 4.20 |
|
|
MI | 30,346 | 304 | 650 | 6.51 | 0.41 | 0.49 | 2.09 |
|
|
MN | 9467 | 168 | 633 | 11.23 | –0.06 | 0.09 | 2.85 |
|
|
MO | 9888 | 161 | 1086 | 17.69 | –0.61 | –3.11 | 6.20 |
|
|
NE | 2498 | 129 | 220 | 11.37 | –0.73 | –1.56 | 3.89 |
|
|
ND | 1584 | 208 | 184 | 24.16 | –0.06 | –1.09 | 4.91 |
|
|
OH | 22035 | 189 | 931 | 7.96 | 0.03 | 0.35 | 2.40 |
|
|
SD | 1141 | 129 | 143 | 16.18 | –0.24 | –0.69 | 2.85 |
|
|
WI | 8511 | 146 | 708 | 12.17 | –0.54 | –0.73 | 3.51 |
| Region 3 |
|
|
|
|
||||
|
|
AL | 10,749 | 219 | 947 | 19.31 | –0.95 | 5.15 | –1.33 |
|
|
AR | 6236 | 207 | 558 | 18.50 | –1.41 | 1.77 | –1.12 |
|
|
DE | 1637 | 168 | 63 | 6.51 | 0.18 | 0.44 | –0.83 |
|
|
DC | 3313 | 469 | 53 | 7.49 | –0.10 | 0.69 | –0.61 |
|
|
FL | 28,001 | 130 | 3879 | 18.06 | –0.54 | 1.09 | –1.74 |
|
|
GA | 23,802 | 224 | 2417 | 22.76 | –0.18 | 1.58 | –1.77 |
|
|
KY | 5339 | 119 | 602 | 13.47 | 2.81 | 2.87 | –0.89 |
|
|
LA | 15,107 | 325 | 718 | 15.44 | 0.13 | 4.65 | –1.29 |
|
|
MD | 12927 | 214 | 556 | 9.19 | 0.14 | 1.09 | –0.72 |
|
|
MS | 2015 | 68 | 823 | 27.64 | 1.18 | 1.88 | –1.53 |
|
|
NC | 21,975 | 210 | 1452 | 13.84 | 0.31 | 0.59 | –0.77 |
|
|
OK | 8220 | 208 | 689 | 17.41 | 0.08 | –0.13 | –1.11 |
|
|
SC | 6362 | 124 | 784 | 15.24 | 0.22 | 1.22 | –1.08 |
|
|
TN | 26,836 | 393 | 1461 | 21.40 | –0.22 | 0.12 | –1.48 |
|
|
TX | 32,712 | 113 | 5994 | 20.67 | –1.16 | –2.07 | –1.56 |
|
|
VA | 16,720 | 196 | 897 | 10.51 | –0.07 | –0.14 | –0.71 |
|
|
WV | 5836 | 338 | 101 | 5.85 | –0.17 | 0.03 | –0.46 |
| Region 4 |
|
|
|
|
||||
|
|
AK | 3704 | 506 | 71 | 9.73 | –0.82 | –0.94 | 0.28 |
|
|
AZ | 8414 | 116 | 652 | 8.96 | –1.32 | –1.46 | 0.31 |
|
|
CA | 106,128 | 269 | 6015 | 15.22 | –0.40 | –0.22 | 0.59 |
|
|
CO | 10,060 | 175 | 292 | 5.07 | –0.01 | 0.32 | 0.15 |
|
|
HI | 2412 | 170 | 219 | 15.45 | 0.02 | –0.49 | 0.36 |
|
|
ID | 2008 | 112 | 312 | 17.47 | –0.09 | 2.06 | 0.57 |
|
|
MT | 1261 | 118 | 97 | 9.07 | –0.51 | –0.88 | 0.26 |
|
|
NV | 3824 | 124 | 614 | 19.92 | –0.77 | –0.24 | 0.57 |
|
|
NM | 5696 | 272 | 143 | 6.81 | 0.44 | 0.50 | 0.19 |
|
|
OR | 4432 | 105 | 239 | 5.67 | –0.06 | 0.00 | 0.16 |
|
|
UT | 3758 | 117 | 352 | 10.98 | –0.13 | 0.07 | 0.27 |
|
|
WA | 11,587 | 152 | 419 | 5.50 | –0.17 | –0.08 | 0.17 |
|
|
WY | 685 | 118 | 42 | 7.23 | –0.57 | –1.11 | 0.14 |
Table 4.
Surveillance metrics for the week of August 24-30, 2020.
| State | Tests per day, n (weekly average) | Daily tests per 100K people, n (daily average for the week) | Positives, n (reported number of new positive test results or confirmed cases per day per 100K people, weekly average) | Speed, n (daily positives per 100K people, weekly average) | Acceleration (day-to-day change in the number of positives per day, weekly average, per 100K people) | Jerk (week-over-week change in acceleration, per 100K people) | 7-day persistence effect on speed (number of new cases per day per 100K people) | ||||||||
| Region 1 |
|
|
|
|
|||||||||||
|
|
CT | 21,027 | 590 | 195 | 5.48 | 0.81 | 0.33 | 0.55 | |||||||
|
|
ME | 3994 | 297 | 25 | 1.88 | 0.05 | 0.12 | 0.28 | |||||||
|
|
MA | 24,300 | 353 | 410 | 5.95 | 0.41 | 1.04 | 0.69 | |||||||
|
|
NH | 1890 | 139 | 21 | 1.54 | –0.07 | –0.15 | 0.19 | |||||||
|
|
NJ | 26,762 | 301 | 302 | 3.40 | 0.05 | –0.34 | 0.51 | |||||||
|
|
NY | 82,233 | 423 | 623 | 3.20 | 0.09 | 0.12 | 0.48 | |||||||
|
|
PA | 13,769 | 108 | 637 | 4.97 | 0.06 | 0.10 | 0.79 | |||||||
|
|
RI | 4963 | 469 | 60 | 5.66 | –1.19 | –1.36 | 1.57 | |||||||
|
|
VT | 1890 | 303 | 8 | 1.35 | 0.16 | 0.21 | 0.15 | |||||||
| Region 2 |
|
|
|
|
|||||||||||
|
|
IL | 44,719 | 353 | 1923 | 15.18 | 0.11 | –0.26 | 5.35 | |||||||
|
|
IN | 12,508 | 186 | 1054 | 15.66 | 0.56 | 0.82 | 3.92 | |||||||
|
|
IA | 5017 | 159 | 921 | 29.18 | 1.95 | 2.38 | 5.83 | |||||||
|
|
KS | 7366 | 253 | 838 | 28.78 | 7.63 | 1.00 | 6.82 | |||||||
|
|
MI | 30,189 | 302 | 817 | 8.18 | 0.90 | 0.48 | 2.18 | |||||||
|
|
MN | 8822 | 156 | 801 | 14.20 | 0.54 | 0.60 | 3.75 | |||||||
|
|
MO | 8486 | 138 | 1226 | 19.97 | 1.51 | 2.11 | 5.92 | |||||||
|
|
NE | 2798 | 145 | 282 | 14.57 | 1.20 | 1.93 | 3.80 | |||||||
|
|
ND | 1297 | 170 | 261 | 34.23 | 1.46 | 1.52 | 8.08 | |||||||
|
|
OH | 30,424 | 260 | 1066 | 9.12 | 0.35 | 0.32 | 2.66 | |||||||
|
|
SD | 1270 | 144 | 292 | 33.04 | 3.86 | 4.10 | 5.41 | |||||||
|
|
WI | 8464 | 145 | 728 | 12.50 | 0.22 | 0.76 | 4.07 | |||||||
| Region 3 |
|
|
|
|
|||||||||||
|
|
AL | 8485 | 173 | 1454 | 29.65 | 2.38 | 3.33 | 0.09 | |||||||
|
|
AR | 6712 | 222 | 612 | 20.27 | 0.49 | 1.90 | 0.08 | |||||||
|
|
DE | 1831 | 188 | 66 | 6.81 | –0.98 | –1.16 | 0.03 | |||||||
|
|
DC | 3149 | 446 | 53 | 7.47 | –0.45 | –0.34 | 0.03 | |||||||
|
|
FL | 24,425 | 114 | 3002 | 13.98 | –0.26 | 0.28 | 0.08 | |||||||
|
|
GA | 22,229 | 209 | 2146 | 20.21 | –0.69 | –0.51 | 0.10 | |||||||
|
|
KY | 9483 | 212 | 643 | 14.40 | –2.59 | –5.40 | 0.06 | |||||||
|
|
LA | 14,987 | 322 | 703 | 15.13 | 1.23 | 1.10 | 0.07 | |||||||
|
|
MD | 12,335 | 204 | 527 | 8.72 | –0.19 | –0.34 | 0.04 | |||||||
|
|
MS | 4988 | 168 | 683 | 22.95 | 0.10 | –1.08 | 0.13 | |||||||
|
|
NC | 23,543 | 224 | 1573 | 15.00 | –0.57 | –0.88 | 0.06 | |||||||
|
|
OK | 7438 | 188 | 694 | 17.53 | 0.36 | 0.29 | 0.08 | |||||||
|
|
SC | 7220 | 140 | 905 | 17.58 | 1.06 | 0.84 | 0.07 | |||||||
|
|
TN | 20,545 | 301 | 1311 | 19.20 | –2.13 | –1.91 | 0.10 | |||||||
|
|
TX | 36,669 | 126 | 4688 | 16.17 | –0.28 | 0.87 | 0.09 | |||||||
|
|
VA | 14,649 | 172 | 969 | 11.35 | 0.07 | 0.15 | 0.05 | |||||||
|
|
WV | 4990 | 289 | 120 | 6.92 | 0.46 | 0.63 | 0.03 | |||||||
| Region 4 |
|
|
|
|
|||||||||||
|
|
AK | 2771 | 379 | 75 | 10.31 | –0.25 | 0.57 | 3.92 | |||||||
|
|
AZ | 6939 | 95 | 508 | 6.98 | 0.33 | 1.65 | 3.61 | |||||||
|
|
CA | 98,685 | 250 | 5177 | 13.10 | –0.26 | 0.14 | 6.14 | |||||||
|
|
CO | 9257 | 161 | 308 | 5.35 | –0.07 | –0.06 | 2.05 | |||||||
|
|
HI | 2536 | 179 | 255 | 17.99 | 0.25 | 0.23 | 6.23 | |||||||
|
|
ID | 2435 | 136 | 288 | 16.11 | 0.00 | 0.09 | 7.04 | |||||||
|
|
MT | 5130 | 480 | 130 | 12.17 | 0.48 | 0.99 | 3.66 | |||||||
|
|
NV | 3065 | 100 | 472 | 15.34 | –0.39 | 0.37 | 8.03 | |||||||
|
|
NM | 6766 | 323 | 125 | 5.97 | –0.48 | –0.93 | 2.75 | |||||||
|
|
OR | 4789 | 114 | 231 | 5.48 | 0.12 | 0.17 | 2.29 | |||||||
