Abstract
COVID-19 pandemic posed an unprecedented threat to global public health and economies. There is no effective treatment of the disease, hence, scaling up testing for rapid diagnosis of SARS-CoV-2 infected patients and quarantine them from healthy individuals is one the best strategies to curb the pandemic. Establishing globally accepted easy-to-access diagnostic tests is extremely important to understanding the epidemiology of the present pandemic. While nucleic acid based tests are considered to be more sensitive with respect to serological tests but present gold standard qRT-PCR-based assays possess limitations such as low sample throughput, requirement for sophisticated reagents and instrumentation. To overcome these shortcomings, recent efforts of incorporating LAMP-based isothermal detection, and minimizing the number of reagents required are on rise. CRISPR based novel techniques, when merge with isothermal and allied technologies, promises to provide sensitive and rapid detection of SARS-CoV-2 nucleic acids. Here, we discuss and present compilation of state-of-the-art detection techniques for COVID-19 using CRISPR technology which has tremendous potential to transform diagnostics and epidemiology.
Keywords: SARS-CoV-2, COVID-19, CRISPR diagnostics, Molecular testing, Diagnostic advancements
1. Introduction: SARS-CoV-2 detection
A group of patients showing presence of pneumonia of an unknown etiology raised concern in Wuhan, China, during December 2019. Sample sequencing of these patients revealed the presence of previously unknown beta-coronavirus named as 2019-nCoV later renamed as SARS-CoV-2. SARS-CoV-2 classified as clade within the subgenus sarbecovirus, Orthocoronavirinae [1], [2]. Recent sequencing alignment revealed 88% to 96% similarity between bat coronavirus and SARS-CoV-2 hypothesized bat as the most possible host of the SARS-CoV-2 before transmitted to humans [3], [4]. Subsequent studies show pangolins are potential intermediate host for SARS-CoV-2 virus [4]. Highly transmissible SARS-CoV-2 virus resulted in increasing number of cases throughout the world, that leads to pandemic declaration by WHO (World Health Organization). Since the declaration of the COVID-19 pandemic on 11th of March 2020 by the WHO, more than 5.6 million people worldwide have been infected with SARS-CoV-2, resulting in more than 356,254 fatalities till 29th May 2020 [5]. An accurate view of the epidemiology of this disease requires rapid testing and identification to inform followed by contact tracing and quarantine strategies necessary to slow the rate of transmission [6]. Complicating the epidemiology of COVID-19 is the finding that Pre-symptomatic carriers are capable to shed/transmit SARS-CoV-2 prior to onset of symptoms [7].
At present, two general types of tests are available for COVID-19, namely, serological tests and nucleic acid-based tests. While serological detection has the advantages of being easier to conduct, without need for sophisticated instruments, these tests depend on antibody detection, which requires seroconversion to occur in patients prior to administration of the test. During the incubation period of 5–14 days prior to symptom onset, serological tests may suffer from a high rate of false-negative reporting due to low titer of IgM or IgG antibodies in blood samples [8]. Different methods are summarized in Table 1 .
Table 1.
Different non CRISPR detection used for COVID-19 testing.
| SN | Test | Description | References |
|---|---|---|---|
| 1 | Serological immunoassays | 1) Rapid diagnostic test (RDT) (detection in 10 to 30 min) 2) Enzyme linked immunosorbent assay (ELISA) (detection in 1 to 5 h) 3) Neutralization assay (detection in 3 to 5 days). |
[8], [12] |
| 2 | Real time RT-PCR | Real time RT-PCR detection is more sensitive and advantageous over reverse transcriptase (RT-PCR) | [13], [14] |
| 3 | Super high-speed qRT-PCR (SHRT-PCR) | Special rotating disk used for rotating samples to different set temperatures specific to each step of PCR, reducing the time required for temperature ramp up and ramp down time | [15] |
| 4 | TaqMan-based RT-PCR | TaqMan probes improve the sensitivity of assays | [13], [16] |
| 5 | Dual TaqMan probe based RT-PCR | 2 probes specific for different sequences used for reducing the detection time with ultra-sensitivity | [17] |
| 6 | Multiplex RT-PCR | Four mismatch-tolerant molecular beacons targeting four gene used for detection to overcome the issue of rapid mutation in sequences | [13], [18] |
| 7 | RT-LAMP | Reverse transcription followed by as loop-mediated isothermal amplification. Abolishes the need of sophisticated instruments like thermal cycler as reaction is possible at single temperature by using water bath. | [10], [19] |
| 8 | RT-RPA | Reverse transcription followed by Recombinase polymerase amplification (RPA) which is also an isothermal amplification | [11] |
| 9 | Microarray based detection | Complementary probes coated on wells of plate utilized for detection | [20] |
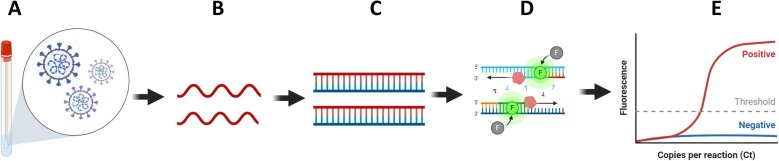
So far, qRT-PCR is considered to be gold standard for SARS-CoV-2 detection. qRT-PCR involves RNA isolation from patient sample (nasal, nasopharyngeal or oropharyngeal swabs, sputum, lower respiratory tract aspirates) and cDNA synthesis followed by amplification of target regions of viral genome detected using double strand specific SYBR green or TaqMan-based fluorescent probes (Fig. 1 ) [9]. Usually N/Nucleocapsid gene, ORF1b, RDRP and E gene of the SARS-CoV-2 is amplified for detection (Table 2 ). Multiplexing i.e. amplification of part of two or more genes at a time is also possible in this technique for further confirmatory detection.
Fig. 1.
Flow diagram demonstrating steps of qRT PCR based detection of viral nucleic acid in sample. A) Nasopharyngeal swab collection from infected patients B) RNAs extracted from samples (red strand) C) Reverse transcription for cDNA synthesis (green strand) and amplification (RT-PCR) conducted by using target specific primers (eg. SARS-CoV-2N gene or RDRP gene) D) Quantitative real time PCR (qRT-PCR) conducted to identify positive samples.E) Final result. Amplification of sample monitored in real time based on fluorescence generated by incorporation of dyes like SYBR green in double stranded DNA. Further TaqMan probes specific to target sequence increases the specificity and sensitivity. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 2.
