Abstract
Background
Zinc finger and BTB domain-containing 4 (ZBTB4), which is a transcriptional regulator, has been identified as a tumor suppressor in several human carcinomas. So far, however, the expression of ZBTB4 and its possible clinical significance in colorectal cancer (CRC) remain unknown.
Materials and Methods
The mRNA and protein expressions of ZBTB4 in five CRC cell lines were respectively detected by performing qRT-PCR and Western Blotting. ZBTB4 expression in colorectal tissue specimens was determined, and subsequently its relationship with clinical prognosis was examined.
Results
The mRNA and protein expressions of ZBTB4 were significantly decreased in all the five CRC cell lines compared with normal colonic epithelial cells. Consistent with the cell data, immunohistochemical results showed that as compared with the normal colorectal tissue samples, ZBTB4 protein expression was clearly lower in the CRC tissue samples, especially in CRC patients with liver metastasis. In addition, low-expressed ZBTB4 was found associated with tumor metastasis stage (P=0.0003) and level of carcinoembryonic antigen (CEA) (P=0.0004). The overall survival (OS) and recurrence-free survival (RFS) in the ZBTB4-low group were significantly lower than those in the ZBTB4-high group (P=0.0007 and P=0.0077).
Conclusion
The current findings showed that patients with high-expressed ZBTB4 in CRC tissues may develop a better prognosis, and ZBTB4 could serve as a potential therapeutic target for CRC treatment.
Keywords: colorectal cancer, ZBTB4 expression, immunohistochemical, clinical significance, prognosis
Introduction
Colorectal cancer (CRC) occurs at the colorectal mucosa and glands. It is one of the most common malignancies, with a high morbidity and mortality in the world.1 Approximately 1.2 million new CRC cases are diagnosed annually, and globally, the incidence of CRC, noticeably more young cases, is gradually increasing.2,3 Carcinogenesis is a highly complex process, involving many factors and stages. Studies on tumorigenesis mechanism and gene functions found that the activation of proto-oncogenes and inactivation of tumor suppressor genes can promote the development and progression of CRC.4 Investigation of the molecular mechanism of CRC has long been the focus of cancer prevention and treatment.
Zinc finger and BTB domain-containing protein is the important member of C2H2 Zinc finger protein family, which contains C2H2 Zinc fingers and a BTB/POZ domain. It can selectively combine with specific target structures to regulate the growth, proliferation and differentiation of both normal cells and tumor cells, and is closely related to the occurrence of many diseases.5–7 In recent years, increasing attention has been paid to the role of ZBTB family in tumors. ZBTB4, alternatively known as KAISO-L1 or ZNF903, belongs to the ZBTB protein family, and functions as a transcriptional repressor.8 Previous studies have reported that ZBTB4 expression is down-regulated in several types of cancers, including in breast cancer, prostate cancer, and Ewing sarcoma.9–11 ZBTB4 can suppress the growth and invasion of breast cancer cells in vitro by inhibiting the expressions of specific protein transcription factors.10 In addition, the up-regulation of Sp repressor ZBTB4 induced by CDODA-Me through targeting miRNAs has been found to inhibit the cell growth of prostate cancer, thus, ZBTB4 overexpression is considered as a prognostic factor for prostate cancer.11 Recent study demonstrated that ZBTB4 inhibits cell growth of Ewing sarcoma, showing pronounced antineoplastic effect by reducing Survivin expression.9 The above findings indicate that ZBTB4 functions as a tumor suppressor in several human carcinomas. So far, however, there is a lack of study on the expression and role of ZBTB4 in CRC.
In this study, the expression of ZBTB4 in CRC cell lines and tissue specimens was determined. Subsequently, the possible clinical significance of ZBTB4 was examined by analyzing clinicopathological data. Our findings demonstrated that ZBTB4 was down-regulated in CRC and that its expression was positively correlated with clinical prognosis of CRC patients. Thus, as a novel prognostic factor, ZBTB4 may be a potential therapeutic target for CRC treatment.
Materials and Methods
Cell Culture
The human normal colonic epithelial cell line NCM460 and the human CRC cell lines (HT-29, HCT-116, SW480, CaCO2 and SW620) were purchased from the American Type Culture Collection (ATCC; USA). All the cell lines were grown in RPMI-1640 medium containing 10% of FBS, glutamine and antibiotics (penicillin/streptomycin) in a humidified incubator at 37°C with 5% CO2/95% air.