|
|
UT | 4382 | 137 | 391 | 12.20 | 0.66 | 0.79 | 4.42 | |||||||
|
|
WA | 11,760 | 154 | 380 | 4.99 | 1.80 | 1.96 | 2.22 | |||||||
|
|
WY | 1486 | 257 | 34 | 5.95 | 0.00 | 0.57 | 2.92 | |||||||
The innovation of this study is the novel metrics we derived to measure how COVID-19 is spreading and changes in terms of transmission rates. These measures should be considered in combination with traditional static numbers including transmission rates and death rates. These novel metrics measure how fast the rates are changing, accounting for their data limitations.
As an example, we tracked the transmission of COVID-19 for the state of Illinois for the week from August 17 to 23, 2020. Illinois had a weekly average of 48,181 COVID-19 tests daily, also expressed as a weekly average of 380 tests per 100,000 population per day. Illinois had a weekly average of 2026 positive tests per day. The speed of the COVID-19 transmission is measured as an increase of 15.99 persons infected per 100,000 population per day. For the week of August 17 to 23 in Illinois, COVID-19 acceleration was 0.37, which means that every day there were .37 more new cases per 100,000 than the day before, or 2.6 more cases per day per 100,000 over the course of the week. The jerk is 0.17, which means that acceleration was increasing: this increased acceleration accounted for 1.4 of the 2.6 additional cases per day per 100,000. Finally, the 7-day lag effect for speed is 3.58, which means that persistence or echo effects accounted for 3.58 or 22% of the 15.99 new daily positive cases per 100,000, which indicates an important but moderate persistence or echo effect for the week of August 17.
We see significant differences in COVID-19 transmission the following week (August 24-30, 2020). Illinois experienced a decrease in weekly average tests to 44,719 daily COVID-19 tests, also expressed as a weekly average of 353 tests per 100,000 population per day. This is 27 fewer tests per 100,000 population from last week. Illinois had a weekly average of 1923 positive tests per day, a decrease from the prior week, also expressed as a speed of 15.18 persons newly infected per day per 100,000 population. During the week of August 24-30, the acceleration decreased from the previous week to 0.11 and the jerk was negative (–0.26), indicating a leveling off of growth in new cases. Finally, the 7-day lag effect on speed is 5.35, which means that the persistence or echo effects accounted for 5.35 or over one-third of the 15.18 new daily positive cases per 100,000. The increased importance of echo effects rather than new cases from other (new) causes is consistent with a leveling off of COVID-19 growth in Illinois during the week of August 24-30.
In summary, the week of August 17-23 showed an increasing COVID-19 speed with positive acceleration and jerk. The week of August 24-30 exhibited a moderation in speed with lower acceleration and negative jerk. This is indicative of a leveling off or an inflection point: the pandemic in Illinois may be starting to decline, or this could be simply a pause before a continued increase in COVID-19 speed.
Discussion
Principal Findings
The dynamic panel data model is a statistically validated analysis of reported COVID-19 transmissions and an important addition to the epidemiological toolkit for understanding the progression of the pandemic. It is important to recognize that surveillance systems require a variety of metrics. Systematic surveillance with standardized measures of decreases and increases in COVID-19 transmission coupled with health policies and guidelines add a critical tool to the epidemiologic arsenal to combat COVID-19.
The specific findings of the modeling exercise confirm that SARS-CoV-2 infection rates are persistent but changeable, and for most states increasing during the period between June 13-19, 2020. We find that for every 100 new COVID-19 cases from June 13-19, the following day would result in 26 new cases, meaning there is a significant reduction each day. However, it is important to recognize that this is an average across states and that state and local experiences will vary, which we measured. From June 20-26, on average in the United States, every 100 new cases on Monday was associated with 65 new cases on Tuesday, indicating the contagion increased 2.5-fold the rate from the prior week. The American pandemic has been ramping up in the past 2 weeks.
Remarkably, the US states diverged into three distinct patterns: (1) decline, (2) constant, and (3) increases consistent with outbreaks. In the 30 states with increasing cases, over the course of 2 weeks, there was a 3.6-fold increase in new infections while the states that had sustained declines in cases decreased by 2.5-fold. Again, these are averages among the three classifications of decline, constant, and increases, but these data could be further refined to show how much each state contributed to increases and decreases. Further investigation could usefully model state and local differences in infection rates, as well as ascertain quickly whether the pandemic will continue to re-emerge in the United States, or whether infection rates will reverse track and decline again even though states reopen.
The strengths of this study are the derived new metrics of the transmission of COVID-19. The limitation of this proof-of-concept surveillance system is that it includes only dynamic cases of COVID-19 infections; a full surveillance system should also include static cases. For example, Table 2 refers only to dynamic, new infections.
Based on the empirical evidence that our metrics of the COVID-19 contagion is a good standardization of increases and decreases for public health surveillance purposes, our future work will focus on the surveillance of 195 countries in eight global regions as defined by the World Bank. When possible, we will provide subcountry-level metrics of the COVID-19 contagion beginning with US states and Canadian provinces. Our surveillance system will include estimates of speed, acceleration, and jerk in acceleration along with traditional surveillance metrics.
Footnotes
Conflicts of Interest: None declared.