Different techniques and subsequent SARS-CoV-2 virus gene targets.
| SN | Test | sgRNA designing target sequence** | Cas | Reference |
|---|---|---|---|---|
| 1 | qRT-PCR | SARS-CoV-2N gene, RdRP gene, & E gene | NA* | [9] |
| 2 | SHERLOCK | SARS-CoV-2 S gene & Orf1ab gene | Cas13a | [26] |
| 3 | DETECTR | SARS-CoV-2N gene & E gene | Cas12a | [7] |
| 4 | AIOD-CRISPR | SARS-CoV-2N gene | Cas12a | [27] |
| 5 | ENHANCE | SARS-CoV-2N gene | Cas12a | [28] |
| 6 | CASdetec | SARS-CoV-2 RdRp gene | Cas12b | [24] |
| 7 | FELUDA | SARS-CoV-2 NSP8 and N gene | Cas9 | [25] |
*NA: Not applicable, **N gene: nucleocapsid protein, S gene: spike protein, Orf1ab gene: replicase polyprotein, E gene: envelope protein, RdRp: RNA dependent RNA polymerase, NSP8: non-structural protein 8.
There are several drawbacks to qRT-PCR-based tests, such as the requirement for a centralized laboratory, trained technical staff, sophisticated instrumentation, and expensive reagents that may not be available at the point of care. Additionally, the overall turnaround time may be quite long. Although detection assays themselves may only require 3–4 h, collection, transport, and data analyses may extend reporting to greater than 24 h.
The other promising nucleic acid based test is RT-LAMP (Loop-mediated isothermal amplification) and its refined version known as RT-RPA (Recombinase Polymerase amplification) [10], [11]. Both the methods are alternatives to qRT-PCR method. Since these are isothermal amplification techniques, a simple heating block can replace expensive thermocycler machines. Elimination of thermocycling helps in reduction of overall time needed for the tests and reduces the cost per assay.
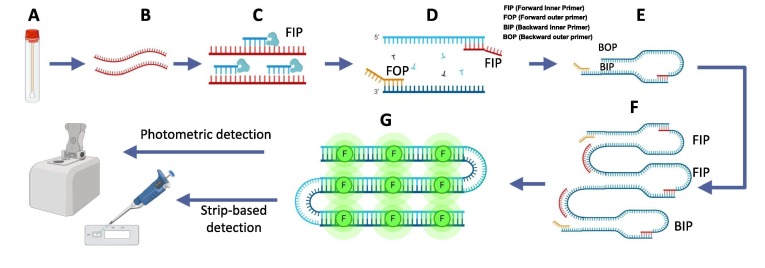
The LAMP technique involves targeting of 6 target sequences with 4 primers along with application of a strand-displacing DNA polymerase [10]. Targeting of several sequences simultaneously makes the assay highly selective for SARS-CoV-2. The reaction endpoint can be determined using double-stranded DNA binding dyes or colorimetric pH indicator dyes (Fig. 2 ). Alternatively, RPA utilizes the key enzymes like recombinase, single stranded DNA-binding protein (SSB) and strand displacing polymerase. The reaction starts with pairing of primer to double-stranded DNA by displacing one strand with the help of recombinase enzyme. Single-stranded DNA stabilized by SSB proteins and then amplification of DNA is carried out by a strand-displacing polymerase akin to PCR amplification. In order to detect RNA targets, amplification occurs after reverse transcription. The advantage of RPA is its optimal temperature ranges between 37 °C and 42 °C, which allows it to be conducted in simple water bath. Within 10 min, few viral copies could be converted into detectable amount [10]. Further addition of TwistAmp exo probes and exonuclease III leads to generation of signal by cleavage of exo-probe functioning as real time detection [11]. Fig. 2, Fig. 3 depicts overall steps in SARS CoV2 detection assays which includes qRT-PCR and CRISPR based assays.
Fig. 2.
Schematic representations of LAMP for detection of viruses in samples. A) Sample (Nasopharyngeal swab) collection, B) RNA extracted from samples (green strand), C) Reverse transcription for cDNA synthesis (red strand) and amplification by strand displacement reaction proceeds at a constant temperature, D - F) Shows LAMP reaction: Target sequence is amplified using four sets of primers with a highly efficient polymerase. Four primer used are specific to target regions involves Forward Inner Primer (FIP), Forward Outer Primer (FOP), Backward Inner Primer (BIP) and Backward Outer Primer (BOP). FOP amplifies the one strand of DNA and displaces the other strand. Displaced strand forms loop and serves as a template for BIP and complementary strand is formed. BOP amplifies the complementary strand and form a dumbbell shape structure. Amplification is achieved by repeating the extensions by respective primers. G) Amplified nucleic acids further can be detected by binding with fluorescent dye, qRT-PCR or paper strip based detection methods (Amplification also observed in real time by using pH sensitive dyes in reaction mixture and monitoring the color change. Due to accumulation of H + ions in the reaction mixture, which leads to increased pH). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
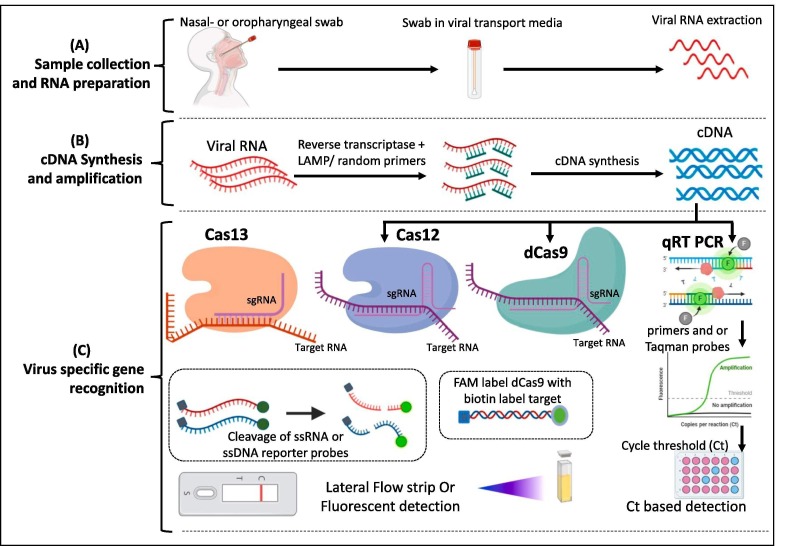
Fig. 3.