Bioinformatics Analysis
To assess the role of ZBTB4 in CRC, relevant data were downloaded from The Cancer Genome Atlas (TCGA) database, and the mRNA expression of ZBTB4 in CRC tissues and normal colorectal tissues were obtained and carefully analyzed.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
The total RNA of the cells (NCM460, HT-29, HCT-116, SW480, CaCO2 and SW620) was extracted by TRIZOL reagent (TaKaRa, Japan). The RNA samples were reverse-transcribed into cDNA using a ReverTra Ace qPCR RT kit (Toyobo, Japan). The primer sequences used were as follows: ZBTB4, forward, 5′-AGGAAGTACCCCTGCCGCTA-3′; reverse, 5′-TTGTAGCCTCCATTGGGTGT-3′. β-actin, forward, 5′-GGACTTCGAGCAAGAGATGG-3′; reverse, 5′-AGCACTGTGTTGGCGTACAG-3′. ZBTB4 expression was determined by quantitative real-time PCR using Power SYBR Green PCR master mix (Applied Biosystems, USA). PCR parameters were as follows: 95°C for 3min, 40 cycles of 95°C for 10s, and 58°C for 30s. The relative mRNA expressions were calculated by the Ct method, and β-actin was chosen as the control. Each sample was run in triplicate.
Western Blotting
The cells were lysed in 1xRIPA buffer (CST, Danvers, MA, USA) containing 1 mM phenylmethylsulfonyl fluoride (PMSF), and the protein concentration was quantified by the Bradford protein assay. A total of 30 μg protein lysates from each sample were separated in 10% SDS-PAGE and then transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). After blocking with 5% non‑fat milk, the membranes were incubated overnight on ice with the primary antibodies against ZBTB4 (polyclonal; Santa Cruz Biotechnology, CA, USA) and GAPDH (all with 1:1000 dilution). After washing, the membranes were probed with horseradish peroxidase-conjugated goat anti-rabbit or anti-rat secondary antibody FluorChem E system (Protein Simple, Santa Clara, CA, USA) was applied for the signal detection.
Patients and Tissue Specimens
A total of 66 CRC tissue samples were collected from the follow-up database of primary CRC patients who underwent surgical resection in our hospital between January 2015 and June 2015. All the patients had complete clinicopathologic data and full follow-up data, and did not undergo preoperative neoadjuvant chemoradiotherapy prior to the study. The diagnosis of CRC was independently confirmed by at least two board-certified pathologists, and tumor staging was decided based on the American Joint Committee on Cancer (AJCC) guidelines. All the procedures of this study were approved by the Clinical Research Ethics Committee of the First Affiliated Hospital of Zhejiang University.
Immunohistochemistry and Evaluation
Immunohistochemical (IHC) staining was performed to detect the expression of ZBTB4 in paraffin-embedded tissue samples from the CRC patients. Briefly, formalin-fixed, paraffin-embedded tissue sections were consecutively cut into 4-μm slices, which were incubated with diluted (1:400) primary ZBTB4 antibody (Santa Cruz Biotechnology, CA, USA) for 1 h at room temperature. Antigen retrieval was conducted by heat treatment in a citrate buffer. After washing with phosphate-buffered saline (PBS), the slices were incubated with second antibody for 20 min, followed by PBS washing. Finally, the slices were visualized with diaminobenzidine and nuclei were counterstained with hematoxylin. The primary antibody was excluded from negative control group.
The IHC staining results were independently evaluated by two pathologists blinded to the clinical-pathological data. The staining of ZBTB4 protein was graded as follows: Low expression is defined when the percentage of ZBTB4 positive cells is less than 50%, and high expression is defined when the percentage of ZBTB4 positive cells is more than 50%.
Statistical Analysis
GraphPad Prism 7 was used for statistical analysis. Student’s t-test was conducted to compare the expression of ZBTB4 between the normal colonic epithelial and CRC cell lines. The relationships between ZBTB4 expression and the CRC clinical features were analyzed using the chi-square test. The Kaplan-Meier method was plotted for the survival analysis. P<0.05 was considered as statistically significant.
