Abstract
The COVID-19 pandemic has affected all sectors of society, from health and economics to socialization and travel. The level and extent of this impact are unprecedented. Although the cause of COVID-19 was quickly identified to be a new coronavirus (SARS-CoV-2), the world was poorly prepared for preventing its spread. One important pillar of preparedness is surveillance of the sources of emerging pathogens and responding appropriately to prevent their spread in the human population. The ever-increasing interaction between humans and animals is one leading factor in facilitating the emergence of new pathogens. In this viewpoint, we discuss the possibility of the zoonotic origin of SARS-CoV-2, highlight the importance of understanding human-animal interaction to improve preparedness for future outbreaks, and outline recommendations for prevention.
Keywords: zoonosis, human-animal interface, COVID-19, SARS-CoV-1, outbreak, virus, transmission, pathogen, emergence, reservoir
Introduction
Currently, the world is battling a pandemic caused by a virus previously undocumented in humans. The new virus, SARS-CoV-2, was identified as the cause of COVID-19. The infection presented with a broad spectrum of symptoms ranging from mild disease to organ failure and death. Similar to severe acute respiratory syndrome coronavirus (SARS-CoV-1) and the Middle East respiratory syndrome coronavirus (MERS-CoV), SARS-CoV-2 is a positive-sense, single-stranded ribonucleic acid (+ssRNA) member of the β-coronavirus genus. However, compared to SARS-CoV-1, SARS-CoV-2 appears to be more efficient in human-to-human transmission [1,2]. The fact that the global spread of the virus outpaced infection control measures has raised concerns about our preparedness to face new viral threats. In this viewpoint, we discuss the potential zoonotic origin of SARS-CoV-2 and revisit the main elements that promote the emergence and spread of new infectious diseases. We also highlight the factors that instigate the rise in new human pathogens and outline recommendations for prevention.
SARS-CoV-2 and the Possibility of an Animal-to-Human Spillover Event
Despite the uncertainty about where and when SARS-CoV-2 originated, the genome sequencing of isolates from early cases indicate an animal origin, with bats being suggested as the most probable source [3-6]. In silico evaluation of a SARS-CoV-2 receptor, angiotensin-converting enzyme 2 (ACE2), indicates that the current pandemic may be caused by a virus with a broad range of hosts, including swine, civets, cats, cows, buffalo, sheep, pigeon, and pangolins [7]. The susceptibility and permissibility of these animals, however, are yet to be tested and proven. One or more domesticated and wild animals have been proposed as an intermediate host for SARS-CoV-2, aiding the spillover to humans [8-11]. Determination of the seroprevalence of SARS-CoV-2 and experimental infection in these animals will help to clarify which, if any, may be involved in the emergence of SARS-CoV-2 [5,7,12]. Moreover, it is also speculated that SARS-CoV-2 could be a result of the recombination of two viruses that circulate in animals living or placed in close proximity [13,14].
If SARS-CoV-2 is the result of a cross-species spillover, the conditions that facilitated the adaptation of SARS-CoV-2 to humans remain unknown [15]. If prejump adaptation occurred in an intermediate host, then SARS-CoV-2 would have undergone genetic refinement in one or more animal species that were spatiotemporally aggregated and in constant contact with humans. The intermediate species is likely to carry an ACE2 receptor that closely clusters with that of humans. If viral adaptation occurred after the spillover event, then continuous passage of the virus from person to person would provide the necessary natural selection opportunities. This would be a realistic assumption given the relatively long incubation (and infectiousness) period, which allows for unnoticeable transmission via a respiratory route. The postadaptation progeny virus can be somewhat genetically distinct from the original virus that made the initial animal-to-human jump [4]. For instance, examining cases of the 2002 severe acute respiratory syndrome (SARS) outbreak revealed that SARS-CoV-1 isolates from early cases (2002) differed from viral isolates obtained from later cases (2003). The viral spike (S) protein involved in binding to the ACE2 receptor showed reduced binding affinity in later viral isolates compared to early isolates [16]. Additionally, a sequence of 29 nucleotides in the C-terminus of the viral genome was deleted in viral isolates from later cases, and this deletion is believed to have occurred through viral adaptation to a human host [17].