Fundamental steps of various CRISPR based COVID-19 detection tests. (A) Collection of sample, transport in Viral transport medium and isolation of RNA (B) Conversion of RNA to cDNA by Reverse transcription. This amplification step could be conducted at isothermal conditions by following RT-RPA of RT-LAMP method. (C) Amplified nucleic acid of viral sample need to be identified by specific viral gene amplification. Conventional method (qRT PCR) utilizes viral gene specific primers to amplify the specific gene segment and further these segments detected by double strand intercalating SYBR dye or fluorescent probes. Recent advancements in CRISPR based diagnostics (CRISPR-Dx) enabled utilization of different Cas enzymes for detection with high sensitivity either from RNA (Cas-13 based detection) or from cDNA (Cas12, Cas9 based etc) followed by color reaction due to the cleavage of reporter probes by the activated Cas enzymes.
Although Isothermal reactions promises cheaper and easier alternative to RT-PCR but optimization of primers and reaction parameters still remains an issue. Therefore, sensitivity and accuracy are challenging for isothermal techniques.
Coupling isothermal reactions with CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) based nucleases found to be an advanced effective tool for nucleic acid based detection.
2. CRISPR associated nuclease based methods
Discovery of CRISPR mediated adaptive immunity and a range of CRISPR-associated proteins (Cas) has not only led transformative advances in genome editing but also in the space of developing next-gen nucleic acid-based diagnostics of disease and pathogen. Integration of lateral-flow chemistry in CRISPR diagnostics opened windows of opportunities towards development of rapid, reliable, specific, and cheap diagnostic kits. CRISPR system has been used for development of molecular diagnostics kits for detection of Dengue virus (DENV), human papillomavirus (HPV) and Zika virus (ZIKV) in human samples [21].
In 2018, Feng Zhang and team developed a method for multiplexed and portable nucleic acid detection platform which used Cas13, Cas12a, and Csm6, called SHERLOCK (specific high-sensitivity enzymatic reporter unlocking) [22] and have successfully used the system for detection of human viruses. In addition to SHERLOCK, there are other CRISPR – based detection techniques published recently (Table-2). One of such techniques is DECTECTR, developed by Chen et al. (2018) [23]. DECTECTR is Cas12a based detection method that involves Cas12a activation after binding with SARS-CoV-2-cDNA and cuts reporter probes (oligonucleotide sequences labelled with the reporter molecules 6-carboxyfluorescein (FAM) and biotin) which confirms detection of virus. Similarly, another Cas12b-based technique CASdetec has been successfully developed, where gRNA has been modified to improve the accuracy further. DECTECTR and CASdetec were reported to detect the viral RNA genome samples at a level of 104 – 105 copies per ml of sample [24]. In addition to SARS-CoV-2 CASdetec has been successfully used for detection of other coronaviruses (SARS-CoV, MERS-CoV, CoV-HKU1, CoV-229E, CoV-OC43, and CoV-NL63) known to cause respiratory diseases with high sensitivity and specificity [24]. Other CRISPR- assisted nucleases such as fnCAS9, Cas12a have also been used for COVID-19 detection, and of these fnCAS9 with its higher mismatch specificity exhibited very high level of accuracy for faster detection [24], [25]. Table-2 shows genes of COVID-19 amplified during different CRISPR-based detection assays. Recently, Sherlock Biosciences (Cambridge, USA), received approval from US Food and Drug Administration’s (FDA) for employing CRISPR-based diagnostic kit for screening COVID-19 cases. The company is working on programming the machinery, which could extract SARS-CoV-2 genetic material in a swab samples from nose, mouth, or throat of patients, or in fluid from the lungs [7].
2.1. CRISPR-Cas13a (C2C2) enzyme based COVID-19 detection
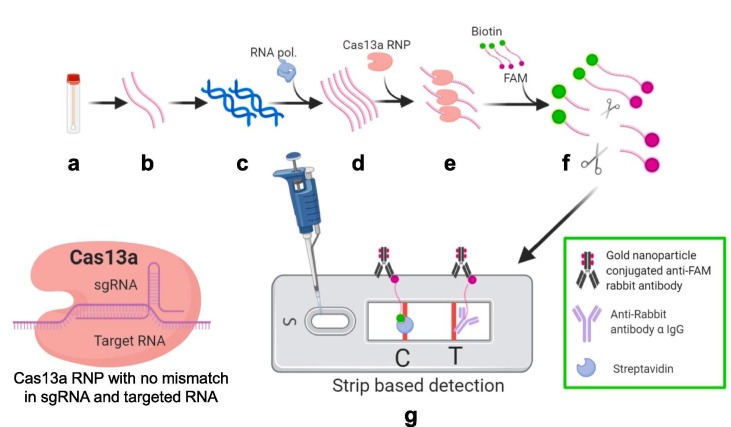
Gootenberg et al. utilized the Cas13a instead of Cas9 considering its ability to bind and cleave RNA rather than DNA substrate [29]. This potential leverage this method to be utilized for viral RNA detection directly. Further contrasting to Cas9, Cas13a becomes an enzymatically active entity instead of returning to inactive state. Active Cas13a cleaves surrounding RNAs regardless of homology. Additionally, this group of scientists developed a technology which provides potential of detection rapidly with high sensitivity, single base specificity and all this on a portable platform. This technology called as SHERLOCK (Specific High-sensitivity Enzymatic Reporter un-LOCKing). which utilizes the recombinase polymerase amplification (RPA) as it an isothermal nucleic acid amplification substitutes need of sophisticated PCR machine. SHERLOCK directly works on RNA virus detection and could also be useful in DNA detection by incorporating T7 RNA polymerase in reaction to convert DNA to RNA. Amplified RNA molecules are subjected to Cas13a nuclease with a guide RNA specific to sequence of interest. After binding to complementary sequence of guide RNA to sample RNA, Cas13a get activated. Active Cas13a shows nonspecific collateral cleavage activity. This collateral cleavage activity reports the signal by cleavage of short nucleotide sequence that is coupled to a fluorescent reporter and a quencher. Cas13a proved its ability to detect RNA as low as 2% of total serum in reactions [29]. There was confirmed differentiation between Zika virus (ZIKV) and dengue virus (DENV) at minimum concentration of 2000 copies of viral genome per mL (3.2aM) [29]. Myhrvold et al. introduced HUDSON technique which offer convenient and simpler RNA extraction from various samples [30]. HUDSON is a method of RNA extraction by lysis of viral particles with inactivation of RNases found in body fluids by using heat and chemical reduction. Advantage of using HUDSON extraction is, the samples after RNA extraction could be directly added to RPA reactions without dilution or purification (to avoid solidification during heating, blood products diluted 1:3). HUDSON combined with SHERLOCK enables detection from range of samples (urine, whole blood, plasma, serum, or saliva) with high sensitivity [30]. In the most recent version of the SHERLOCK technique (SHERLOCKv2) (Fig. 4 ). Gootenberg et al. achieved four-channel multiplexing with 3.5 times increased signal sensitivity to measure as low as 2 aM concentration (2 × 10−18 M viral RNA) [22]. They also discussed application of lateral flow strip-based detection by using FAM-RNA-Biotin reporter. They reported use of HybriDetect strips from Milenia as strip based convenient assay. Addition of CRISPR type-III effector nuclease Csm6 to this reaction for amplified signal detection. Property of synergistic activation of Csm6 by Cas13 leads to enhancement of signal [22].
