Abstract
Oxazolidinones are one of the most important antimicrobials potentially active against glycopeptide- and β-lactam-resistant Gram-positive pathogens. Linezolid—the first oxazolidinone to be approved for clinical use in 2000 by the US Food and Drug Administration—and the newer molecule in the class, tedizolid, inhibit protein synthesis by suppressing the formation of the 70S ribosomal complex in bacteria. Over the past two decades, transferable oxazolidinone resistance genes, in particular cfr and optrA, have been identified in Firmicutes isolated from healthcare-related infections, livestock, and the environment. Our goals in this study were to investigate the genetic contexts and the transferability of the cfr and optrA genes and examine genomic features, such as antimicrobial resistance genes, plasmid incompatibility types, and CRISPR-Cas defenses of a linezolid-resistant Enterococcus faecalis isolated in feces from a healthy pig during an antimicrobial surveillance program for animal production in Brazil. The cfr gene was found to be integrated into a transposon-like structure of 7,759 nt flanked by IS1216E and capable of excising and circularizing, distinguishing it from known genetic contexts for cfr in Enterococcus spp., while optrA was inserted into an Inc18 broad host-range plasmid of >58 kb. Conjugal transfer of cfr and optrA was shown by filter mating. The coexistence of cfr and optrA in an E. faecalis isolated from a healthy nursery pig highlights the need for monitoring the use of antibiotics in the Brazilian swine production system for controlling spread and proliferation of antibiotic resistance.
Keywords: oxazolidinones, resistance, Enterococcus faecalis, cfr gene, optrA gene, livestock
Introduction
Few drugs remain available for treating infections caused by antibiotic-resistant bacteria. Oxazolidinone antimicrobials, including linezolid and tedizolid, are among the few last-line therapies effective for multidrug-resistant (MDR) Gram-positive pathogens. Linezolid inhibits protein synthesis by targeting the peptidyl transferase center of the 50S subunit of bacterial ribosomes, blocking the binding of aminoacyl-tRNA to the A-site of the peptidyl transferase center (PTC), and also affecting the positioning of fMet-tRNA at the P-site, which prevents formation of the initiation complex (1–4). Over the past two decades, however, mutations in domain V of the 50S ribosomal subunit of 23S rRNA or in the ribosomal proteins L3 and L4 (5, 6) and the transferable resistance genes cfr, optrA, and poxtA (7–9) have driven the spread of oxazolidinone resistance in Gram-positive bacteria in healthcare and animal agriculture settings.
The spread of the multiresistance gene cfr has raised concern since its first report in a bovine Staphylococcus sciuri isolate (10). The cfr gene initially reported occurred in Enterococcus spp. from healthcare-related infections in Thailand (11) and from livestock in China (12). So far, a BLASTn search of the GenBank database identifies 3 cfr homologs in enterococci. While cfr has been found in both human and animal isolates of Enterococcus faecalis (Thailand, China) (11–13), Enterococcus faecium (Italy, Ireland, U.S.) (14–16), Enterococcus casseliflavus, and Enterococcus thailandicus (China) (17), the cfr(B) variant has been detected only in clinical isolates of E. faecalis (Japan) (18) and E. faecium (U.S., Germany, Netherlands) (19, 20). The most recently described cfr(D) variant has only four entries so far recorded in NCBI's databases, all E. faecium (France, Ireland, Netherlands) (21).
A Cfr-mediated adenosine modification A2503 in the PTC of 23S rRNA, which confers resistance to the oxazolidinone, phenicol, lincosamide, pleuromutilin, streptogramin A, and 16-member-ring macrolide antimicrobials (22), was until 2015 the only known transferable oxazolidinone resistance mechanism. Since then, the ATP-binding cassette (ABC)-F protein OptrA (23) has also been identified as conferring resistance to oxazolidinones, including the newer molecule in the class, tedizolid (24). optrA was identified in both E. faecalis and E. faecium of human and animal origins (8, 25, 26), as well as in E. thailandicus and Enterococcus gallinarum isolated from hospitals in China (27). Elsewhere in Asia (18, 28), Europe (29, 30), and America (16, 31), optrA has been found in E. faecalis and E. faecium of both human and animal origins. In Africa, optrA-positive E. faecalis isolated from humans (32), urban wastewater (33), and food-producing animals (34) were also reported. The cfr and optrA genes can be either plasmid or chromosomally encoded, and the co-location of both in the same plasmid has already been described in a porcine Staphylococcus sciuri isolate in China (35) and in E. faecium and E. faecalis recovered from hospitalized patients and livestock from Europe and the US (14–16).
In this study, we investigated the genetic contexts and the transferability of the cfr and optrA genes from the linezolid-resistant (LR) E. faecalis strain L9 (CP018004.1), which was isolated from a rectal swab collected from a healthy piglet in a surveillance study of antimicrobial susceptibility in Brazil's swine production system (36). Antimicrobial resistance genes, plasmid incompatibility types, epidemiology, and CRISPR-Cas (Clustered Regularly Interspaced Short Palindromic Repeat) defenses of LR E. faecalis L9 were also examined. Whole-genome sequencing (WGS) analysis revealed the presence of cfr associated with a transposable element capable of excision and formation of an intracellular circular intermediate flanked by IS1216E (CP041775.1), which is different from all previously known genetic contexts in Enterococcus spp. from human and animal sources. Further, the core araC-hp-optrA was found to be inserted into a conjugative Inc18 broad host-range plasmid of >58 kb (CP041776.1) in LR E. faecalis L9.