References
- 1.Lai C, Shih T, Ko W, Tang H, Hsueh P. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int J Antimicrob Agents. 2020 Mar;55(3):105924. doi: 10.1016/j.ijantimicag.2020.105924. http://europepmc.org/abstract/MED/32081636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yuen K, Ye Z-, Fung S, Chan C, Jin D. SARS-CoV-2 and COVID-19: The most important research questions. Cell Biosci. 2020 Mar 16;10(1):40. doi: 10.1186/s13578-020-00404-4. https://cellandbioscience.biomedcentral.com/articles/10.1186/s13578-020-00404-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumari T, Shukla V. COVID-19: Towards confronting an unprecedented pandemic. IJBI. 2020;02(01):01–10. doi: 10.46505/ijbi.2020.2101. [DOI] [Google Scholar]
- 4.Yan Z. Unprecedented pandemic, unprecedented shift, and unprecedented opportunity. Hum Behav Emerg Technol. 2020 Apr 06;2(2):110–112. doi: 10.1002/hbe2.192. http://europepmc.org/abstract/MED/32427197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y, Liu B, Cui J. COVID-19 Evolves in Human Hosts. Preprints. 2020:e. doi: 10.20944/preprints202003.0316.v1. https://www.preprints.org/manuscript/202003.0316/v1. [DOI] [Google Scholar]
- 6.Velavan TP, Meyer CG. The COVID-19 epidemic. Trop Med Int Health. 2020 Mar 16;25(3):278–280. doi: 10.1111/tmi.13383. doi: 10.1111/tmi.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heymann DL, Shindo N. COVID-19: what is next for public health? The Lancet. 2020 Feb;395(10224):542–545. doi: 10.1016/s0140-6736(20)30374-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salathé Marcel, Althaus CL, Neher R, Stringhini S, Hodcroft E, Fellay J, Zwahlen M, Senti G, Battegay M, Wilder-Smith A, Eckerle I, Egger M, Low N. COVID-19 epidemic in Switzerland: on the importance of testing, contact tracing and isolation. Swiss Med Wkly. 2020 Mar 09;150:w20225. doi: 10.4414/smw.2020.20225. https://doi.emh.ch/10.4414/smw.2020.20225. [DOI] [PubMed] [Google Scholar]
- 9.Cohen J, Kupferschmidt K. Countries test tactics in 'war' against COVID-19. Science. 2020 Mar 20;367(6484):1287–1288. doi: 10.1126/science.367.6484.1287. [DOI] [PubMed] [Google Scholar]
- 10.La V, Pham T, Ho M, Nguyen M, P. Nguyen K, Vuong T, Nguyen HT, Tran T, Khuc Q, Ho M, Vuong Q. Policy Response, Social Media and Science Journalism for the Sustainability of the Public Health System Amid the COVID-19 Outbreak: The Vietnam Lessons. Sustainability. 2020 Apr 07;12(7):2931. doi: 10.3390/su12072931. [DOI] [Google Scholar]
- 11.Park S, Choi GJ, Ko H. Information Technology-Based Tracing Strategy in Response to COVID-19 in South Korea-Privacy Controversies. JAMA. 2020 Jun 02;323(21):2129–2130. doi: 10.1001/jama.2020.6602. [DOI] [PubMed] [Google Scholar]
- 12.Hellewell J, Abbott S, Gimma A, Bosse NI, Jarvis CI, Russell TW, Munday JD, Kucharski AJ, Edmunds WJ, Funk S, Eggo RM, Sun F, Flasche S, Quilty BJ, Davies N, Liu Y, Clifford S, Klepac P, Jit M, Diamond C, Gibbs H, van Zandvoort K. Feasibility of controlling COVID-19 outbreaks by isolation of cases and contacts. The Lancet Global Health. 2020 Apr;8(4):e488–e496. doi: 10.1016/s2214-109x(20)30074-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Z, McGoogan JM. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72 314 Cases From the Chinese Center for Disease Control and Prevention. JAMA. 2020 Apr 07;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 14.Parodi SM, Liu VX. From Containment to Mitigation of COVID-19 in the US. JAMA. 2020 Apr 21;323(15):1441–1442. doi: 10.1001/jama.2020.3882. [DOI] [PubMed] [Google Scholar]
- 15.Watkins J. Preventing a covid-19 pandemic. BMJ. 2020 Feb 28;368:m810. doi: 10.1136/bmj.m810. [DOI] [PubMed] [Google Scholar]
- 16.Del Rio Carlos, Malani PN. COVID-19-New Insights on a Rapidly Changing Epidemic. JAMA. 2020 Apr 14;323(14):1339–1340. doi: 10.1001/jama.2020.3072. [DOI] [PubMed] [Google Scholar]
- 17.Bedford J, Enria D, Giesecke J, Heymann DL, Ihekweazu C, Kobinger G, Lane HC, Memish Z, Oh M, Sall AA, Schuchat A, Ungchusak K, Wieler LH. COVID-19: towards controlling of a pandemic. The Lancet. 2020 Mar;395(10229):1015–1018. doi: 10.1016/s0140-6736(20)30673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng VCC, Wong S, Chen JHK, Yip CCY, Chuang VWM, Tsang OTY, Sridhar S, Chan JFW, Ho P, Yuen K. Escalating infection control response to the rapidly evolving epidemiology of the coronavirus disease 2019 (COVID-19) due to SARS-CoV-2 in Hong Kong. Infect Control Hosp Epidemiol. 2020 May 05;41(5):493–498. doi: 10.1017/ice.2020.58. http://europepmc.org/abstract/MED/32131908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spinelli A, Pellino G. COVID-19 pandemic: perspectives on an unfolding crisis. Br J Surg. 2020 Jun 23;107(7):785–787. doi: 10.1002/bjs.11627. http://europepmc.org/abstract/MED/32191340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grech V. Unknown unknowns - COVID-19 and potential global mortality. Early Hum Dev. 2020 May;144:105026. doi: 10.1016/j.earlhumdev.2020.105026. http://europepmc.org/abstract/MED/32247898. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Xu S, Li Y. Beware of the second wave of COVID-19. The Lancet. 2020 Apr;395(10233):1321–1322. doi: 10.1016/s0140-6736(20)30845-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo T, Fan Y, Chen M, Wu X, Zhang L, He T, Wang H, Wan J, Wang X, Lu Z. Cardiovascular Implications of Fatal Outcomes of Patients With Coronavirus Disease 2019 (COVID-19) JAMA Cardiol. 2020 Jul 01;5(7):811–818. doi: 10.1001/jamacardio.2020.1017. http://europepmc.org/abstract/MED/32219356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Modes of transmission of virus causing COVID-19: implications for IPC precaution recommendations. World Health Organization. 2020. Mar, [2020-09-17]. https://www.who.int/news-room/commentaries/detail/modes-of-transmission-of-virus-causing-covid-19-implications-for-ipc-precaution-recommendations.
- 24.Ather A, Patel B, Ruparel NB, Diogenes A, Hargreaves KM. Reply to "Coronavirus Disease 19 (COVID-19): Implications for Clinical Dental Care". J Endod. 2020 Sep;46(9):1342. doi: 10.1016/j.joen.2020.08.005. http://europepmc.org/abstract/MED/32810475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Connors J, Levy J. COVID-19 and its implications for thrombosis and anticoagulation. Blood. 2020 Jun 04;135(23):2033–2040. doi: 10.1182/blood.2020006000. http://europepmc.org/abstract/MED/32339221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao X. COVID-19: immunopathology and its implications for therapy. Nat Rev Immunol. 2020 May 9;20(5):269–270. doi: 10.1038/s41577-020-0308-3. http://europepmc.org/abstract/MED/32273594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicola M, Alsafi Z, Sohrabi C, Kerwan A, Al-Jabir A, Iosifidis C, Agha M, Agha R. The socio-economic implications of the coronavirus pandemic (COVID-19): A review. Int J Surg. 2020 Jun;78:185–193. doi: 10.1016/j.ijsu.2020.04.018. http://europepmc.org/abstract/MED/32305533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenhalgh T, Koh GCH, Car J. Covid-19: a remote assessment in primary care. BMJ. 2020 Mar 25;368:m1182. doi: 10.1136/bmj.m1182. [DOI] [PubMed] [Google Scholar]
- 29.Chew M, Ong L, Koh F, Ng A, Tan Y, Ong B-C. Guest post: Lessons in preparedness? The response to the COVID-19 pandemic by a surgical department in Singapore. BJS. 2020. Mar, [2020-09-17]. https://cuttingedgeblog.com/2020/03/29/guest-post-lessons-in-preparedness-the-response-to-the-covid-19-pandemic-by-a-surgical-department-in-singapore/
- 30.Barro R, Ursúa J, Weng J. The Coronavirus and the Great Influenza Pandemic: Lessons from the “spanish flu” for the coronavirus’s potential effects on mortality and economic activity. National Bureau of Economic Research. 2020 Apr;:e. doi: 10.3386/w26866. https://www.nber.org/papers/w26866. [DOI] [Google Scholar]
- 31.Baker S, Bloom N, Davis S, Kost K, Sammon M, Viratyosin T. The unprecedented stock market impact of COVID-19. National Bureau of Economic Research. 2020 Jun;:e. doi: 10.3386/w26945. https://www.nber.org/papers/w26945. [DOI] [Google Scholar]
- 32.Bradbury-Jones Caroline, Isham L. The pandemic paradox: The consequences of COVID-19 on domestic violence. J Clin Nurs. 2020 Jul 22;29(13-14):2047–2049. doi: 10.1111/jocn.15296. http://europepmc.org/abstract/MED/32281158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taub A. A New Covid-19 Crisis: Domestic Abuse Rises Worldwide. The New York Times. 2020. Apr, [2020-09-17]. https://www.nytimes.com/2020/04/06/world/coronavirus-domestic-violence.html.