Fig. 4.
Schematic representation of Specific High-sensitivity Enzymatic Reporter un-LOCKing (SHERLOCK). a) Sample collection (nasopharyngeal swabs) b) RNA extracted from samples by using HUDSON method (red strand) c) Reverse transcription for cDNA synthesis and amplification d) RNA amplification by using Recombinase Polymerase amplification (RT-RPA) and T7 polymerase under isothermal conditions (green strand) e) Cas13a CRISPR ribonucleoprotein (RNP) complex with target specific crRNA added to amplified samples f) RNP complex get activated due to binding with target RNA sequence of positive samples and cleaves the ssRNA reporter probe (Fluorophore FAM with biotin at respective ends) g) Lateral flow strip based detection. Cleaved FAM bearing part of reporter probe present only in positive samples gets accumulated at test band (T) and further get visible due to gold nanoparticle conjugated anti-FAM antibody accumulation. Uncut reporters get accumulated at control band (C). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.2. CRISPR-Cas12a (Cpf1) enzyme for detection
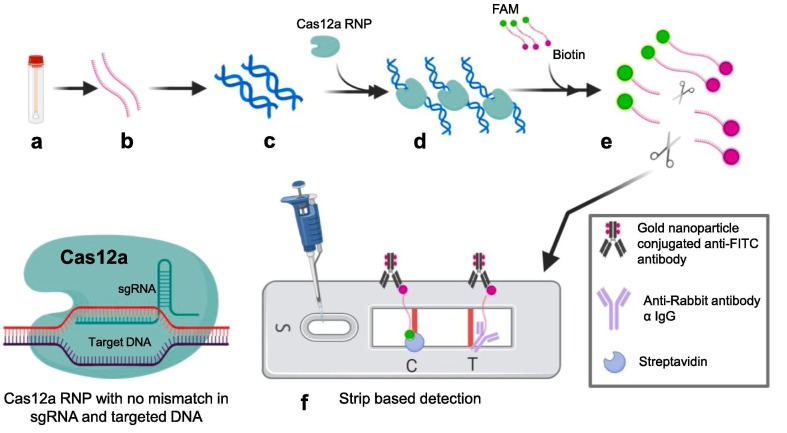
The Doudna group at the University of California Berkley utilized Cas12a in a CRISPR-based detection assay, DNA Endonuclease Targeted CRISPR Trans Reporter (DETECTR). Chen et al. reported remarkable phenomenon about Cas12a. When target sequence complementing at least 15 nucleotides of crRNA, Cas12a cleaves double stranded DNA activating collateral ssDNA cleavage. Activated Cas12a shows trans-ssDNA collateral cleavage activity and cleaves single stranded DNA without any specificity known as indiscriminate single-stranded DNase activity [23]. Mammoth Biosciences Inc. (San Francisco, California, USA) has reconfigured the DETECTR platform for the detection of SARS-CoV-2 within 30 min time from sample to read-out by using lateral flow strip format [7]. RNAs extracted from sample simultaneously subjected to reverse transcription and isothermal amplification by using loop-mediated amplification (RT-LAMP). RT-LAMP reaction takes 20 min and conducted at 62 °C. Further Cas12a binds specifically predefined SARS-CoV-2 gene sequences and get activated. This activated Cas12a cleaves the reporter probes which in turn serves readout in lateral flow strip detection (Fig. 5 ). Cas12a can perform 88 detections in 10 min at 37 °C within 10 min [7].
Fig. 5.
Schematic representation of DNA Endonuclease Targeted CRISPR Trans Reporter (DTECTR) a) Sample collection (nasopharyngeal swab) b) RNA extracted from sample c) Reverse transcription for cDNA synthesis and amplification by using Loop mediated isothermal amplification (RT-LAMP) at 62 °C for 20 min. (green strands with yellow loops) d) Target specific crRNA CRISPR ribonucleoprotein (RNP) complex mixed with amplified products e) Cas12a of RNP complex get activated after binding with target sequences present in positive samples and cleaves ssDNA reporter probes (FAM and biotin at respective ends) f) Lateral flow strip based detection: Cleaved FAM bearing part of reporter probe present only in the positive samples get accumulated at test band and further get visible due to gold nanoparticle conjugated anti-FITC antibody accumulation. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
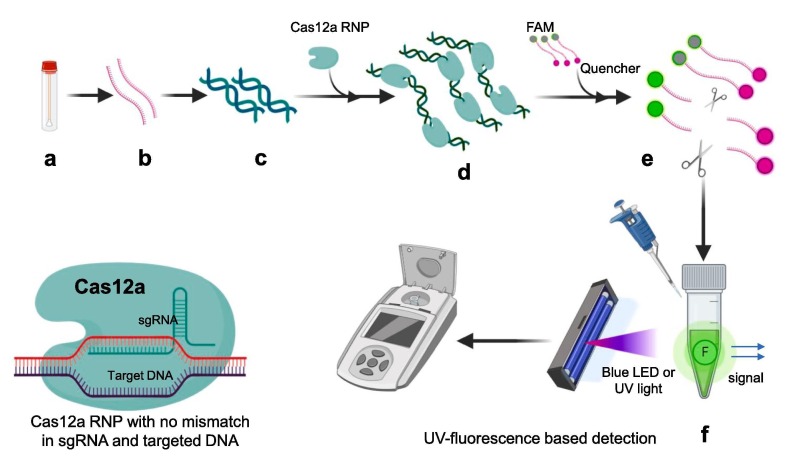
2.3. All-in-One Dual CRISPR-Cas12a (AIOD-CRISPR) assay
Ding et al. utilized two individual crRNAs to generate pair of Cas12a-crRNA which binds to two different sites of target sequence near to RPA primer binding sites [27]. At 37 °C RPA amplification initiates first, and displacement of strands exposes the Cas12a-crRNA binding sites of target sequence. Binding of Cas12a-crRNA on target site activates its endonuclease activity which cleaves the ssDNA-FQ reporter (single stranded DNA sequences labelled with the reporter molecules 6-carboxyfluorescein (FAM) and quencher) to produce fluorescence. This same process repeated continuously on amplified products of RPA. This gives leverage to amplify the signal of detection. Once the complementary sequence binding happens in positive samples, Cas12a cleaves other ssDNA-FQ reporters in proximity by indiscriminate single-stranded DNase activity producing. This process is possible in single tube gives advantage by eliminating need of separate preamplification and amplified product transferring. Further the detection is possible by naked eyes. They also reported that there was no significant difference in positive and negative sample fluorescence when the tubes are kept at room temperature even after 10 min. At 37 °C within one minute there was notable difference in fluorescence of positive sample give further advantage to control the reaction at temperature (Fig. 6 ) [27].