Revisiting the Four Levels of Emerging Infectious Diseases
The dramatic rise in human activities in animal farming, the livestock and poultry industries, live-animal markets, and bushmeat has dangerously increased human-animal and animal-animal contact rates [18]. As a result, and combined with careless practices of animal handling, pathogen attack rates have surged substantially. Animals (wildlife and domestic) have gained much attention as candidate reservoirs for numerous, often serious, emerging viral diseases including monkeypox, Ebola, HIV, West Nile virus, rabies, and A/H5N1. The complex link between humans, animals, and emerging pathogens can be simplified in a 4-level model (Figure 1).
Figure 1.
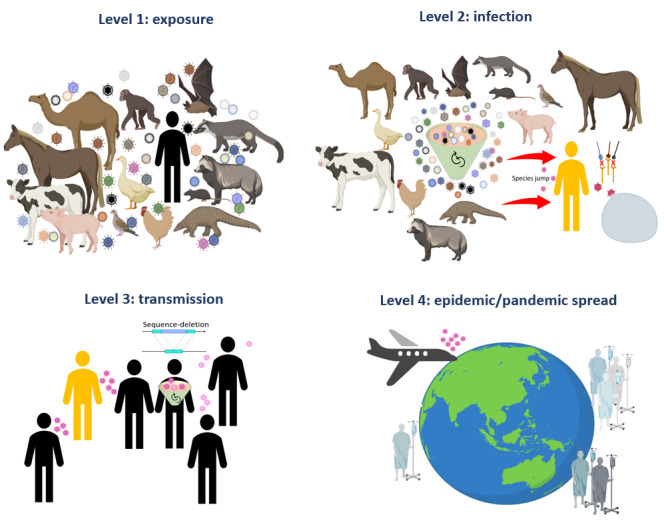
The four-level model of pathogen emergence. Bushmeat production, poultry and livestock industries, and live-animal markets warrant frequent close animal-human and animal-animal contact and increase exposure to pathogens circulating in different species. Frequent exposure (level 1) can turn into a productive infection (level 2), given the necessary viral adaptation has taken place to make humans susceptible. By passaging through humans, the new virus may undergo further adaptation. Pandemic emergence necessitates an efficient human-to-human transmission for sustained propagation. Fast human movement across the globe amplifies the chain of transmission. Efforts should be directed at preventing level 1 (exposure) to significantly decrease the chances of pathogen emergence.
At level 1 (or exposure), the human host encounters the “new” pathogen (eg, virus). Handling animals promotes such encounters. This initial encounter can happen independently of the human’s susceptibility to the virus. Susceptibility means that the host has suitable receptor(s) for viral attachment. The number of viruses to which we are exposed is unknown, but given the profound interaction we have with our environment, we can only assume that the exposure is much higher than we can possibly recognize. At level 2 (or infection), the human-virus encounter becomes fruitful. The human host becomes susceptible and part of the viral lifecycle. Over a thousand pathogenic species have successfully achieved this level. In fact, within the last four decades, new human pathogens have been identified at a rate of more than 3 annually. The majority of these pathogens were viruses [19]. This predominance of viruses as causative agents of emerging diseases is partially attributed to enhanced approaches in viral detection and increased viral mutation rates occurring at relatively short spans [20]. Moreover, almost three-quarters of new human viral infections are attributed to viruses of animal origin that crossed the species barrier [21,22]. Indeed, viruses of nonhuman primates and bats are associated with a high relative risk of emergence [23]. At level 3 (or transmission), the new virus can spread from one person to another. This is generally indicated by the basic reproduction number (R0), a measure that estimates the number of secondary infections in “naïve” susceptible population [24]. A pathogen with an R0 >1 has a better chance of widespread transmission leading to level 4. At level 4 (epidemic/pandemic spread), efficient human-to-human transmission generates sustained propagation. This usually happens without the need for new animal-to-human cross-species transmission events [25].