- 34.Usher K, Bhullar N, Durkin J, Gyamfi N, Jackson D. Family violence and COVID-19: Increased vulnerability and reduced options for support. Int J Ment Health Nurs. 2020 Aug 07;29(4):549–552. doi: 10.1111/inm.12735. http://europepmc.org/abstract/MED/32314526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boserup B, McKenney M, Elkbuli A. Alarming trends in US domestic violence during the COVID-19 pandemic. Am J Emerg Med. 2020 Apr 28;:e. doi: 10.1016/j.ajem.2020.04.077. http://europepmc.org/abstract/MED/32402499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.COVID-19 and violence against women: what the health sector/system can do. World Health Organization. 2020. Apr, [2020-09-17]. https://www.who.int/reproductivehealth/publications/vaw-covid-19/en/
- 37.Leslie E, Wilson R. Sheltering in Place and Domestic Violence: Evidence from Calls for Service during COVID-19. SSRN Journal. 2020 May;:e. doi: 10.2139/ssrn.3600646. doi: 10.2139/ssrn.3600646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfefferbaum B, North CS. Mental Health and the Covid-19 Pandemic. N Engl J Med. 2020 Aug 06;383(6):510–512. doi: 10.1056/nejmp2008017. [DOI] [PubMed] [Google Scholar]
- 39.Yao H, Chen J, Xu Y. Patients with mental health disorders in the COVID-19 epidemic. The Lancet Psychiatry. 2020 Apr;7(4):e21. doi: 10.1016/s2215-0366(20)30090-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holmes Ea, O'Connor Rc, Perry Vh, Tracey I, Wessely S, Arseneault L, Ballard C, Christensen H, Cohen Silver R, Everall I, Ford T, John A, Kabir T, King K, Madan I, Michie S, Przybylski Ak, Shafran R, Sweeney A, Worthman Cm, Yardley L, Cowan K, Cope C, Hotopf M, Bullmore E. Multidisciplinary research priorities for the COVID-19 pandemic: a call for action for mental health science. The Lancet Psychiatry. 2020 Jun;7(6):547–560. doi: 10.1016/S2215-0366(20)30168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greenberg N, Docherty M, Gnanapragasam S, Wessely S. Managing mental health challenges faced by healthcare workers during covid-19 pandemic. BMJ. 2020 Mar 26;368:m1211. doi: 10.1136/bmj.m1211. [DOI] [PubMed] [Google Scholar]
- 42.Mental health and psychosocial considerations during the COVID-19 outbreak, 18 March 2020. World Health Organization. 2020. Mar, [2020-09-17]. https://www.who.int/docs/default-source/coronaviruse/mental-health-considerations.pdf.
- 43.Duan L, Zhu G. Psychological interventions for people affected by the COVID-19 epidemic. The Lancet Psychiatry. 2020 Apr;7(4):300–302. doi: 10.1016/s2215-0366(20)30073-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Lancker W, Parolin Z. COVID-19, school closures, and child poverty: a social crisis in the making. The Lancet Public Health. 2020 May;5(5):e243–e244. doi: 10.1016/s2468-2667(20)30084-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.The Lancet India under COVID-19 lockdown. The Lancet. 2020 Apr;395(10233):1315. doi: 10.1016/s0140-6736(20)30938-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gostin LO, Hodge JG, Wiley LF. Presidential Powers and Response to COVID-19. JAMA. 2020 Apr 28;323(16):1547–1548. doi: 10.1001/jama.2020.4335. [DOI] [PubMed] [Google Scholar]
- 47.Coronavirus disease 2019 (COVID-19) Situation Report – 72. World Health Organization. 2020. Apr, [2020-09-17]. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200401-sitrep-72-covid-19.pdf?sfvrsn=3dd8971b_2.
- 48.Shahid Z, Kalayanamitra R, McClafferty B, Kepko D, Ramgobin D, Patel R, Aggarwal CS, Vunnam R, Sahu N, Bhatt D, Jones K, Golamari R, Jain R. COVID-19 and Older Adults: What We Know. J Am Geriatr Soc. 2020 May 20;68(5):926–929. doi: 10.1111/jgs.16472. http://europepmc.org/abstract/MED/32255507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nikolich-Zugich J, Knox KS, Rios CT, Natt B, Bhattacharya D, Fain MJ. Correction to: SARS-CoV-2 and COVID-19 in older adults: what we may expect regarding pathogenesis, immune responses, and outcomes. GeroScience. 2020 May 3;42(3):1013–1013. doi: 10.1007/s11357-020-00193-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jordan R, Adab P, Cheng K. Covid-19: risk factors for severe disease and death. BMJ. 2020 Mar 26;368:m1198. doi: 10.1136/bmj.m1198. [DOI] [PubMed] [Google Scholar]
- 51.Garnier-Crussard Antoine, Forestier E, Gilbert T, Krolak-Salmon Pierre. Novel Coronavirus (COVID-19) Epidemic: What Are the Risks for Older Patients? J Am Geriatr Soc. 2020 May 12;68(5):939–940. doi: 10.1111/jgs.16407. http://europepmc.org/abstract/MED/32162679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lloyd-Sherlock P, Ebrahim S, Geffen L, McKee M. Bearing the brunt of covid-19: older people in low and middle income countries. BMJ. 2020 Mar 13;368:m1052. doi: 10.1136/bmj.m1052. [DOI] [PubMed] [Google Scholar]
- 53.Ahmed F, Ahmed N, Pissarides C, Stiglitz J. Why inequality could spread COVID-19. The Lancet Public Health. 2020 May;5(5):e240. doi: 10.1016/s2468-2667(20)30085-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sumner A, Hoy C, Ortiz-Juarez E. Estimates of the Impact of COVID-19 on Global Poverty. United Nations University / UNU-WIDER. 2020 Apr;:1–14. doi: 10.35188/unu-wider/2020/800-9. https://www.wider.unu.edu/publication/estimates-impact-covid-19-global-poverty. [DOI] [Google Scholar]
- 55.Laurencin CT, McClinton A. The COVID-19 Pandemic: a Call to Action to Identify and Address Racial and Ethnic Disparities. J Racial Ethn Health Disparities. 2020 Jun 18;7(3):398–402. doi: 10.1007/s40615-020-00756-0. http://europepmc.org/abstract/MED/32306369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mogaji E. Financial Vulnerability During a Pandemic: Insights for Coronavirus Disease (COVID-19) SSRN Journal. 2020:e. doi: 10.2139/ssrn.3564702. [DOI] [Google Scholar]
- 57.Webb Hooper M, Nápoles Anna María, Pérez-Stable Eliseo J. COVID-19 and Racial/Ethnic Disparities. JAMA. 2020 Jun 23;323(24):2466–2467. doi: 10.1001/jama.2020.8598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khunti K, Singh A, Pareek M, Hanif W. Is ethnicity linked to incidence or outcomes of covid-19? BMJ. 2020 Apr 20;369:m1548. doi: 10.1136/bmj.m1548. [DOI] [PubMed] [Google Scholar]
- 59.Chowkwanyun M, Reed AL. Racial Health Disparities and Covid-19 — Caution and Context. N Engl J Med. 2020 Jul 16;383(3):201–203. doi: 10.1056/nejmp2012910. [DOI] [PubMed] [Google Scholar]
- 60.Yancy CW. COVID-19 and African Americans. JAMA. 2020 May 19;323(19):1891–1892. doi: 10.1001/jama.2020.6548. [DOI] [PubMed] [Google Scholar]
- 61.Platt L, Warwick R. Are some ethnic groups more vulnerable to COVID-19 than others. Institute for Fiscal Studies / Nuffield Foundation. 2020. May, [2020-06-19]. https://www.ifs.org.uk/inequality/chapter/are-some-ethnic-groups-more-vulnerable-to-covid-19-than-others/