Fig. 6.
Figure elucidating various steps for AIOD-CRISPR assay. a) Sample collection (nasopharyngeal swab) b) RNA extracted from sample (c) Reverse transcription for cDNA synthesis and amplification by using Recombinase Polymerase amplification (RT-RPA) at 37 °C all in one incubation in single tube d) Cas12a CRISPR ribonucleoprotein (RNP) complex with distinct forward and reverse crRNAs added to amplified samples e) RNP complex get activated due to binding with target sequence of positive samples and cleaves the ssDNA-FQ reporter probe (Fluorophore FAM with quencher molecule) f) Due to cleavage of reporter probe quencher and fluorophore get separated and fluorescence could be detected under blue or UV light in positive samples after 40 min in same single tube. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
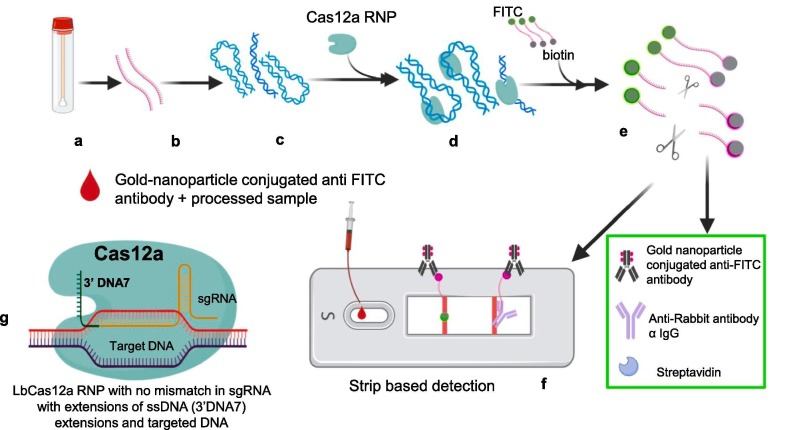
2.4. Enhanced analysis of nucleic acids with crRNA extensions (CRISPR-ENHANCE)
In this technique optimization of CRISPR-based detection assay was achieved. This technique relies on earlier observations about improvement of Cas12a activity due to extended secondary DNA on the guide crRNA or a hairpin RNA structure added to the sgRNA or chemically modified Cas12a guided RNA. Nguyen et al. developed the CRISPR-ENHANCE (Enhanced analysis of nucleic acids with crRNA extensions) technique by using engineered crRNA + 3′DNA7 [28]. Observations also revealed that normal system detects RNA efficiently only when target strand for crRNA is DNA rather than RNA in a heteroduplex. But crRNA + 3′DNA7 is more efficient in enzymatic collateral activity on DNA/RNA heteroduplex compared to wild type crRNA. For detection of SARS-CoV-2 crRNAs were designed to target nucleocapsid phosphoprotein encoding N gene. Engineered crRNA + 3′DNA7 proved higher sensitivity than wild type crRNA of SARS-CoV-2 for detection within 30 min. Integration of paper based lateral flow assay with FITC-ssDNA-Biotin reporter reduced detection time to 20 min and detection of SARS-CoV-2 cDNA to 1 nM without any target amplification. Reverse transcription along with loop mediated isothermal amplification (RT-LAMP) when used the detection limit further reduced to 3–300 copies of RNA by wild type crRNA as well as engineered crRNA + 3′DNA7. But wild type crRNA showed more darker control band (due to insufficient cutting of reporter probes) compared to engineered crRNA + 3′DNA7 on paper strip (due to more efficient cutting of reporter probes). When band intensity compared, ENHANCE showed average 23-fold intensity ratio of positive band to control band compared to wild type crRNA’s average 7-fold ratio at SARS-CoV-2 RNA concentration between 1 nM and 1 pM. Also, in case of ENHANCE visible confirmation was started to get achieved sooner i.e. within 30 s. Thus, in this technique optimization of CRISPR-based detection assay was achieved (Fig. 7 ) [28].
Fig. 7.
Diagrammatic representation of CRISPR-ENHANCE assay. Steps of ENHANCE assay a) Samples like nasopharyngeal swabs collected b) RNAs extracted from samples (red strand) c) Reverse transcription for cDNA synthesis and amplification by using Loop mediated isothermal amplification (RT-LAMP) at 62 °C for 20 min, d) Engineered crRNAs Cas12a RNPs mixed with amplified products e) Cas12a of RNP complex get activated after binding with target sequences present in positive samples and cleaves ssDNA reporter probes (FITC and biotin at respective ends) f) Lateral flow strip based detection: Cleaved FITC bearing part of reporter probe present only in positive samples get accumulated at test band and further get visible due to gold nanoparticle conjugated anti-FITC antibody accumulation. g) Engineered crRNA Cas12a RNPs developed by extended secondary DNA on the guide crRNA. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
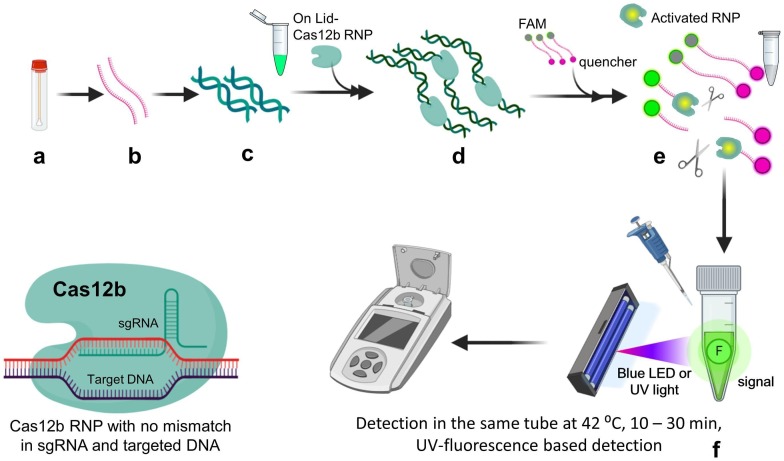
2.5. CRISPR-Cas12b-mediated DNA detection (CASdetec)
CASdetec technique evolved by combining sample treatment protocol with amplification methods and detection of viral nucleic acid by CDetection, which is Cas12b enzyme based. For extraction of viral genome two methods were tested i.e. 1) virus genome extraction kits (spin column) and 2) lysis buffer. Out of which spin column method provide lower detection limit of 10 copies/µL of viral genome but lysis buffer offers convenience at point-of-care testing (POCT). It was observed that 3-fold increase in sgRNA concentration enhances the rate of reaction along with improved fluorescence signal and less background noise by increasing molecular collision between CRISPR and target sequence. For further increment in fluorescence signal incorporation of poly-T fluorescence-quenchers were used. More specifically 7 nucleotide length poly-T reporter was utilized which provided highest signal in shortest time along with sgRNA around RdRp locus. Considering the average viral load representing less than 1 to about 1000 copies per microliter of RNA efficient amplification method is needed. Recombinase-aided amplification (RAA) was used to amplify the substrate 1010 times (from aM to 10 nM) within 10–30 min at constant temperature between 37 °C and 42 °C. It is better to perform amplification RT-RAA and CDetection of viral nucleic acid in single tube, to prevent aerosol contamination chances which is possible when that sample is exposed to environment during testing steps. But when both reactions conducted in single tube it drastically reduces the sensitivity. To overcome this issue, Guo et al. executed RT-RAA reaction within the tube and CDetection reagents were kept in the lid of the tube [24]. Then after amplification the reagents present in lid were spun down into the tube for detection. Single tube operations solved the problem of false positive results due to contamination during transfer and exposure to environment aerosols. To make detection more convenient for POCT, they designed portable dark box fitted with blue LED. Blue LED exposure generates visual confirmation of positive samples in single tube itself (Fig. 8 ). Report suggest detection limit as low as 10 copies/µL without cross contamination [24].