Factors That Spark Animal-to-Human Spillover
The cascade of events that led to the emergence of SARS-CoV-2 are likely to be complicated, but there seems to be a belief that humans at some point were exposed to the new virus, probably directly from bats (the reservoir?) [26], or indirectly through an intermediate and/or amplifying host(s) (wild and/or farm animals) [5,27,28]. Indeed, increased aggregation of people, wildlife, and domestic animals in spatiotemporal closeness, and the various ways of handling animal products, could have increased the exposure rate (level 1). In order to understand what could have precipitated this cross-species transmission (level 2), it is critical to identify the reservoir and intermediate hosts. The intermediate host can act as a biological bridge between the reservoir and humans. Despite active infection, the intermediate host may appear healthy or asymptomatic long enough for the virus to jump unnoticed to humans. The intermediate host offers the virus the additional advantage of a suitable environment for rapid evolution priming for the jump from animal species to humans. Postjump changes in the virus also tend to occur in the human host, and the significance of these changes is yet to be unraveled [29].
In addition to intermediate hosts, the reservoir is a key player in pathogen emergence. Fruit bats have been a common source for a broad range of viruses associated with emerging human infections [30,31]. In the beginning of the 1990s, humans who came in close contact with the blood or body fluids of sick, infected horses (intermediate) acquired a new virus known as Hendra virus. Later, fruit bats were found to be the source for this paramyxovirus. Since then, Hendra virus has caused some isolated outbreaks of serious and often fatal infections in horses and humans [32]. Moreover, in a single outbreak in 1997, Menangle virus, another paramyxovirus circulating in bats, spilled over to pigs. Humans working in pig farms were directly exposed to the virus shed in the secretions of infected pigs, or indirectly through contaminated surfaces [33]. Fruit bats appear to harbor viruses with a broad range of hosts; viruses that utilize cellular receptors that are highly conserved across species have better chances of breaching species barrier [34,35]. For example, Nipah virus was a new human pathogen recognized in 1998 in association with pig farming and cases of fatal encephalitis. In addition to humans and pigs, the virus could naturally infect other species including horses, dogs, and cats [36]. Thus, these animals also became hosts for the virus. Although pigs were the intermediate host in the early outbreaks of Nipah in Malaysia and Singapore, more recent outbreaks in India and Bangladesh suggest direct transmission from bats to humans through contact with fruits contaminated with infectious bat fluids (eg, urine, saliva) [37]. Interestingly, numerous RNA viruses circulate in bats. RNA viruses are a predominant cause of newly emerging zoonotic diseases, such as Ebola and SARS-CoV-2. Accelerated rates of mutation and presence of several variants of a given strain provide RNA viruses with unmatched capacity for rapid adaptation in a short time [20].
Collectively, the above mentioned factors explain how activities pertaining to human-animal interaction pose an increasingly high risk of pathogen emergence. We may not have substantial evidence for the involvement of human-animal contact in the emergence of SARS-CoV-2, but the new outbreaks in mink farms in the Netherlands [38,39] certainly highlight how cross-species transmission could occur. Indeed, this possibly bidirectional spread between humans and animals necessitates policies that regulate human-animal contact. Furthermore, the expanding human population has led to more wildlife hunting, bush meat trade and consumption, live animal markets, densely populated livestock and poultry industries, exotic species trading, and rapid animal transportation to meet the growing demand for such products. This has created continuous and intense contact between humans and potential viral reservoirs, vectors, and intermediate hosts. Additionally, with increased human movement at exceptionally accelerated speed, pathogens can be introduced to previously uninfected areas, which are geographically distant from the source of infection, leading to disastrous pandemics across the globe. Given that we do not have a vaccine or immunity against most emerging viral diseases, resources directed at level 1 (ie, exposure) remain our best hope for preventing costly consequences. It is believed that emerging zoonotic diseases involve a complex interaction between multiple anthropogenic factors that modify the structure and dynamics of wildlife [40].