- 62.Pan A, Liu L, Wang C, Guo H, Hao X, Wang Q, Huang J, He N, Yu H, Lin X, Wei S, Wu T. Association of Public Health Interventions With the Epidemiology of the COVID-19 Outbreak in Wuhan, China. JAMA. 2020 May 19;323(19):1915–1923. doi: 10.1001/jama.2020.6130. http://europepmc.org/abstract/MED/32275295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Raoofi A, Takian A, Akbari Sari A, Olyaeemanesh A, Haghighi H, Aarabi M. COVID-19 Pandemic and Comparative Health Policy Learning in Iran. Arch Iran Med. 2020 Apr 01;23(4):220–234. doi: 10.34172/aim.2020.02. [DOI] [PubMed] [Google Scholar]
- 64.Anderson RM, Heesterbeek H, Klinkenberg D, Hollingsworth TD. How will country-based mitigation measures influence the course of the COVID-19 epidemic? The Lancet. 2020 Mar;395(10228):931–934. doi: 10.1016/s0140-6736(20)30567-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee V, Chiew C, Khong W. Interrupting transmission of COVID-19: lessons from containment efforts in Singapore. J Travel Med. 2020 May 18;27(3):e. doi: 10.1093/jtm/taaa039. http://europepmc.org/abstract/MED/32167146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nkengasong JN, Mankoula W. Looming threat of COVID-19 infection in Africa: act collectively, and fast. The Lancet. 2020 Mar;395(10227):841–842. doi: 10.1016/s0140-6736(20)30464-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dave D, Friedson A, Matsuzawa K, Sabia J. When Do Shelter-in-Place Orders Fight COVID-19 Best? Policy Heterogeneity Across States and Adoption Time. National Bureau of Economic Research. 2020 Aug 03;:e. doi: 10.1111/ecin.12944. http://europepmc.org/abstract/MED/32836519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Loayza N, Pennings S. Macroeconomic policy in the time of COVID-19: A primer for developing countries. Washington, DC: World Bank; 2020. Mar, [Google Scholar]
- 69.Adalja AA, Toner E, Inglesby TV. Priorities for the US Health Community Responding to COVID-19. JAMA. 2020 Apr 14;323(14):1343–1344. doi: 10.1001/jama.2020.3413. [DOI] [PubMed] [Google Scholar]
- 70.Raifman J, Nocka K, Jones D, Bor J, Lipson S, Jay J, Galea S. COVID-19 US State Policy Database. Ann Arbor, MI: Inter-university Consortium for Political and Social Research; 2020. Sep, [Google Scholar]
- 71.Fauci A, Lane H, Redfield R. Covid-19 - Navigating the Uncharted. N Engl J Med. 2020 Mar 26;382(13):1268–1269. doi: 10.1056/NEJMe2002387. http://europepmc.org/abstract/MED/32109011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Haffajee RL, Mello MM. Thinking Globally, Acting Locally — The U.S. Response to Covid-19. N Engl J Med. 2020 May 28;382(22):e75. doi: 10.1056/nejmp2006740. [DOI] [PubMed] [Google Scholar]
- 73.Liu M, Thomadsen R, Yao S. Forecasting the Spread of COVID-19 under Different Reopening Strategies. SSRN Journal. 2020:e. doi: 10.2139/ssrn.3607977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hodge JG. Federal vs. State Powers in Rush to Reopen Amid the Coronavirus Pandemic. SSRN Journal. 2020:e. doi: 10.2139/ssrn.3587011. [DOI] [Google Scholar]
- 75.Harris J. Reopening Under COVID-19: What to Watch For. National Bureau of Economic Research. 2020:e. doi: 10.3386/w27166. https://www.nber.org/papers/w27166. [DOI] [Google Scholar]
- 76.Alhaery M. A COVID-19 Reopening Readiness Index: The Key to Opening up the Economy. medRxiv. 2020:e. doi: 10.1101/2020.05.22.20110577. https://www.medrxiv.org/content/10.1101/2020.05.22.20110577v2. [DOI] [Google Scholar]
- 77.Koonin L. Novel coronavirus disease (COVID-19) outbreak: Now is the time to refresh pandemic plans. J Bus Contin Emer Plan. 2020 Jan 01;13(4):1–15. [PubMed] [Google Scholar]
- 78.Gostic K, Gomez A, Mummah R, Kucharski A, Lloyd-Smith J. Estimated effectiveness of symptom and risk screening to prevent the spread of COVID-19. eLife. 2020;9:e55570. doi: 10.7554/elife.55570. https://elifesciences.org/articles/55570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lurie N, Saville M, Hatchett R, Halton J. Developing Covid-19 Vaccines at Pandemic Speed. N Engl J Med. 2020 May 21;382(21):1969–1973. doi: 10.1056/nejmp2005630. [DOI] [PubMed] [Google Scholar]
- 80.Ahn D, Shin H, Kim M, Lee S, Kim H, Myoung J, Kim B, Kim S. Current Status of Epidemiology, Diagnosis, Therapeutics, and Vaccines for Novel Coronavirus Disease 2019 (COVID-19) J Microbiol Biotechnol. 2020 Mar 28;30(3):313–324. doi: 10.4014/jmb.2003.03011. http://www.jmb.or.kr/journal/view.html?doi=10.4014/jmb.2003.03011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miller A, Reandelar M, Fasciglione K, Roumenova V, Li Y, Otazu G. Correlation between universal BCG vaccination policy and reduced morbidity and mortality for COVID-19: an epidemiological study. medRxiv. 2020:e. doi: 10.1101/2020.03.24.20042937. https://www.medrxiv.org/content/10.1101/2020.03.24.20042937v2. [DOI] [Google Scholar]
- 82.Hennessy CH, Moriarty DG, Zack MM, Scherr PA, Brackbill R. Measuring health-related quality of life for public health surveillance. Public Health Rep. 1994;109(5):665–72. http://europepmc.org/abstract/MED/7938388. [PMC free article] [PubMed] [Google Scholar]
- 83.Woodall WH. The Use of Control Charts in Health-Care and Public-Health Surveillance. Journal of Quality Technology. 2018 Feb 05;38(2):89–104. doi: 10.1080/00224065.2006.11918593. [DOI] [Google Scholar]
- 84.Wharton M, Chorba T, Vogt R, Morse D, Buehler J. Case definitions for public health surveillance. MMWR Recomm Rep. 1990 Oct 19;39(RR-13):1–43. http://www.cdc.gov/mmwr/preview/mmwrhtml/00025629.htm. [PubMed] [Google Scholar]
- 85.Teutsch SM, Churchill RE, editors. Principles and Practice of Public Health Surveillance. 2nd ed. New York, NY: Oxford University Press; 2000. [Google Scholar]
- 86.Lee LM, Teutch SM, Thacker SB, St Louis ME, editors. New York, NY: Oxford University Press; 2010. [Google Scholar]
- 87.Thacker S, Berkelman R. Public health surveillance in the United States. Epidemiol Rev. 1988;10(1):164–90. doi: 10.1093/oxfordjournals.epirev.a036021. [DOI] [PubMed] [Google Scholar]
- 88.German R, Lee L M, Horan J M, Milstein B, Pertowski C, Waller M N, Guidelines Working Group Centers for Disease ControlPrevention (CDC) Updated guidelines for evaluating public health surveillance systems: recommendations from the Guidelines Working Group. MMWR Recomm Rep. 2001 Jul 27;50(RR-13):1–35; quiz CE1. [PubMed] [Google Scholar]
- 89.Declich S, Carter AO. Public health surveillance: historical origins, methods and evaluation. Bull World Health Organ. 1994;72(2):285–304. http://europepmc.org/abstract/MED/8205649. [PMC free article] [PubMed] [Google Scholar]
- 90.Mittal R, Ni R, Seo J. The flow physics of COVID-19. J. Fluid Mech. 2020 May 01;894:e. doi: 10.1017/jfm.2020.330. [DOI] [Google Scholar]
- 91.Cheng KK, Lam TH, Leung CC. Wearing face masks in the community during the COVID-19 pandemic: altruism and solidarity. The Lancet. 2020 Apr;:e. doi: 10.1016/s0140-6736(20)30918-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cheng VC, Wong S, Chuang VW, So SY, Chen JH, Sridhar S, To KK, Chan JF, Hung IF, Ho P, Yuen K. The role of community-wide wearing of face mask for control of coronavirus disease 2019 (COVID-19) epidemic due to SARS-CoV-2. J Infect. 2020 Jul;81(1):107–114. doi: 10.1016/j.jinf.2020.04.024. http://europepmc.org/abstract/MED/32335167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dalton C, Corbett S, Katelaris A. Pre-Emptive Low Cost Social Distancing and Enhanced Hygiene Implemented before Local COVID-19 Transmission Could Decrease the Number and Severity of Cases. SSRN Journal. 2020:e. doi: 10.2139/ssrn.3549276. [DOI] [Google Scholar]