Fig. 8.
Diagrammatic representation of the CASdetec assay. a) Sample collection (nasopharyngeal swab) b) RNA extracted from samples by using spin column-based kit or lysis buffer c) Reverse transcription for cDNA synthesis and amplification by using recombinase aided amplification (RT-RAA) at 42 °C for 30 min (green strands) d) Cas12b CRISPR ribonucleoprotein (RNP) complex stored in lid of tube mixed after amplification e) RNP complex get activated due to binding with target sequence of positive samples and cleaves the ssDNA-FQ 7nt poly-T reporter probe (Fluorophore with quencher molecule) f) Due to cleavage of reporter probe quencher and fluorophore get separated and fluorescence could be detected under blue or UV light in positive samples. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.6. CRISPR-Cas9 enzyme based detection technique
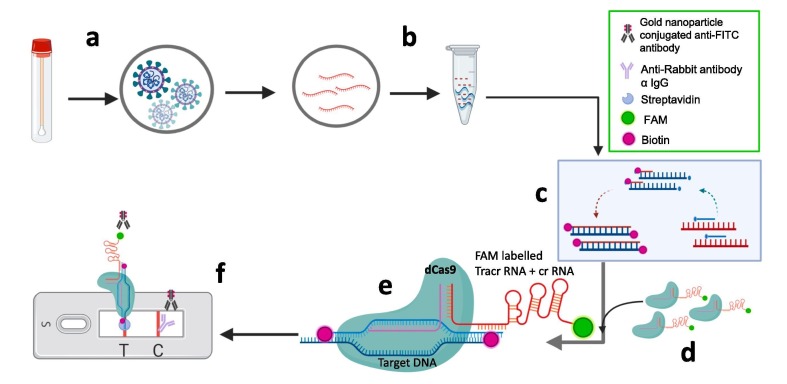
Cas9 enzyme is one of the ubiquitously used enzyme for genetic editing [31]. Cas9 bind specifically to a DNA sequence complementary to guide RNA and serves the endonuclease activity at target site. In case of utilization of this enzyme in CRISPRDx, Azhar et al. reported FnCas9 Editor Linked Uniform Detection Assay (FELUDA) [25]. Cas9 ortholog from Francisella novicida named as FnCas9 used in this assay as it shows high mismatch sensitivity. FnCas9 do not bind to a sequence which harbor mismatch. RNA from viral samples amplified by using biotinylated primers using Recombinase Polymerase Amplification (RPA) method. RPA provides advantage of conducting this technique at isothermal conditions which crucial for point-of-care (POC) where sophisticated setup is not available. CRISPR ribonucleoprotein (RNP) FnCas9 complex labelled with FAM and the biotin labelled amplicons are utilized for detection by using commercially available paper strips. Test line of strip is populated with streptavidin. The biotin labeled amplicons specific to SARS-CoV-2 going to bind at test line along with FAM labelled RNP, subsequently bind to anti-FAM antibody linked gold nanoparticles flow through applied buffer. In absence of biotin labelled amplicons in the sample, the FAM labelled RNP will not bind at streptavidin test band line, Anti-Rabbit antibody is coated at control line, where unbound gold nanoparticle conjugated anti-FAM antibody get bind and shows color band (Fig. 9 ) [16].
Fig. 9.
Schematic representation of paper strip based FnCas9 Editor Linked Uniform Detection Assay (FELUDA): a) Sample (Nasopharyngeal swab) collection b) RNAs extracted from samples c) Reverse transcription for cDNA synthesis and amplification by using biotin labelled primers (red strand) d) FAM labelled dFnCas9 ribonucleoprotein (RNP) complex (FAM labelled tracrRNA is compatible with crRNA) e) RNP complex get activated due to binding with target sequence of positive samples showing no mismatch in sgRNA and target sequences of positive samples f) Lateral flow strip based detection. FAM labelled RNP bound to biotinylated target sequence of positive samples get accumulated at streptavidin test band and further get visible due to gold nanoparticle conjugated anti-FAM antibody accumulation. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3. Summary
Point of care, easy to perform tests are extremely important for diagnosis of COVID-19, as laboratory facilities may not be accessible in poor countries or even places of greater risk of infection such as airports, ports, emergency departments etc. Instrument free, field deployable test kits with higher accuracy and specificity would be ideal for containment of such disease. CRISPR based diagnosis tools promises such accuracy and much shorter turnaround time vis-à-vis other nucleic acid-based methods.