Materials and Methods
Bacterial Isolation
LR E. faecalis L9 comes from a collection of 13 LR E. faecalis (linezolid MIC of 8 mg/L) that were screened from 245 MDR E. faecalis isolated from rectal swabs from healthy piglets (45 days old) in different states of Brazil (36). These 13 optrA-positive E. faecalis, epidemiologically unrelated (ST29, ST330, ST591, ST710, ST711), were recovered from different pigs found to be distributed in 6 out of the 7 states chosen for sample collection. Three LR E. faecalis isolated in the same state (DF) harbored both optrA and cfr (ST591 and ST29), but conjugal transfer of these resistance genes to an enterococcal recipient was achieved only using the ST29 E. faecalis strain L9 as donor in our previous filter mating assays. Therefore, here we investigated the mobile element types that enabled horizontal transfer of cfr and optrA.
Whole-Genome Sequencing and Data Analysis
LR E. faecalis L9 was grown in brain heart infusion (BHI) broth at 37°C (24 h). Genomic DNA was isolated using the QIAGEN DNeasy Blood & Tissue Kit, and quantified using Qubit dsDNA HS. Sequencing libraries were prepared with the Illumina Nextera XT DNA kit and sequenced on a MiSeq instrument (Illumina Inc., USA) at the Massachusetts Eye and Ear Infirmary (MEEI) Ocular Genomics Institute, as 250 nt paired-end reads. De novo assembling was performed using CLC Genomics Workbench 8.0.3. For genome annotations, both the RAST server (Rapid Annotation using Subsystem Technology) and the Prokaryotic Genome Annotation Pipeline (NCBI PGAP) were used. Genome data analysis was performed using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and Center for Genomic Epidemiology (http://www.genomicepidemiology.org) online tools. ResFinder (https://cge.cbs.dtu.dk/services/ResFinder/) was used to identify acquired antimicrobial resistance genes, and PlasmidFinder (https://cge.cbs.dtu.dk/services/PlasmidFinder/) was used to determine plasmid incompatibility types. For detection of the oxazolidinone resistance determinants, LRE-Finder (https://cge.cbs.dtu.dk/services/LRE-finder/) was used as well. Multilocus sequence typing (MLST) loci were assigned by the MLST database (https://pubmlst.org/efaecalis/), and the presence of CRISPR-cas defenses was identified by CRISPRfinder (https://crisprcas.i2bc.paris-saclay.fr).
Filter Mating Assay
Conjugation by filter mating as described previously by Jaworski and Clewell (37) was performed using LR E. faecalis L9 as donor, and the E. faecalis strain OG1RF as recipient. Donor and recipient were grown overnight in BHI broth at 37°C. One milliliter from donor culture plus 1 ml from recipient culture were inoculated in 3 ml of phosphate buffered saline (PBS) solution, filtered through a sterile 25-mm-diameter, 0.22-μm-pore-size membrane filter, and subsequently incubated on BHI agar at 37°C for 24–48 h. PBS (5 ml) was used to wash the filters, and 500 μl of this solution was spread on BHI agar plates (100 × 15 mm Petri plates) containing 25 μg/ml of fusidic acid, 25 μg/ml of rifampicin to select for the OG1RF chromosomal markers, and 25 or 10 μg/ml of chloramphenicol (CHL) to select for oxazolidinone and phenicol resistance genes; linezolid (LZD) (4 μg/ml) instead of chloramphenicol was also tested to select for cfr and optrA. Conjugation efficiency (CFU/ml of transconjugants per CFU/ml of donors) was calculated as previously described (38). PCR using primer sets specific for optrA, cfr, poxtA, fexA, and cat genes (36) and Sanger sequencing were carried out to detect these resistances in OG1RF transconjugants. Minimum inhibitory concentrations (MIC's) of chloramphenicol, florfenicol, linezolid, and tedizolid were determined by broth microdilution testing according to the guidelines of the Clinical Laboratory Standards Institute (CLSI). E. faecalis ATCC 29212 was used as a control for antimicrobial susceptibility testing.
Results and Discussion
Enterococcus faecalis is a commensal bacterium of the gut microbiota of humans and various animal species and also a cause of infections in critically ill patients (39, 40). Besides being an important hospital pathogen, E. faecalis has emerged as a potential reservoir of oxazolidinone resistance genes in animal agriculture settings worldwide (8, 31, 34, 36, 41). It is of substantial concern that antibiotics used in food-producing animals may be selecting for the proliferation of MDR E. faecalis lineages in which cfr and optrA coexist. Cfr rRNA methyltransferase confers resistance to six important antimicrobial classes that target the 50S ribosomal subunit (22), while the ATP-binding cassette (ABC)-F protein OptrA confers resistance to phenicol and oxazolidinone, including resistance to the new oxazolidinone tedizolid (23). The spread of cfr and optrA inter-species/genera has been driven by plasmids containing other important resistance determinants (14–16, 35). Therefore, oxazolidinone resistance can be co-selected by antimicrobials that have been largely used in swine production, such as phenicol, macrolide, lincosamide, and pleuromutilin.