Results
ZBTB4 Expression Was Down-Regulated in Human CRC Tissues and Cells
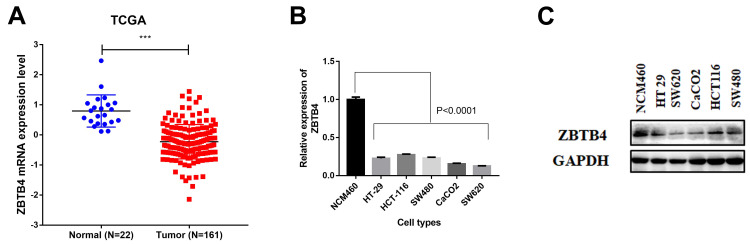
To examine the role of ZBTB4 in CRC, we analyzed the expression of ZBTB4 in TCGA cohort datasets (Figure 1A). From the TCGA cohort, we found that ZBTB4 mRNA expression was significantly down-regulated in CRC tissues (P<0.0001). Then, the mRNA and protein expressions of ZBTB4 were detected in the five CRC cells (HT-29, HCT-116, SW480, CaCO2 and SW620) and normal intestinal epithelial cells (NCM460) respectively by qRT-PCR and Western Blotting. From qRT-PCR, we observed significantly lower mRNA expression of ZBTB4 in all the CRC cell lines than that in the normal intestinal epithelial cells (P<0.0001, Figure 1B). Furthermore, CaCO2 and SW620 exhibited relatively lower expression. Consistent with the above findings, Western Blotting demonstrated that ZBTB4 protein expression was noticeably lower in all the five CRC cell lines than in the normal intestinal epithelial cell line (Figure 1C).
Figure 1.
ZBTB4 expression was down-regulated in colorectal carcinoma (CRC). (A) Relative mRNA expression of ZBTB4 in CRC tissues and their related normal tissues from the TCGA database. P value was calculated by Student’s t-test. The mRNA (B) and protein (C) expressions of ZBTB4 in CRC cell lines (HT-29, HCT-116, SW480, CaCO2 and SW620) and normal colonic epithelial cell lines (NCM460). ***P < 0.001.
Clinical Significance of ZBTB4 in CRC Specimens
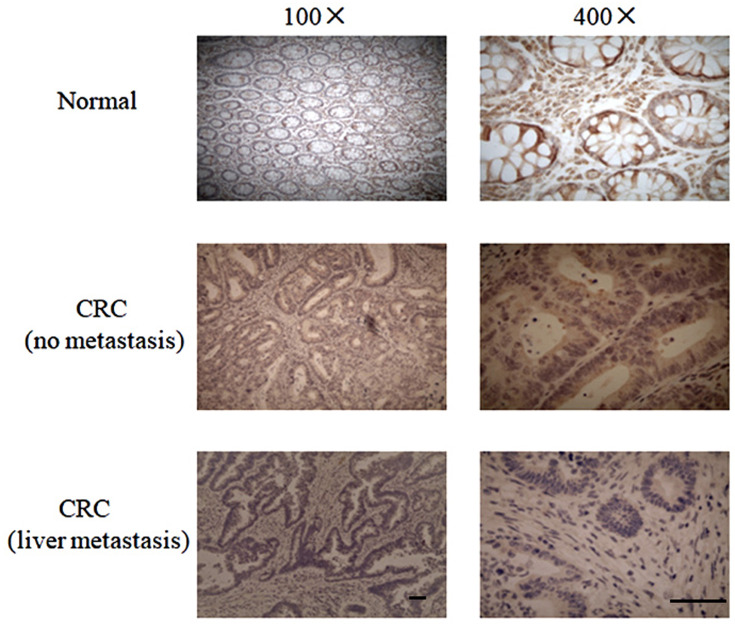
The ZBTB4 protein expression in the paraffin-embedded CRC tissue samples was determined by IHC staining. Representative images of ZBTB4 IHC staining were shown in Figure 2. The results showed that ZBTB4 expression was down-regulated in CRC and was negatively associated with metastasis. Thirty-nine patients (59.09%) out of a total of 66 CRC patients showed negative staining, while the rest 27 patients (40.91%) had positive staining. Negative staining was defined as low-expressed ZBTB4, while positive staining was defined as high-expressed ZBTB4. All the patients were divided into ZBTB4-low group (N=39) or ZBTB4-high group (N=27) according to their IHC staining results. As shown in Table 1, clinical association analysis by chi-squared test demonstrated that ZBTB4 expression were evidently lower in CRC patients with liver metastasis stage (P=0.0003) and high CEA level (P=0.0004). However, there was no correlation between ZBTB4 and other clinicopathological factors, including patient sex (P=0.347), age (P=0.253), differentiation grade (P=0.887), T stage (P=0.233), N stage (P=0.226), tumor location (P=0.461) or vascular invasion (P=0.086).
Figure 2.
IHC staining of ZBTB4 in normal tissues and cancer tissues with or without liver metastasis from CRC patients. Original magnification, ×100 and ×400. Scale bar = 20 μm.
Table 1.