Recommendations
Recognizing the importance of these factors has led to intensive collaboration between different disciplines, including veterinary medicine, public health, environmental sciences, and social sciences, etc. This holistic approach, known as One Health, has shown promising results in curbing the spread of pathogens between humans and animals and between animals and animals [41]. For instance, the integration of veterinary sciences, epidemiology, and virology has led not only to the identification of the different animal reservoirs for rabies but also to the establishment of surveillance systems for monitoring outbreaks and the development of an effective vaccine [42]. These outcomes led to significant improvements in the control of rabies in both animals and humans. Similarly, a vaccine against Hendra virus helped control infection in horses and hence prevented transmission of the virus to humans [43]. Another example of fruitful multidisciplinary work is monitoring climate change to identify temperatures that favor pathogen transmission. For example, changes in climate, which in turn can affect mosquito movement, have been used in prediction models to monitor the emergence of West Nile virus [44].
Although in principle the One Health approach has been shown to be effective in controlling and preventing infectious diseases outbreaks, in reality, implementing it has proven to be challenging. Factors such as limited investments, lack of collaboration, and suboptimal surveillance systems have all hindered the achievement of optimal outcomes [45]. However, continuous progress in the One Health approach and supporting policies remain an essential shield in the face of future outbreaks [42]. This entails serious efforts to improve and implement:
Continuous surveillance of animals and vectors (eg, bats, rats, wild birds, primates, arthropods) for the emergence of new pathogens. This should help predict and prepare for the next pathogen threat and define regions susceptible to outbreaks and tools necessary to interrupt route of transmission. Additionally, further modeling, analyzing, and estimating the risk of emergence can help prepare suitable diagnostic tools for the rapid capture of a new outbreak, and frame essential mitigation protocols [46].
International cooperation to establish pathogen sequence libraries and databases updated with biological characterization of new pathogens. This can facilitate efforts in diagnosis, contact tracing, and potential vaccine development.
Studying and monitoring human behavior associated with increased risk of emerging diseases such as wildlife trade and bush meat consumption. Social sciences are important to help introduce behavioral modifications that can be beneficial and easily adhered to by communities. This also requires education and increase in public awareness to the dangers of high-risk behaviors.
Monitoring alterations in land use, climate change, and air quality, as well as introducing policies to reduce and/or mitigate ecological consequences [42]. Zoonosis is greatly impacted by changes in land use. These changes impact the natural habitat of wild animals and their biodiversity, influence their breeding sites, and determine the exposure rate of humans to pathogens in wild animals. Maintaining the biodiverse pool of wildlife and the natural habitat of wild animals should be a priority when planning landscapes [47].
Monitoring and implementing better, safer, and sustainable animal agricultural practices. Overcrowding, mixing of animals, overuse of antibiotics, and methods of transport are some of the drivers of pathogen emergence and re-emergence.
Conclusion
Humans share a close relationship with animals and their environment. Hence, the risk of emergence of new pathogens that can spill over into the human population cannot be completely eliminated. However, understanding the importance of the human-animal-ecosystem interface and tackling the diverse factors that influence the emergence of new pathogens are essential for better prevention, control, and mitigation. The One Health approach could have a significant positive impact in combatting the emergence of infectious diseases. However, this requires national and international cooperation, sharing of data, funding, implementation of policies and legislation, and political will.
Abbreviations
- +ssRNA
positive-sense, single-stranded RNA
- ACE2
angiotensin-converting enzyme 2
- MERS-CoV
Middle East respiratory syndrome coronavirus
- R0
reproduction number
- RNA
ribonucleic acid
- S
spike
- SARS
severe acute respiratory syndrome
- SARS-CoV-1
severe acute respiratory syndrome coronavirus
Footnotes
Conflicts of Interest: None declared.