Antimicrobial Resistance Determinants of LR E. faecalis L9
ResFinder identified that LR E. faecalis L9 carries the lsa(A) gene, which is responsible for intrinsic LSAP resistance in E. faecalis, and acquired resistance genes for aminoglycoside (str), phenicol fex(A), phenicol and oxazolidinone (optrA), and tetracycline [tet(L), tet(M), tet(S)], in addition to the multiresistance gene cfr. LRE-Finder confirmed the presence of cfr and optrA (CP041775.1 and CP041776.1, respectively), but the phenicol–oxazolidinone–tetracycline resistance gene poxtA was not found. 23S rRNA mutations were not detected in LR E. faecalis L9, nor were they identified in ribosomal protein genes rplC, rplD, and rplV (L3, L4, and L22, respectively).
Genetic Context of cfr in the Porcine LR E. faecalis Isolate L9
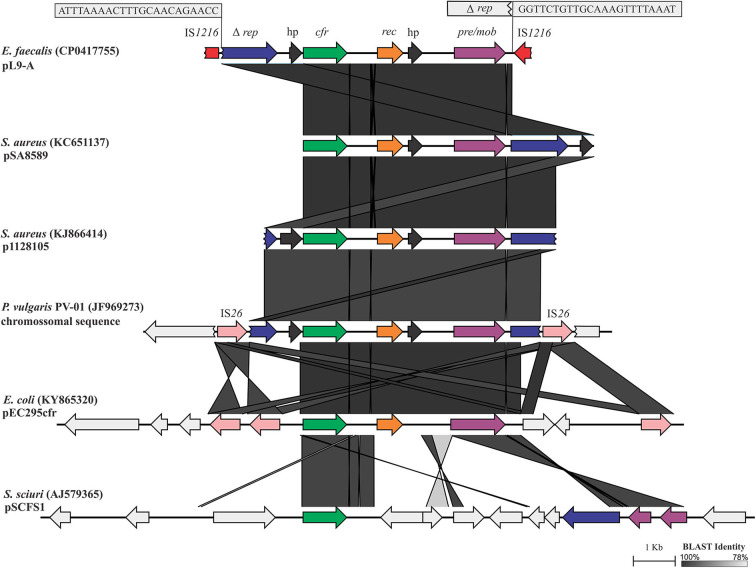
A cfr-carrying DNA segment of 7,759 nt, pL9-A (CP041775.1), was found to be inserted into LR E. faecalis L9 (Figure 1). The cfr gene was flanked upstream by the Tn554-related ΔtnpB gene. Further upstream of ΔtnpB, a gene coding for RepUS18 was detected that was disrupted by the integration of an IS1216E. The repUS18 gene is often found in Inc18 broad host-range plasmids, which have been related to antimicrobial resistance gene transfer in enterococci. Downstream, the cfr gene was flanked by a recombinase rec gene, a gene coding for a hypothetical protein, and a plasmid recombination/mobilization pre/mob gene. In silico predictions indicated that the IS1216E-flanked segment pL9-A could excise and exist within the cell as a non-replicating circular intermediate in LR E. faecalis L9, which was confirmed by PCR and Sanger sequencing using the primers 5′AGGTTTAGAATAATCTCCCGA3′ and 5′GCTGACAACATATCTAATATCTCAA3′.
Figure 1.
Linear comparison of the 7,759-bp cfr-carrying DNA segment pL9-A (CP041775.1) generated by EasyFig. The boxes zoom in on the 23-bp inverted repeats (IR) at the ends of an IS1216 that was inserted into repUS18. The 6,956-bp segment between the IRs shows high DNA identity to the corresponding stretches in pSA8589 (KC651137), p1128105 (KJ866414), and P. vulgaris PV-01 chromosomal sequence (JF969273). Alignment of these sequences revealed only a deletion of 7 bp in repUS18 from pL9-A and an insertion of 10 bp in the hypothetical protein on the flank 5′ of cfr in p1128105, which is represented by the slightly lighter shade of identity over this region.
pL9-A possesses 100% DNA identity over 93% of its length to a chromosomal DNA sequence from Proteus vulgaris PV-01 (JF969273) isolated from a pig nasal swab in China (42), and 99.98% and 99.83% DNA identity to the pSA8589 and p1128105 from Staphylococcus aureus 1900 (43) and S. aureus 1128105 (44) of human origin in the US (KC561137 and KJ866414, respectively), highlighting its very broad-range horizontal transfer capabilities. IS6 insertion sequence family elements, which have been commonly associated with antibiotic resistance genes, appear to be also involved in transposition events of the core cfr-rec-pre/mob in Gram-positive and Gram-negative bacteria.
The cfr-carrying mobile element pL9-A was distinctly different from all known genetic contexts of cfr in Enterococcus spp. from human and animal sources. The similarity of pL9-A to the cfr-carrying segments previously identified in bacteria of other genera indicates that it has most likely been acquired horizontally from other bacteria or, alternatively, could be intrinsic in some lineages of E. faecalis and then transferred to other bacteria. The IS1216E element appears to be involved in the acquisition and dispersal of pL9-A in LR E. faecalis L9. In Brazil, the cfr gene has been reported to date only in an ST398 MSSA strain of human origin (45) in a genetic context other than that observed in the porcine LR E. faecalis isolate L9.