Association of ZBTB4 Expression and Clinical Characteristics in CRC Patients
| Characteristic | N | ZBTB4 Low (N=39) | Expression High (N=27) | χ2value | P value |
|---|---|---|---|---|---|
| Sex | 0.88 | 0.347 | |||
| Male | 37 | 20 | 17 | 4 | |
| Female | 29 | 19 | 10 | ||
| Age | 1.306 | 0.253 | |||
| ≥60 | 36 | 19 | 17 | ||
| <60 | 30 | 20 | 10 | ||
| Differentiation grade | 0.241 | 0.887 | |||
| Well | 2 | 1 | 1 | ||
| Moderate | 41 | 28 | 13 | ||
| Poor | 23 | 17 | 6 | ||
| T stage | 4.275 | 0.233 | |||
| T1 | 1 | 0 | 1 | ||
| T2 | 10 | 4 | 6 | ||
| T3 | 34 | 20 | 14 | ||
| T4 | 21 | 15 | 6 | ||
| N stage | 1.476 | 0.226 | |||
| N0 | 26 | 13 | 13 | ||
| N1+N2 | 40 | 26 | 14 | ||
| M stage | 13.371 | 0.0003 | |||
| M0 | 36 | 14 | 22 | ||
| M1 | 30 | 25 | 5 | ||
| Tumor location | 0.543 | 0.461 | |||
| Rectal | 38 | 21 | 17 | ||
| Colon | 28 | 18 | 10 | ||
| Vascular invasion | 2.948 | 0.086 | |||
| No | 62 | 35 | 27 | ||
| Yes | 4 | 4 | 0 | ||
| CEA(ng/mL) | 12.541 | 0.0004 | |||
| ≥5 | 39 | 30 | 9 | ||
| <5 | 27 | 9 | 18 |
Down-Regulation of ZBTB4 Correlated with an Unfavorable Prognosis
To investigate the clinical significance of ZBTB4 in CRC patients, we examined the relationship between ZBTB4 expression and the patients’ prognosis. Kaplan-Meier analysis was plotted to compare the survival of the two groups, and the results showed that the OS (P=0.0007, Figure 3A) and RFS (P=0.0077, Figure 3B) in the ZBTB4-low group were significantly lower when compared with those in the ZBTB4-high group.
Figure 3.
Kaplan–Meier survival curves for Overall Survival (A) and Recurrence Free Survival (B) in CRC patients in relation to ZBTB4 protein expression.
Discussion
Colorectal cancer (CRC) is a complex disease caused by both genetic and environmental factors. By the time of initial diagnosis, 10% ~ 25% of CRC patients have already developed liver metastasis, which imposes great challenge to surgical treatment. Moreover, the postoperative recurrence rate is high, and about 57% of the patients show local or distant metastasis by the time of treatment.12 Metastasis is a leading cause of cancer-related deaths, thus, the prevention and treatment of tumor metastasis is a basic requirement to the improvement of clinical prognosis.13 To improve the treatment outcome for CRC patients, it is an urgent need to identify the effective biomarkers or therapeutic targets to facilitate individualization of treatment. In this study, we first detected the mRNA and protein expressions of ZBTB4 in CRC cell lines, and the data revealed that the mRNA and protein expressions of ZBTB4 were significantly lower in the cancer cell lines than that in the normal colonic epithelial cell line. Then, ZBTB4 protein expression in the CRC tissue samples derived from 66 patients was determined, and the results showed that the overall positive expression rate was only 40.9%. The above findings indicate that ZBTB4 downregulation is associated with the development and progression of CRC, suggesting that ZBTB4 may be a novel biomarker for CRC.
Many reports have shown that ZBTB4 can regulate multiple cellular processes of human cancers. Our research found that ZBTB4 was down-regulated in CRC tissues and cells, which was consistent with the results of previous studies. Weber et al observed that compared with normal tissues, the expression of ZBTB4 is suppressed in prostate carcinoma and glioblastoma. ZBTB4 can inhibit P21CIP1 expression and cell cycle arrest in response to p53 activation.14 Similarly, Kim et al showed that ZBTB4 was relatively low-expressed in DU145 and PC3 human prostate cancer cells, and the overexpression of ZBTB4 in patients with prostate cancer was a prognostic factor indicative of a longer survival.15 In addition, ZBTB4 has been found down-regulated in breast cancer patients, and its low expression is correlated with shorter relapse-free patient survival.10 In breast carcinoma, previous research showed that ZBTB4 was inversely associated with Sp1 and EZH2.16 Moreover, Yu et al showed that significantly low-expressed ZBTB4 in ES cells and tissues is correlated with shorter survival of ES patients. Functionally, ZBTB4 acts as a tumor suppressor by regulating proliferation, cell cycle progression and apoptosis of ES cells.9 Other studies demonstrated that lower ZBTB4 expression is correlated with higher genome instability in many cancers.8 Our data revealed that ZBTB4 was involved in the progression and metastasis of CRC, and that low-expressed ZBTB4 was associated with poorer prognosis. Taken together, these data suggest that ZBTB4 may be an effective therapeutic target for the treatment of malignant tumors, including CRC.