References
- 1.Chauhan S. Comprehensive review of coronavirus disease 2019 (COVID-19) Biomed J. 2020 Jun 01; doi: 10.1016/j.bj.2020.05.023. https://linkinghub.elsevier.com/retrieve/pii/S2319-4170(20)30087-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harapan H, Itoh N, Yufika A, Winardi W, Keam S, Te H, Megawati Dewi, Hayati Zinatul, Wagner Abram L, Mudatsir Mudatsir. Coronavirus disease 2019 (COVID-19): A literature review. J Infect Public Health. 2020 May;13(5):667–673. doi: 10.1016/j.jiph.2020.03.019. https://linkinghub.elsevier.com/retrieve/pii/S1876-0341(20)30432-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020 Feb 22;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. 2020 Apr 17;26(4):450–452. doi: 10.1038/s41591-020-0820-9. http://europepmc.org/abstract/MED/32284615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lam TT, Jia N, Zhang Y, Shum MH, Jiang J, Zhu H, Tong Y, Shi Y, Ni X, Liao Y, Li W, Jiang B, Wei W, Yuan T, Zheng K, Cui X, Li J, Pei G, Qiang X, Cheung WY, Li L, Sun F, Qin S, Huang J, Leung GM, Holmes EC, Hu Y, Guan Y, Cao W. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020 Jul 26;583(7815):282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 6.Choudhury A, Mukherjee S. In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. J Med Virol. 2020 May 08; doi: 10.1002/jmv.25987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qiu Y, Zhao Y, Wang Q, Li J, Zhou Z, Liao C, Ge X. Predicting the angiotensin converting enzyme 2 (ACE2) utilizing capability as the receptor of SARS-CoV-2. Microbes Infect. 2020 May;22(4-5):221–225. doi: 10.1016/j.micinf.2020.03.003. http://europepmc.org/abstract/MED/32199943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu P, Jiang J, Wan X, Hua Y, Li L, Zhou J, Wang X, Hou F, Chen J, Zou J, Chen J. Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)? PLoS Pathog. 2020 May 14;16(5):e1008421. doi: 10.1371/journal.ppat.1008421. https://dx.plos.org/10.1371/journal.ppat.1008421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi J, Wen Z, Zhong G, Yang H, Wang C, Huang B, Liu R, He X, Shuai L, Sun Z, Zhao Y, Liu P, Liang L, Cui P, Wang J, Zhang X, Guan Y, Tan W, Wu G, Chen H, Bu Z. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020 May 29;368(6494):1016–1020. doi: 10.1126/science.abb7015. http://europepmc.org/abstract/MED/32269068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmitt CA, Bergey CM, Jasinska AJ, Ramensky V, Burt F, Svardal H, Jorgensen MJ, Freimer NB, Grobler JP, Turner TR. ACE2 and TMPRSS2 variation in savanna monkeys (Chlorocebus spp.): Potential risk for zoonotic/anthroponotic transmission of SARS-CoV-2 and a potential model for functional studies. PLoS One. 2020 Jun 23;15(6):e0235106. doi: 10.1371/journal.pone.0235106. https://dx.plos.org/10.1371/journal.pone.0235106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhai X, Sun J, Yan Z, Zhang J, Zhao J, Zhao Z, Gao Q, He W, Veit M, Su S. Comparison of Severe Acute Respiratory Syndrome Coronavirus 2 Spike Protein Binding to ACE2 Receptors from Human, Pets, Farm Animals, and Putative Intermediate Hosts. J Virol. 2020 Jul 16;94(15) doi: 10.1128/jvi.00831-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wassenaar T, Zou Y. 2019_nCoV/SARS-CoV-2: rapid classification of betacoronaviruses and identification of Traditional Chinese Medicine as potential origin of zoonotic coronaviruses. Lett Appl Microbiol. 