- 94.Klompas M, Morris CA, Sinclair J, Pearson M, Shenoy ES. Universal Masking in Hospitals in the Covid-19 Era. N Engl J Med. 2020 May 21;382(21):e63. doi: 10.1056/nejmp2006372. [DOI] [PubMed] [Google Scholar]
- 95.Javid B, Weekes M, Matheson N. Covid-19: should the public wear face masks? BMJ. 2020 Apr 09;369:m1442. doi: 10.1136/bmj.m1442. [DOI] [PubMed] [Google Scholar]
- 96.Mesa Vieira C, Franco OH, Gómez Restrepo Carlos, Abel T. COVID-19: The forgotten priorities of the pandemic. Maturitas. 2020 Jun;136:38–41. doi: 10.1016/j.maturitas.2020.04.004. https://linkinghub.elsevier.com/retrieve/pii/S0378-5122(20)30234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Greenhalgh T, Schmid MB, Czypionka T, Bassler D, Gruer L. Face masks for the public during the covid-19 crisis. BMJ. 2020 Apr 09;369:m1435. doi: 10.1136/bmj.m1435. [DOI] [PubMed] [Google Scholar]
- 98.Eikenberry SE, Mancuso M, Iboi E, Phan T, Eikenberry K, Kuang Y, Kostelich E, Gumel AB. To mask or not to mask: Modeling the potential for face mask use by the general public to curtail the COVID-19 pandemic. Infect Dis Model. 2020;5:293–308. doi: 10.1016/j.idm.2020.04.001. https://linkinghub.elsevier.com/retrieve/pii/S2468-0427(20)30011-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fairchild A, Gostin L, Bayer R. Vexing, Veiled, and Inequitable: Social distancing and the "rights" divide in the age of COVID-19. Am J Bioeth. 2020 Jul 19;20(7):55–61. doi: 10.1080/15265161.2020.1764142. [DOI] [PubMed] [Google Scholar]
- 100.Bernstein J, Hutler B, Rieder T, Faden R, Han H, Barnhill A. An Ethics Framework for the COVID-19 Reopening Process. Johns Hopkins University. 2020. May, [2020-07-20]. https://bioethics.jhu.edu/wp-content/uploads/2019/10/FINAL-SNF-Agora-Covid-19.pdf.
- 101.Halow B. Five Challenges To A Smooth Post-COVID Reopening. 2020. [2020-07-20]. https://www.bashhalow.com/five-challenges-to-a-smooth-post-covid-reopening/
- 102.O'Dowd Adrian. Covid-19: Papers justifying government's plans to reopen schools are "inconclusive," say union bosses. BMJ. 2020 May 27;369:m2108. doi: 10.1136/bmj.m2108. [DOI] [PubMed] [Google Scholar]
- 103.Bhatia R, Jeffrey KM. A Step-by-Step Plan to Reopen California. The Bold Italic. 2020. [2020-07-20]. https://thebolditalic.com/covid-19-next-steps-for-california-c01632c8e8b6.
- 104.Teutsch SM. Considerations in Planning a Surveillance System. In: Lee LS, Teutsch SM, Thacker SB, St. Louis ME, editors. New York, NY: Oxford University Press; 2010. pp. 18–28. [Google Scholar]
- 105.Ren X. Pandemic and lockdown: a territorial approach to COVID-19 in China, Italy and the United States. Eurasian Geography and Economics. 2020 May 08;:1–12. doi: 10.1080/15387216.2020.1762103. [DOI] [Google Scholar]
- 106.Maier BF, Brockmann D. Effective containment explains subexponential growth in recent confirmed COVID-19 cases in China. Science. 2020 May 15;368(6492):742–746. doi: 10.1126/science.abb4557. http://europepmc.org/abstract/MED/32269067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Peng F, Tu L, Yang Y, Hu P, Wang R, Hu Q, Cao F, Jiang T, Sun J, Xu G, Chang C. Management and Treatment of COVID-19: The Chinese Experience. Can J Cardiol. 2020 Jun;36(6):915–930. doi: 10.1016/j.cjca.2020.04.010. http://europepmc.org/abstract/MED/32439306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tang B, Xia F, Bragazzi N. Lessons drawn from China and South Korea for managing COVID-19 epidemic: insights from a comparative modeling study. medRxiv. 2020:e. doi: 10.2471/blt.20.257238. https://www.medrxiv.org/content/10.1101/2020.03.09.20033464v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Salzberger B, Glück Thomas, Ehrenstein B. Successful containment of COVID-19: the WHO-Report on the COVID-19 outbreak in China. Infection. 2020 Apr;48(2):151–153. doi: 10.1007/s15010-020-01409-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chudik A, Pesaran M, Rebucci A. Voluntary and Mandatory Social Distancing: Evidence on COVID-19 Exposure Rates from Chinese Provinces and Selected Countries. National Bureau of Economic Research. 2020. [2020-06-10]. https://www.nber.org/papers/w27039.
- 111.Saez M, Tobias A, Varga D, Barceló Maria Antònia. Effectiveness of the measures to flatten the epidemic curve of COVID-19. The case of Spain. Sci Total Environ. 2020 Jul 20;727:138761. doi: 10.1016/j.scitotenv.2020.138761. http://europepmc.org/abstract/MED/32330703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kenyon C. Flattening-the-curve associated with reduced COVID-19 case fatality rates - an ecological analysis of 65 countries. J Infect. 2020 Jul;81(1):e98–e99. doi: 10.1016/j.jinf.2020.04.007. http://europepmc.org/abstract/MED/32305488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.De Leo S. Covid-19 testing strategies and lockdowns: the European closed curves, analysed by "skew-normal" distributions, the forecasts for the UK, Sweden, and the USA, and the ongoing outbreak in Brazil. medRxiv. 2020:e. doi: 10.1101/2020.06.01.20119461. https://www.medrxiv.org/content/10.1101/2020.06.01.20119461v1. [DOI] [Google Scholar]
- 114.Åslund A. Responses to the COVID-19 crisis in Russia, Ukraine, and Belarus. Eurasian Geography and Economics. 2020 Jun 16;:1–14. doi: 10.1080/15387216.2020.1778499. [DOI] [Google Scholar]
- 115.Emerald Expert Briefings. Bingley, United Kingdom: Emerald Publishing Limited; 2020. Apr, Russia's governors seem set up to fail on COVID-19. [Google Scholar]
- 116.Forsyth IC. COVID-19: China’s Chernobyl, China’s Berlin Airlift, or Neither? Washington Headquarters Services. 2020. Jun, [2020-07-20]. https://www.whs.mil/News/News-Display/Article/2210962/covid-19-chinas-chernobyl-chinas-berlin-airlift-or-neither/
- 117.Mækelæ M, Reggev N, Dutra N, Tamayo Rm, Silva-Sobrinho Ra, Klevjer K, Pfuhl G. Perceived efficacy of COVID-19 restrictions, reactions and their impact on mental health during the early phase of the outbreak in six countries. R Soc Open Sci. 2020 Aug 12;7(8):200644. doi: 10.1098/rsos.200644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Emerald Expert Briefings. Bingley, United Kingdom: Emerald Publishing Limited; 2020. Apr, Belarus leader politically isolated by COVID-19 denial. [Google Scholar]
- 119.Emerald Expert Briefings. Bingley, United Kingdom: Emerald Publishing Limited; 2020. May, Ukraine and Belarus seek loans to ride out COVID-19. [Google Scholar]
- 120.Emerald Expert Briefings. Bingley, United Kingdom: Emerald Publishing Limited; 2020. Mar, COVID-19 forces difficult political choices on Ukraine. [Google Scholar]
- 121.CDC Provisional COVID-19 Death Counts by Week. National Center for Health Statistics. 2020. [2020-07-20]. https://www.cdc.gov/nchs/nvss/vsrr/covid19/index.htm.