In conclusion, our study showed that ZBTB4 was significantly low-expressed in CRC cell lines and tissue samples, and this was closely associated with tumor metastasis status and CEA level. CRC patients with higher expression of ZBTB4 tended to develop a more favorable prognosis. The current findings suggest that ZBTB4 could potentially serve as a new therapeutic target or clinical biomarker for CRC treatment, especially for metastatic CRC patients. However, there are still some limitations in this study, specifically the sample size was small and the underlying molecular mechanism of ZBTB4 down-regulation in CRC cells remains to be further investigated.
Acknowledgments
We are grateful to the patients for their participation, and the record room and pathology department staff at The First Affiliated Hospital of Zhejiang University for their assistance.
Informed Consent
Written informed consent for the information to be stored and used in the hospital database was obtained from each patient.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed on the journal to which the article will be submitted; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59(4):225–249. doi: 10.3322/caac.20006 [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 3.Chen W. Cancer statistics: updated cancer burden in China. Chin J Cancer Res. 2015;27(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slattery ML, Herrick JS, Mullany LE, et al. The co-regulatory networks of tumor suppressor genes, oncogenes, and miRNAs in colorectal cancer. Genes Chromosomes Cancer. 2017;56(11):769–787. doi: 10.1002/gcc.22481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Roy FM, McCrea PD. A role for kaiso-p120ctn complexes in cancer? Nat Rev Cancer. 2005;5(12):956–964. doi: 10.1038/nrc1752 [DOI] [PubMed] [Google Scholar]
- 6.Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Prive GG. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6(10):R82. doi: 10.1186/gb-2005-6-10-r82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamada D, Perez-Torrado R, Filion G, et al. The human protein kinase HIPK2 phosphorylates and downregulates the methyl-binding transcription factor ZBTB4. Oncogene. 2009;28(27):2535–2544. doi: 10.1038/onc.2009.109 [DOI] [PubMed] [Google Scholar]
- 8.Roussel-Gervais A, Naciri I, Kirsh O, et al. Loss of the methyl-CpG-binding protein ZBTB4 alters mitotic checkpoint, increases aneuploidy, and promotes tumorigenesis. Cancer Res. 2017;77(1):62–73. doi: 10.1158/0008-5472.CAN-16-1181 [DOI] [PubMed] [Google Scholar]
- 9.Yu Y, Shang R, Chen Y, et al. Tumor suppressive ZBTB4 inhibits cell growth by regulating cell cycle progression and apoptosis in ewing sarcoma. Biomed Pharmacother. 2018;100:108–115. doi: 10.1016/j.biopha.2018.01.132 [DOI] [PubMed] [Google Scholar]
- 10.Kim K, Chadalapaka G, Lee SO, et al. Identification of oncogenic microRNA-17-92/ZBTB4/specificity protein axis in breast cancer. Oncogene. 2012;31(8):1034–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ross-Adams H, Lamb AD, Dunning MJ, et al. Integration of copy number and transcriptomics provides risk stratification in prostate cancer: a discovery and validation cohort study. Ebiomedicine. 2015;2(9):1133–1144. doi: 10.1016/j.ebiom.2015.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meissner HI, Breen N, Klabunde CN, Vernon SW. Patterns of colorectal cancer screening uptake among men and women in the United States. Cancer Epidemiol Biomarkers Prev. 2006;15(2):389–394. doi: 10.1158/1055-9965.EPI-05-0678 [DOI] [PubMed] [Google Scholar]
- 13.Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127(4):679–695. doi: 10.1016/j.cell.2006.11.001 [DOI] [PubMed] [Google Scholar]
- 14.Weber A, Marquardt J, Elzi D, et al. Zbtb4 represses transcription of P21CIP1 and controls the cellular response to p53 activation. EMBO J. 2008;27(11):1563–1574. doi: 10.1038/emboj.2008.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim K, Chadalapaka G, Pathi SS, et al. Induction of the transcriptional repressor ZBTB4 in prostate cancer cells by drug-induced targeting of microRNA-17-92/106b-25 clusters. Mol Cancer Ther. 2012;11(9):1852–1862. doi: 10.1158/1535-7163.MCT-12-0181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang WS, Chadalapaka G, Cho SG, et al. The transcriptional repressor ZBTB4 regulates EZH2 through a MicroRNA-ZBTB4-specificity protein signaling axis. Neoplasia. 2014;16(12):1059–1069. doi: 10.1016/j.neo.2014.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]