2020 May;70(5):342–348. doi: 10.1111/lam.13285. http://europepmc.org/abstract/MED/32060933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji W, Wang W, Zhao X, Zai J, Li X. Cross–species transmission of the newly identified coronavirus 2019–nCoV. J Med Virol. 2020 Feb 19;92(4):433–440. doi: 10.1002/jmv.25682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lau SK, Luk HK, Wong AC, Li KS, Zhu L, He Z, Fung J, Chan TT, Fung KS, Woo PC. Possible Bat Origin of Severe Acute Respiratory Syndrome Coronavirus 2. Emerg Infect Dis. 2020 Jul;26(7):1542–1547. doi: 10.3201/eid2607.200092. doi: 10.3201/eid2607.200092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Holmes EC. A Genomic Perspective on the Origin and Emergence of SARS-CoV-2. Cell. 2020 Apr;181(2):223–227. doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W, Zhang C, Sui J, Kuhn JH, Moore MJ, Luo S, Wong S, Huang I, Xu K, Vasilieva N, Murakami A, He Y, Marasco WA, Guan Y, Choe H, Farzan M. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005 Apr 20;24(8):1634–43. doi: 10.1038/sj.emboj.7600640. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chinese SARS Molecular Epidemiology Consortium Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004 Mar 12;303(5664):1666–9. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- 18.Lloyd-Smith JO, George D, Pepin KM, Pitzer VE, Pulliam JRC, Dobson AP, Hudson PJ, Grenfell BT. Epidemic dynamics at the human-animal interface. Science. 2009 Dec 04;326(5958):1362–7. doi: 10.1126/science.1177345. http://europepmc.org/abstract/MED/19965751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woolhouse M, Gaunt E. Ecological origins of novel human pathogens. Crit Rev Microbiol. 2007 Oct 11;33(4):231–42. doi: 10.1080/10408410701647560. [DOI] [PubMed] [Google Scholar]
- 20.Domingo E. Rapid evolution of viral RNA genomes. J Nutr. 1997 May;127(5 Suppl):958S–961S. doi: 10.1093/jn/127.5.958S. [DOI] [PubMed] [Google Scholar]
- 21.Morse SS, Mazet JA, Woolhouse M, Parrish CR, Carroll D, Karesh WB, Zambrana-Torrelio C, Lipkin WI, Daszak P. Prediction and prevention of the next pandemic zoonosis. The Lancet. 2012 Dec;380(9857):1956–1965. doi: 10.1016/s0140-6736(12)61684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woolhouse ME. Population biology of emerging and re-emerging pathogens. Trends in Microbiology. 2002 Oct;10(10):s3–s7. doi: 10.1016/s0966-842x(02)02428-9. [DOI] [PubMed] [Google Scholar]
- 23.Cleaveland S, Haydon D, Taylor L. Overviews of pathogen emergence: which pathogens emerge, when and why? Curr Top Microbiol Immunol. 2007;315:85–111. doi: 10.1007/978-3-540-70962-6_5. http://europepmc.org/abstract/MED/17848062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dietz K. The estimation of the basic reproduction number for infectious diseases. Stat Methods Med Res. 1993 Jul 02;2(1):23–41. doi: 10.1177/096228029300200103. [DOI] [PubMed] [Google Scholar]