- 122.Dorn AV, Cooney RE, Sabin ML. COVID-19 exacerbating inequalities in the US. The Lancet. 2020 Apr;395(10232):1243–1244. doi: 10.1016/s0140-6736(20)30893-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.McKibbin W, Fernando R. The Global Macroeconomic Impacts of COVID-19: Seven Scenarios. Asian Economic Papers. 2020 Aug 21;:1–55. doi: 10.1162/asep_a_00796. [DOI] [Google Scholar]
- 124.Baker S, Bloom N, Davis S, Terry S. Covid-induced economic uncertainty. National Bureau of Economic Research; 2020:e. doi: 10.3386/w26983. https://www.nber.org/papers/w26983.pdf. [DOI] [Google Scholar]
- 125.Omer SB, Malani P, Del Rio Carlos. The COVID-19 Pandemic in the US: A Clinical Update. JAMA. 2020 May 12;323(18):1767–1768. doi: 10.1001/jama.2020.5788. [DOI] [PubMed] [Google Scholar]
- 126.Brzezinski A, Deiana G, Kecht V, Van DD. The COVID-19 pandemic: government vs community action across the United States. Covid Economics. 2020:7–156. https://www.researchgate.net/publication/340607113_The_COVID-19_Pandemic_Government_vs_Community_Action_Across_the_United_States. [Google Scholar]
- 127.Ganz S. Does your state have 14-days of declining COVID-19 cases? Finding straightforward answers to a surprisingly tricky question. American Enterprise Institute. 2020 May;:e. https://www.jstor.org/stable/resrep24601?seq=1#metadata_info_tab_contents. [Google Scholar]
- 128.Fox G, Trauer J, McBryde E. Modelling the impact of COVID-19 upon intensive care services in New South Wales. Med J Aust. 2020:e. doi: 10.5694/mja2.50606. https://www.mja.com.au/journal/2020/modelling-impact-covid-19-upon-intensive-care-services-new-south-wales. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Choi S, Ki M. Estimating the reproductive number and the outbreak size of COVID-19 in Korea. Epidemiol Health. 2020 Mar 12;42:e2020011. doi: 10.4178/epih.e2020011. doi: 10.4178/epih.e2020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lauer SA, Grantz KH, Bi Q, Jones FK, Zheng Q, Meredith HR, Azman AS, Reich NG, Lessler J. The Incubation Period of Coronavirus Disease 2019 (COVID-19) From Publicly Reported Confirmed Cases: Estimation and Application. Annals of Internal Medicine. 2020 May 05;172(9):577–582. doi: 10.7326/m20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Calafiore G, Novara C, Possieri C. A modified sir model for the COVID-19 contagion in Italy. arXiv. 2020:e. doi: 10.1016/j.arcontrol.2020.10.005. https://arxiv.org/abs/2003.14391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Korolev I. Identification and estimation of the SEIRD epidemic model for COVID-19. J Econom. 2020 Jul 30;:e. doi: 10.1016/j.jeconom.2020.07.038. http://europepmc.org/abstract/MED/32836680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Pindyck R. COVID-19 and the welfare effects of reducing contagion. National Bureau of Economic Research. 2020:e. doi: 10.3386/w27121. https://www.nber.org/papers/w27121. [DOI] [Google Scholar]
- 134.Stock J. Data gaps and the policy response to the novel coronavirus. National Bureau of Economic Research. 2020:e. doi: 10.3386/w26902. https://www.nber.org/papers/w26902. [DOI] [Google Scholar]
- 135.He S, Tang S, Rong L. A discrete stochastic model of the COVID-19 outbreak: Forecast and control. Math Biosci Eng. 2020:17–2804. doi: 10.3934/mbe.2020153. https://www.aimspress.com/article/10.3934/mbe.2020153. [DOI] [PubMed] [Google Scholar]
- 136.Gandhi M, Yokoe D, Havlir D. Asymptomatic Transmission, the Achilles' Heel of Current Strategies to Control Covid-19. N Engl J Med. 2020 May 28;382(22):2158–2160. doi: 10.1056/NEJMe2009758. http://europepmc.org/abstract/MED/32329972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mani N, Budak J, Lan K, Bryson-Cahn Chloe, Zelikoff Allison, Barker Gwendolyn E C, Grant Carolyn W, Hart Kristi, Barbee Carrie J, Sandoval Marissa D, Dostal Christine L, Corcorran Maria, Ungerleider Hal M, Gates Jeff O, Olin Svaya V, Bryan Andrew, Hoffman Noah G, Marquis Sara R, Harvey Michelle L, Nasenbeny Keri, Mertens Kathleen, Chew Lisa D, Greninger Alexander L, Jerome Keith R, Pottinger Paul S, Dellit Timothy H, Liu Catherine, Pergam Steven A, Neme Santiago, Lynch John B, Kim H Nina, Cohen Seth A. Prevalence of COVID-19 Infection and Outcomes Among Symptomatic Healthcare Workers in Seattle, Washington. Clin Infect Dis. 2020 Jun 16;:2020. doi: 10.1093/cid/ciaa761. http://europepmc.org/abstract/MED/32548613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dodds C, Fakoya I. Covid-19: ensuring equality of access to testing for ethnic minorities. BMJ. 2020 May 29;369:m2122. doi: 10.1136/bmj.m2122. [DOI] [PubMed] [Google Scholar]
- 139.Schmitt-Grohé S, Teoh K, Uribe M. Testing Inequality in New York City. National Bureau of Economic Research. 2020:e. doi: 10.3386/w27019. https://www.nber.org/papers/w27019#:~:text=In%20particular%2C%20the%20test%20distribution,percent%20of%20the%20city's%20income. [DOI] [Google Scholar]
- 140.Voo T, Clapham H, Tam C. Ethical Implementation of Immunity Passports During the COVID-19 Pandemic. J Infect Dis. 2020 Aug 04;222(5):715–718. doi: 10.1093/infdis/jiaa352. http://europepmc.org/abstract/MED/32582943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Beeching N, Fletcher T, Beadsworth M. Covid-19: testing times. BMJ. 2020 Apr 08;369:m1403. doi: 10.1136/bmj.m1403. [DOI] [PubMed] [Google Scholar]
- 142.West CP, Montori VM, Sampathkumar P. COVID-19 Testing: The Threat of False-Negative Results. Mayo Clin Proc. 2020 Jun;95(6):1127–1129. doi: 10.1016/j.mayocp.2020.04.004. http://europepmc.org/abstract/MED/32376102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kumleben N, Bhopal R, Czypionka T, Gruer L, Kock R, Stebbing J, Stigler F. Test, test, test for COVID-19 antibodies: the importance of sensitivity, specificity and predictive powers. Public Health. 2020 Aug;185:88–90. doi: 10.1016/j.puhe.2020.06.006. http://europepmc.org/abstract/MED/32590234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Cassaniti I, Novazzi F, Giardina F, Salinaro F, Sachs M, Perlini S, Bruno R, Mojoli F, Baldanti F, Members of the San Matteo Pavia COVID-19 Task Force Performance of VivaDiag COVID-19 IgM/IgG Rapid Test is inadequate for diagnosis of COVID-19 in acute patients referring to emergency room department. J Med Virol. 2020 Mar 30;:e. doi: 10.1002/jmv.25800. http://europepmc.org/abstract/MED/32227490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Esbin MN, Whitney ON, Chong S, Maurer A, Darzacq X, Tjian R. Overcoming the bottleneck to widespread testing: a rapid review of nucleic acid testing approaches for COVID-19 detection. RNA. 2020 Jul 01;26(7):771–783. doi: 10.1261/rna.076232.120. http://rnajournal.cshlp.org/cgi/pmidlookup?view=long&pmid=32358057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Patel J, Nielsen F, Badiani A, Assi S, Unadkat V, Patel B, Ravindrane R, Wardle H. Poverty, inequality and COVID-19: the forgotten vulnerable. Public Health. 2020 Jun;183:110–111. doi: 10.1016/j.puhe.2020.05.006. http://europepmc.org/abstract/MED/32502699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ji Y, Ma Z, Peppelenbosch MP, Pan Q. Potential association between COVID-19 mortality and health-care resource availability. The Lancet Global Health. 2020 Apr;8(4):e480. doi: 10.1016/s2214-109x(20)30068-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cafer A, Rosenthal M. COVID-19 in the Rural South: A Perfect Storm of Disease, Health Access, and Co-Morbidity. APCRL Policy Briefs. 2020;2:e. https://egrove.olemiss.edu/apcrl_policybriefs/2/ [Google Scholar]