Plasmid-Borne optrA-Carrying Partial Sequence (pL9) in the Porcine LR E. faecalis Isolate L9
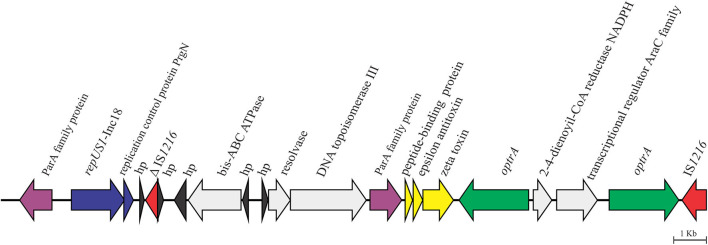
We recently reported that the core araC-hp-optrA of 3,453 nt in length, which was composed of genes coding for a hypothetical protein and an AraC family transcriptional regulator at the 5′ of optrA, was inserted upstream of an IS1216E element into a plasmid of >58 kb, which was not closed during de novo assembly of the high quality draft sequence (CP041776.1) (36). On the flank 5′ of the core araC-hp-optrA, LR E. faecalis L9 showed in silico a duplication of optrA, which was confirmed by PCR and Sanger sequencing using the primers 5′TTGAGTGAAATACCTGTGCG3′ and 5′TGATGGTAATATGGTGTTGGAA3′. Further analysis of pL9 showed the presence of genes coding for the zeta–epsilon–delta (ω-ε-ζ) toxin–antitoxin (TA) system upstream of the duplication of optrA (Figure 2). The ω-ε-ζ TA module, a post-segregational killing system which acts at cell division eliminating progeny that fails to inherit plasmid copy, has been found in various MDR Gram-positive bacteria, including the cfr-carrying conjugative plasmids pW9-2 from E. faecalis, and pW3 and p3-38 from E. thailandicus isolated from sewage in swine farm contexts in China (12) and pEF12-0805 from E. faecium isolated from human blood in Italy (14). pL9 harbored a gene coding for the plasmid replication protein repUS1, which is found in Inc18 broad host-range plasmids. On the 5′ flank of repUS1, a 912-nt open reading frame (ORF) for the partitioning protein ParA which mediates plasmid segregation was found. These mechanisms ensure the maintenance of plasmids that exist in low-copy numbers in a bacterial population, such as Inc18 family plasmids. At the 3′ flank of repUS1, a 288-nt ORF for the replication control protein PrgN is present. Upstream to this region, an 1,494-nt ORF for ATP-binding cassette domain-containing protein came to our attention due to the very few entries so far recorded in NCBI's databases, as it only matches nucleotide sequences from 6 Lactococcus garvieae, which causes fatal hemorrhagic septicaemia in fish (South Korea and Japan), and 1 Lactococcus petauri isolated from human feces in China. This ORF codes for the ATP-binding cassette domain-containing protein (E. faecalis WP_155282194.1), which has 81.74% DNA identity over 99% of its length to the Lactococcus ABC-F-type ribosomal protection protein (WP_019291880.1).
Figure 2.
Genetic context of the 58,593-bp optrA-carrying partial sequence pL9 (CP041776.1) in the porcine LR E. faecalis isolate L9.
pL9 was found to be inserted into a conjugative Inc18 plasmid of >58 kb. Inc18 broad host-range plasmids have been associated with a variety of antibiotic resistances in enterococci, including the high-level vanA glycopeptide resistance carried by Tn1546, which can be transferred to MRSA lineages (46). Inc18 plasmids can play a crucial role in the oxazolidinone resistance emergence, as they are widespread in enterococci, streptococci, and staphylococci in both clinical and environmental settings (47, 48). Moreover, most Inc18 plasmids carry locus coding for stabilization systems, such as the post-segregation killing (PSK) system (49), which has already been implicated in the persistence of the Tn1546-mediated vanA resistance in E. faecium (50). The presence of ORF's adjacent to optrA in pL9 that matched few or no DNA sequences available in GenBank indicates that further investigation is required to understand how new conjugative Inc18 plasmid mosaics are evolving and how that might favor the spread of oxazolidinone resistance in animal agriculture settings.
Transferability of cfr, optrA, and Other Resistance Determinants in LR E. faecalis L9
Filter mating assays were carried out to determine the potential for conjugal transfer of cfr and optrA at different CHL and LZD concentrations (Table 1). optrA/fexA/tet(S)-carrying OG1RF-L9 transconjugants were selected at a frequency of 4 × 10−7 transconjugant cells per donor cell using 25 μg/ml CHL, but conjugation experiments failed to transfer cfr at 25 μg/ml CHL. Decreasing CHL concentration from 25 to 10 μg/ml, countless small colonies of optrA/cfr/fexA/tet(S)-carrying OG1RF-L9 transconjugants could be selected. Linezolid could select only countless small colonies of optrA-positive OG1RF-L9 transconjugants; no cfr-positive OG1RF-L9 transconjugant was obtained, indicating that optrA is responsible for linezolid resistance, and cfr, for a lower-level chloramphenicol resistance phenotype in LR E. faecalis L9.
Table 1.
Conjugation efficiency of cfr and optrA from E. faecalis L9 to E. faecalis OG1RF transconjugants.
| Recipient straina | Conjugation efficiencyb | Resistance genesc | MIC (μg/ml) | |||
|---|---|---|---|---|---|---|
| LZD | TZD | CHL | FFC | |||
| OG1RF-L9 (25 μg/ml CHL) | 4 × 10−7 | optrA, fex(A), tet(S) | 8 | 0.5 | 128 | 64 |
| OG1RF-L9 (10 μg/ml CHL) | ND | optrA, fex(A), tet(S), cfr | 8 | 0.5 | 128 | 64 |
| OG1RF-L9 (8 μg/ml LZD) | ND | optrA, fex(A), tet(S) | 8 | 0.5 | 128 | 64 |
| OG1RF-L9 (4 μg/ml LZD) | ND | optrA, fex(A), tet(S) | 8 | 0.5 | 128 | 64 |
Graded levels of CHL and LZD were tested to select for oxazolidinone and phenicol resistance genes, in addition to 25 μg/ml fusid acid, and 25 μg/ml rifampicin to select for the OG1RF chromosomal markers.