- 25.Wolfe ND, Dunavan CP, Diamond J. Origins of major human infectious diseases. Nature. 2007 May 17;447(7142):279–83. doi: 10.1038/nature05775. http://europepmc.org/abstract/MED/17507975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou P, Yang X, Wang X, Hu B, Zhang L, Zhang W, Si H, Zhu Y, Li B, Huang C, Chen H, Chen J, Luo Y, Guo H, Jiang R, Liu M, Chen Y, Shen X, Wang X, Zheng X, Zhao K, Chen Q, Deng F, Liu L, Yan B, Zhan F, Wang Y, Xiao G, Shi Z. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 Feb 03;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao K, Zhai J, Feng Y, Zhou N, Zhang X, Zou J, Li N, Guo Y, Li X, Shen X, Zhang Z, Shu F, Huang W, Li Y, Zhang Z, Chen R, Wu Y, Peng S, Huang M, Xie W, Cai Q, Hou F, Chen W, Xiao L, Shen Y. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 2020 May 7;583:286–289. doi: 10.1038/s41586-020-2313-x. https://www.nature.com/articles/s41586-020-2313-x. [DOI] [PubMed] [Google Scholar]
- 28.Han G. Pangolins Harbor SARS-CoV-2-Related Coronaviruses. Trends Microbiol. 2020 Jul;28(7):515–517. doi: 10.1016/j.tim.2020.04.001. http://europepmc.org/abstract/MED/32544437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grubaugh ND, Petrone ME, Holmes EC. We shouldn't worry when a virus mutates during disease outbreaks. Nat Microbiol. 2020 Apr 18;5(4):529–530. doi: 10.1038/s41564-020-0690-4. http://europepmc.org/abstract/MED/32071422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Field HE. Bats and emerging zoonoses: henipaviruses and SARS. Zoonoses Public Health. 2009 Aug;56(6-7):278–84. doi: 10.1111/j.1863-2378.2008.01218.x. [DOI] [PubMed] [Google Scholar]
- 31.Letko M, Seifert SN, Olival KJ, Plowright RK, Munster VJ. Bat-borne virus diversity, spillover and emergence. Nat Rev Microbiol. 2020 Aug 11;18(8):461–471. doi: 10.1038/s41579-020-0394-z. http://europepmc.org/abstract/MED/32528128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Field HE. Hendra virus ecology and transmission. Curr Opin Virol. 2016 Feb;16:120–125. doi: 10.1016/j.coviro.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 33.Bowden T, Bingham J, Harper J, Boyle D. Menangle virus, a pteropid bat paramyxovirus infectious for pigs and humans, exhibits tropism for secondary lymphoid organs and intestinal epithelium in weaned pigs. J Gen Virol. 2012 May;93(Pt 5):1007–1016. doi: 10.1099/vir.0.038448-0. [DOI] [PubMed] [Google Scholar]
- 34.Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. Bats: Important Reservoir Hosts of Emerging Viruses. CMR. 2006 Jul;19(3):531–545. doi: 10.1128/cmr.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yinda C, Ghogomu S, Conceição-Neto Nádia, Beller L, Deboutte W, Vanhulle E, Maes Piet, Van Ranst Marc, Matthijnssens Jelle. Cameroonian fruit bats harbor divergent viruses, including rotavirus H, bastroviruses, and picobirnaviruses using an alternative genetic code. Virus Evol. 2018 Jan;4(1):vey008. doi: 10.1093/ve/vey008. http://europepmc.org/abstract/MED/29644096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chua K, Bellini W, Rota P, Harcourt B, Tamin A, Lam S, Ksiazek T G, Rollin P E, Zaki S R, Shieh W, Goldsmith C S, Gubler D J, Roehrig J T, Eaton B, Gould A R, Olson J, Field H, Daniels P, Ling A E, Peters C J, Anderson L J, Mahy B W. Nipah virus: a recently emergent deadly paramyxovirus. Science. 2000 May 26;288(5470):1432–5. doi: 10.1126/science.288.5470.1432. [DOI] [PubMed] [Google Scholar]