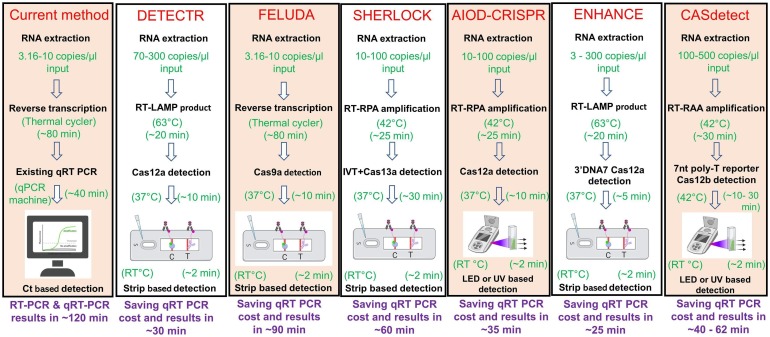
While sherlock provides a quick diagnostic with the help of CRISPR-Cas13a based reporter targeting and recently got approval from CDC USA, other modified version of sherlock such as HUDSON or Sherlock 2.0 aids in simpler RNA extraction/extraction from body fluid directly and higher signal detection respectively. Mammoth bioscience’s DETECTOR technique exploits genome editing capability of Cas12a and its simultaneous single stranded DNA targeting while activated. Dual AIDO CRISPR system further simplifies Cas12a based detection by offering single tube reaction and fluorescence based detection without use of strips. Similarly, CRISPR ENHANCE improves Cas12a based detection even in a femtomolar range of viral nucleic acid by modifying/improving crRNA and other parameters. CDetection platform of Guo et al. exploits editing capability of Cas12b (C2C1) and provides another option for using class-2-V CRISPR system [24]. So far only one report is there for the most popular DNA targeting Class 2, CRISPR Nuclease Cas9, which is FELUDA technology based on dead Fn Cas9 and detects cDNA of viral origin by enzymatic readout, without editing any separate reporter molecule. Fig. 10 shows a comparative analysis of workflow of different CRISPR based systems and total time needed to perform the tests. We believe that with this continuing effort of developing diverse testing methods using CRISPR system further advances the faster COVID-19 detection at the point of care.
Fig. 10.
Workflow comparison of RT -PCR and different CRISPR based assyas for SARS CoV2 detection.
4. Discussion
Current methods like nucleic acid-based diagnostic requires expensive and sophisticated machinery. Sample processing should be minimum and less time consuming considering the situations like pandemic outbreak of virus. Antigen based testing process satisfy the above requisites, but it also brings limitations like less sensitivity and selectivity [30]. In situations like sudden outbreak of SARS-CoV-2, rapid and portable tests which requires minimum sophistication and low cost are the most important and first armor of defense in healthcare system. Recent advancements in CRISPR created new domain of CRISPR diagnostics. Cas9 of type II CRISPR system is most explored protein and due to its high specificity towards target sequence, Cas9 has potential to be used for development of new nucleic acid detection methods. There are several reports about detection methods based on typical double-stranded DNA (dsDNA) cleavage activity of Cas9 for example detection of Zika viruses. Further catalytically deactivated Cas9 (dCas9), has been explored for its potential applicability in nucleic acid detection methods for diagnostic purposes [32]. However, in recent years CRISPR/Cas proteins with collateral cleavage activity made remarkable advancements in CRISPR diagnostics. Initial advancement revealed Cas13a of type III CRISPR based SHERLOCK system demonstrated promising results with its collateral cleavage activity for single-stranded RNA (ssRNA) [26]. Further advancements explored Cas12a (DETECTR, AIOD CRISPR, ENHANCE) and Cas12b (CAS Detec) with its collateral cleavage activity for single-stranded DNA (ssDNA) [7], [24], [27], [28]. CRISPR diagnostic represented attomolar sensitivity based on amplification of collateral cleavage activity. These methods also show remarkable specificity for single base and reduction in total assay time. Further all these methods could be amenable for detection of various viruses by simple changes in target sequences complement.
All these methods are compatible to be leveraged as a point of care diagnostics based on strip and lateral flow based detections. The rapidly spreading pandemic generated requirement of expansion of current capabilities by using applicability of sensors and readout devices. Advancement of SHERLOCK denoted as SHERLOCKv2 has been developed for demonstrating multiplexed detection of target sequences at attomolar concentration in a single reaction [22]. Four channel multiplexing of SHERLOCKv2 involves LwaCas13a, PsmCas13b, CcaCas13b, and AsCas12a in FAM, TEX, Cy5, and HEX channels, respectively which specifically targets orthogonal dinucleotide motifs with specific fluorescence reporters [3]. Ackerman et al. developed Combinaterial Arrayed Reactions for Multiplexed Evaluation of Nucleic Acids (CARMEN) [33]. CARMEN leverages miniaturized and self-organizing microfluidic technology for massive multiplexing. CARMEN involves collection of inputs as droplet emulsions and arrangement of droplets in respective wells. Prior to introduction of microarray chip the amplified sample combined with Cas13 detection mix (solution based fluorescent color code) in microtiter plate. Emulsified solution makes fluorous oil to yield 1-nl droplets which is further loaded into a microwell-array chip. These droplets each containing sample and detection solution get arranged pairwise in chip. Under exposure of an electric field both droplets get merged, which initiates detection reaction. Further implication of computation power of machine learning and image analysis approach improved the analysis with improved accuracy and sensitivity [4]. Current pandemic and future outbreaks generated desperate need for development of new diagnostic approaches. CRISPR diagnostic opened a future avenue for development of low cost, rapid, sensitive, accurate and suitable point of care diagnostics. Future tests will leverage the computational power for better diagnostics.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
All the figures have been created by Biorender.com. Authors also acknowledge Reliance Industries Ltd for funding.
References
- 1.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020;109:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N.a., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu D.i., Wu T., Liu Q., Yang Z. The SARS-CoV-2 outbreak: What we know. Int. J. Infect. Dis. 2020;94:44–48. doi: 10.1016/j.ijid.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Who.int, Coronavirus. (2020). https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed May 29, 2020).