Conjugation efficiency corresponds to the number of CFU transconjugants per CFU donors; ND (not determined): only small bacterial colonies (countless) were obtained, all optrA/fexA-positive.
optrA could be efficiently transferred by conjugation at a frequency of 4 × 10−7 per donor cell using 25 μg/ml of CHL as previously reported (36), and also decreasing CHL concentration or using LZD instead of CHL.
Tn558, which harbors the chloramphenicol/florfenicol efflux MFS transporter fexA gene and the tetracycline resistance ribosomal protection gene tet(S), was also transferred to OG1RF-L9 in a yet to be determined genetic context, as pL9 could not be closed during de novo assembly. A 204-nt fragment of a gene for a conjugal transfer protein 5′ of tet(S) that appears to be involved in its mobilization is identical to homologs occurring in Firmicutes as identified in a BLASTn search. A 210 nt ORF 3′ flank to Tn558 matched a gene coding for the replication-associated protein RepB identified in Listeria monocytogenes (KY613776.1 and KY613741.1) isolated from food in Canada, in Carnobacterium divergens (LT984411.1) from beef carpaccio in France, and in an optrA-carrying conjugative plasmid from the Enterococcaceae strain E508 (MK425645.1) in China. Another 606-nt ORF encoding a hypothetical protein at the extreme 3′ end of repB also matched ORF's from the L. monocytogenes, C. divergens, and Enterococcaceae isolates mentioned above, and the optrA-carrying Enterococcus avium isolate C674 (MH018573.1) from an asymptomatic healthy human in China. At 5′, Tn558 is flanked by a 210-nt ORF for a hypothetical protein that only matches sequences from the optrA gene cluster from E. avium C674 and pStrcfr from Streptococcus suis S10 (KF129409.1). Further upstream, a 1,272-nt ORF for a Y-family DNA polymerase is present, but no match was found for this nucleotide sequence.
Bacterial Immunity of cfr/optrA-Carrying E. faecalis L9
Genome defenses for porcine LR E. faecalis L9 were investigated using CRISPRfinder. Clustered, regularly interspaced short palindromic repeat (CRISPR) loci provide an important defense against parasitic mobile element entry. MDR, hospital-adapted enterococcal lineages lack CRISPR defenses, which are thought to enhance the facility with which they acquire antibiotic resistances on mobile elements (39, 51). A CRISPR-related loci consisting of 9 spacers and direct repeat sequences of 36 bp was found in the L9 chromosome (1,708,044 to 1,708,673 bp), but genes coding for Cas proteins were not identified. A BLASTn search revealed that the L9 CRISPR-related loci possess 95.12% DNA identity over 100% of its length to the corresponding chromosomal DNA sequence from E. faecalis SGAir0397 (CP039434.1), which was recovered from air in Singapore. L9 CRISPR spacer sequences only matched to sequences from E. faecalis FDAARGOS_324 (CP028285.1) isolated from a human eye in the US, and the cyanobacterium Geminocystis sp. isolate NIES-3708 (AP014815.1) from Japan, besides E. faecalis SGAir0397. LR E. faecalis L9 lacked the E. faecalis CRISPR1 locus (a CAS-TypeIIA cluster consisting of Csn2_0_IIA, Cas2_0_I-II-III, Cas1_0_II, Cas9_1_II) typically located between genes EF0672 and EF0673 (51).
To the best of our knowledge, this is the first report of the coexistence of optrA and cfr in a bacterial isolate in Brazil. The fact that LR E. faecalis L9 came from a pool of 13 LR E. faecalis collected from healthy piglets in swine herds distributed across 7 Brazilian States highlights the need for monitoring the use of antibiotics in the country's swine production system in order to preserve the few remaining last-line antibiotics to treat infections caused by MDR pathogens.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, CP018004.1, https://www.ncbi.nlm.nih.gov/genbank/, CP041775.1, https://www.ncbi.nlm.nih.gov/genbank/, CP041776.1.
Ethics Statement
The study was approved by the Ethics Committee of Faculdade de Medicina Veterinária e Zootecnia- Universidade de São Paulo, under number CEUA N.8026060214.
Author Contributions
LA and MG planned the study and wrote the manuscript with suggestions from AG, PB, and FL. PF and AM provided a collection of 245 MDR E. faecalis strains isolated from swine. LA, JS, RS, and IB-J carried out the experimental work and genome sequencing. LA, FZ, AG, PB, and FL contributed to the bioinformatic analyses. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This project has been funded by the Harvard-wide Program on Antibiotic Resistance NIH/NIAID AI083214, and FAPESP scholarship 2014/ 27267-0.