- 37.Ang BSP, Lim TCC, Wang L. Nipah Virus Infection. J Clin Microbiol. 2018 May 25;56(6):e01875-17. doi: 10.1128/jcm.01875-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oreshkova N, Molenaar R, Vreman S, Harders F, Oude Munnink Bas B, Hakze-van der Honing Renate W, Gerhards Nora, Tolsma Paulien, Bouwstra Ruth, Sikkema Reina S, Tacken Mirriam Gj, de Rooij Myrna Mt, Weesendorp Eefke, Engelsma Marc Y, Bruschke Christianne Jm, Smit Lidwien Am, Koopmans Marion, van der Poel Wim Hm, Stegeman Arjan. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Euro Surveill. 2020 Jun;25(23):25. doi: 10.2807/1560-7917.ES.2020.25.23.2001005. http://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2020.25.23.2001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Enserink M. Coronavirus rips through Dutch mink farms, triggering culls. Science. 2020 Jun 12;368(6496):1169–1169. doi: 10.1126/science.368.6496.1169. [DOI] [PubMed] [Google Scholar]
- 40.Daszak P, Cunningham A, Hyatt A. Anthropogenic environmental change and the emergence of infectious diseases in wildlife. Acta Tropica. 2001 Feb;78(2):103–116. doi: 10.1016/s0001-706x(00)00179-0. [DOI] [PubMed] [Google Scholar]
- 41.Gibbs EPJ. The evolution of One Health: a decade of progress and challenges for the future. Vet Rec. 2014 Jan 25;174(4):85–91. doi: 10.1136/vr.g143. [DOI] [PubMed] [Google Scholar]
- 42.Cunningham AA, Daszak P, Wood JLN. One Health, emerging infectious diseases and wildlife: two decades of progress? Philos Trans R Soc Lond B Biol Sci. 2017 Jul 19;372(1725):20160167. doi: 10.1098/rstb.2016.0167. http://europepmc.org/abstract/MED/28584175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Middleton D, Pallister J, Klein R, Feng Y, Haining J, Arkinstall R, Frazer L, Huang J, Edwards N, Wareing M, Elhay M, Hashmi Z, Bingham J, Yamada M, Johnson D, White J, Foord A, Heine HG, Marsh GA, Broder CC, Wang L. Hendra virus vaccine, a one health approach to protecting horse, human, and environmental health. Emerg Infect Dis. 2014 Mar;20(3):372–9. doi: 10.3201/eid2003.131159. doi: 10.3201/eid2003.131159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rabinowitz PM, Kock R, Kachani M, Kunkel R, Thomas J, Gilbert J, Wallace R, Blackmore C, Wong D, Karesh W, Natterson B, Dugas R, Rubin C, Stone Mountain One Health Proof of Concept Working Group Toward proof of concept of a one health approach to disease prediction and control. Emerg Infect Dis. 2013 Dec;19(12) doi: 10.3201/eid1912.130265. doi: 10.3201/eid1912.130265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gebreyes WA, Dupouy-Camet J, Newport MJ, Oliveira CJB, Schlesinger LS, Saif YM, Kariuki S, Saif LJ, Saville W, Wittum T, Hoet A, Quessy S, Kazwala R, Tekola B, Shryock T, Bisesi M, Patchanee P, Boonmar S, King LJ. The global one health paradigm: challenges and opportunities for tackling infectious diseases at the human, animal, and environment interface in low-resource settings. PLoS Negl Trop Dis. 2014 Nov 13;8(11):e3257. doi: 10.1371/journal.pntd.0003257. https://dx.plos.org/10.1371/journal.pntd.0003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Christaki E. New technologies in predicting, preventing and controlling emerging infectious diseases. Virulence. 2015 Jun 11;6(6):558–65. doi: 10.1080/21505594.2015.1040975. http://europepmc.org/abstract/MED/26068569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patz JA, Daszak P, Tabor GM, Aguirre AA, Pearl M, Epstein J, Wolfe ND, Kilpatrick AM, Foufopoulos J, Molyneux D, Bradley DJ, Working Group on Land Use ChangeDisease Emergence Unhealthy landscapes: Policy recommendations on land use change and infectious disease emergence. Environ Health Perspect. 2004 Jul;112(10):1092–8. doi: 10.1289/ehp.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]