- 6.Wilder-Smith A., Freedman D.O. Isolation, quarantine, social distancing and community containment: pivotal role for old-style public health measures in the novel coronavirus (2019-nCoV) outbreak. J. Travel Med. 2020;27:1–4. doi: 10.1093/jtm/taaa020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nature Biotechnology. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruning A.H.L., Leeflang M.M.G., Vos J.M.B.W., Spijker R., De Jong M.D., Wolthers K.C., et al. Rapid tests for influenza, respiratory syncytial virus, and other respiratory viruses: A systematic review and meta-Analysis. Clin. Infect. Dis. 2017;65:1026–1032. doi: 10.1093/cid/cix461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2019;3:1–8. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thi H., Thai C., Le M.Q., Vuong C.D., Parida M., Minekawa H., et al. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus development and evaluation of a novel loop-mediated isothermal amplification method for rapid detectio. J. Clin. Microbiol. 2004;42:1956–1961. doi: 10.1128/JCM.42.5.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aebischer A., Wernike K., Hoffmann B., Beer M. Rapid genome detection of schmallenberg virus and bovine viral diarrhea virus by use of isothermal amplification methods and high-speed real-time reverse transcriptase PCR. J. Clin. Microbiol. 2014;52(6):1883–1892. doi: 10.1128/JCM.00167-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ben-Selma W., Harizi H., Boukadida J. Immunochromatographic IgG/IgM test for rapid diagnosis of active tuberculosis. Clin. Vaccine Immunol. 2011;18(12):2090–2094. doi: 10.1128/CVI.05166-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hadjinicolaou A.V., Farcas G.A., Demetriou V.L., Mazzulli T., Poutanen S.M., Willey B.M., Low D.E., Butany J., Asa S.L., Kain K.C., Kostrikis L.G. Development of a molecular-beacon-based multi-allelic real-time RT-PCR assay for the detection of human coronavirus causing severe acute respiratory syndrome (SARS-CoV): a general methodology for detecting rapidly mutating viruses. Arch. Virol. 2011;156(4):671–680. doi: 10.1007/s00705-010-0906-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noh J.Y., Yoon S.-W., Kim D.-J., Lee M.-S., Kim J.-H., Na W., Song D., Jeong D.G., Kim H.K. Simultaneous detection of severe acute respiratory syndrome, Middle East respiratory syndrome, and related bat coronaviruses by real-time reverse transcription PCR. Arch. Virol. 2017;162(6):1617–1623. doi: 10.1007/s00705-017-3281-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakurai A., Nomura N., Nanba R., Sinkai T., Iwaki T., Obayashi T., Hashimoto K., Hasegawa M., Sakoda Y., Naito A., Morizane Y., Hosaka M., Tsuboi K., Kida H., Kai A., Shibasaki F. Rapid typing of influenza viruses using super high-speed quantitative real-time PCR. J. Virol. Methods. 2011;178(1-2):75–81. doi: 10.1016/j.jviromet.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Q., Xu Z., Wei T., Zeng H., Li J., Gang H., Sun M., Jiang F., Wang X., Dong W., Yang L., Wang J. Development of taqman RT-nested PCR system for clinical SARS-CoV detection. J. Virol. Methods. 2004;119(1):17–23. doi: 10.1016/j.jviromet.2004.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yip S. P L.W.L., To S.S.T., Leung P.H., Cheung T.S., Cheng P.K. Use of dual TaqMan probes to increase the sensitivity of 1-step quantitative reverse transcription-PCR: application to the detection of SARS coronavirus. Clin. Chem. 2005;51:1885–1888. doi: 10.1373/clinchem.2005.054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu X.-F., Pan J.-C., Ye R., Xiang H.-Q., Kou Y., Huang Z.-C. Preparation of armored RNA as a control for multiplex real-time reverse transcription-PCR detection of influenza virus and severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 2008;46(3):837–841. doi: 10.1128/JCM.01904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.da Silva S.J.R., Paiva M.H.S., Guedes D.R.D., Krokovsky L., de Melo F.L., da Silva M.A.L., et al. Development and validation of reverse transcription loop-mediated isothermal amplification (RT-LAMP) for rapid detection of ZIKV in mosquito samples from Brazil. Sci. Rep. 2019;9(1) doi: 10.1038/s41598-019-40960-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hardick J., Metzgar D., Risen L., Myers C., Balansay M., Malcom T., Rothman R., Gaydos C. Initial performance evaluation of a spotted array Mobile Analysis Platform (MAP) for the detection of influenza A/B, RSV, and MERS coronavirus. Diagn. Microbiol. Infect. Dis. 2018;91(3):245–247. doi: 10.1016/j.diagmicrobio.2018.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chertow D.S. Next-generation diagnostics with CRISPR. Science. 2018;360(6387):381–382. doi: 10.1126/science.aat4982. [DOI] [PubMed] [Google Scholar]
- 22.Gootenberg J.S., Abudayyeh O.O., Kellner M.J., Joung J., Collins J.J., Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360(6387):439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., Doudna J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360(6387):436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo L.u., Sun X., Wang X., Liang C., Jiang H., Gao Q., et al. SARS-CoV-2 detection with CRISPR diagnostics. Cell Discov. 2020;6(1) doi: 10.1038/s41421-020-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.S.M. Mohd. Azhar, Rhythm Phutela, Asgar Hussain Ansari, Dipanjali Sinha, Namrata Sharma, Manoj Kumar, Meghali Aich, Saumya Sharma, Khushboo Singhal, Harsha Lad, Pradeep K. Patra, Govind Makharia, Giriraj Ratan Chandak, Debojyoti Chakraborty, Rapid, field-deployable nucleobase detection and identification using FnCas9, BioRxiv. (2020). doi:: https://doi.org/10.1101/2020.04.07.028167.
- 26.Zhang F., Abudayyeh O.O., Gootenberg J.S., Sciences C., Mathers L. A protocol for detection of COVID-19 using CRISPR diagnostics. Bioarchive. 2020:1–8. [Google Scholar]
- 27.Ding X., Yin K., Li Z., Lalla R., Ballesteros E., Sfeir M, Liu C. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 2020;2020(18) doi: 10.1038/s41467-020-18575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen L.T., Smith B.M., Jain P.K. Enhancement of trans-cleavage activity of Cas12a with engineered crRNA enables amplified nucleic acid detection. Nat. Commun. 2020;11(4906) doi: 10.1038/s41467-020-18615-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356(6336):438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myhrvold C., Freije C.A., Gootenberg J.S., Abudayyeh O.O., Metsky H.C., Durbin A.F., et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360(6387):444–448. doi: 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knott G.J., Doudna J.A. CRISPR-Cas guides the future of genetic engineering. Science. 2018;361(6405):866–869. doi: 10.1126/science.aat5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu X., Luo T., Gao J., Lin N., Li W., Xia X., et al. CRISPR-assisted DNA detection, a novel dCas9-based DNA detection technique. BioRxiv. 2020 doi: 10.1101/2020.05.13.093062. [DOI] [PubMed] [Google Scholar]
- 33.Ackerman C.M., Myhrvold C., Thakku S.G., Freije C.A., Metsky H.C., Yang D.K., et al. Massively multiplexed nucleic acid detection with Cas13. Nature. 2020;582(7811):277–282. doi: 10.1038/s41586-020-2279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]