References
- 1.Long KS, Munck C, Andersen TMB, Schaub MA, Hobbie SN, Böttger EC, et al. Mutations in 23S rRNA at the peptidyl transferase center and their relationship to linezolid binding and cross-resistance. Antimicrob Agents Chemother. (2010) 54:4705–13. 10.1128/AAC.00644-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Long KS, Vester B. Resistance to linezolid caused by modifications at its binding site on the ribosome. Antimicrob Agents Chemother. (2012) 56:603–12. 10.1128/AAC.05702-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ippolito JA, Kanyo ZF, Wang D, Franceschi FJ, Moore PB, Steitz TA, et al. Crystal structure of the oxazolidinone antibiotic linezolid bound to the 50S ribosomal subunit. J Med Chem. (2008) 51:3353–6. 10.1021/jm800379d [DOI] [PubMed] [Google Scholar]
- 4.Wilson DN, Schluenzen F, Harms JM, Starosta AL, Connell SR, Fucini P. The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc Natl Acad Sci USA. (2008) 105:13339–44. 10.1073/pnas.0804276105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mendes RE, Hogan PA, Streit JM, Jones RN, Flamm RK. Zyvox® Annual Appraisal of Potency and Spectrum (ZAAPS) program: report of linezolid activity over 9 years (2004-12). J Antimicrob Chemother. (2014) 69:1582–8. 10.1093/jac/dkt541 [DOI] [PubMed] [Google Scholar]
- 6.Pfaller MA, Mendes RE, Streit JM, Hogan PA, Flamm RK. Five-year summary of in vitro activity and resistance mechanisms of linezolid against clinically important gram-positive cocci in the United States from the LEADER surveillance program (2011 to 2015). Antimicrob Agents Chemother. (2017) 61:e00609–17. 10.1128/AAC.00609-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen J, Wang Y, Schwarz S. Presence and dissemination of the multiresistance gene cfr in gram-positive and gram-negative bacteria. J Antimicrob Chemother. (2013) 68:1697–706. 10.1093/jac/dkt092 [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Lv Y, Cai J, Schwarz S, Cui L, Hu Z, et al. A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J Antimicrob Chemother. (2015) 70:2182–90. 10.1093/jac/dkv116 [DOI] [PubMed] [Google Scholar]
- 9.Antonelli A, D'Andrea MM, Brenciani A, Galeotti CL, Morroni G, Pollini S, et al. Characterization of poxtA, a novel phenicol-oxazolidinone-tetracycline resistance gene from an MRSA of clinical origin. J Antimicrob Chemother. (2018) 73:1763–9. 10.1093/jac/dky088 [DOI] [PubMed] [Google Scholar]
- 10.Schwarz S, Werckenthin C, Kehrenberg C. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob Agents Chemother. (2000) 44:2530–3. 10.1128/AAC.44.9.2530-2533.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diaz L, Kiratisin P, Mendes RE, Panesso D, Singh KV, Arias CA. Transferable plasmid-mediated resistance to linezolid due to cfr in a human clinical isolate of Enterococcus faecalis. Antimicrob Agents Chemother. (2012) 56:3917–22. 10.1128/AAC.00419-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, Wang Y, Schwarz S, Li Y, Shen Z, Zhang Q, et al. Transferable multiresistance plasmids carrying cfr in Enterococcus spp. from swine and farm environment Antimicrob Agents Chemother. (2013) 57:42–8. 10.1128/AAC.01605-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang LX, Duan JH, Chen MY, Deng H, Liang HQ, Xiong YQ, et al. Prevalence of cfr in Enterococcus faecalis strains isolated from swine farms in China: predominated cfr-carrying pCPPF5-like plasmids conferring “non-linezolid resistance” phenotype. Infect Genet Evol. (2018) 62:188–92. 10.1016/j.meegid.2018.04.023 [DOI] [PubMed] [Google Scholar]
- 14.Morroni G, Brenciani A, Antonelli A, D'Andrea MM, Di Pilato V, Fioriti S, et al. Characterization of a multiresistance plasmid carrying the optrA and cfr resistance genes from an Enterococcus faecium clinical isolate. Front Microbiol. (2018) 9:2189. 10.3389/fmicb.2018.02189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lazaris A, Coleman DC, Kearns AM, Pichon B, Kinnevey PM, Earls MR, et al. Novel multiresistance cfr plasmids in linezolid-resistant methicillin-resistant Staphylococcus epidermidis and vancomycin-resistant Enterococcus faecium (VRE) from a hospital outbreak: co-location of cfr and optrA in VRE. J Antimicrob Chemother. (2017) 72:3252–7. 10.1093/jac/dkx292 [DOI] [PubMed] [Google Scholar]
- 16.Tyson GH, Sabo JL, Hoffmann M, Hsu C-H, Mukherjee S, Hernandez J, et al. Novel linezolid resistance plasmids in Enterococcus from food animals in the USA. J Antimicrob Chemother. (2018) 73:3254–8. 10.1093/jac/dky369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, Wang Y, Dai L, Wu C, Shen J. First report of multiresistance gene cfr in Enterococcus species casseliflavus and gallinarum of swine origin. Vet Microbiol. (2014) 170:352–7. 10.1016/j.vetmic.2014.02.037 [DOI] [PubMed] [Google Scholar]