- 149.Tsai J, Wilson M. COVID-19: a potential public health problem for homeless populations. The Lancet Public Health. 2020 Apr;5(4):e186–e187. doi: 10.1016/s2468-2667(20)30053-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Harlem G. Corrigendum to: Descriptive analysis of social determinant factors in urban communities affected by COVID-19. J Public Health (Oxf) 2020 Sep 07;:2020. doi: 10.1093/pubmed/fdaa166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Vetter P, Eckerle I, Kaiser L. Covid-19: a puzzle with many missing pieces. BMJ. 2020 Feb 19;368:m627. doi: 10.1136/bmj.m627. [DOI] [PubMed] [Google Scholar]
- 152.Giamello J, Abram S, Bernardi S, Lauria G. The emergency department in the COVID-19 era. Who are we missing? Eur J Emerg Med. 2020 Aug;27(4):305–306. doi: 10.1097/MEJ.0000000000000718. http://europepmc.org/abstract/MED/32345851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Centers for Disease Control (CDC) Guidelines for evaluating surveillance systems. MMWR Suppl. 1988 May 06;37(5):1–18. https://www.cdc.gov/mmwr/preview/mmwrhtml/00001769.htm. [PubMed] [Google Scholar]
- 154.Post LA, Balsen Z, Spano R, Vaca FE. Bolstering gun injury surveillance accuracy using capture-recapture methods. J Behav Med. 2019 Aug 1;42(4):674–680. doi: 10.1007/s10865-019-00017-4. [DOI] [PubMed] [Google Scholar]
- 155.Piltch-Loeb R, Kraemer J, Lin KW, Stoto MA. Public Health Surveillance for Zika Virus: Data Interpretation and Report Validity. Am J Public Health. 2018 Oct;108(10):1358–1362. doi: 10.2105/ajph.2018.304525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Gilman M, Green R. The surveillance gap: The harms of extreme privacy and data marginalization. Faculty Publications. 2018:253–307. https://scholarship.law.wm.edu/facpubs/1883/ [Google Scholar]
- 157.Li Q, Guan X, Wu Peng, Wang Xiaoye, Zhou Lei, Tong Yeqing, Ren Ruiqi, Leung Kathy S M, Lau Eric H Y, Wong Jessica Y, Xing Xuesen, Xiang Nijuan, Wu Yang, Li Chao, Chen Qi, Li Dan, Liu Tian, Zhao Jing, Liu Man, Tu Wenxiao, Chen Chuding, Jin Lianmei, Yang Rui, Wang Qi, Zhou Suhua, Wang Rui, Liu Hui, Luo Yinbo, Liu Yuan, Shao Ge, Li Huan, Tao Zhongfa, Yang Yang, Deng Zhiqiang, Liu Boxi, Ma Zhitao, Zhang Yanping, Shi Guoqing, Lam Tommy T Y, Wu Joseph T, Gao George F, Cowling Benjamin J, Yang Bo, Leung Gabriel M, Feng Zijian. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med. 2020 Mar 26;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. http://europepmc.org/abstract/MED/31995857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lipsitch M, Cohen Ted, Cooper Ben, Robins James M, Ma Stefan, James Lyn, Gopalakrishna Gowri, Chew Suok Kai, Tan Chorh Chuan, Samore Matthew H, Fisman David, Murray Megan. Transmission dynamics and control of severe acute respiratory syndrome. Science. 2003 Jun 20;300(5627):1966–70. doi: 10.1126/science.1086616. http://www.sciencemag.org/cgi/pmidlookup?view=long&pmid=12766207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kucharski A, Russell T, Diamond C. Early dynamics of transmission and control of COVID-19: a mathematical modelling study. medRxiv. 2020:e. doi: 10.1101/2020.01.31.20019901. https://www.medrxiv.org/content/10.1101/2020.01.31.20019901v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jones JH. Models of Infectious Disease. Stanford Spring Workshop in Formal Demography / Stanford University. 2008. [2020-06-19]. https://web.stanford.edu/~jhj1/teachingdocs/Jones-Epidemics050308.pdf.
- 161.Martcheva M. An Introduction to Mathematical Epidemiology. New York, NY: Springer; 2015. [Google Scholar]
- 162.Braveman PA, Egerter SA, Woolf SH, Marks JS. When do we know enough to recommend action on the social determinants of health? Am J Prev Med. 2011 Jan;40(1 Suppl 1):S58–66. doi: 10.1016/j.amepre.2010.09.026. [DOI] [PubMed] [Google Scholar]
- 163.Guidelines: Opening Up America Again. White House / Centers for Disease Control and Prevention. 2020. [2020-09-21]. https://www.whitehouse.gov/openingamerica/
- 164.The COVID Tracking Project. The Atlantic. 2020. [2020-08-31]. https://covidtracking.com/
- 165.Brownstein JS, Freifeld CC, Madoff LC. Digital Disease Detection — Harnessing the Web for Public Health Surveillance. N Engl J Med. 2009 May 21;360(21):2153–2157. doi: 10.1056/nejmp0900702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.COVID 19 Tracking. GitHub. 2020. [2020-08-31]. https://github.com/COVID19Tracking/covid-tracking-data.
- 167.Oehmke J, Oehmke T, Singh L, Post L. Dynamic Panel Estimates of SARS-CoV-2 Infection Rates: Health Surveillance Informs Public Health Policy. J Med Internet Res. 2020 Sep 09;:e. doi: 10.2196/20924. doi: 10.2196/20924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hansen LP, Singleton KJ. Generalized Instrumental Variables Estimation of Nonlinear Rational Expectations Models. Econometrica. 1982 Sep;50(5):1269. doi: 10.2307/1911873. [DOI] [Google Scholar]
- 169.Hansen LP. Large Sample Properties of Generalized Method of Moments Estimators. Econometrica. 1982 Jul;50(4):1029. doi: 10.2307/1912775. [DOI] [Google Scholar]
- 170.Roodman D. xtabond2: Stata module to extend xtabond dynamic panel data estimator. Statistical Software Components S435901, Boston College Department of Economics. 2018. [2020-07-20]. https://ideas.repec.org/c/boc/bocode/s435901.html.
- 171.Roodman D. How to do Xtabond2: An Introduction to Difference and System GMM in Stata. The Stata Journal. 2018 Nov 19;9(1):86–136. doi: 10.1177/1536867x0900900106. [DOI] [Google Scholar]
- 172.Blundell R, Bond S. GMM Estimation with persistent panel data: an application to production functions. Econometric Reviews. 2000 Jan;19(3):321–340. doi: 10.1080/07474930008800475. [DOI] [Google Scholar]
- 173.Bond S, Hoeffler A, Temple J. GMM estimation of empirical growth models. Economics Papers 2001-W21, Economics Group, Nuffield College, University of Oxford. 2001. [2020-07-20]. https://ideas.repec.org/p/nuf/econwp/0121.html.
- 174.Arellano M. Dynamic Panel Data Estimation Using DPD: A Guide for Users. Great Britain: Institute for Fiscal Studies London; 1988. [Google Scholar]
- 175.COVID-19 Dashboard. Johns Hopkins University. 2020. [2020-09-18]. https://coronavirus.jhu.edu/us-map.


