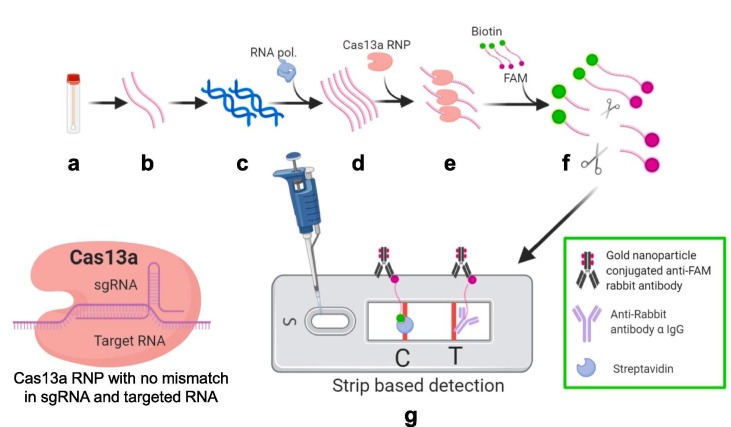
Fig. 4.
Schematic representation of Specific High-sensitivity Enzymatic Reporter un-LOCKing (SHERLOCK). a) Sample collection (nasopharyngeal swabs) b) RNA extracted from samples by using HUDSON method (red strand) c) Reverse transcription for cDNA synthesis and amplification d) RNA amplification by using Recombinase Polymerase amplification (RT-RPA) and T7 polymerase under isothermal conditions (green strand) e) Cas13a CRISPR ribonucleoprotein (RNP) complex with target specific crRNA added to amplified samples f) RNP complex get activated due to binding with target RNA sequence of positive samples and cleaves the ssRNA reporter probe (Fluorophore FAM with biotin at respective ends) g) Lateral flow strip based detection. Cleaved FAM bearing part of reporter probe present only in positive samples gets accumulated at test band (T) and further get visible due to gold nanoparticle conjugated anti-FAM antibody accumulation. Uncut reporters get accumulated at control band (C). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)