- 18.Kuroda M, Sekizuka T, Matsui H, Suzuki K, Seki H, Saito M, et al. Complete genome sequence and characterization of linezolid-resistant Enterococcus faecalis clinical isolate KUB3006 carrying a cfr(B)-transposon on its chromosome and optrA-Plasmid. Front Microbiol. (2018) 9:2576. 10.3389/fmicb.2018.02576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deshpande LM, Ashcraft DS, Kahn HP, Pankey G, Jones RN, Farrell DJ, et al. Detection of a new cfr-Like Gene, cfr(B), in Enterococcus faecium isolates recovered from human specimens in the United States as part of the SENTRY antimicrobial surveillance program. Antimicrob Agents Chemother. (2015) 59:6256–61. 10.1128/AAC.01473-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bender JK, Fleige C, Klare I, Fiedler S, Mischnik A, Mutters N, et al. Detection of a cfr(B) variant in German Enterococcus faecium Clinical Isolates and the impact on linezolid resistance in Enterococcus spp. PLoS ONE. (2016) 11:e0167042. 10.1371/journal.pone.0167042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guerin F, Sassi M, Dejoies L, Zouari A, Schutz S, Potrel S, et al. Molecular and functional analysis of the novel cfr(D) linezolid resistance gene identified in Enterococcus faecium. J Antimicrob Chemother. (2020) 75:1699–703. 10.1093/jac/dkaa125 [DOI] [PubMed] [Google Scholar]
- 22.Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. The Cfr rRNA methyltransferase confers resistance to Phenicols, Lincosamides, Oxazolidinones, Pleuromutilins, and Streptogramin A antibiotics. Antimicrob Agents Chemother. (2006) 50:2500–5. 10.1128/AAC.00131-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharkey LKR, O'Neill AJ. Antibiotic resistance ABC-F proteins: bringing target protection into the limelight. ACS Infect Dis. (2018) 4:239–46. 10.1021/acsinfecdis.7b00251 [DOI] [PubMed] [Google Scholar]
- 24.Fala L. Sivextro (Tedizolid Phosphate) approved for the treatment of adults with acute bacterial skin and skin-structure infections. Am Health Drug Benefits. (2015) 8:111–5. [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Dong G, Li J, Chen L, Liu H, Bi W, et al. A high incidence and coexistence of multiresistance genes cfr and optrA among linezolid-resistant enterococci isolated from a teaching hospital in Wenzhou, China. Eur J Clin Microbiol Infect Dis. (2018) 37:1441–8. 10.1007/s10096-018-3269-8 [DOI] [PubMed] [Google Scholar]
- 26.Shang Y, Li D, Shan X, Schwarz S, Zhang S-M, Chen Y-X, et al. Analysis of two pheromone-responsive conjugative multiresistance plasmids carrying the novel mobile optrA locus from Enterococcus faecalis. Infect Drug Resist. (2019) 12:2355–62. 10.2147/IDR.S206295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai J, Wang Y, Schwarz S, Lv H, Li Y, Liao K, et al. Enterococcal isolates carrying the novel oxazolidinone resistance gene optrA from hospitals in Zhejiang, Guangdong, and Henan, China, 2010-2014. Clin Microbiol Infect. (2015) 21:1095.e1–4. 10.1016/j.cmi.2015.08.007 [DOI] [PubMed] [Google Scholar]
- 28.Chien J-Y, Mendes RE, Deshpande LM, Hsueh P-R. Empyema thoracis caused by an optrA-positive and linezolid-intermediate Enterococcus faecalis strain. J Infect. (2017) 75:182–4. 10.1016/j.jinf.2017.05.003 [DOI] [PubMed] [Google Scholar]
- 29.Sassi M, Guérin F, Zouari A, Beyrouthy R, Auzou M, Fines-Guyon M, et al. Emergence of optrA-mediated linezolid resistance in enterococci from France, 2006-16. J Antimicrob Chemother. (2019) 74:1469–72. 10.1093/jac/dkz097 [DOI] [PubMed] [Google Scholar]
- 30.Bender JK, Fleige C, Lange D, Klare I, Werner G. Rapid emergence of highly variable and transferable oxazolidinone and phenicol resistance gene optrA in German Enterococcus spp. clinical isolates Int J Antimicrob Agents. (2018) 52:819–27. 10.1016/j.ijantimicag.2018.09.009 [DOI] [PubMed] [Google Scholar]
- 31.Cavaco LM, Bernal JF, Zankari E, Léon M, Hendriksen RS, Perez-Gutierrez E, et al. Detection of linezolid resistance due to the optrA gene in Enterococcus faecalis from poultry meat from the American continent (Colombia). J Antimicrob Chemother. (2017) 72:678–83. 10.1093/jac/dkw490 [DOI] [PubMed] [Google Scholar]
- 32.Said HS, Abdelmegeed ES. Emergence of multidrug resistance and extensive drug resistance among enterococcal clinical isolates in Egypt. Infect Drug Resist. (2019) 12:1113–25. 10.2147/IDR.S189341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Freitas AR, Elghaieb H, León-Sampedro R, Abbassi MS, Novais C, Coque TM, et al. Detection of optrA in the African continent (Tunisia) within a mosaic Enterococcus faecalis plasmid from urban wastewaters. J Antimicrob Chemother. (2017) 72:3245–51. 10.1093/jac/dkx321 [DOI] [PubMed] [Google Scholar]
- 34.Elghaieb H, Freitas AR, Abbassi MS, Novais C, Zouari M, Hassen A, et al. Dispersal of linezolid-resistant enterococci carrying poxtA or optrA in retail meat and food-producing animals from Tunisia. J Antimicrob Chemother. (2019) 74:2865–9. 10.1093/jac/dkz263 [DOI] [PubMed] [Google Scholar]
- 35.Li D, Wang Y, Schwarz S, Cai J, Fan R, Li J, et al. Co-location of the oxazolidinone resistance genes optrA and cfr on a multiresistance plasmid from Staphylococcus sciuri. J Antimicrob Chemother. (2016) 71:1474–8. 10.1093/jac/dkw040 [DOI] [PubMed] [Google Scholar]
- 36.Almeida LM, Lebreton F, Gaca A, Bispo PM, Saavedra JT, Calumby RN, et al. Transferable resistance gene optrA in Enterococcus faecalis from Swine in Brazil. Antimicrob Agents Chemother. (2020) 64:e00142–20. 10.1128/AAC.00142-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaworski DD, Clewell DB. Evidence that coupling sequences play a frequency-determining role in conjugative transposition of Tn916 in Enterococcus faecalis. J Bacteriol. (1994) 176:3328–35. 10.1128/JB.176.11.3328-3335.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jett BD, Hatter KL, Huycke MM, Gilmore MS. Simplified agar plate method for quantifying viable bacteria. BioTechniques. (1997) 23:648–50. 10.2144/97234bm22 [DOI] [PubMed] [Google Scholar]
- 39.Gilmore MS, Lebreton F, van Schaik W. Genomic transition of enterococci from gut commensals to leading causes of multidrug-resistant hospital infection in the antibiotic era. Curr Opin Microbiol. (2013) 16:10–6. 10.1016/j.mib.2013.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Tyne D, Gilmore MS. Friend turned foe: evolution of enterococcal virulence and antibiotic resistance. Annu Rev Microbiol. (2014) 68:337–56. 10.1146/annurev-micro-091213-113003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morroni G, Brenciani A, Simoni S, Vignaroli C, Mingoia M, Giovanetti E. Commentary: nationwide surveillance of novel oxazolidinone resistance gene optrA in Enterococcus isolates in China from 2004 to 2014. Front Microbiol. (2017) 8:1631. 10.3389/fmicb.2017.01631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, Wang Y, Wu CM, Schwarz S, Shen Z, Zhang W, et al. Detection of the staphylococcal multiresistance gene cfr in Proteus vulgaris of food animal origin. J Antimicrob Chemother. (2011) 66:2521–6. 10.1093/jac/dkr322 [DOI] [PubMed] [Google Scholar]
- 43.Mendes RE, Deshpande LM, Bonilla HF, Schwarz S, Huband MD, Jones RN, et al. Dissemination of a pSCFS3-like cfr-carrying plasmid in Staphylococcus aureus and Staphylococcus epidermidis clinical isolates recovered from hospitals in Ohio. Antimicrob Agents Chemother. (2013) 57:2923–8. 10.1128/AAC.00071-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Locke JB, Zuill DE, Scharn CR, Deane J, Sahm DF, Denys GA, et al. Linezolid-resistant Staphylococcus aureus strain 1128105, the first known clinical isolate possessing the cfr multidrug resistance gene. Antimicrob Agents Chemother. (2014) 58:6592–8. 10.1128/AAC.03493-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gales AC, Deshpande LM, de Souza AG, Pignatari ACC, Mendes RE. MSSA ST398/t034 carrying a plasmid-mediated Cfr and Erm(B) in Brazil. J Antimicrob Chemother. (2015) 70:303–5. 10.1093/jac/dku366 [DOI] [PubMed] [Google Scholar]
- 46.Kohler V, Vaishampayan A, Grohmann E. Broad-host-range Inc18 plasmids: occurrence, spread and transfer mechanisms. Plasmid. (2018) 99:11–21. 10.1016/j.plasmid.2018.06.001 [DOI] [PubMed] [Google Scholar]
- 47.Freitas AR, Coque TM, Novais C, Hammerum AM, Lester CH, Zervos MJ, et al. Human and swine hosts share vancomycin-resistant Enterococcus faecium CC17 and CC5 and Enterococcus faecalis CC2 clonal clusters harboring Tn1546 on indistinguishable plasmids. J Clin Microbiol. (2011) 49:925–31. 10.1128/JCM.01750-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu W, Murray PR, Huskins WC, Jernigan JA, McDonald LC, Clark NC, et al. Dissemination of an Enterococcus Inc18-Like vanA plasmid associated with vancomycin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. (2010) 54:4314–20. 10.1128/AAC.00185-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meinhart A, Alonso JC, Sträter N, Saenger W. Crystal structure of the plasmid maintenance system epsilon/zeta: functional mechanism of toxin zeta and inactivation by epsilon 2 zeta 2 complex formation. Proc Natl Acad Sci USA. (2003) 100:1661–6. 10.1073/pnas.0434325100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sletvold H, Johnsen PJ, Hamre I, Simonsen GS, Sundsfjord A, Nielsen KM. Complete sequence of Enterococcus faecium pVEF3 and the detection of an omega-epsilon-zeta toxin-antitoxin module and an ABC transporter. Plasmid. (2008) 60:75–85. 10.1016/j.plasmid.2008.04.002 [DOI] [PubMed] [Google Scholar]
- 51.Palmer KL, Gilmore MS. Multidrug-resistant enterococci lack CRISPR-cas. mBio. (2010) 1:e00227–10. 10.1128/mBio.00227-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, CP018004.1, https://www.ncbi.nlm.nih.gov/genbank/, CP041775.1, https://www.ncbi.nlm.nih.gov/genbank/, CP041776.1.